SUMMARY
Tau accumulation is a major pathological hallmark of Alzheimer’s disease (AD) and other tauopathies, but the mechanism(s) of tau aggregation remains unclear. Taking advantage of the identification of tau filament cores by cryoelectron microscopy, we demonstrate that the AD tau core possesses the intrinsic ability to spontaneously aggregate in the absence of an inducer, with antibodies generated against AD tau core filaments detecting AD tau pathology. The AD tau core also drives aggregation of full-length wild-type tau, increases seeding potential, and templates abnormal forms of tau present in brain homogenates and antemortem cerebrospinal fluid (CSF) from patients with AD in an ultrasensitive real-time quaking-induced conversion (QuIC) assay. Finally, we show that the filament cores in corticobasal degeneration (CBD) and Pick’s disease (PiD) similarly assemble into filaments under physiological conditions. These results document an approach to modeling tau aggregation and have significant implications for in vivo investigation of tau transmission and biomarker development.
In brief
Carlomagno et al. demonstrate the AD tau filament core is able to spontaneously aggregate and recruit full-length wild-type tau to filaments. They also show that abnormal tau species in brain and CSF from AD patients template the AD core in a real-time QuIC assay.
Graphical Abstract

INTRODUCTION
Alzheimer’s disease (AD) is characterized pathologically by the presence of amyloid-beta (Aβ) plaques and tau neurofibrillary tangles (NFTs), with tau NFT burden correlating better with neurodegeneration and clinical symptoms than Aβ plaque pathology in patients with AD (Arriagada et al., 1992; Josephs et al., 2008). As such, significant research efforts have been devoted to understanding the mechanism(s) by which tau polymerizes into paired helical filaments (PHFs) and straight filaments (SFs), the building blocks of NFTs in AD. Recent studies utilizing cryoelectron microscopy (cryo-EM) to resolve the structures of PHFs and SFs have discovered that both are derived from the same filament core encompassing residues 304/306–378/380 of the tau protein (Falcon et al., 2018b; Fitzpatrick et al., 2017). Moreover, despite the clinical and pathological heterogeneity associated with AD, the filament core structure is remarkably consistent (Falcon et al., 2018b), suggesting common underlying mechanism(s) may drive tau filament formation in AD.
As tau pathology has been shown to spread transsynaptically in vivo (de Calignon et al., 2012; Wu et al., 2016) and follow a stereotypical pattern of progression through the brain along anatomically connected pathways in AD (Adams et al., 2019; Braak and Del Tredici, 2016), abnormal forms of tau are believed to act as seeds to recruit and convert normal tau to a pathologic species in a prion-like manner. This idea is supported by studies injecting AD brain homogenates (Ahmed et al., 2014; Boluda et al., 2015; Clavaguera et al., 2013; Guo et al., 2016) or synthetic tau filaments (Iba et al., 2013, 2015) into mice and demonstrating propagation of tau pathology through the brain from the site of injection. Moreover, this unique self-propagating prion-like property of abnormal tau has been exploited for developing a diagnostic biomarker using an ultrasensitive real-time quaking-induced conversion (QuIC) assay (Saijo et al., 2017). While recent studies have demonstrated that real-time QuIC is able to differentiate abnormal from normal tau by enhancement of thioflavin positivity, the assays rely on a His-tagged recombinant tau substrate and the presence of heparin as an inducer (Kraus et al., 2019a; Saijo et al., 2017), which may influence filament structure and negatively impact assay sensitivity (Falcon et al., 2015; Zhang et al., 2019).
In the current study, to characterize and evaluate potential seeding activity of the AD tau filament core sequence (referred to hereafter as the AD tau core), we first generated recombinant protein and demonstrated enhanced aggregation propensity relative to full-length tau. Notably, we discovered that the AD tau core exhibits a potent ability to spontaneously form filaments that is not observed with full-length tau. The AD tau core was also able to recruit normal, full-length tau to filaments to create a pathogenic species with enhanced seeding capability. Taking advantage of the unique assembly properties of the recombinant AD tau core, we generated an antibody against AD tau core filaments that detects tau pathology in postmortem brain tissue from patients with AD, as well as other tauopathies. We also explored diagnostic potential by using the recombinant AD tau core as a substrate to seed onto abnormal tau species present in postmortem brain tissue and antemortem cerebrospinal fluid (CSF) from patients with AD. Finally, we generated the filament core from the primary tauopathies corticobasal degeneration (CBD) and Pick’s disease (PiD) and demonstrated that similar to the AD tau core, the CBD and PiD tau cores also spontaneously form filaments. In characterizing the assembly properties and seeding capacity of the AD tau filament core, demonstrating that antibodies produced from AD tau core filaments recognize AD tau pathology, and revealing the CBD and PiD tau cores can also form filaments under physiological conditions, the current study uncovers an approach to model tau seeding in vitro and investigate disease pathogenesis in tauopathies.
RESULTS
The AD tau filament core exhibits unique assembly properties
To investigate the inherent aggregation properties of the AD tau filament core (corresponding to amino acids 306–378 of full-length 4R2N tau isoform), we cloned this sequence into a recombinant protein vector and optimized the purification protocol to obtain a highly pure protein yield of the AD tau core (Figure S1A, lane 6). Following purification, we assessed polymerization and filament assembly of the AD tau core, with full-length tau included as a positive control for aggregation. Using either dextran sulfate or heparin to induce assembly, we observed a significant increase in aggregation of the AD tau core relative to full-length tau as revealed by incorporation of thioflavin (Figures S1B, S1E, and S1G–S1J), a shift from the soluble to the pellet fractions in ultracentrifugation analysis (Figures S1C and S1F), and filament formation by EM (Figure S1D). Notably, similar results were observed with an equal concentration (0.4 μg/μL; Figures S1B–S1F) or molar ratio (10 μM; Figures S1G–S1J) of the AD tau core and full-length tau. While these results indicate that the AD tau core is very aggregation prone, given that heparin-induced tau filaments have been shown to be structurally distinct from AD brain-derived tau filaments (Zhang et al., 2019), the standard approach to stimulate tau filament assembly and model aggregation in vitro with the presence of an inducer reduces physiological relevance.
As such, we next tested various incubation conditions in the absence of an inducer. Remarkably, we discovered that continuous agitation of the recombinant AD tau core protein at 37°C leads to spontaneous filament assembly. We confirmed this finding with two different preparations of the recombinant AD tau core protein and demonstrated that no filaments are detected if the protein is incubated at 37°C in the absence of shaking (Figure S2A). In addition, we found that while agitation is required to promote filament assembly, the incubation temperature is more flexible, with filaments detected when shaking was performed at either room temperature or 37°C (Figure S2B). Moreover, AD tau core filaments are also stable following assembly, with storage at 4°C or frozen at −80°C for 48 h having no significant impact on filament formation (Figure S2B). Finally, while AD tau core filaments typically clump together, we demonstrate that sonication can be utilized to disrupt the large clumps and break the long filaments into smaller, shorter filaments (Figure S2C).
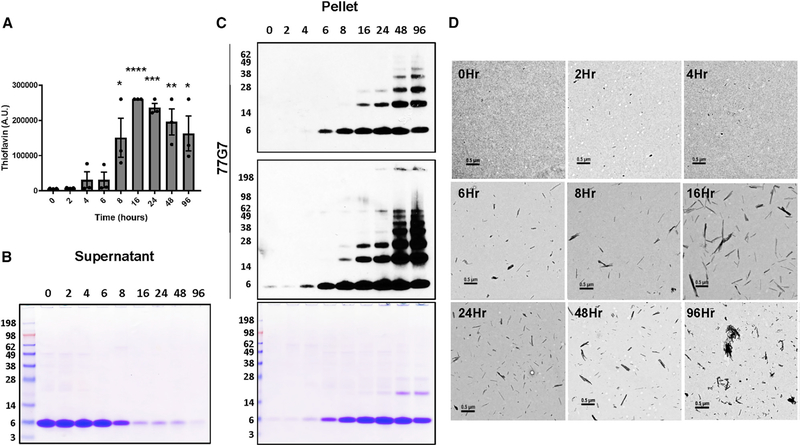
Following the initial characterization and determination of ideal assay and storage conditions, we then wanted to determine the time course of polymerization and filament formation. To do so, we incubated the AD tau core at 37°C with continuous agitation and examined aggregation by thioflavin, ultracentrifugation analysis, and EM at the indicated time points (Figures 1 and S3A). With three different preparations of the recombinant AD tau core protein, a slight increase in thioflavin was detected as early as 4–6 h, with a robust increase observed at 8 h (Figure 1A). Similarly, ultracentrifugation analysis revealed a subtle increase in the amount of AD tau core in the pellet at 4–6 h, with a striking shift from the soluble supernatant fraction to the insoluble pellet fraction at 8 h (Figures 1B and S3A). The pellet fraction was also evaluated by immunoblotting with an antibody that recognizes the AD tau core (77G7), which mirrored the Coomassie-blue-stained pellet fraction, and revealed the gradual increase in higher molecular weight tau species across time (Figure 1C). In addition, we verified the presence of filaments as early as 6–8 h with EM (Figure 1D), in agreement with the thioflavin and ultracentrifugation analysis. Finally, we observed a dose-dependent increase in lactate dehydrogenase (LDH) release from M17 neuroblastoma cells treated with increasing concentrations of AD tau core filaments, indicative of toxicity (Figures S3B and S3C). Collectively, these results demonstrate that the recombinant AD tau core possesses a remarkable ability to polymerize and form toxic filaments in the absence of an inducer, which may increase physiological relevance.
Figure 1. Time-dependent aggregation of AD tau core with no inducer.
(A) The recombinant AD tau core was incubated at 37°C with constant agitation, and thioflavin fluorescence was used to monitor assembly in the reaction mixtures at the indicated time points. Each point represents the average thioflavin signal (measured in triplicate) of three different preparations of recombinant protein (see ultracentrifugation data in Figure S3A). Data are represented as mean ± SEM. *p = 0.01; **p < 0.01; ***p < 0.001; ****p < 0.0001.
(B and C) Assembly reactions were pelleted at the indicated time points, and the supernatant and pellet fractions were evaluated by SDS-PAGE followed by Coomassie blue staining (B). The pellet fraction was also evaluated by immunoblotting with 77G7, an antibody that detects the AD tau core (C).
(D) Filament formation was evaluated at the indicated time points by electron microscopy (EM). Scale bar represents 0.5 μm.
See also Figures S1–S3.
AD core antibodies detect tau pathology in AD
To develop an antibody to detect the AD tau filament core, we utilized our optimized assembly paradigm to generate filaments derived from the AD tau core in the absence of an inducer, which was subsequently used as an immunogen. Serum collected from immunized rabbits (called ADC1 and ADC2) was first examined for the presence of AD tau core antibodies by western blotting (Figure S4A). As the AD tau core sequence is also present in full-length tau, the detection of both recombinant full-length tau and the AD tau core by ADC1 and ADC2 confirms the presence of AD tau core antibodies in serum from both rabbits, while the lack of immunoreactivity toward recombinant alpha-synuclein was assessed for specificity. Moreover, as the AD tau core is resistant to pronase digestion of AD tau filaments (Fitzpatrick et al., 2017), we treated the sarkosyl-insoluble tau fraction from 3 different patients with AD with pronase and examined ADC1 and E1 (N-terminal epitope/antibody that detects full-length tau, but not the AD tau core) immunoreactivity (Figure S4B). Unlike E1, which detects an N-terminal epitope that is located on the fuzzy coat of tau filaments in AD and is sensitive to pronase treatment (Figure S4B, right), the ADC1 epitope is completely resistant to pronase (Figure S4B, left). We also confirmed that similar to the sarkosyl-insoluble fraction from patients with AD, polymerized recombinant AD tau core is resistant to pronase treatment, while monomeric AD tau core is sensitive to pronase digestion (Figure S4B, left). Next, we examined ADC1 and ADC2 immunoreactivity in the sarkosyl-insoluble fraction from AD postmortem brain tissue by western blotting (Figure S4C), which revealed the accumulation of various tau core-containing species across different patients.
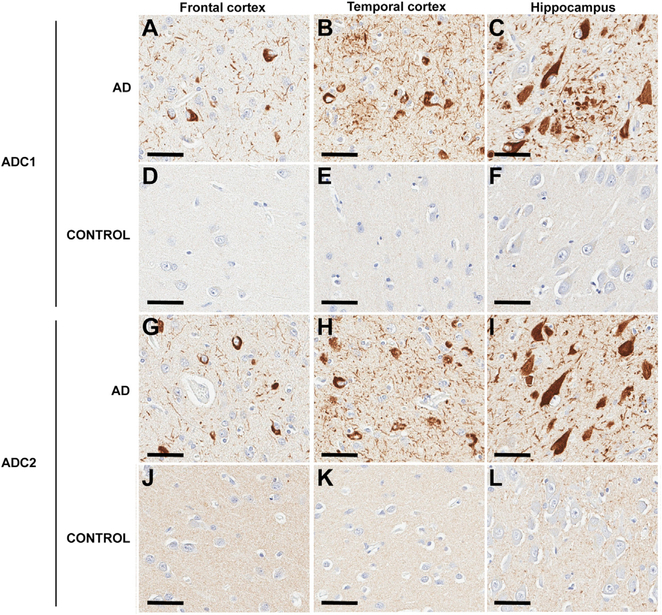
In order to evaluate whether antibodies generated against recombinant AD tau core filaments detect tau pathology in paraffin-embedded tissue sections from patients with AD, we performed immunohistochemistry. Consistently across brain regions, aberrant tau accumulation was labeled by ADC1 and ADC2 in patients with AD (Figures 2A–2C and 2G–2I; Table S1), but not in healthy controls (Figures 2D–2F and 2J–2L), confirming specificity for pathological forms of tau. In addition, ADC1 and ADC2 detected pathologies characteristic of AD tau, including NFTs, neuropil threads, and neuritic plaques. Moreover, we also verified colocalization with the well-validated tau antibody PHF1 (specific for phosphorylation on serine 396/404) and thioflavin using confocal microscopy (Figures S5A and S5B). To determine the ability of ADC1 and ADC2 to recognize early versus late forms of tau pathology in AD, as well as to compare with the well-characterized pathological tau antibody CP13 (detects tau phosphorylated on serine 202), we stained hippocampal sections ranging in disease severity (Figures 3A–3Dd; Table S1). Notably, while CP13 labeled pretangles in the CA4 field of the hippocampus (Figures 3A and 3B), this early form of tau pathology was not detected by ADC1 (Figures 3F and 3G) or ADC2 (Figures 3K and 3L). By contrast, as the CA4 becomes more affected in Braak stages IV–VI, ADC1 (Figures 3H–3J) and ADC2 (Figures 3M–3O) detect tau pathology that is not recognized by CP13 (Figures 3C–3E), consistent with previous observations that CP13 preferentially labels early, but not late, forms of tau pathology (Espinoza et al., 2008). Similar results were obtained in the entorhinal cortex, with the increasingly robust pathology across Braak stage detected by ADC1 (Figures 3U–3Y) and ADC2 (Figures 3Z–3Dd), but not recognized by CP13 (Figures 3P–3T).
Figure 2. AD tau core antibodies detect AD pathology.
(A–L) Immunohistochemical analysis with the AD core 1 (ADC1; A–F) and AD core 2 (ADC2; G–L) antibodies in paraffin-embedded tissue sections from a control (D–F and J–L) and AD (A–C and G–I) case. Representative images are presented from the frontal cortex (A, D, G, and J), temporal cortex (B, E, H, and K), and hippocampus (C, F, I, and L) as indicated. Nuclei are counterstained with hematoxylin. Scale bar represents 50 μm.
Figure 3. AD tau core antibodies recognize late-stage AD pathology.
(A–O) Immunohistochemistry with CP13 (A–E), ADC1 (F–J), and ADC2 (K–O) was performed using paraffin-embedded hippocampal tissue sections from patients with different Braak stages, as indicated. Representative images from the CA4 field of the hippocampus are presented, with nuclei counterstained with hematoxylin.
(P–Dd) Immunohistochemical analysis with CP13 (P–T), ADC1 (U–Y), and ADC2 (Z–Dd) was performed using paraffin-embedded hippocampal tissue sections from patients with different Braak stages, as indicated, with representative images presented from the entorhinal cortex. Nuclei are counterstained with hematoxylin.
Scale bar represents 100 μm.
Finally, to examine whether ADC1 and ADC2 are selective for AD tau pathology, we performed immunohistochemistry on brain tissue from patients with progressive supranuclear palsy (PSP) and PiD. Similar to CP13, both antibodies detected neuronal inclusions and oligodendroglia coiled bodies in the basal ganglia in PSP (Figures S5C, S5E, and S5G) as well as Pick bodies in the frontal cortex in PiD (Figures S5D, S5F, and S5H). Collectively, these results indicate that antibodies generated against AD tau core filaments detect a pronase-resistant late stage of pathology in AD, with the filament core epitopes for ADC1 and ADC2 common across tauopathies.
Full-length tau increases aggregation and seeding potential of the AD tau core
Given that the AD tau core spontaneously assembles into higher ordered structures that are seeding competent and antibodies generated against AD tau core filaments detect tau pathology in AD, we wanted to further assess potential disease relevance by elucidating whether the AD tau filament core possesses the ability to stimulate polymerization of full-length wild-type tau in the absence of an inducer. To do so, we utilized the assembly protocol we developed and incubated recombinant full-length wild-type tau (4R0N isoform) in the presence or absence of the AD tau core. Following 3 days of shaking at 37°C, we evaluated aggregation by ultracentrifugation analysis (Figure 4A) and incorporation of thioflavin (Figure 4B). Full-length tau and associated degradation products (due to the prolonged incubation at 37°C) remained localized to the soluble fraction when incubated in the absence of the AD tau core, as detected by E1, ADC1, and Coomassie blue staining (Figure 4A). By contrast, the AD tau core incubated alone remained predominantly localized to the pellet fraction by ADC1 immunoreactivity and Coomassie blue staining (Figure 4A). However, the AD tau core was able to stimulate the transition of full-length tau from the soluble to the pellet fraction as revealed by E1, ADC1, and Coomassie blue staining (Figure 4A). Moreover, a dramatic increase in thioflavin signal was observed when full-length tau was incubated with the AD tau core (Figure 4B), indicating that the AD tau core promotes aggregation of full-length wild-type tau. To confirm that full-length wild-type tau forms filaments in the presence of the AD tau core, we performed immuno-EM using E1 and MC1 (conformational antibody that recognizes a discontinuous epitope formed when tau’s N terminus interacts with the third microtubule-binding repeat domain [Jicha et al., 1997, 1999]). Notably, we observed filaments positive for both E1 (Figure 4C) and MC1 (Figure 4D) in reactions that contained both full-length wild-type tau and the AD tau core, indicating that the AD tau core acts as a seed to promote aggregation and filament formation of full-length wild-type tau.
Figure 4. AD tau core seeds polymerization of full-length wild-type tau.
(A) The AD tau core and full-length tau were incubated alone or together for 3 days at 37°C with constant agitation. The assembly reactions were centrifuged to obtain the supernatant (S) and pellet (P) fractions, which were evaluated by SDS-PAGE followed by Coomassie blue staining or immunoblotting for the tau antibodies E1 or ADC1.
(B) Following the 3-day incubation, aggregation was monitored by thioflavin positivity in the assembly reactions. Data are shown as the mean ± SEM and were analyzed by one-way ANOVA with Tukey’s multiple comparisons test. *p < 0.05; ****p < 0.0001.
(C and D) Following co-incubation of the AD tau core with full-length wild-type tau, assembly reactions were evaluated by immuno-EM. Filaments are labeled with E1 (C) and MC1 (D) detected with gold particles. Scale bar represents 0.2 μm.
(E) The sarkosyl-insoluble fraction obtained from the frontal cortex of a patient with AD (AD P3) was added to the tau biosensor cell line, and the induction of FRET relative to buffer-treated cells was quantified by flow cytometry. Data are represented as mean ± SEM.
(F) Recombinant AD tau core assembled by incubating at 37°C with constant agitation (220 rpm) for the indicated time points was added to the tau biosensor cell line. After 3 days, cells were collected, and the induction of FRET was measured by flow cytometry. Data are represented as mean ± SEM.
(G–L) Confocal microscopy was utilized to examine inclusion formation of the YFP-tagged tau construct expressed in the biosensor cell line following treatment with assembly mixtures after 0 (G–I) or 3 (J–L) days of incubation. Assembly mixtures contained full-length 4R0N tau alone (G and J), AD tau core alone (H and K), or full-length 4R0N and the AD tau core (I and L). Scale bar represents 10 μm.
(M) Tau biosensor cells were exposed to assembly mixtures on day 0 or following the 3-day incubation, with cell lysates collected 48 h later and FRET quantified by flow cytometry. Data are shown as the mean ± SEM and were analyzed by two-way ANOVA with Sidak’s multiple comparisons test. ****p < 0.0001.
Next, we examined tau seeding potential in a well-characterized biosensor cell line, in which stable expression of cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP)-tagged tau fragments (amino acids 244–372) containing the P301S mutation enable quantification of tau seeding activity by monitoring inclusion formation and measuring fluorescence resonance energy transfer (FRET) (Holmes et al., 2014; Sanders et al., 2014). Consistent with previous reports (Holmes et al., 2014; Sanders et al., 2014), we observed a significant increase in FRET signal in biosensor cells exposed to the sarkosyl-insoluble fraction prepared from postmortem brain tissue from a patient with AD (AD P3) (Figure 4E). Following development of assay conditions that detect seeding activity in AD P3 material, we assessed seeding potential in the pellet fraction from the AD tau core assembled across time. While no seeding activity was observed in the AD tau core pellet at or prior to 8 h of assembly, a sudden increase in FRET was detected in cells exposed to the AD tau core pellet assembled for 16 h, indicative of tau seeding (Figure 4F). Notably, as higher molecular weight tau species are detected by immunoblotting (Figure 1C) and robust filament assembly is observed by EM (Figure 1D) at this time point, these features may be linked to seeding competency of the AD tau core.
To quantitate seeding activity of AD core-induced full-length tau filaments and determine whether the presence of full-length wild-type tau alters the seeding potential of homogeneous AD tau core filaments (Figure 4F), we exposed tau biosensor cells to assembly reactions following 0 (monomeric protein) or 3 days of incubation. While no inclusions were detected in cells treated with assembly reactions from day 0 (Figures 4G–4I), there was a marked increase in inclusion formation upon treatment with day 3 reactions containing both AD core and full-length tau compared with either protein alone (Figures 4J–4L). Consistent with observations by confocal microscopy, although there was an increase in FRET signal detected in cells exposed to aggregated AD core compared with full-length tau alone, there was a dramatic increase in FRET following exposure to day 3 assembly reactions containing both AD core and full-length tau (Figure 4M). By contrast, there was no FRET signal detected in cells treated with the same concentrations of AD core and/or full-length tau protein prior to the 3-day assembly reaction (Figure 4M; day 0), indicating that polymerization into higher-ordered structures rather than just monomeric protein is required for FRET and seeding activity.
Pathologic, brain-, and CSF-derived AD tau species seed the AD tau core
Considering that the AD tau core can recruit wild-type tau to filaments and possesses potent seeding activity, we wanted to determine whether the recombinant AD tau core could be utilized as a substrate for a real-time QuIC assay with enhanced sensitivity and diagnostic potential for AD. We first identified the optimal concentration of AD tau core to enable template-driven seeding detection and quantification. In particular, as continuous agitation at 37°C leads to self-assembly of the AD tau core (100 μM) within 6–8 h as reflected by the incorporation of thioflavin (Figure 1), new assay conditions were required to ensure minimal assembly of the AD tau core in the absence of test samples (e.g., brain homogenates, patient biofluids, etc.). Therefore, we monitored aggregation by thioflavin positivity with reduced concentrations of AD tau core, which revealed that while increased thioflavin is detected with the AD tau core alone at 25 μM, no thioflavin signal is observed at 12.5 μM (Figure S6A). Following identification of 12.5 μM as the optimal concentration of recombinant AD tau core for the assay, we next tested different dilutions of brain homogenates to determine the ideal conditions. We prepared brain lysates from the frontal cortex of people with AD and controls (Table S1) and evaluated templating of the AD tau core in dilutions ranging from 10−1 to 10−9 (Figure S6B). While increased aggregation of the recombinant AD tau core was observed with both control and AD samples in concentrated brain homogenates, there was a clear separation between AD and controls at more dilute concentrations (10−8–10−9) in the real-time QuIC assay (Figure S6B). After confirmation that no thioflavin signal is observed in brain homogenates diluted to 10−9 in the absence of the recombinant AD tau core (Figure S6C), we selected a concentration of 12.5 μM for the recombinant AD tau core and a dilution of 10−9 for brain homogenates as the ideal assay conditions for the real-time QuIC reaction. We then assessed the ability of our optimized real-time QuIC assay to differentiate between control and AD brain homogenates. Notably, we observed a significant increase in thioflavin positivity with brain lysates from the affected frontal cortex of patients with AD, with minimal thioflavin signal in frontal cortex lysates from controls (Figure 5A). Given that the cerebellum is unaffected by tau pathology in AD, we also evaluated brain lysates prepared from the cerebellum of people with AD as an additional control. As expected, we observed no increase in thioflavin positivity upon incubation of cerebellar AD brain lysates with the recombinant AD tau core (Figure 5A), indicating that the presence of pathologic AD tau species is required to stimulate aggregation of the recombinant AD tau core in our assay. In addition, we observed a significantly reduced thioflavin signal in brain lysates from patients with the tauopathies CBD, PSP, and chronic traumatic encephalopathy (CTE), demonstrating specificity of the assay for AD (Figure 5B; Table S1).
Figure 5. The AD tau core seeds pathologic, brain-derived AD tau species.
(A) Real-time QuIC analysis was performed with the AD tau core alone, or with brain homogenates diluted 10−9. Trace for the AD tau core alone was performed in triplicate. Lines corresponding to AD (frontal cortex or cerebellum) or control (frontal cortex) represent the average of 10 cases (each case performed in triplicate; see Table S1 for case information), with error bars depicting SEM. Significant differences between AD and control frontal cortex samples were first detected at 7.5 h (p = 0.02), reaching p < 0.0001 by 8.5+ h. (B) Real-time QuIC analysis was performed with brain homogenates prepared from the frontal cortex and diluted 10−9. Lines corresponding to each group represent the average of all cases (AD n = 13, CBD n = 4, PSP n = 4, CTE n = 3, control n = 2), with each case tested in triplicate, and errors bars depicting SEM. AD samples were significantly different from CBD, PSP, CTE, and control cases (p < 0.0001).
(C) Real-time QuIC analysis using the AD tau core and full-length tau mixture was performed with antemortem CSF from control or AD patients. Line graph represents the average of 10 control or 10 AD cases (see Table S2 for case information), with each case measured in quadruplicate, and errors bars depicting SEM. AD samples were significantly different from control samples by 16 h (p = 0.04), reaching p < 0.0001 by 19 h.
In order to assess biomarker potential, we modified the real-time QuIC assay to include both recombinant AD tau core and full-length tau (4R0N isoform) as a substrate given our finding that the AD core induces aggregation of full-length tau (Figure 4), as well as previous reports that tau species in antemortem CSF are truncated and lack the AD tau core sequence (Barthélemy et al., 2019; Sato et al., 2018). Following optimization of the ideal protein concentrations that showed no amplification of thioflavin signal in the absence of CSF, we determined that a volume of 2.5 μL of CSF per reaction provided the greatest separation between control and AD CSF samples (Figures S6D and S6E). We then tested a collection of 10 antemortem CSF samples from people with AD compared with 10 age- and sex-matched controls (Table S2), which revealed a significant increase in thioflavin signal in AD CSF compared with control CSF (Figure 5C). These results indicate that seeding-competent tau species are present in AD CSF and are capable of templating mixed full-length:AD tau core filaments.
The CBD and PiD tau filament cores spontaneously form filaments under physiological conditions
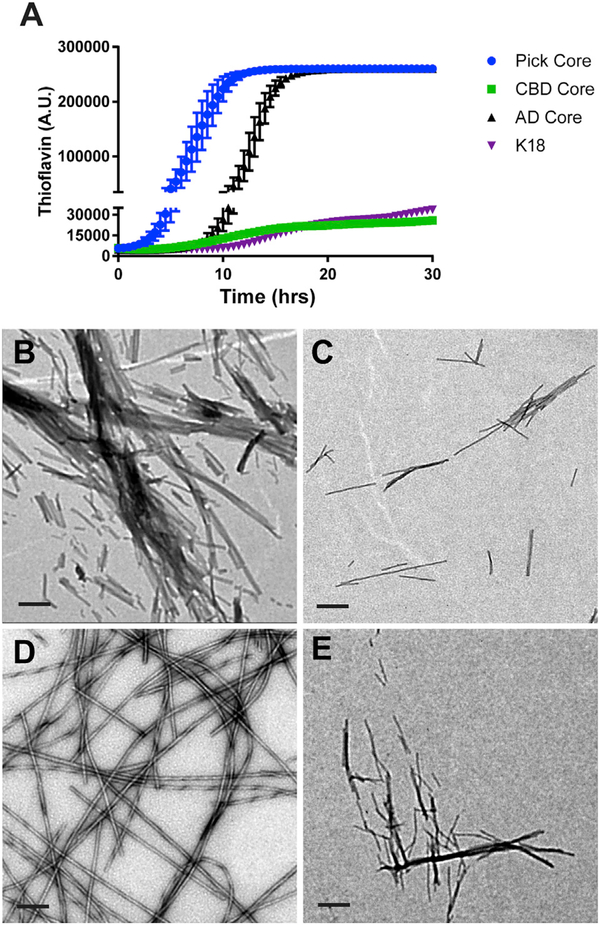
Following characterization of the AD tau core, we wanted to determine whether the filament core in the primary tauopathies CBD and PiD would exhibit a similar tendency to assemble into filaments with constant agitation. To do so, we cloned the CBD core (amino acids 274–380 corresponding to full-length 4R2N tau isoform) (Arakhamia et al., 2020; Zhang et al., 2020) and PiD core (amino acids 254–274; 306–378 corresponding to full-length 4R2N isoform; PiD core is 3R, so residues 275–305 encoded by exon 10 are missing) (Falcon et al., 2018a) into a recombinant protein vector, and we optimized the purification protocol for each core protein. In addition, we also purified the K18 tau fragment (amino acids 244–368 of full-length 4R2N isoform; corresponds to the microtubule-binding domain of 4R tau) to determine how filament assembly compares with the CBD and PiD cores, as the K18 fragment is widely used to model tau aggregation (Barré and Eliezer, 2013; Holmes et al., 2014; Iba et al., 2013; Kaniyappan et al., 2020; Karikari et al., 2017, 2019; Sanders et al., 2014; Shammas et al., 2015; Weismiller et al., 2018; Zhang et al., 2019). We first monitored thioflavin signal with shaking at 37°C to assess the kinetics of aggregation of the different core proteins (Figure 6A). While the assembly of the PiD core was faster than the AD core, the CBD core and K18 fragment exhibited a much slower rate of aggregation (Figure 6A). We also used EM to monitor filament formation after shaking reactions for 24 h (Figures 6B–6E). Consistent with the thioflavin data, all tau proteins polymerized into filaments to a varying degree, with the AD (Figure 6B) and PiD (Figure 6D) cores driving robust filament assembly, while filaments derived from the K18 fragment (Figure 6C) or CBD core (Figure 6E) were sparsely distributed across the grid. These results indicate that tau aggregation and filament assembly with the complete microtubule-binding domain of tau are less efficient than the shorter tau core proteins under physiological conditions.
Figure 6. Filament assembly of different tau fragments.
(A) The recombinant K18 fragment and AD, CBD, and PiD tau cores were incubated at 37°C with constant agitation, and assembly was monitored by thioflavin fluorescence at the indicated time points. The experiment was performed in triplicate, with the data represented as mean ± SEM. The PiD core was significantly different from other cores at 5 h (p = 0.04), reaching p < 0.0001 by 6 h. The difference between the PiD and AD cores was no longer significant by 15.5 h. The AD core was significantly different from the K18 and CBD cores by 11.5 h (p < 0.002) and 12 h (p < 0.0001).
(B–E) Filament formation by recombinant AD tau core (B), K18 fragment (C), PiD tau core (D), and the CBD tau core (E) following shaking at 220 rpm for 24 h was evaluated by EM. Scale bar represents 0.2 μm.
DISCUSSION
In this study, we demonstrate that the potent aggregation propensity of the AD tau filament core drives spontaneous self-assembly into filaments, even in the absence of an inducer to facilitate aggregation. We also reveal that antibodies generated against recombinant AD tau core filaments detect AD tau pathology in patients. Providing support for the idea that the AD tau core can act to seed pathology, we show that the AD tau core recruits full-length wild-type tau to filaments, generating an abnormal tau species with enhanced seeding activity. Capitalizing on the unique aggregation propensity and seeding potential of the AD tau core, we demonstrate that abnormal forms of tau present in dilute AD brain homogenates and CSF template the AD tau core (combined with full-length tau for CSF) in an ultrasensitive real-time QuIC assay. Finally, we reveal that similar to the AD tau core, the CBD and PiD tau cores also spontaneously assemble into filaments albeit with different characteristics, while the K18 tau fragment exhibits minimal assembly under similar conditions. Collectively, our findings have significant implications for modeling tauopathy in vitro and in vivo, providing a different approach to produce tau filaments in vitro under more physiological conditions, which may also generate the ideal tau seed to evaluate propagation in vivo.
Since the initial resolution of AD tau filament structure and identification of the core sequence cryo-EM (Fitzpatrick et al., 2017), the field has rapidly evolved with subsequent discoveries, including the development of structural models for tau filaments in the primary tauopathies PiD (Falcon et al., 2018a), CBD (Arakhamia et al., 2020; Zhang et al., 2020), and CTE (Falcon et al., 2019). Therefore, while the current study has primarily focused on defining the aggregation and seeding propensity of the AD tau filament core, future studies will need to expand on this initial characterization of the CBD and PiD cores and assess how the three filament cores compare. As tau has been shown to undergo liquid-liquid phase separation (LLPS) following phosphorylation or in the presence of RNA (Wegmann et al., 2018; Zhang et al., 2017), it would also be intriguing in future studies to assess the interaction between each tau core with RNA, as well as the impact on LLPS dynamics. In addition, it will be particularly exciting to evaluate whether each tau core exhibits preferential amplification of brain- and/or CSF-derived tau species from its corresponding tauopathy. Considering that filaments formed by inducing tau aggregation with heparin do not accurately recapitulate brain-derived tau filaments (Fichou et al., 2018; Zhang et al., 2019), it will also be important to determine whether the protocol developed in the current study to generate AD core-stimulated filaments (either homogeneous or heterogeneous filaments with full-length wild-type tau) more closely resemble the structure of AD brain-derived tau filaments. Of note, although the AD tau core spontaneously assembles into filaments within 6–8 h, it is interesting that only SFs are detected, despite the fact that the same sequence forms the core of both straight and PHFs in AD (Fitzpatrick et al., 2017). Given that aggregated tau has been shown to be hyperphosphorylated and ubiquitinated in AD (Arakhamia et al., 2020; Cripps et al., 2006), and posttranslational modifications (PTMs) have been implicated in the regulation of tau filament structure (Arakhamia et al., 2020), the lack of PTMs on the AD tau core in the current study may favor SF over PHF formation. In addition, as a longer tau fragment consisting of the microtubule-binding domain of 3R tau (244–274; 306–394) has been shown to form PHFs in vitro (Schweers et al., 1995), it is possible the additional flanking regions may also be required to support the formation of PHFs. Resolution of these key questions could illuminate critical epitopes to target to prevent PHF formation, as well as provide the means to generate PHFs in a high-throughput and reproducible manner for screening purposes. This would significantly advance the field, providing a simple yet accurate in vitro model of tau filaments to guide future research efforts, such as PET ligand development, small-molecule identification and binding affinity optimization, and generation of new antibodies with an increased focus on conformational epitopes for biomarker and immunotherapy approaches. Moreover, as studies investigating tau propagation in vivo rely on either brain-derived insoluble tau or heparin-induced tau filaments (both full-length [Iba et al., 2013, 2015] and the K18 tau fragment [Iba et al., 2013] have been used), our results suggest the tau filament cores provide an alternative approach to generate abnormal tau species for in vivo propagation studies without the confounding effect of heparin. Given the recent identification of the low-density lipoprotein receptor-related protein 1 (LRP1) as a master regulator of tau endocytosis (Rauch et al., 2020), future studies will need to assess whether the filament cores also act as substrates for LRP1 similar to full-length tau and the K18 fragment, which could reveal the ideal form of tau to use for investigating in vivo propagation (i.e., AD tau core alone, or AD tau core-induced full-length filaments).
While tau pathology is implicated in the functional deficits associated with AD given the correlation with cognitive impairment (Arriagada et al., 1992; Holtzman et al., 2011; Josephs et al., 2008), the pathogenic mechanism(s) to explain the onset of tauopathy in AD remains to be determined. Here, our discovery that the recombinant AD tau core can act as a seed to recruit full-length wild-type tau to filaments may have potential implications for disease pathogenesis. In particular, although it is unknown whether a fragment corresponding to the AD tau core is produced in vivo, the detection of several proteolytic cleavage sites near both the 306 and 378 residues (Derisbourg et al., 2015; Quinn et al., 2018) raises the distinct possibility that the AD tau core fragment could be generated as a result of aberrant proteolysis, incomplete degradation, or another yet-to-be-identified mechanism. In fact, a recent study demonstrated that while proteolytic cleavage of tau normally occurs within the AD tau core sequence, conformational changes in the tau protein lead to altered cleavage sites, leaving the AD tau core intact in patients with AD (Chen et al., 2018). Our findings illustrate how failure to degrade tau within the AD core sequence could enhance aggregation, recruiting normal tau to inclusions and facilitating propagation in disease. In future studies, it will be important to assess how the flanking regions at the N- or C-terminal side of the AD tau core sequence impact aggregation propensity or seeding activity, which could identify specific tau fragments likely to be most pathogenic.
The approach utilized in the current study to model tau aggregation and filament assembly in vitro also provides the means to drive assembly of full-length wild-type tau, even in the absence of a pathogenic mutation. This increases significance and relevance to AD, which unlike primary tauopathies is not associated with pathogenic mutations in tau. In addition, our findings provide clarity for a controversy surrounding the tau biosensor cell line. In particular, a recent study demonstrated that GFP interferes with the in vitro assembly of recombinant tau due to steric hindrance (Kaniyappan et al., 2020), leading the authors to speculate that the presence of fluorophore tags would similarly perturb tau assembly in cell models, questioning the interpretation of changes in FRET signal in the biosensor cell line used in the current study. Moreover, the authors note the inefficient induction of FRET by purified tau assemblies in comparison with tissue lysates indicate that factors other than tau may be responsible for inducing FRET in biosensor cells, such as cytokines (Kaniyappan et al., 2020). However, our observation that purified AD tau core filaments and AD tau core-induced full-length tau filaments both promote significant increases in FRET positivity in the tau biosensor cell line clearly indicates the response is mediated by tau, with a higher FRET signal associated with more aggregation-prone forms of tau, consistent with the idea that FRET is an indication of seeding activity in this cell model. While the resulting structure of the aggregated forms of tau that are FRET positive is unlikely to recapitulate brain-derived tau filaments, as heparin-induced tau filaments also do not recapitulate the structure of brain-derived tau filaments (Fichou et al., 2018; Zhang et al., 2019), the effect of GFP on heparin-induced tau filament formation in vitro (Kaniyappan et al., 2020) may be physiologically irrelevant.
In the AD tau core real-time QuIC assay developed in this report, we detected robust amplification in thioflavin signal in the presence of AD brain homogenates consistent with previous observations (Kraus et al., 2019a), but with a higher signal and greater separation between AD and disease controls at the lower dilutions (10−8 and 10−9) in the current study. The increased sensitivity could be due to the use of an untagged AD tau core substrate, the absence of heparin, or a combination of these two factors. In addition, our modified real-time QuIC assay for CSF, which incorporates a mixture of AD tau core and full-length tau as a substrate for pathogenic tau seeds, provided remarkable sensitivity and specificity to differentiate antemortem CSF samples collected from patients with AD from age-matched healthy controls. While a recent study also observed a signal by real-time QuIC assay in CSF collected from a subset of people with AD (Saijo et al., 2020), as the assay conditions were optimized for primary tauopathies with low sensitivity for AD tau pathology in brain homogenates, our AD CSF-optimized real-time QuIC assay appears to improve the detection of abnormal tau species in AD CSF. Consistent with these results, seeding-competent tau species have been detected in AD CSF (Skachokova et al., 2019). In addition, tau seeding activity in postmortem brain tissue from patients with AD has recently been shown to correlate with the aggressiveness of the disease course, indicating that alterations in tau seeding capacity may explain the significant clinical heterogeneity observed across the AD spectrum (Dujardin et al., 2020). These findings support the idea that our development of a quantitative assay reliant on tau’s seeding capacity will be capable of providing diagnostic and more importantly prognostic insight, which will be further evaluated in future studies.
In conclusion, we present evidence that the AD tau filament core possesses the capability to spontaneously assemble into filaments in the absence of an inducer, with antibodies generated against AD tau core filaments exhibiting immunoreactivity for tau pathology in postmortem brain tissue from patients with AD and other tauopathies. The AD tau core also promotes aggregation of full-length wild-type tau into filaments positive for the MC1 conformational epitope, generating an aberrant tau species with enhanced seeding capacity. Finally, development of real-time QuIC assays that utilize either the AD tau core alone (brain) or AD tau core combined with full-length tau (CSF) as a substrate is capable of differentiating patients with AD from controls in both diluted brain homogenates (10−9) and CSF samples. Taken together, this study describes the development of reagents, assays, and an approach to modeling tau filament assembly in vitro that is more physiologically relevant than standard approaches that rely on an inducer to support tau filament formation.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Leonard Petrucelli (petrucelli.leonard@mayo.edu).
Materials availability
All unique and stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Data and code availability
The published article includes all datasets generated or analyzed during this study.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cloning and construct generation
Human cDNAs encoding full-length wild-type tau (4R0N isoform), AD tau core, CBD tau core, PiD tau core, K18 tau fragment, and alpha-synuclein were all cloned into the pET30a vector (EMD Millipore) and transformed into Rosetta 2(DE3) pLysS competent cells (EMD Millipore) for protein expression. All constructs were sequence-verified using ABI3730 with Big Dye chemistry following manufacturer’s protocols (Applied Biosystems, Foster City, CA, USA).
Human samples
Human postmortem brain tissue was provided by the Mayo Clinic Florida Brain Bank. Information on all human postmortem patients is provided in Table S1, with neuropathological diagnosis on all cases determined by a single neuropathologist (D.W.D.). Written informed consent was obtained before study entry from all subjects or their legal next of kin if they were unable to give written consent, and biological samples were obtained with Mayo Clinic Institutional Review Board (IRB) approval. CSF samples from patients with AD were purchased from Precision Medicine, Inc (Solana Beach, CA). CSF samples from age- and sex-matched control individuals were obtained, based on availability, from a collection of subjects participating in the Mayo Clinic Florida Ataxia Program, the Mayo Clinic Florida ALS Program, and from Massachusetts General Hospital and Washington University through their Dominantly Inherited ALS (DIALS) Network study. Information for all patients with antemortem CSF is provided in Table S2.
METHOD DETAILS
Protein purification
Cultures were grown to saturation overnight, and were subsequently utilized to inoculate larger cultures in the morning (1:100 dilution). Cells were grown in Luria Broth on a shaker (220rpm, 37°C) until the OD600 reached 0.5 absorbance units. Protein expression was then induced with 0.5mM IPTG for 4 hours, and the cells collected by centrifugation and washed in 1X TBS before storing pellets at −80°C.
To purify full-length tau, as well as the Pick’s core and K18 fragment, the cell pellets were resuspended in lysis buffer (100mM Tris [pH8], 50mM NaCl, 1mM MgCl2, 1mM PMSF, 1mM dithiothreitol, 21U/mL DNAase) and lysed with three freeze-thaw cycles using liquid nitrogen and a tepid water bath. The lysates were then centrifuged to eliminate cell debris, and 500mM NaCl was added to supernatants, which were heated (80°C, 10 minutes) and subsequently cooled on ice (10 minutes). Precipitates were removed by centrifugation, and the resulting supernatant was diluted 1:10 with cold water and loaded onto a 5mL HiTrap Mono S column (GE Healthcare Life Sciences, Pittsburgh, PA) using the ÄKTA FPLC system (GE Healthcare Life Sciences). The column was washed with 10 column volumes of wash buffer (10 mM HEPES [pH 7.4], 150 mM NaCl), and tau proteins were eluted with a linear gradient of NaCl at 20mM/min. Fractions collected between 250mM to 350mM NaCl were used for CSF RT-QuIC analysis following dialysis (10mM HEPES pH7.4 and 100mM NaCl), or further purified by HPLC (Shimadzu System) using a C8 column (PrincetonSPHER-300 C8 300A 5u 150×10 mm) for recombinant assembly evaluation. Fractions containing the highly-pure protein were lyophilized and stored at −80C until use.
The AD and CBD tau cores were purified by resuspending cell pellets in lysis buffer (10mM HEPES [pH7.4], 50mM NaCl, 1mM MgCl2, 1mM PMSF, 1mM dithiothreitol, 21U/mL DNAase), followed by sonication (40% power for 30 s on, 30 s off for a total of 3 minutes) and three freeze-thaw cycles using liquid nitrogen and a tepid water bath. Lysates were then centrifuged to eliminate cell debris. For the CBD tau core, 500mM NaCl was added to supernatants, which were heated (80°C, 5 minutes), cooled on ice (10 minutes), and precipitates removed by centrifugation. For both cores, the supernatant was diluted with cold water (1:10 for the AD tau core, 1:20 for the CBD tau core) and loaded onto a 5mL HiTrap Mono S column (GE Healthcare Life Sciences, Pittsburgh, PA) using the ÄKTA FPLC system (GE Healthcare Life Sciences). The column was washed with 10 column volumes of wash buffer (10 mM HEPES [pH 7.4], 50 mM NaCl for the AD tau core, 10 mM HEPES [pH 7.4], 100 mM NaCl for the CBD tau core), and the protein eluted with a step gradient (100mM NaCl for the AD tau core, 250mM NaCl for the CBD tau core). Fractions were collected and stored until use at −80C.
Alpha-synuclein was purified by resuspending cell pellets in lysis buffer (10mM Tris-HCl [pH8] 50mM NaCl, 1mM MgCl2, 1mM PMSF, 1mM dithiothreitol, 21U/mL DNAase), and lysing by sonication (40% power for 30 s on, 30 s off for a total of 3 minutes). The inclusion body fraction was isolated by centrifugation and resuspended in buffer (10mM Tris-HCl [pH8], 0.75M NaCl, 1mM dithiothreitol), followed by heating (90°C, 10 minutes) and subsequently cooling on ice (10 minutes). Precipitates were removed by centrifugation, and the resulting supernatant was diluted 1:10 with cold water and loaded onto a 5mL HiTrap Mono Q column (GE Healthcare Life Sciences, Pittsburgh, PA) using the ÄKTA FPLC system (GE Healthcare Life Sciences). The column was washed with 10 column volumes of wash buffer (10 mM HEPES [pH 7.4], 150 mM NaCl), and alpha-synuclein protein was eluted with a linear gradient of NaCl at 7.5 mM/min. Fractions of approximately 1mL were collected and analyzed by SDS-PAGE and Coomassie blue staining. Fractions that contained high concentrations of purified protein were then pooled and dialyzed against 200 volumes of 10mM HEPES buffer at pH 7.4, and protein was subsequently frozen and stored at −80 until use. For all protein, a BCA protein assay kit (Pierce Biotechnology, Rockford, IL) was used to determine protein concentration before use.
Tau assembly with inducer
The assembly reaction was performed with 0.4 μg/μL (Figure S1B–F) or 10 μM (Figure S1G–J) of protein solubilized in assembly buffer (10mM HEPES [pH7.4], 100mM NaCl, 1mM MgCl, 0.3mM dithiothreitol, 0.1mM EDTA, 2mM EGTA, 5 μM ThT) containing 0.04mg/mL dextran sulfate or heparin at 37°C with constant agitation at 330 rpm. ThT signal was measured by FLUOstar Omega (BMG) in a 96 well plate.
Tau assembly reaction with no inducer
To investigate assembly of the recombinant AD core, protein was incubated in Assembly Buffer (10 mM HEPES [pH 7.4], 100 mM NaCl, 1mM TCEP) at 37°C (unless otherwise indicated) at a concentration of 100μM. At the indicated time point, 15 uL of the reaction was incubated with 105uL of buffer (10 mM HEPES [pH 7.4], 100 mM NaCl, 7 μM ThT), and ThT signal measured by FLUOstar Omega (BMG). To evaluate co-polymerization of the AD tau core with full-length tau (4R0N), each protein were incubated at a concentration of 25 μM (1 μg/μL 4R0N and 0.2 μg/μL AD tau core) in Assembly Buffer, with the assembly reaction incubated at 37°C shaking (220rpm) for 3 days. ThT signal was evaluated as described above. To compare the K18 fragment and different tau core species, each protein was incubated at a concentration of 100 μM in Assembly Buffer (10 mM HEPES [pH 7.4], 100 mM NaCl, 1mM TCEP, 5 μM ThT) at 37°C with constant agitation at 330 rpm. ThT signal was measured by FLUOstar Omega (BMG) in a 96 well plate.
Electron Microscopy
Polymerized tau reactions were diluted 1:4 in TBS (pH7.4), and absorbed onto carbon/formvar-coated 400 mesh copper grids (Electron Microscopy Sciences, Hatfield, PA) for 60 s. Grids were then stained with 2% uranyl acetate (Electron Microscopy Sciences, Hatfield, PA) for 60 s, followed by three washes in 20uL TBS. Grids were examined with a Philips 208S electron microscope (Philips, Hillsboro, OR).
Immuno-Electron Microscopy
As above, polymerized tau reactions were diluted 1:4 in TBS (pH. 7.4) and absorbed onto carbon/formvar-coated 400 mesh copper grids for 60 s. Non-specific binding was blocked by incubating grids with blocking buffer (1mg/mL of BSA in TBS) for 30 minutes at room temperature in a humidified chamber, followed by a 60-minute incubation in primary antibody (diluted 1:200 in blocking buffer) at room temperature. After three washes in blocking buffer (40uL drops), grids were incubated with gold-labeled secondary antibody (diluted 1:20 in blocking buffer, 60 minute incubation in the same humidified chamber at room temperature). Grids were washed six times with TBS (40uL drops), stained with 2% uranyl acetate (Electron Microscopy Sciences, Hatfield, PA) for 60 s, and examined with a Philips 208S electron microscope (Philips, Hillsboro, OR).
Ultracentrifugation analysis
Following the polymerization reaction (40uL), samples were centrifuged at 266,100 × g for 40 minutes (TLA110 70000 rpm) at 4°C to separate soluble and insoluble fractions. The pellets containing aggregated and polymerized tau were resuspended in 47 μL 1× SDS sample buffer, while 7uL of 6× SDS sample buffer was added to the soluble fraction. The samples were boiled at 95°C for 10 minutes, separated by SDS-PAGE on a 10% Bis-tris gel, and subsequently visualized by staining with Coomassie blue (Imperial Protein Staining). Immunoblotting for 77G7, ADC1 and E1 were performed as described below.
Sample preparation and immunoblotting procedure
Recombinant protein (50ng each of full-length tau, AD tau core and alpha-synuclein) was diluted in dH2O, 2× Tris-glycine SDS sample buffer (Life Technologies), and 5% beta-mercaptoethanol (Sigma Aldrich), heat-denatured for 5 minutes at 95°C, and run on SDS-PAGE Tris-glycine gels (Life Technologies). Gels to evaluate antibody specificity, as well as gels from ultracentrifugation analysis (described above) were transferred to PVDF membrane (Millipore). Membranes were blocked in 5% non-fat dry milk in TBS/0.1% Triton X-100, and incubated overnight in primary antibody rocking at 4°C. Membranes were incubated in HRP-conjugated secondary antibodies (1:5000; Jackson ImmunoResearch, West Grove, PA) for 1 hour at room temperature, and detected by ECL (PerkinElmer, Waltham, MA).
AD core toxicity in M17 neuroblastoma cells
M17 cells were treated with different concentrations of recombinant AD core polymerized by shaking for 24 hours in the absence of inducer. 72 hours following treatment, media was collected for an LDH assay following manufacturer’s protocols (Promega). Cells were washed with PBS, treated with trypsin for 2 minutes, and collected by ultracentrifugation. Cell pellets were washed in PBS, and subsequently lysed in cell lysis buffer (50 mM Tris, 274 mM NaCl, 5 mM KCl, 5mM EDTA, 1% Triton X-100, 1% SDS, 1 mM PMSF, and protease and phosphatase inhibitor cocktails).
Pronase treatment
Sarkosyl-insoluble material was purified from 150mg of AD postmortem brain tissue and resuspended in 250uL TBS. Recombinant AD tau core (polymerized or monomeric) was diluted to 0.125ug/uL. 40uL of each sample was incubated in the presence or absence of 0.4mg/mL pronase (Millipore) at 37°C. Protease inhibitor cocktail was then added to stop the reaction at either 1, 2 or 5 minutes as indicated. Pronase-treated and untreated samples were then spotted onto a nitrocellulose membrane to evaluate immunoreactivity for ADC1 and E1 tau antibodies. Briefly, dot blots were washed in TBS plus 0.1% Triton-X (TBS-T), and incubated overnight at 4C with the indicated primary antibodies. The following day, blots were washed in TBS-T, incubated with donkey anti-rabbit IgG secondary antibodies conjugated to horseradish peroxidase (1:5000; Jackson ImmunoResearch) for 1 hour, and again washed with TBS-T. Protein expression was then visualized by enhanced chemiluminescence treatment and exposure to film.
Seeding in FRET biosensor cells
The tau FRET biosensor cell line (ATCC® CRL-3275) was purchased from ATCC (Manassas, VA). Cells were seeded at 1.5×10^5 cells/well in a 24-well plate, and 24 hours following plating were treated with different mixtures of recombinant material or sarkosyl-insoluble (P3) brain fraction using 0.5uL/well of Lipofectamine 2000 (Invitrogen). For the AD tau core time course experiment, 1uM (5uL of the assembly reaction) was added to each well. For the AD tau core-induced full-length tau copolymerization experiment, 0.25uM (5uL of the assembly reaction) was added to each well to test the seeding activity. Three days following treatment, cells were harvested and fixed with 2% paraformaldehyde for 10 minutes at room temperature. Fixed cells were subsequently resuspended in flow cytometry buffer (Hank’s Balanced Salt Solution buffer with 2% fetal bovine serum) and run on the Attune NxT Flow Cytometer (Thermo Fisher), with the technician blinded to sample information. CFP and FRET signals were excited with the 405 nm laser and detected with 460/50nm and 525/50nm filters, respectively. YFP signal was excited with the 488nm laser and detected with a 540/30nm filter. Gating and compensation strategies were performed as described(Holmes et al., 2014). Integrated FRET density was calculated by multiplying the median fluorescence intensity of FRET-positive cells by the percentage of cells that are positive for FRET, as described(Holmes et al., 2014). For confocal microscopy, cells were plated on poly-D-lysine-coated coverslips in a 24-well plate, and treated as above. Following fixation, Hoescht 33258 (1 μg/mL, H3569, Thermo Fisher Scientific) was used to stain cellular nuclei. Images were obtained on a Zeiss LSM 880 laser scanning confocal microscope equipped with 405nm and 488nm excitation.
Immunohistochemistry
Paraffin-embedded tissue blocks were sectioned at 5 microns, mounted on positively-charged glass slides, and dried overnight in a 60°C oven. Tissue sections were then deparaffinized in xylene, and rehydrated in a graded series of alcohols. Slides were first incubated with 98% formic acid (EMD Chemicals Catalog, #FX0440–6) for 30 minutes, and following several washes, antigen retrieval was performed by steaming in distilled water for 30 minutes. Slides were then incubated with 3% hydrogen peroxide to quench endogenous peroxidase activity, and subsequently rinsed in distilled water prior to a 5-minute incubation in TBS with 0.05% tween 20. Sections were then immunostained using the DAKO Autostainer (DAKO North America, Carpinteria, CA) and the DAKO EnVision + HRP system to detect ADC1 (1:2500) and ADC2 (1:5000), which were generated by immunizing two separate rabbits with AD tau core filaments (polymerized by shaking at 220RPM for 24 hours, with filament formation verified by EM prior to injection). The stained slides were then counterstained with hematoxylin, dehydrated, coverslipped, and scanned with the Leica Aperio AT2 Slide Scanner (Aperio, Vista, CA).
Immunofluorescence and thioflavin S staining
Paraffin-embedded (5 μm) of human hippocampal brain sections were deparaffinized and rehydrated as above. Thioflavin S staining was performed immediately following rehydration by incubating slides in 1% Thioflavin S (in dH2O) for 7 minutes, and subsequently washed in water. To colocalize ADC2 and PHF1, slides were incubated in 98% formic acid for 30 minutes following rehydration. Slides were then steamed for 30 minutes in dH2O, blocked with Dako Protein Block for 1 hour, and incubated with primary antibody overnight in a humidified chamber at 4°C. The next day, slides were washed in PBS, and incubated with the corresponding Alexa Fluor 488- or 568-conjugated donkey anti-species (1:500, Molecular Probes) for 2 hours. Hoechst 33258 (1 μg/ml, Thermo Fisher Scientific) was used to stain cellular nuclei. Images were obtained on a Zeiss LSM 880 laser scanning confocal microscope.
RT-QuIC assay
The RT-QuIC assay was performed as previously described(Manne et al., 2017, 2019, 2020) with minor modifications. In brief, the RT-QuIC assay was carried out using a 96-well clear-bottom plate (CORNING Costar® Assay Plate) with the reaction mixture consisting of final concentrations of 100 mM NaCl, 10 mM HEPES, 5 μM ThT, 1 mM TCEP and 12.5μM (10μg) of recombinant AD tau core for brain homogenates. For CSF, the reaction mixture contained 6.25μM recombinant AD tau core along with 6.25μM of full-length (4R0N isoform) and two 0.8-mm silica beads (OPS Diagnostics) in each well. Brain tissues were homogenized to 10% (w/v) using 1X PBS with phosphatase and protease inhibitors and further diluted 10-fold with 1X PBS as described previously(Kondru et al., 2017). Each reaction consisted of 2 μL of 10−9 dilution of brain tissue homogenates as a seed (each sample was tested in triplicate) and 98 μL of AD core RT-QuIC reaction mixture of a 96-well plate, with the technician blinded to sample classification. For CSF samples, 2.5 μL of undiluted CSF was used as a seed and mixed with 97.5 μL of the reaction mixture containing both recombinant AD tau core and full-length tau, with the technician again blinded to sample information. The plates were sealed with a plate sealer (CORNING Thermowell™ Sealing Tape) and incubated at 30°C in a FLUOstar Omega (BMG) plate reader with continuous shaking (double orbital, 300 rpm) and the gain was set to 1300 units for brain homogenates and 1550 for CSF samples. ThT fluorescence readings were recorded every 30 min at excitation and emission wavelengths of 450 and 480 nm, respectively, with the maximum fluorescence set at 260,000. The RT-QuIC reaction was conducted for 20–48 hours, and the results were then analyzed to identify positive samples. A sample was considered positive if it crossed the threshold fluorescence value, and it could be calculated by averaging fluorescence readings over the 2–7 hour time window plus 100 standard deviations.
QUANTIFICATION AND STATISTICAL ANALYSIS
GraphPad Prism was utilized to perform statistical analyses. Differences among groups were analyzed using unpaired Student’s t test (Figures 4E, S1B, S1E, S1G, and S1I), one-way ANOVA, with posthoc analysis using Tukey’s multiple comparisons tests (Figure 4B) or Dunnett’s multiple comparisons test (Figures 1A, 4F, 5B, and S3C). In addition, statistical differences were also assessed by two-way ANOVA with Sidak’s multiple comparisons test (Figure 4M) or two-way ANOVA with repeated-measures with Sidak’s multiple comparisons test (Figures 5A, 5C, and 6A). p < 0.05 was considered statistically-significant.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-tau (77G7, 316–355) | Biolegend | Cat#816701; RRID:AB_2564801 |
| Rabbit polyclonal anti-AD core filaments (ADC1) | This paper | ADC1; RRID:AB_2888919 |
| Rabbit polyclonal anti-AD core filaments (ADC2) | This paper | ADC2; RRID:AB_2888920 |
| Rabbit polyclonal anti-Tau (Human-specific) | L. Petrucelli, Mayo Clinic; Jacksonville, FL; USA | E1; RRID: 2819185 |
| Mouse monoclonal anti-tau (conformational epitope 5–15, 312–322) | P. Davies, Albert Einstein College of Medicine; New York, NY; USA | MC1; RRID:AB_2314773 |
| Mouse monoclonal anti-GAPDH | Meridian Life Science | Cat#H86504M; RRID:AB_151542 |
| Mouse monoclonal anti-alpha-synuclein | BD Biosciences | Cat#610787; RRID:AB_398108 |
| Mouse monoclonal anti-pTau (phosphorylated at Ser202) | P. Davies, Albert Einstein College of Medicine; New York, NY; USA | CP13; RRID: AB_2314223 |
| Mouse monoclonal anti-pTau (phosphorylated at Ser396 and Ser404) | P. Davies, Albert Einstein College of Medicine; New York, NY; USA | PHF1; RRID: AB_2315150 |
| peroxidase-conjugated donkey anti-rabbit | Jackson ImmunoResearch | Cat#711–036–152; RRID: AB_2340590 |
| peroxidase-conjugated donkey anti-mouse | Jackson ImmunoResearch | Cat#715–036–150; RRID: AB_2340773 |
| 18nm colloidal gold-conjugated goat anti-mouse | Jackson ImmunoResearch | Cat#115–215–146; RRID: AB_2338738 |
| 6nm colloidal gold-conjugated goat anti-rabbit | Jackson ImmunoResearch | Cat#111–195–144; RRID: AB_2338015 |
| Donkey anti-rabbit Alexa Fluor 488 | Invitrogen | A-21206; RRID:AB_2535792 |
| Donkey anti-mouse Alexa Fluor 568 | Invitrogen | A10037; RRID:AB_2534013 |
| Donkey anti-rabbit Alexa Fluor 568 | Invitrogen | A10042; RRID:AB_2534017 |
| Bacterial and virus strains | ||
| Rosetta 2(DE3)pLysS competent cells | Millipore/Sigma | 71403 |
| Biological samples | ||
| Human postmortem brain tissue (see Table S1) | Mayo Clinic Brain Bank | http://www.mayo.edu/research/departments-divisions/departmentneuroscience-florida/brain-banks/mayo-clinic-brain-bank |
| CSF samples from AD patients (see Table S2) | Precision Medicine | https://www.precisionmed.com/services/csf-samples/ |
| CSF samples from control patients (see Table S2) | Mayo Clinic Florida, Massachusetts General Hospital and Washington University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DNase | Sigma-Aldrich | D5025 |
| dextran sulfate (MW6500–10000Da) | Sigma-Aldrich | D8906 |
| heparin | Sigma-Aldrich | H4784 |
| Thioflavin T | Sigma-Aldrich | T3516 |
| beta-mercaptoethanol | Sigma-Aldrich | M3148 |
| dithiothreitol (DTT) | Sigma-Aldrich | D9779 |
| isopropyl 1-thio-D-galactopyranoside (IPTG) | Thermo Fisher Scientific | BP1755 |
| Imperial protein stain (Coomassie blue) | Thermo Fisher Scientific | 24615 |
| protease inhibitor cocktail | EMD Millipore | 539131 |
| PMSF | Thermo Fisher Scientific | 36978 |
| phosphatase inhibitor cocktails A and B | Bimake | B15001 |
| pronase | EMD Millipore | 537088 |
| Western Lightning Plus-ECL | PerkinElmer | NEL105001EA |
| Thioflavin S | Sigma-Aldrich | T1892 |
| Hoescht 33258 | Thermo Fisher Scientific | H3569 |
| recombinant AD tau core | This paper | N/A |
| recombinant full-length human tau (4R0N) | This paper | N/A |
| recombinant Pick tau core | This paper | N/A |
| recombinant CBD tau core | This paper | N/A |
| recombinant K18 tau fragment | This paper | N/A |
| recombinant alpha-synuclein | This paper | N/A |
| BigDye Terminator v3.1 | Applied Biosystems | 4337455 |
| HiDi formamide | Thermo Fisher Scientific | 4311320 |
| TCEP | Sigma-Aldrich | 646547 |
| lipofectamine 2000 | Invitrogen | 11668500 |
| formic acid | EMD Chemicals Catalog | FX0440–6 |
| uranyl acetate | Electron Microscopy Sciences | 22400 |
| TFA | Sigma-Aldrich | T6508 |
| Kanamycin sulfate | VWR | 0408–25 |
| Chloramphenicol | Millipore | 220551 |
| LB Broth Miller | Thermo Fisher Scientific | BP1426–2 |
| Acetonitrile | Thermo Fisher Scientific | A9906–4 |
| Critical commercial assays | ||
| BCA protein assay | Thermo Fisher Scientific | 23227 |
| cytotox 96 non-radioactive cytotoxicity assay | Promega | G1780 |
| EnVision+HRP system for Rabbit antibodies | Dako | K401111 |
| EnVision+HRP system for Mouse antibodies | Dako | K400711 |
| Deposited data | ||
| raw data | This paper | see raw data file |
| Experimental Models: Cell Lines | ||
| Tau RD P301S FRET biosensor | ATCC | CRL-3275 |
| Recombinant DNA | ||
| AD tau core | This paper | N/A |
| Pick’s tau core | This paper | N/A |
| CBD tau core | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 7 | GraphPad | https://www.graphpad.com/ |
| Omega Data Analysis | BMG | https://www.bmglabtech.com/ |
| Other | ||
| formvar/carbon copper grids 400 mesh | Electron Microscopy Services | FCF400-Cu |
| HiTrap SP HP column | GE Healthcare Life Sciences | 17–1152–01 |
| HiTrap QP HP column | GE Healthcare Life Sciences | 17–1154–01 |
| PrincetonSPHER-300 C8 300A 5u 150 ×10 mm column | Princeton | 150100–08502 |
| 96-well Costar Assay plate; clear-bottom | Corning | 3631 |
| 0.8mm silica beads | OPS Diagnostics | BMBG800–200–02 |
| Thermowell Sealing Tape | Corning | 6570 |
Highlights.
The AD tau filament core spontaneously aggregates in the absence of an inducer
Aggregation and seeding of full-length wild-type tau are enhanced by the AD tau core
The AD tau core templates abnormal tau in brain and antemortem CSF from AD patients
Tau filament cores from the tauopathies CBD and PiD also spontaneously aggregate
ACKNOWLEDGMENTS
We would like to thank the patients and their families for their participation in this work. Human postmortem brain samples and associated data were provided by the Mayo Clinic Brain Bank. We would also like to thank Katie Vicari for the artwork depicted in the graphical abstract. This work was supported by Mayo Clinic Foundation (L.P. and D.W.D.); The Liston Family Foundation (L.P.); Mayo Clinic Center for Regenerative Medicine (Z.K.W.); Mayo Clinic Neuroscience-Focused Research Team (Z.K.W.); National Institutes of Health/National Institute on Aging (R01AG063780 [C.N.C.], ADRC 2 P50 AG016574-16 [L.P. and D.W.D.], and R01AG062171 [L.P.]); National Institutes of Health/National Institute of Neurological Disorders and Stroke (U54NS100693 [L.P., D.W.D., and C.N.C.], R35NS097273 [L.P.], and U01NS110438 [A.W.P.F. and L.P.]); and Cure Alzheimer’s Fund (L.P. and A.W.P.F.). In addition, this work was also supported by gifts from The Sol Goldman Charitable Trust (Z.K.W.), the Donald G. and Jodi P. Heeringa Family (Z.K.W.), the Haworth Family Professorship in Neurodegenerative Diseases fund (Z.K.W.), and The Albertson Parkinson’s Research Foundation (Z.K.W.).
Footnotes
DECLARATION OF INTERESTS
The authors have no potential conflicts of interest to disclose.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108843.
REFERENCES
- Adams JN, Maass A, Harrison TM, Baker SL, and Jagust WJ (2019). Cortical tau deposition follows patterns of entorhinal functional connectivity in aging. eLife 8, e49132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Z, Cooper J, Murray TK, Garn K, McNaughton E, Clarke H, Parhizkar S, Ward MA, Cavallini A, Jackson S, et al. (2014). A novel in vivo model of tau propagation with rapid and progressive neurofibrillary tangle pathology: the pattern of spread is determined by connectivity, not proximity. Acta Neuropathol. 127, 667–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakhamia T, Lee CE, Carlomagno Y, Duong DM, Kundinger SR, Wang K, Williams D, DeTure M, Dickson DW, Cook CN, et al. (2020). Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains. Cell 180, 633–644.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriagada PV, Growdon JH, Hedley-Whyte ET, and Hyman BT (1992). Neurofibrillary tangles but not senile plaques parallel duration and severity of Alzheimer’s disease. Neurology 42, 631–639. [DOI] [PubMed] [Google Scholar]
- Barré P, and Eliezer D (2013). Structural transitions in tau k18 on micelle binding suggest a hierarchy in the efficacy of individual microtubule-binding repeats in filament nucleation. Protein Sci. 22, 1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthélemy NR, Mallipeddi N, Moiseyev P, Sato C, and Bateman RJ (2019). Tau Phosphorylation Rates Measured by Mass Spectrometry Differ in the Intracellular Brain vs. Extracellular Cerebrospinal Fluid Compartments and Are Differentially Affected by Alzheimer’s Disease. Front. Aging Neurosci. 11, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boluda S, Iba M, Zhang B, Raible KM, Lee VM, and Trojanowski JQ (2015). Differential induction and spread of tau pathology in young PS19 tau transgenic mice following intracerebral injections of pathological tau from Alzheimer’s disease or corticobasal degeneration brains. Acta Neuropathol. 129, 221–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, and Del Tredici K (2016). Potential Pathways of Abnormal Tau and α-Synuclein Dissemination in Sporadic Alzheimer’s and Parkinson’s Diseases. Cold Spring Harb. Perspect. Biol. 8, a023630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Liu P, Auger P, Lee SH, Adolfsson O, Rey-Bellet L, Lafrance-Vanasse J, Friedman BA, Pihlgren M, Muhs A, et al. (2018). Calpain-mediated tau fragmentation is altered in Alzheimer’s disease progression. Sci. Rep. 8, 16725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavaguera F, Akatsu H, Fraser G, Crowther RA, Frank S, Hench J, Probst A, Winkler DT, Reichwald J, Staufenbiel M, et al. (2013). Brain homogenates from human tauopathies induce tau inclusions in mouse brain. Proc. Natl. Acad. Sci. USA 110, 9535–9540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps D, Thomas SN, Jeng Y, Yang F, Davies P, and Yang AJ (2006). Alzheimer disease-specific conformation of hyperphosphorylated paired helical filament-Tau is polyubiquitinated through Lys-48, Lys-11, and Lys-6 ubiquitin conjugation. J. Biol. Chem. 281, 10825–10838. [DOI] [PubMed] [Google Scholar]
- de Calignon A, Polydoro M, Suárez-Calvet M, William C, Adamowicz DH, Kopeikina KJ, Pitstick R, Sahara N, Ashe KH, Carlson GA, et al. (2012). Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron 73, 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derisbourg M, Leghay C, Chiappetta G, Fernandez-Gomez FJ, Laurent C, Demeyer D, Carrier S, Buée-Scherrer V, Blum D, Vinh J, et al. (2015). Role of the Tau N-terminal region in microtubule stabilization revealed by new endogenous truncated forms. Sci. Rep. 5, 9659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin S, Commins C, Lathuiliere A, Beerepoot P, Fernandes AR, Kamath TV, De Los Santos MB, Klickstein N, Corjuc DL, Corjuc BT, et al. (2020). Tau molecular diversity contributes to clinical heterogeneity in Alzheimer’s disease. Nat. Med. 26, 1256–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza M, de Silva R, Dickson DW, and Davies P (2008). Differential incorporation of tau isoforms in Alzheimer’s disease. J. Alzheimers Dis. 14, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon B, Cavallini A, Angers R, Glover S, Murray TK, Barnham L, Jackson S, O’Neill MJ, Isaacs AM, Hutton ML, et al. (2015). Conformation determines the seeding potencies of native and recombinant Tau aggregates. J. Biol. Chem. 290, 1049–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon B, Zhang W, Murzin AG, Murshudov G, Garringer HJ, Vidal R, Crowther RA, Ghetti B, Scheres SHW, and Goedert M (2018a). Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 561, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon B, Zhang W, Schweighauser M, Murzin AG, Vidal R, Garringer HJ, Ghetti B, Scheres SHW, and Goedert M (2018b). Tau filaments from multiple cases of sporadic and inherited Alzheimer’s disease adopt a common fold. Acta Neuropathol. 136, 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R, Crowther RA, Newell KL, Ghetti B, Goedert M, and Scheres SHW (2019). Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature 568, 420–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichou Y, Vigers M, Goring AK, Eschmann NA, and Han S (2018). Heparin-induced tau filaments are structurally heterogeneous and differ from Alzheimer’s disease filaments. Chem. Commun. (Camb.) 54, 4573–4576. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, and Scheres SHW (2017). Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JL, Narasimhan S, Changolkar L, He Z, Stieber A, Zhang B, Gathagan RJ, Iba M, McBride JD, Trojanowski JQ, and Lee VM (2016). Unique pathological tau conformers from Alzheimer’s brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 213, 2635–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes BB, Furman JL, Mahan TE, Yamasaki TR, Mirbaha H, Eades WC, Belaygorod L, Cairns NJ, Holtzman DM, and Diamond MI (2014). Proteopathic tau seeding predicts tauopathy in vivo. Proc. Natl. Acad. Sci. USA 111, E4376–E4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Morris JC, and Goate AM (2011). Alzheimer’s disease: the challenge of the second century. Sci. Transl. Med. 3, 77sr1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba M, Guo JL, McBride JD, Zhang B, Trojanowski JQ, and Lee VM (2013). Synthetic tau fibrils mediate transmission of neurofibrillary tangles in a transgenic mouse model of Alzheimer’s-like tauopathy. J. Neurosci. 33, 1024–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba M, McBride JD, Guo JL, Zhang B, Trojanowski JQ, and Lee VM (2015). Tau pathology spread in PS19 tau transgenic mice following locus coeruleus (LC) injections of synthetic tau fibrils is determined by the LC’s afferent and efferent connections. Acta Neuropathol. 130, 349–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jicha GA, Bowser R, Kazam IG, and Davies P (1997). Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J. Neurosci. Res. 48, 128–132. [DOI] [PubMed] [Google Scholar]
- Jicha GA, Berenfeld B, and Davies P (1999). Sequence requirements for formation of conformational variants of tau similar to those found in Alzheimer’s disease. J. Neurosci. Res. 55, 713–723. [DOI] [PubMed] [Google Scholar]
- Josephs KA, Whitwell JL, Ahmed Z, Shiung MM, Weigand SD, Knopman DS, Boeve BF, Parisi JE, Petersen RC, Dickson DW, and Jack CR Jr. (2008). Beta-amyloid burden is not associated with rates of brain atrophy. Ann. Neurol. 63, 204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniyappan STK, Biernat J, Chandupatla RR, Hubschmann S, Irsen S, Bicher S, Klatt C, Mandelkow EM, and Mandelkow E (2020). FRET-based Tau seeding assay does not represent prion-like templated assembly of Tau fibers. Mol. Neurodegener. 15, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikari TK, Turner A, Stass R, Lee LC, Wilson B, Nagel DA, Hill EJ, and Moffat KG (2017). Expression and purification of tau protein and its frontotemporal dementia variants using a cleavable histidine tag. Protein Expr. Purif. 130, 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karikari TK, Nagel DA, Grainger A, Clarke-Bland C, Crowe J, Hill EJ, and Moffat KG (2019). Distinct Conformations, Aggregation and Cellular Internalization of Different Tau Strains. Front. Cell. Neurosci. 13, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondru N, Manne S, Greenlee J, West Greenlee H, Anantharam V, Halbur P, Kanthasamy A, and Kanthasamy A (2017). Integrated Organotypic Slice Cultures and RT-QuIC (OSCAR) Assay: Implications for Translational Discovery in Protein Misfolding Diseases. Sci. Rep. 7, 43155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus A, Saijo E, Metrick MA 2nd, Newell K, Sigurdson CJ, Zanusso G, Ghetti B, and Caughey B (2019a). Seeding selectivity and ultrasensitive detection of tau aggregate conformers of Alzheimer disease. Acta Neuropathol. 137, 585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne S, Kondru N, Nichols T, Lehmkuhl A, Thomsen B, Main R, Halbur P, Dutta S, and Kanthasamy AG (2017). Ante-mortem detection of chronic wasting disease in recto-anal mucosa-associated lymphoid tissues from elk (Cervus elaphus nelsoni) using real-time quaking-induced conversion (RT-QuIC) assay: A blinded collaborative study. Prion 11, 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne S, Kondru N, Hepker M, Jin H, Anantharam V, Lewis M, Huang X, Kanthasamy A, and Kanthasamy AG (2019). Ultrasensitive Detection of Aggregated α-Synuclein in Glial Cells, Human Cerebrospinal Fluid, and Brain Tissue Using the RT-QuIC Assay: New High-Throughput Neuroimmune Biomarker Assay for Parkinsonian Disorders. J. Neuroimmune Pharmacol. 14, 423–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manne S, Kondru N, Jin H, Anantharam V, Huang X, Kanthasamy A, and Kanthasamy AG (2020). α-Synuclein real-time quaking-induced conversion in the submandibular glands of Parkinson’s disease patients. Mov. Disord. 35, 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JP, Corbett NJ, Kellett KAB, and Hooper NM (2018). Tau Proteolysis in the Pathogenesis of Tauopathies: Neurotoxic Fragments and Novel Biomarkers. J. Alzheimers Dis. 63, 13–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch JN, Luna G, Guzman E, Audouard M, Challis C, Sibih YE, Leshuk C, Hernandez I, Wegmann S, Hyman BT, et al. (2020). LRP1 is a master regulator of tau uptake and spread. Nature 580, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo E, Ghetti B, Zanusso G, Oblak A, Furman JL, Diamond MI, Kraus A, and Caughey B (2017). Ultrasensitive and selective detection of 3-repeat tau seeding activity in Pick disease brain and cerebrospinal fluid. Acta Neuropathol. 133, 751–765. [DOI] [PubMed] [Google Scholar]
- Saijo E, Metrick MA 2nd, Koga S, Parchi P, Litvan I, Spina S, Boxer A, Rojas JC, Galasko D, Kraus A, et al. (2020). 4-Repeat tau seeds and templating subtypes as brain and CSF biomarkers of frontotemporal lobar degeneration. Acta Neuropathol. 139, 63–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, Barker SJ, Foley AC, Thorpe JR, Serpell LC, et al. (2014). Distinct tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C, Barthélemy NR, Mawuenyega KG, Patterson BW, Gordon BA, Jockel-Balsarotti J, Sullivan M, Crisp MJ, Kasten T, Kirmess KM, et al. (2018). Tau Kinetics in Neurons and the Human Central Nervous System. Neuron 97, 1284–1298.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweers O, Mandelkow EM, Biernat J, and Mandelkow E (1995). Oxidation of cysteine-322 in the repeat domain of microtubule-associated protein tau controls the in vitro assembly of paired helical filaments. Proc. Natl. Acad. Sci. USA 92, 8463–8467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shammas SL, Garcia GA, Kumar S, Kjaergaard M, Horrocks MH, Shivji N, Mandelkow E, Knowles TP, Mandelkow E, and Klenerman D (2015). A mechanistic model of tau amyloid aggregation based on direct observation of oligomers. Nat. Commun. 6, 7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skachokova Z, Martinisi A, Flach M, Sprenger F, Naegelin Y, SteinerMonard V, Sollberger M, Monsch AU, Goedert M, Tolnay M, and Winkler DT (2019). Cerebrospinal fluid from Alzheimer’s disease patients promotes tau aggregation in transgenic mice. Acta Neuropathol. Commun. 7, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann S, Eftekharzadeh B, Tepper K, Zoltowska KM, Bennett RE, Dujardin S, Laskowski PR, MacKenzie D, Kamath T, Commins C, et al. (2018). Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 37, e98049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismiller HA, Murphy R, Wei G, Ma B, Nussinov R, and Margittai M (2018). Structural disorder in four-repeat Tau fibrils reveals a new mechanism for barriers to cross-seeding of Tau isoforms. J. Biol. Chem. 293, 17336–17348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JW, Hussaini SA, Bastille IM, Rodriguez GA, Mrejeru A, Rilett K, Sanders DW, Cook C, Fu H, Boonen RA, et al. (2016). Neuronal activity enhances tau propagation and tau pathology in vivo. Nat. Neurosci. 19, 1085–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Lin Y, Eschmann NA, Zhou H, Rauch JN, Hernandez I, Guzman E, Kosik KS, and Han S (2017). RNA stores tau reversibly in complex coacervates. PLoS Biol. 15, e2002183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Falcon B, Murzin AG, Fan J, Crowther RA, Goedert M, and Scheres SH (2019). Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. eLife 8, e43584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Tarutani A, Newell KL, Murzin AG, Matsubara T, Falcon B, Vidal R, Garringer HJ, Shi Y, Ikeuchi T, et al. (2020). Novel tau filament fold in corticobasal degeneration. Nature 580, 283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study.