Abstract
Epigenetic mechanisms play an important part in the regulation of gene expression and these alterations may induce long-term changes in gene function and metabolism. They have received extensive attention in bridging the gap between environmental exposures and disease development via their influence on gene expression. DNA methylation is the earliest discovered epigenetic alteration. In this review, we try to examine the role of DNA methylation and histone modification in Age related macular degeneration (AMD) and Diabetic Retinopathy (DR), its vascular complications and recent progress. Given the complex nature of AMD and DR, it is crucial to improve therapeutics which will greatly enhance the quality of life and reduce the burden for millions of patients living with these potentially blinding conditions.
Keywords: Age-related macular degeneration (AMD), Diabetic Retinopathy (DR), Epigenetics, Gene expression
Introduction
Epigenetic modifications are changes in gene expression and phenotype, without alterations in the DNA sequence.1 Of late, epigenetic modifications have been emphasized as chief mechanisms for the advancement of several diseases.2 Epigenetic modifications include DNA methylation, histone modification and non-coding RNAs3,4 and they activate multiple signaling pathways and regulate gene expression in eyes.5 Exploring the interactions between epigenome, genome, and environmental changes will increase the understanding of epigenetic-disease mechanisms.6 DNA methylation is the earliest discovered and most important epigenetic modification, which is exerted by DNMTs at the 5-position of cytosine residues in CpG dinucleotides,7 which are often grouped in clusters, as CpG islands. DNA methylation of promoter CpG islands can control gene expression8 and hence alterations in these processes may induce long-term changes in gene function and metabolism.9 Conventionally, epigenetic changes were static in gene expression regulation, but, this impression is now being reformed and epigenetic marks, including DNA methylation are dynamic. Though the studies are limited, its known that, epigenetic modifications are protective and it can be reversible, this can be used as therapeutic tools targeting vascular complications. In this review, we examined the role of DNA methylation and histone modification in AMD and DR, its vascular complications and recent progress that have significantly accelerated this field.
Environment gene interactions in AMD
Advances in ageing affects molecular, “cellular and physiological levels” which results in AMD.10 About 8.7% of all cases of blindness globally are contributed by AMD and it is the primary cause of visual loss in technologically advanced countries.11 It has been assessed that about 18.9 million will be affected by the year 2040.12 At present, treatments exist only for the exudative or ‘wet’ form of AMD and these rely on angiogenesis inhibitors.13 There is no treatment for atrophic ‘dry’ AMD, characterized by RPE loss and progressive neuroretinal cellular dysfunction. Extensive studies across the world have highlighted the involvement of genomics to the risk of developing AMD.14
Drusen formation leads to visual disturbances characterized by the role of epigenetics in AMD pathogenesis and the epigenetic mechanisms show a promising gap between environmental exposure and disease development, their influence on gene expression.15 Such studies will allow researchers to generate much-needed therapeutics in preventing disease progression and therapeutic improvement, will greatly advance the value of life and cut the disease burden for millions of patients with blinding conditions.16
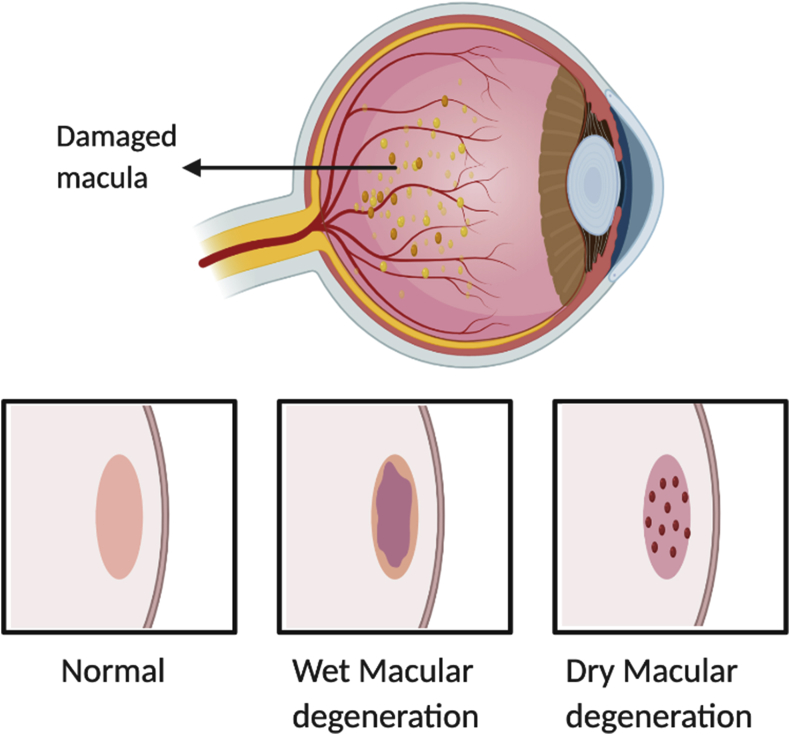
Soft drusen formation between Bruch's membrane and retinal pigmented epithelial (RPE) and loss of RPE cells in the macular region will eventually leads to permanent vision loss.17 Hence, it is crucial to appreciate that cellular phenotype and function are not exclusively decided by genetic information. The non-genetic factors interact with epigenome and alter the expression of gene.18 Therefore all these three factors viz; environment, gene and epigenetics are involved in the AMD pathogenesis. Environmental risk factors, including diet, obesity, smoking, sun exposure, and age, may elicit epigenetic changes that accumulate over a lifetime, eventually resulting in altered gene expression. In AMD, ageing is an important factor for gene silencing due to epigenetic alterations.19 At a molecular level, epigenetic modifications affect protein, metabolic and transcriptomic activities in diseases and can be used as targets for new drugs. These epigenetic modifications are represented in Table 1 and the changes that occur in retina of AMD patients are illustrated in Fig. 1.
Table 1.
Summary of relevant studies on epigenetic modifications in AMD and DR
| S.No. | Disease type | Epigenetic Assay | Marker | Results | Ref |
|---|---|---|---|---|---|
| 1. | DR | DNA methylation | Sirt1 promoter | Hypermethylated Sirt1 promoter | |
| 2. | DR | Histone acetylation - Quantified using Chromatin Immunoprecipitation (ChIP) | MMP-9 promoter | Overexpression of Sirt1 | |
| 3. | Type-I diabetes with proliferative DR | Genome wide analysis (GWAS) and DNA methylation | 349 CpG site - 233 unique genes including TNF, CHI3L1 (also known as YKL-40), CHN2, GIPR, GLRA1, GPX1, AHRR, and BCOR | Decreased DNA methylation | 60 |
| 4. | DR | DNA methylation | MMP-9 promoter | SIRT1 gene and protein expressions were decreased and increased acetylation of NF-κB at MMP-9 promoter and the activity of MMP-9 | |
| 5. | DR | Histone modification - Streptozotocin-induced diabetic rats | MMP-9 promoter | decreased H3K9me2 and increased Ac-H3K9 and p65 at the retinal MMP-9 promoter | 61 |
| 6. | DR | Histone modification - H3k20me3 and H3k9 NF-κB p65 |
Sod2 | Increased H4K20me3, acetyl H3K9, and NF-κB p65 at the promoter and enhancer of retinal sod2 | 64 |
| 7. | DR | DNA methylation | POLG | down-regulated, the D-loop region and Hypermethylation at regulatory region |
58 |
| 8. | DR | Histone Modification – Chromatin Immunoprecipitation (human retinal endothelial cells) | MMP-9 promoter | elevated H3K27me3 levels | |
| 9. | DR | Histone modification – Chromatin immunoprecipitation | H3K4 methylation in Nrf2 binding at Gclc-ARE4 | H3K4me3 and H3K4me1 decreased | 45 |
| 10. | DR | Histone modification – chromatin-immunoprecipitation and co-immunoprecipitation | MMP-9 promoter | hyperacetylaation | 56 |
| 11. | DR | Global DNA methylation - 5-methylcytosine content was assessed by reversed-phase high-pressure liquid chromatography of peripheral blood leukocytes | Global DNA methylation | increasing trend in global DNA methylation levels | 61 |
| 12. | DR | DNA methylation | D- loop | Methylation in retinal capillary cells | 45 |
| 13. | DR | DNA methylation | Cyt b region | Increased methylation | 45 |
| 14. | DR | Histone modification - Methylation enriched H3k4me1 and activated setD7 | Keap1 promoter | Keap1 promoter increased expression | 45 |
| 15. | DR | Histone methylation | H3k4 by a lysine specific histone demethylase (LSD-1) | Transcriptional repression | |
| 16. | DR | DNA methylation | POLG | Hypermethylation of CpG sites at the regulatory regions | 63 |
| 17. | AMD RPE cells |
DNA methylation | GSTM1 and GSTM5 promoters | GSTM1 promoter hypermethylation | 30 |
| 18. | AMD | DNA methylation | IL17RC promoter | Demethylation | 27 |
| 19. | AMD -RPE cells | Histone modification | SIRT1 induced deacetylation of the p53 protein | Hypomethylation | 15,39 |
| 20. | DR and AMD [Human retinal endothelial cells (HRECs) and Retinal Pigmented epithelial cells (ARPE-19)] | miRNA analysis - Hcy induced BRB dysfunction | HDAC, DNMT |
MiRNA - HDAC and DNMT activation | 37 |
| 21. | AMD | DNA methylation | IL17RC promoter | Hypomethylation | 27 |
| 22. | AMD | DNA methylation | IL17RC promoter | Hypermethylation | 28,29 |
| 23. | AMD | DNA methylation | GSTM1 and GSTM2 | Hypermethylation | 31 |
| 24. | AMD (in retinas and peripheral blood) | DNA methylation | IL17RC | Hypomethylation | 12 |
Table shows the list of evidence of recent studies on Epigenetic modifications (DNA methylation and histone modifications) in Age related macular degeneration (AMD) and Diabetic Retinopathy (DR).
Figure 1.
Illustrates the retina structure in normal eye and in the pathogenesis of AMD.
DNA methylation status in AMD
Both genetic and environmental elements contribute to the risk of AMD, it is less clear how these 2 systems interact. One type of epigenetic mark that has been explored in a number of studies is the difference in DNA methylation patterns between cases and controls. Currently, only inadequate studies have been reported in ocular diseases related to epigenetics which include age-related macular degeneration (AMD), diabetic retinopathy, cataract, pterygium, retinoblastoma, uveal melanoma, glaucoma, keratitis, and uveitis.20, 21, 22 In this context, DNMTs activity was found to be higher in AMD patients than in controls. AMD patients also exhibited up-regulation of DNMT1 and DNMT3B expression. It has been observed that expression of HIF1a is downregulated by HDAC1 and expression of VEGF is downregulated by HDAC7 and promotes the angiogenesis genes.23,24 Epigenetic changes involved in inflammatory cytokines and T cells leads to another hallmark of AMD25 and Interleukin (IL)-17 receptor C promoter demethylation enhances inflammatory response.26 A study on methylation status revealed an elevated level of mRNA expression in response to IL17RC promoter hypomethylation in retina and peripheral blood of AMD patients and this epigenetic modification may impact the proinflammatory activity of monocytes in AMD pathogenesis, although further research is needed to confirm these results.12 Wei et al26 assessed methylation changes at the IL17RC promoter in discordant siblings for AMD as well as in an AMD case–control study and evaluated IL17RC expression in the eyes and blood of AMD patients and showed that hypomethylation of the IL17RC promoter led to an elevated expression of its protein and mRNA, suggesting that the DNA methylation pattern and expression of IL17RC may potentially serve as a biomarker for the diagnosis of AMD. In contrast, Oliver et al27 could not find any evidence for differential methylation between AMD and controls, therefore concluding that hypomethylation of the promoter region will not be an applicable biomarker for AMD diagnosis, further pin-pointing the requirement for more studies in this area. Similarly, Oxidative damage induces decreased mRNA and protein levels of detoxification enzymes, such as glutathione S-transferase (GSTM1 and GSTM5) in RPE cells correlated with GSTM1 promoter hypermethylation.28 In precise, GSTM1 and GSTM5 were subjected to hypermethylation leading to lower amounts of corresponding mRNA and proteins in RPE and choroid which increases the susceptibility to oxidative stress in patients with AMD.29 Interestingly, both mitochondrial and nuclear DNA from RPE cells of patients with AMD showed increased oxidative damage, thereby indicating the role of imbalanced redox enzymes in the pathogenesis of AMD.30
Histone modifications and chromatin remodeling in AMD
Chromatin is the complex of chromosomal DNA and proteins and DNA is packaged in chromatin around the histone protein units called nucleosomes, which are highly alkaline, and positively charged amine groups31 which help histone proteins to interact with and bind to the negatively charged phosphate backbone. During acetylation, amines on the histone change into amides; thereby, positive charges on the histone become neutralized and subsequently the capability of the histones to bind with DNA is reduced32 and this results in expansion of chromatin allowing genetic transcription. The histone deacetylation is catalyzed when histone deacetylases removing acetyl groups and increase the positively charged amine groups of histones and facilitate high-affinity binding between the histones and the phosphate backbone.33
Vavilala et al,34 showed that pure honokiol inhibits the HIF pathway and hypoxia-mediated expression of pro-angiogenic genes in retinal pigmented epithelial (RPE) cell lines using chromatin immunoprecipitation assays, demonstrating that honokiol inhibits binding of HIF to hypoxia-response elements present on VEGF promoter, making it an ideal therapeutic agent for the treatment of ocular neovascular diseases. Subsequently, miRNA pathway analysis has shown their involvement in HDAC and DNMT activation and oxidative stresses, inflammation, hypoxia, and angiogenesis pathways. Hcy-induced epigenetic modifications may be involved in retinopathies associated with HHcy, such as age related macular degeneration and diabetic retinopathy.35 Previous evidence supports the role of aberrant epigenetic modifications with significant increase in mRNA expression of HDAC1, HDAC3, HDAC6, DNMT1 and DNMT3a in RPE cells of mice with excessive iron levels and, thus, are at a higher risk for AMD.36 Cultured RPE cells derived from patients with AMD showed hypomethylation of the clusterin promoter as well.37 Recently, aberrant epigenetic modifications have been identified in the pathogenesis of AMD. This has led to the development of alternative therapies that can alter aberrant chromatin-remodelling processes involved in AMD. These novel therapeutic agents could help to ameliorate the challenges in current treatment. However, epigenetic based treatments are young and more pre-clinical studies are needed to evaluate their mechanism in AMD therapeutics.38
Epigenetic changes in diabetic retinopathy (DR)
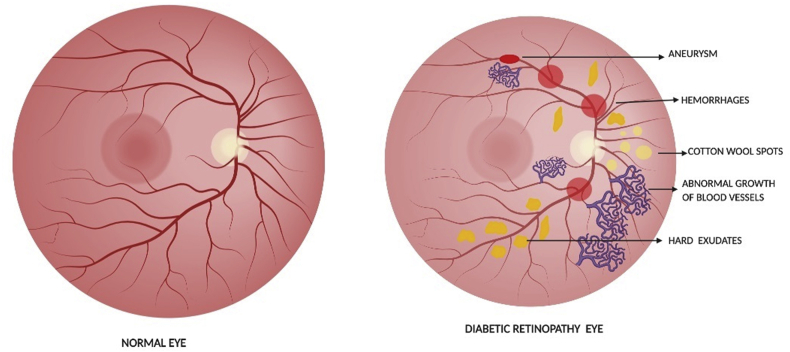
Diabetic Retinopathy (DR) is a microvascular complication in diabetes and its major initiator is hyperglycemia.39 Development of DR occurred even after hyperglycemia was replaced by normal glycemia.40, 41, 42, 43, 44, 45, 46 Circulation of high glucose is considered as the major instigator for changes in metabolic and deleterious functions.39,43,47 DNA is highly dynamic and responds to the environmental stimuli by modifying its properties48 and changes in DNA methylation can lasts for several years.49 Hyperglycemia initiates metabolic abnormalities which induces genetic alterations in the retina and continues to progress even after hyperglycemia termination.43,50,51 The epigenetic changes are depicted in Table 1 and the alterations that occur in DR are represented in Fig. 2.
Figure 2.
Illustrates the normal retina and in DR pathogenesis.
Role of DNA methylation in DR
Reversal of normal glycemia after a period does not reverse the epigenetic changes and mitochondria continues to be dysfunctional.52,53 In diabetic patients, the DNMT activity is increased,52 and the maintenance enzyme DNMT1 is upregulated.54 Tewari et al52 also demonstrated a significant increase in DNMT activity in the retinal nuclear fraction of the rodents with DR. Differential DNA methylation patterns are observed in the blood samples of patients with proliferative DR as well.55 Oxidative stress has been known to play a role in DR. The mtDNA hypermethylation plays an important role for the continued imbalance in mitochondrial homeostasis. Diabetes increases Tet activity in the retina and this plays a significant role in the MMP-9 upregulation, an enzyme implicated in mitochondrial damage. Increased binding of Tet2 results in promoter hypomethylation of MMP-9 which results in activation of transcription.56 Regulation of DNMT1 inhibits glucose-induced mtDNA methylation in retinal endothelial cells, and ameliorates its transcription.54 Hypermethylation at the regulatory region of DNA polymerase gamma (POLG1) was hypermethylated in DR57 leads to impairment of the mitochondrial DNA replication system and subsequent apoptosis of retinal capillary cells. Recent studies58,59 states that diabetic environment favors methylation of CpG dinucleotides forming 5-methylcytosine (5 mC). Continued hypermethylation of the CpG sites at the regulatory region of POLG affects its binding to the mtDNA, compromising the transcriptional activity. Modulation of DNA methylation using pharmaceutic or molecular means could help maintain mitochondria homeostasis, and prevent further progression of DR61. These studies show a strong association between altered methylation in mtDNA and mitochondrial homeostasis.54,56
Histone modifications in DR and its therapeutic interventions
Diabetic environment also brings about epigenetic modifications in the retina, and enzymes responsible for modifications of histones and DNA are altered.43 Hyperglycaemia leads to a significant epigenetic alteration of Sod2 in H4K20me3 and H3K9 in a diabetic rodent model.61 Reversal of hyperglycemia fails to provide any benefit to epigenetic modifications of Keap1 promoter, suggesting its role in DR.43 Effect of high glucose on monomethyl H3K4 (H3K4me1), dimethyl H3K4 (H3K4me2), and lysine-specific demethylase-1 (LSD1) was quantified at Sod2 by chromatin immunoprecipitation and histone methylation of retinal Sod2 has an important role in the development of DR and in the metabolic memory phenomenon associated with its continued progression. Similarly, the effect of high glucose on the binding of transcriptional factor Sp1 at Keap1 promoter and histone methylation status was investigated in retinal endothelial cells. Epigenetic modifications at Keap1 promoter by SetD7 facilitate its binding with Sp1, increasing its expression. Polymorphisms in H3K9 and DNMT have also been observed in diabetic vascular complications62 and the enzyme important for trimethylation of H4K20, is elevated in DR.63 Epigenetic modifications play an important role in regulation of molecular mechanisms associated with DR and histone modifications in MMP-9. 53 Epigenetic alterations in the MMP-9 promoter region have been identified in human donor eyes with DR53 and diabetic C57BL/6J mice.61 Interestingly, experimental evidence has shown that, alterations in histone modifications affects maintenance of mitochondrial homeostasis in DR.43 Mitochondrial superoxide levels are elevated in the diabetic retina and the dysfunctional mitochondria accelerates retinal cell apoptosis43,64, which precedes the histopathology in DR, incriminating it in DR. The Sirt1 regulation in diabetes further indicates the role of epigenetic influence on transcriptional suppression.65 These studies show the significant link between histone modifications and DR. These kinds of studies are warranted in future to identify a suitable potential biomarker for DR. Analyzing the epigenetic changes and the outcome of the results helps in sensible evaluation of their mechanism of action, specificity and adverse effects. An increased understanding of the complex relationships between genetic, epigenetic, and environmental factors involved in the development of complex diseases, such as degenerative retinal diseases, preventative and therapeutic interventions will reduce the severity and complications arising from such diseases, thereby improving the quality of life for older patients.
Conclusions and future perspectives
As AMD and DR have a complex nature, it is crucial to establish a database of aberrant methylation and acetylation patterns. Eventually, this will allow clinicians to devise individualized therapies and promise patients ‘healthy ageing’. DNA methylation plays a critical role in the pathogenesis of retinal disease complications and better understanding of the role and mechanism of DNA methylation and ocular complications can inspire critical implications for the early prevention of these diseases and provide unique opportunities to develop novel therapeutic approaches. However, there are only limited studies related to histone modifications in eye diseases and these kinds of studies are warranted in future. Furthermore, increasing evidence shows that epigenetic modifications are not static, which are dynamic and even reversible and therefore, in this view of epigenetic mechanisms, intervention with pharmaceuticals or other interventions during early course of the disease may ameliorate its complications in later life.
Conflict of Interests
The authors declare no conflict of interests.
Acknowledgements
We thank Vision Research Foundation, Sankara Nethralaya, Chennai for supporting necessary facilities to carry out this review.
Footnotes
Peer review under responsibility of Chongqing Medical University.
References
- 1.Feinberg A.P. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 2.Willyard C. The saving switch. Nat Med. 2010;16(1):18–21. doi: 10.1038/nm0110-18. [DOI] [PubMed] [Google Scholar]
- 3.Skinner M.K., Manikkam M., Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. TEM (Trends Endocrinol Metab) 2010;21(4):214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu W., Wang F., Yu Z., Xin F. Epigenetics and cellular metabolism. Genet Epigenet. 2016;8:43–51. doi: 10.4137/GEG.S32160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchi C., Miccoli R., Del Prato S. Hyperglycemia and vascular metabolic memory: truth or fiction? Curr Diabetes Rep. 2013;13(3):403–410. doi: 10.1007/s11892-013-0371-2. [DOI] [PubMed] [Google Scholar]
- 6.Pons D., de Vries F.R., van den Elsen P.J., Heijmans B.T., Quax P.H., Jukema J.W. Epigenetic histone acetylation modifiers in vascular remodelling: new targets for therapy in cardiovascular disease. Eur Heart J. 2009;33(3):266–277. doi: 10.1093/eurheartj/ehn603. [DOI] [PubMed] [Google Scholar]
- 7.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 8.Reik W., Dean W. DNA methylation and mammalian epigenetics. Electrophoresis. 2001;22(14):2838–2843. doi: 10.1002/1522-2683(200108)22:14<2838::AID-ELPS2838>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 9.Tonna S., El-Osta A., Cooper M.E., Tikellis C. Metabolic memory and diabetic nephropathy: potential role for epigenetic mechanisms. Nat Rev Nephrol. 2010;6(6):332–341. doi: 10.1038/nrneph.2010.55. [DOI] [PubMed] [Google Scholar]
- 10.Sharma K., Sharma N.K., Anand A. Why AMD is a disease of ageing and not of development: mechanisms and insights. Front Aging Neurosci. 2014;6 doi: 10.3389/fnagi.2014.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Health Organization Age Related Macular Degeneration: Priority eye diseases. http://www.who.int/blindness/causes/priority/en/index8.html
- 12.Sobrin L., Seddon J.M. Nature and nurture- genes and environment- predict onset and progression of macular degeneration. Prog Retin Eye Res. 2014;40:1–15. doi: 10.1016/j.preteyeres.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muthiah M.N., Keane P.A., Zhong J. Adaptive optics imaging shows rescue of macula cone photoreceptors. Ophthalmol Times. 2014;121(1):430–431. doi: 10.1016/j.ophtha.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 14.Bharti K., Rao M., Hull S.C. Developing cellular therapies for retinal degenerative diseases. Invest Ophthalmol Vis Sci. 2014;55(2):1191–1202. doi: 10.1167/iovs.13-13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suuronen T., Nuutinen T., Ryhanen T., Kaarniranta K., Salminen A. Epigenetic regulation of clusterin/apolipoprotein J expression in retinal pigment epithelial cells. Biochem Biophys Res Commun. 2007;357(2):397–401. doi: 10.1016/j.bbrc.2007.03.135. [DOI] [PubMed] [Google Scholar]
- 16.Pennington K.L., DeAngelis M.M. Epigenetic mechanisms of the aging human retina. J ExpNeurosc. 2015;9(S2):51–79. doi: 10.4137/JEN.S25513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan K.N., Mahroo O.A., Khan R.S. Differentiating drusen: drusen and drusen-like appearances associated with ageing, age-related macular degeneration, inherited eye disease and other pathological processes. Prog Retin Eye Res. 2016;53:70–106. doi: 10.1016/j.preteyeres.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein B.E., Meissner A., Lander E.S. The mammalian epigenome. Cell. 2007;128(4):669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 19.Sedivy J.M., Banumathy G., Adams P.D. Aging by epigenetics: a consequence of chromatin damage? Exp Cell Res. 2008;314(9):1909–1917. doi: 10.1016/j.yexcr.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauthier A.C., Liu J. Epigenetics and signaling pathways in glaucoma. BioMed Res Int. 2017;2017 doi: 10.1155/2017/5712341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma A., Stei M.M., Frohlich H., Holz F.G., Loeffler K.U., Herwig-Carl M.C. Genetic and epigenetic insights into uveal melanoma. Clin Genet. 2018;93(5):952–961. doi: 10.1111/cge.13136. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X., Zhao L., Hambly B., Bao S., Wang K. Diabetic retinopathy: reversibility of epigenetic modifications and new therapeutic targets. Cell Biosci. 2017;7(1) doi: 10.1186/s13578-017-0167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M.S., Kwon H.J., Lee Y.M. Histone deacetylase induces angiogenesis by negative regulation of tumour suppressor genes. Nat Med. 2001;7(4):437–443. doi: 10.1038/86507. [DOI] [PubMed] [Google Scholar]
- 24.Wang S., Li X., Parra M., Verdin E., Rhonda B.D., Olson E.N. Control of endothelial cell proliferation and migration by VEGF signaling to histone deacetylase 7. Proc Natl Acad Sci Unit States Am. 2008;105(22):7738–7743. doi: 10.1073/pnas.0802857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He S., Li X., Chan N., Hinton D.R. Epigenetic mechanisms in ocular disease. Mol Vis. 2013;19:665–674. [PMC free article] [PubMed] [Google Scholar]
- 26.Wei L., Liu B., Tuo J. Hypomethylation of the IL17RC promoter associates with age related macular degeneration. Cell Rep. 2012;2(5):1151–1158. doi: 10.1016/j.celrep.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliver V.F., Franchina M., Jaffe A.E. Hypomethylation of the IL17RC promoter in peripheral blood leukocytes is not a hallmark of age-related macular degeneration. Cell Rep. 2013;5(6):1527–1535. doi: 10.1016/j.celrep.2013.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hunter A., Spechler P.A., Cwanger A. DNA methylation is associated with altered gene expression in AMD. Invest Ophthalmol Vis Sci. 2012;53(4):2089–2105. doi: 10.1167/iovs.11-8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gemenetzi M., Lotery A.J. The role of epigenetics in age-related macular degeneration. Eye. 2014;28(12):1407–1417. doi: 10.1038/eye.2014.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarrett S.G., Lin H., Godley B.F., Boulton M.E. Mitochondrial DNA damage and its potential role in retinal degeneration. Prog Retin Eye Res. 2008;27(6):596–607. doi: 10.1016/j.preteyeres.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Fedorova E., Zink D. Nuclear architecture and gene regulation. Biochim Biophys Acta. 2008;1783(11):2174–2184. doi: 10.1016/j.bbamcr.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 32.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Nan X., Ng H.H., Johnson C.A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 34.Vavilala D.T., Ponnaluri V.K.C., Kanjilal D., Mukherji M. Evaluation of anti-HIF and anti- angiogenic properties of honokiol for the treatment of ocular neovascular diseases. PloS One. 2014;9(11) doi: 10.1371/journal.pone.0113717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elmasry K., Riyaz M., Isha S. Epigenetic modifications in hyperhomocysteinemia: potential role in diabetic retinopathy and age-related macular degeneration. Oncotarget. 2018;9(16):12562–12590. doi: 10.18632/oncotarget.24333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gnana-Prakasam J.P., Veeranan-Karmegam R., Coothankandaswamy V. Loss of Hfe leads to progression of tumor phenotype in primary retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2013;54(1):63–71. doi: 10.1167/iovs.12-10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suuronen T., Nuutinen T., Ryhanen T., Kaarniranta K., Salminen A. Epigenetic regulation of clusterin/apolipoprotein J expression in retinal pigment epithelial cells. Biochem Biophys Res Commun. 2007;357(2):397–401. doi: 10.1016/j.bbrc.2007.03.135. [DOI] [PubMed] [Google Scholar]
- 38.Faith A.A., Kwa1, Thilini R.T. Epigenetic modifications as potential therapeutic targets in age-related macular degeneration and diabetic retinopathy. Drug Discov Today. 2014;19(9):1387–1393. doi: 10.1016/j.drudis.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 39.Frank R.N. Diabetic retinopathy. N Engl J Med. 2004;350:48–58. doi: 10.1056/NEJMra021678. [DOI] [PubMed] [Google Scholar]
- 40.Chilelli N.C., Burlina S., Lapolla A. AGEs, rather than hyperglycemia are responsible for microvascular complications in diabetes: a ‘‘glycoxidation-centric’’ point of view. Nutr Metabol Cardiovasc Dis. 2013;23(10):913–919. doi: 10.1016/j.numecd.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Kowluru R.A., Zhong Q., Kanwar M. Metabolic memory and diabetic retinopathy: role of inflammatory mediators in retinal pericytes. Exp Eye Res. 2010;90(5):617–623. doi: 10.1016/j.exer.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Madsen-Bouterse S.A., Mohammad G., Kanwar M., Kowluru R.A. Role of mitochondrial DNA damage in the development of diabetic retinopathy and the metabolic memory phenomenon associated with its progression. Antioxidants Redox Signal. 2010;13(6):797–805. doi: 10.1089/ars.2009.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mishra M., Kowluru R.A. Retinal mitochondrial DNA mismatch repair in the development of diabetic retinopathy and its continued progression after termination of hyperglycemia. Invest Ophthalmol Vis Sci. 2014;55(10):6960–6967. doi: 10.1167/iovs.14-15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos J.M., Kowluru R.A. Role of mitochondria biogenesis in the metabolic memory associated with the continued progression of diabetic retinopathy and its regulation by lipoic acid. Invest Ophthalmol Vis Sci. 2011;52(12):8791–8798. doi: 10.1167/iovs.11-8203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santos J.M., Kowluru R.A. Impaired transport of mitochondrial transcription factor A (TFAM) and the metabolic memory phenomenon associated with the progression of diabetic retinopathy. Diabetes/Metab Res Rev. 2013;29(3):204–213. doi: 10.1002/dmrr.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos J.M., Mishra M., Kowluru R.A. Posttranslational modification of mitochondrial transcription factor A in impaired mitochondria biogenesis: implications in diabetic retinopathy and metabolic memory phenomenon. Exp Eye Res. 2014;121:168–177. doi: 10.1016/j.exer.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kowluru R.A., Kowluru A., Mishra M., Kumar B. Oxidative stress and epigenetic modifications in the pathogenesis of diabetic retinopathy. Prog Retin Eye Res. 2015;48:40–61. doi: 10.1016/j.preteyeres.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kowluru R.A. Diabetic retinopathy, metabolic memory and epigenetic modifications. Vis Res. 2017;139:30–38. doi: 10.1016/j.visres.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 49.Zawia N.H., Lahiri D.K., Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic Biol Med. 2009;46(9):1241–1249. doi: 10.1016/j.freeradbiomed.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White N.H., Sun W., Cleary P.A. Effect of prior intensive therapy in type 1 diabetes on 10-year progression of retinopathy in the DCCT/EDIC: comparison of adults and adolescents. Diabetes. 2010;59(5):1244–1253. doi: 10.2337/db09-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddy M.A., Zhang E., Natarajan R. Epigenetic mechanisms in diabetic complications and metabolic memory. Diabetologia. 2015;58(3):443–455. doi: 10.1007/s00125-014-3462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tewari S., Zhong Q., Santos J.M., Kowluru R.A. Mitochondria DNA replication and DNA methylation in the metabolic memory associated with continued progression of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2012;53(8):4881–4888. doi: 10.1167/iovs.12-9732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong Q., Kowluru R.A. Epigenetic modification of Sod 2 in the development of diabetic retinopathy and in the metabolic memory: role of histone methylation. Invest Ophthalmol Vis Sci. 2013;54(1):244–250. doi: 10.1167/iovs.12-10854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mishra M., Kowluru R.A. Epigenetic modification of mitochondrial DNA in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2015;56(9):5133–5142. doi: 10.1167/iovs.15-16937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agardh E., Lundstig A., Perfilyev A. Genome-wide analysis of DNA methylation in subjects with type 1 diabetes identifies epigenetic modifications associated with proliferative diabetic retinopathy. BMC Med. 2015;13 doi: 10.1186/s12916-015-0421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shan Y., Kowluru R. Dynamic epigenetic modifications of retinal matrix metalloproteinase-9 in the development of diabetic retinopathy. Lab Invest. 2016;96:1040–1049. doi: 10.1038/labinvest.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tewari S., Santos J.M., Kowluru R.A. Damaged mitochondrial DNA replication system and the development of diabetic retinopathy. Antioxidants Redox Signal. 2012;17(3):492–504. doi: 10.1089/ars.2011.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maghbooli Z., Larijani B., Emamgholipour S., Amini M., Keshtkar A., Pasalar P. Aberrant DNA methylation patterns in diabetic nephropathy. J Diabetes Metab Disord. 2014;13 doi: 10.1186/2251-6581-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zou L., Yan S., Guan X., Pan Y., Qu X. Hypermethylation of the prkcz gene in type 2 diabetes mellitus. J Diabetes Res. 2013;2013 doi: 10.1155/2013/721493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santos J.M., Tewari S., Kowluru R.A. A compensatory mechanism protects retinal mitochondria from initial insult in diabetic retinopathy. Free Radic Biol Med. 2012;53(9):1729–1737. doi: 10.1016/j.freeradbiomed.2012.08.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kowluru R.A., Shan Y., Mishra M. Dynamic DNA methylation of matrix metalloproteinase-9 in the development of diabetic retinopathy. Lab Invest. 2016;96(10):1040–1049. doi: 10.1038/labinvest.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Syreeni A., Aei-Osta A., Forsblom C., Sandholm N., Parkkonen M., Tarnow L. Genetic examination of SETD7 and SUV39H1/H2 methyltransferases and the risk of diabetes complications in patients with type 1 diabetes. Diabetes. 2011;60(11):3073–3080. doi: 10.2337/db11-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhong Q., Kowluru R.A. Epigenetic changes in mitochondrial superoxide dismutase in the retina and the development of diabetic retinopathy. Diabetes. 2011;60(4):1304–1313. doi: 10.2337/db10-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kowluru R.A., Abbas S.N. Diabetes-induced mitochondrial dysfunction in the retina. Invest Ophthalmol Vis Sci. 2003;44(12):5327–5334. doi: 10.1167/iovs.03-0353. [DOI] [PubMed] [Google Scholar]
- 65.Mishra M., Duraisamy A.J., Kowluru R.A. Sirt1-a guardian of the development of diabetic retinopathy. Diabetes. 2018;67(4):745–754. doi: 10.2337/db17-0996. [DOI] [PMC free article] [PubMed] [Google Scholar]