Abstract
Circular RNAs are a large class of noncoding RNAs. Smad5 functions in cell differentiation, cell proliferation and metastasis. It has been reported that lnc-Smad5 can inhibit the proliferation of diffuse large B cell lymphoma. However, the function of circ-Smad5 has not yet been reported. Lentivirus vectors were constructed to establish circ-Smad5 upregulated and circ-Smad5 downregulated cell models. A CCK-8 assay was used to detect the proliferation of JB6 cells. FACS was used to analyze the cell cycle in the cell models. Western blot, immunofluorescence staining and TOP/FOP flash dual luciferase activity assays were used to determine the activity of the Wnt signaling pathway. The results revealed that the expression level of circ-Smad5 in JB6 cells was significantly lower than the expression level of linearized-Smad5. Compared with the control group, the percentage of S phase cells and the expression level of cyclin D1 protein were significantly higher in the sh-circ-Smad5 group. In the sh-circ-Smad5 group, β-catenin and LEF-1 were significantly increased, p-β-catenin was significantly decreased, and the relative activity of the TOP/FOP reporter gene was higher compared to the control group levels. These phenomena could be reversed by treating with Wnt signaling inhibitor PNU-74654. We conclude that the circ-Smad5 retards the proliferation and the cell cycle progression of JB6 cells. Thus, circ-Smad5 may function by inhibiting the activation of Wnt/β-catenin/Lef 1 signaling, which inhibits the expression of cyclin D1. To the best of our knowledge, we are the first to report the function of circ-Smad5.
Keywords: Cell cycle, Circ-Smad5, Circular RNA, Wnt signaling, Cell proliferation
Introduction
Circular RNAs are a large class of noncoding RNAs that were first reported decades ago. In recent years, research on circular RNAs has arisen again with the advancement of sequencing techniques. Circular RNAs are the products of backsplicing. In eukaryotic cells, they are found both in the cell plasma and in the cell nucleus.1 Several biological functions of circular RNA have already been revealed. Some circular RNAs such as CiRS-7 and circ-HIPK3 can function as microRNA sponges.2,3 Some circular RNAs such as circ-ABCC4 and circ-PABPN1 can function as ceRNAs to regulate the formation of corresponding mRNAs.4, 5, 6 Some circular RNAs can even encode proteins,7,8 which arises a new question. Should circular RNA be classed as non-coding RNAs? Recently, circ-Foxo 3 was found to regulate the cell cycle by forming ternary complexes with p21 and CDK2.9 This identifies a new area of circular RNA.
The cell cycle consists of G1, S, G2 and M phases. The cell cycle is involved in all the life activities of a cell, including cell proliferation, cell differentiation, cell migration, cell senescence and cell death.10,11 The epidermis is the largest defensive barrier of the body. Epidermal cells are renewing all the time through proliferation and differentiation. Any factor that affects the cell cycle of epidermal cells will affect the function of the epidermis.12,13 Cyclins function as the regulatory units in cell cycle progression.14,15
Both circular Smad5 (circ-Smad5) and linearized Smad5 (Smad5 mRNA) are encoded by the Smad5 gene. Smad5 is a critical molecule in the TGF-β signaling pathway. It functions in mesenchymal stem cell differentiation, cell proliferation and metastasis.16, 17, 18 It has been reported that lnc-Smad5-AS1 can inhibit the proliferation of diffuse large B cell lymphoma.19 However, the function of circ-Smad5 has not yet been reported. Epidermal cells quickly proliferate in vivo. Thus, epidermal cell lines are good models for research on the cell cycle. Our study showed that circ-Smad5 functions as an inhibitor of cell cycle progression in epidermal cells.
Materials and methods
Cell culture
JB6 and 293T cell lines were cultured as previously reported.20 The growth medium for the JB6 cell line was high-glucose DMEM (HyClone, USA) with 10% fetal bovine serum (HyClone, USA). The growth medium for 293T cells was high-glucose DMEM (HyClone, USA) with 10% fetal bovine serum (Everygreen, China).
Construction of vectors and stable cell lines
The siRNAs were synthesized by Oligobio (Beijing, China). The sequences are as follows: msi-1 GCCACAATGAACCGCACAT, msi-2 GCCCAAGAATGAGGCTGTT. msi-3 GGTGTACGCTGAGTGTCTT. The short hairpin RNA (shRNA) vector (pwslv-sh07-mCherry-circSmad5) was constructed by Synbio Tech (Suzhou, China). The target sequence of shRNA is TCCAGCAGAGGACTTGCACTA. The ectopic expression vector PLCDH was purchased from Geneseed Biotechnology (Guangzhou, China). Pwslv-sh07-circSmad5, psPAX2 and pMD2.G plasmids were simultaneously transfected into 293T cells using Lipofectamine 2000 (Invitrogen, USA) for preparation of the lentivirus suspension. Then JB6 cells were infected with the lentivirus suspension. Puromycin selection and fluorescence microscopy were used to screen the successfully infected cells. The same treatment was performed with the PLCDH-circSmad5 plasmid to generate circSmad5 overexpressed cells.
RT-PCR
The total RNA of JB6 cells was extracted with an Eastep Super Total RNA Extraction Kit (Promega, China) according to the manufacturer's protocol. The concentrations and purity of the isolated RNA were measured with a NanoDrop One instrument (Thermo Fisher Scientific, USA). cDNA was synthesized with a First Strand cDNA Synthesis Kit (ToYoBo, Japan). Then, 2× Taq Master Mix (Novoprotein, China) was used to amplify the target genes. Target bands were detected by 1.5% agarose gel electrophoresis. Circular primers were designed to detect the expression of circular RNA Smad 5, linearized primers were designed to detect the expression of Smad5 mRNA, and all-Smad5 primers were designed to detect the expression of both circular RNA Smad5 and mRNA Smad5 (Fig. 1B). The sequences of the primers were as follows: 5′-TCAGACCATGCCCAGCATATC-3’ (sense), and 5′-AGATACCAAAAGCGTGGCTC-3’ (antisense) for circ-Smad5; 5′-CCATGGATTCGAGGCTGTGT-3’ (sense) and 5′-AAATGGGGTTCAGCGGAGAG-3’ (antisense) for linearized-Smad5; 5′-ACAAGGTGACGAGGAAGA-3’ (sense) and 5′-ACTAATACTGGAGGTAAGACTG-3’ (antisense) for all-Smad5; 5′-TGCTGTCCCTGTATGCCTCT-3’ (sense) and 5′-TTGATGTCACGCACGATTTC-3’ (antisense) for β-actin.
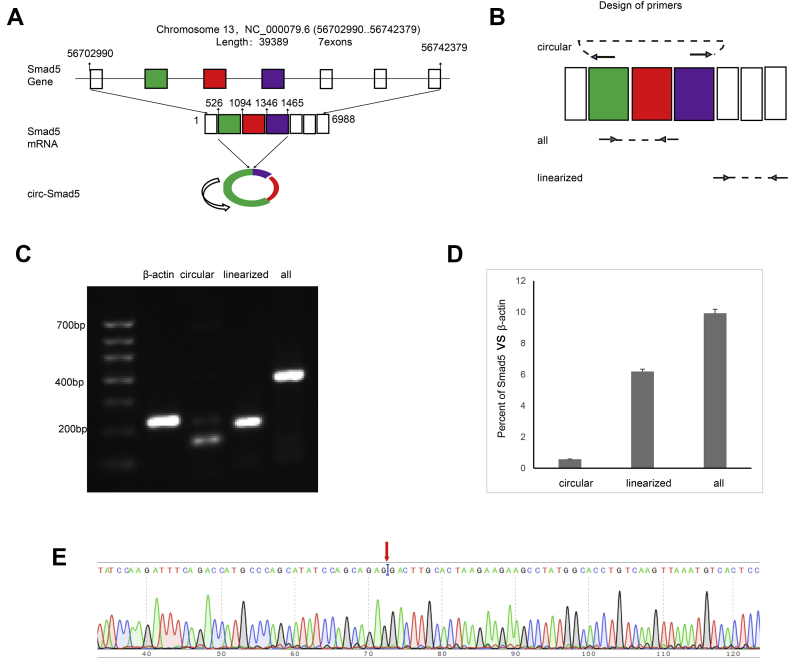
Figure 1.
The expression of circ-Smad5 in JB6 cells. (A) Information about circ-Smad5 is shown. It is the backsplicing product of exon 2 to exon 4 (B) The design of primers used to detect circ-Smad5, linearized-Smad5 and all-Smad5 are shown (C) RT-PCR results for the RNAs detected in JB6 cells are shown (D) QRT-PCR results are shown for the RNAs detected in JB6 cells. The expression levels of Smad5 RNAs were standardized with the expression level of β-actin (E) Sequencing results are shown for the circ-Smad5. The red arrow shows the backsplicing site.
Cell proliferation assay
Cultured JB6 cells were resuspended to a concentration of 2 × 104/mL. Cells (100 μL) were seeded into a 96-well plate. Three replicate wells were plated for each sample. At 24 h, 48 h, 72 h and 96 h after seeding, optical density at 450 nm (OD450) was measured with a microplate reader. At 1 h, 2 h or 4 h before measurement, 10 μL of CCK-8 (Beyotime, China) was added to the culture medium. The proliferation rate of each group was calculated based on the value of OD450.
Cell cycle assay
At 24 or 48 h after seeding, JB6 cell pellets were collected following treatment with 0.25% trypsin. The cells were washed with prechilled PBS, fixed with 70% ethanol and stained with propidium iodide staining solution (Beyotime, China) at 37 °C for 30 min in the dark. Red fluorescence was detected by flow cytometry. FlowJo 7.0 software was used for DNA content and light scattering analysis.
Western blot analysis
The JB6 cell line was used. At 24 h after seeding, PNU-74654 (Selleck, China), LiCl (Sigma, USA) or DMSO (Sigma, USA) was added into the culture medium. At 48 h after seeding, the cell pellets were collected following treatment with 0.25% trypsin. The total protein from the cell pellets were extracted respectively. SDS-PAGE was performed with 0.8% polyacrylamide gels. Proteins were then transferred to PVDF membranes (Millipore, USA), and membranes were blocked with 5% nonfat dry milk, in TBST (Tris-buffered saline containing 0.1% Tween-20). The PVDF membranes were then incubated with diluted primary antibodies overnight at 4 °C, washed with TBST, incubated with the HRP-labeled secondary antibodies (Sangon Biotech, China), and washed with TBST again. Finally, ECL (Bio-Rad, USA) was added to the PVDF membrane. The results were observed and recorded in a gel imager (Bio-Rad, USA). Primary antibodies against the following proteins were used: CyclinD1 (1:500, Sangon Biotech, China), β-Catenin (1:1000, Santa Cruz, USA), p-β-Catenin (1:5000, CST, USA), LEF1 (1:1000, Santa Cruz, USA). Anti-Smad 5 (1:1000, proteintech, China) was used to detect the expression of Smad5 protein after treatments.
Immunofluorescence staining
JB6 cells were seeded onto coverslips. When the confluence reached 80%, the coverslips containing the cells were gently rinsed with PBS and fixed with 4% paraformaldehyde solution for 15 min at room temperature. Then, the coverslips were washed with PBS, blocked with blocking solution for 1 h at room temperature and incubated with diluted primary antibodies at 4 °C overnight. Primary antibodies against the following proteins were used: CyclinD1 (1:25, Sangon Biotech, China), β-Catenin (1:100, Santa Cruz, USA), p-GSK3β (1:100, CST, USA), LEF1 (1:100, Santa Cruz, USA). Then, the samples were washed with PBS and incubated with the diluted secondary antibodies (1:500, Beyotime, China) at room temperature for 1 h in the dark. DAPI (1:10000, Beyotime, China) was added to the coverslips, which was followed by 10 min of incubation in the dark. At last, the coverslips were washed, mounted with an anti-fluorescence quenching solution (Beyotime, China), and observed under a fluorescence microscope.
Dual-luciferase activity assay
JB6 cells were seeded into 24-well plates and cultured overnight. PTK plasmid (expressing Renilla luciferase, Promega, USA) and TOP flash plasmid (expressing firefly luciferase, Promega, USA) or PTK plasmid and FOP flash plasmid (expressing firefly luciferase, Promega, USA) were simultaneously transfected into JB6 cells using Lipofectamine 2000 (Invitrogen, USA) according to the manufacturers' protocol. Specifically, 0.04 μg of PTK plasmid and 4 μg of TOP flash or FOP flash were transfected into the cells of every well. Lysis buffer was added to 24-well plate after 24 h of normal culture to prepare cell lysate, debris was removed by centrifugation, and 20 μL of supernatant was used for the detection of luciferase values. The firefly luciferase values (RLU1 and RLU2) were detected with a dual-luciferase reporter assay system (Promega, USA) according to the manufacturer's protocol. The TOP flash relative value = RLU1/RLU2, the FOP flash relative value = RLU1/RLU2, the final ratio = TOP flash relative value/FOP flash relative value. The final ratio was used as an indicator of the activation of the Wnt/β-catenin signaling pathway.
Statistical analysis
GraphPad 7.0 software was used for statistical analyses. All data were presented as the mean ± SD. The cell proliferation assay was analyzed by two-way ANOVA. All other assays were analyzed by unpaired t-tests. P < 0.05 was considered to be statistically significant.
Results
Circ-Smad5 is expressed in epidermal cells
The mouse Smad5 gene is located on chromosome 13 and contains 39389 base pairs. Its mRNA consists of 6988 base pairs and has 7 exons. Circ-Smad5 is formed by the backsplicing of exons 2, 3, and 4 (Fig. 1A). Divergent primers between exon 2 and exon 4 were designed to detect circ-Smad5. The convergent primers between exon 2 and exon 4 were designed to detect both linearized and circular Smad5 RNAs. The convergent primers outside of exon 2 to exon 4 were designed to detect the linearized mRNA of Smad5 (Fig. 1B). RT-PCR analysis showed that circ-Smad5 was expressed in JB6 cell lines (Fig. 1C). Quantitative RT-PCR analysis showed that the expression level of all-Smad5 was higher than the summation of circ-Smad5 and linearized-Smad5 (Fig. 1D). The expression of circ-Smad5 was confirmed by sequencing of PCR products (Fig. 1E).
Silencing of circ-Smad5 promotes cell cycle progression
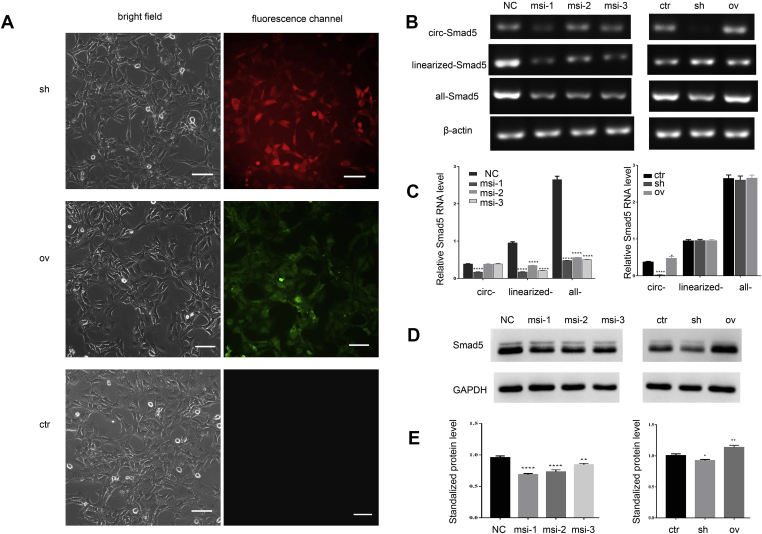
To research the function of circ-Smad5 in the epidermal cells, we constructed stable cell lines that overexpress circ-Smad5 or express circ-Smad5 shRNA. MCherry expression demonstrated that the sh-circ-Smad5 cell line was successfully constructed, whereas copGFP expression demonstrated that the ov-circ-Smad5 cell line was successfully constructed (Fig. 2A). RT-PCR results showed that the expression of linearized Smad5 and all-Smad5 in the siRNA-treated groups were all significantly lower than they were in the negative control group. The target sequence of msi-1 was also included in the circ-Smad5, so the expression level of circ-Smad5 in the msi-1 treated group was significantly lower than it was in the msi-2 and msi-3 treated groups. The expression level of circ-Smad5 in the silenced group was significantly lower than it was in the control group. The expression level of circ-Smad5 in the overexpression group was not significantly higher than that in the control group (Fig. 2B and C). Western blot results showed that the expression of Smad5 protein in the siRNA-treated groups or the silenced group was significantly lower than it was in the control groups or overexpressed group. The expression of Smad5 protein in the overexpressed group was significantly higher than it was in the control group (Fig. 2D and E). On one hand, as a circular RNA, the endogenous expression of circ-Smad5 was sufficiently high, the research on the knockdown of circ-Smad5 is more important. On the other hand, the efficiency of the overexpression of circ-Smad5 was not high enough. We focused on the effects of circ-Smad5 knockdown in the following experiments.
Figure 2.
The validation of the overexpression and silencing of circ-Smad5 in JB6 cells (A) Expression of mCherry or copGFP is shown with the expression of sh-circ-Smad5 or ov-circ-Smad5. Scale bar = 25 μm (B) RT-PCR results show the expression of Smad5-related RNAs in the treated cells. β-actin was used as the internal control (C) The relative expression levels of Smad5-related RNAs in (B) were standardized with β-actin (D) Western blot results show the expression of Smad5 in the treated cells. GAPDH was used as the internal control (E) The relative expression levels of Smad5 in (D) was standardized with GAPDH. OV: overexpression of circ-Smad5, sh: silencing of circ-Smad5, ctr: control, NC: negative control, msi: siRNA target to the mRNA of Smad5. N = 3, *P < 0.05.
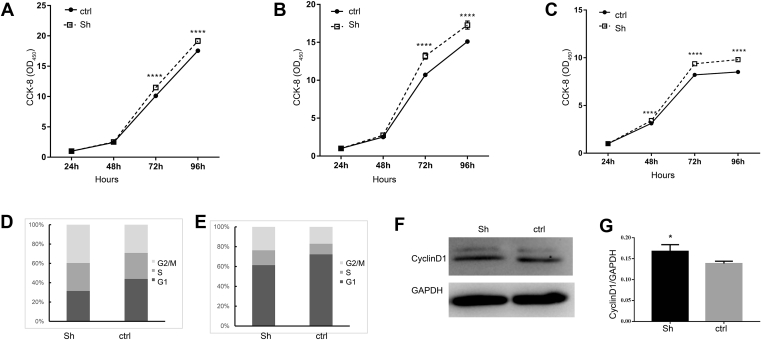
To investigate the biological function of circ-Smad5 in epidermal cells, cell proliferation was detected by CCK-8 assays. The OD450 displays the relative amount of proliferating cells from the addition of CCK-8 to the time of detection. When the period was 1 or 2 h, the relative number of proliferating cells in the sh-circ-Smad5 group was significantly greater than it was 72 h after seeding. When the period was prolonged to 4 h, the relative number of proliferating cells in the sh-circ-Smad5 group was significantly greater than it was 48 h after seeding (Fig. 3A–C). The cell cycle was analyzed by flow cytometry. At 24 h after seeding, the percentage of cells in the G1, S and G2 phases was 31.46%, 28.91% and 22.13% for the sh-circ-Smad5 group, whereas 43.90%, 17.69% and 26.98% for the control group, respectively (Fig. 3D). At 48 h after seeding, the percentage of cells in the G1, S and G2 phases was 61.39%, 14.96% and 13.90% for the sh-circ-Smad5 group, whereas it was 72.26%, 10.74% and 12.41% for the control group, respectively (Fig. 3E). The percentage of cells in the S phase of the sh-circ-Smad5 group was higher than it was in the control group. Western blot results showed that the expression level of cyclinD1 protein in the sh-circ-Smad5 group was significantly higher than it was in the control group (Fig. 3F and G). These results indicate that circ-Smad5 is involved in the cell cycle progression of epidermal cells.
Figure 3.
Circ-Smad 5 inhibits the proliferation of JB6 cells. (A–C) The relative OD450 value was analyzed at 24 h, 48 h, 72 h and 96 h after seeding. The CCK-8 reagent was added at 1 h (A), 2 h (B) or 4 h (C) before detection (D–E) The cell cycle was detected by FACS at 24 h (D) or 48 h (E) after seeding (F) The expression of cyclin D1 was detected by Western blot. GAPDH was used as the internal control (G) The relative expression level of cyclin D1 was standardized with GAPDH. sh: silencing of circ-Smad5, ctr: control. N = 3, *P < 0.05.
Silencing of circ-Smad5 activates Wnt/β-catenin/Lef1 signaling
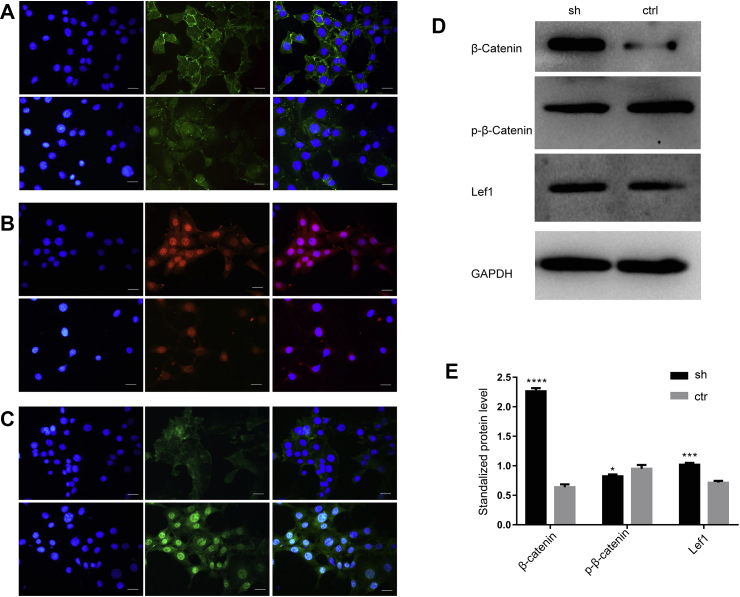
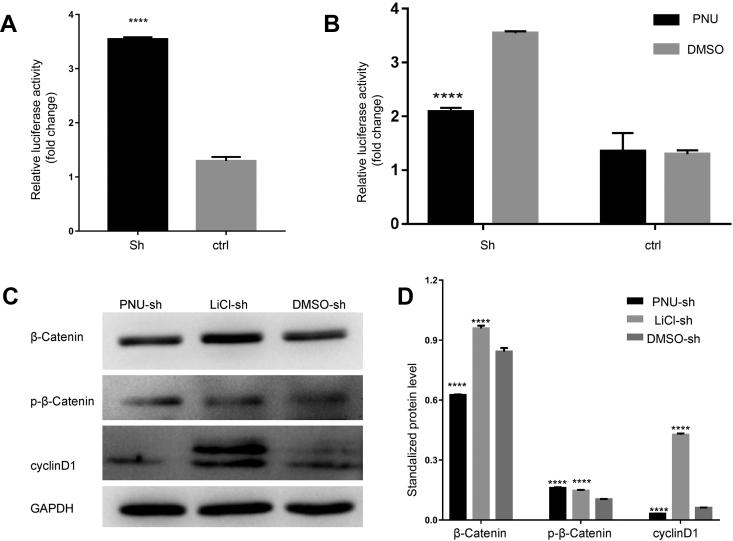
The Wnt signaling pathway is a major signaling pathway reported to regulate the proliferation of cells. Therefore, we analyzed the expression of key factors from the Wnt signaling pathway in sh-circ-Smad5 JB6 cells. Compared with the control group, the expression of β-catenin was enhanced both in the cell plasma and nucleus of the sh-circ-Smad5 group (Fig. 4A). The expression of Lef 1 was enhanced (Fig. 4B) and the expression of p-GSK3β was reduced in the sh-circ-Smad5 group (Fig. 4C). Western blot results showed that β-catenin, and LEF-1 were significantly increased, while p-β-catenin was significantly decreased in the sh-circ-Smad5 group (Fig. 4D and E). The relative luciferase activity of the TOP/FOP reporter system was significantly higher in the sh-circ-Smad5 group than it was in the control group (Fig. 5A). Collectively, these results revealed that sh-circ-Smad5 activates the Wnt/β-catenin/Lef1 signaling pathway.
Figure 4.
Expression of key molecules in the Wnt signaling pathway in sh-circ-Smad5 cells. (A–C) Expression of β-catenin (A), Lef 1 (B) and p-GSK3β (C) was determined by immunofluorescence. Upper: sh-circ-Smad5 group, lower: control group, left panel: DAPI, middle panel: target molecules, right panel: the merge of left two panels. Scale bar = 10 μm (D) Expression of key molecules in the Wnt signaling pathway was detected by Western blot. GAPDH was used as an internal control (E) The relative expression levels of the molecules were standardized with GAPDH. sh: silencing of circ-Smad5, ctr: control. N = 3, *P < 0.05.
Figure 5.
Wnt signaling pathway activity in sh-circ-Smad5 cells. (A) The activity of the Wnt signaling pathway in sh-circ-Smad5 cells was detected by the TOP/FOP flash reporter system (B) The activity of the Wnt signaling pathway in PNU-74654 treated sh-circ-Smad5 cells was detected by the TOP/FOP flash reporter system (C) Expression of Wnt signaling pathway key molecules in PNU-74654-treated or LiCl-treated sh-circ-Smad5 cells was detected by Western blot. GAPDH was used as an internal control (D) The expression levels of the molecules in Fig. 5C were standardized with GAPDH. DMSO was used as a control. PNU: PNU-74654, sh: silencing of circ-Smad5, ctr: control. N = 3, *P < 0.05.
PNU-74654 treatment reverses activation of the Wnt/β-catenin/cyclin D1 pathway by sh-circ-Smad5
To verify the effect of sh-circ-Smad5, Wnt activator LiCl and Wnt inhibitor PNU-74654 were added into the culture medium of sh-circ-Smad5-treated JB6 cells.21 At 24 h after addition, the relative luciferase activity of the TOP/FOP flash reporter system was significantly lower in the PNU-treated group than it was in the DMSO-treated group (Fig. 5B). Compared with the DMSO-treated group, the protein expression levels of β-catenin and cyclinD1 were significantly decreased in the PNU-treated group. The protein expression level of p-β-catenin was significantly increased in the PNU-treated group (Fig. 5C and D). Taken together, these data suggest that sh-circ-Smad5 induced cell cycle progression by regulating the Wnt/β-catenin signaling pathway.
Discussion
Circular RNAs were discovered decades ago, and they were thought to be a byproducts of splicing.22 However, these RNAs have recently been found to have many biological functions.23 This finding has become a new hot issue of noncoding RNAs.24 To identify the circular RNAs, researchers usually design convergent primers and divergent primers to differentiate between circular RNA and linearized mRNA. Here we designed an extra pair of primers (all-Smad5) that detect both the circular RNA and linearized mRNA. The use of the three pairs of primers can be a new method to identify circular RNAs. If the expression level of all-RNA equals to the expression level of circular-RNA plus linearized-RNA, then we can infer that there is only one kind of circ-RNA. If the expression level of circular-RNA plus linearized-RNA exceeds the expression level of all-RNA, then we can infer that additional circ-RNAs produced from splicing at different sites from the target circ-RNA may exist. If the expression level of circular-RNA plus linearized-RNA is less than the expression level of all-RNA, then we can infer that additional circ-RNAs produced from splicing at the same sites as the target circ-RNA may exist. Here, the expression level of circular-Smad5 plus linearized-Smad5 is less than the expression level of all-RNA, thus we infer that there are other circ-Smad5s. There are other additional bands in the RT-PCR results, which further support this (Fig. 1C). However, the target circ-Smad5 has the highest expression level, and we consider the target circular Smad5 to be circ-Smad5 in this paper. We are the first to propose a three-primer system to identify circular RNAs.
CCK-8 assays are commonly used to measure cell proliferation and cell viability. Compared with the control group, cells in the sh-circ-Smad5 group had an increased rate of proliferation. Cell cycle analysis suggests that the cell cycle change in the sh-circ-Smad5 group was an important factor for the change in cell proliferation. The cell cycle is regulated by a group of CDKs. The activity of CDKs depends on cyclins. Cyclin D1 is a major cyclin that regulates the cell cycle transition from G1 to S.25,26 The expression of cyclin D1 was higher in the sh-circ-Smad5 group than in the control group. These data demonstrate that circ-Smad5 may regulate the proliferation of JB6 cells by regulating the expression cyclin D1.
Cyclin D1 is the major executor of the cell cycle. Usually, cyclin D1 is not directly regulated by a molecule. It has been reported that the expression of cyclin D1 and cyclin D2 is regulated by LEF/TCF, which are transcription factors downstream of the canonical Wnt signaling pathway.27 The canonical Wnt signaling pathway is also named Wnt/β-catenin signaling pathway. When Wnt ligands bind to the Frizzled receptor and coreceptors, β-catenin becomes dephosphorylated, translocates from the cell plasma to nucleus, binds with TCF/LEF and regulates the expression of downstream molecules.28 In sh-circ-Smad5 JB6 cells, the expression of β-catenin, lef 1 and cyclin D1 was upregulated, whereas the expression of p-β-catenin was downregulated. Combining with the upregulation of the relative dual luciferase values noted above, we conclude that Wnt/β-catenin/Lef 1 signaling was activated. Interestingly, the knockdown of circ-Smad5 increased the expression of β-catenin both in the cell plasma and nucleus. In addition to its function in the Wnt signaling pathway, β-catenin also function in cell adhesion. This phenomenon implies that circ-Smad5 may also function in cell adhesion. In addition, the activation of the Wnt signaling pathway was blocked by Wnt signaling inhibitor PNU-74654. These data demonstrate that cyclin D1 is upregulated by Wnt/β-catenin/Lef1 signaling in sh-circ-Smad5 JB6 cells.
Our data revealed that circ-Smad5 could function by inhibiting the Wnt signaling pathway. However, other mechanisms may also be involved. Since circ-Smad5 is formed through the backsplicing of Smad5 exons, its production may impact the function of Smad5 by some unknown mechanisms. For example, the Smad5-related TGFβ signaling pathway plays important roles in cell zebrafish posterior lateral line formation,29 and we can speculate that circ-Smad5 may also have some kind of function in this process. We also revealed an interesting phenomenon: neither overexpression nor silencing of circ-Smad5 changed the expression level of Smad5 mRNAs, but they all changed the expression level of Smad5 protein. This implies that circ-Smad5 is not a competitive RNA to linearized-Smad5 and circ-Smad5 may impact the expression of Smad5 protein indirectly. Wnt/β-catenin signaling pathway may be one of the signaling pathway between them. From our data, we speculate that circ-Smad5 may function through sponging microRNAs or binding with other regulators. The mechanism that how circ-Smad5 impact Wnt/β-catenin signaling pathway should be revealed in future.
Conclusions
In summary, we report that circ-Smad5 inhibits the proliferation of JB6 cells. The function of circ-Smad5 has not been reported before; thus, we revealed a new function for circ-Smad5. We also found one possible mechanism by which circ-Smad5 regulates cell proliferation: inhibiting the activation of Wnt/β-catenin/Lef 1 signaling, thus inhibiting the expression of cyclinD1 and the progression of the cell cycle.
Conflict of Interests
The authors declare no conflict of interests.
Acknowledgements
This work was supported by the National Natural Science Foundation of China [grant number 81472895], the Natural Science Foundation of Chongqing [grant number cstc2018jcyjAX0053] and Key Talents Support Plan of Army Medical University (2019). We thank Min Gao and Yizhan Xing at the Army Medical University for technical support.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Yuhong Li, Email: liyuhongtmmu@hotmail.com.
Yiming Zhang, Email: zhangyiming@tmmu.edu.cn.
References
- 1.Yang Q., Wu J., Zhao J. Circular RNA expression profiles during the differentiation of mouse neural stem cells. BMC Syst Biol. 2018;12(Suppl 8):e128. doi: 10.1186/s12918-018-0651-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zheng Q., Bao C., Guo W. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:e11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng L., Yuan X.Q., Li G.C. The emerging landscape of circular RNA ciRS-7 in cancer (Review) Oncol Rep. 2015;33(6):2669–2674. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y.H., Song J., Shen L., Shao J. Systematic identification of lncRNAs and circRNAs-associated ceRNA networks in human lumbar disc degeneration. Biotech Histochem. 2019;94(8):606–616. doi: 10.1080/10520295.2019.1622782. [DOI] [PubMed] [Google Scholar]
- 5.Huang C., Deng H., Wang Y. Circular RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23(9):6112–6119. doi: 10.1111/jcmm.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdelmohsen K., Panda A.C., Munk R. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y., Fan X., Mao M. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pamudurti N.R., Bartok O., Jens M. Translation of CircRNAs. Mol Cell. 2017;66(1):9–21. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du W.W., Yang W., Liu E. Foxo 3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phua S.C., Chiba S., Suzuki M. Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell. 2017;168(1–2):264–279. doi: 10.1016/j.cell.2016.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mohamed T.M.A., Ang Y.S., Radzinsky E. Regulation of cell cycle to stimulate adult cardiomyocyte proliferation and cardiac regeneration. Cell. 2018;173(1):104–116. doi: 10.1016/j.cell.2018.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying Z., Sandoval M., Beronja S. Oncogenic activation of PI3K induces progenitor cell differentiation to suppress epidermal growth. Nat Cell Biol. 2018;20(11):1256–1266. doi: 10.1038/s41556-018-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joost S., Jacob T., Sun X. Single-cell transcriptomics of traced epidermal and hair follicle stem cells reveals rapid adaptations during wound healing. Cell Rep. 2018;25(3):585–597. doi: 10.1016/j.celrep.2018.09.059. [DOI] [PubMed] [Google Scholar]
- 14.Tsytlonok M., Sanabria H., Wang Y. Dynamic anticipation by Cdk 2/Cyclin A-bound p27 mediates signal integration in cell cycle regulation. Nat Commun. 2019;10(1):e1676. doi: 10.1038/s41467-019-09446-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen G., Ding X.F., Bouamar H. Everolimus induces G1 cell cycle arrest through autophagy-mediated protein degradation of cyclin D1 in breast cancer cells. Am J Physiol Cell Physiol. 2019;317(2):C244–C252. doi: 10.1152/ajpcell.00390.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang W., Chen L., Wu J. Long noncoding RNA TUG1 inhibits osteogenesis of bone marrow mesenchymal stem cells via Smad 5 after irradiation. Theranostics. 2019;9(8):2198–2208. doi: 10.7150/thno.30798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z., Wang J., Zeng X. Two-stage study of lung cancer risk modification by a functional variant in the 3'-untranslated region of SMAD5 based on the bone morphogenetic protein pathway. Mol Clin Oncol. 2018;8(1):38–46. doi: 10.3892/mco.2017.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Q., Gan H., Song W. MicroRNA-145 promotes esophageal cancer cells proliferation and metastasis by targeting SMAD5. Scand J Gastroenterol. 2018;53(7):769–776. doi: 10.1080/00365521.2018.1476913. [DOI] [PubMed] [Google Scholar]
- 19.Zhao C.C., Jiao Y., Zhang Y.Y. Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/beta-catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell Death Dis. 2019;10(4):e252. doi: 10.1038/s41419-019-1479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y.H., Zhang K., Yang K. Adenovirus-mediated Wnt10b overexpression induces hair follicle regeneration. J Investig Dermatol. 2013;133(1):42–48. doi: 10.1038/jid.2012.235. [DOI] [PubMed] [Google Scholar]
- 21.Duong H.Q., Nemazanyy I., Rambow F. The endosomal protein CEMIP links WNT signaling to MEK1-ERK1/2 activation in selumetinib-resistant intestinal organoids. Cancer Res. 2018;78(16):4533–4548. doi: 10.1158/0008-5472.CAN-17-3149. [DOI] [PubMed] [Google Scholar]
- 22.Song X., Zhang N., Han P. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44(9) doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 24.Qu S., Yang X., Li X. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Wu H., Ploeger J.M., Kamarajugadda S. Evidence for a novel regulatory interaction involving cyclin D1, lipid droplets, lipolysis, and cell cycle progression in hepatocytes. Hepatol Commun. 2019;3(3):406–422. doi: 10.1002/hep4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun F., Li N., Tong X. Ara-c induces cell cycle G1/S arrest by inducing upregulation of the INK4 family gene or directly inhibiting the formation of the cell cycle-dependent complex CDK4/cyclin D1. Cell Cycle. 2019;18(18):2293–2306. doi: 10.1080/15384101.2019.1644913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye B., Li L., Xu H. Opposing roles of TCF7/LEF1 and TCF7L2 in cyclin D2 and Bmp 4 expression and cardiomyocyte cell cycle control during late heart development. Lab Investig. 2019;99(6):807–818. doi: 10.1038/s41374-019-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macdonald B.T., Semenov M.V., He X. SnapShot: Wnt/beta-catenin signaling. Cell. 2007;131(6):e1204. doi: 10.1016/j.cell.2007.11.036. [DOI] [PubMed] [Google Scholar]
- 29.Xing C., Gong B., Xue Y. TGFbeta1a regulates zebrafish posterior lateral line formation via Smad 5 mediated pathway. J Mol Cell Biol. 2015;7(1):48–61. doi: 10.1093/jmcb/mjv004. [DOI] [PubMed] [Google Scholar]