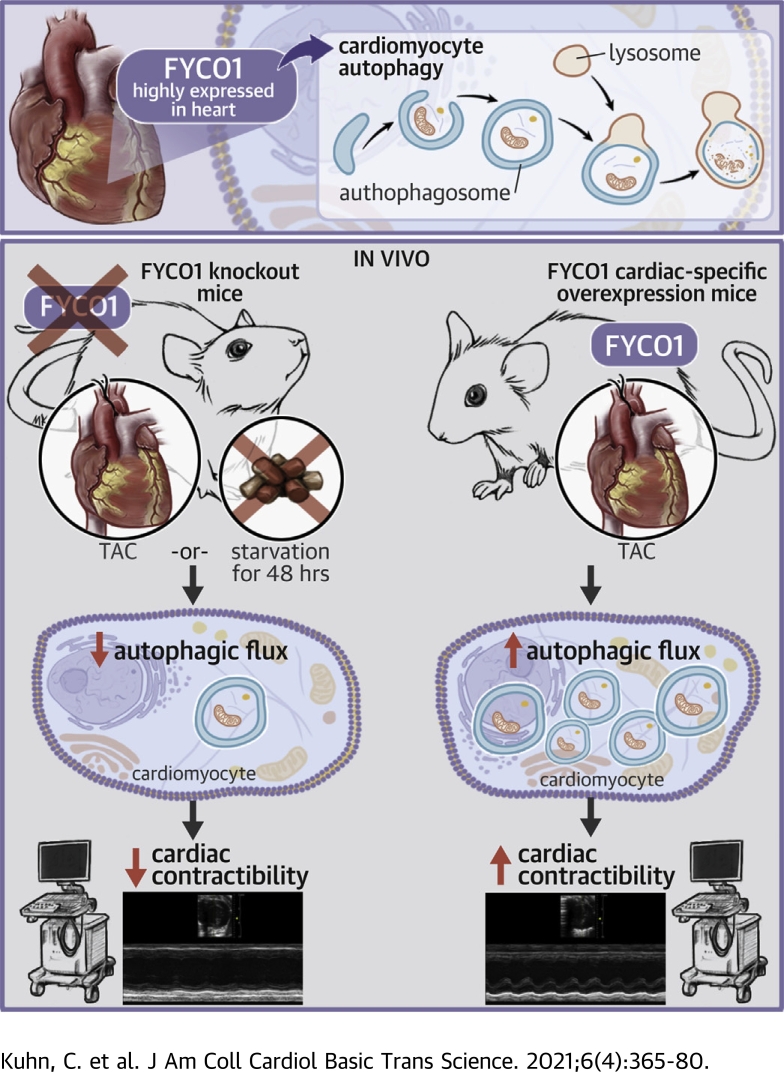
Visual Abstract
Key Words: autophagy, FYCO1, heart failure
Abbreviations and Acronyms: BFA, bafilomycin A1; CSA, cell surface area; GFP, green fluorescent protein; KO, knockout; MHC, myosin heavy chain; microRNA, micro–ribonucleic acid; mRNA, messenger ribonucleic acid; NRCM, neonatal rat cardiomyocytes; RFP, red fluorescent protein; TAC, transverse aortic constriction; TG, transgenic; WT, wild-type
Highlights
-
•
FYCO1, a component of the autophagic machinery, is highly expressed in the heart and a potent inducer of cardiomyocyte autophagy.
-
•
Loss of FYCO1 in vivo inhibits adaptation to starvation or biomechanical stress of the heart by an abrogated increase of autophagic flux and results in contractile dysfunction.
-
•
Heart specific overexpression of FYCO1 improves autophagic flux and rescues contractile dysfunction following pressure overload.
Summary
Autophagy is a cellular degradation process that has been implicated in diverse disease processes. The authors provide evidence that FYCO1, a component of the autophagic machinery, is essential for adaptation to cardiac stress. Although the absence of FYCO1 does not affect basal autophagy in isolated cardiomyocytes, it abolishes induction of autophagy after glucose deprivation. Likewise, Fyco1-deficient mice subjected to starvation or pressure overload are unable to respond with induction of autophagy and develop impaired cardiac function. FYCO1 overexpression leads to induction of autophagy in isolated cardiomyocytes and transgenic mouse hearts, thereby rescuing cardiac dysfunction in response to biomechanical stress.
Heart failure is a major cause of morbidity and mortality worldwide (1). Several cardiovascular diseases, such as myocardial infarction, inherited cardiomyopathy, and arterial hypertension, can result in the clinical syndrome of heart failure. The pathophysiology of heart failure and cardiomyopathy is complex and still incompletely understood. Despite its heterogeneous etiology, heart failure typically involves cardiac remodeling, which is characterized by progressive changes in cellular size, structure, and function along with molecular alterations (2), subsequently prompting altered systolic and diastolic contractile function.
Only recently, protein homeostasis came into the focus of attention as an additional mechanism contributing to the pathogenesis of heart failure and cardiomyopathy. Besides the well-established ubiquitin-proteasome system, autophagy is another major mechanism to regulate protein turnover (3). Autophagy is a lysosome-dependent degradation mechanism, and 3 types of autophagy can be distinguished: macroautophagy, microautophagy and chaperone-mediated autophagy (4). Macroautophagy is characterized by the inclusion of cytosolic material, from proteins to organelles, into double-membrane vesicles termed autophagosomes. Autophagosomes fuse with lysosomes, where their cargo is degraded and finally released for intracellular recycling. Thereby, autophagy supports the cellular adaptation to environmental stress.
Previously we conducted a bioinformatics search for cardiac enriched genes in expressed sequence tag databases to identify novel genes that are involved in cardiac hypertrophy and failure (5). In this screen we identified several sequences corresponding to the gene FYCO1 (FYVE and coiled-coil domain-containing protein 1). As predicted, FYCO1 is highly enriched in human cardiac and skeletal muscle (6). FYCO1 directly interacts with LC3, Rab7, and phosphatidylinositol-3-phosphate, key players in autophagy (7). However, the functional role of FYCO1 in the heart, where it is predominantly expressed, still remains elusive.
Methods
Isolation and culture of neonatal rat cardiomyocytes
Neonatal rat cardiomyocytes (NRCMs) were isolated as described previously (8). NRCMs were infected at a multiplicity of infection of 25 with an adenovirus 24 h after isolation. For glucose deprivation, we applied glucose-free Dulbecco’s modified Eagle’s medium without serum supplementation for 24 h. Bafilomycin A1 (BFA; Sigma-Aldrich, St. Louis, Missouri) was resolved in dimethyl sulfoxide and used at a final concentration of 50 nmol/l for 4 h.
Cloning and generation of adenoviruses
Cloning of human FYCO1 into a pDonR201 gateway vector (Life Technologies, Carlsbad, California) was carried out using standard polymerase chain reaction techniques. An adenovirus encoding full-length human FYCO1 complementary deoxyribonucleic acid with a C-terminal V5 tag was generated using the ViraPower Adenoviral Expression System (Life Technologies). A galactosidase V5–encoding adenovirus served as control (Life Technologies).
Oligonucleotides encoding synthetic micro–ribonucleic acids (microRNAs) specifically targeting FYCO1 were designed using OligoPerfect software (Thermo Fisher Scientific, Waltham, Massachusetts). Sequences are outlined in the Supplemental Appendix. Oligonucleotides were cloned into the pcDNA6.2-GW/miR vector. As a negative control, we used the pcDNA6.2-GW/miR plasmid, which can form a hairpin structure and is consecutively processed into a mature microRNA yet is predicted not to target any known mammalian gene. Adenoviruses encoding synthetic microRNAs were generated using the ViraPower Adenoviral Expression System.
Immunoblotting
Cells were harvested in a lysis buffer containing 20 mmol/l Tris, pH 7.5, 12.5% (v/v) glycerol, 10 mmol/l dithiothreitol, 500 mmol/l NaCl, and 1% (v/v) Nonidet P 40 (Sigma-Aldrich), supplemented with protease inhibitor cocktail tablets (Roche, Basel, Switzerland) as well as phosphatase inhibitor cocktail 2 and 3 (Sigma-Aldrich). After up to 3 brief freeze-and-thaw cycles and a centrifugation step, whole-cell lysate was obtained.
Heart tissue was harvested, immediately transferred into lysis buffer, and homogenized using an Ultra-Turrax T25 tissue separator (Janke & Kunkel, Staufen im Breisgau, Germany). Whole-cell extracts and heart homogenates were resolved using sodium dodecyl sulfate polyacrylamide-gel electrophoresis and transferred to a nitrocellulose or polyvinylidene difluoride membrane. Application of the primary antibody was followed by a horseradish peroxidase-coupled or Cy3-coupled secondary antibody (Supplemental Table 1).
Tissue processing and immunochemistry
For histological staining, whole mouse hearts were embedded in Tissue-Tek O.C.T. Compound (Sakura, Osaka, Japan). Serial 7-μm-thick cryosections were cut with a refrigerated microtome (CryoStar NX70, Thermo Fisher Scientific). Cryosections and NRCMs were washed with phosphate-buffered saline and then fixed in 4% (w/v) paraformaldehyde (Sigma-Aldrich) in phosphate-buffered saline for 10 min at room temperature. After permeabilization, blocking solution was applied, followed by incubation with primary and secondary antibodies. All images were captured at room temperature using the BZ-9000E HS all-in-one fluorescence microscope (Keyence, Osaka, Japan) with the BZ-II Viewer version 2.1 (Keyence). Bright-field macro images of whole-heart sections were taken in Z-sync and AutoRangePhoto mode with a 20% overlap between neighboring pictures and merged automatically using BZ-II Analyzer version 2.1 (Keyence). A detailed description can be found in the Supplemental Appendix.
Electron microscopy
Electron microscopy was carried out as described previously (9). The heart was perfused with 1% procaine in 0.1 mol/l phosphate-buffered saline and fixed with 6% glutaraldehyde in 0.1 mol/l phosphate-buffered saline by transcardial vascular perfusion to harvest papillary muscles. Tissue blocks were post-fixed with 2% osmium tetroxide and embedded in araldite. Ultrathin sections were processed with uranyl acetate and lead citrate and viewed using a Zeiss EM 900 microscope (Zeiss, Oberkochen, Germany).
Generation of Fyco1-knockout and transgenic mice
For generation of Fyco1-deficient mice, exons 4 and 5 were flanked by loxP, neomycin selection marker by FRT, and puromycin by F3 sites. Constitutive knockouts (KOs) were produced by Cre-mediated deletion of exons 4 and 5. Moreover, deletion of exons 4 and 5 resulted in frame shift by splicing from exon 3. Fyco1-KO mice were generated in cooperation with TaconicArtemis (Leverkusen, Germany) in the background of C57BL/6N.
To generate transgenic (TG) mice, a complementary deoxyribonucleic acid encoding full-length human FYCO1 was cloned into a plasmid containing the α-myosin heavy chain (α-MHC) promoter, a carboxyterminal Flag tag, and a human growth hormone 1 polyadenylation signal, which was a kind gift of Jeffrey Robbins (10). Microinjections of the linearized construct into pronuclei of fertilized oocytes and implantation of the microinjected eggs into pseudopregnant female mice were performed at Interfakultäre Biomedizinische Forschungseinrichtung at the University of Heidelberg. Oocytes and surrogate mothers had the same C57BL/6N genetic background.
TG and KO offspring were identified by polymerase chain reaction analysis of genomic DNA isolated from tail biopsies.
Animal experiments
For starvation experiments, female mice (8 to 9 weeks old) had unrestricted access to water but did not receive food for 48 h. Animals were examined by echocardiography and killed for further analyses directly after the starvation period.
For analyses of autophagic flux in vivo, we applied chloroquine 50 mg/kg or vehicle (NaCl 0.9%) intraperitoneally. The animals were starved for 48 h and were injected 4 h before the end of the experiment.
Transverse aortic constriction (TAC) was performed in female KO mice and their wild-type (WT) littermates (11 to 13 weeks old) for 1 and 2 weeks. TAC was performed in male TG mice and their WT littermates for 2 weeks. A detailed description can be found in the Supplemental Appendix.
Statistical analysis
All results are shown as mean ± SEM unless stated otherwise. Statistical analyses of the data were carried out using 1-way or 2-way analysis of variance followed by Student-Newman-Keuls post hoc tests. If appropriate, Student’s t-test (2 sided) or a rank sum test was used. P values <0.05 were considered to indicate statistical significance. For statistical analyses, we applied SigmaPlot version 13.0 (Systat Software, San Jose, California) and GraphPad Prism version 8.4.3 (GraphPad Software, La Jolla, California).
Study approval
Animal handling was performed according to the institutional guidelines of the University Medical Center Schleswig-Holstein as well as of the state of Schleswig-Holstein, Germany. All animal experiments were approved by the state of Schleswig-Holstein (V312-72241.121-4 [12-1/12] and V242-7224.121-4 [64-6/14]). Human left ventricular myocardial tissue was taken from explanted hearts with end-stage heart failure. Healthy donor hearts that could not be transplanted were used as controls. The study conformed with the principles outlined in the Declaration of Helsinki and was reviewed and approved by the ethics committee of the University Hospital Hamburg (Az. 532/116/9.7.1991).
Results
Overexpression of FYCO1 induces autophagy in cardiomyocytes in vitro
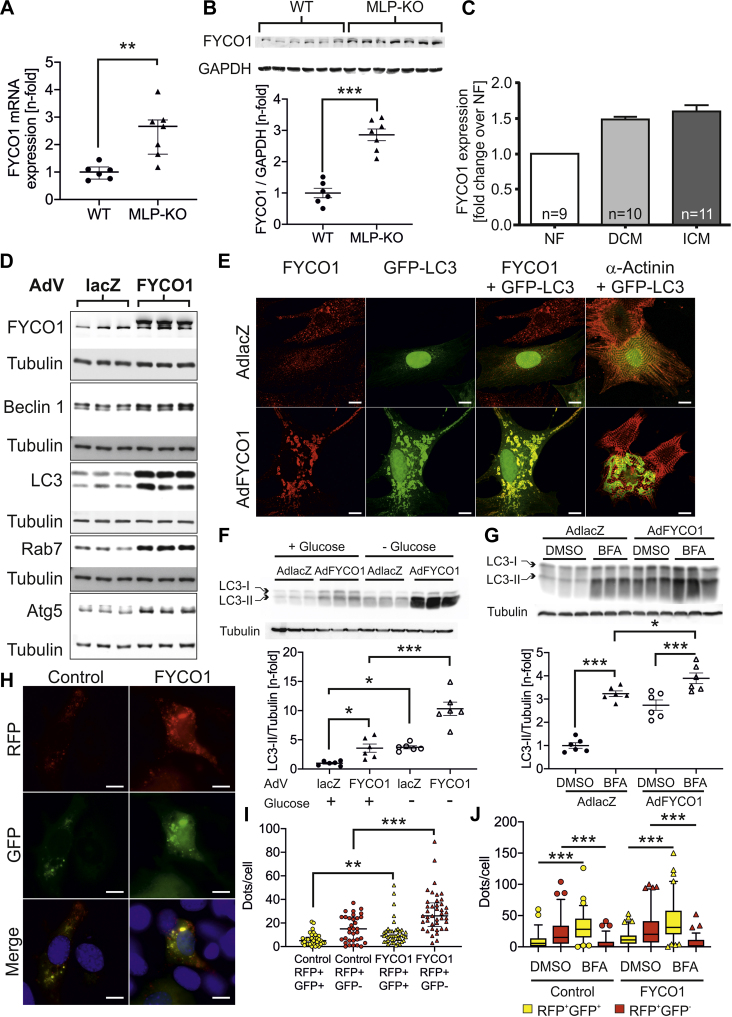
As FYCO1 messenger ribonucleic acid (mRNA) is highly enriched in heart muscle (Supplemental Figures 1A to 1C) (6) and in cardiomyocytes (Supplemental Figure 1D), we first examined the expression of FYCO1 under cardiac disease conditions. In muscle LIM protein–KO mice, an established model of dilated cardiomyopathy (11), we observed induction of Fyco1 mRNA and protein (Figures 1A and 1B). Likewise, FYCO1 mRNA is up-regulated in human dilated and ischemic cardiomyopathy (Figure 1C). However, in contrast to these results, FYCO1 protein is reduced in the heart of human patients with dilated and ischemic cardiomyopathy (Supplemental Figure 1E).
Figure 1.
Overexpression of FYCO1 Induces Autophagy In Vitro
(A) FYCO1 (FYVE and coiled-coil domain-containing protein 1) messenger RNA (mRNA) (data are presented as dot plots with median and 25th and 75th percentiles; Mann-Whitney rank sum test) and (B) protein (data are presented as dot plots with mean ± SEM; 2-sided Student’s t-test) are induced in hearts of muscle LIM protein (MLP)–knockout (KO) mice, a model of dilated cardiomyopathy (DCM), as indicated by quantitative polymerase chain reaction and immunoblotting. (C) FYCO1 mRNA expression in human DCM and ischemic cardiomyopathy (ICM) using nCounter technology (NanoString, Seattle, Washington). (D) Immunoblots for autophagic proteins after adenoviral overexpression of FYCO1 (multiplicity of infection [MOI] = 25) in neonatal rat cardiomyocytes (NRCMs). An adenovirus (AdV) encoding β-galactosidase (lacZ, MOI = 25) serves as control. (E) Immunocytochemistry of NRCMs that overexpress FYCO1. Immunostaining was performed with green fluorescent protein (GFP)–fused LC3 (green) and antibodies against FYCO1 or α-actinin (red). (F) LC3-II expression in NRCMs that overexpress FYCO1 and have been starved by glucose deprivation for 24 h. (G) LC3-II expression in NRCMs that overexpress FYCO1 and have been treated with bafilomycin A1 (BFA) for 4 h. Data are presented as dot plots with mean ± SEM (2-way analysis of variance [ANOVA] followed by Student-Newman-Keuls post hoc tests). (H,I) Quantification of autophagosomes/autolysosomes in C2C12 myoblasts labeled by a tandem fluorescent (red fluorescent protein [RFP], GFP) LC3. C2C12 cells overexpress FYCO1 simultaneously (data are presented as dot plots with median and 25th and 75th percentiles; Mann-Whitney rank sum test). (J) Quantification of autophagosomes/autolysosomes in C2C12 myoblasts labeled by a tandem fluorescent (RFP, GFP) LC3. C2C12 cells overexpress FYCO1 simultaneously and are treated with BFA. Data are presented as box plots with median and 5th and 95th percentile whiskers (Kruskal-Wallis ANOVA on ranks followed by Dunn’s post hoc tests). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Scale bar, 10 μm. DMSO = dimethyl sulfoxide; GAPDH = glyceraldehyde 3-phosphate dehydrogenase; NF = non-failing hearts; WT = wild-type.
To study the effects of FYCO1 overexpression on cardiomyocyte autophagy, we generated an adenovirus encoding FYCO1. An adenovirus encoding lacZ served as control. Overexpression of FYCO1 in NRCMs was accompanied by significantly higher levels of beclin-1, LC3-II, Rab7, and Atg5 (Figure 1D). Furthermore, overexpression of FYCO1 in cardiomyocytes resulted in an accumulation of large vesicles labeled by green fluorescent protein (GFP)–fused LC3 (12) and FYCO1 throughout the cells (Figure 1E). Starvation of NRCMs by glucose deprivation for 24 h led to increased LC3-II levels in control cells. LC3-II levels were further augmented by FYCO1 overexpression in starved NRCMs (Figure 1F). Next, we used BFA, a pharmacological inhibitor of autophagosome lysosome fusion, to examine the effects of FYCO1 overexpression on the autophagic flux. First, we observed the expected accumulation of LC3-II in control NRCMs treated with BFA (Figure 1G). Although FYCO1 overexpression raised the content of LC3-II compared with control NRCMs treated with vehicle (dimethyl sulfoxide), BFA treatment exaggerated LC3-II accumulation caused by FYCO1 overexpression, consistent with an induction of the autophagic flux by FYCO1. Because FYCO1 is expressed in skeletal muscle at high levels, we also analyzed the autophagic flux in C2C12 myoblasts. Transfection of a tandem fluorescent (red fluorescent protein [RFP] and GFP) LC3 (13) enabled the pH-dependent visualization of autophagosomes and autophagolysosomes. FYCO1 overexpression caused an increase in red (RFP+GFP−) and yellow (RFP+GFP+) dots per cell, suggesting increased autophagic flux rather than a block of degradation (Figures 1H and 1I). As predicted, treatment with BFA led to an accumulation of yellow dots and a dramatic decrease of red-only dots in control cells. Overexpression of FYCO1 produced a very similar regulation (Figure 1J). Taken together, these data indicate that autophagosome accumulation in response to FYCO1 overexpression is caused by an increased autophagic flux rather than by a block at late stages of autophagosome maturation.
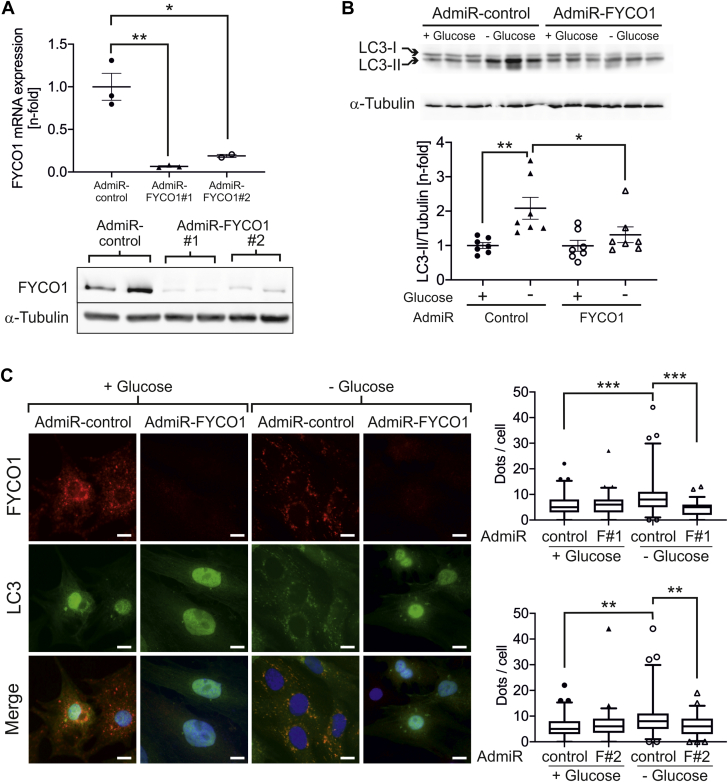
Knockdown of Fyco1 inhibits cardiomyocyte autophagy
To analyze potential opposite effects on cardiomyocyte autophagy by Fyco1 down-regulation, we generated adenoviruses encoding 2 different synthetic microRNAs targeting Fyco1. Both of these constructs produced a significant knockdown of Fyco1 at the mRNA and protein levels (Figure 2A) in NRCMs. Glucose deprivation of control NRCMs, transduced with a microRNA that has no target, resulted in elevated LC3-II levels. In contrast, knockdown of Fyco1 almost completely inhibited this effect (Figure 2B). Using a GFP-tagged LC3 adenovirus, we were able to visualize autophagosomes in cardiomyocytes. Glucose-deprived cells that had been treated with a control microRNA showed formation of autophagosomes labeled by LC3 and Fyco1. Yet significantly fewer autophagosomes could be detected in starved cardiomyocytes after Fyco1 knockdown caused by either synthetic microRNA (Figure 2C).
Figure 2.
Knockdown of FYCO1 Inhibits Cardiomyocyte Autophagy
(A) FYCO1 mRNA and protein expression in NRCMs after adenoviral transduction (MOI = 25) with 2 different synthetic micro–ribonucleic acids (microRNAs) that target FYCO1. Data are presented as dot plots with mean ± SEM (2-sided Student’s t-test). (B) LC3-II expression in NRCMs transduced with synthetic microRNA against FYCO1 and treated with glucose deprivation for 24 h. Data are presented as dot plots with mean ± SEM (2-way ANOVA followed by Student-Newman-Keuls post hoc tests). (C) Quantification of autophagosomes labeled by GFP-LC3 in NRCMs that overexpress a microRNA against FYCO1 and have been starved by glucose deprivation for 24 h. Data are presented as box plots with median and 5th and 95th percentile whiskers (Kruskal-Wallis ANOVA on ranks followed by Wilcoxon rank sum tests). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗ p < 0.001. Scale bar, 10 μm. Abbreviations as in Figure 1.
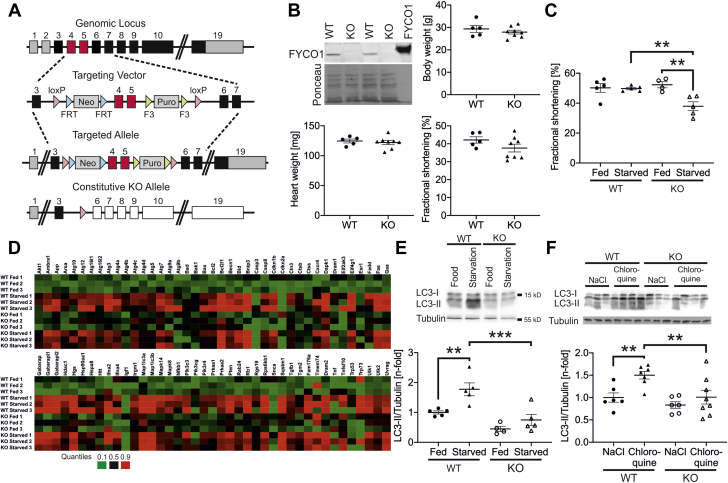
Fyco1 deficiency prevents induction of autophagy in vivo
To gain further insight into the regulation of autophagy in the heart by Fyco1 in vivo, we next generated Fyco1-deficient mice (KO) by targeted deletion of exons 4 and 5 as well as by introducing a frame shift in the downstream exons (Figure 3A). The complete absence of Fyco1 in homozygous KO mice was confirmed at the protein level (Figure 3B). KO mice were born in expected Mendelian ratios and developed normally until adulthood. At the age of 4 months (Supplemental Figures 2A to 2C) and at the age of 1 year, Fyco1-deficient mice did not display differences in body weight, heart weight, or cardiac function (Figure 3B and Supplemental Table 2).
Figure 3.
Fyco1 Deficiency Prevents Induction of Autophagy Upon Starvation In Vivo
(A) Targeting strategy of FYCO1. (B) Fyco1 expression in hearts from homozygous Fyco1-KO mice. Body weight, heart mass, and echocardiographic analysis of fractional shortening in female KO mice at the age of 1 year. Data are presented as dot plots with mean ± SEM (2-sided Student’s t-test). (C) Echocardiographic analysis of fractional shortening in Fyco1-KO mice that have been starved for 48 h. (D) Heatmap of the expression profile of autophagic genes of fed and starved WT and Fyco1-KO mice, which was assessed using real-time polymerase chain reaction arrays (n-fold regulation on the basis of fed WT: black, 50th percentile; green, 10th percentile; red, 90th percentile). (E) LC3-II expression in hearts from Fyco1-KO mice after starvation for 48 h. (F) LC3-II expression in hearts from Fyco1-KO mice after starvation for 48 h and treatment with chloroquine (50 mg/kg intraperitoneally for 4 h). Data are presented as dot plots with mean ± SEM (2-way ANOVA followed by Student-Newman-Keuls post hoc tests). ∗∗p < 0.01 and ∗∗∗p < 0.001. Neo = neomycin; Puro = puromycin; other abbreviations as in Figure 1.
Autophagic flux is active in cardiomyocytes under basal conditions and is usually induced by stress conditions such as starvation (14). In the heart tissue, we did not observe a significant reduction of LC3-II under baseline conditions. We thus challenged Fyco1-KO mice by food deprivation. Fyco1-KO animals and WT controls had unrestricted access to water but did not receive food for 48 h. Whereas starved WT mice showed normal contractile function despite food deprivation, KO mice revealed significantly impaired fractional shortening as assessed by echocardiography (Figure 3C and Supplemental Table 3). Systematic analysis of 82 autophagy-related genes revealed an induction of the majority of genes in starved WT as well as in starved KO animals (Figure 3D). Yet whereas starvation of WT mice led to significant accumulation of LC3-II protein in the heart, Fyco1-deficient mice were resistant to this effect. Although Map1lc3a (n = 3 per group) and Map1lc3b (n = 3 per group) encoding LC3 were induced in heart tissue of KO animals, LC3-II protein remained unchanged in starved KO mice (Figure 3E). Consistently, in cardiomyocytes isolated from neonatal WT mice, we observed an increase of LC3-II levels after glucose deprivation, which was absent in cardiomyocytes isolated from KO animals (Supplemental Figure 2D). To analyze autophagic flux in vivo, we starved additional WT and KO mice for 48 h. These animals were treated with either chloroquine (50 mg/kg) or vehicle (NaCl 0.9%) intraperitoneally 4 h before the end of the experiment. Treatment with chloroquine resulted in a 1.5 ± 0.1-fold accumulation of LC3-II in starved WT mice compared with starved WT animals that received vehicle only. In contrast, Fyco1-KO mice were resistant to LC3-II accumulation after starvation and chloroquine treatment (Figure 3F), consistent with impaired autophagic flux.
Fyco1-deficient mice are resistant to autophagy induced by pressure overload
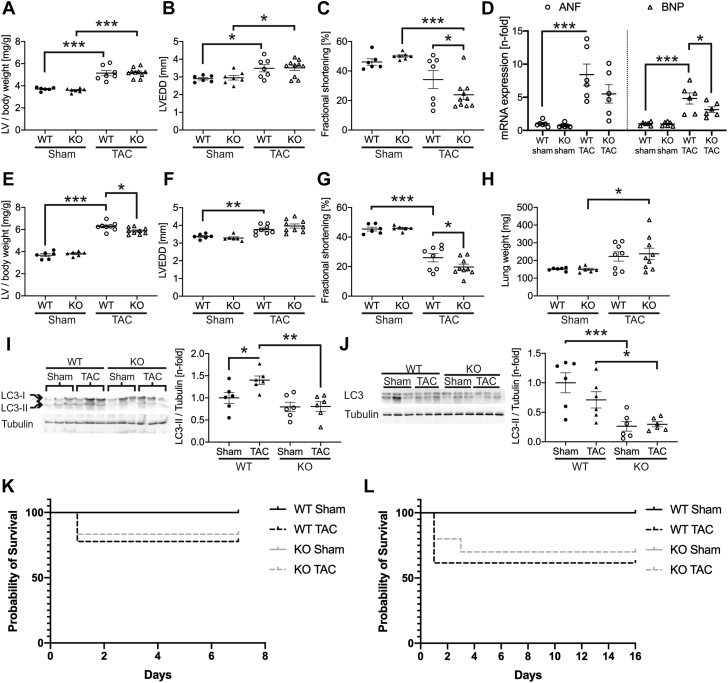
As FYCO1 protein is up-regulated in response to pressure overload in mice (Supplemental Figures 3A and 3B) we subjected Fyco1-deficient mice to severe TAC. After 1 week of pressure overload, WT and KO mice developed left ventricular hypertrophy and ventricular dilation to a similar degree (Figures 4A and 4B). Yet fractional shortening in Fyco1-KO mice that had undergone TAC was found to be significantly impaired compared with banded WT animals (Figure 4C and Supplemental Table 4). Whereas atrial natriuretic factor/Nppa and brain natriuretic peptide/Nppb, members of the hypertrophic gene program, were expressed at higher levels in WT mice after TAC, expression of Nppb was significantly lower, and Nppa trended toward lower levels in banded KO animals (Figure 4D). After 2 weeks of TAC, WT and KO animals developed hypertrophy and left ventricular dilation to the same extent (Figures 4E and 4F). Cardiac function of banded WT mice worsened, but the impairment of fractional shortening was still more pronounced in KO mice (Figure 4G and Supplemental Table 5). Both banded WT and KO mice showed signs of pulmonary congestion, measured by increased lung weights (Figure 4H). Moreover, cardiac hypertrophy after 1 week was accompanied by elevated levels of LC3-II in WT TAC mice, while accumulation of LC3-II was absent in Fyco1-deficient mice (Figure 4I), consistent with inhibition of autophagy upon pressure overload in vivo. Interestingly, after 2 weeks of severe TAC, we did not observe elevated LC3-II levels in banded WT mice anymore, but we still detected lower levels of LC3-II in sham-operated and banded KO mice (Figure 4J). Survival after TAC was not affected by genotype (Figures 4K and 4L).
Figure 4.
Fyco1-Deficient Mice Are Resistant to Autophagy Induced by Pressure Overload
Fyco1-KO mice and WT littermates were subjected to transverse aortic constriction (TAC). Ratio of left ventricular (LV) mass to body weight (A) and echocardiographic analyses of LV end-diastolic diameter (LVEDD) (B) and fractional shortening (C) after 1 week of TAC. (D) Quantification of Nppa/ANF (atrial natriuretic factor) and Nppb/BNP (brain natriuretic peptide) expression after 1 week of TAC. (E) LV mass/body weight and echocardiographic analyses of (F) LVEDD and (G) fractional shortening after 2 weeks of TAC. (H) Lung weight after 2 weeks of TAC. LC3-II protein expression in the left ventricle after 1 week (I) and after 2 weeks (J) of severe TAC. All datasets are presented as dot plots with mean ± SEM (2-way ANOVA followed by Student-Newman-Keuls post hoc tests). Kaplan-Meier survival curves of WT and KO mice during (K) 1 week and during (L) 2 weeks of TAC. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Abbreviations as in Figure 1.
Cardiac-specific overexpression of FYCO1 in vivo induces autophagy and causes mild hypertrophy of the heart
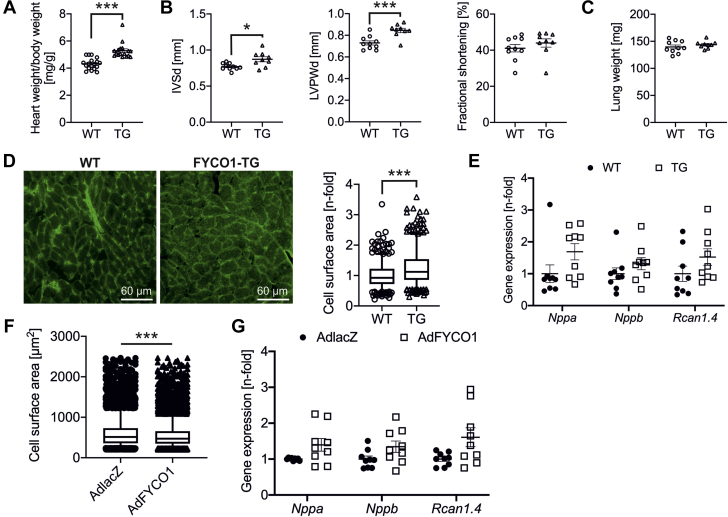
Next, we generated a TG mouse model that overexpresses human FYCO1 under control of an α-MHC promoter (10). We established 6 founder lines (lines A to F), 2 of which revealed significant FYCO1 protein expression. The overexpression of FYCO1 in 3-month-old male mice was moderate in line A (13.1 ± 1.3-fold) and high in line F (20.1 ± 2.4-fold) (Supplemental Figure 4A). The phenotypes of these 2 TG mouse lines were similar under basal conditions. Here we present the data of line F. Morphometric and echocardiographic data for TG mouse line A are shown in Supplemental Table 6. TG mice did not differ from WT littermates with respect to body weight but developed a significantly higher ratio of heart weight to body weight, indicating mild cardiac hypertrophy (Figure 5A). Likewise, echocardiography revealed a slightly thickened septum and left ventricular posterior wall in TG mice, while fractional shortening was unchanged (Figure 5B and Supplemental Table 7). Lung weight as a surrogate marker of cardiac dysfunction did not vary in TG animals (Figure 5C). Cardiomyocyte cell surface area (CSA) was significantly higher in FYCO1 TG mice than in control animals, while the expression of hypertrophic marker genes (Nppa, Nppb, and Rcan1.4) did not significantly differ (Figures 5D and 5E). Overexpression of FYCO1 in NRCMs was associated with 8% smaller CSAs than in control cells that overexpressed β-galactosidase (Figure 5F). As in TG animals, overexpression of FYCO1 did not result in an induction of Nppa, Nppb, and Rcan1.4 either (Figure 5G). Interestingly, treatment with the hypertrophic agent phenylephrine did not change the subcellular localization of autophagosomes in FYCO1-overexpressing NRVCMs compared with starved cells (Supplemental Figure 4B).
Figure 5.
FYCO1-TG Mice Reveal Mild Cardiac Hypertrophy Without Induction of Maladaptive Hypertrophic Genes
(A) Heart/body weight ratios of transgenic (TG) mice with cardiac-specific overexpression of FYCO1 (TG) and their WT littermates. (B) Echocardiographic analyses of interventricular septal thickness in diastole (IVSd), left ventricular posterior wall thickness in diastole (LVPWd), and fractional shortening. (C) Lung weight of TG and WT mice. Data are presented as dot plots with mean ± SEM (2-sided Student’s t test). (D) Quantification of cardiomyocyte CSAs by staining of TG and WT heart sections with wheat germ agglutinin lectin. Data are presented as box plots with median and 5th and 95th percentile whiskers (2-tailed Mann-Whitney rank sum test). (E) Gene expression of Nppa, Nppb, and Rcan1.4 by quantitative polymerase chain reaction (qPCR). Data are presented as dot plots with mean ± SEM (2-sided Student’s t-test). (F) Quantification of CSA in isolated NRCMs that overexpress FYCO1. Data are presented as box plots with median and 5th and 95th percentile whiskers (2-tailed Mann-Whitney rank sum test). (G) Expression of hypertrophic genes Nppa, Nppb, and Rcan1.4 by qPCR in NRCMs after overexpression of FYCO1 (MOI = 50). Data are presented as dot plots with mean ± SEM (2-sided Student’s t-test). ∗p < 0.05 and ∗∗∗p < 0.001. Abbreviations as in Figure 1.
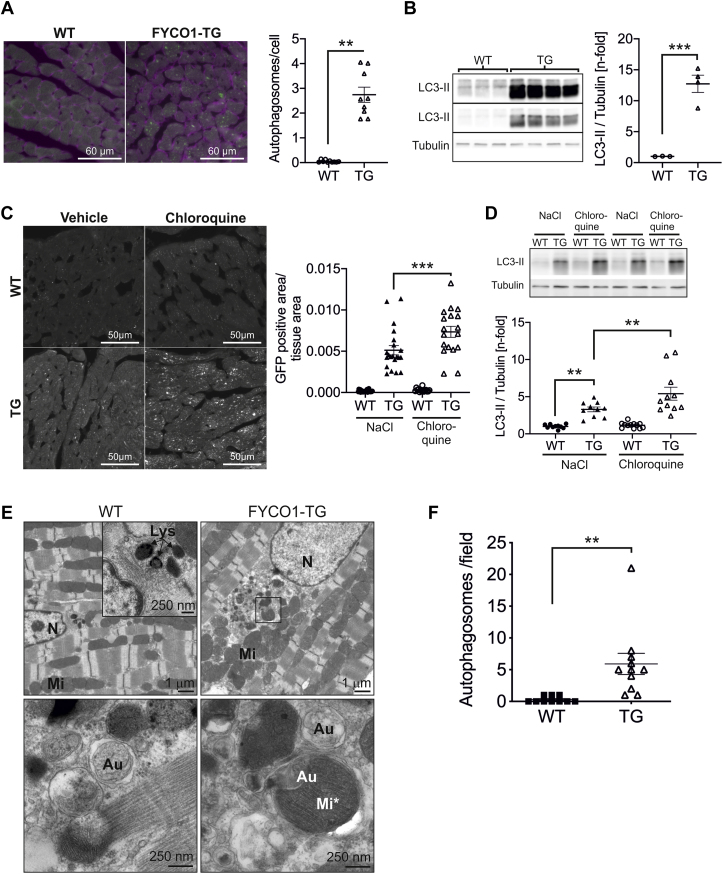
To analyze autophagy in FYCO1-TG mice, we generated double-TG mice that overexpressed FYCO1 and GFP-LC3 (14) simultaneously and determined the number of autophagosomes. Mice that overexpressed only GFP-LC3 served as a control group. Indeed, we found more autophagosomes in FYCO1-TG mice than in control animals (Figure 6A). Accordingly, overexpression of FYCO1 in vivo led to significant accumulation of LC3-II protein (Figure 6B). Next, we applied chloroquine (50 mg/kg) or vehicle (NaCl 0.9%) intraperitoneally for 4 h to analyze autophagic flux in TG mice. Animals were starved for 48 h. Although the accumulation of autophagosomes and LC3-II was mild and not significant in WT mice after chloroquine treatment, in FYCO1-TG mice, the number of autophagosomes and LC3-II was higher in vehicle-treated animals and rose further with chloroquine treatment (Figure 6C). Immunoblotting for LC3-II produced similar results (Figures 6D). Electron microscopy revealed a normal ultrastructure of autophagosomes in TG mice compared with WT mice (Figure 6E), with an increase of autophagosomes in TG (Figure 6F). The sarcomere structure did not show signs of disarray, which might accompany pathological cardiomyocyte hypertrophy.
Figure 6.
FYCO1 Overexpression Causes Induction of Autophagy In Vivo
(A) Quantification of cardiac autophagosomes per cell in double-TG mice that overexpress FYCO1 and GFP-LC3. Mice that overexpress GFP-LC3 only served as control (3 animals per group, 2-sided Student’s t-test). Cell surface area was determined by wheat germ agglutinin lectin. (B) LC3-II protein expression in FYCO1 transgenic mice (LC3-II with shorter and longer exposure time). (C) Quantification of autophagosomes labeled by GFP-LC3 in heart sections of TG mice that have been starved for 48 h and treated with either chloroquine (50 mg/kg) or vehicle alone (NaCl 0.9%) intraperitoneally for 4 h (3 animals per group, 18–25 images per animal). (D) LC3-II protein expression in starved TG mice after chloroquine treatment. Data are presented as dot plots with mean ± SEM (2-way ANOVA followed by Student-Newman-Keuls post hoc tests). (E) Analysis of papillary muscles by electron microscopy. Scale bar as indicated. (F) Quantification of autophagosomes/autolysosomes per visual field at 3,000× magnification. Organelles were validated at 12,000× and 20,000× magnification (WT: 10 fields from 3 mice; TG: 11 fields from 2 mice). Data are presented as dot plots with mean ± SEM (2-sided Student’s t-test). ∗∗p < 0.01 and ∗∗∗p < 0.001. Au = autophagosome; Lys = lysosome; Mi = mitochondria; Mi∗ = mitochondrium in autophagsome; N = nucleus; other abbreviations as in Figure 1.
FYCO1 overexpression in vivo blunts cardiac dysfunction because of pressure overload
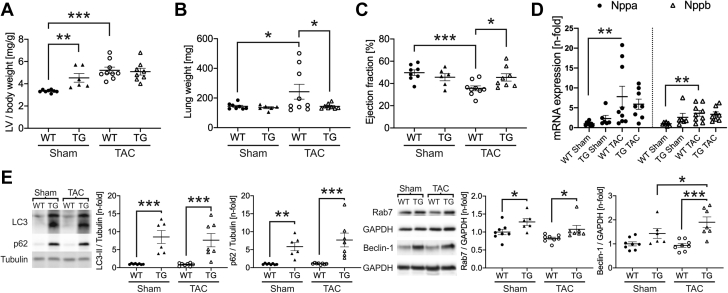
As the loss of FYCO1 is detrimental under cardiac stress conditions, we next asked whether cardiac-specific overexpression of FYCO1 might be beneficial under pressure overload. Thus, we subjected TG mice and their WT littermates to TAC. Banded WT mice developed left ventricular hypertrophy after 2 weeks, as expected. Similar to the baseline data described earlier, sham-operated TG mice revealed mild cardiac hypertrophy. Interestingly, the ratio of left ventricular mass to body weight of TG and WT mice following TAC was identical, implying that the additional hypertrophy due to TAC was less pronounced in TG mice (Figure 7A). Although lung weight as a marker of congestive heart failure was elevated in banded WT mice, banded TG mice did not develop pulmonary congestion (Figure 7B). As expected, WT mice exhibited cardiac dysfunction with reduced ejection fraction and fractional shortening after 2 weeks of severe TAC (Supplemental Table 8). In contrast, the ejection fraction of banded TG animals was markedly better and almost completely preserved (Figure 7C). The hypertrophic phenotype of banded WT animals was accompanied by a significant induction of hypertrophic marker genes (Nppa, Nppb) (Figure 7D). In TG mice, the induction of these genes by TAC was milder and not statistically different compared with sham-operated animals. The autophagic marker proteins LC3-II, p62, and Rab7 were found up-regulated in both sham-operated FYCO1 TG mice and TAC operated TG animals. Only beclin-1 was further induced by severe TAC after 2 weeks in TG mice (Figure 7E).
Figure 7.
FYCO1 Overexpression Blunts Cardiac Dysfunction Due to Pressure Overload
FYCO1 TG and WT mice were subjected to transverse aortic constriction (TAC) for 2 weeks. (A) LV mass/body weight and (B) lung weight as a marker of congestive heart failure after TAC. (C) Echocardiographic analysis of ejection fraction after TAC. (D) Expression of Nppa/ANF and Nppb/BNP by qPCR in left ventricles of banded TG and WT mice. (E) Expression of autophagic proteins LC3-II, p62, Rab7, and beclin-1 in left ventricles of banded TG and WT animals. All datasets are presented as dot plots with mean ± SEM (2-way ANOVA followed by Student-Newman-Keuls post hoc tests). ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001. Abbreviations as in Figures 1, 4, and 5.
To further examine the underlying mechanism that is responsible for the contractile phenotype of FYCO1 deficient and TG mice after TAC, we additionally performed a yeast 2-hybrid screen with FYCO1 as bait. From this screen, we identified several clones encoding beta-MHC (β-MHC/Myh7) as a potential interacting protein. Coimmunoprecipitation of overexpressed Myh7 with FYCO1 confirmed the protein interaction (Supplemental Figure 5A). Myh7 is a prototypical member of the “fetal gene program,” up-regulated in the context of pathological hypertrophy (15). Consistently, Myh7 mRNA was induced in WT mice after TAC. The induction of Myh7 in banded KO or TG mice did not differ significantly from banded WT animals (Supplemental Figure 5B). However, although there was only a trend toward higher Myh7 protein levels in banded KO mice, overexpression of FYCO1 in vivo resulted in significantly mitigated induction of Myh7 protein in banded TG mice (Supplemental Figure 5C), which might contribute to preservation of contractile function in FYCO1 TG mice subjected to increased biomechanical stress. To further test the effect of FYCO1 overexpression in a cell model, we overexpressed FYCO1 in isolated cardiomyocytes. Myh7 expression was significantly reduced by FYCO1 overexpression, while the expression of Myh6 remained unchanged (Supplemental Figure 5D). To examine the role of the autophagic flux potentially modulating Myh7, overexpression of FYCO1 was combined with glucose deprivation and BFA treatment. Again, Myh7 expression was reduced by FYCO1 overexpression. Starvation and inhibition of autophagic flux by BFA exaggerated the suppression by FYCO1 (Supplemental Figure 5E).
In summary, the functional and molecular data support the concept that FYCO1 overexpression is beneficial under circumstances of increased biomechanical stress due to pressure overload.
Discussion
FYCO1 modulates cardiac autophagy
Autophagy is a lysosome-dependent cellular degradation process that has been implicated in diverse disease processes. FYCO1 has recently been linked to autophagy by its interaction with key members of the autophagic machinery: LC3, Rab7, and phosphatidylinositol-3-phosphate (7). Although FYCO1 is highly expressed in striated muscle tissue, its role so far has not been investigated in isolated cardiomyocytes or in the heart in vivo. Thus we aimed to establish its cellular function in cardiomyocytes under basal and “stress” conditions such as glucose deprivation and increased biomechanical stress. The absence of FYCO1 is accompanied by a reduction of the autophagic flux in human embryonic kidney 293 (HEK293) cells (16). Consistently, we found an even more pronounced inhibition of autophagic flux after starvation in cardiomyocytes with reduced or absent FYCO1 levels. Accordingly, Fyco1-deficient mice also revealed impaired autophagic flux, as the treatment with chloroquine did not result in LC3-II accumulation.
Under stress conditions, such as starvation, autophagy is increased in the heart (14). Therefore, inhibition of autophagic flux might be detrimental and maladaptive. Consistent with this notion, inhibition of autophagy by BFA in starved mice is associated with cardiac dysfunction (17). Likewise, mice deficient for FoxO1 or haploinsufficient for beclin-1, genetic modifications that both result in suppression of autophagy, develop contractile dysfunction after 48 h of starvation (18). We here provide evidence that induction of autophagic flux is completely prevented upon FYCO1 deficiency, both in hearts from Fyco1-deficient mice subjected to starvation and in cardiomyocytes isolated from Fyco1-KO mice or in NRCMs with Fyco1 knockdown in a glucose-free medium.
Interestingly, heart-specific deletion of Atg5 by Cre recombinase under the control of a myosin light chain 2v promoter results in animals that lack the ability to increase autophagic activity and experience left ventricular dilation and impaired contractile function 1 week after “moderate” pressure overload due to TAC, while the hypertrophic response is unchanged (19). In contrast, “severe” TAC causes induction of autophagy as early as 24 h after TAC, and autophagy remains elevated for at least 2 weeks (20). Partial inhibition of autophagy (e.g., due to beclin-1 haploinsufficiency) results in improved cardiac function, while overexpression of beclin-1 is associated with reduced fractional shortening following TAC (20). We noticed elevated LC3-II levels 1 week after TAC, when mice already showed contractile dysfunction. After 2 weeks of pressure overload, mice developed severe contractile dysfunction but did not exhibit elevated LC3-II levels anymore. A possible explanation might be that we used a severe TAC model resulting in an ultra-rapid transition to heart failure, consistent with recently published data by other groups (21). Of note, we did not observe an induction of autophagy after 1 or 2 weeks of TAC in Fyco1-deficient mice. The inability of Fyco1-deficient cardiomyocytes to respond to cardiac stress conditions with induction of the autophagic flux due to impaired autophagosome maturation might cause the observed heart failure phenotype, which closely resembles the Atg5 conditional KO model (19).
Overexpression of FYCO1 in cardiomyocytes results in the formation of relatively large vesicles positive for LC3 and FYCO1, which resembles the effects of FYCO1 overexpression in human HEK293 cells, probably because of homotypic fusion (16). We show that, in vitro, overexpression of FYCO1 causes an accumulation of autophagosomes under nutrient-rich conditions. Likewise, autophagic flux is also markedly increased in TG mice that overexpress FYCO1 in vivo. Under TAC conditions, this animal model reacts with improved adaptation to increased biomechanical stress, resulting in preservation of contractile function. Our present data as well as findings by other groups (19,22) support the concept that the tightly regulated increase of autophagy is an adaptive response to cellular stress, while its inhibition is detrimental for cardiac function.
Interestingly, overexpression of FYCO1 is associated with increased myocardial content of Rab7. Rab7 is necessary for autophagosome maturation (23,24), and knockdown of Rab7 results in impaired autophagic flux (25). Similarly, Rab7 is decreased in hearts of aged mice that exhibit disturbed autophagic flux and respond to starvation with cardiac dysfunction (26). Contrarily, treatment of aged mice with the ALDH2 activator alda-1 results in restoration of autophagic flux, which is accompanied by an induction of Rab7 and prevention of starvation-induced cardiac dysfunction (26).
FYCO1 affects cardiac function and hypertrophy
Nevertheless, can additional mechanisms explain the protective effects of FYCO1? Given higher ratios of heart to body weight and thickened left ventricular walls in FYCO1-TG mice, one might expect that FYCO1 overexpression influences cardiac hypertrophy. However, the overexpression of FYCO1 does not affect the expression of typical members of the hypertrophic gene program, neither in TG animals nor in isolated cardiomyocytes. Although CSA is increased in TG mice, overexpression of FYCO1 in NRCMs results in smaller CSAs. Of note, the ratio of left ventricular mass to body weight is identical in banded WT and TG mice, which implies less pathological hypertrophy due to TAC in TG mice. Moreover, we established an interaction of FYCO1 with β-MHC/Myh7, a component of the thick filament and contractile apparatus. FYCO1 overexpression attenuates the induction of “fetal” Myh7 protein, which typically accompanies pathological cardiac hypertrophy in mice and rats that had undergone TAC (15,27). Of note, replacement of α-MHC (Myh6) by β-MHC (Myh7) in a TG mouse model leads to impaired contractility (28) and more pronounced left ventricular dilation and dysfunction after myocardial infarction (29). This suggests that under stress conditions, a minimized up-regulation or increased degradation of Myh7 by FYCO1 overexpression might contribute to improved contractile function, as observed in our animal models. Moreover, interaction with other sarcomeric proteins, which had been found in the yeast 2-hybrid screen using FYCO1 as bait (e.g., TPM1 to TPM3), might affect their stoichiometry, thereby altering cardiac growth and function, as observed in some forms of hypertrophic cardiomyopathy (30). In summary, morphometric data do not necessarily reflect pathological cardiac hypertrophy by FYCO1 overexpression on the molecular level. A speculative hypothesis is that the observed phenotype in TG mice might resemble physiological hypertrophy, which we will investigate in future experiments (e.g., by subjecting FYCO1 TG/Fyco1-KO mice to forced and voluntary physical exercise).
Study limitations
We provide evidence that FYCO1 expression levels significantly affect cardiomyocyte autophagy, especially under stress conditions. As we do not provide data, for example, for pharmacological inhibition of the autophagic flux following aortic banding, we cannot exclude that other factors beyond inhibition of cardiac autophagy may contribute to contractile dysfunction under pressure overload in Fyco1-deficient mice. Conversely, the preservation of contractile function following TAC in FYCO1-overexpressing mice is probably not caused by exaggerated autophagy alone, as, for example, Myh7 expression is differentially regulated. Although the degree of pressure overload is similar in TG and WT mice, because the gradients over the stenosis are nearly identical (peak velocity: WT TAC, 2,366 ± 250 mm/s; TG TAC, 2,452 ± 174 mm/s), we cannot prove that this is true for Fyco1-KO experiments as well. These caveats should be considered when interpreting the results.
Conclusions
In this study we introduce FYCO1 as a crucial regulator of cardiac autophagy. Although knockdown of FYCO1 reduces autophagy in isolated cardiomyocytes, overexpression of FYCO1 leads to increased autophagic flux in vitro. The absence of FYCO1 in vivo results in suppression of autophagy following starvation and pressure overload and finally contractile dysfunction. In contrast, cardiac-specific overexpression of FYCO1 leads to increased autophagy and restoration of cardiac function following TAC. Our data support the notion that FYCO1 is necessary to maintain cardiac function under conditions of starvation and hemodynamic stress in vivo.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: FYCO1 is necessary for the autophagic response in the heart to diverse stress stimuli. FYCO1 expression modulates contractile function and heart failure via autophagic flux.
TRANSLATIONAL OUTLOOK: As overexpression of FYCO1 protects from cardiac dysfunction in response to pressure overload, we propose that enhancing autophagic flux via FYCO1 overexpression might be a promising therapeutic strategy to treat or prevent heart failure.
Funding Support and Author Disclosures
The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
The authors thank Janine Eisenmann, Michaela Schmack, Wiona Burmeister, Katharina Stiebeling, Christin Tannert, and Dagmar Niemeier for their excellent technical assistance. Furthermore, the authors thank N. Mizushima for kindly providing GFP-LC3 TG mice and J.A. Hill for providing the GFP-LC3 adenovirus. The authors thank S. Billmann-Born for laser scanning microscopy. The authors thank Maksymilian Prondzynski and Elisabeth Krämer (Universitätsklinikum Hamburg-Eppendorf, Hamburg) for their help with the NanoString experiments. This work was supported by the German Centre for Cardiovascular Research and the Federal Ministry of Education and Research.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental tables and figures, please see the online version of this paper.
Appendix
References
- 1.Roger V.L. Epidemiology of heart failure. Circ Res. 2013;113:646–659. doi: 10.1161/CIRCRESAHA.113.300268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kehat I., Molkentin J.D. Molecular pathways underlying cardiac remodeling during pathophysiological stimulation. Circulation. 2010;122:2727–2735. doi: 10.1161/CIRCULATIONAHA.110.942268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizushima N., Klionsky D.J. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Will R.D., Eden M., Just S. Myomasp/LRRC39, a heart- and muscle-specific protein, is a novel component of the sarcomeric M-band and is involved in stretch sensing. Circ Res. 2010;107:1253–1264. doi: 10.1161/CIRCRESAHA.110.222372. [DOI] [PubMed] [Google Scholar]
- 6.Kiss H., Yang Y., Kiss C. The transcriptional map of the common eliminated region 1 (C3CER1) in 3p21.3. Eur J Hum Genet. 2002;10:52–61. doi: 10.1038/sj.ejhg.5200758. [DOI] [PubMed] [Google Scholar]
- 7.Pankiv S., Alemu E.A., Brech A. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol. 2010;188:253–269. doi: 10.1083/jcb.200907015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn C., Frank D., Dierck F. Cardiac remodeling is not modulated by overexpression of muscle LIM protein (MLP) Basic Res Cardiol. 2012;107:262. doi: 10.1007/s00395-012-0262-8. [DOI] [PubMed] [Google Scholar]
- 9.Rangrez A.Y., Eden M., Poyanmehr R. Myozap deficiency promotes adverse cardiac remodeling via differential regulation of mitogen-activated protein kinase/serum-response factor and beta-catenin/GSK-3beta protein signaling. J Biol Chem. 2016;291:4128–4143. doi: 10.1074/jbc.M115.689620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gulick J., Subramaniam A., Neumann J., Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J Biol Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- 11.Arber S., Hunter J.J., Ross J., Jr. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 12.Tannous P., Zhu H., Nemchenko A. Intracellular protein aggregation is a proximal trigger of cardiomyocyte autophagy. Circulation. 2008;117:3070–3078. doi: 10.1161/CIRCULATIONAHA.107.763870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kimura S., Noda T., Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 14.Mizushima N., Yamamoto A., Matsui M., Yoshimori T., Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercadier J.J., Lompre A.M., Wisnewsky C. Myosin isoenzyme changes in several models of rat cardiac hypertrophy. Circ Res. 1981;49:525–532. doi: 10.1161/01.res.49.2.525. [DOI] [PubMed] [Google Scholar]
- 16.Olsvik H.L., Lamark T., Takagi K. FYCO1 contains a C-terminally extended, LC3A/B-preferring LC3-interacting region (LIR) motif required for efficient maturation of autophagosomes during basal autophagy. J Biol Chem. 2015;290:29361–29374. doi: 10.1074/jbc.M115.686915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanamori H., Takemura G., Maruyama R. Functional significance and morphological characterization of starvation-induced autophagy in the adult heart. Am J Pathol. 2009;174:1705–1714. doi: 10.2353/ajpath.2009.080875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hariharan N., Maejima Y., Nakae J., Paik J., Depinho R.A., Sadoshima J. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakai A., Yamaguchi O., Takeda T. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 20.Zhu H., Tannous P., Johnstone J.L. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007;117:1782–1793. doi: 10.1172/JCI27523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang B., Nie J., Wu L. AMPKalpha2 protects against the development of heart failure by enhancing mitophagy via PINK1 phosphorylation. Circ Res. 2018;122:712–729. doi: 10.1161/CIRCRESAHA.117.312317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhuiyan M.S., Pattison J.S., Osinska H. Enhanced autophagy ameliorates cardiac proteinopathy. J Clin Invest. 2013;123:5284–5297. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jager S., Bucci C., Tanida I. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 24.Gutierrez M.G., Munafo D.B., Beron W., Colombo M.I. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 25.Su H., Li F., Ranek M.J., Wei N., Wang X. COP9 signalosome regulates autophagosome maturation. Circulation. 2011;124:2117–2128. doi: 10.1161/CIRCULATIONAHA.111.048934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu B., Yu L., Wang Y. Aldehyde dehydrogenase 2 activation in aged heart improves the autophagy by reducing the carbonyl modification on SIRT1. Oncotarget. 2016;7:2175–2188. doi: 10.18632/oncotarget.6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dorn G.W., II, Robbins J., Ball N., Walsh R.A. Myosin heavy chain regulation and myocyte contractile depression after LV hypertrophy in aortic-banded mice. Am J Physiol. 1994;267:H400–H405. doi: 10.1152/ajpheart.1994.267.1.H400. [DOI] [PubMed] [Google Scholar]
- 28.Krenz M., Sanbe A., Bouyer-Dalloz F. Analysis of myosin heavy chain functionality in the heart. J Biol Chem. 2003;278:17466–17474. doi: 10.1074/jbc.M210804200. [DOI] [PubMed] [Google Scholar]
- 29.Krenz M., Robbins J. Impact of beta-myosin heavy chain expression on cardiac function during stress. J Am Coll Cardiol. 2004;44:2390–2397. doi: 10.1016/j.jacc.2004.09.044. [DOI] [PubMed] [Google Scholar]
- 30.van Dijk S.J., Dooijes D., dos Remedios C. Cardiac myosin-binding protein C mutations and hypertrophic cardiomyopathy: haploinsufficiency, deranged phosphorylation, and cardiomyocyte dysfunction. Circulation. 2009;119:1473–1483. doi: 10.1161/CIRCULATIONAHA.108.838672. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.