Abstract
Exhausted CD8+ T (Tex) cells are dysfunctional due to persistent antigen exposure in chronic viral infection and tumor contexts. A stem cell-like Tex (Tex-stem) subset can self-renew and differentiate into terminally exhausted (Tex-term) cells. Here, we show that ectopic Tcf1 expression potently promoted the generation of Tex-stem cells in both a chronic viral infection and preclinical tumor models. Tcf1 overexpression diminished coinhibitory receptor expression and enhanced polycytokine-producing capacity while retaining a heightened responses to checkpoint blockade, leading to enhanced viral and tumor control. Mechanistically, ectopically expressed Tcf1 exploited existing and novel chromatin accessible sites as transcriptional enhancers or repressors and modulated the transcriptome by enforcing pre-existing expression patterns in Tex-stem cells, such as enhanced suppression of Blimp1 and Bim and acquisition of new downstream genes, including Mx1, Tox2, and Runx3. These findings reveal a pronounced impact of ectopic Tcf1 expression on Tex functional restoration and highlight the therapeutic potential of harnessing Tcf1-enforced transcriptional programs.
Keywords: T cell exhaustion, Tcf1, Stem cell features
Subject terms: Lymphocyte differentiation, Immunogenetics
Introduction
CD8+ T lymphocytes are essential players in protective cellular immune responses against pathogens and malignantly transformed cells. Depending on the nature of the challenge, CD8+ T cells are equipped with plasticity to maximize the reduction in pathogen load or tumor growth.1,2 The most productive CD8+ T cell responses are found in acute viral or intracellular bacterial infections, for which fully functional, cytotoxic effector CD8+ T cells facilitate the clearance of pathogens, followed by formation of memory CD8+ T cells, which provide accelerated and heightened protection against reinfection by the same pathogens.3 On the other hand, in a chronic viral infection context or intratumor microenvironment, effector CD8+ T cells become ‘functionally exhausted’ or ‘hypofunctional’ due to antigen persistence, resulting in incomplete pathogen or tumor control.4,5 The ‘exhausted’ CD8+ T cells are nonetheless critical in curtailing viral replication and tumor growth, and these critical functions can be enhanced by blocking coinhibitory receptors/pathways (known as checkpoint blockade).6,7 However, the epigenetic stability of Tex cells limits the durability of checkpoint blockade-elicited functional enhancement.8,9 It remains unknown whether Tex cells can be intrinsically reprogrammed to achieve more stable functional restoration.
‘Exhausted’ CD8+ T (Tex) cells have distinct phenotypes with relatively high expression of coinhibitory receptors such as PD-1 and LAG-3, and their ability to produce cytokines, exert cytotoxic effects, and proliferate in response to cognate antigens is progressively impaired.4,5 It is now recognized that Tex cells represent a transcriptomic and epigenetic state that differs from that of effector and memory CD8+ T cells elicited by acute infection.10,11 Recent studies have clarified the heterogeneity of Tex cells elicited by chronic viral infection, revealing at least two distinct subsets. One subset is a terminally exhausted population (Tex-term) associated with the PD-1hiTim3hi phenotype, which retains the ability to perform cytotoxic functions but fails to persist in the long term. The other subset has stem cell-like features (termed Tex-stem herein) and a PD-1intTim3loCXCR5+ phenotype.12–14 Tex-stem cells persist longer due to their self-renewal capacity, differentiate into Tex-term cells, and exhibit increased proliferative capacity in response to checkpoint blockade. Furthermore, tumor-infiltrating CD8+ T cells (TILs), detected in mouse models or patient tumors, share a remarkably similar molecular signature with Tex cells, and the Tex-stem equivalent population in TILs is marked by the expression of Slamf6 instead of CXCR5.15–17 Tex-term and Tex-stem cells share characteristics of the common molecular program that leads to T cell exhaustion18, and both subsets show strong dependence on Nfat,19 Irf4,20 and Tox transcription factors.21–25 On the other hand, the Tex subsets have distinct transcriptional features, with Tcf1, Bcl6, and Id3 promoting and Blimp1, Runx3, and Id2 antagonizing Tex-stem cell formation.12–14,26,27 There is an urgent need to exploit these underlying transcriptional circuits in Tex cell subsets for improved control of infection and/or tumors.
The transcription factor Tcf1 (encoded by Tcf7) is critical for T cell development and mature T cell differentiation.28–30 In CD8+ lineage T cells, Tcf1 is necessary for establishing CD8+ T cell identity,31 and it promotes the longevity and recall responses of the memory CD8+ T cells elicited by acute infection.32,33 In the context of chronic viral infection and TILs, Tex-stem cells express higher levels of Tcf1 than Tex-term cells and are often referred to as Tcf1+ Tex cells.5 Importantly, Tex-stem cells depend on Tcf1 for generation and persistence.12,14,15,26,34 Recent studies suggest that the divergence of Tex-stem and Tex-term cells may occur rapidly after CD8+ T cells are activated by chronic viral infection, before they even reach peak responses or show detectable signs of dysfunctional cytotoxicity.23,24 In addition, the generation of precursors to Tex-stem cells appears to depend on Tcf1.35 Remarkably, Tcf1 is identified as a single transcription factor that is positively correlated with favorable prognosis in melanoma patients after checkpoint blockade therapy,36 highlighting the requirement for Tcf1 in preserving CD8+ T cell functionality upon exposure to persisting viral or tumor antigens.
An equally important question to address is whether Tcf1 is sufficient to confer functional advantages to Tex cells, where an affirmative answer will provide a strong rationale for utilizing the Tcf1-dependent regulatory circuit in therapeutic applications. Several recent reports used retroviruses to deliver the Tcf7 gene into preprimed CD8+ T cells, which were subsequently infected with lymphocytic choriomeningitis virus clone 13 (LCMV-Cl13) to generate a chronic viral infection.26,35,37 These studies observed that the Tcf7- expressing retrovirus partially promoted Tex-stem phenotypes, for example, by inducing CXCR5 or repressing Tim3 when observed on day 8 post infection. As explained above, CD8+ T cells activated by chronic viral infection do not exhibit dysfunction, such as diminished cytokine production or reduced expression of cytotoxic molecules, in the early stage. Because T cell exhaustion is a progressive differentiation process, it is critical to assess the impact of ectopic Tcf1 expression in the long term and on truly dysfunctional Tex cells. This evaluation is technically challenging for retrovirus-delivered genes because of the progressive silencing by epigenetic mechanisms during long-term cell propagation.38 Another disadvantage of retrovirus-mediated gene delivery is associated with the need for prepriming CD8+ T cells outside the context of chronic viral infection to increase transduction efficiency. When retrovirus-transduced cells are transferred into new hosts pre- or reinfected with LCMV-Cl13, the LCMV-specific TCRs are restimulated in the already activated CD8+ T cells. These stimuli may have an unwanted impact on the activation of the Tex program and the generation of Tex-stem cells, particularly in light of recent findings that suggest the early bifurcation of Tex-stem and Tex-term cell fates.23,24 These challenges have contributed to the incomplete understanding of the full-spectrum impact of ectopic Tcf1 expression on Tex cell phenotype, function, fate divergence, and the underlying transcriptomic and epigenetic mechanisms.
To overcome the limitations of a retroviral approach, we employed a Tcf1 transgenic mouse strain to achieve ectopic expression of full-length Tcf1. We previously showed that transgenic expression of Tcf1, together with its coactivator β-catenin in a constitutively active form, expands the memory CD8+ T cell pool in response to acute infections.39 In the current work, we extended this approach to chronic viral infection and preclinical tumor models and demonstrated that ectopic Tcf1 expression promoted Tex cells to predominantly adopt the Tex-stem phenotype while preserving their enhanced functionality, leading to more effective viral or tumor control. The stably expressed Tcf1 transgene also facilitated a mechanistic investigation into its impact on Tex cells at an advanced stage of CD8+ T cell exhaustion, and we found that ectopically expressed Tcf1 exploits novel and existing genomic regulatory regions and target genes to solidify and enforce the stem cell-like program of Tex cells.
Results
Ectopic expression of full-length Tcf1 promotes the generation of Tex-stem cells
Tcf1 is expressed as multiple isoforms in T cells, with the full-length protein of 45 kDa (known as p45 Tcf1). By interfering with exon 1 to 2 splicing, we specifically abrogated the expression of p45 Tcf1 while preserving the expression of the shorter isoforms (called p45–/– herein).40,41 To induce the ectopic expression of p45 Tcf1, we utilized a Tcf1 transgene driven by the H-2Kb promoter (called p45-Tg).42 In CD8+ T cells, we confirmed that the Tcf1 expression was elevated by 3–4-fold in the p45-Tg cells (Fig. S1a) and that the ectopic Tcf1 expression did not detectably alter the naïve state or cause the aberrant expression of coinhibitory receptors such as Tim3, PD-1, 2B4, LAG-3 or key transcription factors such as T-bet, Eomes and Tox (Fig. S1b–d). The N-terminal domain of p45 Tcf1 interacts with β-catenin. In response to Wnt ligand stimulation, β-catenin is stabilized, and is translocated into the nucleus to function as a coactivator of Tcf/Lef family transcription factors.29 To investigate a potential synergistic effect resulting from the Tcf1 and β-catenin interaction, we employed a nondegradable (hence constitutively active) β-catenin transgene driven by the Cd4 promoter (called βCat-Tg).43 We crossed these genetically modified strains to a transgenic P14 TCR, which specifically recognizes the glycoprotein 33–41 epitope (GP33) of LCMV. We then adoptively transferred naïve P14 CD8+ T cells with various genotypes into CD45-disparate recipients, followed by infection with LCMV-Cl13 to elicit chronic viral infection (Fig. 1a).
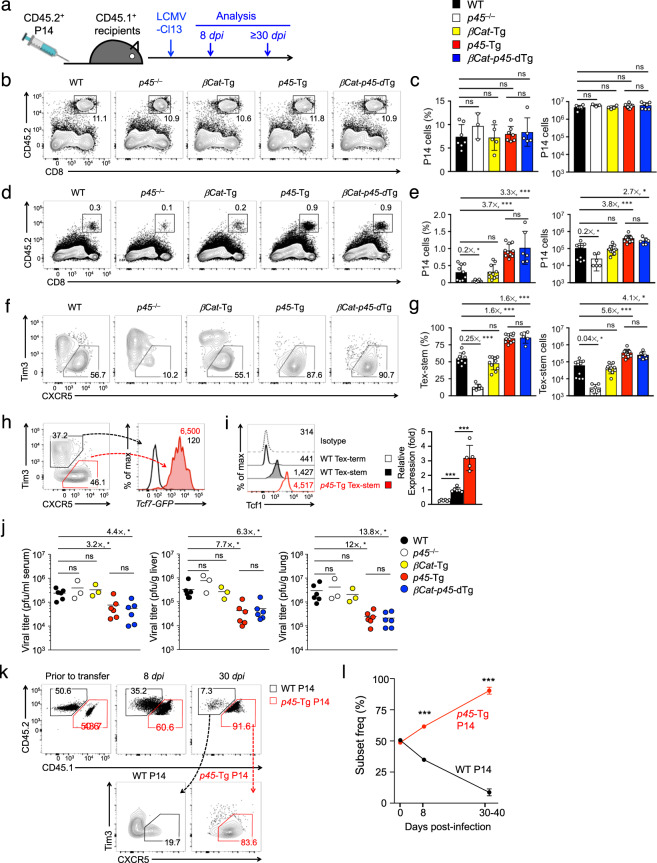
Fig. 1.
Ectopic Tcf1 expression promotes the generation of Tex-stem cells in response to chronic viral infection. a Experimental design. Detection of P14 Tex cells with the indicated genotypes on day 8 (b, c) and days 28–35 (d, e) post infection, with representative contour plots in (b) and (d). Cumulative data on cell frequency (left) and numbers (right panels) in (c) and (e) are the means ± S.D. from 2 to 4 independent experiments. Detection of the Tex-stem subset in the CD45.2+CD8+ cells based on the CXCR5+Tim3lo phenotype at 28–35 dpi, with representative data in (f) and cumulative data in (g). h Detection of Tcf7-GFP reporter activity in Tex-stem and Tex-term cells at 30 days after adoptive transfer of the Tcf7GFP/+ P14 cells and subsequent LCMV-Cl13 infection. i Detection of Tcf1 protein in the Tex-stem and Tex-term cells as determined by intranuclear staining on 30 dpi. Representative half-stack histograms are in the left panel, with values denoting geometric mean fluorescence intensity (gMFI). The right panel shows cumulative data from 2 to 3 experiments, with the Tcf1 gMFI in the WT Tex-stem cells set at 1 and its relatively expression in other subsets/genotypes normalized for each independent experiment. j Viral titers in the sera, livers and lungs of the recipient mice at 28–35 days post infection. Note that recipients of the WT and p45–/– P14 cells had similar viral loads despite low numbers of p45–/– P14 cells, which was likely the result of endogenous immune responses in the recipients. k–l Tracking WT and p45-Tg P14 Tex cells under competitive conditions. CD45.2+ WT and CD45.1+CD45.2+ p45-Tg P14 Tex cells were mixed at a 1:1 ratio (prior to transfer) and adoptively transferred into the same CD45.1+ recipients. On 8 and 30 dpi, both cell types were detected in the spleens, with a representative contour plot in (k) and cumulative data in (l). Significance of the differences among multiple groups was assessed by one-way ANOVA, and p values of the differences between specific pairs were determined by Tukey’s test. Student’s t test was used for comparisons between two groups only (such as in (l)). ns, not significant; *p < 0.05; **p < 0.01; and ***p < 0.001. Values denote fold changes compared with WT P14 cells. This annotation applies to all figures in this work
At the peak response on day 8 post infection (8 dpi), the expansion of the early P14 Tex cells was similar regardless of the gain or loss of p45 Tcf1 expression or ectopic expression of β-catenin alone or with p45 Tcf1 (Fig. 1b, c). In contrast, after 28 dpi, when T cell dysfunction reached an advanced stage with pronounced phenotypic and functional alterations, the frequency and number of the P14 Tex cells with the p45–/– genotype were greatly decreased but those with the p45-Tg genotype were substantially increased, compared with the wild-type (WT) cells (Fig. 1d, e). On the other hand, the number of βCat-Tg-expressing Tex cells was similar to the number of WT cells, and the β-catenin and p45 Tcf1 double transgene (called βCat-p45-dTg) Tex cells did not exceed the number of p45-Tg cells (Fig. 1d, e). These data indicate that the number of virus-specific Tex cells after chronic infection depended on p45 Tcf1 and could be enhanced by the ectopic expression of p45 Tcf1, even without additional β-catenin stabilization.
Further analysis of the Tex cells at the advanced exhaustion stage revealed that the CXCR5+Tim3lo Tex-stem population was greatly diminished in frequency and numbers in the p45–/– P14 cells compared with those of WT P14 cells (Fig. 1f, g), a finding consistent with previous reports,12,14,26 highlighting the strong dependence of Tex-stem cells on the full-length Tcf1 protein. In fact, a knock-in Tcf7-GFP reporter44 clearly demarcated the Tcf1+ Tex-stem from the Tcf1– Tex-term cells (Fig. 1h). Intranuclear detection validated this observation and revealed an ~3-fold increase in Tcf1 protein levels in the Tex-stem cells upon ectopic expression (Fig. 1i). Although expression of βCat-Tg alone did not lead to discernable advantages, we found a strong impact of p45-Tg, which expanded Tex-stem cells to become the predominant population among the Tex cells, and this impact was not detectably enhanced by βCat-p45-dTg (Fig. 1f, g). Notwithstanding endogenous CD8+ T cell responses in the recipients, which should contribute similarly to the viral control in all groups, the increase in total P14 and Tex-stem cell numbers in recipients of p45-Tg or βCat-p45-dTg P14 cells further reduced the viral titers in the sera, livers and lungs (Fig. 1j). These observations indicated that ectopic Tcf1 expression alone, in cooperation with endogenous β-catenin and/or other coactivators, promotes the accumulation of Tim3loCXCR5+ Tex-stem cells to be the predominant CD8+ T cell subset after chronic viral infection and improves viral control.
To exclude the possibility that the observed difference in viral load may have contributed to the phenotypic changes of the Tex cells, CD45.2+ WT P14 and CD45.1+CD45.2+ p45-Tg P14 CD8+ T cells were cotransferred at a 1:1 ratio into CD45.1+ recipients followed by LCMV-Cl13 infection. Under this condition, the p45-Tg and WT CD8+ T cells were exposed to the same cytokine milieu and antigen load. At 8 dpi, the p45-Tg Tex cells started to exhibit a proliferative advantage over the WT Tex cells, and at more than 30 dpi, the p45-Tg Tex cells were at least 10-fold more abundant than the WT Tex cells (Fig. 1k, l). Whereas ~20% of the WT Tex cell population had a Tex-stem cell phenotype, more than 80% of the p45-Tg Tex cell population had a Tex-stem phenotype (Fig. 1k). These data thus support the notion that the skewing of Tex cells toward a stem-like phenotype by p45-Tg directly resulted from the intrinsic impact of ectopic Tcf1 expression and was not a secondary effect caused by differences in antigen load and/or inflammation.
Ectopic Tcf1 expression ameliorates CD8 T cell exhaustion
We then investigated whether ectopic Tcf1 expression exerts broader impacts on molecular and functional aspects of Tex cells in addition to promoting a Tex-stem phenotype. Consistent with previous reports,12,14 WT Tex-stem cells expressed lower levels of coinhibitory receptors, including PD-1 and 2B4, than did the WT Tex-term cells, although the LAG-3 expression was similar in both cell types (Fig. 2a, b). Interestingly, PD-1 expression, but not 2B4 or LAG-3 expression, was reduced in the p45-Tg Tex-stem cells (Fig. 2a, b). T-bet and Eomes are both essential for sustaining the Tex cell pool for the control of chronic infection,45 and Tox family TFs are indispensable for activating the Tex-specific transcriptional program.21–25 We observed a lower expression of T-bet but a higher expression of Eomes and Tox in WT Tex-stem cells than in WT Tex-term cells (Fig. 2c, d). The p45-Tg Tex-stem cells showed modestly suppressed Eomes and Tox expression with no detectable effects on T-bet levels (Fig. 2c, d). In addition, the p45-Tg Tex-stem cells retained other features of the WT Tex-stem cells, expressing lower levels of FasL but higher levels of Fas, IL-7Rα, ICOS and Bcl6 than did the WT Tex-term cells (Fig. S2). The dysfunction of Tex cells manifests as diminished capacity of IFN-γ production, especially coproduction with IL-2 and TNF-α.46 Because direct ex vivo stimulation diminished the surface expression of Tim3 and CXCR5, we used the combination of CD39 and Slamf6 to differentiate the Tex-term and Tex-stem cells12 (Fig. 2e), because the expression of CD39 and Slamf6 remained more clearly observed in assays measuring cytokine production. Although WT Tex-stem and Tex-term cells showed a similar capacity for producing IFN-γ, larger numbers of WT Tex-stem cells retained the ability to coproduce IL-2 and/or TNF-α (Fig. 2f, g). Importantly, the p45-Tg Tex-stem cells exhibited an increase in polycytokine production (Fig. 2f, g). These data demonstrate that ectopic Tcf1 expression not only allows Tex cells to assume a Tex-stem phenotype but also to adopt enhanced intrinsic programs that are characteristic of Tex-stem cells, such as suppression of key coinhibitory receptor(s) and expansion of polycytokine production capacity.
Fig. 2.
Ectopic Tcf1 expression promotes phenotypic and functional features characteristic of Tex-stem cells. Detection of key proteins in Tex cells by surface staining (a–b) or intranuclear staining (c–d). WT or p45-Tg P14 Tex cells were detected in recipient spleens at 28 dpi or later, and the CXCR5–Tim3hi Tex-term and CXCR5+Tim3lo Tex-stem cells were analyzed for the indicated proteins. Representative half-stack histograms are shown in (a) and (c), with values denoting gMFI. Cumulative data on relative protein expression were from 2–3 experiments (b, d), with the gMFI of each protein in the WT Tex-stem cells set at 1 and its relative expression in other subsets/genotypes normalized for each independent experiment. e CD39 and Slamf6 proteins were used as alternative markers to identify Tex-stem and Tex-term in the P14 cells on 30 dpi. f–g Detection of cytokines in the Tex cells. WT and p45-Tg P14 Tex cells were stimulated with PMA and ionomycin for 5 h, surface stained to identify CD39hiSlamf6– Tex-term and Slamf6+ Tex-stem subsets and then intracellularly stained for the indicated cytokines. Representative contour plots are shown in (f), and cumulative data on polycytokine-producing Tex cell numbers were from 2 independent experiments (g)
We next determined the extent to which ectopic Tcf1 expression affects the kinetics of Tex cell responses to chronic viral infection. During the contraction phase (e.g., 12 dpi), WT Tex-term cells showed more evident apoptosis than did the WT Tex-stem cells, with the p45-Tg Tex-stem cells showing a trend toward attenuated apoptosis (Fig. 3a, b), which may partly account for their enhanced persistence over time (Fig. 3c). The number of WT P14 Tex cells showed progressive reduction during the 90-day observation period, while the p45-Tg P14 Tex cells persisted at higher numbers (Fig. 3c). Tex-stem cells show a stronger proliferative burst in response to PD-1 blockade than did the Tex-term cells.12 Because PD-1 expression was diminished in the p45-Tg Tex-stem cells compared with the WT Tex-stem cells (Fig. 2a), we determined their proliferative capacity before and after treatment with anti-PD-L1 by measuring BrdU incorporation (Fig. 3d). Because Tex-stem cells are continuously differentiating into Tex-term cells with or without checkpoint blockade, the Tex-term cell pool at a given time point consists of at least two sources, a pre-existing subset that undergoes turnover and a newly generated subset derived from Tex-stem cells. With this potential caveat in mind, we observed that the WT Tex-stem cells showed reduced proliferation compared with the WT Tex-term cells in the untreated mice, and the p45-Tg Tex-stem cells exhibited an even lower rate of proliferation (Fig. 3e, f). The relative quiescence of the WT and p45-Tg Tex-stem cells was consistent with their stem cell-like features, which are similar to those of hematopoietic stem cells in that they are maintained in a quiescent state for their functional preservation.47 Whereas all the cell types showed increased BrdU incorporation upon anti-PD-L1 treatment, both the WT and p45-Tg Tex-stem cells showed a greater increase in proliferative capacity than did the Tex-term cells (Fig. 3e, f). As a result, the total number of P14 Tex cells was elevated by the anti-PD-L1 treatment in both groups, with the p45-Tg P14 Tex cells maintaining the numerical advantage (Fig. 3g). These data indicate that ectopic Tcf1 expression confers a survival advantage to Tex cells while preserving their enhanced proliferative capacity in response to checkpoint blockade.
Fig. 3.
Ectopic Tcf1 expression promotes Tex cell persistence by enhancing survival and quiescence. Detection of cell survival in the WT and p45-Tg P14 Tex subsets by Annexin V and 7-AAD staining on 12 dpi in recipient spleens, with representative contour plots shown in (a) and cumulative data on live cells shown in (b). c Longitudinal tracking of the number of P14 Tex cells after LCMV-Cl13 infection. d Experimental design to determine Tex cell cycling status and response to PD-1 blockade. e–f. Detection of the cycling status of Tex cells in the resting state (untreated) or in response to anti-PD-L1 treatment, determined by measuring BrdU incorporation on 30 dpi. Representative contour plots are shown in (e), and cumulative data on BrdU+ cells are shown in (f). g Cumulative data on the numbers of WT and p45-Tg P14 Tex cells before and after anti-PD-L1 blockade. All data are from 2 to 3 experiments. h Experimental design to determine Tex-stem cell function on a per cell basis. i Expansion of WT and p45-Tg Tex-stem cells in response to secondary challenge. Sorted WT and p45-Tg Tex-stem cells (5000 each) were transferred into TCRα–/– mice followed by LM-GP33 infection with or without anti-PD-L1 treatment. On day 5 after LM infection, Vα2+CD8+ cells were enumerated in recipient spleens. j Detection of the bacterial load in the livers and lungs of the recipients on day 5 after LM infection. CFU colony-forming unit; L.O.D. limit of detection
To further determine whether ectopic Tcf1 expression enhanced Tex-stem cell functions on a per cell basis, the WT and p45-Tg P14 Tex-stem cells were sort-purified, and the same number of each type of cell was adoptively transferred into TCRα–/– recipients in which endogenous T cell responses were abrogated (Fig. 3h), followed by rechallenge with Listeria monocytogenes expressing the GP33 epitope (LM-GP33). Although the WT and p45-Tg Tex-stem cells expanded similarly in response to the recall stimulation, the bacterial burden in the livers and lungs in recipients of the p45-Tg cells was reduced by approximately two orders of magnitude compared with that in the recipients of the WT cells (Fig. 3i, j). In parallel, a cohort of WT P14 Tex-stem cell recipients was given two doses of anti-PD-L1 treatment (Fig. 3h), which were sufficient to increase the number of P14 cells responding to the LM-GP33 rechallenge and diminish the bacterial loads (Fig. 3i, j). Collectively, these data suggest that ectopic Tcf1 expression confers functional enhancement to Tex-stem cells even in the absence of numerical advantage, leading to enhanced control of an infection.
Ectopic Tcf1 expression enforces the Tex-stem transcriptome profile
A major difference between Tex-stem and Tex-term cells is their differential Tcf1 expression, with the Tex-stem cells being Tcf1+ (Fig. 1h, i),12,26,34 and therefore, their transcriptomic differences are, at least partly, ascribed to Tcf1 expression itself. Expression profiling of the Tcf1+ Tex-stem and Tcf1– Tex-term cells, using a Tcf7-GFP reporter BAC transgene, led to the identification of 1400 downregulated and 1166 upregulated genes in the Tcf7-GFP+ compared with the Tcf7-GFP– Tex cells.34 To mechanistically understand the strong impact of ectopic Tcf1 expression on Tex cell phenotype and function, we analyzed the transcriptomes of the WT and p45-Tg Tex-stem cells by RNA-seq. The down- and upregulated genes in the Tcf7-GFP+ Tex cells exhibited a strong tendency to further decrease or increase in the p45-Tg Tex-stem compared with WT Tex-stem cells, respectively (Fig. 4a). This global trend suggests that ectopic Tcf1 expression enforces gene expression patterns that have been established in Tcf7-GFP+ Tex-stem cells by endogenous Tcf1 expression.
Fig. 4.
Ectopic Tcf1 expression modulates the Tex cell transcriptome. a Boxplot showing concordant gene expression trends between endogenous and ectopic Tcf1 expression in Tex cells. WT and p45-Tg P14 Tex-stem cells, each in duplicate, were sort-purified on 30 dpi and subjected to RNA-seq analysis. Transcriptomic data on the Tcf7-GFP+ and Tcf7-GFP– Tex cells were retrieved from Ref. 34 to define the upregulated, downregulated, or not significantly different (ns) genes due to endogenous Tcf1 expression. Genes in each subset were assessed for global expression trends in Tex-stem cells with the ectopic expression of Tcf1. *p < 1e–10, as determined by Wilcoxon rank sum test. b Scatter plots comparing the DEGs of the p45-Tg/WT Tex-stem cells and those of the Tcf7-GFP+/Tcf7-GFP– Tex cells. Different groups of DEGs are marked as g1–g6, with the g1 and g4 genes showing concordant expression changes and g2 and g5 genes showing unique changes regulated by ectopic Tcf1 expression. The genes in g3 and g6 with discordant expression changes were not analyzed in detail because these genes may not account for the stem cell-like features of the Tcf7-GFP+ Tex cells, as their expression was reversed by ectopic Tcf1 expression. Heatmaps showing select downregulated (c) and upregulated (d) genes in the p45-Tg compared with WT Tex-stem cells in the indicated functional categories; the data on the Tcf7-GFP+ and Tcf7-GFP– Tex cells are included for direct comparison. Gene symbols in bold and color (blue/red for downregulated/upregulated genes, respectively) mark the genes highlighted in Figs. 5 and 6 that are associated with differential chromatin accessibility and/or Tcf1 binding peaks
To define specific genes affected by ectopic Tcf1 expression, we identified differentially expressed genes (DEGs) in the p45-Tg and WT Tex-stem cells using the DESeq2 algorithm48 with the criteria of ≥1.5-fold expression changes and FDR < 0.1. The DEGs were then stratified by their status as down- and upregulated genes in the Tcf7-GFP+ Tex compared with Tcf7-GFP– Tex cells. Genes in groups 1 and 4, as displayed on a scattered plot, showed concordant expression changes in the p45-Tg Tex-stem and Tcf7-GFP+ Tex cells, as commonly repressed and activated genes, respectively (Fig. 4b). These genes were likely Tcf1-dependent genes in the WT Tex-stem cells, and their expression was enforced by ectopic Tcf1 expression. On the other hand, the genes in groups 2 and 5, which were not differentially expressed in the Tcf7-GFP+ and Tcf7-GFP– Tex cells, were repressed and induced in the p45-Tg Tex-stem cells, respectively (Fig. 4b). These genes likely represented novel targets acquired upon ectopic Tcf1 expression. These analyses suggest that ectopically expressed Tcf1 can enforce characteristic genes in the Tex-stem cells and/or antagonize genes in the Tex-term cells, and further explore novel downstream target genes to solidify a Tex-stem cell identity.
The functional annotation of downregulated genes in p45-Tg compared with WT Tex-stem cells (in groups 1 and 2) led to the identification of several key transcription and epigenetic regulators. For example, Prdm1 in group 1 (encoding the Blimp1 transcription factor, known to promote a Tex-term fate49) and Runx3 in group 2 (encoding the Runx3 transcription factor that antagonizes the generation of Tex-stem cells27) were repressed in the p45-Tg over WT Tex-stem cells (Fig. 4c). Other important functional categories included receptors and signaling molecules. Among these, Klrg1 in group 1 (encoding killer cell lectin-like receptor G1, known to be associated with the terminal differentiation of effector CD8+ T cells) and Pdcd1 in group 2 (encoding the PD-1 coinhibitory receptor) showed diminished expression upon ectopic Tcf1 expression (Fig. 4c). In addition, Bcl2l11 (encoding the pro-apoptotic Bim protein) and a number of cell cycle regulators were repressed in the p45-Tg Tex-stem cells (Fig. 4c), consistent with their improved survival and increased stem cell-like quiescence (Fig. 3). In line with these findings, gene set enrichment analysis (GSEA), which was used to assess the behavior of a given gene set without preset thresholds, showed negative enrichment of the ‘KEGG_CELL_ CYCLE’ gene set in the WT Tex-stem cells (i.e., they had diminished expression in the p45-Tg Tex-stem cells) (Fig. S3a).
The upregulated genes in the p45-Tg Tex-stem cells (in groups 4 and 5) included Irf7 and Zbtb32 (encoding members of IRF and POK family transcription factors, respectively) and Mapk11 (encoding p38 MAP kinase β) (Fig. 4d), which all have active roles in antiviral CD8+ T cell responses.50–52 In addition, genes in ‘immune system process’ and ‘cell adhesion,’ such as interferon-responsive Ifit3, Ifit3b, Oas2, Oasl2, and Mx1 and integrins, including Itga7 and Itga4, showed elevated expression in the p45-Tg Tex-stem cells (Fig. 4d). GSEA also revealed substantial enrichment of several IFN-β1-, IFN-α, or TNF-α-induced gene sets in the p45-Tg Tex-stem cells (Fig. S3b–d). These findings were consistent with a recent single-cell RNA-seq-based observation indicating that Tcf1 is coexpressed with interferon-stimulated genes in early precursors of the Tex-stem cells.35 Collectively, these analyses suggest that ectopic Tcf1 expression modulates the Tex cell transcriptome to an enhanced Tex-stem state.
Ectopic Tcf1 expression modulates chromatin accessibility to enforce Tex-stem cell identity
To further define the impact of ectopic Tcf1 expression on Tex cells, we measured chromatin accessibility using ATAC-seq with the p45-Tg Tex-stem, WT Tex-stem (Tcf1+) and WT Tex-term (Tcf1–) cells, which were segregated into distinct clusters by principal component analysis (PCA) (Fig. 5a). Among a total of 57,340 ATAC-seq peaks identified in all three cell states, 14,686 peaks (25.6%) exhibited significantly different signal strengths for each pairwise comparison, with the cutoff at a 2-fold change and FDR < 10–5, and were thus defined as differential chromatin accessibility (Diff ChrAcc) sites. Compared with the WT Tex-term cells, both the WT and p45-Tg Tex-stem cells showed extensive ChrAcc changes, with substantial overlap among the Diff ChrAcc sites (as shown in the a2 + a5 subclusters among the decreased ChrAcc sites and b2 + b5 subclusters among the increased ChrAcc sites; Fig. 5b). On the other hand, ectopic Tcf1 expression resulted in distinct ChrAcc changes compared with WT Tex-term cells (a1 + a4 and b1 + b4 subclusters, Fig. 5b). A comparison of the p45-Tg with WT Tex-stem cells also demonstrated that ectopic Tcf1 expression not only enforced the existing ChrAcc changes established in the WT Tex-stem cells (a5 and b5 subclusters) but also exploited novel chromatin sites to modulate their accessibility (a4 + a6 and b4 + b6 subclusters, Fig. 5b). These data indicate that ectopically expressed Tcf1 has substantial impact on the ChrAcc landscape in Tex cells.
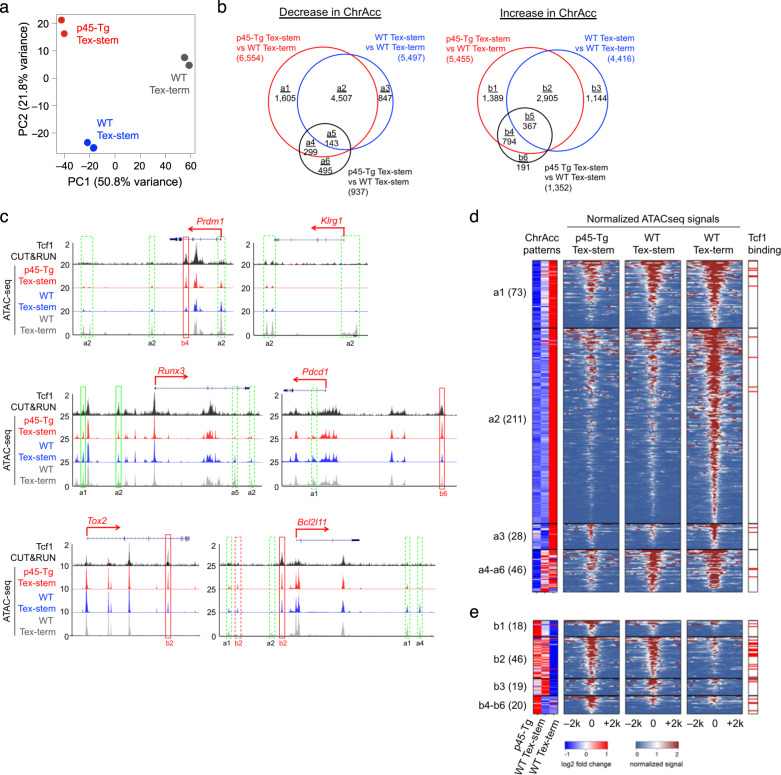
Fig. 5.
Ectopic Tcf1 expression modulates Tex cell chromatin accessibility for target gene repression. a Principal component analysis (PCA) of the ATAC-seq signals of the p45-Tg Tex-stem cells, WT Tex-stem and Tex-term cells, with each analyzed in duplicate on 30 dpi. Normalized average ATAC-seq signals of the merged ATAC-seq peaks were extracted for all replicates. The variance levels of the top two principal components are marked. b Venn diagrams showing the distribution of decreased (left) or increased (right) ChrAcc sites based on the indicated pairwise comparisons. All Diff ChrAcc sites are allocated to subclusters a1–a6 and b1–b6, and the values denote the number of sites in each subcluster. Diff ChrAcc sites in the p45-Tg and WT Tex-stem cell comparison did not overlap with the a3 or b3 clusters, which is indicative of strong concordant ChrAcc changes with ectopic or endogenous Tcf1 expression in these cell types. c ATAC-seq tracks of all three cell types and Tcf1 CUT&RUN tracks of the p45-Tg Tex-stem cells for select downregulated gene loci. The structure and transcription direction are marked on top, and the rectangles denote individual or groups of Diff ChrAcc sites, with their subcluster association annotated below. Green and red rectangles denote concordant and discordant changes in ChrAcc and gene expression, respectively. Diff ChrAcc sites overlapping with high-confidence Tcf1 binding peaks are marked with solid rectangles, while those lacking Tcf1 binding are marked with dotted rectangles. Heatmaps showing concordant (d) and discordant (e) ChrAcc site changes associated with downregulated genes in the p45-Tg Tex-stem cells. Far left, relative ChrAcc change patterns. Middle three panels, normalized ATAC-seq signals at the center of the Diff ChrAcc sites and their ±2 kb neighboring regions. Far right, presence of high-confidence Tcf1 binding event as marked with a red line at each corresponding Diff ChrAcc site. Diff ChrAcc site subclusters are labeled, and the number of Diff ChrAcc sites in each subcluster is in parentheses. Subclusters b4–b6 and a4–a6 are clustered together to highlight the unique effects of ectopic Tcf1 expression
To connect ChrAcc changes to the enhanced functional output in Tex cells upon ectopic Tcf1 expression, we used a gene-centric approach. Each ChrAcc site was assigned to a gene located within the gene body and ±50 kb flanking sequences. DEGs derived from the comparison of the p45-Tg and WT Tex-stem transcriptomes (Fig. 4b) were associated with more Diff ChrAcc sites than were the non-DEGs (Fig. S4a). The downregulated genes in the p45-Tg Tex-stem cells were more frequently associated with a decrease in ChrAcc sites than were the non-DEGs, as determined by both site distribution frequency (Fig. S4b) and site counts per gene (Fig. S4c). Similarly, the upregulated genes in the p45-Tg Tex-stem cells were more frequently associated with an increase in ChrAcc sites than were the non-DEGs (Fig. S4b, d). These observations validated our focus on DEG-associated Diff ChrAcc sites.
The 100 downregulated genes in the p45-Tg Tex-stem cells (combining genes in groups 1 and 2, Fig. 4b) were associated with 461 unique Diff ChrAcc sites, with 39 sites found in promoter regions (defined as transcription start site, TSS, and ±3 kb flanking sequences) of 29 genes (gene list in Fig. S5a and Table S1). Thirty-five of the 39 promoter-associated Diff ChrAcc sites showed diminished signals in the p45-Tg and WT Tex-stem cells compared with those of the WT Tex-term cells, showing a concordant pattern with gene downregulation. For example, the Prdm1 TSS was less accessible, while the Klrg1 promoter became completely inaccessible in the p45-Tg and WT Tex-stem cells (Fig. 5c). On the other hand, the remaining 422 Diff ChrAcc sites in distal, nonpromoter regulatory regions were associated with 91 downregulated genes (Fig. S5a, Table S1), and most of them (76%) showed concordant changes with gene expression, appearing in the a1–a6 subclusters (Fig. 5d). For example, decreased ChrAcc sites were found in the upstream and/or downstream regions of Prdm1, Klrg1, Runx3, and Bcl2l11 and intronic regions of Runx3 and Pdcd1 (Fig. 5c). Notably, several sites were particularly decreased in the p45-Tg Tex-stem cells, such as those belonging to the a1, a4, or a5 subclusters, as observed in the Runx3, Pdcd1, and Bcl2l11 gene loci (Fig. 5c), highlighting the unique impact of ectopic Tcf1 expression on enforcing a Tex-stem-characteristic ChrAcc landscape.
Another significant observation was that 49 of the 100 downregulated genes in p45-Tg Tex-stem cells were also associated with sites showing an increase in ChrAcc sites, as in the b1–b6 subclusters (Fig. 5e). Such sites were observed in the introns of Prdm1 and Tox2 and upstream regions of the Pdcd1 and Bcl2l11 genes (marked by red rectangles in Fig. 5c). In addition to increased ChrAcc sites common to both the p45-Tg and WT Tex-stem cells (as in the b2 subclusters, such as those in the Bcl2l11 and Tox2 loci), ectopic Tcf1 expression did result in a uniquely strong increase in ChrAcc in the p45-Tg Tex-stem cells, as observed in the Prdm1 intron 4 (b4 subcluster) and the Pdcd1 upstream region (b6 subcluster) (Fig. 5c). We posit that the sites showing increased ChrAcc may function as transcriptional silencers (or negative regulatory elements) and cooperatively contribute to target gene repression in Tex-stem cells (also refer to the next section).
The 74 upregulated genes in the p45-Tg Tex-stem cells (combining genes in groups 4 and 5, Fig. 4b) were associated with 195 Diff ChrAcc sites, with 26 sites found in the promoter regions of 24 genes (gene list in Fig. S5b and Table S2). Twenty-two of the 26 promoter-associated Diff ChrAcc sites showed increased signals in the p45-Tg or WT Tex-stem compared with the WT Tex-term cells, as observed in Tshz2, Hopx, and Itga7 TSSs (Fig. 6a). The remaining 169 Diff ChrAcc sites in distal regulatory regions were associated with 60 upregulated genes (Fig. S5b, Table S2), with most of them (57%) showing concordant changes with gene expression, as evident in the b1–b6 subclusters (Fig. 6b). For example, increased ChrAcc sites were found in the intronic regions of the Tshz2, Itga7, Mapk11, and Mx1, upstream of Hopx, and downstream of Mx1 genes (Fig. 6a). In addition to an increase in ChrAcc sites common to both the p45-Tg and WT Tex-stem cells (as evident in b2 subclusters), ectopic Tcf1 expression caused a unique increase in ChrAcc in the p45-Tg Tex-stem cells, as observed in the Tshz2 intron (b5 subcluster) and Mx1 intronic region (b4 subcluster) (Fig. 6a). We also found that the upregulated genes in the p45-Tg and WT Tex-stem cells were associated with sites showing a decrease in ChrAcc (Fig. 6c), as seen in an upstream region of Mx1 (Fig. 6a), suggesting a possible abrogation of silencers/negative regulatory elements by Tcf1 expression. Collectively, these analyses revealed that ectopic Tcf1 expression modulates the ChrAcc landscape in Tex cells by stabilizing/enforcing existing ChrAcc features in Tex-stem cells as well as exploiting novel ChrAcc sites.
Fig. 6.
Ectopic Tcf1 expression modulates Tex cell chromatin accessibility for target gene induction. a ATAC-seq and Tcf1 CUT&RUN tracks for select genes that were upregulated in the p45-Tg Tex-stem cells, with the same format as that in Fig. 5c. Heatmaps showing concordant (b) and discordant (c) ChrAcc changes associated with upregulated genes in the p45-Tg Tex-stem cells, with the same format as that in Fig. 5d, e
Tcf1 expression favors increasing ChrAcc for target gene regulation
To investigate potential regulatory mechanisms critical for the Diff ChrAcc sites upon ectopic Tcf1 expression, we performed a systematic motif analysis for all the Diff ChrAcc sites derived from the p45-Tg and WT Tex-stem cell comparison (a4–6 and b4–6 subclusters in Fig. 5). We defined an enrichment score by using the area under the receiver-operating (AUC) to quantify the association of each motif with Diff ChrAcc sites. In this method, motifs with enrichment scores over 0.5, as determined by the AUC, were considered to be preferably enriched in sites with increased ChrAcc, while those with scores under 0.5 were enriched in sites with decreased ChrAcc in the p45-Tg Tex-stem cells. Strikingly, the TCF7 and LEF1 motifs were the most enriched motifs associated with increased ChrAcc sites (Fig. 7a). It is also noteworthy that the NFKB1, NFKB2, RELB, and REL motifs were strongly associated with decreased ChrAcc sites, implying a possible scenario in which ectopic Tcf1 expression diminishes responsiveness to NF-κB pathway activation (Fig. 7a). By applying the same approach to all Diff ChrAcc sites derived from the WT Tex-stem and Tex-term cell comparison (a2, 3, 5 and b2, 3, 5 subclusters), we found that the TCF7 and LEF1 motifs remained the top 2 most enriched sequences in association with increased ChrAcc sites in the Tcf1+ Tex-stem cells (Fig. S6a). These data suggest that ectopic and endogenous Tcf1 expression (in the p45-Tg and WT Tex-stem cells, respectively) may have potent and direct roles in enforcing an open chromatin state and/or exploiting novel accessible sites.
Fig. 7.
Ectopic Tcf1 expression directly increases ChrAcc on a genome-wide scale and at DEG loci. a Motif analysis of Diff ChrAcc sites in the p45-Tg and WT Tex-stem cells. The top 10 enriched motifs associated with increased (red bars) and those with decreased (blue bars) Diff ChrAcc sites are displayed together with their motif logos. b Genome-wide mapping of Tcf1 binding locations in the p45-Tg Tex-stem cells using the CUT&RUN approach. Displayed are heatmap of all high-confidence Tcf1 peaks, centered on peak summits and their ±2 kb flanking sequences (left panel) and heatmap of ATAC-seq signals at corresponding genomic locations. c The top 10 motifs associated with increased and decreased ChrAcc sites (as shown in a) were assessed for the likelihood of direct binding by Tcf1 and their statistical significance, as displayed in volcano plots, where only motifs with positive log2 (odds ratio) values are labeled. The occurrence of a given motif among the corresponding Diff ChrAcc sites is denoted by circle size, and the occurrence of Tcf1 binding peaks at the corresponding Diff ChrAcc sites containing a given motif is denoted by circle colors. d Venn diagrams showing the co-occurrence of Tcf1 binding and Diff ChrAcc at the promoter or distal regions of the downregulated (left) and upregulated (right panel) genes derived from the p45-Tg and WT Tex-stem cell comparison. e Summation of the frequency of Tcf1 binding to Diff ChrAcc sites associated with DEGs, as quantitative presentation of data in Figs. 5d, e and 6b, c
To further develop this notion and discern a direct regulatory effect of Tcf1 in Tex cells, we employed the CUT&RUN approach to map global Tcf1 occupancies in the p45-Tg Tex-stem cells. By focusing the analysis on p45-Tg cells, we identified Tcf1 binding events that could be extrapolated to genome-wide binding sites occupied by endogenous Tcf1 in the WT Tex-stem cells as well as those newly acquired upon ectopic Tcf1 expression. By setting a fivefold enrichment and p value < 10–5 threshold in MACS algorithm,53 we identified 11,055 Tcf1 binding peaks with high confidence, with 54.8% distributed in the promoters and the others in distal regulatory regions. By stratifying these data with ATAC-seq profiles, we found that over 99% of Tcf1 binding peaks overlapped with ATAC-seq peaks detected in the p45-Tg Tex-stem cells (Fig. 7b), indicating that Tcf1-bound chromatin regions were mostly accessible. Stratifying the Tcf1 binding peaks with increased ChrAcc sites in the p45-Tg compared with the WT Tex-stem cells revealed that >25% of Tcf1 motif+ sites overlapped with high-confidence Tcf1 peaks (Fig. 7c). We used an odds-ratio score to quantify the co-occurrence of a motif and Tcf1 peak(s) at the Diff ChrAcc sites. This approach revealed that the TCF7 and LEF1 motif-associated sites with increased ChrAcc exhibited the highest likelihood of actual Tcf1 binding (Fig. 7c, left panel), supporting a direct role of Tcf1 in increasing ChrAcc in Tex cells. By applying this approach to other highly enriched motifs, shown in Fig. 7a, we demonstrated the likely association of Tcf1 binding to NANOG/SOX and GATA family TF motifs (Fig. 7c, left panel), suggesting potential cooperativity of Tcf1 with these factors. On the other hand, Tcf1 peaks were found in only <5% of the decreased ChrAcc sites containing the NF-κb family TF motifs, despite the reasonably strong positive odds ratios (Fig. 7c, right panel). These global analyses collectively indicate a preferable association of Tcf1 binding with increased over decreased ChrAcc sites, implying that a predominant effect of ectopic Tcf1 expression is to increase chromatin accessibility in Tex cells.
In addition to the global trends observed above, we focused on the DEGs derived from the p45-Tg and WT Tex-stem cell comparison. Tcf1 peaks were found in 26 and 17 promoters of the 100 downregulated and 74 upregulated genes in the p45-Tg Tex-stem cells, respectively (Fig. 7d). Tcf1 also bound to distal regulatory regions of the DEGs, with 171 sites associated with 80 downregulated genes, 129 sites associated with 59 upregulated genes, with a portion of these genes containing Tcf1 binding in both promoters and distal regions (Fig. 7d, gene lists in Fig. S6b, c, Tables S1 and S2). These data suggest that Tcf1 exerts a direct regulatory effect on more than 80% of the DEGs upon ectopic expression. Furthermore, some Tcf1 peaks overlapped with the Diff ChrAcc sites in both promoters and distal regions of the DEGs (Fig. 7d), and these Tcf1 peaks were more frequently found at increased ChrAcc sites in the p45-Tg Tex-stem cells, regardless of whether the associated genes were up- or downregulated in the p45-Tg Tex-stem cells (Fig. 7e, summation of Figs. 5d, e and 6b, c).
Specifically, for concordant changes between the gene expression and ChrAcc sites (as in Figs. 5d and 6b), Tcf1 peaks were less frequently observed for downregulated gene-associated and decreased ChrAcc sites in the p45-Tg Tex-stem cells (Figs. 5d and 7e), with the two upstream regions of Runx3 among the few examples (Fig. 5c). In contrast, Tcf1 binding was more frequently found at the upregulated gene-associated and increased ChrAcc sites (Figs. 6b and 7e); these events were observed upstream of Hopx, downstream of Mx1, and in introns of Tshz2 and Itga7 (Fig. 6a), suggesting that ectopic Tcf1 expression directly increases the ChrAcc at these sites to function as transcriptional enhancers.
Transcriptional silencers are known to be chromatin accessible. Indeed, strong ATAC-seq signals were detected in the Tex subsets for the well-characterized Cd4 silencer and a recently defined Foxp3 CNS0 element, where Tcf1 binds and contributes to silencing these genes in naïve CD8+ T cells31,54 (Fig. S6d). In line with this established concept, Tcf1 peaks were also frequently found at the increased ChrAcc sites associated with downregulated genes in the p45-Tg Tex-stem cells (Fig. 7e), such as those in the introns of Prdm1 and Tox2 and the upstream regions of Pdcd1 and Bcl2l11 (Fig. 5c). In such a gene context, these Tcf1-bound regions likely function as transcriptional repressors. These data collectively suggest that a predominant effect of ectopic Tcf1 expression is to increase chromatin accessibility at existing or novel sites for target gene activation or repression, solidifying the Tex-stem transcriptional program.
Ectopic Tcf1 expression enhances tumor control
Recent studies have revealed that CD8+ tumor-infiltrating lymphocytes (TILs) in both preclinical tumor models and patients are heterogeneous and contain self-renewing stem cell-like and more dysfunctional subsets.16,17,36,55,56 Similar to the Tex-stem cells elicited by chronic viral infection, stem cell-like CD8+ TILs (called Tex-stem TILs for consistency) exhibit a distinct Tim3loSlamf6+ phenotype and depend on Tcf1 for generation and persistence.15,16 To investigate the impact of ectopic Tcf1 expression on the CD8+ TILs, we adoptively transferred WT or p45-Tg P14 CD8+ T cells into CD45.1+ congenic recipient mice, followed by subcutaneous implantation of B16 melanoma cells expressing the LCMV GP33 epitope (B16-GP33)15,57,58 (Fig. 8a). When the tumors became palpable, usually 10 days after implantation, the recipients were given a therapeutic vaccine, i.e., GP33 peptide-keyhole limpet hemocyanin (GP33-KLH) conjugates27 mixed with poly(I:C) adjuvant (Fig. 8a). Consistent with previous reports,15,57 without vaccination, neither the WT nor p45-Tg P14 cells showed curtailed aggressive tumor growth; however, tumor growth was impeded in the vaccinated mice that received WT P14 cells and was further diminished and/or delayed in the vaccinated mice that received p45-Tg P14 cells (Fig. 8b).
Fig. 8.
Ectopic Tcf1 expression promotes Tex-stem features in the TILs exposed to the tumor microenvironment. a Experimental design. b Tracking tumor growth in the presence of the WT or p45-Tg P14 cells with or without GP33-KLH vaccination. c Total counts of P14 TILs at 14 days after vaccination. d–e Detection of Tim3loSlamf6+ Tex-stem cells in the P14 TILs. In representative contour plots in (d), the values denote percentages of Tex-stem TILs. Cumulative data on the frequency (left) and number (right) of the Tex-stem TILs are from three independent experiments (e). Detection of key proteins in the TILs, as determined by surface staining (f–g) or intranuclear staining (h–i). WT and p45-Tg P14 TILs were observed at 14 days after vaccination, and the Tim3hiSlamf6– Tex-term and Tim3loSlamf6+ Tex-stem TILs were analyzed for the indicated proteins. Representative half-stack histograms are shown in (f) and (h), with values denoting the gMFI. Cumulative data on the relative protein expression were from 2–3 experiments (g, i), with the gMFI of each protein in the WT Tex-stem TILs set at 1 and its relative expression in other subsets/genotypes normalized for each independent experiment. j–l Detection of cytokine production in TILs. Fourteen days after vaccination, WT or p45-Tg P14 TILs were surface stained to identify the CD39hiSlamf6− Tex-term and CD39loSlamf6+ Tex-stem cells and then intracellularly stained to detect the indicated cytokines. Representative contour plots are shown in (j), and cumulative data on the number of IFN-γ− or polycytokine-producing Tex-stem TILs (k) and total TILs (l) are from three independent experiments
In line with reduced tumor sizes, the number of p45-Tg P14 TILs was significantly increased compared with that of the WT P14 TILs after therapeutic vaccination (Fig. 8c). Compared with WT P14 TILs, the p45-Tg P14 TILs showed clear increase in frequency and numbers of Tim3loSlamf6+ Tex- stem cells (Fig. 8d, e). Similar to what was observed for the WT and p45-Tg Tex-stem cells elicited by chronic viral infection, the expression of PD-1 and 2B4 coinhibitory receptors and the T-bet transcription factor was lower in the WT Tex-stem TILs than in the Tex-term TILs and was further reduced in the p45-Tg Tex-stem TILs (Fig. 8f–i). On the other hand, the WT and p45-Tg Tex-stem TILs had some features that differed from their LCMV-Cl13-elicited Tex counterparts, showing progressively diminished expression of LAG-3 and Tox, without detectable impact on Eomes (compare Fig. 8f–i with 2a–d). In contrast to the TILs, the WT and p45-Tg P14 cells in the secondary lymphoid organs did not show elevated expression of coinhibitory receptors such as Tim3, PD-1, 2B4, or LAG-3 with or without the vaccination (Fig. S7), and such phenomena are likely due to their minimal exposure to persisting antigens outside the tumor microenvironment. Consistent with a previous report,15 polycytokine production was better retained in WT Tex-stem than in the Tex-term TILs and was further enhanced in the p45-Tg Tex-stem TILs (Fig. 8j). As a consequence, we observed a >10-fold increase in the number of polyfunctional p45-Tg TILs compared with the WT Tex-stem TILs, as well as a substantial increase in polyfunctional total P14 TILs (Fig. 8k, l). Collectively, these observations demonstrate that ectopic Tcf1 expression has the capacity to enforce Tex-stem transcriptional programs in CD8+ TILs and hence improve tumor control.
Discussion
Recent advances reveal the heterogeneity of exhausted CD8+ T cells that have been exposed to persistent antigen stimulation, where the Tex-stem subset shows stem/progenitor cell-like features and enhanced responsiveness to checkpoint blockade therapy.5,12,18 Expanding Tex-stem cells has clear therapeutic value in enhancing viral and tumor immunity; however, a durable impact is infrequently achieved by checkpoint blockade due to epigenetic stability of Tex cells.8,9 In this study, we demonstrated that ectopic Tcf1 expression not only skewed most Tex cells to the Tex-stem phenotype with a numerical increase but also showed a pronounced impact on global chromatin accessibility and the transcriptome in Tex cells. These ‘Tcf1-enhanced’ cells exhibited prolonged survival and enhanced functionality on a per cell basis, resulting in better control of chronic viral infection and preclinical tumors. Importantly, Tex-stem cells with ectopic Tcf1 expression retained their responsiveness to anti-PD-L1 treatment despite decreased expression of PD-1. These findings highlight the Tcf1-enforced transcriptional program as an attractive therapeutic target, alone or in combination with checkpoint blockade, to durably expand cells with stem cell-like features.
Several previous studies used retrovirus to deliver the Tcf7 gene into preprimed CD8+ T cells followed by in vivo transfer and observed an increased frequency of Tex-stem precursors or cells with some Tex-stem markers at the early stage of response to chronic viral infection.26,35,37 Despite the early divergence of Tex-stem and Tex-term fate decisions, as recently suggested,24,35 Tex cells do not manifest dysfunction at this early stage. In this work, we used a genetic approach to stably express Tcf1, which facilitated investigation of its impact on Tex cells at advanced stages of exhaustion, longitudinal tracking of Tex cell persistence, and evaluation of conserved beneficial effects in tumor-infiltrating Tex cells. Among the advantages, for example, we consistently observed repression of PD-1 by transgenic Tcf1 in Tex-stem cells in both chronic viral infection and tumor models; this effect, however, was not evident in Tex cells with retroviral Tcf1 expression at day 22 post infection,35 which might be due to insufficient elevation of Tcf1 protein and/or silencing of the retrovirally delivered Tcf7 gene during extensive CD8+ T cell propagation. Inhibitors of glycogen synthesis kinase 3β (GSK-3β), such as TWS119 and BIO, have been used to stabilize β-catenin in CD8+ T cells. When used in vitro, TWS119 caused transient induction of Tcf1, which peaked at 8 h and then returned to basal levels within one day but arrested CD8+ T cell division and IFN-γ production.59 A more recent study testing BIO in vivo demonstrated a modest induction of Tcf1 and enhanced IFN-γ production in early Tex cells without suppressing cell proliferation.37 Notably, neither retrovirally delivered Tcf1 nor GSK-3β inhibitor-induced Tcf1 conferred numerical advantages to the targeted CD8+ T cell population. The revelation that a stably expressed Tcf1 expands the Tex-stem pool with evident functional restoration may constitute a reasonably strong rationale for therapeutic considerations.
Stable expression of Tcf1 by a transgene also facilitated in-depth mechanistic investigations. Naturally occurring WT Tex-stem cells expressed higher levels of Tcf1 than Tex-term cells. It is therefore not surprising that ectopic Tcf1 expression enforced existing transcriptomic and chromatin accessibility changes in WT Tex-stems over Tex-term cells. Tex-stem (p45-Tg or WT) and Tex-term cell comparisons revealed strong diversification of the transcriptome and ChrAcc landscape,34,60 reflecting the impact by the presence versus absence of Tcf1 expression in these subsets. On the other hand, the p45-Tg and WT Tex-stem cell comparisons represent the impact by ectopic versus endogenous Tcf1 expression, and it is therefore not unexpected that these two subsets had smaller numbers of differential gene expression and ChrAcc sites. It should be noted, however, that Tcf1 exploited novel ChrAcc sites and new downstream genes upon ectopic expression. In fact, the ChrAcc and transcriptomic changes, enforced or newly acquired through ectopic Tcf1 expression, showed meaningful links to Tex cell biology, as also manifested in phenotypic and functional changes of Tex-stem cells in vivo. In-depth data mining further showed that a predominant effect of ectopic Tcf1 expression was to increase chromatin opening at existing and novel sites, which could function as positive or negative regulatory elements. In line with our findings, it was reported that forced expression of Tcf1 in bone marrow progenitors and even fibroblasts increased T-lineage gene expression and chromatin openings at associated gene loci, hence promoting T cell fate commitment.61,62 These observations collectively point to an interesting capacity of ectopic Tcf1 expression to reprogram target cell types to confer favorable T cell-associated functional output.
In addition to a direct impact on chromatin accessibility, ectopically expressed Tcf1 may employ multiple mechanisms in directing Tex cells to a Tex-stem fate. We observed direct Tcf1 binding to the promoter regions of DEGs, and these Tcf1-bound promoters were not always (in fact, they were quite infrequently) associated with chromatin accessibility changes. At the promoter regions, Tcf1 may coopt coactivator or corepressor proteins to modulate the stability and/or activity of the basic transcriptional machinery complex to control transcriptional output. β-catenin is a known coactivator for Tcf/Lef TFs, and we previously reported that ectopic expression of both p45 Tcf1 and constitutively active β-catenin has synergistic effects on expanding the memory CD8+ T cell pool in response to acute infection by L. monocytogenes39; however, this effect was not observed in the Tex cells, especially Tex-stems, elicited by chronic viral infection. It is now recognized that Tex cells represent a molecularly and functionally cell lineage distinct from effector or memory CD8+ T cells generated in response to acute infections. The differences in molecular circuitry may have thus determined their sensitivity to the exogenous input of active β-catenin. In the context of follicular helper T cell differentiation that is elicited by acute infection, we recently discovered that Ezh2, in its phosphorylated form at the serine 21 residue, functions as a coactivator of Tcf1 to induce the expression of Bcl6 and Icos.63 For target gene repression, Tcf1 utilizes intrinsic histone deacetylase activity and/or cooperates with Groucho/Tle corepressors to establish CD8+ T cell identity during thymic development.31,64 The interplay of Tcf1 with its cooperating pathways warrants in-depth molecular dissection in Tex-stem cells to maximize its therapeutic potential.
Tex cells, generated in response to chronic viral infection or in the tumor microenvironment, share similar functional impairments and, to a certain extent, similar molecular signatures.17 For example, the expression of Tcf1, Eomes, and Tox transcription factors largely showed concordant changes starting from the early stage of response to chronic viral infection.21,22,35 We observed higher expression of Tox and Eomes in naturally occurring Tcf1+ Tex-stem cells than in their Tcf1– Tex-term counterparts elicited by chronic viral infection. On the other hand, distinct molecular wiring between antiviral and TIL Tex cells has also been recognized.5 In an inducible liver cancer model, Tcf1 expression in TILs was negatively correlated with that of Tox.22 In line with this result, we found that Tcf1+ WT Tex-stem TILs expressed lower amounts of Tox but similar levels of Eomes compared with Tex-term TILs. Despite these differences, ectopically expressed Tcf1 invariably repressed Tox and a few coinhibitory receptors in both antiviral and TIL Tex-stem cells, and these changes appeared to be compatible with a favorable functional output in both model systems. Although using transgenic expression in human T cells is not a feasible approach, the functional and molecular advantages of ectopic Tcf1 expression revealed in this work imply that the Tcf1-enforced transcriptional program could hold the key to broadly boosting antiviral and antitumor immunity. With the advent of CRISPR/Cas9 technology, the genome of human cells is more amenable to manipulation, and one can envision a site-directed insertion of the Tcf1 expression cassette into a ubiquitously expressed gene locus such as Rosa26 to achieve stable expression without involving viral components.
Accession numbers
The RNA-seq, ATAC-seq and Tcf1 CUT&RUN data are deposited at the Gene Expression Omnibus under SuperSeries GSE 139058.
Supplementary information
Acknowledgements
We thank Werner Held (University of Lausanne, Switzerland) and Zuoming Sun (City of Hope) for providing p45 Tcf1 transgenic and β-catenin transgenic mice, respectively; Ananda Goldrath (UCSD) for sharing the B16-GP33 melanoma cells; Jian Zhang (U of Iowa) for sharing TCRα–/– mice; the University of Iowa Flow Cytometry Core facility for cell sorting, the Genomics Division of Iowa Institute of Human Genetics and Admera Health for next-generation sequencing. This study was supported by grants from the NIH (AI112579, AI121080 and AI139874 to H.-H.X., GM133712 to C.Z., and GM113961, AI147064 and AI114543 to V.P.B.), and the Veteran Affairs BLR&D Merit Review Program (BX002903) to H.-H.X.
Author contributions
Q.S., with assistance from X.C. and D.B.D., performed the experiments and analyzed the data; S.H. analyzed the high-throughput sequencing data under the supervision of C.Z.; V.P.B., C.Z., and H.H.X. designed and supervised the study.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Qiang Shan, Sheng’en Hu
Contributor Information
Chongzhi Zang, Email: zang@virginia.edu.
Hai-Hui Xue, Email: haihui.xue@hmh-cdi.org.
Supplementary information
The online version of this article (10.1038/s41423-020-0436-5) contains supplementary material.
References
- 1.Omilusik KD, Goldrath AW. Remembering to remember: T cell memory maintenance and plasticity. Curr. Opin. Immunol. 2019;58:89–97. doi: 10.1016/j.coi.2019.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Youngblood B, Hale JS, Ahmed R. Memory CD8 T cell transcriptional plasticity. F1000Prime Rep. 2015;7:38. doi: 10.12703/P7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin MD, Badovinac VP. Defining memory CD8 T cell. Front. Immunol. 2018;9:2692. doi: 10.3389/fimmu.2018.02692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLane LM, Abdel-Hakeem MS, Wherry EJ. CD8 T Cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 2019;37:457. doi: 10.1146/annurev-immunol-041015-055318. [DOI] [PubMed] [Google Scholar]
- 5.Blank CU, et al. Defining ‘T cell exhaustion’. Nat. Rev. Immunol. 2019;19:665. doi: 10.1038/s41577-019-0221-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel SA, Minn AJ. Combination cancer therapy with immune checkpoint blockade: mechanisms and strategies. Immunity. 2018;48:417–433. doi: 10.1016/j.immuni.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribas A, Wolchok JD. Cancer immunotherapy using checkpoint blockade. Science. 2018;359:1350–1355. doi: 10.1126/science.aar4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pauken KE, et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 2016;354:1160–1165. doi: 10.1126/science.aaf2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sen DR, et al. The epigenetic landscape of T cell exhaustion. Science. 2016;354:1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doering TA, et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity. 2012;37:1130–1144. doi: 10.1016/j.immuni.2012.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philip M, et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 2017;545:452–456. doi: 10.1038/nature22367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Im SJ, et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537:417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He R, et al. Follicular CXCR5- expressing CD8(+) T cells curtail chronic viral infection. Nature. 2016;537:412–428. doi: 10.1038/nature19317. [DOI] [PubMed] [Google Scholar]
- 14.Leong YA, et al. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 2016;17:1187–1196. doi: 10.1038/ni.3543. [DOI] [PubMed] [Google Scholar]
- 15.Siddiqui I, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 2019;50:195–211 e110. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 16.Kurtulus S, et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1(−)CD8(+) tumor-infiltrating T cells. Immunity. 2019;50:181–194 e186. doi: 10.1016/j.immuni.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller BC, et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 2019;20:326–336. doi: 10.1038/s41590-019-0312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kallies A, Zehn D, Utzschneider DT. Precursor exhausted T cells: key to successful immunotherapy? Nat. Rev. Immunol. 2019;20:128. doi: 10.1038/s41577-019-0223-7. [DOI] [PubMed] [Google Scholar]
- 19.Martinez GJ, et al. The transcription factor NFAT promotes exhaustion of activated CD8(+) T cells. Immunity. 2015;42:265–278. doi: 10.1016/j.immuni.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Man K, et al. Transcription factor IRF4 promotes CD8(+) T cell exhaustion and limits the development of memory-like t cells during chronic infection. Immunity. 2017;47:1129–1141 e1125. doi: 10.1016/j.immuni.2017.11.021. [DOI] [PubMed] [Google Scholar]
- 21.Alfei F, et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 2019;571:265–269. doi: 10.1038/s41586-019-1326-9. [DOI] [PubMed] [Google Scholar]
- 22.Scott AC, et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 2019;571:270–274. doi: 10.1038/s41586-019-1324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan O, et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature. 2019;571:211–218. doi: 10.1038/s41586-019-1325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao C, et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8(+) T cell persistence in chronic infection. Nat. Immunol. 2019;20:890–901. doi: 10.1038/s41590-019-0403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo H, et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc. Natl Acad. Sci. USA. 2019;116:12410–12415. doi: 10.1073/pnas.1905675116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu T, et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 2016;1:eaai8593. doi: 10.1126/sciimmunol.aai8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shan Q, et al. The transcription factor Runx3 guards cytotoxic CD8(+) effector T cells against deviation towards follicular helper T cell lineage. Nat. Immunol. 2017;18:931–939. doi: 10.1038/ni.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinke FC, Xue HH. From inception to output, Tcf1 and Lef1 safeguard development of T cells and innate immune cells. Immunol. Res. 2014;59:45–55. doi: 10.1007/s12026-014-8545-9. [DOI] [PubMed] [Google Scholar]
- 29.Xue HH, Zhao DM. Regulation of mature T cell responses by the Wnt signaling pathway. Ann. N.Y. Acad. Sci. 2012;1247:16–33. doi: 10.1111/j.1749-6632.2011.06302.x. [DOI] [PubMed] [Google Scholar]
- 30.Raghu D, Xue HH, Mielke LA. Control of lymphocyte fate, infection, and tumor immunity by TCF-1. Trends Immunol. 2019;40:1149–1162. doi: 10.1016/j.it.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 31.Xing S, et al. Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat. Immunol. 2016;17:695–703. doi: 10.1038/ni.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, et al. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity. 2010;33:229–240. doi: 10.1016/j.immuni.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeannet G, et al. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc. Natl Acad. Sci. USA. 2010;107:9777–9782. doi: 10.1073/pnas.0914127107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Utzschneider DT, et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity. 2016;45:415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. 2019;51:840–855 e845. doi: 10.1016/j.immuni.2019.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sade-Feldman M, et al. Defining T cell states associated with response to checkpoint immunotherapy in Melanoma. Cell. 2018;175:998–1013 e1020. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, et al. The transcription factor TCF1 preserves the effector function of exhausted CD8 T cells during chronic viral infection. Front. Immunol. 2019;10:169. doi: 10.3389/fimmu.2019.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 39.Zhao DM, et al. Constitutive activation of Wnt signaling favors generation of memory CD8 T cells. J. Immunol. 2010;184:1191–1199. doi: 10.4049/jimmunol.0901199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Z, et al. Cutting edge: beta-catenin-Interacting Tcf1 isoforms are essential for thymocyte survival but dispensable for thymic maturation transitions. J. Immunol. 2017;198:3404–3409. doi: 10.4049/jimmunol.1602139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gullicksrud JA, et al. Differential requirements for Tcf1 long isoforms in CD8(+) and CD4(+) T cell responses to acute viral infection. J. Immunol. 2017;199:911–919. doi: 10.4049/jimmunol.1700595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ioannidis V, Beermann F, Clevers H, Held W. The beta-catenin–TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat. Immunol. 2001;2:691–697. doi: 10.1038/90623. [DOI] [PubMed] [Google Scholar]
- 43.Xie H, Huang Z, Sadim MS, Sun Z. Stabilized beta-catenin extends thymocyte survival by up-regulating Bcl-xL. J. Immunol. 2005;175:7981–7988. doi: 10.4049/jimmunol.175.12.7981. [DOI] [PubMed] [Google Scholar]
- 44.Yang Q, et al. TCF-1 upregulation identifies early innate lymphoid progenitors in the bone marrow. Nat. Immunol. 2015;16:1044–1050. doi: 10.1038/ni.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paley MA, et al. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 2012;338:1220–1225. doi: 10.1126/science.1229620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wherry EJ. T cell exhaustion. Nat. Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 47.Yu S, et al. Hematopoietic and leukemic stem cells have distinct dependence on Tcf1 and Lef1 transcription factors. J. Biol. Chem. 2016;291:11148–11160. doi: 10.1074/jbc.M116.717801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin H, et al. A role for the transcriptional repressor Blimp-1 in CD8(+) T cell exhaustion during chronic viral infection. Immunity. 2009;31:309–320. doi: 10.1016/j.immuni.2009.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou S, Cerny AM, Fitzgerald KA, Kurt-Jones EA, Finberg RW. Role of interferon regulatory factor 7 in T cell responses during acute lymphocytic choriomeningitis virus infection. J. Virol. 2012;86:11254–11265. doi: 10.1128/JVI.00576-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shin HM, et al. Transient expression of ZBTB32 in anti-viral CD8+ T cells limits the magnitude of the effector response and the generation of memory. PLoS Pathog. 2017;13:e1006544. doi: 10.1371/journal.ppat.1006544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Conze D, et al. Activation of p38 MAP kinase in T cells facilitates the immune response to the influenza virus. Mol. Immunol. 2000;37:503–513. doi: 10.1016/s0161-5890(00)00078-x. [DOI] [PubMed] [Google Scholar]
- 53.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinke FC, et al. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat. Immunol. 2014;15:646–656. doi: 10.1038/ni.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, et al. Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. 2019;176:775–789 e718. doi: 10.1016/j.cell.2018.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brummelman J, et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8(+) T cells infiltrating human tumors. J. Exp. Med. 2018;215:2520–2535. doi: 10.1084/jem.20180684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Danahy DB, et al. Sepsis-induced state of immunoparalysis is defined by diminished CD8 T cell-mediated antitumor immunity. J. Immunol. 2019;203:725–735. doi: 10.4049/jimmunol.1900435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Milner JJ, et al. Runx3 programs CD8(+) T cell residency in nonlymphoid tissues and tumours. Nature. 2017;552:253–257. doi: 10.1038/nature24993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gattinoni L, et al. Wnt signaling arrests effector T cell differentiation and generates CD8+ memory stem cells. Nat. Med. 2009;15:808–813. doi: 10.1038/nm.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jadhav RR, et al. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc. Natl Acad. Sci. USA. 2019;116:14113–14118. doi: 10.1073/pnas.1903520116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weber BN, et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature. 2011;476:63–68. doi: 10.1038/nature10279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnson JL, et al. Lineage-determining transcription factor TCF-1 initiates the epigenetic identity of T Cells. Immunity. 2018;48:243–257 e210. doi: 10.1016/j.immuni.2018.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li F, et al. Ezh2 programs TFH differentiation by integrating phosphorylation-dependent activation of Bcl6 and polycomb-dependent repression of p19Arf. Nat. Commun. 2018;9:5452. doi: 10.1038/s41467-018-07853-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xing S, et al. Tle corepressors are differentially partitioned to instruct CD8(+) T cell lineage choice and identity. J. Exp. Med. 2018;215:2211–2226. doi: 10.1084/jem.20171514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.