Abstract
Our recent work identified a genetic variant of the α345 hexamer of the collagen IV scaffold that is present in patients with glomerular basement membrane diseases, Goodpasture’s disease (GP) and Alport syndrome (AS), and phenocopies of AS in knock-in mice. To understand the context of this “Zurich” variant, an 8-amino acid appendage, we developed a construct of the WT α345 hexamer using the single-chain NC1 trimer technology, which allowed us to solve a crystal structure of this key connection module. The α345 hexamer structure revealed a ring of 12 chloride ions at the trimer–trimer interface, analogous to the collagen α121 hexamer, and the location of the 170 AS variants. The hexamer surface is marked by multiple pores and crevices that are potentially accessible to small molecules. Loop-crevice-loop features constitute bioactive sites, where pathogenic pathways converge that are linked to AS and GP, and, potentially, diabetic nephropathy. In Pedchenko et al., we demonstrate that these sites exhibit conformational plasticity, a dynamic property underlying assembly of bioactive sites and hexamer dysfunction. The α345 hexamer structure is a platform to decipher how variants cause AS and how hypoepitopes can be triggered, causing GP. Furthermore, the bioactive sites, along with the pores and crevices on the hexamer surface, are prospective targets for therapeutic interventions.
Keywords: extracellular matrix, collagen, X-ray crystallography, atomic force microscopy (AFM), genetic disease
Abbreviations: AS, Alport syndrome; DN, diabetic nephropathy; GBM, glomerular basement membrane; GP, Goodpasture’s disease; LCL, loop-crevice-loop
Prominent diseases of the glomerular basement membrane (GBM), a specialized form of the extracellular matrix, are diabetic nephropathy (DN), Alport syndrome (AS), and Goodpasture’s disease (GP). The morphological abnormalities in the GBM involve structural alterations in collagen IVα345 scaffold, the major GBM component (1, 2, 3). The mechanisms whereby collagen IV enables normal GBM function or causes GBM abnormalities and dysfunction in disease are unknown.
In Pokidysheva et al. (4), we found that the α345 hexamer, a key connection module within the collagen IVα345 scaffold, is a focal point of bioactivity within the GBM, based on investigation of the Zurich variant that caused AS. To understand how variants, including the Z-variant, in AS cause renal dysfunction, knowledge of the 3D structure of the α345 hexamer is critical. Moreover, this knowledge is also critical to understanding renal dysfunction in GP and DN and development of therapies. Therefore, we solved a crystal structure of the α345NC1 hexamer, a goal that has been pursued by scientists for several decades. The crystal structure revealed features critical for GBM function and in pathogenesis of AS and GP, and, potentially, DN, thus providing a framework for the development of therapies.
Results
After decades of attempts to isolate and crystallize the α345 hexamer, we developed a recombinant single-chain NC1 trimer technology (5) and used it to define the arrangement of chains and solve the crystal structure of the recombinant α345 hexamer.
Composition and arrangement of chains within the α345 hexamer
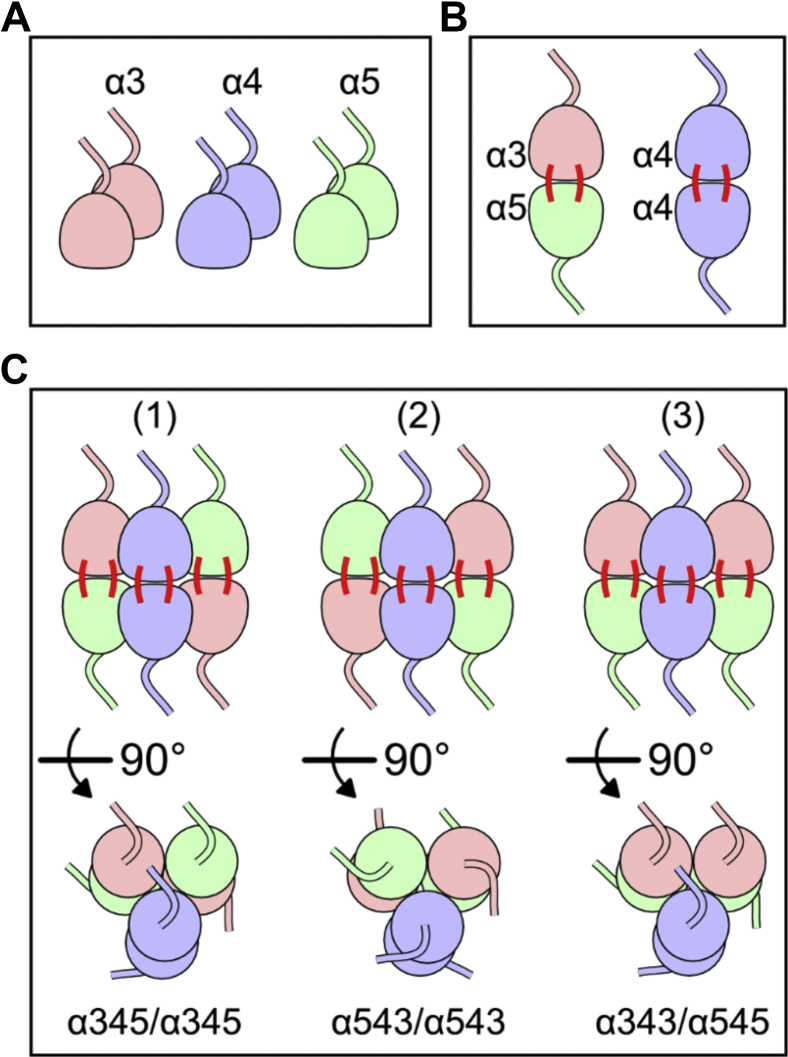
The collagen IVα345 scaffold, composed of the α3, α4, and α5 chains, is a major constituent of the GBM. The α345 hexamer can be extracted from the GBM using collagenase treatment (6). We previously determined the equimolar composition of α3, α4, and α5 chains in the α345 hexamers isolated from the GBM (7) (Fig. 1A). In the same study, we also found the presence of α3-α5 and α4-α4 covalently linked dimers (7), where chain monomers from opposite trimers were connected by sulfilimine bonds (8) (Fig. 1B). However, the exact chain arrangement within the hexamer is unknown. Taking into account our previous findings, there are only three possible chain arrangements in the α345 hexamers: (1) α345-to-α345, (2) α543-to-α543, and (3) α343-to-α545 (Fig. 1C).
Figure 1.
Possible chain orientations within the α345 hexamer. We previously found that the GBM NC1α345 hexamer contains two copies of the α3 (light red), α4 (light blue), and α5 (light green) NC1 monomers (A) and sulfilimine crosslinking of opposite chains forming two types of dimers, α3-α5 and α4-α4 (B) (7). Taking into account the composition and sulfilimine crosslinking pattern, three chain orientations are possible within the NC1α345 hexamer (C). GBM, glomerular basement membrane.
We tested these possibilities experimentally using the single-chain NC1 trimer technology (5) developed and verified for the α121 hexamer. This technology allows to define composition and orientation of the chains in the trimer (Fig. 2). The α345, α543, α343, and α545 NC1 single-chain NC1 trimers carrying the signal peptide for secretion (Fig. S1) were transiently expressed in expiCHO cells. Total cell lysates and media were analyzed for the presence of protein of interest using Western blotting (Fig. S2). Only α345 and α545 constructs were detected in the media, whereas α543 and α343 were exclusively trapped within the cells, indicating misfolding problem. Although α545 was partially secreted to the medium, the required partner, α343, was trapped within the cells. Coexpression of α343 and α545 did not rescue secretion of α343. Collectively, the single-chain α345 NC1 trimer represents native composition and orientation of chains. This is also supported by previous studies where association of individual α4 or α5 NC1 monomers with the α3 chain was selectively blocked by the mAbs (7).
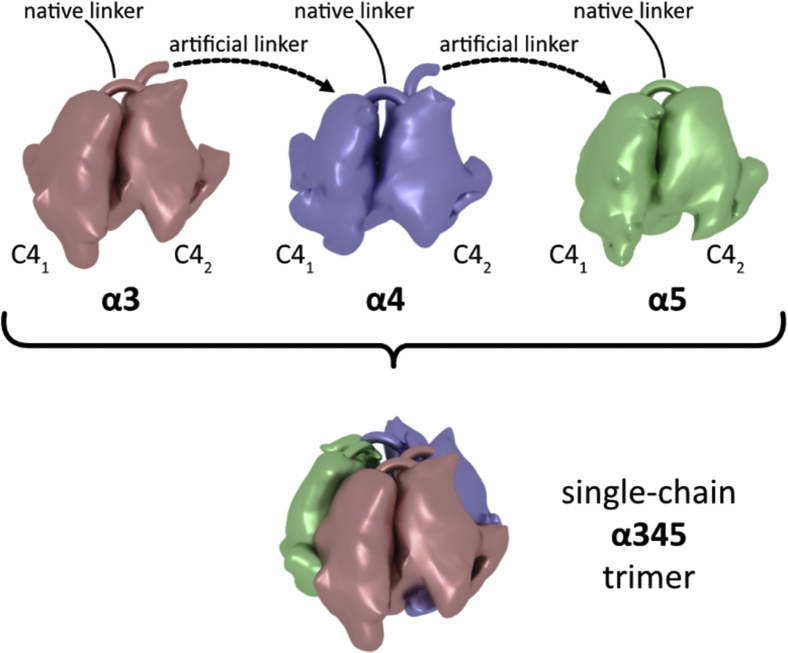
Figure 2.
Schematic presentation of the single-chain technology used for generating the α345 NC1 trimer. A single polypeptide combines NC1 monomers of α3, α4, and α5 chains via artificial linkers analogous to native linkers between C4 subdomains within each monomer. Short linkers restrict orientation of monomers in a way that the C42 subdomain of the α3 chain will interface with the C41 subdomain of the α4 chain and the C42 subdomain of the α4 chain will interface with the C41 subdomain of the α5 chain. Finally, the C42 subdomain of the α5 chain will interface with the C41 subdomain of the α3 chain in the folded single-chain α345 NC1 trimer. The same strategy was applied to generate α543, α343, and α545 forms of the single-chain NC1 trimer.
Crystal structure of the α345 hexamer
The single-chain α345 NC1 trimer produced recombinantly in stably transfected HEK293 cells was purified to homogeneity (Fig. S3) and crystallized in the presence of sodium chloride. The crystal structure was solved at the 1.76 Å resolution with a single polypeptide chain per asymmetric unit (Table S1). It has the designed orientation of α chains in the order of α3-to-α4-to-α5 (Figs. 2 and 3). The atomic structure is homologous to the crystal structure of the α121 NC1 domain (5) (Fig. 3), which is also homologous to all reported crystal structures of tissue extracted from the human and bovine α121 NC1 domain (9, 10, 11). Least-square superpositions of whole α345 and α121 trimers and individual chains revealed no significant variations between corresponding Cα atoms (overall r.m.s.d. 0.67 Å) (Table S2). Remarkably, α4 chain has the highest r.m.s.d. value of 2.03 Å when superposed with the α2 chain, although still in the range for highly homologous proteins (12). Most of the structural difference is due to presence of two extra residues, which is unique for α4 chain, within the bottom loop, making a contact with the adjacent α5 chain (Fig. 3). Superposition of α3 and α5 chains with corresponding α1 chains has only 0.55 Å and 0.54 Å r.m.s.d. values, which are typical even for identical proteins (12). Structures of α3 and α5 chains in the α345 NC1 trimer are also identical to the crystal structures observed in α3 and α5 homotrimers/homohexamers, r.m.s.d. of 0.55 to 0.60 Å for α3 and 0.58 Å for α5 (Table S3). Nevertheless, NC1 domains demonstrate sufficient plasticity by forming α2 homotetramer/homo-octamer and α4 homohexamer/homododecamer upon crystallization (13), although for the price of ∼20% and ∼10% of their inner sequences being unstructured. Superposition of the α4 structure of the α345 trimer with the structured part of α4 in the artefactual α4 homohexamer/homododecamer has r.m.s.d. values in the range 4.50 to 4.74 Å, pointing to significant variations even for the structured part (Table S3).
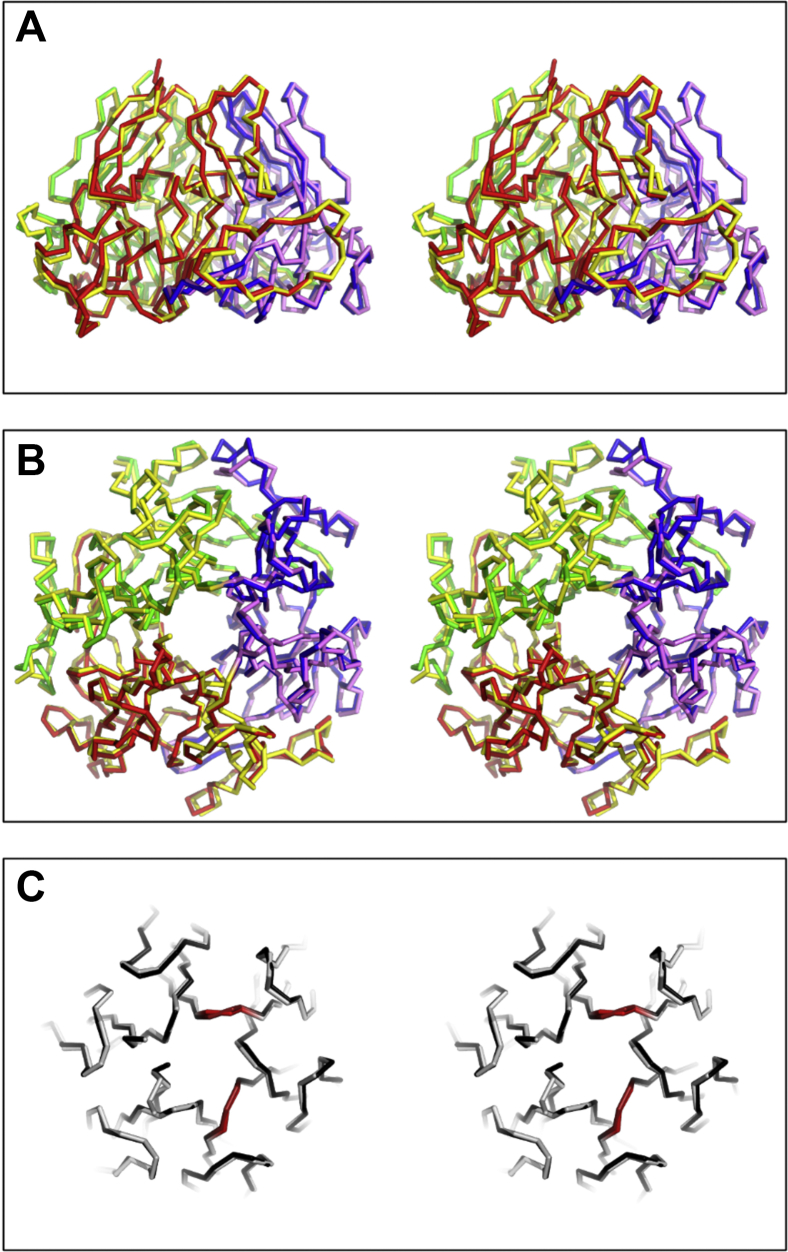
Figure 3.
Crystal structure comparison of the single-chain α345 (this study) and α121 NC1 trimers (PDB code: 6mpx) (5). The side view (A) and top view (B) of the superposition of α345 and α121 shown as stereopairs with the wireframe backbone. The α345 NC1 trimer coloring is red for α3, blue for α4, and green for α5. The α121 NC1 trimer coloring is yellow for α1 and violet for α5. C, the top slab of the top view showing the superposition of α345 (white) and α121 (black) shown as a stereopair with a wireframe backbone. Artificial linkers between chains are shown in red.
High homology and identity of the structures of α3, α4, and α5 chains in the α345 NC1 trimer with the α1 and α2 chains in the NC1 trimer and with α3 and α5 chains in homotrimers/homohexamers also verifies the correct design of the artificial linkers connecting α3-to-α5 and α4-to-α5. All C4 subdomain linkers, native and artificially introduced, are well structured (Fig. 3C), related by a pseudohexagonal symmetry and have comparable atomic displacement factors (Fig. S8 in Supporting Section 3), which further verifies the design of artificial linkers.
Analysis of the crystal structure of the single-chain α345 NC1 trimer reveals an unexpected pairing of chains at the hexamer interface. In the crystal structures of the α121 NC1, the pairing follows the rule even–even and odd–odd, that is, α2–α2 and α1–α1, which is consistent with covalent sulfilimine cross-linking of these pairs (8). Based on sulfilimine cross-linking analysis of the GBM α345 hexamer, the expected chain pairs were α3–α5 and α4–α4 (7), although in the crystal structure, we observed α3–α3 and α4–α5 pairs. Thus, the present crystal structure of the α345 hexamer demonstrates a labile nature of a trimer–trimer orientation in the hexamer before sulfilimine cross-linking. Presumably, during crystallization process, only one particular form was selected as compatible with a given crystal packing. In support of this labile nature of the hexamer are the crystal structures of α3 and α5 homohexamers, which demonstrated alternative pairs of chains in hexamers (13). How nature selects one particular orientation before covalent cross-linking by sulfilimine bonds remains to be explored. In conclusion, our single-chain NC1 trimer method allowed for determination of the α345 hexamer crystal structure, a major goal that has been pursued by scientists for decades.
The chloride ions at the NC1 α345 trimer–trimer interface
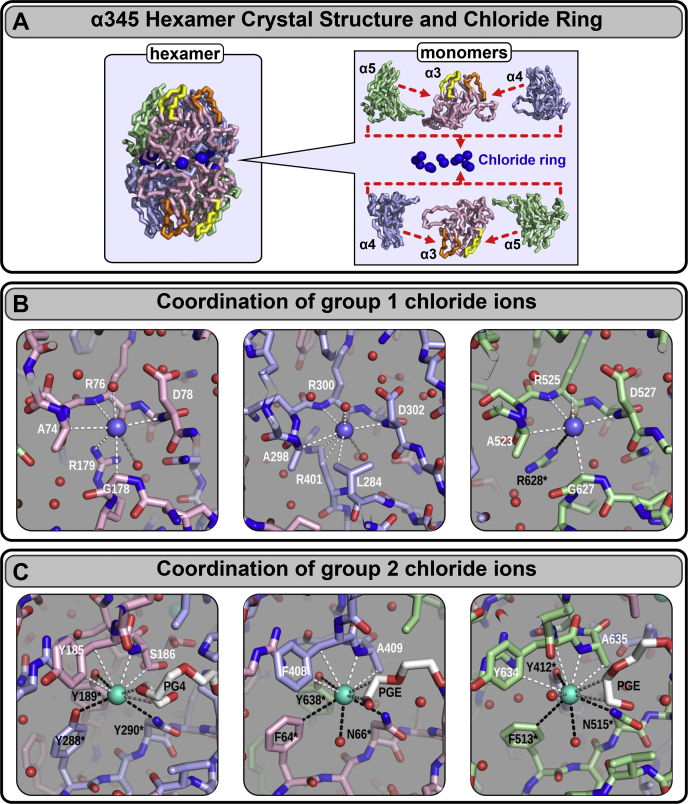
Despite rotational mismatch of the crystallized hexamer, we discovered a set of twelve Cl- ions at the trimer–trimer interface (Fig. 4) having the same geometry as in the α121 hexamer (5). All 12 Cl- at the trimer–trimer interface have comparable electron densities (Fig. S4) and atomic displacement factors in the range from 19.1 to 19.7 Å2, like the core residues of the polypeptide chain (Fig. 8). Together, the 12 ions form a chloride ring at the hexamer interface (Fig. 5). Analogously to the Cl- ions in the α121 hexamer, these ions form two structurally different groups (Figs. 5, S4, and Table S4). Geometry and residue specificity of Cl- coordination are identical between α345 and α121, with only one exception, that is, two-thirds of group 1 ions (four ions per hexamer) are not coordinated by the salt bridge to the arginine residue of the opposite trimer, rather identical arginine within the same trimer coordinates respective ions (Cl- ions #1–2). Potentially, the absence of these salt bridges might impact the hexamer stability of the crystallized form. The other two Cl- ions (#3) in group 1 have ‘classical’ geometry observed in the α121 hexamer with the arginine residues from the opposite trimer involved in the coordination (5).
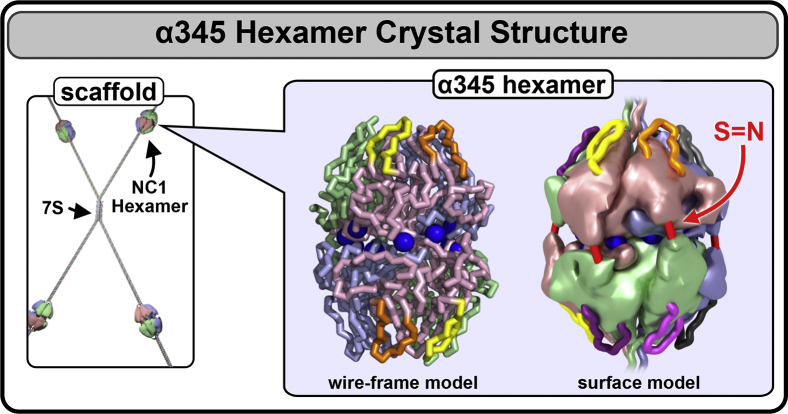
Figure 4.
The crystal structure of the collagen IVα345hexamer. In the GBM, collagen IVα345 scaffold is built from α345 protomers, where the NC1 hexameric complex is a key assembly unit (left). The crystal structure of the α345 hexamer is shown as a backbone wire-frame (middle) depicting loops (GP hypoepitopes) and the chloride ring at the trimer–trimer interface. The corrected surface model (right) depicts the appropriate chain pairing and sulfilimine crosslinks (S = N; shown in red). GBM, glomerular basement membrane.
Figure 8.
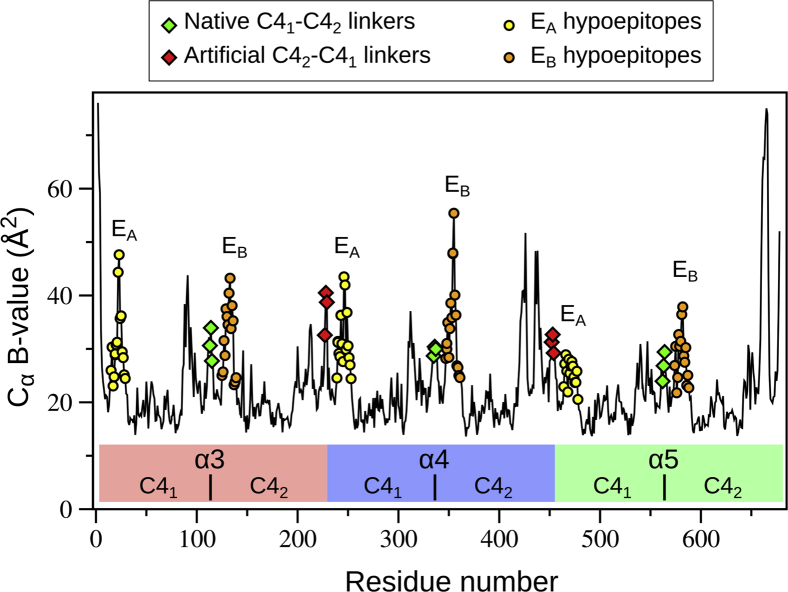
Mean square displacement (B values) of Cα atoms. B values for native and artificial linkers are comparable as depicted by green and red diamonds, respectively. B values for EA and EB hypoepitopes are shown as yellow and orange circles. Residue positions are marked with colored bars: light red for α3, light blue for α4, and light green for α5 chains. Borders between C41 and C42 subdomains are depicted as vertical lines.
Figure 5.
The α345 hexamer crystal structure reveals a ring of chloride ions that coordinate the trimer–trimer interface.A, the crystal structure of the α345 hexamer is shown as a backbone wireframe (left). Individual chains are indicated by different colors as shown on the right. EA and EB loops in the α3 chain are shown in yellow and orange, respectively. 12 chloride ions (blue spheres) form a Cl- ring at the interface between two α345 trimers (left and right). B, chloride ion coordination for group 1 chloride. Group 1 chloride are responsible for introducing intramolecular salt bridges utilizing chloride coordinating Arg (R76, R300, and R525 in α3, α4, and α5 respectively). Coloring for group 1 chloride is blue shown as blue spheres, while carbon atom coloring in NC1 chains is light red for α3, light blue for α4, and light green for α5. C, chloride ion coordination for group 2 chloride. Group 2 chloride are responsible for directly bridging trimeric protomers in the collagen IV hexamer, and unlike group 1 chloride, are available for interaction with PEG molecules (PG4 and PGE). Coloring for group 2 chloride is cyan, while carbon atom coloring in PEG molecules (PG4 and PGE) is white. Chloride interactions from one NC1 molecule are shown as white dashes, while interactions from the opposite molecule are shown as black dashes. Carbon atom coloring for NC1 chains is the same as above.
Chloride ions of group 2 are located in pockets and also interact with PEG molecules used for crystallization (Fig. 6C and Table S4). Although those interactions are nonspecific and weak, they point to possibility to modulate chloride binding and the hexamer assembly by specifically developed agents, which might become drugs. In summary, like the α121 hexamer, the α345 hexamer possesses two groups of chloride ions at the trimer–trimer interface forming a 12-ion ring critical for hexamer assembly and stability.
Figure 6.
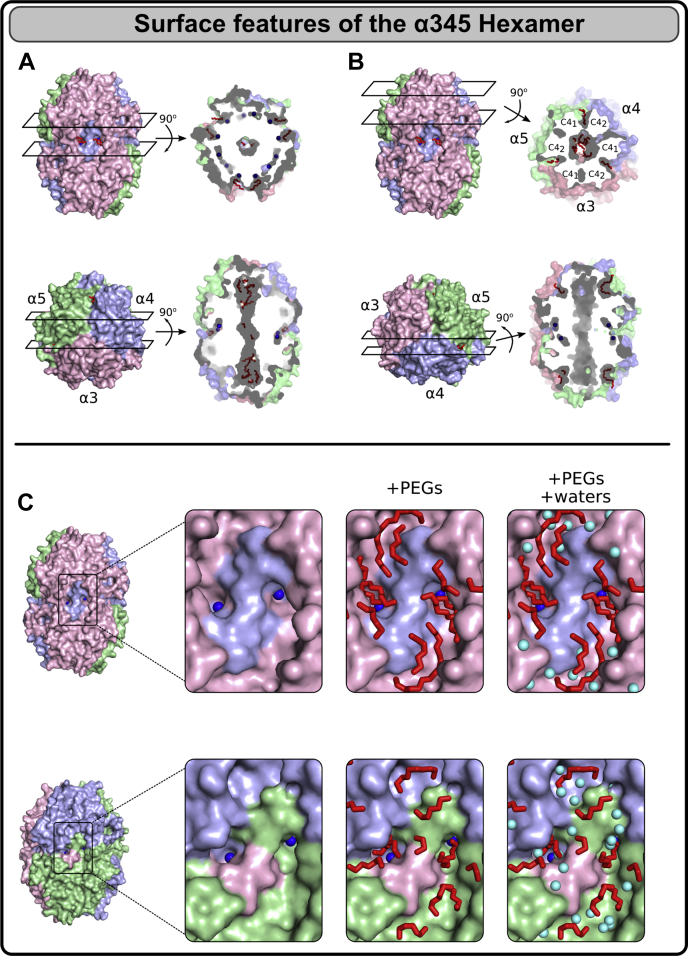
The α345 hexamer crystal structure reveals crevices, pockets, and cavities along the surface and are solvent accessible.A, equatorial and meridian slices through the surface of the hexamer reveal a big central cavity going from one trimer to the other, small inner cavities encapsulating six chloride ions of group 1, and pockets accommodating six chloride ions of group 2. The central cavity contains multiple structured PEG molecules. Pockets also contain parts of PEG molecules. B, the same orientation of slices but shifted toward the crevices between chains and C4 subdomains. Interchain crevices are occupied by PEG molecules. Outside surface of the α345 hexamer is colored in light red for α3, light blue for α4, and light green for α5. The inner surface is colored in gray. Chloride ions of both group 1 and group 2 are shown as blue spheres. PEG molecules are shown as red wireframes. C, PEG molecules penetrate and surround the pockets with chloride ions. Coloring of the NC1 chains is light red for α3, light blue for α4, and light green for α5. Chloride ions of group 2 are shown as blue spheres. Structured PEG molecules are shown as red wireframes. Structured water molecules are shown as cyan spheres.
Crevices, pockets, and inner cavities in the α345 hexamer
The crystal structure of the α345 hexamer reveals several crevices, pockets, and inner cavities, which are large enough to accommodate small molecules (Fig. 6, A and B). Under crystallization conditions used, we observe not only chloride ions, which are physiologically relevant and critical for the hexamer assembly, but also multiple PEG molecules. As discussed earlier, the chloride ions of group 2 are sitting at the bottom of pockets, which are also occupied by PEG molecules. Chloride ions of group 2 are localized in small inner cavities, which additionally contain several structured water molecules (Fig. 6C). The central inner cavity going from one trimer to another through the hexamer interface accommodates multiple structured PEG molecules but would accommodate much larger molecules if present during protein folding or the hexamer assembly. We also found crevices between chains and between C4 subdomains within each chain. The crevices between chains are wider and occupied by PEG molecules (Fig. 6B). The crevices are close enough to the inner cavity and potentially there is a communication between these structures under physiological conditions. In support of this molecular channel is the presence of PEG molecules in the inner cavity, although the protein used for crystallization has been already in the hexamer form (addressed below). Thus, even for the fully assembled hexamer there is a mechanism of penetration of ligands into the inner cavity.
The outer surface of the α345 hexamer represents a complex landscape with multiple hills and valleys. We found multiple PEG molecules interacting with the surface and some of them having contacts with two adjacent chains (Fig. S5).
GP hypoepitope loops on the surface of the α345 hexamer
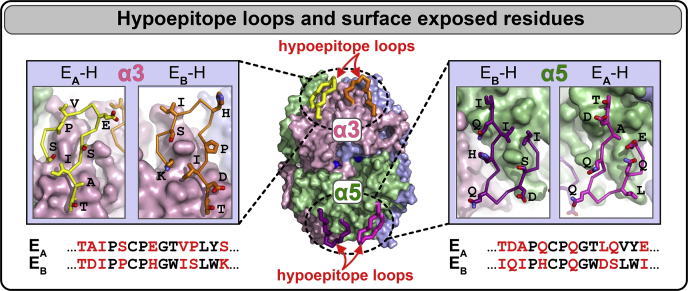
There are surface-exposed loops on the α345 hexamer (Fig. 7) that encompass the EA and EB regions of GP immunoreactivity (14, 15). The loops are designated herein as hypoepitopes as they are not recognized by GP autoantibodies but can undergo a conformational transition into neoepitopes that bind the antibodies (16, 17). The EA and EB loops (Fig. 7) demonstrate elevated mean square displacement values (Fig. 8), which reflect increased dynamic mobility of the loops. The EA loop of α5 chain is involved in crystal packing, thus having relatively lower B values (Fig. 8). Mutation analysis showed evolutionary pressure on the loop sequences, particularly on the EA loop in α2-α5 chains (Fig. S6), supporting the functional importance of these loops. One of the crevices is located between EA and EB hypoepitope loops forming loop-crevice-loop (LCL) regions at the apexes of the α345 hexamer and is juxtaposed with the T-cell receptor epitopes (18, 19, 20).
Figure 7.
The α345 hexamer crystal structure reveals the EAand EBloops on the surface of the α3 and α5 NC1 domains within the native α345NC1 hexamer. The loops are presented in different colors as indicated. Insets show the side-chain geometry for surface residues available for signaling and binding with other partners. The corresponding loop sequences are shown at the bottom with surface residues highlighted in red font.
Z-appendage location within the α345 hexamer
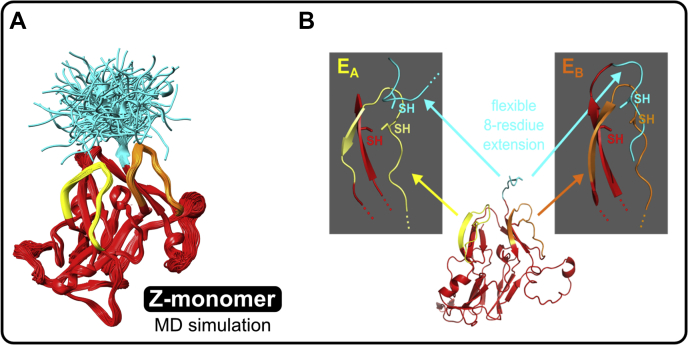
The Z-appendage is an 8-residue C-terminal extension of the native primary structure of the α3 chain of collagen IV. It is located at the apex of the α3NC1 monomer, in juxtaposition with EA and EB hypoepitope loops (Fig. 9). To assess Z-appendage flexibility, a molecular dynamics (MD) simulation was performed on the appendage in the context of a model of the α3 chain NC1 monomer. To sample all possible orientations of the appendage, 1000 initial conformations were originally generated by high-temperature MD. Each of those was extended for an additional 1 ns of simulation time at the physiological temperature, resulting in 1 μs of total MD sampling. Cysteines were reduced. Clustering analysis of the Z-appendage residues from all 1000 trajectories (using a 7 Å r.m.s.d. cutoff) revealed 134 conformational families. Within two of those 134 clusters, we found multiple conformations of the Z-appendage that had the cysteine residue positioned adjacent to the EA or EB loop disulfides. Two such conformations are shown in Figure 9, B and C. In these two conformations, the mutant cysteine is near the WT EA/EB cysteines that form an intraloop disulfide. These conformations suggest that the appendage cysteine residue may form alternative disulfides. Interference with disulfide formation and disturbing other interactions within the monomer can lead to a conformational change of the monomer and influence assembly of the hexamer. In conclusion, the Z-appendage can assume multiple conformations and its free thiol group can participate in a number of reactions including those with EA and EB epitope loops.
Figure 9.
Molecular dynamic (MD) simulations predict the Z-appendage can assume multiple conformations.A, a molecular dynamics simulation (1000 independent runs) (cyan) was performed to sample conformations of the Zurich variant. B, MD simulation analysis revealed two clusters of conformants of the 8-residue extension particularly close to the EA or EB regions. This confirms that free cysteine residue within the extension can form alternative disulfide bridges and disturb other interactions within the monomer. Interference with disulfide formation can lead to conformational changes of the EA and EB regions leading to appearance of immunogenic neoepitopes.
Analysis of known Alport variants in COL4A3, COL4A4, and COL4A5
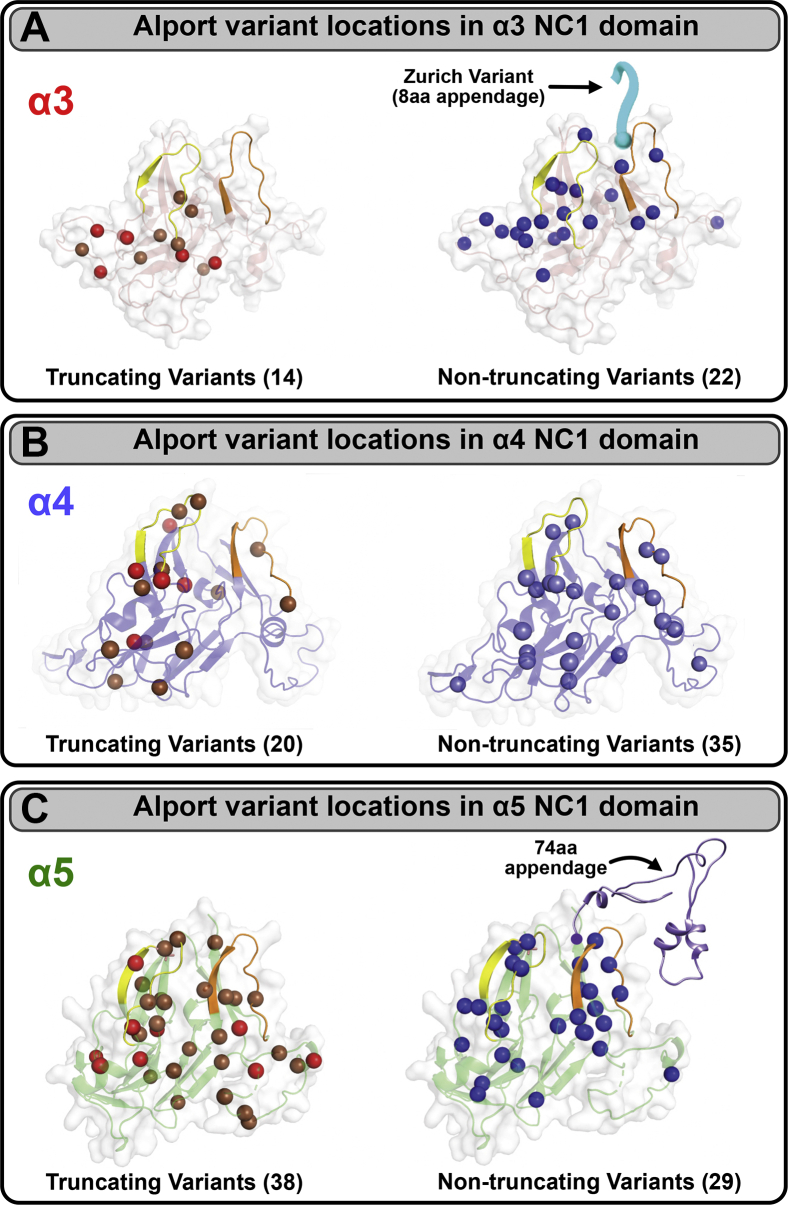
Solving the crystal structure of the α345 hexamer allowed for 3D mapping of known Alport variants. The maps of Alport variants in the α3, α4, and α5 chains of collagen IV and their localizations within 3D structures of the α3, α4, and α5NC1 domains are shown in Figure 10 (for additional details, see Figs. S7–S9). The analysis reveals two classes of NC1 variants, that is, truncating and nontruncating. Potentially, both classes are amenable to protein replacement therapy and the nontruncating class also presents a possibility for development of small-molecule therapies. The descriptions for each variant are provided in the top part of Figs. S7–S9, according to the human genome variation society nomenclature (21). The Zurich variant of the α3NC1 domain stands out among other Alport variants as it results in a C-terminal extension of protein polypeptide chain producing an 8-amino acid Z-appendage as shown in Fig. S7; a variant analogous to the Zurich variant producing a 74-amino acid appendage has been identified in α5NC1 (Fig. S9).
Figure 10.
Crystal structure of the α345 hexamer reveals the location of known Alport variants in the α3, α4, and α5 NC1 domains.A, distinct pathogenic Alport variants were mapped within the 3D structure of the α3 NC1 domain (source: HGMD 2020.1). The total number of truncating (small deletion, brown; nonsense, red) and nontruncating (missense variants, blue) variants are shown in parentheses. Truncating variants result in the premature stop codon and expression of the truncated form of COL4A3, which does not incorporate into the GBM (left). Although nontruncating missense variants (blue) do not affect the overall length of the NC1 domain (right), they may result in conformational changes of crucial regions within NC1. These nontruncating variants can incorporate into the GBM but are functionally defective. The Zurich variant (cyan) is a combination of a small deletion and insertion, resulting in an 8-aa appendage to the NC1 domain. Therefore, it belongs to “nontruncating” variant subgroup (right). B, distinct pathogenic Alport variants were mapped within the 3D structure of the α4 NC1 domain (source: HGMD 2020.1). Color codes are as in panel A. C, distinct pathogenic Alport variants were mapped within the 3D structure of the α5 NC1 domain (source: HGMD 2020.1). A distinct Alport variant, 74-amino acid appendage at the C-terminus of α5 chain, is depicted on the right. Color codes are as in panel A. For an expanded description of the genetic identity of the α3, α4, and α5 variants, see Figs. S12–S14. GBM, glomerular basement membrane.
Analysis of potential glycoxidation sites on the surface of the α345 hexamer relevant to DN
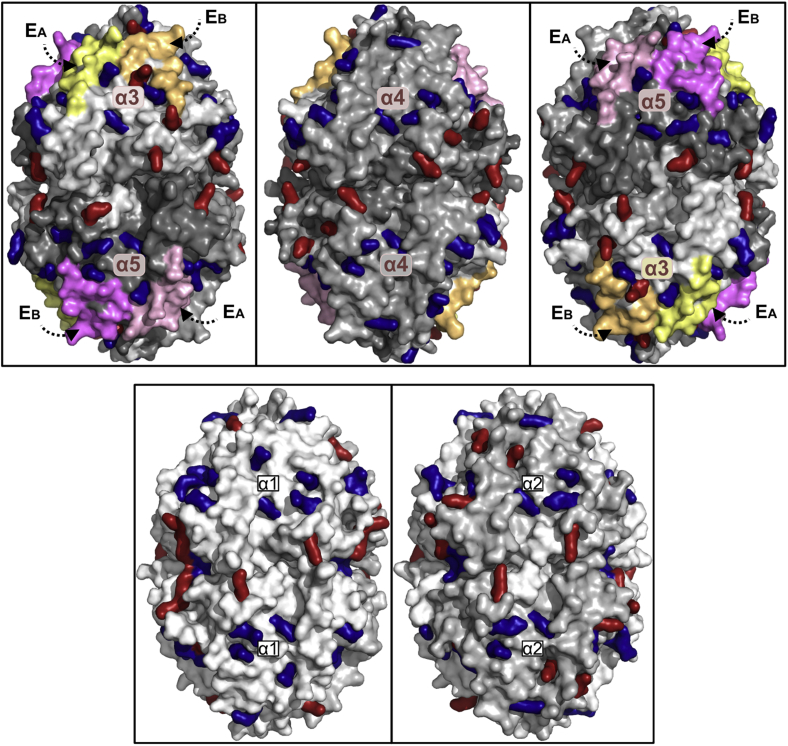
The α345 hexamer possesses multiple surface-exposed lysine (Lys) and arginine (Arg) residues (Fig. 11) that can be targeted by hyperglycemia-derived reactive carbonyl products to form stable adducts that underlie DN pathogenesis, including Lys–Lys and Lys–Arg crosslinks (22). There are 78 surface-exposed Lys and Arg side chains in the hexamer. Importantly, six of these residues in the α3NC1 domain and four in the α5NC1 domain are adjacent to or located on the respective EA and EB hypoepitopes (Fig. 11).
Figure 11.
The surface-exposed lysine (Lys) and arginine (Arg) residues of collagen IV hexamers of α345 (top) and α121 (bottom) GBM scaffolds that can be adducted by glucose and glucose-derived reactive carbonyl products in diabetes.Top panel: The 3D structure of α345 hexamer is shown in three different projections rotated by 120° about the vertical axis. The labels indicate individual α3, α4, and α5 NC1 domains within the hexamer structure and locations of EA and EB hypoepitopes. The surface-exposed Lys and Arg side chains are shown in red and blue colors, respectively. Bottom panel: the 3D structure of α121 hexamer in two projections (120° rotation about the vertical axis) showing the location of the surface-exposed Lys and Arg side chains using the same color coding as in the top panel. GBM, glomerular basement membrane.
Discussion
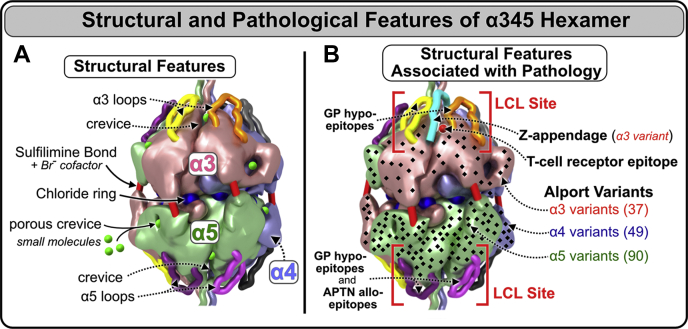
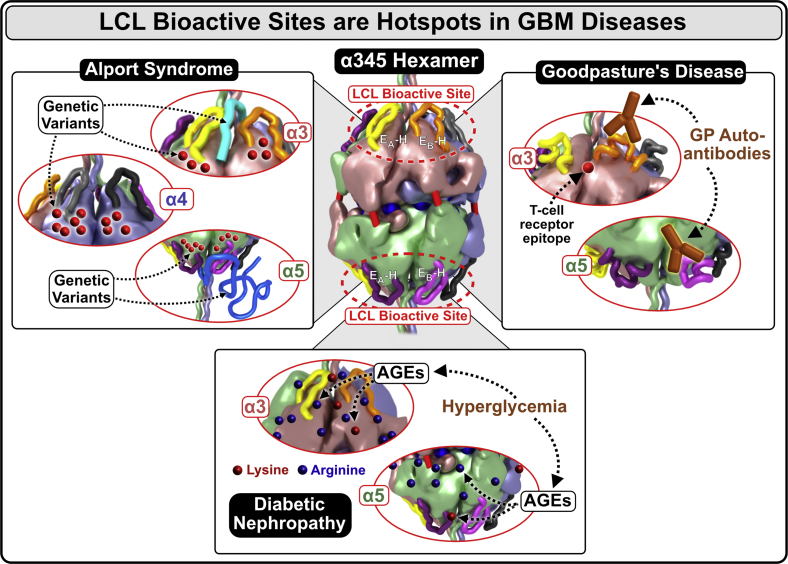
The crystal structure of the α345 hexamer provided a framework to interpret a role that the Z-appendage, a representative AS variant, played in AS and as a possible structural risk factor for GP, as described in Pokidysheva et al. (4). The crystal structure of the hexamer revealed a ring of 12 chloride ions that, together with up to six sulfilimine bonds, stabilizes the hexamer structure (Fig. 12A). The α345 hexamer harbors a number of structural features associated with pathology, which are located within the LCL sites where pathogenic mechanisms of AS and GP converge, and potentially DN. Within the LCL sites, there are multiple Alport-associated variants in the α3, α4, and α5 NC1 domains including the Z-appendage, which is juxtaposed with the GP hypoepitopes (Fig. 12B). In addition, GP hypoepitope loops and a T-cell receptor epitope (18, 19, 20) are located within the LCL sites (Fig. 12B).
Figure 12.
Structural and pathological features of the α345 hexamer.A, the α345 hexamer is a key connection module within the collagen IVα345 scaffold. The hexamer structure features a ring of 12 chloride ions, required for hexamer assembly, at the interface of the two trimers. The assembled hexamer also features up to six sulfilimine bonds between the protomers that reinforce the stability of the scaffold. The surface of α345 hexamer is marked by multiple pores and crevices that are accessible to small molecules. B, the α345 hexamer harbors a number of features involved in pathogenesis of Alport syndrome and Goodpasture's diseases. Multiple Alport-associated variants occur within α3, α4, and α5 NC1 domains (black dots) including the Zurich variant of α3 NC1, which produces an 8-amino acid Z-appendage shown in cyan. In juxtaposition to the Z-appendage, there are Goodpasture's disease (GP) hypoepitope loops and a T-cell receptor epitope located at the bottom of the crevice between the loops. Together, these features constitute a loop-crevice-loop (LCL) site where pathogenic mechanism of Alport syndrome and Goodpasture diseases converge (top red square brackets). The analogous LCL site is located within the α5 NC1 domain (bottom red square brackets) and within the α4 NC1 domain (not shown).
Furthermore, the 3D structure provided the framework for designing hexamer assembly studies in Pedchenko et al. (23), which demonstrated that the LCL sites have conformational plasticity. This plasticity along with the structural features of the LCL sites indicate bioactive functions that may include signaling and organizing macromolecular complexes. These functions can be perturbed by the Z-appendage and other genetic variants that occur in the hexamer in AS, endogenous and exogenous triggers in GP, and hyperglycemia in DN (Fig. 13). Moreover, because a significant number of Alport variants occurs throughout the hexamer structure (Fig. 12), the multiple pores, crevices, and cavities on the surface of the hexamer can be potential targets for therapeutic interventions such as small-molecule drugs and protein replacement.
Figure 13.
The crystal structure of the α345 hexamer reveals common “hotspots” of bioactivity, where pathogenic mechanisms converge. The Z-appendage and the GP hypoepitopes are located at the same sites of α3 and α5 subunits near the apices of the α345 hexamer which are called loop-crevice-loop (LCL) bioactive sites (α4 LCL site located on back side of the hexamer). This indicates that the pathogenic mechanisms of GP and AS converge at these sites, thus revealing “hotspots” of bioactivity. In Alport syndrome, a number of pathogenic variants including the Zurich variant are located within the LCL sites (top left). In addition, there are numerous other hypomorph variants on the surface that can affect LCL function (see Figs. S12–S14). In familial GP disease, the Zurich variant within the LCL sites could predispose the site for a trigger of autoantibody production (top right). In sporadic GP, the same LCL site is vulnerable to endogenous and exogenous triggers that elicit the immune response. Similarly, in diabetic nephropathy, this site is vulnerable to hyperglycemia-derived modifications of Lys and Arg residues at the hexamer surface. These modifications are in fact equivalent to genetic variants that cause structural perturbation and dysfunction. They can cause GBM thickening, a hallmark feature of diabetic nephropathy (35). AS, Alport syndrome; GBM, glomerular basement membrane; GP, Goodpasture’s disease.
Experimental procedures
Design, expression, and purification of single-chain NC1 trimers
Four combinations, α345, α543, α343, and α545, of human DNA sequences encoding residues of α3, α4, and α5 of collagen IV NC1 domain were cloned in-frame with the BM-40 signal peptide and the FLAG tag of the pRc-X vector (14) between restriction sites NheI and BspDI using the previously developed strategy (5). Constructs were transiently expressed (and, in case of α343 and α545, coexpressed) in ExpiCHO cells. For the single-chain α345 NC1 trimer construct, a stable clone of HEK293 cells was developed as described (5) and used for bulk production of the α345 NC1 trimer for assembly studies and crystallization. The recombinant protein fused with an N-terminal FLAG tag was purified using FLAG-affinity resin as described (6). Size-exclusion chromatography using Superdex 200 Increase 10/300GL column (GE Healthcare) was used for the final purification step.
Single-chain α345 NC1 trimer crystallization and structure determination
The single-chain α345 NC1 trimer was crystallized in the tetragonal form (space group, P41212) using the hanging-drop vapor diffusion method. The protein solution (∼10 mg/ml) in 5-mM Tris HCl, pH 7.5, and 150-mM NaCl was mixed for the drop solution in a 1:1 proportion with a reservoir solution of 100-mM Tris HCl, pH 8.5, and 56% PEG 200. The crystals grew to a final size of ∼0.2 × 0.15 × 0.15 mm after 45 days at 22 °C. The crystals were flash-frozen in liquid nitrogen. Data collection was performed remotely on crystals cryocooled to 100 K at the Life Sciences Collaborative Access Team beamline 21-ID-G at the Advanced Photon Source, Argonne National Laboratory. Data extending to 1.85 Å resolution were indexed using iMOSFLM (24) and then scaled and merged using Scala (25). Amplitudes were converted to structure factors using CTRUNCATE (26). Five percent of the data were set aside to monitor Rfree. Initial phases were obtained by molecular replacement using Phaser-MR (27) and the previously solved single-chain α112 NC1 trimer (PDB code: 6MPX) (5) as the search model. One single-chain NC1 polypeptide was found per asymmetric unit (VM = 2.82 Å3/Da; solvent content = 56.4% (28)). Refinement was carried out using Phenix (29) with translation–libration–screw-rotation model restraints. The models were manually adjusted between each refinement cycle using Coot (30). Model geometry assessed using MolProbity (31) showed 97.5% of the residues in the favored region and 2.5% in the additionally allowed region, with none in the outlier regions. The final data collection and refinement statistics are shown in Table S6. Model superimpositions were performed using LSQ Superpose function in Coot (30).
Structural modeling of the NC1 domain: MD simulations
To examine the conformational space accessible by the mutant residues, we performed 1000 1-ns MD simulations of the mutant α3 monomer using the graphics processing unit-enabled codes in the AMBER18 suite of molecular mechanics programs (32). We began by extracting the monomer structure from a previously published α345 model (33) and from the structure solved in the present study. The mutant residues, in an extended strand-like conformation, were appended to the WT residues. We then heated this extended mutant to 1000K while holding the WT residues close to their starting position using restraints and modeling the cysteine residues in a reducing environment. We used the generalized born (GB) implicit solvent model to maximize conformational kinetics by removing friction with solvent molecules while still providing a good approximation of solvent-shielding effects (34). We then performed a 10-ns restrained MD simulation at 1000K from which we captured a snapshot every 10 ps. The resulting 1000 snapshots were then cooled to 310 K over 50 ps and used as the starting structures for 1000 x 1-ns unrestrained MD simulations at 310 K, also using the GB approximation. The resulting 1 μs worth of MD conformational samples were then clustered and analyzed with the CPPTRAJ program, to visualize representative conformational accessibility and to analyze distances between the mutant cysteine and the WT cysteine CG atoms.
Data availability
All data described in this article are available in the main text or supporting information. The atomic coordinates and structure factors (code 6WKU) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Supporting information
This article contains supporting information (13, 36)
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Neonila Danylevych and Mohamed Rafi for their technical assistance, Dr Julie Hudson for editing, and Dr Aaron Fidler for figure production, writing, and coordination in manuscript preparation. We also thank Vanderbilt Center for Structural Biology for the use of Protein Characterization and Biomolecular Crystallography facilities.
This research used resources of the Advanced Photon Source, a U.S. Department of Energy Office of Science User Facility, operated by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. The use of the LS-CAT Sector 21 was supported by a grant (085P1000817) from Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor.
Author contributions
S. P. B. and B. G. H. conceived the study and conceptualized and wrote the manuscript. S. P. B. and R. B. performed the experiments. J. S. performed the in silico modeling and analysis. S. P. B., R. B., S. V. C., S. I., P. A. V., and B. G. H. analyzed the data. S. P. B., P. A. V., and B. G. H. edited the manuscript.
Funding and additional information
This work was supported by Grants R01DK18381-50 and a supplement R01DK018381-49S (to B. G. H.), R24DK103067 (to B. G. H.), R01DK065138 (to B. G. H. and P. A. V.) from the National Institute of Diabetes and Digestive and Kidney Diseases. Additional support was provided by the Aspirnaut program from the Center for Matrix Biology at Vanderbilt University Medical Center. S. P. B. was supported, in part, by start-up funding from Department of Medicine, Division of Nephrology, Vanderbilt University Medical Center.
Edited by Gerald Hart
Supporting information
References
- 1.Hudson B.G., Reeders S.T., Tryggvason K. Type IV collagen: Structure, gene organization, and role in human diseases. Molecular basis of goodpasture and Alport syndromes and diffuse leiomyomatosis. J. Biol. Chem. 1993;268:26033–26036. [PubMed] [Google Scholar]
- 2.Hudson B.G., Tryggvason K., Sundaramoorthy M., Neilson E.G. Alport's syndrome, Goodpasture's syndrome, and type IV collagen. N. Engl. J. Med. 2003;348:2543–2556. doi: 10.1056/NEJMra022296. [DOI] [PubMed] [Google Scholar]
- 3.Naylor R.W., Morais M., Lennon R. Complexities of the glomerular basement membrane. Nat. Rev. Nephrol. 2021;17:112–127. doi: 10.1038/s41581-020-0329-y. [DOI] [PubMed] [Google Scholar]
- 4.Pokidysheva E.N., Seeger H., Pedchenko V., Chetyrkin S., Bergmann C., Abrahamson D., Cui Z.W., Delpire E., Fervenza F.C., Fidler A.L., Fogo A.B., Gaspert A., Grohmann M., Gross O., Haddad G. Collagen IVα345 dysfunction in glomerular basement membrane diseases. I. Discovery of a COL4A3 variant in familial Goodpasture’s and Alport diseases. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100590. 100590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedchenko V., Bauer R., Pokidysheva E.N., Al-Shaer A., Forde N.R., Fidler A.L., Hudson B.G., Boudko S.P. A chloride ring is an ancient evolutionary innovation mediating the assembly of the collagen IV scaffold of basement membranes. J. Biol. Chem. 2019;294:7968–7981. doi: 10.1074/jbc.RA119.007426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boudko S.P., Danylevych N., Hudson B.G., Pedchenko V.K. Basement membrane collagen IV: Isolation of functional domains. Methods Cel. Biol. 2018;143:171–185. doi: 10.1016/bs.mcb.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borza D.B., Bondar O., Todd P., Sundaramoorthy M., Sado Y., Ninomiya Y., Hudson B.G. Quaternary organization of the goodpasture autoantigen, the alpha 3(IV) collagen chain. Sequestration of two cryptic autoepitopes by intrapromoter interactions with the alpha4 and alpha5 NC1 domains. J. Biol. Chem. 2002;277:40075–40083. doi: 10.1074/jbc.M207769200. [DOI] [PubMed] [Google Scholar]
- 8.Vanacore R., Ham A.J., Voehler M., Sanders C.R., Conrads T.P., Veenstra T.D., Sharpless K.B., Dawson P.E., Hudson B.G. A sulfilimine bond identified in collagen IV. Science. 2009;325:1230–1234. doi: 10.1126/science.1176811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sundaramoorthy M., Meiyappan M., Todd P., Hudson B.G. Crystal structure of NC1 domains. Structural basis for type IV collagen assembly in basement membranes. J. Biol. Chem. 2002;277:31142–31153. doi: 10.1074/jbc.M201740200. [DOI] [PubMed] [Google Scholar]
- 10.Than M.E., Henrich S., Huber R., Ries A., Mann K., Kuhn K., Timpl R., Bourenkov G.P., Bartunik H.D., Bode W. The 1.9-A crystal structure of the noncollagenous (NC1) domain of human placenta collagen IV shows stabilization via a novel type of covalent Met-Lys cross-link. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6607–6612. doi: 10.1073/pnas.062183499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanacore R.M., Shanmugasundararaj S., Friedman D.B., Bondar O., Hudson B.G., Sundaramoorthy M. The alpha1.alpha2 network of collagen IV. Reinforced stabilization of the noncollagenous domain-1 by noncovalent forces and the absence of Met-Lys cross-links. J. Biol. Chem. 2004;279:44723–44730. doi: 10.1074/jbc.M406344200. [DOI] [PubMed] [Google Scholar]
- 12.Kufareva I., Abagyan R. Methods of protein structure comparison. Methods Mol. Biol. 2012;857:231–257. doi: 10.1007/978-1-61779-588-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casino P., Gozalbo-Rovira R., Rodriguez-Diaz J., Banerjee S., Boutaud A., Rubio V., Hudson B.G., Saus J., Cervera J., Marina A. Structures of collagen IV globular domains: Insight into associated pathologies, folding and network assembly. IUCrJ. 2018;5:765–779. doi: 10.1107/S2052252518012459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Netzer K.O., Leinonen A., Boutaud A., Borza D.B., Todd P., Gunwar S., Langeveld J.P., Hudson B.G. The Goodpasture autoantigen. Mapping the major conformational epitope(s) of alpha3(IV) collagen to residues 17-31 and 127-141 of the NC1 domain. J. Biol. Chem. 1999;274:11267–11274. doi: 10.1074/jbc.274.16.11267. [DOI] [PubMed] [Google Scholar]
- 15.Hellmark T., Burkhardt H., Wieslander J. Goodpasture disease. Characterization of a single conformational epitope as the target of pathogenic autoantibodies. J. Biol. Chem. 1999;274:25862–25868. doi: 10.1074/jbc.274.36.25862. [DOI] [PubMed] [Google Scholar]
- 16.Pedchenko V., Bondar O., Fogo A.B., Vanacore R., Voziyan P., Kitching A.R., Wieslander J., Kashtan C., Borza D.B., Neilson E.G., Wilson C.B., Hudson B.G. Molecular architecture of the Goodpasture autoantigen in anti-GBM nephritis. N. Engl. J. Med. 2010;363:343–354. doi: 10.1056/NEJMoa0910500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calvete J.J., Revert F., Blanco M., Cervera J., Tarrega C., Sanz L., Revert-Ros F., Granero F., Perez-Paya E., Hudson B.G., Saus J. Conformational diversity of the Goodpasture antigen, the noncollagenous-1 domain of the alpha3 chain of collagen IV. Proteomics. 2006;6(Suppl 1):S237–S244. doi: 10.1002/pmic.200500495. [DOI] [PubMed] [Google Scholar]
- 18.Ooi J.D., Petersen J., Tan Y.H., Huynh M., Willett Z.J., Ramarathinam S.H., Eggenhuizen P.J., Loh K.L., Watson K.A., Gan P.Y., Alikhan M.A., Dudek N.L., Handel A., Hudson B.G., Fugger L. Dominant protection from HLA-linked autoimmunity by antigen-specific regulatory T cells. Nature. 2017;545:243–247. doi: 10.1038/nature22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phelps R.G., Rees A.J. The HLA complex in Goodpasture's disease: A model for analyzing susceptibility to autoimmunity. Kidney Int. 1999;56:1638–1653. doi: 10.1046/j.1523-1755.1999.00720.x. [DOI] [PubMed] [Google Scholar]
- 20.Xie L.J., Cui Z., Chen F.J., Pei Z.Y., Hu S.Y., Gu Q.H., Jia X.Y., Zhu L., Zhou X.J., Zhang H., Liao Y.H., Lai L.H., Hudson B.G., Zhao M.H. The susceptible HLA class II alleles and their presenting epitope(s) in Goodpasture's disease. Immunology. 2017;151:395–404. doi: 10.1111/imm.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.den Dunnen J.T., Dalgleish R., Maglott D.R., Hart R.K., Greenblatt M.S., McGowan-Jordan J., Roux A.F., Smith T., Antonarakis S.E., Taschner P.E. HGVS Recommendations for the description of sequence variants: 2016 Update. Hum. Mutat. 2016;37:564–569. doi: 10.1002/humu.22981. [DOI] [PubMed] [Google Scholar]
- 22.Thorpe S.R., Baynes J.W. Maillard reaction products in tissue proteins: New products and new perspectives. Amino Acids. 2003;25:275–281. doi: 10.1007/s00726-003-0017-9. [DOI] [PubMed] [Google Scholar]
- 23.Pedchenko V., Boudko S.P., Barber M., Mikhailova T., Saus J., Harmange J.-C., Hudson B.G. Collagen IVα345 dysfunction in glomerular basement membrane diseases. III. A functional framework for α345 hexamer assembly. J. Biol. Chem. 2021;296 doi: 10.1016/j.jbc.2021.100592. 100592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battye T.G., Kontogiannis L., Johnson O., Powell H.R., Leslie A.G. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans P. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 26.Padilla J.E., Yeates T.O. A statistic for local intensity differences: Robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr. D Biol. Crystallogr. 2003;59:1124–1130. doi: 10.1107/s0907444903007947. [DOI] [PubMed] [Google Scholar]
- 27.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matthews B.W. Solvent content of protein crystals. J. Mol. Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 29.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C. Phenix: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen V.B., Arendall W.B., 3rd, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., Richardson D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Case D.A., Cheatham T.E., Darden T., Gohlke H., Luo R., Merz K.M., Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanacore R.M., Ham A.J., Cartailler J.P., Sundaramoorthy M., Todd P., Pedchenko V., Sado Y., Borza D.B., Hudson B.G. A role for collagen IV cross-links in Conferring immune Privilege to the goodpasture autoantigen: Structural basis for the Crpticity of B cell epitopes. J. Biol. Chem. 2008;283:22737–22748. doi: 10.1074/jbc.M803451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salomon-Ferrer R., Gotz A.W., Poole D., Le Grand S., Walker R.C. Routine Microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent Particle Mesh Ewald. J. Chem. Theor. Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 35.Mauer S.M., Steffes M.W., Ellis E.N., Sutherland D.E., Brown D.M., Goetz F.C. Structural-functional relationships in diabetic nephropathy. J. Clin. Invest. 1984;74:1143–1155. doi: 10.1172/JCI111523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silk M., Petrovski S., Ascher D.B. MTR-Viewer: identifying regions within genes under purifying selection. Nucleic Acids Res. 2019;47:W121–W126. doi: 10.1093/nar/gkz457. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data described in this article are available in the main text or supporting information. The atomic coordinates and structure factors (code 6WKU) have been deposited in the Protein Data Bank (http://wwpdb.org/).