Abstract
Objective
Arcuate nucleus neuropeptide Y/agouti-related peptide (NPY/AgRP) neurons drive ingestive behavior. The M-current, a subthreshold non-inactivating potassium current, plays a critical role in regulating NPY/AgRP neuronal excitability. Fasting decreases while 17β-estradiol increases the M-current by regulating the mRNA expression of Kcnq2, 3, and 5 (Kv7.2, 3, and 5) channel subunits. Incorporating KCNQ3 into heteromeric channels has been considered essential to generate a robust M-current. Therefore, we investigated the behavioral and physiological effects of selective Kcnq3 deletion from NPY/AgRP neurons.
Methods
We used a single adeno-associated viral vector containing a recombinase-dependent Staphylococcus aureus Cas9 with a single-guide RNA to selectively delete Kcnq3 in NPY/AgRP neurons. Single-cell quantitative measurements of mRNA expression and whole-cell patch clamp experiments were conducted to validate the selective knockdown. Body weight, food intake, and locomotor activity were measured in male mice to assess disruptions in energy balance.
Results
The virus reduced the expression of Kcnq3 mRNA without affecting Kcnq2 or Kcnq5. The M-current was attenuated, causing NPY/AgRP neurons to be more depolarized, exhibit a higher input resistance, and require less depolarizing current to fire action potentials, indicative of increased excitability. Although the resulting decrease in the M-current did not overtly alter ingestive behavior, it significantly reduced the locomotor activity as measured by open-field testing. Control mice on a high-fat diet exhibited an enhanced M-current and increased Kcnq2 and Kcnq3 expression, but the M-current remained significantly attenuated in KCNQ3 knockdown animals.
Conclusions
The M-current plays a critical role in modulating the intrinsic excitability of NPY/AgRP neurons that is essential for maintaining energy homeostasis.
Keywords: Neuropeptide Y, Agouti-related peptide, 17β-estradiol, KCNQ channels, High-fat diet, Energy balance
Highlights
-
•
SaCas9 CRISPR vector selectively reduces Kcnq3 expression in NPY/AgRP neurons.
-
•
Knockdown of Kcnq3 significantly attenuates the M current.
-
•
While ingestive behavior appears normal, locomotion is significantly reduced.
-
•
A high-fat diet in conjunction with Kcnq3 knockdown promotes weight gain.
-
•
A high-fat diet increases Kcnq3 mRNA and the M current in control NPY/AgRP neurons.
1. Introduction
In the hypothalamic arcuate nucleus (ARH), two distinct neuronal cell types differentially modulate energy homeostasis: the proopiomelanocortin (POMC) and neuropeptide Y/agouti-related peptide (NPY/AgRP) neurons. NPY and AgRP are co-expressed in the same ARH neurons, and AgRP is the endogenous antagonist for the melanocortin (MC4) receptor, thus blocking the anorectic effects of the POMC peptide, α-melanocyte-stimulating hormone [1,2]. Ablation of NPY/AgRP neurons in adults causes rapid starvation [3,4], while injecting NPY into the paraventricular nucleus stimulates food intake [5]. Optogenetic and chemogenetic stimulation of NPY/AgRP neurons swiftly increases food consumption [6,7], whereas stimulation of POMC neurons attenuates food intake [6,8]. In vivo measurements of calcium activity have shown that NPY/AgRP neurons integrate anticipatory sensory cues with the current energy state to generate appropriate feeding behavior [9]. In fact, plasticity and responsiveness of ARH circuits are critical for maintaining energy balance.
In their orexigenic role, NPY/AgRP neurons respond to many peripheral and central hormonal signals that control energy homeostasis [[10], [11], [12], [13], [14]]. NPY/AgRP neurons are also the major CNS targets of the anorectic hormone 17β-estradiol (E2) [15,16], where, for example, E2 suppresses NPY expression in the ARH [17,18]. Conversely, NPY/AgRP neurons are more active after fasting, and leptin reduces their excitability [13]. Leptin and other peripheral signals (for example, insulin and gastrin-releasing peptide) modulate NPY/AgRP excitability by targeting calcium and potassium (K+) channels [14,[19], [20], [21]]. One such conductance, the M-current, is a sub-threshold, non-inactivating, voltage-dependent, outward potassium (K+) current. This conductance modulates neuronal excitability, action potential kinetics, and burst firing [22,23]. The M-current is itself regulated by numerous neurotransmitters and neuropeptides that modulate neuronal excitability through G-protein-coupled receptors including acetylcholine, serotonin, substance P, and gonadotropin-releasing hormone [22,24].
The M-current amplitude is mediated by KCNQ (Kv1-7) channels [25,26]. NPY/AgRP neurons highly express transcripts for the subunits KCNQ 2, 3, and 5, but not 4, with most individual cells expressing detectable levels of two or more of these subunits [27] to form hetero-tetrameric [28] and possibly homo-tetrameric complexes [29]. Global knockout of Kcnq2 is perinatally lethal [30], unlike deletion of Kcnq3 [29] or Kcnq5 [31]. When Kcnq2 or Kcnq3 is individually expressed in Xenopus oocytes, the M-current is negligible, but equimolar injections of both transcripts produce a significantly higher current [25], suggesting that these heteromers contribute the bulk of the native conductance or that KCNQ3 co-expression aids in trafficking to the membrane [[32], [33], [34]]. However, these assumptions have been questioned using conditional knockout mice. When KCNQ subunits are individually deleted from cortical pyramidal cells, only Kcnq2 knockout produces a robust increase in neuronal excitability, suggesting that KCNQ2 is solely obligatory [35]. It is currently unknown whether this represents a general rule for KCNQ channels or is cell-type specific. The role and importance of KCNQ5 in the M-current is not as well-characterized. KCNQ5 subunits are thought to exist in homomeric or KCNQ3/5 heteromeric complexes [36,37]. In the hippocampus, the expression of Kcnq5 has been suggested to decrease the M-current due to competition for KCNQ3 subunits to the detriment of KCNQ2/3 heteromer formation [31]. In the ARH, however, elevated Kcnq5 expression appears to contribute to the E2 potentiation of the M-current [27]. Surprisingly, Kcnq2 KO mice also have a 40% loss of KCNQ5 protein, although the underlying cause remains unknown [35]. Previously studied KCNQ3 knockouts have been global [[29], [30], [31]] or present during development [35,38], and therefore, the influence of compensatory mechanisms cannot be dismissed. Cre-dependent CRISPR/Cas9 vectors enable region and cell-type knockout specificity in adult animals [[39], [40], [41]]. This strategy has recently been improved upon by creating a single adeno-associated viral vector [42]. Using this novel approach, we tested the hypothesis that knockdown (KD) of Kcnq3 in NPY/AgRP neurons of adult animals would increase their excitability, causing disruptions in the regulation of energy balance.
2. Materials and methods
2.1. Animals and treatments
All the animal procedures described in this study were performed in accordance with institutional guidelines based on National Institutes of Health standards and approved by the Institutional Animal Care and Use Committee at Oregon Health and Science University or the University of Washington, Seattle.
2.2. Mice
DATCre mice [43] were bred and housed at the University of Washington, Seattle. NPYhrGFP (JAX #006417) [14] and AgRPCre (JAX #012899) [44] transgenic mice (breeders provided by Dr. Brad Lowell, Harvard University, Cambridge, MA, USA) were selectively bred at OHSU. All the mice were maintained under controlled temperature (25 °C) and photoperiod conditions (lights on at 6 a.m. and off at 6 p.m.) with a standard chow diet (CD, 5L0D, Laboratory Diets, St. Louis, MO, USA) and water ad libitum unless otherwise stated. For high-fat diet experiments, the mice were instead fed a high-fat diet (HFD, D12451, 45% kcal from fat, Research Diets, New Brunswick, NJ, USA) either at weaning (mRNA expression) or four weeks after stereotactic injections when the mice were typically 3–4 months old (behavior and electrophysiology). For single-cell reverse transcription polymerase chain reaction (scRT-PCR) and electrophysiology, female and male mice were used, whereas behavior was measured solely in males. Groups were arranged to balance starting weight and fat mass as determined by an EchoMRI Whole-Body Composition Analyzer (Echo Medical Systems, Houston, TX, USA).
2.3. Visualized whole-cell patch recordings
Coronal brain slices (240 μm) containing the ARH from gonadectomized female or intact male mice were produced in an ice-cold sucrose cutting solution (see recipe) and stored in a bubbled (95% 02 and 5% CO2) chamber containing artificial cerebrospinal fluid (aCSF; see recipe). Whole-cell patch recordings were produced from YFP/mCherry-positive neurons using an Olympus BX51W1 upright microscope with an Exfo X-Cite 120 Series fluorescence light source, epifluorescence (FITC and mCherry filters), and video-enhanced infrared-differential interference contrast. Patch pipettes were filled with K+-gluconate internal solution (in mM): 128 potassium gluconate, 10 NaCl, 1 Mg Cl2, 11 EGTA, 10 HEPES, 3 ATP, and 0.25 GTP (pH was adjusted to 7.3–7.4 with 1N KOH, 290–300 mOsm). Pipette resistances ranged from 3 to 5 MΩ in whole-cell configurations. Fluorescent cells in the AgRPCre mice were targeted for recordings. Cells were included in the analysis only if their input resistance was above 800 MΩ and access resistance remained less than 20 MΩ throughout the experiment. I–V relationships of currents were examined with voltage steps.
2.4. Electrophysiology data analysis
Electrophysiological signals were digitized with Digidata 1440A (Molecular Devices), amplified with an Axopatch 200B (Molecular Devices), and analyzed using ClampFit version 10 software (RRID:SCR_011323, v10.3, Molecular Devices). Subsequently, all the electrophysiology data were transferred to Sigma Plot 13 (Jandel Scientific, San Rafael, CA, USA) or Graph Pad Prism 6/7 (La Jolla, CA. USA) for statistical analysis. The liquid junction potential was corrected for the data analysis. All the values are expressed as mean ± SEM. Comparisons between two groups were made using un-paired Student's t-tests or between multiple groups using ANOVA (with post hoc comparisons) with p values < 0.05 considered significant. When variances differed significantly, the Mann–Whitney test was used instead.
2.5. Electrophysiological solutions and drugs
A sucrose solution was used during Vibratome slicing (in mM): 2 KCl, 1 MgCl2–6H2O, NaH2PO4, 10 HEPES, 10 glucose, 208 sucrose, 26 NaHCO3, 2 MgSO4–7H2O, and 1 CaCl2. For harvesting and electrophysiological recordings, a standard artificial cerebrospinal fluid was used (in mM): 124 NaCl, 5 KCl, 1.44 NaH2PO4, 5 HEPES, 10 glucose, 26 NaHCO3, 2 MgSO4–7H2O, and 2 CaCl2. Tetrodotoxin (1 μM, Alomone Labs, Jerusalem, Israel) was added to the bath for all of the recordings except when measuring the rheobase. Tetraethylammonium (TEA, 5 mM, Sigma–Aldrich, St. Louis, MO, USA), a non-specific K+ channel blocker, and XE 991 (40 μM, Tocris, Bristol, UK), a KCNQ channel antagonist, were also used.
2.6. AAV delivery
Viruses were prepared at the University of Washington according to published methods [42,45]. While incorporating the smaller SaCas9 sequence allowed the inclusion of the sgRNA in a single vector, there was not sufficient room remaining in the cassette for a fluorophore sequence. To visualize the quality and location of the injection/infection, the CRISPR/SaCas9 vector was spiked with a high titer virus encoding mCherry or YFP. The resulting mixture allowed a single injection of both viruses at a similar titer. Co-injected viruses were always the same serotype (AAV1) so as not to affect the transduction efficiency [42]. Four to sixteen weeks prior to each experiment, the AgRPCre mice (>60 days old) received bilateral ARH injections of Cre-dependent adeno-associated (AAV, serotype 1) viral vectors encoding yellow fluorescent protein, YFP (AAV1-EF1α-YFP), or mCherry mCh (AAV1-EF1α-mCh) either alone or co-injected with an AAV1 designed to encode SaCas9 and a single-guide RNA (sgRNA). See the SaCas9 section for specifics on the sgRNA design. Using aseptic techniques, anesthetized (1–1.5% isoflurane/O2) mice received a medial skin incision to expose the surface of the skull. A glass pipette (#3-000-203-G/X, Drummond Scientific, Broomall, PA, USA) with a beveled tip (diameter = 45 μm) was filled with mineral oil and loaded with an aliquot of AAV using a Nanoject II (Drummond Scientific). ARH injection coordinates were anteroposterior (AP): −1.15 mm, mediolateral (ML): ±0.30 mm, and dorsoventral (DV): −5.80 and −5.70 (surface of the brain z = 0.0 mm); 250 nl of the AAV (2 × 1012 particles/ml) was injected (100 nl/min) at each position, and the pipette was left in place for 10 min post-injection and then slowly retracted from the brain. The skin incision was closed using Vetbond (3M), and each mouse received analgesia (Rimadyl, 4–5 mg/kg, sc).
2.7. Gonadectomy
Male gonads were left intact, and all the females were subjected to ovariectomy (OVX) at least 7 days prior to each experiment. Rimadyl (4–5 mg/kg, sc) was given immediately after surgery to relieve post-operative pain. For the quantitative measurement of CRISPR gene editing, we used fed OVX female mice that were treated with 17β-estradiol benzoate (EB). We selected this model in females to enhance the expression of Kcnq 2, 3, and 5 [27] with the goal of maximizing the sensitivity to detect reductions in transcripts and currents. Females received an injection of a low (0.25 μg) and then a high (1.5 μg) dose the following day of EB administered in the morning on the two days preceding the experiments. We have documented that this regimen induces a preovulatory surge of luteinizing hormone in females, replicating proestrus [46,47]. Circulating levels of E2 were verified by uterine weights (>95 mg) at the time of hypothalamic slice preparation (between 8:30 and 10:30 am).
2.8. Generation and validation of AAV1-FLEX-SaCas9-U6-sgKcnq3
The sgRNA for targeting the Kcnq3 locus was designed as previously described [42]. The following oligos (Sigma) were used to clone into pAAV-FLEX-SaCas9-U6-sgRNA (Addgene 124844). Kcnq3 forward: CACCGCCACCGCCGAGTCCCCTCCAG; Kcnq3 reverse: AAACCTGGAGGGGACTCGGCGGTGGC. Targeted deep sequencing of Kcnq3 locus: AAV1-FLEX-SaCas9-U6-sgKcnq3 and AAV1-FLEX-EGFP-KASH were co-injected into the VTA of the DAT-Cre mice. Nuclei were isolated and FACS for EGFP-KASH-positive and -negative nuclei was performed, followed by targeted deep sequencing of whole-genome amplified (WGA) DNA as previously described [42]. Four weeks after surgery, tissue punches of the ventral midbrain were pooled from 3 mice into a single group and homogenized in 2 mL of homogenization buffer containing (in mM): 320 sucrose (sterile filtered), 5 CaCl2 (sterile filtered), 3 Mg(Ac)2 (sterile filtered), 10 Tris pH 7.8 (sterile filtered), 0.1 EDTA pH 8 (sterile filtered), 0.1% NP40, 0.1 Protease Inhibitor Cocktail (PIC, Sigma), and β-mercaptoethanol. Homogenization was performed using 2 mL glass dounces (Sigma Cat#: D8938-1SET) 25 times with pestle A and then 25 times with pestle B. The volume of the homogenate was transferred to a 15 mL conical tube and increased to 5 mL using homogenization buffer, mixed, and incubated on ice for 5 min. Then 5 mL of 50% Optiprep density gradient medium (Sigma Cat#: D1556-250 ML) containing (in mM): 5 CaCl2 (sterile filtered), 3 Mg(Ac)2 (sterile filtered), 10 Tris pH 7.8 (sterile filtered), 0.1 PIC, and 1 β-mercaptoethanol was added to the homogenate and mixed by inversion. The suspension was layered onto 10 mL of 29% iso-osmolar Optiprep solution in a 1 × 3.5 Beckman centrifuge tube (SW32 Ti rotor) and centrifuged at 7500 RPM for 30 min at 4 °C. Floating cell debris was removed, and the supernatant was gently poured out. The pellets were resuspended in sterile 1 x PBS. Then 500 GFP-positive and 500 GFP-negative nuclei were sorted directly into 3 μL of REPLI-g Advanced Storage Buffer (Qiagen Cat#: 150365) in a PCR tube strip (Genessee Cat#: 24-706) using a BD AriaFACS III. Whole-genome amplification (WGA) was performed directly following FACS using a REPLI-g Advanced DNA Single-Cell kit (Qiagen Cat#: 150365) according to the manufacturer's instructions. To generate amplicons, 1 μl of WGA DNA was diluted to 1:50 and amplified (PCR 1) with Phusion High-Fidelity Polymerase (Thermo Fisher Scientific Cat#: F530L) using the following thermocycler protocol: initial denaturation (30 s, 95 °C); denaturation (10 s, 95 °C), annealing (20 s, 69 °C), and extension (10 s, 72 °C); the cycle was repeated 34 times until the final extension (5 min, 72 °C). For PCR 2, 1 uL of PCR 1 was amplified with a second set of primers using the same thermocycler protocol. The 205-bp amplicon from PCR 2 was gel extracted using a MinElute gel extraction kit (Qiagen Cat#: 28606) and sent to Genewiz for Amplicon-EZ targeted deep sequencing and Sanger sequencing. The primers used to generate the amplicons were as follows: PCR 1 forward: CAAGTGCTCCTACTTCCC reverse: GTCTTTGCCAGGAGCCCGAT, PCR 2 forward: AGATGGGTCTCAAGGCTC, reverse: AGGGTCCCGTCTTTGTCG.
2.9. Single-cell harvesting
Ovariectomized (OVX) EB-treated female mice were rapidly decapitated and coronal brain sections (240 μm) were cut using a Vibratome (VT-1000S, Leica, Wetzlar, Germany). The ARH was microdissected from basal hypothalamic slices (3-4 slices per mouse). Gentle trituration following incubation with papain (Sigma–Aldrich) was used to dissociate the ARH neurons before dispersion onto a glass-bottomed dish where the healthy cells settled and adhered to the glass bottom. After 15 min, the aCSF was removed and fresh artificial cerebrospinal fluid (aCSF) was added to the plate. This washing procedure was repeated two times. Throughout the dispersion and harvesting procedure, a constant flow (2 ml/min) of oxygenated aCSF circulated into the plate while the effluent circulated out using a peristaltic pump. The aCSF flow helped ensure that fresh oxygenated media was reaching the cells and assisted in clearing out unhealthy cells and debris from the trituration. The cells harvested were those observed to be fully intact, with one to three processes and smooth cell membranes as visualized using an inverted microscope (DMIL, Leica) equipped with a fluorescent LED light source (X-Cite 110LED, Excelitas Technologies Corp., Waltham, MA, USA). Individual neurons were patched and then harvested with gentle suction into pipettes using a XenoWorks Micromanipulator/Microinjector system (Sutter Instrument Company, Novato, CA) and expelled into a siliconized 0.65 ml microcentrifuge tube containing 5X Superscript III buffer (Invitrogen, Carlsbad, CA, USA), 15 U of RNasin (Promega, Madison, WI, USA), 10 mM of dithiothreitol (DTT), and diethylpyrocarbonate (DEPC)-treated water in a total of 8 μl for quantitative real-time PCR (qPCR) (5 or 10 cell pools/tube). Collected pools of cells were stored at −80 °C until immediately prior to reverse transcription.
2.10. Quantitative RT-PCR and single-cell RT-PCR and primer design
cDNA synthesis was performed in a reaction volume of 25 μl (5- or 10-cell pools) containing dNTPs (0.5 mM, Promega), random primers (100 ng/per tube, Promega), anchored oligo(dT)20 primers (400 ng/tube, Invitrogen), Superscript III reverse-transcriptase (100 U/per tube, Invitrogen), RNAsin (15 U), DTT (6 mM), and DEPC-treated water according to established protocols [46] and stored at −20 °C. Primers for genes that encode for Kcnq2 (Kv7.2), Kcnq3 (Kv7.3), Kcnq5 (Kv7.5), and β-actin were designed to cross at least one intron-exon boundary and are listed as follows: Kcnq2 (accession number NM_010611) 92 bp PCR product, forward primer 1079–1098 nt reverse primer 1151–1170 nt, Kcnq3 (accession number NM_152923) 94 bp PCR product, forward primer 879–896 nt, reverse primer 955–972 nt, Kcnq5 (accession number NM_023872) 98 bp PCR product, forward primer 1277–1297 nt, reverse primer 1354–1374 nt, Actb (β-actin) (accession number NM_007393) 110 bp PCR product, forward primer 446–465 nt, and reverse primer 535–555 nt. Of note, our first choice for primer location for Kcnq3 was to span the PAM and sgRNA sites for a more accurate determination of Kcnq3 mRNA expression knockdown. However, because the PAM and sgRNA sites were found at nt 488–514 (exon1) of the 5’ end of the Kcnq3 mRNA sequence (NM_152923.3), and primers spanning these sites would not cross exon-intron boundaries, we designed a highly efficient set of downstream primers as previously noted (forward nt 879–896 and reverse nt 885–886) producing a 94 bp product spanning the intron-exon boundary between exons 2 and 3.
qPCR was performed on a QuantStudio 7 Flex Real-Time PCR System (Thermo Fisher Scientific) using Power Sybr Green Master Mix (Thermo Fisher Scientific). The efficiencies of the target and reference gene amplifications were determined by serially diluting cDNAs (1:50 to 1:12,800) from mouse basal hypothalamus to construct standard curves. The efficiency was calculated according to the following formula: E = 10(−1/m)-1 and m = slope [48,49] for Kcnq2 m = −3.367, r2 = 0.98; Kcnq3 m = −3.432, and Kcnq5 m = −3.487, and r2 = 0.99. Based on the high efficiencies, the comparative ΔΔCT method was used to determine values from duplicate samples of 4 μl for the target genes (Kcnq2, Kcnq3, and Kcnq5) and 2 μl for the reference gene (Actb). The relative linear quantity was determined using the 2− ΔΔCT equation [46]. To determine the relative expression levels of mRNA in neurons obtained from the control and KCNQ3 knockdown mice, the mean ΔCT for the target gene from the control samples was used as the calibrator, and the data expressed as an n-fold change in gene expression were normalized to the reference gene Actb relative to the calibrator. A two-tailed unpaired t-test was used for analysis, and the data are expressed as mean ± SEM, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001.
2.11. Open field
The animals were tested during their relative light phase between zeitgeiber 6–8 at least 1 h before testing to allow the mice to acclimate in a dimly lit room adjacent to the testing apparatus in the presence of white noise. The open field test was conducted in 40 × 40 cm opaque white boxes cleaned with acetic acid at the beginning and end of testing days and 70% ethanol between subjects, allowing time for any odors to dissipate. The room light was adjusted to 200 lux with white noise also present. The mice were tracked using Ethovision XT 10 (Noldus, Leesburg, VA, USA) for 10 min. The distance (cm) traveled and average velocity (cm/s) were calculated in addition to the percent time spent and frequency of crossing into the center, with the latter two considered measures of anxiety.
2.12. Lickometer
Med Associates operant chambers (ENV-307W) equipped with dual lickometers contained in sound-attenuating cubicles (ENV-022V) were used to measure vanilla Ensure (Abbott Nutrition) consumption. The mice were given access to two sipper tubes, either for Ensure (36 g in 100 ml of tap water) or tap water. The house light was on throughout the 30-min testing period. The day prior to start of training, a small dish containing Ensure was placed in the mouse home cages to prevent neophobia. Chambers were controlled, and data were collected with MedPC 5 software (Med Associates) and exported to Excel before analysis using GraphPad Prism 6/7.
2.13. Food intake measurements
The mice were singly housed in clean cages with added enrichment (crinkle paper or tubes) and ramekin bowls containing standard food pellets with ad libitum access to water. After three days of habituation to single housing and two days to the ramekin, all the food was removed from the hopper and 9–14 g of chow was placed in the ramekin. In the morning on the following two days, the remaining pellets and crumbs were weighed, and the difference was marked as baseline food consumed. After fasting, the mice were moved to clean cages (to prevent coprophagy) with water but no food for an overnight fast. After 16 h, food was restored and food intake was measured for an additional day (refeed) [50].
3. Results
3.1. CRISPR editing of Kcnq3 significantly decreased Kcnq3 expression
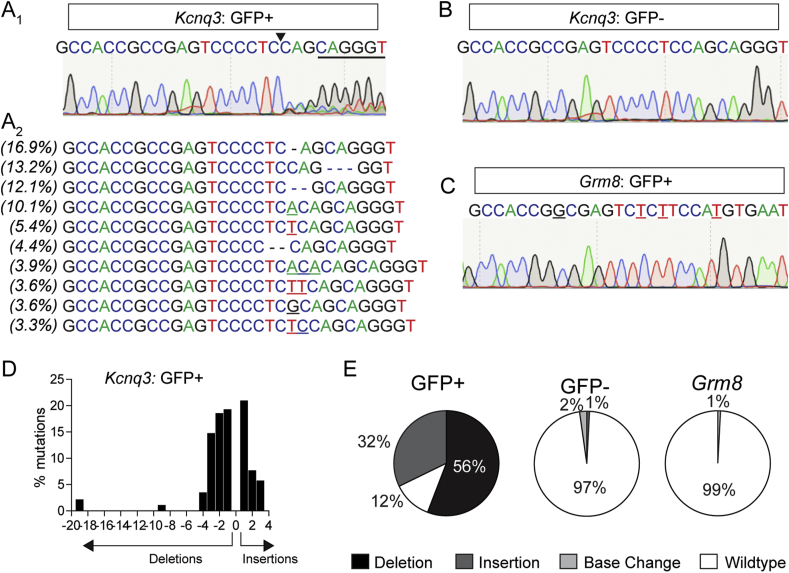
The CRISPR/Cas9 system is recognized as an efficient means of generating insertion/deletion (indel) mutations to cause a loss of function in targeted genes [[51], [52], [53]]. However, due to the large size of Staphylococcus pyogenes, there is insufficient capacity in the expression cassette of adeno-associated viruses to also include the single-guide RNA (sgRNA) necessary for gene targeting. This limitation requires using a separate viral vector [39,54] or incorporating a Cre-dependent SpCas9 transgenic line [55]. We recently developed a single viral vector for conditional expression of the smaller Staphylococcus aureus (SaCas9) and sgRNA that yields high-efficiency mutagenesis in specific cell types [42]. To inactivate Kcnq3, we generated a guide targeting exon 1, which is conserved across all splice variants. Because of the small size of the ARH and the relatively low number of AgRP neurons, we confirmed the efficacy of SaCas9 mutagenesis of Kcnq3 in midbrain dopamine neurons as previously described [42]. Briefly, the DAT-Cre mice were co-injected with AAV1-FLEX-SaCas9-U6-sgKcnq3 and AAV1-FLEX-EGFP-KASH and tissue was harvested four weeks following surgery. EGFP-positive and -negative nuclei were isolated by FACS and whole-genome amplification (WGA) was performed followed by targeted deep sequencing of a PCR amplicon containing the targeted region of Kcnq3 and CRISPOR-predicted [56] off-target Grm8 (Figure 1A–C). SaCas9 generated numerous insertions and deletions (indels) centered at 3 base pairs upstream of the protospacer adjacent motif (PAM) (Figure 1A and D-E). A small number of base changes (2%) and deletions (1%) were observed in GFP-negative nuclei and a similarly small number of base changes were observed in Grm8, likely reflecting the sequencing fidelity (Figure 1B,C and E). Of the sequenced GFP+ cells, only 12% retained the wild-type sequence. Therefore, the majority of GFP+ cells likely had a loss-of-function Kcnq3 mutation.
Figure 1.
Analysis of targeted KCNQ3 mutagenesis in DATCremice. Analysis of FACS-sorted GFP+ (A) and GFP- (B) nuclei from mice co-injected with AAV1-FLEX-SaCas9-U6-sgKCNQ3 and AAV1-FLEX-EGFP-KASH into the VTA. (A1) sgKcnq3 sequence with PAM underlined and SaCas9 cut site indicated by black arrow. Chromatogram results display multiple peaks beginning at the cut site. (A2) Top 10 mutations at the cut site with the percent of total reads for which they occur on the left. Base changes: bolded. Insertions: underlined. Deletions: marked with a “-” (dash). (B) Chromatogram of GFP- nuclei sequencing displaying no evidence of mutations at the SaCas9 cut site. (C) Chromatogram of sgKcnq3 predicted off-target in Grm8 displaying no evidence of mutations at the SaCas9 cut site. (D) Frequency distribution of insertions and deletions in Kcnq3 from GFP+ nuclei. (E) Percent of wild-type, deletions, insertions, and base changes as percent of total reads for Kcnq3 in GFP+ (left) and GFP- nuclei (middle) and the predicted off-target Grm8 (right).
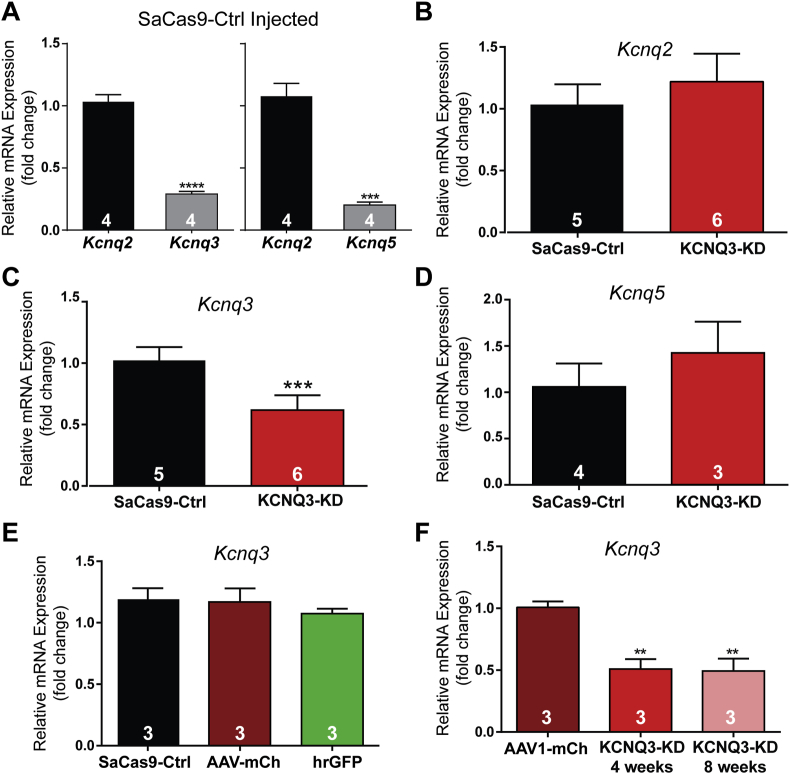
Based on the promising results of Kcnq3 editing by the SaCas9/sgRNA vector in the DATCre mice, a cohort of AgRPCre mice was given bilateral stereotaxic injections in the ARH of either AAV1-FLEX-SaCas9-sgKcnq3 or a control virus containing the Kcnq3 guide with three base pairs in the mutated seed region (SaCas9-control) as previously described [42]. An additional Cre-dependent virus with the same serotype (AAV1) that drove the expression of a fluorophore (YFP or mCherry) was co-administered to visualize the injection quality and facilitate cell harvesting. Female mice were chosen for the initial cohort as estradiol increases the M-current [27], which could help in detecting the current attenuation. After three weeks, the mice underwent OVX and were subsequently given EB treatment on the two days prior to sacrifice. Brain slices were prepared, and cells were harvested as previously described [57] and analyzed with qPCR. Of note, the relative expression of Kcnq2 mRNA was much higher than that of Kcnq3 mRNA (Figure 2A, left) or Kcnq5 mRNA (Figure 2A, right). The Kcnq2 mRNA expression was not changed in NPY/AgRP neurons in response to Kcnq3 editing (Figure 2B), but the KCNQ3-KD group did display a reduction in the relative expression of Kcnq3 compared to the control group (Figure 2C). The Kcnq5 mRNA expression was also unaffected (Figure 2D). Hence, the qPCR data corroborated the sequencing data in the sgKcnq3-targeted mice (Figure 1A), indicating that we selectively reduced the Kcnq3 gene expression in the targeted cells.
Figure 2.
Quantitative RT-PCR of KCNQ channel subunits in NPY/AgRP neurons from OVX EB-treated female mice. (A) The Kcnq2 transcript (1.03 ± 0.06) was significantly more abundant than Kcnq3 (0.29 ± 0.02) using an unpaired t-test (t(6) = 11.32). Similarly, Kcnq5 (0.20 ± 0.02) was expressed at a much lower relative level to Kcnq2 (1.03 ± 0.06) according to an unpaired t-test (t(6) = 8.147). Only data from the control-injected animals are shown. (B) The relative expression of Kcnq2 was unaffected by the control (1.03 ± 0.07) or sgKcnq3 SaCas9 virus (1.22 ± 0.23) as determined by an unpaired t-test (t(9) = 1.535, p > 0.05). (C) Relative expression of Kcnq3 (10 cell pools, 4 per animal) normalized to the control average (1.02 ± 0.05) was significantly reduced in the knockdown animals (0.62 ± 0.05) using an unpaired t-test (t(9) = 5.811). (D) Relative expression of Kcnq5 (10 cell pools, 4 per animal) normalized to the control average. There was no significant difference between the control (1.06 ± 0.13) and sgKcnq3 (1.4 ± 0.20) virus-injected mice (unpaired t-test, t(5) = 1.649). (E) The relative expression of Kcnq3 mRNA (5 cell pools, 4 per animal) was no different between several control groups that included the AgRPCre animals injected with the SaCas9-control (0.97 ± 0.05) and AAV-DIO-mCh, AAV-DIO-mCh alone (0.91 ± 0.05), or uninjected NPYhrGFP mice (0.97 ± 0.07) using one-way ANOVA (F(2, 6) = 0.195). (F) Knockdown of Kcnq3 (5 cell pools, 4 per animal) was stable between 4 weeks (0.49 ± 0.1, unpaired t-test, t(4) = 5.370) and 8 weeks (0.51 ± 0.14, unpaired t-test, t(4) = 4.626) post-injection compared to 4-week fluorophore controls (1.01 ± 0.05). ∗∗p < 0.01 and ∗∗∗p < 0.001. Unless stated otherwise for qRT-PCR, 10 cell pools with 4 pools per animal were used. The number of animals per group is indicated in each panel. Error bars indicate SEM.
We then tested whether the procedure itself affected the Kcnq3 mRNA expression compared to the control SaCas9 virus. The NPYhrGFP and AgRPCre lines were bred to congenicity with C57BL/6J, and in the ARH, NPY and AgRP neurons were functionally identical. We expected their relative expression of Kcnq3 to be similar. However, the Cre-expression, stereotactic surgery, or viral infection could have affected cell health and transcription. Therefore, the AgRPCre female mice were given ARH injections of AAV1-EF1α-DIO-mCh, and after three weeks, the mice underwent OVX and were subsequently given EB treatment on the two days prior to sacrifice. After 4 weeks post-injection, the animals were sacrificed, and tissue processed for qRT-PCR as previously described. Age-matched NPYhrGFP female mice were also selected and processed. As expected, there was no significant difference between any of the groups (Figure 2E). Together these data indicated that the observed reduction in Kcnq3 mRNA expression was solely due to specific CRISPR gene editing by the vector with the correctly targeted sgRNA.
We wanted to determine the extent and stability of the knockdown. This was an important detail to develop a time frame in which to conduct behavioral and electrophysiological experiments. Therefore, six female AgRPCre mice were once again prepared as previously described. At 4–5 weeks, three of the mice were euthanized and their fluorescent ARH cells harvested. The remaining three mice were processed 8–10 weeks after injection. Kcnq3 relative expression was reduced at both the 4-week and 8-week marks compared to the mice injected with only the fluorophore virus, and there was no difference between the two time points (Figure 2F). These data indicated that the full effect of the single-vector CRISPR/SaCas9 gene editing was achieved by four weeks and remained stable, at least within this window, to allow a reasonable amount of time to test the effects of KCNQ3 knockdown.
3.2. Selective deletion of Kcnq3 reduced the M-current in NPY/AgRP neurons
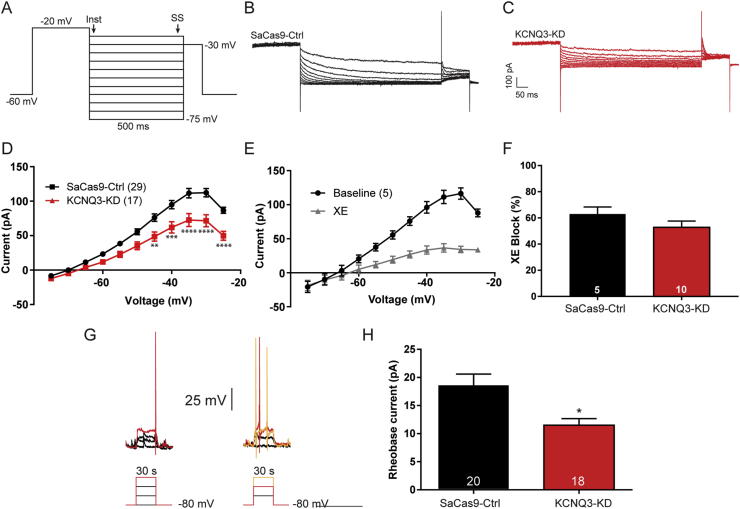
The M-current is a subthreshold K+ current that influences the resting membrane potential to regulate the firing of a neuron. Fasting and loss of E2 reduce the amplitude of the M-current in NPY/AgRP neurons, increasing their excitability [27] and presumably underlying, in part, the hyperphagia associated with those conditions. Therefore, we initially measured the effects of CRISPR-mediated Kcnq3 deletion in NPY/AgRP neurons from the fed EB-treated OVX females. Most patch clamp recordings were taken from slices adjacent to those used for the qPCR experiments, enabling gene transcription and electrophysiology data to be collected from the same animal. Although there was significantly higher input resistance in the KCNQ3-KD NPY/AgRP neurons (Ctrl 1.2 ± 0.1 GΩ, n = 31 vs KCNQ3-KD 1.6 ± .0.1 GΩ, n = 54; unpaired t-test, t(50) = 3.358; p < 0.01), the membrane capacitance (Ctrl 13.9 ± 0.6 pF vs KCNQ3-KD 14.7 ± 0.5 pF; unpaired t-test, t(67) = 0.9767, p > 0.05) and resting membrane potential (Ctrl −50.8 ± 1.7 mV, n = 31 vs KCNQ3-KD -47.4 ± 1.6 mV, n = 54; unpaired t-test, t(83) = 1.349, p > 0.05) were no different.
The M-current contributes to whole-cell K+ currents and can be elucidated in voltage clamps through established protocols [24,58]. Therefore, the deactivation or relaxation was measured as the difference between the steady state (>400 ms) and instantaneous (5–15 ms after voltage step) current (Figure 3A). The membrane potential was stepped from −60 (Vhold) to −20 mV to open the maximal number of KCNQ channels before stepping down to more hyperpolarized voltages (−25 to −75 mV). The current relaxation represents the deactivation of the KCNQ channels with greater “sags,” indicating that a higher M-current is present [27]. Current responses to this protocol were measured in the presence of tetrodotoxin (1 μM, TTX) to prevent sodium-driven action potentials (Figure 3B,C). In the KCNQ3-KD group, the IV plot of the M-current was significantly reduced at more depolarized voltages (Figure 3D, −45 to −25 mV). As this is a subthreshold current, the amplitude of the M-current should be greatest in this range of membrane potentials [23], which would further support the selective nature of CRISPR gene editing. XE 991 (40 μm) is a selective and potent blocker of KCNQ channels that inhibits the M-current (Figure 3E) [27]. XE displayed equivalent efficacy, blocking the M-current in the control and KCNQ3-KD animals (Figure 3F), which is also seen when recording from hippocampal neurons in KCNQ3-knockout mice [29].
Figure 3.
CRISPR gene editing of Kcnq3 decreased the M-current in OVX EB-treated female mice. (A) The deactivation protocol in voltage clamps started by holding the cell at −60 mV before a voltage step to −20 mV (300 ms), followed by 5 mV incremental steps (−25 to −75 mV, 500 ms). The current measured during the steady state (ss) was subtracted from the instantaneous (Inst) current. TTX (1 μM) was present in the bath. (B and C) The sag seen following the voltage step was the deactivation of the M-current, which was higher in the control animals (B) compared to the KCNQ3-KD animals (C). (D) Graphed as an I–V plot, the M-current was smaller across a range of voltages in the KCNQ3-KD mice (two-way ANOVA, main effect of group, F(1, 484) = 93.40) with significant post hoc differences between −45 and −25 mV (Bonferroni's test). (E) The M-current was strongly inhibited after 10 min of bath application of 40 μM XE 991, a selective blocker of KCNQ channels. (F) The area under the curve for the deactivation protocol was calculated before and after XE application to determine the percent block of the current. The XE sensitive current was the M-current with the residual current due to other K+ conductance. There was no difference in the effectiveness of XE between the control (62.9 ± 5.5%) and KCNQ3-KD (52.8 ± 4.9%) cells (unpaired t-test, t(13) = 1.360). Error bars indicate SEM. (G) In current clamps, the rheobase was assessed in both Cas9-control and Cas9-KCNQ3. The membrane potential was held at −80 mV before small amounts of current were injected (∼6 pA/step, 500 ms). The red current trace represents the minimum current necessary to elicit an action potential. (H) The amount of current required to elicit a spike was significantly greater in the controls than KCNQ3-KD cells (18.4 ± 2.2 vs 11.5 ± 1.2 pA, n = 20/18) even when normalized for capacitance (1.3 ± 0.1 vs 0.9 ± 0.1 pA/pS). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001. Number of cells per group is indicated in each panel. Error bars indicate SEM.
To further test for an increase in neuronal excitability, the rheobase was measured in current clamps [27]. Each cell was held in a hyperpolarized state at −80 mV while incremental current steps were applied to the cell (Figure 3G, bottom). The amount of injected current required to fire an action potential (rheobase current) was noted and compared between the AgRPCre mice injected with either the Cas9-Ctrl or KCNQ3-KD virus. The KCNQ3-KD mice required significantly less rheobase current to fire action potentials (Figure 3H), and this difference was significant even after normalizing to the cell capacitance. These results indicated that the deletion of Kcnq3 increased the excitability of NPY/AgRP neurons.
3.3. CRISPR knockdown of Kcnq3 did not affect food consumption
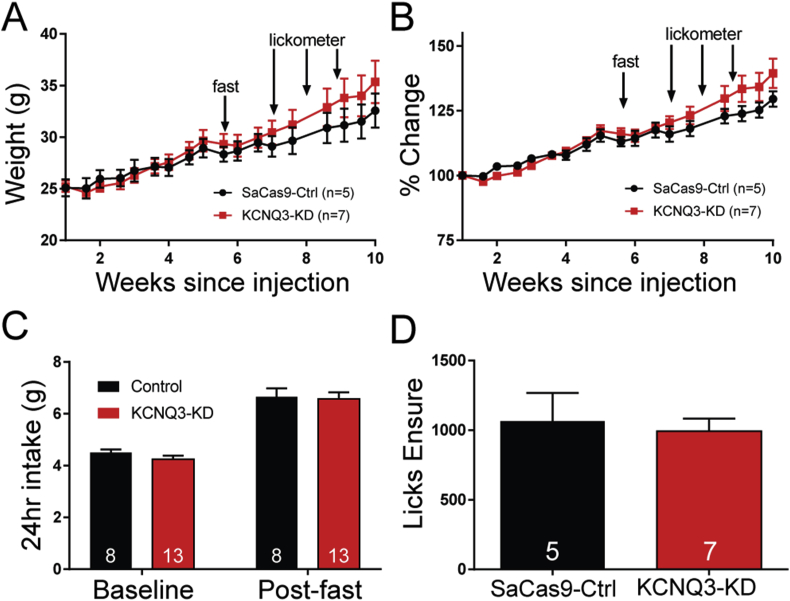
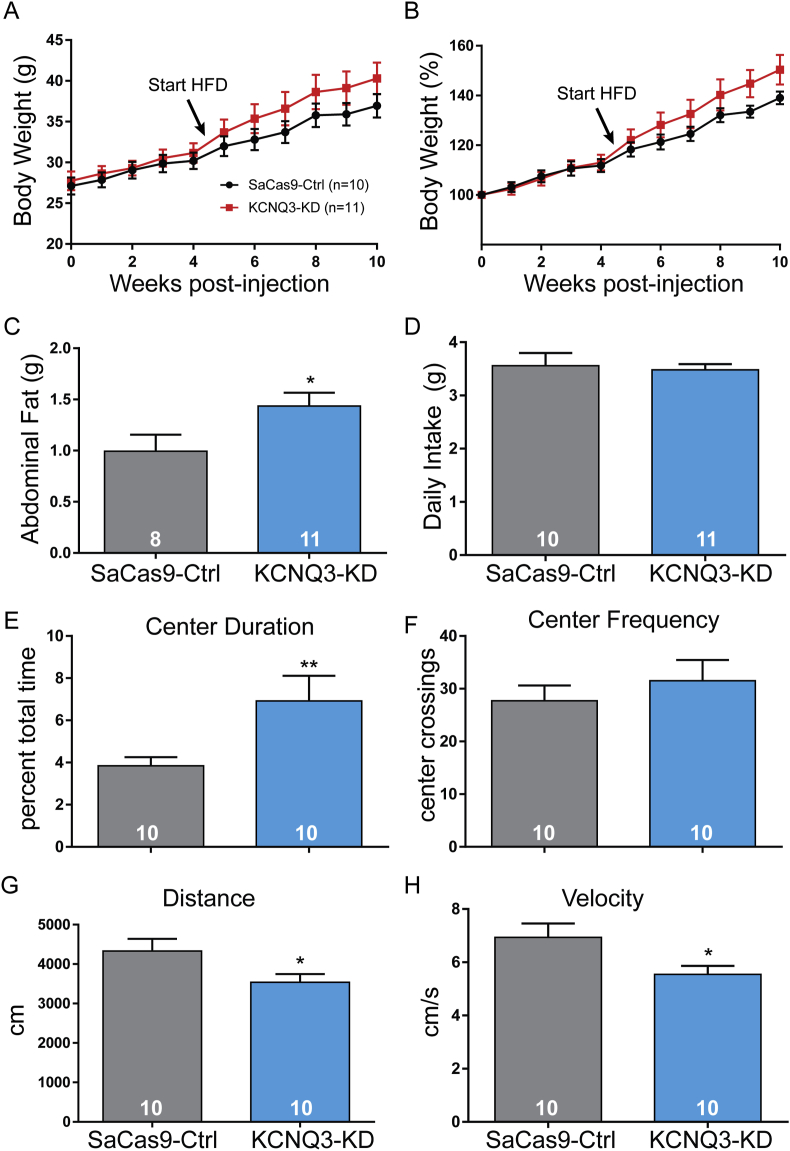
Females were used for the previously described PCR and initial electrophysiological experiments. However, we found estradiol can protect females from the effects of other manipulations such as a high-fat diet (HFD) [59]. We suspected that males might be more sensitive to KCNQ3 knockdown since they lacked the protective effects of estradiol, which enhances the expression of Kcnq5 mRNA and amplitude of the M-current [27]. Unlike in females, the resting membrane potential was significantly depolarized in KCNQ3-KD NPY/AgRP neurons (Ctrl −52.6 ± 1.3 mV, n = 32; KCNQ3-KD -43.3 ± 1.7 mV, n = 39; unpaired t-test, t(69) = 4.641, p < 0.0001). In global KCNQ3-knockout mice, the M-current is significantly diminished in cell types known to sparsely express Kcnq5 [29]. Therefore, we evaluated feeding behavior in the male mice. Weight was tracked for 10 weeks, but no difference was seen over this period (Figure 4A,B). This finding suggested that food consumption was the same between the groups. To test this, the mice were individually housed, and their daily food intake measured (Figure 4C), and as suspected, no difference was apparent between the control (4.5 ± 0.1 g) and KCNQ3-KD (4.2 ± 0.1 g) mice. However, KCNQ subunit expression responds to the animal's energy state, and following an overnight fast, relative Kcnq2 and Kcnq3 expression is reduced by almost half with a diminished M-current and increased food intake [27]. If Kcnq3 expression is already at a nadir, the elevated consumption could be attenuated, as both groups displayed a similar increase in post-fasting food intake (Ctrl 6.7 ± 0.3 g vs KCNQ3-KD 6.6 ± 0.3 g), perhaps due to a ceiling effect.
Figure 4.
Effects of Kcnq3 knockdown on ingestive behavior in intact male mice. (A) AgRPCre male mice underwent an EchoMRI and the measurements were used to balance the group assignment. After bilateral ARH injections of either SaCas9-Ctrl or KCNQ3-KD virus, the mice were weighed regularly. At five weeks post-injection prior to the onset of behavioral testing, the average weight of each group was similar. (B) Even after normalization to the initial weight, there was no difference in the average percent change between the groups. Interestingly, a trend of greater weight gain in the KCNQ3-KD mice began to emerge after overnight fasting and subsequent exposure to Ensure, a palatable liquid diet (see arrows). (C) The mice were individually housed and their daily intake of standard chow was evaluated (unpaired t-test, t(19) = 1.259, p > 0.05). The mice were then placed in clean cages with no food for 16 h. Once food was returned, as expected, the mice overconsumed compared to the baseline intake. This paradigm is known to decrease the expression of both Kcnq2 and Kcnq3. However, there was no difference in post-fast chow consumption (unpaired t-test, t(19) = 0.1763, p > 0.05) which would suggest a ceiling effect. (D) The mice were trained to drink from lickometers containing Ensure, a highly palatable nutritionally complete diet. After four days of training over a 30-min access period, there were no significant differences between the number of Ensure licks (Ctrl 1065 ± 202 vs KCNQ3-KD 993 ± 90; unpaired t-test, t(10) = 0.36, p > 0.05). The number of animals per group is indicated in each panel. Error bars indicate SEM.
Palatability is another factor that can drive food overconsumption [60]. Ensure (Abbott Nutrition) is a nutritionally complete liquid diet often used in feeding and reward studies. Rodents readily consume Ensure with minimal training, even increasing their daily energy intake in a sustained manner [61]. Therefore, ingestive behavior was also measured using a short-term access paradigm. Fed mice during their relative light phase were placed in an operant chamber (Med Associates) equipped with dual lickometers for 30 min on four training days. One bottle contained tap water and the other vanilla Ensure (36 g in 100 ml of tap water). However, both groups registered a similar number of licks with no statistically significant difference (Figure 4D).
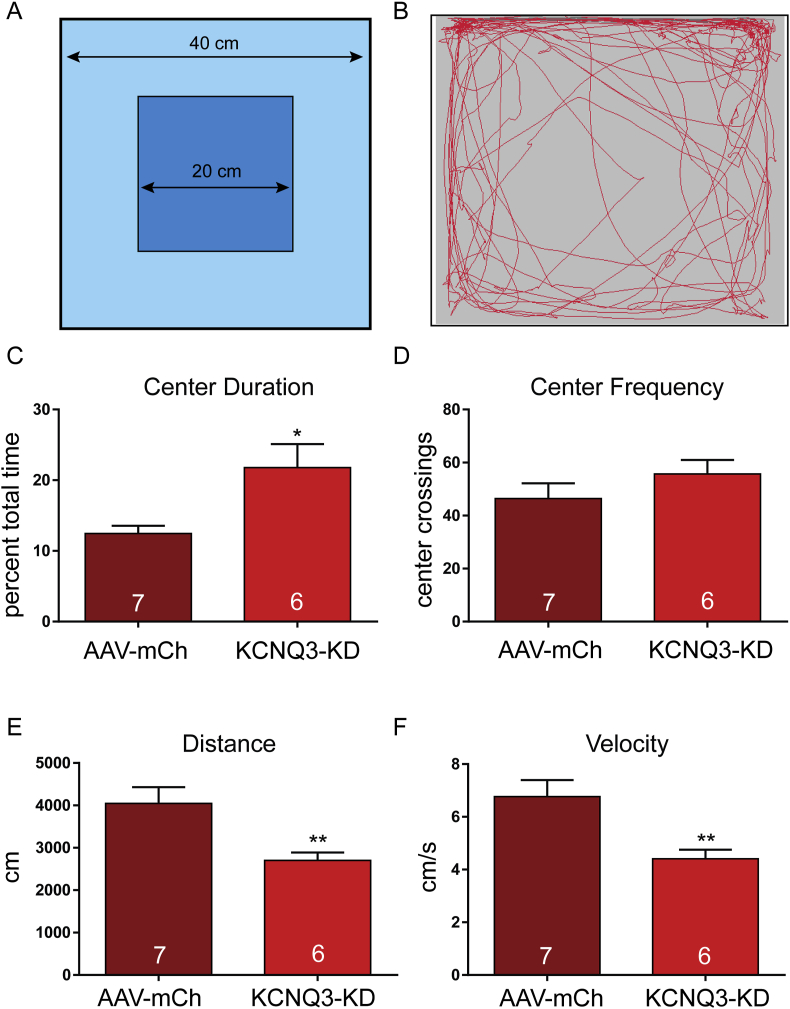
3.4. CRISPR knockdown of Kcnq3 affected open field behavior
NPY/AgRP activity is not just associated with ingestive behavior. Brain infusion of NPY agonists [62,63] and manipulations that increase NPY/AgRP activity [64,65] tend to produce anxiolytic effects. The Open Field Test (OFT, Figure 5A,B) is a well-established assay for comparing differences in anxiety between groups [66,67] based on time spent in the center (Figure 5C) and frequency of center crossing (Figure 5D) as well as a general assessment of locomotor activity [68,69]. Because the surgical control (AAV-mCh) and control vector (SaCas9-Ctrl) injected mice expressed the same levels of Kcnq3 mRNA (Figure 2E), we used the AAV-mCh injected mice for these OFT studies. Interestingly, there was a clear behavioral difference between groups. The KCNQ3-KD mice spent more time in the center, suggesting reduced anxiety. Although not as useful as home cage observations or as definitive as metabolic chamber measurements, the total distance traveled and average velocity (Figure 5E,F) provide insight into energy expenditure [70,71]. Again, the KCNQ3-KD mice were different than the control animals with significantly lower distance and velocity. With the center duration higher but center frequency the same, it was not surprising that the KCNQ3-KD animals exhibited lower total distances traveled. At the end of the behavioral studies, the mice were euthanized, and the quality of the injections was confirmed before subjects were included in the final analysis. Of the eight KCNQ3-KD animals, four had bilateral injections and three had unilateral injections. One had no apparent labeling and was excluded from the final analysis. The mice with better injections tended to show a more robust behavioral phenotype. For example, decreased distance and velocity correlated with the injection quality (r2 = 0.6, p < 0.05, and n = 7). Therefore, while the KCNQ3-KD animals did not display differences in consumption of standard chow, the KCNQ3-KD mice began to accelerate their weight gain following overnight fasting and access to palatable food. Coupled with indications of diminished physical activity, animals with KCNQ3-KD specifically in NPY/AgRP neurons may be more susceptible to developing obesity [72,73].
Figure 5.
Consequence of Kcnq3 knockdown on open field behavior in intact male mice. (A) Open field test (OFT) was conducted in 40 cm × 40 cm opaque white boxes over 10 min. (B) Movement was tracked using a video analysis. (C) KCNQ3 KD increased the time spent in the center during the OFT (control 12.5 ± 1.1% vs KCNQ3 KD 21.8 ± 3.3%, unpaired t-test, t(11) = 2.851, p < 0.01), indicating an anxiolytic effect. (D) The frequency of crossing between the center and periphery was the same between the groups (control 46.4 ± 5.7 vs KCNQ3 KD 55.7 ± 5.3, unpaired t-test, t(6) = 1.353, p > 0.05). (E) The KCNQ3-KD animals exhibited lower total distances traveled (control 4046 ± 384 cm vs KCNQ3 KD 2703 ± 185 cm, unpaired t-test, t(12) = 3.147, p < 0.01) and (F) lower velocity (control 6.8 ± 0.6 cm/s vs KCNQ3 KD 4.4 ± 0.8 cm/s, unpaired t-test, t(12) = 3.147, p < 0.01). The number of animals per group is indicated in each panel. Error bars indicate SEM.
3.5. KCNQ3-KD promoted weight gain on a high-fat diet
The rapidity at which the energy and reproductive state of the animal modulates KCNQ subunit expression and M-current activity in NPY/AgRP neurons indicates an important role in maintaining homeostasis. If fasting decreases the M-current through reduced Kcnq2 and Kcnq3 expression, one would predict that overconsumption or a high-fat diet (HFD) would cause the reverse. Furthermore, a more calorically dense diet typically drives weight gain in C57BL/6J mice [74], which could help clarify group differences within the time frame in which we established KCNQ3-KD was stable. Therefore, a new cohort of AgRPCre male mice was injected with AAV-DIO-YFP and either AAV-DIO-SaCas9-Ctrl or -Kcnq3 viruses into the ARH. At four weeks post-injection, when the CRISPR-mediated knockdown appeared to be complete and stable (Figure 2F), the mice were switched from standard chow to a high-fat diet (Research Diets, D12451). Both groups immediately began to accelerate their gain weight, albeit at different rates (Figure 6A), which was more evident when normalized as a percent of the starting weight (Figure 6B). The percent of starting weight values was analyzed with a two-way ANOVA, which indicated a main effect of weeks post-injection (F(10, 190) = 90.32 and p < 0.0001) and an interaction with viral treatment (F(10, 190) = 2.224, p < 0.05). A post hoc analysis found the difference in weights in weeks 10 and 11 approached but did not reach significance. A post mortem analysis revealed that the KCNQ3-KD animals had more abdominal fat (1.4 ± 0.1 g) than the controls (1.0 ± 0.2 g) (Figure 6C). Still, the average daily intake was not different between the groups (Figure 6D, Ctrl 3.6 ± 0.2; KCNQ3-KD 3.5 ± 0.1 g), which was similar to the observations with a standard chow diet. Therefore, a group difference in weight gain could not be attributed to greater consumption. We again utilized the OFT to assess differences in locomotor activity and, indirectly, energy expenditure. Consistent with the regular chow-fed mice, KCNQ3-KD in NPY/AgRP neurons was associated with increased time spent in the center (Figure 6E, Ctrl 3.8 ± 0.4% vs KCNQ3-KD 6.9 ± 1.2%), which reduced the total distance traveled (Figure 6G, 4333 ± 307 vs 3538 ± 209 cm, p < 0.05) and lowered the average velocity (Figure 6H, 6.9 ± 0.5 vs 5.5 ± 0.3 cm/s, p < 0.05).
Figure 6.
Weight gain from high-fat diet was exacerbated by Kcnq3 knockdown in NPY/AgRP neurons in intact male mice. (A) Body weight was tracked for 10 weeks from the day of injection in the male mice. After 4 weeks, the mice (arrow) were switched to high-fat chow (45% kcal from fat) and maintained on this diet until sacrifice. The KCNQ3-KD group tended to gain weight faster. (B) When normalized to starting measurements, the divergence in body weights became more apparent. A two-way ANOVA found that the HFD produced a main effect of time post-injection (F(10, 190) = 90.32) and an interaction with viral injection (F(10, 190) = 2.224). (C) Post mortem examination also found a significant increase in abdominal fat (unpaired t-test, t(17) = 2.14, p < 0.05). (D) The weight gain did not appear to be due to overconsumption of the HFD as both groups displayed the same average daily intake (3.6 ± 0.2 g vs 3.5 ± 0.1 g, t(19) = 0.308, p > 0.05). (E) At approximately 11 weeks post-injection and after 5–6 weeks of the HFD, the mice were tested in an open field. While the percent time spent in the center was lower than for the chow-fed group (Ctrl 3.8 ± 0.4 vs KCNQ3-KD 6.9 ± 1.2, unpaired t-test t(18) = 2.531, p < 0.01), the relative proportions remained approximately the same such that the KCNQ3-KD mice tended to spend more time in the center (t(18) = 2.53, p < 0.05). (F) As previously observed with a chow diet, knockdown of KCNQ3 did not affect the frequency of center crossings (t(18) = 0.77, p > 0.05). (G) As seen with the chow-fed group, KCNQ3 knockdown decreased the total distance traveled during a 10-min open field test (unpaired t-test, t(18) = 2.138, p < 0.05). (H) KCNQ3 knockdown also led to a diminished average velocity (unpaired t-test, t(18) = 2.251, p < 0.05). The number of animals per group is indicated in each panel. Error bars indicate SEM.
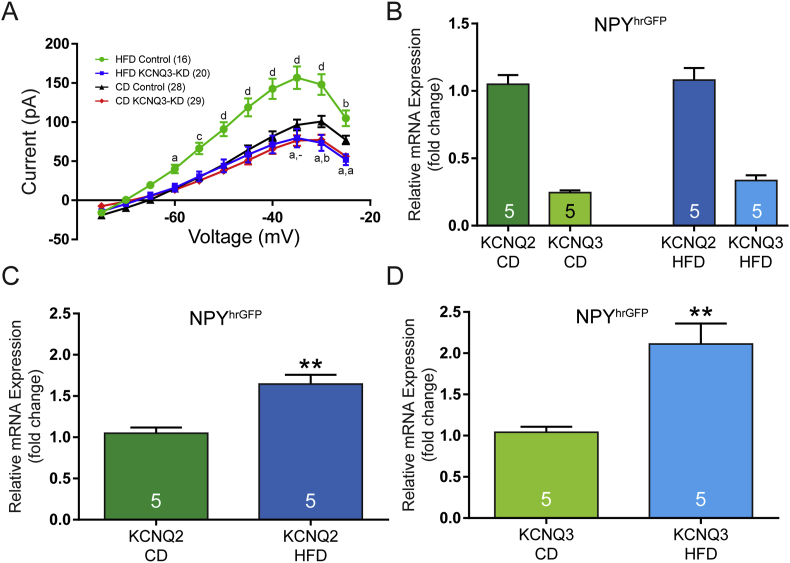
3.6. High-fat diet enhanced the M-current
The mice were euthanized, and the M-current was measured using the deactivation protocol. As expected, the M-current was smaller in KCNQ3-KD NPY/AgRP neurons compared to the CD and HFD controls (Figure 7A). Importantly, there was a dramatic increase in the M-current in NPY/AgRP neurons from the control and HFD mice. As this phenomenon was not obvious in the control diet mice, males or females, we hypothesized that the enhanced M-current was a consequence of the HFD. Therefore, we introduced a group of male mice to the HFD during weaning at 3 weeks of age while age-matched controls were maintained on the control diet. The NPYhrGFP mice were used as their expression levels were no different from the injected controls (Figure 2E) and did not require surgeries/injections. After 10–13 weeks, these mice along with the controls were processed for qRT-PCR to measure Kcnq2 and Kcnq3 as previously described (Section 3.1). Regardless of diet, Kcnq2 was still expressed at roughly four times that of Kcnq3 (Figure 7B). The HFD did lead to an almost 60% increase in the expression of Kcnq2 (Figure 7C) and doubled the expression of Kcnq3 (Figure 7D).
Figure 7.
Long-term high-fat diet enhanced the M-current in intact male mice. (A) The M-current was measured in vitro using whole-cell recordings from YFP+ AgRPCre neurons. Compared to recordings from the chow-fed (CD) SaCas9-control injected mice, CRISPR editing of Kcnq3 mRNA decreased the M-current to a similar extent in both the CD and high-fat diet (HFD) groups (two-way ANOVA, F(3, 979) = 89). The KCNQ3-KD (CD and HFD) groups differed from the CD control-injected NPY/AgRP neurons at more depolarized voltages. In addition, these three groups had significantly lower currents than those measured in cells from the HFD control-injected animals, in which the M-current was enhanced by the HFD (two-way ANOVA, post hoc comparisons made to the CD control). (B) As previously seen with the control diet, the Kcnq2 transcript (1.05 ± 0.07, n = 5) was significantly more abundant than Kcnq3 (0.25 ± 0.02) using an unpaired t-test (t(8) = 11.33). A similar expression ratio was seen with the HFD in which Kcnq2 (1.08 ± 0.09) was more highly expressed than Kcnq3 (0.34 ± 0.04) as determined by an unpaired t-test (t(8) = 7.63). (C) Normalized to the relative expression of Kcnq2 (1.05 ± 0.07) in the control diet group, the HFD led to a significant increase in the expression of Kcnq2 (1.65 ± 0.11) based on an unpaired t-test (t(8) = 4.53). (D) Normalized to the relative expression of Kcnq3 (1.04 ± 0.07, n = 5) in the control diet group, the HFD doubled the expression of Kcnq3 (2.11 ± 0.25) according to an unpaired t-test (t(8) = 4.17). The number of animals per group is indicated in each panel. For qRT-PCR experiments, 10 cell pools with 4 pools per animal were used. a, p < 0.05; b, p < 0.01; c, p < 0.001; and d, p < 0.0001. Error bars indicate SEM.
4. Discussion
4.1. Single-vector CRISPR gene editing
Using a newly developed CRISPR strategy, we showed that this single-vector approach reduced the expression of Kcnq3 in NPY/AgRP neurons, resulting in molecular, cellular, and behavioral changes. While this approach was previously validated [42], we extended this finding using a stringent method of harvesting healthy cells paired with a quantitative measurement of the relative gene expression [57]. We confirmed that this change was not due to the surgery, SaCas9-alone, or fluorophore. The single-vector SaCas9 system enabled a stable (at least 10 weeks) region and cell-specific knockdown of a gene in adult animals, avoiding potential developmental compensation or fetal lethality. Sanger sequencing found that only 12% of GFP+ cells retained a wild-type sequence while most cells exhibited an insertion or deletion that likely formed a frameshifted mRNA and loss-of-function mutation (Figure 1E) [42]. The current construct has PAM, sgRNA, and more than 50 nucleotides from an exon junction in the 5′ to 3′ prime direction, which support rapid nonsense-mediated decay of the mutated transcript [75]. The premature stop codon is generated at the start of the sequence before the first exon junction complex, meaning that the decay process is likely efficient [76]. As we selected fluorescent cells for the mRNA expression analysis, the remaining transcripts detected (Figure 2B) were probably edited, non-translatable Kcnq3 transcripts resistant to nonsense-mediated decay pathways [77,78] in addition to the small fraction of cells retaining a wild-type sequence. Still, SaCas9 gene editing is likely to produce milder phenotypes than traditional deletion methods due to incomplete coverage/infection by the virus and gene deletion.
A final innovation of the current approach is the targeting of ion channel subunits such as Kcnn3 in Hunker et al. [42]. Unlike the suprathreshold stimulus resulting from optogenetic or chemogenetic stimulation [6,7], CRISPR deletion of Kcnq3 increased the overall excitability of NPY/AgRP neurons without directly driving action potential firing, but still produced a behavioral phenotype. Therefore, this is a powerful tool for revealing the functional significance of channels in a specific neuronal population in adult animals. The ability to target subunits is a particularly exciting strategy. Many, if not most, receptors and channels lack subunit selective agonists and antagonists. The “gold standard” antagonists for studying the M-current, XE 991 and linopirdine, are broadly effective. The small-molecule UCL2077 [79] exhibits some subtype selectivity, strongly blocking KCNQ1-2, slightly inhibiting KCNQ3-4, and enhancing KCNQ5 currents [80]. However, heteromeric permutations complicate pharmacological manipulations. For example, while KCNQ subunits present differing sensitivity to tetraethylammonium (TEA), a non-selective K+ channel blocker [81], the most common heteromeric complexes are insensitive to the compound. Therefore, other than in heterologous expression systems, pharmacologically untangling the naturally occurring subpopulations of KCNQ channels is a daunting task. Herein we found that deletion of Kcnq3 attenuated the magnitude of the M-current, but the subunit was clearly not obligatory. In fact, the relative expression of Kcnq3 was less than half of Kcnq2 (Figure 2D). A rough estimate of the stoichiometry would therefore suggest that wild-type channels were predominantly KCNQ2 homomers. While KCNQ2/5 heteromers were possible, it was not clear how strongly KCNQ5 interacted with KCNQ2 [37]. One would expect that deleting Kcnq2 in NPY/AgRP neurons would produce more robust behavioral alterations, and deletion of Kcnq5 could reduce or eliminate the previously described sex differences [27]. However, the much lower abundance of Kcnq5 suggested that increased expression was unlikely to be the sole reason for the E2-enhanced M-current [27]. Other E2-driven mechanisms must regulate the M-current such as altered coupling between ghrelin receptors and KCNQ channels in ARH kisspeptin neurons [82]. However, the strategy we outlined can easily be adapted to study other KCNQ subunits to more fully explicate the M-current in future experiments.
4.2. The M-current controlled resting membrane potential and was suppressed during fasting in NPY neurons
Arcuate NPY/AgRP neurons are involved in the control of food intake and are affected by multiple central and peripheral signals that reflect energy states such as satiety and fasting (reviewed in [11,83,84]). In fed animals, NPY/AgRP neurons are less active and exhibit reduced firing activity [13], whereas NPY expression and release increase during periods of food restriction or deprivation [[85], [86], [87]]. In our previous studies, we demonstrated that NPY/AgRP neurons in fed animals were relatively quiescent and in a more hyperpolarized state compared to the fasted state [27]. The depolarized state reported in fasted mice was probably due to a combination of presynaptic inputs, changes in peripheral hormonal signals such as leptin [88,89], and modulation of endogenous K+ conductance such as KATP channels [90,91] and KCNQ channels [27]. For example, fasting greatly reduced the M-current while increasing the neuronal excitability of the NPY/AgRP neurons [27]. This effect on M-current activity was partially restored 24 h after refeeding, but not by a shorter 2 h refeeding. The suppression of the M-current by fasting may involve peripheral signals such as serum leptin or ghrelin concentrations. For example, ghrelin increases the excitability of NPY/AgRP neurons by inhibiting the M-current [92,93]. Therefore, in addition to changes in subunit expression, the coupling between metabotropic receptors and KCNQ channels can alter the magnitude of the M-current.
Most arcuate NPY/AgRP neurons express multiple KCNQ subunits and exhibit a robust M-current [27]. One of the primary functions of the M-current is to control action potential firing, spike-frequency adaptation, depolarizing afterpotentials, and afterhyperpolarization (AHP) currents [26,29,94,95]. We previously examined the effects of XE991, a selective blocker of KCNQ channels, on the excitability of NPY/AgRP neurons from fed males. We found that by blocking the M-current, quiescent NPY/AgRP neurons became depolarized and started firing continuously with a concomitant reduction in the rheobase current [27]. However, action potential firing did not occur in the presence of glutamatergic and GABAergic blockers although the membrane potential was similarly depolarized. The data suggest that the M-current in hypothalamic NPY/AgRP neurons is important for controlling the resting membrane potential. The M-current has a role in action potential generation in other neuronal cell types (for example, hippocampal CA1 pyramidal neurons) [96,97]. In arcuate NPY/AgRP neurons, the rheobase current was significantly reduced (Figure 3H), which in combination with presynaptic glutamatergic inputs generated “more perfect” synaptic integration [98]. Therefore, any alteration in M-current activity could have significantly affected the excitability of NPY/AgRP neurons.
4.3. Behavioral consequences of KCNQ3 knockdown in NPY/AgRP neurons
NPY/AgRP neurons are critical for the homeostatic regulation of energy balance. To fulfill this role, they position to detect changes in energy reserves [[99], [100], [101], [102]] and drastically alter their connectivity and excitability within hours [88]. The M-current is one mechanism thought to underlie this rapid adaptability [27]. At the start of these studies, it was uncertain whether attenuation of the M-current would be sufficient to promote weight gain. Suprathreshold activation, as seen with opto- and chemogenetic approaches, drives rapid and robust food consumption [6,7]. Furthermore, alterations that chronically increase NPY/AgRP activity, such as deletion of the leptin receptor or expression of the bacterial sodium channel, lead to hyperphagia and weight gain [103,104]. Yet, in the present study, neither daily consumption of standard chow nor weight gain was different between the groups despite elevated neuronal excitability as evidenced by the change in the rheobase. This led us to hypothesize that challenges might be necessary to draw out an ingestive phenotype. First, we measured intake following overnight fast access and found no difference. Next, the mice were given brief access to a vanilla Ensure as in vivo studies have shown that the palatability affects the degree to which NPY/AgRP neurons respond to food presentation [105]. However, neither measure of consumption was significantly different between the groups. Interestingly, the group weights and change in weight began to diverge following measures of ingestive behavior. While by no means conclusive, results from the OFT suggested that the weight gain might have been due to decreased energy expenditure. However, the additional time spent in the center could have reflected a less anxious phenotype. In the future, additional tests such as the elevated plus maze, running wheel activity, or metabolic chambers could be used to more precisely define the metabolic and behavioral phenotype of KCNQ3-KD mice.
Interestingly, by six months of age, global KCNQ3-knockout males gain weight, exhibit reduced activity, and breed poorly compared to controls and female knockouts (personal communication of Dr. Anastasios Tzingounis, University of Connecticut). These observations led us to question whether we could accelerate the phenotype development in our conditional KCNQ3-KD mice through a more calorically dense diet. A HFD is frequently used to induce obesity [106], and male mice in particular quickly become obese and glucose intolerant [107]. Indeed, the group body weights immediately began to diverge after introduction to the HFD (Figure 6A,B), and at time of sacrifice, the KCNQ3-KD mice had more abdominal fat. No differences were observed between the groups in average daily food intake, which was consistent with what we saw with control diet consumption. There was a reduction in food intake from earlier control diet measurements (Figure 4C), but likely reflected a homeostatic compensation to account for the greater caloric density of the HFD (3.36 kcal/g vs 4.7 kcal/g). These findings led us to again employ the OFT as an indirect measure of energy expenditure. Consistent with previous results, KCNQ3-KD in NPY/AgRP neurons led to decreased locomotor activity (Figure 6E,F). Therefore, the weight gain that followed KCNQ3-KD likely stemmed from a slightly positive energy balance due to decreased energy expenditure. However, further testing will be required to confirm this hypothesis and fully distinguish between anxiety and locomotor behavioral phenotypes as the center duration was reduced by KCNQ3-KD by both diets (Figure 5, Figure 6E).
We used whole-cell recordings in voltage clamps to confirm that the M-current was reduced in the KCNQ3-KD group, and although this was the case, we were surprised to see that HFD substantially increased the M-current in the control group (Figure 7A). As stated earlier, fasting is known to dramatically decrease the expression of Kcnq2 and Kcnq3 [27,108] with E2 increasing Kcnq5 expression and the magnitude of the M-current [27]. In addition, our findings of a greatly augmented M-current with the HFD suggested that the M-current was highly involved in modulating the excitability of NPY/AgRP neurons and ultimately the homeostatic energy balance. Interestingly, a long-term HFD (>8 weeks) is associated with increased NPY/AgRP neuronal excitability, although this effect was thought to involve Kv2.1 but not KCNQ channels [91,109]. Perhaps a longer exposure to a HFD would eventually result in a diminished M-current. Alternatively, an enhanced M-current could be a homeostatic response to counteract the attenuation of this leptin-sensitive K+ current [91]. Still, the average food intake reduced with the HFD to compensate for greater caloric density compared to the CD. The lack of an augmented M-current in the CRISPR-edited animals suggested the effect relied on KCNQ3, whose expression was sensitive to the energy state of the animals, and this conjecture appeared borne out by the qRT-PCR data (Figure 7C,D).
NPY/AgRP neurons make extensive projections throughout the brain to regions not directly involved in food intake [110]. The influence of increased NPY/AgRP activity, therefore, could also manifest as alterations in related but non-ingestive behaviors [111]. Chronic chemogenetic activation of NPY/AgRP neurons causes both hyperphagia and reduced physical activity [7], but can also diminish anxiety-related behaviors [65], although this could be due to lowered risk aversion rather than an anxiolytic effect [112]. Lacking available food, the animal may instead display displacement behaviors such as grooming rather than continue foraging [65]. Therefore, the decreased distance traveled in the KCNQ3-KD mice could have reflected lower levels of physical activity (distance traveled), a change in anxiety, or displacement behaviors in the absence of food. It should be noted that we measured this decreased activity during the relative light phase when the animals would normally be at rest. Future experiments can examine whether KCNQ3-KD affects dark phase behavior that is more sensitive to disruption by KCNQ3-KD. Nevertheless, our data demonstrated that the M-current is an important cellular mechanism by which NPY/AgRP activity is regulated in response to the animal's energy state. Significantly, loss or impairment of this channel subunit appear to cause behavioral alterations that predispose an organism to developing obesity.
Funding
This study was supported by United States National Institutes of Health Grants DK 68098 (MJK and OKR), NS 43330 (OKR), NS 38809 (MJK), and MH 104450 (LZ).
Acknowledgments
We thank Dr. Anastasios Tzioungounis (University of Connecticut) for insightful discussion and sharing unpublished findings.
Contributor Information
Todd L. Stincic, Email: stincic@ohsu.edu.
Martin J. Kelly, Email: kellym@ohsu.edu.
Conflict of interest
None declared.
References
- 1.Haskell-Luevano C., Cone R.D., Monck E.K., Wan Y.-P. Structure activity studies of the Melanocortin-4 receptor by in Vitro mutagenesis: indentification of agouti-related protien (AGRP), melanocortin agonist and synthetic peptide antagonist interaction determinants. Biochemistry. 2001;40(20):6164–6179. doi: 10.1021/bi010025q. [DOI] [PubMed] [Google Scholar]
- 2.Lu X.Y., Nicholson J.R., Akil H., Watson S.J. Time course of short-term and long-term orexigenic effects of Agouti-related protein (86-132) NeuroReport. 2001;12(6):1281–1284. doi: 10.1097/00001756-200105080-00045. [DOI] [PubMed] [Google Scholar]
- 3.Gropp E., Shanabrough M., Borok E., Xu A.W., Janoschek R., Buch T. Agouti-related peptide-expressing neurons are mandatory for feeding. Nature Neuroscience. 2005;8(10):1289–1291. doi: 10.1038/nn1548. [DOI] [PubMed] [Google Scholar]
- 4.Luquet S., Perez F.A., Hnasko T.S., Palmiter R.D. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science. 2005;310(5748):683–685. doi: 10.1126/science.1115524. [DOI] [PubMed] [Google Scholar]
- 5.Stanley B.G., Leibowitz S.F. Neuropeptide Y injected in the paraventricular hypothalamus: a powerful stimulant of feeding behavior. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(11):3940–3943. doi: 10.1073/pnas.82.11.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aponte Y., Atasoy D., Sternson S.M. AGRP neurons are sufficient to orchestrate feeding behavior rapidly and without training. Nature Neuroscience. 2011;14(3):351–355. doi: 10.1038/nn.2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krashes M.J., Koda S., Ye C., Rogan S.C., Adams A.C., Cusher D.S. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. Journal of Clinical Investigation. 2011;121(4):1424–1428. doi: 10.1172/JCI46229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei Q., Krolewski D.M., Moore S., Kumar V., Li F., Martin B. Uneven balance of power between hypothalamic peptidergic neurons in the control of feeding. Proceedings of the National Academy of Sciences. 2018;115(40):E9489–E9498. doi: 10.1073/pnas.1802237115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y., Knight Z.A. Making sense of the sensory regulation of hunger neurons. BioEssays. 2016;38(4):316–324. doi: 10.1002/bies.201500167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elmquist J.K. Hypothalamic pathways underlying the endocrine, autonomic, and behavioral effects of leptin. Physiology & Behavior. 2001;74:703–708. doi: 10.1016/s0031-9384(01)00613-8. [DOI] [PubMed] [Google Scholar]
- 11.Gao Q., Horvath T.L. Neurobiology of feeding and energy expenditure. Annual Review of Neuroscience. 2007;30:367–398. doi: 10.1146/annurev.neuro.30.051606.094324. [DOI] [PubMed] [Google Scholar]
- 12.Kohno D., Nakata M., Maekawa F., Fujiwara K., Maejima Y., Kuramochi M. Leptin suppresses ghrelin-induced activation of neuropeptide Y neurons in the arcuate nucleus via phosphatidylinositol 3-kinase- and phosphodiesterase 3-mediated pathway. Endocrinology. 2007;148(5):2251–2263. doi: 10.1210/en.2006-1240. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi K.A., Cone R.D. Fasting induces a large, leptin-dependent increase in the intrinsic action potential frequency of orexigenic arcuate nucleus neuropeptide Y/Agouti-related protein neurons. Endocrinology. 2005;146(3):1043–1047. doi: 10.1210/en.2004-1397. [DOI] [PubMed] [Google Scholar]
- 14.Van den Pol A.N., Yao Y., Fu L.-Y., Foo K., Huang H., Coppari R. Neuromedin B and gastrin-releasing peptide excite arcuate nucleus neuropeptide Y neurons in a novel transgenic mouse expressing stong renilla green fluorescent protein in NPY neurons. Journal of Neuroscience. 2009;29(14):4622–4639. doi: 10.1523/JNEUROSCI.3249-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rønnekleiv O.K., Kelly M.J. Diversity of ovarian steroid signaling in the hypothalamus. Frontiers in Neuroendocrinology. 2005;26(2):65–84. doi: 10.1016/j.yfrne.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Micevych P.E., Kelly M.J. Membrane estrogen receptor regulation of hypothalamic function. Neuroendocrinology. 2012;96(2):103–110. doi: 10.1159/000338400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crowley W.R., Tessel R.E., O'Donohue T.L., Adler B.A., Kalra S.P. Effects of ovarian hormones on the concentrations of immunoreactive neuropeptide Y in discrete brain regions of the female rat: correlation with serum luteinizing hormone (LH) and median eminence LH-releasing hormone. Endocrinology. 1985;117(3):1151–1155. doi: 10.1210/endo-117-3-1151. [DOI] [PubMed] [Google Scholar]
- 18.Pelletier G., Li S., Luu-The V., Labrie F. Oestrogenic regulation of pro-opiomelanocortin, neuropeptide Y and corticotrophin-releasing hormone mRNAs in mouse hypothalamus. Journal of Neuroendocrinology. 2007;19(6):426–431. doi: 10.1111/j.1365-2826.2007.01548.x. [DOI] [PubMed] [Google Scholar]
- 19.Yang M.J., Wang F., Wang J.H., Wu W.N., Hu Z.L., Cheng J. PI3K integrates the effects of insulin and leptin on large-conductance Ca2+-activated K+ channels in neuropeptide Y neurons of the hypothalamic arcuate nucleus. American Journal of Physiology: Endocrinology and Metabolism. 2010;298(2):E193–E201. doi: 10.1152/ajpendo.00155.2009. [DOI] [PubMed] [Google Scholar]
- 20.Wang J.-H., Wang F., Yang M.-J., Yu D.-F., Wu W.-N., Liu J. Leptin regulated calcium channels of NPY and POMC neurons by activation of different signal pathways. Neuroscience. 2008;156(1):89–98. doi: 10.1016/j.neuroscience.2008.04.079. [DOI] [PubMed] [Google Scholar]
- 21.Qiu J., Zhang C., Borgquist A., Nestor C.C., Smith A.W., Bosch M.A. Insulin excites anorexigenic proopiomelanocortin neurons via activation of canonical transient receptor potential channels. Cell Metabolism. 2014;19(4):682–693. doi: 10.1016/j.cmet.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delmas P., Brown D.A. Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nature Reviews Neuroscience. 2005;6(11):850–862. doi: 10.1038/nrn1785. [DOI] [PubMed] [Google Scholar]
- 23.Robbins J. KCNQ potassium channels: physiology, pathophysiology, and pharmacology. Pharmacology & Therapeutics. 2001;90(1):1–19. doi: 10.1016/s0163-7258(01)00116-4. [DOI] [PubMed] [Google Scholar]
- 24.Xu C., Roepke T.A., Zhang C., Rønnekleiv O.K., Kelly M.J. Gonadotropin-releasing hormone (GnRH) activates the M-current in GnRH neurons: an autoregulatory negative feedback mechanism? Endocrinology. 2008;149(5):2459–2466. doi: 10.1210/en.2007-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang H.S., Pan Z., Shi W., Brown B.S., Wymore R.S., Cohen I.S. KCNQ2 and KCNQ3 potassium channel subunits: molecular correlates of the M-channel. Science. 1998;282(5395):1890–1893. doi: 10.1126/science.282.5395.1890. [DOI] [PubMed] [Google Scholar]
- 26.Peters H.C., Hu H., Pongs O., Storm J.F., Isbrandt D. Conditional transgenic suppression of M channels in mouse brain reveals functions in neuronal excitability, resonance and behavior. Nature Neuroscience. 2005;8(1):51–60. doi: 10.1038/nn1375. [DOI] [PubMed] [Google Scholar]
- 27.Roepke T.A., Qiu J., Smith A.W., Rønnekleiv O.K., Kelly M.J. Fasting and 17β-estradiol differentially modulate the M-current in neuropeptide Y neurons. Journal of Neuroscience. 2011;17(33):11825–11835. doi: 10.1523/JNEUROSCI.1395-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tatulian L., Delmas P., Abogadie F.C., Brown D.A. Activation of expressed KCNQ potassium currents and native neuronal M-type potassium currents by the anti-convulsant drug retigabine. Journal of Neuroscience. 2001;21(15):5535–5545. doi: 10.1523/JNEUROSCI.21-15-05535.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tzingounis A.V., Nicoll R.A. Contribution of KCNQ2 and KCNQ3 to the medium and slow afterhyperpolarization currents. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(50):19974–19979. doi: 10.1073/pnas.0810535105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe H., Nagata E., Kosakai A., Nakamura M., Yokoyama M., Tanaka K. Disruption of the epilepsy KCNQ2 gene results in neural hyperexcitability. Journal of Neurochemistry. 2000;75(1):28–33. doi: 10.1046/j.1471-4159.2000.0750028.x. [DOI] [PubMed] [Google Scholar]
- 31.Tzingounis A.V., Heidenreich M., Kharkovets T., Spitzmaul G., Jensen H.S., Nicoll R.A. The KCNQ5 potassium channel mediates a component of the afterhyperpolarization current in mouse hippocampus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(22):10232–10237. doi: 10.1073/pnas.1004644107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwake M., Pusch M., Kharkovets T., Jentsch T.J. Surface expression and single channel properties of KCNQ2/KCNQ3, M-type K+ channels involved in epilepsy. Journal of Biological Chemistry. 2000;275(18):13343–13348. doi: 10.1074/jbc.275.18.13343. [DOI] [PubMed] [Google Scholar]
- 33.Etxeberria A., Santana-Castro I., Regalado M.P., Aivar P., Villarroel A. Three mechanisms underlie KCNQ2/3 heteromeric potassium M-channel potentiation. Journal of Neuroscience. 2004;24(41):9146–9152. doi: 10.1523/JNEUROSCI.3194-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasmussen H.B., Frøkjær-Jensen C., Jensen C.S., Jensen H.S., Jørgensen N.K., Misonou H. Requirement of subunit co-assembly and ankyrin-G for M-channel localization at the axon initial segment. Journal of Cell Science. 2007;120(6):953–963. doi: 10.1242/jcs.03396. [DOI] [PubMed] [Google Scholar]
- 35.Soh H., Pant R., LoTurco J.J., Tzingounis A.V. Conditional deletions of epilepsy-associated KCNQ2 and KCNQ3 channels from cerebral cortex cause differential effects on neuronal excitability. Journal of Neuroscience. 2014;34(15):5311–5321. doi: 10.1523/JNEUROSCI.3919-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lerche C., Scherer C.R., Seebohm G., Derst C., Wei A.D., Busch A.E. Molecular cloning and functional expression of KCNQ5, a potassium channel subunit that may contribute to neuronal M-current diversity. Journal of Biological Chemistry. 2000;275(29):22395–22400. doi: 10.1074/jbc.M002378200. [DOI] [PubMed] [Google Scholar]
- 37.Schroeder B.C., Hechenberger M., Weinreich F., Kubisch C., Jentsch T.J. KCNQ5, a novel potassium channel broadly expressed in brain, mediates M-type currents. Journal of Biological Chemistry. 2000;275(31):24089–24095. doi: 10.1074/jbc.M003245200. [DOI] [PubMed] [Google Scholar]
- 38.Gorski J.A., Talley T., Qiu M., Puelles L., Rubenstein J.L.R., Jones K.R. Cortical excitatory neurons and Glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. Journal of Neuroscience. 2002;22(15):6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swiech L., Heidenreich M., Banerjee A., Habib N., Li Y., Trombetta J. In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nature Biotechnology. 2015;33(1):102–106. doi: 10.1038/nbt.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiyama J., Mikuni T., Yasuda R. Virus-mediated genome editing via homology-directed repair in mitotic and postmitotic cells in mammalian brain. Neuron. 2017;96(4):755–768. doi: 10.1016/j.neuron.2017.10.004. e755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bak R.O., Dever D.P., Porteus M.H. CRISPR/Cas9 genome editing in human hematopoietic stem cells. Nature Protocols. 2018;13(2):358–376. doi: 10.1038/nprot.2017.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hunker A.C., Soden M.E., Krayushkina D., Heymann G., Awatramani R., Zweifel L.S. Conditional single vector CRISPR/SaCas9 viruses for efficient mutagenesis in the adult mouse nervous system. Cell Reports. 2020;30(12):4303–4316. doi: 10.1016/j.celrep.2020.02.092. e4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhuang X., Masson J., Gingrich J.A., Rayport S., Hen R. Targeted gene expression in dopamine and serotonin neurons of the mouse brain. Journal of Neuroscience Methods. 2005;143(1):27–32. doi: 10.1016/j.jneumeth.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 44.Tong Q., Ye C.-P., Jones J.E., Elmquist J.K., Lowell B.B. Synaptic release of GABA by AgRP neurons is required for normal regulation of energy balance. Nature Neuroscience. 2008;11(9):998–1000. doi: 10.1038/nn.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gore B.B., Soden M.E., Zweifel L.S. Manipulating gene expression in projection-specific neuronal populations using combinatorial viral approaches. Current Protocols in Neuroscience. 2013;65:1–20. doi: 10.1002/0471142301.ns0435s65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bosch M.A., Tonsfeldt K.J., Rønnekleiv O.K. mRNA expression of ion channels in GnRH neurons: subtype-specific regulation by 17β-Estradiol. Molecular and Cellular Endocrinology. 2013;367(1–2):85–97. doi: 10.1016/j.mce.2012.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C., Tonsfeldt K.J., Qiu J., Bosch M.A., Kobayashi K., Steiner R.A. Molecular mechanisms that drive estradiol-dependent burst firing of Kiss1 neurons in the rostral periventricular preoptic area. American Journal of Physiology: Endocrinology and Metabolism. 2013;305(11):E1384–E1397. doi: 10.1152/ajpendo.00406.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 49.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jensen T., Kiersgaard M., Sørensen D., Mikkelsen L. Fasting of mice: a review. Laboratory Animals. 2013;47(4):225–240. doi: 10.1177/0023677213501659. [DOI] [PubMed] [Google Scholar]
- 51.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, N.Y.) 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471(7340):602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, N.Y.) 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Duan Y., Ma G., Huang X., D'Amore P.A., Zhang F., Lei H. The clustered, regularly interspaced, short palindromic repeats-associated endonuclease 9 (CRISPR/Cas9)-created MDM2 T309G mutation enhances vitreous-induced expression of MDM2 and proliferation and survival of cells. Journal of Biological Chemistry. 2016;291(31):16339–16347. doi: 10.1074/jbc.M116.729467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. doi: 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Concordet J.-P., Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Research. 2018;46(W1):W242–W245. doi: 10.1093/nar/gky354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stincic T.L., Grachev P., Bosch M.A., Rønnekleiv O.K., Kelly M.J. Estradiol drives the anorexigenic activity of proopiomelanocortin neurons in female mice. eNeuro. 2018;5(4):e0103–e0118. doi: 10.1523/ENEURO.0103-18.2018. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schweitzer P. Cannabinoids decrease the K+ M-current in hippocampal CA1 neurons. Journal of Neuroscience. 2000;20(1):51–58. doi: 10.1523/JNEUROSCI.20-01-00051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu J., Bosch M.A., Zhang C., Rønnekleiv O.K., Kelly M.J. Estradiol protects neuropeptide Y/agouti-related peptide neurons against insulin resistance in females. Neuroendocrinology. 2020;110(1–2):105–118. doi: 10.1159/000501560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Erlanson-Albertsson C. How palatable food disrupts appetite regulation. Basic and Clinical Pharmacology and Toxicology. 2005;97(2):61–73. doi: 10.1111/j.1742-7843.2005.pto_179.x. [DOI] [PubMed] [Google Scholar]
- 61.Archer Z.A., Brown Y.A., Rayner D.V., Stubbs R.J., Mercer J.G. Effect of flavour of liquid Ensure diet supplement on energy intake in male SD rats. Physiology & Behavior. 2006;89(3):414–419. doi: 10.1016/j.physbeh.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 62.Heilig M., Söderpalm B., Engel J.A., Widerlöv E. Centrally administered neuropeptide Y (NPY) produces anxiolytic-like effects in animal anxiety models. Psychopharmacology. 1989;98(4):524–529. doi: 10.1007/BF00441953. [DOI] [PubMed] [Google Scholar]
- 63.Kokare D.M., Dandekar M.P., Chopde C.T., Subhedar N. Interaction between neuropeptide Y and alpha-melanocyte stimulating hormone in amygdala regulates anxiety in rats. Brain Research. 2005;1043(1):107–114. doi: 10.1016/j.brainres.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 64.Dietrich M.O., Bober J., Ferreira J.G., Tellez L.A., Mineur Y.S., Souza D.O. AgRP neurons regulate development of dopamine neuronal plasticity and nonfood-associated behaviors. Nature Neuroscience. 2012;15(8):1108–1110. doi: 10.1038/nn.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dietrich M.O., Zimmer M.R., Bober J., Horvath T.L. Hypothalamic Agrp neurons drive stereotypic behaviors beyond feeding. Cell. 2015;160(6):1222–1232. doi: 10.1016/j.cell.2015.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carola V., D'Olimpio F., Brunamonti E., Mangia F., Renzi P. Evaluation of the elevated plus-maze and open-field tests for the assessment of anxiety-related behaviour in inbred mice. Behavioural Brain Research. 2002;134(1):49–57. doi: 10.1016/s0166-4328(01)00452-1. [DOI] [PubMed] [Google Scholar]
- 67.Prut L., Belzung C. The open field as a paradigm to measure the effects of drugs on anxiety-like behaviors: a review. European Journal of Pharmacology. 2003;463(1):3–33. doi: 10.1016/s0014-2999(03)01272-x. [DOI] [PubMed] [Google Scholar]
- 68.Gould T.D., Dao D.T., Kovacsics C.E. The open field test. In: Gould T.D., editor. Mood and anxiety related phenotypes in mice: characterization using behavioral tests. Humana Press; Totowa, NJ: 2009. pp. 1–20. [Google Scholar]
- 69.Kraeuter A.-K., Guest P.C., Sarnyai Z. The open field test for measuring locomotor activity and anxiety-like behavior. In: Guest P.C., editor. Pre-clinical models: techniques and protocols. Springer New York; New York, NY: 2019. pp. 99–103. [DOI] [PubMed] [Google Scholar]
- 70.Lund T.D., Rovis T., Chung W.C., Handa R.J. Novel actions of estrogen receptor-beta on anxiety-related behaviors. Endocrinology. 2005;146(2):797–807. doi: 10.1210/en.2004-1158. [DOI] [PubMed] [Google Scholar]
- 71.Estrada C.M., Ghisays V., Nguyen E.T., Caldwell J.L., Streicher J., Solomon M.B. Estrogen signaling in the medial amygdala decreases emotional stress responses and obesity in ovariectomized rats. Hormones and Behavior. 2018;98:33–44. doi: 10.1016/j.yhbeh.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 72.Blundell J.E., Finlayson G. Is susceptibility to weight gain characterized by homeostatic or hedonic risk factors for overconsumption? Physiology & Behavior. 2004;82(1):21–25. doi: 10.1016/j.physbeh.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 73.Spiegelman B.M., Flier J.S. Obesity and the regulation of energy balance. Cell. 2001;104(4):531–543. doi: 10.1016/s0092-8674(01)00240-9. [DOI] [PubMed] [Google Scholar]
- 74.Collins S., Martin T.L., Surwit R.S., Robidoux J. Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: physiological and molecular characteristics. Physiology & Behavior. 2004;81(2):243–248. doi: 10.1016/j.physbeh.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 75.Neu-Yilik G., Amthor B., Gehring N.H., Bahri S., Paidassi H., Hentze M.W. Mechanism of escape from nonsense-mediated mRNA decay of human β-globin transcripts with nonsense mutations in the first exon. RNA. 2011;17(5):843–854. doi: 10.1261/rna.2401811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lindeboom R.G.H., Supek F., Lehner B. The rules and impact of nonsense-mediated mRNA decay in human cancers. Nature Genetics. 2016;48(10):1112–1118. doi: 10.1038/ng.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garneau N.L., Wilusz J., Wilusz C.J. The highways and byways of mRNA decay. Nature Reviews. Molecular Cell Biology. 2007;8(2):113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 78.Lykke-Andersen S., Jensen T.H. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nature Reviews. Molecular Cell Biology. 2015;16(11):665–677. doi: 10.1038/nrm4063. [DOI] [PubMed] [Google Scholar]
- 79.Shah M.M., Javadzadeh-Tabatabaie M., Benton D.C.H., Ganellin C.R., Haylett D.G. Enhancement of hippocampal pyramidal cell excitablility by the novel selective slow-afterhyperpolarization channel blocker 3-(triphenylmethylaminomethyl)pyridine (UCL2077) Molecular Pharmacology. 2006;70(5):1494–1502. doi: 10.1124/mol.106.026625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soh H., Tzingounis A.V. The specific slow afterhyperpolarization inhibitor UCL2077 is a subtype-selective blocker of the epilepsy associated KCNQ channels. Molecular Pharmocology. 2010;78(6):1088–1095. doi: 10.1124/mol.110.066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hadley J.K., Noda M., Selyanko A.A., Wood I.C., Abogadie F.C., Brown D.A. Differential tetraethylammonium sensitivity of KCNQ1-4 potassium channels. British Journal of Pharmacology. 2000;129(3):413–415. doi: 10.1038/sj.bjp.0703086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Conde K., Roepke T. 17β-estradiol increases arcuate KNDy neuronal sensitivity to ghrelin inhibition of the M-current in female mice. Neuroendocrinology. 2019 doi: 10.1159/000503146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Saper C.B., Chou T.C., Elmquist J.K. The need to feed: homeostatic and hedonic control of eating. Neuron. 2002;36(2):199–211. doi: 10.1016/s0896-6273(02)00969-8. [DOI] [PubMed] [Google Scholar]
- 84.Woods S.C. The control of food intake: behavioral versus molecular perspectives. Cell Metabolism. 2009;9(6):489–498. doi: 10.1016/j.cmet.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brady L.S., Smith M.A., Gold P.W., Herkenham M. Altered expression of hypothalamic neuropeptide mRNA in food-restricted and food-deprived rats. Neuroendocrinology. 1995;52(5):441–447. doi: 10.1159/000125626. [DOI] [PubMed] [Google Scholar]
- 86.Yoshihara T., Honma S., Honma K. Effects of restricted daily feeding on neuropeptide Y release in the rat paraventricular nucleus. American Journal of Physiology. 1996;270(4 Pt 1):E589–E595. doi: 10.1152/ajpendo.1996.270.4.E589. [DOI] [PubMed] [Google Scholar]
- 87.Hahn T.M., Breininger J.F., Baskin D.G., Schwartz M.W. Coexpression of Agrp and NPY in fasting-activated hypothalamic neurons. Nature Neuroscience. 1998;1(4):271–272. doi: 10.1038/1082. [DOI] [PubMed] [Google Scholar]
- 88.Pinto S., Roseberry A.G., Liu H., Diano S., Shanabrough M., Cai X. Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science. 2004;304(5667):110–115. doi: 10.1126/science.1089459. [DOI] [PubMed] [Google Scholar]
- 89.Baskin D.G., Breininger J.F., Bonigut S., Miller M.A. Leptin binding in the arcuate nucleus is increased during fasting. Brain Research. 1999;828(1–2):154–158. doi: 10.1016/s0006-8993(99)01252-4. [DOI] [PubMed] [Google Scholar]
- 90.van den Top M., Lyons D.J., Lee K., Coderre E., Renaud L.P., Spanswick D. Pharmacological and molecular characterization of ATP-sensitive K+ conductances in CART and NPY/AgRP expressing neurons of the hypothalamic arcuate nucleus. Neuroscience. 2007;144(3):815–824. doi: 10.1016/j.neuroscience.2006.09.059. [DOI] [PubMed] [Google Scholar]
- 91.Baver S.B., Hope K., Guyot S., Bjorbaek C., Kaczorowski C., O'Connell K.M. Leptin modulates the intrinsic excitability of AgRP/NPY neurons in the arcuate nucleus of the hypothalamus. Journal of Neuroscience. 2014;34(16):5486–5496. doi: 10.1523/JNEUROSCI.4861-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shi L., Bian X., Qu Z., Ma Z., Zhou Y., Wang K. Peptide hormone ghrelin enhances neuronal excitability by inhibition of Kv7/KCNQ channels. Nature Communications. 2013;4:1435. doi: 10.1038/ncomms2439. [DOI] [PubMed] [Google Scholar]
- 93.Yasrebi A., Hsieh A., Mamounis K.J., Krumm E.A., Yang J.A., Magby J. Differential gene regulation of GHSR signalling pathway in the arcuate nucleus and NPY neurons by fasting, diet-induced obesity, and 17β-estradiol. Molecular and Cellular Endocrinology. 2016;422:42–56. doi: 10.1016/j.mce.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gu N., Vervaeke K., Hu H., Storm J.F. Kv7/KCNQ/M and HCN/h, but not KCa2/SK channels, contribute to the somatic medium after-hyperploarization and excitability control in CA1 hippocampal pyramidal cells. The Journal of Physiology. 2005;566(Pt 3):689–715. doi: 10.1113/jphysiol.2005.086835. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yue C., Yaari Y. KCNQ/M channels control spike afterdepolarization and burst generation in hippocampal neurons. Journal of Neuroscience. 2004;24(19):4614–4624. doi: 10.1523/JNEUROSCI.0765-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leao R.N., Tan H.M., Fisahn A. Kv7/KCNQ channels control action potential phasing of pyramidal neurons during hippocampal gamma oscillations in vitro. Journal of Neuroscience. 2009;29(42):13353–13364. doi: 10.1523/JNEUROSCI.1463-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu H., Vervaeke K., Storm J.F. M-channels (Kv7/KCNQ channels) that regulate synaptic integration, excitability, and spike pattern of CA1 pyrimidal cells are located in the perisomatic region. Journal of Neuroscience. 2007;27(8):1853–1867. doi: 10.1523/JNEUROSCI.4463-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Branco T., Tozer A., Magnus C.J., Sugino K., Tanaka S., Lee A.K. Near-perfect synaptic integration by Nav1.7 in hypothalamic neurons regulates body weight. Cell. 2016;165(7):1749–1761. doi: 10.1016/j.cell.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lam T.K.T., Pocai A., Guiterrez-Juarez R., Obici S., Bryan J., Aguilar-Bryan L. Hypothalamic sensing of circulating fatty acids is required for glucose homeostasis. Nature Medicine. 2005;11(3):320–327. doi: 10.1038/nm1201. [DOI] [PubMed] [Google Scholar]
- 100.Chaudhri O., Small C., Bloom S. Gastrointestinal hormones regulating appetite. Philosophical Transactions of the Royal Society B: Biological Sciences. 2006;361(1471):1187–1209. doi: 10.1098/rstb.2006.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Coll A.P., Farooqi I.S., O'Rahilly S. The hormonal control of food intake. Cell. 2007;129(2):251–262. doi: 10.1016/j.cell.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dietrich M.O., Horvath T.L. Feeding signals and brain circuitry. European Journal of Neuroscience. 2009;30(9):1688–1696. doi: 10.1111/j.1460-9568.2009.06963.x. [DOI] [PubMed] [Google Scholar]
- 103.Xu J., Bartolome C.L., Low C.S., Yi X., Chien C.H., Wang P. Genetic identification of leptin neural circuits in energy and glucose homeostases. Nature. 2018;556(7702):505–509. doi: 10.1038/s41586-018-0049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhu C., Jiang Z., Xu Y., Cai Z.L., Jiang Q., Xu Y. Profound and redundant functions of arcuate neurons in obesity development. Nature Metabolism. 2020;2(8):763–774. doi: 10.1038/s42255-020-0229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen Y., Lin Y.-C., Kuo T.-W., Knight Z.A. Sensory detection of food rapidly modulates arcuate feeding circuits. Cell. 2015;160(5):829–841. doi: 10.1016/j.cell.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang C.-Y., Liao J.K. A mouse model of diet-induced obesity and insulin resistance. Methods in Molecular Biology (Clifton, N.J.) 2012;821:421–433. doi: 10.1007/978-1-61779-430-8_27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qiu J., Bosch M.A., Meza C., Navarro U.V., Nestor C.C., Wagner E.J. Estradiol protects proopiomelanocortin neurons against insulin resistance. Endocrinology. 2018;159(2):647–664. doi: 10.1210/en.2017-00793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Henry F.E., Sugino K., Tozer A., Branco T., Sternson S.M. Cell type-specific transcriptomics of hypothalamic energy-sensing neuron responses to weight-loss. eLife. 2015;4 doi: 10.7554/eLife.09800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wei W., Pham K., Gammons J.W., Sutherland D., Liu Y., Smith A. Diet composition, not calorie intake, rapidly alters intrinsic excitability of hypothalamic AgRP/NPY neurons in mice. Scientific Reports. 2015;5 doi: 10.1038/srep16810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Broberger C., Johansen J., Johansson C., Schalling M., Hokfelt T. The neuropeptide Y/agouti gene-related protein (AGRP) brain circuitry in normal, anorectic, and monosodium glutamate-treated mice. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(25):15043–15048. doi: 10.1073/pnas.95.25.15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sutton A.K., Krashes M.J. Integrating hunger with rival motivations. Trends in Endocrinology and Metabolism. 2020;31(7):495–507. doi: 10.1016/j.tem.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 112.Padilla S.L., Qiu J., Soden M.E., Sanz E., Nestor C.C., Barker F.D. Agouti-related peptide neural circuits mediate adaptive behaviors in the starved state. Nature Neuroscience. 2016;19(5):734–741. doi: 10.1038/nn.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]