Abstract
Background and aims
Realizing the transmission potential and the magnitude of the coronavirus disease 2019 (COVID‐19) aids public health monitoring, strategies, and preparation. Two fundamental parameters, the basic reproduction number (R 0) and case fatality rate (CFR) of COVID‐19, help in this understanding process. The objective of this study was to estimate the R 0 and CFR of COVID‐19 and assess whether the parameters vary in different regions of the world.
Methods
We carried out a systematic review to find the reported estimates of the R 0 and the CFR in articles from international databases between January 1 and August 31, 2020. Random‐effect models and Forest plots were implemented to evaluate the mean effect size of R 0 and the CFR. Furthermore, R 0 and CFR of the studies were quantified based on geographic location, the tests/thousand population, and the median population age of the countries where the studies were conducted. To assess statistical heterogeneity among the selected articles, the I 2 statistic and the Cochran's Q test were used.
Results
Forty‐five studies involving R 0 and 34 studies involving CFR were included. The pooled estimation of R 0 was 2.69 (95% CI: 2.40, 2.98), and that of the CFR was 2.67 (2.25, 3.13). The CFR in different regions of the world varied significantly, from 2.49 (2.08, 2.94) in Asia to 3.40 (2.81, 4.04) in North America. We observed higher mean CFR values for the countries with lower tests (3.15 vs 2.16) and greater median population age (3.13 vs 2.27). However, R 0 did not vary significantly in different regions of the world.
Conclusions
An R 0 of 2.69 and a CFR of 2.67 indicate the severity of the COVID‐19. Although R 0 and CFR may vary over time, space, and demographics, we recommend considering these figures in control and prevention measures.
Keywords: basic reproduction number, case fatality rate, COVID‐19, meta‐analysis, systematic review
1. INTRODUCTION
Since the emergence of SARS‐CoV‐2, the virus causing COVID‐19, in late 2019, there have been more than 35 million confirmed cases, and 950 000 confirmed deaths have been reported as of September 19, 2020. 1 The basic reproduction number (R 0) determines a disease's transmission potential and can be used to inform public health policies. In a susceptible population, the expected number of cases generated by one infected case is specified as R 0. 2 , 3 Outbreaks will continue to propagate until this number falls below 1.0. A timely and accurate assessment of COVID‐19's R 0 notifies COVID is of great significance for assessing the spread of the virus, predicting future trends, and adjusting a series of control measures.
Another important factor to inform is the case fatality rate (CFR), which is the proportion of patients who die from a specified disease compared with the total number of patients identified with the disease within a defined population of interest and is typically presented as a percentage. 4 CFR is an indicator of disease severity and can be used to identify high‐risk populations to inform targeted interventions and the overall public health response. 5
To understand the transmissibility and severity of SARS‐CoV‐2 in populations, there have been several estimates of R 0 and the CFR. Estimates of R 0 and CFR have varied in different countries and different settings. 6 For example, in the Diamond Princess cruise ship, the estimated median R 0 was 2.28 (95% CI: 2.06‐2.52). 7 At the early stages of the epidemic in Wuhan, the value of R 0 was estimated as 2.24 (95% CI: 1.96‐2.55). 8 R 0 has also been estimated for several other countries, including Italy, 3.2 (95% CI: 2.9‐3.5) 9 ; USA, 5.3 (95% CI: 4.35‐6.25) 10 ; and, Japan, 2.6 (95% CI: 2.4‐2.8). 11
Likewise, the estimates of CFRs have varied in different countries and phases of the pandemic. A study found that, at the start of the epidemic, CFR in China was 2%, but on March 5, 2020, it increased to 3.7%. In South Korea, the same study reported the CFR as 0.6% on March 5, 2020. 12 In Europe, during March 2020, Italy, Spain, and France had a CFR of 9.26%, 6.16%, and 4.21%, respectively. 13 Given the considerable variation in estimates of both these parameters, in this current study, our aim was to perform a systematic review and meta‐analysis of the published studies on the COVID‐19 outbreak to provide summary statistics of R 0 and the CFR that are most applicable to other countries or regions.
2. METHODS
We followed the Preferred Reporting Items for Systematic Review and Meta‐Analysis Protocols (PRISMA‐P) 2009. The protocol of the study was registered with the International prospective register of systematic reviews (PROSPERO) (registration number ID: CRD42020182867).
2.1. Search strategy
We systematically searched the major databases: LitCovid (a curated COVID‐19 database of PubMed), PubMed, EMBASE, Scopus, and Web of Science from January 1 to August 31, 2020. We searched articles for “Basic reproduction number (R 0)” and “Case‐fatality rate (CFR)” of the COVID‐19 patients. For basic reproduction number, the search terms were as follows: “2019 novel coronavirus and COVID‐19” AND “Basic reproduction number”, OR “Basic reproduction rate” OR “R 0.” For the case fatality rate, the search terms were “2019 novel coronavirus and COVID‐19” AND “case fatality rate,” OR “case fatality ratio,” OR “CFR” according to the indices of the various databases (Table S1). We also manually scanned the reference lists of the included articles and consulted specialists in the field to ensure a thorough analysis. The records were managed by Mendeley version 1.19.4 software to exclude duplicates.
2.2. Inclusion and exclusion criteria
Articles were selected if they reported CFR and/or R0 with a confidence interval of the patients with COVID‐19. The inclusion criteria for studies were laboratory‐confirmed parents with COVID‐19, reported at sufficient sample size, considered all age‐groups, and published in English and any counties or regions of the world. Studies were excluded if they were pre‐prints, review articles, editorials and case reports, letters to the editor, short communications, and specific to children or pregnant women. However, we did not exclude some special populations representative of the general population in a particular setting (eg, Diamond Princess cruise ship). The steps followed in the literature search are illustrated in Figure 1.
FIGURE 1.
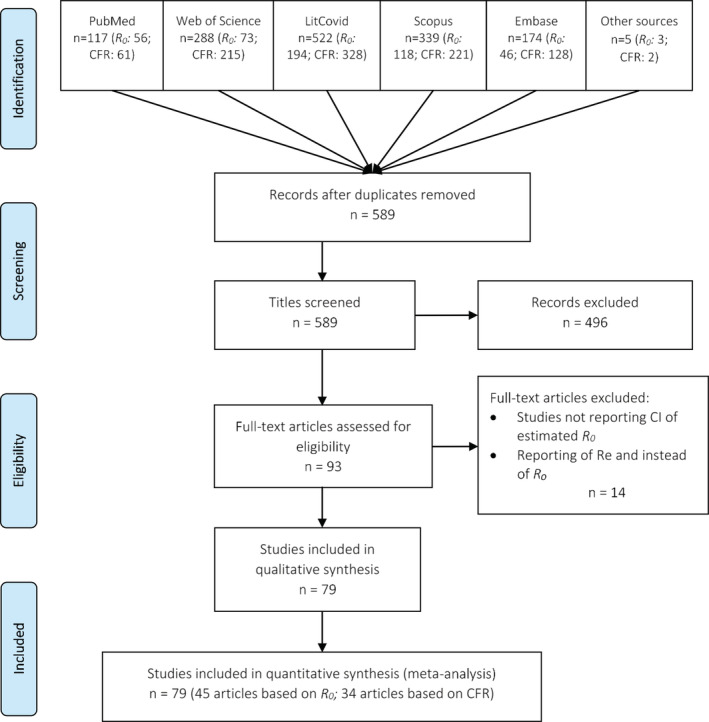
Flow diagram for included studies in the meta‐analysis. CFR, case fatality rate; R 0, basic reproduction number; Re, effective reproduction number
2.3. Data screening and extraction
After excluding duplicate papers, two investigators (TA and AA) independently evaluated the full‐text articles for inclusion after screening the titles and abstracts using the eligibility criterion. The co‐authors helped to solve any conflict in opinions. A standardized form was used to extract data from all eligible studies. For each selected article, publication details [title, first author, publication year, journal name and publisher]; design and population [country, study design, study period, and sample size]; participants' characteristics and major findings [number of deaths, case fatality rate, basic reproduction number, confidence interval, methods used, SD, and other related statistics] were extracted. Forty‐five research articles 7 , 9 , 10 , 11 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 for R 0 and 34 research articles 13 , 18 , 50 , 55 , 56 , 57 , 58 , 59 , 60 , 61 , 62 , 63 , 64 , 65 , 66 , 67 , 68 , 69 , 70 , 71 , 72 , 73 , 74 , 75 , 76 , 77 , 78 , 79 , 80 , 81 , 82 , 83 , 84 , 85 for CFR estimation were selected.
2.4. Critical appraisal and publication bias
The critical appraisal of each selected study was assessed by TA and AA independently using modified risk‐of‐bias (ROB) operational criteria (Tables S2 and S3). 86 A funnel plot was used to determine publication bias, and an Egger's test with P < .05 was an indication of small‐study effects.
2.5. Statistical analysis
We defined R 0 as the number of expected cases directly generated by one infected case in a susceptible population and CFR as the percentage of COVID‐19 cases that result in death. Background statistics of the COVID‐19 patients were recorded through frequency distribution analysis. A random‐effect model was, therefore, used to perform a meta‐analysis. The geo‐location variations for R 0 and geo‐location, tests per thousand population, and population median age variations for CRF were examined using subgroup analysis. However, R 0 estimations for Africa and South America and CFR estimations for Africa and Oceania might not reflect the actual situations of these continents because of the inadequate data used for these estimates. The median values of the frequency distributions were used as cutoff values of the subgroups. Heterogeneity was assessed among the selected studies using Higgin's and Thompson's I 2 statistic and Cochran's Q test. Stata SE version 16.1 (Stata Corporation, College Station, TX, USA 5) was used for all statistical analyses.
3. RESULTS
We identified a total of 490 articles estimating R 0. After considering the duplicates, inclusion, and exclusion criteria, we reviewed these research articles. We searched for titles, abstracts and keywords, methodology, and references and found 59 articles to investigate further (Figure 1). Out of these 59 articles, we found 45 research articles that reported 108 R 0 estimates ranging from 0.27 to 8.21 with related statistics for a meta‐analysis (Table S4). Similarly, for the CFR, after considering filtering criteria, only 34 research articles of 955 articles qualified for quantifying parameter values of CFR (Figure 1), and these 34 articles provided 158 estimates of CFR ranging from 0.00 to 25.9 for use in the meta‐analysis (Table S5). No studies were excluded.
3.1. Pooled estimation of the basic reproduction number (R0)
The pooled R 0 for COVID‐19, in the random‐effects model, was found to be 2.69 (2.40, 2.98), indicating that an infected person with COVID‐19 can transmit the infection to, on average, three susceptible people (Table 1, and Figure 2). There was significant heterogeneity between studies (I 2: 99.96; Table 1).
TABLE 1.
Effect size and test of heterogeneity for the basic reproduction number (R 0) by region
| Statistics | Region | Test for Subgroup differences | Overall | ||||
|---|---|---|---|---|---|---|---|
| Africa | Asia | Europe | North America | South America | |||
| Effect Size (95% CI) |
2.40 (0.96, 3.84) |
2.56 (2.19, 2.94) |
2.70 (2.26, 3.13) |
3.69 (1.46, 5.92) |
2.32 (2.07, 2.57) |
3.81 (P value = .43) |
2.69 (2.40, 2.98) |
| I 2 statistic | 99.43% | 99.96% | 99.95% | 99.98% | 0.00% | 99.96% | |
| Cochran's Q statistics | ‐ | ‐ | ‐ | ‐ | ‐ | 1.6×105 | |
| n (sample = 108) | 3 | 49 | 48 | 6 | 2 | 108 | |
FIGURE 2.
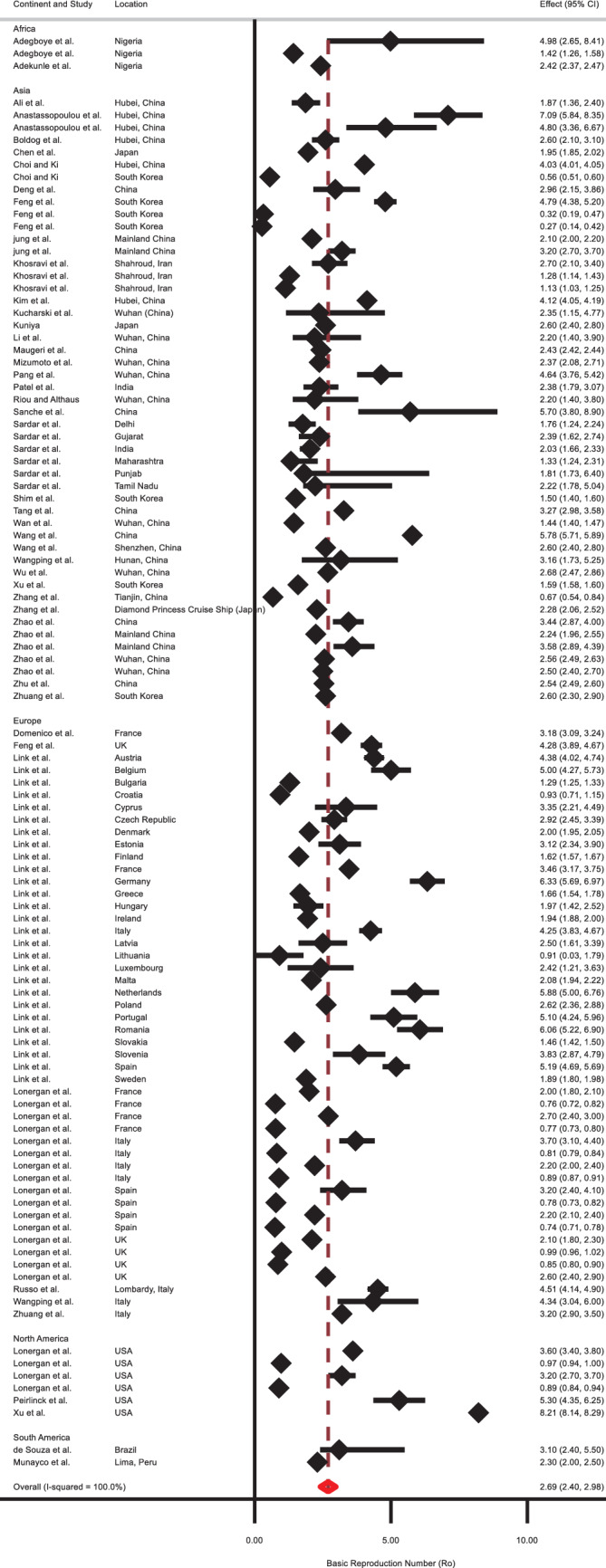
Forest plot of the basic reproduction number (R 0) values based on the random‐effects model
The outcome of the subgroup analysis showed that the R 0 value for Asia was 2.56 (2.19, 2.94), for “Europe” was 2.70 (2.26, 3.13), and for North America was 3.69 (1.46, 5.92; Table 1). There were inadequate studies for Africa (n = 3) and South America (n = 2), and it might not reflect the actual R 0 for these two continents. Moreover, the test of geographic subgroup variations shows that the calculated mean R 0 values across these regions are not significantly different (P = .43). Figure 3 illustrates a funnel plot for assessing publishing bias in R 0 values. The presence of asymmetry and publication bias is indicated by this funnel plot. Additionally, the existence of small‐study effects was indicated by Egger's test (P < .001).
FIGURE 3.
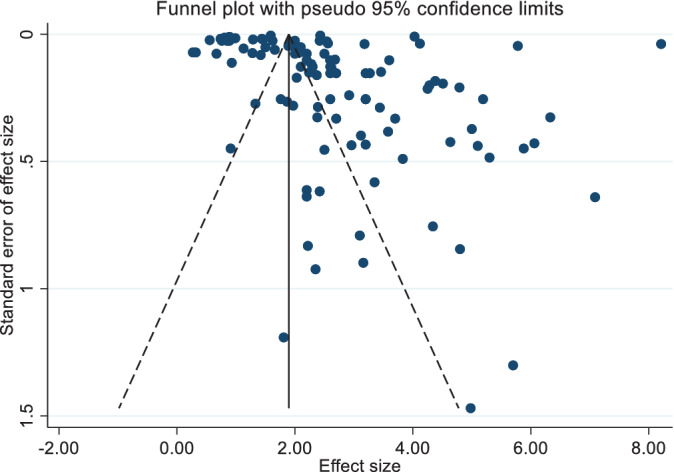
Funnel plot for the basic reproduction number (R 0) values based on the random‐effects model
3.2. Pooled estimation of the case fatality rate
The pooled estimation of the CFR from the random‐effects model was 2.67 (2.25, 3.13). There was significant heterogeneity between studies (I 2: 99.95, Q = 315 324.53 with P = .00; Table 2, and Figure 4). The high value of the I 2 statistic (99.95%) confirms a high level of between‐study heterogeneity.
TABLE 2.
Effect size and test of heterogeneity for the case fatality rate (CFR) by region, population median age (median value), and tests per thousand population (median value)
| Statistics | Region | Test for Subgroup differences | Overall | |||||
|---|---|---|---|---|---|---|---|---|
| Africa | Asia | Europe | North America | Oceania | South America | |||
| Effect Size (95% CI) |
7.11 (6.38, 7.91) |
2.49 (2.08, 2.94) |
2.82 (1.98, 3.81) |
3.40 (2.81, 4.04) |
0.20 (0.02, 0.51) |
2.89 (2.03, 3.89) |
254.44 (P‐value = .00) |
2.67 (2.25, 3.13) |
| I 2 statistic | ‐ | 99.66% | 99.98% | 99.84% | 32.87% | 99.69% | 99.95% | |
| Cochran's Q statistics | ‐ | ‐ | ‐ | ‐ | ‐ | ‐ | 315 324.53 | |
| n (sample = 158) | 1 | 69 | 60 | 15 | 4 | 9 | 158 | |
| Total tests per thousand population | ||||||||
| Less than or equal to 111.16 tests | Greater than 111.16 tests | Overall | ||||||
| Effect size (95% CI) | 3.15 (2.57, 3.79) | 2.16 (1.57, 2.83) |
5.14 (P value = .02) |
2.68 (2.25, 3.14) | ||||
| I 2 statistic | 99.90% | 99.97% | 99.95% | |||||
| Cochran's Q statistics | ‐ | ‐ | 315 324.53 | |||||
| n (sample = 157) | 87 | 70 | 157 | |||||
| Population Median Age Category | ||||||||
| Less than or equal to 38.7 years | Greater than 38.7 years | Overall | ||||||
| Effect size (95% CI) | 2.27 (1.85, 2.74) | 3.13 (2.44, 3.91) |
4.30 (P value = .04) |
2.67 (2.25, 3.13) | ||||
| I 2 statistic | 99.80% | 99.97% | 99.95% | |||||
| Cochran's Q statistics | ‐ | ‐ | 315 324.53 | |||||
| n (sample = 158) | 82 | 76 | 158 | |||||
FIGURE 4.
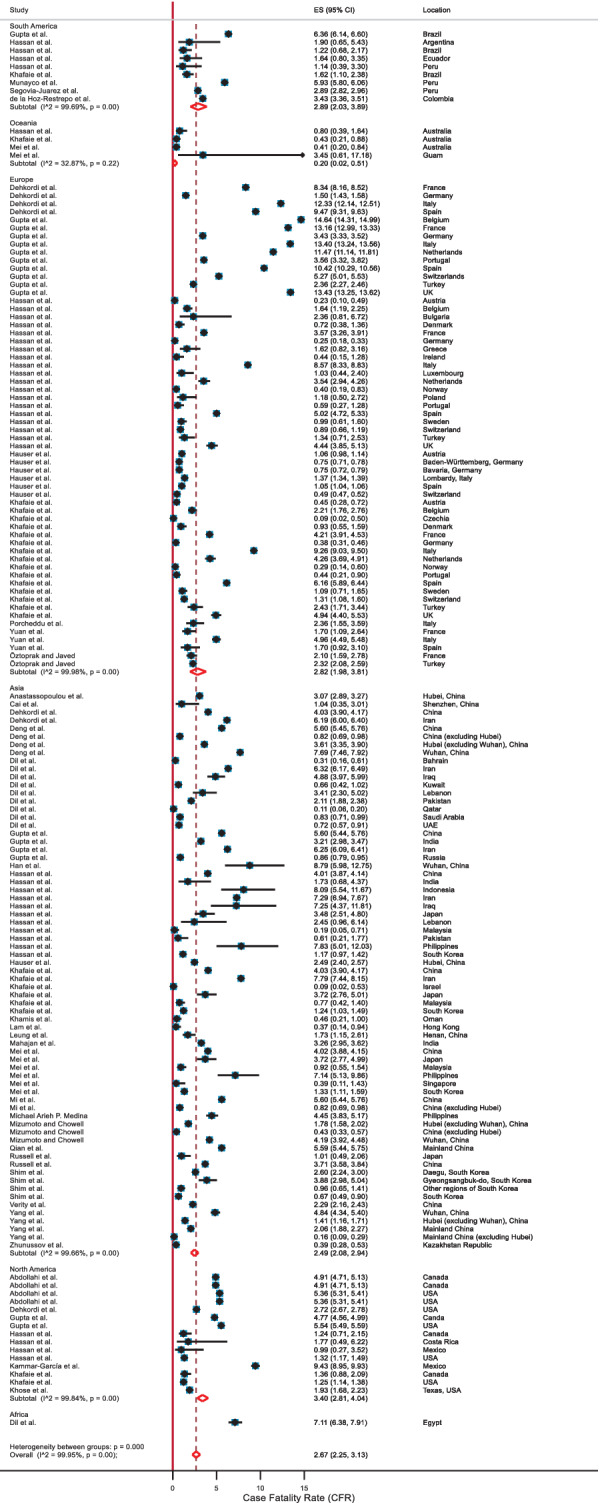
Forest plot of the case fatality rate (CFR) values based on the random‐effects model
The outcome of the subgroup analyses stratified by region showed that the CFR for Asia was 2.49 (2.08, 2.94), for Europe was 2.82 (1.98, 3.81), for North America was 3.40 (2.81, 4.04), and for South America was 2.89 (2.03, 3.89; Table 2). Our estimated CFR might not reflect the actual CFR for Africa and Oceania as there were only one study included for Africa and four studies for Oceania. We observed higher mean CFR values for the countries with lower tests (3.15 vs 2.16) and greater median population age (3.13 vs 2.27). The subgroup difference tests were significant (Table 2), indicating that the calculated mean CFR values for these subgroups were significantly different. The funnel plot for CFR values is shown in Figure 5. Though the funnel plot indicates asymmetry, the Egger test (P = .182) suggests that there was no significant evidence of small‐study effects.
FIGURE 5.
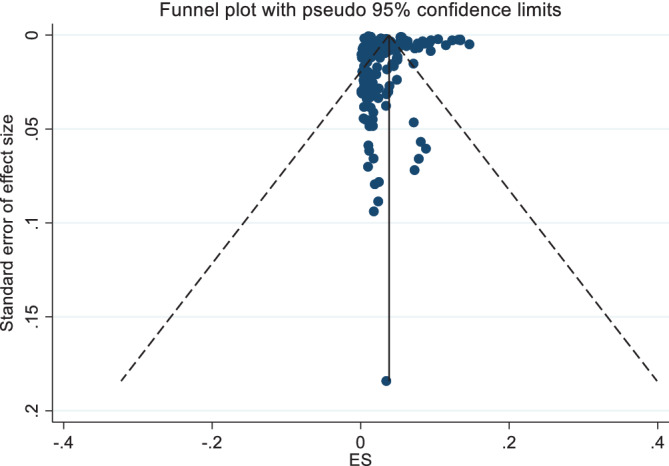
Funnel plot for the case fatality rate (CFR) values based on the random‐effects model
However, if we exclude the extreme CFR values (reported in the included articles), the pooled CFR estimation became 2.63 (2.24, 3.05). Furthermore, if we exclude the cases where naïve CFR (estimated as the ratio of the deaths to the confirmed cases mentioned in the article) >4, the pooled CFR estimation became 1.23 (0.97, 1.53).
4. DISCUSSION
According to our estimation of the R 0 statistic, in the absence of any control measures, one infected individual can transmit the infection to more than two and a half susceptible persons in a naïve population. However, R 0 did not significantly differ across the continents. Among these regions, the highest R 0 was estimated for “North America” (3.69) and the lowest for “Asia” (2.56). We estimated a CFR of above two and a half, and the CFR varied in different continents (highest 3.40 in North America, lowest 2.49 in Asia). We further observed lower CFR values of COVID‐19 in the countries where a higher number of samples (per thousand population) were tested and higher CFR values where the median age of the national population was higher (>38.7 years).
Our estimated overall R 0 is slightly greater than WHO estimates of 1.4 to 2.5 87 but smaller than the results of the other studies, that is, He et al. and Alimohamadi et al. estimated R 0 as 3.15 (95% CI: 2.41‐3.90) and 3.32 (95% CI: 2.81‐3.82), respectively. 88 , 89 Our estimated R 0 value is close to the R 0 values for the severe acute respiratory syndrome epidemic (R 0 = 2‐3) but greater than the Middle East respiratory syndrome in the Middle East (R 0 < 1). 90 However, high R 0 values tend to be estimated in the early stages of epidemics and pandemics due to small sample sizes and incomplete information. However, the total value of R 0 in a population is the average of the R 0 subtypes of that community. Therefore, the probability of transmission in specific subgroups of a population can still be high, even if its overall R0 value is low.
Our estimated value for CFR is similar to the estimates of the Chinese Center for Disease Control and Prevention (China CDC; 2.3%) 91 and He et al. (2.72% with 95% CI:1.29%‐4.16%) 88 but less than the WHO reported estimate of 3.4%. 92 Additionally, our estimated CFR is lower than the CFR values estimated for the other severe coronavirus, severe acute respiratory syndrome epidemic (9.6%), and the Middle East respiratory syndrome (40%), 90 but higher than that of influenza (0·1%). 93 , 94 Although several researchers found higher case fatality rates than ours, usually those are not the actual case fatality rates, as mild, atypical, and asymptomatic cases may not be identified (contrary to deaths), particularly in countries where access to testing is limited. 95 For example, before March 11, 2020 in United States, there was insufficient state‐funded testing. 96 According to a study of 565 Wuhan evacuated Japanese citizens, the CFR would then only be around one‐tenth of what it is now if we consider the underreporting. 97 Our included studies may suffer from underreporting bias, which could increase the pooled CFR estimation. Thus, we further estimated the CFR by excluding the extreme values and the naive CFR greater than four. These gave us lower CFR values close to one.
In our estimation, the CFR varied among different regions, but R 0 did not vary significantly. We found lower CFR in the countries with a higher number of sample testing per thousand population. A higher number of testing allowed the detection of more pre‐/a‐symptomatic and mild cases, which reduced the rate of fatal outcomes. 98 We further found that countries with higher median age had higher CFR values. Older age has been recognized as one of the most common risk factors for a severe outcome of COVID‐19. 99 , 100 , 101 A study conducted with 5484 patients in Italy found the CFR of COVID‐19 was 0.43% for people below 70 years and 10.5% for people aged 70 years and above. 101 Another study showed that an increase of 1% population that is aged 65 years and above increased the risk of death due to COVID‐19 by 10%. 100
An estimate of R 0 is affected by several factors, including the proportion of the susceptible population, population density, and awareness among people. Because SARS‐COV‐2 is a novel virus, we believe the proportion of the susceptible population remains the same across the world; however, other factors (density and awareness) vary. The density of the population is lower in European and North American countries compared with Asian counterparts. Thus, our estimation of R 0 for nonsignificant variation came as a surprise. However, the early phase of the pandemic produces incomplete data. Further studies on R 0 are essential to understand how different lockdown measures, especially face covering, social distancing, and demographic (population density), and environmental factors (temperature, humidity, UV light), affect the transmission dynamic of SARS‐CoV‐2. 102 , 103 , 104 , 105
For developing and populated countries, the R 0 value of 2.69 indicates the rapid spread of the COVID‐19, and the CFR value of 2.67 is an indication of a relatively poor outcome of the treatments. Necessary precautions and strategies such as restricting mass gatherings, limiting the transportation system, and closing educational institutions should continue to prevent the outbreak of this disease until a successful vaccine arrives.
5. LIMITATIONS
An important limitation of our study was that very few studies on Africa (one article on CFR and three articles on R 0), South America (two articles on R 0), and Oceania (four articles on CFR) were available in peer‐reviewed journals. Therefore, our estimated R 0 and CFR estimates for these regions might not be representative. Thus, we were unable to draw any conclusion on CFR or R 0 of these geographical regions. Furthermore, we did not include any preprint articles, although a number of those were published as peer‐reviewed articles and included in this review (those published up until August 31, 2020).
Future research on R 0 and CFR should include more studies of these regions. Moreover, some of the selected studies had limitations because of sample size, missing information on the onset of COVID‐19, and data availability. As a result, the findings, as mentioned earlier, should be interpreted with caution in that regard. Furthermore, future systematic reviews should include Chinese literature since our article was limited to the publications written in English language only. Another important limitation is the case detection and testing heterogeneity. Countries' responses varied, and some countries are facing difficulties in testing enough samples. Furthermore, the quality of the testing samples also varies in different countries. All these have affected the case detection and thus impacted R 0 and CFR. However, we believe the effects are minimum, and data across the world are still comparable.
6. CONCLUSION
The estimated R 0 of SARS‐CoV‐2 was 2.69 (95% CI: 2.40, 2.98) and the CFR of COVID‐19 was 2.67 (95% CI: 2.25, 3.13) based on published literature from January to August 2020. The analysis by subgroups of regions confirmed a significant CFR variation, but the same was not found for R 0. Considering the estimated COVID‐19's R 0 and CFR values, implementing the social distancing program and other non‐pharmaceutical interventions are necessary steps to control the epidemic until an effective vaccine reaches all susceptible populations.
CONFLICT OF INTEREST
The authors declare there is no conflict of interest.
AUTHOR CONTRIBUTIONS
Conceptualization: Najmul Haider, Md. Jamal Uddin
Data curation: Tanvir Ahammed, Aniqua Anjum
Formal analysis: Tanvir Ahammed
Resources: Tanvir Ahammed, Mohammad Meshbahur Rahman, Najmul Haider, Md. Jamal Uddin
Visualization: Tanvir Ahammed, Mohammad Meshbahur Rahman
Supervision: Najmul Haider, Md. Jamal Uddin
Writing ‐ original draft preparation: Tanvir Ahammed, Aniqua Anjum
Writing ‐ review and editing: Najmul Haider, Richard Kock, Md. Jamal Uddin
All authors have read and approved the final version of the manuscript.
Md. Jamal Uddin had full access to all of the data in this study and takes complete responsibility for the integrity of the data and the accuracy of the data analysis.
TRANSPARENCY STATEMENT
Tanvir Ahammed affirms that this manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned (and, if relevant, registered) have been explained.
Supporting information
Appendix S1: Supplementary Information
ACKNOWLEDGMENTS
The authors acknowledge the authors of the selected research articles. NH and RK are members of the Pan‐African Network on Emerging and Re‐Emerging Infections (PANDORA‐ID‐NET – https://www.pandora-id.net/) funded by the European and Developing Countries Clinical Trials Partnership (EDCTP2) program, which is supported under Horizon 2020, the European Union's Framework Programme for Research and Innovation. The authors also acknowledge Laura Macfarlane‐Berry for reviewing and editing the manuscript and Md. Ahadur Rahman for his resource‐related support.
Ahammed T, Anjum A, Rahman MM, Haider N, Kock R, Uddin MJ. Estimation of novel coronavirus (COVID‐19) reproduction number and case fatality rate: A systematic review and meta‐analysis. Health Sci Rep. 2021;4:e274. 10.1002/hsr2.274
DATA AVAILABILITY STATEMENT
The complete list of data and the data entries for all included studies are provided in the paper (Tables S4 and S5). No additional supporting data are available.
REFERENCES
- 1. Worldometer . COVID‐19 coronavirus pandemic; 2020. https://www.worldometers.info/coronavirus/ (Accessed September 11, 2020)
- 2. Fraser C, Donnelly CA, Cauchemez S, et al. Pandemic potential of a strain of influenza a (H1N1): early findings. Science. 2009;324(5934):1557‐1561. 10.1126/science.1176062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Department of Health Australia . The reproduction number; 2006. https://www1.health.gov.au/internet/publications/publishing.nsf/Content/mathematical-models~mathematical-models-models.htm~mathematical-models-2.2.htm (Accessed June 1, 2020)
- 4. Kanchan T, Kumar N, Unnikrishnan B. Mortality: statistics. Encycl Forensic Leg Med. 2016;3:572‐577. https://linkinghub.elsevier.com/retrieve/pii/B9780128000342002974. [Google Scholar]
- 5. Reich NG, Lessler J, Cummings DAT, Brookmeyer R. Estimating absolute and relative case fatality ratios from infectious disease surveillance data. Biometrics. 2012;68(2):598‐606. 10.1111/j.1541-0420.2011.01709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fanelli D, Piazza F. Analysis and forecast of COVID‐19 spreading in China, Italy and France. Chaos, Solitons Fractals. 2020;134:109761 https://linkinghub.elsevier.com/retrieve/pii/S0960077920301636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang S, Diao M, Yu W, Pei L, Lin Z, Chen D. Estimation of the reproductive number of novel coronavirus (COVID‐19) and the probable outbreak size on the Diamond Princess cruise ship: a data‐driven analysis. Int J Infect Dis. 2020;93:201‐204. https://linkinghub.elsevier.com/retrieve/pii/S1201971220300916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao S, Lin Q, Ran J, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019‐nCoV) in China, from 2019 to 2020: a data‐driven analysis in the early phase of the outbreak. Int J Infect Dis. 2020;92:214‐217. https://linkinghub.elsevier.com/retrieve/pii/S1201971220300539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhuang Z, Zhao S, Lin Q, et al. Preliminary estimates of the reproduction number of the coronavirus disease (COVID‐19) outbreak in Republic of Korea and Italy by March 5, 2020. Int J Infect Dis. 2020;95:308‐310. https://linkinghub.elsevier.com/retrieve/pii/S1201971220302599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peirlinck M, Linka K, Sahli Costabal F, Kuhl E. Outbreak dynamics of COVID‐19 in China and the United States. Biomech Model Mechanobiol. 2020;27:1 http://www.ncbi.nlm.nih.gov/pubmed/32342242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuniya T. Prediction of the epidemic peak of coronavirus disease in Japan. J Clin Med. 2020;9(3):789. https://www.mdpi.com/2077-0383/9/3/789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chughtai A, Malik A. Is coronavirus disease (COVID‐19) case fatality ratio underestimated? Glob Biosecurity. 2020;1(3). http://jglobalbiosecurity.com/article/10.31646/gbio.56. [Google Scholar]
- 13. Khafaie MA, Rahim F. Cross‐country comparison of case fatality rates of COVID‐19/SARS‐COV‐2. Osong Public Heal Res Perspect. 2020;11(2):74‐80. 10.24171/j.phrp.2020.11.2.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus‐infected pneumonia. N Engl J Med. 2020;382(13):1199‐1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Riou J, Althaus CL. Pattern of early human‐to‐human transmission of Wuhan 2019 novel coronavirus (2019‐nCoV), December 2019 to January 2020. Euro Surveill. 2020;25(4):2000058. 10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Russo L, Anastassopoulou C, Tsakris A, et al. Tracing day‐zero and forecasting the COVID‐19 outbreak in Lombardy, Italy: A compartmental modelling and numerical optimization approach. Plos one. 2020;15(10):e0240649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanche S, Lin YT, Xu C, Romero‐Severson E, Hengartner N, Ke R. High contagiousness and rapid spread of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(7):1470‐1477. http://wwwnc.cdc.gov/eid/article/26/7/20-0282_article.htm. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shim E, Tariq A, Choi W, Lee Y, Chowell G. Transmission potential and severity of COVID‐19 in South Korea. Int J Infect Dis. 2020;93:339‐344. https://linkinghub.elsevier.com/retrieve/pii/S1201971220301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang B, Xia F, Tang S, et al. The effectiveness of quarantine and isolation determine the trend of the COVID‐19 epidemics in the final phase of the current outbreak in China. Int J Infect Dis. 2020;95:288‐293. https://linkinghub.elsevier.com/retrieve/pii/S1201971220301375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019‐nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395(10225):689‐697. https://linkinghub.elsevier.com/retrieve/pii/S0140673620302609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao S, Musa SS, Lin Q, et al. Estimating the unreported number of novel coronavirus (2019‐nCoV) cases in China in the first half of January 2020: a data‐driven modelling analysis of the early outbreak. J Clin Med. 2020;9(2):388 https://www.mdpi.com/2077-0383/9/2/388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu Y, Chen YQ. On a statistical transmission model in analysis of the early phase of COVID‐19 outbreak. Stat Biosci. 2021;13(1):1‐17. 10.1007/s12561-020-09277-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao S, Stone L, Gao D, et al. Imitation dynamics in the mitigation of the novel coronavirus disease (COVID‐19) outbreak in Wuhan, China from 2019 to 2020. Ann Transl Med. 2020;8(7):448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ali M, Shah STH, Imran M, Khan A. The role of asymptomatic class, quarantine and isolation in the transmission of COVID‐19. J Biol Dyn. 2020;14(1):389‐408. http://www.ncbi.nlm.nih.gov/pubmed/32498655. [DOI] [PubMed] [Google Scholar]
- 25. Deng Y, You C, Liu Y, Qin J, Zhou X. Estimation of incubation period and generation time based on observed length‐biased epidemic cohort with censoring for COVID‐19 outbreak in China. Biometrics. 2020;1‐13. 10.1111/biom.13325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim S, Choi S, Ko Y, Ki M, Jung E. Risk estimation of the SARS‐CoV‐2 acute respiratory disease outbreak outside China. Theor Biol Med Model. 2020;17(1):9 http://www.ncbi.nlm.nih.gov/pubmed/32498721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maugeri A, Barchitta M, Battiato S, Agodi A. Estimation of unreported novel coronavirus (SARS‐CoV‐2) infections from reported deaths: a susceptible‐exposed‐infectious‐recovered‐dead model. J Clin Med. 2020;9(5):1350. http://www.ncbi.nlm.nih.gov/pubmed/32380708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mizumoto K, Kagaya K, Chowell G. Effect of a wet market on coronavirus disease (COVID‐19) transmission dynamics in China, 2019–2020. Int J Infect Dis. 2020;97:96‐101. 10.1016/j.ijid.2020.05.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pang L, Liu S, Zhang X, Tian T, Zhao Z. Transmission dynamics and control strategies of COVID‐19 in Wuhan, China. J Biol Syst. 2020;28(3):543‐560. 10.1142/S0218339020500096. [DOI] [Google Scholar]
- 30. Wan K, Chen J, Lu C, Dong L, Wu Z, Zhang L. When will the battle against novel coronavirus end in Wuhan: a SEIR modeling analysis. J Glob Health. 2020;10(1):011002. http://jogh.org/documents/issue202001/jogh-10-011002.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang K, Lu Z, Wang X, et al. Current trends and future prediction of novel coronavirus disease (COVID‐19) epidemic in China: a dynamical modeling analysis. Math Biosci Eng. 2020;17(4):3052‐3061. 10.3934/mbe.2020173. [DOI] [PubMed] [Google Scholar]
- 32. Wang K, Zhao S, Liao Y, et al. Estimating the serial interval of the novel coronavirus disease (COVID‐19) based on the public surveillance data in Shenzhen, China, from 19 January to February 22, 2020. Transbound Emerg Dis. 2020;67(6):2818‐2822. 10.1111/tbed.13647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wangping J, Ke H, Yang S, et al. Extended SIR prediction of the epidemics trend of COVID‐19 in Italy and compared with Hunan, China. Front Med. 2020;7:169. 10.3389/fmed.2020.00169/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y, Li Y, Wang L, Li M, Zhou X. Evaluating transmission heterogeneity and super‐spreading event of COVID‐19 in a Metropolis of China. Int J Environ Res Public Health. 2020;17(10):3705. http://www.ncbi.nlm.nih.gov/pubmed/32456346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao Y, Wang R, Li J, Zhang Y, Yang H, Zhao Y. Analysis of the transmissibility change of 2019‐novel coronavirus pneumonia and its potential factors in China from 2019 to 2020. Biomed Res Int. 2020;2020:1‐7. https://www.hindawi.com/journals/bmri/2020/3842470/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Z, Yang J, Dai B. Forecast possible risk for COVID‐19 epidemic dissemination under current control strategies in Japan. Int J Environ Res Public Health. 2020;17(11):3872. http://www.ncbi.nlm.nih.gov/pubmed/32486011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Khosravi A, Chaman R, Rohani‐Rasaf M, Zare F, Mehravaran S, Emamian MH. The basic reproduction number and prediction of the epidemic size of the novel coronavirus (COVID‐19) in Shahroud, Iran. Epidemiol Infect. 2020;148:e115. https://www.cambridge.org/core/product/identifier/S0950268820001247/type/journal_article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Patel P, Athotra A, Vaisakh T, Dikid T, Jain S, COVID N. Impact of nonpharmacological interventions on COVID‐19 transmission dynamics in India. Indian J Public Health. 2020;64(6):142. http://www.ijph.in/text.asp?2020/64/6/142/285626. [DOI] [PubMed] [Google Scholar]
- 39. Sardar T, Nadim SS, Rana S, Chattopadhyay J. Assessment of lockdown effect in some states and overall india: A predictive mathematical study on covid‐19 outbreak. Chaos, Solitons & Fractals. 2020;139:110078. 10.1016/j.chaos.2020.110078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu C, Dong Y, Yu X, et al. Estimation of reproduction numbers of COVID‐19 in typical countries and epidemic trends under different prevention and control scenarios. Front Med. 2020;14:613‐622. http://www.ncbi.nlm.nih.gov/pubmed/32468343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Adekunle AI, Adegboye OA, Gayawan E, McBryde ES. Is Nigeria really on top of COVID‐19? Message from effective reproduction number. Epidemiol Infect. 2020;148:e166. https://www.cambridge.org/core/product/identifier/S0950268820001740/type/journal_article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Souza WM, Buss LF, Candido D d S, et al. Epidemiological and clinical characteristics of the COVID‐19 epidemic in Brazil. Nat Hum Behav. 2020;4(8):856‐865. 10.1038/s41562-020-0928-4. [DOI] [PubMed] [Google Scholar]
- 43. Di Domenico L, Pullano G, Sabbatini CE, Boëlle P‐Y, Colizza V. Impact of lockdown on COVID‐19 epidemic in Île‐de‐France and possible exit strategies. BMC Med. 2020;18(1):240. 10.1186/s12916-020-01698-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Adegboye OA, Adekunle AI, Gayawan E. Early transmission dynamics of novel coronavirus (COVID‐19) in Nigeria. Int J Environ Res Public Health. 2020;17(9):3054 https://www.mdpi.com/1660-4601/17/9/3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Feng X, Chen J, Wang K, et al. Phase‐adjusted estimation of the COVID‐19 outbreak in South Korea under multi‐source data and adjustment measures: a modelling study. Math Biosci Eng. 2020;17(4):3637‐3648. 10.3934/mbe.2020205. [DOI] [PubMed] [Google Scholar]
- 46. Feng L‐X, Jing S‐L, Hu S‐K, Wang D‐F, Huo H‐F. Modelling the effects of media coverage and quarantine on the COVID‐19 infections in the UK. Math Biosci Eng. 2020;17(4):3618‐3636. 10.3934/mbe.2020204. [DOI] [PubMed] [Google Scholar]
- 47. Linka K, Peirlinck M, Kuhl E. The reproduction number of COVID‐19 and its correlation with public health interventions. Comput Mech. 2020;66:1035‐1050. 10.1007/s00466-020-01880-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lonergan M, Chalmers JD. Estimates of the ongoing need for social distancing and control measures post‐“lockdown” from trajectories of COVID‐19 cases and mortality. Eur Respir J. 2020;56(1):2001483. 10.1183/13993003.01483-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Munayco CV, Tariq A, Rothenberg R, et al. Early transmission dynamics of COVID‐19 in a southern hemisphere setting: Lima‐Peru: February 29th‐March 30th, 2020. Prepr Serv Heal Sci. 2020;5:338‐345. https://linkinghub.elsevier.com/retrieve/pii/S2468042720300130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anastassopoulou C, Russo L, Tsakris A, Siettos C. Data‐based analysis, modelling and forecasting of the COVID‐19 outbreak. PLoS One. 2020;15(3):e0230405. 10.1371/journal.pone.0230405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boldog P, Tekeli T, Vizi Z, Dénes A, Bartha FA, Röst G. Risk assessment of novel coronavirus COVID‐19 outbreaks outside China. J Clin Med. 2020;9(2):571 https://www.mdpi.com/2077-0383/9/2/571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Choi S, Ki M. Estimating the reproductive number and the outbreak size of COVID‐19 in Korea. Epidemiol Health. 2020;42:e2020011. 10.4178/epih.e2020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jung S, Akhmetzhanov AR, Hayashi K, et al. Real‐time estimation of the risk of death from novel coronavirus (COVID‐19) infection: inference using exported cases. J Clin Med. 2020;9(2):523 https://www.mdpi.com/2077-0383/9/2/523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kucharski AJ, Russell TW, Diamond C, et al. Early dynamics of transmission and control of COVID‐19: a mathematical modelling study. Lancet Infect Dis. 2020;20(5):553‐558. https://linkinghub.elsevier.com/retrieve/pii/S1473309920301444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mizumoto K, Chowell G. Estimating risk for death from coronavirus disease, China, January‐February 2020. Emerg Infect Dis. 2020;26(6):1251‐1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Javed A. Case fatality rate estimation of COVID‐19 for European countries: Turkey's current scenario amidst a global pandemic; comparison of outbreaks with European countries. Eurasian J Med Oncol. 2020;4(2):149‐156. 10.14744/ejmo.2020.60998/. [DOI] [Google Scholar]
- 57. Porcheddu R, Serra C, Kelvin D, Kelvin N, Rubino S. Similarity in case fatality rates (CFR) of COVID‐19/SARS‐COV‐2 in Italy and China. J Infect Dev Ctries. 2020;14(2):125‐128. http://www.ncbi.nlm.nih.gov/pubmed/32146445. [DOI] [PubMed] [Google Scholar]
- 58. Russell TW, Hellewell J, Jarvis CI, et al. Estimating the infection and case fatality ratio for coronavirus disease (COVID‐19) using age‐adjusted data from the outbreak on the Diamond Princess cruise ship. Eurosurveillance. 2020;25(12):2000256. 10.2807/1560-7917.ES.2020.25.12.2000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Verity R, Okell LC, Dorigatti I, et al. Estimates of the severity of coronavirus disease 2019: a model‐based analysis. Lancet Infect Dis. 2020;20(6):669‐677. https://linkinghub.elsevier.com/retrieve/pii/S1473309920302437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yang S, Cao P, Du P, et al. Early estimation of the case fatality rate of COVID‐19 in mainland China: a data‐driven analysis. Ann Transl Med. 2020;8(4):128‐128. http://atm.amegroups.com/article/view/36613/html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yuan J, Li M, Lv G, Lu ZK. Monitoring transmissibility and mortality of COVID‐19 in Europe. Int J Infect Dis. 2020;95:311‐315. https://linkinghub.elsevier.com/retrieve/pii/S120197122030182X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mahajan P, Kaushal J. Epidemic trend of COVID‐19 transmission in India during Lockdown‐1 phase. J Community Health. 2020;45(6):1291‐1300. 10.1007/s10900-020-00863-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Semenova Y, Glushkova N, Pivina L, et al. Epidemiological characteristics and forecast of COVID‐19 outbreak in the Republic of Kazakhstan. J Korean Med Sci. 2020;35(24):e227. 10.3346/jkms.2020.35.e227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gupta S, Kumar Patel K, Sivaraman S, Mangal A. Global epidemiology of first 90 days into COVID‐19 pandemic: disease incidence, prevalence, case fatality rate and their association with population density, urbanisation and elderly population. J Health Manag. 2020;22(2):117‐128. 10.1177/0972063420932762. [DOI] [Google Scholar]
- 65. Dil S, Dil N, Maken ZH. COVID‐19 trends and forecast in the eastern Mediterranean region with a particular focus on Pakistan. Cureus. 2020;12(6):e8582. https://doi.org/10.7759/cureus.8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Deng X, Yang J, Wang W, et al. Case fatality risk of the first pandemic wave of novel coronavirus disease 2019 (COVID‐19) in China. Clin Infect Dis. 2020. 10.1093/cid/ciaa578/5837356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Lam HY, Lam TS, Wong CH, et al. The epidemiology of COVID‐19 cases and the successful containment strategy in Hong Kong–January to may 2020. Int J Infect Dis. 2020;98:51‐58. https://linkinghub.elsevier.com/retrieve/pii/S1201971220304926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Abdollahi E, Champredon D, Langley JM, Galvani AP, Moghadas SM. Temporal estimates of case‐fatality rate for COVID‐19 outbreaks in Canada and the United States. CMAJ. 2020;192(25):E666‐E670. http://www.ncbi.nlm.nih.gov/pubmed/32444481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Munayco C, Chowell G, Tariq A, Undurraga EA, Mizumoto K. Risk of Death by Age and Gender from CoVID‐19 in Peru. Albany NY: Aging; 2020:2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. de la Hoz‐Restrepo F, Alvis‐Zakzuk NJ, de la Hoz‐Gomez JF, et al. Is Colombia an example of successful containment of the COVID‐19 2020 pandemic? A critical analysis of the epidemiological data. Int J Infect Dis. 2020;99:522‐529. https://doi.org/10.1016/j.ijid.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kammar‐Garcia A, Vidal‐Mayo J, Vera‐Zertuche JM, et al. Impact of comorbidities in MEXICAN SARS‐COV‐2‐positive patients: a retrospective analysis in a national cohort. Rev Invest Clin. 2020;72(3):151‐158. http://www.ncbi.nlm.nih.gov/pubmed/32584330. [DOI] [PubMed] [Google Scholar]
- 72. Hauser A, Counotte MJ, Margossian CC, et al. Estimation of SARS‐CoV‐2 mortality during the early stages of an epidemic: a modeling study in Hubei, China, and six regions in Europe. PLOS Med. 2020;17(7):e1003189. 10.1371/journal.pmed.1003189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Segovia‐Juarez J, Castagnetto JMJM, Gonzales GF. High altitude reduces infection rate of COVID‐19 but not case‐fatality rate. Respir Physiol Neurobiol. 2020;281:103494 https://linkinghub.elsevier.com/retrieve/pii/S156990482030152X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Qian J, Zhao L, Ye R‐Z, Li X‐J, Liu Y‐L. Age‐dependent gender differences of COVID‐19 in mainland China: comparative study. Clin Infect Dis. 2020;71(9):2488‐2494. https://doi.org/10.1093/cid/ciaa683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Cai Q, Huang D, Ou P, et al. COVID‐19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy. 2020;75(7):1742‐1752. 10.1111/all.14309. [DOI] [PubMed] [Google Scholar]
- 76. Shim E, Mizumoto K, Choi W, Chowell G. Estimating the risk of COVID‐19 death during the course of the outbreak in Korea, February‐May 2020. J Clin Med. 2020;9(6):1641. https://doi.org/10.3390/jcm9061641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Khose S, Moore JX, Wang HE. Epidemiology of the 2020 pandemic of COVID‐19 in the State of Texas: the first month of community spread. J Community Heal. 2020;45(4):696‐701. 10.1007/s10900-020-00854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Hassan MM, El Zowalaty ME, Khan SA, Islam A, Nayem MRK, Järhult JD. Role of environmental temperature on the attack rate and case fatality rate of coronavirus disease 2019 (COVID‐19) pandemic. Infect Ecol Epidemiol. 2020;10(1):1792620. 10.1080/20008686.2020.1792620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Khamis F, Al Rashidi B, Al‐Zakwani I, Al Wahaibi AH, Al Awaidy ST. Epidemiology of COVID‐19 infection in Oman: analysis of the first 1304 cases. Oman Med J. 2020;35(3):e145‐e145. http://omjournal.org/articleDetails.aspx?coType=1&aId=2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hoseinpour Dehkordi A, Alizadeh M, Derakhshan P, Babazadeh P, Jahandideh A. Understanding epidemic data and statistics: a case study of COVID‐19. J Med Virol. 2020;92(7):868‐882. 10.1002/jmv.25885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Han H, Xie L, Liu R, et al. Analysis of heart injury laboratory parameters in 273 COVID‐19 patients in one hospital in Wuhan, China. J Med Virol. 2020;92(7):819‐823. http://www.ncbi.nlm.nih.gov/pubmed/32232979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Leung K, Wu JT, Liu D, Leung GM. First‐wave COVID‐19 transmissibility and severity in China outside Hubei after control measures, and second‐wave scenario planning: a modelling impact assessment. Lancet. 2020;395(10233):1382‐1393. https://linkinghub.elsevier.com/retrieve/pii/S0140673620307467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mei Y, Hu J. Preparedness is essential for Western Pacific Islands during the COVID‐19 pandemic. Disaster Med Public Heal Prep. 2020;16:1‐5. https://www.cambridge.org/core/product/identifier/S1935789320001020/type/journal_article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mi Y, Huang T, Zhang J, et al. Estimating the instant case fatality rate of COVID‐19 in China. Int J Infect Dis. 2020;97:1‐6. https://linkinghub.elsevier.com/retrieve/pii/S120197122030271X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Medina MA. Preliminary estimate of COVID‐19 case fatality rate in The Philippines using linear regression analysis. SSRN Electron J. 2020. https://dx.doi.org/10.2139/ssrn.3569248. [Google Scholar]
- 86. Hoy D, Brooks P, Woolf A, et al. Assessing risk of bias in prevalence studies: modification of an existing tool and evidence of interrater agreement. J Clin Epidemiol. 2012;65(9):934‐939. 10.1016/j.jclinepi.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 87. Liu Y, Gayle AA, Wilder‐Smith A, Rocklöv J. The reproductive number of COVID‐19 is higher compared to SARS coronavirus. J Travel Med. 2020;13(2):taaa021. 10.1093/jtm/taaa021/5735319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. He W, Yi GY, Zhu Y. Estimation of the basic reproduction number, average incubation time, asymptomatic infection rate, and case fatality rate for COVID‐19: meta‐analysis and sensitivity analysis. J Med Virol. 2020;92(11):2543‐2550. http://www.ncbi.nlm.nih.gov/pubmed/32470164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Alimohamadi Y, Taghdir M, Sepandi M. Estimate of the basic reproduction number for COVID‐19: a systematic review and meta‐analysis. J Prev Med Public Heal. 2020;53(3):151‐157. 10.3961/jpmph.20.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. The Lancet Infectious Diseases . MERS—an uncertain future. Lancet Infect Dis. 2015;15(10):1115 https://linkinghub.elsevier.com/retrieve/pii/S1473309915003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Epidemiology Working Group for NCIP Epidemic Response Chinese Center for Disease Control and Prevention . The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID‐19) in China. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(2):145‐151. http://www.ncbi.nlm.nih.gov/pubmed/32064853. [DOI] [PubMed] [Google Scholar]
- 92. World Health Organization (WHO) . WHO Director‐General's opening remarks at the media briefing on COVID‐19 ‐ 3 March 2020. https://www.who.int/director‐general/speeches/detail/who‐director‐general‐s‐opening‐remarks‐at‐the‐media‐briefing‐on‐covid‐19‐‐‐3‐march‐2020 (Accessed cited June 3, 2020).
- 93. De Wit E, van Doremalen N, Falzarano D, Munster VJ. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523‐534. http://www.ncbi.nlm.nih.gov/pubmed/27344959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fauci AS, Lane HC, Redfield RR. Covid‐19 ‐ navigating the uncharted. N Engl J Med. 2020;382(13):1268‐1269. http://www.ncbi.nlm.nih.gov/pubmed/32109011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. German Network for Evidence‐based Medicine . COVID‐19: Where's the Evidence? 2020. https://www.ebm-netzwerk.de/de/veroeffentlichungen/covid-19. (Accessed June 3, 2020)
- 96. Centers for Disease Control and Prevention . Coronavirus Disease 2019 (COVID‐19); 2020 https://www.cdc.gov/coronavirus/2019-ncov/testing-in-us.html (Accessed June 27, 2020)
- 97. Nishiura H, Kobayashi T, Yang Y, et al. The rate of under ascertainment of novel coronavirus (2019‐nCoV) infection: estimation using Japanese passengers data on evacuation flights. J Clin Med. 2020;9(2):419 https://www.mdpi.com/2077-0383/9/2/419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Liang L‐L, Tseng C‐H, Ho HJ, Wu C‐Y. Covid‐19 mortality is negatively associated with test number and government effectiveness. Sci Rep. 2020;10(1):12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rhodes J, Dunstan F, Laird E, Subramanian S, Kenny RA. COVID‐19 mortality increases with northerly latitude after adjustment for age suggesting a link with ultraviolet and vitamin D. BMJ Nutr Prev Heal. 2020;3(1):118‐120. 10.1136/bmjnph-2020-000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Haider N, Yavlinsky A, Chang Y‐M, et al. The Global Health security index and joint external evaluation score for health preparedness are not correlated with countries' COVID‐19 detection response time and mortality outcome. Epidemiol Infect. 2020;148:e210. https://www.cambridge.org/core/product/identifier/S0950268820002046/type/journal_article. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Poletti P, Tirani M, Cereda D, et al. Age‐specific SARS‐CoV‐2 infection fatality ratio and associated risk factors, Italy, February to April 2020. Eurosurveillance. 2020;25(31):2001383. 10.2807/1560-7917.ES.2020.25.31.2001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Bukhari Q, Jameel Y. Will Coronavirus Pandemic Diminish by Summer? SSRN Electron J. 2020; https://www.ssrn.com/abstract=3556998. [Google Scholar]
- 103. Silberstein M. Vitamin D: A simpler alternative to tocilizumab for trial in COVID‐19? Med Hypotheses. 2020;140:109767. 10.1016/j.mehy.2020.109767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. University of Cambridge . Seasonal immunity: Activity of thousands of genes differs from winter to summer. ScienceDaily. 2015. https://www.sciencedaily.com/releases/2015/05/150512112356.htm (Accessed June 26, 2020)
- 105. Jüni P, Rothenbühler M, Bobos P, et al. Impact of climate and public health interventions on the COVID‐19 pandemic: a prospective cohort study. Can Med Assoc J. 2020;192(21):E566‐E573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supplementary Information
Data Availability Statement
The complete list of data and the data entries for all included studies are provided in the paper (Tables S4 and S5). No additional supporting data are available.


