Abstract
Scuticociliates are dangerous parasitic pathogens causing systemic tissue destruction and high mortality in marine fish worldwide. In this study, the first identification of Uronema marinum (Ciliophora, Scuticociliatida) from cultured turbot (Psetta maxima) larvae using mitochondrial cytochrome c oxidase 1 (cox1) gene sequence as well as species-specific primers was reported. The mean prevalence values of infected fish were calculated, and partial sequencing obtained from the mitochondrial cox1 gene region was also compared with isolates registered in the Genbank database. The sequence comparison showed 93.00% identity to U. marinum, and the parasite has also been deposited in the GenBank database. This study is the first case of U. marinum infection in Turkish marine aquaculture, contributing to the systematics and molecular epidemiology of scuticociliate in Turkey.
Key Words: Cytochrome c oxidase 1, Parasite, Polymerase chain reaction, Turbot, Uronema marinum
Introduction
The aquatic environment may offer proper conditions for sustaining parasites’ life cycles, and both wild and cultured fish species may be suitable hosts for the parasites.1 Because of the numerous stress factors, parasitic infections’ prevalence and adverse effects are usually more severe in cultured fish.2
The ciliates belonging to the scuticociliates class are obligate parasites causing significant economic losses in aquatic animals. These ciliates in fish tissues has been associated with various pathological changes, including anemia, ascites, hemorrhages, ulcers, muscular necrosis, and encephalitis in the brain.3,4 In the early stages of infection, the ciliates are encountered in the connective tissue of skin and fins and nervous tissue. During the following stages, the whole organism can get infected.5,6
Turbot is a carnivorous flatfish living at or near the bottom of the marine environments from North Africa to the Atlantic. On the other hand, the Black Sea turbot (Psetta maxima) is an endemic subspecies having potential for aquaculture in the Black Sea region.7 For the last two decades, the Black Sea turbot has been cultured by Central Fisheries Research Institute (CFRI) in Turkey in partnership with the Japan International Cooperation Agency. About 20,000 larvae are produced annually in a seawater based recirculation system, and juvenile turbot is tagged and released into the Black Sea coast of Turkey.8
Ciliates present in cultured fish species in Turkey are mostly unknown. Miamensis avidus, a scuticociliate species, has only been isolated from reared common dentex (Dentex dentex).2 Also, presumptive identification of Philasterides dicentrarchi in cultured juvenile turbot (P. maxima), based on a parasitological examination of the skin and fins, has been reported.9 The P. dicentrarchi was the source of severe internal and external clinical signs in farmed turbot Scophthalmus maximus in Spain.4
As an alternative to or support the microscopy-based identification methods, mitochondrial cytochrome c oxidase 1 (cox1) gene sequencing has been recommended as a new and additional taxonomic approach scuticociliate species identification. This technique is a sensitive, rapid, and specific diagnostic tool10 and has been used to identify scuticociliates named U. marinum, Pseudocohnilembus persalinus, P. longisetus, and M. avidus in various fish species, including olive flounder (Paralichthys olivaceus) and black rockfish (Sebastes schlegelii).11 As a result, in this study, we report the first identification of U. marinum (scuticociliatida) from cultured turbot (P. maxima) larvae using mitochondrial cox1 gene sequencing.
Materials and Methods
Examined fish. Turbot is cultured in the hatchery of CFRI, Turkey. About 20,000 larvae are produced annually in a seawater-based system. All examined fish were collected from CFRI Turbot Hatchery, Trabzon, Turkey. Fish were randomly sampled. Sampling was taken place in late October 2018. The water temperature, pH, and salinity recorded at the sampling time were 15.00 ˚C, 7.20, and 18.00 ppt, respectively. Thirty turbot larvae (2.00 - 4.00 g) were sampled for parasitic examination after a suspected disease. All sampled fish were examined externally and internally.
Ciliate isolation. Scuticociliatosis parasites cause many fish mortality (20.00 - 25.00% annually) in turbot hatchery. Turbot larva is especially sensitive to the pathogen. The presumptive identification of cliates was accomplished by microscopic observation of affected fish. Skin and fin tissues were dissected from moribund turbot larvae and examined via light microscopy (E400; Nikon, Tokyo, Japan). Morphological studies were also carried out using the wet Chatton-Lwoff silver nitrate impregnation method described by Foissner.12 Live ciliates from all affected fish were cultured in seawater containing 0.18% NaCl at hatchery temperature (15.00 ˚C) for ten days. The ciliates were concentrated by centrifugation at 1000 g for 5 min, and the supernatant was removed.4 In this study, the prevalence of ciliata was also calculated in sampled fish.13
DNA extraction. Fresh tissue samples of 13 ciliata affected fish were mixed in a tube, and DNA extraction and polymerase chain reaction (PCR) assays were conducted. According to the manufacturer's guidelines, ciliata genomic DNA was extracted by the DNA extraction kit (Qiagen, Hilden, Germany). The concentration and quality of DNA were assessed by a Nanodrop (ND 8000; Thermo Fisher Scientific, Waltham, USA), and the final DNA concentration was adjusted to 20.00 ng μL-1.
PCR amplification of cox 1 and sequence analysis. The presumptive identification of ciliates was made by microscopic observation of fish. Molecular identification of ciliates was performed by amplifying the mitochondrial cox1 gene by species-specific primers (Table 1) and sequence analysis of amplified fragments.11 The DNA amplification was conducted using AmpliTaq Gold Master Mix (Thermo Fisher Scientific) in a thermal cycler (Applied Biosystems, Foster City, USA). The PCR reaction components were used according to the manufacturer's instructions. The following PCR conditions were used for all primer sets: Pre-denaturation at 95.00 ˚C for 10 min, 35 cycles of 95.00 ˚C for 30 sec, 52.00 ˚C for 45 sec and 72.00 ˚C for 30 sec and a final extension at 72.00 ˚C for 7 min. Analysis of amplification product was performed using 1.00% agarose gel containing SYBR Green. DNA fragment size was evaluated using the 100-bp DNA ladder (BioBasic Inc., Ontario, Canada). The expected sizes of the strong PCR products for U. marinum, M. avidus, P. persalinus, and P. longisetus were 285, 422, 229, and 341 bp, respectively (Table 1). The sequencing reaction was performed by BigDye Terminator Cycle Sequencing kit (version 3.1; Applied Biosystems), according to the manufacturer's instructions and sequences were determined in an ABI PRISM 3500 Genetic Analyzer and ABI Prism DNA Sequencing Analysis Software (version 5.1; Applied Biosystems). The nucleotide sequences were compared with previously published data in GenBank (www.ncbi.nlm.nih.gov). Uronema marinum, Cyclidium glaucoma, and Tetrahymena sp. nucleotide sequences were aligned by CLUSTALW Multiple Sequence Alignment Program (UCD, Dublin, Ireland), and the phylo-genetic tree was constructed on MEGA software (version 10.0; Biodesign Institute, Tempe, USA).14 When designing the phylogenetic tree, Tamura-Nei substitution model,15 gamma-distributed with invariant sites for rates among sites, partial deletion for gap/missing data treatment was chosen. The phylogenetic tree was constructed by maximum likelihood method with 1000 replicates.16
Table 1.
Primers used for this study
| Organism | Primer | Sequence (5′–3′) | Size (bp) |
|---|---|---|---|
| Uronema marinum | UM-F UM-R |
AACATAGAGCATATAGAGAGTACTCTAA TTCATCCAGCTGTTGTTAATGT |
285 |
| Miamiensis avidus | MA-F MA-R |
AGTAATAATAGAACATTTAACGAATTTAATAACAC CGTCTTGTAATTAATAAATTTGTAAACGATAC |
422 |
| Pseudocohnilembus persalinus | PP-F PP-R |
TAAATCTAATCATCGTAATAATAGAGAATTGTTAG CTTATCGATACGACTAACTGCAT |
229 |
| Pseudocohnilembus longisetus | PL-F PL-R |
AAATCAAATCATAGAAATAATAGAGAATTTTTAAATG GCTCCAACACCAGTATATTTAATG |
341 |
Results
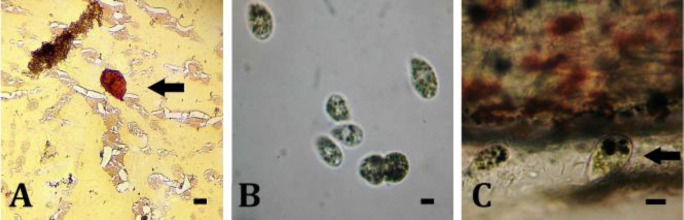
Skin lesions were observed as the most common finding in larvae with scuticociliates infections. The affected fish displayed petechial hemorrhages on various locations such as skin, operculum, and around the mouth. They also exhibited anorexia, lethargy, and darkened skin. Also, ascites in the internal organs were seen. The existence of infection caused by U. marinum, a pathogenic parasite, was identified in 13 out of 30 specimens. The mean prevalence of infected fish in this facility was calculated as 43.33%. The scuticociliates isolated from the naturally-infected fish larvae were identified by the traditional technique of light microscopy to evaluate morphological characters. The ciliate's morphological characteristics included its distinct size of approximately 40.00 - 50.00 × 20.00 - 30.00 μm and pear-like shape (Fig. 1).
Fig. 1.
Morphological characteristics of Uronema marinum (arrow) from turbot. A) Silver nitrate stain; B) Ciliates observed through light microscopy; C) A ciliata in sub-dermal layer. (Scale bars = 10.00 μm)
An expected PCR fragment size of about 285-bp was obtained (not shown), and the 251 bp mitochondrial cox1 gene sequence was aligned to the GenBank database. The sequence comparison showed 93.00% identity to the sequences of U. marinum (Accession no.: MG372368.1). Thus, based on the sequence identity, the ciliata isolated from the diseased turbot were confirmed as U. marinum.
The sequencing results obtained from the mitochondrial cox1 gene regions were deposited in GenBank database and compared with isolates from different countries registered in the database.
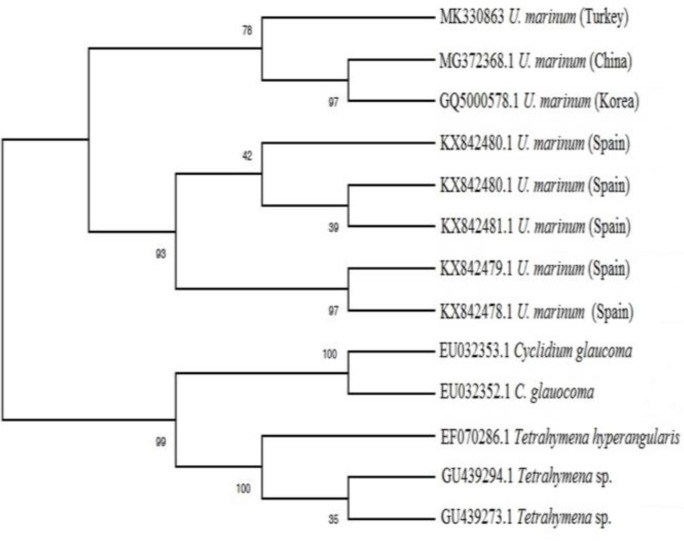
The parasite has also been deposited in GenBank databases (Accession no: MK330863). The phylogenetic tree showed U. marinum, C. glaucoma, and Tetrahymena sp. came from a common ancestor, and then U. marinum separated from its relatives. The U. marinum strains from Spain are under the same clade, while the U. marinum strain from Turkey separated from U. marinum strains belonging to Korea and China (Fig. 2).
Fig. 2.
Phylogenetic tree based on mitochondrial cytochrome cox1 gene sequence comparison, obtained by the MEGAX package program based on the maximum-likelihood method with 1000 bootstraps showing the Uronema marinum strains from different countries
Discussion
Uronema is a genus under scuticociliatida found world-wide in both freshwater and marine environments.11,17 In a previous study, new Urenema species, including Uronemella binucleata, U. filificum, and U. cymruensis have been reported from China's marine waters.18 In this study, U. marinum was identified as an etiological agent of the disease outbreak in cultured turbot in the CFRI, the first Uronema species reported in Turkey's marine waters.
Pleuronema coronatum, Cohnilembus verminus, Philaste-rides armatalis, Porpostoma notatum, Cyclidium varibonneti, Ancistrum crissum,19 Philasterides dicentrarchi,4 Uronemella filificum, U. binucleata, U. cymruensis, Pseudocohnilembus hargisi, Cyclidium citrullus,18 Miamiensis avidus,10 Pseudo-cohnilembus persalinus, P. longisetus, Uronema marinum,11 Pleuronema elegans, P. setigerum, P. grolierei and U. orientalis 17 are important ciliata species belonging to the orders scuticociliatida. The existence of ciliata with the capacity to destroy cells and tissues of fish has been known for many years. The typical features of all ciliata are tissue damage and high mortality in aquatic animals. Similarly, the results detailed in previous reports, hemorrhage in the external and internal organs, skin color changes, abnormal swimming behavior, anorexia, and lethargy were detected in affected fish. The cumulative mortality was approximately reached 25.00%.
The mitochondrial cox1 gene appears to be a new candidate for use as a DNA barcode. The particular regions of the cox1 gene are highly conserved. This case allows the design of universal primers. The cox1 gene also contains regions of the hypervariable sequence, which allows generating species-specific primers.20 In our study, U. marinum was identified based on the mitochondrial cox1 gene sequence in cultured turbot larvae. Phylogenetic tree displayed strain from Turkey diverged from strains belonging to Spain, and it is closer to strains from Korea and China. The divergence between strains can be caused due to different geographical conditions. This result indicates that the hypervariable sequences of the cox1 gene may represent a useful diagnostic tool for rapid identification of scuticociliate species.
Turbot larva is a susceptible species to stress factors, including water temperature changes, parasitic infections, and handling. Thus, in the case of parasitic infection, high mortalities must be expected in larvae culture.4,8 Various chemicals and chemotherapeutics have been used to treat scuticociliatosis throughout aquaculture production. However, the desired recovery could not be achieved. Therefore, preventive measures such as environmental stress reduction may be essential to decrease the risk of infection. To our knowledge, this study represents the first U. marinum identification in Turkey by targeting the mitochondrial cox1 gene. In our opinion, the results of this study will help to advance the systematics and molecular epidemiology of scuticociliate in Turkey.
Acknowledgments
The author would like to thank Dr. Attila Karsi (Mississippi State University, USA) and Recep Parlak (CFRI, Turkey) for their help. This research was supported by CFRI, Turkey. The author followed all applicable international, national, and/or institutional guidelines for the care and use of animals. The research permission for the animal experiment (Protocol No: 42208298-040-04-05) was received from the Animal Experiments Local Ethics Committee of CFRI, Turkey.
Conflict of interest
The author declares that there is no conflict of interest regarding the publication of this article.
References
- 1.Barber I. Parasites, behaviour and welfare in fish. Appl Anim Behav Sci. 2007;104(3-4):251–264. [Google Scholar]
- 2.Turgay E, Steinum TM, Gül AE. et al. An outbreak of scuticociliatosis in cultured common dentex (Dentex dentex) in Turkey. Bull Eur Assoc Fish Pathol. 2015;35(3):104–111. [Google Scholar]
- 3.Bradbury PC. Pathogenic ciliates. In: Hausmann K, Bradbury PC, editors. Ciliates: cells as organisms. Deer-field Beach, USA: Gustav Fischer Verlag; 1996. pp. 463–477. [Google Scholar]
- 4.Iglesias R, Paramá A, Alvarez MF, et al. Philasterides dicentrarchi (Ciliophora, Scuticociliatida) as the causative agent of scuticociliatosis in farmed turbot Scophthalmus maximus in Galicia (NW Spain) Dis Aquat Organ. 2001;46(1):47–55. doi: 10.3354/dao046047. [DOI] [PubMed] [Google Scholar]
- 5.Sterud E, Hansen MK, Mo TA. Systemic infection with Uronema like ciliates in farmed turbot, Scophthalmus maximus (L. ). J Fish Dis. 2000;23(1):33–37. [Google Scholar]
- 6.Du G, Qu L, Shang K, et al. Ciliate Uronema marinum is the causative agent of scuticociliatosis in farm raised turbot Scophthalmus maximus. J Oceanol Limnol. 2019;37:1726–1735. [Google Scholar]
- 7.Sahin T. Larval rearing of the Black Sea turbot, Scophthalmus maximus (Linnaeus, 1758), under laboratory conditions. Turk J Zool. 2001;25:447–452. [Google Scholar]
- 8.Aksungur N, Aksungur M, Akbulut B, et al. Effects of stocking density on growth performance, survival andfood conversion ratio of turbot (Psetta maxima) in the net cages on the southeastern coast of the Black Sea. Turkish J Fish Aquat Sci. 2007;7:147–152. [Google Scholar]
- 9.Kayis S, Yandi I, Altinok I, et al. Treatment by vinegar of Philasterides dicentrarchi (Ciliophora: Scuticociliatida) infestation in cultured juvenile turbot (Psetta maxima) Isr J Aquac. 2011;63:627–631. [Google Scholar]
- 10.Jung SJ, e al. Small subunit ribosomal RNA and mitochondrial cytochrome c oxidase subunit 1 gene sequences of 21 strains of the parasitic scuticociliate Miamiensis avidus (Ciliophora, Scuticociliatia) Parasitol Res. 2011;108(5):1153–1161. doi: 10.1007/s00436-010-2157-7. [DOI] [PubMed] [Google Scholar]
- 11.Whang I, Kang HS, Lee J. Identification of scuticociliates (Pseudocohnilembus persalinus, P longisetus, Uronema marinum and Miamiensis avidus) based on the cox1 sequence. Parasitol Int. 2013;62(1):7–13. doi: 10.1016/j.parint.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 12.Foissner W. Basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa. Eur J Protistol. 1991;27(4):313–330. doi: 10.1016/S0932-4739(11)80248-8. [DOI] [PubMed] [Google Scholar]
- 13.Er A, Kayış Ş. Intensity and prevalence of some crustacean fish parasites in Turkey and their molecular identification. Turk J Zool. 2015;39(6):1142–1150. [Google Scholar]
- 14.Kumar S, Stecher G, Li M, et al. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 16.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 17.Pan X, Huang J, Fan X, et al. Morphology and phylogeny of four marine scuticociliates (Protista, ciliophora), with descriptions of two new species: Pleuronema elegans spec nov and Uronema orientalis spec nov. Acta Protozool. 2015;54:31–43. [Google Scholar]
- 18.Song W, Wilbert N. Reinvestigations of three “well-known” marine scuticociliates: Uronemella filificum (Kahl, 1931) nov gen nov comb Pseudocohnilembus hargisi Evans & Thompson, 1964 and Cyclidium citrullus Cohn 1865, with description of the new genus Uronemella (Protozoa, Ciliophora, Scuticociliatida) Zool Anz. 2002;241(4):317–331. [Google Scholar]
- 19.Song W. Morphological and taxonomical studies on some marine scuticociliates from China Sea, with description of two new species, Philasteridesarmatalis sp and Cyclidium varibonneti sp (Protozoa: Ciliophora: Scuticociliatida) Acta Protozool. 2000;39:295–322. [Google Scholar]
- 20.Folmer O, Black M, Hoeh W, et al. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–299. [PubMed] [Google Scholar]