Abstract
Sulindac has shown significant clinical benefit in preventing colorectal cancer progression, but its mechanism of action has not been fully elucidated. We have found that sulindac sulfide (SS) is able to inhibit cell cycle progression in human colorectal cancer cells, particularly through G1 arrest. To understand the underlying mechanisms of sulindac inhibitory activity, we have demonstrated that Cyclin G2 up-regulation upon SS treatment can substantially delay cell cycle progression by enhancing the transcriptional activity of FOXO3a in human colorectal tumor cells. MiR-182, an oncogenic microRNA known to inhibit FOXO3a gene expression, is also involved in the suppressive effect of SS on cell cycle progression. This process begins with the down-regulation of miR-182, followed by the enhancement of FOXO3a transcriptional activity and the up-regulation of Cyclin G2. To further determine the clinical utility of this axis, we analyzed the expression of miR-182/FOXO3a/Cyclin G2 in human colorectal tumor samples. Our results show not only that there are significant differences in miR-182/FOXO3a/Cyclin G2 between tumors and normal tissues, but also that the synergetic effect of miR-182 and FOXO3a is associated with predicting tumor progression. Our study demonstrates a novel mechanistic axis consisting of miR-182/FOXO3a/Cyclin G2 that mediates sulindac inhibition of cell cycle progression.
Keywords: Cell cycle, Colorectal cancer, Gene regulation, miRNA, Sulindac
Introduction
Colorectal cancer (CRC) remains one of the leading causes of cancer-related death. In 2020, the United States will have approximately 53,200 new deaths from CRC, along with 147,950 newly diagnosed cases.1 Such statistics show an unmet need to further develop novel treatment strategies that are not only efficacious but also have fewer adverse side effects when compared to standard chemotherapy regimens. Nonsteroidal anti-inflammatory drugs (NSAIDs) have been extensively reported to be effective as chemopreventative agents, in both the control and management of cancer progression.2 Of significance, prescription-strength sulindac shows compelling efficacy to reduce pre-cancerous adenomas in individuals with familial adenomatous polyposis (FAP).3 It is notable that the anti-neoplastic activity of NSAIDs is generally believed to result from anti-proliferative and pro-apoptotic effects via inhibiting cyclooxygenase-2 (COX-2) activity. This, in turn, reduces the synthesis of prostaglandin 2 (PGE2), which is known to block apoptosis and stimulate angiogenesis and tumor invasion.4 Unfortunately, long-term administration of high doses of NSAIDs for cancer intervention was approached with certain precautions, due to the fact that COX/PGE2 inhibition is related to the possibility of gastrointestinal, renal, and cardiovascular toxicities.5,6 The recent efforts examining the discovery of novel, less toxic, non-COX inhibitory derivatives of NSAIDs support that COX-inhibition may not be fully responsible for the anti-cancer activities of NSAIDs. With chemical modifications, NSAID derivatives showed similar, and in many cases, improved anti-cancer activities; however, cellular cytotoxicity was significantly reduced. For example, several non-COX inhibitory derivatives of sulindac were reported for compelling efficacy and improved potency compared to the parent compounds for the inhibition of CRC cell growth.7,8
The mammalian cell cycle is a highly organized and systematically controlled process consisting of four sequential phases: Gap phase 1 (G1), DNA synthesis (S), Gap phase 2 (G2), and Mitosis (M) phase. Dysregulation of cell cycle control, such as unscheduled proliferation, is regarded as one of the key drivers for both genomic and chromosomal instability that facilitate tumorigenesis.9 The cell cycle is tightly controlled by a subfamily of cyclin-dependent kinases (CDKs), and their activity is commonly induced by cyclins, or suppressed by CDK inhibitors (CDKIs), such as p21wif1/cip1 and p27kip1, or balanced by the interactions between CDKs and CDKIs.9
Most of cyclins have been shown to promote cell cycle progression, but Cyclin G2 appears to be an “outlier,” together with Cyclin G1 and Cyclin I, which are mainly involved in the maintenance of cell quiescent status by inducing cell cycle arrest.10 There is accumulating evidence that supports the notion of Cyclin G2 functioning as a tumor suppressor gene, through inhibiting tumor cell proliferation, epithelial-to-mesenchymal transition (EMT) and metastasis.11,12 It was also reported as a putative prognostic marker that is correlated with disease progression in different tumor types.13, 14, 15, 16, 17
In this study, we first determined that Cyclin G2 could be up-regulated by sulindac sulfide (SS) in human CRC cell lines, HCT116 and HT29. Then, we found that Cyclin G2 was involved in G1 arrest in CRC cells upon SS treatment through a validated mechanism mediated by a tumor suppressor gene, FOXO3a, that can regulate the expression of Cyclin G2 at the transcriptional level. Of interest, FOXO3a is specifically targeted by miR-182, an oncogenic miRNA that can be targeted by SS. Therefore, our study demonstrates a new mechanistic axis consisting of miR-182/FOXO3a/Cyclin G2 that mediates sulindac inhibition of cell cycle progression.
Materials and methods
Tissue culture and reagents
Human CRC cell lines (HCT116 and HT29) and normal colon epithelial cell line (CCD841-CoN) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and incubated with McCoy's 5A medium (Gibco, Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Atlanta Biologicals, Flowery Branch, GA, USA) in 5% CO2 at 37 °C in a humidified incubator. HCT116 cells have low to a null expression of COX-2, and HT29 cells have intact level of COX-2.18 Sulindac sulfide (SS) was purchased from Thermo Fisher Scientific (Waltham, MA, USA). Thereafter, only those materials and reagents that were not purchased from Thermo Fisher Scientific will be marked. The vendor information for all antibodies used in this study were exhibited in Table S1.
Cell viability assay
Cells were seeded in 96-well microtiter plates at a density of 5000 cells per well, and then treated with SS at different concentrations for 36 h. The relative cell viability was measured using the Cell Titer Glo Assay following the instruction specified by the manufacturer (Promega, WI, USA). The half maximal inhibitory concentration (IC50) was determined and calculated using GraphPad Prism 7.
Cell cycle analysis
For cell cycle analysis, HCT116 and HT29 cells were seeded in 6-cm dishes, with a cell number of 1 × 105 per dish. We utilized serum starvation to induced cell cycle synchronization. After 24 h of serum starvation, HCT116 and HT29 cells were passaged and released into cell cycle by adding serum, and then treated with SS in a time-course experiment. When harvesting, cells were washed with the ice-cold phosphate-buffered solution (PBS) and fixed with ice-cold 70% ethanol overnight. RNase (Promega) at 10 μg/ml was added to the cells before they were stained with 50 μg/ml of Propidium iodide (MP Biomedicals, OH, USA). DNA contents were determined by flow cytometry. In brief, we selected the forward scatter and side scatter to gate total cells population, then utilzed the channel FL3 PEAK (pulse processing) to gate the single cell and exclude cell doublets.
Quantitative real-time PCR
Total RNA was extracted from cells using Trizol reagent as reported previously.19 Two micrograms of RNA were reverse-transcribed using a high capacity cDNA reverse transcriptase kit. PCR was then performed on cDNA with gene-specific primers. The protocol for the detection of mature miRNAs using a stem-loop gene-specific reverse transcription primer was performed as described previously.19 SYBR Green Master Mix was used for PCR reactions that were performed on the ABI 7500 Real-Time PCR System.
Western blot
Cells were harvested and washed twice with ice-cold PBS and lysed with RIPA buffer containing protease inhibitor cocktail (1%). The lysates were vortexed on ice for 15 s every 10 min, for a total of 40 min, then centrifuged at 12,000g for 20 min at 4 °C. Protein was quantified by DC protein assay (Bio-Rad, Hercules, CA, USA). Cell lysates containing equivalent amounts of protein were separated and concentrated with the sodium dodecyl sulfate (SDS) polyacrylamide gel electrophoresis, then transferred onto a PVDF membrane (Bio-Rad) and probed with the primary antibodies (1:1000), followed by secondary antibodies (Bio-Rad 1:1000). Equal loading was confirmed with primary antibodies against α-tubulin (whole-cell lysates) or HDAC-1 (nuclear preparation). Image lab software (Bio-Rad) was utilized to quantify the densitometry of Western Blot results.
RNA interference
Cyclin G2 gene was knocked down using siRNAs that were purchased from Dharmacon™ (Lafayette, CO). Lipofectamine RNAiMAX was used for the transfection of siRNAs targeting Cyclin G2 (SMARTpool) or a non-targeting control (Pool #2). The process followed a published protocol.20
Sub-cellular fractionation
To isolate the nuclear fraction, cells were harvested and then fractionated using the Nuclear/Cytoplasmic Fractionation Kit. Briefly, 5 × 106 cells/sample were washed with PBS and centrifuged. The cell pellets were then re-suspended with cold cytoplasmic extraction reagent 1 (CER1), followed by a vigorous vortex for 15 s. Then, ice-cold CER II was added to the tube, which was vortexed for 15 s every 10 min, for a total of 40 min. The cytosolic fraction (supernatant) was collected after centrifuging at 16,000×g for 5 min. Being re-suspened with ice-cold nuclear extraction reagent (NER), the pellets were vortexed and then centrifuged at 16,000×g for 10 min to release the nuclear fraction (supernatant). The purity of fractions was tested by immunoblotting with antibodies specific for the cytoplasmic (tubulin) or the nuclear (HDAC-1) extractions.
Immunofluorescence assay
To detect the cellular distribution of FOX03a, cells were seeded in Fluoro-dishes (World Precision Instruments, FL, USA) with a number of 5 × 104 per dish overnight at 37 °C and then treated with 100 μM SS or the same volume of 0.1% DMSO. The cells were fixed with 4% formaldehyde for 30 min and permeabilized with 1% Triton X-100 (Sigma–Aldrich) for 15 min, and then blocked with 2.5% BSA for 30 min, before incubating with the primary antibodies at 4 °C overnight. After washing with PBS, the cells were incubated with the fluorescent secondary antibody for 1 h at room temperature. Cells were then washed and counterstained with DAPI at a concentration of 5 ng/ml. All images were taken using confocal microscopy.
Chromatin immunoprecipitation assay (ChIP)
ChIP analysis was performed with EZ-Magna ChIP kit from Millipore (Billerica, MA, USA). In brief, a total of 1 × 107 CRC cells were placed in a 15-cm culture dish and treated with SS for 12 h before cross-linking with 1% formaldehyde. The cell lysates were sonicated to shear DNA to sizes of 300-1000 bp. Equal aliquots of chromatin supernatants were separated and incubated with the antibodies against FOXO3a, or IgG overnight at 4 °C with continuous rotation. After reverse cross-link of protein/DNA complexes to free DNA, PCR was performed for 30 cycles consisting of denaturing for 20 s at 94 °C, annealing for 3 s at 59 °C and extension for 30 s at 72 °C. Finally, PCR products were electrophoresed with a 2% agarose gel.
CRISPR/cas9 gene editing
The sgRNAs were designed by the CRISPR Design Tool (http://crispr.mit.edu/; sgRNAs for FOXO3a: Forward: CACCGGGCGGCGGGAGCGGCACGCT; Reverse: AAACAGCGTGCCGCTCCCGCCGCCC). The synthesized sgRNA oligos were cloned into the lentiCRISPRv2 vectors (Addgene), referring to the published protocol.21 The constructed plasmids were transfected into HCT116 cells using Lipofectamine and incubated for 24 h, followed by the selection with puromycin (2 μg/ml). The surviving cells were expanded and used in the following studies.
Luciferase report assay
We established the wildtype and mutated elements of 3′-UTR of FOXO3a containing miR-182 binding sites, and then cloned them into the pMIR-REPORT vectors. The luciferase assay was performed with HCT116 cells, as described previously.22 Briefly, cells were co-transfected with 400 ng of pMIR-REPORT plasmids, 10 ng of pRL-TK renilla plasmid (Promega), and 150 nM miR-182 precursor or negative control mimic on 24-well plates. After incubation for 24 h, cells were analyzed by using a luminometer (Promega).
Studies of human bio-specimens
Twenty paired, archival biopsy tumor samples from CRC patients were collected through the Louisiana Cancer Research Center (LCRC) tissue biorepository (IRB # 9814). Tissues were thawed on ice and then transferred to 1.5 ml tubes pre-filled with beads (Omni International, Inc.). Next, TRIzol (1 ml) was added into the tubes, and tissue was shredded using the Omni Bead Ruptor 4 (Omni International, Inc.). Finally, total RNA were extracted from the suspension following the protocol as previously reported.19
For qRT-PCR analyses of 20 paired colorectal tumor and adjacent normal tissues, quantitative data for each sample was collected, and the means of the triplicates were calculated. The relative expressions of miR-182, FOXO3a, Cyclin G2, and COX-2 in tumor tissues, compared to their adjacent normal tissues, were calculated using the 2-ΔΔct method, as reported previously.23 The expression differences between the tumor and adjacent normal tissues (ΔΔCt) were tested using the paired t-test. The 95% confidence interval of relative expression was calculated based on the confidence interval of ΔΔCt.24
We also calculated the relative expressions of these four biomarkers for their association with tumor progression (based upon clinical tumor stage and lymph node metastasis). All markers were grouped to the low- and high-expression groups based upon the cut-point of median relative expressions. CRC stage was categorized as either low (T1/T2) or high (T3/T4) stage, respectively, with the metastatic status dichotomized to the low (N0) and high (N1) metastasis group. The associations between the expressed status (high/low) of these biomarkers with tumor stage or metastasis were then analyzed using the Fisher's exact test. SAS v.9.4 was utilized for statistical analsyis.
Results
SS inhibits cell cycle progression through up-regulating Cyclin G2 in human CRC cells
Utilizing the cell viability assay, we determined the IC50 values of SS in human CRC cell lines, HCT116 and HT29. The human normal colon epithelial cells (CCD841-CoN) are included as the control to determine the selectivity of SS in normal cells. As shown in Fig. 1A, the IC50s of SS in HCT116 and HT29 cells are 93 μM and 102 μM, respectively, which are nearly 50% lower than that in CCD841-CoN cells (147 μM), demonstrating that SS has a selective inhibition in tumor cells. We chose 100 μM as a standard dose of SS in the following experiments. First, we compared the inhibitory effect of 100 μM SS on the growth of HCT116, HT29, and CCD841-CoN cells with a time-course study. As seen in Fig. 1B, HCT116 and HT29 cells displayed a similar pattern responding to SS treatment at 60 h with an inhibition rate of over 75%; however, only 25% CCD841-CoN cells were suppressed, showing an apparent resistance to SS treatment. These results support the notion that SS has a selective inhibition in CRC cells, indicating that different targets and pathways could be involved in SS inhibitory activity in tumor cells versus normal cells. In this study, we will focus on the inhibitory activity of SS in CRC cells.
Figure 1.
Sulindac sulfide (SS) inhibits cell cycle progression in human CRC cells. (A) Cell viability assay result demonstrates that SS inhibits the growth of CRC HCT116 and HT29 cells versus human normal colon epithelial CCD841-CoN cells. Cells were treated with SS at different concentrations for 36 h. (B) SS at 100 μM significantly inhibits HCT116 and HT29 cell proliferation compared to CCD841-CoN cells in a time-course study. Serum starvation was utilized to induce cell cycle synchronization. After 24 h of serum starvation, HCT116 and HT29 cells were passaged and released into cell cycle by adding serum, and then treated with SS in a time-course experiment. (C) HCT116 cells show G1 arrest at 12 h after 100 μM SS treatment, as determined by the flow cytometry analysis. (D) HT29 cells show G1 arrest at 16 h after 100 μM SS treatment as determined by flow cytometry analysis. Error bars represent mean ± SD from 3 replicates.
Given that the doubling time of HCT116 and HT29 cells is ~18 h and 24 h, respectively, we analyzed their cell cycle progression after they were treated with SS at 100 μM for 36 h and 48 h, which are considered as a close approximation of roughly two cell cycles for HCT116 and HT29 cells. We utilized serum starvation to induce cell cycle synchronization. After starvation, HCT116 and HT29 cells were passaged and released into cell cycle by adding serum. Re-feeding control HCT116 and HT29 cells with serum caused a substantial enrichment in S phase compared to SS-treated counterparts. In HCT116 cells, the average percentages of cells in the S phase are 87.8% (Ctrl) vs. 34.7% (SS); in HT29 cells, they are 55.1% (Ctrl) vs. 21.8% (SS). Of significance, the average percentages of cells in the G1 phase are 10.4% (Ctrl) vs. 59.3% (SS) in HCT116 cells, and 28.4% (Ctrl) vs.71.7% (SS) in HT29 cells (Fig. 1C and D). These results demonstrate that SS treatment can lead to an apparent G1 arrest in HCT116 and HT29 cells. Notablely, it has been reported that HCT116 cells have low to a null expression of COX-2, and HT29 cells have intact level of COX-2.18 In our results, G1 arrest by SS treatment shows a similar pattern in both cell lines, which implies that COX-2 inhibition might not play a significant role in this activity.
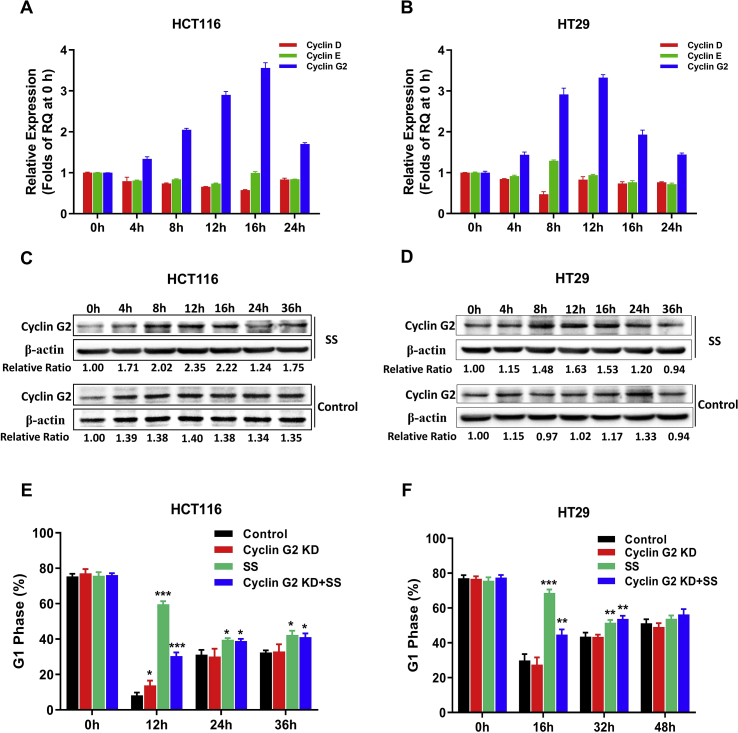
To determine the underlying molecular mechanism of SS inhibiting cell cycle progression, we also examined Cyclin D, Cyclin E, and Cyclin G2, which are associated with G1 arrest in accordance with the previous study.9 Our results show that, upon SS treatment, Cyclin G2 elevated more than 3 folds steadily within a course of 24 h in both cell lines, while Cyclin D and Cyclin E were not altered significantly in HCT116 cells. Only in HT29 cells, Cyclin D showed a decrease of ~50% at 8 h (Fig. 2A and B). We further validated the protein expression of Cyclin G2 using Western Blot analysis (Fig. 2C and D), showing an increase up to 2.35 folds in HCT116 cells and 1.63 folds in HT29 cells within a course of 36 h. These results are consistent with the transcriptional expression of Cyclin G2, as shown in Fig. 2A and B.
Figure 2.
Cyclin G2 was involved in SS inhibition of cell cycle progression. Cyclin D, Cyclin E, and Cyclin G2 were evaluated in HCT116 (A) and HT29 (B) cells upon SS treatment with qRT-PCR relative quantitation (RQ) analysis. Fold-change was utilized to indicate the alterations of select cyclins, which were calculated using the relative quantitation (RQ) values of different time points to divide the RQ value at 0 h. Western blot analysis of Cyclin G2 in HCT116 (C) and HT29 (D) cells after SS treatment for indicated times. The protein signal of Cyclin G2 for each sample was normalized by comparing with the signal of β-actin individually. G1 arrest caused by SS treatment could be diminished by Cyclin G2 knockdown with siRNA in both HCT116 (E) and HT29 (F) cells, as determined by flow cytometry. All samples were compared to the control cells at different time points. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, by t-test. Error bars represent mean ± SD from 3 replicates.
After successfully knocking down Cyclin G2 gene utilizing siRNA in both HCT116 and HT29 cells (Fig. S1), we analyzed their cell cycle progression upon SS treatment. As shown in Fig. 2E and F, Cyclin G2 knockdown (KD) did not significantly affect cell cycle progression. However, when treated with SS, these cells showed a significant reduction in G1 arrest compared to the parent cells (HCT116: 49% vs. HT29: 35%). These results support the notion that Cyclin G2 absence could compromise SS inhibitory effect on cell cycle progression in CRC cells.
SS inducing Cyclin G2 through regulating FOXO3a
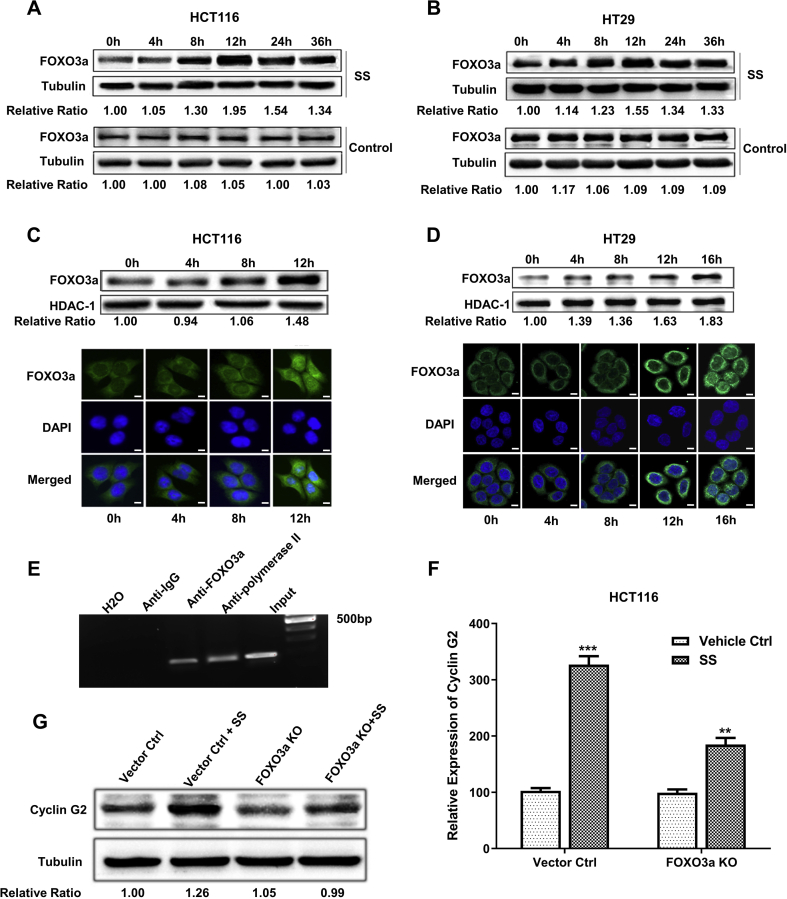
FOXO3a has been previously reported as a tumor suppressor gene that is involved in numerous cellular processes including apoptosis and suppression of cell proliferation.25 FOXO3a is also known to function as a transcription factor that has the capacity to regulate a number of genes related to cell cycle control, such as Cyclin G2.26 In order to determine the mechanism of SS regulating Cyclin G2, we examined the expression of FOXO3a in HCT116 and HT29 cells after the treatment of SS at 100 μM in a serial time course of up to 36 h. We found that FOXO3a was up-regulated by SS up to 1.95 times in HCT116 cells and 1.55 times in HT29 cells (Fig. 3A and B), which is consistent with the expression pattern of Cyclin G2 in response to SS treatment, as shown in Fig. 2C and D.
Figure 3.
Cyclin G2 regulated by FOXO3a is involved in SS inhibition of cell cycle progression in human CRC tumor cells. FOXO3a was examined in HCT116 cells (A) and HT29 cells (B) upon 100 μM SS treatment at different time points using Western Blot. The protein signal of FOXO3a for each sample was normalized by comparing to the signal of tubulin individually. The nuclear translocation of FOXO3a upon SS treatment (100 μM) was also examined in HCT116 cells (C) and HT29 cells (D) using Western Blot and confocal microscopy imaging (40x). Green (anti-FOXO3a Ab) indicates FOXO3a distribution, with blue (DAPI) indicating the location of the nucleus. Scale bar: 10 μm. The protein signal of nuclear FOXO3a in Western Blot was normalized by comparing to the signal of HDAC-1 individually. (E) ChIP assay was performed to determine if FOXO3a protein could bind to the promoter of Cyclin G2. Anti-IgG and anti-polymerase II were used as negative and positive controls, respectively. The efficacy of SS treatment (100 μM) for up-regulating Cyclin G2 is decreased in HCT116-FOXO3a KO cells compared to the vector control cells, as determined by qRT-PCR (F) and Western Blot (G). Some blots were cropped from different parts of the same gel and incubated with different antibodies. The protein signal of Cyclin G2 for each sample was normalized by comparing to the signal of tubulin individually. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, by t-test. Error bars represent mean ± SD from 3 replicates. KO: knockout.
Through sub-cellular fractionation, we examined the distribution of FOXO3a by Western Blot analysis, finding that the nuclear FOXO3a was increased by SS treatment in a time-dependent manner. In HCT116 and HT29 cells upon SS treatment for up to 12 h and 16 h, FOXO3a expression in the nuclease increased 48% and 83%, respectively. We further confirmed these results with confocal microscopy imaging (Fig. 3C and D). Our results show that SS is able to enhance the transcriptional activity of FOXO3a in the nucleus, resulting in up-regulation of its target genes, such as Cyclin G2. We further performed the chromatin immunoprecipitation (ChIP) assay in order to “pull down” the mixture of FOXO3a protein binding to Cyclin G2 promoter fragments with a specific FOXO3a antibody. As shown in Fig. 3E, the DNA fragment of the Cyclin G2 promoter was detected from the pull-down mixture by PCR. We also established an in vitro FOXO3a knockout (KO) model utilizing CRISPR/cas9 technology in HCT116 cells (Fig. S2) to further examine the levels of Cyclin G2 expression upon SS treatment. As shown in Fig. 3F and G, SS was not capable of up-regulating the expression of Cyclin G2 to a significant extent in HCT116-FOXO3a-KO cells, at both mRNA and protein levels. These results strongly support our hypothesis that SS up-regulates Cyclin G2 through a FOXO3a-mediated transcriptional regulation.
MiR-182 mediates SS regulation of FOXO3a
Our previous study has shown that a panel of miRNAs could be regulated by SS in CRC tumor cells.27 Herein, we were interested in further examining if miRNA may be involved in the regulation of FOXO3a by SS. Utilizing several different online algorithms, such as TargetScan, Pictar, and miRANDA, we identified several miRNAs that can putatively target FOXO3a for further investigation. These included miR-302a, miR-27b, miR-29b, and miR-182, but only miR-182 was found to be dominantly down-regulated by SS treatment in a time-dependent fashion (Fig. 4A and B). We further compared the expression patterns of miR-182, versus FOXO3a, in both HCT116 and HT29 cells treated with SS in a time-course experiment. We found that the trend of miR-182 expression was opposite to that seen with FOXO3a (Fig. 4C and D), which implies that up-regulation of FOXO3a might be due to the SS repression placed upon miR-182.
Figure 4.
MiR-182 is involved in SS induction of FOXO3a. Select miRNAs that potentially target FOXO3a were screened in HCT116 cells (A) and HT29 cells (B) treated with 100 μM SS in a time-course study. The relative expression of individual miRNA upon SS treatment at different time points was calculated by comparing it to the controls at 0 h. The expression patterns of miR-182 and FOXO3a were analyzed by qRT-PCR in HCT116 cells (C), and HT29 cells (D) treated with 100 μM SS in a time-course study. (E) Luciferase reporter assay was used to determine the direct binding of miR-182 to the 3′-UTR of FOXO3a. WT: wild type; MT: mutated; PC: positive control that is a synthesized oligo with the equal length to mature miR-182 and complement to the sequence of the 3′-UTR of FOXO3a. (F) QRT-PCR analysis shows the expression of miR-182 in HCT116 cells after transient transfection of miR-182 mimics and inhibitors, respectively, and Western Blot analysis indicates that FOXO3a expression is negatively correlated to miR-182 expression in HCT116 cells, supporting that FOXO3a is one of the targets of miR-182. (G) SS can not induce FOXO3a in HCT116 cells with miR-182 knockout (KO) by CRISPR/cas9. (H) MiR-182 is able to attenuate the inductive effect of SS on FOXO3a in HCT116 cells. MiR-182 mimics were transiently transfected to HCT116 cells to overexpress miR-182. DMSO is the vehicle of SS. Some blots were cropped from different parts of the same gel and incubated with different antibodies. The protein signal of FOXO3a for each sample was normalized by comparing to the signal of tubulin individually. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, by t-test. Error bars represent mean ± SD from 3 replicates.
We further studied the interaction between miR-182 and FOXO3a, utilizing the luciferase reporter assay. MiR-182 mimics (mature miR-182) was co-transfected with different formats of 3′-UTR of FOXO3a (wildtype and mutated) and the positive control containing complementary sequences to miR-182. As shown in Fig. 4E, miR-182 is able to interact with the wildtype 3′-UTR of FOXO3a with a reduction of 30% relative luciferase activity but not the sequence containing the mutations. When we artificially altered the expression of miR-182 with its specific mimics and inhibitors, FOXO3a expression was changed at the protein levels with an opposite trend of miR-182 (Fig. 4F). Furthermore, using CRISPR/cas9 technology, we established miR-182 knockout (KO) models with HCT116 cells, in which FOXO3a is up-regulated to 20% comparable to the induction of SS treatment. However, SS treatment was unable to up-regulate FOXO3a in miR-182-KO cells (Fig. 4G). We then transfected miR-182 mimics to the parent HCT116 cells, finding that the enrichment of miR-182 could attenuate the induction of FOXO3a by SS (Fig. 4H). These results support the concept that SS up-regulating FOXO3a is primarily through the suppression of miR-182.
Predictive capacity of miR-182, FOXO3a, and Cyclin G2 in CRC progression
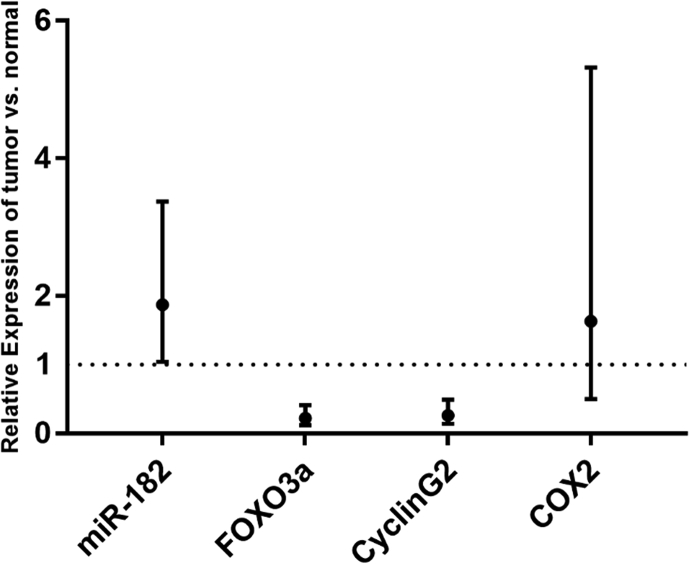
We collected 20 human CRC tumor specimens paired with “normal” adjacent, non-neoplastic tissues, correlating them with the clinical information of patients, as shown in Table S2. After isolation of total RNA from these samples, we analyzed the expression of miR-182, FOXO3a, Cyclin G2, and COX-2 using qRT-PCR. As shown in Fig. 5, the expression of miR-182 in tumors is significantly higher than that in the adjacent normal tissues (paired t-test, P=0.038). Additionally, the expressions of FOXO3a and Cyclin G2 in the tumor tissues were much lower than that in the adjacent normal tissues (P=4.3×10-5 and P=2.9×10-4, respectively). The relative gene expressions of the tumor vs. matched normal tissues are 1.87 [95% confidence interval (CI)=1.04-3.37], 0.22 (95% CI=0.12-0.41), and 0.26 (95% CI=0.14-0.49) for miR-182, FOXO3a, and Cyclin G2, respectively. We found no significant difference in COX-2 expression between the tumor and matched normal tissues (P=0.402), with the relative expressions of 1.63 (95% CI=0.5-5.32).
Figure 5.
The predictive capacity of miR-182, FOXO3a, and Cyclin G2 in CRC progression. QRT-PCR analysis of miR-182, FOXO3a, Cyclin G2, and COX-2 expression with the means and 95% confidence intervals of relative expression in tumor vs. normal tissues. Paired t-test P-values are 0.038 for miR-182, 4.3 × 10-5 for FOXO3a, 2.9 × 10-4 for CyclinG2, and 0.402 for COX-2.
The expressed status (high/low) of miR-182, FOXO3a, Cyclin G2, and COX-2 were also defined, based upon the medians of the relative expressions (2.02, 0.21, 0.33 and 0.99, respectively). There was no significant difference identified between all four biomarkers with the metastasis status (N1). For tumor stage, Cyclin G2 and COX-2 were not significantly associated with colon tumor stage, but miR-182 and FOXO3a were marginally associated with a higher tumor stage (Fisher's exact P=0.070 and 0.070, respectively). As shown in Table 1, the patients with relatively low expressions of miR-182 and FOXO3a tended to have a high stage of CRC (100% high tumor stage vs. 55% overall). The Fisher's exact test P-value is 0.024 for comparing the tumor stage for the four sub-groups of the miR-182 and FOXO3a combinations.
Table 1.
Synergetic effect of miR-182 and FOXO3a on predicting CRC progression.
| FOXO3aa | miR-182a |
||
|---|---|---|---|
| Low % of Stage III/IV (n) |
High % of Stage III/IV (n) |
Total % of Stage III/IV (n) |
|
| Low | 100% (6) | 50% (4) | 80% (10) |
| High | 50% (4) | 16.7% (6) | 30% (10) |
| Total | 80% (10) | 30% (10) | 55% (20) |
Based on the cut-point of median relative expressions (2.02 for miR-182 and 0.21 for FOXO3a), Fisher's exact test P = 0.024 for testing four sub-groups with tumor stage.
Discussion
One of the hallmarks of cancer is characterized by uncontrolled cell proliferation, owing to genetic or epigenetic changes in either cell cycle regulators or their upstream pathways.28 Previous studies have reported that select cyclins/CDK complexes are involved in the SS-induced cell cycle inhibition, such as Cyclin E,29 Cyclin D1,30,31 p34cdc2,32 and proliferating cell nuclear antigen (PCNA).33 SS has also been reported to induce cell cycle arrest in mouse embryonic fibroblast (MEFs) cells, which is mediated by the retinoblastoma tumor suppressor protein (Rb) and the cyclin kinase inhibitor p21waf1/cip1. Of interest, MEF cells with deficient p21waf1/cip1, or Rb, were found to be sensitive to SS treatment, implying that SS could be more effective in those pre-malignant cells with cell cycle checkpoint deficits.34
In this study, we demonstrate that SS treatment can induce G1 arrest in both HCT116 and HT29 cells. After analyzing several cyclins that are involved in G1 arrest, we found that Cyclin G2 exhibited a time-dependent response to SS treatment, with a pattern tightly correlated to cell cycle progression. Previous studies have reported that up-regulation of Cyclin G2 can cause G1-phase growth arrest.12,35,36 In our loss-of-function study, Cyclin G2 knockdown can significantly attenuate the SS inhibitory effect on cell cycle progression with G1 arrest.
We further explored the molecular mechanisms involved in the regulation of Cyclin G2 by SS. FOXO3a is a member of the forkhead box family, and it has been reported to regulate Cyclin G2 at the transcriptional level.26 Our results show that SS can not only induce the expression of FOXO3a in a time-dependent manner, but also promote its translocation and accumulation in the nucleus, resulting in up-regulation of Cyclin G2. We further determined the regulatory effect of FOXO3a on Cyclin G2 at the transcriptional level utilizing ChIP assay and loss-of-function studies with CRISPR/cas9 technology. All of these results consistently support our hypothesis that Cyclin G2's elevation upon the treatment of SS is through the increased transcriptional activity of FOXO3a.
MicroRNAs (miRNAs) are a set of endogenous, small, non-coding RNA molecules that are able to regulate gene expression at the post-transcriptional and translational levels. Given the short length (18–22 nt), a single miRNA has the capacity to trigger hundreds, or even thousands, of putative target genes by binding to their 3′-untranslated regions (UTRs). Many of these genes are shown to be involved with diversified cellular and molecular processes, such as the cell cycle, proliferation, differentiation, apoptosis, invasion, and metastasis, to name but a few.37 Therefore, targeting an individual or panel of miRNAs will provide valuable insight as to the numerous genes that are co-targeted by these miRNAs, but additionally involved in different pathways or cellular events.
Recently, we reported that select oncogenic miRNAs could be down-regulated by SS through the suppression of NF-κB signaling, supporting a novel mechanism distinct from COX-inhibition.27 In the current study, we examined several miRNAs that putatively target FOXO3a, but only miR-182 was validated to inhibit FOXO3a in CRC cells. Moreover, we found that miR-182 could mediate the inductive effect of SS on FOXO3a, in both HCT116 and HT29 cells in a time-dependent manner. MiR-182 expression levels were also found to be highly correlated with the patterns of cell cycle growth arrest, as well as FOXO3 alteration upon SS treatment. In particular, we found that miR-182 mimics were able to attenuate the inductive effect of SS on FOXO3a, which supports the notion that miR-182 mediates SS regulation of FOXO3a. Although miR-182 was reported as an oncogenic miRNA previously,38 very few studies have investigated its impact upon the activity of NSAIDs. For the first time, our study demonstrates its involvement in sulindac inhibitory activity against CRC cells.
Furthermore, we expanded our observation on the clinical utility of miR-182/FOXO3a/Cyclin G2, evaluating their predictive capacity as biomarkers using CRC patient speciems. We analyzed the expression levels of miR-182, FOXO3a, and Cyclin G2 in 20 CRC samples that were paired with adjacent non-neoplastic tissues. Our results showed that their dysregulation dominantly existed within tumor tissues. More significantly, miR-182 and FOXO3a displayed a synergistic effect upon the prediction of tumor progression, and their dysregulation had a strong correlation with advanced tumor stages. COX-2 was assessed in parallel, but no significant difference was identified between tumor and non-neoplastic control tissues.
Sulindac has non-selective COX-1 and COX-2 inhibitory effects, with the latter regarded as one of the key mechanisms accounting for the anti-cancer activity of sulindac. However, there are significant side effects that have limited the clinical utility of sulindac, such as heart and kidney toxicity secondary to PGE2 suppression, which is derived from COX-2 inhibition.39,40 It is no doubt that the generic NSAIDs, such as sulindac, have unique advantages to be re-purposed as a more affordable and effective option for cancer control and prevention. In particular, decreasing the potential side effect profile or other toxicity would greatly benefit the patient. In our study, we used two distinct human CRC cell line models with different expressions of COX-2 activity. HCT116 cells have low to a null expression of COX-2 gene, with HT29 cells having a completely intact level of COX-2 activity.18 However, G1 arrest by SS treatment showed a similar pattern in both cell lines, which implies that COX-2 inhibition might not play a significant role in this activity.
In summary, we demonstrate a new mechanistic axis consisting of miR-182, FOXO3a, and Cyclin G2. This axis is involved in the inhibitory effect of sulindac sulfide (SS) on cell cycle progression and arrest of the G1 phase in human CRC cells. The activity of SS is able to down-regulate the expression of oncogenic miR-182. FOXO3a is the target of miR-182, and down-regulation of miR-182 promotes the transcriptional activity of this tumor suppressor gene. Ultimately, FOXO3a is able to up-regulate Cyclin G2, which plays a key role in the regulation of cell cycle progression, in particular for G1 arrest. Therefore, our study not only highlights an innovative insight into the discovery of novel drugs that can trigger Cyclin G2 for the treatment of CRC, but also provides a new mechanism of miR-182/FOXO3a/Cyclin G2 to understand the preventative advantage of sulindac in CRC occurrence.
Conflicts of Interests
Authors declare no conflict of interest.
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2020.11.006.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Smalley W., Ray W.A., Daugherty J. Use of nonsteroidal anti-inflammatory drugs and incidence of colorectal cancer: a population-based study. Arch Intern Med. 1999;159(2):161–166. doi: 10.1001/archinte.159.2.161. [DOI] [PubMed] [Google Scholar]
- 3.Giardiello F.M., Hamilton S.R., Krush A.J. Treatment of colonic and rectal adenomas with sulindac in familial adenomatous polyposis. N Engl J Med. 1993;328(18):1313–1316. doi: 10.1056/NEJM199305063281805. [DOI] [PubMed] [Google Scholar]
- 4.De Silva B., Banney L., Uttley W. Pseudoporphyria and nonsteroidal antiinflammatory agents in children with juvenile idiopathic arthritis. Pediatr Dermatol. 2000;17(6):480–483. doi: 10.1046/j.1525-1470.2000.01827.x. [DOI] [PubMed] [Google Scholar]
- 5.Mukherjee D., Nissen S.E., Topol E.J. Risk of cardiovascular events associated with selective COX-2 inhibitors. J Am Med Assoc. 2001;286(8):954–959. doi: 10.1001/jama.286.8.954. [DOI] [PubMed] [Google Scholar]
- 6.Smolinske S.C., Hall A.H., Vandenberg S.A. Toxic effects of nonsteroidal anti-inflammatory drugs in overdose. An overview of recent evidence on clinical effects and dose-response relationships. Drug Saf. 1990;5(4):252–274. doi: 10.2165/00002018-199005040-00003. [DOI] [PubMed] [Google Scholar]
- 7.Piazza G.A., Keeton A.B., Tinsley H.N. A novel sulindac derivative that does not inhibit cyclooxygenases but potently inhibits colon tumor cell growth and induces apoptosis with antitumor activity. Canc Prev Res. 2009;2(6):572–580. doi: 10.1158/1940-6207.CAPR-09-0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitt J.D., Li N., Tinsley H.N. A novel sulindac derivative that potently suppresses colon tumor cell growth by inhibiting cGMP phosphodiesterase and beta-catenin transcriptional activity. Canc Prev Res. 2012;5(6):822–833. doi: 10.1158/1940-6207.CAPR-11-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malumbres M., Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Canc. 2009;9(3):153–166. doi: 10.1038/nrc2602. [DOI] [PubMed] [Google Scholar]
- 10.Bernaudo S., Khazai S., Honarparvar E. Epidermal growth factor promotes cyclin G2 degradation via calpain-mediated proteolysis in gynaecological cancer cells. PloS One. 2017;12 doi: 10.1371/journal.pone.0179906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernaudo S., Salem M., Qi X. Cyclin G2 inhibits epithelial-to-mesenchymal transition by disrupting Wnt/beta-catenin signaling. Oncogene. 2016;35(36):4816–4827. doi: 10.1038/onc.2016.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennin D.A., Don A.S., Brake T. Cyclin G2 associates with protein phosphatase 2A catalytic and regulatory B' subunits in active complexes and induces nuclear aberrations and a G1/S phase cell cycle arrest. J Biol Chem. 2002;277(30):27449–27467. doi: 10.1074/jbc.M111693200. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y., Shintani S., Kohno Y. Cyclin G2 dysregulation in human oral cancer. Canc Res. 2004;64(24):8980–8986. doi: 10.1158/0008-5472.CAN-04-1926. [DOI] [PubMed] [Google Scholar]
- 14.Choi M.G., Noh J.H., An J.Y. Expression levels of cyclin G2, but not cyclin E, correlate with gastric cancer progression. J Surg Res. 2009;157(2):168–174. doi: 10.1016/j.jss.2008.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Hasegawa S., Nagano H., Konno M. Cyclin G2: a novel independent prognostic marker in pancreatic cancer. Oncol Lett. 2015;10(5):2986–2990. doi: 10.3892/ol.2015.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zimmermann M., Arachchige-Don A.P.S., Donaldson M.S. Cyclin G2 promotes cell cycle arrest in breast cancer cells responding to fulvestrant and metformin and correlates with patient survival. Cell Cycle. 2016;15(23):3278–3295. doi: 10.1080/15384101.2016.1243189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun G.G., Zhang J., Hu W.N. CCNG2 expression is downregulated in colorectal carcinoma and its clinical significance. Tumor Biol. 2013;35(4):3339–3346. doi: 10.1007/s13277-013-1440-5. [DOI] [PubMed] [Google Scholar]
- 18.Cianchi F., Cortesini C., Schiavone N. The role of cyclooxygenase-2 in mediating the effects of histamine on cell proliferation and vascular endothelial growth factor production in colorectal cancer. Clin Canc Res. 2005;11(19 Pt 1):6807–6815. doi: 10.1158/1078-0432.CCR-05-0675. [DOI] [PubMed] [Google Scholar]
- 19.Xi Y., Nakajima G., Gavin E. Systematic analysis of microRNA expression of RNA extracted from fresh frozen and formalin-fixed paraffin-embedded samples. RNA. 2007;13(10):1668–1674. doi: 10.1261/rna.642907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xi Y., Nakajima G., Hamil T. Association of insulin-like growth factor binding protein-3 expression with melanoma progression. Mol Canc Therapeut. 2006;5(12):3078–3084. doi: 10.1158/1535-7163.MCT-06-0424. [DOI] [PubMed] [Google Scholar]
- 21.Chang H., Yi B., Ma R. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci Rep. 2016;6 doi: 10.1038/srep22312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li X., Zhang J., Gao L. MiR-181 mediates cell differentiation by interrupting the Lin28 and let-7 feedback circuit. Cell Death Differ. 2011;19(3):378–386. doi: 10.1038/cdd.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Yuan J.S., Reed A., Chen F. Statistical analysis of real-time PCR data. BMC Bioinf. 2006;7 doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu Z., Tindall D.J. FOXOs, cancer and regulation of apoptosis. Oncogene. 2008;27(16):2312–2319. doi: 10.1038/onc.2008.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Gac L., Marques M., Garcia Z. Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead. Mol Cell Biol. 2004;24(5):2181–2189. doi: 10.1128/MCB.24.5.2181-2189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., Gao L., Cui Q. Sulindac inhibits tumor cell invasion by suppressing NF-kappaB-mediated transcription of microRNAs. Oncogene. 2012;31(48):4979–4986. doi: 10.1038/onc.2011.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hanahan D., Weinberg Robert A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Qiao L., Shiff S.J., Rigas B. Sulindac sulfide alters the expression of cyclin proteins in HT-29 colon adenocarcinoma cells. Int J Canc. 1998;76(1):99–104. doi: 10.1002/(sici)1097-0215(19980330)76:1<99::aid-ijc16>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 30.Li H., Liu L., David M.L. Pro-apoptotic actions of exisulind and CP461 in SW480 colon tumor cells involve beta-catenin and cyclin D1 down-regulation. Biochem Pharmacol. 2002;64(9):1325–1336. doi: 10.1016/s0006-2952(02)01345-x. [DOI] [PubMed] [Google Scholar]
- 31.Boon E.M., Keller J.J., Wormhoudt T.A. Sulindac targets nuclear beta-catenin accumulation and Wnt signalling in adenomas of patients with familial adenomatous polyposis and in human colorectal cancer cell lines. Br J Canc. 2004;90(1):224–229. doi: 10.1038/sj.bjc.6601505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldberg Y., Nassif, Pittas A. The anti-proliferative effect of sulindac and sulindac sulfide on HT-29 colon cancer cells: alterations in tumor suppressor and cell cycle-regulatory proteins. Oncogene. 1996;12(4):893–901. [PubMed] [Google Scholar]
- 33.Qiao L., Shiff S.J., Rigas B. Sulindac sulfide induces several subpopulations of colon cancer cells, defined by PCNA/Ki-67 and DNA strand breaks. Biochim Biophys Acta. 1997;1359(3):222–232. doi: 10.1016/s0167-4889(97)00102-x. [DOI] [PubMed] [Google Scholar]
- 34.Jung B., Barbier V., Brickner H. Mechanisms of sulindac-induced apoptosis and cell cycle arrest. Canc Lett. 2005;219(1):15–25. doi: 10.1016/j.canlet.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 35.Le X.F., Arachchige-Don A.S., Mao W. Roles of human epidermal growth factor receptor 2, c-jun NH2-terminal kinase, phosphoinositide 3-kinase, and p70 S6 kinase pathways in regulation of cyclin G2 expression in human breast cancer cells. Mol Canc Therapeut. 2007;6(11):2843–2857. doi: 10.1158/1535-7163.MCT-07-0109. [DOI] [PubMed] [Google Scholar]
- 36.Horne M.C., Donaldson K.L., Goolsby G.L. Cyclin G2 is up-regulated during growth inhibition and B cell antigen receptor-mediated cell cycle arrest. J Biol Chem. 1997;272(19):12650–12661. doi: 10.1074/jbc.272.19.12650. [DOI] [PubMed] [Google Scholar]
- 37.Esquela-Kerscher A., Slack F.J. Oncomirs - microRNAs with a role in cancer. Nat Rev Canc. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 38.Song L., Liu L., Wu Z. TGF-beta induces miR-182 to sustain NF-kappaB activation in glioma subsets. J Clin Invest. 2012;122(10):3563–3578. doi: 10.1172/JCI62339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fitzgerald G.A. Coxibs and cardiovascular disease. N Engl J Med. 2004;351(17):1709–1711. doi: 10.1056/NEJMp048288. [DOI] [PubMed] [Google Scholar]
- 40.Sheng H., Shao J., Kirkland S.C. Inhibition of human colon cancer cell growth by selective inhibition of cyclooxygenase-2. J Clin Invest. 1997;99(9):2254–2259. doi: 10.1172/JCI119400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.