Highlights
-
•
Delta like canonical Notch ligand 3 (DLL3) is an independent prognostic factor for invasive breast cancer (IBC) and represents a worse prognosis.
-
•
DLL3 may be the messenger of the Notch and P53 signaling pathways.
-
•
DLL3 may promote tumor progression by affecting the infiltration of B cells, neutrophils and T cells in tumor microenvironment.
-
•
DLL3 may be used as an immunoadjuvant checkpoint or a marker of immunotherapy effect.
Keywords: Delta like canonical Notch ligand 3, Invasive breast cancer, Prognosis biomarker, P53 signaling pathway, Immune infiltration
Abstract
Background
Previous studies have shown the prognostic value of delta like canonical Notch ligand 3 (DLL3) in patients with different types of tumors, but the role and predictive value of DLL3 in invasive breast cancer (IBC) have not been reported. In this study, we explored the prognostic ability and potential ways of DLL3 in IBC patients.
Methods
We retrospectively enrolled 130 IBC patients from a single institution from 2004 to 2019 for bioinformatics and statistical analysis. The Cancer Genome Atlas breast invasive carcinoma (TCGA-BRCA) cohort was used for verification.
Results
High expression of DLL3 was associated with overall survival (OS) in IBC patients (P = 0.023). Multivariate analysis further showed that DLL3 expression was an independent prognostic factor (hazard ratio [HR]: 1.08; 95% confidence interval [CI]: 1.01–1.15; P = 0.017). Time-dependent receiver operating characteristic (ROC) with the area under the curve (0.786) demonstrated that DLL3 expression can predict the survival outcome of IBC patients. Furthermore, the expression of DLL3 was related to a variety of tumor infiltrating immune cells (TIICs), particularly T cells regulatory (Tregs). Gene set enrichment analysis (GSEA) and immunohistochemistry (IHC) results indicated that DLL3 was closely related to p53 signaling pathway.
Conclusions
High expression of DLL3 was associated with poor prognosis and immune cell infiltration in IBC patients. Moreover, P53 signaling pathway may be the key pathway.
Introduction
Breast cancer is among the most commonly diagnosed cancer and the leading cause of cancer death [1]. There are 1.6 million new breast cancer cases in the world every year, with about 500,000 deaths [2]. Despite recent advances in diagnosis and therapy, the prognosis of this malignancy remains a challenge [3,4]. Therefore, it is necessary to determine the specific biomarkers and therapeutic targets of breast cancer and their molecular mechanisms.
The Notch signaling pathway is evolutionarily conserved and operated by four Notch receptors (Notch1–4) and five ligands (Jag1, Jag2, DLL1, DLL3 and DLL4) of the Jagged/Serrate and Delta families, respectively [5,6]. Gamma secretase inhibitors (GSIs) are drugs that inhibit Notch signaling and can successfully in controlling cancer cell growth in conjunction with standard chemotherapy, but its application is limited by a large number of side effects [[7], [8]–9]. Therefore, drugs targeting specific Notch receptors and ligands may be more effective and accurate.
The abnormal activation of Notch signal is related to the occurrence of breast cancer [10]. Cbx4 upregulates carcinogenesis of breast cancer through Notch1 Signaling Pathway [11], while anomalous Notch1 signal transduction leads to apoptosis of breast cancer cells [12]. Notch3 maintains luminal phenotype and suppresses tumorigenesis and metastasis of breast cancer via trans-activating estrogen receptor-α [13]. DLL1-mediated Notch signaling promotes luminal breast cancer [6]. These studies show that receptors and ligands of NOTCH pathway play an important role in the development of breast cancer. Delta-like ligand 3 (DLL3) is an inhibitory Notch pathway ligand that is aberrantly expressed in small cell lung cancer (SCLC) and other high-grade neuroendocrine tumors [[14], [15]–16].
However, the implications of DLL3 in IBC pathogenesis and tumor progression, as well as its value as a promising biomarker for patient stratification, are not yet characterized. Here, we reported that up-regulation of DLL3 expression in IBC was closely related to patient's age and distant metastasis. In addition, DLL3 was an independent prognostic factor in patients with IBC, and its high expression phenotype was significantly enriched in the p53 signaling pathway. Furthermore, CIBERSORT [17], a widely accepted evaluation algorithm, was performed to analyze whether DLL3 is related to the relative abundance of TIICs in the tumor microenvironment. At the same time, we discussed the potential mechanism of DLL3.
Patients and methods
Patient information collection and sorting
Between July 2004 and February 2019, we admitted 130 patients with IBC in Huashan Hospital of Fudan University. The clinical research ethics committee of Fudan University approved this study with the approval number 2018-Y016 and each patient included in the cohort obtained informed consent. The specimens were independently review by two or more pathologists from Huashan Hospital, and a consensus diagnosis was reached with reference to the 2012 WHO diagnostic criteria for breast tumors [18].
The gene expression data and corresponding clinical information were downloaded from TCGA official website (https://portal.gdc.cancer.gov/). A total of 914 patients with complete data were retained and further analyzed.
DLL3 expression and survival analysis
Formalin-fixed tissue paraffin block made 3 mm white plate for IHC analysis. We used anti-DLL3 antibody (1:200; Proteintech), anti-BID antibody (1:200; Proteintech), anti-CDK4 antibody (1:200; Proteintech) and anti-P53 (1:100; Dako) for tissue staining. Three pathologists used a semi-quantitative immune response scoring algorithm to evaluate DLL3 staining independently. The semiquantitative immunoreactivity score (IRS) is between 0 and 30, based on the increase in IHC staining intensity (0, negative; 1, weak; 2, middle; 3, strong) and the percentage of positive tumor cells (one point per 10% increase, the percentage of positive tumor cells 1–10) [19]. The critical value of p53 staining was 10% (≤ 10% of tumor cells were negative and > 10% were positive) [20,21]. Disagreements were resolved by three pathologists negotiated under a multi- lens.
Wilcoxon test and paired analysis between normal tissue and tumor tissue in the same patient were used to analyze the expression of DLL3. Kaplan-Meier curve was performed to analyze the survival of DLL3.
Analysis of the correlation between TIICs and DLL3
CIBERSORT, a deconvolution algorithm to provide an estimation of the abundances of member cell types in a mixed cell population, can recognize the types of immune cells sensitively and accurately[22,23]. The algorithm analyzed the relative proportion of 22 kinds of immune cells between the high expression group and the low expression group of DLL3. At the same time, we used Spearman method to compare the correlation between DLL3 and immune cells. Furthermore, the GEPIA 2 "correlation" model further confirmed the relationship between the expression of DLL3 and the possible gene markers of TIICs.
Functional enrichment analysis and verification
Pearson correlation analysis (| R | ≥ 0.3) was used to extract genes related to DLL3 expression. A total of 375 DLL3 related genes were identified. Gene ontology (GO) analysis was performed to analyze the related genes via ‘clusterProfiler’ R package [24]. The gene set variation analysis (GSVA) R package [25] was used to KEGG pathways between low and high DLL3 expression groups. Gene terms with |logFC| ≥ 0.2 and P < 0.05 were considered statistically significant. Gene set enrichment analysis [26] was employed to verify the biological processes in the two groups stratified as described above. Significant enrichment criteria: the absolute value of NES was greater than 1, the nom p-value was less than 0.05, and the FDR q-value was less than 0.25.
Statistical analysis
All statistical analyses were conducted using R (v.3.6.2). The relationship between clinicopathologic features and DLL3 were analyzed with the Wilcoxon test and logistic regression. Receptor-operating characteristic (ROC) curve was performed to evaluate the predictive accuracy of DLL3 expression. P value < 0.05 was regarded as statistically significant.
Results
High expression of DLL3 indicates poor prognosis of IBC patients
Table 1 summarized the clinicopathological information of 130 patients. In our cohort, the median age was 56 years (21–88 years), of which 80 cases (61.5%) were younger than 60 years old. In histological grading, G2 stage accounted for the most (66.9%). According to AJCC staging, 46 (35.4%) patients had lymph node involvement and 9 (6.9%) patients had distal metastasis. Among 130 patients, 18 (13.8%) were luminal A, 20 (15.5%) were luminal B, 41 (31.5) were Basal-like and 51 (39.2%) were HER-2 overexpression.
Table 1.
Patients’ characteristics.
| Clinical characteristics | Total | % | |
|---|---|---|---|
| Age at diagnosis | 56 (21–88) | ||
| Age < 60 | 80 | 61.5 | |
| Age ≥ 60 | 50 | 38.5 | |
| Grade | |||
| G1 | 5 | 3.8 | |
| G2 | 87 | 66.9 | |
| G3 | 38 | 29.2 | |
| Tumor status | |||
| T1 | 68 | 52.3 | |
| T2 | 58 | 44.6 | |
| T3 | 4 | 0.9 | |
| Remote metastasis | |||
| M0 | 121 | 93.1 | |
| M1 | 9 | 6.9 | |
| Lymphatic invasion | |||
| N0 | 84 | 64.6 | |
| N1 | 46 | 35.4 | |
| Tumor classification | |||
| HER-2 overexpression | 51 | 39.2 | |
| Basal-like | 41 | 31.5 | |
| Luminal-A | 18 | 13.8 | |
| Luminal-B | 20 | 15.5 |
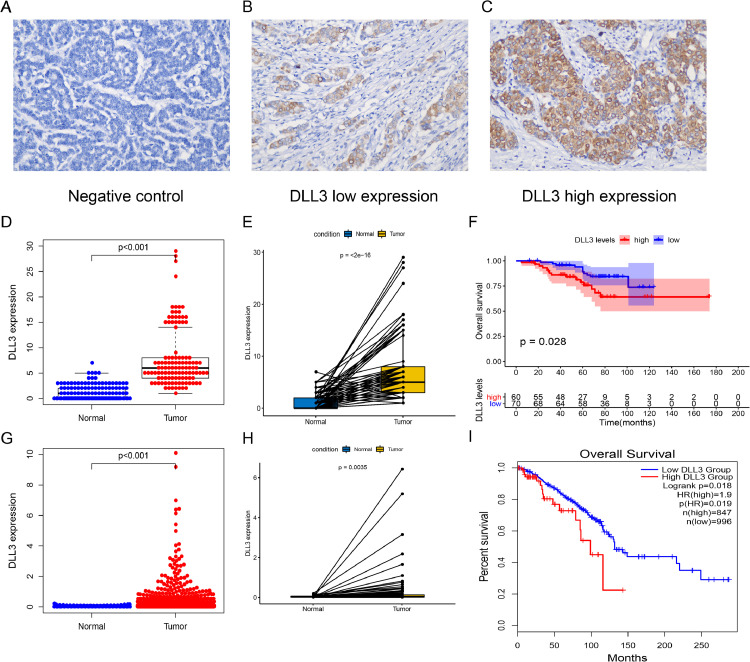
The expression of DLL3 was detected by immunohistochemistry in 130 patients. DLL3 was mainly expressed in the cytoplasm (Fig. 1B,C). Fig. 1A was the negative control. DLL3 was divided into high and low expression groups according to the median value (median: 5; IRS range: 1–29). The expression of DLL3 in IBC was significantly higher than that in normal tissues (P < 0.001; Fig. 1D). Paired analysis showed that the expression of DLL3 was increased in the same patient's tumor tissue (P < 0.001; Fig. 1E). In patients with breast cancer subtypes, DLL3 also has higher expression levels in cancer tissues (all P < 0.001; Fig. S1A). Kaplan Meier survival analysis showed that high expression of DLL3 was associated with poor prognosis (P = 0.028; Fig. 1F). We further explored the prognostic ability of DLL3 in different breast cancer subtypes, and the results showed that patients with high DLL3 expression in luminal A (P = 0.007) and HER2 overexpression (P = 0.009) subtypes had worse survival, there was no statistically significant difference between luminal B and Basal like (Fig.S1B).We validated our results through the TCGA-BRCA cohort. As shown in Fig. 1G–I, high expression of DLL3 in tumor tissue represented poor prognosis (all P < 0.05).
Fig. 1.
Expression and survival analysis of DLL3. (A–C) IHC staining of DLL3 in IBC samples (200 × magnification) including (A) Negative control of IHC performed on the IBC tissue using nonimmune IgG instead of the primary antibody; (B) Low DLL3 expression in the tumor tissue; (C) High DLL3 expression in the tumor tissue. In our cohort: (D) DLL3 expression in normal and tumor tissues; (E) Paired analysis; (F) Kaplan–Meier survival analysis of IBC patients with OS based on DLL3 expression. In TCGA–BRCA cohort: (G) DLL3 expression in normal and tumor tissues; (H) Paired analysis; (I) Kaplan–Meier survival analysis.
Relationship between DLL3 expression and clinicopathological characteristic
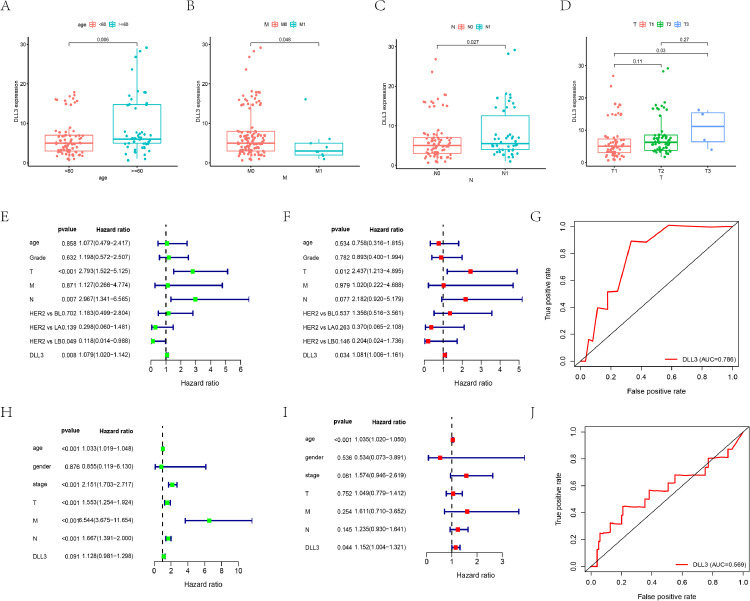
DLL3 expression data of a total of 130 IBC samples were analyzed from our cohort. As showed in Fig. 2A–D, increased expression of DLL3 correlated significantly with the patients’ age (P = 0.006). Patients in high DLL3 group tended to have more lymphatic invasion (P = 0.027), advanced T stages (P = 0.03) and less metastatic sites (P = 0.048). Univariate analysis using logistic regression revealed that DLL3 expression as a categorical dependent variable (based on median expression value of 6) was associated with poor prognostic patients’ age (Table S1; OR = 2.18 for age ≥ 60 vs. age < 60; P = 0.033). Similarly, we found that the expression of DLL3 is a protective factor for distant metastasis (OR = 0.31 for M1 vs. M0; P = 0.045).
Fig. 2.
Correlation analyses of the DLL3 expression with various clinicopathological characteristics of the IBC patients. The analysis compares the expression of DLL3 in our IBC cohort from stratified according to (A) Age; (B) Remote metastasis; (C) Lymphatic invasion and (D) Tumor status. Estimation of the prognostic accuracy of the DLL3 expression in the IBC patients. (E) Univariate, (F) Multivariate Cox regression analysis of the correlation between OS and various clinicopathological characteristics and DLL3; (G) ROC curve analysis shows the prognostic accuracy of DLL3 expression in our cohort. (H–J) Verify the conclusion in TCGA-BRCA cohort, including (H) Univariate Cox regression analysis; (I) Multivariate Cox regression analysis; (J) ROC curve analysis. HER2:HER-2 overexpression; BL: Basal-like; LA: luminal A; LB: luminal B.
The expression of DLL3 was an independent prognostic factor
We performed univariate and multivariate Cox regression analyses to determine if the DLL3 expression was an independent prognostic factor for patients with IBC (Table S2). Univariate analyses showed that T stage (P < 0.001), lymphatic invasion (P = 0.007) and DLL3 expression level (P = 0.008) were significantly associated with OS in IBC patients (Fig. 2E). Luminal B subtypes had longer survival time compared with HER-2 overexpression subtype (P = 0.049). Multivariate analyses manifested that T stage (P = 0.012) and DLL3 expression level (P = 0.034) were independent prognostic factors of IBC (Fig. 2F). The area under the ROC curve for DLL3 expression was 0.786, which proved its good prediction performance (Fig. 2G). Then we verified our results in the TCGA-BRCA cohort. As shown in Fig. 2H–J, the expression of DLL3 was independently associated with OS in 914 IBC samples (P < 0.05). ROC curve showed that DLL3 expression could predict the survival of IBC patients.
DLL3 influences the proportion of infiltrating immune cells in IBC
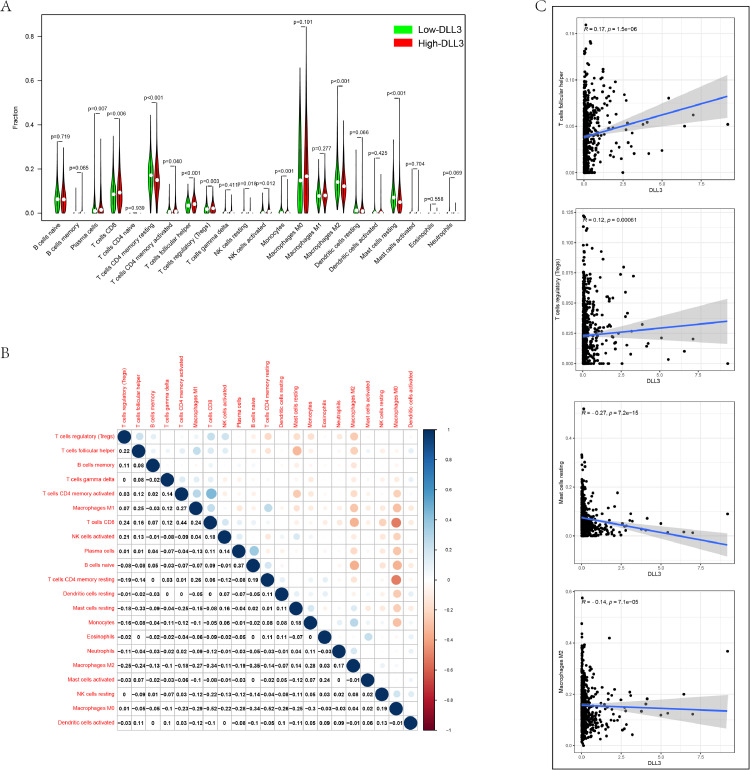
Genomic and transcriptome data from tumor samples have been used to study the tumor microenvironment (TME) [27]. Using gene expression characteristics, T cell receptors and B cell receptor systems to analyze the TME to determine the immune target of neoantigens, which provides rich information for many cancer types and has prognostic value [28]. Therefore, we tried to find out whether the expression of DLL3 was related to the immune invasion of IBC. The results were shown in Fig. 3A. The proportion of Plasma cells (P < 0.001), T cells CD8 (P = 0.002), T cells CD4 memory activated (P < 0.001), T cells follicular helper (P < 0.001), Tregs (P < 0.001), Macrophages M0 (P < 0.001) in high expression group was higher than the low expression group and T cells CD4 memory resting, Monocytes, Macrophages M2, Dendritic cells resting, Mast cells resting were significantly reduced (all P < 0.001).
Fig. 3.
Association between DLL3 expression and composition of TIICs. (A) The ratio of 22 immune cells in IBC tissues in the DLL3 high and low expression groups. (B) Correlation degree matrix of the relative proportion of immune cells in the microenvironment. (C) Correlation analysis between DLL3 and immune cells.
Cytotoxic T lymphocytes play a central role in eliminating tumor cells [29]. The heat map reflected the correlation between different TIIC subgroups (Fig. 3B). We have seen that CD8 T cells were negatively correlated with Macrophages M0 (R = - 0.52), while CD8 T cells were significantly positively correlated with T cells CD4 memory activated (R = 0.44). Further Spearman correlation analysis showed that DLL3 was positively correlated with Tregs, T cells follicular helper, while DLL3 was negatively correlated with macrophages M2, mast cells resting (|R| < 0.1; P < 0.001) (Fig. 3C). Meanwhile, we analyzed the relationship between DLL3 expression and cell surface markers (Table S3). Gene markers affected by DLL3 expression include: CD8B of CD8 + T cells, CD19, and CD79A of B cell, KIR2DL1, KIR2DL3 of Natural killer cell, CD11b of Neutrophils, PD-1, CTLA4, LAG3 and TIM-3 of T cell exhaustion, TPSB2, TPSAB2, CPA3 and MS4A2 of Mast cells (P < 0.05).
DLL3-related signaling pathways in IBC
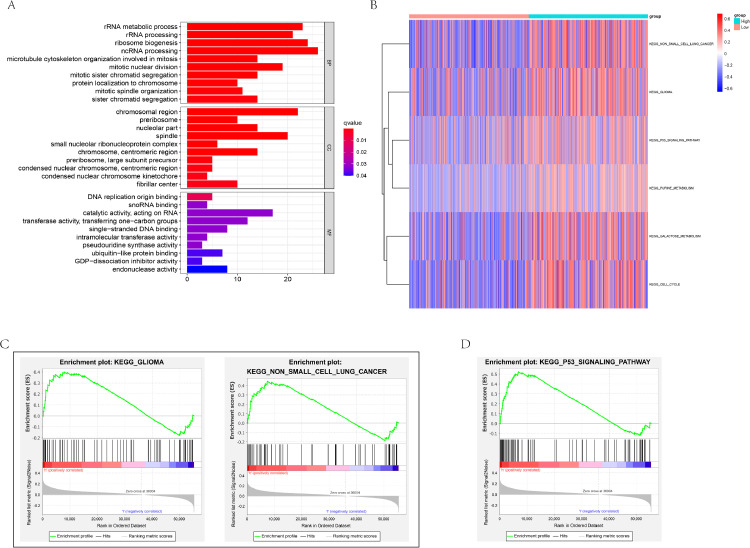
To better understand the function role of DLL3 in IBC, we conducted GO for the 375 related genes. The results showed that DLL3 was involved in mitotic nuclear division, mitotic sister chromatid segregation, rRNA processing, rRNA metabolic process (Fig. 4A). In addition, GSVA showed the pathways involved in High-DLL3 group were different in non-small cell lung cancer, glioma, P53 signaling pathway, and cell cycle (Fig. 4B). GSEA has reached a similar conclusion, including glioma, non-small cell lung cancer, P53 signaling pathway (Fig. 4C,D).
Fig. 4.
DLL3-related biological signatures and signal pathways in IBC. (A) DLL3-related biological processes in IBC by GO analysis; DLL3-related KEGG pathways via (B) GSVA and GSEA verified that the high DLL3 group was in the (C) cancer related pathway and (D) P53 signaling pathway.
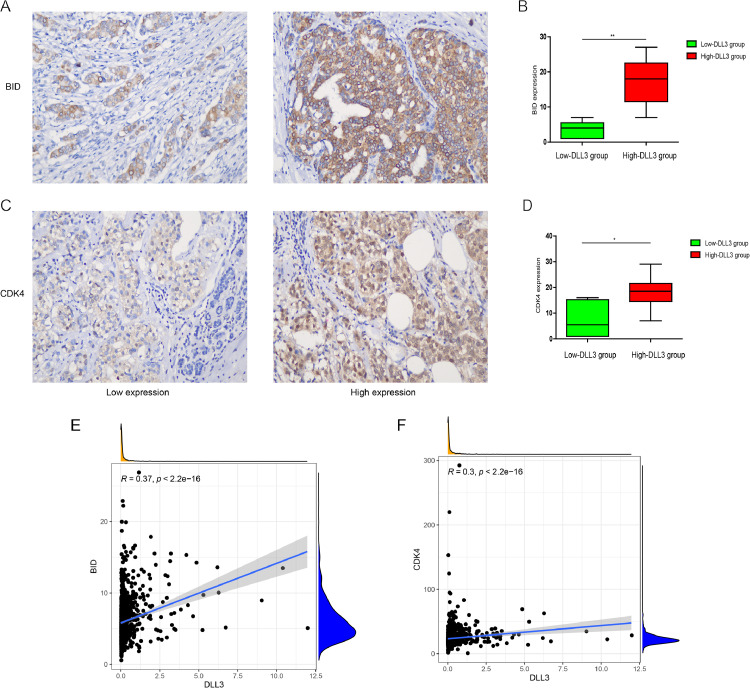
The role of the p53 protein in tumor suppression relies on the ability of p53 to regulate the transcription of genes that are important in cell-cycle arrest and in apoptosis. Previous reports have shown that bid is a me regulated by p5330. Cyclin-dependent kinase 4 (CDK4) is a key promoter of cell proliferation, and the regulation of CDK4 by P53 is also an important way to regulate the cell cycle [30,31]. In order to further verify the conclusion that DLL3 was related to p53 signaling pathway. We first analyzed the relationship between DLL3 and different P53 states and found that DLL3 expression was higher in P53-positive patients (Fig. S2A; P = 0.0056). Further hierarchical analysis showed that there was a positive correlation between the expression of p53 and DLL3 in luminal A (P = 0.0048) and HER2 overexpression (P = 0.0015) subtypes (Figs. S2C,D). We used Spearman analysis and IHC to detect the correlation of marker molecules of P53 signaling pathway. As shown in Fig. 5A–D, the expression of BID and CDK4 in IBC patients with high or low DLL3 groups were consistent with that of DLL3 expression level (P < 0.05). Correlation analysis demonstrated that DLL3 was positively correlated with BID and CDK4 expression (R > 0.3; P < 0.001) (Fig. 5E,F).
Fig. 5.
DLL3 positively regulated P53 signaling pathway. IHC staining of (A) BID and (C) CDK4 in IBC samples (200 × magnification). IHC scores for (B) BID and (D) CDK4 were compared between the low-DLL3 group and high-DLL3 group. n = 3 randomly selected fields of 10 samples, *P < 0.05 **P < 0.01. (E-F) Correlation analysis.
Discussion
In previous studies, the DLL3 is mainly associated with SCLC and other Neuroendocrine tumors. DLL3 is highly expressed in SCLC but not in normal lung tissue and it promoted tumor growth, migration and invasion by modulating Snail [32,33]. Loredana et al. found that DLL3 was expressed in neuroendocrine prostate cancer and was associated with neuroendocrine marker expression and invasive clinical characteristics. Rovalpituzumab teserine, a DLL3 targeted antibiotic drug regulate, has been used in clinical trials of SCLC and neuroendocrine prostate cancer [16,34]. These studies suggest that DLL3 is associated with the proliferation and migration of neuroendocrine tumors and is a promising therapeutic target. Here, we found that DLL3 was highly expressed in IBC, but little or no expression was found in normal breast tissues. High expression of DLL3 was associated with age and advanced T stage and represents a poorer prognosis for IBC. Therefore, DLL3 may be an effective therapeutic target for IBC. In addition, our results proved that DLL3 was an independent prognostic factor for IBC, which can accurately predict the survival outcome of IBC patients.
The proportion of immune cells in the TME plays an important role in tumor cell survival and tumor patient prognosis [[35], [36]–37]. In our study, a noteworthy conclusion was that the expression of DLL3 is associated with the level of immune infiltration in IBC. DLL3 significantly affected the infiltration of B cells, Neutrophils and T cells in TME. We verified these results by the correlation between the gene markers of different immune cells and the expression of DLL3. These correlations indicated that DLL3 may have a potential regulatory effect on B and T cells in IBC and affect the survival of tumor cells. Then, we explored another important aspect. Recently, immune checkpoint inhibitors (ICI) have been approved for use in patients with IBC, which has been shown to improve OS in patients [38]. But not all patients can benefit from ICI [39,40]. It was urgent to find more immune targets or immune mechanisms to assist in the treatment of IBC. Our study found that DLL3 was positively correlated with the expression of PD-1 and CTLA4, suggested that DLL3 may be used as an immunoadjuvant checkpoint or a marker of immunotherapeutic effect.
Stylianou et al. demonstrated an accumulation of Notch1 ICD in breast cancer cells compared with normal tissue and found that in overexpressing Notch1 ICD cell lines, the TP53 mediated DNA damage pathway was blocked to prevent cells from completing their apoptotic process [10]. However, the interaction between Notch and p53 in breast cancer remains unclear. Our study found that the high DLL3 group was enriched in the p53 signaling pathway, which indicated that DLL3 may affect tumor development through p53 signaling pathway. Tumor suppressor protein p53 plays an important role in maintaining genome integrity [41]. It can induce cell cycle arrest and apoptosis in response to cell stress [42]. P53 increases the expression of proapoptotic Bcl-2 family members, including BAX, BID, NOXA and Puma in response to DNA damage [43]. Hyperactivation of CDK4/6–cyclin D1 complexes is central regulators of the G1-S transition of the cell cycle in breast cancer [44]. Targeting CDK4/6 activity has long been considered a promising approach for cancer treatment. These inhibitors, combined with endocrine therapy, have been approved by the FDA for the treatment of breast cancer [45,46]. Our study found that DLL3 is more highly expressed in P53-positive patients, and it is positively correlated with BID and CDK4, the main markers of the p53 signaling pathway. High DLL3 group was significantly enriched in the cell cycle pathway. It suggested that DLL3 may regulate cell cycle and inhibit apoptosis through p53 pathway, leading to carcinogenesis. Therefore, targeting DLL3 or combined targeting may be an effective treatment strategy. Furthermore, we noticed that over activated Notch signaling attenuates mitochondrial activity and induce glycolysis in a p53 dependent manner in breast cancer [47]. Our results also demonstrated that the galactose metabolism pathway was activated in high DLL3 group. This may be one of the ways that Notch pathway regulates the energy homeostasis of cells.
The current study has limitations. Firstly, the work was only performed in our cohort and TCGA-BRCA cohort and we found DLL3 has a stronger predictive role in HER-2overexpression and luminal A patients. This conclusion needs to be further verified in more external cohorts and more samples. Secondly, we will further standardize every step in preanalytic, analytic, and postanalytic phases during IHC to achieve reproducible and reliable immunohistochemistry test results for DLL3 staining. Finally, further experimental validation should be performed to prove the biologic impact of DLL3.
In conclusions, DLL3 expression may be a potential prognostic molecular marker of poor survival in IBC. Moreover, the p53 signaling pathway may be the key pathway regulated by DLL3 in IBC.
Contributions
Chong Yuan: Conceptualization, Methodology, Writing - Original Draft, Data Curation
Kaili Chang: Software, Validation, Visualization
Chenyue Xu: Investigation, Formal analysis
Funding
This work was supported by the National Nature Science Foundation of China (Grant no. 81874064 and 82072692).
Ethical approval and consent to participate
The clinical research ethics committee of Fudan University approved this study with the approval number 2018-Y016 and each patient included in the cohort obtained informed consent.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Thanks to three doctors (Du, Liu, Chen) in the pathology department of Huashan Hospital Affiliated to Fudan University for reading the film.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101080.
Contributor Information
Qingquan Li, Email: 061101040@fudan.edu.cn.
Zunguo Du, Email: duzunguo@126.com.
Appendix. Supplementary materials
Reference
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Soerjomataram I., Dikshit R. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Kan Z., Ding Y., Kim J. Multi-omics profiling of younger Asian breast cancers reveals distinctive molecular signatures. Nat. Commun. 2018;9(1):1725. doi: 10.1038/s41467-018-04129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Corona S.P., Sobhani N., Ianza A. Advances in systemic therapy for metastatic breast cancer: future perspectives. Med. Oncol. 2017;34(7):119. doi: 10.1007/s12032-017-0975-5. [DOI] [PubMed] [Google Scholar]
- 5.Bray S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016;17(11):722–735. doi: 10.1038/nrm.2016.94. [DOI] [PubMed] [Google Scholar]
- 6.Kumar S., Srivastav R.K., Wilkes D.W. Estrogen-dependent DLL1-mediated Notch signaling promotes luminal breast cancer. Oncogene. 2019;38(12):2092–2107. doi: 10.1038/s41388-018-0562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krop I., Demuth T., Guthrie T. Phase I pharmacologic and pharmacodynamic study of the gamma secretase (Notch) inhibitor MK-0752 in adult patients with advanced solid tumors. J. Clin. Oncol. 2012;30(19):2307–2313. doi: 10.1200/JCO.2011.39.1540. [DOI] [PubMed] [Google Scholar]
- 8.Samon J.B., Castillo-Martin M., Hadler M. Preclinical analysis of the gamma-secretase inhibitor PF-03084014 in combination with glucocorticoids in T-cell acute lymphoblastic leukemia. Mol. Cancer Ther. 2012;11(7):1565–1575. doi: 10.1158/1535-7163.MCT-11-0938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Capaccione K.M., Pine S.R. The Notch signaling pathway as a mediator of tumor survival. Carcinogenesis. 2013;34(7):1420–1430. doi: 10.1093/carcin/bgt127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stylianou S., Clarke R.B., Brennan K. Aberrant activation of notch signaling in human breast cancer. Cancer Res. 2006;66(3):1517–1525. doi: 10.1158/0008-5472.CAN-05-3054. [DOI] [PubMed] [Google Scholar]
- 11.Zeng J.S., Zhang Z.D., Pei L. CBX4 exhibits oncogenic activities in breast cancer via Notch1 signaling. Int. J. Biochem. Cell Biol. 2018;95:1–8. doi: 10.1016/j.biocel.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Simmons M.J., Serra R., Hermance N., Kelliher M.A. NOTCH1 inhibition in vivo results in mammary tumor regression and reduced mammary tumorsphere-forming activity in vitro. Breast Cancer Res. 2012;14(5):R126. doi: 10.1186/bcr3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dou X.W., Liang Y.K., Lin H.Y. Notch3 maintains luminal phenotype and suppresses tumorigenesis and metastasis of breast cancer via trans-activating estrogen receptor-alpha. Theranostics. 2017;7(16):4041–4056. doi: 10.7150/thno.19989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koshkin V.S., Garcia J.A., Reynolds J. Transcriptomic and protein analysis of small-cell bladder cancer (SCBC) identifies prognostic biomarkers and DLL3 as a relevant therapeutic target. Clin. Cancer Res. 2019;25(1):210–221. doi: 10.1158/1078-0432.CCR-18-1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spino M., Kurz S.C., Chiriboga L. Cell surface notch ligand DLL3 is a therapeutic target in isocitrate dehydrogenase-mutant glioma. Clin. Cancer Res. 2019;25(4):1261–1271. doi: 10.1158/1078-0432.CCR-18-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Puca L., Gavyert K., Sailer V. Delta-like protein 3 expression and therapeutic targeting in neuroendocrine prostate cancer. Sci. Transl. Med. 2019;11(484) doi: 10.1126/scitranslmed.aav0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newman A.M., Liu C.L., Green M.R. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank G.A., Danilova N.V., Andreeva I., Nefedova N.A. [WHO classification of tumors of the breast, 2012] Arkh. Patol. 2013;75(2):53–63. [PubMed] [Google Scholar]
- 19.Qu Y., Lin Z., Qi Y. PAK1 expression determines poor prognosis and immune evasion in metastatic renal cell carcinoma patients. Urol. Oncol. 2020;38(4):293–304. doi: 10.1016/j.urolonc.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Zlobec I., Steele R., Michel R.P., Compton C.C., Lugli A., Jass J.R. Scoring of p53, VEGF, Bcl-2 and APAF-1 immunohistochemistry and interobserver reliability in colorectal cancer. Mod. Pathol. 2006;19(9):1236–1242. doi: 10.1038/modpathol.3800642. [DOI] [PubMed] [Google Scholar]
- 21.Yamashita H., Toyama T., Nishio M. p53 protein accumulation predicts resistance to endocrine therapy and decreased post-relapse survival in metastatic breast cancer. Breast Cancer Res. 2006;8(4):R48. doi: 10.1186/bcr1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yin X., Zhang X., Liu Z. Assessment for prognostic value of differentially expressed genes in immune microenvironment of clear cell renal cell carcinoma. Am. J. Transl. Res. 2020;12(9):5416–5432. [PMC free article] [PubMed] [Google Scholar]
- 23.Frey B., Ruckert M., Weber J. Hypofractionated irradiation has immune stimulatory potential and induces a timely restricted infiltration of immune cells in colon cancer tumors. Front. Immunol. 2017;8:231. doi: 10.3389/fimmu.2017.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A., Tamayo P., Mootha V.K. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charoentong P., Finotello F., Angelova M. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18(1):248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Thorsson V., Gibbs D.L., Brown S.D. The immune landscape of cancer. Immunity. 2019;51(2):411–412. doi: 10.1016/j.immuni.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 29.Masugi Y., Abe T., Ueno A. Characterization of spatial distribution of tumor-infiltrating CD8(+) T cells refines their prognostic utility for pancreatic cancer survival. Mod. Pathol. 2019;32(10):1495–1507. doi: 10.1038/s41379-019-0291-z. [DOI] [PubMed] [Google Scholar]
- 30.O'Leary B., Finn R.S., Turner N.C. Treating cancer with selective CDK4/6 inhibitors. Nat. Rev. Clin. Oncol. 2016;13(7):417–430. doi: 10.1038/nrclinonc.2016.26. [DOI] [PubMed] [Google Scholar]
- 31.Ewen M.E. p53-dependent repression of cdk4 synthesis in transforming growth factor-beta-induced G1 cell cycle arrest. J. Lab. Clin. Med. 1996;128(4):355–360. doi: 10.1016/s0022-2143(96)80006-0. [DOI] [PubMed] [Google Scholar]
- 32.Regzedmaa O., Li Y., Li Y. Prevalence of DLL3, CTLA-4 and MSTN expression in patients with small cell lung cancer. Onco. Targets Ther. 2019;12:10043–10055. doi: 10.2147/OTT.S216362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furuta M., Kikuchi H., Shoji T. DLL3 regulates the migration and invasion of small cell lung cancer by modulating Snail. Cancer Sci. 2019;110(5):1599–1608. doi: 10.1111/cas.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudin C.M., Pietanza M.C., Bauer T.M. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol. 2017;18(1):42–51. doi: 10.1016/S1470-2045(16)30565-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flister M.J., Bergom C. Genetic modifiers of the breast tumor microenvironment. Trends Cancer. 2018;4(6):429–444. doi: 10.1016/j.trecan.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gordon B., Gadi V.K. The role of the tumor microenvironment in developing successful therapeutic and secondary prophylactic breast cancer vaccines. Vaccines. 2020;8(3) doi: 10.3390/vaccines8030529. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bainer R., Frankenberger C., Rabe D., An G., Gilad Y., Rosner M.R. Gene expression in local stroma reflects breast tumor states and predicts patient outcome. Sci. Rep. 2016;6:39240. doi: 10.1038/srep39240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adams S., Gatti-Mays M.E., Kalinsky K. Current landscape of immunotherapy in breast cancer: a review. JAMA Oncol. 2019 doi: 10.1001/jamaoncol.2018.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmid P., Adams S., Rugo H.S. Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. N. Engl. J. Med. 2018;379(22):2108–2121. doi: 10.1056/NEJMoa1809615. [DOI] [PubMed] [Google Scholar]
- 40.Adams S., Loi S., Toppmeyer D. Pembrolizumab monotherapy for previously untreated, PD-L1-positive, metastatic triple-negative breast cancer: cohort B of the phase II KEYNOTE-086 study. Ann. Oncol. 2019;30(3):405–411. doi: 10.1093/annonc/mdy518. [DOI] [PubMed] [Google Scholar]
- 41.Ryan K.M., Ernst M.K., Rice N.R., Vousden K.H. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404(6780):892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 42.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 43.Shankar E., Sivaprasad U., Basu A. Protein kinase C epsilon confers resistance of MCF-7 cells to TRAIL by Akt-dependent activation of Hdm2 and downregulation of p53. Oncogene. 2008;27(28):3957–3966. doi: 10.1038/onc.2008.39. [DOI] [PubMed] [Google Scholar]
- 44.AbuHammad S., Cullinane C., Martin C. Regulation of PRMT5-MDM4 axis is critical in the response to CDK4/6 inhibitors in melanoma. Proc. Natl. Acad. Sci. USA. 2019;116(36):17990–18000. doi: 10.1073/pnas.1901323116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Finn R.S., Crown J.P., Lang I. The cyclin-dependent kinase 4/6 inhibitor palbociclib in combination with letrozole versus letrozole alone as first-line treatment of oestrogen receptor-positive, HER2-negative, advanced breast cancer (PALOMA-1/TRIO-18): a randomised phase 2 study. Lancet Oncol. 2015;16(1):25–35. doi: 10.1016/S1470-2045(14)71159-3. [DOI] [PubMed] [Google Scholar]
- 46.Cristofanilli M., Turner N.C., Bondarenko I. Fulvestrant plus palbociclib versus fulvestrant plus placebo for treatment of hormone-receptor-positive, HER2-negative metastatic breast cancer that progressed on previous endocrine therapy (PALOMA-3): final analysis of the multicentre, double-blind, phase 3 randomised controlled trial. Lancet Oncol. 2016;17(4):425–439. doi: 10.1016/S1470-2045(15)00613-0. [DOI] [PubMed] [Google Scholar]
- 47.Landor S.K., Mutvei A.P., Mamaeva V. Hypo- and hyperactivated Notch signaling induce a glycolytic switch through distinct mechanisms. Proc. Natl. Acad. Sci. USA. 2011;108(46):18814–18819. doi: 10.1073/pnas.1104943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.