Abstract
Accumulation of amyloid beta peptides is thought to initiate the pathogenesis of Alzheimer's disease. However, the precise mechanisms mediating their neurotoxicity are unclear. Our microarray analyses show that, in Drosophila models of amyloid beta 42 toxicity, genes involved in the unfolded protein response and metabolic processes are upregulated in brain. Comparison with the brain transcriptome of early-stage Alzheimer's patients revealed a common transcriptional signature, but with generally opposing directions of gene expression changes between flies and humans. Among these differentially regulated genes, lactate dehydrogenase (Ldh) was up-regulated by the greatest degree in amyloid beta 42 flies and the human orthologues (LDHA and LDHB) were down-regulated in patients. Functional analyses revealed that either over-expression or inhibition of Ldh by RNA interference (RNAi) slightly exacerbated climbing defects in both healthy and amyloid beta 42-induced Drosophila. This suggests that metabolic responses to lactate dehydrogenase must be finely-tuned, and that its observed upregulation following amyloid beta 42 production could potentially represent a compensatory protection to maintain pathway homeostasis in this model, with further manipulation leading to detrimental effects. The increased Ldh expression in amyloid beta 42 flies was regulated partially by unfolded protein response signalling, as ATF4 RNAi diminished the transcriptional response and enhanced amyloid beta 42-induced climbing phenotypes. Further functional studies are required to determine whether Ldh upregulation provides compensatory neuroprotection against amyloid beta 42-induced loss of activating transcription factor 4 activity and endoplasmatic reticulum stress. Our study thus reveals dysregulation of lactate dehydrogenase signalling in Drosophila models and patients with Alzheimer's disease, which may lead to a detrimental loss of metabolic homeostasis. Importantly, we observed that down-regulation of ATF4-dependent endoplasmic reticulum-stress signalling in this context appears to prevent Ldh compensation and to exacerbate amyloid beta 42-dependent neuronal toxicity. Our findings, therefore, suggest caution in the use of therapeutic strategies focussed on down-regulation of this pathway for the treatment of Alzheimer's disease, since its natural response to the toxic peptide may induce beneficial neuroprotective effects.
Keywords: Alzheimer's disease, Drosophila, ATF4, Ldh, UPR
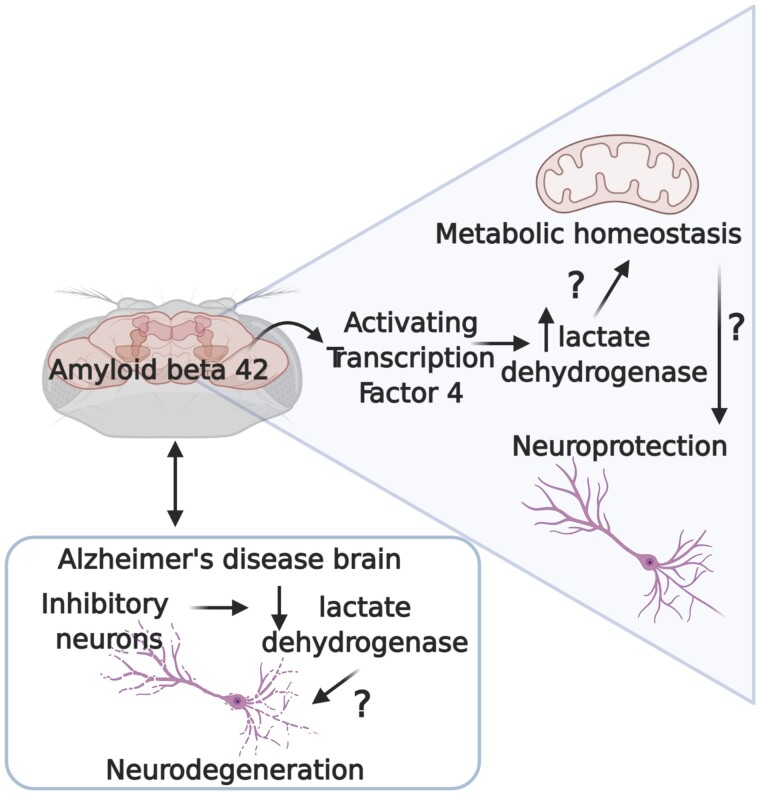
Niccoli et al. report, in Drosophila, that neuronal lactate dehydrogenase, through activating transcription factor 4, is increased in response to Aβ42. Conversely, lactate dehydrogenase is reduced in inhibitory neurons, which are vulnerable to neurodegeneration in Alzheimer's disease patients. It is therefore possible that upregulation of Ldh maintains neuronal homeostasis.
Graphical Abstract
Graphical Abstract.
Introduction
Alzheimer's disease is the most common form of dementia, and affects over 50 million people worldwide.1 The main risk factor for Alzheimer's disease is advancing age,2 with incidence increasing from 0.6% at age 60–65 to over 8% for people aged over 85.3 Although the age-specific incidence of Alzheimer's disease has declined in many parts of the world in recent years,4 human lifespan has increased steadily over the last decades.5 Alzheimer's disease is therefore becoming one of the most common causes of disability and death, with no effective preventative measures or cures yet available.
Alzheimer's disease is characterized by widespread neurodegeneration, but how this is mediated is still unclear. Pathologically, brains of Alzheimer's disease patients display an intracellular accumulation of neurofibrillary tangles, composed of Tau protein, and a substantial increase in extracellular amyloid plaques composed of amyloid beta (Aß) peptides, derived from the mis-processing of the amyloid precursor protein (APP). The most widely accepted model of Alzheimer's disease aetiology is the amyloid hypothesis, first postulated in 1992,6 and based on the observation that all early onset, dominantly inherited forms of the disease are caused by mutations that lead to the abnormal-processing of APP. The amyloid hypothesis states that Alzheimer's disease is initiated by the accumulation of toxic Aß peptides,6 which induce a downstream cascade of events, ultimately resulting in neuronal cell death. Yet, the mechanisms by which Aß accumulation leads to neuronal dysfunction remain to be resolved.
Efforts to identify pathways leading to neuronal cell death have been driven forward by recent advances in single-cell sequencing. This has facilitated the identification of cell-type-specific responses to accumulation of toxic entities and formulation of a more precise picture of the cellular phase of Alzheimer's disease pathogenesis,7 during which specific neuronal responses to the accumulation of toxic proteins ultimately lead to the demise of several neuronal populations. A recent study using single-cell sequencing of Alzheimer's disease patient brains has shown that different cell types show distinct and sometimes opposing transcriptional responses to disease.8 Whether these transcriptional events play a causal role in disease progression, or whether they reflect bystander responses to proteotoxicity, requires further study.
Model organisms play a key role in investigations where genes highlighted by human studies can be manipulated to determine whether they affect disease development.9Drosophila models are excellent for uncovering the molecular mechanisms of disease, thanks to their powerful genetic toolkit developed during the century that flies have been used in research.10 Moreover, 75% of human disease genes have homologues in flies.11 Additionally, their short lifespan enables the assessment of pathological responses to toxic insults across the lifespan of a complex organism. Drosophila Aß toxicity models are widely used in neurodegeneration research 12 and display a range of pathologies, including neuronal dysfunction, behavioural decline and early death in response to amyloid accumulation in the fly brain. We have used an inducible model to express pathogenic Arctic Aß42, tagged with an endoplasmic reticulum (ER) export signal peptide,13 specifically in neurons of the adult fly, thereby removing any confounding developmental effects. These flies show shortened lifespan, climbing defects and neurodegeneration phenotypes.14
Fly models have shown cellular responses to Aß accumulation similar to those seen in Alzheimer's disease patients. For example, the ER stress response is induced in fly models of Aß toxicity15 and in human Alzheimer's disease brain.16,17 Functional genomic approaches using Drosophila have confirmed that this is a protective response, since up-regulation of specific components of the Unfolded Protein Response (UPR), including Xbp1 and BiP,15,18 can protect against Aß toxicity. Similarly, Nrf2, a transcriptional activator of cell protection genes, has been shown to be altered in Alzheimer's disease patients,19 with some studies finding up-regulation and others down-regulation, possibly due to looking at different disease stages. Our studies in flies have shown that restoring Nrf2 activity can ameliorate Aß toxicity by promoting degradation of the amyloid peptide and increasing resistance to cellular stress.20 Therefore, Drosophila appear to mount an evolutionarily conserved response to Aß accumulation in the brain and, due to the ease of genetic manipulation, flies present a powerful tool to identify molecular mediators of Alzheimer's disease pathogenesis and thus potential targets for drug development and clinical translation.
We set out to identify potential age-specific responses to Aß accumulation in the Drosophila brain and to identify conserved transcriptional responses to amyloid toxicity between fly and human AD. Given that Alzheimer's disease is a late-onset disorder, we monitored transcriptional responses at different ages. However, the majority of transcriptional alterations in response to Aß42 in older fly brains overlapped with those in young flies, suggesting that these represent general brain-specific responses to amyloid early in disease pathogenesis. Comparing microarray analysis of fly brains to single-cell transcriptomics of brains of Alzheimer's disease patients, we identified several genes that are differentially expressed, although reciprocally regulated, in flies and humans, including genes implicated in metabolism, ER-stress, proteostasis and cell cycle. We further identified Ldh as a conserved gene mis-expressed in the presence of Aß downstream of ATF4-dependent UPR activation. Activation of the UPR could potentially modulate a protective metabolic response in Alzheimer's disease that warrants further investigation for potential therapeutic benefit.
Materials and methods
Drosophila microarray analyses
Upstream Activating Sequence UAS-ArcAß42/+; elavGS/+ flies were treated, for 7 days, with standard SY medium containing 200 µM Mifepristone (RU486) or medium containing carrier alone (−RU) from either 5 or 20 days of age and brains dissected one week after withdrawal of induction, at 19 or 34 days of age, respectively. Treatments for each replicate were staggered and brains dissected on consecutive days over a 2-hour period to circumvent circadian effects. Frozen fly heads from each replicate were used to measure Aß42 peptide concentrations by enzyme-linked immunosorbent assay (ELISA).
Brain tissues used for microarray analyses were stored in Allprotect tissue reagent (Qiagen # 76405) at −80°C and, for each array, RNA extracted from 25 brains using RLT buffer + 0.01% β-mercaptoethanol and purified with RNeasy columns (Qiagen, Valencia, CA, USA) following the manufacturer's instructions. The quality and concentration of RNA was confirmed using an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA), and further procedures followed the standard Affymetrix protocol. All samples were hybridized to the Drosophila Genome 2.0 Genechip. In total, 5 biological replicates of each condition (−RU 19d, +RU [d 5–12] 19d, −RU 34d and +RU [d 20–27] 34d) were performed.
Differential expression analysis of Drosophila brain data-sets
Differential gene expression was determined as previously described.20 Briefly, raw data (cel files) were processed to correct for probe-sequence biases using gcrma (http://www.bioconductor.org) in R (http://www.r-project.org) and presence of target transcripts, with a P-value <0.111, determined using Affymetrix's MicroArray Suite 5.0 (bioconductor's package affy).21 Raw data were summarized and normalized using the Robust Multichip Average (rma) function (bioconductor's package affy22). A linear model was fitted and differential expression of genes was assessed using the empirical Bayes moderated t-statistic in R's limma package.23P-values were adjusted for multiple hypothesis testing by applying the Benjamini and Hochberg correction for false discovery rate. Summarized probe-sets were mapped to transcripts using R's package ‘drosophila2.db’. Transcripts not mapping to any known or predicted genes were excluded from further analysis.
Gene ontology analysis of Drosophila brain data-sets
Pathway analyses were performed as previously described.20 The Wilcoxon rank sum test, as implemented in Catmap,24 was used to determine significant enrichment of Gene Ontology (GO) categories. FlyBase (http://flybase.org) gene identifiers were mapped to Gene Ontology identifiers (FlyBase version FB2014_01). Ranks of genes were based on the P-value derived from the Bayes t-statistic for differential expression. To account for multiple hypothesis testing, an enrichment of GO terms was deemed statistically significant if the P-value derived from the Wilcoxon rank sum test was ≤1.0 × 10−05. Gene lists were sorted by log-fold change and P-value. For all microarray experiments, two sets of lists were derived; a gene list comprising most differentially up-regulated (log-fold change > 0) genes at the top of the list and most differentially down-regulated genes (log-fold change < 0) at the bottom of the list (termed up-to-down) and vice versa (termed down-to-up). If a GO category was found to be statistically significant in the up-to-down list, this GO was referred to as up-regulated, and conversely if statistically significant in the down-to-up list, this GO was referred to as down-regulated, meaning that a large enough proportion of genes in these categories were found to be up or down-regulated respectively. Statistical significance of overlaps of GOs between age-groups was determined using Fisher's exact test. To account for multiple hypothesis testing, a P-value cut-off of ≤1.0 × 10−05 was used.
Comparison of human Alzheimer’s disease single-cell transcriptional changes versus Drosophila models
Human genes differentially expressed between early and no Alzheimer pathology in six cell types were obtained from Supplementary Table 2 of Mathys et al. Human genes were considered significant according to the author’s definition (column ‘DEGs.Ind.Model’). Fly genes were defined as differentially expressed if their adjusted P-value was <0.05. To analyse sharing in candidate genes between Drosophila and humans, we first transformed human candidate genes to fly orthologues using a table with human to Drosophila orthologue mappings from the Alliance of Genome Resources (AIG) (Alliance of Genome Resources), which employs the Drosophila RNAi Screening Center Integrative Ortholog Prediction Tool. Only genes for which the forward (human to fly orthologues) and reverse orthologue search (fly orthologues to human genes) resulted in the same top gene hits were included (i.e. columns ‘BestForward’ and ‘BestReverse’ were filtered for ‘yes’). We included all matching orthologue hits when one human gene mapped to multiple fly orthologues. Where different human genes mapped to the same fly orthologue, the orthologue hit was only considered once. Genes that were not in common between the above Drosophila brain and human single-cell datasets were removed from the analysis, so that the background size varied between 4,464 and 5,757 genes dependent on the human cell type. Shared candidate genes between the Drosophila and human cell type datasets were then obtained and SuperExactTest (Wang et al.) used to assess whether the number of overlapping genes is significantly different than expected by chance. Performing the analysis based on human genes resulted in qualitatively similar results (not shown).
Fly husbandry and stocks
All flies were reared at 25°C on a 12-h:12-h light:dark cycle at constant humidity and on standard sugar-yeast-agar (SYA) medium (agar, 15 g/l; sugar, 50 g/l; autolyzed yeast, 100 g/l; nipagin, 30 ml/l (of 10% solution in ethanol) and propionic acid, 2 ml/l). For induction with RU486, 24–48 h after eclosion, the female flies carrying a heterozygous copy of elavGS and at least one UAS construct were fed SYA medium supplemented with 200 µM mifepristone (RU486) to induce transgene expression. ElavGS, derived from the original elavGS 301.2 line25 was a gift from Dr H. Tricoire (CNRS), the UAS-Aß42Arc stock was a gift from Dr D. Crowther (University of Cambridge). The UAS-Ldh stock was generated by PCR amplifying the genomic locus with primers (ATGGCCGCCATTAAGGACAGTCTGTTGGC and TTAGAACTTCAGACCAGCCTGGACATCGGA), and gateway cloned into an entry vector and transferred into a gateway compatible pUASTattB vector according to standard protocols and inserted into the attP40 locus. LdhRNAi stock: y1 v1; P{TRiP.HMS00039}attP2, P{lacW}simaj11B7 and ATF4 RNAi stock, P{TRiP.JF02007}attP2 were all from Bloomington stock centre. All flies were back-crossed six times into a w1118 (for over-expression lines) or a v1w+ background (for RNAi lines) line to ensure homogenous back-ground.
Lifespan analysis
Flies were raised at a uniform density in 200 ml bottles. After eclosion, flies were allowed to mate for 24–48 h. At least 110–150 females of the appropriate genotype were split into groups of 15 and housed in vials containing SYA medium and either carrier alone or RU486. Deaths were scored and flies tipped onto fresh food three times a week. All lifespans were performed at 25°C.
Climbing assay
The climbing assays for Aß expressing flies were performed as previously described.14 Briefly, 15 flies were placed in a 25 cm pipette, tapped to the bottom, and allowed to climb for 45 s. The number of flies in the top 5 cm, centre, and bottom 3 cm was scored. A performance index was calculated for each time point and plotted. For flies expressing Ldh and LdhRNAi in neurons climbing assays were performed according to Woodling et al.26 Briefly, flies allowed to climb in a vertical 20 cm column formed by two plastic vials, each fly height was scored in ImageJ and used for statistical analysis. Climbing assays were performed every 3–4 days and at least 100 flies were used per condition.
Quantitative PCR
Total RNA was extracted from 15 to 20 fly heads using Trizol (Invitrogen) and subsequently treated with DNAse I (Ambion) for DNA digestion. The RNA was then reverse transcribed using Superscript II (Invitrogen) with oligo(dT) primers. Quantitative gene expression analysis was performed on a 7900HT real-time PCR system (Applied Biosystems) using SYBR-green technology (ABI). Relative quantities of transcripts were determined using the relative standard curve method normalized to eIF. Primer sequences can be found in Table 1.
Table 1.
Primers for quantitative real-time PCR analysis
| Gene | Forward Primer | Reversed primer |
|---|---|---|
| Aß | CGATCCTTCTCCTGCTAACC | CACCATCAAGCCAATAATCG |
| Atf-4 | TCGATGCTTACAAACAGGCG | AAAGTTAAAGGGCGTGGCAG |
| eIF | ATCAGCTCCGAGGAT | GCGGAGACAGACGTT |
| LDH | GGTATCGGGACTGTA | GCAGCACGGCTCCAACTTTC |
| Sima | CACCTTCAAGAGCGTGCTGA | CGTGGCCTGGCTAAGAATC |
LDH assay
To measure LDH activity, 10 fly heads were homogenized in 62.5 µl of 0.2 M NaPO4, pH 6.5 plus 0.2% phenylthiourea. The samples were centrifuged at 21,000 g for 5 min at 4°C, the clear supernatant was taken to measure the activity of LDH. Protein extracts were quantified using the Bradford protein assay (Bio-Rad protein assay reagent; Bio-Rad laboratories Ltd (UK)). For the measurement in the direction pyruvate to lactate, the assay mixture contained 0.05 M sodium phosphate, pH 6.5, 1 mM sodium pyruvate, and 0.2 mM NADH. For the measurement in the direction lactate to pyruvate, the assay mixture contained 0.1 M sodium phosphate, pH 7.5, 100 mM D, L-lithium lactate, and 4.13 mM NAD+. Blank samples contained everything but NADH or NAD+. 10 µg of the protein extracts were added to the reaction mixture. The activity of LDH was measured spectrophotometrically at 25°C at 340 nm on a Tecan Infinite M200 platereader. The LDH activity is represented as a slope of the reaction.
Lactate pyruvate levels
To measure lactate and pyruvate levels, 15 fly heads were homogenized in 30 µl 4% cold Trichloroacetic acid. The samples were centrifuged for 15 min at 11,500 g for 15 min at 4°C. 20 µl of the clear supernatant was neutralized with 180 µl of 1:10 dilution of 1 M Tris-HCL pH8. Lactate and pyruvate levels were measured using the Lactate assay kit (Sigma) and the Pyruvate assay kit (Sigma). The samples were diluted 1:10 in the assay buffer, then the pyruvate and lactate levels were performed according to the manufacturers’ instructions. The pellet was resuspended in 75 µl of 10 mM Tris pH 10.4 to extract the proteins. Protein extracts were quantified using the Bradford protein assay (Bio-Rad protein assay reagent; Bio-Rad laboratories Ltd (UK)). The amount of lactate and pyruvate in each sample is expressed as the ratio of the total protein content (µg/g total protein).
Aß42 ELISA
Total Aß42 was extracted from fly heads in GnHCl buffer (5 M Guanidinium HCl, 50 mM Hepes pH 7.3, protease inhibitor cocktail (Sigma, P8340) and 5 mM EDTA), as previously described (Sofola O et al.). Aß42 levels were then measured using the High Sensitivity Human Amyloid Aß42 ELISA kit (Millipore), according to the manufacturers’ instructions. Protein extracts were quantified using the Bradford protein assay (Bio-Rad laboratories Ltd, UK) and the amount of Aß42 in each sample expressed as a ratio of the total protein content (pmoles/g total protein).
Fluorescence-activated cell sorting of neurons
Green Fluorescent Protein (GFP) labelled neurons were sorted according to DeSalvo et al.27 Briefly, 10 brains per sample were dissected in Schneider’s medium containing 1% Bovine Serum Albumin and quickly transferred into the same medium on ice. Samples were dissociated as described previously and GFP positive neurons were fluorescence-activated cell sorting (FACS) sorted straight into lysis medium, ready for RNA extraction.
Statistical analyses
Microarray analyses are described above. For climbing assays, data were analysed by ordinal logistic regression or linear regression in R (http://www.r-project.org), using the individual heights for each fly as data points. For lifespans, data are presented as cumulative survival curves and survival rates were compared using log-rank tests in Excel. For all other experiments, data are presented as means ± SEM, from a minimum of 3 independent biological repeats, and were analysed by unpaired student’s t-test or one-way ANOVA and Tukey’s post-hoc analyses using Graphpad Prism 8.0 (https://www.graphpad.com/scientific-software/prism/).
Data availability
The raw microarray data generated in this study are deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress) with identifier E-MTAB-8865.
Results
To understand how Aß toxicity is mediated in neurons, and whether age influences this response, we measured RNA expression in the brains of flies expressing Aß42 peptide either early or later in life (Supplementary Fig. 1A and B). The lifespan curves associated with these induction patterns have been published elsewhere (Fig. 2C in the study by Rogers et al.28) 224 genes were differentially expressed between uninduced controls and Aß42-expressing brains at the two ages (Supplementary Tables 1 and 2), 41 of which were in common between the young and older flies, a significant enrichment (Fig. 1A, Supplementary Fig. 1C and Table 3). A larger number of genes were altered in response to Aß42 in young flies (Fig. 1A), possibly because young flies can activate a more robust response, which might also explain why older individuals are more susceptible to Aß toxicity. Most genes with altered expression at older age were also altered in the young brains, suggesting that the majority of these represented generic responses to Aß42, albeit with magnitudes that varied slightly with age (Fig. 1A, Supplementary Fig. 1C).
Figure 2.
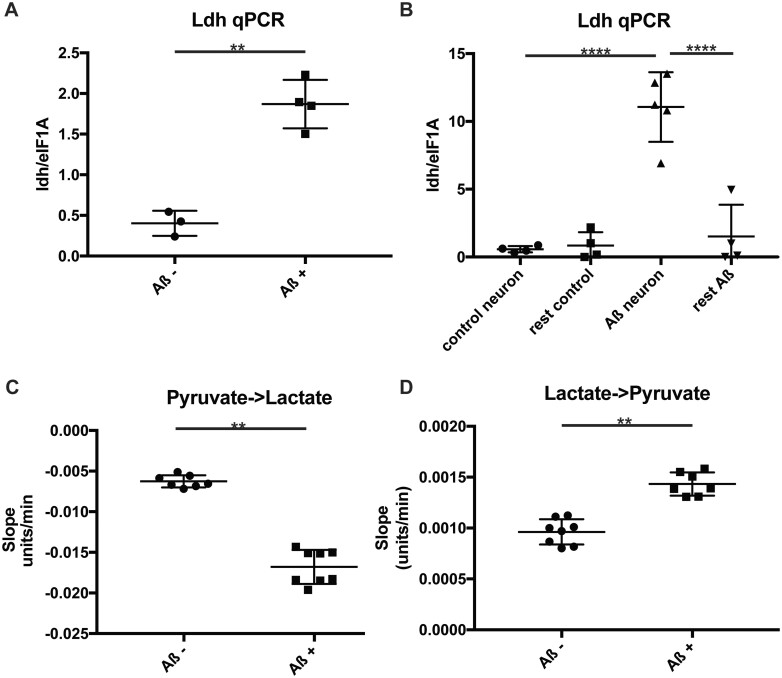
Ldh is upregulated in Aß expressing fly brains. (A) Ldh qPCR analysis of brains expressing Aß (Aß+) and uninduced controls (Aß−). Genotypes: UAS-Aß; elav GS. (B) Ldh qPCR analysis of FACS sorted GFP expressing neurons and other cells, expressing Aß (Aß) and driver alone (controls). Genotypes: UAS-Aß/UAS-eGFP; elav GS and UAS-eGFP; elav GS. (C and D) Ldh enzymatic assay on brain extracts expressing Aß (Aß+) and uninduced controls (Aß−). Assayed in the direction of lactate (C) and pyruvate production (D). Values shown are the slopes generated by the enzymatic reaction. Lactate production generates a negative slope, so a lower negative value signifies a greater activity. Genotypes: UAS-Aß; elav GS. A, C and D were compared by t-test, B by one-way ANOVA followed by Tukey’s post-hoc test. ** P < 0.01, ****P < 0.0001, N = 3–5 per condition for qPCR; N = 7–8 per condition for LDH activity.
Figure 1.
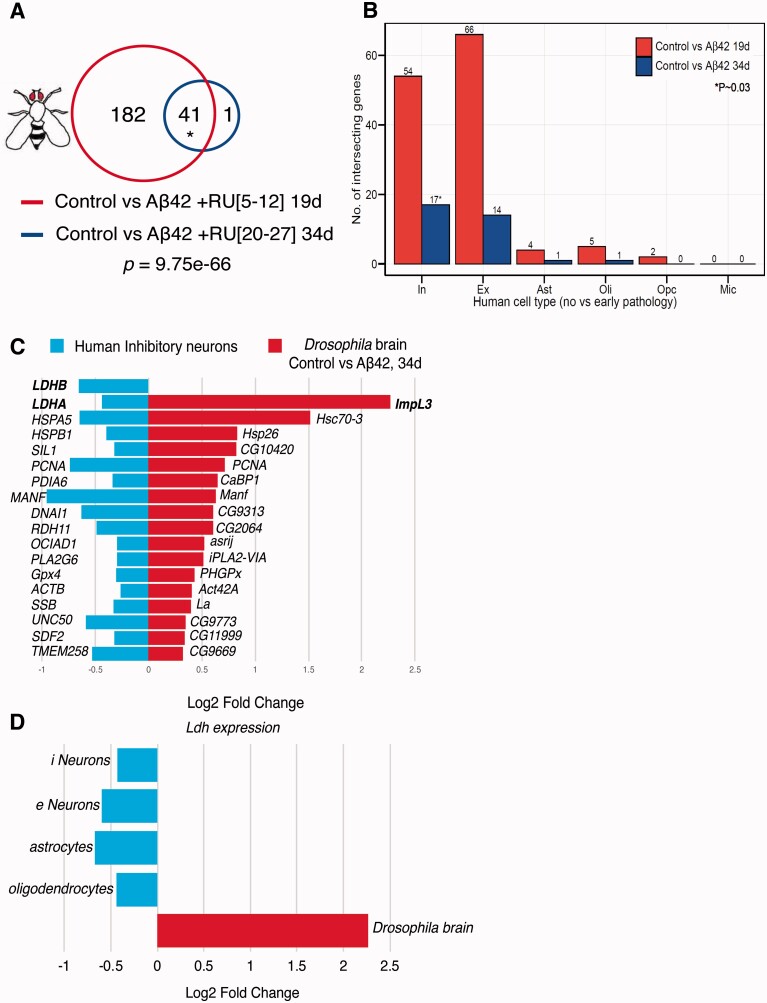
Overlapping transcriptional responses to early AD pathology in aged Drosophila and human brain. (A) Significant overlaps in differentially expressed (DE) genes were observed between young (19d) and old (34d) fly brains in response to Aß42 expression (41 of 224 DE genes analysed; p = 9.75e-66, Bayes moderated t-statistic and Benjamini and Hochberg FDR correction), and individual genes are detailed in Supplementary Fig. 1C. (total gene number n = 8566). (B) Plot depicts the number of intersecting genes that are differentially expressed in no versus early pathology in human AD brain, across various cell types, and control versus Aß42 expression in fly brain. Red represents overlaps between human genes and alterations in young flies and blue overlaps with old flies. Significant overlaps (P = 0.03; SuperExactTest) were observed only when comparing no versus early pathology in human inhibitory neurons and control versus Aß42+ 34d (old flies). Abbreviations: In, inhibitory neurons; Ex, excitatory neurons; Ast, astrocytes; Oli, oligodendrocytes; Opc, oligodendrocyte precursor cell; Mic, microglia. (C) Plot showing the 17 genes from the significant overlap in B (*). Genes are upregulated in flies (red) and downregulated in humans (blue). (D) Ldh was the principal upregulated gene in response to Aß42 in aged Drosophila brain and its human orthologue LDH was significantly downregulated in inhibitory (i) neurons (LDH A&B). LDHB expression was also downregulated in human neurons excitatory (e), astrocytes and oligodendrocytes in brain tissue from early-stage AD patients compared with controls. The fly image in this figure was originally created by Dr Fiona Kerr and produced in Kerr et al.56
A recent single-cell study also found that transcriptional changes in Alzheimer’s disease patients early and later in disease development were quite similar, suggesting that there is a strong transcriptional signature early in response to Aß42 that does not greatly change with disease progression.8 To determine if the changes in gene expression in Drosophila brains were conserved in human Alzheimer's disease brains, we compared our young and old fly data-sets to single-cell sequencing of early and late stage Alzheimer's disease patients versus healthy controls8 (Fig. 1B, Supplementary Table 8). Significant overlaps were observed only between control versus Aß42 34d in flies and no pathology versus early Alzheimer's disease pathology in inhibitory neurons in humans (Fig. 1B), with 17 genes commonly regulated between Alzheimer's disease flies and patients (Fig. 1C). Of these, lactate dehydrogenase (Ldh; Drosophila ImpL3) was up-regulated to the greatest degree in Aß42 fly brains (Fig. 1C and D), while both LDHA and LDHB isoforms were down-regulated in inhibitory patient neurons. LDHB was also significantly down-regulated in excitatory neurons, astrocytes and oligodendrocytes of Alzheimer's disease patients with early pathology (Fig. 1D).
Moreover, several genes involved in the Unfolded Protein Response (UPR) including Hsc70, CG10420 and CaBP1 were significantly up-regulated in Aß42 fly brain, and their orthologues HSPA5/BiP, SIL1 and PDIA6 down-regulated in inhibitory neurons of Alzheimer's disease patients (Fig. 1C), consistent with ER-stress associated responses under Alzheimer's disease conditions in flies and humans. Supporting this observation, GO pathway analyses in Drosophila (Supplementary Tables 4–7) confirmed that UPR, ER and Golgi processes were significantly enriched in differentially expressed upregulated genes in both young and old Aß42 fly brain (Supplementary Tables 4 and 6). These data suggest that our fly model represents early stages of Alzheimer's disease pathogenesis, but with mainly opposing effects on expression of the same genes, including defects in LDH and UPR levels. This discrepancy in the direction of change requires further investigation, but may represent cell-type specific effects which are not detectable using a whole-brain approach to transcriptional analyses in flies compared to human studies.
LDH catalyses the conversion of lactate into pyruvate and vice-versa and is a key enzyme in the glycolytic cascade. Its activity is increased in Alzheimer's disease patients' brains,29 and mRNA levels of LDHA are higher in fibroblasts derived from late-onset Alzheimer's disease patients versus controls.30 However, it is unclear whether LDH plays a direct role in Alzheimer's disease pathogenesis. Up-regulation of glycolysis in neurons, and specifically of key enzymes, including LDHA, confers resistance to Aß toxicity.31 We, therefore, sought to understand whether the increase in LDH activity observed in flies is itself a compensatory response to Aß42, or whether instead it contributes to disease development.
We confirmed by qPCR that expression of Aß in fly neurons leads to up-regulation of Ldh RNA in fly brains (Fig. 2A), and this upregulation does not occur in driver alone flies treated with RU (Supplementary Fig. 2A). This is accompanied by increased enzymatic activity in both directions (Fig. 2C and D). However, this increase in enzymatic activity did not result in an increase in either lactate or pyruvate (Supplementary Fig. 2B and C). This was, possibly, because flux through the pathway was increased in both directions, and the steady-state metabolite concentrations were hence maintained. Ldh is expressed both in neurons and in glia, where it contributes to the neuronal lactate shuttle. In glia, Ldh catalyses the conversion of pyruvate to lactate, which is then shuttled to neurons via monocarboxylate transporters where Ldh converts it back to pyruvate which then enters glycolysis for rapid energy production.32 Whereas in mammalian systems these two reactions are catalysed by different isoenzymes, composed of different subunits, in flies there is a single Ldh gene (Impl3), with the same enzyme catalysing the reaction in both directions. We performed the above enzymatic analysis of Ldh using whole head lysates, hence further studies were required to determine whether the increase in LDH activity occurred in neurons or in glia.
To identify which cell-types were responsible for the increase in LDH activity in Alzheimer's disease flies, we next measured transcript levels in isolated neurons versus non-neuronal cells in the fly. We FACS sorted GFP positive cells, using control or Aß42 flies co-expressing GFP specifically in neurons. We confirmed that the GFP sorted cell fractions were indeed enriched for neurons by assessing the levels of the neuronal marker elav (Supplementary Fig. 2D). We then measured Ldh mRNA levels on FACS sorted cells, and found that Ldh increased in neurons (Fig. 2B) but not in the other cell types in the brain.
To test whether an increase in Ldh was protective, we generated flies over-expressing Ldh in adult neurons and confirmed the over-expression (Supplementary Fig. 3).
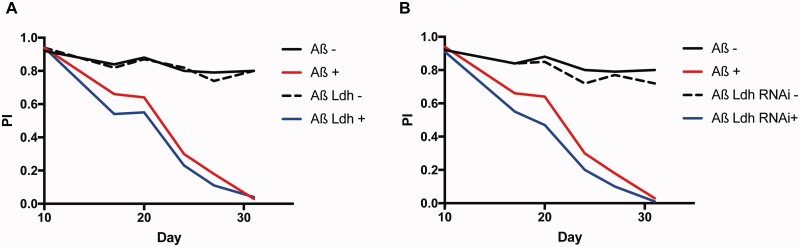
We then over-expressed Ldh in adult neurons that also expressed Aß42, and found an exacerbation of the impaired climbing phenotype observed in these flies, suggesting that Ldh up-regulation may contribute to Aß42-induced neuronal toxicity (Fig. 3A). qPCR analysis of Ldh transcripts showed that, even in the −RU condition, the UAS-Aß; UAS-Ldh fly lines showed increased Ldh expression (Supplementary Fig. 3A), leading to increased Ldh enzymatic (Supplementary Fig. 3B and C). The increase in transcript and enzyme activity in the UAS line in the un-induced condition most likely reflects the leakiness of the elavGS driver.33 There was a trend towards increased lactate levels (which did not reach significance) and there was a slight decrease in pyruvate in the over-expression line (Supplementary Fig. 3D and E), possibly indicating that excessive Ldh activity in neurons is driving a higher rate of pyruvate consumption by the TCA cycle.
Figure 3.
Manipulation of Ldh leads to exacerbation of climbing defects. (A) Plot of Performance index for a negative geotaxis assay of flies expressing Aß (Aß+) alone compared to flies expressing Aß and Ldh (Aß Ldh+), and their un-induced controls (Aß− and Aß Ldh−). Genotypes: UAS-Aß/UAS-Ldh; elav GS, UAS-Aß; elavGS. Aß UAS Ldh was not significantly different to Aß alone (P = 0.139 by ordinal logistics regression). (B) Plot of Performance index for a climbing assay of flies expressing Aß (Aß+) alone compared to flies expressing Aß and Ldh RNAi (Aß Ldh RNAi +), and their un-induced controls (Aß− and Aß Ldh RNAi−). Genotypes: UAS-Aß/UAS-Ldh RNAi; elav GS, UAS-Aß; elavGS. Aß Ldh RNAi genotype was significantly worse than Aß alone (P = 0.00552 by ordinal logistics regression). Note experiments were run in parallel with Aß+ and Aß− curves in Fig. 3a, but depicted separately for clarity.
To test this further, we down-regulated Ldh using RNAi, in flies over-expressing Aß42, but this also led to a worsening of the climbing phenotype (Fig. 3B). Again, qPCR analysis, showed that Ldh transcripts were down-regulated, even in the non-induced conditions, leading to changes in enzymatic activity and metabolite levels both in the induced and un-induced condition. This is consistent with previous studies showing that RNAi lines can display strong knock-down even in the un-induced condition.33 Pyruvate and lactate levels were very low in the presence of Ldh RNAi, however, suggesting that Ldh enzyme is affecting the production of both metabolites under our experimental conditions (Supplementary Fig. 3), possibly because of a compensatory down-regulation of the whole pathway, but this remains to be determined.
Over-expression or knock-down of Ldh, however, also negatively affected climbing behaviour in healthy flies (Supplementary Fig. 3F and G) suggesting that its expression must be finely-controlled to maintain metabolic homeostasis and neuronal function. As further genetic manipulation of Ldh also induced detrimental effects in Aß42-expressing flies, these findings could indicate that its natural up-regulation in response to Aß42 production represents a compensatory mechanism to maintain optimal levels of gene expression and protection against metabolic dysfunction under these conditions.
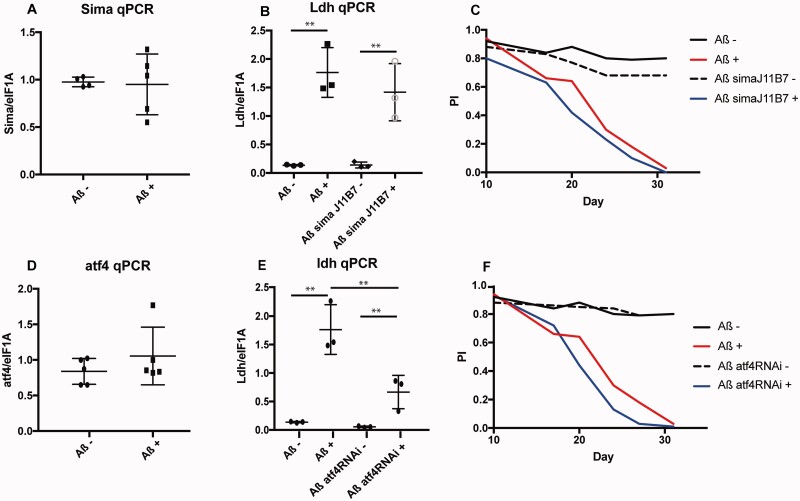
We next investigated the potential molecular mechanisms responsible for regulation of Ldh in response to Aß42. Ldh levels are regulated by hypoxia inducible factor 1 (HIF1)34,35 in mammalian systems and by the Hif1 homologue, similar (sima), in flies.36 In Aß-resistant human neurons, elevation of Hif1 levels is thought to lead to increased LDHA31,37 and Hif1 protein levels are increased in Alzheimer's disease mouse models.31 To investigate whether sima was responsible for the up-regulation of Ldh in Alzheimer's disease fly brain in response to Aß, we first examined the levels of sima transcript, and observed no significant difference. However, sima can be regulated post-transcriptionally, so this does not rule out its involvement. We down-regulated sima genetically, using a heterozygous null mutant,38 but this had no effect on Ldh expression in brains of Aß over-expressing flies (Fig. 4B), suggesting that sima does not regulate Ldh levels in response to Aß42. Down-regulation of sima exacerbated the negative geotaxis phenotype of both Aß and control flies, suggesting that it is generally detrimental to fly climbing but not specifically to Aß toxicity.
Figure 4.
Sima does not but ATF4 does induce Ldh. (A) qPCR for Sima in fly heads expressing Aß (Aß+) and un-induced controls (Aß−). Genotype: UAS-Aß; elav GS. (B) qPCR of Ldh in heads from sima mutant flies expressing Aß (Aß sima J11B7+) and their un-induced controls (Aß−, Aß sima J11B7−) Genotypes: UAS-Aß; elav GS/sima J11B7, UAS-Aß; elavGS. **P < 0.01, N = 3–5 per condition. (C) Plot of Performance index for a climbing assay of flies expressing Aß alone (Aß+), together with mutant sima (Aß sima J11B7+) as well as their uninduced controls (Aß−, Aß sima J11B7−). Genotypes as above. UAS-Aß/sima J11B7 genotypes were significantly worse than Aß alone (p = 4.03e-06 respectively by ordinal logistics regression) but there was no interaction with RU, suggesting the sima mutation affects control and Aß expressing flies similarly. Note experiments were run in parallel with Aß+ and Aß− curves in Fig. 3a, but depicted separately for clarity. (D) qPCR for ATF4 in fly heads expressing Aß (Aß+) and un-induced controls (Aß−). Genotype: UAS-Aß; elav GS. (E) qPCR for Ldh in heads from flies expressing Aß together with RNAi for ATF4. Genotypes: UAS-Aß; elavGS/UAS-ATF4 RNAi and UAS-Aß; elav GS. **P < 0.01, N = 3–5 per condition. (F) Plot of Performance index for a climbing assay of flies expressing Aß alone (Aß+), together with RNAi for ATF4 (Aß ATF4RNAi +) and their uninduced controls (Aß− and Aß ATF4RNAi−). Genotypes as above. The Aß ATF4RNAi genotype displayed a significantly worse response to RU over time relative to Aß alone (P = 0.03379 for a three-way interaction of RU, genotype and day by ordinal logistics regression). Note experiments were run in parallel with Aß+ and Aß− curves in Fig. 3a, but depicted separately for clarity.
ATF4 is an effector of the UPR, induced downstream of protein kinase R-like endoplasmic reticulum kinase (PERK) and eukaryotic Initiation Factor 2 alpha (eIF2alpha),39 and has been shown to regulate glucose homeostasis and energy expenditure.40 In particular, in flies it has been shown to up-regulate glycolytic enzymes, including Ldh in response to ER stress.41 The UPR is up-regulated in response to Aß in patients,42 animal models43 and flies.18ATF4 is also induced in Alzheimer's disease patient brains44 and in animal models of Alzheimer's disease.45 Given that both Ldh and ER-stress associated genes were altered in response to Aß42 in our flies, we therefore further explored potential connections between these processes by assessing ATF4 level and its potential functional role in mediating neuronal damage in AD. ATF4 transcript was unaltered in Aß42 expressing flies compared to uninduced controls (Fig. 4D). However, as ATF4 is translationally regulated,39 alterations in mRNA may not be expected. Indeed, down-regulation of ATF4 by RNAi dampened the increased expression of Ldh in response to Aß42 (Fig. 4E), suggesting that ATF4 does contribute, at least partially, to the regulation of Ldh under these conditions. Aß transcripts, on the other hand, were unaltered (Supplementary Fig. 4), suggesting the effect is specific to Ldh. ATF4 down-regulation further decreased the climbing ability of Aß expressing flies (Fig. 4F), providing correlative evidence to suggest that activation of ATF4 could contribute to a protective response to Aß42 accumulation, possibly via its up-regulation of Ldh. More experiments will be required to prove this is the case.
Discussion
We have shown that flies mount a conserved transcriptional response to the accumulation of Aß42, and that this response is similar in young and old flies. Similarly, in humans, it was found that the ageing signature was orthogonal to the disease response in Alzheimer's disease patients.8 Our findings highlight Ldh, as well as several ER-stress associated genes, as a major transcriptional responder to early Aß42 toxicity in both young and old fly brain. Greater transcriptional responses were detected in response to Aß42 in the brain of young flies, but further studies are required to confirm whether these represent protective responses that are lost with age.
Consistent with our observation that both Ldh and ER-associated genes are altered in both Aß fly models and Alzheimer's disease patient brain, we have also shown that expression of Aß42 induces Ldh expression, and subsequent activity, in fly brain via ATF4, a downstream effector of the UPR. The induction of Ldh could potentially be neuroprotective since both down-regulating Ldh and blocking the Aß42-induced increase in Ldh expression using ATF4 RNAi in neurons are detrimental. Up-regulation of Ldh has been observed in Alzheimer's disease patients' brains.29 Elevated CSF Ldh activity is also used as an indicator of neuronal damage,46 although the exact source of this increase is not clear46 and could be due to both cellular and blood–brain barrier damage releasing the enzyme into the CSF or a direct increase in enzymatic activity of CSF-expressed Ldh. Our findings in the fly, are consistent with other studies showing that up-regulation of Ldh37 and other glycolytic enzymes31 increases resistance to Aß toxicity in cortical neurons, however, further experiments will be required to directly prove this, for example by blocking the ATF4 induced increase in Ldh and checking whether this has a detrimental effect.
We show for the first time that the Aß42 peptide directly induces Ldh expression in an animal model.
It is interesting that the fly response appears reciprocal to the one discovered in inhibitory human neurons.47 Inhibitory neurons are extremely susceptible to Aß toxicity. Severe loss of inhibitory GABAergic neurons has also been observed in several animal models of Alzheimer's disease and in patients,47,48 where their degeneration appears to precede that in other cell types, at least in early disease stages.49 It is possible, therefore, that fly neurons can mount a protective response to Aß, whereas inhibitory neurons display a loss of protective pathways, thus making them selectively susceptible to disease. Alternatively, by measuring transcriptional responses in heterogeneous neuronal and glial cell types using a whole-brain approach, our study may have excluded detection of cell-type specific transcriptional changes in our fly model. Hence, further work is required to investigate these functional connections using human neuronal models of Aβ toxicity.
LDH upregulation in mammalian systems has been shown to be regulated by a variety of mechanisms,46 one of the most prevalent being Hif1. Hif1 is responsible for the up-regulation of glycolysis in cancer cells, leading to the switch from oxidative to glycolytic metabolism, through the Warburg effect, which promotes their survival.46 It has been proposed that survival of neurons in Alzheimer's disease can also be promoted by the Warburg effect,37 and that this is also mediated by Hif1. However, in our in vivo model of Aß42 toxicity the transcription factor ATF4, and not sima (the fly Hif1 homologue), is responsible for the induction of Ldh expression in response to Aß42.
These findings suggest that Ldh upregulation is downstream of UPR activation. The UPR is mediated by 3 effectors: PERK, Iris and Atf6.50 PERK phosphorylates eIF2alpha to inhibit canonical translation and induce the translation of specific factors, such as ATF4. The UPR is induced in the brain of Alzheimer's disease patients and in animal models,51 however, whether this is pathological or protective is controversial.51 In particular, up-regulation of ATF4 has been observed in Alzheimer’s disease patients' brains,44 downstream of PERK activation and eIF2alpha phosphorylation.44 The phosphorylation of eIF2alpha is usually considered pathological in neurodegenerative conditions52–54 and its pharmacological or genetic inhibition has been shown to protect animal models of frontotemporal dementia55 and Alzheimer's disease.54 Functionally, however, its role in early pathogenesis of Alzheimer's disease is far from clear, with studies in cells suggesting that the up-regulation of eIF2alpha phosphorylation and ATF4 activation contributes to cellular resistance to Aß toxicity.44 Our in vivo data using a fly model of Aß42 toxicity agree with this finding and further suggest that ATF4 might also play a protective role, potentially by contributing to Ldh induction. However, formally demonstrating that this is indeed the case would require deleting the binding sites for ATF4 in the Ldh endogenous promoter and showing that this abrogates Ldh induction resulting in a detrimental effect in the presence of Aß.
This work requires further investigation, however, it lends a word of caution towards therapies geared purely towards down-regulating the UPR as a therapy for Alzheimer's disease, since part of its endogenous response may be beneficial.
Supplementary material
Supplementary material is available at Brain Communications online.
Supplementary Material
Acknowledgements
The authors thank Dr Herve Tricoire (CNRS, France) for providing the elavGS line and Dr Damian Crowther (University of Cambridge) for the UAS-Arc Aß42 line used in this study. The authors also thank Sebastian Grönke and Jacqueline Dols for injecting the plasmid to generate the Ldh fly lines. Graphical abstract Created with BioRender.com.
Glossary
- Aß =
amyloid ß
- AD =
Alzheimer's disease
- APP =
amyloid precursor protein
- ATF4 =
activating transcription factor 4
- Elav=
embryonic lethal abnormal vision
- eIF2alpha =
eukaryotic initiation factor 2 alpha
- ELISA =
enzyme-linked immunosorbent assay
- ER =
endoplasmic reticulum
- FACS =
fluorescence-activated cell sorting
- GFP =
green fluorescent protein
- GO =
gene ontology
- GS =
gene-switch
- Hif1 =
hypoxia-inducible factor 1
- Ldh =
lactate dehydrogenase
- NADH =
nicotinamide adenine dinucleotide
- PERK =
protein kinase R-like endoplasmic reticulum kinase
- qRTPCR =
quantitative reverse transcription polymerase chain reaction
- RNAi =
RNA interference
- RU =
Mifepristone
- sima =
similar
- SYA =
sugar-yeast-agar
- TCA =
trichloroacetic acid
- UAS =
upstream activating sequence
- UPR =
unfolded protein response
Funding
T.N., I.S., J.A., D.F., D.I. and J.T. were funded by a Wellcome Trust Strategic Award to L.P. (grant number WT098565/Z/12/Z to L.P.) and F.K. by an Alzheimer’s Research UK (ARUK) project grant awarded to L.P. (ART-2009–4; F.K.). I.S. was supported by the Erasmus Student Exchange Programme. O.A.-S. was funded by an Alzheimer’s Society Senior Fellowship. T.N. is currently funded by an ARUK fellowship (ARUK-SRF2018A-003) and F.K. by a Glasgow Caledonian University Research Fellowship.
Competing interests
The authors report no competing interests.
References
- 1.Alzheimer's Disease International. World Alzheimer’s report 2019, Attitudes to dementia. 2019.
- 2. Niccoli T, Partridge L.. Ageing as a risk factor for disease. Curr Biol. 2012;22(17):R741–R752. [DOI] [PubMed] [Google Scholar]
- 3. Hebert LE, Scherr PA, Beckett LA, et al. Age-specific incidence of Alzheimer's disease in a community population. JAMA. 1995;273(17):1354–1359. https://www.ncbi.nlm.nih.gov/pubmed/7715060. [PubMed] [Google Scholar]
- 4. Langa KM. Is the risk of Alzheimer's disease and dementia declining? Alzheimers Res Ther. 2015;7(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dong X, Milholland B, Vijg J.. Evidence for a limit to human lifespan. Nature. 2016;538(7624):257–259. [DOI] [PubMed] [Google Scholar]
- 6. Hardy JA, Higgins GA.. Alzheimer's disease: The amyloid cascade hypothesis. Science. 1992;256(5054):184–185. [DOI] [PubMed] [Google Scholar]
- 7. De Strooper B, Karran E.. The cellular phase of Alzheimer's disease. Cell. 2016;164(4):603–615. [DOI] [PubMed] [Google Scholar]
- 8. Mathys H, Davila-Velderrain J, Peng Z, et al. Single-cell transcriptomic analysis of Alzheimer's disease. Nature. 2019;570(7761):332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wangler MF, Yamamoto S, Chao HT, et al. Members of the Undiagnosed Diseases Network (UDN). Model organisms facilitate rare disease diagnosis and therapeutic research. Genetics. 2017;207(1):9–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bellen HJ, Tong C, Tsuda H.. 100 years of drosophila research and its impact on vertebrate neuroscience: A history lesson for the future. Nat Rev Neurosci. 2010;11(7):514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cheng L, Baonza A, Grifoni D.. Drosophila models of human disease. Biomed Res Int. 2018;2018:7214974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McGurk L, Berson A, Bonini NM.. Drosophila as an in vivo model for human neurodegenerative disease. Genetics. 2015;201(2):377–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Crowther DC, Kinghorn KJ, Miranda E, et al. Intraneuronal abeta, non-amyloid aggregates and neurodegeneration in a drosophila model of Alzheimer's disease. Neuroscience. 2005;132(1):123–135. [DOI] [PubMed] [Google Scholar]
- 14. Sofola O, Kerr F, Rogers I, et al. Inhibition of gsk-3 ameliorates abeta pathology in an adult-onset drosophila model of Alzheimer's disease. PLoS Genet. 2010;6(9):e1001087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Niccoli T, Cabecinha M, Tillmann A, et al. Increased glucose transport into neurons rescues abeta toxicity in drosophila. Curr Biol. 2016;26(18):2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van der Harg JM, Nolle A, Zwart R, et al. The unfolded protein response mediates reversible tau phosphorylation induced by metabolic stress. Cell Death Dis. 2014;5:e1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoozemans JJ, van Haastert ES, Nijholt DA, Rozemuller AJ, Eikelenboom P, Scheper W.. The unfolded protein response is activated in pretangle neurons in Alzheimer's disease hippocampus. Am J Pathol. 2009;174(4):1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Casas-Tinto S, Zhang Y, Sanchez-Garcia J, Gomez-Velazquez M, Rincon-Limas DE, Fernandez-Funez P.. The er stress factor xbp1s prevents amyloid-beta neurotoxicity. Hum Mol Genet. 2011;20(11):2144–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fao L, Mota SI, Rego AC.. Shaping the nrf2-are-related pathways in Alzheimer's and parkinson's diseases. Ageing Res Rev. 2019;54:100942. [DOI] [PubMed] [Google Scholar]
- 20. Kerr F, Sofola-Adesakin O, Ivanov DK, et al. Direct keap1-nrf2 disruption as a potential therapeutic target for Alzheimer's disease. PLoS Genet. 2017;13(3):e1006593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ling D, Salvaterra PM.. Robust rt-qpcr data normalization: Validation and selection of internal reference genes during post-experimental data analysis. PLoS One. 2011;6(3):e17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gautier L, Cope L, Bolstad BM, Irizarry RA.. Affy–analysis of affymetrix genechip data at the probe level. Bioinformatics. 2004;20(3):307–315. [DOI] [PubMed] [Google Scholar]
- 23. Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Breslin T, Eden P, Krogh M.. Comparing functional annotation analyses with catmap. BMC Bioinformatics. 2004;5(1):193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Osterwalder T, Yoon KS, White BH, Keshishian H.. A conditional tissue-specific transgene expression system using inducible gal4. Proc Natl Acad Sci U S A. 2001;98(22):12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Woodling NS, Aleyakpo B, Dyson MC, et al. The neuronal receptor tyrosine kinase alk is a target for longevity. Aging Cell. 2020;19(5):e13137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. DeSalvo MK, Hindle SJ, Rusan ZM, et al. The drosophila surface glia transcriptome: Evolutionary conserved blood-brain barrier processes. Front Neurosci. 2014;8:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rogers I, Kerr F, Martinez P, Hardy J, Lovestone S, Partridge L.. Ageing increases vulnerability to abeta42 toxicity in drosophila. PLoS One. 2012;7(7):e40569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bigl M, Bruckner MK, Arendt T, Bigl V, Eschrich K.. Activities of key glycolytic enzymes in the brains of patients with Alzheimer's disease. J Neural Transm (Vienna). 1999;106(5-6):499–511. [DOI] [PubMed] [Google Scholar]
- 30. Sonntag KC, Ryu WI, Amirault KM, et al. Late-onset Alzheimer's disease is associated with inherent changes in bioenergetics profiles. Sci Rep. 2017;7(1):14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Soucek T, Cumming R, Dargusch R, Maher P, Schubert D.. The regulation of glucose metabolism by hif-1 mediates a neuroprotective response to amyloid beta peptide. Neuron. 2003;39(1):43–56. [DOI] [PubMed] [Google Scholar]
- 32. Mason S. Lactate shuttles in neuroenergetics-homeostasis, allostasis and beyond. Front Neurosci. 2017;11:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scialo F, Sriram A, Stefanatos R, Sanz A.. Practical recommendations for the use of the geneswitch gal4 system to knock-down genes in drosophila melanogaster. PLoS One. 2016;11(8):e0161817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Firth JD, Ebert BL, Ratcliffe PJ.. Hypoxic regulation of lactate dehydrogenase a. Interaction between hypoxia-inducible factor 1 and camp response elements. J Biol Chem. 1995;270(36):21021–21027. [DOI] [PubMed] [Google Scholar]
- 35. Semenza GL, Jiang BH, Leung SW, et al. Hypoxia response elements in the aldolase a, enolase 1, and lactate dehydrogenase a gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol Chem. 1996;271(51):32529–32537. [DOI] [PubMed] [Google Scholar]
- 36. Wang CW, Purkayastha A, Jones KT, Thaker SK, Banerjee U.. In vivo genetic dissection of tumor growth and the Warburg effect. Elife. 2016;5:e18126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Newington JT, Pitts A, Chien A, Arseneault R, Schubert D, Cumming RC.. Amyloid beta resistance in nerve cell lines is mediated by the Warburg effect. PLoS One. 2011;6(4):e19191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Doronkin S, Djagaeva I, Nagle ME, Reiter LT, Seagroves TN.. Dose-dependent modulation of hif-1alpha/sima controls the rate of cell migration and invasion in drosophila ovary border cells. Oncogene. 2010;29(8):1123–1134. [DOI] [PubMed] [Google Scholar]
- 39. Blais JD, Filipenko V, Bi M, et al. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol. 2004;24(17):7469–7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Seo J, Fortuno ES 3rd, Suh JM, et al. Atf4 regulates obesity, glucose homeostasis, and energy expenditure. Diabetes. 2009;58(11):2565–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee JE, Oney M, Frizzell K, Phadnis N, Hollien J.. Drosophila melanogaster activating transcription factor 4 regulates glycolysis during endoplasmic reticulum stress. G3 (Bethesda). 2015;5(4):667–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hoozemans JJ, Veerhuis R, Van Haastert ES, et al. The unfolded protein response is activated in Alzheimer's disease. Acta Neuropathol. 2005;110(2):165–172. [DOI] [PubMed] [Google Scholar]
- 43. Soejima N, Ohyagi Y, Nakamura N, et al. Intracellular accumulation of toxic turn amyloid-beta is associated with endoplasmic reticulum stress in Alzheimer's disease. Curr Alzheimer Res. 2013;10(1):11–20. [PubMed] [Google Scholar]
- 44. Lewerenz J, Maher P.. Basal levels of eif2alpha phosphorylation determine cellular antioxidant status by regulating atf4 and xct expression. J Biol Chem. 2009;284(2):1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baleriola J, Walker CA, Jean YY, et al. Axonally synthesized atf4 transmits a neurodegenerative signal across brain regions. Cell. 2014;158(5):1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Valvona CJ, Fillmore HL, Nunn PB, Pilkington GJ.. The regulation and function of lactate dehydrogenase A: Therapeutic potential in brain tumor. Brain Pathol. 2016;26(1):3–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Vico Varela E, Etter G, Williams S.. Excitatory-inhibitory imbalance in Alzheimer's disease and therapeutic significance. Neurobiol Dis. 2019;127:605–615. [DOI] [PubMed] [Google Scholar]
- 48. Sanchez-Mejias E, Nunez-Diaz C, Sanchez-Varo R, et al. Distinct disease-sensitive gabaergic neurons in the perirhinal cortex of Alzheimer's mice and patients. Brain Pathol. 2020;30(2):345–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mikkonen M, Alafuzoff I, Tapiola T, Soininen H, Miettinen R.. Subfield- and layer-specific changes in parvalbumin, calretinin and calbindin-d28k immunoreactivity in the entorhinal cortex in Alzheimer's disease. Neuroscience. 1999;92(2):515–532. [DOI] [PubMed] [Google Scholar]
- 50. Hetz C, Mollereau B.. Disturbance of endoplasmic reticulum proteostasis in neurodegenerative diseases. Nat Rev Neurosci. 2014;15(4):233–249. [DOI] [PubMed] [Google Scholar]
- 51. Gerakis Y, Hetz C.. Emerging roles of er stress in the etiology and pathogenesis of Alzheimer's disease. FEBS J. 2018;285(6):995–1011. [DOI] [PubMed] [Google Scholar]
- 52. Chang RC, Wong AK, Ng HK, Hugon J.. Phosphorylation of eukaryotic initiation factor-2alpha (eif2alpha) is associated with neuronal degeneration in Alzheimer's disease. Neuroreport. 2002;13(18):2429–2432. [DOI] [PubMed] [Google Scholar]
- 53. Kim HS, Choi Y, Shin KY, et al. Swedish amyloid precursor protein mutation increases phosphorylation of eif2alpha in vitro and in vivo. J Neurosci Res. 2007;85(7):1528–1537. [DOI] [PubMed] [Google Scholar]
- 54. Ma T, Trinh MA, Wexler AJ, et al. Suppression of eif2alpha kinases alleviates Alzheimer's disease-related plasticity and memory deficits. Nat Neurosci. 2013;16(9):1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Halliday M, Radford H, Zents KAM, et al. Repurposed drugs targeting eif2α-p-mediated translational repression prevent neurodegeneration in mice. Brain. 2017;140(6):1768–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kerr F, Bjedov I, Sofola-Adesakin O. Molecular Mechanisms of Lithium Action: Switching the Light on Multiple Targets for Dementia Using Animal Models. Front. Mol. Neurosci. 2018;11:297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw microarray data generated in this study are deposited in ArrayExpress (http://www.ebi.ac.uk/arrayexpress) with identifier E-MTAB-8865.