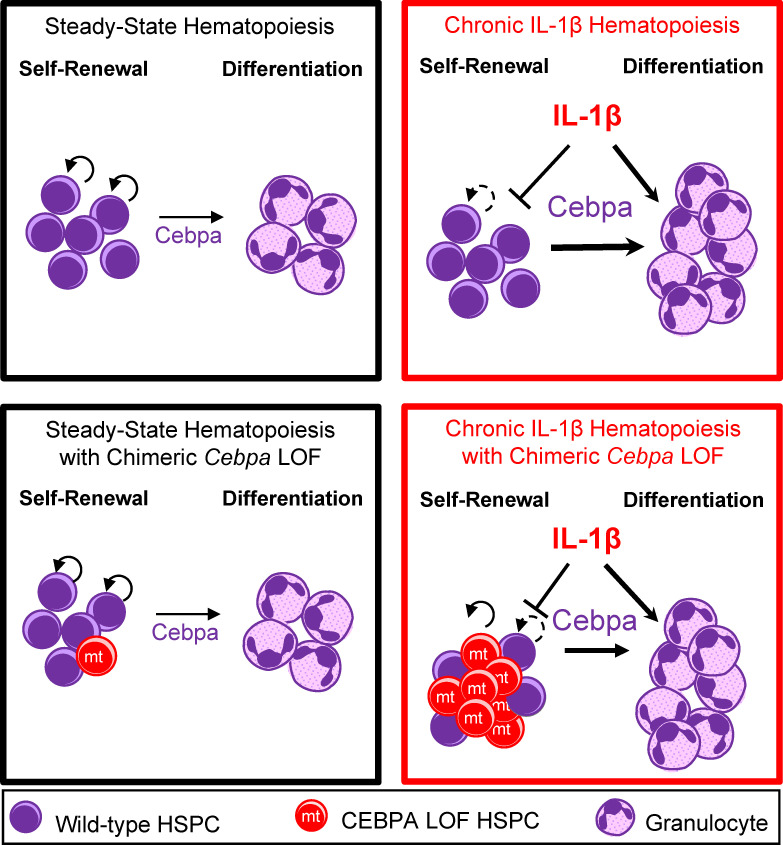
Higa et al. show that chronic interleukin-1β exposure primes multipotent hematopoietic progenitor cells for myelopoiesis by upregulating myeloid differentiation programs and repressing stem gene programs in a Cebpa-dependent manner. Consequently, interleukin-1 potently selects for Cebpa loss in hematopoietic stem and progenitor cells.
Abstract
The early events that drive myeloid oncogenesis are not well understood. Most studies focus on the cell-intrinsic genetic changes and how they impact cell fate decisions. We consider how chronic exposure to the proinflammatory cytokine, interleukin-1β (IL-1β), impacts Cebpa-knockout hematopoietic stem and progenitor cells (HSPCs) in competitive settings. Surprisingly, we found that Cebpa loss did not confer a hematopoietic cell–intrinsic competitive advantage; rather chronic IL-1β exposure engendered potent selection for Cebpa loss. Chronic IL-1β augments myeloid lineage output by activating differentiation and repressing stem cell gene expression programs in a Cebpa-dependent manner. As a result, Cebpa-knockout HSPCs are resistant to the prodifferentiative effects of chronic IL-1β, and competitively expand. We further show that ectopic CEBPA expression reduces the fitness of established human acute myeloid leukemias, coinciding with increased differentiation. These findings have important implications for the earliest events that drive hematologic disorders, suggesting that chronic inflammation could be an important driver of leukemogenesis and a potential target for intervention.
Graphical Abstract
Introduction
Hematologic disorders such as clonal hematopoiesis, myelodysplastic syndrome (MDS), myeloproliferative neoplasm (MPN), and acute myeloid leukemia (AML) are thought to originate from multipotent hematopoietic stem and progenitor cells (HSPCs), which are responsible for lifelong regeneration of the blood system. Understanding the earliest events that drive these disorders is thus important to understand the etiology of the malignancy and to devise potential intervention strategies.
Risk factors associated with hematologic disorders include age and prior exposure to radiation or chemotherapy, conditions characterized by underlying inflammation (Bowman et al., 2018). Prior studies have shown that aging, radiation, chronic infection, and chronic inflammation promote HSPC differentiation at the expense of self-renewal (Fleenor et al., 2015a; Matatall et al., 2016; Mejia-Ramirez and Florian, 2020; Pietras, 2017). The proinflammatory cytokine IL-1 is important in regulating hematopoiesis and the immune response in the acute setting, such as by promoting emergency granulopoiesis in response to infection, but can drive pathology in the chronic setting (Mantovani et al., 2019). For instance, IL-1 can promote AML expansion in ex vivo culture (Carey et al., 2017), and increased expression of IL-1 receptor accessory protein is associated with poor prognosis in AML (Barreyro et al., 2012).
Many oncogenic mutations associated with AML are known to block differentiation and/or enhance self-renewal (Corces et al., 2017; Kelly and Gilliland, 2002; Ley et al., 2013). CCAAT/enhancer-binding protein α (C/EBPα) is a bZIP transcription factor that regulates myeloid lineage commitment in HSPCs. Its function is frequently altered in MDS/MPN and AML by loss-of-function (LOF) mutations or by (epi)genetic alterations that suppress CEBPA expression, such as mutations in NPM1 or RUNX1, FLT3-ITD, AML1-ETO, or BCR-ABL, together accounting for ∼70% of molecular alterations in de novo AML (Ernst et al., 2010; Gu et al., 2018; Guo et al., 2012; Ley et al., 2013; Pabst et al., 2001a; Perrotti et al., 2002; Zheng et al., 2004). Further, Cebpa LOF leads to a block in myeloid differentiation, contributing to AML-like diseases in mouse models (Bereshchenko et al., 2009; Kirstetter et al., 2008; Pabst et al., 2001b; Porse et al., 2005; Zhang et al., 1997; Zhang et al., 2004). Therefore, CEBPA LOF represents a common mechanism of hematopoietic dysregulation across hematologic disorders. Yet we lack insight into the mechanisms driving selection for the CEBPA LOF phenotype in hematologic disorders.
Analysis of steady-state hematopoiesis suggests that multipotent progenitors (MPPs) derived from hematopoietic stem cells (HSCs) are major daily contributors to blood production and are long-lived under unperturbed conditions in mice (Busch et al., 2015; Säwén et al., 2016; Sun et al., 2014). Thus, in order to understand the origin of hematologic disorders, it is important to consider both HSCs and MPPs, collectively referred to as HSPCs, as potential oncogenic reservoirs, and to appreciate how they interact with their microenvironment.
We hypothesized that because chronic exposure to IL-1 drives precocious differentiation, it may drive selection for adaptive phenotypes that counteract HSPC differentiation as achieved by Cebpa LOF. To test this hypothesis, we modeled the impact of chronic inflammation on competition between WT and Cebpa-knockout HSPCs in vivo, by treating chimeric mice with IL-1β for 20 d. Cebpa-knockout HSPCs are resistant to the prodifferentiative effects of IL-1β, thereby providing a fitness advantage in this setting, and leading to their preferential expansion in the bone marrow (BM). These data illustrate the potential for inflammation to alter the intrinsic transcriptional landscape of HSPCs and to drive the selective fitness of potentially oncogenic HSPCs in the BM. Finally, we explored how genes regulated by IL-1β and dependent on Cebpa are altered in AMLs and the impact of restored C/EBPα activity on AML fitness.
Results
HSPC-specific Cebpa knockout causes expansion of MPPs
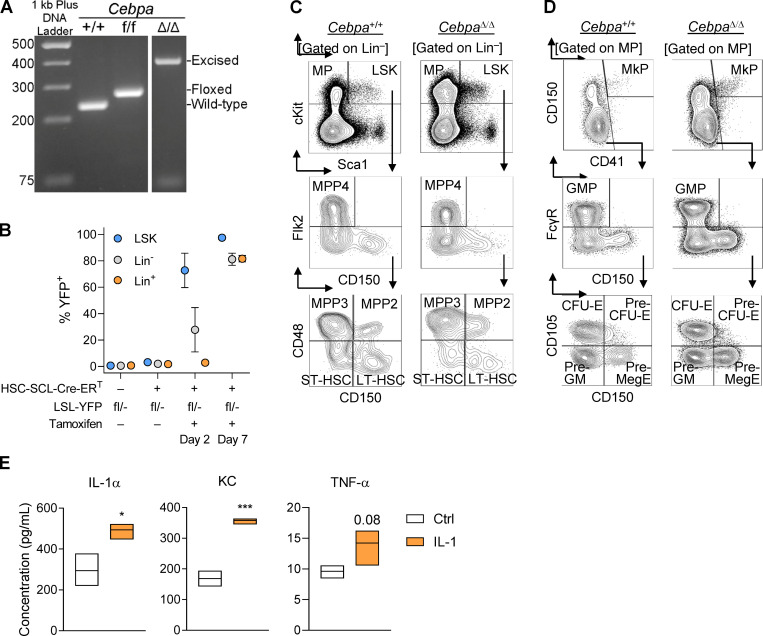
To understand how Cebpa disruption impacts HSPC fitness in a context-dependent manner, we crossed Cebpaf/f mice (Zhang et al., 2004) with R26R-LSL-EYFP and HSC-SCL-Cre-ERT transgenic mice (Göthert et al., 2005) to achieve traceable, tamoxifen-inducible, HSPC-specific knockout of Cebpa (herein referred to as CebpaΔ/Δ). To induce Cebpa excision, we treated mice with 2.5 mg tamoxifen for 3 d and analyzed mice 7 d later (Fig. 1 A). We confirmed that YFP accurately reports Cebpa excision by genotyping individual colonies formed on methylcellulose from YFP+ HSPC-enriched Lineage−Sca1+cKit+ (LSK) cells at 95% specificity (Fig. S1 A and not depicted). 2 d after tamoxifen treatment, the LSK population was highly and specifically labeled, followed by the appearance of YFP+ cells within more differentiated populations by day 7 (Fig. S1 B).
Figure 1.
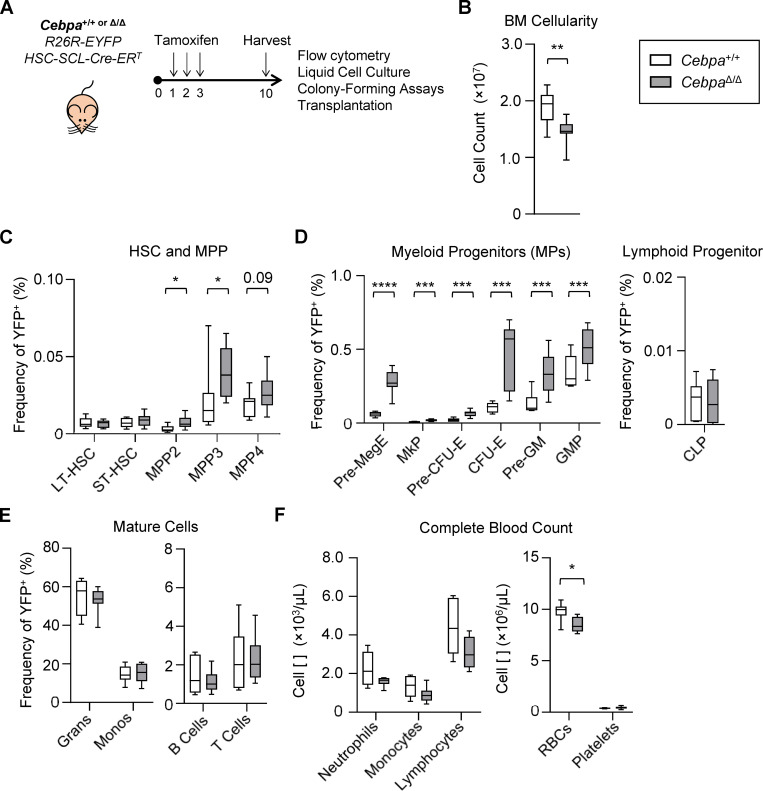
HSPC-specific Cebpa knockout causes expansion of MPPs. (A–F) Hematopoietic characterization of Cebpa+/+ and CebpaΔ/Δ mice 7 d after deletion. (A) Schematic to achieve traceable, tamoxifen-inducible, HSPC-specific knockout of Cebpa. (B) BM cellularity from one femur and one tibia per mouse. (C) HSPC frequency. (D) Committed progenitor frequency. (E) Mature cell frequency. (F) Complete blood count. CLP, common lymphoid progenitor; LT, long-term; ST, short-term. Data are from three separate experiments with n = 3 per group (n = 9 per genotype), presented as mean ± SD, and analyzed by unpaired Mann–Whitney U test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Figure S1.
Characterization of primary Cebpa-knockout mice. (A) PCR products to detect WT (235-bp), floxed (269-bp), or excised allele (361-bp) run on a 2% agarose gel with 1 kb Plus DNA ladder (Thermo Fisher Scientific). (B) Kinetics of tamoxifen-induced YFP expression within Lin−Sca1+cKit+ (LSK), Lin−, and Lin+ BM fractions. Tamoxifen did not influence HSPC behavior in cell culture or IL-1 levels in BM fluid (data not shown). (C and D) Representative flow cytometry plots to characterize HSPC populations (C) and myeloid progenitors (D) from Cebpa+/+ and CebpaΔ/Δ mice. (E) Cytokine analysis of BM fluid from vehicle or chronic IL-1β–treated mice (n = 3 mice per group; representative of two experiments). Data are presented as mean ± SD and analyzed by unpaired t test (in E). *, P < 0.05; ***, P < 0.001.
To assess the impact of Cebpa knockout on hematopoietic lineage distribution in this model, we analyzed BM and peripheral blood from Cebpa+/+ and CebpaΔ/Δ mice (Fig. S1, C and D; and Fig. 1, B–F). BM cellularity was significantly reduced in CebpaΔ/Δ mice (Fig. 1 B). Cebpa knockout did not significantly affect the frequency of phenotypically defined long-term HSC (LT-HSC) or short-term HSC (ST-HSC), but significantly increased the frequency of megakaryocyte/erythroid (MegE)-biased MPP2s and granulocyte/monocyte (GM)-biased MPP3s, with a trending increase in lymphoid-biased MPP4s (Fig. 1 D). CebpaΔ/Δ mice also had a significant buildup of MegE-lineage progenitors and GM-lineage progenitors, suggestive of a block in myeloid lineage differentiation at this level. We did not observe differences in common lymphoid progenitors (Fig. 1 D). CebpaΔ/Δ mice had no significant changes in the frequency of mature BM cell populations (Fig. 1 E) or mature peripheral blood cells, except for a significant reduction in RBC counts (Fig. 1 F). The lack of changes in mature BM cells or in LT-HSCs or ST-HSCs differs from previous studies (Ye et al., 2013; Zhang et al., 2004). This is likely due to using an HSPC-specific Cre rather than Mx1-Cre, in which the effect of a differentiation block is delayed due to the slower turnover of HSCs and MPPs, and also performing analyses 7 d after deletion. This mouse model thus corroborates previous findings that C/EBPα acts during myeloid lineage specification upstream of granulocyte-monocyte progenitors (GMPs; Zhang et al., 2004) and preGMs (Pundhir et al., 2018).
HSPC-specific Cebpa knockout enhances MPP colony-forming potential
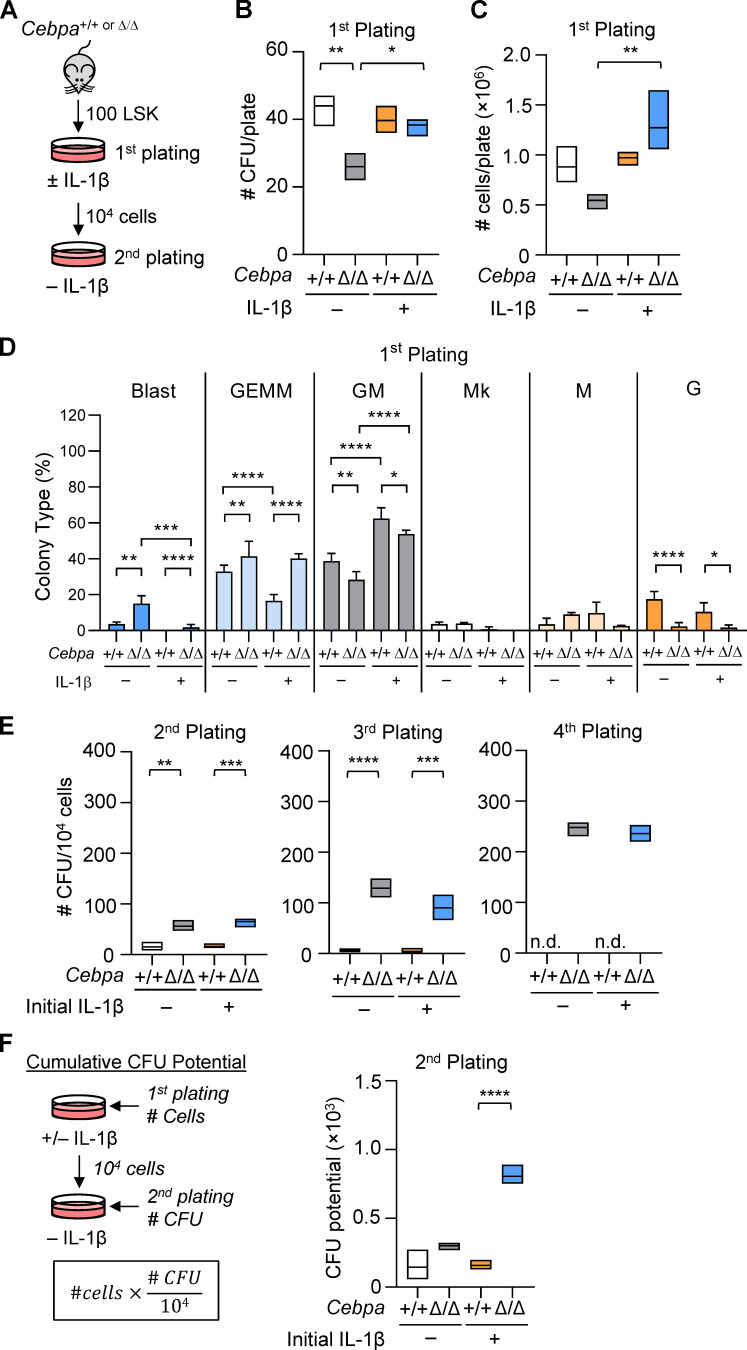
IL-1β promotes precocious myeloid differentiation with concomitant loss of HSC self-renewal (Pietras et al., 2016). Because Cebpa knockout blocks myeloid differentiation upstream of GMPs and preGMs, we hypothesized that CebpaΔ/Δ LSK cells would retain serial replating activity when exposed to IL-1β. We sorted 100 LSK cells from Cebpa+/+ or CebpaΔ/Δ mice and performed methylcellulose CFU assays with or without IL-1β (Fig. 2 A). We assessed colony types and numbers using both morphologic scoring and flow cytometry (data not shown). While IL-1β did not affect the number of CFUs or total cell numbers from Cebpa+/+ LSK cells on initial plating (Fig. 2, B and C), it did skew output toward significantly more differentiated CFU-GM colonies (both Mac1lowGr1+ and Mac1highGr1+ by flow cytometry), with significantly fewer CFU-GEMM (mixed lineage of granulocyte, erythroid, macrophage, megakaryocyte; Lin−Sca1−cKit+ by flow cytometry) colonies (Fig. 2 D), consistent with previous studies using purified HSCs (Pietras et al., 2016). In the absence of IL-1β, CebpaΔ/Δ LSK cells formed significantly fewer and smaller CFUs, producing fewer mature CFU-G (Mac1lowGr1+ by flow cytometry) than Cebpa+/+ LSK cells while retaining significantly more immature CFU-blast (Lin−Sca1+cKit+ by flow cytometry; Fig. 2, B–D). In contrast, IL-1β significantly increased the clonogenic potential of CebpaΔ/Δ LSK cells (Fig. 2 B), while having no significant impact on the clonogenic potential of Cebpa+/+ LSK cells. Even in the presence of IL-1β, CebpaΔ/Δ LSK cells maintained significantly more multipotent CFU-GEMM and produced fewer mature CFU-GM and CFU-G than that of Cebpa+/+ (Fig. 2 D).
Figure 2.
HSPC-specific Cebpa knockout enhances MPP activity. (A) Experimental design for LSK CFU assay. (B–F) LSK CFU assay data. (B) CFU counts per plate for initial plating. (C) Total cell counts per plate for initial plating. (D) Frequencies of CFU types for initial plating. Mk, megakaryocyte. (E) CFU counts per plate on replating 104 cells. n.d., not done because too few cells were recovered. (F) Calculation and data for cumulative CFU potential for second plating. Data are representative of three separate experiments with n = 3 per group, presented as mean ± SD, and analyzed by two-way ANOVA with Tukey’s multiple comparisons test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Could not perform statistics for fourth replating due to missing data points.
To functionally assess HSPC clonogenic potential, we serially replated 104 cells from the initial plating onto methylcellulose without IL-1β (Fig. 2 A). On the second plating, CebpaΔ/Δ cells formed significantly more CFUs than Cebpa+/+ cells, regardless of prior IL-1β exposure. Cebpa+/+ cells were not maintained past the third replating, whereas CebpaΔ/Δ cells continued to expand in number up to a fourth replating (Fig. 2 E). Since IL-1β significantly increased both the number of CFU and total cells produced from CebpaΔ/Δ LSK cells in the initial plating (Fig. 2, B and C), we accounted for this initial expansion difference in the overall clonogenic activity of CebpaΔ/Δ LSK cells by calculating a “cumulative CFU potential.” We multiplied the total number of cells from the first plating by the CFUs formed per 104 cells at replating (Fig. 2 F). Using this approach, we found that IL-1β significantly increased the cumulative CFU potential of CebpaΔ/Δ LSK cells but not Cebpa+/+ LSK cells (Fig. 2 F). Together, these data indicate that the clonogenic potential of CebpaΔ/Δ HSPCs is potentiated by IL-1β without inducing precocious myeloid differentiation as in Cebpa+/+ cells.
Cebpa knockout confers a fitness advantage in the context of chronic IL-1β
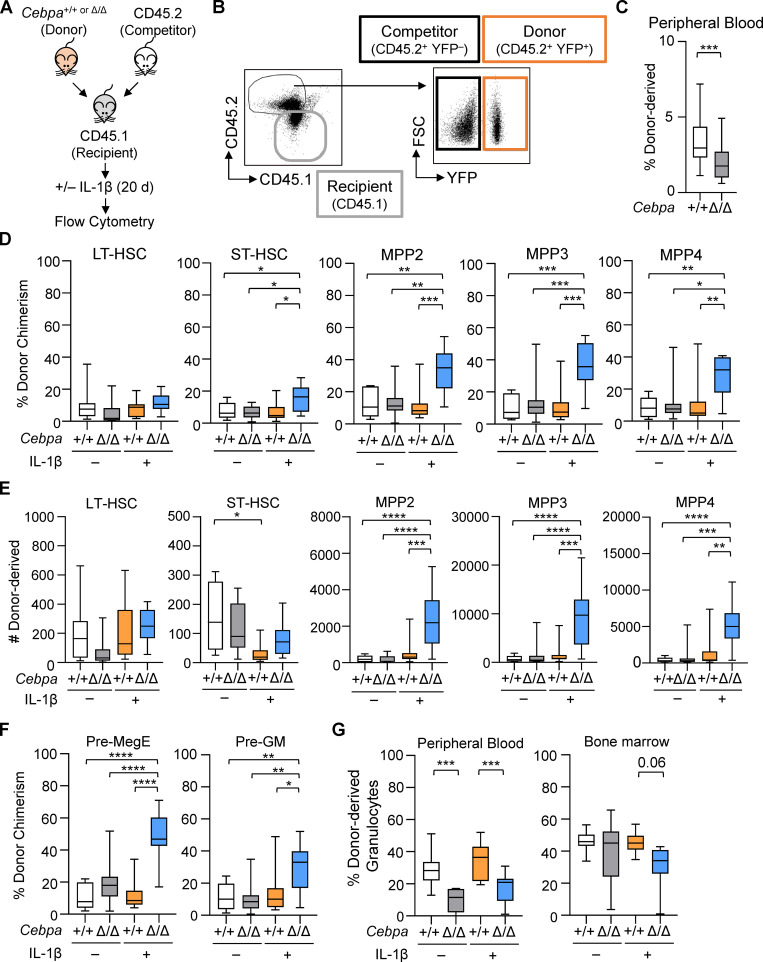
The IL-1β–enhanced colony-forming activity we observed led us to hypothesize that IL-1β may engender a fitness advantage for CebpaΔ/Δ LSK cells in vivo. To test this concept, we competitively transplanted sorted LSK cells from donor Cebpa+/+ or CebpaΔ/Δ mice with C57BL/6 (CD45.2) competitor BM into congenic Boy/J (CD45.1) recipients, allowing us to track the donor, competitor, and recipient populations by flow cytometry (Fig. 3, A and B). To isolate the effects of chronic IL-1β in the transplant setting, we used mild busulfan conditioning, which we previously showed to be less inflammatory and less disruptive to the hematopoietic system than irradiation (Henry et al., 2015). Because busulfan does not significantly deplete mature hematopoietic cells, we also treated recipients with CD4 and CD8 T cell–depleting antibodies to prevent rejection of Cebpa+/+ or CebpaΔ/Δ donor cells expressing YFP, a potential foreign antigen. At 3 wk after transplant, we found that peripheral blood chimerism ranged between 1 and 7% and was significantly lower for CebpaΔ/Δ-derived cells (Fig. 3 C). Thus, this system models early stages of leukemogenesis whereby rare oncogenically initiated cells must compete with more abundant normal progenitors.
Figure 3.
Cebpa knockout confers a fitness advantage in the context of chronic IL-1β in competitive transplants. (A) Experimental design for competitive BM transplants. (B) Representative flow cytometry plots to identify donor (CD45.2+YFP+), competitor (CD45.2+YFP−), and recipient (CD45.1+) populations. FSC, forward scatter. (C) Initial engraftment in the periphery was assessed by frequency of donor-derived (CD45.2+YFP+) leukocytes 3 wk after transplant. (D–F) Following 20 d of treatment with vehicle or IL-1β, BM was analyzed as follows. (D) Donor chimerism presented as percentage CD45.2+YFP+ of each indicated HSPC population. LT, long-term; ST, short-term. (E) Absolute numbers of donor-derived (CD45.2+YFP+) HSPCs in the BM from one femur and one tibia per mouse. (F) Donor chimerism presented as percentage CD45.2+YFP+ of each indicated myeloid progenitor population. (G) Granulocyte derived from donors in peripheral blood and BM. Data are representative of three separate experiments with n = 10 per group, presented as mean ± SD, and analyzed by unpaired Mann–Whitney U test (C) and two-way ANOVA with Tukey’s multiple comparisons test (D–G). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
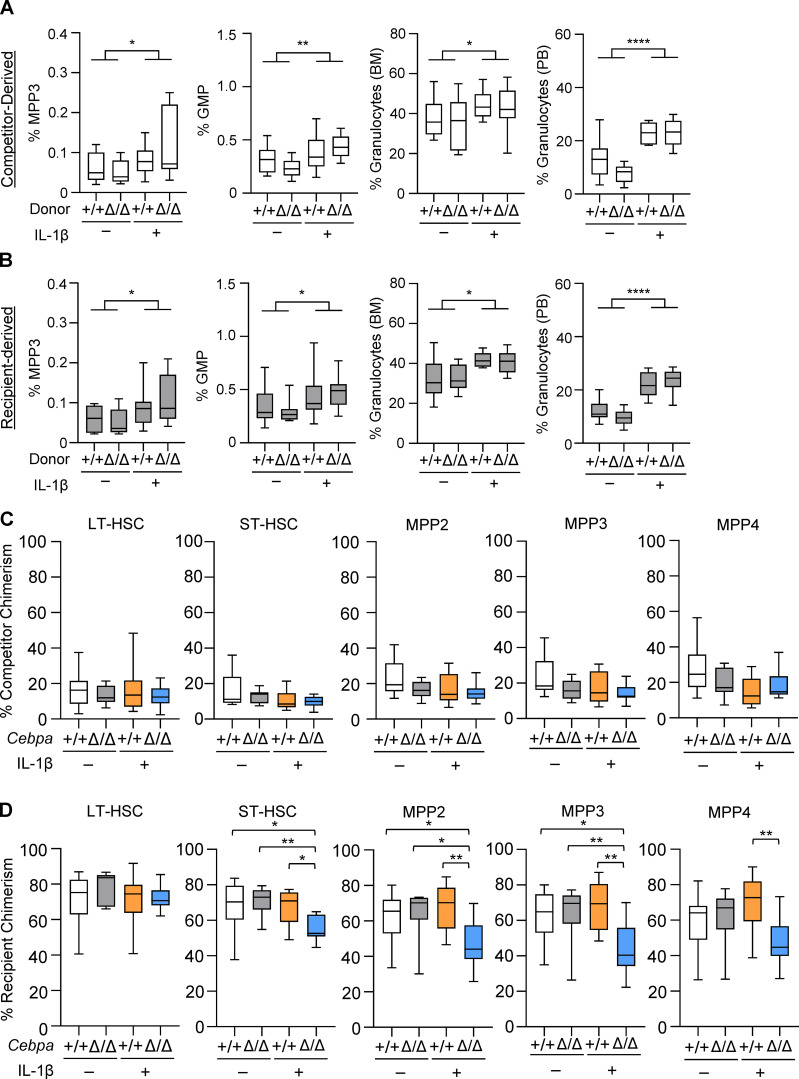
We subsequently treated the recipient mice ± IL-1β for 20 d (Fig. 3 A). As anticipated, chronic IL-1β treatment increased levels of inflammatory cytokines in the BM, in particular IL-1α, TNF-α, and the chemokine KC (Fig. S1 E). To confirm that our transplant conditions did not alter hematopoietic responses to IL-1β, we first assessed the hematopoietic output of WT competitor and recipient populations. As previously reported, chronic IL-1β increased production of myeloid-biased MPP3s, GMPs, and granulocytes (Fig. S2, A and B; Pietras et al., 2016). We next evaluated competition dynamics within HSCs, MPPs, and myeloid progenitors by assessing donor, recipient, and competitor chimerism within each compartment (Fig. 3, D and F; and Fig. S2, C and D). Importantly, in the absence of chronic IL-1β, Cebpa+/+ and CebpaΔ/Δ donor chimerism was equivalent, suggesting that the CebpaΔ/Δ LSK population does not possess a hematopoietic cell intrinsic competitive advantage (Fig. 3 D). In contrast, chronic IL-1β administration triggered selective expansion of donor-derived CebpaΔ/Δ ST-HSC, MPP2, MPP3, and MPP4 compartments (Fig. 3 D). This expansion was also reflected in the absolute number of donor-derived CebpaΔ/Δ MPPs, with the largest numerical expansion occurring within the MPP3 fraction (Fig. 3 E). Chronic IL-1β–dependent expansion of donor-derived CebpaΔ/Δ cells likewise occurred in downstream preMegE and preGM populations (Fig. 3 F). On the other hand, at this time point we observed significant deficits in CebpaΔ/Δ donor-derived granulocytes in the periphery (Fig. 3 G), consistent with a block in myeloid differentiation. Hence, by using less inflammatory methods for transplant, we did not see any evidence of cell-intrinsic competitive advantage in CebpaΔ/Δ LSK cells. On the other hand, chronic inflammatory signaling via IL-1β triggered selective expansion of CebpaΔ/Δ HSPCs in our model.
Figure S2.
Myeloid differentiation and chimerism in competitor and recipient HSPCs. (A and B) Frequency of MPP3, GMP, and granulocytes derived from competitors (A) and recipients (B) in competitive transplants. Graphs in A and B show the percentage of MPP3 within CD45.1+ or CD45.2+ YFP− cell gates, respectively. PB, peripheral blood. (C) Recipient chimerism within indicated HSPC population. (D) Competitor chimerism within indicated HSPC population. LT, long-term; ST, short-term. Data are representative of three separate experiments with n = 10 per group, presented as mean ± SD, and analyzed by two-way ANOVA with Tukey’s multiple comparisons test. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
Withdrawal of IL-1β eliminates the fitness advantage of Cebpa-knockout HSPC
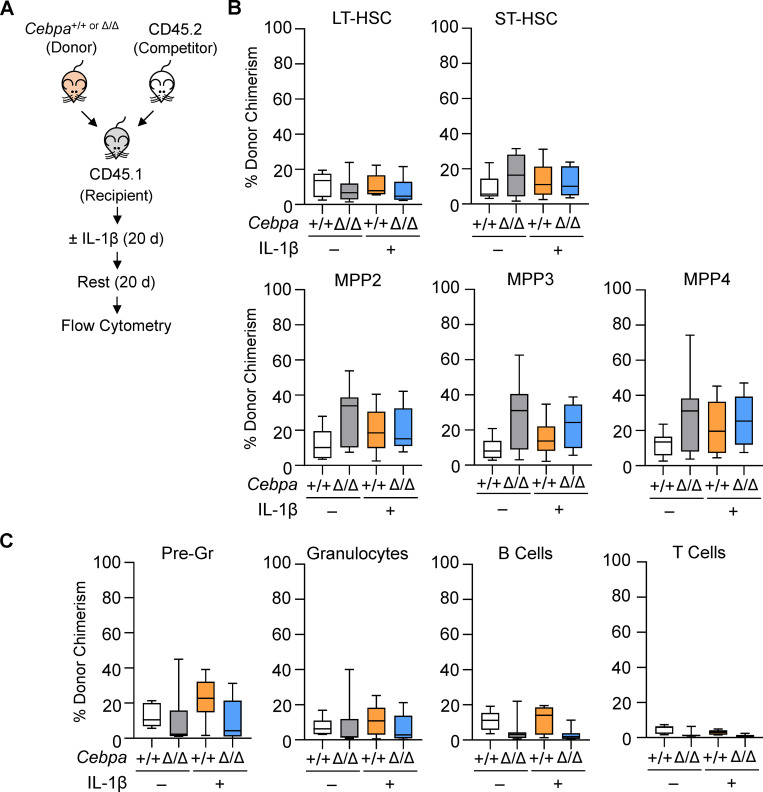
The effects of IL-1β on HSCs are reversible, as HSC self-renewal and balanced myeloid and lymphoid differentiation are recovered upon withdrawal of IL-1β (Pietras et al., 2016). Thus, we hypothesized that the competitive advantage of Cebpa-knockout HSPCs would likewise be dampened upon withdrawal of IL-1β. To test this, we performed competitive transplants and treated recipient mice with IL-1β for 20 d as described above, and then allowed the recipient mice to rest without treatment for an additional 20 d (Fig. 4 A). Consistent with our prediction, we found no difference in Cebpa+/+ and CebpaΔ/Δ chimerism within the LT-HSC, ST-HSC, MPP2, MPP3, or MPP4 compartments or within mature cell compartments after withdrawal of IL-1β (Fig. 4, B and C). These data suggest that withdrawal of IL-1β eliminates the fitness advantage of Cebpa-knockout HSPCs, supporting a model in which selective expansion of these cells requires ongoing exposure to inflammatory signals.
Figure 4.
Withdrawal of IL-1β eliminates the fitness advantage of Cebpa-knockout HSPCs. (A) Experimental design for competitive BM transplant. (B and C) After treatment with vehicle or IL-1β for 20 d and subsequent withdrawal of treatment for 20 d, BM was analyzed as follows. (B) Donor chimerism presented as percentage CD45.2+YFP+ of each indicated HSPC population. LT, long-term; ST, short-term. (C) Donor chimerism presented as percentage CD45.2+YFP+ of each indicated mature cell population. Data are from two separate experiments with n = 3–5 per group, presented as mean ± SD, and analyzed by two-way ANOVA with Tukey’s multiple comparisons test. No comparisons were significant.
Chronic IL-1β–driven myeloid gene programs are Cebpa dependent
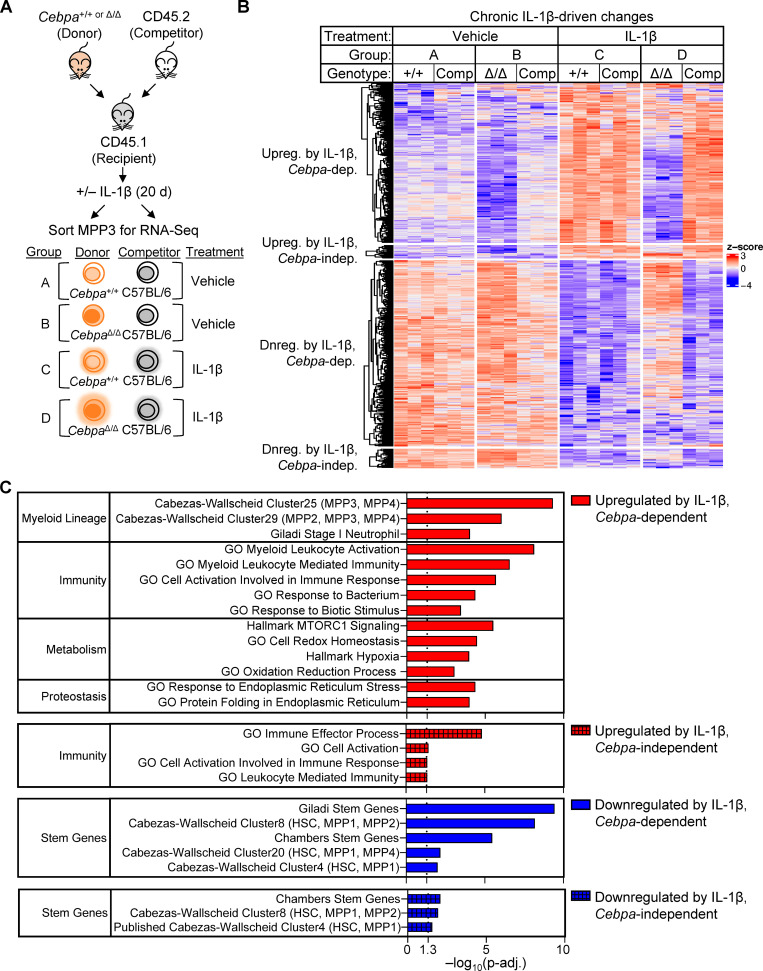
As shown above in Fig. 3, the CebpaΔ/Δ MPP3 compartment exhibited the most striking expansion during chronic IL-1β exposure. Because chronic IL-1β and Cebpa knockout are likely to exert opposing effects on myeloid lineage output, we chose to investigate the molecular mechanism by which CebpaΔ/Δ confers a fitness advantage on the GM-biased MPP3 compartment in the context of chronic IL-1β exposure. Thus, we isolated both donor- and competitor-derived MPP3s from recipient mice and performed RNA sequencing (RNA-seq; Fig. 5 A). After validating Cebpa knockout by assessing its transcript levels in donor-derived MPP3s (Fig. S3 A), we determined the gene expression consequences of Cebpa deficiency in MPP3s (Fig. S3 B). There were 494 differentially expressed genes (adjusted P < 0.05) between CebpaΔ/Δ and Cebpa+/+ MPP3s isolated from vehicle-treated mice (Fig. S3 B; gene list in Table S1). Chronic IL-1β had little effect on the relative expression of these genes between CebpaΔ/Δ and Cebpa+/+ MPP3s, suggesting that loss of Cebpa is epistatic to chronic IL-1β (Fig. S3 B). We then focused on chronic IL-1β–driven gene expression changes by comparing gene expression in Cebpa+/+ MPP3s isolated from recipient mice treated with or without chronic IL-1β. There were 555 differentially expressed genes (adjusted P < 0.05). Interestingly, chronic IL-1β failed to induce most gene expression changes in Cebpa-knockout MPP3s, suggesting that Cebpa is required for the majority of chronic IL-1β–induced gene programs in MPP3s (Fig. 5 B). Only a small subset of chronic IL-1β–induced gene expression changes were independent of Cebpa (Fig. 5 B). Il1r1 expression was unchanged in CebpaΔ/Δ MPP3s, ruling out the lack of IL-1 signaling as an explanation for the attenuated transcriptional signature (Fig. S3 A).
Figure 5.
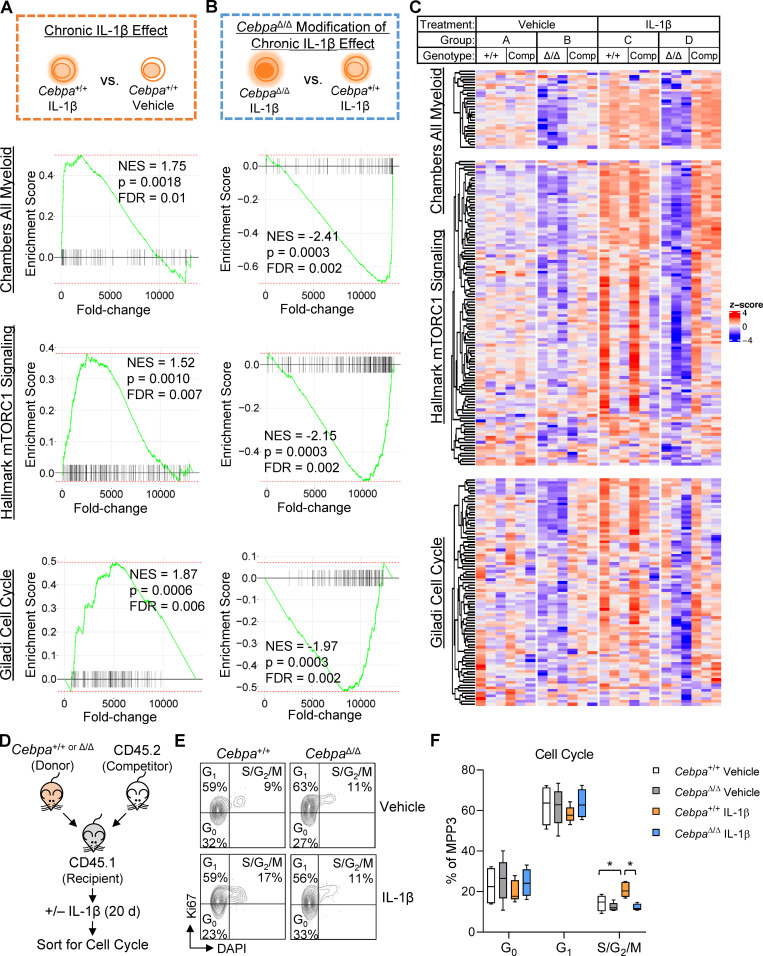
Cebpa mediates many of the chronic IL-1β gene expression changes. (A) Experimental design for RNA-seq (n = 3 of 3–4 mice pooled to obtain 2,000 donor [CD45.2+YFP+] or competitor [CD45.2+YFP−] MPP3s). (B) Heatmap of all genes significantly changed (adjusted P < 0.05) by chronic IL-1β, grouped based on pattern of expression: up-regulated (upreg.) with IL-1β, Cebpa-dependent; upreg. with IL-1β, Cebpa-independent; down-regulated (dnreg.) with IL-1β, Cebpa-dependent; dnreg. with IL-1β, Cebpa-independent (genes for heatmap are listed in Table S1). (C) Table showing select significant pathways (adjusted P < 0.05) from ORA.
Figure S3.
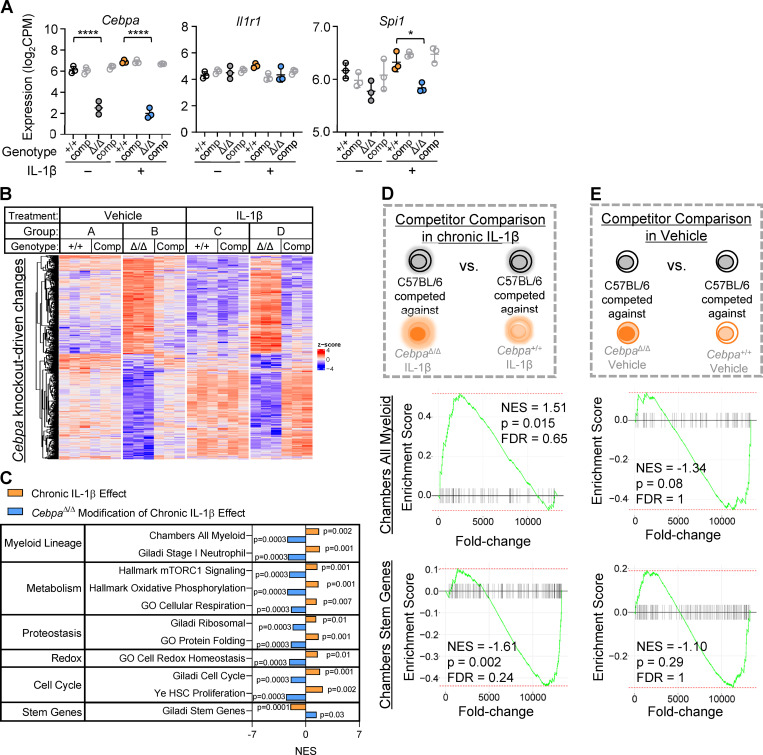
Cebpa knockout confers a fitness advantage in the context of chronic IL-1β in vivo. (A) Expression of Cebpa, Il1r1, and Spi1 plotted as log2(counts per million) from RNA-seq (n = 3 of three to four mice pooled). (B) Heatmap of all genes significantly changed (adjusted P < 0.05) by Cebpa knockout (genes for heatmap are listed in Table S1). (C) Table summarizing the GSEA normalized enrichment score (NES) for select gene sets for “chronic IL-1β effect” and “CebpaΔ/Δ modification of chronic IL-1β effect” pairwise comparisons. (D and E) GSEA with “Chambers all myeloid” and “Chambers stem genes” gene sets. (D) Schematic and enrichment plots for pairwise comparison between C57BL/6 MPP3 competed against CebpaΔ/Δ or Cebpa+/+ from chronic IL-1β–treated mice. (E) Schematic and enrichment plots for pairwise comparison between C57BL/6 MPP3 competed against CebpaΔ/Δ or Cebpa+/+ from vehicle-treated mice. FDR, false discovery rate; NES, normalized enrichment score. Data are presented as mean ± SD and analyzed by two-way ANOVA with Tukey’s multiple comparisons test (in A). *, P < 0.05; ****, P < 0.0001.
To gain insight into the biological impact of these gene expression changes, we performed overrepresentation analysis (ORA) on genes grouped based on whether they were significantly up- or down-regulated by chronic IL-1β and whether this change was Cebpa dependent or independent (Fig. 5 C). From the chronic IL-1β up-regulated genes that are dependent on Cebpa, 61 pathways were overrepresented with adjusted P < 0.05 (Fig. 5 C), with many related to myeloid lineage or immunity. In accordance with ORA, pairwise gene set enrichment analyses (GSEA) revealed that Cebpa knockout prevented activation of myeloid gene programs despite chronic IL-1β exposure (Fig. 6, A–C; and Fig. S3 C). These data are consistent with the failure of donor-derived CebpaΔ/Δ to overproduce granulocytes in response to chronic IL-1β (Fig. 3 G).
Figure 6.
Cebpa knockout counteracts chronic IL-1β–driven transcriptional programs. (A–C) GSEA using “Chambers all myeloid,” “hallmark mTORC1 signaling,” and “Giladi cell cycle” gene sets. (A) Schematic and enrichment plots for pairwise comparison between Cebpa+/+ MPP3 isolated from chronic IL-1β–treated versus vehicle-treated mice (“chronic IL-1β effect”). (B) Schematic and enrichment plots for pairwise comparison between CebpaΔ/Δ and Cebpa+/+ MPP3s isolated from chronic IL-1β–treated mice (“CebpaΔ/Δ modification of chronic IL-1β effect”). (C) Heatmaps for leading edge genes from B (genes for heatmap are listed in Table S1). (D) Experimental design for cell cycle analysis (n = 5 per group). (E) Representative flow cytometry plots for Ki67/DAPI cell cycle analysis. (F) Graph of frequency of MPP3 in G0, G1, S/G2/M. FDR, false discovery rate; NES, normalized enrichment score. In D–F, cell cycle data are from one experiment with n = 5 mice per group, presented as mean ± SD, and analyzed by two-way ANOVA with Tukey’s multiple comparisons test. *, P < 0.05; **, P < 0.01.
Additional pathways that chronic IL-1β up-regulates in a Cebpa-dependent manner included those related to metabolism, protein synthesis, and redox homeostasis (Fig. 5 C; Fig. 6, A–C; and Fig. S3 C). Multiple studies have shown that tight regulation of these pathways is required for HSC function (García-Prat et al., 2017; Ito and Suda, 2014; Signer et al., 2014). For instance, mTORC1 signaling, which is an important regulator of cell growth by regulating protein synthesis and cellular metabolism, has been shown to be important for myeloid cell differentiation (Peng et al., 2018; Zhang et al., 2019), and its hyperactivation can lead to HSC exhaustion (Gan and DePinho, 2009). HSPC differentiation is likewise associated with increased metabolic and biosynthetic demands, so it seems appropriate that chronic IL-1β priming of MPP3s for myeloid differentiation is associated with up-regulation of the cellular processes required for differentiation. These data underscore the role of Cebpa as a master regulator coordinating not only myeloid gene programs, but also gene programs for cellular processes required to support differentiation.
Chronic IL-1β–induced Cebpa-knockout MPP3 expansion is cell cycle independent
IL-1β can stimulate HSPC proliferation (Pietras et al., 2016; Ueda et al., 2009; Weisser et al., 2016). Conversely, C/EBPα inhibits cell cycle entry through direct interaction with cyclin-dependent kinases and E2F (Johnson, 2005), and Cebpa-knockout HSCs exhibit loss of quiescence (Hasemann et al., 2014; Ye et al., 2013). Thus, we reasoned that chronic IL-1β may directly activate increased CebpaΔ/Δ MPP3 proliferation, resulting in their selective expansion. Unexpectedly, Cebpa knockout actually prevented chronic IL-1β–driven up-regulation of mTORC1 and cell cycle genes associated with cell proliferation (Fig. 6, A–C). Consistent with our gene expression data, cell cycle analysis of donor-derived Cebpa+/+ and CebpaΔ/Δ MPP3s isolated from recipient mice treated with or without chronic IL-1β revealed a modest but significant increase in the frequency of Cebpa+/+ MPP3s in S/G2/M phase following treatment with chronic IL-1β, whereas the cell cycle status of CebpaΔ/Δ MPP3s was unchanged relative to controls (Fig. 6, D–F). Together, these data suggest that the competitive advantage of CebpaΔ/Δ MPP3s in the presence of IL-1β is not explained by changes in cell cycle activity.
Chronic IL-1β triggers a self-renewal gene program in Cebpa-knockout MPP3s
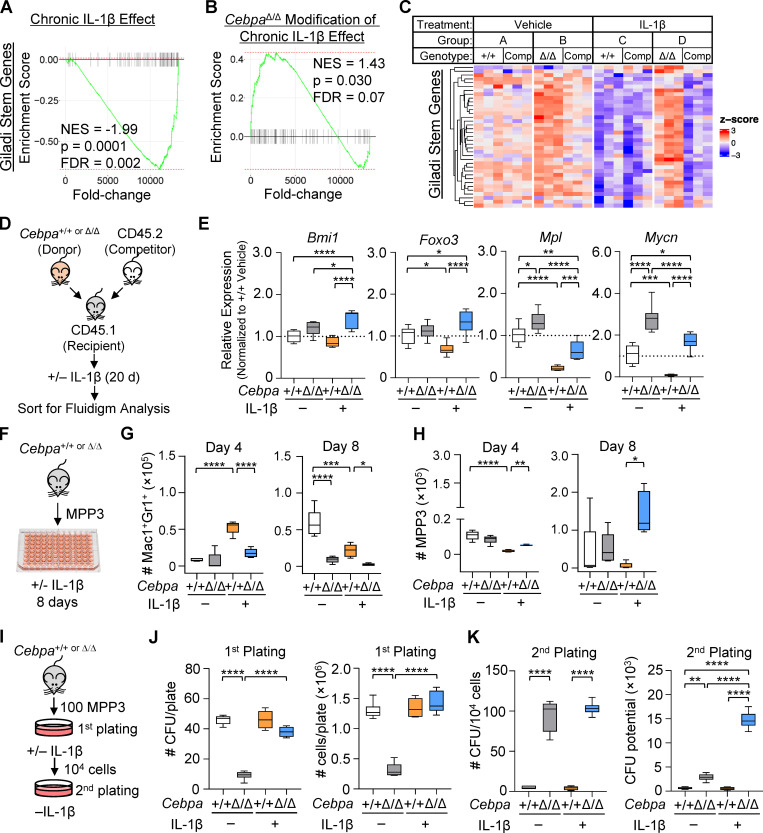
To further assess the molecular basis for selective expansion of CebpaΔ/Δ MPP3s in vivo, we analyzed genes down-regulated by IL-1β in Cebpa+/+ but not CebpaΔ/Δ MPP3s. ORA revealed that pathways with this expression pattern included those related to stemness (Fig. 5 C), suggesting that chronic IL-1β represses stem cell gene programs in MPP3 via a C/EBPα-dependent mechanism. GSEA supported these findings, showing enrichment for stem cell gene signatures in CebpaΔ/Δ MPP3s from chronic IL-1β–treated recipient mice (Fig. 7, A–C; and Fig. S3 C). To validate these findings, we isolated donor-derived Cebpa+/+ and CebpaΔ/Δ MPP3s from recipient mice treated with or without chronic IL-1β and performed quantitative RT-PCR analysis using a custom Fluidigm gene expression array (Fig. 7 D). We found that expression of genes essential for HSC self-renewal, such as Bmi1, Foxo3, Mpl, and Mycn (Iwama et al., 2004; Laurenti et al., 2008; Miyamoto et al., 2007; Nitta et al., 2020; Yoshihara et al., 2007), were elevated in CebpaΔ/Δ MPP3s as compared with Cebpa+/+ MPP3s from chronically IL-1β–exposed mice (Fig. 7 E). Thus, the fitness advantage in the context of chronic IL-1β may be attributed to a coordinate increase in self-renewal–associated genes in CebpaΔ/Δ MPP3s.
Figure 7.
Chronic IL-1β triggers aberrant expansion potential of Cebpa-knockout MPP3. (A–C) GSEA using “Giladi stem genes” gene set. (A) Schematic and enrichment plot for pairwise comparison between Cebpa+/+ MPP3 isolated from chronic IL-1β–treated versus vehicle-treated mice (“chronic IL-1β effect”). (B) Schematic and enrichment plot for pairwise comparisons between CebpaΔ/Δ versus Cebpa+/+ MPP3 isolated from chronic IL-1β–treated mice (“CebpaΔ/Δ modification of chronic IL-1β effect”). (C) Heatmap for leading edge genes from B (genes for heatmap are provided in Table S1). (D) Experimental design for Fluidigm Biomark gene expression analysis (one experiment independent from RNA-Seq, eight replicates from n = 5 pooled mice per group). (E) Relative expression of self-renewal genes (Bmi1, Foxo3, Mpl, and Mycn) calculated by first normalizing to Gusb within each group, then normalizing to Cebpa+/+ vehicle group. (F–H) Liquid culture data representative of three separate experiments with n = 3–5 per group. (F) Experimental design for MPP3 liquid culture. (G) Time course of Mac1+Gr1+ absolute numbers. (H) Time course of MPP3 absolute numbers. (I–K) CFU assay representative of two experiments with n = 3 or 6 per group. (I) Experimental design for MPP3 CFU. (J) CFU counts per plate and total cell counts per plate from initial plating. (K) CFU counts per plate upon replating 104 cells and calculated cumulative CFU potential from second plating. FDR, false discovery rate; NES, normalized enrichment score. Data are presented as mean ± SD and analyzed by two-way ANOVA with Tukey’s multiple comparisons test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, except in G and H, in which symbols for different comparisons are indicated.
IL-1β triggers aberrant expansion potential of Cebpa-knockout MPP3s
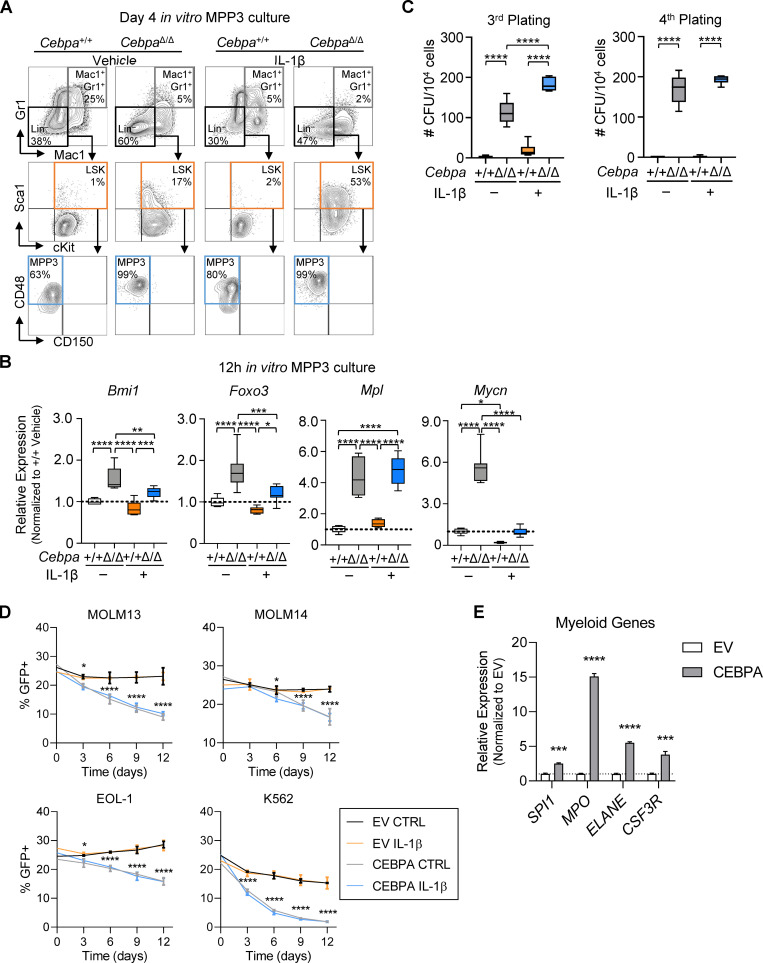
To assess the direct impact of IL-1β on Cebpa+/+ or CebpaΔ/Δ MPP3 expansion, we sorted MPP3s from primary Cebpa+/+ or CebpaΔ/Δ mice for liquid culture with or without IL-1β and assessed MPP3 maintenance and differentiation by flow cytometry (Figs. 7 F and S4 A). Similar to studies performed with purified HSCs (Pietras et al., 2016), IL-1β induced precocious differentiation of Cebpa+/+ MPP3s and early accumulation of Mac1+Gr1+ cells (Fig. 7 G). This was associated with a concomitant early depletion of MPP3s, resulting in significantly reduced differentiation potential at day 8 (Fig. 7, G and H). Despite IL-1β treatment, CebpaΔ/Δ MPP3s formed very few differentiated Mac1+Gr1+ cells over the 8-d culture (Fig. 7 G). Consistent with our in vivo data, there was no difference in the number of phenotypic Cebpa+/+ or CebpaΔ/Δ MPP3 cells after 8 d of culture without IL-1β. However, IL-1β induced selective expansion of CebpaΔ/Δ MPP3s in culture (Fig. 7 H). CebpaΔ/Δ MPP3s cultured with IL-1β also expressed elevated levels of HSC self-renewal genes identified in our in vivo studies, including Bmi1, Foxo3, Mpl, and Mycn (Fig. S4 B).
Figure S4.
CEBPA regulates expression of self-renewal and myeloid genes and cell competition in culture. (A and B) MPP3 liquid culture (data are representative of two experiments with n = 3 per group). (A) Representative flow cytometry plots for MPP3 cell culture at day 4. (B) Relative expression of self-renewal genes (Bmi1, Foxo3, Mpl, and Mycn) calculated by first normalizing to Gusb within each group, then normalizing to Cebpa+/+ vehicle group from MPP3 cell culture at 12 h. (C) MPP3 CFU assay (data are representative of two experiments with n = 3 or 6 per group): third and fourth plating CFU counts per plate upon replating 104 cells. (D and E) Cell culture of AML cell lines MOLM-13, MOLM-14, EOL-1, and K562 (representative of two experiments with n = 3 per group). (D) Cell competition dynamics. (E) Gene expression analysis of CEBPA target genes from sorted GFP+ MOLM-13 cells at day 6. EV, empty vector. Data are presented as mean ± SD and analyzed by two-way ANOVA with Tukey’s multiple comparisons test (B–D) or unpaired t test (E). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We also analyzed serial CFU activity of Cebpa+/+ or CebpaΔ/Δ MPP3s with or without IL-1β (Fig. 7 I). As seen with LSK cells, IL-1β did not affect the total number of CFUs or cells formed by Cebpa+/+ MPP3s on the initial round of plating. While CebpaΔ/Δ MPP3s formed fewer CFUs and had fewer total cells in the absence of IL-1β, their clonogenic activity increased significantly when cultured with IL-1β (Fig. 7 J). Upon replating, Cebpa+/+ cells formed very few CFUs, but CebpaΔ/Δ cells formed significantly more CFUs regardless of whether previously plated with or without IL-1β (Fig. 7 K), and CebpaΔ/Δ MPP3s retained clonogenic potential up to the third and fourth replatings (Fig. S4 C). Again, we calculated the cumulative CFU potential at replating and found that IL-1β significantly increased the cumulative CFU potential of CebpaΔ/Δ MPP3s (Fig. 7 K). This demonstration of IL-1β–specific CebpaΔ/Δ MPP3 expansion in cell culture recapitulates our in vivo data and further isolates the fitness advantage as reliant specifically on IL-1β. In addition, these data suggest that CebpaΔ/Δ MPP3s expand following IL-1β exposure via triggering aberrant self-renewal activity, which is consistent with up-regulation of key stem genes in vivo.
Cebpa knockout exacerbates chronic IL-1β gene programs in competitor MPP3s
Somatic cell fitness describes how cell-intrinsic properties (Cebpa knockout) interact with microenvironmental factors (IL-1β) and other cells within its niche. Cell competition dynamics are appreciated to be an important factor in tumorigenesis (Bowling et al., 2019). We hypothesized that WT competitor MPP3 would exhibit differential gene expression in competition against a more fit population (versus CebpaΔ/Δ MPP3 in chronic IL-1β) versus in neutral competition (versus Cebpa+/+ MPP3 in chronic IL-1β), even though they are identical by genotype and treatment. We performed GSEA pairwise comparisons between WT competitor MPP3s transplanted with Cebpa+/+ or CebpaΔ/Δ in the context of chronic IL-1β (Fig. S3 D). Interestingly, WT MPP3s that were competed against CebpaΔ/Δ MPP3s had enrichment for myeloid gene programs above that of WT MPP3s that were competed against Cebpa+/+ MPP3s in the context of chronic IL-1β. Further, WT MPP3s that were competed against CebpaΔ/Δ MPP3s exhibited reduced expression of stem gene programs as compared with WT MPP3s competed against Cebpa+/+ MPP3s in the context of chronic IL-1β (Fig. S3 D). When the same comparison was made between competitors isolated from vehicle-treated mice, we observed no enrichment for myeloid genes and much less enrichment for stem genes in the WT MPP3s competed against CebpaΔ/Δ MPP3s (Fig. S3 E). This suggests that WT competitors differ in gene expression only when in the presence of a fitness differential. These data provide evidence at the molecular level that competition in the presence of CebpaΔ/Δ MPP3s exacerbated chronic IL-1β activation of myeloid gene programs and repression of stem gene programs.
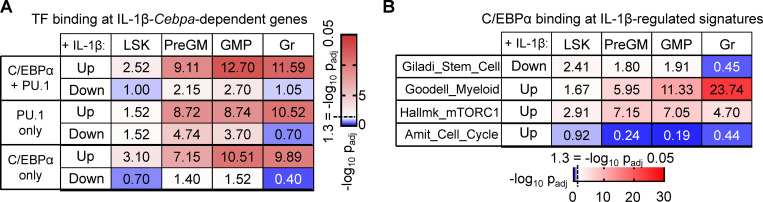
Cebpa-dependent IL-1 target genes are enriched for C/EBPα and PU.1 binding sites
C/EBPα act both independently and cooperatively with PU.1 to regulate GM-lineage gene programs in HSPCs and can regulate PU.1 levels (Pundhir et al., 2018). To better understand how Cebpa-dependent genes are regulated in our system, we mined publicly available C/EBPα and PU.1 chromatin immunoprecipitation sequencing (ChIP-seq) data from LSK, preGM, GMP, and granulocytes from Cebpa+/+ and CebpaΔ/Δ mice (Pundhir et al., 2018). We first determined the enrichment between the genes in each binding category and genes that are up- or down-regulated by chronic IL-1β in a Cebpa-dependent manner. We found that genes up-regulated by chronic IL-1β in a Cebpa-dependent manner, which were largely related to myeloid lineage or immunity (Fig. 5, B and C), were highly enriched for C/EBPα-, PU.1-, and dual-bound regions in LSK, preGM, GMP, and granulocytes (Fig. 8 A). Genes down-regulated by chronic IL-1β in a C/EBPα-dependent manner, enriched in stem gene pathways (Fig. 5, B and C), were significantly enriched for C/EBPα-, PU.1-, and dual bound regions in preGM and GMP, but not granulocytes (Fig. 8 A). On the other hand, these genes were significantly enriched for PU.1 in LSK, preGM, and GMP (Fig. 8 A), consistent with reduced Spi1 expression in IL-1–exposed CebpaΔ/Δ MPP3s (Fig. S3 A). These data support a model in which loss of Cebpa may (at least in part) indirectly prevent differentiation-associated down-regulation of stem cell genes via failure to induce appropriate levels of PU.1 expression. When we further analyzed genes regulated by IL-1β in a C/EBPα-dependent manner based on functional categories, we observed significant enrichment for C/EBPα binding to promoters of stem cell, myeloid, and mTORC1 pathway genes across the differentiation spectrum (except for stem cell genes in granulocytes), while binding to cell cycle promoters was not evident, suggesting indirect regulation of these genes (Fig. 8 B). Taken together, these data suggest that the Cebpa-dependent IL-1β–regulated genes are broadly regulated by direct binding by C/EBPα, PU.1, or both.
Figure 8.
IL-1β-Cebpa–dependent genes are highly enriched for C/EBPα and PU.1 binding sites. (A) Table summarizing C/EBPα-, PU.1-, and dual-bound regions at genes that are up- or down-regulated by chronic IL-1β in a Cebpa-dependent manner in LSK, preGM, GMP, and granulocytes. (B) Table summarizing C/EBPα binding for IL-1β-Cebpa–dependent gene within indicated gene sets in LSK, preGM, GMP, and granulocytes.
The Cebpa-dependent IL-1β–regulated gene signature is enriched in previously irradiated (IRP) HSPCs
Our laboratory had previously shown that months after γ-irradiation exposure, HSPCs exhibit precocious differentiation and impaired self-renewal, and that prior irradiation selects for loss of Cebpa within the HSPC compartment (Fleenor et al., 2015b). Because the IRP HSPC phenotype is similar to HSPCs exposed to chronic IL-1β, we hypothesized that the Cebpa-dependent IL-1β–regulated gene signature would be enriched in IRP HSPCs. We performed ORA on RNA-seq performed on cultured LSK cells isolated from IRP or unirradiated control mice (Fleenor et al., 2015b). We found that Cebpa-dependent IL-1β–down-regulated genes (e.g., stemness genes) were overrepresented in genes significantly down-regulated in IRP LSK cells (P = 2.79 × 10−6), and Cebpa-dependent IL-1β–up-regulated genes (e.g., myeloid differentiation and cell cycle genes) were overrepresented in genes significantly up-regulated in IRP LSK cells (P = 5.0 × 10−4). These data suggest that the IRP phenotype in HSPCs may in part be driven by a Cebpa-dependent IL-1β–regulated gene program.
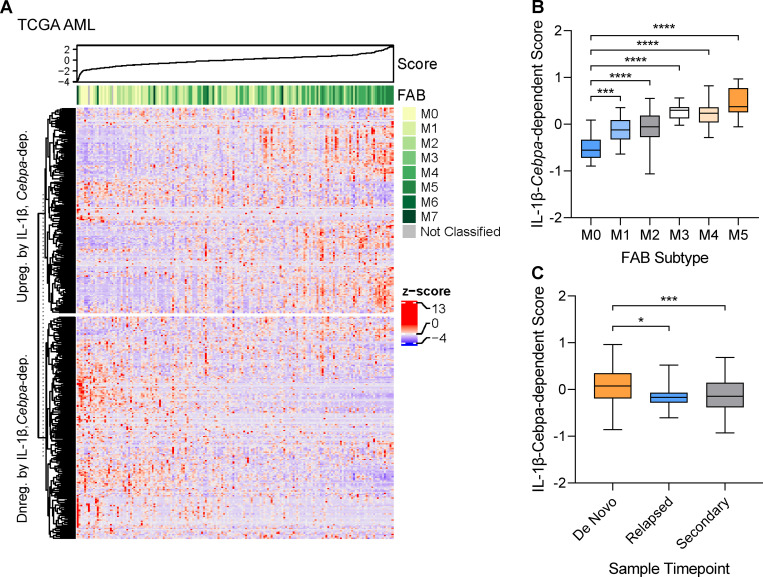
The Cebpa-dependent IL-1β gene signature is associated with AML subtype
CEBPA LOF occurs frequently in hematologic malignancies. As our data indicate that Cebpa is crucial for IL-1β–mediated HSPC differentiation, we hypothesized that the Cebpa-dependent IL-1β–regulated gene signature we identified would correlate with the differentiation status of AML. Historically, AML has been classified using the French–American–British (FAB) classification scheme, where M0 represents the most undifferentiated AML and M5 represents the most differentiated, monocytic-type AML (Bennett et al., 1985). Importantly, CEBPA mutations are commonly associated with primitive M1 and M2 FAB subtypes (Green et al., 2010; Dufour et al., 2012). Thus, we mined publicly available gene expression databases of AML samples from The Cancer Genome Atlas (TCGA; Cerami et al., 2012) and Beat AML (Tyner et al., 2018) and assigned each AML a Cebpa-dependent IL-1β–regulated gene score, where a higher score is determined by greater expression of Cebpa-dependent IL-1β–up-regulated genes (e.g., myeloid differentiation and cell cycle genes) and lesser expression of Cebpa-dependent IL-1β–down-regulated genes (e.g., stemness genes; Fig. 9 A). Interestingly, and consistent with the association between Cebpa mutations and primitive FAB subtypes, we found that the primitive M0–M2 subtypes were significantly associated with a lower Cebpa-dependent IL-1β–regulated gene score, whereas the M5 differentiated subtype corresponded to a higher score (Fig. 9 B). Similar results were seen for AML samples from Beat AML (data not shown).
Figure 9.
The IL-1β-Cebpa–dependent signature is associated with AML subtype. (A) Heatmap of IL-1β-Cebpa–dependent gene score for AML samples from TCGA. (B) Graph of IL-1β-Cebpa–dependent gene score for AML samples from TCGA by FAB subtype. (C) Graph of IL-1β-Cebpa–dependent gene score for de novo, relapse, or secondary to MDS or MPN AML samples from Beat AML. Data are presented as mean ± SD and analyzed by one-way ANOVA with Tukey’s multiple comparisons test. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
Previous studies have shown that AML at relapse is enriched for leukemia stem cells, as demonstrated via limiting dilution transplantation experiments (Ho et al., 2016). Analyses of the Beat AML datasets reveals that the Cebpa-dependent IL-1β–regulated gene score is higher in AMLs at diagnosis than at relapse, consistent with enrichment for stem cell activity (Fig. 9 C). The score was similarly lower for AMLs secondary to MDS or MPN (Fig. 9 C). While speculative at present, the inflammation associated with therapy or prior MDS/MPN may select for Cebpa loss, contributing to the more primitive phenotypes of relapse and secondary AMLs.
Ectopic CEBPA expression induces differentiation and impairs fitness of AML cells
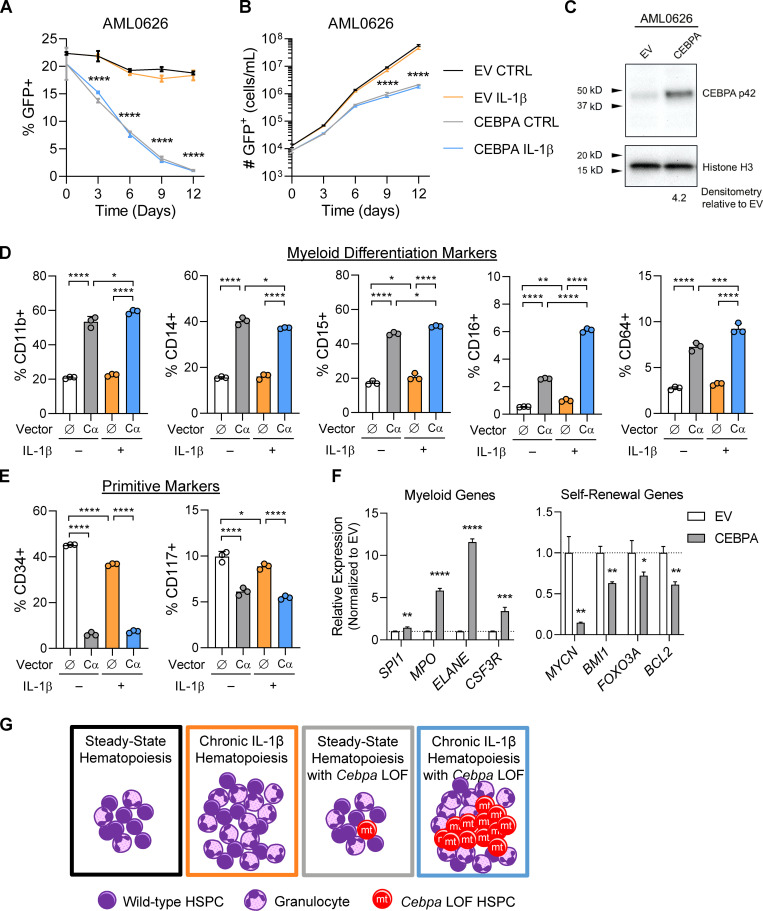
Because a low Cebpa-dependent IL-1β gene score correlated with primitive AML, we hypothesized that ectopic expression of CEBPA in AML cells would induce differentiation and impair fitness. Overexpression of CEBPA has been shown to induce neutrophilic differentiation in U937 cells (Radomska et al., 1998) and in a mouse model of acute promyelocytic leukemia (Truong et al., 2003) and to override epigenetic reprogramming mediated by RUNX1 fusions in AMLs (Loke et al., 2018). To investigate CEBPA LOF in this context, we used a primary AML (AML0626) characterized as M0/M1/M2; AML cell lines MOLM-13 and EOL-1, which harbor CEBPA mutations (in-frame insertion and deletion, respectively) according to the Cancer Cell Line Encyclopedia (Ghandi et al., 2019); chronic myelogenous leukemia line K562 (no detectable expression of CEBPA; Radomska et al., 1998); and MOLM-14, which is from the same patient as MOLM-13 and exhibits a more differentiated phenotype (Matsuo et al., 1997). We set up competitive cell cultures using cells transduced with the empty vector or a CEBPA-expressing vector, both expressing GFP, versus nontransduced cells to obtain a culture containing ∼25% GFP+ cells and cultured with and without IL-1β (Fig. 10, A and B; and Fig. S4 D). Analyses of transduced cells via Western blot demonstrated that ectopic expression roughly quadrupled C/EBPα levels in AML0626 (Fig. 10 C). We assessed the percentage of GFP+ cells and cell numbers at days 3–12. Across the multiple AML cell lines and primary cultures, we found that ectopic expression of CEBPA reduced the fraction of GFP+ cells and absolute number of GFP+ cells maintained in culture, indicative of a competitive disadvantage (Fig. 10, A and B; and Fig. S4 D). This was regardless of whether cells were cultured with or without IL-1β, suggesting that C/EBPα is epistatic to IL-1β (consistent with our gene expression data; Figs. S3 B and 5 B). Ectopic expression of CEBPA increased the frequency of AML0626 cells expressing myeloid differentiation markers CD11b, CD14, CD15, CD16, and CD64 and reduced the frequency of cells expressing primitive cell surface markers CD34 and CD117 (Fig. 10, D and E). Notably, IL1β treatment boosted the ability of ectopic C/EBPα to increase expression of some myeloid markers such as CD16 (Fig. 10 D). Finally, GFP+ cells were sorted for gene expression analysis, and ectopic expression of CEBPA did indeed cause up-regulation of myeloid genes and down-regulation of self-renewal genes (Figs. 10 F and S4 E). Thus, the maintenance of low C/EBPα activity appears important for AML cell fitness, by reducing differentiation and increasing self-renewal.
Figure 10.
Ectopic CEBPA expression induces differentiation and impairs fitness in AML cells. (A) Cell competition dynamics of primary AML sample AML0626 in cell culture. EV, empty vector. (B) Cell expansion of primary AML sample AML0626 in cell culture. (C) Western blot of sorted GFP+ cells at day 6. (D) Flow cytometric analysis of differentiation markers on GFP+ cells at day 6. (E) Flow cytometric analysis of primitive markers on GFP+ cells at day 6. (F) Gene expression analysis of CEBPA target genes from sorted GFP+ cells at day 6. (G) Model of inflammation-driven HSPC differentiation that selects for phenotypes such as with Cebpa LOF that prevent HSPC differentiation and/or promote self-renewal. For A–F, n = 3 per group, presented as mean ± SD, and analyzed by two-way ANOVA with Tukey’s multiple comparisons test (A and B, D and E) and unpaired t test (F). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. Data are representative of three experiments.
Discussion
Using a mouse model with HSPC-specific Cebpa deletion and less-inflammatory competitive transplant methods, we show that Cebpa-knockout HSPCs do not have a hematopoietic cell intrinsic competitive advantage relative to WT HSPCs. Strikingly, chronic IL-1β exposure triggered a fitness advantage for Cebpa-knockout MPPs. While chronic IL-1β remodels the HSPC compartment by expanding myeloid-biased MPPs to increase myeloid output, C/EBPα blocks myeloid differentiation. Together, these phenotypes cooperate and ultimately result in chronic IL-1β–dependent expansion of CebpaΔ/Δ cells (Fig. 10 G). While prior studies had shown an inherent competitive advantage of CebpaΔ/Δ HSCs, these investigations used polyinosinic:polycytidylic acid to induce Cre recombination and lethal irradiation to condition transplant recipients (Ye et al., 2013; Zhang et al., 2004). Both approaches are highly inflammatory and impair BM function. In addition, Mx1-Cre–mediated Cebpa deletion rapidly depleted granulocytes, which can activate MPP proliferation (Säwén et al., 2016) and confound functional studies of HSPCs. Our data demonstrate that an inflammatory environment is required to reveal the competitive advantage of Cebpa-knockout HSPCs.
Transcriptional analyses revealed that Cebpa knockout prevents chronic IL-1β–driven activation of myeloid differentiation, as well as metabolic and proteostatic gene programs important for differentiation. Despite the preexisting molecular priming of MPP3s toward myeloid lineage differentiation (Cabezas-Wallscheid et al., 2014; Pietras et al., 2015), our data show that chronic IL-1β further activates these pathways, consistent with the capacity of myeloid lineage progenitors to overproduce myeloid cells in response to “emergency” cues (Hérault et al., 2017; Kang et al., 2020; Manz and Boettcher, 2014). The dependence of these gene programs on C/EBPα is consistent with its role as a key master regulator that initiates myeloid lineage priming in the LSK population (Pundhir et al., 2018). It is interesting that chronic IL-1β has been shown to induce precocious myeloid differentiation via PU.1 (Pietras et al., 2016). C/EBPα can regulate PU.1 expression and enhancer binding in preGMs (Pundhir et al., 2018). In fact, PU.1 expression was significantly reduced in CebpaΔ/Δ MPP3s in the context of chronic IL-1β (Fig. S3 A). Collectively, these data suggest that Cebpa is important for GM-lineage specification in MPP3s and that chronic IL-1β primes GM-lineage output via Cebpa-dependent gene programs in MPP3s. In addition, Cebpa knockout prevents chronic IL-1β–driven repression of stem cell self-renewal genes, Bmi1, Foxo3, Mpl, and Mycn (Iwama et al., 2004; Laurenti et al., 2008; Miyamoto et al., 2007; Nitta et al., 2020; Yoshihara et al., 2007), and C/EBPα has been demonstrated to directly bind to the promoters of Mycn (Ye et al., 2013) and Foxo3 (Hasemann et al., 2014). Mining of publicly available ChIP-seq datasets revealed that the Cebpa-dependent IL-1β–regulated genes are directly regulated by C/EBPα and/or PU.1, further supporting a model in which C/EBPα integrates inflammatory cues to regulate gene expression programs, either independently or in concert with PU.1.
While steady-state hematopoiesis results in balanced lineage output, chronic IL-1β expands the MPP compartment to increase myeloid output. However, IL-1β–induced differentiation gene programs likely serve as a limiting mechanism that prevents aberrant accumulation of HSPCs. Our data support a model wherein aberrant expansion of HSPCs with Cebpa LOF is an emergent property triggered by inflammatory signals such as chronic IL-1β. Our findings suggest that Cebpa LOF HSPCs fail to differentiate and instead activate expression of stem cell self-renewal genes, thereby promoting their selective expansion, while WT HSPCs differentiate in response to chronic IL-1β (Fig. 10 G). Still, this selective expansion is reversed upon resolution of inflammation (Fig. 4), suggesting that inflammation must be sufficiently chronic to provide a window of opportunity to favor Cebpa LOF and thus leukemia initiation. As such, IL-1β promotion of emergency granulopoiesis, dependent on C/EBPα, is able to play its critical role in host defense (essential for host fitness). These programs carry the risk of promoting leukemogenesis, although this risk is largely relegated to older ages or rare circumstances (evolutionarily, a minor fitness cost). Still, in these contexts, chronic inflammation and its downstream effectors may represent a therapeutic target that could slow leukemogenesis.
Finally, we showed that primitive M0–M2 AML subtypes, relapsed AML, and AML secondary to MDS or MPN were associated with a lower Cebpa-dependent IL-1β–regulated gene score, corresponding to greater expression of Cebpa-dependent IL-1β–down-regulated genes (e.g., stemness genes). The Cebpa-dependent IL-1β–regulated gene score may provide additional information on the phenotype of an AML and aid in predicting prognosis and therapeutic response in AML. We further show that the maintenance of low CEPBA activity by AMLs is important for their fitness, as ectopic expression of CEBPA conferred a competitive disadvantage on AMLs, coinciding with increased differentiation and reduced expression of stemness genes. AMLs may need to evolve reduced CEPBA activity to convert IL-1β from a negative regulator of HSPC fitness (Pietras et al., 2016; Chavez et al., 2021) into a positive contributor to AML fitness (Carey et al., 2017).
There is growing evidence of context-specific expansion of oncogenically initiated hematopoietic progenitors: BCR-ABL or Ppm1d deficiency with chemotherapy (Bilousova et al., 2005; Hsu et al., 2018; Kahn et al., 2018); Notch activation, Tp53-deficiency, or Cebpa-deficiency following γ-irradiation (Bondar and Medzhitov, 2010; Fleenor et al., 2015b; Marusyk et al., 2009; Marusyk et al., 2010); BCR-ABL, Myc, NRasV12, or AML1-ETO in aging (Henry et al., 2015; Henry et al., 2010; Vas et al., 2012); JakV617F with TNF-α exposure (Fleischman et al., 2011); Tet2-knockout with LPS challenge; microbe-induced inflammation (Cai et al., 2018; Meisel et al., 2018); or DNMT3A mutations in the setting of bacteria-induced IFNγ signaling (Hormaechea-Agulla et al., 2021). Here, we show that chronic IL-1β is sufficient to drive selective expansion of Cebpa-knockout MPPs, directly connecting an inflammatory signaling pathway to a transcriptional fate network. These findings have important implications for the earliest events that could drive clonal hematopoiesis with indeterminate potential, MDS, MPN, or AML. Initially, IL-1 drives selection for mutations in HSPCs that provide resistance to the prodifferentiative effects of IL-1. This expansion will proportionally increase the odds of successive mutation accumulation in the same clone, increasing the possibility of leukemic transformation. Once transformed, IL-1 can then act as a growth factor for AML blasts (Carey et al., 2017) and contribute to more aggressive disease (Barreyro et al., 2012). Therefore, inflammation could be an important driver of leukemogenesis and could represent a potential target for intervention.
Materials and methods
Mice
CebpaloxP/loxP mice (kind gift of Daniel G. Tenen) were previously described (Zhang et al., 2004). HSC-SCL-Cre-ERT transgenic mice were generously provided by Joachim Göthert (Göthert et al., 2005). R26R-EYFP transgenic reporter mice were generously provided by Dennis Roop (Srinivas et al., 2001). All mice were backcrossed for at least five generations on a C57BL/6 background and bred to be heterozygous for R26R-EYFP and heterozygous for HSC-SCL-Cre-ERT, while each litter contained a mixture of Cebpa genotypes, such that Cebpa+/+ and CebpaΔ/Δ littermates were used in experiments. To induce recombination, mice were treated i.p. with 2.5 mg of tamoxifen (prepared at 10 mg/ml in corn oil) daily for 3 d and were used for experiments 7 d after the final dose of tamoxifen. Genotyping of mice and detection of recombined product were as previously described (Zhang et al., 2004). C57BL/6 (Ly5.2) and congenic (Ly5.1) mice were ordered from Charles River Laboratories. All procedures were performed in accordance with the University of Colorado Anschutz Medical Campus Institutional Animal Care and Use Committee–approved animal protocols.
Lentiviral vectors
The CEBPA-expressing vector was generated by subcloning the full-length ORF (Sino Biological) into the GFP-expressing lentiviral vector pUltra (Addgene). Lentiviral particles where produced by triple transfection of HEK293FT cells with pUltra-CEBPA, pSPAX2, and pMD2.G. The supernatant was harvested after 24 h, and the virus was concentrated by ultracentrifugation at 40,000 ×g for 2 h at 4°C over a 20% sucrose cushion. Lentiviral transduction was performed by plating the cells at 500,000–1,000,000/ml with the lentiviral supernatant in the presence of 4 µg/ml polybrene and centrifuging the plate at 1,000 ×g for 1 h at 32°C followed by incubation for 24 h at 37°C, at the end of which the medium was replaced with fresh medium. C/EBPα expression was determined by Western blot.
Cell preparation and flow cytometry
Peripheral blood for flow cytometry analysis or complete blood count was collected in heparin (50 U/ml) from the submandibular vein of live mice or via cardiac puncture from terminal mice. Complete blood count analyses were run on a Heska HT5 blood analyzer. For flow cytometry analyses, RBCs were hemolyzed with ammonium-chloride-potassium (ACK) lysis buffer, and cells were washed and filtered. Cell were stained for mature immune cells using the following antibodies: rat IgG for blocking, Mac1 (M1/70) Brilliant Ultraviolet 395 (BUV395), Gr1 (Rb6-8C5) Pacific Blue, CD45.2 (104) biotin, streptavidin Brilliant Violet (BV) 605, B220 (RA3-6B2) BV786, CD45.1 (A20) PerCP-Cy-5.5, CD8 (53-6.7) PE, Ter-119 (TER-119) PE-Cy5, CD4 (GK1.5) PE-Cy7, IgM (RMM-1) APC, CD3 (17A2) Alexa Fluor 700, and CD19 (1D3) APC-Cy7. BM for flow analysis was obtained by flushing cells from one tibia and one femur per mouse. RBCs were hemolyzed with ACK lysis buffer, and cells were washed and filtered. Cell counts were collected on the Guava easyCyte System (Millipore). BM cells were stained for mature immune cells as with peripheral blood and for HSPCs with the following antibodies: rat IgG, Mac1 BUV395, Sca1 (D7) BV421, CD41 (MWReg30) BV510, CD45.2 biotin, streptavidin BV605, CD105 (MJ7/18) BV711, CD150 (TC15-12F12.2) BV785, CD45.1 PerCP-Cy.5.5, Flk2 (A2F10) PE, lineage cocktail (B220, CD3, CD4, CD5 [53-7.3], CD8, Gr1, Ter119) conjugated with PE-Cy5, FcgR (93) PE-Cy7, CD34 (RAM34) eFluor(e)660, CD48 (HM48-1) Alexa Fluor 700, and cKit (2B8) APC-Cy7. Data were collected on the ZE5 cell analyzer (Bio-Rad) or the LSRFortessa (BD Biosciences) and analyzed with FlowJo software v10. BM for cell sorting was obtained by crushing all long bones and spines from each mouse. RBCs were hemolysed with ACK lysis buffer, and cells were washed and filtered. BM cells were lineage depleted using Mouse Lineage Cell Depletion Kit (Miltenyi Biotec). Cells were stained to identify HSPCs with the following antibodies: rat IgG, CD48 e450, CD45.2 biotin, streptavidin BV605, CD150 BV786, CD45.1 PerCP-Cy5.5, Flk2 PE, cKit PE-Cy7, Sca1 APC, and lineage cocktail conjugated with APC-Cy7. Cell sorting was performed with the 100-µm nozzle on the FACSAria Fusion (BD Biosciences). Flow cytometry analysis was performed on cells from liquid culture and on individual colonies from methylcellulose with the following antibodies: rat IgG, Mac1 BUV395, Gr1 Pacific Blue, Sca1 APC, and cKit PE-Cy7. Data were collected on the ZE5 cell analyzer (Bio-Rad) and analyzed with FlowJo v10. AML cells were immunophenotyped with the following antibodies: CD11b BUV395, CD117 BV421, CD64 BV510, CD16 BV605, CD38 PE, CD15 PE-Cy5, CD45 PE-Cy7, CD34 APC, and CD14 APC-Cy7, and analyzed on the LSRFortessa (BD Biosciences; van Lochem et al., 2004).
Cell culture
For liquid cultures of murine primary cells, 200 YFP+ LSK cells were sorted from Cebpa+/+ or CebpaΔ/Δ mice into wells of a 96-well culture plate. Cells were grown in IMDM (Gibco) with 20% heat-inactivated FBS (HyClone), 50 µM β-mercaptoethanol, 100 ng/ml mIL-6, 100 ng/ml mFlt3-ligand, 100 ng/ml murine stem cell factor (SCF), and 10 ng/ml IL-11, as previously described (Varnum-Finney et al., 2003). For CFU assays, 100 YFP+ LSK cells were sorted from Cebpa+/+ or CebpaΔ/Δ mice and plated in methylcellulose (HSC006; R&D Systems; 1 ml in 3-cm dish) containing IMDM, antibiotic-antimycotic (Gibco), 25 ng/ml SCF, 25 ng/ml Flt3-liter, 25 ng/ml IL-11, 10 ng/ml IL-3, 10 ng/ml GM-CSF, 25 ng/ml thrombopoietin, and 4 U/ml erythropoietin with or without 25 ng/ml IL-1β. Colonies were counted and scored under a dissecting scope after 7 d. For serial replating, 104 cells were replated in fresh methylcellulose with or without IL-1β. All cytokines were obtained from Peprotech. We calculated cumulative CFU potential with the following equation:
AML cell lines MOLM-13, MOLM-14, and EOL-1 and CML cell line K562 were maintained in IMDM with 2 mM glutamine, 10% heat-inactivated FBS, and 100 U/ml penicillin/streptomycin. Primary AML sample AML0626 (derived from a relapsed patient, FAB = M0/1/2, with IDH1-R132 and KIT-D816V mutations) was generously provided by Craig Jordan (University of Colorado Anschutz Medical Campus, Aurora, CO) and was maintained in IMDM, 20% BSA, insulin, and transferrin (BIT; Stem Cell Technologies), LDL 1:300 (Calbiochem), 50 µM β-mercaptoethanol, 100 U/ml penicillin/streptomycin, 10 ng/ml SCF 10 ng/ml, 10 ng/ml IL-3, and 10 ng/ml FLT3L. For AML cell competition assays, cells were transduced with the empty vector or CEBPA-expressing vector and then mixed with nontransduced cells to obtain a population containing ∼25% GFP+ cells. MOLM13, MOLM14, and EOL-1 were plated at 100,000 cells/ml, K562 were plated at 50,000 cells/ml, and AML0626 were plated at 200,000 cells/ml with or without 25 ng/ml IL-1β. Every 3 d, cells were counted, the percentage of GFP+ cells was tested with the Guava easyCyte flow cytometer (Millipore), and cells were replated at the same initial density.
Competitive transplants
To obtain a starting donor chimerism of ∼5%, 2 × 103 LSK cells were sorted and transplanted with 2 × 106 competitor whole BM cells into mildly conditioned congenic mice. Mild conditioning was achieved by treating mice with 20 mg/kg of busulfan 4 d before transplant and 30 µg of anti-CD4 antibody, clone GK1.5 (BioXcell), and 30 µg of anti-CD8 antibody, clone 2.43 (Bio-X-Cell), 2 d before transplant. Busulfan was prepared fresh at 20 mg/ml in DMSO and diluted in saline to 2 mg/ml, while kept warm and protected from light. CD4 and CD8 T cell–depleting antibodies were prepared at 30 µg per 0.1 ml of PBS. Initial chimerism was assessed 3 wk after transplant, and mice were subsequently treated daily for 20 d with 0.1% BSA/PBS or 0.5 µg IL-1β (Peprotech) in 0.1% BSA/PBS via i.p. injection.
Fluidigm Biomark gene expression analysis
Cells were cultured for 12 h as described above. Live cells were subsequently sorted to purity into 96-well plates at 50 cells/well in 5 µl CellsDirect 2× Reaction Buffer (Invitrogen). A panel of 96 target genes were preamplified for 20 rounds with Superscript III (Invitrogen) using a custom-designed set of target-specific primers. Preamplified cDNA was treated with Exonuclease I (New England Biolabs) to remove excess primers, diluted in DNA suspension buffer (Clontech), loaded with custom-designed primer sets onto a Fluidigm 96.96 DynamicArray IFC, and run on a BioMark HD System (Fluidigm) using SsoFast Sybr Green for detection (Bio-Rad). Data were analyzed by the cycle threshold (ΔΔCt) method using Fluidigm software, and gene expression values were normalized to Gusb expression.
RNA-seq
Cells were sorted directly into RLT Buffer, and RNA was extracted using RNeasy Micro Kit (Qiagen). Library construction was performed using the SMARTer Stranded Total RNA-Seq Kit v2, Pico Input Mammalian (Takara), and paired-end sequencing was performed on the NovaSeq 6000 instrument (Illumina) by the University of Colorado Cancer Center Genomics Shared Resource. Illumina adapters were removed using BBDuk (Bushnell, 2019), and reads <50 bp after trimming were discarded. Reads were aligned and quantified using STAR v2.6.0a (Dobin et al., 2013) to the Ensembl mouse transcriptome (GRCm38.p6, release 96). Normalization and differential expression were calculated using the limma R package (Ritchie et al., 2015). Differentially expressed genes (adjusted P < 0.05) comparing the donors of group C versus the donors of group A were determined to be influenced by IL-1β (Fig. 3 A). These genes were further determined to be Cebpa dependent or independent based on whether they were significantly altered when comparing the donors of group D to the donors of group B. ORA was performed on these genes using the ClusterProfiler R package (Yu et al., 2012) with Hallmark and GO Biological Processes gene sets from the Molecular Signatures Database (Liberzon et al., 2011), along with select published gene sets (Cabezas-Wallscheid et al., 2014; Chambers et al., 2007; Giladi et al., 2018; Ye et al., 2013). GSEA was performed on the fold change of the genes for each comparison using the fGSEA R package with 10,000 permutations (Sergushichev, 2016 Preprint) with the same gene sets. Heatmaps were generated with the ComplexHeatmap R package (Gu et al., 2016) following z-score transformation. The genes in the pathway heatmaps were selected because of their presence in the leading edge as determined by fGSEA for the indicated comparison. Raw and processed RNA-seq data are deposited in GEO under accession no. GSE166629 (Edgar et al., 2002).
Cell cycle analysis
Cells were sorted and fixed into FACS buffer, and then immediately fixed in BD Cytofix/Cytoperm. Cells were restained with surface stain, and intracellular staining for Ki67 (SolA15) APC was performed with the Fixation/Permeabilization Solution Kit (BD Bioscience) and as previously described (Jalbert and Pietras, 2018). Data were collected on the ZE5 cell analyzer (Bio-Rad) and analyzed with FlowJo v10.
RNA-seq data mining
The fgsea R package was used to conduct pathway analysis on the effect of in vitro radiation in LSK (GEO accession no. GSE61602; Fleenor et al., 2015b).
ChIP-seq data mining
C/EBPα and PU.1 ChIP binding data in LSK, preGM, GMP, and granulocytes were downloaded from GEO (accession no. GSE89767; Pundhir et al., 2018). The clusterProfiler R package was used to determine the enrichment between Cebpa-dependent IL1 up- and down-regulated genes and the genes in each binding category.
AML data mining
TCGA AML (Firehose Legacy; Ley et al., 2013) transcripts per million–normalized and Tyner AML (Tyner et al., 2018) counts per million–normalized gene expression data were downloaded from cBioPortal on June 19, 2020 (Cerami et al., 2012). Genes with mean transcripts per million <1 in the TCGA were removed. The genes of each dataset were z-score transformed and separated into Cebpa-dependent IL1 up- and down-regulated genes. The difference in the sample sum of z-scores for each gene grouping was calculated and subsequently z-score transformed. A higher signature score corresponds to greater similarity to the Cebpa-dependent changes observed upon IL-1 treatment.
Statistics
Unpaired Mann-Whitney U test or unpaired Student's t-test (for bivariate comparison) or two-way ANOVA with Tukey’s multiple comparisons test (for multivariate comparison) was performed in Prism software v8 (GraphPad); *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001.
Online supplemental material
Fig. S1 shows characterization of primary Cebpa-knockout mice. Fig. S2 shows supporting analyses of competitive transplants. Fig. S3 shows that Cebpa knockout is epistatic to chronic IL-1β, by preventing up-regulation of myeloid differentiation programs and down-regulation of stem gene programs, and that gene expression of competitors is affected by the donor genotype they were competed against. Fig. S4 shows supporting MPP3 cell culture data and cell competition in AML cell lines. Table S1 lists genes from heatmaps presented in Figs. 5 B, 6 C, 7 C, and S3 B.
Supplementary Material
lists gene names from heatmaps in Figs. 5 B, 6 C, 7 C, and S3 B.
Acknowledgments
We thank Patricia Ernst and Craig Jordan for critical review of the manuscript and Pavel Davizon-Castillo for performing complete blood counts. We thank Emmanuelle Passegue for providing BM fluid cytokine array data from IL-1–treated mice. We thank the Human Immune Monitoring Shared Resource and the Clinimmune Flow Core. We thank the University of Colorado Cancer Center Shared Resources (Flow Cytometry, Genomics, Cell Technologies and Bioinformatics/Biostatistics), supported by National Cancer Institute grant P30-CA046934.
These studies were supported by grants from the National Institutes of Health (R01-AG066544 and R01-AG067584 to J. DeGregori; R01-DK119394 to E.M. Pietras; R35-CA197697 and P01-HL131477 to D.G. Tenen; F30-CA210383 and T32-AG000279 to K.C. Higa; F31-HL138754 to J.L. Rabe); the Cleo Meador and George R. Scott Endowed Chair of Medicine in Hematology and Department of Medicine Outstanding Early Career Scholar Program award to E.M. Pietras; Leukemia and Lymphoma Society SCOR grant 7020-19 to J. DeGregori; and the Courtenay C. and Lucy Patten Davis Endowed Chair in Lung Cancer Research to J. DeGregori.
Author contributions: K.C. Higa performed all experiments with assistance from J.S. Chavez, V. Zaberezhnyy, J.L. Rabe, M. De Dominici, and E.M. Pietras. K.C. Higa, A. Goodspeed, and E.P. Danis performed bioinformatic analyses. D.G. Tenen provided resources and interpreted results. K.C. Higa, E.M. Pietras, and J. DeGregori designed research studies, interpreted results, and wrote the manuscript.
References
- Barreyro, L., Will B., Bartholdy B., Zhou L., Todorova T.I., Stanley R.F., Ben-Neriah S., Montagna C., Parekh S., Pellagatti A., et al. 2012. Overexpression of IL-1 receptor accessory protein in stem and progenitor cells and outcome correlation in AML and MDS. Blood. 120:1290–1298. 10.1182/blood-2012-01-404699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett, J.M., Catovsky D., Daniel M.T., Flandrin G., Galton D.A., Gralnick H.R., and Sultan C.. 1985. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann. Intern. Med. 103:620–625. 10.7326/0003-4819-103-4-620 [DOI] [PubMed] [Google Scholar]
- Bereshchenko, O., Mancini E., Moore S., Bilbao D., Månsson R., Luc S., Grover A., Jacobsen S.E., Bryder D., and Nerlov C.. 2009. Hematopoietic stem cell expansion precedes the generation of committed myeloid leukemia-initiating cells in C/EBPalpha mutant AML. Cancer Cell. 16:390–400. 10.1016/j.ccr.2009.09.036 [DOI] [PubMed] [Google Scholar]
- Bilousova, G., Marusyk A., Porter C.C., Cardiff R.D., and DeGregori J.. 2005. Impaired DNA replication within progenitor cell pools promotes leukemogenesis. PLoS Biol. 3:e401. 10.1371/journal.pbio.0030401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondar, T., and Medzhitov R.. 2010. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell. 6:309–322. 10.1016/j.stem.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowling, S., Lawlor K., and Rodríguez T.A.. 2019. Cell competition: the winners and losers of fitness selection. Development. 146:dev167486. 10.1242/dev.167486 [DOI] [PubMed] [Google Scholar]
- Bowman, R.L., Busque L., and Levine R.L.. 2018. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell. 22:157–170. 10.1016/j.stem.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch, K., Klapproth K., Barile M., Flossdorf M., Holland-Letz T., Schlenner S.M., Reth M., Höfer T., and Rodewald H.R.. 2015. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 518:542–546. 10.1038/nature14242 [DOI] [PubMed] [Google Scholar]
- Bushnell, B. 2019. BBTools:BBMap. https://sourceforge.net/projects/bbmap/
- Cabezas-Wallscheid, N., Klimmeck D., Hansson J., Lipka D.B., Reyes A., Wang Q., Weichenhan D., Lier A., von Paleske L., Renders S., et al. 2014. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 15:507–522. 10.1016/j.stem.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Cai, Z., Kotzin J.J., Ramdas B., Chen S., Nelanuthala S., Palam L.R., Pandey R., Mali R.S., Liu Y., Kelley M.R., et al. 2018. Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem Cell. 23:833–849.e5. 10.1016/j.stem.2018.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey, A., Edwards D.K. V, Eide C.A., Newell L., Traer E., Medeiros B.C., Pollyea D.A., Deininger M.W., Collins R.H., Tyner J.W., et al. 2017. Identification of interleukin-1 by functional screening as a key mediator of cellular expansion and disease progression in acute myeloid leukemia. Cell Rep. 18:3204–3218. 10.1016/j.celrep.2017.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami, E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers, S.M., Boles N.C., Lin K.Y., Tierney M.P., Bowman T.V., Bradfute S.B., Chen A.J., Merchant A.A., Sirin O., Weksberg D.C., et al. 2007. Hematopoietic fingerprints: an expression database of stem cells and their progeny. Cell Stem Cell. 1:578–591. 10.1016/j.stem.2007.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez, J.S., Rabe J.L., Loeffler D., Higa K.C., Hernandez G., Mills T.S., Ahmed N., Gessner R.L., Ke Z., Idler B.M., et al. 2021. PU.1 enforces quiescence and limits hematopoietic stem cell expansion during inflammatory stress. J. Exp. Med. 218:e20201169. 10.1084/jem.20201169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces, M.R., Chang H.Y., and Majeti R.. 2017. Preleukemic hematopoietic stem cells in human acute myeloid leukemia. Front. Oncol. 7:263. 10.3389/fonc.2017.00263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufour, A., Schneider F., Hoster E., Benthaus T., Ksienzyk B., Schneider S., Kakadia P.M., Sauerland M.C., Berdel W.E., Büchner T., et al. AML CG study group . 2012. Monoallelic CEBPA mutations in normal karyotype acute myeloid leukemia: independent favorable prognostic factor within NPM1 mutated patients. Ann. Hematol. 91:1051–1063. 10.1007/s00277-012-1423-4 [DOI] [PubMed] [Google Scholar]
- Edgar, R., Domrachev M., and Lash A.E.. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, T., Chase A., Zoi K., Waghorn K., Hidalgo-Curtis C., Score J., Jones A., Grand F., Reiter A., Hochhaus A., and Cross N.C.. 2010. Transcription factor mutations in myelodysplastic/myeloproliferative neoplasms. Haematologica. 95:1473–1480. 10.3324/haematol.2010.021808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleenor, C.J., Higa K., Weil M.M., and DeGregori J.. 2015a. Evolved cellular mechanisms to respond to genotoxic insults: Implications for radiation-induced hematologic malignancies. Radiat. Res. 184:341–351. 10.1667/RR14147.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleenor, C.J., Rozhok A.I., Zaberezhnyy V., Mathew D., Kim J., Tan A.C., Bernstein I.D., and DeGregori J.. 2015b. Contrasting roles for C/EBPα and Notch in irradiation-induced multipotent hematopoietic progenitor cell defects. Stem Cells. 33:1345–1358. 10.1002/stem.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischman, A.G., Aichberger K.J., Luty S.B., Bumm T.G., Petersen C.L., Doratotaj S., Vasudevan K.B., LaTocha D.H., Yang F., Press R.D., et al. 2011. TNFα facilitates clonal expansion of JAK2V617F positive cells in myeloproliferative neoplasms. Blood. 118:6392–6398. 10.1182/blood-2011-04-348144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan, B., and DePinho R.A.. 2009. mTORC1 signaling governs hematopoietic stem cell quiescence. Cell Cycle. 8:1003–1006. 10.4161/cc.8.7.8045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Prat, L., Sousa-Victor P., and Muñoz-Cánoves P.. 2017. Proteostatic and metabolic control of stemness. Cell Stem Cell. 20:593–608. 10.1016/j.stem.2017.04.011 [DOI] [PubMed] [Google Scholar]
- Ghandi, M., Huang F.W., Jané-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. III, Barretina J., Gelfand E.T., Bielski C.M., Li H., et al. 2019. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. 569:503–508. 10.1038/s41586-019-1186-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giladi, A., Paul F., Herzog Y., Lubling Y., Weiner A., Yofe I., Jaitin D., Cabezas-Wallscheid N., Dress R., Ginhoux F., et al. 2018. Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat. Cell Biol. 20:836–846. 10.1038/s41556-018-0121-4 [DOI] [PubMed] [Google Scholar]
- Göthert, J.R., Gustin S.E., Hall M.A., Green A.R., Göttgens B., Izon D.J., and Begley C.G.. 2005. In vivo fate-tracing studies using the Scl stem cell enhancer: embryonic hematopoietic stem cells significantly contribute to adult hematopoiesis. Blood. 105:2724–2732. 10.1182/blood-2004-08-3037 [DOI] [PubMed] [Google Scholar]
- Green, C.L., Koo K.K., Hills R.K., Burnett A.K., Linch D.C., and Gale R.E.. 2010. Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J. Clin. Oncol. 28:2739–2747. 10.1200/JCO.2009.26.2501 [DOI] [PubMed] [Google Scholar]
- Gu, X., Ebrahem Q., Mahfouz R.Z., Hasipek M., Enane F., Radivoyevitch T., Rapin N., Przychodzen B., Hu Z., Balusu R., et al. 2018. Leukemogenic nucleophosmin mutation disrupts the transcription factor hub that regulates granulomonocytic fates. J. Clin. Invest. 128:4260–4279. 10.1172/JCI97117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Z., Eils R., and Schlesner M.. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 32:2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Guo, H., Ma O., Speck N.A., and Friedman A.D.. 2012. Runx1 deletion or dominant inhibition reduces Cebpa transcription via conserved promoter and distal enhancer sites to favor monopoiesis over granulopoiesis. Blood. 119:4408–4418. 10.1182/blood-2011-12-397091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasemann, M.S., Lauridsen F.K., Waage J., Jakobsen J.S., Frank A.K., Schuster M.B., Rapin N., Bagger F.O., Hoppe P.S., Schroeder T., and Porse B.T.. 2014. C/EBPα is required for long-term self-renewal and lineage priming of hematopoietic stem cells and for the maintenance of epigenetic configurations in multipotent progenitors. PLoS Genet. 10:e1004079. 10.1371/journal.pgen.1004079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, C.J., Casás-Selves M., Kim J., Zaberezhnyy V., Aghili L., Daniel A.E., Jimenez L., Azam T., McNamee E.N., Clambey E.T., et al. 2015. Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. J. Clin. Invest. 125:4666–4680. 10.1172/JCI83024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, C.J., Marusyk A., Zaberezhnyy V., Adane B., and DeGregori J.. 2010. Declining lymphoid progenitor fitness promotes aging-associated leukemogenesis. Proc. Natl. Acad. Sci. USA. 107:21713–21718. 10.1073/pnas.1005486107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérault, A., Binnewies M., Leong S., Calero-Nieto F.J., Zhang S.Y., Kang Y.A., Wang X., Pietras E.M., Chu S.H., Barry-Holson K., et al. 2017. Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature. 544:53–58. 10.1038/nature21693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, T.C., LaMere M., Stevens B.M., Ashton J.M., Myers J.R., O’Dwyer K.M., Liesveld J.L., Mendler J.H., Guzman M., Morrissette J.D., et al. 2016. Evolution of acute myelogenous leukemia stem cell properties after treatment and progression. Blood. 128:1671–1678. 10.1182/blood-2016-02-695312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hormaechea-Agulla, D., Matatall K.A., Le D.T., Kain B., Long X., Kus P., Jaksik R., Challen G.A., Kimmel M., and King K.Y.. 2021. Chronic infection drives Dnmt3a-loss-of-function clonal hematopoiesis via IFNγ signaling. Cell Stem Cell.:S1934-5909(21)00108-9. 10.1016/j.stem.2021.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, J.I., Dayaram T., Tovy A., De Braekeleer E., Jeong M., Wang F., Zhang J., Heffernan T.P., Gera S., Kovacs J.J., et al. 2018. PPM1D mutations drive clonal hematopoiesis in response to cytotoxic chemotherapy. Cell Stem Cell. 23:700–713.e6. 10.1016/j.stem.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K., and Suda T.. 2014. Metabolic requirements for the maintenance of self-renewing stem cells. Nat. Rev. Mol. Cell Biol. 15:243–256. 10.1038/nrm3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwama, A., Oguro H., Negishi M., Kato Y., Morita Y., Tsukui H., Ema H., Kamijo T., Katoh-Fukui Y., Koseki H., et al. 2004. Enhanced self-renewal of hematopoietic stem cells mediated by the polycomb gene product Bmi-1. Immunity. 21:843–851. 10.1016/j.immuni.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Jalbert, E., and Pietras E.M.. 2018. Analysis of Murine Hematopoietic Stem Cell Proliferation During Inflammation. In Cellular Quiescence: Methods and Protocols. Lacorazza H.D., editor. Springer New York, New York, NY. pp. 183–200. 10.1007/978-1-4939-7371-2_14 [DOI] [PubMed] [Google Scholar]
- Johnson, P.F. 2005. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J. Cell Sci. 118:2545–2555. 10.1242/jcs.02459 [DOI] [PubMed] [Google Scholar]
- Kahn, J.D., Miller P.G., Silver A.J., Sellar R.S., Bhatt S., Gibson C., McConkey M., Adams D., Mar B., Mertins P., et al. 2018. PPM1D-truncating mutations confer resistance to chemotherapy and sensitivity to PPM1D inhibition in hematopoietic cells. Blood. 132:1095–1105. 10.1182/blood-2018-05-850339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y.A., Pietras E.M., and Passegué E.. 2020. Deregulated Notch and Wnt signaling activates early-stage myeloid regeneration pathways in leukemia. J. Exp. Med. 217:e20190787. 10.1084/jem.20190787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, L.M., and Gilliland D.G.. 2002. Genetics of myeloid leukemias. Annu. Rev. Genomics Hum. Genet. 3:179–198. 10.1146/annurev.genom.3.032802.115046 [DOI] [PubMed] [Google Scholar]
- Kirstetter, P., Schuster M.B., Bereshchenko O., Moore S., Dvinge H., Kurz E., Theilgaard-Mönch K., Månsson R., Pedersen T.A., Pabst T., et al. 2008. Modeling of C/EBPalpha mutant acute myeloid leukemia reveals a common expression signature of committed myeloid leukemia-initiating cells. Cancer Cell. 13:299–310. 10.1016/j.ccr.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Laurenti, E., Varnum-Finney B., Wilson A., Ferrero I., Blanco-Bose W.E., Ehninger A., Knoepfler P.S., Cheng P.F., MacDonald H.R., Eisenman R.N., et al. 2008. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 3:611–624. 10.1016/j.stem.2008.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley, T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J. Jr., Laird P.W., Baty J.D., et al. Cancer Genome Atlas Research Network . 2013. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368:2059–2074. 10.1056/NEJMoa1301689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon, A., Subramanian A., Pinchback R., Thorvaldsdóttir H., Tamayo P., and Mesirov J.P.. 2011. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 27:1739–1740. 10.1093/bioinformatics/btr260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loke, J., Chin P.S., Keane P., Pickin A., Assi S.A., Ptasinska A., Imperato M.R., Cockerill P.N., and Bonifer C.. 2018. C/EBPα overrides epigenetic reprogramming by oncogenic transcription factors in acute myeloid leukemia. Blood Adv. 2:271–284. 10.1182/bloodadvances.2017012781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani, A., Dinarello C.A., Molgora M., and Garlanda C.. 2019. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity. 50:778–795. 10.1016/j.immuni.2019.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz, M.G., and Boettcher S.. 2014. Emergency granulopoiesis. Nat. Rev. Immunol. 14:302–314. 10.1038/nri3660 [DOI] [PubMed] [Google Scholar]
- Marusyk, A., Casás-Selves M., Henry C.J., Zaberezhnyy V., Klawitter J., Christians U., and DeGregori J.. 2009. Irradiation alters selection for oncogenic mutations in hematopoietic progenitors. Cancer Res. 69:7262–7269. 10.1158/0008-5472.CAN-09-0604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk, A., Porter C.C., Zaberezhnyy V., and DeGregori J.. 2010. Irradiation selects for p53-deficient hematopoietic progenitors. PLoS Biol. 8:e1000324. 10.1371/journal.pbio.1000324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matatall, K.A., Jeong M., Chen S., Sun D., Chen F., Mo Q., Kimmel M., and King K.Y.. 2016. Chronic infection depletes hematopoietic stem cells through stress-induced terminal differentiation. Cell Rep. 17:2584–2595. 10.1016/j.celrep.2016.11.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo, Y., MacLeod R.A., Uphoff C.C., Drexler H.G., Nishizaki C., Katayama Y., Kimura G., Fujii N., Omoto E., Harada M., and Orita K.. 1997. Two acute monocytic leukemia (AML-M5a) cell lines (MOLM-13 and MOLM-14) with interclonal phenotypic heterogeneity showing MLL-AF9 fusion resulting from an occult chromosome insertion, ins(11;9)(q23;p22p23). Leukemia. 11:1469–1477. [DOI] [PubMed] [Google Scholar]
- Meisel, M., Hinterleitner R., Pacis A., Chen L., Earley Z.M., Mayassi T., Pierre J.F., Ernest J.D., Galipeau H.J., Thuille N., et al. 2018. Microbial signals drive pre-leukaemic myeloproliferation in a Tet2-deficient host. Nature. 557:580–584. 10.1038/s41586-018-0125-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejia-Ramirez, E., and Florian M.C.. 2020. Understanding intrinsic hematopoietic stem cell aging. Haematologica. 105:22–37. 10.3324/haematol.2018.211342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, K., Araki K.Y., Naka K., Arai F., Takubo K., Yamazaki S., Matsuoka S., Miyamoto T., Ito K., Ohmura M., et al. 2007. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 1:101–112. 10.1016/j.stem.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Nitta, E., Itokawa N., Yabata S., Koide S., Hou L.B., Oshima M., Aoyama K., Saraya A., and Iwama A.. 2020. Bmi1 counteracts hematopoietic stem cell aging by repressing target genes and enforcing the stem cell gene signature. Biochem. Biophys. Res. Commun. 521:612–619. 10.1016/j.bbrc.2019.10.153 [DOI] [PubMed] [Google Scholar]
- Pabst, T., Mueller B.U., Harakawa N., Schoch C., Haferlach T., Behre G., Hiddemann W., Zhang D.-E., and Tenen D.G.. 2001a. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat. Med. 7:444–451. 10.1038/86515 [DOI] [PubMed] [Google Scholar]
- Pabst, T., Mueller B.U., Zhang P., Radomska H.S., Narravula S., Schnittger S., Behre G., Hiddemann W., and Tenen D.G.. 2001b. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-α (C/EBPalpha), in acute myeloid leukemia. Nat. Genet. 27:263–270. 10.1038/85820 [DOI] [PubMed] [Google Scholar]
- Peng, H., Kasada A., Ueno M., Hoshii T., Tadokoro Y., Nomura N., Ito C., Takase Y., Vu H.T., Kobayashi M., et al. 2018. Distinct roles of Rheb and Raptor in activating mTOR complex 1 for the self-renewal of hematopoietic stem cells. Biochem. Biophys. Res. Commun. 495:1129–1135. 10.1016/j.bbrc.2017.11.140 [DOI] [PubMed] [Google Scholar]
- Perrotti, D., Cesi V., Trotta R., Guerzoni C., Santilli G., Campbell K., Iervolino A., Condorelli F., Gambacorti-Passerini C., Caligiuri M.A., and Calabretta B.. 2002. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat. Genet. 30:48–58. 10.1038/ng791 [DOI] [PubMed] [Google Scholar]
- Pietras, E.M. 2017. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 130:1693–1698. 10.1182/blood-2017-06-780882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Mirantes-Barbeito C., Fong S., Loeffler D., Kovtonyuk L.V., Zhang S., Lakshminarasimhan R., Chin C.P., Techner J.M., Will B., et al. 2016. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat. Cell Biol. 18:607–618. 10.1038/ncb3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietras, E.M., Reynaud D., Kang Y.A., Carlin D., Calero-Nieto F.J., Leavitt A.D., Stuart J.M., Göttgens B., and Passegué E.. 2015. Functionally distinct subsets of lineage-biased multipotent progenitors control blood production in normal and regenerative conditions. Cell Stem Cell. 17:35–46. 10.1016/j.stem.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porse, B.T., Bryder D., Theilgaard-Mönch K., Hasemann M.S., Anderson K., Damgaard I., Jacobsen S.E., and Nerlov C.. 2005. Loss of C/EBP alpha cell cycle control increases myeloid progenitor proliferation and transforms the neutrophil granulocyte lineage. J. Exp. Med. 202:85–96. 10.1084/jem.20050067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pundhir, S., Bratt Lauridsen F.K., Schuster M.B., Jakobsen J.S., Ge Y., Schoof E.M., Rapin N., Waage J., Hasemann M.S., and Porse B.T.. 2018. Enhancer and transcription factor dynamics during myeloid differentiation reveal an early differentiation block in Cebpa null progenitors. Cell Rep. 23:2744–2757. 10.1016/j.celrep.2018.05.012 [DOI] [PubMed] [Google Scholar]
- Radomska, H.S., Huettner C.S., Zhang P., Cheng T., Scadden D.T., and Tenen D.G.. 1998. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 18:4301–4314. 10.1128/MCB.18.7.4301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie, M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., and Smyth G.K.. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Säwén, P., Lang S., Mandal P., Rossi D.J., Soneji S., and Bryder D.. 2016. Mitotic history reveals distinct stem cell populations and their contributions to hematopoiesis. Cell Rep. 14:2809–2818. 10.1016/j.celrep.2016.02.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergushichev, A.A. 2016. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. BioRxiv. (Preprint posted June 20, 2016.)
- Signer, R.A., Magee J.A., Salic A., and Morrison S.J.. 2014. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 509:49–54. 10.1038/nature13035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas, S., Watanabe T., Lin C.-S., William C.M., Tanabe Y., Jessell T.M., and Costantini F.. 2001. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1:4. 10.1186/1471-213X-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J., Ramos A., Chapman B., Johnnidis J.B., Le L., Ho Y.J., Klein A., Hofmann O., and Camargo F.D.. 2014. Clonal dynamics of native haematopoiesis. Nature. 514:322–327. 10.1038/nature13824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong, B.T., Lee Y.J., Lodie T.A., Park D.J., Perrotti D., Watanabe N., Koeffler H.P., Nakajima H., Tenen D.G., and Kogan S.C.. 2003. CCAAT/Enhancer binding proteins repress the leukemic phenotype of acute myeloid leukemia. Blood. 101:1141–1148. 10.1182/blood-2002-05-1374 [DOI] [PubMed] [Google Scholar]
- Tyner, J.W., Tognon C.E., Bottomly D., Wilmot B., Kurtz S.E., Savage S.L., Long N., Schultz A.R., Traer E., Abel M., et al. 2018. Functional genomic landscape of acute myeloid leukaemia. Nature. 562:526–531. 10.1038/s41586-018-0623-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda, Y., Cain D.W., Kuraoka M., Kondo M., and Kelsoe G.. 2009. IL-1R type I-dependent hemopoietic stem cell proliferation is necessary for inflammatory granulopoiesis and reactive neutrophilia. J. Immunol. 182:6477–6484. 10.4049/jimmunol.0803961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lochem, E.G., van der Velden V.H., Wind H.K., te Marvelde J.G., Westerdaal N.A., and van Dongen J.J.. 2004. Immunophenotypic differentiation patterns of normal hematopoiesis in human bone marrow: reference patterns for age-related changes and disease-induced shifts. Cytometry B Clin. Cytom. 60B:1–13. 10.1002/cyto.b.20008 [DOI] [PubMed] [Google Scholar]
- Varnum-Finney, B., Brashem-Stein C., and Bernstein I.D.. 2003. Combined effects of Notch signaling and cytokines induce a multiple log increase in precursors with lymphoid and myeloid reconstituting ability. Blood. 101:1784–1789. 10.1182/blood-2002-06-1862 [DOI] [PubMed] [Google Scholar]
- Vas, V., Senger K., Dörr K., Niebel A., and Geiger H.. 2012. Aging of the microenvironment influences clonality in hematopoiesis. PLoS One. 7:e42080. 10.1371/journal.pone.0042080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser, M., Demel U.M., Stein S., Chen-Wichmann L., Touzot F., Santilli G., Sujer S., Brendel C., Siler U., Cavazzana M., et al. 2016. Hyperinflammation in patients with chronic granulomatous disease leads to impairment of hematopoietic stem cell functions. J. Allergy Clin. Immunol. 138:219–228.e9. 10.1016/j.jaci.2015.11.028 [DOI] [PubMed] [Google Scholar]
- Ye, M., Zhang H., Amabile G., Yang H., Staber P.B., Zhang P., Levantini E., Alberich-Jordà M., Zhang J., Kawasaki A., and Tenen D.G.. 2013. C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat. Cell Biol. 15:385–394. 10.1038/ncb2698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara, H., Arai F., Hosokawa K., Hagiwara T., Takubo K., Nakamura Y., Gomei Y., Iwasaki H., Matsuoka S., Miyamoto K., et al. 2007. Thrombopoietin/MPL signaling regulates hematopoietic stem cell quiescence and interaction with the osteoblastic niche. Cell Stem Cell. 1:685–697. 10.1016/j.stem.2007.10.020 [DOI] [PubMed] [Google Scholar]
- Yu, G., Wang L.-G., Han Y., and He Q.-Y.. 2012. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, D.-E., Zhang P., Wang N.D., Hetherington C.J., Darlington G.J., and Tenen D.G.. 1997. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc. Natl. Acad. Sci. USA. 94:569–574. 10.1073/pnas.94.2.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, M., Liu F., Zhou P., Wang Q., Xu C., Li Y., Bian L., Liu Y., Zhou J., Wang F., et al. 2019. The MTOR signaling pathway regulates macrophage differentiation from mouse myeloid progenitors by inhibiting autophagy. Autophagy. 15:1150–1162. 10.1080/15548627.2019.1578040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., Iwasaki-Arai J., Iwasaki H., Fenyus M.L., Dayaram T., Owens B.M., Shigematsu H., Levantini E., Huettner C.S., Lekstrom-Himes J.A., et al. 2004. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 21:853–863. 10.1016/j.immuni.2004.11.006 [DOI] [PubMed] [Google Scholar]
- Zheng, R., Friedman A.D., Levis M., Li L., Weir E.G., and Small D.. 2004. Internal tandem duplication mutation of FLT3 blocks myeloid differentiation through suppression of C/EBPalpha expression. Blood. 103:1883–1890. 10.1182/blood-2003-06-1978 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists gene names from heatmaps in Figs. 5 B, 6 C, 7 C, and S3 B.