Visual Abstract
Key Words: bacterial interactions, cardiovascular disease, enteric bacterial microflora, intestinal barrier function, intestinal microbiology
Abbreviations and Acronyms: ANOVA, analysis of variance; CHD, congenital heart disease; CPB, cardiopulmonary bypass; DNA, deoxyribonucleic acid; EBD, epithelial barrier dysfunction; FABP2, fatty acid binding protein 2; LCOS, low–cardiac output syndrome; NPO, nil per os; OTU, operational taxonomic unit; PGE2, prostaglandin E2; RA, relative abundance; rRNA, ribosomal ribonucleic acid
Highlights
-
•
Little is understood regarding the connection between congenital heart disease, the intestinal Microbiome, and how this could influence systemic inflammation.
-
•
To date, no study has evaluated the microbiome in congenital heart disease and the effects of cardiopulmonary bypass.
-
•
There is significant dysbiosis in patients with congenital heart disease and this is exacerbated following cardiopulmonary bypass.
-
•
Cardiopulmonary bypass induces intestinal barrier dysfunction which was not seen in control surgeries.
-
•
This data improves our understanding of intestinal dysbiosis, intestinal barrier dysfunction, and intestinal inflammatory eicosanoids and their interactions with cardiopulmonary bypass.
Summary
There are no data evaluating the microbiome in congenital heart disease following cardiopulmonary bypass. The authors evaluated patients with congenital heart disease undergoing cardiopulmonary bypass and noncardiac patients undergoing surgery without bypass. Patients with congenital heart disease had differences in baseline microbiome compared with control subjects, and this was exacerbated following surgery with bypass. Markers of barrier dysfunction were similar for both groups at baseline, and surgery with bypass induced significant intestinal barrier dysfunction compared with control subjects. This study offers novel evidence of alterations of the microbiome in congenital heart disease and exacerbation along with intestinal barrier dysfunction following cardiopulmonary bypass.
Evidence continues to mount reinforcing the relationship of the intestinal microbiome and its impact on systemic health (1). Recent evidence suggests that changes to this diverse microbiome drive pathology in the host (2). Additionally, certain disease states can alter the microbiome and produce intestinal dysbiosis. Intestinal dysbiosis is a disruption of the normal microorganism flora that exists within the intestinal tract (3). Systemic inflammatory processes, such as those seen following cardiopulmonary bypass (CPB), can exacerbate intestinal inflammation by promoting a reduction in gut health promoting bacteria and an increase in proinflammatory bacteria (4, 5, 6). These bacteria interact with the intestinal epithelium to secrete cytokines. These cytokines can activate key inflammatory pathways and perpetuate critical illness (7, 8, 9).
There are also data suggesting that cytokines released following CPB are similar to those released during other types of systemic inflammation, such as sepsis and trauma (10,11). CPB is well documented to promote systemic inflammation, manifested by clinical findings such as hypotension, tachycardia, and capillary leak (12, 13, 14). It thus stands to reason that CPB may be associated with gut microbial changes as seen in other systemic inflammatory conditions. Available research exploring this relationship, however, is sparse; thus the aim of this study was to rigorously investigate this relationship.
Intestinal epithelial barrier dysfunction (EBD) evidenced by changes in epithelial integrity, epithelial function, and paracellular integrity is known to be caused by CPB (15). Intestinal barrier dysfunction has been identified in states of physiological stress (16,17). Although Typpo et al. (15) evaluated intestinal EBD during CPB, its correlation with the intestinal microbiome in patients with cardiac defects has yet to be evaluated. This would be new information to build upon our understanding of the effects of congenital heart disease (CHD) and CPB on the intestinal microbiome and intestinal EBD. No data currently exist comparing surgery with CPB versus surgery without CPB to evaluate the effects of CHD and CPB on the microbiome and EBD in a controlled fashion.
CHD affects roughly 1% of all pediatric patients: 40,000 births annually (18). The majority of these patients require cardiac surgery to correct defects. This involves CPB, a primary cause of systemic inflammation experienced by these patients. The inflammatory process also leads to a second major contributor of morbidity and mortality post-operatively, low–cardiac output syndrome (LCOS), which occurs in roughly 20% to 25% of all patients, with the majority occurring in neonates and infants. Among neonates and infants, almost half appear to have some degree of LCOS (19,20). CPB is a necessary technology needed to support patients during correction of CHD. It is important to understand how the inflammation associated with CPB affects the body and if the inflammatory response can be modified to promote healing and improved outcomes following CPB. Serum markers for EBD have not been comprehensively evaluated in CBP. This could provide key cross-sectional data into the severity of gut injury following surgery with CPB. If cardiac surgery with CPB induces changes to the intestinal microbiome similar to other systemic inflammatory conditions, we may be able to identify methods to minimize the effect on the microbiome and promote growth of healthy intestinal bacteria, which could reduce EBD and improve the outcomes of patients following CPB. Only limited studies have evaluated the microbiome in CHD (21, 22, 23). Although there are a small number of adult data investigating the microbiome in cardiac surgery (24,25), no study has evaluated the role of the microbiome in CHD as it relates to cardiac surgery or CPB. This study was thus undertaken to investigate the intestinal microbiome in patients with CHD undergoing CPB, its effects on EBD, and correlations with the microbiome and EBD. Our hypothesis was that surgery with CPB would result in reduced bacterial biodiversity and bacterial richness as well as intestinal barrier dysfunction compared with surgery without CPB.
Methods
Patients
Institutional Review Board approval was obtained from Saint Louis University (#28431). Patients meeting eligibility scheduled for surgery at Cardinal Glennon Children’s Hospital in Saint Louis were approached and consented in pre-anesthesia, as per the inclusion and exclusion criteria, from November 2017 to January 2019 (Table 1). The comparison arm procedures were selected to control for perioperative variables (e.g., surgical duration, perioperative antibiotics, nil per os [NPO] time, and anesthetic agents). The control group consisted of patients without CHD. Exclusion criteria were selected to eliminate variables of noncardiac chronic disease, which can influence the gastrointestinal microbiome.
Table 1.
Inclusion and Exclusion Criteria
| Inclusion Criteria | Exclusion Criteria |
|---|---|
| Age 1 month to 18 yrs | Previous surgery in past 3 months |
| Hospital stay >48 h | Gastrointestinal pathology or intestinal surgery, excluding gastrostomy tube |
| Elective surgery | Liver disease |
| Cardiac surgery | Dialysis-dependent kidney disease |
| Expected surgical duration >90 min | Antibiotics within 3 months |
| Previous chemotherapy | |
| Continuous enteral feeds prior to surgery |
Surgery
The operative course and anesthetic regimens were per standard of care. All patients, in both study groups, received perioperative and post-operative intravenous cefazolin for 3 doses except in case of documented penicillin allergy or documented methicillin-resistant Staphylococcus aureus. All patients received similar anesthetic agents in both study arms. For surgery with CPB, blood prime was used for patients weighing <10 kg and clear-primed for those weighing more than 10 kg. Modified ultrafiltration and cell-saver techniques were used to minimize exogenous transfusions. Mild hypothermia was used in all CPB cases, and moderate hypothermia with regional perfusion was used in procedures involving complex intracardiac or aortic arch reconstruction. No deep hypothermia or deep hypothermic cardiac arrest procedures were included in this cohort.
Stool collection
A pre-surgery stool sample was collected either from a spontaneous bowel movement or via sterile fecal swab inserted into the rectum in the operating room prior to surgery. A second stool sample was collected post-operatively. This was collected anywhere between 2 and 4 days post-operatively, as per previously published research (19). Samples were stored in a freezer at −80°C until deoxyribonucleic acid (DNA) extraction was performed.
Blood sample collection
Blood samples for measurement of fatty acid binding protein 2 (FABP2), claudin-3, and citrulline were collected from indwelling catheters following induction with anesthesia prior to surgery. Post-operatively, blood samples were collected from indwelling catheters or peripheral venous puncture at 24 and 48 h. Samples were immediately placed on ice and stored at 4°C. The samples were spun at 3,500 rpm for 15 min within 2 h of collection, and the serum was stored at −80°C. All samples were collected with mean arterial blood pressure within 1 SD of normal for age.
Extraction of DNA from stool
DNA from the stool samples was extracted using the Qiagen PowerFecal DNA Extraction Kit (Qiagen, Hilden, Germany). Extracted DNA was quantified using a Qubit 2.0 fluorometer and Quant-iT dsDNA broad range assay kits (Thermo Fisher Scientific, Waltham, Massachusetts). DNA was then diluted to a standard volume and concentration for 16S ribosomal ribonucleic acid (rRNA) library preparation.
16S rRNA library preparation and sequencing
Analysis of the microbiome was performed only in patients in whom pre-surgery and post-surgery stool could be collected. Extracted fecal DNA was processed at the University of Missouri DNA Core Facility. Bacterial 16S rRNA amplicons were constructed via amplification of the V4 region of the 16S rRNA gene with universal primers (U515F/806R) developed against the V4 region, flanked by Illumina standard adapter sequences (Illumina, San Diego, California) (26,27). Oligonucleotide sequences were available at probeBase (28). Dual-indexed forward and reverse primers were used in all reactions. Polymerase chain reaction was performed in 50 μl reactions containing 100 ng metagenomic DNA, primers (0.2 μmol/l each), deoxyribonucleotide triphosphates (200 μmol/l each), and Phusion high-fidelity DNA polymerase (1U) (New England Biolabs, Ipswich, Massachusetts). Amplification parameters were 98°C(3:00) + [98°C(0:15) + 50°C(0:30) + 72°C(0:30)] × 25 cycles + 72°C(7:00). Amplicon pools (5 μl/reaction) were combined, thoroughly mixed, and then purified by addition of Axygen AxyPrep MagPCR clean-up beads (Thermo Fisher Scientific) to an equal volume of 50 μl of amplicons and incubated for 15 min at room temperature. Products were then washed multiple times with 80% ethanol, and the dried pellet was resuspended in 32.5 μl EB buffer, incubated for 2 min at room temperature, and then placed on the magnetic stand for 5 min. The final amplicon pool was evaluated using the Advanced Analytical Fragment Analyzer automated electrophoresis system (Agilent, Santa Clara, California), quantified using quant-iT HS dsDNA reagent kits (Thermo Fisher Scientific), and diluted according to Illumina’s standard protocol for sequencing on the MiSeq instrument.
Analysis of FABP2 and claudin-3
Serum samples were thawed and used to determine the concentrations of FABP2 and claudin-3 by enzyme-linked immunosorbent assay (catalog #RAB0537-1KT, Sigma-Aldrich, St. Louis, Missouri; catalog #SEF293Hu, Cloud-Clone, Houston, Texas). Both kits were used according to manufacturer instructions. The plates were developed and analyzed using a spectrophotometer (EL-800, BioTek Instruments, Winooski, Vermont) at 450 nm.
Analysis of citrulline
Serum samples were deproteinized with 35% sulfosalicylic acid. Using the Hitachi L-8800 Amino Acid Analyzer (Hitachi, Tokyo, Japan), the samples were injected onto an ion-exchange column (4.6 mm inside diameter × 60 mm, packed with Hitachi custom ion exchange resin). Amino acids were selectively eluted from the column by buffers of increasing pH and a rise in temperature. Ninhydrin was mixed with the buffer amino acid solution, heated to a variation in temperature between 20° C and 70°C (the column is at 60°C for citrulline), and read at 570 nm for amino acids. The amount of citrulline was calculated using the area under the peak.
Metabolomics
Metabolomic eicosanoid analysis was performed when sufficient remaining samples was available for analysis. These samples consisted of both pre-surgery and post-surgery stool in both groups. The stool samples were standardized for dehydration and mass prior to analysis. A Shimadzu Nexera UPLC system (Shimadzu Scientific Instruments, Columbia, Maryland) equipped with 2 pumps (LC-30 AD), a column oven (CTO-30AS), and an auto-sampler (SIL-30AC) were used. Mass spectrometric detection was performed on an LCMS-8060 system (Shimadzu Scientific Instruments), equipped with a dual ion source operated in negative electrospray ionization mode. All chromatographic separations were performed using a Shim-pack XR-ODSIII (150 × 2.00 mm, 2.2 μm) as the analytic columns equipped with a Shim-pack C18 guard column (Shimadzu Scientific Instruments). The mobile phase consisted of 0.1% acetic acid in water (mobile phase A; 80%) and acetonitrile (20%) (mobile phase B), at a total flow rate of 0.37 ml/min. Chromatographic separation was achieved using a 25-min gradient elution and separated eicosanoids.
All stool eicosanoids were detected in negative ionization mode with the following instrument-dependent mass spectrometer parameters: nebulizer gas, 2.2 l/min; heating gas, 10 l/min; drying gas, 10 l/min; interface temperature, 300°C; desolvation line temperature, 250°C; heat block temperature, 400°C; and interface temperature, 300°C.
Analytes were extracted from stool sample, and calibration standards were performed on the stool samples by solid phase extraction using Oasis HLB 3-ml, 60-mg SPE cartridges (Waters, Milford, Massachusetts). Each stool sample was accurately weighed and then homogenized with triple-distilled deionized water at a 10- fold dilution factor using a TissueLyserII (Qiagen Life Sciences, Louisville, Kentucky). The resultant homogenate sample (200 μl) was spiked with IS (20 μl) and diluted with 5% acetic acid in water (1,500 μl), vortexed for 30 s, and then loaded onto SPE cartridges pre-conditioned with methanol (MeOH; 2 mL), followed by 0.1% acetic acid in water (2 ml). Loaded cartridges were washed with 5% MeOH (2 ml) and eluted with MeOH (2 ml). For all standards and samples, eluates were evaporated under vacuum at room temperature and reconstituted with 50% acetonitrile in water (100 μl). All unlabeled eicosanoids standards and stable deuterated isotope-labeled internal standards, including prostaglandin E2 (PGE2)–d4, thromboxane B2–d4, arachidonic acid–d8, 15-hydroxyeicosatetraenoic acid–d8, leukotriene B4–d8, and resolvin D1–d5, were purchased from Cayman Chemicals (Ann Arbor, Michigan).
Data analysis
Subject demographics, clinical characteristics, and laboratory values were analyzed by study group. Data were tested for normality using the Shapiro-Wilk method. Normally distributed continuous variables are expressed as mean ± SD. Medians and interquartile ranges are reported for continuous data not normally distributed. Categorical variables are expressed as counts and percentages. The experimental group was compared with the control group using chi-square tests for categorical variables, independent-samples Student’s t-tests for normally distributed continuous variables, and the Mann-Whitney U test for variables not normally distributed. Feeding intolerance was calculated using a feeding intolerance score in a similar study, with a cumulative score over the previous 24 h (use of an antiemetic agent, abdominal distention, emesis, diarrhea, and gastrointestinal bleed). The variables were scored if present or absent from the previous 24 h, with a maximum score of 5 (15). Feeding intolerance was considered present for a score ≥2 in 24 h or ≥1 for 2 consecutive days. Data were analyzed using SPSS version 26 (IBM, Armonk, New York) at a significance level of 0.05.
Read merging, clustering, and annotation of DNA sequences were performed at the University of Missouri Informatics Research Core Facility. Paired DNA sequences were merged using FLASH software (29) and removed if found to be far from the expected length of 292 bases after trimming for base quality of 31. Cutadapt (30) (https://github.com/marcelm/cutadapt) was used to remove the primers at both ends of the contig and cull contigs that did not contain both primers. The usearch (31) fastq_filter command (http://drive5.com/usearch/manual/cmd_fastq_filter.html) was used for quality trimming of contigs, rejecting those for which the expected number of errors was >0.5. All contigs were trimmed to 248 bases, and shorter contigs were removed. The QIIME (32) 1.9 command split_libraries_fastq.py was used to demultiplex the samples. The outputs for all samples were combined into a single file for clustering. The UPARSE (33) method (http://www.drive5.com/uparse/) was used to both cluster contigs with 97% identity and remove chimeras. Taxonomy was assigned to selected operational taxonomic units (OTUs) using BLAST (34) against the SILVA database (version 128 [35]) of 16S rRNA sequences and taxonomy.
For univariate outcomes (e.g., α-diversity, or the relative abundance [RA] of a specific taxon), data were first tested for normality and equal variance using the Shapiro-Wilk and Brown-Forsyth methods, respectively. Non-normally distributed data were tested using the Kruskal-Wallis test, while traditional 2-way analysis of variance (ANOVA) was performed on normally distributed data. These analyses were performed using SigmaPlot version 14.0 (Systat Software, Chicago, Illinois). Multivariate statistical comparisons performed on the microbiome analysis, such as testing for differences in β-diversity between groups of samples, were made via permutational multivariate ANOVA of nontransformed data using Past version 3.25 (36). Repeated-measures ANOVA was performed on FABP2, claudin-3, and citrulline, adjusting for experimental group.
Analysis of eicosanoids was performed at the University of Nebraska Medical Center College of Pharmacy. The raw data obtained were converted to concentration data (nanograms per gram) using Lab Solution software version 5.8 (Shimadzu Scientific Instruments). Metabolomic data analysis was performed using the MetaboAnalyst version 4.0 online platform. The Mann-Whitney U test was used to evaluate differences in eicosanoid levels between the CPB group both pre- and post-surgery.
Results
Enrollment
After Institutional Review Board approval was obtained, a total of 86 patients were screened (Figure 1). After excluding those who did not meet criteria, we approached a total of 66 patients for consent. Of these, 18 patients refused or were unable to provide consent, leaving us with a total of 48 patients for our study. In the CPB arm, 12 patients were removed from study after enrollment because of an inability to obtain an adequate stool sample (n = 5), return to the operating room (n = 2), and discharge home prior to collection of a post-surgery stool sample (n = 5). In the control arm, 6 patients were removed from study after enrollment because of return to the operating room (n = 1), discharge home prior to collection of a post-surgery stool sample (n = 4), and diagnosis of a previously unidentified gastrointestinal pathology (n = 1). Only 1 patient in the CPB arm received clindamycin. All other patients in both arms received cefazolin.
Figure 1.
Enrollment Process
Patients reviewed and those excluded according to criteria. A number refused to provide consent. Between the 2 groups, patients were removed from study because of inability to collect pre-operative sample (n = 5), post-operative stool sample (n = 9), return to the operating room (n = 3), and identification of gastrointestinal (GI) pathology (n = 1). CPB = cardiopulmonary bypass.
Patient characteristics
Demographic data and details of the surgery and post-operative characteristics are listed in Table 2. Maximum vasoactive-inotropic score was calculated with a multiplier on the basis of vasoactive agent and dose and reported for patients in the CPB group using a standardized calculation (37). No patients in the control group required inotropic support. There were no statistical differences in age, sex, duration NPO prior to surgery, use of antibiotics or steroids, and surgical time between groups. The type of feeding differed between the 2 groups, favoring oral intake in the control group and more nasogastric feeds initiated in the CPB group. The incidence of feeding intolerance was greater in the CPB group. The CPB group had a longer post-operative intensive care unit and hospital length of stay compared with the control group. Further perioperative details for each patient in both groups are outlined in Supplemental Table 1.
Table 2.
Patient Demographic Data
| CPB Patients (n = 17) | Control Subjects (n = 12) | p Value | |
|---|---|---|---|
| Age (months) | 19 (7.0–128.0) | 34 (13.3–115.0) | 0.49 |
| Male | 10 (55.6) | 4 (33.3) | 0.23 |
| NPO time (h) | 6 (3) | 7 (2) | 0.32 |
| Surgical time (min) | 310 (247.5–378.0) | 242.5 (226.3–265.0) | 0.08 |
| Bypass time (min) | 138.9 (59.2) | — | — |
| Cross-clamp time (min) | 54.1 (44.7) | — | — |
| Ventilator days (days) | 1.25 (0.0–5.0) | — | — |
| Inotrope days (days) | |||
| Milrinone | 2 (1.0–4.5) | — | — |
| Epinephrine | 0 (0.0–1.8) | — | — |
| Dopamine | 1.25 (0.5–2.3) | — | — |
| Vasopressin | 0 | — | — |
| Maximum vasoactive-inotropic score | 10.6 (3.6) | — | — |
| PICU LOS (days) | 4 (3.0–6.3) | 1 (1–1) | <0.001 |
| Hospital LOS (days) | 7(6–9) | 3 (3–4) | <0.001 |
| Time to start feeds (h) | 37 (8.0–50.3) | 5.5 (4.0–8.5) | 0.001 |
| Feeding type | |||
| Regular diet | 7 (41.2) | 10 (83.3) | 0.035 |
| Nasogastric feeds | 10 (58.8) | 2 (16.7) | 0.027 |
| Feeding intolerance | 5 (27.8) | 0 (0.0) | <0.001 |
| Steroids | 1 (5.8) | 3 (25.0) | 0.14 |
| Antibiotics | 17 (100) | 12 (100) | |
| Cefazolin | 16 (94) | 12 (100) | 0.84 |
| Clindamycin | 1 (6) | 0 (0) |
Values are median (interquartile range) or n (%). Data were compared using Student’s t-test, the Mann-Whitney U test, or the chi-square test.
CBP = cardiopulmonary bypass; LOS = length of stay; NPO = nil per os; PICU = pediatric critical care unit.
The diagnoses within the CPB group and surgical procedures performed are listed in Table 3. A total of 14 patients had mild hypothermia and were cooled to 32°C to 34°C. Three patients underwent moderate hypothermia to 28°C with regional cerebral perfusion. The control group did not have CHD and had diagnoses consisting of lumbar and thoracic scoliosis, craniosynostosis, and posterior fossa tumor. Control group procedures included spinal fusion (n = 2), Chiari malformation repair (n = 3), craniosynostosis repair (n = 4), and posterior fossa tumor resection (n = 3). At the time of each blood sample collection, subjects in both groups were within 1 SD of age-appropriate mean arterial blood pressures.
Table 3.
Cardiopulmonary Bypass Arm Surgical Diagnoses and Operations Performed
| Diagnosis | n | Surgery Performed | n |
|---|---|---|---|
| Septal defects | 5 | Septal repair | 5 |
| TOF | 4 | TOF repair | 4 |
| HLHS | 3 | Fontan revision | 1 |
| AV canal defects | 3 | Bidirectional Glenn shunt | 3 |
| DORV | 1 | Aortic arch reconstruction | 3 |
| Aortic stenosis | 1 | DKS procedure | 2 |
| Pulmonary stenosis/atresia | 5 | Ross procedure | 1 |
| Coarctation of aorta/hypoplastic arch | 3 | Aortic valve repair | 1 |
| RV-PA conduit | 4 | ||
| Rastelli procedure | 1 | ||
| AV canal repair | 1 |
The total number of surgical diagnoses and operations performed exceeds 17, as there were patients with multiple defects repaired. This is further detailed in Supplemental Table 1.
AV = atrioventricular; DKS = Damus-Kaye-Stansel; DORV = double-outlet right ventricle; HLHS = hypoplastic left heart syndrome; PA = pulmonary artery; RV = right ventricle; TOF = tetralogy of Fallot.
Microbiome
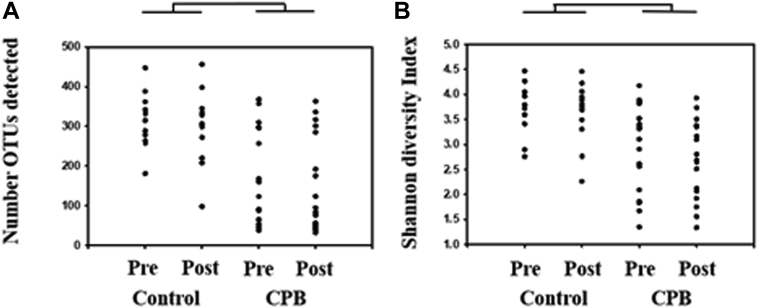
Sequencing of 16S rRNA amplicon libraries generated using fecal DNA resulted in a total of 5.37 million high-quality sequences, and a mean of 92,520 ± 16,581 sequences per sample. For comparisons of richness and α-diversity, samples were subsampled to a uniform sequence number. Richness, or the number of distinct OTUs detected in each sample, differed significantly between CPB and control subjects (p < 0.001), with CPB subjects harboring incredibly sparse fecal communities (Figure 2A). The community structure was also compared for α-diversity, reflecting richness as well as evenness of distribution, revealing a similar disparity with greater Shannon diversity values in control subjects, relative to CPB patients (Figure 2B). In both comparisons, there was no significant effect of time.
Figure 2.
Bacterial Richness and Diversity
(A) The number of operational taxonomic units (OTUs) or bacterial richness present in each sample. (B) The alpha-diversity, a representation of bacterial richness and evenness of bacterial species within each sample. Statistically significant reductions in bacterial richness (p < 0.001) and alpha-diversity (p < 0.001) existed in the cardiopulmonary bypass (CPB) group compared with the control group.
Stacked bar charts demonstrate the taxonomies present in each group at the level of phylum and genus (Figure 3). Even at this coarse taxonomic resolution, there were differences between CPB patients and control subjects in the RA of several bacterial species. Specifically, CPB patients harbored a greater RA of Proteobacteria (p < 0.001) and Actinobacteria (p = 0.028), with no significant effects of time or interactions between group and time in either phylum. This was offset by a lower RA of Bacteroides in CPB subjects (p = 0.004), which actually decreased from pre-surgery to post-surgery in the CPB group (p = 0.032) but not the control subjects (p = 0.853) (Supplemental Figures 1A to 1C). Although control subjects harbored a greater mean RA of Firmicutes compared with CPB subjects, the difference did not reach statistical significance (p = 0.064) (Supplemental Figure 1D).
Figure 3.
Taxonomy of Bacteria
(A) The bacteria at the phylum level in each sample pre- and post-surgery for the control group and the cardiopulmonary bypass (CPB) group. There was a significant difference in the pro-inflammatory bacteria (red bars) between the CPB group and the control group in both pre- and post-surgery samples (p < 0.001). (B) The genus-level bacteria in each sample. Red bars indicate pro-inflammatory bacteria. Blue, green, yellow, and tan bars indicate healthy gut–promoting bacteria.
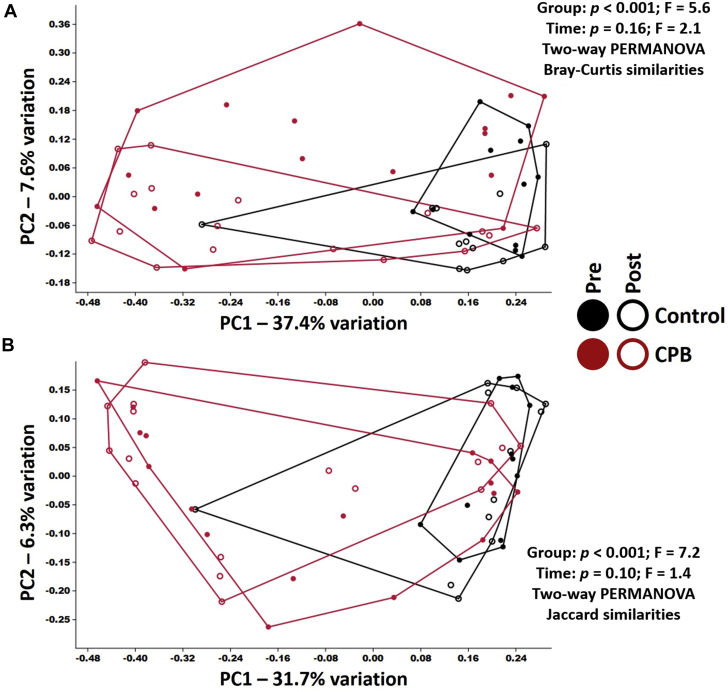
Overall β-diversity (i.e., intersubject differences in community composition) was visualized using principal coordinate analysis and tested statistically using permutational multivariate ANOVA, allowing us to compare groups using more than 1 measure of similarity. For the sake of comparison, we opted to use the Bray-Curtis and Jaccard similarities, which place more and less emphasis, respectively, on differences in the RA of shared taxa when calculating intersample similarity. Using the relatively weighted Bray-Curtis similarities, significant differences were detected between control and CPB (pre-surgery, p = 0.001; post-surgery, p < 0.001), with significant changes in composition pre-surgery to post-surgery within CPB subjects (p = 0.04) but not pre- to post-surgery in control subjects (p = 0.14), on the basis of 1-way permutational multivariate ANOVA. Two-factor permutational multivariate ANOVA indicated a group effect (p < 0.001) and a time-dependent effect (p = 0.016), indicating that the compositional differences between control and experimental samples outweigh the time-dependent differences in both groups (Figure 4A). Comparison using the unweighted Jaccard similarities revealed a very similar pattern, including a significant difference between control and CPB at both time points (pre-surgery, p = 0.0008; post-surgery, p = 0.001) but no significant time-dependent changes in composition in either group (control, p = 0.55; CPB, p = 0.19) on the basis of 1-way permutational multivariate ANOVA (Figure 4B). Two-factor ANOVA indicated a group effect (p = 0.0001) but no time-dependent effect (p = 0.10), suggesting that the subtle time-dependent compositional differences detected previously (using the Bray-Curtis similarity) are predicated primarily on changes in RA of bacteria present at both time points, rather than introduction or loss of taxa between sampling.
Figure 4.
Beta-Diversity in Patients Undergoing CPB and Control Subjects
(A) The Bray-Curtis beta-diversity similarities between patients undergoing cardiopulmonary bypass (CPB) and control subjects. (B) The Jaccard beta-diversity similarities between patients undergoing CPB and control subjects. Red indicates patients undergoing CPB and black indicates control subjects. Closed circles represent pre-surgery samples for each group, and open circles represent post-surgery samples for each group. PC = principal component; PERMANOVA = permutational multivariate analysis of variance.
To identify specific taxonomic differences associated with group (CPB vs. control) or time of collection, serial 2-factor ANOVA was performed on all taxa detected in >15% of either group, resulting in a total of 319 OTUs tested. The p values resulting from serial testing for differences in the RA of detected sequences were adjusted to control the false discovery rate associated with multiple tests, using the method of Benjamini and Hochberg (38) with a conservative false discovery rate of 10% (39). Of the 14 OTUs yielding time-dependent p values < 0.05, none withstood correction for multiple testing. Conversely, of the 81 OTUs yielding group-dependent p values <0.05, 56 withstood correction (Supplemental Table 2). Of those 56 OTUs, 49 were detected at an increased RA in control relative to CPB, and 7 were at an increased RA in CPB patients. Taxa present at greater RA in samples from control patients comprised bacteria from the phyla Actinobacteria (3 OTUs), Bacteroidetes (6 OTUs), and Firmicutes (40 OTUs). The OTUs within phylum Firmicutes were predominantly members of families Ruminococcaceae and Lachnospiraceae, historically referred to as clostridium clusters IV and XIVa, respectively, and known to contain many butyrate-producing genera, including taxa identified as underrepresented in CPB patients, including Faecalibacterium sp., Agathobacter sp., Eubacterium sp., and Roseburia sp. (40). In contrast, the 7 OTUs compensating for these differences, and thus detected at greater RA in CPB patients compared with control subjects, comprised 3 OTUs from phylum Firmicutes and 4 OTUs from the family Enterobacteriaceae within the phylum Proteobacteria.
Serum FABP2, claudin-3, and citrulline
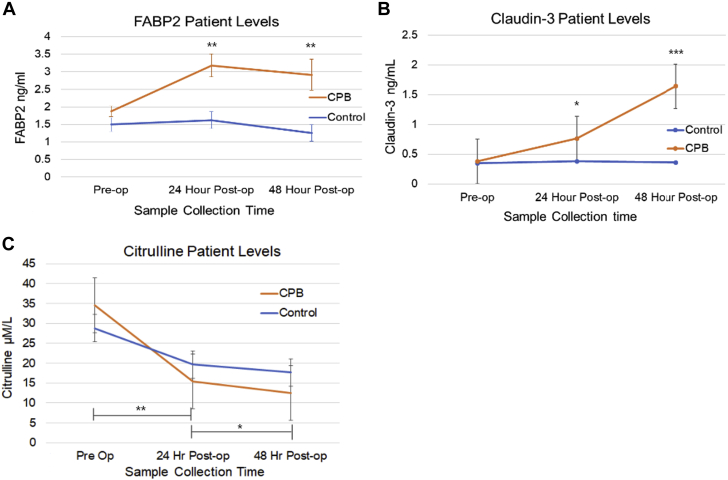
Serum markers of intestinal EBD were added to the analysis after enrollment of the first 3 patients had occurred, giving a total of 14 for the CPB group and 9 for the control group. For 3 patients in each group, we were not able to collect the 48-h blood sample. Patients in both arms had FABP2, claudin-3, and citrulline levels measured pre-surgery and then 24 and 48 h post-surgery. Figure 5 outlines the markers of intestinal barrier dysfunction. All patients started with similar levels of FABP2. FABP2 is a cytosolic enzyme found in mature enterocytes within the small intestines (41). There was a statistically significant increase in FABP2 levels in the CPB group compared with control subjects at 24 h post-surgery (p = 0.001) and at 48 h post-surgery (p = 0.012), most evident at 24 h post-surgery. FABP2 showed no appreciable change between pre- and post-surgery levels in the control group. Claudin-3 is one of a group of proteins involved in maintaining the tight junction barrier properties of the intestinal epithelial cells (42,43). Claudin-3 levels were noted to increase significantly at the 48-h sample collection, which is similar to previously published data of Typpo et al. (15). This was statistically significant compared with control subjects (p = 0.002). No change of claudin-3 levels at any time point was noted within the control group. Citrulline levels were not statistically different at baseline in either group, but baseline and the 24-h post-surgery sample collection time and baseline and the 48-h post-surgery sample collection time in the CPB group compared with control subjects (Δ 24 h, p = 0.001; Δ 48 h, p = 0.035). These results are summarized in Table 4. Variables, such as CPB duration, cross-clamp duration, antibiotic agents administered post-surgery, and vasoactive-inotropic score did not influence FABP2, claudin-3, and citrulline levels in the CPB group. The effect of mild hypothermia, moderate hypothermia with regional cerebral perfusion, or whether CPB was blood-primed or clear-primed did not have an effect on the degree of intestinal EBD via measurements of FABP2.
Figure 5.
Changes in Markers of Intestinal Epithelial Barrier Dysfunction
(A) Significant increase of fatty acid binding protein 2 (FABP2) in patients undergoing CPB versus control subjects (p = 0.001). (B) Significant increase of claudin-3 in patients undergoing CPB versus control subjects (p < 0.001). (C) Significant reduction of citrulline in patients undergoing CPB versus control subjects (p = 0.001). Independent Student’s t-test for means at each time point. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Table 4.
Patient Levels of Markers of Intestinal Barrier Dysfunction
| CPB Patients |
Control Subjects |
p Value∗ | |||
|---|---|---|---|---|---|
| n | Value | n | Value | ||
| FABP2 pre-surgery | 14 | 1.80 (1.32 to 2.46) | 9 | 1.58 (0.97 to 1.98) | 0.12 |
| FABP2 24 h post-surgery | 14 | 3.08 (2.14 to 4.29) | 9 | 1.65 (1.20 to 2.14) | 0.001 |
| FABP2 48 h post-surgery | 11 | 2.27 (1.85 to 3.93) | 6 | 1.20 (0.80 to 1.74) | 0.002 |
| FABP2 Δ 24 h | 14 | 1.06 (0.55 to 2.18) | 9 | 0.14 (0.08 to 0.28) | <0.001 |
| FABP2 Δ 48 h | 11 | 0.88 (−0.12 to 1.84) | 6 | −0.21 (−0.97 to −0.08) | 0.022 |
| Claudin-3 pre-surgery | 14 | 0.35 (0.21 to 0.45) | 9 | 0.37 (0.17 to 0.45) | 0.93 |
| Claudin-3 24 h post | 14 | 0.55 (0.47 to 1.00) | 9 | 0.43 (0.20 to 0.53) | 0.028 |
| Claudin-3 48 h post-surgery | 11 | 1.62 (1.24 to 1.78) | 6 | 0.22 (0.20 to 0.59) | <0.001 |
| Claudin-3 Δ 24 h | 14 | 0.26 (0.09 to 0.57) | 9 | 0.06 (0.02 to 0.08) | 0.004 |
| Claudin-3 Δ 48 h | 11 | 1.04 (0.89 to 1.67) | 6 | 0.06 (0.01 to 0.21) | <0.001 |
| Citrulline pre-surgery | 14 | 34.5 ± 8.9 | 9 | 28.8 ± 3.6 | 0.08 |
| Citrulline 24 h post-surgery | 14 | 15.4 ± 4.2 | 9 | 19.7 ± 5.0 | 0.041 |
| Citrulline 48 h post-surgery | 11 | 12.5 ± 3.2 | 6 | 17.7 ± 6.9 | 0.043 |
| Citrulline Δ 24 h | 14 | −19.1 ± 7.4 | 9 | −9.1 ± 4.3 | 0.001 |
| Citrulline Δ 48 h | 11 | −22.1 ± 9.5 | 6 | −12.0 ± 5.9 | 0.035 |
Values are median (interquartile range) or mean ± SD.
CPB = cardiopulmonary bypass; FABP2 = fatty acid binding protein 2.
Student’s t-test and the Mann-Whitney U was test were used for normally and non-normally distributed data, respectively.
Repeated-measures ANOVA was performed on FABP2, claudin-3, and citrulline. FABP2 showed a difference over time that varied by experimental group (p = 0.005). The differences by experimental group over time are seen between the pre-surgery baseline and 24-h post-surgery samples (p = 0.009) and between the pre-surgery baseline and 48-h post-surgery samples (p = 0.011). Claudin-3 demonstrated a difference over time that varied by experimental group (p < 0.001). The differences by experimental group over time are seen between the pre-surgery baseline and 48-h post-surgery samples (p = 0.001) and between the 24-h post-surgery and 48-h post-surgery samples (p = 0.001). Citrulline exhibited a difference over time, and this difference varied by experimental group (p = 0.037). The difference by experimental group over time is seen between the pre-surgery baseline and 24-h post-surgery samples (p = 0.009). The difference between the pre-surgery baseline and 48-h post-surgery sample was significant at p < 0.001, but this did not vary by experimental group (p = 0.25). On the basis of multiple comparisons for markers of intestinal EBD, we used the Bonferroni correction, which resulted in an adjustment of significance to p = 0.017.
Metabolomics
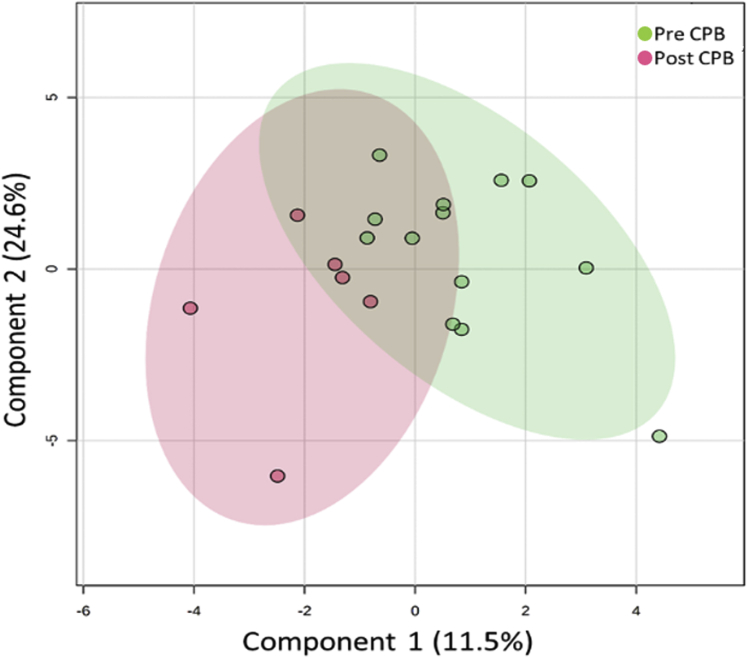
Eicosanoid concentrations, associated with pro-inflammatory signaling, were determined in stool samples from patients using a validated liquid chromatographic/tandem mass spectrometric method. A total of 67 eicosanoids were detected, and their metabolites were detected above the limit of detection in stool samples; the 35 largest differences are presented in Supplemental Table 3. There was substantial variability in eicosanoid concentrations in stool samples with concentrations of PGE2 varying over a 10-fold range. Among stool prostaglandins, prostaglandin D2 had the highest concentrations (12.03 ng/g), whereas prostaglandin E3 (0.01 ng/g) was found in the lowest concentration. The 5 eicosanoids with the highest degrees of variation from pre-surgery levels to post-surgery levels in the CPB group are presented in Figure 6. We observed about a 2-fold increase in the stool levels of PGE2 and its various metabolites. There was also substantial variability in many of these metabolites in the CPB group and the control group. Principal-component analysis was performed on the CPB group comparing pre-CPB and post-CPB levels of eicosanoids, shown in Figure 7.
Figure 6.
Stool Eicosanoid Levels Distribution in the CPB Group
(A) Levels of prostaglandin E2 (PGE2). (B) Levels of 11-hydroxy-12(E),14(Z)-eicosadienoic acid (11-HEDE). (C) Levels of 12(13)Di-HOME. (D) Levels of 11-dehydro-thromboxane E3 (11-DeTXB3). (E) Levels of prostaglandin E3 (PGE3). Stool eicosanoids measured in pre-surgical cardiopulmonary bypass (CPB) (n = 6) and post-surgical CPB (n = 12) patients. On the x-axis, pre-CPB refers to the pre-surgical samples and post-CPB to the post-surgical samples. A total of 67 eicosanoids were analyzed, and the 35 largest differences are shown. The Mann-Whitney U test was used to assess differences between the 2 groups.
Figure 7.
Principal-Component Analysis for Stool Eicosanoids in the CPB Group
Principal-component analysis was performed with pre–cardiopulmonary bypass (CPB) (red; n = 6) and post-CPB (green; n = 12). Two-dimensional principal-component score plots are separated from other groups, indicating a distinct metabolite composition between the 2 groups. Component 1 indicates the degree of variation between the groups on the basis of their total metabolite content. Component 2 indicates the differences within groups. Measuring distances between samples with partial least squares discriminate analysis revealed p = 0.09.
Discussion
The goal of this study was to map the intestinal microbiome in patients with CHD undergoing surgery with CPB and determine if the changes observed were similar to other disease states with systemic inflammation. We found novel evidence that pediatric patients with CHD have a disrupted microbiome at baseline, which includes significantly large proportions of pro-inflammatory, lipopolysaccharide-expressing bacteria and a reduction of gut health–promoting bacteria. CPB further exacerbated these derangements. Additionally, our data demonstrated no evidence of intestinal EBD in either group prior to surgery. Moreover, only patients within the CPB group developed EBD, indicating that the development of a “leaky gut” was exclusive to this group. This would indicate that intestinal EBD is more related to CPB than to the surgery itself and that, despite abnormal amounts of pro-inflammatory bacteria at baseline, CHD itself does not result in measurable EBD. The possibility of minimal EBD below the threshold level of our detection method occurring pre-operatively in patients with CHD cannot be ruled out. Additionally, it is possible patients with CHD may experience EBD following any surgical intervention, which may be due to the baseline dysbiosis identified in these patients.
Patients with CHD had significant dysbiosis prior to surgery, which was exacerbated by CPB and could influence outcomes post-operatively. CPB led to an average increase of 17% to 27% in pro-inflammatory bacteria following surgery, which only further exacerbated the dysbiosis. Surprisingly, patients in the control arm experienced a relatively unchanged intestinal microbiome, which signifies that surgery itself may not significantly alter the intestinal microbiome, but larger studies are needed to confirm these results. We believe that dysbiosis influenced the post-operative course predominantly through modification of inflammatory responses. However, the possibility that ventilation, post-operative antibiotics, inotropic support, method of feeding, and length of time spent in the intensive care unit could also contribute to the worsened dysbiosis in the post-surgery CPB group cannot be ruled out completely. Although our analysis of antibiotic duration between patients in the CPB group did not demonstrate a statistical difference in the degree of dysbiosis, a larger sample size may help verify whether this can be a contributing factor. The degree of dysbiosis did not correlate with the degree of EBD following CPB. We were unable to evaluate correlation between the duration of CPB, cross-clamp time, total duration of surgery, or vasoactive-inotropic score with increases in FABP2 and claudin-3 or a reduction in citrulline. A larger sample size may identify correlations among these variables and the degree of intestinal EBD. Antibiotic duration also did not appear to influence the degree of dysbiosis or EBD. Overall, patients with CHD were more likely to have dysbiosis at baseline, and the systemic inflammation following surgery with CPB exacerbated these microbial disturbances and also induced intestinal EBD, which could be a result of the underlying dysbiosis and inflammatory insult of CPB. Collectively, the comparisons of fecal bacterial richness, α-diversity, β-diversity, and the RA of individual taxa from fecal samples collected pre- and post-surgery from pediatric patients undergoing surgical intervention in the presence or absence of CPB shows that the greatest source of variability within the dataset is a pre-existing difference between the 2 groups that is maintained post-surgery. This suggests that the differences observed in the CPB group are the result of an underlying morbidity. Although multivariate analysis is best performed when evaluating the beta-diversity, a small sample size limits interpretation. Limited data are available regarding the effect of probiotics on the microbiome in CHD and on cytokine levels in the pre-operative period. One strain of Bifidobacterium infantis has been shown to not influence circulating cytokine concentrations (22), and increased understanding of the microbiome is needed. In particular, which strains are up-regulated or down-regulated will provide information on how probiotics influence the growth of healthy bacteria in the gut. In a study evaluating a synbiotic agent and the effect on sepsis, reductions in culture-proven sepsis, necrotizing enterocolitis, and mortality were all observed (21). Mechanisms as to how a probiotic or synbiotic agent influences systemic inflammation in the post-operative period have yet to be evaluated.
LCOS is a clinical diagnosis and exists on a spectrum of concern for cardiac dysfunction and systemic inflammation post-operatively. Scoring systems have been attempted in the past, but significant bias and limitations have prevented their use in the clinical management of children (44). In children with CHD, there is the added feature in some patients of hypoxia and/or reduced gut perfusion, which many have influences on the composition of the microbiome, as well as mild degrees of ongoing intestinal EBD. Moreno-Indias et al. (45) showed that intermittent hypoxia induces cyclic hypoxia and reoxygenation, affecting oxygen delivery to the intestinal epithelium. Chronic hypoxia associated with CHD likely has similar effects on epithelial (and periepithelial) oxygenation. This may in turn influence the microbiota that develops in the intestinal tract of these patients. Additionally, time spent in the neonatal intensive care unit prior to discharge or surgical correction influences the development of the intestinal microbiome, and the full effects of this are not yet understood (45, 46, 47).
Patients undergoing surgery with CPB develop systemic inflammation and intestinal EBD. The influence of EBD on patient outcomes has yet to be studied in detail; however, the markers used in the present study have been associated with feeding intolerance, clinical severity, and fluid overload (15). Although strong data support EBD as being associated with the inflammatory process of CPB, it remains unclear whether this is directly related to CPB or whether the existing dysbiosis and the increasing amounts of proinflammatory bacteria in the gut play a vital role in the development of EBD following the inflammatory insult of CPB. FABP2 has been used as a marker evaluating intestinal ischemia and has shown a correlation with epithelial injury and ischemia-reperfusion injury (48, 49, 50). Claudin expression and cellular distribution have been associated with a variety of intestinal inflammatory diseases, and dysregulation in the normal levels of claudin proteins correlates with tight junction loss, signaling increased intestinal permeability (41,51,52). In addition, citrulline has been used as a marker to determine the functional enterocyte mass in pediatric short-bowel syndrome (53, 54, 55). Reductions in butyrate via consumption by proinflammatory bacteria and reduced production via lesser amounts of butyrate-producing healthy bacteria could be a contributing factor to the development of intestinal EBD in this patient population after surgery with CPB (56,57). The degree of systemic inflammation may significantly alter nutritional absorption and the development of reactive oxygen species (58). Genetic expression of DUOX2, a gene encoding dual oxidase 2, has been shown to correlate with the expansion of Proteobacteria in the microbiome (58). Selective decontamination could also influence the development of inflammation through endotoxemia and cytokine activation following CPB (59). It will be important to evaluate this with larger populations to determine if there is a correlation with changes to the microbiome following CPB and the development, or severity, of EBD.
Following completion of the primary analysis, stool samples were evaluated for eicosanoids. Many important eicosanoids were found to be elevated post-surgery in the stool samples of patients undergoing CPB. Many inflammatory markers were also elevated pre-surgery in the CPB group compared with control subjects. This indicates an ongoing inflammatory process within the intestinal tract that is exacerbated by CPB. Although the difference between pre-surgery samples and post-surgery samples was not statistically significant, we believe that a larger sample size would demonstrate significant differences in the measured eicosanoids. Many of the eicosanoids elevated in patients with CHD undergoing CPB are involved in platelet aggregation and neutrophil activation. Intestinal permeability is present, and the question remains if these metabolites are originating in the intestinal tract or migrating from the systemic circulation because of inflammation into the intestinal tract. Although our data do not answer this question, we believe the eicosanoids are originating in the stool from proinflammatory bacteria producing them and/or activating intestinal epithelial cell production. Identifying up-regulation of the prostaglandins in the post-CPB samples was a very interesting finding, as it has well-known involvement in inflammation and intestinal barrier integrity. Understanding the role of inflammatory mediators in activation of both local and systemic inflammation through up-regulation of these mediators by intestinal epithelial cells and the intestinal microbiota will be very important to our understanding of this process. Future studies to evaluate both serum and stool levels of eicosanoids and other inflammatory mediators, as well as mRNA and transcription factors for these metabolites, will be important to understand the origin, signaling, and causal role in the systemic inflammation process.
This study showed that the 2 groups had variability in their gut microbiomes. This makes analysis comparing the effects of surgery with CPB compared with surgery without CPB difficult, despite our statistical corrections, and poses a relative limitation. Some patients did not have all blood samples collected because of either parental refusal without a line to draw blood or venipuncture that would have been needed only to collect a research related sample. Although these nuances pose limitations, they also highlight the pragmatic nature of this study. There were differences between the CPB group and the control group at the time of post-surgery stool collection. All of the control group patients were out of the intensive care unit, off the ventilator, on no inotropic support, and on feeds at the time of post-surgery stool collection. The CPB group had 47% of patients off the ventilator, 65% of patients off inotropic support, 76% on feeds, and 41% had left the intensive care unit by the time of the post-surgery stool collection. It is possible that in addition to CPB, these variables may have also influenced the degree of dysbiosis noted in the CPB group post-surgery compared with the control group.
Although surgical duration was not statistically different between the 2 groups in this cohort, a larger sample size may reveal other potential factors that could have an influence on changes to both the microbiome and markers of intestinal EBD. There were no statistically significant differences in age, sex, NPO duration, anesthetic agents, or steroid use between the 2 groups in this study. Additional examination of a large cohort is warranted to confirm that these factors do not influence outcomes. Taken together, limitations of this study include the small sample size and the presence of CHD in 1 group and not the other. The design of this study focused on perioperative variables, and a study to examine cardiac surgery on CHD with and without CPB is needed. This will likely require a multicenter study over an extended duration to be sufficiently powered.
Many factors exist to influence the outcome measures of this study. We focused on variables associated with perioperative management. Dietary patterns, probiotics, and other environmental and genetic factors have potential to influence results. Further studies controlling for the variables need to be performed and are part of our ongoing studies. Additionally, many patients with CHD spend time in the neonatal intensive care unit prior to discharge home. There are data (44, 45, 46) showing that time spent in a critical care unit influences the development of the intestinal microbiome, and the full effects of this are not yet understood.
The results of this study show novel evidence of the dysbiosis that exists in patients with CHD and that is exacerbated following surgery with CPB. These data offer an important addition to the existing knowledge regarding the causes and factors influencing the systemic inflammatory process following CPB. With this new evidence, further studies aimed to reduce the degree of intestinal dysbiosis should be performed with the aim of improving outcomes following cardiac surgery with CPB. Animal models to evaluate the microbiome with CPB can offer insight into mechanisms and potential interventions to improve this dysbiosis, which could improve outcomes for patients requiring surgery with CPB to correct their cardiac defects. A model of animal CPB to evaluate interventions on the microbiome, such as probiotics, fecal microbiome transplantation, and decontamination, and its effects on systemic inflammation is warranted for future investigations into mechanisms. An intervention to determine if beneficial effects can be achieved by manipulation of the intestinal microbiota to reduce systemic inflammation, as has been observed after critical illness (60,61), is required in this patient population. Additionally, recent findings support the involvement of micro–ribonucleic acid in both the development and exacerbation of CHD as well as influences on the microbiome (62, 63, 64, 65, 66). Targeting micro–ribonucleic acid may also yield therapeutic interventions. Signaling pathways, such as hypoxia-inducing factor 1α and nuclear factor κB, are influenced by hypoxia, inflammation, and the microbiome. How these pathways are regulated in CHD as it relates to changes in the microbiome may also improve our understanding of systemic inflammation. This may be a key component for patients who experience LCOS post-operatively to reduce the degree of systemic inflammation and improve outcomes for this high-risk population.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: This study offers novel evidence to support the influence of CHD and CPB on the microbiome and intestinal EBD. This is the first study to attempt to account for perioperative variables to determine changes to the microbiome. Future studies to expand upon this pilot study and evaluate changes to the microbiome with markers of systemic inflammation could yield knowledge of the microbiome’s role in the inflammatory process and healing following surgery with CPB.
TRANSLATIONAL OUTLOOK: The translational outlook will involve evaluating large populations of patients with CHD to determine the microbiome composition that may be cardiac lesion specific. Animal studies to evaluate interventions on the microbiome and its effect on systemic inflammation will also be important prior to clinical translation. Future therapeutic interventions may involve prebiotic, probiotic, and synbiotic agents guided by disease to improve the microbial composition prior to cardiac interventions aimed at improving outcomes following cardiac surgery.
Funding Support and Author Disclosures
This work was supported by an internal Fleur de Lis $25,000 grant from Saint Louis University School of Medicine. Dr. Lindsey is a Stokes-Shackleford Professor at the University of Nebraska Medical Center. This research was supported by funding from the National Institutes of Health under awards DK121046-01, DK124950-01, DK98623-01A1, DK124095-01A1, HL129823, HL137319, and OD019924-01 and from the U.S. Department of Veterans Affairs Office of Research and Development under awards, and 5I01BX000505. The content is solely the responsibility of the authors and does not necessarily represent the official views of any funding agencies. All authors have reviewed and approved this article. The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental tables and a figure, please see the online version of this paper.
Appendix
References
- 1.Kamada N., Chen G.Y., Inohara N., Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–690. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohland C.L., Jobin C. Microbial activities and intestinal homeostasis: a delicate balance between health and disease. Cell Mol Gastroenterol Hepatol. 2015;1:28–40. doi: 10.1016/j.jcmgh.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bull M.J., Plummer N.T. Part 1: the human gut microbiome in health and disease. Integr Med (Encinitas) 2014;13:17–22. [PMC free article] [PubMed] [Google Scholar]
- 5.Wang H., Wei C.-X., Min L., Zhu L.-Y. Good or bad: gut bacteria in human health and diseases. Biotechnol Biotechnolog Equip. 2018;32:1075–1080. [Google Scholar]
- 6.Cabrera-Perez J., Badovinac V.P., Griffith T.S. Enteric immunity, the gut microbiome, and sepsis: rethinking the germ theory of disease. Exp Biol Med (Maywood) 2017;242:127–139. doi: 10.1177/1535370216669610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu K., Ogura H., Hamasaki T. Altered gut flora are associated with septic complications and death in critically ill patients with systemic inflammatory response syndrome. Dig Dis Sci. 2011;56:1171–1177. doi: 10.1007/s10620-010-1418-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alverdy J.C., Krezalek M.A. Collapse of the microbiome, emergence of the pathobiome, and the immunopathology of sepsis. Crit Care Med. 2017;45:337–347. doi: 10.1097/CCM.0000000000002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hakansson A., Molin G. Gut microbiota and inflammation. Nutrients. 2011;3:637–682. doi: 10.3390/nu3060637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gogos C.A., Drosou E., Bassaris H.P., Skoutelis A. Pro- versus anti-inflammatory cytokine profile in patients with severe sepsis: a marker for prognosis and future therapeutic options. J Infect Dis. 2000;181:176–180. doi: 10.1086/315214. [DOI] [PubMed] [Google Scholar]
- 11.Halter J., Steinberg J., Fink G. Evidence of systemic cytokine release in patients undergoing cardiopulmonary bypass. J Extra Corpor Technol. 2005;37:272–277. [PMC free article] [PubMed] [Google Scholar]
- 12.Sinclair D.G., Haslam P.L., Quinlan G.J., Pepper J.R., Evans T.W. The effect of cardiopulmonary bypass on intestinal and pulmonary endothelial permeability. Chest. 1995;108:718–724. doi: 10.1378/chest.108.3.718. [DOI] [PubMed] [Google Scholar]
- 13.Hamada Y., Kawachi K., Tsunooka N., Nakamura Y., Takano S., Imagawa H. Capillary leakage in cardiac surgery with cardiopulmonary bypass. Asian Cardiovasc Thorac Ann. 2004;12:193–197. doi: 10.1177/021849230401200303. [DOI] [PubMed] [Google Scholar]
- 14.Cremer J., Martin M., Redl H. Systemic inflammatory response syndrome after cardiac operations. Ann Thorac Surg. 1996;61:1714–1720. doi: 10.1016/0003-4975(96)00055-0. [DOI] [PubMed] [Google Scholar]
- 15.Typpo K.V., Larmonier C.B., Deschenes J., Redford D., Kiela P.R., Ghishan F.K. Clinical characteristics associated with postoperative intestinal epithelial barrier dysfunction in children with congenital heart disease. Pediatr Crit Care Med. 2015;16:37–44. doi: 10.1097/PCC.0000000000000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karl J.P., Margolis L.M., Madslien E.H. Changes in intestinal microbiota composition and metabolism coincide with increased intestinal permeability in young adults under prolonged physiological stress. Am J Physiol Gastrointest Liver Physiol. 2017;312:G559–G571. doi: 10.1152/ajpgi.00066.2017. [DOI] [PubMed] [Google Scholar]
- 17.Leclercq S., Matamoros S., Cani P.D. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci U S A. 2014;111:E4485–E4493. doi: 10.1073/pnas.1415174111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention Data and statistics on congenital heart defects. https://www.cdc.gov/ncbddd/heartdefects/data.html Available at:
- 19.Wessel D.L. Managing low cardiac output syndrome after congenital heart surgery. Crit Care Med. 2001;29:S220–S230. doi: 10.1097/00003246-200110001-00004. [DOI] [PubMed] [Google Scholar]
- 20.Chandler H.K., Kirsch R. Management of the low cardiac output syndrome following surgery for congenital heart disease. Curr Cardiol Rev. 2016;12:107–111. doi: 10.2174/1573403X12666151119164647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dilli D., Aydin B., Zenciroğlu A., Özyazici E., Beken S., Okumuş N. Treatment outcomes of infants with cyanotic congenital heart disease treated with synbiotics. Pediatrics. 2013;132:e932–e938. doi: 10.1542/peds.2013-1262. [DOI] [PubMed] [Google Scholar]
- 22.Ellis C.L., Bokulich N.A., Kalanetra K.M. Probiotic administration in congenital heart disease: a pilot study. J Perinatol. 2013;33:691–697. doi: 10.1038/jp.2013.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ellis C.L., Rutledge J.C., Underwood M.A. Intestinal microbiota and blue baby syndrome: probiotic therapy for term neonates with cyanotic congenital heart disease. Gut Microbes. 2010;1:359–366. doi: 10.4161/gmic.1.6.14077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aardema H., Lisotto P., Kurilshikov A. Marked changes in gut microbiota in cardio-surgical intensive care patients: a longitudinal cohort study. Front Cell Infect Microbiol. 2019;9:467. doi: 10.3389/fcimb.2019.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ding W., Liu J., Zhou X. Clinical multi-omics study on the gut microbiota in critically ill patients after cardiovascular surgery combined with cardiopulmonary bypass with or without sepsis (MUL-GM-CSCPB study): a prospective study protocol. Front Med (Lausanne) 2020;7:269. doi: 10.3389/fmed.2020.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walters W.A., Caporaso J.G., Lauber C.L., Berg-Lyons D., Fierer N., Knight R. PrimerProspector: de novo design and taxonomic analysis of barcoded polymerase chain reaction primers. Bioinformatics. 2011;27:1159–1161. doi: 10.1093/bioinformatics/btr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caporaso J.G., Lauber C.L., Walters W.A. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A. 2011;108(suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loy A., Maixner F., Wagner M., Horn M. probeBase—an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 2007;35:D800–D804. doi: 10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 31.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 32.Kuczynski J., Stombaugh J., Walters W.A., González A., Caporaso J.G., Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics. 2011 doi: 10.1002/0471250953.bi1007s36. Chapter 10:Unit 10.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 34.Altschul S.F., Madden T.L., Schäffer A.A. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pruesse E. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammer O., Harper D.A.T., Ryan P.D. PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica. 2001;4:1–9. [Google Scholar]
- 37.Gaies M.G., Gurney J.G., Yen A.H. Vasoactive-inotropic score as a predictor of morbidity and mortality in infants after cardiopulmonary bypass. Pediatr Crit Care Med. 2010;11:234–238. doi: 10.1097/PCC.0b013e3181b806fc. [DOI] [PubMed] [Google Scholar]
- 38.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B Methodol. 1995;57:289–300. [Google Scholar]
- 39.Jiang L., Amir A., Morton J.T., Heller R., Arias-Castro E., Knight R. Discrete false-discovery rate improves identification of differentially abundant microbes. mSystems. 2017;2:e00092–e00117. doi: 10.1128/mSystems.00092-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Louis P., Young P., Holtrop G., Flint H.J. Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol. 2010;12:304–314. doi: 10.1111/j.1462-2920.2009.02066.x. [DOI] [PubMed] [Google Scholar]
- 41.Derikx J.P., Luyer M.D., Heineman E., Buurman W.A. Non-invasive markers of gut wall integrity in health and disease. World J Gastroenterol. 2010;16:5272–5279. doi: 10.3748/wjg.v16.i42.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu Z., Ding L., Lu Q., Chen Y.H. Claudins in intestines: Distribution and functional significance in health and diseases. Tissue Barriers. 2013;1 doi: 10.4161/tisb.24978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel R.M., Myers L.S., Kurundkar A.R., Maheshwari A., Nusrat A., Lin P.W. Probiotic bacteria induce maturation of intestinal claudin 3 expression and barrier function. Am J Pathol. 2012;180:626–635. doi: 10.1016/j.ajpath.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alten J.A., Gaies M. Defining low cardiac output syndrome: an ode to Justice Potter Stewart. Pediatr Crit Care Med. 2017;18:85–87. doi: 10.1097/PCC.0000000000000989. [DOI] [PubMed] [Google Scholar]
- 45.Moreno-Indias I., Torres M., Montserrat J.M. Intermittent hypoxia alters gut microbiota diversity in a mouse model of sleep apnoea. Eur Respir J. 2015;45:1055–1065. doi: 10.1183/09031936.00184314. [DOI] [PubMed] [Google Scholar]
- 46.Rogers M.B., Firek B., Shi M. Disruption of the microbiota across multiple body sites in critically ill children. Microbiome. 2016;4:66. doi: 10.1186/s40168-016-0211-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hayakawa M., Asahara T., Henzan N. Dramatic changes of the gut flora immediately after severe and sudden insults. Dig Dis Sci. 2011;56:2361–2365. doi: 10.1007/s10620-011-1649-3. [DOI] [PubMed] [Google Scholar]
- 48.Holmes J.H.I.V., Lieberman J.M., Probert C.B. Elevated intestinal fatty acid binding protein and gastrointestinal complications following cardiopulmonary bypass: a preliminary analysis. J Surg Res. 2011;100:192–196. doi: 10.1006/jsre.2001.6237. [DOI] [PubMed] [Google Scholar]
- 49.Kano H., Takahashi H., Inoue T., Tanaka H., Okita Y. Transition of intestinal fatty acid-binding protein on hypothermic circulatory arrest with cardiopulmonary bypass. Perfusion. 2017;32:200–205. doi: 10.1177/0267659116667807. [DOI] [PubMed] [Google Scholar]
- 50.Pathan N., Burmester M., Adamovic T. Intestinal injury and endotoxemia in children undergoing surgery for congenital heart disease. Am J Respir Crit Care Med. 2011;184:1261–1269. doi: 10.1164/rccm.201104-0715OC. [DOI] [PubMed] [Google Scholar]
- 51.Moore S.A., Nighot P., Reyes C. Intestinal barrier dysfunction in human necrotizing enterocolitis. J Pediatr Surg. 2016;51:1907–1913. doi: 10.1016/j.jpedsurg.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Catalioto R.M., Maggi C.A., Giuliani S. Intestinal epithelial barrier dysfunction in disease and possible therapeutical interventions. Curr Med Chem. 2011;18:398–426. doi: 10.2174/092986711794839179. [DOI] [PubMed] [Google Scholar]
- 53.Rhoads J.M., Plunkett E., Galanko J. Serum citrulline levels correlate with enteral tolerance and bowel length in infants with short bowel syndrome. J Pediatr. 2005;146:542–547. doi: 10.1016/j.jpeds.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 54.Merlin E., Minet-Quinard R., Pereira B. Non-invasive biological quantification of acute gastrointestinal graft-versus-host disease in children by plasma citrulline. Pediatr Transplant. 2013;17:683–687. doi: 10.1111/petr.12128. [DOI] [PubMed] [Google Scholar]
- 55.Gosselin K.B., Feldman H.A., Sonis A.L. Serum citrulline as a biomarker of gastrointestinal function during hematopoietic cell transplantation in children. J Pediatr Gastroenterol Nutr. 2014;58:709–714. doi: 10.1097/MPG.0000000000000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vital M., Karch A., Pieper D.H. Colonic butyrate-producing communities in humans: an overview using omics data. mSystems. 2017;2:e00130–e00217. doi: 10.1128/mSystems.00130-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riviere A., Selak M., Lantin D., Leroy F., De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baumler A.J., Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. 2016;535:85–93. doi: 10.1038/nature18849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez-Pellus A.E., Merino P., Bru M. Can selective digestive decontamination avoid the endotoxemia and cytokine activation promoted by cardiopulmonary bypass? Crit Care Med. 1993;21:1684–1691. doi: 10.1097/00003246-199311000-00017. [DOI] [PubMed] [Google Scholar]
- 60.Lankelma J.M., van Vught L.A., Belzer C. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: a pilot study. Intensive Care Med. 2017;43:59–68. doi: 10.1007/s00134-016-4613-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fox A.C., McConnell K.W., Yoseph B.P. The endogenous bacteria alter gut epithelial apoptosis and decrease mortality following Pseudomonas aeruginosa pneumonia. Shock. 2012;38:508–514. doi: 10.1097/SHK.0b013e31826e47e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He W., Che H., Jin C., Ge S. Effects of miR-23b on hypoxia-induced cardiomyocytes apoptosis. Biomed Pharmacother. 2017;96:812–817. doi: 10.1016/j.biopha.2017.09.148. [DOI] [PubMed] [Google Scholar]
- 63.Li J., Cao Y., Ma X.J. Roles of miR-1-1 and miR-181c in ventricular septal defects. Int J Cardiol. 2013;168:1441–1446. doi: 10.1016/j.ijcard.2012.12.048. [DOI] [PubMed] [Google Scholar]
- 64.Li J.W., He S.Y., Feng Z.Z. MicroRNA-146b inhibition augments hypoxia-induced cardiomyocyte apoptosis. Mol Med Rep. 2015;12:6903–6910. doi: 10.3892/mmr.2015.4333. [DOI] [PubMed] [Google Scholar]
- 65.Liu S., da Cunha A.P., Rezende R.M. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe. 2016;19:32–43. doi: 10.1016/j.chom.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu S., Weiner H.L. Control of the gut microbiome by fecal microRNA. Microb Cell. 2016;3:176–177. doi: 10.15698/mic2016.04.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.