Abstract
Cowpea (Vigna unguiculata (L.) Walp) is an important multipurpose legume crop grown in arid and semi-arid areas of sub-Saharan Africa. The crop associates with a wide diversity of high ecological value rhizobia bacteria, improving biological soil fertility and crop production. Here, we evaluated the symbiotic efficiency (SE) and genetic diversity of native rhizobia isolated from root nodules of cowpea genotypes cultivated in semi-arid areas of lower Eastern Kenya. Rhizobia trapping and SE experiments were done in the greenhouse while genetic diversity was evaluated based on 16S rRNA gene sequencing. Twenty morphologically distinct isolates representing a total of 94 isolates were used for genetic analysis. After 16S rRNA gene sequencing, the isolates closely resembled bacteria belonging to the genus Rhizobium, Paraburkholderia and non-rhizobial endophytes (Enterobacter, Strenotrophomonas and Pseudomonas). This study also reports for the first time the presence of an efficient native cowpea nodulating Beta-Rhizobia (Paraburkholderia phenoliruptrix BR3459a) in Africa. Symbiotic efficiency of the native rhizobia isolates varied (p < 0.0001) significantly. Remarkably, two isolates, M2 and M3 recorded higher SE of 82.49 % and 72.76 % respectively compared to the commercial strain Bradyrhizobium sp. USDA 3456 (67.68 %). Our results form an important step in the development of efficient microbial inoculum and sustainable food production.
Keywords: Cowpea, Genetic diversity, Rhizobia, Symbiotic efficiency, Kenya
Cowpea, Genetic diversity, Rhizobia, Symbiotic efficiency, Kenya.
1. Introduction
Worldwide, an estimated 3.3 million tons of cowpea (Vigna unguiculata (L.) Walp.) are produced annually out of which 64 % is produced in Africa [1]. Many countries in sub-Saharan Africa are characterized by rapid growth of human population and they rely on the production of cowpea as one of the measures to curb food insecurity [2]. Predominantly cultivated by smallholder farmers, the legume serves as a main source of food in semiarid parts of Kenya, where the leaves are used as vegetables and the grains as a protein source. The crop is also a source of income and animal fodder for the low-income rural populations [3]. In sustainable agricultural systems, cowpea plays an important role in nutrient cycling particularly biological nitrogen fixation in arid and semi-arid regions. The legume also acts as a cover crop and helps overcome pollution associated with transport of sediment into surface and ground water sources [4]. Despite its significance, cowpea yields remain low in Kenya making it too risky an investment to many growers. Some of the variables linked to decline in yields are low soil fertility, low technology cultivation techniques and extensive use of inorganic fertilizers [5]. Adoption of bio-fertilizers which promote environmental and socio-economic stability of agriculture has been suggested as an alternative method with a potential to scale up cowpea yields. Rhizobia inoculants are some of the commonly recommended bio-fertilizers in production of cowpea [6].
Inoculation of cowpea with rhizobia is a viable alternative that can improve nitrogen fixation, soil fertility, and increase legume yields [7]. The practice is especially necessary in soils where the native rhizobia are incompatible with the host plants, the population is low or is inefficient in N fixation [8]. Indigenous rhizobia isolates have shown superiority in performance and therefore are recommended over exotic strains in legume production [9]. Majority of the commercial inoculants utilized particularly in Kenya contain exotic cultures that may not be well adapted to the local conditions and this may affect their competitiveness and efficiency [10]. Additionally, subtle differences in host genotype at the sub-species level have been reported to shape rhizobial populations in the soil and also influence their symbiotic efficiency [11]. Evaluating the symbiotic efficiency (SE) and diversity of rhizobia is critical in family farming systems for enhanced cowpea production and assured food security. So far, very few trials have been conducted on rhizobia isolation and molecular characterization from the semi-arid regions of Eastern Kenya and considering cowpea cultivation offers environmental sustainability and economic growth [5], studies are necessary to genetically identify rhizobia strains in the soils capable of establishing efficient symbiotic relationships with locally grown cowpea genotypes. Therefore, the objective of this study was to evaluate the symbiotic efficiency and genetic diversity of rhizobia isolated from agricultural soils in lower Eastern Kenya by using different cowpea genotypes as the trap plant. Results of this study forms a basis upon which cheap and affordable bio-fertilizers alternatives can be developed.
2. Materials and methods
2.1. Study sites description
The study was conducted in smallholder farms in the semi-arid areas of Embu (Mbeere South sub-county) latitude 1o 10′S longitude 37o 47′E and Kitui (Kitui West sub-county) latitude 0o46′S longitude 37o39′E counties in Eastern Kenya. Ten farms with no history of rhizobia inoculation and cultivated for more than ten years were selected (five farms per county). The areas are hot and dry with temperatures ranging from 14 °C to 34 °C and they receive an annual rainfall between 700-900 mm [12]. The areas represent some of the typical semi-arid areas in Kenya predominated by smallholder farmers where agriculture is the main stay. Key crops grown for food and cash generation in these regions include cowpeas, green grams, common beans, pigeon peas and maize. Greenhouse bioassays and laboratory experiments were carried out at Kenyatta University, Nairobi, Kenya.
2.2. Soil sampling
Twenty kilograms of soil were obtained from each farm at a depth of 5–30 cm after clearing soil debris from the soil surface. The soils from each farm were then mixed thoroughly to obtain a homogenous soil sample, packed separately and transported to Kenyatta University for greenhouse experiments. Soil samples that were not used immediately were stored at 4 °C.
2.3. Greenhouse rhizobia trapping
Rhizobia trapping experiment was done in an even span greenhouse which received 12 h of sunlight while the temperature and humidity ranged from 21 to 28 °C and 60–78 % respectively. Three kilograms of homogenous soil samples obtained from each farm were distributed into plastic pots, conical in shape with a breadth of 17.8 cm and height 17.8 cm. The pots were sterilized before being used by washing them in 3 % sodium hypochlorite and later after drying they were swabbed with 70 % ethanol. The 3 kgs of soil was constituted as follows; 1.25 kg of sites soil was added to 1.75 kg of sterilized sand to make 3 kgs of soil per pot. The sand had been oven sterilized at 200 °C for three consecutive days. Cowpea genotypes (K80, M66, KVU 27-1, and Kikamba, which is a preferred cowpea landrace by farmers in the study area) were used in the trapping experiment. The genotypes are recommended by Kenya Agricultural and Livestock Research Organization (KALRO) for ASAL areas in Kenya. The cowpea seeds were sterilized in 3 % sodium hypochlorite for 5 min and rinsed in 6 changes of sterilized distilled water [13]. Three seedlings were planted pot−1 and later thinned to two after 5 days of germination. The pots were arranged in a complete randomized block design. Watering was carried out as needed where approximately 100 ml of water was added to the plants per day. Each treatment (farm soil) was replicated four times with two plants in each pot (4 cowpea varieties x 4 replicates) making a total of 160 pots representing the ten farms. Harvesting was done after 45 days. The roots and shoots were separated. The roots were carefully washed in a stream of running water after which the nodules were carefully detached. The detached root nodules for each treatment were separated and preserved by wrapping them in absorbent paper towels and left to dry at room temperature as described by Muthini et al. [13]. Shoot, root dry weights and nodulation were also determined.
2.4. Isolation and purification of nodule isolates
Nodules representing treatment were immersed in sterile distilled water and allowed to imbibe for 2 h. They were then dipped in 70 % ethanol (v/v) for 30 s to remove air bubbles from the tissues and reduce surface tension. The nodules were then dipped in 3 % sodium hypochlorite (v/v) for 3 min for further sterilization and then rinsed in six changes of sterile distilled water [13]. The nodules were later crushed in a drop of distilled water with a sterile glass rod. A loop full of the suspension was streaked onto Yeast Extract Mannitol Agar (YEMA) plates containing 0.025 mg/l of Congo red and incubated at 27 °C in the dark [13]. Colonies emerged after three days and after five days well isolated colonies were streaked on YEMA with Congo red [13].
2.5. Morphological and biochemical grouping of nodule isolates
Morphological characteristics that include colour change, colony elevation, shape, colony size, exo-polysaccharide gum, transparency and mucosity were used for presumptive identification of the rhizobia. Biochemical identification, Bromothymol Blue Test (BTB) and Gram staining procedures were carried out as guided by Beck et al. [14]. The isolates were tested for acid/alkali production by growing them on YEMA with BTB (0.025 mg/L) indicator at pH 6.8. The cultures were then incubated at 28 °C in a rotating orbital shaker for up to 5 days. The isolates were allowed to grow and then grouped as acidproducing, alkali producing or neutral, depending on colour changes observed in the media [15].
2.6. Authentication of representative rhizobia isolates
The nodule forming ability of the rhizobia isolates was assessed in the even span greenhouse described in sub-topic 2.3 above. Leonard jar assemblies were prepared and they comprised of a modified plastic cup with a diameter of 8 cm (brim) and a bottom diameter of 4 cm. A rectangular hole 1.5 cm2 was made at the bottom of the cup and fit with a 20 cm long wick. Prior to fitting the cups were swabbed with 70 % ethanol while the wick was sterilized in 3 % sodium hypochlorite. A larger plastic vessel was decontaminated using 70 % ethanol and used to suspend the cup assembly [13].
Procedures described by Muthini et al. [16] were used to prepare the sterile nitrogen–free plant nutrient medium. Five stock solutions were later autoclaved at 121 °C for 15 min. The rooting medium used was sterilized vermiculite. The vermiculite was soaked in water overnight then thoroughly washed for two days. For the final rinse, distilled water was used. The vermiculite was then autoclaved and later packed into the cups of the Leornard jar assemblies which were later covered with sterilized aluminum foil to maintain the sterile conditions of the assembly. The jars were then put in khaki bags for insulation.
Cowpea seeds of uniform size and shape were selected and surface sterilized in 3 % (v/v) sodium hypochlorite for 5 min then rinsed in six changes of sterile distilled water [13]. The seeds were then pre-germinated on sterile moist vermiculite packed in Kilner jars for 3 days at 28 °C. Three seedlings were transplanted into sterilized Leonard jars using sterile forceps and later thinned to two. Eight days after transplanting, the seedlings were inoculated with (1 ml) broth culture of each representative rhizobia isolates. The rhizobia isolates were cultured in Yeast Extract Mannitol Broth (YEMB) for three days (to exponential phase) before inoculation. Leonard jars inoculated with commercial Bradyrhizobium sp. strain USDA 3456 were used as positive controls. Jars of un-inoculated seedlings were used as negative controls. The experiment was laid out in a randomized complete block design and each treatment replicated four times. The nitrogen free media was replenished every week.
After 45 days the plants were harvested. Vermiculite and liquid medium were emptied out of the cup and Leonard jars. Plant roots were washed under running tap water to rinse off vermiculite and the attached wick removed. The plants were scored for presence and absence of nodules. The presence of a single nodule in a Leonard jar for any plant was viewed as evidence that the isolate was rhizobia [16].
2.7. Determination of symbiotic efficiency of representative rhizobia isolates
This experiment was also performed in the even span greenhouse described in sub-section 2.3 above. Sterilization, pre-germination, transplantation and thinning of cowpea seeds in Leonard jars was done as described in the authentication experiment above. After eight days, one milliliter (1 ml) broth culture of each authenticated rhizobia isolate and a reference strain Bradyrhizobium sp. strain USDA 3456 were inoculated onto the seedlings. Plants inoculated with the reference strain (Bradyrhizobium sp. strain USDA 3456) and those in Leonard jars supplied with nitrogen (sterile 0.05 % KNO3 solution) were used as positive controls. Non-inoculated plants in Leonard jars supplied with nitrogen free media were used as negative controls. The experiment was arranged in a randomized complete block design and each treatment was replicated five times. Nutrient medium in the jars was replenished every week. Harvesting was done after 45 days using procedures proposed by Beck et al. [14]. Shoots were separated from the roots after which the roots were carefully washed with tap water. All the nodules were detached and counted. Apart from nodule number (NN), nodule dry weight (NDW), shoot dry weight (SDW), and root dry weight (RDW) were recorded. Symbiotic efficiency (SE %) was calculated by (dividing the total dry weight of the inoculated plants with the total dry weight of non-inoculated control plants supplemented with nitrogen (0.05 % KNO3) × 100 [16].
2.8. Genomic DNA extraction and 16S rRNA gene amplification
Rhizobia cultures were re-suspended in eppendorf tubes containing 400 μl of normal saline to remove polymerase chain reaction (PCR) inhibitors like exopolysaccharides (EPS). The mixture was vortexed for 20 s then centrifuged at 13,000 rpm for 10 min. The supernatant was poured out leaving the cell pellets. These pellets were washed four times with normal saline, then harvested and re-suspended in 400 μl of genomic lysis buffer. This mixture was incubated in a water bath set at 65 °C for 30 min. This was followed by centrifugation at 13,000 rpm for 5 min and the supernatant transferred into another sterilized eppendorf tubes. Four hundred microliters of isopropanol was then added to the supernatant and samples incubated at -20 °C. The samples were then centrifuged at 13,000 rpm for 3 min and the isopropanol discarded. Thereafter, 400 μl of DNA buffer (70 % alcohol) was added to the pellet, centrifuged at 13,000 rpm for 1 min and the liquid phase decanted out gently. The pellets were then air-dried followed by dissolving the DNA in 50 μl of elution buffer (TE). The quality and quantity of DNA was determined by resolving SYBR green stained DNA in 0.8 % agarose gel and observing the DNA bands in a UV trans illuminator. Extracted DNA was stored at -20 °C [9].
PCR was carried out in a 25 μl reaction volume containing 9.0 μl sterile PCR water, 1.25 μl of 10 μM primer 1492 R and 1.25 μl of 10 μM primer 27 F, 12.5 μl One Taq 2X master mix with standard buffer (Biolabs) and 1.0 μl of DNA template. Sequence for the forward primer was (27F 5′-AGAGTTTGATCCTGGCTCAG-3′) while the reverse primer was (1492R 5′-GGTTACCTTGTTACGACTT-3′) [17]. The PCR reaction was carried out in a Techgene thermocycler, FTGENE5D model (Techne UK). The PCR conditions for amplification were as follows: An Initial DNA denaturation at 94 °C for 2 min then denaturation at 94 °C for 45 s, annealing at 62 °C for 45 s and extension at 72 °C for 2 min (35 cycles). Final extension was carried out at 72 °C for 5 min. Amplified DNA was held at 4 °C [9].
The PCR products were separated by gel electrophoresis in 1.4 % agarose in 0.5X TBE buffer at 80 V for 1 h and stained with SYBR-Green. A 1kb DNA ladder (Biolabs) was used to estimate the molecular sizes of the bands. Gel visualization was done using a UV trans-illuminator and photographed using a digital camera.
2.9. 16S rRNA gene sequencing and analysis
The PCR products were sent to Inqaba Biotech in (Pretoria) South Africa for purification and Sanger sequencing using a ABI 3730 DNA sequencer (Applied Biosystems, USA). Base calling for sequenced data was done using Finch Tv software [18]. Consensus sequences were created using BioEdit software [19] after which the sequences obtained were compared with sequences in the National Centre for Biotechnological Information (NCBI) GenBank database using Basic Local Arrangement Search Tool (BLAST) program. The sequence identities that were closely related to the sequences of the study isolates were retrieved from the NCBI sequence data repository. The contig and corresponding retrieved sequences were then aligned using clustal W. The evolutionary history of the isolates was then presented in the form of a phylogenetic tree by neighbor joining method using Molecular Evolutionary Genetics Analysis (MEGA) 7 software [20].
2.10. Statistical analyses
For cowpea genotypes, data on the number of nodules, nodule dry weight, shoot dry weight and root dry weight was analyzed using two-way analysis of variance (ANOVA). Means were separated by Tukey's HSD test at 5 % probability level. All ANOVAs and post hoc tests were carried out using SAS software version 9.2 [21]. Wherever feasible, data was log (x+1) transformed to fulfill the assumptions of ANOVA.
3. Results
3.1. Morphological characteristics of representative rhizobia isolates
All the cowpea genotypes exhibited bacterial communities in their root nodules. A total of 94 root nodule isolates were obtained and placed into 20 groups based on biochemical and morphological characteristics as published in Njeru et al. [22]. The 20 groups were code-named M1 to M20.
3.2. 16S rRNA gene characterization of rhizobia isolates
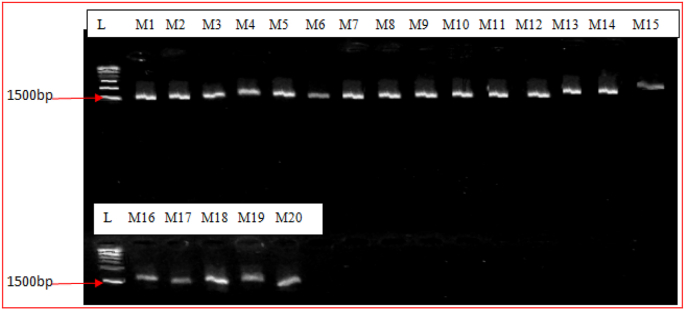
The 16S rRNA gene sequencing showed that the study isolates belong to 20 different bacterial strains including Rhizobium and other non rhizobial endophytes. The amplified region of the PCR products showed definite and appropriately sized band in all lanes at approximately at 1500 base pairs (Figure 1) when resolved in 1.4 % agarose gel at 80 V for 1 h.
Figure 1.
Gel electrophoresis image of PCR amplified 16S rRNA gene of 20 bacterial isolates on 1.4 % agarose gel. L, 1 kb DNA ladder from New England Biolabs, USA; size is indicated on the left hand margin. Lane M1-M20, bacterial isolates from cowpea root nodules.
Alignment and sequence comparison of the 16S rRNA gene study sequences with the obtained known bacterial sequences in the NCBI database showed that the study isolates were closely related to known bacterial lineages (Table 1). The genus Rhizobium made up 45 % of the total number of isolates. BLAST revealed the presence of Rhizobium alamii strain S10001 (99.71 % 16S rRNA gene sequence similarity with isolate M1); Rhizobium mesosinicum strain CCBAU 25010 (99.09 % 16S rRNA gene sequence similarity with isolate M3); Rhizobium sp. strain 1616 (99.93 % 16S rRNA gene sequence similarity with isolate M5);Rhizobium sp. L120T (100 % 16S rRNA gene sequence similarity with isolate M6); Rhizobium tropici strain SY137 (91.16 % 16S rRNA gene sequence similarity with isolate M11); Rhizobium pusense strain CFBP5875 (99.85 % 16S rRNA gene sequence similarity with isolate M17); Rhizobium pusense strain WTB176 (99.78 % 16S rRNA gene sequence similarity with isolate M18) and Rhizobium sp. strain CM-CNRG 562 (99.85 % 16S rRNA gene sequence similarity with M20).
Table 1.
Bacterial isolates from cowpea nodules showing sequence similarity (%) with NCBI database bacterial strains after 16s rRNA gene sequencing.
| Laboratory designation | Species/strain identification | Genbank Accession number | 16S rRNA gene similarity (%) | Sizes of sequences |
|---|---|---|---|---|
| M1 | Rhizobium alamii strain S10001 (MF977610.1) | MT775434 | 99.71 | 1350 |
| M2 | Paraburkholderia phenoliruptrix BR3459a (CP003864.1) | MT775435 | 99.86 | 1390 |
| M3 | Rhizobium mesosinicum strain CCBAU 25010 (NR043548.1) | MT775436 | 99.09 | 1330 |
| M4 | Enterobacter sp. strain PB-1121-E (MK208589.1) | MT775437 | 98.73 | 1410 |
| M5 | Rhizobium sp. strain 1616 (MK280695.1) | MT775438 | 99.93 | 1350 |
| M6 | Rhizobium sp. L120T (KM894194.1) | MT775439 | 100 | 1350 |
| M7 | Enterobacter cloacae strain IAE252 (MK414959.1) | MT775440 | 99.56 | 1410 |
| M8 | Enterobacter hormaechei strain E11 (KF145192.1) | MT775441 | 99.71 | 1410 |
| M9 | Enterobacter cloacae strain VITPSSJ (KP305908.1) | MT775442 | 99.93 | 1400 |
| M10 | Pseudomonas putida strain AR4 (KX343951.1) | MT775443 | 99 | 1480 |
| M11 | Rhizobium tropici strain SY137 (KP687380.1) | MT775444 | 91.16 | 1270 |
| M12 | Enterobacter oryziphilus strain REICA_084 (JF795012.1) | MT775445 | 99.64 | 1410 |
| M13 | Enterobacter sp. JFZ-10 (KT446410.1) | MT775446 | 99.9 | 1400 |
| M14 | Stenotrophomonas maltophilia strain W9-4 (MG905303.1) | MT775447 | 99.86 | 1400 |
| M15 | Enterobacteriaceae sp. strain HXDJ-2S1 (MH135802.2:1) | MT775448 | 87.34 | 1410 |
| M16 | Enterobacter ludwigii strain OS5.4 (KX242269.1) | MT775449 | 94.96 | 1410 |
| M17 | Rhizobium pusense strain CFBP5875 (CPO39894.1) | MT775450 | 99.85 | 1350 |
| M18 | Rhizobium pusense strain WTB176 (MK734334.1) | MT775451 | 99.78 | 1350 |
| M19 | Enterobacter sp. HK169 (CPO17087.1) | MT775452 | 99.93 | 1390 |
| M20 | Rhizobium sp. strain CM-CNRG 562 (MK108017.1) | MT775453 | 99.85 | 1360 |
The 16S rRNA nucleotide sequences for the isolates in this study (M1-M20) have been deposited in NCBI Genbank database under accession numbers MT775434-MT775453. They can be accessed through: http://www.ncbi.nlm.nih.gov/nuccore/?term=MT775434:MT775453%5baccn].
Apart from rhizobia, there were associative endophytic bacterial strains which represented 55 % of the total isolates (Table 1). Nine of the isolates (M4, M7, M8, M9, M12, M13, M15, M16, M19) had 87 %–100 % sequence similarity with members of genus Enterobacter. Isolate M2 had 99.86 % sequence similarity with Paraburkholderia phenoliruptrix BR3459a. Isolate M14 had a sequence similarity of 99.86 % with Strenotrophomonas maltophila while isolate M10 had a sequence similarity of 99 % with Pseudomonas putida strain AR4 (Table 1).
Evolution relatedness of the isolated rhizobia and other non rhizobial endophytes was evaluated based on the 16S rRNA gene. The phylogenetic tree, based on the neighbour joining method clustered the strains into four main clusters (Clusters A, B, C and D) (Figure 2). The grouping was based on the strains’ similarity in relation to their 16S rRNA gene composition. Cluster A, supported by a bootstrap value of 100 % comprised of isolates M1, M3, M5, M17, M18, M20, M6, and M11. The isolates in this cluster were closely related to members of genus Rhizobium (Figure 2) and all strains showed potential to nodulate cowpea. Cluster B comprised of isolates M2 and M14. This cluster was supported by a bootstrap value of 81 % with only isolate M2 eliciting nodulation in cowpea. Members for this cluster were closely related to members of the genera Paraburkholderia and Strenotrophomonas (Figure 2). Cluster C exclusively contained Pseudomonas putida strain AR4. This cluster was supported by bootstrap value of 100 %. Cluster D supported with a bootstrap value of 100 % comprised of eleven isolates (M4, M12, M13, M8, M9, M19, M15, M7 and M16) belonging to the Enterobacter spp. (Figure 2).
Figure 2.
A dendogram showing the phylogenetic relationship of 20 bacterial isolates from cowpea plants cultivated in lower Eastern Kenya. The tree was inferred using the Neighbor-Joining method after 16s rRNA gene sequencing of the bacterial isolates. The dendogram was constructed using MEGA 7 software. Percentage bootstrap values for 1000 iterations are shown at the nodes. Only bootstrap values ≥40 % are shown. The evolutionary distances were computed using Maximum Composite Likelihood method. The 20 isolates designated M1-M20 formed four clusters which were labeled A, B, C and D.
3.3. Authentication and symbiotic efficiency of representative isolates
The ability of nodule isolates to induce nodule formation in the host plant authenticates the isolates as rhizobia. The results in this study confirmed that only nine isolates namely (M1, M2, M3, M5, M6, M11, M17, M18 and M20) were rhizobia and they nodulated the host plant. Eleven isolates (M4, M7, M8, M9, M10, M12, M13, M14, M15, M16, and M19) did not elicit nodulation and were established to be non-rhizobial endophytes (NRE). The nine rhizobia isolates were tested for their effectiveness and the results showed that there were significant differences in nodule number (p < 0.0001), nodule dry weight (p < 0.0001), root dry weight (p = 0.0006) and shoot dry weight (p < 0.0001) of cowpea (Table 2). Cowpea plants inoculated with isolate M3 recorded the highest mean nodule number (46.00 ± 7.25) which was statistically similar to isolate M2 and REF while plants inoculated with isolate M5 had the lowest mean number of nodules (2.50 ± 2.50). As expected, the un-inoculated control and the nitrogen supplemented control plants did not nodulate. Isolate M2 registered the highest nodule dry weight (0.22 ± 0.09) which was statistically different from all other isolates except isolates M3, M6, and REF. Inoculation with the different isolates augmented shoot dry weight (SDW) of the plants. Isolate M2 recorded the highest shoot dry weight (0.85 ± 0.08) which was statistically different from all other isolates except M3, REF and PC while isolate M1 recorded the lowest shoot dry weight (0.13 ± 0.03). There was a significant difference (p < 0.0001) in symbiotic efficiency of representative isolates (Table 2). However, none of the isolates had symbiotic effectiveness (SE) above 100 % in comparison with the nitrogen supplemented control (PC). Isolate M2 had the highest SE of 82.49 % followed by isolate M3 (72.76 %). However, the SE of isolate M2 and M3 was not statistically different from that of the reference strain (REF) and PC. Isolates with SE > 80 % were ranked as most effective (isolate M2), while those with SE between 51-80 % were listed as effective (isolate M3) and the rest of the isolates with SE < 50 % were classified as moderately effective.
Table 2.
Symbiotic efficiency of native rhizobia isolates inoculated on cowpea plants and their effect on nodule number, shoot, root and nodule dry weight.
| Isolate | Nodule Number | Nodule Dry Weight (g plant−1) | Shoot Dry Weight (g plant−1) | Root Dry Weight (g plant−1) | Symbiotic Efficiency (%) |
|---|---|---|---|---|---|
| M1 | 4.50(4.50)c | 0.01(0.01)bc | 0.13(0.03)d | 0.07(0.02)c | 12.85(2.90)d |
| M2 | 38.50(9.98)ab | 0.22(0.09)a | 0.85(0.08)ab | 0.30(0.03)a | 82.49(7.64)ab |
| M3 | 46.00(7.25)a | 0.08(0.02)abc | 0.75(0.07)ab | 0.16(0.01)abc | 72.76(6.38)ab |
| M5 | 2.50(2.50)c | 0.00(0.00)c | 0.17(0.04)d | 0.16(0.05)abc | 16.49(4.26)d |
| M6 | 21.25(8.75)bc | 0.11(0.04)abc | 0.49(0.07)bcd | 0.09(0.01)bc | 47.59(7.08)bcd |
| M11 | 6.25(2.43)c | 0.01(0.00)bc | 0.21(0.03)d | 0.16(0.00)abc | 20.77(2.76)d |
| M17 | 8.50(4.97)c | 0.02(0.01)bc | 0.25(0.06)cd | 0.08(0.02)c | 24.76(5.85)cd |
| M18 | 3.50(3.50)c | 0.01(0.01)c | 0.18(0.04)d | 0.17(0.02)abc | 17.06(3.82)d |
| M20 | 4.50(4.50)c | 0.01(0.01)bc | 0.21(0.05)d | 0.13(0.05)abc | 20.27(4.97)d |
| REF | 34.50(9.10)ab | 0.14(0.07)ab | 0.70(0.15)abc | 0.22(0.04)abc | 67.68(14.60)abc |
| PC | 0.00(0.00)c | 0.00(0.00)c | 1.03(0.34)a | 0.29(0.11)ab | 100.00(32.61)a |
| NC |
0.00(0.00)c |
0.00(0.00)c |
0.24(0.03)cd |
0.09(0.03)bc |
22.86(3.25)cd |
| P value | p < 0.0001 | p < 0.0001 | p < 0.0001 | p = 0.0006 | p < 0.0001 |
Values followed by the same letters within the column are not significantly different from each other according to Tukey's Honest Significant Difference (HSD) at 5 % level. NC-Non inoculated plants, REF- Rhizobia reference strain (Bradyrhizobium sp. strain USDA 3456), PC-un-inoculated control plants supplemented with nitrogen (0.05 % KNO3). SE % = (Total dry weight of inoculated plant/Total dry weight of PC-un-inoculated control plants) x 100. Standard error is indicated in parentheses.
4. Discussion
4.1. Morphological characteristics of indigenous rhizobia isolates
In the present study, we identified rhizobial and non rhizobial bacterial isolates that inhabit cowpea nodules and their potential to stimulate nodulation and growth of the plants. Full data or information on morphological characteristics of the 20 isolates in this study was reported in the publication Njeru et al. [22].
4.2. Symbiotic nitrogen fixation efficiency of the native rhizobia isolates
A substantial increase in dry weights and nodule numbers was noted in plants inoculated with native rhizobia strains in comparison with the un-inoculated control. Conclusions made from studies by Yusif et al. [23] and Boddey et al. [24] collaborate these findings. The likelihood of the rhizobia-legume association being synergistic and benefiting the host has been shown to be determined by the effectiveness of the rhizobia isolates used [25]. Absence of nodules in the un-inoculated and nitrogen supplemented control demonstrated absence of external contamination [15]. The variation in symbiotic efficiency observed among the rhizobia isolates in this study was similarly reported in native rhizobia population nodulating cowpea in Kenya [25]. According to Girija et al. [26] differences in symbiotic efficiency among rhizobia might be due to host specificity. In the present study SE ranged from 12.85 % to 82.49 %. Kawaka et al. [27] conducted a similar study on 16 native rhizobia isolates from common bean nodules and reported SE of the isolates ranged from 67 % to 164 %. Two isolates (M2 and M3) exhibited higher performance in terms of nodulation, SDW and SE. This reveals the existence of ablest native rhizobia isolates from Kenyan semi-arid regions and it can be suggested that these isolates are highly adapted to the functions required for effective bacterial survival and colonization within the rhizosphere in response to plant signals [28]. Isolate M2 had an SE of 82.49 % and based on Lalande et al. [29] guidelines on symbiotic efficiency, isolates are considered highly effective only if they record symbiotic efficiency >80 %. This resonates the potential of isolate M2 to be formulated into a cost effective inoculum for cowpea and adapted by the small holder farmers in the arid and semi-arid regions.
The commercial inoculant, Bradyrhizobium sp. strain USDA 3456 showed better performance in terms of nodulation, SDW and SE than most of the isolates. Similar results were reported in a study conducted by Kyei-Boahen et al. [6]. The authors noted that cowpea plants inoculated with Bradyrhizobium sp. strain USDA 3456 registered higher nodulation, SDW and symbiotic efficiency when compared to those treated indigenous Bradyrhizobium spp. Additionally some Bradyrhizobia species have been reported to possess plant growth promoting traits such as phosphorus solubilization and production of IAA hormone that expedites cell elongation and differentiation therefore, making them superior to others [30].
4.3. 16S rRNA gene characterization of nodule isolates
In the present study, analysis of 16S rRNA gene was done to investigate the diversity of bacterial communities associated with nodules from cowpea plants grown in the semi-arid regions of Kenya. In our study eight different bacteria strains from the class Alphaproteobacteria, genus Rhizobium were detected. These findings are supported by previous studies by Jaramillo et al. [31], Castro et al. [32] and Ngeno et al. [33] who demonstrated the ability of cowpea to trap rhizobia from soils under different agricultural systems. Our findings also support the “promiscuous” nature of cowpea to nodulate with a mélange of rhizobia isolates which also aids the plant in thriving in diverse environs where other legumes may not be able to survive [32]. However, these findings contradict previous conclusions made that Bradyrhizobium is the primary symbiont of cowpea [34]. Low numbers of Bradyrhizobium strains have been reported to nodulate legumes in arid and semi-arid areas when compared to other genera [35]. Factors such as soil-climatic conditions have been suggested to affect Bradyrhizobia abundance and diversity in sub-Saharan Africa [36].
Based on results on symbiotic efficiency all the rhizobia strains showed potential to nodulate cowpea. However, phylogenetically not all the rhizobia strains were closely related. This suggests that rhizobia strains, from the study area, with the potential to nodulate cowpea are not restricted to a phylogenetic group [37]. Therefore, these strains might have divergently evolved to colonize root nodules of different cowpea genotypes but retaining their critical genes that code for the nodulation of cowpea. Lack of phenotypic and genotypic correlation in rhizobia isolates nodulating cowpeas in Indian soils has been reported before by Arora et al. [37]. According to Gratten et al. [38] genetic changes can cause pleiotropic effects on several traits simultaneously thus affecting the resulting phenotypes. Isolate M2, identified as Paraburkholderia phenoliruptrix BR3459a was unique as it showed ability to nodulate cowpea and recorded high efficiency. To our knowledge, we report for the first time a Beta-Rhizobia in the genus Parabulkholdreia that nodulates cowpea in Kenya and Africa. This strain could be developed into an effective cowpea inoculum after evaluating its genetic stability, its potential to nodulate cowpea in presence of background rhizobia and its ability to survive in inoculum carrier material. There are several works documenting nodulation of legumes in the sub-family Papilionoideae such as cowpea by members of β-Proteobacteria in Brazil [32] and Venezuela [39]. In Africa Beta-rhizobia genus Paraburkholderia with symbiotic properties have been isolated from legumes such as Aspalathus linearis in South Africa [40]. So far there has been no citing of Beta-Rhizobia nodulating cowpea in Africa. Majority of the studies done have reported members of genera Bradyrhizobium, Rhizobium, Ensifer and Mesorhizobium as microsymbionts of cowpea [41]. Occurrence of cowpea nodulating Paraburkholderia spp. in Kenyan soils could be attributed to horizontal gene transfer between α and β proteobacteria [39]. Genus Paraburkholderia have also been reported to tolerate and dominate in environments with high aluminium contents, low soil fertility and low soil pH and this can form a basis for utilization of these bacteria as bio fertilizers [42].
The non-rhizobial endophytes detected in the cowpea nodules in this study belonged to class Gammaproteobacteria and genus Pseudomonas, Strenotrophomonas and Enterobacter. These findings support previous reports by Chidebe et al. [43] and Leite et al. [44] who documented on diversity of non-rhizobial endophytes (NRE) associated with cowpea root nodules. These non-rhizobial bacterial strains have been previously isolated from Vigna unguiculata nodules in Brazil [44] and from Phaesolus vulgaris [45] in Western Kenya. Occurrence of these NRE in the present study may be attributed to compatibility of the microbe with the host plant and that each microbe occupies a different ecological niche in the root nodule [46]. Generally, most NRE are nonpathogenic however some for instance Enterobacter, Strenotrophomonas, and Burkholderia have been profiled as mammalian pathogens [47]. None of the NREs in this study caused nodulation in cowpea plants. Their presence in cowpea rhizosphere is not accidental and similar findings have been documented by Castro et al. [32].These NREs are capable of entering nodule infection threads and have been reported to coexist with rhizobia strains as antagonists or they can cause synergistic effects between host-microbial associations [48]. Through production of iron chelating siderophores, providing intrinsic resistance to heavy metals, and increasing nitrogen content in plants these NREs have been reported to enhance symbiosis between rhizobia and host plants [49, 50, 51].
5. Conclusion
This study demonstrated great diversity of bacterial isolates that nodulate different cowpea genotypes cultivated in Kenya. Documented for the first time in Africa is an efficient cowpea nodulating Beta-Rhizobia (Paraburkholderia phenoliruptrix BR3459a) coded isolate M2. Isolate M3 (Rhizobium mesosinicum strain CCBAU 25010) also depicted high nodulation and SE. These two isolates can be used for the development of low cost native microbial inocula after subjecting them to further field trials so as to establish their competitiveness and genetic stability under different environmental conditions.
Declarations
Author contribution statement
Mercy Martha Muindi, Morris Muthini: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Ezekiel Mugendi Njeru, John Maingi: Conceived and designed the experiments; Wrote the paper.
Funding statement
This work was supported by The World Academy of Science (TWAS) (Grant number 16-180; RG/BIO/AF/AC_I), The Future Leaders –African Independent Researchers (FLAIR) Fellowship Programme, which is a partnership between the African Academy of Sciences and the Royal Society funded by the UK Government's Global Challenges Research Fund (Grant number FLR∖R1∖190944); and, Kenyatta University Vice-Chancellor's Research Grant.
Data availability statement
Data associated with this study has been deposited at NCBI database (http://www.ncbi.nlm.nih.gov/nuccore/?term=MT775434:MT775453%5baccn) under the accession numbers MT775434-MT775453. The data supporting this study can be accessed at Dryad Digital Repository. Dryad for review URL: https://datadryad.org/stash/resources/82332/review. Alongside is the Dryad DOI: https://doi.org/10.5061/dryad.c866t1g4x [dataset [52].
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
The authors wish to thank farmers from Kitui and Embu counties who participated in the field work.
References
- 1.Faostat F. Publisher: FAO (Food and Agriculture Organization of the United Nations); Rome, Italy: 2017. FAOSTAT Statistical Database.http://www.fao.org/3/b-i6407e.pdf Accessed on. [Google Scholar]
- 2.Conti M.V., Campanaro A., Coccetti P., De Giuseppe R., Galimberti A., Labra M., Cena H. Potential role of neglected and underutilized plant species in improving women’s empowerment and nutrition in areas of sub-Saharan Africa. Nutr. Rev. 2019;77(11):817–828. doi: 10.1093/nutrit/nuz038. [DOI] [PubMed] [Google Scholar]
- 3.De Jager I., Borgonjen-Van Den Berg K.J., Giller K.E., Brouwer I.D. Current and potential role of grain legumes on protein and micronutrient adequacy of the diet of rural Ghanaian infants and young children: using linear programming. Nutr. J. 2019;18(1):1–12. doi: 10.1186/s12937-019-0435-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tani E., Abraham E., Chachalis D., Travlos I. Molecular, genetic and agronomic approaches to utilizing pulses as cover crops and green manure into cropping systems. Int. J. Math. Stat. 2017;18(6):1202. doi: 10.3390/ijms18061202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oruru M.B., Njeru E.M., Pasquet R., Runo S. Response of a wild-type and modern cowpea cultivars to arbuscular mycorrhizal inoculation in sterilized and non-sterilized soil. J. Plant Nutr. 2018;41(1):90–101. [Google Scholar]
- 6.Kyei-Boahen S., Savala C.E., Chikoye D., Abaidoo R. Growth and yield responses of cowpea to inoculation and phosphorus fertilization in different environments. Front. Plant Sci. 2017;8:646. doi: 10.3389/fpls.2017.00646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi R. Tripartite plant microbe and nutrients interaction in the below ground soil ecosystems. J. Med. Plants Stud. 2018;6(4):36–40. [Google Scholar]
- 8.Yanni Y., Zidan M., Dazzo F., Rizk R., Mehesen A., Abdelfattah F., Elsadany A. Enhanced symbiotic performance and productivity of drought stressed common bean after inoculation with tolerant native rhizobia in extensive fields. Agric. Ecosyst. Environ. 2016;232:119–128. [Google Scholar]
- 9.Koskey G., Mburu S.W., Kimiti J.M., Ombori O., Maingi J.M., Njeru E.M. Genetic characterization and diversity of Rhizobium isolated from root nodules of mid-altitude climbing bean (Phaseolus vulgaris L.) varieties. Front. Microbiol. 2018;9:968. doi: 10.3389/fmicb.2018.00968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ondieki D.K., Nyaboga E.N., Wagacha J.M., Mwaura F.B. Morphological and genetic diversity of Rhizobia nodulating cowpea (Vigna unguiculata L.) from agricultural soils of lower eastern Kenya. Internet J. Microbiol. 2017:1–9. doi: 10.1155/2017/8684921. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Westhoek A., Field E., Rehling F., Mulley G., Webb I., Poole P.S., Turnbull L.A. Policing the legume-Rhizobium symbiosis: a critical test of partner choice. Sci. Rep. 2017;7(1):1–10. doi: 10.1038/s41598-017-01634-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kisaka O.M., Mucheru-Muna M., Ngetich K.F., Mugwe N.J., Mugendi D., Mairura F. Rainfall variability, drought characterization, and effficacy of rainfall data reconstruction: case of Eastern Kenya. Adv. Meteorol. 2015;16:380404. [Google Scholar]
- 13.Muthini M., Maingi J.M., Muoma J.O., Amoding A., Mukaminega D., Osoro N., Mgutu A., Ombori O. Morphological assessment and effectiveness of indigenous rhizobia isolates that nodulate P. vulgaris in water hyacinth compost testing field in Lake Victoria basin. BJAST. 2014;4:718–738. [Google Scholar]
- 14.Beck D.P., Materon L.A., Afandi F. Practical Rhizobium-legume technology manual. Practical Rhizobium-legume technology manual. ICARDA Aleppo (Syria) 1993;19:389. [Google Scholar]
- 15.Mwenda B., Kiambi D., Kungu J., Van De Gevel J., Farda C., Morimoto Y. Vulnerability and adaptation strategies to drought and erratic rains as key extreme events: insights from small scale farming households in mixed crop agro ecosystems of semi-arid eastern Kenya. Afr. J. Agric. 2019;14(15):712–728. [Google Scholar]
- 16.Somasegaran P., Hoben H.J. Handbook for Rhizobia. Springer; New York, NY: 1994. Quantifying the growth of rhizobia; pp. 47–57. [Google Scholar]
- 17.Lane D.J. 16S/23S rRNA sequencing. In: Stackebrandt E., Goodfellow M., editors. Nucleic Acid Techniques in Bacterial Systematic. John Wiley Sons; New York: 1991. pp. 115–175. [Google Scholar]
- 18.Treves D.S. Review of three DNA analysis applications for use in the microbiology or genetics classroom. JM&BE. 2010;11(2):186. doi: 10.1128/jmbe.v11i2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall T.A. Nucleic Acids Symp. Ser. Vol. 41. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; pp. 95–98. [Google Scholar]
- 20.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. Mega X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan Y. Multiple imputations using SAS software. J. Stat. Software. 2011;45(6):1–25. [Google Scholar]
- 22.Njeru E.M., Morris M., Mercy M.M., Omwoyo O., Shem B.N., Steve R., John M.M. Climate Impacts on Agricultural and Natural Resource Sustainability in Africa. Springer; Cham: 2020. Exploiting arbuscular mycorrhizal fungi-rhizobia-legume symbiosis to increase smallholder farmers’ crop production and resilience under a changing climate; pp. 471–488. [Google Scholar]
- 23.Yusif S.A., Muhammad I., Hayatu N.G., Sauwa M.M., Tafinta I.Y., Mohammed M.A., Lukman S.A., Abubakar G.A., Hussain A.M. Effects of biochar and rhizobium inoculation on nodulation and growth of groundnut in Sokoto State, Nigeria. JALSI. 2016;9:1–9. [Google Scholar]
- 24.Boddey R.M., Fosu M., Atakora W.K., Miranda C.H., Boddey L.H., Guimaraes A.P., Ahiabor B.D. Cowpea (Vigna unguiculata) crops in Africa can respond to inoculation with rhizobium. Ex agric. 2017;53(4):578–587. [Google Scholar]
- 25.Nyaga J.W., Njeru E.M. Potential of native rhizobia to improve cowpea growth and production in semiarid regions of Kenya. Front. Agron. 2020;2:606293. [Google Scholar]
- 26.Girija D., Panchami P.S., Jose P.E., Saeed T., Nair S.S. Isolation and characterization of native cowpea rhizobia from Wayanad India. Legume Research-An International Journal. 2020;43(1):126–133. [Google Scholar]
- 27.Kawaka F., Dida M.M., Opala P.A., Ombori O., Maingi J., Osoro N., Muthini M., Amoding A., Mukaminega D., Muoma J. Symbiotic efficiency of native rhizobia nodulating common bean (Phaseolus vulgaris L.) in soils of Western Kenya. Int. Sch. Res. Notices. 2014;8:1–8. doi: 10.1155/2014/258497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barret M., Morrissey J.P., O’gara F. Functional genomics analysis of plant growth-promoting rhizobacterial traits involved in rhizosphere competence. Biol. Fertil. Soils. 2011;47(7):729–743. [Google Scholar]
- 29.Lalande R., Bigwaneza P.C., Antoun H. Symbiotic effectiveness of strains of Rhizobium leguminosarum biovar phaseoli isolated from soils of Rwanda. Plant Soil. 1990;121(1):41–46. [Google Scholar]
- 30.Badawi F.S.F., Biomy A.M.M., Desoky A.H. Peanut plant growth and yield as influenced by co-inoculation with Bradyrhizobium and some rhizo-microorganisms under sandy loam soil conditions. Ann. Agric. Sci. (Cairo) 2011;56(1):17–25. [Google Scholar]
- 31.Jaramillo P.M.D., Guimarães A.A., Florentino L.A., Silva K.B., Nóbrega R.S.A., Moreira F.M.D.S. Symbiotic nitrogen-fixing bacterial populations trapped from soils under agroforestry systems in the Western Amazon. Sci. Agric. 2013;70(6):397–404. [Google Scholar]
- 32.Castro J.L.D., Souza M.G., Rufini M., Guimarães A.A., Rodrigues T.L., Moreira F.M.D.S. Diversity and efficiency of rhizobia communities from iron mining areas using cowpea as a trap plant. Rev. Bras. Ciênc. Solo. 2017;41 [Google Scholar]
- 33.Ngeno J.K., Chemining’wa G.N., Hutchinson M.J. Symbiotic efficiency of commercial and native rhizobia of Cowpea (Vigna unguiculata L.) in South Western Kenya. AJAAR. 2018;8:1–17. [Google Scholar]
- 34.Sena P.T.S., do Nascimento T.R., Lino J de O.S., Oliveira G.S., Ferreira Neto R.A., de Freitas A.D.S. Molecular, physiological, and symbiotic characterization of cowpea rhizobia from soils under different agricultural systems in the semiarid region of Brazil. J. Soil Sci. Plant Nutr. 2020:1–15. [Google Scholar]
- 35.Li M., Li Y., Chen W.F., Sui X.H., Li Y., Jr., Li Y., Wang E.T., Chen W.X. Genetic diversity, community structure and distribution of rhizobia in the root nodules of Caragana spp. from arid and semi-arid alkaline deserts, in the north of China. Syst. Appl. Microbiol. 2012;35(4):239245. doi: 10.1016/j.syapm.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 36.Grönemeyer J.L., Reinhold-Hurek B. Diversity of Bradyrhizobia in sub-sahara Africa: a rich resource. Front. Microbiol. 2018;9:2194. doi: 10.3389/fmicb.2018.02194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arora N.K., Khare E., Singh S., Tewari S. Phenetic, genetic diversity and symbiotic compatibility of rhizobial strains nodulating pigeon pea in Northern India. 3 Biotech. 2018;8(1):52. doi: 10.1007/s13205-017-1074-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gratten J., Visscher P.M. Genetic pleiotropy in complex traits and diseases: implications for genomic medicine. Genome Med. 2016;8(1):78. doi: 10.1186/s13073-016-0332-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramírez M.D.A., España M., Lewandowska S., Yuan K., Okazaki S., Ohkama-Ohtsu N., Yokoyama T. Phylogenetic analysis of symbiotic bacteria associated with two Vigna species under different agro-ecological conditions in Venezuela. Microb. Environ. 2020;35(1):ME19120. doi: 10.1264/jsme2.ME19120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassen A.I., Bopape F.L., Habig J., Lamprecht S.C. Nodulation of rooibos (Aspalathus linearis Burm.f), an indigenous South African legume by members of both the α-Proteobacteria and β-Proteobacteria. Biol. Fertil. Soils. 2012;48:295–303. [Google Scholar]
- 41.Valdez-Nuñez R.A., Castro-Tuanama R., Castellano-Hinojosa A., Bedmar E.J., Ríos-Ruiz W.F. Microbial Probiotics for Agricultural Systems. Springer; Cham: 2019. PGPR Characterization of non-nodulating bacterial endophytes from root nodules of Vigna unguiculata (L.) Walp; pp. 111–126. [Google Scholar]
- 42.Dall'Agnol R.F., Plotegher F., Souza R.C., Mendes I.C., dos Reis Junior F.B., Béna G., Moulin L., Hungria M. Paraburkholderia nodosa is the main N2-fixing species trapped by promiscuous common bean (Phaseolus vulgaris L.) in the Brazilian ‘Cerradão’. FEMS Microbiol. Ecol. 2016;92(8):fiw108. doi: 10.1093/femsec/fiw108. [DOI] [PubMed] [Google Scholar]
- 43.Chidebe I.N., Jaiswal S.K., Dakora F.D. Distribution and phylogeny of microsymbionts associated with cowpea (Vigna unguiculata) nodulation in three agroecological regions of Mozambique. Appl. Environ. Microbiol. 2018;84(2):e01712–e01717. doi: 10.1128/AEM.01712-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leite J., Fischer D., Rouws L.F., Fernandes-Júnior P.I., Hofmann A., Kublik S., Schloter M., Xavier G.R., Radl V. Cowpea nodules harbor non-rhizobial bacterial communities that are shaped by soil type rather than plant genotype. Front. Plant Sci. 2017;7:2064. doi: 10.3389/fpls.2016.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kawaka F., Makonde H., Dida M., Opala P., Ombori O., Maingi J., Muoma J. Genetic diversity of symbiotic bacteria nodulating common bean (Phaseolus vulgaris) in western Kenya. PloS One. 2018;13(11) doi: 10.1371/journal.pone.0207403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Meyer S.E., De Beuf K., Vekeman B., Willems A. A large diversity of non-rhizobial endophytes found in legume root nodules in Flanders (Belgium) Soil Biol. Biochem. 2015;83:1–11. [Google Scholar]
- 47.Martínez-Hidalgo P., Hirsch A.M. The nodule microbiome: N2-fixing rhizobia do not live alone. Phytobiomes. 2017;1(2):70–82. [Google Scholar]
- 48.Wekesa C.S., Okun D., Juma K., Shitabule D., Okoth P., Nyongesa P., Katoo A., Mulamu S., Wamalwa E., Mahalo C., Koyo M. Abundance and symbiotic potential of common bean (Phaseolus vulgaris) nodule associated bacteria in western Kenya soil. J. Agric. Sci. 2016;1:1–9. [Google Scholar]
- 49.Ryan R.P., Monchy S., Cardinale M., Taghavi S., Crossman L., Avison M.B., Berg G., Van Der Lelie D., Dow J.M. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 2009;7:514–525. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 50.Singh R.P., Jha P.N. The PGPR Stenotrophomonas maltophilia SBP-9 augments resistance against biotic and abiotic stress in wheat plants. Front. Microbiol. 2017;8:1945. doi: 10.3389/fmicb.2017.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H.B., Singh R.K., Singh P., Song Q.Q., Xing Y.X., Yang L.T., Li Y.R. Genetic diversity of nitrogen-fixing and plant growth promoting Pseudomonas species isolated from sugarcane rhizosphere. Front. Microbiol. 2017;8:1268. doi: 10.3389/fmicb.2017.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muindi M.M., Muthini M., Njeru E.M., Maingi J. 2020. Symbiotic Efficiency and Genetic Characterization of Bacteria Associated with Cowpea Genotypes Grown in Semiarid Tropics of Kenya. (dataset) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data associated with this study has been deposited at NCBI database (http://www.ncbi.nlm.nih.gov/nuccore/?term=MT775434:MT775453%5baccn) under the accession numbers MT775434-MT775453. The data supporting this study can be accessed at Dryad Digital Repository. Dryad for review URL: https://datadryad.org/stash/resources/82332/review. Alongside is the Dryad DOI: https://doi.org/10.5061/dryad.c866t1g4x [dataset [52].