Abstract
Migratory shorebirds have many unique life history characteristics, such as long‐distance travel between breeding sites, stopover sites, and wintering sites. The physiological challenges for migrant energy requirement and immunity may affect their gut microbiome community. Here, we reviewed the specific features (e.g., relatively high proportion of Corynebacterium and Fusobacterium) in the gut microbiome of 18 migratory shorebirds, and the factors (e.g., diet, migration, environment, and phylogeny) affecting the gut microbiome. We discussed possible future studies of the gut microbiome in migratory shorebirds, including the composition and function of the spatial‐temporal gut microbiome, and the potential contributions made by the gut microbiome to energy requirement during migration.
Keywords: composition and function, energy requirement, migratory shorebirds, the gut microbiome
We synthesised published 16S rRNA gene data in the migratory shorebirds (Cho & Lee, 2020; Grond et al., 2019, 2020; Risely et al., 2017, 2018), and found the predominant phyla of these 13 shorebird species include Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria.

1. INTRODUCTION
Bird migration is the regular seasonal movement between breeding and wintering sites, and one common route is between north and south (Alerstam et al., 2003; Berthold, 2001). Out of these migratory birds, the shorebirds perform some of the longest and fasters migrations ever record (Gill et al., 2009). The shorebirds refer to species in the suborder (Charadrii) (such as plovers or sandpipers) that frequent seashore areas. Most shorebirds (e.g., Calidris ruficollis) travel long distances between breeding sites, stopover sites, and wintering sites (Gill et al., 2009). During long‐distance migration, they face specific physiological challenges, such as energy requirement, environmental pressure, and immunity (Buehler & Piersma, 2008; Klaassen, 1996; Weber, 2009). These extreme physiological challenges (e.g., body mass loss and partial atrophy of the gastrointestinal tract [McWilliams & Karasov, 2001; Piersma et al., 1999]) may affect the host–microbe interactions and lead the changes in the gut microbiome community (e.g., Cho & Lee, 2020; Grond et al., 2019; Risely et al., 2017, 2018).
The gut microbiome is defined as a combination of all the genomes of microorganisms in a specified environment (Grond et al., 2018). The gut microbiome plays an important role in the activities of the host (Kohl, 2012). Understanding the importance of the gut microbiome to bird host physiology and health is important, and studies have focused on the formation and maintenance of the gut microbiome and its effects on host fitness (Grond et al., 2018; Rooks et al., 2014; Waite & Taylor, 2015). Thus, the current studies on the gut microbiome of the migratory shorebirds mainly focus on the composition and specific microbes associated with the extreme physiological challenges during migration. Here, we reviewed the specific features of the gut microbiome of 18 migratory shorebirds (Table 1) and discuss possible future studies of the gut microbiome in migratory shorebirds.
TABLE 1.
The information of the published gut microbiome data in 18 migratory shorebirds
| Species | Sampling site | Location | Sampling time | Sample type | 16s rRNA sequencing | References |
|---|---|---|---|---|---|---|
| Himantopus himantopus | Wintering grounds | Tagus Estuary, Portugal | 2008–2009 | Cloacal | 16s full length, clone | Santos et al. (2012) |
| Limosa limosa | Wintering grounds | Tagus Estuary, Portugal | 2008–2009 | Cloacal | 16s full length, clone | Santos et al. (2012) |
| Tringa totanus | Wintering grounds | Tagus Estuary, Portugal | 2008–2009 | Cloacal | 16s full length, clone | Santos et al. (2012) |
| Arenaria interpres | Stopover site | Delaware Bay, USA | 2011 | Feces | V1‐V4, clone | Ryu et al. (2014) |
| Calidris canutus | Stopover site | Delaware Bay, USA | 2011 | Feces | V1‐V4, clone | Ryu et al. (2014) |
| Calidris pusilla | Stopover site | Delaware Bay, USA | 2011 | Feces | V1‐V4, clone | Ryu et al. (2014) |
| Arenaria interpres | Wintering grounds | Australia | 2015–2016 | Cloacal | V1‐V3, high throughput | Risely et al. (2017, 2018) |
| Calidris acuminata | Wintering grounds | Australia | 2015–2016 | Cloacal | V1‐V3, high throughput | Risely et al. (2017, 2018) |
| Calidris ferruginea | Wintering grounds | Australia | 2015–2016 | Cloacal | V1‐V3, high throughput | Risely et al. (2017, 2018) |
| Calidris ruficollis | Wintering grounds | Australia | 2015–2016 | Cloacal | V1‐V3, high throughput | Risely et al. (2017, 2018) |
| Calidris alpina | Breeding grounds | Arctic region | 2011–2014 | Feces | V3‐v4, high throughput | Grond et al. (2019) |
| Calidris mauri | Breeding grounds | Arctic region | 2011–2014 | Feces | V3‐v4, high throughput | Grond et al. (2019) |
| Calidris melanotos | Breeding grounds | Arctic region | 2011–2014 | Feces | V3‐v4, high throughput | Grond et al. (2019) |
| Calidris pusilla | Breeding grounds | Arctic region | 2011–2014 | Feces | V3‐v4, high throughput | Grond et al. (2019) |
| Limnodromus scolopaceus | Breeding grounds | Arctic region | 2011–2014 | Feces | V3‐v4, high throughput | Grond et al. (2019) |
| Phalaropus fulicarius | Breeding grounds | Arctic region | 2011–2014 | Feces | V3‐v4, high throughput | Grond et al. (2019) |
| Phalaropus lobatus | Breeding grounds | Arctic region | 2011–2014 | Feces | V3‐v4, high throughput | Grond et al. (2019) |
| Pluvialis dominica | Breeding grounds | Arctic region | 2011–2014 | Feces | V3‐v4, high throughput | Grond et al. (2019) |
| Calidris alpina US | Breeding grounds | Delaware Bay, USA | 2018 | Gut contents | V3‐v4, high throughput | Grond et al. (2019) |
| Calidris pusilla US | Breeding grounds | Delaware Bay, USA | 2018 | Gut contents | V3‐v4, high throughput | Grond et al. (2019) |
| Calidris alba | Breeding grounds | Northern Greenland | 2017 | Feces | V3‐v4, high throughput | Cho and Lee (2020) |
| Actitis hypoleucos | Unclear | Equatorial Guinea | 2016 | Gut contents | V3‐v4, high throughput | Capunitan et al. (2020) |
2. CORYNEBACTERIUM ENRICHED IN THE GUT MICROBIOME OF THE RECENTLY ARRIVED MIGRATORY SHOREBIRDS POTENTIALLY RELATED TO THE ENERGY REQUIREMENT
We synthesized published 16S rRNA gene data (Table 1, 506 samples using high‐throughput sequencing technology) in the migratory shorebirds (Cho & Lee, 2020; Grond et al., 2019, 2020; Risely et al., 2017, 2018) and found the predominant phyla of these 13 shorebird species include Firmicutes, Bacteroidetes, Proteobacteria, Fusobacteria, and Actinobacteria (Figure 1). Firmicutes is also a dominant phylum in the other seven migratory shorebirds (Table 1): Clone sequencing: Himantopus himantopus, Limosa limosa, and Tringa totanus (Santos et al., 2012); clone sequencing: Arenaria interpres, Calidris canutus, and Calidris pusilla (Ryu et al., 2014); one sample: Actitis hypoleucos (Capunitan et al., 2020). Risely et al. (2017), Risely et al. (2018) revealed that the recent Calidris migrants (e.g., Calidris ruficollis) harbor a relatively higher proportion of Corynebacterium in the gut microbiome than that of the residents (Risely et al., 2017, 2018). Corynebacterium, one type of Gram‐negative bacteria, is a potential pathogen group in the human and animal gut (Bernard, 2012; Khamis et al., 2004), and the increased abundance of this genus may reflect the host immune response (Dorella et al., 2006; Rooks et al., 2014). The bacterial endotoxins (produced by some Gram‐negative bacteria) or exotoxins can lead to host inflammatory responses that increase fat deposition and energy harvest (Boulangé et al., 2016; Cani et al., 2009; Zhao, 2013). Thus, a high proportion of Corynebacterium in the recently arrived migrants may enable migrating shorebirds to maximize fat deposition (Risely et al., 2017, 2018). This specific feature in the migrant gut microbiome may also be associated with the physiological changes during the migration or an intestinal immune response (Risely et al., 2017, 2018). However, the strain level and real function of Corynebacterium in the migratory shorebird gut still not know and need to be investigated in the experiments (e.g., germ‐free animal models).
FIGURE 1.
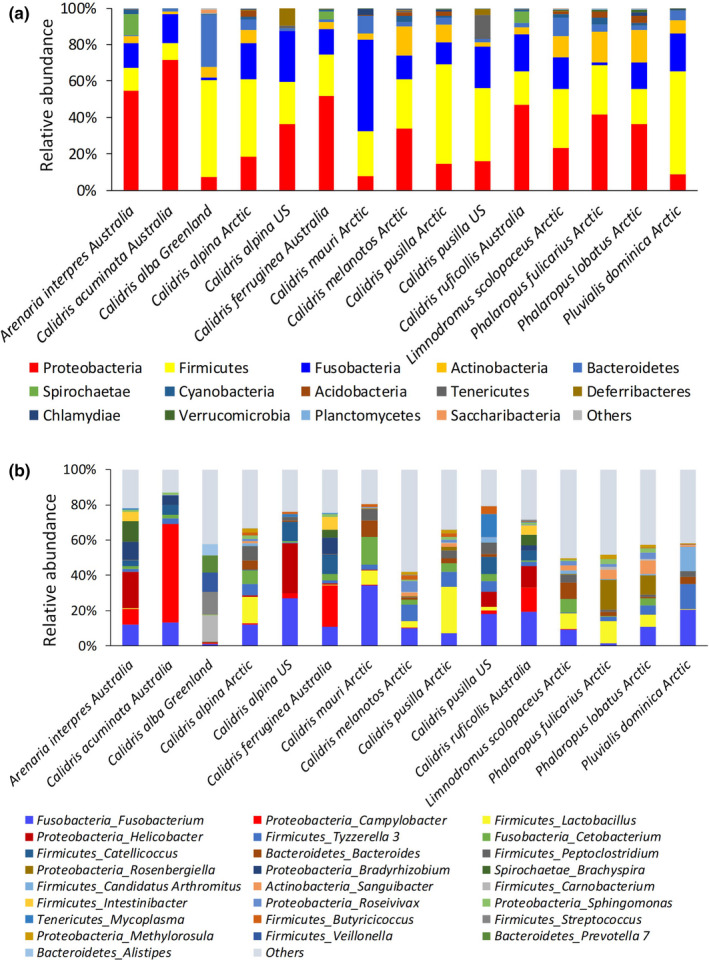
The analysis of the published bacterial 16S ribosomal RNA gene data (using high‐throughput sequencing) of the migratory shorebirds. (a) Phylum level. (b) Genus level. Details of the published data are shown in Table 1. The trimmomatic function was used for quality control on these raw datasets (Bolger et al., 2014). Operational taxonomic units (OTU) were defined as sharing >97% sequence identity by searching clean sequences against the SILVA132 database by pick‐up‐closed‐OTU methods (Christian et al., 2013). Taxon summary was created using the OTUs table (2,000 sequences per sample) in QIIME 1.9 (Caporaso et al., 2010)
Moreover, the analysis of the published 16S rRNA gene data of the migratory shorebirds showed Corynebacterium was mainly enriched in Calidris shorebirds in Australia (Figure 2a). The abundance of Corynebacterium in the Calidris shorebirds in other regions (e.g., Arctic, Greenland, and US regions) was low (Figure 2a). Considering Corynebacterium was rare in the living environment (e.g., sediment [Risely et al., 2017]), we speculated that the different physiological stages (e.g., recently arrived migrants vs. breeding stage) might explain this difference in the abundance of Corynebacterium among these geographic populations. However, the difference in the dietary composition and living environment would also affect the migrant gut microbiome composition and function. Thus, we still need to investigate that whether this finding was the common pattern in the recently arrived migratory shorebirds along with different geographic populations.
FIGURE 2.
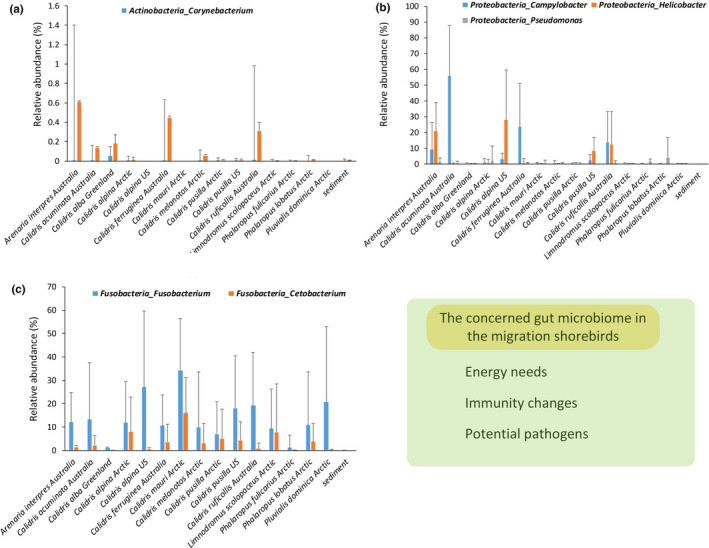
The specific features in the gut microbiome of the migratory shorebirds based on the analysis of the published bacterial 16S Ribosomal RNA gene data (using high‐throughput sequencing). (a) Corynebacterium. (b) Fusobacterium and Cetobacterium (c) Campylobacter and Helicobacter. Details of the published data are shown in Table 1
3. A HIGH PROPORTION OF FUSOBACTERIUM IN THE GUT MICROBIOME OF THE MIGRATORY SHOREBIRDS POTENTIALLY RELATED TO THE CARNIVOROUS DIET
A high proportion of Fusobacteria (e.g., Fusobacterium) was identified in the gut microbiome of the migratory shorebirds in the Arctic region (Grond et al., 2019). The analysis on the published 16S rRNA gene data of the migratory shorebirds revealed another specific feature that most of the migratory shorebirds across different geographic populations (e.g., Australia, US, and Arctic regions) harbored a high proportion of Fusobacteria (mean abundance about 20 percent) (Figure 2b). Fusobacteria is the Gram‐negative and non‐spore‐forming anaerobes and may produce endotoxins (Tan et al., 1996; Warner et al., 1975). Fusobacterium is the common members of the gut microbiome in birds (Hird et al., 2015; Waite & Taylor, 2015), especially in the vulture (mean abundance about 10 percent) (Roggenbuck et al., 2014). A high proportion of Fusobacterium in the shorebirds' gut microbiome may result from the diet‐associated microbiome (e.g., biofilm comprised of microphytobenthos and bacteria) (Grond et al., 2019). Interestingly, a high proportion of Fusobacterium of the gut microbiome is also found in the alligators (mean abundance over 50 percent) (Keenan et al., 2013) and the meat‐eat carnivorans (mean abundance about 30 percent) (Zhu et al., 2018). For example, the high proportion of Fusobacterium in the vulture gut microbiome may indicate that vultures can benefit from the bacterial breakdown of carrion while tolerating bacterial toxins (Roggenbuck et al., 2014). Therefore, the common feature of the enriched Fusobacteria in the gut microbiome of these vertebrates might partially be related to their carnivorous or carrion diet.
4. A HIGH PROPORTION OF OPPORTUNISTIC PATHOGENS IN THE GUT MICROBIOME OF THE MIGRATORY SHOREBIRDS
Several bacteria, considered to be opportunistic pathogens, can cause bird morbidity and mortality (Kohl, 2012). The potentially pathogenic bacteria include Gram‐negative Campylobacter and Helicobacter in the migratory shorebird species (e.g., Calidris species) (Grond et al., 2020; Risely et al., 2018; Ryu et al., 2014; Santos et al., 2012). The analysis uncovered that several migratory shorebird's gut microbiome harbors a high proportion of Campylobacter and Helicobacter among geographic populations (Australia and US regions) (Figure 2c). The mean abundance of migratory Calidris acuminate in Australia was over 50 percent. As mentioned by Ryu et al. (2014), the migratory shorebirds are the important reservoirs of these opportunistic pathogens. Campylobacter lives in the digestive tract of animals such as wild birds, pigs, rodents, cats, and dogs (Acheson & Allos, 2001; Kaakoush et al., 2015; Moore et al., 2005). A high proportion of Campylobacter might reflect the high immune pressure in these migratory shorebirds under the physiological changes or the different environment during migration. However, the negative effect on the health in the migratory shorebirds is still unclear. In addition, given that specially enriched Gram‐negative bacteria (e.g., Corynebacterium, Fusobacterium, and Campylobacter) and their contradictory (pathogenic potential and energy harvest), the investigation on the function of migratory shorebirds' gut microbiome should focus on the interaction between the microbes and the host–microbe interaction.
5. HOST PHYLOGENY AND POTENTIAL SOCIAL CONTACT AFFECTING THE GUT MICROBIOME COMMUNITY OF THE MIGRATORY SHOREBIRDS DURING SYMPATRIC CLUSTERING
Most of the current research of the bird gut microbiome focus on nonmigratory birds, and in these species, host system development is the main driving force for the diversity of intestinal microbes (Grond et al., 2018; Waite & Taylor, 2015). The microbiome is a trait of the host species, and the evolutionary relationship between them and the potential shorebird phylogeny is assessed by comparative phylogenetic methods (Capunitan et al., 2020). The gut microbiome community may differ among different species of migratory shorebirds. Here, we re‐analyzed the raw dataset of eight migratory shorebird species in the Arctic region (Grond et al., 2019) and found a significant difference in the abundance of some gut microbiome among these species (Figure 3a). For example, Actinobacteria was enriched in Phalaropus lobatus, and Proteobacteria was enriched in Calidris melanotos. However, NDMS analysis using unweighted UniFrac distance displayed the mixed pattern in the gut microbiome community among these eight species (Figure 3b). These findings indicated the partial or weak effect on the gut microbiome by host phylogeny, and other factors might also affect the gut microbiome community.
FIGURE 3.
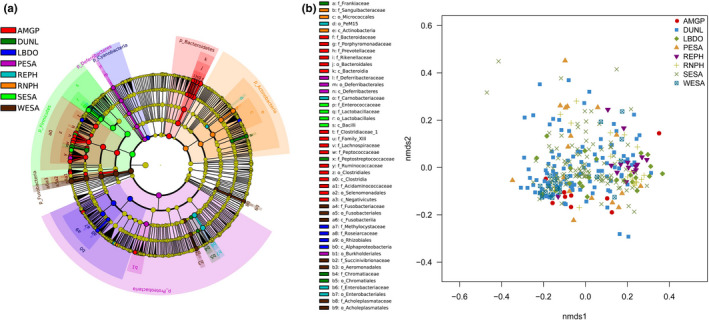
The re‐analysis on the gut microbiome community of eight migratory shorebirds (Grond et al., 2019). (a) Nonmetric multidimensional scaling (NMDS) using unweighted UniFrac distance displayed the mixed pattern in the gut microbiome among eight migratory shorebirds. (b) LEfSe (linear discriminant analysis effective size) determined the significant difference in the abundance of the gut microbial taxon among eight migratory shorebirds. AMGP, Pluvialis dominica; DUNL, Calidris alpine; LBDO, Limnodromus scolopaceus; PESA, Calidris melanotos; REPH, Phalaropus fulicarius; RNPH, Phalaropus lobatus; SESA, Calidris pusilla; WESA, Calidris mauri. The raw dataset of sediment microbiome came from Risely et al. (2017)
The impact of shared habitat on the microbiota may be manifested in the sympatric clusters of migratory birds (Lewis et al., 2017). Social contact among intraspecies and interspecies may lead to the gut microbiome transmission and affect the gut bacterial communities in individual animals (Antwis et al., 2018; Archie & Tung, 2015; Grond et al., 2014; Kwong & Moran, 2016; Ryu et al., 2014). Different species of migratory shorebirds have direct or indirect contact because they share habitat and food during their migrations or breeding (Cho & Lee, 2020; Grond et al., 2019; Risely et al., 2017, 2018; Ryu et al., 2014; Santos et al., 2012). Therefore, we speculated that the effects of social contact on the gut microbiome community of the migratory shorebirds might also modify the effects contributed by host phylogeny.
6. THE POTENTIAL EFFECT BY ENVIRONMENT MICROBIOME
Risely et al. (2017) found red‐necked stints (Calidris ruficollis) acquired little environmental microbiome (about 0.1% of gut microbiome putatively from foraging sediment) (Risely et al., 2017). This indicated that these migratory shorebirds did not acquire microbes from the sediment during migration (Risely et al., 2017). This migratory shorebird species may resist the invasion of environmental microorganisms, which allows them to maintain a stable gut microbiome during migration from the breeding area to wintering sites (Risely et al., 2017). This study raised an interesting question on the gut microbiome transmission between the living environment (e.g., food source and living habitat source) in the migratory shorebirds during migration. The natural environment of the breeding site of some wild migratory shorebirds (such as the western sandpiper [Calidris mauri] and semipalmated sandpiper [Calidris pusilla]) can have a greater impact on the composition of intestinal microbes than phylogeny (Grond et al., 2019). The migratory shorebirds, during migration, would face different environments (e.g., winter sites, stopover sites, and breeding sites) (Gill et al., 2009). Currently, only one pilot study has tried to evaluate the contribution of the sediment‐source microbiome to the gut microbiome of Calidris ruficollis in winter sites (Risely et al., 2017). Thus, the relationship between the potential dynamics of migratory shorebirds' gut microbiome during long‐distance migration and the environment‐source microbial contribution is still unknown.
7. CONCLUSION
Here, we reviewed that the current studies mainly reveal the community in the migratory shorebird gut microbiome, which was potentially associated with the physiological challenges and energy requirement during migration. Thus, the next main scientific question related to common patterns and the function of the gut microbiome will be important and necessary.
8. FUTURE PERSPECTIVES
8.1. Spatial–temporal patterns of migratory bird gut microbiomes
Currently, it is hard to conclude which factors were mainly lead to the difference in the gut microbiome community of the migratory shorebirds during migration due to the differences in sampling time (not in the same migration season), geographic regions (not in the sample migration route), and sample types (Figure 4). The global distribution of sampling sites for migratory shorebirds research shows that data from Asia, Africa, and South America need to be supplemented (Figure 5). Therefore, to complete the global shorebirds data, these regions are key for future research, especially the migration route from Siberia to the eastern coast of China, which is significant for studying the community dynamics of the gut microbiome of migratory shorebirds. A large project would involve tracking changes in the composition of migratory shorebird gut microbiome and their living environment microbiome during migration, including premigration, migration, and end of the migration. This will be an international project, and it will face many challenges. The study will provide information on the relationship between the changes in the specific gut microbiota and migration. Studying the spatial–temporal changes of the gut microbiome during migration is significant for understanding the response of the dramatic physiological challenges and the survival status of migratory shorebird populations.
FIGURE 4.
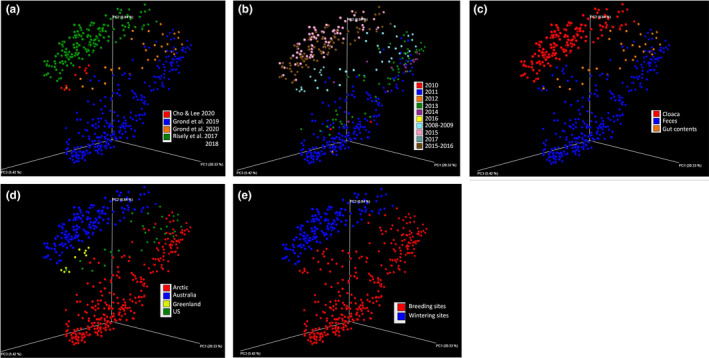
The PCoA analysis of the published bacterial 16S ribosomal RNA gene data of the migratory shorebirds using unweighted UniFrac distance. (a) The data source; (b) sampling time; (c) sample type; (d) study area; (e) sampling site. Details of the published data are shown in Table 1
FIGURE 5.
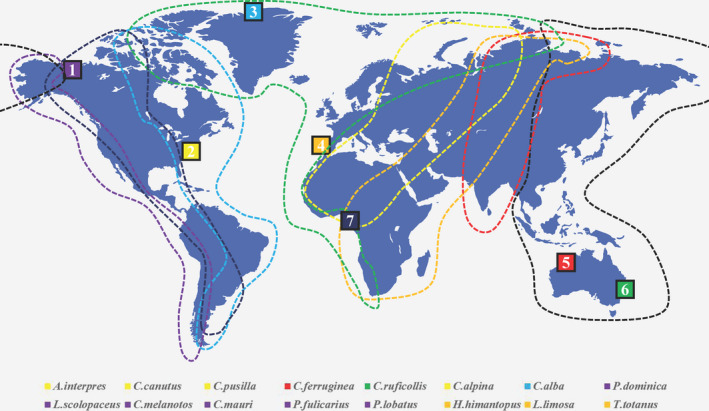
The main migration route worldwide (The sample location data were referred to published data: Cho & Lee, 2020; Grond et al., 2020; Grond et al., 2019; Risely et al., 2018; Risely et al., 2017; Ryu et al., 2014; Santos et al., 2012. The number represents the research location: 1 = MacKenzie River Delta in Alaska and Canada; 2 = Delaware Bay, USA; 3 = Sirius Passet; North Greenland; 4 = Tagus estuary, Portugal; 5 = Broome, Western Australia; 6 = Victoria, Eastern Australia. The migration route is redrawn according to the content of the URL https://www.sovon.nl/ and https://www.wwt.org.uk/. The black dotted line indicates the East Asia–Australia migration route. The red dotted line indicates the Central Asia–India migration route. The orange dotted line indicates the migration route from West Asia to East Africa. The yellow dotted line indicates the Mediterranean–Black Sea migration route. The green dotted line indicates the migration route in the eastern Atlantic Ocean. The cyan dotted line indicates the migration route of the western Atlantic Ocean. The blue dotted line indicates the Mississippi migration route. The purple dotted line indicates the migration route in the eastern Pacific Ocean)
8.2. The function of the gut microbiome in migratory shorebirds (e.g., energy requirements)
The function of the gut microbiome is particularly important for migratory shorebirds, as they face extraordinary metabolic, nutritional, and immune challenges during migration (Altizer et al., 2011; Grond et al., 2019; Risely et al., 2018; Wikelski et al., 2003). Before migration, dunlin and semipalmated sandpipers will quickly gain weight and deposit fat (Grond et al., 2020). The trade‐off between microbial colonization and migratory bird energy requirement remains to be explored. Moreover, the function of gut microbiome (e.g., significant changes in some microbial group during migration) in the migratory shorebird gut needs to be deeply investigated in the experiments (e.g., germ‐free animals).
CONFLICT OF INTEREST
The authors declared no conflicts of interest relevant to this manuscript.
AUTHOR CONTRIBUTIONS
Zheng Zhang: Writing‐original draft (equal). Zhisong Yang: Writing‐original draft (equal). Lifeng Zhu: Conceptualization (equal); Formal analysis (equal); Funding acquisition (equal); Writing‐original draft (equal).
ACKNOWLEDGMENTS
This study is supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Zhang Z, Yang Z, Zhu L. Gut microbiome of migratory shorebirds: Current status and future perspectives. Ecol Evol. 2021;11:3737–3745. 10.1002/ece3.7390
DATA AVAILABILITY STATEMENT
Not applicable.
REFERENCES
- Acheson, D. , & Allos, B. M. (2001). Campylobacter jejuni infections: Update on emerging issues and trends. Clinical Infectious Diseases, 32, 1201–1206. 10.1086/319760 [DOI] [PubMed] [Google Scholar]
- Alerstam, T. , Hedenström, A. , & Åkesson, S. (2003). Long‐distance migration: Evolution and determinants. Oikos, 103, 247–260. 10.1034/j.1600-0706.2003.12559.x [DOI] [Google Scholar]
- Altizer, S. , Bartel, R. , & Han, B. A. (2011). Animal migration and infectious disease risk. Science, 331, 296–302. 10.1126/science.1194694 [DOI] [PubMed] [Google Scholar]
- Antwis, R. E. , Lea, J. M. D. , Unwin, B. , & Shultz, S. (2018). Gut microbiome composition is associated with spatial structuring and social interactions in semi‐feral Welsh Mountain ponies. Microbiome, 6, 1–11. 10.1186/s40168-018-0593-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archie, E. A. , & Tung, J. (2015). Social behavior and the microbiome. Current Opinion in Behavioral Sciences, 6, 28–34. 10.1016/j.cobeha.2015.07.008 [DOI] [Google Scholar]
- Bernard, K. (2012). The genus Corynebacterium and other medically relevant coryneform‐like bacteria. Journal of Clinical Microbiology, 50, 3152–3158. 10.1128/JCM.00796-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold, P. (2001). Bird migration: A general survey. Demand: Oxford University Press. [Google Scholar]
- Bolger, A. M. , Lohse, M. , & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulangé, C. L. , Neves, A. L. , Chilloux, J. , Nicholson, J. K. , & Dumas, M.‐E. (2016). Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Medicine, 8, 1–12. 10.1186/s13073-016-0303-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehler, D. M. , & Piersma, T. (2008). Travelling on a budget: Predictions and ecological evidence for bottlenecks in the annual cycle of long‐distance migrants. Philosophical Transactions of the Royal Society B: Biological Sciences, 363, 247–266. 10.1098/rstb.2007.2138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani, P. D. , Possemiers, S. , Van de Wiele, T. , Guiot, Y. , Everard, A. , Rottier, O. , Geurts, L. , Naslain, D. , Neyrinck, A. , Lambert, D. M. , Muccioli, G. G. , & Delzenne, N. M. (2009). Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP‐2‐driven improvement of gut permeability. Gut, 58, 1091–1103. 10.1136/gut.2008.165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, G. , Kuczynsk, I. J. , Stombaugh, J. , Bittinge, R. K. , Bushman, F. , Costello, E. , Costello, E. K. , Fierer, N. , Peña, A. G. , Goodrich, J. K. , Gordon, J. I. , Huttley, G. A. , Kelley, S. T. , Knights, D. , Koenig, J. E. , Ley, R. E. , Lozupone, C. A. , McDonald, D. , Muegge, B. D. , … Knight, R. (2010). QIIME allows integration and analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capunitan, D. C. , Johnson, O. , Terrill, R. S. , & Hird, S. M. (2020). Evolutionary signal in the gut microbiomes of 74 bird species from Equatorial Guinea. Molecular Ecology, 29, 829–847. 10.1111/mec.15354 [DOI] [PubMed] [Google Scholar]
- Cho, H. , & Lee, W. Y. (2020). Interspecific comparison of the fecal microbiota structure in three Arctic migratory bird species. Ecology and Evolution, 10, 5582–5594. 10.1002/ece3.6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian, Q. , Elmar, P. , Pelin, Y. , Jan, G. , Timmy, S. , Pablo, Y. , Jörg, P. , & Frank Oliver, G. C. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web‐based tools. Nucleic Acids Research, 41, 590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorella, F. A. , Pacheco, L. G. C. , Oliveira, S. C. , Miyoshi, A. , & Azevedo, V. (2006). Corynebacterium pseudotuberculosis: Microbiology, biochemical properties, pathogenesis and molecular studies of virulence. Veterinary Research, 37, 201–218. [DOI] [PubMed] [Google Scholar]
- Gill, R. E. Jr , Tibbitts, T. L. , Douglas, D. C. , Handel, C. M. , Mulcahy, D. M. , Gottschalck, J. C. , Warnock, N. , McCaffery, B. J. , Battley, P. F. , & Piersma, T. (2009). Extreme endurance flights by landbirds crossing the Pacific Ocean: Ecological corridor rather than barrier? Proceedings of the Royal Society B: Biological Sciences, 276, 447–457. 10.1098/rspb.2008.1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grond, K. , Guilani, H. , & Hird, S. M. (2020). Spatial heterogeneity of the shorebird gastrointestinal microbiome. Royal Society Open Science, 7, 191609. 10.1098/rsos.191609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grond, K. , Ryu, H. , Baker, A. J. , Santo Domingo, J. W. , & Buehler, D. M. (2014). Gastro‐intestinal microbiota of two migratory shorebird species during spring migration staging in Delaware Bay, USA. Journal of Ornithology, 155, 969–977. 10.1007/s10336-014-1083-3 [DOI] [Google Scholar]
- Grond, K. , Sandercock, B. K. , Jumpponen, A. , & Zeglin, L. H. (2018). The avian gut microbiota: Community, physiology and function in wild birds. Journal of Avian Biology, 49, e01788. [Google Scholar]
- Grond, K. , Santo Domingo, J. W. , Lanctot, R. B. , Jumpponen, A. , Bentzen, R. L. , Boldenow, M. L. , Brown, S. C. , Casler, B. , Cunningham, J. A. , Doll, A. C. , Freeman, S. , Hill, B. L. , Kendall, S. J. , Kwon, E. , Liebezeit, J. R. , Pirie‐Dominix, L. , Rausch, J. , & Sandercock, B. K. (2019). Composition and drivers of gut microbial communities in arctic‐breeding shorebirds. Frontiers in Microbiology, 10, 2258. 10.3389/fmicb.2019.02258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird, S. M. , Sánchez, C. , Carstens, B. C. , & Brumfield, R. T. (2015). Comparative gut microbiota of 59 neotropical bird species. Frontiers in Microbiology, 6, 1403. 10.3389/fmicb.2015.01403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush, N. O. , Castaño‐Rodríguez, N. , Mitchell, H. M. , & Man, S. M. (2015). Global epidemiology of Campylobacter infection. Clinical Microbiology Reviews, 28, 687–720. 10.1128/CMR.00006-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan, S. W. , Engel, A. S. , & Elsey, R. M. (2013). The alligator gut microbiome and implications for archosaur symbioses. Scientific Reports, 3, 1–7. 10.1038/srep02877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamis, A. , Raoult, D. , & La Scola, B. (2004). rpoB gene sequencing for identification of Corynebacterium species. Journal of Clinical Microbiology, 42, 3925–3931. 10.1128/JCM.42.9.3925-3931.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen, M. (1996). Metabolic constraints on long‐distance migration in birds. Journal of Experimental Biology, 199, 57–64. [DOI] [PubMed] [Google Scholar]
- Kohl, K. D. (2012). Diversity and function of the avian gut microbiota. Journal of Comparative Physiology B, 182, 591–602. 10.1007/s00360-012-0645-z [DOI] [PubMed] [Google Scholar]
- Kwong, W. K. , & Moran, N. A. (2016). Gut microbial communities of social bees. Nature Reviews Microbiology, 14, 374–384. 10.1038/nrmicro.2016.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, W. B. , Moore, F. R. , & Wang, S. (2017). Changes in gut microbiota of migratory passerines during stopover after crossing an ecological barrier. The Auk, 134, 137–145. 10.1642/AUK-16-120.1 [DOI] [Google Scholar]
- McWilliams, S. R. , & Karasov, W. H. (2001). Phenotypic flexibility in digestive system structure and function in migratory birds and its ecological significance. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology, 128, 577–591. 10.1016/S1095-6433(00)00336-6 [DOI] [PubMed] [Google Scholar]
- Moore, J. E. , Corcoran, D. , Dooley, J. S. , Fanning, S. , Lucey, B. , Matsuda, M. , McDowell, D. A. , Mégraud, F. , Millar, B. C. , & O'Mahony, R. (2005). Campylobacter. Veterinary Research, 36, 351–382. [DOI] [PubMed] [Google Scholar]
- Piersma, T. , Gudmundsson, G. A. , & Lilliendahl, K. (1999). Rapid changes in the size of different functional organ and muscle groups during refueling in a long‐distance migrating shorebird. Physiological and Biochemical Zoology, 72, 405–415. 10.1086/316680 [DOI] [PubMed] [Google Scholar]
- Risely, A. , Waite, D. W. , Ujvari, B. , Hoye, B. J. , & Klaassen, M. (2018). Active migration is associated with specific and consistent changes to gut microbiota in Calidris shorebirds. Journal of Animal Ecology, 87, 428–437. [DOI] [PubMed] [Google Scholar]
- Risely, A. , Waite, D. , Ujvari, B. , Klaassen, M. , & Hoye, B. (2017). Gut microbiota of a long‐distance migrant demonstrates resistance against environmental microbe incursions. Molecular Ecology, 26, 5842–5854. 10.1111/mec.14326 [DOI] [PubMed] [Google Scholar]
- Roggenbuck, M. , Schnell, I. B. , Blom, N. , Bælum, J. , Bertelsen, M. F. , Sicheritz‐Pontén, T. , Sørensen, S. J. , Gilbert, M. T. P. , Graves, G. R. , & Hansen, L. H. (2014). The microbiome of New World vultures. Nature Communications, 5, 1–8. 10.1038/ncomms6498 [DOI] [PubMed] [Google Scholar]
- Rooks, M. G. , Veiga, P. , Wardwell‐Scott, L. H. , Tickle, T. , Segata, N. , Michaud, M. , Gallini, C. A. , Beal, C. , van Hylckama‐Vlieg, J. E. T. , Ballal, S. A. , Morgan, X. C. , Glickman, J. N. , Gevers, D. , Huttenhower, C. , & Garrett, W. S. (2014). Gut microbiome composition and function in experimental colitis during active disease and treatment‐induced remission. The ISME Journal, 8, 1403–1417. 10.1038/ismej.2014.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu, H. , Grond, K. , Verheijen, B. , Elk, M. , Buehler, D. M. , & Santo Domingo, J. W. (2014). Intestinal microbiota and species diversity of Campylobacter and Helicobacter spp. in migrating shorebirds in Delaware Bay. Applied and Environment Microbiology, 80, 1838–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, S. S. , Pardal, S. , Proenca, D. N. , Lopes, R. J. , Ramos, J. A. , Mendes, L. , & Morais, P. V. (2012). Diversity of cloacal microbial community in migratory shorebirds that use the Tagus estuary as stopover habitat and their potential to harbor and disperse pathogenic microorganisms. FEMS Microbiology Ecology, 82, 63–74. 10.1111/j.1574-6941.2012.01407.x [DOI] [PubMed] [Google Scholar]
- Tan, Z. , Nagaraja, T. , & Chengappa, M. (1996). Fusobacterium necrophorum infections: Virulence factors, pathogenic mechanism and control measures. Veterinary Research Communications, 20, 113–140. 10.1007/BF00385634 [DOI] [PubMed] [Google Scholar]
- Waite, D. W. , & Taylor, M. (2015). Exploring the avian gut microbiota: Current trends and future directions. Frontiers in Microbiology, 6, 673. 10.3389/fmicb.2015.00673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, J. , Fales, W. , Sutherland, R. , & Teresa, G. (1975). Endotoxin from Fusobacterium necrophorum of bovine hepatic abscess origin. American Journal of Veterinary Research, 36, 1015–1019. [PubMed] [Google Scholar]
- Weber, J.‐M. (2009). The physiology of long‐distance migration: Extending the limits of endurance metabolism. Journal of Experimental Biology, 212, 593–597. 10.1242/jeb.015024 [DOI] [PubMed] [Google Scholar]
- Wikelski, M. , Tarlow, E. M. , Raim, A. , Diehl, R. H. , Larkin, R. P. , & Visser, G. H. (2003). Costs of migration in free‐flying songbirds. Nature, 423, 704. 10.1038/423704a [DOI] [PubMed] [Google Scholar]
- Zhao, L. (2013). The gut microbiota and obesity: From correlation to causality. Nature Reviews Microbiology, 11, 639–647. 10.1038/nrmicro3089 [DOI] [PubMed] [Google Scholar]
- Zhu, L. , Wu, Q. , Deng, C. , Zhang, M. , Zhang, C. , Chen, H. , Lu, G. , & Wei, F. (2018). Adaptive evolution to a high purine and fat diet of carnivorans revealed by gut microbiomes and host genomes. Environmental Microbiology, 20, 1711–1722. 10.1111/1462-2920.14096 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


