Abstract
F-box proteins β-TrCP1 and β-TrCP2 are paralogs present in the human genome. They control several cellular processes including cell cycle and DNA damage signaling. Moreover, it is reported that they facilitate DNA damage-induced accumulation of p53 by directing proteasomal degradation of MDM2, a protein that promotes p53 degradation. However, the individual roles of β-TrCP1 and β-TrCP2 in the genotoxic stress-induced activation of cell cycle checkpoints and DNA damage repair remain largely unknown. Here, using biochemical, molecular biology, flow cytometric, and immunofluorescence techniques, we show that β-TrCP1 and β-TrCP2 communicate during genotoxic stress. We found that expression levels of β-TrCP1 are significantly increased while levels of β-TrCP2 are markedly decreased upon induction of genotoxic stress. Further, our results revealed that DNA damage-induced activation of ATM kinase plays an important role in maintaining the reciprocal expression levels of β-TrCP1 and β-TrCP2 via the phosphorylation of β-TrCP1 at Ser158. Phosphorylated β-TrCP1 potently promotes the proteasomal degradation of β-TrCP2 and MDM2, resulting in the activation of p53. Additionally, β-TrCP1 impedes MDM2 accumulation via abrogation of its lysine 63-linked polyubiquitination by β-TrCP2. Thus, β-TrCP1 helps to arrest cells at the G2/M phase of the cell cycle and promotes DNA repair upon DNA damage through attenuation of β-TrCP2. Collectively, our findings elucidate an intriguing posttranslational regulatory mechanism of these two paralogs under genotoxic stress and revealed β-TrCP1 as a key player in maintaining the genome integrity through the attenuation of β-TrCP2 levels in response to genotoxic stress.
Keywords: DNA damage, p53, polyubiquitination, cell cycle
Abbreviations: β-TrCP, β-transducin repeat-containing protein; Cul1, Cullin1; NS, non-silencing; SCF complex, SKP1, Cul1 and F-box protein; UPS, ubiquitin-proteasome system
The ubiquitin-proteasome system (UPS) is mainly responsible for ubiquitination of proteins. Ubiquitination is an important posttranslational modification and is a process of conjugation of ubiquitin to protein (1). Ubiquitination of proteins is carried out through three major steps by a sequential activity of three classes of enzymes. In the first step, ubiquitin is activated by ubiquitin-activating enzyme E1 in the presence of ATP. The activated ubiquitin is then transferred to the ubiquitin-conjugating enzyme E2, and finally the activated ubiquitin is transferred to the substrate proteins through E3 ubiquitin ligase enzyme (2).
SCF E3 ubiquitin ligases are the largest class of E3 ubiquitin ligases. They play a crucial role in cell cycle progression, cell proliferation, DNA damage, apoptosis, and many other cellular functions (3). SCF complex is a multisubunit complex comprising three invariable components SKP1, Cullin1 (Cul1), Rbx1, and a variable component F-box protein (4). F-box protein β-transducin repeat-containing protein (β-TrCP) is a component of SCF (SKP1, Cul1, and F-box protein) E3 ubiquitin ligase complex. It is well established that β-TrCP recognizes substrates having consensus sequence DSGXXS degron to promote their proteasomal degradation (5). In the human genome, β-TrCP has two paralogs (namely β-TrCP1 and β-TrCP2) (6). Previous study showed that β-TrCP1 knockout mice are viable with reduced fertility (7). It is also reported that MEFs from β-TrCP1 knockout mice display mitotic abnormalities with centrosome overduplication and chromosome misalignment indicating that it plays important role in progression of mitosis (7). In contrast, β-TrCP2 knockout mice died before embryonic day 10.5 indicating that these two paralogs have distinct cellular function (8).
Several studies have shown that cellular functions of β-TrCP1 and β-TrCP2 are indistinguishable and therefore maximum studies cited them as β-TrCP (5, 9, 10). Hence, it was difficult to understand whether the study is related to β-TrCP1 or β-TrCP2. β-TrCP1 and β-TrCP2 form homo and heterodimer by utilizing their dimerization domain; however, homodimer is more functional in promoting the degradation of its substrate (11). β-TrCP is known to regulate many cellular processes such as NF-κB signaling (12), β-catenin signaling (13), TGF-β signaling (10), apoptosis (14), autophagy (15), and cell cycle (9, 16).
β-TrCP is involved in diseases such as cancer. It was reported that β-TrCP can function as either tumor suppressor or oncogene or both in a context-dependent manner (5, 9, 17). A recent study showed that β-TrCP1 may function as tumor suppressor and β-TrCP2 as an oncogene (18). Generally, perturbation of function of tumor suppressor and/or oncogene results in impairment of DNA damage response/repair that gives rise to chromosomal instability and cancer predisposition (19, 20). Previous studies showed that β-TrCP plays important roles in DNA damage response and maintains the genome integrity by controlling multiple substrates (17, 21, 22, 23). For instance, β-TrCP promotes CReP degradation upon DNA damage to shut down the translational machinery through inactivation of eIF2α (24). It is also reported that β-TrCP downregulates Cdc25A to maintain intra-S-phase checkpoint (16) while SCFβTrCP-mediated Claspin degradation is important for checkpoint recovery during genotoxic stress (25). Recent studies showed that β-TrCP directs the degradation of MDM2 in response to DNA damage to initiate DNA damage response pathway through activation of p53 (21, 22); however, it is not clear whether β-TrCP1 or β-TrCP2 is involved in DNA damage-induced activation of p53. β-TrCP also promotes the degradation of pro-caspase-3 to protect the cells from DNA damage-induced apoptotic cell death (26). Though numerous studies have been done to understand the role of β-TrCP in DNA damage response; however, distinct role of individual β-TrCP1 and β-TrCP2 in DNA damage response and repair is poorly understood. Our study established that tumor suppressor β-TrCP1 promotes the degradation of β-TrCP2 following genotoxic stress to facilitate the activation of cell cycle checkpoint and DNA damage repair to maintain the genomic stability.
Results
β-TrCP1 is accumulated, whereas β-TrCP2 is declined at the protein level upon genotoxic stresses
β-TrCP functions as a tumor suppressor/oncogene in a context-dependent manner. Tumor suppressor as well as oncogene plays key role during genotoxic stress. Several studies showed that β-TrCP plays a crucial role in DNA damage response. However, during genotoxic stress, role of β-TrCP1 and β-TrCP2 at the individual level is largely unknown. Toward this, first we examined the levels of β-TrCP1 and β-TrCP2 following exposure to genotoxic stress by either radiation (UV and IR) or oxidative stress (H2O2) in MCF7 cells. Interestingly, we observed that β-TrCP1 levels were significantly increased while β-TrCP2 levels were markedly declined following DNA damage induction (Fig. 1A). Similar results were also observed in HCT116 cells, indicating that DNA damage-induced differential expression of β-TrCP1 and β-TrCP2 is a general phenomenon (Fig. 1B). Interestingly, the converse correlation of their expression was observed in single-strand break (UV radiation and H2O2) and double-strand break (ionizing radiation) DNA damage induction, indicating that the observed phenomenon is also not specific to a particular type of DNA damage (Fig. 1, A and B). To further support our observation, we examined the levels of β-TrCP1 and β-TrCP2 following exposure to different doses of IR. Results showed a dose-dependent ablation of β-TrCP2 levels with concomitant increased levels of β-TrCP1 (Fig. 1, C and D).
Figure 1.
Expression levels of β-TrCP1 are increased while expression levels of β-TrCP2 are decreased following genotoxic stresses.A, MCF7 cells were treated with 5 Gy ionizing radiation (IR) for 4 h or 10 mJ/m2 UV for 4 h or 0.05% H2O2 for 2 h. Whole-cell protein lysates (WCL) were immunoblotted for the indicated proteins. p53 was used as positive control for induction of DNA damage and tubulin was used as loading control. Data are representative of three independent experiments. B, HCT116 cells were treated with 5 Gy ionizing radiation (IR) for 4 h or 10 mJ/m2 UV for 4 h or 0.05% H2O2 for 2 h. WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. C, MCF7 cells were treated with increasing doses of IR for 4 h. WCL were immunoblotted for the indicated proteins. Data are representative of three independent experiments. D, HCT116 cells were treated with increasing doses of IR for 4 h. WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. E, MCF7 cells were treated with 5 Gy IR and cells were collected at the indicated time of post IR and WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. F, MCF7 cells were treated with 5 Gy IR and cells were collected at the indicated time of post IR for immunofluorescence analysis using β-TrCP1 and β-TrCP2 antibodies. Data are representative of three independent experiments. G, quantification of expression levels of indicated proteins as in Figure 1F. Random fields were selected and the fluorescence intensity was measured using Image J software.
To prevent the deleterious effects of DNA damage and to maintain the genome integrity, cells attempt to repair their genome. We therefore asked whether there is any correlation of differential expression of β-TrCP1 and β-TrCP2 during DNA damage response and repair. To address this, first we examined the expression levels of β-TrCP1 and β-TrCP2 for a period of 24 h post irradiation. Immunoblotting results showed that levels of β-TrCP1 were increased with decreased levels of β-TrCP2 till 6 h post IR and then β-TrCP1 was decreasing with concomitant increasing levels of β-TrCP2 to restore their cellular levels at 24 h post irradiation (Fig. 1E). To further support this observation, we performed immunofluorescence study. In agreement with the immunoblotting data, immunofluorescence results also showed a converse correlation in the expression levels of β-TrCP1 and β-TrCP2 following genotoxic stress (Fig. 1, F and G). Collectively, our results demonstrated that β-TrCP1 and β-TrCP2 levels are differentially regulated upon DNA damage.
DNA damage-induced differential regulation of β-TrCP1 and β-TrCP2 is at the proteasomal level
Next, we were interested to understand the mechanism of the genotoxic-stress-induced differential expression of β-TrCP. The observed differential expression of β-TrCP1 and β-TrCP2 following genotoxic stresses could be either at the transcriptional level or at the posttranslational level. We, therefore, first monitored the mRNA levels of β-TrCP1 and β-TrCP2 following DNA damage. The real-time RT-PCR results showed that mRNA levels of β-TrCP1 and β-TrCP2 remained unaltered following genotoxic stress (Fig. 2, A and B), indicating that DNA damage-induced differential expression of β-TrCP1 and β-TrCP2 could be at the posttranscriptional level. To explore this, we have irradiated cells in the absence and presence of proteasomal inhibitor MG132. Immunoblotting results showed that IR-induced ablation of β-TrCP2 was markedly blocked following inhibition of proteasome by MG132 at 4 h post irradiation (Fig. 2C). Interestingly, ablation of β-TrCP1 was also significantly inhibited following treatment of MG132 at 24 h post irradiation (Fig. 2C). In addition, we performed cycloheximide chase assay and results revealed that half-life of β-TrCP2 is shortened following genotoxic stress (Fig. 2, D and E). As expected, the half-life of β-TrCP1 was extended following irradiation (Fig. 2, D and E). To further authenticate the proteasomal regulation of β-TrCP2, we examined the polyubiquitinated levels of β-TrCP2 following genotoxic stress. Immunoblotting of immunoprecipitates showed that the levels of polyubiquitinated β-TrCP2 were significantly increased at 4 h post irradiation (Fig. 2F). Collectively, these results suggest that β-TrCP2 undergoes proteasomal degradation at the early time point of genotoxic stress induction.
Figure 2.
DNA damage-induced alteration of β-TrCP1 and β-TrCP2 expression levels occurs through proteasome mediated pathway.A and B, MCF7 cells were treated with 5 Gy IR as indicated and cell were collected at 4 h post radiation exposure. Then, total RNA was extracted using TRIzol reagent, cDNA prepared followed by real-time RT-PCR to examine the mRNA levels of β-TrCP1(A)/β-TrCP2(B). Data are representative of three independent experiments. C, MCF7 cells were treated with 5 Gy IR as indicated and cells were collected at the indicated time point of post IR in the absence or presence of MG132. WCL were immunoblotted for the indicated proteins. Data are representative of three independent experiments. D, MCF7 cells were exposed to 5 Gy IR and allowed to grow for 1 h. Then, cycloheximide (40 μg/ml) was added to control and irradiated cells for the indicated time. WCL were immunoblotted for the indicated proteins. Data are representative of three independent experiments. E, quantification of levels of β-TrCP1 and β-TrCP2 as in Figure 2D. Protein levels were quantified using Image J software. Expression levels of β-TrCP1 and β-TrCP2 were normalized with tubulin and then set to 1 for β-TrCP1 and β-TrCP2 at 0 h. The levels at other time points were calculated with respect to 0 h. F, MCF7 cells expressing His-ubiquitin were exposed to 5 Gy IR and irradiated cells were grown for indicated time periods. WCL were pulled down with Ni-NTA beads. Pulled fractions and inputs WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. CHX, cycloheximide.
β-TrCP1 directs polyubiquitination-mediated degradation of β-TrCP2 upon induction of DNA damage
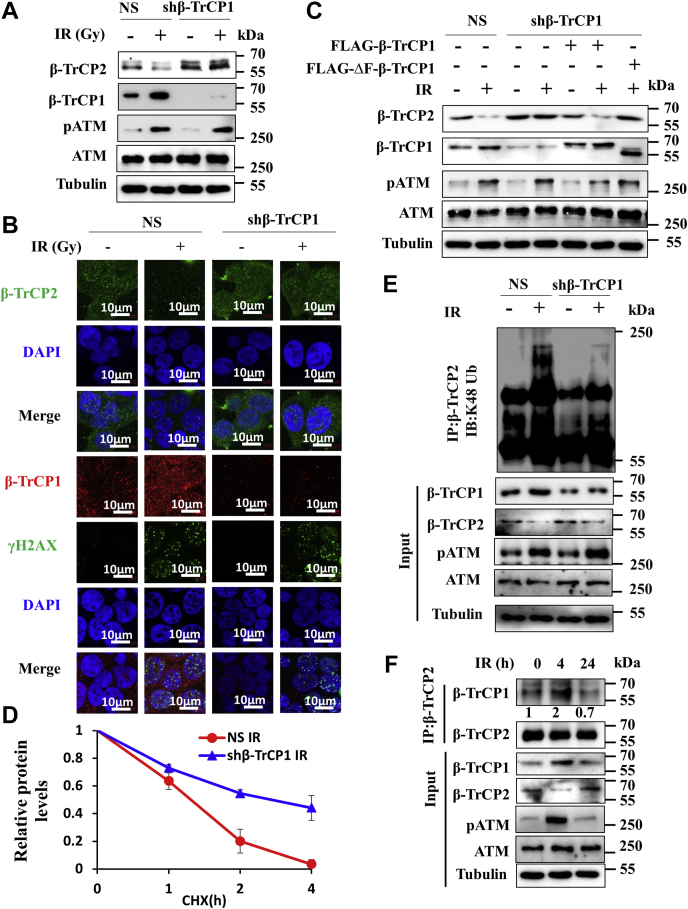
The preceding results demonstrate that β-TrCP1 is accumulated and β-TrCP2 is degraded upon induction of DNA damage (Figs. 1 and 2). A previous study showed that β-TrCP1 and β-TrCP2 regulate each other at the proteasomal level (18). We therefore posit that β-TrCP1 might be involved in degradation of β-TrCP2 following induction of DNA damage. To test this, β-TrCP2 levels were monitored in MCF7 cells expressing either NS (non-silencing shRNA) or β-TrCP1 shRNA following exposure to IR. Immunoblotting results revealed that DNA damage-induced β-TrCP2 ablation was markedly blocked in β-TrCP1 knockdown cells, indicating that β-TrCP1 plays an important role in proteasomal attenuation of β-TrCP2 upon induction of DNA damage (Fig. 3A). To support our observation, we examined the levels of β-TrCP2 in HCT116 cells following depletion of β-TrCP1. Results revealed that DNA damage-induced degradation of β-TrCP2 was significantly blocked in β-TrCP1-depleted HCT116 cells (Fig. S1A). To support our immunoblotting results, we performed immunofluorescence study. In agreement with the immunoblotting results, we found that DNA damage-induced ablation of TrCP2 is markedly blocked in β-TrCP1 depleted cells (Fig. 3B and Fig. S1B). To further authenticate the involvement of β-TrCP1 in DNA damage-induced ablation of β-TrCP2, we examined the levels of β-TrCP2 following ectopic expression of the wild type and the F-box motif deleted β-TrCP1 mutant (ΔF-β-TrCP1) in β-TrCP1-depleted cells. Immunoblotting results showed that DNA damage-induced ablation of β-TrCP2 in β-TrCP1 depleted cells was resumed following ectopic expression of the wild-type β-TrCP1 but not with the mutant ΔF-β-TrCP1 (Fig. 3C). Next we examined how the regulation of β-TrCP2 by β-TrCP1 is happening through SCF complex under normal condition. We found that, unlike β-TrCP1, ΔF-β-TrCP1 could not promote the degradation of β-TrCP2 (Fig. S1C). Taken together, these results suggest that β-TrCP1 directs the DNA damage-induced degradation of β-TrCP2 through SCF complex. To authenticate the SCF complex-dependent event, expression levels of β-TrCP2 were examined following depletion of Cul1 upon genotoxic stress. Immunoblotting result showed that DNA damage-induced degradation of β-TrCP2 was impaired upon Cul1 depletion (Fig. S1D). Further, cycloheximide chase immunoblotting assay was performed to examine the stability of β-TrCP2. Result revealed that DNA damage-induced turnover of β-TrCP2 is declined in β-TrCP1 knockdown cells (Fig. 3D and Fig. S1E). Next, polyubiquitinated levels of β-TrCP2 were examined in β-TrCP1 knockdown cells under genotoxic stress. We found that DNA damage-induced polyubiquitinated levels of β-TrCP2 were sharply declined following depletion of β-TrCP1 (Fig. 3E). Finally, we investigated whether the degradation of β-TrCP2 following DNA damage is due to its enhanced interaction with β-TrCP1, which further leads to polyubiquitination-mediated degradation. To test this, we performed co-immunoprecipitation experiment. Immunoblotting of immunoprecipitates revealed that the interaction of β-TrCP1 with β-TrCP2 was significantly increased at 4 h and then declined to basal level interaction at 24 h post irradiation (Fig. 3F). Collectively, these results showed that accumulated β-TrCP1 directs the proteasomal degradation of β-TrCP2 under genotoxic stress.
Figure 3.
β-TrCP1 directs ablation of β-TrCP2 upon DNA damage.A, MCF7 cells expressing either NS or β-TrCP shRNA were treated with 5 Gy IR for 4 h as indicated. WCL were immunoblotted for the indicated proteins. pATM was used as DNA damage sensing marker. Data are representative of two independent experiments. B, NS or β-TrCP1 knockdown MCF7 cells were treated with 5 Gy IR and cells were collected for immunofluorescence analysis using β-TrCP1 and β-TrCP2 antibodies. γH2AX was used as positive control. Data are representative of three independent experiments. C, NS and β-TrCP1-depleted MCF7 cells were transfected with either vector control or indicated plasmids for 36 h. Transfected NS and β-TrCP1 knockdown cells were then treated with 5 Gy IR for 4 h as indicated. WCL were immunoblotted for the indicated proteins. pATM was used as DNA damage marker. Data are representative of three independent experiments. D, quantification of levels of β-TrCP2 as in Fig. S1E. Protein levels were quantified using Image J software. Expression levels of β-TrCP2 were normalized with tubulin and then set to 1 for β-TrCP2 at 0 h. The levels at other time points were calculated with respect to 0 h. E, NS or β-TrCP1 knockdown MCF7 cells were either untreated or treated with 5 Gy IR for 4 h and WCL were immunoprecipitated with anti-β-TrCP2 antibody. Immunoprecipitates and inputs WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. F, MCF7 cells were exposed to 5 Gy IR and irradiated cells were collected at the indicated time points. WCL were immunoprecipitated with anti-β-TrCP2 antibody. Immunoprecipitates and inputs WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. CHX, cycloheximide.
ATM phosphorylates and stabilizes β-TrCP1 to promote β-TrCP2 degradation upon DNA damage
The above described results prompted us to examine how β-TrCP1 is empowered to direct the degradation of β-TrCP2 under genotoxic stresses. We speculated that DNA damage signaling pathways might play crucial role in this process. Among the DNA damage signaling pathways, ATM kinase plays a pivotal role in double-strand DNA damage response and repair processes (27, 28). It is reported that ATM phosphorylates approximately 700 substrates having SQ/TQ site (28). Interestingly, analysis of the β-TrCP1 and β-TrCP2 amino acid sequences revealed the presence of one putative ATM phosphorylation site present in β-TrCP1 at Ser-158 position, which is conserved among the vertebrates (Fig. 4A). In contrast, β-TrCP2 does not have ATM phosphorylation site. We therefore reasoned that ATM might be involved in the accumulation as well as empowerment of β-TrCP1 to facilitate the degradation of β-TrCP2 upon DNA damage. To test this possibility, we performed a series of experiments. First, the cells were exposed to ionizing radiation in the absence and presence of an ATM inhibitor. The results showed that the DNA damage-induced accumulation of β-TrCP1 as well as downregulation of β-TrCP2 expression is significantly abrogated upon inactivation of ATM (Fig. 4B).
Figure 4.
ATM phosphorylates and stabilizes β-TrCP1 upon DNA damage.A, schematic shows the conservation of Ser-158 residue in β-TrCP1 among the vertebrates. B, MCF7 cells were exposed to 5 Gy IR. Irradiated cells were grown in the absence or presence of KU-55933 for 4 h as indicated and WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. C, MCF7 cells expressing His-ubiquitin were treated with 5 Gy IR in the absence or presence of KU-55933 for 4 h and WCL were pulled down with Ni-NTA beads. Pulled fractions and inputs WCL were then immunoblotted for the indicated proteins. Data are representative of two independent experiments. D, MCF7 cells expressing either wild type or S158A mutant of β-TrCP1 were irradiated in the absence or presence of KU-55933. WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. E, WCL of MCF7 cells expressing either wild type or S158D mutant of β-TrCP1 in the absence or presence of myc-β-TrCP2 were immunoblotted for the indicated proteins. Data are representative of two independent experiments. F, WCL of MCF7 cells transfected with either myc-β-TrCP2 alone or with wild type, S158A or S158D mutants of β-TrCP1 were immunoprecipitated with anti-FLAG antibody. Immunoprecipitates and inputs WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments.
Proteasomal degradation of protein warrants its polyubiquitination. We therefore examined the levels of polyubiquitinated β-TrCP2 following induction of genotoxic stress in the absence and presence of ATM inhibitor. Immunoblotting of immunoprecipitates demonstrated that DNA damage-induced polyubiquitinated β-TrCP2 levels were markedly declined following inactivation of ATM (Fig. 4C). To understand how ATM inactivation abrogates DNA damage-induced polyubiquitination of β-TrCP2, we asked whether ATM plays any role in the interaction of substrate (β-TrCP2) and receptor (β-TrCP1). Immunoblotting of immunoprecipitates revealed that the interaction of β-TrCP1 and β-TrCP2 was sharply increased following induction of DNA damage (Fig. S2A) and was sharply diminished following inactivation of ATM (Fig. S2A). These results suggest that ATM-mediated phosphorylation of β-TrCP1 at Ser158 position might be playing an important role for its accumulation as well as interaction between β-TrCP1 and β-TrCP2. To test this possibility, we generated the wild-type, phosphorylation mimetic (S158D) and phosphorylation defective (S158A) mutants of β-TrCP1. Immunoblotting results demonstrated that ectopically expressed β-TrCP1 is significantly accumulated following induction of genotoxic stress, which was abrogated following inactivation of ATM (Fig. 4D). In contrast, phosphorylation defective β-TrCP1 mutant failed to accumulate following induction of DNA damage (Fig. 4D), indicating that ATM plays role in accumulation of β-TrCP1 upon genotoxic stress. To further support our observation, we examined phosphorylated levels of the wild type and the mutant (S158A) β-TrCP1 following irradiation. Results revealed that phospho-serine levels of the wild-type β-TrCP1 were significantly increased upon induction of genotoxic stress, which was significantly declined following inhibition of ATM (Fig. S2B). In contrast, we did not observe any delectable change in phospho serine levels of the mutant (S158A) β-TrCP1 following induction of DNA damage, indicating that ATM phosphorylates β-TrCP1 at Ser158 position (Fig. S2B).
Next, we investigated why the phosphorylated form of β-TrCP1 is accumulated upon DNA damage. Is it because of incompetency of β-TrCP2 to target the phosphorylated β-TrCP1? To explore this possibility, we examined the levels of β-TrCP1 phosphomimetic S158D mutant following ectopic expression β-TrCP2. Result showed that unlike the wild-type β-TrCP1, the β-TrCP1(S158D) mutant is resistant toward degradation by β-TrCP2, indicating that ATM-mediated phosphorylation at Ser158 protects β-TrCP1 from β-TrCP2-mediated degradation upon DNA damage (Fig. 4E). Further, co-immunoprecipitation results demonstrated that inability of β-TrCP2 to degrade β-TrCP1 was neither due to disruption of β-TrCP1 and β-TrCP2 interaction nor due to preferential homodimer formation of β-TrCP1 (Fig. S2, C and D). Finally, we examined the polyubiquitinated levels of β-TrCP1 wild type and S158D mutant following expression of β-TrCP2. Results revealed that β-TrCP2 facilitates the polyubiquitination of the wild-type β-TrCP1, as expected, but failed to do so for the β-TrCP1(S158D) mutant (Fig. 4F). Thus, ATM protects β-TrCP1 from the E3 SCFβ-TrCP2-mediated proteasomal degradation under genotoxic stress.
Degron motif in β-TrCP2 is important for its degradation by β-TrCP1
F-box proteins are the substrate receptor in SCF complex. Our preceding results showed that β-TrCP1 directs proteasomal degradation of β-TrCP2 upon genotoxic stress. We then asked whether β-TrCP1 targets the single form of β-TrCP2 (non-SCF complex form). To address this we generated F-box motif deleted β-TrCP2 mutant (Fig. 5A). Immunoblotting results showed that the F-box motif deleted β-TrCP2 undergoes degradation upon genotoxic stress (Fig. 5B). We also found that DNA damage-induced degradation of the F-box motif deleted β-TrCP2 is inhibited in β-TrCP1 knockdown cells indicating that β-TrCP1 targets non-complex form of β-TrCP2 for directing degradation (Fig. 5C).
Figure 5.
DNA damage-induced β-TrCP1-mediated β-TrCP2 degradation is degron sequence dependent.A, schematic shows wild-type β-TrCP2, ΔF-β-TrCP2, and degron mutant β-TrCP2. B, MCF7 cells expressing β-TrCP2, ΔF-β-TrCP2 were exposed to 5 Gy IR for 4 h. WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. C, NS and β-TrCP1 knockdown MCF7 cells expressing ΔF-β-TrCP2 were exposed to 5 Gy IR for 4 h. WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. D, wild type and degron mutant β-TrCP2 expressing MCF7 cells were exposed to 5 Gy IR for 4 h. WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. E, wild type and degron mutant β-TrCP2 expressing MCF7 cells were exposed to 5 Gy IR and allowed to grow for 1 h. Then, cycloheximide (40 μg/ml) was added to control and irradiated cells for the indicated time. WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. F, quantification of levels of β-TrCP2 as in Figure 5E. Protein levels were quantified using Image J software. Expression levels of β-TrCP2 were normalized with tubulin and then set to 1 for β-TrCP2 at 0 h. The levels at other time points were calculated with respect to 0 h. G, wild type and degron mutant β-TrCP2 expressing MCF7 cells were exposed to 5 Gy IR for 4 h. WCL were immunoprecipitated with anti-myc antibody. Immunoprecipitates and inputs WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. CHX, cycloheximide.
β-TrCP recognizes phospho-degron motif (DSGxxS) for directing degradation of the substrate (5). We then asked whether β-TrCP1 recognizes phospho-degron motif in β-TrCP2 to promote its degradation upon genotoxic stress. To address this, we generated β-TrCP2 degron mutant (DM) and examined the expression following genotoxic stress (Fig. 5A). Immunoblotting results showed that DM is resistant to DNA damage-induced degradation (Fig. 5D). In agreement, we found that turnover of DM is slowed down as compared with the wild-type β-TrCP2 (Fig. 5, E and F). Finally, we examined the polyubiquitinated levels of the DM following genotoxic stress. Results showed that the DM is not polyubiquitinated following genotoxic stress (Fig. 5G). Collectively, these results suggest that degron motif is important for the degradation of β-TrCP2 under genotoxic stress.
β-TrCP1 assists G2/M cell cycle arrest upon DNA damage by facilitating degradation of β-TrCP2 and MDM2
Next, we were interested to understand why β-TrCP1 directs degradation of β-TrCP2 under genotoxic stresses. It is reported that β-TrCP plays an important role in proteasomal degradation of MDM2 to stabilize p53 (21, 22). However, whether β-TrCP1 or β-TrCP2 facilitates the proteasomal degradation of MDM2 was not addressed. Interestingly, β-TrCP1 is accumulated under genotoxic stresses (Fig. 1). We therefore hypothesized that β-TrCP1 might be involved in degradation of MDM2 during genotoxic stress. Indeed, we found that DNA damage-induced MDM2 attenuation and p53 stabilization are inhibited in β-TrCP1-depleted cells (Fig. 6A, compare lane 2 and lane 4). We then speculated that accumulated β-TrCP2 in β-TrCP1-depleted cells might impede the DNA damage-induced degradation of MDM2. To test this possibility, we examined the levels of MDM2 in β-TrCP1 and β-TrCP2 double-knockdown cells following genotoxic stress. Interestingly, we found that MDM2 is degraded following DNA damage induction in β-TrCP1-β-TrCP2 double-knockdown cells (Fig. 6A). This observation prompted us to examine whether β-TrCP2 has any role in protecting MDM2 from degradation during genotoxic stress. Immunoblotting results revealed that DNA damage-induced degradation of MDM2 is blocked following ectopic expression of β-TrCP2 (Fig. 6B). Interestingly, as expected, genotoxic stress-mediated accumulation of p53 is also severely compromised following ectopic expression of β-TrCP2 (Fig. 6B), indicating that β-TrCP2 may protect MDM2 from degradation by facilitating degradation defective polyubiquitination. To test this possibility, we examined the turnover of MDM2 in MCF7 cells expressing shRNA for either NS or β-TrCP1 or β-TrCP2 or both β-TrCP1-β-TrCP2 by cycloheximide chase assay. Immunoblotting results revealed that turnover of MDM2 is significantly increased upon depletion of β-TrCP2 (Fig. 6C and Fig. S3A). To strengthen our observation, polyubiquitinated levels of MDM2 were examined upon β-TrCP2 expression in the absence and presence of irradiation. Results revealed that β-TrCP2 promoted K63-linked polyubiquitination of MDM2, which is declined upon DNA damage (Fig. 6D). In a complementary approach, we examined the K63-linked polyubiquitinated levels of MDM2 upon depletion of either β-TrCP1 or β-TrCP2 or both. Results revealed that levels of K63-linked polyubiquitination of MDM2 were significantly decreased upon β-TrCP2 depletion. Interestingly, K63-linked polyubiquitination was increased following depletion of β-TrCP1 and noticeably decreased upon co-depletion of β-TrCP2 in β-TrCP1-depleted cells (Fig. 6E). In addition, we found that ectopically expressed β-TrCP1 promotes K48-linked polyubiquitination, whereas β-TrCP2 directs K63-linked polyubiquitination of MDM2 (Fig. S3B). To further authenticate, polyubiquitination assay of MDM2 by β-TrCP1/β-TrCP2 was performed in vitro. In agreement with in vivo polyubiquitination data, in vitro results confirmed that β-TrCP1 promotes K48-linked ubiquitination, whereas β-TrCP2 directs K63-linked ubiquitination of MDM2 (Fig. S3, C and D). Finally, we examined the direct interaction of β-TrCP1/β-TrCP2 with MDM2 using recombinant proteins. Immunoblotting of immunoprecipitates showed that MDM2 interacts with both β-TrCP1 and β-TrCP2 (Fig. S3E). In addition, we found that β-TrCP1 and MDM2 interaction was increased following genotoxic stress (Fig. S3F). Taken together, these results suggest that, during genotoxic stress condition, β-TrCP1 negatively regulates MDM2 by promoting K48-linked polyubiquitination, and in parallel it inhibits K63-linked ubiquitination of MDM2 by promoting β-TrCP2 degradation. Thus, β-TrCP1-mediated attenuation of MDM2 and β-TrCP2 is important for the activation of p53 during genotoxic stress.
Figure 6.
β-TrCP1-mediated β-TrCP2 degradation activates G2/M checkpoint upon DNA damage through ablation of MDM2.A, MCF7 cells expression indicated that shRNA was exposed to 5 Gy IR for 4 h and WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. B, MCF7 cells were transfected with indicated plasmid for 42 h. Transfected cells were then exposed to IR (5 Gy) as indicated. Cells were then collected at 4 h post irradiation and WCL were immunoblotted for the indicated proteins. Data are representative of two independent experiments. C, quantification of levels of MDM2 as in Fig. S3A. Protein levels were quantified using Image J software. Expression levels of MDM2 were normalized with tubulin and then set to 1 for MDM2 at 0 h. The levels at other time points were calculated with respect to 0 h. D, MCF7 cells expressing His K63-only ubiquitin mutant, myc-MDM2, and FLAG-β-TrCP2 as indicated were either untreated or treated with 5 Gy IR for 4 h. Cells were then harvested and whole-cell lysates were incubated with Ni-NTA beads. The pulled down fractions and inputs were immunoblotted for the indicated proteins. Data are representative of three independent experiments. E, WCL of NS, β-TrCP1 knockdown, β-TrCP2 knockdown, or β-TrCP1 and β-TrCP2 double-knockdown MCF7 cells were immunoprecipitated with anti-MDM2 antibody. The immunoprecipitates and inputs were immunoblotted for the indicated proteins. Data are representative of two independent experiments. F, quantification of cell population at the G2/M phase from the Fig. S3G. Data represent the mean (±SD) of three independent experiments. CHX, cycloheximide.
Genotoxic-stress-induced MDM2 degradation facilitates p53 accumulation (21, 29). It is known that accumulated p53 helps to arrest the cells at G2/M phase of the cell cycle during genotoxic stress (30). We found that β-TrCP1 helps in accumulation of p53 under genotoxic stress (Fig. 6A). We therefore examined whether DNA damage-induced β-TrCP1 accumulation has any role in G2/M phase cell cycle arrest. FACS analysis revealed that β-TrCP1-depleted cells are defective in arresting cells at the G2/M phase (Fig. 6F and Fig. S3G). In contrast, co-depletion of β-TrCP2 in β-TrCP1 depleted cells restores the DNA damage-induced G2/M cell cycle arrest (Fig. 6F and Fig. S3G). Collectively, our results demonstrated that β-TrCP1 plays an important role in p53 accumulation-mediated cell cycle arrest upon genotoxic stress via facilitating ablation of β-TrCP2 as well as MDM2.
DNA damage-induced attenuation of β-TrCP2 by β-TrCP1 is important to promote cell survival
DNA damage response pathways activate multiple downstream processes that arrest cell cycle to allow the repair of damaged DNA (31, 32, 33, 34). Our results show that β-TrCP1 helps to arrest the cells at the G2/M phase through facilitating p53 accumulation. We then examined whether β-TrCP1-mediated attenuation of β-TrCP2 has any role in DNA damage repair process. We monitored the DNA repair by assessing the response of H2A histone family member X (H2AX) phosphorylation (γH2AX), a marker of DNA double-strand breaks (35). Results revealed that β-TrCP1-depleted cells are defective in DNA damage repair (Fig. 7, A and B and Fig. S4A). For instance, noticeable number of γH2AX foci was detected even after 24 h of post DNA damage in β-TrCP1-depleted cells (Fig. 7, A and B). Interestingly, depletion of β-TrCP2 in β-TrCP1-depleted cells resumed the DNA damage repair (Fig. 7, A and B). In a complementary approach, we examined γH2AX foci following ectopic expression of either β-TrCP1 or ΔF- β-TrCP1 mutant in β-TrCP1-depleted cells. Results revealed that DNA damage repair defect in β-TrCP1-depleted cells was resolved upon ectopic expression of the wild type β-TrCP1 but not with the ΔF- β-TrCP1 mutant (Fig. 7, A and B). Further, DNA damage repair was assessed by comet assay and result showed the prevalence of distinct comet tails even at 24 h of post DNA damage in β-TrCP1-depleted cells (Fig. 7C and Fig. S4B). Interestingly, overexpression of the wild-type β-TrCP1 but not the ΔF- β-TrCP1 in β-TrCP1-depleted cells resolved the DNA damages (absence of comet tail at post 24 h of irradiation) (Fig. 7C and Fig. S4B).
Figure 7.
β-TrCP1 regulates DNA repair and cell survival upon DNA damage through attenuating β-TrCP2.A, MCF7 cells stably expressing NS, β-TrCP1 or coexpressing β-TrCP1 and β-TrCP2 shRNA were transfected with either vector control or WT- β-TrCP1 or ΔF-β-TrCP1 as indicated for 36 h. Cells were then exposed to 5 Gy IR for 0, 4, and 24 h. Cells were stained for γH2AX (cyan) and DNA (blue) and γH2AX foci were counted in total 110 cells from ten random fields. Data are representative of three independent experiments. B, quantification of γH2AX foci per cell as in panel A. Random ten fields were observed and counted number of γH2AX per cell. C, MCF7 cells stably expressing NS, β-TrCP1 or coexpressing β-TrCP1 and β-TrCP2 shRNA were transfected with either vector control or WT- β-TrCP1 or ΔF-β-TrCP1 as indicated for 36 h. Cells were then exposed to 5 Gy IR for 0, 4, and 24 h. Cell were then trypsinized and subjected for comet assay. Comet positive cells were counted from ten random fields and plotted. Data are representative of three independent experiments. D, MCF7 cells stably expressing NS, β-TrCP1 or coexpressing β-TrCP1 and β-TrCP2 shRNA were transfected with either vector control or WT- β-TrCP1 or ΔF-β-TrCP1 as indicated for 36 h. Cells were then exposed to 5 Gy IR and allowed to form colonies. Colonies were then stained with crystal violet dye. Colonies were counted and colony number in NS untreated condition was normalized to 100% and the number of colonies for others were determined with respect to NS untreated cells. Data are representative of three independent experiments.
Typically, DNA damage repair defective cells are more prone to cell death upon genotoxic cell death. We therefore examined the survival of wild-type cells, β-TrCP1-depleted cells, and β-TrCP1- β-TrCP2 double-knockdown cells with and without irradiation. In agreement with the previous study, we observed an increased colony-forming ability of β-TrCP1 knockdown cells under normal condition (Fig. 7D) (18). The long-term colony formation assay also revealed that irradiated β-TrCP1-depleted cells form lesser number of colonies as compared with the wild-type cells (Fig. 7D). However, knockdown of β-TrCP2 in β-TrCP1-depleted cells increased the survival of the irradiated cells and formed similar number of colonies as in control cells (Fig. 7D). Collectively, these results demonstrated that β-TrCP1 and β-TrCP2 axis is important to activate DNA damage response and repair pathway to maintain the genome integrity.
Discussion
Paralogs β-TrCP1 and β-TrCP2 are known to regulate numerous biological processes including cell cycle progression, cellular signaling, development, DNA damage response, and their deregulation is associated with pathological conditions such as cancer. Previous studies have shown that β-TrCP1 can function as a tumor suppressor while β-TrCP2 can function as an oncogene (17, 18, 36, 37, 38, 39, 40, 41, 42). It is also reported that tumor suppressors play important role in DNA damage response (43, 44, 45) and β-TrCP is known to be involved in DNA damage (16, 21, 22, 23). However, their distinct role as well as regulation during DNA damage response and repair has remained elusive. In this study, we unravel the functional importance of β-TrCP1 and β-TrCP2 regulation in the context of DNA damage response. Our study reveals that β-TrCP1 attenuates expression levels of β-TrCP2 to facilitate p53 activation and cell fitness upon DNA damage (Figs. 6 and 7). For the first time, we elucidated the molecular insight of cross-regulation of β-TrCP1 and β-TrCP2 during DNA damage response (Fig. 8).
Figure 8.
Model depicting the proteasomal regulation β-TrCP2 by β-TrCP1 following DNA damage.
β-TrCP1 activates DNA damage response signal by directing the ablation of β-TrCP2. β-TrCP1 and β-TrCP2 form homodimer and heterodimer by utilizing their dimerization domain. Interestingly, homodimer is more potent than heterodimer in promoting the degradation of the substrates (11). It was thought that β-TrCP1-β-TrCP2 heterodimer also targets the substrate for proteasomal degradation. It was a long-standing question why E3 ubiquitin ligase activity of β-TrCP1- β-TrCP2 heterodimer is less potent than the homodimer. We have demonstrated the molecular insights responsible for functionality of homo- and heterodimer under genotoxic stress. A recent study (18) and our present study demonstrate that heterodimer is formed to degrade each other. Under glucose deprivation or serum starvation, heterodimer is formed to promote degradation of β-TrCP1 by SCF-β-TrCP2. Here we showed that heterodimer formation results in degradation of β-TrCP2 by β-TrCP1 under genotoxic stress. Thus, heterodimer formation is facilitated by stress conditions, but degradation of paralogs is dependent on the cell fate decision. It appears that if cells need to survive glucose deprivation or serum starvation, SCF- β-TrCP2 is empowered to degrade β-TrCP1 to prevent cell death, and conversely SCF- β-TrCP1 is empowered to direct degradation of β-TrCP2 to facilitate DNA damage response and repair process.
ATM is the central upstream kinase activated upon genotoxic-stress-induced DNA double-strand breaks (46). Upon activation, ATM phosphorylates approximately 700 cellular proteins having SQ/TQ consensus motif (28). We found that β-TrCP1 significantly accumulated and directs the degradation of β-TrCP2 in ATM-dependent manner. Further, we showed that ATM-mediated phosphorylation of β-TrCP1 at Ser158 is important for its protection from proteasomal degradation by β-TrCP2. ATM plays a critical role in empowering the ubiquitin ligase activity of SCF-β-TrCP1 over SCF-β-TrCP2. Thus, ATM is the key player in blocking the degradation of tumor suppressor β-TrCP1 to facilitate the inactivation of oncogene β-TrCP2 during DNA damage response. Therefore, our study suggests that β-TrCP1 may play a critical role in inactivating oncogenic function of β-TrCP2 to activate DNA damage checkpoints during genotoxic-stress-inducing cancer chemotherapeutic drugs. In contrast to genotoxic stress, previous study showed that glucose deprivation or serum starvation empowers SCF-β-TrCP2 to target β-TrCP1 for proteasomal degradation to inactivate SCF-β-TrCP1 ubiquitin ligase activity in AMPK kinase dependent manner (18). Interestingly, both β-TrCP1 and β-TrCP2 have phosphorylation site for AMPK kinase. However, AMPK selectively phosphorylates β-TrCP1 at Ser82 to mark it for proteasomal degradation by β-TrCP2. Interestingly, between β-TrCP1 and β-TrCP2, only β-TrCP1 has putative phosphorylation of ATM. Our data suggest that ATM phosphorylates β-TrCP1 at Serine-158 position, which makes it resistant for β-TrCP2-mediated degradation but enhances heterodimerization of β-TrCP1 and β-TrCP2. It is also possible that phosphorylation at Serine-158 might alter the conformational change in the degron motif in β-TrCP1 and make it resistant to β-TrCP2. We found that β-TrCP1 directs the degradation of β-TrCP2 through recognizing degron sequence in β-TrCP2. It might be possible that other DNA damage-related kinases may phosphorylate β-TrCP2 to direct its proteasomal degradation by β-TrCP1 under genotoxic stress. It is also possible that other type of posttranslational modifications might help to empower β-TrCP1 to direct β-TrCP2 degradation. Thus, kinase pathways play a pivotal role in determining the stability of paralogs under different stress conditions.
It is reported that potency of cancer chemotherapeutic drugs depends on the activation of p53 (47). Activation of p53 is blocked through proteasomal degradation by MDM2 in cancer. Therefore, activation of p53 requires degradation of MDM2. A previous study showed that β-TrCP1 and β-TrCP2 interact with MDM2 (21). However, it was unclear whether β-TrCP1 or β-TrCP2 or both were involved in proteasomal degradation of MDM2 since both β-TrCP1 and β-TrCP2 were silenced simultaneously in all the experiments. Moreover, it was shown that depletion of either β-TrCP1 or β-TrCP2 results in accumulation of MDM2 (22). Our study, for the first time, revealed that β-TrCP1 facilitates the activation of p53 through promoting the degradation of both MDM2 and β-TrCP2 during genotoxic stress. β-TrCP1 directs K48-linked polyubiquitination of MDM2 (Fig. S3B top panel and Fig. S3D); in contrast, β-TrCP2 facilitates K63-linked polyubiquitination of MDM2 and thereby prevents its degradation (Fig. 6D, Fig. S3B bottom panel and Fig. S3C). Our study suggests that β-TrCP1 increases the turnover of MDM2 through two parallel pathways following induction of genotoxic stress; prevents K63-linked polyubiquitination of MDM2 by β-TrCP2 and directly facilitates the degradation-specific polyubiquitination of MDM2.
During genotoxic stress, halting the cells at different cell cycle phases is important to provide sufficient time to repair the genome. Halting the cells at different phases requires activation of checkpoints. Our study showed that β-TrCP1 plays a vital role in halting the genotoxic stress exposed cells at the G2/M phase of the cell cycle through activation of p53. A previous study showed that cells defective in activation of G2/M phase checkpoint undergo cell death following irradiation-induced genotoxic stress (48). Indeed, we found that β-TrCP1-depleted cells are defective in arresting at the G2/M phase of the cell cycle and these cells are hypersensitive to cell death following genotoxic stress (Figs. 6F and 7D).
Typically, tumor suppressors play vital role in DNA damage repair to maintain the genome integrity. Our results demonstrated that β-TrCP1-depleted cells are defective in resolving the DNA damage due to accumulation of β-TrCP2 (Fig. 7). Accumulated β-TrCP2 in β-TrCP1 knocked down cells facilitates augmentation of MDM2 levels through promoting K63-linked polyubiquitination to prevent activation of p53. Genotoxic stress-induced p53 monitors DNA damage repair through activating nonhomologous end joining and homologous recombination process (49). Therefore, p53 induction deficient β-TrCP1-depleted cells are defective in DNA damage repair as evident from the presence of γH2AX foci even after 24 h post irradiation. As a result, β-TrCP1-depleted cells are more prone for cell death upon genotoxic stress. Thus, β-TrCP1 may play a critical role in barring normal cells from generation of genomically unstable cells as well as protect the cells from cell death during radiation therapy. On the other hand, underexpression of β-TrCP1 in cancer cells may facilitate the accumulation of mutant cells that have the potential to fuel cancer development. Overall, our study deciphers the molecular insights of context-dependent function of β-TrCP, which reveals that β-TrCP1 could function as tumor suppressor, whereas β-TrCP2 could function as an oncogene following genotoxic stresses and therefore β-TrCP1-mediated attenuation of β-TrCP2 could be exploited for effective cancer chemotherapy.
Experimental procedures
Cell culture
HEK-293T, MCF7, HCT116 cells were kind gift from Prof. Michael R. Green, University of Massachusetts Medical School. Cells were cultured in either DMEM or RPMI supplemented with 10% FBS containing streptomycin and penicillin under humid condition at 37 °C.
Plasmids and shRNAs
The cDNAs of β-TrCP2 was cloned in mammalian expression vector pCMV6-Entry having C-terminal FLAG and myc-tag. GST-SKP1 was a kind gift from Ashutosh Kumar (IIT Bombay, India). Ubc13 was from kind gift from Dr Ranabir Das (NCBS, India). His-ubiquitin and K63 (ubiquitin mutant has only K63 lysine, rest lysine residues are mutated to alanine)-only ubiquitin mutants were the kind gift from Dr Wuhan Xiao (Chinese Academy of Science, China). cDNA of β-TrCP1 was a kind gift from Jae U. Jung (University of Southern California, USA). β-TrCP1 was subcloned in p3XFLAG-CMV-14 and pCMV-myc-C vectors. FLAG-F-box deleted β-TrCP1 (ΔF-β-TRCP1), S158A-β-TrCP1 and S158D-β-TrCP1 were cloned in p3XFLAG-CMV-14 vector (Sigma). FLAG-β-TrCP2 and FLAG-F-box deleted β-TrCP2 (FLAG-ΔF-β-TrCP2) were also cloned in p3XFLAG-CMV-14 vector (Sigma). β-TrCP2 and degron mutant-β-TrCP2 (DM) were also cloned in pCMV-myc-C vector. β-TrCP1 and β-TrCP2 were also cloned in PET-28a vector. Primers used in this study are listed in Table S1. Short hairpin RNAs (shRNAs) and packaging plasmids (pPAX2 and pMD2.G), GST-MDM2 and GST-MDM2 (C464A) were kind gifts from Prof. Michael R. Green (University of Massachusetts Medical School, USA). shRNAs sequences are mentioned in Table S2.
Generation of stable knockdown cells
Stable knockdown cells were generated as described previously (50). Briefly, lentivirus shRNA along with viral packaging vectors (shRNA:pPAX2:pMD2.G = 1:1:0.5 weight ratio) was transfected in HEK-293T cells using polyethyleneimine for 48 h. Virus-containing media was collected, filtered through 0.45 μm syringe filter, and host cells were transduced with filtered virus in the presence of 8 μg/ml polybrene for 48 h. Infected cells were then grown in the presence of 1 μg/ml puromycin for 7 days to select lentivirus-expressing cells. Scramble NS shRNA-expressing cells were used as wild-type cells throughout our study. Knockdown efficiency was determined by immunoblotting.
Antibodies
Anti-β-TrCP1 (sc-390629), anti-His (sc-8036), anti-HA (sc-7392), anti-Ubiquitin (sc-8017), anti-p53 (sc126), anti-ATM (sc-53173), anti-GST (sc-138), and anti-MDM2 (sc-813) were purchased from Santa Cruz Biotechnology. anti-pATM (13050) and anti-K63-linked ubiquitin (5621), anti-γH2AX (9718), and anti-K48-linked ubiquitin antibodies (8081) were purchased from Cell Signaling Technology. We purchased anti-β-TrCP2 (PA5-29878) from Thermo Fisher Scientific, anti-γH2AX (ab26350) from Abcam and anti-myc (11667149001) from Roche. Antibody against Cullin1 (100-401-A01) was procured from Rockland. We obtained anti-FLAG (F1804) and anti-Tubulin (T5168) antibodies from Sigma-Aldrich.
SDS-PAGE and western blotting
Cells were harvested, washed with ice-cold PBS, and lysed with lysis buffer (50 mM Tris pH7.4, 250 mM NaCl, 5 mM EDTA, 50 mM NaF, 1 mM Na3VO4, 0.5% Triton X-100, and protease inhibitor cocktail) in ice for 30 min. Lysates were then spun down at 16,000g at 4 °C for 20 min and supernatants were collected. The protein concentration was measured by Bradford assay using bovine serum albumin as a standard (51). Protein samples were prepared using SDS sample buffer (50 mM Tris pH 6.8, 2% SDS, 10% glycerol 1% β-mercaptoethanol, 0.024% bromophenol blue) and boiled for 5 min and stored at −80 °C. Samples were run on SDS-PAGE to resolve the proteins according to their molecular weight and transferred onto the PVDF membrane. After blocking with 3% skimmed milk/5% BSA, the membrane was incubated with respective primary antibody for overnight with gentle rocking. Next day, membrane was washed with TBST buffer and incubated with HRP-conjugated respective secondary antibody for 1 h at 25 °C. Finally, the membrane was washed thrice with TBST buffer and developed using chemiluminescence substrates (Pierce) in GE Amersham Imager 600.
In vivo ubiquitination assay
The in vivo ubiquitination assays were performed as described previously (50). Briefly, plasmids were transfected in MCF7 cells with indicated combination for 36 h. Transfected cells were then grown in the presence of 5 μM MG132 for additional 6 h. Cells were then harvested and protein extracts were prepared as described above. Protein extracts (600–800 μg per immunoprecipitation reaction) were then immunoprecipitated with 2 to 3 μg primary antibody. The immunoprecipitates and input protein extracts were resolved by SDS-PAGE and immunoblotted for the indicated proteins.
In addition, we also performed polyubiquitination assay under denaturing condition as described previously (52). Briefly, cells were harvested and lysed in 1% SDS lysis buffer (50 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1% SDS, 1 mM DTT), and boiled for 5 min. Lysates were clarified by centrifugation at 16,000g for 20 min. Supernatant was diluted ten times and the proteins concentration was measured using Bradford method. The diluted proteins (800 μg/pull-down experiment) were then incubated with Ni-NTA beads overnight at 4 °C. After washing thrice, the beads were incubated with 1X SDS dye for 10 min at RT followed by boiling in water bath for 5 min. Elutes were loaded on SDS-PAGE and polyubiquitinated proteins were detected by immunoblotting.
Protein purification
Recombinant proteins were purified from BL21 DE3 strain as described previously (53). Briefly, the transformed cells were grown at 37 °C in LB broth containing ampicillin/kanamycin until they reached early log phase (A 600 ∼ 0.4–0.6), and then protein expression was induced by the addition of 0.5 mM IPTG to the culture and was allowed to grow for an additional 6 h. Cells were harvested by centrifugation at 3000g at 4 °C for 10 min and suspended in lysis buffer (50 mM Tris pH 8, 100 mM NaCl, 1 mM phenyl-methylsulfonyl fluoride, 0.1% (v/v) β-mercaptoethanol, and 10 mM MgCl2). Lysozyme (0.4 mg/ml) was added to the cell suspension, incubated on ice for 1 h, and then the suspension was sonicated for six cycles with 20 s pulse. The insoluble debris was removed by centrifugation at 12,000g at 4 °C for 30 min. The supernatant was allowed to bind to respective agarose beads, and the protein was eluted with respective elution buffer. Protein concentrations were determined by the Bradford method (51).
In vitro protein–protein interaction
GST-MDM2, His-β-TrCP1, and His-β-TrCP2 were expressed in bacterial system and purified by affinity chromatography. For in vitro interaction, purified GST or GST-MDM2 protein was incubated with His-β-TrCP1 or His-β-TrCP2 as indicated for 6 h at 4 °C. His-β-TrCP1 and His-β-TrCP2 were pulled down using Ni-NTA beads. Finally, beads were washed three times and proteins were eluted by adding 1X SDS-dye to the reaction mixture and boiled for 5 min. The reaction mixtures were resolved by SDS-PAGE, transferred onto PVDF membrane, and blots were probed with the indicated antibodies.
In vitro ubiquitination assay
For in vitro ubiquitination assay, β-TrCP1, β-TrCP2, SKP1, Ubiquitin, E1, Ubc13, and Cdc34 were affinity purified using bacterial expression system. The E3 ligase complex together with purified E1, E2, Rbx1, ubiquitin, and ATP was incubated as indicated with purified GST-MDM2 (C464A) in ubiquitylation buffer (50 mM Tris8, 5 mM MgCl2, 0.1% Tween 20, 1 mM β–mercaptoethanol) at 25 °C for 1 h. The reactions were stopped by the addition of 5XSDS sample loading buffer, boiled for 5 min, and run on SDS-PAGE for immunoblotting.
Real-time RT-PCR
Total RNA was extracted using Trizol (Invitrogen) according to the manufacturer’s instructions and 1 μg of extracted total RNA was used to synthesize cDNA using kit from Takara. Real-time PCR was performed using SYBR green reagent from Takara. mRNA levels of actin were used as internal control to normalize the mRNA levels of β-TrCP1 and β-TrCP2 and the ratio of β-TrCP1:actin or β-TrCP2:actin was set to 1 for control condition. The primers used for real-time RT-PCR are listed in Table S3.
Immunofluorescence
Cells were fixed with 3.7% formaldehyde and permeabilized with ice-cold methanol. Cells were stained with primary antibodies for 3 h at room temperature. Then, the cells were incubated with fluorophore-conjugated secondary antibody (Alexa fluor 488 or Alexa fluor 594) for 2 h, and then DNA was stained with Hoechst-33258. After each step of staining, cells were washed thrice in PBS. Finally, stained cells were mounted, observed under confocal microscope (Zeiss LSM 880). Data was analyzed using ZEISS ZEN software quantified using ImageJ software.
Comet assay
Comet assay was performed as described previously (54). Briefly, cells were embedded in 0.65% low-melting agarose on a glass slide. Cells were incubated in lysis buffer (2.5 M NaCl, 0.1 M EDTA, 10 mM Tris, 1% SDS, 1% (v/v) Triton X-100, pH-10) containing 10% DMSO overnight at 4 °C. Slides were then incubated with alkaline electrophoresis buffer (10 M NaOH and 200 mM EDTA, pH-13) for 30 min to allow the DNA to unwind. Subsequently, electrophoresis was carried out for 30 min at 300 mA. Next, the slides were neutralized with 0.4 M Tris buffer pH-7.5 and then immersed in 70% ethanol for 5 min, air-dried, stained with ethidium bromide, and were observed under an epifluorescence microscope (Olympus 1X71 microscope). Comet positive cells were counted and plotted.
Long-term colony formation assay
For colony formation assay, NS, shβ-TrCP1, and shβ-TrCP1-shβ-TRCP2 expressing cells (5 × 103 cells per 35 mm dish) were seeded and grown for 3 days. Next, colonies were exposed to 5 Gy IR and then allowed to form colonies. Finally, cells were fixed with 3.7% formaldehyde and colonies were stained with crystal violet (Sigma-HT90132) for 15 min. Stained cells were then washed with PBS to remove residual crystal violet solution. Images were acquired using GE Amersham Imager 600.
Genotoxic stress induction
Genotoxic stresses were induced by either radiation exposure or chemical treatments. For exposure of cells to UV irradiation, media was removed, washed two times with PBS, and then cells were exposed to UV radiation (10 mJ/m2) using a Hoefer Scientific UV cross-linker. For exposure of cells to ionizing radiation, cells were irradiated with 5 Gy using a Co60 irradiator. Irradiated cells were then kept in the CO2 incubator and collected at the indicated time periods. For H2O2, cells were grown in the press of 0.05% H2O2 for 2 h. Collected cells were then processed for either immunoblotting or immunoflourescence or comet assay or FACS analysis.
FACS analysis
Cells were irradiated and collected by trypsinization at the indicated time. After washing with PBS, cells were fixed with 95% ethanol and stored at −20 °C overnight. Cells were washed with PBS, treated with RNase A, labeled with propidium iodide, and analyzed by BD FACS Calibur for cell cycle.
Statistical analysis
Each experiment was repeated at least three times. Values are shown as mean ± SD. Statistical significance was tested using Student’s t-test (Microsoft Excel) or ANOVA. p values of ∗∗∗(<0.001), ∗∗ (<0.01), and ∗(<0.05) were considered significant.
Data availability
All data presented in the paper are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Prof. Michael R. Green (University of Massachusetts Medical School, USA) for providing cell lines and shRNAs used in this study. We would like to thank Prof. Edward Yeh (University of California, USA), Dr Wuhan Xiao (Chinese Academy of Science, China), and Prof. Jae U. Jung (University of Southern California, USA) for providing cDNA constructs. We thank Dr Jayati Mullick for editorial assistance. This work was partly supported by National Centre for Cell Science and partly by Department of Biotechnology, Government of India (BT/HRD/NBA/39/01/2018-19). S. I. is senior DBT research fellow and P. D., O. S. are Senior UGC research fellows.
Author contributions
Conception and design: S. I., M. K. S. Development of methodology: S. I., M. K. S. Acquisition of data: S. I., P. D., O. S. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S. I., M. K. S. Writing the manuscript: S. I., P. D., M. K. S. Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): M. K. S. Study supervision: M. K. S.
Edited by Patrick Sung
Supporting information
References
- 1.Glickman M.H., Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A., Heller H., Elias S., Ciechanover A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 1983;258:8206–8214. [PubMed] [Google Scholar]
- 3.Zheng N., Wang Z., Wei W. Ubiquitination-mediated degradation of cell cycle-related proteins by F-box proteins. Int. J. Biochem. Cell Biol. 2016;73:99–110. doi: 10.1016/j.biocel.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng N., Schulman B.A., Song L., Miller J.J., Jeffrey P.D., Wang P., Chu C., Koepp D.M., Elledge S.J., Pagano M., Conaway R.C., Conaway J.W., Harper J.W., Pavletich N.P. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 5.Frescas D., Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: Tipping the scales of cancer. Nat. Rev. Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki H., Chiba T., Kobayashi M., Takeuchi M., Suzuki T., Ichiyama A., Ikenoue T., Omata M., Furuichi K., Tanaka K. IκBα ubiquitination is catalyzed by an SCF-like complex containing Skp1, cullin-1, and two F-box/WD40-repeat proteins, βTrCP1 and βTrCP2. Biochem. Biophys. Res. Commun. 1999;256:127–132. doi: 10.1006/bbrc.1999.0289. [DOI] [PubMed] [Google Scholar]
- 7.Guardavaccaro D., Kudo Y., Boulaire J., Barchi M., Busino L., Donzelli M., Margottin-Goguet F., Jackson P.K., Yamasaki L., Pagano M. Control of meiotic and mitotic progression by the F box protein β-Trcp1 in vivo. Dev. Cell. 2003;4:799–812. doi: 10.1016/s1534-5807(03)00154-0. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa T., Araki T., Nakagawa M., Hirao A., Unno M., Nakayama K. S6 kinase- and β-TrCP2-dependent degradation of p19 Arf is required for cell proliferation. Mol. Cell. Biol. 2015;35:3517–3527. doi: 10.1128/MCB.00343-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs S.Y., Spiegelman V.S., Kumar K.G.S. The many faces of β-TrCP E3 ubiquitin ligases: Reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- 10.Ray D., Terao Y., Nimbalkar D., Chu L.-H., Donzelli M., Tsutsui T., Zou X., Ghosh A.K., Varga J., Draetta G.F., Kiyokawa H. Transforming growth factor facilitates beta-TrCP-mediated degradation of Cdc25A in a Smad3-dependent manner. Mol. Cell. Biol. 2005;25:3338–3347. doi: 10.1128/MCB.25.8.3338-3347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki H., Chiba T., Suzuki T., Fujita T., Ikenoue T., Omata M., Furuichi K., Shikama H., Tanaka K. Homodimer of two F-box proteins betaTrCP1 or betaTrCP2 binds to IkappaBalpha for signal-dependent ubiquitination. J. Biol. Chem. 2000;275:2877–2884. doi: 10.1074/jbc.275.4.2877. [DOI] [PubMed] [Google Scholar]
- 12.Hatakeyama S., Kitagawa M., Nakayama K., Shirane M., Matsumoto M., Hattori K., Higashi H., Nakano H., Okumura K., Onoé K., Good R.A., Nakayama K.I. Ubiquitin-dependent degradation of IκBα is mediated by a ubiquitin ligase Skp1/Cul 1/F-box protein FWD1. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3859–3863. doi: 10.1073/pnas.96.7.3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang J., Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F- box/WD40-repeat protein Slimb. Nature. 1998;391:493–496. doi: 10.1038/35154. [DOI] [PubMed] [Google Scholar]
- 14.Ding Q., He X., Hsu J.-M., Xia W., Chen C.-T., Li L.-Y., Lee D.-F., Liu J.-C., Zhong Q., Wang X., Hung M.-C. Degradation of Mcl-1 by β-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol. Cell. Biol. 2007;27:4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z., Zhong J., Gao D., Inuzuka H., Liu P., Wei W. DEPTOR ubiquitination and destruction by SCF (β-TrCP) Am. J. Physiol. Endocrinol. Metab. 2012;303:E163–E169. doi: 10.1152/ajpendo.00105.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busino L., Donzelli M., Chiesa M., Guardavaccaro D., Ganoth D., Dorrello N.V., Hershko A., Pagano M., Draetta G.F. Degradation of Cdc25A by β-TrCP during S phase and in response to DNA damage. Nature. 2003;426:87–91. doi: 10.1038/nature02082. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z., Liu P., Inuzuka H., Wei W. Roles of F-box proteins in cancer. Nat. Rev. Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui D., Dai X., Shu J., Ma Y., Wei D., Xiong X., Zhao Y. The cross talk of two family members of β-TrCP in the regulation of cell autophagy and growth. Cell Death Differ. 2020;27:1119–1133. doi: 10.1038/s41418-019-0402-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dasika G.K., Lin S.C.J., Zhao S., Sung P., Tomkinson A., Lee E.Y.H.P. DNA damage-induced cell cycle checkpoints and DNA strand break repair in development and tumorigenesis. Oncogene. 1999;18:7883–7899. doi: 10.1038/sj.onc.1203283. [DOI] [PubMed] [Google Scholar]
- 20.Halazonetis T.D., Gorgoulis V.G., Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 21.Inuzuka H., Tseng A., Gao D., Zhai B., Zhang Q., Shaik S., Wan L., Ang X.L., Mock C., Yin H., Stommel J.M., Gygi S., Lahav G., Asara J., Xiao Z.X.J. Phosphorylation by casein kinase I promotes the turnover of the Mdm2 oncoprotein via the SCFβ-TRCP ubiquitin ligase. Cancer Cell. 2010;18:147–159. doi: 10.1016/j.ccr.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Inuzuka H., Zhong J., Fukushima H., Wan L., Liu P., Wei W. DNA damage-induced activation of ATM promotes β-TRCPmediated Mdm2 ubiquitination and destruction. Oncotarget. 2012;3:1026–1035. doi: 10.18632/oncotarget.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J.M., Jin J. CRL ubiquitin ligases and DNA damage response. Front. Oncol. 2012;2:29. doi: 10.3389/fonc.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loveless T.B., Topacio B.R., Vashisht A.A., Galaang S., Ulrich K.M., Young B.D., Wohlschlegel J.A., Toczyski D.P. DNA damage regulates translation through β-TRCP targeting of CReP. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mailand N., Bekker-Jensen S., Bartek J., Lukas J. Destruction of claspin by SCFβTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell. 2006;23:307–318. doi: 10.1016/j.molcel.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 26.Tan M., Gallegos J.R., Gu Q., Huang Y., Li J., Jin Y., Lu H., Sun Y. SAG/ROC-SCFβ-TrCP E3 ubiquitin ligase promotes pro-caspase-3 degradation as a mechanism of apoptosis protection. Neoplasia. 2006;8:1042–1054. doi: 10.1593/neo.06568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khanna K.K., Lavin M.F., Jackson S.P., Mulhern T.D. ATM, a central controller of cellular responses to DNA damage. Cell Death Differ. 2001;8:1052–1065. doi: 10.1038/sj.cdd.4400874. [DOI] [PubMed] [Google Scholar]
- 28.Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., Hurov K.E., Luo J., Bakalarski C.E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S.P., Elledge S.J. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 29.Pereg Y., Shkedy D., De Graaf P., Meulmeester E., Edelson-Averbukh M., Salek M., Biton S., Teunisse A.F.A.S., Lehmann W.D., Jochemsen A.G., Shiloh Y. Phosphorylation of Hdmx mediates its Hdm2- and ATM-dependent degradation in response to DNA damage. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5056–5061. doi: 10.1073/pnas.0408595102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor W.R., Stark G.R. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 31.Khanna K.K., Jackson S.P. DNA double-strand breaks: Signaling, repair and the cancer connection. Nat. Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 32.Helt C.E., Wang W., Keng P.C., Bambara R.A. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle. 2005;4:529–532. doi: 10.4161/cc.4.4.1598. [DOI] [PubMed] [Google Scholar]
- 33.Branzei D., Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 34.O’Connor M.J. Targeting the DNA damage response in cancer. Mol. Cell. 2015;60:547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 35.Kuo L.J., Yang L.X. γ-H2AX- a novel biomaker for DNA double-strand breaks. In Vivo. 2008;22:305–309. [PubMed] [Google Scholar]
- 36.Yang Q., Li K., Huang X., Zhao C., Mei Y., Li X., Jiao L., Yang H. lncRNA SLC7A11-AS1 promotes chemoresistance by blocking SCFβ-TRCP-mediated degradation of NRF2 in pancreatic cancer. Mol. Ther. Nucleic Acids. 2020;19:974–985. doi: 10.1016/j.omtn.2019.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L., Feng W., Yang X., Yang F., Wang R., Ren Q., Zhu X., Zheng G. Fbxw11 promotes the proliferation of lymphocytic leukemia cells through the concomitant activation of NF-κB and β-catenin/TCF signaling pathways. Cell Death Dis. 2018;9:427. doi: 10.1038/s41419-018-0440-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q., Zheng J., Liu L. The long noncoding RNA PCGEM1 promotes cell proliferation, migration and invasion via targeting the miR-182/FBXW11 axis in cervical cancer. Cancer Cell Int. 2019;19:304. doi: 10.1186/s12935-019-1030-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang J., Wang W.F., Xie S., Zhang X.L., Qi W.F., Zhou X.P., Hu J.X., Shi Q., Yu R.T. β-transducin repeat-containing E3 ubiquitin protein ligase inhibits migration, invasion and proliferation of glioma cells. Oncol. Lett. 2017;14:3131–3135. doi: 10.3892/ol.2017.6533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He N., Li C., Zhang X., Sheng T., Chi S., Chen K., Wang Q., Vertrees R., Logrono R., Xie J. Regulation of lung cancer cell growth and invasiveness by β-TRCP. Mol. Carcinog. 2005;42:18–28. doi: 10.1002/mc.20063. [DOI] [PubMed] [Google Scholar]
- 41.Kim S.E., Yoon J.Y., Jeong W.J., Jeon S.H., Park Y., Yoon J.B., Park Y.N., Kim H., Choi K.Y. H-Ras is degraded by Wnt/β-catenin signaling via β-TrCP-mediated polyubiquitylation. J. Cell Sci. 2009;122:842–848. doi: 10.1242/jcs.040493. [DOI] [PubMed] [Google Scholar]
- 42.Bufalieri F., Infante P., Bernardi F., Caimano M., Romania P., Moretti M., Lospinoso Severini L., Talbot J., Melaiu O., Tanori M., Di Magno L., Bellavia D., Capalbo C., Puget S., De Smaele E. ERAP1 promotes Hedgehog-dependent tumorigenesis by controlling USP47-mediated degradation of βTrCP. Nat. Commun. 2019;10:3304. doi: 10.1038/s41467-019-11093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meek D.W. Tumour suppression by p53: A role for the DNA damage response? Nat. Rev. Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 44.Wu J., Lu L.Y., Yu X. The role of BRCA1 in DNA damage response. Protein Cell. 2010;1:117–123. doi: 10.1007/s13238-010-0010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shiloh Y., Ziv Y. The ATM protein kinase: Regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 2013;14:197–210. [PubMed] [Google Scholar]
- 46.Bakkenist C.J., Kastan M.B. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421:499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 47.O’Connor P.M., Jackman J., Bae I., Myers T.G., Fan S., Mutoh M., Scudiero D.A., Monks A., Sausville E.A., Weinstein J.N., Friend S., Fornace A.J., Kohn K.W. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–4300. [PubMed] [Google Scholar]
- 48.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 49.Janic A., Valente L.J., Wakefield M.J., Di Stefano L., Milla L., Wilcox S., Yang H., Tai L., Vandenberg C.J., Kueh A.J., Mizutani S., Brennan M.S., Schenk R.L., Lindqvist L.M., Papenfuss A.T. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat. Med. 2018;24:947–953. doi: 10.1038/s41591-018-0043-5. [DOI] [PubMed] [Google Scholar]
- 50.Santra M.K., Wajapeyee N., Green M.R. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature. 2009;459:722–725. doi: 10.1038/nature08011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 52.Nishikawa H., Ooka S., Sato K., Arima K., Okamoto J., Klevit R.E., Fukuda M., Ohta T. Mass spectrometric and mutational analyses reveal Lys-6-linked polyubiquitin chains catalyzed by BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 2004;279:3916–3924. doi: 10.1074/jbc.M308540200. [DOI] [PubMed] [Google Scholar]
- 53.Beuria T.K., Krishnakumar S.S., Sahar S., Singh N., Gupta K., Meshram M., Panda D. Glutamate-induced assembly of bacterial cell division protein FtsZ. J. Biol. Chem. 2003;278:3735–3741. doi: 10.1074/jbc.M205760200. [DOI] [PubMed] [Google Scholar]
- 54.Olive P.L., Banáth J.P. The comet assay: A method to measure DNA damage in individual cells. Nat. Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data presented in the paper are contained within the article.