Abstract
A novel coronavirus, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), caused a worldwide pandemic. Our aim in this study is to produce new fusion inhibitors against SARS-CoV-2, which can be the basis for developing new antiviral drugs. The fusion core comprising the heptad repeat domains (HR1 and HR2) of SARS-CoV-2 spike (S) were used to design the peptides. A total of twelve peptides were generated, comprising a short or truncated 24-mer (peptide #1), a long 36-mer peptide (peptide #2), and ten peptide #2 analogs. In contrast to SARS-CoV, SARS-CoV-2 S-mediated cell-cell fusion cannot be inhibited with a minimal length, 24-mer peptide. Peptide #2 demonstrated potent inhibition of SARS-CoV-2 S-mediated cell-cell fusion at 1 µM concentration. Three peptide #2 analogs showed IC50 values in the low micromolar range (4.7-9.8 µM). Peptide #2 inhibited the SARS-CoV-2 pseudovirus assay at IC50=1.49 µM. Given their potent inhibition of viral activity and safety and lack of cytotoxicity, these peptides provide an attractive avenue for the development of new prophylactic and therapeutic agents against SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, Fusion inhibitors, Antiviral drugs
INTRODUCTION
The SARS-CoV-2 pandemic that began in 2019 has posed a significant threat worldwide. In the past two decades, three coronaviruses have emerged and endangered public health, including the severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome CoV (MERS-CoV), and SARS-CoV-2 (Peeri et al., 2020). Until the moment, there is no approved drug for treating SARS-CoV-2 infection. Therefore, discovery of new potential chemotherapeutics is an urgent demand.
The viral CoV genome encodes four structural proteins: spike, membrane (M), envelope (E), and nucleocapsid (N). Viral membrane fusion is an essential step of virus replication, which is accomplished by the viral spike and leads to the fusion of the viral and cell membranes (Bosch et al., 2003). The CoV S protein is composed of two subunits, S1 and S2. S1 binds the host cell angiotensin-converting enzyme-2 (ACE2) receptor (Song et al., 2018; Hoffmann et al., 2020). Cleavage of S1 by host cell proteases exposes a highly hydrophobic membrane-binding domain of the S2 subunit (Kirchdoerfer et al., 2016). The S2 subunit contains two domains, heptad repeat domain 1 (HR1) and heptad repeat domain 2 (HR2). HR1 forms a homotrimer exposing three hydrophobic pockets on its surface, which host the HR2 domain during the active fusion process (Xia et al., 2014). An HR domain is composed of tandem repeat motifs of seven residues (named a-g). Of the seven residues, the first (a) and fourth (d) are predominantly hydrophobic or bulky (Gao et al., 2013).
From a therapeutic perspective, fusion inhibitors have been successfully used in inhibiting viral infections as they harbor intrinsic ability to prevent the fusion of viral and cellular membranes. In this study, we investigated a current knowledge gap in the discovery of anti-SARS-CoV-2 fusion drugs. First, the inhibitory properties of a truncated 24-mer peptide (peptide #1) and a 36-mer peptide (peptide #2) were compared. The results indicated strong inhibition of S-mediated cell-cell fusion and SARS-CoV-2 pseudovirus activity. In addition, ten peptide #2 analogs were developed. The structural and molecular aspects of the associated inhibitory properties are discussed.
MATERIALS AND METHODS
Fusion peptides
Previous studies of SARS-CoV, MERS-CoV, and other viruses such as HIV and Ebola showed that HR1 analogs are usually weaker inhibitors than HR2-derived peptides (Liu et al., 2004; Lu et al., 2014; Shabane et al., 2019). Furthermore, our previous work on developing MERS-CoV peptides showed that 36-amino acids HR2 analogs strongly inhibited MERS-CoV replication (Patent pending, USPTO application no. 16/857136). Previous studies on MERS-CoV peptides showed that a 19-mer peptide did not show any antiviral activity, and the 45-mer peptide demonstrated marginal activity (Lu et al., 2014). Based on these data, we designed shorter 24-amino acid (peptide #1) and 36-amino acid (peptide #2) HR2 analogs for potential anti-SARS-CoV2 activity. The concluded sequence for peptide synthesis was generated after alignment of SARS-CoV, SARS-CoV-2, and MERS-CoV HR2 sequences. The peptides were designed to include the central helix and residues on the extended N-terminal region (Fig. 1). In addition, a series of single or multiple modified peptide #2 analogs were produced by introducing a single, double, or triple amino acid mutation (Fig. 1). The mutations were based on the improvement of free energy of binding after amino acid mutations, as previously described (Dehouck et al., 2013; Brender and Zhang, 2015). Each mutant residue was estimated to improve the binding energy by 0.1-0.5 kcal/mol. Peptide synthesis orders were sent to Apeptides (Shanghai, China) and Biomatik Inc (Ontario, Canada).
Fig. 1.
The structure of SARS-CoV-2 fusion core and sequence of peptides. (A) Schematic representation of SARS-CoV-2 Spike. The S1 subunit contains the N-terminal domain (NTD) and the receptor-binding domain (RBD). The S2 subunit comprises the fusion protein (FP) and two heptad repeat domains, HR1 and HR2. (B) HR1 and HR2 monomers during the fusion state showing the regions comprising the synthesized. (C) The position of peptides #1 and 2 on HR2. The side chains of amino acids is presented in green dots. (D) The sequences of peptides #1-12, the conserved sequences were highlighted with the same colour.
Molecular dynamics simulations (MDs)
MDs were used to investigate the stability of the complex between the HR2-modified peptides #2-12 with HR1. The mutations were generated by Schrodinger Maestro package version 12 and saved as PDB files. MDs were performed as previously described with slight modifications (Kandeel et al., 2018). GROMACS version 5.5.5 and GROMOS96 43a1 force field were used (Van Der Spoel et al., 2005). The simulation continued for 10 ns to determine the root mean square deviation (RMSD) of the structure and the root mean square fluctuation (RMSF) of the residues. Snapshots were taken every 10 ps, providing 1000 snapshots for every complex.
Cell lines and plasmids
HEK293T is an immortalized cell line derived from a human fetal kidney. A pair of previously described 293FT-based reporter cell lines that constitutively express individual split proteins (DSP1-7 and DSP8-11 proteins) (Wang et al., 2014) were used and maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and 1 g/mL puromycin. Calu-3 cells (ATCC HTB-55) were maintained in Eagle’s minimum essential medium (EMEM) containing 10% fetal bovine serum (FBS). For the construction of transient transfection vectors, a synthetic DNA corresponding to the S gene of SARS-CoV-2 (NC_045512.2) was cloned into a lentiviral transfer plasmid (CD500B-1, SBI, Palo Alto, CA, USA) and the VSV(Vesicular stomatitis virus)-G gene was cloned into pCAG plasmid.
DSP assay to monitor membrane fusion
The DSP assay using 293FT cells was performed as described previously (Yamamoto et al., 2020) to monitor SARS-CoV-2-S-mediated membrane fusion. Briefly, effector cells expressing SARS-CoV-2-S protein with DSP8-11, target cells expressing ACE2, and transmembrane serine protease 2 (TMPRSS2) with DSP1-7 were seeded in 10 cm culture dishes (4×106 cells/10 mL) one day before the assay. Two hours before the DSP assay, cells were treated with 6 µM EnduRen (Promega, Madison, WI, USA), a substrate for Renilla luciferase, to activate EnduRen. One microliter of each peptide dissolved in dimethyl sulfoxide (DMSO) was added to the 384-well plates (Greiner Bioscience, Frickenhausen, Germany). Next, 50 µL of each single-cell suspension (effector and target cells) was added to the wells using a Multidrop dispenser (Thermo Fisher Scientific, Waltham, MA, USA). After incubation at 37°C for 4 h, luciferase activity was measured using a Centro xS960 luminometer (Berthold, Bad-Wildbad, Germany).
Preparation of pseudotyped VSV viral particles and infection experiments
293T cells were transfected with an expression plasmid for SARS-CoV-2-S, VSV-G, or control expression plasmid by calcium-phosphate precipitation. At 16 h posttransfection, the cells were inoculated with a replication-deficient VSV, VSV-ΔG-Luciferase, which lacks the VSV-G gene and encodes firefly luciferase, at an MOI=1 as described previously (Tani et al., 2010). After 2 h of incubation, cells were washed with DMEM and further incubated for 16 h before supernatants containing the pseudotyped viral particles were harvested. Cellular debris was removed from the supernatants using a syringe filter with a 0.2 µm size pore (Millipore, Bedford, MA, USA).
For an infection assay, target Calu-3 cells were seeded in 96-well plates (5×104 cells/100 µL) one day before the assay. Cells were pre-treated with peptides for 1 h before infection. Pseudotyped viral particles were added to cells with the peptides. After 2 h of incubation, the culture supernatant was removed, and cells were washed with EMEM. Cells were further incubated in EMEM containing 10% FBS without peptides and pseudotyped viral particles. At 16 h postinfection, luciferase activity was measured using the Bright-Glo Luciferase Assay System (Promega) and Centro xS960 luminometer (Berthold).
Statistical analysis
Statistically significant differences between the mean values were determined using a two-tailed Student’s t-test. All data represent three independent experiments, and values represent the mean ± standard deviation (SD), with p<0.05 considered statistically significant. Several correlation runs were conducted to evaluate the relationship between the obtained activity and the molecular and structural descriptors of the peptides. Pearson’s correlation coefficient and the degree of significance were estimated in GraphPad Prism software (GraphPad Software, San Diego, CA, USA).
RESULTS
The peptides sequences
The peptide design comprised testing the hypothesis of potential inhibition of SARS-CoV fusion by truncated or short peptide, a 24-mer (peptide #1). Furthermore, the 36-mer peptides were designed based on our previous research on MERS-CoV fusion as well as several fusion inhibition reports. All peptides contained the central helix of HR2 (Fig. 1). Peptides #2-12 contained several mutations of the WT 36-mer peptide #2 (Fig. 1). The inserted mutations were calculated to increase the binding free energy by ~0.5 kcal/mol.
DSP assay for SARS-CoV-2 S-mediated cell-cell fusion
We evaluated the effects of the twelve different peptides on SARS-CoV-2 S protein-mediated membrane fusion by DSP assay, which we previously established (Yamamoto et al., 2020). Four peptides (#2, #3, #5 and #10) significantly reduced luciferase activity at 10 µM in the DSP assay (Fig. 2A). Two peptides (#7 and #11) also reduced luciferase activity at 10 µM with statistical meaning, although their inhibitory activities were much less than those of the four peptides listed above. The other six peptides did not affect luciferase activity. None of the twelve peptides affected luciferase activity in cells carrying the preformed DSP1-7/DSP8-11 reporter complex, indicating that luciferase activity under the treatment of each peptide in Fig. 2A reflects its effect on SARS-CoV-2 S mediated cell-cell fusion (Fig. 2B). Further analysis of the four peptides that strongly inhibited membrane fusion showed that Peptide #2 suppressed luciferase activity in a dose-response manner and exhibited the greatest inhibitory activity on the four peptides with a minimum IC50 value at 1.0 µM. (Fig. 2C, 2D). The IC50 values of #3, #5, and #10 were 7.2, 4.7, and 8.9 µM, respectively, indicating that the inhibitory activities of the three peptides were approximately 4- to 9-fold less than that of peptide #2 (Fig. 2D). Therefore, we chose peptide #2 for further analysis.
Fig. 2.
The effect of peptides on SARS-CoV-2 S-mediated membrane fusion. (A) The effect of each peptides on the coculture fusion assay using DSP as a reporter. Peptides were tested at different concentrations, and the additional proteins other than reporters (DSPs) transduced into the effector and target cells are indicated below the graph. The relative cell fusion was represented as the DSP value (RL activity measured in RLU) normalized to that of the control assay with DMSO alone. (B) The effect of each peptides on RL measurement. Each peptides was added to cells co-expressing DSP1-7 and DSP8-11 to evaluate its direct inhibitory effects on RL. The relative DSP signal is indicated in the vertical axis by setting the control value with DSP alone as 100%. (C) Dose-response analysis for 5 peptides. The relative cell fusion was represented as the DSP value normalized to that of the control assay with DMSO alone. (D) Calculated IC50 value in figure (C). *p<0.05, **p<0.01 vs. control assay with DMSO.
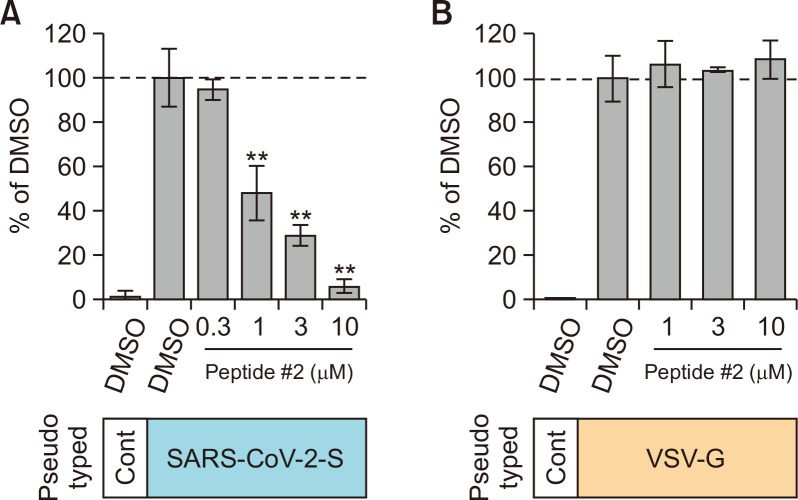
The effect of peptide #2 on SARS-CoV-2-S pseudotyped VSV infection
We then examined the effect of peptide #2 on infection of human lung epithelial cells, Calu-3 cells with SARS-CoV-2-S, or VSV-G pseudotyped VSV viral particles. To analyze the effect of peptide #2 on the virus entry step, we treated Calu-3 cells with peptide #2 for 1 h before infection and for an additional 2 h during infection. After the pseudotyped viral particles were removed, cells were incubated for 16 h without the peptide to induce luciferase expression. The luciferase assay showed that the infectious activity of SARS-CoV-2-S pseudotyped viral particles was significantly suppressed by the treatment with peptide #2 (IC50=1.49 µM) (Fig. 3A), while infection with VSV-G pseudotyped viral particles was not affected by peptide #2 (Fig. 3B). These results indicate that peptide #2 potently inhibits infection with SARS-CoV-2-S pseudotyped viral particles by preventing viral-cell fusion, possibly through binding of peptide #2 to the SARS-CoV-2 Spike.
Fig. 3.
The effect of peptide #2 on SARS-CoV-2 S-mediated viral infection. (A) The effect of peptide #2 on infection of Calu-3 cells with SARS-CoV-2 S pseudotyped VSV viral particles. The relative infectivity was represented as the RLU normalized to that of the control assay with DMSO alone. (B) The effect of peptide #2 on infection of Calu-3 cells with VSV-G pseudotyped VSV viral particles. The relative infectivity was represented as the RLU normalized to that of the control assay with DMSO alone. **p<0.01.
Stability of peptides-HR1 complexes
The initial structure of the fusion complex contained a trimer of the 6-helix coiled-coil bundle of the HR1-HR2 fusion core. HR1 comprised the residues 912-989 of SARS-CoV S1, and HR2 contained 38 residues (1164-1202). MDs were used to evaluate the structural stability of the designed peptides and HR1 complexes. The RMSD of peptides-HR1 complexes is provided in Fig. 4A. Table 1 summarizes the average RMSD for every peptide-HR1 complex. The notable feature is the stability of the peptide complexes with low RMSD values (<0.2 nm). Overall, the results indicate tight binding of the peptides complexes. RMSF values revealed that all peptides had a similar profile of low RMSF values, and the peak of high fluctuations corresponds to the loop connecting HR1 and HR2 (residues no. 70-80, Fig. 4B), as previously observed (Kandeel et al., 2018).
Fig. 4.
Molecular dynamics simulation of the peptides #2-12 complexes with HR1. (A) RMSD plot showing the changes of the α-carbon atom over the simulation time. (B) RMSF plot showing the fluctuations of the peptidesHR2 complex residues.
Table 1.
The average RMSD values for peptides #2-12 after 10 ns molecular dynamics simulation
| Peptide # | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RMSD (nm) | 0.192 | 0.169 | 0.208 | 0.189 | 0.195 | 0.179 | 0.194 | 0.176 | 0.181 | 0.176 | 0.193 |
The sequence and molecular descriptors of the composition of the peptides were analyzed to understand the observed inhibitory activity of the peptides on a molecular basis. In addition, multiple correlation analysis was adopted by investigating the relationship of each molecular descriptor with the corresponding activity (Table 2). Within the examined descriptors, only hydrophilic substitution showed a significant correlation with the obtained S-mediated cell-cell fusion % to DMSO (r=–0.63, p<0.05) (Table 3).
Table 2.
The protein structure statistics of the used peptides
| ID | α-helix (residues) |
α-helix % | Frequency of residues | Half-life in mammals (h) | %cell-cell fusion activity | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Negative charge | Positive charge | Other | Hydrophobic | Hydrophilic | Other | |||||
| # 2 | 7-34 | 75 | 0.194 | 0.083 | 0.722 | 0.444 | 0.28 | 0.28 | 1.1 | 3.078 |
| # 3 | 7-34 | 75 | 0.194 | 0.083 | 0.722 | 0.444 | 0.28 | 0.28 | 1.1 | 13.694 |
| # 4 | 7-34 | 75 | 0.167 | 0.111 | 0.722 | 0.444 | 0.28 | 0.28 | 1.1 | 108.542 |
| # 5 | 7-34 | 75 | 0.167 | 0.083 | 0.75 | 0.472 | 0.278 | 0.25 | 1.1 | 7.609 |
| # 6 | 7-34 | 75 | 0.167 | 0.083 | 0.75 | 0.5 | 0.25 | 0.25 | 1.1 | 85.133 |
| # 7 | 7-25, 28-33 | 66.7 | 0.167 | 0.083 | 0.75 | 0.5 | 0.25 | 0.25 | 1.1 | 67.81 |
| # 8 | 7-34 | 75 | 0.222 | 0.25 | 0.306 | 0.444 | 0.25 | 0.306 | 1.1 | 93.096 |
| # 9 | 7-34 | 75 | 0.194 | 0.083 | 0.722 | 0.472 | 0.25 | 0.278 | 1.1 | 94.155 |
| # 10 | 5-34 | 80.6 | 0.222 | 0.083 | 0.694 | 0.417 | 0.278 | 0.306 | 1.1 | 31.54 |
| # 11 | 7-34 | 75 | 0.194 | 0.083 | 0.722 | 0.472 | 0.25 | 0.278 | 1.1 | 72.85 |
| # 12 | 7-25, 27-34 | 69.4 | 0.222 | 0.083 | 0.694 | 0.472 | 0.222 | 0.306 | 1.1 | 89.708 |
Table 3.
Correlation statistics of the obtained cell-cell fusion inhibition properties of the peptides with their sequence and molecular descriptors
| Activity% vs. Alfa-helix % |
Activity% vs. Negative charge |
Activity% vs. Positive charge |
Activity% vs. other charge |
Activity% vs. hydrophobic |
Activity% vs. hydrophilic |
Activity% vs. other |
|
|---|---|---|---|---|---|---|---|
| Pearson r | –0.2782 | 0.02148 | 0.3442 | –0.282 | 0.3148 | –0.6326 | 0.1641 |
| 95% confidence interval | –0.7525 to 0.3861 | –0.5859 to 0.6135 | –0.3222 to 0.7825 | –0.7543 to 0.3826 | –0.3514 to 0.7694 | –0.8934 to –0.05273 | –0.4834 to 0.6955 |
| R squared | 0.07737 | 0.0004614 | 0.1184 | 0.07953 | 0.09911 | 0.4002 | 0.02692 |
| p value | |||||||
| p (two-tailed) | 0.4075 | 0.9500 | 0.3000 | 0.4008 | 0.3457 | 0.0367 | 0.6298 |
| p value summary | ns | ns | ns | ns | ns | * | ns |
| Significant? (alpha=0.05) | No | No | No | No | No | Yes | No |
| Number of XY pairs | 11 | 11 | 11 | 11 | 11 | 11 | 11 |
DISCUSSION
The discovery of new therapeutic agents against emerging viruses is the most effective tool to fight disease and decrease morbidity and mortality. In this context, fusion inhibitors have demonstrated substantial promise for both prophylaxis and treatment of viral infections. In this study, we introduced the first generation of small chemical molecules with anti-MERS-CoV fusion activity (Kandeel et al., 2020b). Given the lack of approved anti-SARS-CoV-2 drugs, this study was conducted to initiate a drug discovery campaign by targeting the SARS-CoV-2 fusion step. Fusion inhibitors are commonly used for treating HIV. Currently, there are approximately 11 completed and one recruiting clinical trials for treating HIV patients with fusion inhibitors (https://www.clinicaltrials.gov). Fusion inhibition has been found a useful strategy to inhibit SARS-CoV (Liu et al., 2004; Sainz et al., 2006; Chu et al., 2008; Liu et al., 2009) and MERS-CoV (Lu et al., 2014).
The virus entry inhibitors comprise two important steps of the virus replication cycle: attachment and fusion. The viral receptor-binding domain (RBD) is an important attachment target for developing specific antiviral antibodies or vaccines. However, this target is prone to frequent mutation (Xia et al., 2019). Therefore, fusion inhibitors target a highly conserved region in viral proteins.
Previous report revealed that CoVs can infect the cells via two entry mechanisms, direct membrane fusion or endocytosis (Qinfen et al., 2004). SARS-CoV-2 entry can also be accomplished by both plasma membrane and endocytic pathways (Seyedpour et al., 2020). Both mechanisms depend on the activation of the viral spike after cleavage, specifically, by a TMPRSS2 for plasma membrane pathway and the pH-dependent cysteine protease cathepsin L for the endocytic pathway. The pathways used for each cell line have been identified; for example, SARS-CoV-2 infects Calu-3 cells which endogenously express TMPRSS2 by the plasma membrane pathway and infects Vero cells which lacks expression of TMPRSS2 by the endocytic pathway (Hoffmann et al., 2020). Since recent analysis of single-cell RNA-seq datasets from human tissues revealed that TMPRSS2 is expressed in a cell type specific manner (Sungnak et al., 2020; Ziegler et al., 2020). It is likely that the entry pathway also depends on the cell type. TMPRSS2-knockout resulted in reduced spread of SARS-CoV and MERS-CoV in the airways accompanied by reduced severity of lung pathology in a mouse model (Iwata-Yoshikawa et al., 2019). Therefore, TMPRSS2-dependent plasma membrane pathway is likely crucial for SARS-CoV-2 spread and disease development in vivo, and targeting plasma membrane pathway is likely to be an effective strategy to cure COVID-19. In this study, the S-mediated cell-cell fusion assay in TMPRSS2-expressing 293FT cells and pseudovirus assays in Calu-3 cells were dependent on TMPRSS2. Therefore, our results suggest that the peptides may inhibit the TMPRSS2-dependent plasma membrane pathway of the membrane fusion process.
In comparing the HR1-HR2 complexes, the SARS-CoV-2 complex has stronger binding energy than SARS-CoV, determined by the higher α–helicity of HR1 (Zhu et al., 2020). All peptides showed similar α-helical content of 75% except #7 and #12, which demonstrated 66.7 and 69.4%, respectively (Table 1). A previous study determined that the enhanced α-helical content of fusion peptides is associated with higher antiviral efficacy (Sainz et al., 2006).
Both SARS and SARS-CoV-2 proteins demonstrate a high identity rate (Kandeel et al., 2020a; Kandeel and Al-Nazawi, 2020). Experience from SARS-CoV fusion inhibitors shows that truncated peptides with a minimal length of 23-mer peptides can inhibit the virus, which showed inhibition of fusion at IC50 of 1.04 µM (Liu et al., 2009). In this study, the truncated peptide #1 did not show significant inhibition. A 36-mer peptide of an HR analog strongly inhibited MERS-CoV S-mediated cell-cell fusion at 0.93 µM (Lu et al., 2014). In this study, the 36-mer peptide #2 demonstrated potent inhibition of both SARS-CoV-2 S-mediated cell-cell fusion and pseudovirus activity at 1 and 1.4 µM, respectively. Novel antiviral compounds with IC50 in the low micromolar range are expected to be promising lead structures (Liu et al., 2004). Furthermore, the tested peptides were lacking any cytotoxicity suggesting their potential use in future drug discovery optimizations.
The use of peptide as chemotherapeutic agents has been widely applied in treating viral diseases e.g. HIV (Yao et al., 2012), Zika virus (Jackman et al., 2018), African swine fever virus (Hakobyan et al., 2018), dengue virus (De La Guardia and Lleonart, 2014), influenza virus (Kadam et al., 2017) and feline immune deficiency virus (Mizukoshi et al., 2009). Modifications of peptides had led to improvement of their systemic activity. For instance, amphipathic alfa helical peptides and cell penetrating peptides had improved the penetration of cell membranes and body barriers as blood brain barrier (Stalmans et al., 2015). Engineering of peptides by using dextrorotary (D)-amino acids improved the peptide stability and efficiency in delivery to remote organs (Garton et al., 2018). The noninvasive methods of peptides delivery is now attracting the researchers’ attention by oral, transdermal or nasal routes. Conjugation of peptides as insulin with polyarginines resulted in a dramatic increase in the systemic delivery after oral administration (Morishita et al., 2007). Huge market potential for peptide-conjugates is expected, as well as there are several peptide drugs are now undergoing clinical trials (Vhora et al., 2015). More recently, conjugation of peptides with polymeric, lipid and magnetic nanoparticles improved the systemic their delivery (Bashyal et al., 2016). In the field of antiviral therapy, the antiviral fusion peptide enfuvirtide had been approved by FDA for treating HIV. This parent peptide has been used as a target for improved potency, stability, pharmacokinetics and drug delivery e.g. PEG conjugation (Cheng et al., 2016), antithrombin-binding carrier pentasaccharide conjugates (Huet et al., 2010) and transdermal delivery through low-frequency, low-Power ultrasound transducer patch (Snook et al., 2019). Therefore, the promising advances in peptides therapy and delivery support the premise of using peptides in the treatment of SARS-CoV-2 infections.
In brief, this study investigated the discovery of new peptide inhibitors against SARS-CoV-2 fusion. Peptides # 2 inhibited S-mediated cell-cell fusion at 1 and 4.4 µM, respectively. This suggests the potent inhibition of SARS-CoV-2, probably by inhibition of the membrane fusion mechanism. Peptide #1 can be regarded as a lead compound for further antiviral optimization.
ACKNOWLEDGMENTS
The authors acknowledge the financial support from the Administration of International Cooperation and Knowledge Exchange (ICKEA), King Faisal University, Project No. IC-01-20. This work was supported in part by grants-in-aid from the Japanese Society for the Promotion of Science (16H06575 to J. I., 20K07610 to M. Y.), a Program of Japan Initiative for Global Research Network on Infectious Diseases (JGRID) from AMED (JP20wm0125002 to Y. K.). The authors acknowledge the computational facilities at the college of veterinary medicine. King Faisal University.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Bashyal S., Noh G., Keum T., Choi Y. W., Lee S. Cell penetrating peptides as an innovative approach for drug delivery; then, present and the future. J. Pharm. Investig. 2016;46:205–220. doi: 10.1007/s40005-016-0253-0. [DOI] [Google Scholar]
- Bosch B. J., Van, der Zee R., De Haan C. A., Rottier P. J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brender J. R., Zhang Y. Predicting the effect of mutations on protein-protein binding interactions through structure-based interface profiles. PLoS Comput. Biol. 2015;11:e1004494. doi: 10.1371/journal.pcbi.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Wang Y., Zhang Z., Lv X., Gao G. F., Shao Y., Ma L., Li X. Enfuvirtide-PEG conjugate: a potent HIV fusion inhibitor with improved pharmacokinetic properties. Eur. J. Med. Chem. 2016;121:232–237. doi: 10.1016/j.ejmech.2016.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L. H. M., Chan S. H., Tsai S. N., Wang Y., Cheng C. H. K., Wong K. B., Waye M. M. Y., Ngai S. M. Fusion core structure of the severe acute respiratory syndrome coronavirus (SARS-CoV): in search of potent SARS-CoV entry inhibitors. J. Cell. Biochem. 2008;104:2335–2347. doi: 10.1002/jcb.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Guardia C., Lleonart R. Progress in the identification of dengue virus entry/fusion inhibitors. BioMed Res. Int. 2014;2014:825039. doi: 10.1155/2014/825039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehouck Y., Kwasigroch J. M., Rooman M., Gilis D. BeAtMuSiC: prediction of changes in protein-protein binding affinity on mutations. Nucleic Acid Res. 2013;41:W333–W339. doi: 10.1093/nar/gkt450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Lu G., Qi J., Li Y., Wu Y., Deng Y., Geng H., Li H., Wang Q., Xiao H. Structure of the fusion core and inhibition of fusion by a heptad repeat peptide derived from the S protein of Middle East respiratory syndrome coronavirus. J. Virol. 2013;87:13134–13140. doi: 10.1128/JVI.02433-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garton M., Nim S., Stone T. A., Wang K. E., Deber C. M., Kim P. M. Method to generate highly stable D-amino acid analogs of bioactive helical peptides using a mirror image of the entire PDB. Proc. Natl. Acad. Sci. U.S.A. 2018;115:1505–1510. doi: 10.1073/pnas.1711837115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakobyan A., Galindo I., Nañez A., Arabyan E., Karalyan Z., Chistov A. A., Streshnev P. P., Korshun V. A., Alonso C., Zakaryan H. Rigid amphipathic fusion inhibitors demonstrate antiviral activity against African swine fever virus. J. Gen. Virol. 2018;99:148–156. doi: 10.1099/jgv.0.000991. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T. S., Herrler G., Wu N. H., Nitsche A., Müller M. A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huet T., Kerbarh O., Schols D., Clayette P., Gauchet C., Dubreucq G., Vincent L., Bompais H., Mazinghien R., Querolle O., Salvador A., Lemoine J., Lucidi B., Balzarini J., Petitou M. Long-lasting enfuvirtide carrier pentasaccharide conjugates with potent anti-human immunodeficiency virus type 1 activity. Antimicrob. Agents Chemother. 2010;54:134–142. doi: 10.1128/AAC.00827-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J. Virol. 2019;93:e01815–18. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman J. A., Costa V. V., Park S., Real A., Park J. H., Cardozo P. L., Ferhan A. R., Olmo I. G., Moreira T. P., Bambirra J. L., Queiroz V. F., Queiroz-Junior C. M., Foureaux G., Souza D. G., Ribeiro F. M., Yoon B. K., Wynendaele E., De Spiegeleer B., Teixeira M. M., Cho N. J. Therapeutic treatment of Zika virus infection using a brain-penetrating antiviral peptide. Nat. Mater. 2018;17:971–977. doi: 10.1038/s41563-018-0194-2. [DOI] [PubMed] [Google Scholar]
- Kadam R. U., Juraszek J., Brandenburg B., Buyck C., Schepens W. B., Kesteleyn B., Stoops B., Vreeken R. J., Vermond J., Goutier W. Potent peptidic fusion inhibitors of influenza virus. Science. 2017;358:496–502. doi: 10.1126/science.aan0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M., Abdelrahman A. H. M., Oh-Hashi K., Ibrahim A., Venugopala K. N., Morsy M. A., Ibrahim M. A. A. Repurposing of FDA-approved antivirals, antibiotics, anthelmintics, antioxidants, and cell protectives against SARS-CoV-2 papain-like protease. J. Biomol. Struct. Dyn. 2020a doi: 10.1080/07391102.2020.1784291. doi: 10.1080/07391102.2020.1784291 [Online ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M., Al-Nazawi M. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Sci. 2020;251:117627. doi: 10.1016/j.lfs.2020.117627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M., Al-Taher A., Li H., Schwingenschlogl U., Al-Nazawi M. Molecular dynamics of Middle East Respiratory Syndrome Coronavirus (MERS CoV) fusion heptad repeat trimers. Comput. Biol. Chem. 2018;75:205–212. doi: 10.1016/j.compbiolchem.2018.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandeel M., Yamamoto M., Al-Taher A., Watanabe A., Oh-Hashi K., Park B. K., Kwon H. J., Inoue J. I., Al-Nazawi M. Small molecule inhibitors of Middle East respiratory syndrome coronavirus fusion by targeting cavities on heptad repeat trimers. Biomol. Ther. (Seoul) 2020b;28:311–319. doi: 10.4062/biomolther.2019.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchdoerfer R. N., Cottrell C. A., Wang N., Pallesen J., Yassine H. M., Turner H. L., Corbett K. S., Graham B. S., McLellan J. S., Ward A. B. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu I. J., Kao C. L., Hsieh S. C., Wey M. T., Kan L. S., Wang W. K. Identification of a minimal peptide derived from heptad repeat (HR) 2 of spike protein of SARS-CoV and combination of HR1-derived peptides as fusion inhibitors. Antiviral Res. 2009;81:82–87. doi: 10.1016/j.antiviral.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Xiao G., Chen Y., He Y., Niu J., Escalante C. R., Xiong H., Farmar J., Debnath A. K., Tien P., Jiang S. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet. 2004;363:938–947. doi: 10.1016/S0140-6736(04)15788-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Liu Q., Zhu Y., Chan K. H., Qin L., Li Y., Wang Q., Chan J. F., Du L., Yu F., Ma C., Ye S., Yuen K. Y., Zhang R., Jiang S. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat. Commun. 2014;5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizukoshi F., Baba K., Goto Y., Setoguchi A., Fujino Y., Ohno K., Oishi S., Kodera Y., Fujii N., Tsujimoto H. Antiviral activity of membrane fusion inhibitors that target gp40 of the feline immunodeficiency virus envelope protein. Vet. Microbiol. 2009;136:155–159. doi: 10.1016/j.vetmic.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Morishita M., Kamei N., Ehara J., Isowa K., Takayama K. A novel approach using functional peptides for efficient intestinal absorption of insulin. J. Control. Release. 2007;118:177–184. doi: 10.1016/j.jconrel.2006.12.022. [DOI] [PubMed] [Google Scholar]
- Peeri N. C., Shrestha N., Rahman M. S., Zaki R., Tan Z., Bibi S., Baghbanzadeh M., Aghamohammadi N., Zhang W., Haque U. The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned? Int. J. Epidemiol. 2020;49:717–726. doi: 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qinfen Z., Jinming C., Xiaojun H., Huanying Z., Jicheng H., Ling F., Kunpeng L., Jingqiang Z. The life cycle of SARS coronavirus in Vero E6 cells. J. Med. Virol. 2004;73:332–337. doi: 10.1002/jmv.20095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz B., Jr, Mossel E. C., Gallaher W. R., Wimley W. C., Peters C. J., Wilson R. B., Garry R. F. Inhibition of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) infectivity by peptides analogous to the viral spike protein. Virus Res. 2006;120:146–155. doi: 10.1016/j.virusres.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedpour S., Khodaei B., Loghman A. H., Seyedpour N., Kisomi M. F., Balibegloo M., Nezamabadi S. S., Gholami B., Saghazadeh A., Rezaei N. Targeted therapy strategies against SARS-CoV-2 cell entry mechanisms: a systematic review of in vitro and in vivo studies. J. Cell. Physiol. 2020 doi: 10.1002/jcp.30032. doi: 10.1002/jcp.30032 [Online ahead of print] [DOI] [PubMed] [Google Scholar]
- Shabane P. S., Izadi S., Onufriev A. V. General purpose water model can improve atomistic simulations of intrinsically disordered proteins. J. Chem. Theory Comput. 2019;15:2620–2634. doi: 10.1021/acs.jctc.8b01123. [DOI] [PubMed] [Google Scholar]
- Snook K. A., Van Ess R., 2nd, Werner J. R., Clement R. S., Ocon-Grove O. M., Dodds J. W., Ryan K. J., Acosta E. P., Zurlo J. J., Mulvihill M. L. Transdermal delivery of enfuvirtide in a porcine model using a low-frequency, low-power ultrasound transducer patch. Ultrasound Med. Biol. 2019;45:513–525. doi: 10.1016/j.ultrasmedbio.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Gui M., Wang X., Xiang Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018;14:e1007236. doi: 10.1371/journal.ppat.1007236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stalmans S., Bracke N., Wynendaele E., Gevaert B., Peremans K., Burvenich C., Polis I., De Spiegeleer B. Cell-penetrating peptides selectively cross the blood-brain barrier in vivo. PLoS ONE. 2015;10:e0139652. doi: 10.1371/journal.pone.0139652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., Worlock K. B., Yoshida M., Barnes J. L. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H., Shiokawa M., Kaname Y., Kambara H., Mori Y., Abe T., Moriishi K., Matsuura Y. Involvement of ceramide in the propagation of Japanese encephalitis virus. J. Virol. 2010;84:2798–2807. doi: 10.1128/JVI.02499-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A. E., Berendsen H. J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Vhora I., Patil S., Bhatt P., Misra A. Chapter one - proteinand peptide-drug conjugates: an emerging drug delivery technology. In: Donev R., editor. Advances in Protein Chemistry and Structural Biology. Vol. 98. Academic Press; 2015. pp. 1–55. [DOI] [PubMed] [Google Scholar]
- Wang H., Li X., Nakane S., Liu S., Ishikawa H., Iwamoto A., Matsuda Z. Co-expression of foreign proteins tethered to HIV-1 envelope glycoprotein on the cell surface by introducing an intervening second membrane-spanning domain. PLoS ONE. 2014;9:e96790. doi: 10.1371/journal.pone.0096790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Liu Q., Wang Q., Sun Z., Su S., Du L., Ying T., Lu L., Jiang S. Middle East respiratory syndrome coronavirus (MERS-CoV) entry inhibitors targeting spike protein. Virus Res. 2014;194:200–210. doi: 10.1016/j.virusres.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Yan L., Xu W., Agrawal A. S., Algaissi A., Tseng C. K., Wang Q., Du L., Tan W., Wilson I. A., Jiang S., Yang B., Lu L. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci. Adv. 2019;5:eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Kiso M., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Imai M., Takeda M., Kinoshita N., Ohmagari N., Gohda J., Semba K., Matsuda Z., Kawaguchi Y., Kawaoka Y., Inoue J. I. The anticoagulant nafamostat potently inhibits SARS-CoV-2 S protein-mediated fusion in a cell fusion assay system and viral infection in vitro in a cell-type-dependent manner. Viruses. 2020;12:629. doi: 10.3390/v12060629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X., Chong H., Zhang C., Waltersperger S., Wang M., Cui S., He Y. Broad antiviral activity and crystal structure of HIV-1 fusion inhibitor sifuvirtide. J. Biol. Chem. 2012;287:6788–6796. doi: 10.1074/jbc.M111.317883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Yu D., Yan H., Chong H., He Y. Design of potent membrane fusion inhibitors against SARS-CoV-2, an emerging coronavirus with high fusogenic activity. J. Virol. 2020;94:e00635–20. doi: 10.1128/JVI.00635-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler C. G. K., Allon S. J., Nyquist S. K., Mbano I. M., Miao V. N., Tzouanas C. N., Cao Y., Yousif A. S., Bals J., Hauser B. M., Feldman J., Muus C., Wadsworth M. H., 2nd, Kazer S. W., Hughes T. K., Doran B., Gatter G. J., Vukovic M., Taliaferro F., Mead B. E., Guo Z., Wang J. P., Gras D., Plaisant M., Ansari M., Angelidis I., Adler H., Sucre J. M. S., Taylor C. J., Lin B., Waghray A., Mitsialis V., Dwyer D. F., Buchheit K. M., Boyce J. A., Barrett N. A., Laidlaw T. M., Carroll S. L., Colonna L., Tkachev V., Peterson C. W., Yu A., Zheng H. B., Gideon H. P., Winchell C. G., Lin P. L., Bingle C. D., Snapper S. B., Kropski J. A., Theis F. J., Schiller H. B., Zaragosi L. E., Barbry P., Leslie A., Kiem H. P., Flynn J. L., Fortune S. M., Berger B., Finberg R. W., Kean L. S., Garber M., Schmidt A. G., Lingwood D., Shalek A. K., Ordovas-Montanes J. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]