Abstract
Background
In in vitro fertilisation (IVF) with or without intracytoplasmic sperm injection (ICSI), selection of the most competent embryo(s) for transfer is based on morphological criteria. However, many women do not achieve a pregnancy even after 'good quality' embryo transfer. One of the presumed causes is that such morphologically normal embryos have an abnormal number of chromosomes (aneuploidies). Preimplantation genetic testing for aneuploidies (PGT‐A), formerly known as preimplantation genetic screening (PGS), was therefore developed as an alternative method to select embryos for transfer in IVF. In PGT‐A, the polar body or one or a few cells of the embryo are obtained by biopsy and tested. Only polar bodies and embryos that show a normal number of chromosomes are transferred.
The first generation of PGT‐A, using cleavage‐stage biopsy and fluorescence in situ hybridisation (FISH) for the genetic analysis, was demonstrated to be ineffective in improving live birth rates. Since then, new PGT‐A methodologies have been developed that perform the biopsy procedure at other stages of development and use different methods for genetic analysis.
Whether or not PGT‐A improves IVF outcomes and is beneficial to patients has remained controversial.
Objectives
To evaluate the effectiveness and safety of PGT‐A in women undergoing an IVF treatment.
Search methods
We searched the Cochrane Gynaecology and Fertility (CGF) Group Trials Register, CENTRAL, MEDLINE, Embase, PsycINFO, CINAHL, and two trials registers in September 2019 and checked the references of appropriate papers.
Selection criteria
All randomised controlled trials (RCTs) reporting data on clinical outcomes in participants undergoing IVF with PGT‐A versus IVF without PGT‐A were eligible for inclusion.
Data collection and analysis
Two review authors independently selected studies for inclusion, assessed risk of bias, and extracted study data. The primary outcome was the cumulative live birth rate (cLBR). Secondary outcomes were live birth rate (LBR) after the first embryo transfer, miscarriage rate, ongoing pregnancy rate, clinical pregnancy rate, multiple pregnancy rate, proportion of women reaching an embryo transfer, and mean number of embryos per transfer.
Main results
We included 13 trials involving 2794 women. The quality of the evidence ranged from low to moderate. The main limitations were imprecision, inconsistency, and risk of publication bias.
IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses
Polar body biopsy
One trial used polar body biopsy with array comparative genomic hybridisation (aCGH). It is uncertain whether the addition of PGT‐A by polar body biopsy increases the cLBR compared to IVF without PGT‐A (odds ratio (OR) 1.05, 95% confidence interval (CI) 0.66 to 1.66, 1 RCT, N = 396, low‐quality evidence). The evidence suggests that for the observed cLBR of 24% in the control group, the chance of live birth following the results of one IVF cycle with PGT‐A is between 17% and 34%. It is uncertain whether the LBR after the first embryo transfer improves with PGT‐A by polar body biopsy (OR 1.10, 95% CI 0.68 to 1.79, 1 RCT, N = 396, low‐quality evidence). PGT‐A with polar body biopsy may reduce miscarriage rate (OR 0.45, 95% CI 0.23 to 0.88, 1 RCT, N = 396, low‐quality evidence). No data on ongoing pregnancy rate were available. The effect of PGT‐A by polar body biopsy on improving clinical pregnancy rate is uncertain (OR 0.77, 95% CI 0.50 to 1.16, 1 RCT, N = 396, low‐quality evidence).
Blastocyst stage biopsy
One trial used blastocyst stage biopsy with next‐generation sequencing. It is uncertain whether IVF with the addition of PGT‐A by blastocyst stage biopsy increases cLBR compared to IVF without PGT‐A, since no data were available. It is uncertain if LBR after the first embryo transfer improves with PGT‐A with blastocyst stage biopsy (OR 0.93, 95% CI 0.69 to 1.27, 1 RCT, N = 661, low‐quality evidence). It is uncertain whether PGT‐A with blastocyst stage biopsy reduces miscarriage rate (OR 0.89, 95% CI 0.52 to 1.54, 1 RCT, N = 661, low‐quality evidence). No data on ongoing pregnancy rate or clinical pregnancy rate were available.
IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis
Eleven trials were included in this comparison. It is uncertain whether IVF with addition of PGT‐A increases cLBR (OR 0.59, 95% CI 0.35 to 1.01, 1 RCT, N = 408, low‐quality evidence). The evidence suggests that for the observed average cLBR of 29% in the control group, the chance of live birth following the results of one IVF cycle with PGT‐A is between 12% and 29%. PGT‐A performed with FISH probably reduces live births after the first transfer compared to the control group (OR 0.62, 95% CI 0.43 to 0.91, 10 RCTs, N = 1680, I² = 54%, moderate‐quality evidence). The evidence suggests that for the observed average LBR per first transfer of 31% in the control group, the chance of live birth after the first embryo transfer with PGT‐A is between 16% and 29%. There is probably little or no difference in miscarriage rate between PGT‐A and the control group (OR 1.03, 95%, CI 0.75 to 1.41; 10 RCTs, N = 1680, I² = 16%; moderate‐quality evidence). The addition of PGT‐A may reduce ongoing pregnancy rate (OR 0.68, 95% CI 0.51 to 0.90, 5 RCTs, N = 1121, I² = 60%, low‐quality evidence) and probably reduces clinical pregnancies (OR 0.60, 95% CI 0.45 to 0.81, 5 RCTs, N = 1131; I² = 0%, moderate‐quality evidence).
Authors' conclusions
There is insufficient good‐quality evidence of a difference in cumulative live birth rate, live birth rate after the first embryo transfer, or miscarriage rate between IVF with and IVF without PGT‐A as currently performed. No data were available on ongoing pregnancy rates. The effect of PGT‐A on clinical pregnancy rate is uncertain.
Women need to be aware that it is uncertain whether PGT‐A with the use of genome‐wide analyses is an effective addition to IVF, especially in view of the invasiveness and costs involved in PGT‐A. PGT‐A using FISH for the genetic analysis is probably harmful.
The currently available evidence is insufficient to support PGT‐A in routine clinical practice.
Plain language summary
Preimplantation genetic testing for abnormal chromosome numbers for couples undergoing in vitro fertilisation
Review question
Does preimplantation genetic testing for abnormal chromosome numbers improve the chances of a pregnancy followed by a live‐born baby?
Background
In in vitro fertilisation (IVF) with or without intracytoplasmic sperm injection (ICSI), the selection of the best embryo(s) for transfer is mainly based on morphological assessment of the embryos, which includes the number of cells, the regularity of cells, and the presence of cell fragments. Unfortunately, almost two‐thirds of couples do not get pregnant even after transfer of ‘good quality’ embryos. One of the presumed causes is that such embryos have an abnormal number of chromosomes (aneuploidy). Preimplantation genetic testing for aneuploidy (PGT‐A) is a technique used to analyse the number of chromosomes present in IVF embryos. In PGT‐A, a polar body (a waste product of maternal meiosis),or one or a few cells of the embryo are obtained by biopsy and tested. Only polar bodies or embryos with a normal number of chromosomes in each cell, so‐called 'euploid embryos', are transferred into the uterus. The idea is that this will increase the live birth rate per started IVF cycle. Previous studies on PGT‐A that used a genetic analysis technique called fluorescence in situ hybridisation (FISH) found PGT‐A to be ineffective in improving live birth rates. Since then, new methodologies and techniques in PGT‐A have been developed that perform the procedure on polar bodies or other stages of embryo development and use different methods of genetic analysis (array comparative genomic hybridisation (aCGH) or next‐generation sequencing (NGS)).
We compared the benefits and risks of IVF with and without PGT‐A, performed with different techniques at different stages: polar body or other stage of embryo development.
Study characteristics
We included 13 randomised controlled trials (a type of study in which participants are assigned to one of two or more treatment groups using a random method) involving a total of 2794 women. The evidence is current to September 2019.
Key results
IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses
Polar body biopsy
There was not enough evidence to determine whether there is any difference in the cumulative live birth rate (cLBR) or live birth rate (LBR) after the first embryo transfer with the addition of PGT‐A using polar body biopsy to IVF. There may be a reduction in the miscarriage rate with the addition of PGT‐A. No studies reported on ongoing pregnancy rate. It is also uncertain whether the addition of PGT‐A with polar body biopsy to IVF leads to more clinical pregnancies.
Blastocyst stage biopsy
No studies reported on cLBR after blastocyst stage biopsy. It is uncertain if the addition of PGT‐A with biopsy in the blastocyst stage improves LBR after the first embryo transfer or reduces the miscarriage rate. No studies reported on ongoing pregnancy rate or clinical pregnancy rate.
IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis
The addition of PGT‐A by FISH does not increase cLBR where FISH is used for the genetic analysis. Live birth rate after the first embryo transfer is probably reduced by the addition of PGT‐A. There is probably little or no difference in miscarriage rates between IVF with and without PGT‐A using FISH. PGT‐A using FISH may reduce ongoing pregnancies and probably reduces clinical pregnancies.
Quality of the evidence
The quality of the evidence ranged from low to moderate. The main limitations of the evidence were the limited number of studies and events, inconsistency in the estimates between studies, and indications that results may be biased because not all eligible studies have been published.
Summary of findings
Background
Description of the condition
One in six couples experience subfertility, defined as the failure to conceive after one year of unprotected intercourse, at least once during their reproductive lifetime (Dyer 2016). In vitro fertilisation (IVF) with and without intracytoplasmic sperm injection (ICSI) has evolved as the intervention of choice to help these couples conceive.
In IVF, selection of the most competent embryo(s) for transfer is mainly based on morphological criteria, such as the number of pronuclei, the number and regularity of blastomeres, and the percentage of fragmentation in cleavage stage embryos, and assessment of trophectoderm, inner cell mass, and expansion in the blastocyst stage of embryo development. Even so, two‐thirds of women do not reach pregnancy, whilst some others might result in a miscarriage, after transfer of embryos that are morphologically of good quality (De Geyter 2018).
One of the suggested causes is that such morphologically normal embryos contain an abnormal number of chromosomes (aneuploidies). Those embryos have been regarded as the main reason for implantation failure, miscarriage, or recurrent miscarriages and prolonged time to pregnancy in IVF.
Since the early 1980s, many reports have been published showing numerical chromosome abnormalities in morphologically normal human cleavage stage embryos (Angell 1983; Munné 1993; Benadiva 1996; Delhanty 1997). Since aneuploid embryos are not expected to develop to term, preimplantation genetic testing for aneuploidies (PGT‐A) was introduced in 1995 to improve live birth rates (Verlinsky 1995; Sermon 2016).
Description of the intervention
The previous terminology of 'preimplantation genetic screening' (PGS) has been replaced by 'preimplantation genetic testing', or PGT, following a revision of terminology used in infertility care (Zegers‐Hochschild 2017). PGT is defined as a test performed to analyse the DNA from mature oocytes (polar bodies) or embryos (cleavage or blastocyst stage) for determining genetic abnormalities. This includes PGT for chromosomal structural rearrangements (PGT‐SR), PGT for monogenic/single‐gene defects (PGT‐M), and PGT for aneuploidies (PGT‐A). Although the technology used in PGT‐A and PGT‐SR is nearly identical, PGT‐A and PGT‐SR have completely different indications. PGT‐A aims to improve treatment outcome in subfertile couples undergoing an IVF treatment, whereas PGT‐M and PGT‐SR aim to prevent the birth of affected children in fertile couples with a high risk of transmitting genetic disorders.
In PGT‐A, embryos created in vitro are analysed for aneuploidies, and only those that show a normal number of chromosomes, that is those that are euploid for the chromosomes tested, are transferred into the uterine cavity. There are different approaches to obtain nuclear material for this genetic analysis.
One approach is aspiration of the first and second polar body from the unfertilised oocyte or the zygote (Verlinsky 1995; Montag 2009; Geraedts 2016; Verpoest 2018). Polar bodies are a waste product of maternal meiosis. Mitotic errors and paternally derived meiotic errors and mutations cannot be detected from polar bodies. Polar body biopsy can be an alternative to embryo biopsy due to regulations that prohibit embryo biopsy in specific regions. Another approach is removal of one or two blastomeres from embryos at the early cleavage stage (Handyside 1989). A third approach is removal of trophectoderm cells at the blastocyst stage (Dokras 1990). The amount of DNA is higher since multiple cells are analysed. Finally, new sources of embryonic genetic material can be obtained by blastocyst fluid aspiration, Gianaroli 2014; Capalbo 2018, or spent embryo culture medium analysis, where cell‐free genomic DNA is obtained (in a non‐invasive way) from the embryo culture medium, may potentially be used for genetic testing (Hammond 2017; Vera‐Rodriquez 2018; Belandres 2019; Leaver 2020).
The method of genetic analysis can be limited to a certain number of chromosomes, mostly when using fluorescence in situ hybridisation (FISH) for the analysis, or it can encompass almost all or all chromosomes in genome‐wide technologies. FISH technology is based on the use of specific DNA probes that are labelled with distinctive fluorochromes; following hybridisation, results are visualised via fluorescence microscopy. Multiple genome‐wide technologies are described; array‐based comparative genomic hybridisation (aCGH) (Wells 2008; Geraedts 2011), single nucleotide polymorphism (SNP) array (van Uum 2012), or next generation sequencing (NGS) (Treff 2013). The principle of aCGH is a molecular cytogenetic technique for the detection of chromosomal copy number changes. aCGH compares the genome against a reference DNA sample, utilising the same principles as FISH. The application of NGS for the detection of copy number changes differs from aCGH by using direct reads of genomic sequencing fragments and their quantification according to sequence read numbers instead of a signal intensity comparison between fluorescently labelled test and reference DNA samples.
PGT‐A was first recommended and carried out for the following indications: advanced maternal age (Gianaroli 1999; Munné 1999; Kahraman 2000; Obasaju 2001; Munné 2003; Montag 2004; Platteau 2005), repeated IVF failure (Gianaroli 1999; Kahraman 2000; Munné 2003; Pehlivan 2003; Wilding 2004), recurrent miscarriage (Pellicer 1999; Rubio 2003; Munné 2005; Rubio 2005), and severe male factor (Silber 2003; Platteau 2004). Later PGT‐A was also offered to younger women with a good prognosis for a pregnancy, as high aneuploidy rates were found in their embryos as well (Baart 2006; Goossens 2009).
How the intervention might work
Initially PGT‐A was introduced based on the hypothesis that selection of an aneuploid embryo from a cohort of embryos available for transfer in an IVF treatment would lead to better clinical outcomes (Verlinsky 1995). Part of the embryos available in IVF were reported to be aneuploid, and aneuploidies were expected to result in arrested development, implantation failure, or miscarriage (Wilton 2002). Nowadays, the main reason for choosing to use PGT‐A is the hypothesis that PGT‐A could reduce the chance of miscarriage and shorten time to pregnancy leading to live birth (Sermon 2016; Cimadomo 2020). PGT‐A can never increase the cumulative live birth rate, as it selects out the aneuploid embryos that would likely not have resulted in a pregnancy (Paulson 2017).
First‐generation PGT‐A was characterised by biopsy at the cleavage stage of embryo development using fluorescence in situ hybridisation (FISH) for the genetic analysis; however, it was demonstrated to be ineffective in improving IVF live birth rates and reducing miscarriage rates (Twisk 2006; Mastenbroek 2011). The mosaic nature of human preimplantation embryos, which means that not all cells of an embryo have the same chromosomal constitution, and the limited capability and accuracy of the FISH analysis, are considered the main reasons for this failure (Scriven 2010; van Echten‐Arends 2011; Mastenbroek 2014; Taylor 2014). Consequently, PGT‐A at the blastocyst stage with the use of genome‐wide analyses became common practice, as mosaicism was claimed to be less of a problem at later stages of embryo development, or could at least be noticed as multiple cells are available for the analysis at the blastocyst stage, and genome‐wide analyses were claimed to be more accurate.
PGT‐A and IVF is a controversial subject, as evidence has appeared undermining the rationale of PGT‐A (Rosenwaks 2018; Mochizuki 2020). It was demonstrated that mosaic embryos could also lead to healthy babies (Greco 2015). Subsequently it was proposed that mosaic embryos, which till then were in most cases discarded, could now be transferred to the uterus, although perhaps with a lower priority than euploid embryos (Cram 2019). An assumed reason is that there is a self‐correction process of aneuploidies in preimplantation embryos, allowing mosaic embryos to result in healthy live births, although perhaps with lower efficiency than fully euploid embryos (Bolton 2016; Singla 2020). But next to mosaicism, there is still a debate surrounding the accuracy of analysis methods, not only since healthy live births have been reported after transfer of aneuploid embryos (Patrizio 2019), but also since there is increasing evidence that the genome‐wide analyses currently being used have a high false‐positive rate (Popovic 2018; Lawrenz 2019; Popovic 2020).
Why it is important to do this review
It has been demonstrated that the former approach for PGT‐A, that is cleavage stage biopsy with the use of FISH for genetic analysis, is ineffective in improving IVF live birth rates and in reducing miscarriage rates (Twisk 2006). New approaches of PGT‐A have been developed that perform the procedure on polar bodies or other stages of embryo development and use different methods of genetic analysis.
These new approaches of PGT‐A have been introduced into routine clinical practice and are often offered; however, their effectiveness remains unclear (Mastenbroek 2014). We therefore undertook and updated this systematic review to investigate whether there is a difference in cumulative live birth rate, live birth per first embryo transfer, miscarriage rate, ongoing pregnancy rate, or clinical pregnancy rate. This involved comparing IVF with PGT‐A versus IVF without PGT‐A.
Objectives
To evaluate the effectiveness and safety of PGT‐A in women undergoing an IVF treatment.
Methods
Criteria for considering studies for this review
Types of studies
Randomised controlled trials (RCT) were eligible for inclusion in the review. We excluded non‐randomised studies and quasi‐randomised trials. We also excluded cross‐over trials, which are not appropriate in this context.
Due to bias by design in favour of PGT‐A, we excluded RCTs that only reported results on women reaching a euploid embryo transfer, studies where PGT‐A was not performed on the full cohort of embryos, and studies where multiple oocyte retrievals were allowed before a transfer was attempted without the possibility to extract data per ovum pick‐up.
Types of participants
Women undergoing IVF treatment with or without PGT‐A offered for all suggested indications or a combination of indications, that is:
advanced maternal age (AMA);
repeated IVF failure (RIF);
recurrent miscarriage;
testicular sperm extraction (TESE‐ICSI);
good‐prognosis patients.
Types of interventions
We compared IVF with PGT‐A versus IVF without PGT‐A.
We included the following two randomised comparisons.
IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses
IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis
In these two randomised comparisons, different subgroups exist between the moment of biopsy in embryo development, as follows.
Polar body biopsy
Cleavage stage biopsy
Blastocyst stage biopsy
Types of outcome measures
Primary outcomes
Cumulative live birth rate per woman, defined as the birth of a living child after 20 weeks of gestation, after one IVF cycle, including the results of cryopreserved thawed embryo transfers.
Secondary outcomes
Live birth rate per woman after the first embryo transfer, defined as the birth of a living child after 20 weeks of gestation after the first embryo transfer in an IVF cycle.
Miscarriage rate per woman and per pregnancy.
Ongoing pregnancy per woman, defined as ultrasound‐confirmed evidence of a gestation sac with fetal heart motion at 12 weeks.
Clinical pregnancy per woman, defined by the presence of an intrauterine gestational sac or the presence of intrauterine gestational sac with fetal heartbeat.
Multiple pregnancy per woman and per live birth.
Proportion of women reaching embryo transfer.
Mean number of embryos per transfer.
Search methods for identification of studies
We searched for all published and unpublished RCTs on PGT‐A versus non‐PGT‐A, with no language or date restriction and in consultation with the Cochrane Gynaecology and Fertility Group (CGF) Information Specialist.
Electronic searches
We searched the following electronic databases for relevant trials:
the Cochrane Gynaecology and Fertility Group (CGF) Specialised Register of Controlled Trials; ProCite platform, searched on 9 September 2019 (Appendix 1);
Cochrane Central Register of Controlled Trials (CENTRAL); Ovid platform, searched on 9 September 2019 (Issue August 2019) (Appendix 2);
MEDLINE Epub Ahead of Print, In‐Process & Other Non‐Indexed Citations; Ovid platform, searched from 1946 to 9 September 2019 (Appendix 3);
Embase; Ovid platform, searched from 1980 to 9 September 2019 (Appendix 4);
PsycINFO; Ovid platform, searched from 1806 to 9 September 2019 (Appendix 5);
CINAHL (Cumulative Index to Nursing and Allied Health Literature); EBSCO platform, searched from 1961 to 9 September 2019 (Appendix 6).
We combined the MEDLINE search with the Cochrane Highly Sensitive Search Strategy for identifying randomised trials described in the Cochrane Handbook for Systematic Reviews of Interventions (Chapter 6, 6.4.11; Lefebvre 2011). The Embase, PsycINFO, and CINAHL searches are combined with trial filters developed by the Scottish Intercollegiate Guidelines Network (SIGN) (www.sign.ac.uk/what-we-do/methodology/search-filters/).
We also searched the following other electronic sources of trials.
Trial registers for ongoing and registered trials, searched on 9 September 2019:
US National Institutes of Health Ongoing Trials Register ClinicalTrials.gov (www.clinicaltrials.gov);
World Health Organization International Clinical Trials Registry Platform (www.who.int/trialsearch/Default.aspx).
Note: it is now mandatory for Cochrane Reviews to include searches of trial registers.
LILACS (Latin American and Caribbean Health Science Information database; from 1982 to 9 September 2019) and other Spanish and Portuguese language databases found in the Virtual Health Library Regional Portal (VHL) (bvsalud.org/portal/?lang=en) (the right‐hand drop down box allows you to filter out MEDLINE records).
Searching other resources
We searched the following conference abstracts:
American Society for Reproductive Medicine and Canadian Fertility and Andrology Society (ASRM/CFAS) Conjoint Annual Meeting (2018), Abstracts of the Scientific Oral and Poster Sessions, Program Supplement;
European Society of Human Reproduction and Embryology (ESHRE) Annual Meeting (2018), Abstracts of the Scientific Oral and Poster Sessions, Program Supplement.
We handsearched the references cited in all obtained studies. We searched PubMed and Google for any recent trials that had not yet been indexed in MEDLINE.
We contacted experts in the field to obtain additional data. We contacted original authors for clarification and further data if the trial report was unclear.
We did not perform a separate search for adverse effects of PGT‐A. We considered adverse effects described in the studies only.
Data collection and analysis
Selection of studies
After an initial screen of titles and abstracts retrieved by the search, conducted by the CGF Information Specialist (Marian Showell), two review authors (SC and MZ) retrieved the full texts of all potentially eligible studies. We used the software program Covidence to manage the screening of titles and abstracts and to generate the PRISMA flow diagram (Covidence). Two review authors (SC and MZ) independently examined the full texts against the inclusion criteria to determine their eligibility. In the case of disagreement between review authors, a third review author (SM) was consulted to establish consensus on whether to include the trial or not. We documented the selection process with a PRISMA flow chart (Figure 1).
1.
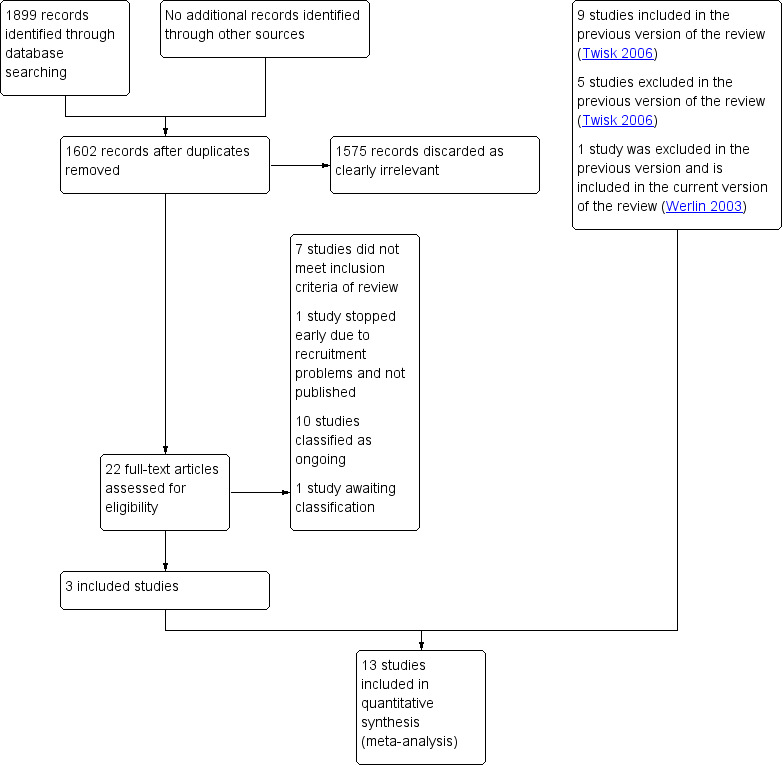
Study flow diagram.
Data extraction and management
Two review authors (SC and MZ) independently extracted the outcome data and information on location, clinical and design details, funding, and participants. In the case of disagreement between review authors, a third review author (SM) was consulted to establish consensus. Any differences were resolved by discussion. Details of the studies are provided in the Characteristics of included studies tables. Studies that appeared to meet the inclusion criteria but were excluded from the review are presented in the Characteristics of excluded studies tables along with the reasons for their exclusion in brief.
Assessment of risk of bias in included studies
Two review authors (SC and MZ) independently assessed the included studies for risk of bias using the Cochrane 'Risk of bias' tool (Higgins 2011). The tool addresses the following domains:
selection bias (random sequence generation, allocation concealment);
performance bias (blinding of participants and personnel);
detection bias (blinding of outcome assessors);
attrition bias (incomplete outcome data);
reporting bias (selective reporting);
other forms of bias such as selective reporting of subgroups, or potential influence from funders.
In case of disagreement between review authors, a third review author (EK) was consulted to establish consensus. We assigned judgements to each of these domains, as recommended in the Cochrane Handbook for Systematic Reviews of Interventions (Higgins 2011). Special attention was given to selective reporting, as it affects the internal validity of an individual study. Studies might not present data per woman starting an IVF cycle. This information is described in the Characteristics of included studies table and Figure 2 and Figure 3 and provides a context for discussing the reliability of the results. Our conclusions are presented in the 'Risk of bias' table.
2.

Risk of bias graph: review authors' judgements about each risk of bias item presented as percentages across all included studies.
3.

Risk of bias summary: review authors' judgements about each risk of bias item for each included study.
Measures of treatment effect
All of our outcomes represented dichotomous data (e.g. live birth rates). We used the numbers of events in the control and intervention groups of each study to calculate Mantel‐Haenszel odds ratios (ORs). We reversed the direction of effect of individual studies if required to ensure consistency across trials. We used mean difference (MD) for continuous outcomes. We presented 95% confidence intervals (CIs) for all outcomes. We assessed whether the estimates calculated in the review for individual studies were compatible in each case with the estimates reported in the study publications.
Unit of analysis issues
We analysed the data per woman randomised. We also analysed per clinical pregnancy for miscarriage. Data that did not permit valid analysis (e.g. per woman or per ovum pick‐up) were excluded. We counted multiple pregnancy as one event; this outcome was also analysed per live birth.
Dealing with missing data
We analysed the data on an intention‐to‐treat (ITT) basis as far as possible (i.e. including all randomised participants in the analysis in the groups to which they had been randomised). We anticipated that trials conducted over 10 years ago might not have data on cumulative live birth rates. When there was insufficient information in the published report, we attempted to contact the authors of the included studies for clarification. If missing data became available, we included these in the analysis. When we were unable to obtain these data, we undertook imputation of individual values for cumulative live birth rate and live birth rate per first embryo transfer only. Live birth was assumed not to have occurred in participants without a reported outcome. For other outcomes, we analysed only the available data.
Assessment of heterogeneity
We considered whether the clinical and methodological characteristics of the included studies were sufficiently similar for meta‐analysis to provide a meaningful summary. We assessed statistical heterogeneity by measuring the I² statistic. We assumed that there was substantial heterogeneity when I² was calculated as greater than 50% (Higgins 2011).
Assessment of reporting biases
We planned that if more than 10 studies were identified, we would produce a funnel plot to evaluate the risk of reporting bias, to explore the possibility of small‐study effects (a tendency for estimates of the intervention effect to be more beneficial in smaller studies) (Chaimani 2013). In view of the difficulty of detecting and correcting for publication bias and other reporting biases, we aimed to minimise their potential impact by ensuring a comprehensive search for eligible studies and by being alert to duplication of data.
Data synthesis
When multiple studies were available for a similar comparison, we used Review Manager 2014 software to perform the meta‐analyses, employing the Mantel‐Haenszel method with a fixed‐effect model; otherwise, results from trials that could not be combined were presented in data tables in a narrative format.
Subgroup analysis and investigation of heterogeneity
We planned that when sufficient data were available, we would perform subgroup analyses of the following subgroups to determine the potential causes of heterogeneity.
Polar body biopsy
Cleavage stage biopsy
Blastocyst stage biopsy
If we detected substantial heterogeneity, we would explore it by employing the random‐effects model. We aimed to take any statistical heterogeneity into account when interpreting the results, especially if there was any variation in the direction of effect.
We did not pool the results per subgroup for the comparison ‘IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses’, since the stages of biopsy are different.
Sensitivity analysis
We conducted sensitivity analyses for the primary outcome. These analyses included consideration of whether the review conclusions would have differed if:
the summary effect measure had been risk ratio rather than odds ratio;
eligibility had been restricted to studies with low risk of bias for randomisation and allocation concealment.
Overall quality of the body of evidence: 'Summary of findings' tables
We prepared 'Summary of findings' tables using GRADEpro GDT and Cochrane methods. These tables evaluate the overall quality of the body of evidence for the main review outcomes (cumulative live birth, live birth rate after the first embryo transfer, miscarriage rate per woman, ongoing pregnancy rate, and clinical pregnancy rate) for the two main review comparisons (IVF with PGT‐A versus IVF without PGT‐A with use of genome‐wide analyses; and IVF with PGT‐A versus IVF without PGT‐A with use of FISH for genetic analysis). We assessed the quality of the evidence using the GRADE criteria: risk of bias, consistency of effect, imprecision, indirectness, and publication bias. Two review authors (SC and MZ) independently made judgements about evidence quality (high, moderate, low, or very low); in the case of disagreement between review authors, a third review author (EK) was consulted to establish consensus. Judgements were justified, documented, and incorporated into the reporting of results for each outcome.
Results
Description of studies
Results of the search
For this update, we screened 1899 titles and identified 22 articles from searching electronic databases and other resources. From these 22 studies, we included three new studies, nine studies were earlier included. One study was excluded in the previous version of the review, but is included in this updated version (Werlin 2003).12 articles were excluded because they did not meet the inclusion criteria of the review; the reasons for their exclusion are shown in Characteristics of excluded studies. One of these trials was stopped early due to recruitment problems and is not published (NCT02265614). Ten other studies are ongoing and documented in the Characteristics of ongoing studies section of the review. One study is awaiting classification (see Characteristics of studies awaiting classification). Consequently, we included 13 studies with a total of 2794 participants in the review. Details of the screening and selection process are shown in the PRISMA study flow diagram (Figure 1).
Included studies
Study design and setting
We included a total of 13 studies in this update review, nine studies from the previous version and three new studies. One earlier excluded study was included in this version of the review (Werlin 2003). This study was previously excluded for the reason that only biochemical pregnancy rates were reported, but it was possible to extract data for one secondary outcome, therefore this study is now included.
A couple was offered one treatment cycle in 10 trials, Werlin 2003; Staessen 2004; Blockeel 2008; Hardarson 2008; Jansen 2008; Staessen 2008; Meyer 2009; Schoolcraft 2009; Rubio 2013; Munné 2019, and a maximum of three cycles in one trial (Mastenbroek 2007). In one trial a non‐randomised treatment cycle was offered to participants in the PGT‐A group if in the first treatment cycle all oocytes were aneuploid (Verpoest 2018). Eleven participants started a second cycle, 10 of which had ICSI in a second cycle, and PGT‐A was performed for nine of them. Of the nine evaluated participants, six did not have any euploid embryos in this second cycle. The authors kindly provided us data about those outcomes, which permitted an analysis per first treatment cycle (Verpoest 2018 [pers comm]). In one trial a couple could participate in the study several times with independent randomisation for each cycle (Debrock 2010). The authors of the Twisk 2006 Cochrane Review provided data for this trial, in which each participant was only included in one treatment group, by personal communication, and we used them again in this update (DeBrock 2010 [pers comm]).
Rubio 2013 published one paper where two studies are described. In the first study patients with AMA were included, and in the second study patients with RIF could participate. We only used the data for women with RIF in this review. In the study group of participants with AMA, a second IVF treatment cycle was offered to participants before randomisation, therefore no data per started cycle could be extracted.
In three trials one embryo was transferred (Jansen 2008; Staessen 2008; Munné 2019); in one trial two embryos were transferred if there were two embryos available (Mastenbroek 2007); in two trials one or two embryos were transferred according to the policy per centre, participant wishes, and the availability of normal embryos (Hardarson 2008; Verpoest 2018); and in one trial up to three embryos were transferred (Blockeel 2008). In one trial the number of embryos transferred depended on the age of the woman, namely up to three blastocysts when the woman was between 37 and 39 years old and up to a maximum of six blastocysts if the woman was 40 years of age or older (Staessen 2004). In one trial a maximum of two to three embryos were transferred before 1 July 2003, and only one embryo after 1 July 2003 in the first IVF attempt in women younger than 36 years (Debrock 2010). In two trials the transfer policy was not described (Meyer 2009; Schoolcraft 2009).
All trials performed fresh embryo transfers, except for one recent trial where a freeze‐all strategy was used (Munné 2019). Data on cryopreservation and pregnancies originating from frozen‐thawed embryos were available in four trials (Mastenbroek 2007; Debrock 2010; Rubio 2013; Verpoest 2018). In one of these trials only one embryo transfer was performed in the PGT‐A group and two in the control group, although many more embryos were cryopreserved (Debrock 2010). In this study, one pregnancy was obtained in the PGT‐A group after a mixed transfer of both a non‐biopsied embryo (from a previous IVF cycle) and a biopsied embryo, so no conclusion regarding the origin of the embryo could be made. In this study no pregnancies were obtained in the control group.
In all of the included studies, in the control group the morphologically best embryos were transferred, and in the intervention group embryos that were found to be chromosomally normal were transferred. In two studies undetermined embryos with good morphologic features were transferred if no chromosomally normal embryos with good morphologic features were available (Mastenbroek 2007; Verpoest 2018); in the other studies undetermined embryos were not transferred. Although in one study, one embryo was transferred in which no result was obtained for chromosomes 16 and 18 due to technical difficulties, and one embryo was transferred in which the chromosomal pattern was only evaluated in one nucleus, whilst this study normally removed and investigated two embryos (Debrock 2010).
The sample size was based on a power calculation in 10 trials (Staessen 2004; Mastenbroek 2007; Blockeel 2008; Hardarson 2008; Jansen 2008; Staessen 2008; Meyer 2009; Debrock 2010; Verpoest 2018; Munné 2019). In four trials the calculated number of inclusions was not reached. In one trial the study was ended prematurely because an interim analysis showed such a lower implantation rate for the PGT‐A group that it was considered unethical to continue (Meyer 2009); in one trial the study was ended prematurely because an interim analysis showed futility (Staessen 2008); and a further trial was ended prematurely because the trend was opposite to that required to disprove the null hypothesis and because in the control group many more cryo‐stored blastocysts were accumulating (Jansen 2008). In one trial the study was ended prematurely because an interim analysis showed a very low conditional power of superiority for the primary outcome (Hardarson 2008). In one trial the targeted sample size was not reached because of suboptimal recruitment; however, the included sample allowed a 90% power to detect the targeted increase (Verpoest 2018). In three studies the power calculation was based on embryos instead of women (Staessen 2004; Blockeel 2008; Debrock 2010).
In the Staessen 2004 study, 400 women were randomised, with 200 assigned to the PGT‐A group and 200 to the control group. In the PGT‐A group one woman did not fulfil the inclusion criteria, whilst in the control group 10 women did not fulfil the inclusion criteria, therefore a total of 199 versus 190 women were correctly assigned to the treatment and control group respectively. In 51 women in the treatment group and 49 women in the control group ovum pick‐up was not performed. In the original article these women were excluded from the analysis, but we included these women in an ITT analysis, therefore we included from this article a total of 199 women in the PGT‐A group and 190 women in the control group.
In the Blockeel 2008 study, 95 women were randomised to the PGT‐A group and 105 women were randomised to the control group. In the PGT‐A group eight women did not fulfil the inclusion criteria, whilst in the control group 10 women did not fulfil the inclusion criteria; we excluded these from the meta‐analysis. The authors excluded nine women in the PGT‐A group and 11 women in the control group because of wrong allocation and a spontaneous pregnancy, therefore a total of 87 versus 95 women were correctly assigned to the treatment and the control group respectively. Nine women in the PGT‐A group and 10 women in the control group were wrongly allocated. These women should have been included in an ITT analysis; however, this was not possible because the results of their treatment are unknown to us. The study authors further excluded six women in the PGT‐A group and 18 women in the control group because of insufficient ovarian response, stop further fertility treatment, and spontaneous pregnancy. These women should have been included in an ITT analysis, but this was not possible for the same reason described above. Consequently, from this article we included a total of 72 women in the PGT‐A group and 67 women in the control group.
In the Meyer 2009 study, four women were excluded from the analysis because no embryo transfer was performed for personal reasons. In the original article these women were excluded from the analysis, but we included these women in an ITT analysis, therefore from this article we included 23 women in the PGT‐A group and 24 women in the control group.
In the Debrock 2010 study, 52 women were randomised to the PGT‐A group and 52 women to the control group. However, women could be included several times with independent randomisation for each cycle, which introduces a bias since these cycles are not independent. After excluding these cycles, that is when each woman could be included only once, 44 women could be included in the PGT‐A group and 50 women in the control group (DeBrock 2010 [pers comm]). Fifteen women in the PGT‐A group and 28 women in the control group did not receive the intended treatment; we included these women in an ITT analysis, therefore from this study we included 44 women in the PGT‐A group and 50 women in the control group.
One study reported only percentages and used transfers and pregnancies as units of analysis, therefore we recalculated the numbers per participant for the various outcomes (Schoolcraft 2009). From this article we included 32 women in the PGT‐A group and 30 women in the control group.
Types of participants
PGT‐A was performed for the indication advanced maternal age in seven studies (Werlin 2003; Staessen 2004; Mastenbroek 2007; Hardarson 2008; Schoolcraft 2009; Debrock 2010; Verpoest 2018); in good‐prognosis patients in five studies (Werlin 2003; Jansen 2008; Staessen 2008; Meyer 2009; Munné 2019); and for the indication repeated IVF failure in three studies (Werlin 2003; Blockeel 2008; Rubio 2013).
We included seven studies for the indication advanced maternal age. Advanced maternal age was defined as 37 years or higher (Staessen 2004), 35 years or higher (Schoolcraft 2009; Debrock 2010), 36 to 40 years (Verpoest 2018), 35 till 41 years (Mastenbroek 2007), 38 years or higher (Hardarson 2008), and 39 years or higher (Werlin 2003). Other inclusion criteria in these studies were: normal karyotype of both partners (Staessen 2004), absence of any type of hereditary condition in the patients or partners personal and family history (Verpoest 2018), body mass index (BMI) between 18 and 30 kg/m² (Verpoest 2018), need for ICSI with motile sperm (Staessen 2004), at least two fertilised oocytes one day after ovum pick‐up (Debrock 2010), at least two 6‐cell stage embryos on day 3 (Debrock 2010), at least five 6‐cell stage embryos with no more than 15% fragmentation on day 3 (Schoolcraft 2009), no previous failed IVF cycles (Mastenbroek 2007), not three or more previous failed IVF cycles (Verpoest 2018), no objection to double embryo transfer (Mastenbroek 2007; Verpoest 2018), and at least three embryos of good morphological quality if double embryo transfer was performed, or at least two embryos of good morphological quality if single embryo transfer was performed (Hardarson 2008).
We included five studies for the indication good‐prognosis patients. Good‐prognosis patients were defined in one trial as patients below 39 years, with normal ovarian reserve, BMI below 30 kg/m², presence of ejaculated sperm, a normal uterus, no more than two previous failed IVF cycles, and at least four embryos containing at least 5 cells with less than 40% fragmentation (Meyer 2009). In another trial, good‐prognosis patients were defined as women below 36 years with the need for ICSI with motile sperm and a normal karyotype of both partners (Staessen 2008). In the third trial, good‐prognosis patients were defined as patients below 38 years, with no objection to single embryo transfer, in their first or second IVF attempt with no cycles cancelled because of poor response. Additional criteria were: no fewer than eight ovarian follicles over 1 cm in diameter at day 8 to 10 of stimulation, at least four embryos with seven or more cells on day 3 of culture, and at least two blastocysts for biopsy on day 5 or 6 (Jansen 2008). In the latest trial, good‐prognosis patients were defined as female age 25 to 40 years undergoing IVF with autologous oocytes with at least two blastocysts of sufficient quality, but the exclusion criteria for this trial included more than two failed IVF‐embryo transfers, more than one miscarriage, or severe male factor (Munné 2019).
We included three studies for the indication repeated IVF failure. In these three studies repeated IVF failure was defined as three or more failed IVF or ICSI attempts with embryos of good morphological quality (Werlin 2003; Blockeel 2008; Rubio 2013). In one study other inclusion criteria were subfertility with need for assisted reproduction with motile spermatozoa, maternal age less than 37 years, and a normal karyotype in both partners (Blockeel 2008). In another study other inclusion criteria were maternal age less than 40 years, fresh embryos in each cycle, and no abnormality in the infertility work‐up (Rubio 2013).
Types of interventions
One study compared PGT‐A with biopsy of the first and second polar body with the use of array comparative genomic hybridisation (aCGH) as genome‐wide analysis technique versus no PGT‐A (Verpoest 2018).
No studies performed PGT‐A with biopsy in cleavage stage with the use of genome‐wide analyses.
One study performed PGT‐A with biopsy at the blastocyst stage with the use of next‐generation sequencing (NGS)–based genome‐wide analyses versus no PGT‐A (Munné 2019).
Ten studies compared PGT‐A with cleavage stage biopsies with the use of FISH for the genetic analysis versus no PGT‐A (Werlin 2003; Staessen 2004; Mastenbroek 2007; Blockeel 2008; Hardarson 2008; Staessen 2008; Meyer 2009; Schoolcraft 2009; Debrock 2010; Rubio 2013).
One study compared PGT‐A with biopsy in the blastocyst stage with the use of FISH for the genetic analysis versus no PGT‐A followed by FISH genetic analysis versus no PGT‐A (Jansen 2008).
In the majority of studies the aneuploidy screening was performed by FISH (Werlin 2003; Staessen 2004; Mastenbroek 2007; Blockeel 2008; Hardarson 2008; Jansen 2008; Staessen 2008; Meyer 2009; Schoolcraft 2009; Debrock 2010; Rubio 2013). In two studies the aneuploidy screening was performed with the use of genome‐wide analyses (Verpoest 2018; Munné 2019). In one study PGT‐A was performed with use of aCGH analysis of both polar bodies (Verpoest 2018). In another study blastocyst trophectoderm biopsy was followed by NGS (Munné 2019).
Types of outcomes
Cumulative live birth rate outcomes could be analysed from two trials (Mastenbroek 2007; Verpoest 2018). The live births were those recorded following embryo transfer, thus including spontaneous pregnancies. In the Verpoest 2018 study, the primary outcome was live birth per participant within one year from the first follicle aspiration after enrolment in the study. The live births were those recorded following embryo transfer, thus excluding deliveries following coincidental spontaneous pregnancies. The results in this review data analysis were per first treatment cycle; the study authors kindly provided live birth data per first treatment cycle on request (Verpoest 2018 [pers comm]). In another trial the results of frozen‐thawed embryos of the first treatment cycle and the spontaneous pregnancies were taken into account (Mastenbroek 2007). Only cumulative ongoing pregnancy rates per treatment cycle were provided. The authors kindly provided us the cumulative live birth rate per first treatment cycle (Mastenbroek 2007 [pers comm]). In this trial two live births were obtained after a frozen‐thawed embryo transfer in the control group, and no pregnancies were obtained in the PGT‐A group (Mastenbroek 2007 [pers comm].
The live birth rate after the first embryo transfer per woman was reported in seven trials (Blockeel 2008; Hardarson 2008; Jansen 2008; Staessen 2008; Meyer 2009; Rubio 2013; Munné 2019). In one trial the live birth rate after the embryo transfer was incomplete since one pregnancy was still ongoing at the time of writing (Blockeel 2008), but the author kindly provided us the outcome of this pregnancy (Blockeel 2018 [pers comm]). In one trial the live birth rate was reported per participant, but the authors kindly provided us the outcome of the live birth rate after the first transfer (Mastenbroek 2007). In one trial live birth rate was not reported, but it was possible to calculate the numbers from the data provided (Schoolcraft 2009). The outcome measure live birth was not defined the same in all studies. Live birth was defined as a live‐born child after 20 weeks of gestation (Blockeel 2018 [pers comm];Staessen 2008), as a live‐born child after 24 weeks of gestation (Mastenbroek 2007), as progression of pregnancy past the 24th week of gestation (Schoolcraft 2009), or it was not defined (Hardarson 2008; Jansen 2008; Meyer 2009; Debrock 2010; Verpoest 2018). In the Munné 2019 study, the primary study outcome ongoing pregnancy rate at 20 weeks gestation reflects the live birth rate, as all ongoing pregnancies continued to live birth in this study cohort.
Miscarriage data were available for all included studies expect one (Werlin 2003). Miscarriage data were confirmed to be loss of a clinical pregnancy (not biochemical) in three studies (Mastenbroek 2007; Meyer 2009; Verpoest 2018). In seven studies the miscarriage data were a mixture of biochemical and clinical pregnancy losses (Staessen 2004; Blockeel 2008; Hardarson 2008; Jansen 2008; Staessen 2008; Schoolcraft 2009; Debrock 2010). In one study miscarriage was not defined (Munné 2019). We have taken the pragmatic view to include all miscarriage data independent of the used definition, as according to the study authors the majority of the pregnancy losses were from clinical pregnancies.
Ongoing pregnancy was reported in five studies (Staessen 2004; Mastenbroek 2007; Blockeel 2008; Debrock 2010; Rubio 2013). A clinical pregnancy was defined as the presence of at least one intrauterine gestational sac on ultrasound exam, Mastenbroek 2007; Meyer 2009; Debrock 2010; Verpoest 2018; Munné 2019, or as fetal heart activity on ultrasound exam. Seven studies reported multiple pregnancy rates (Staessen 2004; Mastenbroek 2007; Hardarson 2008; Staessen 2008; Debrock 2010; Rubio 2013; Verpoest 2018). In nine studies the proportion of women reaching embryo transfer was reported (Staessen 2004; Mastenbroek 2007; Blockeel 2008; Hardarson 2008; Jansen 2008; Meyer 2009; Debrock 2010; Rubio 2013). The mean number of embryos per transfer could be calculated in nine studies (Staessen 2004; Mastenbroek 2007; Blockeel 2008; Hardarson 2008; Meyer 2009; Debrock 2010; Rubio 2013; Verpoest 2018; Munné 2019).
Excluded studies
We excluded 12 studies. Six of these studies performed PGT‐A with the use of genome‐wide analyses (Yang 2012; Forman 2013; Scott 2013; Forman 2014; Rubio 2017; Ozgur 2019), and the other six studies performed PGT‐A with the use of FISH for the genetic analysis (Gianaroli 1997; Gianaroli 1999; Stevens 2004; Mersereau 2008; Moayeri 2016).
Two studies were excluded after retrieving and reading the full text because couples were allocated to the treatment or control group on the basis of their volunteer decision, instead of random allocation (Gianaroli 1997; Gianaroli 1999). One study was excluded because the trial was stopped early and no data published (NCT02265614). One study, Stevens 2004, was excluded because the participants included in this study were also included in another, larger study (Schoolcraft 2009). Moayeri 2016 was excluded because the authors only reported on biochemical pregnancies, and number of participants reaching embryo transfer was not reported. We excluded other studies due to inappropriate design or reporting of the study that did not allow for a fair evaluation of the effect of PGT‐A on IVF treatment outcomes, such as the use of single embryo transfer (SET) in the PGT‐A group versus double embryo transfer (DET) in the control group, Forman 2013; Forman 2014, and only reporting on women that had an embryo transfer, Yang 2012; Scott 2013. In IVF with PGT‐A, compared to IVF without PGT‐A, a relatively large group of women will not have an embryo transfer, as all embryos are considered aneuploid or unsuitable for transfer. If a study only includes women with an embryo transfer or only reports outcomes per embryo transfer, then the study favours PGT‐A by design by leaving these women that do not get pregnant (as they do not have an embryo transfer) out of the equation. These studies do not permit the drawing of any conclusion on the effect of PGT‐A on IVF effectiveness, where treatment outcomes should be calculated per woman (including all women going for treatment) or per started treatment cycle (including all started treatments). One study was excluded because outcome measures were reported as percentages without mentioning the unit of analysis, and it was not possible to calculate the exact numbers (Mersereau 2008). In two other studies multiple ovum pickups were allowed to collect oocytes before a transfer was attempted, and it was not possible to calculate the exact numbers per first ovum pick‐up (Rubio 2013; Rubio 2017). Multiple ovum‐pickups per woman before PGT‐A is performed hampers a fair evaluation of the effect of PGT‐A on IVF treatment outcomes, as such an artificial increase of oocyte number can be expected to benefit the PGT‐A arm more than the control arm, especially when reporting per first transfer only, and it was not possible to correct for this different strategy. In the Rubio 2013 study, data from the AMA group were excluded for this reason. The authors of Ozgur 2019 performed blastocyst stage biopsy in only the best morphologically scoring embryo in case a participant was randomised to the PGT‐A group. Euploid embryos were transferred when available, otherwise a non‐biopsied embryo was chosen for transfer. Consequently, the right unit of analysis (e.g. per woman or per ovum pick‐up) could not be used for evaluating PGT‐A effectiveness.
Risk of bias in included studies
Risk of bias in the included studies is summarised in Figure 2 and Figure 3.
Allocation
Sequence generation
As shown in Figure 2 and Figure 3, seven studies reported adequate methods for random sequence generation and were therefore rated as at low risk of bias for sequence generation (Mastenbroek 2007; Hardarson 2008; Meyer 2009; Schoolcraft 2009; Rubio 2013; Verpoest 2018; Munné 2019). The other six studies did not describe the method used and were rated as at unclear risk of bias for this domain.
Allocation concealment
Four studies described adequate methods for allocation concealment (Mastenbroek 2007; Meyer 2009; Verpoest 2018; Munné 2019). The remaining nine studies did not describe methods of allocation concealment and were scored as at unclear risk of bias for this domain.
Blinding
Blinding of participants and personnel (performance bias)
In three studies participants and clinicians were blinded (Mastenbroek 2007; Verpoest 2018; Munné 2019). Participants and clinicians involved in the study were blinded until after embryo transfer, Verpoest 2018; Munné 2019, or study completion, Mastenbroek 2007.
In six studies participating women and personnel were not blinded (Staessen 2004; Blockeel 2008; Hardarson 2008; Staessen 2008; Meyer 2009; Debrock 2010). Three trials did not mention whether participants and treatment providers were blind to assignment status (Werlin 2003; Jansen 2008; Schoolcraft 2009). We assessed these nine studies as being at high risk of performance bias. None of the included studies blinded the personnel performing PGT‐A, but this would have been impossible.
Blinding of outcome assessors (detection bias)
We judged all 13 studies to be at low risk of detection bias because the outcomes (cumulative live birth, live birth, miscarriage, ongoing pregnancy, and clinical pregnancy) are objective and therefore cannot be influenced by knowledge of the intervention. One study described that blinding for outcome assessors was performed (Mastenbroek 2007). In the remaining studies outcome assessors were not blinded, however we still deemed these studies as having a low risk of bias due to the reason described above.
Incomplete outcome data
In seven trials there were dropouts in both the intervention and the control group (respectively 52/200 and 77/206 and 13/120 and 14/95 and 15/52 and 8/205 and 56/330 in the intervention group and 59/200 and 71/202 and 13/120 and 28/105 and 28/52 and 7/191 and 18/331 in the control group) (Staessen 2004; Mastenbroek 2007; Blockeel 2008; Staessen 2008; Debrock 2010; Verpoest 2018; Munné 2019). Reasons for cancelling the intended treatment cycle were insufficient ovarian response (Staessen 2004; Mastenbroek 2007; Blockeel 2008), no oocytes at ovum pick‐up (Debrock 2010), no fertilisation (Debrock 2010; Verpoest 2018), fewer than two fertilised oocytes available (Debrock 2010), fewer than two embryos with at least six cells available on day 3 (Debrock 2010), no embryo available for biopsy (Debrock 2010), no euploid embryos available for transfer, thaw failure, protocol deviation (Munné 2019), cancer cyst detected (Staessen 2004), no technical support for genetic analysis (Debrock 2010), other medical reasons (Mastenbroek 2007; Verpoest 2018), inability to manage the treatment burden (Staessen 2004; Mastenbroek 2007; Staessen 2008), stop further fertility treatment (Blockeel 2008), spontaneous pregnancy (Staessen 2004; Blockeel 2008; Staessen 2008; Verpoest 2018), not finished at the end of follow‐up (Mastenbroek 2007), participant withdrew (Debrock 2010; Verpoest 2018; Munné 2019), and other reasons (Mastenbroek 2007). Dropouts were included in an ITT analysis in three trials (Mastenbroek 2007; Verpoest 2018; Munné 2019), but they were not included in an ITT analysis in the other trials (Staessen 2004; Blockeel 2008; Staessen 2008; Debrock 2010). The remaining trials did not mention dropouts. In one study, four women declined embryo transfer for reasons not related to the study, two in the PGT‐A treatment group and two in the control group; these women were not included in an ITT analysis (Meyer 2009). Dr Debrock kindly provided the writers of the previous version of this review the information about the dropout cycles so it could be included in an ITT analysis.
Selective reporting
There was no evidence of selective reporting. We considered all studies to be at low risk of reporting bias because they reported and published all outcomes they had set out to investigate. This was confirmed on communication with authors and by referencing against information in online trials registers if available.
Other potential sources of bias
We found no potential sources of within‐study bias in 11 studies and assessed these studies as having a low risk of other bias (Staessen 2004; Mastenbroek 2007; Blockeel 2008; Jansen 2008; Staessen 2008; Meyer 2009; Schoolcraft 2009; Debrock 2010; Rubio 2013; Munné 2019; Verpoest 2018).
We assessed two studies as having a high risk of within‐study bias (Werlin 2003; Hardarson 2008). This was due to the difference in day of embryo transfer between study arms (day 5 for intervention and day 3 for control (Hardarson 2008); or day 3 or 5 for the control group based on physician preference (Werlin 2003)). This difference in maturity of the embryo could have had an impact on the likelihood of an ongoing pregnancy.
Effects of interventions
Summary of findings 1. Preimplantation genetic testing for aneuploidies with the use genome‐wide analyses in in vitro fertilisation.
| Preimplantation genetic testing for aneuploidies with the use genome‐wide analyses in in vitro fertilisation | ||||||
|
Patient or population: couples with an in vitro fertilisation indication
Settings: fertility clinics
Intervention: PGT‐A with the use of genome‐wide analyses Comparison: no PGT‐A | ||||||
| Outcomes per woman | Embryo stage of biopsy | Illustrative comparative risks* (95% CI) | Relative effect (95% CI) | No. of participants (studies) | Quality of the evidence (GRADE) | |
| Assumed risk | Corresponding risk | |||||
| Control | PGT‐A with the use of genome‐wide analyses | |||||
| Cumulative live birth | Polar body biopsy | 236 per 1000 | 245 per 1000 (169 to 338) | OR 1.05 (0.66 to 1.66) | 396 (1 RCT) | ⊕⊕⊝⊝ lowa |
| Live birth rate after the first embryo transfer | Polar body biopsy | 199 per 1000 | 215 per 1000 (144 to 308) |
OR 1.10 (0.68 to 1.79) | 396 (1 RCT) | ⊕⊕⊝⊝ lowa |
| Blastocyst stage biopsy | 435 per 1000 | 417 per 1000 (347 to 494) |
OR 0.93 (0.69 to 1.27) | 661 (1 RCT) | ⊕⊕⊝⊝ lowa | |
| Miscarriage | Polar body biopsy | 141 per 1000 | 69 per 1000 (36 to 127) | OR 0.45 (0.23 to 0.88) | 396 (1 RCT) | ⊕⊕⊝⊝ lowb |
| Blastocyst stage biopsy | 82 per 1000 | 73 per 1000 (44 to 121) |
OR 0.89 (0.52 to 1.54) | 661 (1 RCT) | ⊕⊕⊝⊝ lowa | |
| Ongoing pregnancy | No studies reported on ongoing pregnancy. | |||||
| Clinical pregnancy | Polar body biopsy | 366 per 1000 | 308 per 1000 (224 to 402) | OR 0.77 (0.50 to 1.16) | 396 (1 RCT) | ⊕⊕⊝⊝ lowa |
| *The basis for the assumed risk (e.g. the median control group risk across studies) is provided in footnotes. The corresponding risk (and its 95% confidence interval) is based on the assumed risk in the comparison group and the relative effect of the intervention (and its 95% CI). CI: confidence interval; OR: odds ratio; PGT‐A: preimplantation genetic testing for aneuploidies; RCT: randomised controlled trial | ||||||
|
GRADE Working Group grades of evidence
High quality: further research is very unlikely to change our confidence in the estimate of effect. Moderate quality: further research is likely to have an important impact on our confidence in the estimate of effect and may change the estimate. Low quality: further research is very likely to have an important impact on our confidence in the estimate of effect and is likely to change the estimate. Very low quality: we are very uncertain about the estimate. | ||||||
aDowngraded two levels for imprecision: results based on one study, small number of events; CI crosses the line of no effect. bDowngraded two levels for imprecision: results based on one study, small number of events.
Summary of findings 2. Preimplantation genetic testing for aneuploidies with the use of fluorescence in situ hybridisation (FISH) for the genetic analysis in in vitro fertilisation.
| Preimplantation genetic testing for aneuploidies with the use of fluorescence in situ hybridisation (FISH) for the genetic analysis in in vitro fertilisation | |||||
|
Patient or population: couples with an in vitro fertilisation indication
Settings: fertility clinics
Intervention: PGT‐A with the use of FISH for the genetic analysis Comparison: no PGT‐A | |||||
| Outcomes per woman | Illustrative comparative risks* (95% CI) | Relative effect (95% CI) | No. of participants (studies) | Quality of the evidence (GRADE) | |
| Assumed risk | Corresponding risk | ||||
| Control | PGT‐A with the use of FISH for the genetic analysis | ||||
| Cumulative live birth | 287 per 1000 | 192 per 1000 (124 to 289) | OR 0.59 (0.35 to 1.01) | 408 (1 RCT) | ⊕⊕⊝⊝ lowa |
| Live birth rate after the first embryo transfer | 307 per 1000 | 215 per 1000 (160 to 287) | OR 0.62 (0.43 to 0.91) | 1680 (10 RCTs) | ⊕⊕⊕⊝ moderateb |
| Miscarriage | 105 per 1000 | 108 per 1000 (81 to 143) | OR 1.03 (0.75 to 1.41) | 1680 (10 RCTs) | ⊕⊕⊕⊝ moderatec |
| Ongoing pregnancy | 274 per 1000 | 204 per 1000 (161 to 253) | OR 0.68 (0.51 to 0.90) | 1121 (5 RCTs) | ⊕⊕⊝⊝ lowb,c |
| Clinical pregnancy | 333 per 1000 | 219 per 1000 (174 to 267) | OR 0.56 (0.42 to 0.73) | 1131 (5 RCTs) | ⊕⊕⊕⊝ moderatec |
| *The basis for the assumed risk (e.g. the median control group risk across studies) is provided in footnotes. The corresponding risk (and its 95% confidence interval) is based on the assumed risk in the comparison group and the relative effect of the intervention (and its 95% CI). CI: confidence interval; OR: odds ratio; PGT‐A: preimplantation genetic testing for aneuploidies; RCT: randomised controlled trial | |||||
|
GRADE Working Group grades of evidence
High quality: further research is very unlikely to change our confidence in the estimate of effect. Moderate quality: further research is likely to have an important impact on our confidence in the estimate of effect and may change the estimate. Low quality: further research is very likely to have an important impact on our confidence in the estimate of effect and is likely to change the estimate. Very low quality: we are very uncertain about the estimate. | |||||
aDowngraded two levels for imprecision: results based on one study, small number of events. bDowngraded one level for inconsistency: I² > 50% in results across studies. cDowngraded one level for imprecision: the CI for most studies crosses the line of no effect, or small number of events.
See: Preimplantation genetic testing for aneuploidies with the use genome‐wide analyses in in vitro fertilisation (Table 1) and Preimplantation genetic testing for aneuploidies with the use of fluorescence in situ hybridisation (FISH) for the genetic analysis in in vitro fertilisation (Table 2).
(1) IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses
Two studies undertook this comparison (Verpoest 2018; Munné 2019), with a total of 1057 participants.
Primary outcomes
1.1 Cumulative live birth rate after the first treatment cycle per woman randomised
Polar body biopsy
Only one study provided cumulative live birth data (Verpoest 2018). There were 50 events reported in the women randomised to the PGT‐A arm and 45 events in the 396 women randomised to the control arm. It is unclear whether there is any difference in rate of cumulative live birth per woman between the groups (odds ratio (OR) 1.05, 95% confidence interval (CI) 0.66 to 1.66, 1 RCT, N = 396, low‐quality evidence) (Analysis 1.1, Figure 4). The evidence suggests that if the rate of cumulative live birth in the control groups is 24%, the rate with the use of PGT‐A with genome wide‐analysis would be between 17% and 34%.
1.1. Analysis.

Comparison 1: IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, Outcome 1: Cumulative live birth rate after the first treatment cycle, per woman randomised
4.

Forest plot of comparison: 1 IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, outcome: 1.1 Cumulative live birth rate after the first treatment cycle, per woman randomised.
We did not perform a sensitivity analysis restricted to studies at low risk of selection bias as only one included study was at low risk of selection bias. A sensitivity analysis on the effect measured by risk ratio did not influence this finding substantially.
Blastocyst stage biopsy
No data were provided for this outcome.
Secondary outcomes
1.2 Live birth rate per first embryo transfer per woman randomised
Two trials involving a total of 1057 women reported on live birth rate after the first embryo transfer (Verpoest 2018; Munné 2019). We divided the PGT‐A performed with the use of genome‐wide analyses into subgroups of stage of biopsy.
Polar body biopsy
One trial performed biopsy on polar bodies of the embryo. There were 44 events reported in the women randomised to the PGT‐A arm and 38 events in the 396 women randomised to the control arm. It is unclear whether there is any difference in rate of live birth after the first embryo transfer per woman (OR 1.10, 95% CI 0.68 to 1.79, 1 RCT, N = 396, low‐quality evidence) (Analysis 1.2, Figure 5). The evidence suggests that if the live birth rate after the first embryo transfer in the control groups is 20%, the rate with the use of PGT‐A with the use of genome‐wide analyses would be between 14% and 31%.
1.2. Analysis.

Comparison 1: IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, Outcome 2: Live birth rate after the first embryo transfer per woman randomised
5.

Forest plot of comparison: 1 IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, outcome: 1.2 live birth rate after the first embryo transfer per woman randomised.
Blastocyst stage biopsy
One trial performed biopsy in the blastocyst stage of the embryo. There were 138 events reported in the women randomised to the PGT‐A arm and 144 events in the 661 women randomised to the control arm (OR 0.93, 95% CI 0.69 to 1.27, 1 RCT, N = 661, low‐quality evidence) (Analysis 1.2, Figure 5), indicating that we are uncertain about the effectiveness of the intervention. Translated into absolute risks, this means that for a woman with a 44% chance of achieving a live birth after the first embryo transfer without the performance of PGT‐A, the chance of a live birth after the first embryo transfer with PGT‐A with the use of genome‐wide analyses would be between 35% and 50%.
1.3 Miscarriage rate per woman randomised
One study defined miscarriage data as loss of clinical pregnancy (Verpoest 2018), whilst the other study did not define miscarriage (Munné 2019).
Polar body biopsy
One trial performed biopsy on polar bodies of the embryo (Verpoest 2018). The evidence suggests that PGT‐A performed with the use of genome‐wide analyses may decrease miscarriage rate compared to an IVF treatment without PGT‐A (OR 0.45, 95% CI 0.23 to 0.88, 1 RCT, N = 396, low‐quality evidence) (Analysis 1.3, Figure 6). The evidence suggests that if the miscarriage rate without the addition of PGT‐A is 14%, the rate associated with PGT‐A with the use of genome‐wide analyses would be between 4% and 13%.
1.3. Analysis.

Comparison 1: IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, Outcome 3: Miscarriage rate per woman randomised
6.

Forest plot of comparison: 1 IVF without PGT‐A versus IVF with PGT‐A with the use of genome‐wide analyses, outcome: 1.3 Miscarriage rate per woman randomised.
Blastocyst stage biopsy
One trial performed biopsy in the blastocyst stage of the embryo (Munné 2019). We are uncertain of the effect of PGT‐A performed with the use of genome‐wide analyses on the miscarriage rate compared to IVF treatment without PGT‐A (OR 0.89, 95% CI 0.52 to 1.54, 1 RCT, N = 661, low‐quality evidence) (Analysis 1.3, Figure 6). The evidence suggests that if the miscarriage rate without the addition of PGT‐A is 8.2%, the rate associated with PGT‐A with the use of genome‐wide analyses would be between 4% and 12%.
1.4 Miscarriage rate per pregnancy
Polar body biopsy
When miscarriage rate is expressed per clinical pregnancy (Verpoest 2018), there also may be reduction in the miscarriage rate in the PGT‐A group (OR 0.47, 95% CI 0.22 to 1.00, 1 RCT, N = 136) (Analysis 1.4).
1.4. Analysis.

Comparison 1: IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, Outcome 4: Miscarriage rate per clinical pregnancy
Blastocyst stage biopsy
No data were provided for this outcome.
Ongoing pregnancy rate per woman randomised
No data were provided for this outcome.
1.5 Clinical pregnancy per woman randomised
Polar body biopsy
Only one study reported this outcome (Verpoest 2018). There were 63 clinical pregnancies in the 205 women randomised to the intervention group, and 70 pregnancies in the 191 women randomised to the control group. It is unclear whether there is any difference between interventions in clinical pregnancy rates (OR 0.77, 95% CI 0.50 to 1.16, 1 RCT, N = 396, low‐quality evidence) (Analysis 1.5).
1.5. Analysis.

Comparison 1: IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, Outcome 5: Clinical pregnancy per woman randomised
Blastocyst stage biopsy
No data were provided for this outcome.
1.6 Multiple pregnancy per woman randomised
Polar body biopsy
One study reported this outcome (Verpoest 2018). It is uncertain whether PGT‐A with the use of genome‐wide analyses for embryo selection influences multiple pregnancy rates (OR 0.53, 95% CI 0.20 to 1.37, 1 RCT, N = 396) (Analysis 1.6).
1.6. Analysis.

Comparison 1: IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, Outcome 6: Multiple pregnancy per woman randomised
Blastocyst stage biopsy
No data were provided for this outcome.
1.7 Multiple pregnancy per live birth
Polar body biopsy
It is also uncertain if there is a difference between groups in multiple pregnancy rate per live birth (OR 0.45, 95% CI 0.16 to 1.26, 1 RCT, N = 95) (Analysis 1.7).
1.7. Analysis.

Comparison 1: IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, Outcome 7: Multiple pregnancy rate per live birth
Blastocyst stage biopsy
No data were provided for this outcome.
1.8 Proportion of women reaching embryo transfer
In two studies a total of 423 participants reached an embryo transfer in the PGT‐A group and 484 participants reached an embryo transfer in the control group (Verpoest 2018; Munné 2019).
Polar body biopsy
One trial performed biopsy on polar bodies (Verpoest 2018). The evidence suggests that PGT‐A performed with the use of genome‐wide analyses decreases the proportion of women reaching an embryo transfer (OR 0.31, 95% CI 0.18 to 0.54, 1 RCT, N = 396) (Analysis 1.8).
1.8. Analysis.

Comparison 1: IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, Outcome 8: Proportion of women reaching embryo transfer per woman randomised
Blastocyst stage biopsy
One trial performed biopsy in the blastocyst stage of the embryo (Munné 2019). The evidence suggests that PGT‐A performed with the use of genome‐wide analyses decreases the proportion of women reaching an embryo transfer (OR 0.28, 95% CI 0.16 to 0.49, 1 RCT, N = 661) (Analysis 1.8).
1.9 Mean number of embryos per transfer
Polar body biopsy
In the Verpoest 2018 study a single embryo transfer or double transfer was performed subject to availability of (euploid) embryos, that is if there was only one embryo euploid tested, a single embryo transfer was performed (Verpoest 2018). In the PGT‐A group 177 embryos were transferred with a mean number of 1.4 embryos per transfer. In the control group 249 embryos were transferred with a mean number of 1.8 embryo per transfer. The evidence suggests that PGT‐A with the use of genome‐wide analyses for embryo selection decreases the number of embryos available for transfer, but this finding is unclear as the evidence is of low quality (mean difference (MD) −0.40, 95% CI −0.49 to −0.31, 1 RCT, N = 426) (Analysis 1.9).
1.9. Analysis.

Comparison 1: IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses, Outcome 9: Mean number of embryos transferred per transfer
Blastocyst stage biopsy
In the study of Munné 2019, the standard care was a single embryo transfer, in the control group and the intervention group.
(2) IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis
Primary outcomes
2.1 Cumulative live birth rate per woman
One study provided cumulative live birth rate data (Mastenbroek 2007), with a total of 408 participants. It is uncertain whether there is any difference between the group with addition of PGT‐A and the control group in rate of cumulative live birth (OR 0.59, 95% CI 0.35 to 1.01, 1 RCT, N = 408, low‐quality evidence) (Analysis 2.1, Figure 7). The evidence suggests that for the observed average cumulative live birth rate per woman of 29% in the control group, the chance of live birth following the results of one IVF treatment cycle with PGT‐A is between 12% and 29%. In Mastenbroek 2007 PGT‐A was offered to women who were randomly assigned to undergo three cycles of IVF, with embryo selection based on either PGT‐A or conventional embryo selection. For the outcome cumulative live birth rate only the cryopreserved embryos from the first IVF cycle were taken into account.
2.1. Analysis.

Comparison 2: IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, Outcome 1: Cumulative live birth rate after the first treatment cycle, per woman randomised
7.

Forest plot of comparison: 2 IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, outcome: 2.1 Cumulative live birth rate after the first treatment cycle, per woman randomised.
We did not perform a sensitivity analysis restricted to studies at low risk of selection bias as only one included study was at low risk of selection bias.
Secondary outcomes
2.2 Live birth rate per first embryo transfer per woman randomised
All 10 trials for this comparison provided data on live birth after the first embryo transfer. We found that the intervention, PGT‐A with the use of FISH for the genetic analysis, resulted in fewer live births than in the control group (OR 0.62, 95% CI 0.43 to 0.91, 10 RCTs, N = 1680, I² = 54%, moderate‐quality evidence) (Analysis 2.2, Figure 8). The evidence suggests that for the observed average live birth rate after the first embryo transfer per woman of 31% in the control group, the chance of live birth after the first embryo transfer with PGT‐A is between 16% and 29%.
2.2. Analysis.

Comparison 2: IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, Outcome 2: Live birth rate after the first embryo transfer per woman randomised
8.

Forest plot of comparison: 2 IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, outcome: 2.2 live birth rate after the first embryo transfer per woman randomised.
We divided the PGT‐A performed with use of FISH into subgroups: one trial performed biopsy on blastocyst stage, and nine trials performed biopsy in cleavage stage of embryo development. There may be little or no difference between the subgroups (P = 0.28). The OR was 0.40 (95% CI 0.18 to 0.90, 1 RCT, N = 101) for PGT‐A with blastocyst stage biopsy and 0.66 (95% CI 0.45 to 0.98, 9 RCTs, N = 1579) for PGT‐A with cleavage stage biopsy (Analysis 2.2, Figure 8).
2.3 Miscarriage rate per woman randomised
All 10 studies provided data on miscarriage. Of 855 women randomised to the intervention arm, 92 experienced a miscarriage, whereas of 825 women randomised to the control arm, 87 experienced a miscarriage. There is probably little or no difference in miscarriage rate between PGT‐A and the control group (OR 1.03, 95% CI 0.75 to 1.41, 10 RCTs, N = 1680, I² = 16%, moderate‐quality evidence) (Analysis 2.3).
2.3. Analysis.

Comparison 2: IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, Outcome 3: Miscarriage rate per woman randomised
2.4 Miscarriage rate per clinical pregnancy
When miscarriage rate is expressed per clinical pregnancy (Staessen 2004; Mastenbroek 2007; Blockeel 2008; Jansen 2008; Debrock 2010), there may be a reduction in miscarriage rate in the control group (OR 1.77, 95% CI 1.10 to 2.86, 5 RCTs, N = 288, I² = 45%, moderate‐quality evidence) (Analysis 2.4).
2.4. Analysis.

Comparison 2: IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, Outcome 4: Miscarriage rate per clinical pregnancy
2.5 Ongoing pregnancy rate per woman randomised
Four studies provided data on ongoing pregnancy rate (Staessen 2004; Mastenbroek 2007; Blockeel 2008; Rubio 2013). The intervention, PGT‐A with the use of FISH for the genetic analysis, reduces ongoing pregnancies (OR 0.68, 95% CI 0.51 to 0.90, 5 RCTs, N = 1121, I² = 60%, low‐quality evidence) (Analysis 2.5).
2.5. Analysis.

Comparison 2: IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, Outcome 5: Ongoing pregnancy rate per woman randomised
2.6 Clinical pregnancy rate per woman randomised
Five studies provided clinical pregnancy data (Staessen 2004; Mastenbroek 2007; Blockeel 2008; Jansen 2008; Debrock 2010). There were 131 clinical pregnancies in the 576 women randomised to the PGT‐A arm and 185 clinical pregnancies in the 555 women randomised to the control arm. The intervention, PGT‐A with the use of FISH for the genetic analysis, reduces clinical pregnancies (OR 0.60, 95% CI 0.45 to 0.81, 5 RCTs, N = 1131, I² = 0%, moderate‐quality evidence) (Analysis 2.6).
2.6. Analysis.

Comparison 2: IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, Outcome 6: Clinical pregnancy per woman randomised
2.7 Multiple pregnancy rate per woman randomised
Six studies provided multiple pregnancy data (Staessen 2004; Mastenbroek 2007; Hardarson 2008; Staessen 2008; Debrock 2010; Rubio 2013). There were 20 multiple pregnancies in the 673 women randomised to the PGT‐A arm and 29 multiple pregnancies in the 658 women randomised to the control arm. Due to the wide confidence interval, it is unclear whether there is a difference in the multiple pregnancy rate per woman between the two groups (OR 0.66, 95% CI 0.37 to 1.17, 6 RCTs, N = 1331, I² = 0%) (Analysis 2.7).
2.7. Analysis.

Comparison 2: IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, Outcome 7: Multiple pregnancy rate per woman randomised
2.8 Multiple pregnancy rate per live birth
Six studies provided multiple pregnancy rates per live birth (Staessen 2004; Mastenbroek 2007; Hardarson 2008; Staessen 2008; Debrock 2010; Rubio 2013). There were 20 multiple pregnancies amongst the 164 live births to the PGT‐A arm and 29 multiple pregnancies amongst the 200 live births to the control arm. Due to the wide confidence interval, it is unclear whether there is difference in the multiple pregnancy rate per live birth between the two groups (OR 0.97, 95% CI 0.51 to 1.82, 6 RCTs, N = 364, I² = 0%) (Analysis 2.8).
2.8. Analysis.

Comparison 2: IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, Outcome 8: Multiple pregnancy rate per live birth
2.9 Proportion of women reaching embryo transfer
Nine studies provided data on the proportion of women reaching embryo transfer (Werlin 2003; Staessen 2004; Mastenbroek 2007; Blockeel 2008; Hardarson 2008; Jansen 2008; Meyer 2009; Debrock 2010; Rubio 2013). There were 508 women in the PGT‐A arm and 532 women in the control group reaching an embryo transfer. The proportion of woman reaching embryo transfer probably decreases with the addition of PGT‐A with the use of FISH for the genetic analysis to an IVF treatment (OR 0.54, 95% CI 0.40 to 0.74, 9 RCTs, N = 1286, I2 = 48%) (Analysis 2.9).
2.9. Analysis.

Comparison 2: IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, Outcome 9: Proportion of women reaching embryo transfer per woman randomised
2.10 Mean number of embryos per transfer
Seven studies provided data on the mean number of embryos transferred per transfer (Staessen 2004; Mastenbroek 2007; Blockeel 2008; Hardarson 2008; Meyer 2009; Debrock 2010; Rubio 2013). The mean number of embryos transferred per transfer was decreased in the intervention group, PGT‐A with the use of FISH for the genetic analysis (MD −0.23, 95% CI −0.30 to −0.16, 7 RCTs, I² = 81%) (Analysis 2.10). There was statistical heterogeneity, with I² = 81%. This could be explained in part by the fact that the maximum embryos for transfer was much higher in one study, namely up to six if a woman was 40 years or older (Staessen 2004), as compared to the other studies, where the maximum of embryos for transfer was two or three. However, if this study was excluded from the analysis, the I² was still high, namely 69%.
2.10. Analysis.

Comparison 2: IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis, Outcome 10: Mean number of embryos transferred per transfer
Discussion
Summary of main results
This review compared the effectiveness and safety of PGT‐A in an IVF treatment. We found two trials that compared the addition of PGT‐A with the use of genome‐wide analyses with an IVF treatment control group, and 10 trials that compared the addition of PGT‐A by FISH with an IVF treatment control group. The aim of the comparisons was to assess the potential advantages of selecting embryos by PGT‐A to increase live birth rates and prevent miscarriages.
IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses
All findings for this comparison were uncertain because of the low quality of the evidence due to the limited number of studies. It is unclear whether there is any difference between the addition of PGT‐A by genome‐wide analyses in an IVF treatment versus morphology embryo screening in terms of cumulative live birth or live birth rate after the first embryo transfer. PGT‐A with polar body biopsy may decrease miscarriage rate, but it is uncertain if PGT‐A reduces miscarriage rates with blastocyst stage biopsy. It is unclear whether there is any difference in clinical pregnancy rates.
There is insufficient good‐quality evidence of a difference in cumulative live birth rate, live birth rate after the first embryo transfer, or miscarriage rate to choose between an IVF treatment with or without PGT‐A performed by genome‐wide analyses. No data were available on ongoing pregnancy rates. The effect of PGT‐A on clinical pregnancy rate is uncertain.
IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis
We are uncertain if the addition of PGT‐A with the use of FISH for the genetic analysis leads to a higher cumulative live birth rate. The addition of PGT‐A probably reduces live birth rate after the first embryo transfer; may reduce ongoing pregnancy; and probably reduces clinical pregnancy rate. There is probably little or no difference in miscarriage between the intervention and the control group. The currently available evidence does not support the routine use of PGT‐A with the use of FISH for the genetic analysis.
Overall completeness and applicability of evidence
The comparisons of PGT‐A with the use of genome‐wide analyses and PGT‐A with the use of FISH for the genetic analysis included 1057 and 1680 women, respectively.
In the comparison ‘IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses’, a trophectoderm biopsy in blastocyst stage of the embryo development was performed in one study (Munné 2019), whilst analysis of the first and second polar bodies of the fertilised oocytes was performed in the other study (Verpoest 2018).
It is important to mention that the blastocyst biopsy or trophectoderm biopsy is at present the most widely used technique (Sermon 2016; De Rycke 2017).
The outcome miscarriage in the comparison ‘IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses’ suggests that the miscarriage rate is probably reduced with the use of PGT‐A in the subgroup with polar body biopsy. In this subgroup, only women "with advanced maternal age", aged 36 to 40 years, were included (Verpoest 2018). In the subgroup with blastocyst stage biopsy, women were aged 25 to 40 years (Munné 2019). In this last study a post hoc analysis of overall pregnancy outcomes in women aged 35 to 40 years demonstrated no significant difference in the miscarriage rate.
There was heterogeneity between the two trials included in the comparison ‘IVF with PGT‐A versus IVF without PGT‐A by genome‐wide analyses’. The included participants had different indications for PGT‐A; the timing of the biopsy in embryo development was dissimilar; and the embryo transfer policy ‘fresh and or only frozen’ differed.
For the studies comparing ‘IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis’, the included population represented women with different indications for PGT‐A. This variety adds to the broad applicability of results to clinical practice. Whilst cleavage stage biopsy was the most widely practiced form of embryo biopsy for over a decade (Harton 2011), its clinical use has now decreased.
Cumulative live birth rate is the outcome of choice for patients for multiple reasons. A frozen transfer treatment is less burdensome, and in many settings in the world cheaper than initiating a new IVF treatment. Also, the contribution of cryopreservation programmes to the general IVF treatment effectiveness has increased dramatically in the past decades (Wong 2014; Maheshwari 2015). Both the number of frozen transfers per started IVF cycle, and the results per frozen transfer have increased (Wong 2014). Cumulative live birth rate was therefore the primary outcome in our review. However, we have also reported the live birth rate after the first embryo transfer, as this is common in the PGT‐A literature. It must be noted that this outcome favours PGT‐A by design, and should thus only be considered in the right context. It appears that embryos selected by PGT‐A have in themselves a higher implantation potential, but with PGT‐A fewer embryos will be available for transfer. For a fair comparison, for example to evaluate whether embryos were left out rightfully after PGT‐A, all embryos should be taken into account, which is reflected by the cumulative live birth rate. We have reported both outcomes, cumulative live birth rate and live birth rate after the first transfer, per woman randomised. Reporting rates after the first transfer (including only women who received a transfer) is simply incorrect when evaluating IVF effectiveness, and will favour PGT‐A by design (Griesinger 2016).
Only two studies reported data on cumulative live birth rate (Mastenbroek 2007; Verpoest 2018). The Verpoest 2018 study also reported the outcome "time to pregnancy", which did not show a significant difference between study arms. Future studies should focus on the cumulative live birth rate and time to pregnancy, where a time horizon (e.g. of 12 months after randomisations) could be considered.
According to the applicability, it is important to emphasise that the PGT‐A with the use of FISH for genetic analysis is an outdated technical procedure. More advanced genome‐wide analyses approaches are today widely applied, although we await more recent and robust data to support the evidence.
Quality of the evidence
Using the GRADE approach, we found the evidence for all outcomes comparing the addition of PGT‐A on IVF with the use of genome‐wide analyses versus the control group to be of low quality due to the limited number of studies comparing PGT‐A, inconsistency, imprecision, and the risk of publication bias. As there are four ongoing trials registered (NCT01946945; NCT02032264; NCT02265614; NCT02353364), with a study start before 2015, an estimated completion date in the past, and not published, we need to take publication bias into account.
For the comparison addition of PGT‐A on IVF with the use of FISH for the genetic analysis versus the control group, we assessed the evidence for cumulative live birth rate as low quality due to the limited number of studies comparing PGT‐A. Evidence for the other outcomes in this comparison was of low to moderate quality.
Potential biases in the review process
Given the extensive search strategy, including the electronic database search and handsearching of relevant references, the chance of incomplete identification of studies was low. We aimed to identify all eligible studies for inclusion in this review, and contacted the authors of included studies on many occasions in an effort to include as much information as possible. The authors of most studies were forthcoming with further study information, which helped us to accrue a full picture of the study outcomes, as well as providing information needed to assess and establish risk of bias.
Agreements and disagreements with other studies or reviews
Our results are in line with the outcomes of the previous Cochrane Review, Twisk 2006, in concluding that PGT‐A performed with embryo biopsy in cleavage stage using FISH for the genetic analysis decreases live birth rates per first embryo transfer. In Twisk 2006, all trials used FISH for the genetic analysis. Twisk 2006 did not evaluate the outcome of cumulative live birth rate. There is insufficient evidence as to whether PGT‐A with the use of FISH for the genetic analysis improves cumulative live birth rate.
The European Society for Human Reproduction and Embryology stated very recently that using FISH for genetic analysis is not recommended for PGT‐A, as only a subset of chromosomes can be tested, and better genome‐wide analysing techniques exist (ESHRE PGT‐SR/PGT‐A Working Group 2020).
In line with the results of our review, the American Society of Reproductive Medicine stated there was insufficient evidence to recommend the use of blastocyst biopsy with aneuploidy testing, with the use of genome‐wide analyses and the use of FISH for the genetic analysis (ASRM 2018).
Technologies have been developed in the fields of genomics and biopsy techniques. We added the comparison ‘IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses’ to the review, for which we could only include two published trials. Since the introduction of PGT‐A with these new approaches, there are no published systematic reviews with the same inclusion and exclusion criteria on the topic of PGT‐A with the use of genome‐wide analyses.
Authors' conclusions
Implications for practice.
It is uncertain whether the addition of blastocyst stage preimplantation genetic testing for aneuploidies (PGT‐A) where genome‐wide analyses are used, that is the current standard in routine practice today, improves cumulative live birth rates or reduces miscarriage rate. Evidence based on one trial showed a decrease in miscarriage rate with the use of polar body biopsy. There were no data available on ongoing pregnancy rates. The effect of PGT‐A on the number of clinical pregnancies is also uncertain.
The currently available evidence does not support the routine use of PGT‐A with the use of fluorescence in situ hybridisation (FISH) for the genetic analysis. It is uncertain if PGT‐A with the use of FISH improves cumulative live birth rate. PGT‐A with the use of FISH probably decreases live birth rate after the first embryo transfer. There is probably little or no difference in the number of miscarriages between groups. PGT‐A with the use of FISH may reduce ongoing pregnancies and probably reduces the number of clinical pregnancies.
Women and partners who receive IVF should have access to up‐to‐date information on the uncertainty of the effectiveness of PGT‐A, especially considering the invasiveness and costs, in order to make informed choices regarding their treatment. The currently available evidence is insufficient to support PGT‐A in routine clinical practice.
Implications for research.
Considering the outcomes of our review, new developments should be properly evaluated before their routine clinical application. This involves method assessment studies, pilot‐studies showing a potential benefit in terms of cumulative live birth per woman, followed by randomised controlled trials. New research on the effectiveness of PGT‐A should be specifically directed at retaining the highest cumulative live birth chances whilst preventing miscarriages. Another reason for the need for new research is the low number of new included studies in this update review. Future trials should ideally: randomise the participants before the ovum pick‐up so there is no withdrawal of participants without an embryo transfer; have a clear description of the indication(s) for PGT‐A in the study; include cumulative live birth rate and time to pregnancy as outcomes; and use the right unit of analysis (e.g. per woman, per ovum pick‐up).
New research on PGT‐A with the use of FISH for genetic analysis is no longer necessary.
What's new
| Date | Event | Description |
|---|---|---|
| 16 January 2020 | New citation required but conclusions have not changed | The addition of new studies has not led to a change in our conclusions. |
| 16 January 2020 | New search has been performed | New studies added (Rubio 2013; Verpoest 2018; Munné 2019). All fields opened and format amended. |
History
Protocol first published: Issue 2, 2005 Review first published: Issue 1, 2006
| Date | Event | Description |
|---|---|---|
| 13 June 2008 | Amended | New randomised controlled trials included. Results and Discussion updated. |
| 11 November 2005 | New citation required and conclusions have changed | Substantive amendment |
Acknowledgements
The authors wish to acknowledge the support of the Cochrane Gynaecology and Fertility Group staff.
We acknowledge the contribution of Moniek Twisk, Fulco van Veen, Professor Maas Jan Heineman, and Professor Sjoerd Repping to previous versions of this review.
The review authors are grateful to the following peer reviewers for their time and comments.
Noortje Uphoff (consumer), University of York, United Kingdom
Rui Wang, Monash University, Robinson Research Institute, University of Adelaide, Australia
Evi Vogiatzi, Athens, Greece
Ben Willem Mol, Department of Obstetrics and Gynaecology, Monash University, Clayton, Victoria, Australia
Appendices
Appendix 1. Cochrane Gynaecology and Fertility Specialised Register search strategy
Searched 9 September 2019
PROCITE platform
Keywords CONTAINS "preimplantation genetic diagnosis" or "pre‐implantation genetic diagnosis" or "pre‐implantation genetic screening" or "preimplantation genetic analysis" or "preimplantation genetic diagnosis" or "preimplantation genetic screening" or "genetic analysis" or "genetic screening" or "genetic testing" or "genetic techniques" or "chromosomal abnormalities" or "chromosomes" or "aneuploidy screening" or "PGS" or "PGD" or Title CONTAINS "preimplantation genetic diagnosis" or "pre‐implantation genetic diagnosis" or "pre‐implantation genetic screening" or "preimplantation genetic analysis" or "preimplantation genetic diagnosis" or "preimplantation genetic screening" or "genetic analysis" or "genetic screening" or "genetic testing" or "genetic techniques" or "chromosomal abnormalities" or "chromosomes" or "aneuploidy screening" or "PGS" or "PGD"
(212 records)
Appendix 2. CENTRAL search strategy
Searched 9 September 2019 (Issue August 2019)
Ovid platform
1 exp chromosome aberrations/ or exp aneuploidy/ (518) 2 exp Preimplantation Diagnosis/ (39) 3 (Preimplant* adj3 gene*).tw. (244) 4 aneuploid*.tw. (450) 5 (PGS or PGD).tw. (571) 6 chromosom*.tw. (2292) 7 exp in situ hybridization/ or exp in situ hybridization, fluorescence/ (303) 8 In Situ Hybridi?ation.tw. (990) 9 FISH.tw. (5518) 10 (Preimplant* adj3 Diagnos*).tw. (93) 11 (Preimplant$ adj3 test*).tw. (44) 12 (Preimplant* adj3 screen*).tw. (141) 13 (gene* adj3 screen*).tw. (1204) 14 exp Comparative Genomic Hybridization/ (10) 15 (genom$ adj2 hybridi?ation).tw. (106) 16 CGH.tw. (128) 17 aCGH.tw. (56) 18 exp Polymerase Chain Reaction/ (2195) 19 Quantitative Polymerase.tw. (460) 20 (qPCR or SNP).tw. (2516) 21 Polymorphism, Single Nucleotide/ (1367) 22 single nucleotide polymorphism.tw. (586) 23 Nucleic Acid Amplification Techniques/ (29) 24 nucleic acid amplification.tw. (147) 25 whole genome amplification.tw. (33) 26 WGA.tw. (33) 27 Next generation sequencing.tw. (791) 28 (polar body adj3 biops*).tw. (6) 29 (trophectoderm adj3 biops*).tw. (90) 30 spent culture medi*.tw. (12) 31 (Pre implant* adj3 screen*).tw. (9) 32 (Pre implant* adj3 test*).tw. (6) 33 (Pre implant* adj3 diagnos*).tw. (9) 34 or/1‐33 (16669) 35 exp Fertilization in Vitro/ (2012) 36 exp Embryo Transfer/ (1063) 37 (embryo* or blastocyst* or blastomer*).tw. (6728) 38 in vitro fertili?ation.tw. (2965) 39 (ivf or ICSI).tw. (6362) 40 intracytoplasmic sperm injection*.tw. (1123) 41 poor implantation*.tw. (6) 42 pregnancy fail*.tw. (110) 43 recurrent implantation failure*.tw. (140) 44 recurrent miscarriage*.tw. (254) 45 (advanced adj2 age*).tw. (1481) 46 recurrent pregnancy loss*.tw. (181) 47 or/35‐46 (11746) 48 34 and 47 (639)
Appendix 3. MEDLINE search strategy
Searched from 1946 to 9 September 2019
Ovid platform
1 exp chromosome aberrations/ or exp aneuploidy/ (150579) 2 exp Preimplantation Diagnosis/ (3020) 3 (Preimplant* adj3 gene*).tw. (3324) 4 aneuploid*.tw. (19922) 5 (PGS or PGD).tw. (11332) 6 chromosom*.tw. (344530) 7 exp in situ hybridization/ or exp in situ hybridization, fluorescence/ (92080) 8 In Situ Hybridi?ation.tw. (95705) 9 FISH.tw. (155327) 10 (Preimplant* adj3 Diagnos*).tw. (2735) 11 (Preimplant$ adj3 test*).tw. (461) 12 (Preimplant* adj3 screen*).tw. (560) 13 (gene* adj3 screen*).tw. (31714) 14 exp Comparative Genomic Hybridization/ (5930) 15 (genom$ adj2 hybridi?ation).tw. (10663) 16 CGH.tw. (6462) 17 aCGH.tw. (1630) 18 exp Polymerase Chain Reaction/ (441419) 19 Quantitative Polymerase.tw. (19081) 20 (qPCR or SNP).tw. (87333) 21 Polymorphism, Single Nucleotide/ (109177) 22 single nucleotide polymorphism.tw. (23704) 23 Nucleic Acid Amplification Techniques/ (9982) 24 nucleic acid amplification.tw. (3431) 25 whole genome amplification.tw. (937) 26 WGA.tw. (5203) 27 Next generation sequencing.tw. (26557) 28 (polar body adj3 biops*).tw. (82) 29 (trophectoderm adj3 biops*).tw. (279) 30 spent culture medi*.tw. (346) 31 (Pre implant* adj3 screen*).tw. (77) 32 (Pre implant* adj3 test*).tw. (67) 33 (Pre implant* adj3 diagnos*).tw. (364) 34 or/1‐33 (1237396) 35 exp Fertilization in Vitro/ (34607) 36 exp Embryo Transfer/ (15580) 37 (embryo* or blastocyst* or blastomer*).tw. (345625) 38 in vitro fertili?ation.tw. (22191) 39 (ivf or ICSI).tw. (26262) 40 intracytoplasmic sperm injection*.tw. (6870) 41 poor implantation*.tw. (40) 42 pregnancy fail*.tw. (980) 43 recurrent implantation failure*.tw. (312) 44 recurrent miscarriage*.tw. (2016) 45 (advanced adj2 age*).tw. (21335) 46 recurrent pregnancy loss*.tw. (1833) 47 or/35‐46 (394966) 48 34 and 47 (60634) 49 randomized controlled trial.pt. (488816) 50 controlled clinical trial.pt. (93272) 51 randomized.ab. (453764) 52 randomised.ab. (90509) 53 placebo.tw. (206022) 54 clinical trials as topic.sh. (188234) 55 randomly.ab. (317666) 56 trial.ti. (204296) 57 (crossover or cross‐over or cross over).tw. (81543) 58 or/49‐57 (1299117) 59 exp animals/ not humans.sh. (4615941) 60 58 not 59 (1195133) 61 48 and 60 (539)
Appendix 4. Embase search strategy
Searched from 1980 to 9 September 2019
Ovid platform
1 exp prenatal diagnosis/ (99852) 2 exp chromosome aberration/ (172013) 3 (Preimplant* adj3 gene*).tw. (5414) 4 aneuploid$.tw. (25977) 5 (PGS or PGD).tw. (14996) 6 chromosom*.tw. (378625) 7 exp in situ hybridization/ or exp hybridization/ (315511) 8 In Situ Hybridi?ation.tw. (111385) 9 (Preimplant* adj3 Diagnos*).tw. (3934) 10 (gene* adj3 screen*).tw. (43126) 11 exp comparative genomic hybridization/ (17746) 12 (genom* adj3 hybridi?ation).tw. (15601) 13 CGH.tw. (10850) 14 aCGH.tw. (3568) 15 (preimplant* adj3 screen*).tw. (1494) 16 exp polymerase chain reaction/ (863341) 17 Quantitative Polymerase.tw. (21564) 18 (qPCR or SNP).tw. (145147) 19 exp single nucleotide polymorphism/ (170844) 20 single nucleotide polymorphism.tw. (29777) 21 nucleic acid amplification/ (8579) 22 Nucleic Acid Amplification.tw. (4909) 23 whole genome amplification.tw. (1685) 24 WGA.tw. (5961) 25 Next generation sequencing.tw. (44684) 26 exp next generation sequencing/ (41183) 27 (polar body adj3 biops*).tw. (162) 28 (trophectoderm adj3 biops*).tw. (1096) 29 spent culture medi*.tw. (432) 30 (Pre implant* adj3 diagnos*).tw. (657) 31 (Pre implant* adj3 test*).tw. (137) 32 (Pre implant* adj3 screen*).tw. (241) 33 or/1‐32 (1817829) 34 exp in vitro fertilization/ (66328) 35 (embryo* or blastocyst* or blastomere*).tw. (381802) 36 in vitro fertili?ation.tw. (29209) 37 (ivf or ICSI).tw. (45598) 38 intracytoplasmic sperm injection*.tw. (9229) 39 poor implantation*.tw. (65) 40 pregnancy fail*.tw. (1270) 41 recurrent implantation failure*.tw. (701) 42 recurrent pregnancy loss*.tw. (3197) 43 recurrent miscarriage*.tw. (3502) 44 (advanced adj2 age*).tw. (31235) 45 or/34‐44 (459904) 46 33 and 45 (80839) 47 Clinical Trial/ (952533) 48 Randomized Controlled Trial/ (565068) 49 exp randomization/ (84159) 50 Single Blind Procedure/ (36462) 51 Double Blind Procedure/ (162252) 52 Crossover Procedure/ (60492) 53 Placebo/ (327459) 54 Randomi?ed controlled trial$.tw. (210783) 55 Rct.tw. (33803) 56 random allocation.tw. (1909) 57 randomly.tw. (417169) 58 randomly allocated.tw. (33204) 59 allocated randomly.tw. (2474) 60 (allocated adj2 random).tw. (809) 61 Single blind$.tw. (23300) 62 Double blind$.tw. (195064) 63 ((treble or triple) adj blind$).tw. (1005) 64 placebo$.tw. (290276) 65 prospective study/ (548416) 66 or/47‐65 (2309129) 67 case study/ (64016) 68 case report.tw. (380889) 69 abstract report/ or letter/ (1072000) 70 or/67‐69 (1507016) 71 66 not 70 (2257002) 72 (exp animal/ or animal.hw. or nonhuman/) not (exp human/ or human cell/ or (human or humans).ti.) (5806303) 73 71 not 72 (2100129) 74 46 and 73 (2176)
Appendix 5. PsycINFO search strategy
Searched from 1806 to 9 September 2019
Ovid platform
1 exp Prenatal Diagnosis/ (678) 2 (Preimplant$ adj3 gene$).tw. (115) 3 exp Chromosome Disorders/ (9749) 4 aneuploid$.tw. (227) 5 (Preimplant$ adj2 screen$).tw. (3) 6 (PGS or PGD).tw. (655) 7 chromosom*.tw. (9183) 8 In Situ Hybridi?ation.tw. (3480) 9 (Preimplant* adj3 Diagnos*).tw. (112) 10 (gene* adj3 screen*).tw. (2299) 11 (genom* adj3 hybridi?ation).tw. (228) 12 (CGH or aCGH).tw. (175) 13 Quantitative Polymerase.tw. (493) 14 (qPCR or SNP).tw. (4869) 15 single nucleotide polymorphism.tw. (2493) 16 Nucleic Acid Amplification.tw. (54) 17 whole genome amplification.tw. (3) 18 WGA.tw. (128) 19 Next generation sequencing.tw. (568) 20 spent culture medi*.tw. (1) 21 (Pre implant* adj3 diagnos*).tw. (27) 22 (Pre implant* adj3 test*).tw. (6) 23 (Pre implant* adj3 screen*).tw. (0) 24 or/1‐23 (30228) 25 exp Reproductive Technology/ (1761) 26 exp Infertility/ (2091) 27 (ivf or ICSI).tw. (576) 28 (embryo* or blastocyst* or blastomer*).tw. (10906) 29 in vitro fertili?ation.tw. (724) 30 intracytoplasmic sperm injection*.tw. (56) 31 poor implantation*.tw. (1) 32 pregnancy fail*.tw. (36) 33 recurrent pregnancy loss*.tw. (15) 34 recurrent miscarriage*.tw. (27) 35 (advanced adj2 age*).tw. (2068) 36 or/25‐35 (16302) 37 24 and 36 (868) 38 random.tw. (56139) 39 control.tw. (430242) 40 double‐blind.tw. (22341) 41 clinical trials/ (11428) 42 placebo/ (5346) 43 exp Treatment/ (1012719) 44 or/38‐43 (1397582) 45 37 and 44 (202)
Appendix 6. CINAHL search strategy
Searched from 1961 to 9 September 2019
Ebsco platform
| # | Query | Results |
| S57 | S44 AND S56 | 329 |
| S56 | S45 OR S46 OR S47 OR S48 OR S49 OR S50 OR S51 OR S52 OR S53 OR S54 OR S55 | 1,348,419 |
| S55 | TX allocat* random* | 10,952 |
| S54 | (MH "Quantitative Studies") | 23,227 |
| S53 | (MH "Placebos") | 11,446 |
| S52 | TX placebo* | 58,971 |
| S51 | TX random* allocat* | 10,952 |
| S50 | (MH "Random Assignment") | 56,376 |
| S49 | TX randomi* control* trial* | 174,775 |
| S48 | TX ( (singl* n1 blind*) or (singl* n1 mask*) ) or TX ( (doubl* n1 blind*) or (doubl* n1 mask*) ) or TX ( (tripl* n1 blind*) or (tripl* n1 mask*) ) or TX ( (trebl* n1 blind*) or (trebl* n1 mask*) ) | 1,030,939 |
| S47 | TX clinic* n1 trial* | 250,159 |
| S46 | PT Clinical trial | 86,851 |
| S45 | (MH "Clinical Trials+") | 266,213 |
| S44 | S31 AND S43 | 2,527 |
| S43 | S32 OR S33 OR S34 OR S35 OR S36 OR S37 OR S38 OR S39 OR S40 OR S41 OR S42 | 42,195 |
| S42 | TX recurrent pregnancy loss* | 659 |
| S41 | TX (advanced N2 age*) | 4,923 |
| S40 | TX recurrent miscarriage* | 557 |
| S39 | TX recurrent implantation failure* | 249 |
| S38 | TX pregnancy fail* | 7,850 |
| S37 | TX poor implantation* | 922 |
| S36 | TX (ivf or ICSI) | 4,828 |
| S35 | TX in vitro fertili?ation | 6,852 |
| S34 | TX (embryo* or blastocyst* or blastomer*) | 23,319 |
| S33 | TX intracytoplasmic sperm injection* | 866 |
| S32 | (MM "Fertilization in Vitro") | 3,354 |
| S31 | S1 OR S2 OR S3 OR S4 OR S5 OR S6 OR S7 OR S8 OR S9 OR S10 OR S11 OR S12 OR S13 OR S14 OR S15 OR S16 OR S17 OR S18 OR S19 OR S20 OR S21 OR S22 OR S23 OR S24 OR S25 OR S26 OR S27 OR S28 OR S29 OR S30 | 54,932 |
| S30 | TX (Pre implant* N3 test*) | 202 |
| S29 | TX (Pre implant* N3 diagnos*) | 121 |
| S28 | TX (Pre implant* N3 screen*) | 50 |
| S27 | TX (spent culture medi*) | 705 |
| S26 | TX (trophectoderm N3 biops*) | 102 |
| S25 | TX (polar body N3 biops*) | 8 |
| S24 | TX Next generation sequenc* | 3,502 |
| S23 | TX WGA | 101 |
| S22 | TX whole genome amplification* | 154 |
| S21 | TX Nucleic Acid Amplification Technique* | 1,436 |
| S20 | (MM "Nucleic Acid Amplification Techniques+") | 3,510 |
| S19 | TX single nucleotide polymorphism | 8,513 |
| S18 | (MM "Polymorphism, Genetic") | 14,077 |
| S17 | Polymorphism, Single Nucleotide | 8,444 |
| S16 | TX (qPCR or SNP) | 9,861 |
| S15 | TX Quantitative Polymerase | 7,315 |
| S14 | (MM "Polymerase Chain Reaction+") | 3,016 |
| S13 | TX aCGH | 174 |
| S12 | TX CGH | 465 |
| S11 | TX (genom* N2 hybridi?ation) | 667 |
| S10 | (MM "Nucleic Acid Hybridization") | 83 |
| S9 | TX (gene* N2 screen*) | 14,852 |
| S8 | TX (Preimplant* N2 screen*) | 202 |
| S7 | TX Preimplant* Diagnos* | 896 |
| S6 | (MM "In Situ Hybridization+") OR (MM "In Situ Hybridization, Fluorescence+") | 450 |
| S5 | TX (PGS or PGD‐AS) | 1,296 |
| S4 | TX aneuploid* | 2,332 |
| S3 | TX (Preimplant* N3 gene*) | 758 |
| S2 | (MM "Aneuploidy") | 615 |
| S1 | (MM "Preimplantation Diagnosis") | 500 |
Data and analyses
Comparison 1. IVF with PGT‐A versus IVF without PGT‐A with the use of genome‐wide analyses.
| Outcome or subgroup title | No. of studies | No. of participants | Statistical method | Effect size |
|---|---|---|---|---|
| 1.1 Cumulative live birth rate after the first treatment cycle, per woman randomised | 1 | Odds Ratio (M‐H, Random, 95% CI) | Totals not selected | |
| 1.1.1 Polar body biopsy | 1 | Odds Ratio (M‐H, Random, 95% CI) | Totals not selected | |
| 1.2 Live birth rate after the first embryo transfer per woman randomised | 2 | Odds Ratio (M‐H, Fixed, 95% CI) | Subtotals only | |
| 1.2.1 Polar body biopsy | 1 | 396 | Odds Ratio (M‐H, Fixed, 95% CI) | 1.10 [0.68, 1.79] |
| 1.2.2 Blastocyst stage biopsy | 1 | 661 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.93 [0.69, 1.27] |
| 1.3 Miscarriage rate per woman randomised | 2 | Odds Ratio (M‐H, Fixed, 95% CI) | Subtotals only | |
| 1.3.1 Polar body biopsy | 1 | 396 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.45 [0.23, 0.88] |
| 1.3.2 Blastocyst stage biopsy | 1 | 661 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.89 [0.52, 1.54] |
| 1.4 Miscarriage rate per clinical pregnancy | 1 | Odds Ratio (M‐H, Fixed, 95% CI) | Subtotals only | |
| 1.4.1 Polar body biopsy | 1 | 136 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.47 [0.22, 1.00] |
| 1.5 Clinical pregnancy per woman randomised | 1 | Odds Ratio (M‐H, Random, 95% CI) | Totals not selected | |
| 1.5.1 Polar body biopsy | 1 | Odds Ratio (M‐H, Random, 95% CI) | Totals not selected | |
| 1.6 Multiple pregnancy per woman randomised | 1 | 396 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.53 [0.20, 1.37] |
| 1.6.1 Polar body biopsy | 1 | 396 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.53 [0.20, 1.37] |
| 1.7 Multiple pregnancy rate per live birth | 1 | 95 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.45 [0.16, 1.26] |
| 1.7.1 Polar body biopsy | 1 | 95 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.45 [0.16, 1.26] |
| 1.8 Proportion of women reaching embryo transfer per woman randomised | 2 | Odds Ratio (M‐H, Fixed, 95% CI) | Subtotals only | |
| 1.8.1 Polar body biopsy | 1 | 396 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.31 [0.18, 0.54] |
| 1.8.2 Blastocyst stage biopsy | 1 | 661 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.28 [0.16, 0.49] |
| 1.9 Mean number of embryos transferred per transfer | 2 | Mean Difference (IV, Fixed, 95% CI) | Subtotals only | |
| 1.9.1 Polar body biopsy | 1 | 426 | Mean Difference (IV, Fixed, 95% CI) | ‐0.40 [‐0.49, ‐0.31] |
| 1.9.2 Blastocyst stage biopsy | 1 | 587 | Mean Difference (IV, Fixed, 95% CI) | Not estimable |
Comparison 2. IVF with PGT‐A versus IVF without PGT‐A with the use of FISH for the genetic analysis.
| Outcome or subgroup title | No. of studies | No. of participants | Statistical method | Effect size |
|---|---|---|---|---|
| 2.1 Cumulative live birth rate after the first treatment cycle, per woman randomised | 1 | Odds Ratio (M‐H, Fixed, 95% CI) | Totals not selected | |
| 2.1.1 Cleavage stage biopsy | 1 | Odds Ratio (M‐H, Fixed, 95% CI) | Totals not selected | |
| 2.2 Live birth rate after the first embryo transfer per woman randomised | 10 | 1680 | Odds Ratio (M‐H, Random, 95% CI) | 0.62 [0.43, 0.91] |
| 2.2.1 Cleavage stage biopsy | 9 | 1579 | Odds Ratio (M‐H, Random, 95% CI) | 0.66 [0.44, 0.98] |
| 2.2.2 Blastocyst stage biopsy | 1 | 101 | Odds Ratio (M‐H, Random, 95% CI) | 0.40 [0.18, 0.90] |
| 2.3 Miscarriage rate per woman randomised | 10 | 1680 | Odds Ratio (M‐H, Fixed, 95% CI) | 1.03 [0.75, 1.41] |
| 2.3.1 Cleavage stage biopsy | 9 | 1579 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.99 [0.72, 1.36] |
| 2.3.2 Blastocyst stage biopsy | 1 | 101 | Odds Ratio (M‐H, Fixed, 95% CI) | 4.50 [0.51, 39.99] |
| 2.4 Miscarriage rate per clinical pregnancy | 5 | 288 | Odds Ratio (M‐H, Fixed, 95% CI) | 1.77 [1.10, 2.86] |
| 2.4.1 Cleavage stage biopsy | 4 | 267 | Odds Ratio (M‐H, Fixed, 95% CI) | 1.93 [1.17, 3.19] |
| 2.4.2 Blastocyst stage biopsy | 1 | 21 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.53 [0.08, 3.76] |
| 2.5 Ongoing pregnancy rate per woman randomised | 5 | 1121 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.68 [0.51, 0.90] |
| 2.5.1 Cleavage stage biopsy | 5 | 1121 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.68 [0.51, 0.90] |
| 2.6 Clinical pregnancy per woman randomised | 5 | 1131 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.60 [0.45, 0.81] |
| 2.6.1 Cleavage stage biopsy | 4 | 1030 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.63 [0.46, 0.86] |
| 2.6.2 Blastocyst stage biopsy | 1 | 101 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.47 [0.21, 1.04] |
| 2.7 Multiple pregnancy rate per woman randomised | 6 | 1331 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.66 [0.37, 1.17] |
| 2.7.1 Cleavage stage biopsy | 6 | 1331 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.66 [0.37, 1.17] |
| 2.8 Multiple pregnancy rate per live birth | 6 | 364 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.97 [0.51, 1.82] |
| 2.8.1 Cleavage stage biopsy | 6 | 364 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.97 [0.51, 1.82] |
| 2.9 Proportion of women reaching embryo transfer per woman randomised | 9 | 1286 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.54 [0.40, 0.74] |
| 2.9.1 Cleavage stage biopsy | 8 | 1185 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.55 [0.40, 0.75] |
| 2.9.2 Blastocyst stage biopsy | 1 | 101 | Odds Ratio (M‐H, Fixed, 95% CI) | 0.39 [0.02, 9.82] |
| 2.10 Mean number of embryos transferred per transfer | 7 | 1433 | Mean Difference (IV, Fixed, 95% CI) | ‐0.23 [‐0.30, ‐0.16] |
| 2.10.1 Cleavage stage biopsy | 7 | 1433 | Mean Difference (IV, Fixed, 95% CI) | ‐0.23 [‐0.30, ‐0.16] |
Characteristics of studies
Characteristics of included studies [ordered by study ID]
Blockeel 2008.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between March 2001 and December 2007 in Belgium Sample size: a sample size calculation was performed 134 women randomised |
|
| Participants |
Inclusion criteria: couples with 3 or more failed IVF or ICSI attempts with embryos of good morphological quality, maternal age < 37 years and a normal karyotype in both partners Exclusion criteria: not applicable |
|
| Interventions |
Type of biopsy: cleavage stage biopsy. Oocytes with 2 pronuclei were assessed on day 2 and day 3 after injection for embryonic development, and embryos reaching at least the 5‐cell stage on day 3 of development were biopsied. Genetic analysis: performed by FISH, analysis for chromosomes X, Y, 13, 16, 18, 21, 22 |
|
| Outcomes | Miscarriage rate Ongoing pregnancy rate Clinical pregnancy rate Multiple pregnancy rate Proportion of women reaching embryo transfer Mean number of embryos per transfer |
|
| Notes |
Treatment procedure: ICSI only Embryo transfer policy: maximum 3 embryos for transfer Note: between 2001 and October 2005, 2 blastomeres were removed from those embryos with at least 6 blastomeres. From November 2005 onwards, only 1 blastomere was removed from the embryo. |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Quote: "A computer‐generated list was used for randomization" |
| Allocation concealment (selection bias) | Unclear risk | Judgement comment: the list was not concealed from the physicians |
| Blinding of participants and personnel (performance bias) All outcomes | High risk | Judgement comment: no blinding |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: flow chart of attrition including reasons |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | Low risk | Judgement comment: no sources of bias detected. No funding |
Debrock 2010.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between June 2002 and January 2007 in Belgium Sample size calculation: not stated 94 women randomised |
|
| Participants |
Inclusion criteria: women 35 years or older, with at least 2 fertilised oocytes available on day 1 after oocyte retrieval, and with at least 2 embryos consisting of 6 or more cells at day 3 after oocyte retrieval Exclusion criteria: NA |
|
| Interventions |
Type of biopsy: cleavage stage biopsy. Biopsy on embryos with at least 6 blastomeres on day 3. 2 blastomeres with a nucleus were gently aspirated from each embryo. Genetic analysis: performed by FISH analysis for chromosomes X, Y, 13, 16, 18, 21, 22 |
|
| Outcomes | live birth rate Miscarriage rate Ongoing pregnancy rate Clinical pregnancy rate Multiple pregnancy rate Proportion of women reaching embryo transfer Mean number of embryos per transfer |
|
| Notes |
Treatment procedure: IVF and ICSI Embryo transfer policy: before 1 July 2003 a maximum of 2 to 3 embryos were transferred, after that only 1 embryo was transferred in the first trial in women less than 36 years old. Maximum 3 embryos for transfer |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Judgement comment: method of randomisation with sealed envelopes |
| Allocation concealment (selection bias) | Unclear risk | Judgement comment: "sealed" envelopes, but not stated if opaque |
| Blinding of participants and personnel (performance bias) All outcomes | High risk | Judgement comment: no blinding |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: flow chart of attrition including reasons |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | Low risk | Judgement comment: no sources of bias detected. Unclear whether this was a sponsored trial |
Hardarson 2008.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between November 2003 and February 2007 in Sweden Sample size calculation: a sample size with power calculation was performed 56 and 53 participants were randomised into the PGT‐A and control group, respectively. |
|
| Participants |
Inclusion criteria: couples with infertility of female or male origin, intending to undergo IVF or ICSI, who had signed a written consent form and in which the age of the woman was ≥38 years were eligible for randomisation. The couple had to have at least 3 embryos of GQE, only 2 GQE were required if the participant only wanted 1 embryo back. Exclusion criteria: patients who had previously been randomised to either of the 2 study groups in this trial |
|
| Interventions |
Type of biopsy: cleavage stage biopsy. Biopsy on embryos with at least 6 cells and less than 20% fragmentation. Mostly 1 blastomere removed. Genetic analysis: FISH analysis for chromosomes X, Y, 13, 18, 21 |
|
| Outcomes | live birth rate Miscarriage rate Ongoing pregnancy rate Clinical pregnancy rate Proportion of women reaching embryo transfer Mean number of embryos per transfer |
|
| Notes |
Treatment procedure: IVF or ICSI Embryo transfer policy: maximum 2 embryos for transfer |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Quote: "Randomization into two groups was performed using a computerized randomization program" |
| Allocation concealment (selection bias) | Unclear risk | Judgement comment: concealment of allocation not stated |
| Blinding of participants and personnel (performance bias) All outcomes | High risk | Judgement comment: no blinding |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: number of randomised participants is the number of analysed participants |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | High risk | Judgement comment: high risk of within‐study bias due to the difference in day of embryo transfer between study arms (day 5 for intervention and day 3 for control) Funding: sponsored by the Swedish Medical Research Council and Serono Nordic AB |
Jansen 2008.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between August 2004 and November 2006 in Sydney, Australia Sample size calculation: not performed 101 women randomised |
|
| Participants |
Inclusion criteria: women below 38 years of age in their first or second IVF with no cancelled cycles because of poor response, agreement to elective single embryo transfer, with at least 8 follicles of 1 cm or more on day 8 to 10 of stimulation Exclusion criteria: women with fewer than 4 embryos with eat least 7 cells on day 3, and women with fewer than 2 blastocysts for biopsy were excluded |
|
| Interventions |
Type of biopsy: blastocyst stage biopsy, biopsy of the trophectoderm. Biopsies consisted of 2–9 trophectoderm cells. Genetic analysis: FISH analysis for chromosomes X, Y, 13, 18, 21 |
|
| Outcomes | live birth rate Miscarriage rate Clinical pregnancy rate Proportion of women reaching embryo transfer |
|
| Notes |
Treatment procedure: method for fertilisation not reported Embryo transfer policy: single embryo transfer Note: the trial was suspended and then terminated earlier when it was unable to show an advantage for PGS |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Judgement comment: randomised but no further details |
| Allocation concealment (selection bias) | Unclear risk | Judgement comment: used "sealed envelopes", no details as to whether they were opaque |
| Blinding of participants and personnel (performance bias) All outcomes | High risk | Judgement comment: no blinding |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: all randomised participants analysed. Also data on women who were withdrawn from the study before randomisation because of suboptimal responses to stimulation, and data on women who were eligible but elected not to take part in the study |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | Low risk | Judgement comment: no sources of bias detected Funding: unclear whether this was a sponsored trial |
Mastenbroek 2007.
| Study characteristics | ||
| Methods |
Study design: multicentre, randomised, double‐blind, controlled trial Duration and location of the trial: between May 2003 and November 2005, in the Netherlands Sample size calculation: a sample size with power calculation was performed 408 women randomised An intention‐to‐treat analysis was performed. |
|
| Participants |
Inclusion criteria: women between 35 and 41 years of age with no previous failed IVF cycles Exclusion criteria: women who objected to double embryo transfer were excluded |
|
| Interventions |
Type of biopsy: cleavage stage biopsy. Biopsy on embryos with at least 4 blastomeres and with a maximum of 50% fragmentation, mostly 1 blastomere removed Genetic analysis: FISH analysis for chromosomes X, Y, 1, 13, 16, 17, 18, 21 |
|
| Outcomes | Cumulative live birth rate Miscarriage rate Ongoing pregnancy rate Clinical pregnancy rate Multiple pregnancy rate Proportion of women reaching embryo transfer Mean number of embryos per transfer |
|
| Notes |
Treatment procedure: IVF and ICSI Embryo transfer policy: maximum 2 embryos for transfer |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Judgement comment: randomisation by computer |
| Allocation concealment (selection bias) | Low risk | Judgement comment: used computer randomisation with concealment of allocation |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | Judgement comment: blinding of participants and personnel performed |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: blinding of outcome assessment performed |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: number of randomised participants is the number of analysed participants |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | Low risk | Judgement comment: no suggestion of other bias Funding: supported by grant from the Netherlands Organisation for Health Research and Development (ZonMw) |
Meyer 2009.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between June 2004 and November 2006, private infertility clinic, Illinois, USA Sample size calculation: a sample size with power calculation was performed 47 women randomised An intention‐to‐treat analysis was performed. |
|
| Participants |
Inclusion criteria: women below 39 years of age with a normal ovarian reserve, a body mass index below 30 kg/m², a normal uterus, no more than 2 previous failed IVF cycles with at least 4 embryos containing at least 5 cells with less than 40% fragmentation Exclusion criteria: smoking history, the presence of a hydrosalpinx or uterine anomaly, a history of more than 2 previous spontaneous pregnancy losses, and the presence of severe male factor infertility |
|
| Interventions |
Type of biopsy: cleavage stage biopsy, mostly 1 blastomere removed Genetic analysis: FISH analysis for chromosomes X, Y, 13, 16, 17, 18, 21, 22 |
|
| Outcomes | live birth rate Miscarriage rate Clinical pregnancy rate Proportion of women reaching embryo transfer Mean number of embryos per transfer |
|
| Notes |
Treatment procedure: ICSI only Embryo transfer policy: maximum embryos for transfer not reported |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Judgement comment: computer‐generated randomisation |
| Allocation concealment (selection bias) | Low risk | Judgement comment: sealed, opaque envelopes randomisation |
| Blinding of participants and personnel (performance bias) All outcomes | High risk | Judgement comment: not blinded |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: 4 participants declined transfers for reasons not related to the study, all other participants randomised were analysed |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | Low risk | Judgement comment: no suggestion of other bias Funding: supported in part by Serono |
Munné 2019.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: from October 2014 to January 2016, multinational, multicentre, 34 clinics, 9 laboratories across the USA, Canada, the UK, and Australia Sample size calculation: a sample size with power calculation was performed 661 women randomised |
|
| Participants |
Inclusion criteria: female age 25 to 40 years undergoing IVF with autologous oocytes with at least 2 blastocysts of sufficient quality for biopsy and vitrification by day 6 Exclusion criteria: diminished ovarian reserve, more than 2 previous failed IVF‐ET, more than 1 miscarriage, azoospermia, or severe oligospermia |
|
| Interventions |
Type of biopsy: blastocyst stage biopsy Genetic analysis: comprehensive chromosome screening analysis, using next‐generation sequencing |
|
| Outcomes | live birth rate Ongoing pregnancy rate Clinical pregnancy rate Miscarriage rate |
|
| Notes |
Treatment procedure: IVF Embryo transfer policy: single frozen‐thawed embryo transfer |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Judgement comment: randomisation by computer. An electronic data capture system with a randomisation module was used to randomise participants 1:1 into the control and PGT‐A arms. |
| Allocation concealment (selection bias) | Low risk | Judgement comment: concealment of allocation |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | Quote: "The providers performing the transfer and the study subjects were blinded to the randomisation status and PGT‐A results until after the embryo transfer or pregnancy outcome was known. The genetic laboratory investigators and personnel performing PGS were blinded to the aggregate pregnancy outcomes until study completion." |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: 7/330 and 14/331 participants withdrew during the study from PGT‐A and control groups, respectively |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | Low risk | Judgement comment: no suggestion of other bias Funding: supported by Illumina |
Rubio 2013.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between 2004 and December 2011, private infertility clinic, Valencia, Spain Sample size: a sample size calculation was performed 90 women randomised |
|
| Participants |
Inclusion criteria: FSH levels < 10 IU/mL, E2 < 65 pg/mL, antral follicle count 8 or more, and 5 or more metaphase II (MII) oocytes in previous cycles. Women < 40 years; 3 or more implantation failures in previous IVF/ICSI cycle with transfer of at least 2 good‐quality, fresh embryos in each cycle; and no abnormality in the infertility work‐up performed Exclusion criteria: any abnormality detected in the previous infertility work‐up; hydrosalpinx; previous ectopic pregnancies or miscarriages; presence of myomas, adhesions, or polyps; previous ET with high difficulty or bleeding, or both; patients with other indications for PGD/PGS; and patients with fewer than 5 MII oocytes in a single cycle |
|
| Interventions |
Type of biopsy: cleavage stage biopsy Genetic analysis: genetic analysis with FISH analysis |
|
| Outcomes | live birth rate Miscarriage rate Multiple pregnancy rate Proportion of women reaching embryo transfer Mean number of embryos per transfer |
|
| Notes |
Treatment procedure: ICSI only Embryo transfer policy: maximum embryos for transfer not reported |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Judgement comment: computer‐generated randomisation |
| Allocation concealment (selection bias) | Unclear risk | Judgement comment: concealment of allocation not reported |
| Blinding of participants and personnel (performance bias) All outcomes | High risk | Judgement comment: not blinded |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: number of randomised participants is the number of analysed participants |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | Low risk | Judgement comment: no suggestion of other bias. Funding: unclear whether this was a sponsored trial |
Schoolcraft 2009.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between May 2002 and March 2004, private fertility clinic, New Jersey, USA Sample size: not performed 62 women randomised |
|
| Participants |
Inclusion criteria: women 35 years of age or older with at least 5 embryos consisting of at least 6 cells and less than 15% fragmentation on day 3 Exclusion criteria: ‐ |
|
| Interventions |
Type of biopsy: cleavage stage biopsy Genetic analysis: genetic analysis with FISH analysis |
|
| Outcomes | live birth rate Miscarriage rate Proportion of women reaching embryo transfer |
|
| Notes |
Treatment procedure: IVF and ICSI Embryo transfer policy: maximum number of embryos transferred not reported |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Quote: "eligible couples were assigned randomly by a computer‐generated random number table" |
| Allocation concealment (selection bias) | Unclear risk | Judgement comment: concealment of allocation not reported |
| Blinding of participants and personnel (performance bias) All outcomes | High risk | Judgement comment: not blinded |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: all women randomised were analysed |
| Selective reporting (reporting bias) | Low risk | Judgement comment: a priori outcomes reported |
| Other bias | Low risk | Judgement comment: no suggestion of other bias Funding: unclear whether this was a sponsored trial |
Staessen 2004.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between March 2000 and December 2003 in the University Hospital, Dutch‐speaking Brussels Free University Sample size: not performed 400 women randomised, note: 1 woman in the intervention group and 10 women in the control group did not fulfil the inclusion criteria |
|
| Participants | Inclusion criteria: women 37 years or older receiving ICSI as infertility treatment Exclusion criteria: patients with an abnormal karyotype or non‐motile sperm were excluded | |
| Interventions |
Type of biopsy: cleavage stage biopsy Genetic analysis: genetic analysis with FISH analysis |
|
| Outcomes | live birth rate Miscarriage rate Ongoing pregnancy rate Clinical pregnancy rate Multiple pregnancy rate Mean number of embryos per transfer |
|
| Notes |
Treatment procedure: ICSI only Embryo transfer policy: a maximum of 3 blastocysts were transferred in women between 37 and 39 years of age, and up to a maximum of 6 blastocysts were transferred in women 40 years of age or older |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Judgement comment: randomised, but method of randomisation is not described |
| Allocation concealment (selection bias) | Unclear risk | Judgement comment: concealment of allocation not reported |
| Blinding of participants and personnel (performance bias) All outcomes | High risk | Judgement comment: not blinded |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | High risk | Judgement comment: data on dropouts not reported |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | Low risk | Judgement comment: no suggestion of other bias |
Staessen 2008.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between October 2004 and December 2006 in the University Hospital, Dutch‐speaking Brussels Free University Sample size: 240 women randomised |
|
| Participants |
Inclusion criteria: women below 36 years of age Exclusion criteria: patients with an abnormal karyotype or non‐motile sperm were excluded |
|
| Interventions |
Type of biopsy: cleavage stage biopsy Genetic analysis: genetic analysis with FISH analysis |
|
| Outcomes | live birth rate Miscarriage rate Ongoing pregnancy rate Clinical pregnancy rate Multiple pregnancy rate Proportion of women reaching embryo transfer Mean number of embryos per transfer |
|
| Notes |
Treatment procedure: ICSI only Embryo transfer policy: ICSI only. Single embryo transfer Note: study recruitment was terminated prematurely on basis of interim analysis |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Judgement comment: randomised ‐ no details |
| Allocation concealment (selection bias) | Unclear risk | Judgement comment: concealment of allocation not reported |
| Blinding of participants and personnel (performance bias) All outcomes | High risk | Judgement comment: clinicians and embryologists who performed the embryo transfer were blinded only with respect to the patients' participation in the study |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | High risk | Judgement comment: data on dropouts not reported |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | Low risk | Judgement comment: no suggestion of other bias Funding: grant from the Belgium Fund for Scientific Research Flanders (FWO‐Vlaanderen) |
Verpoest 2018.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between June 2012 and December 2016, multicentre and multinational Sample size: a sample size calculation was performed 396 women randomised |
|
| Participants |
Inclusion criteria: women aged 36 to 40 years who were being treated for infertility through ICSI were eligible Exclusion criteria: couples meeting 1 or more of the following criteria:
|
|
| Interventions |
Type of biopsy: polar body (PB) biopsy Genetic analysis: comprehensive chromosome screening (CCS) with array comparative genomic hybridisation (aCGH) |
|
| Outcomes | Cumulative live birth rate live birth rate Miscarriage rate Clinical pregnancy rate Multiple pregnancy rate Proportion of women reaching embryo transfer Mean number of embryos per transfer |
|
| Notes |
Treatment procedure: ICSI only Embryo transfer policy: transfer of more than 2 embryos was not allowed. If not all embryos produced a result on PB CCS, single embryo transfer (SET) or double embryo transfer was performed subject to availability of genetically transferable embryos, i.e. if there was only 1 embryo euploid on the basis of PB CCS, SET was performed, and supernumerary embryos without diagnosis were cryopreserved. |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Low risk | Ouote: "Web‐based block randomisation was performed with a block size of 6, with stratification for centre and age categories 36‐37 and 38‐40" |
| Allocation concealment (selection bias) | Low risk | Judgement comment: concealment of allocation |
| Blinding of participants and personnel (performance bias) All outcomes | Low risk | Ouote: "Both the patient and clinicians were blinded at the time of enrolment until the day after the intervention" |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: non‐blinding not likely to affect objective outcomes |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: number of randomised participants is the number of analysed participants |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of other bias |
| Other bias | Low risk | Judgement comment: second cycle when no embryo for embryo transfer in PGT‐A group only (heterogeneity) Funding: this study was funded by the European Society of Human Reproduction and Embryology. Illumina provided micro arrays and other consumables necessary for aCGH testing of polar bodies. |
Werlin 2003.
| Study characteristics | ||
| Methods |
Study design: randomised controlled trial Duration and location of the trial: between August 2001 and August 2002, location and centre are not described Sample size: a sample size calculation was not described |
|
| Participants |
Inclusion criteria: patients with recurrent pregnancy loss, defined as 2 spontaneous pregnancy losses; patients of advanced maternal age, defined as > 38 years of age; and patients with repeated failed IVF cycles, defined as > 2 failed cycles Exclusion criteria: not described |
|
| Interventions |
Type of biopsy: cleavage stage biopsy Genetic analysis: genetic analysis with FISH analysis |
|
| Outcomes | Proportion of women reaching embryo transfer | |
| Notes |
Treatment procedure: ICSI only Embryo transfer policy: in the control group, ET was performed on day 3 or 5 after retrieval based on physician preference |
|
| Risk of bias | ||
| Bias | Authors' judgement | Support for judgement |
| Random sequence generation (selection bias) | Unclear risk | Judgement comment: randomised ‐ no details |
| Allocation concealment (selection bias) | Unclear risk | Judgement comment: concealment of allocation not reported |
| Blinding of participants and personnel (performance bias) All outcomes | High risk | Judgement comment: not described |
| Blinding of outcome assessment (detection bias) All outcomes | Low risk | Judgement comment: no blinding of outcome assessment mentioned |
| Incomplete outcome data (attrition bias) All outcomes | Low risk | Judgement comment: number of randomised participants is the number of analysed participants |
| Selective reporting (reporting bias) | Low risk | Judgement comment: no suggestion of selective reporting |
| Other bias | High risk | Judgement comment: high risk of within‐study bias due to the difference in day of embryo transfer between study arms (day 5 for intervention and day 3 or 5 for control) Funding: unclear whether this was a sponsored trial |
ET: embryo transfer
E2: Estradiol
FISH: Fluorescence in situ hybridisation
FSH: Follicle stimulating hormone
GQE: good morphological quality
ICSI: intracytoplasmic sperm injection
IVF: in vitro fertilisation
PGD: Preimplantation genetic diagnosis
PGS: Pre‐implantation genetic screening
Characteristics of excluded studies [ordered by study ID]
| Study | Reason for exclusion |
|---|---|
| Forman 2013 | Inappropriate study design: SET in the PGS group vs DET in the control group |
| Forman 2014 | Inappropriate study design: SET in the PGS group vs DET in the control group |
| Gianaroli 1997 | Allocation to intervention or control group based on volunteer decision. |
| Gianaroli 1999 | Allocation to intervention or control group based on volunteer decision. |
| Mersereau 2008 | Unknown status: ongoing or stopped. Outcome measures reported as percentages without mentioning the unit of analysis, not possible to calculate the exact numbers. |
| Moayeri 2016 | No data beyond biochemical pregnancy |
| NCT02265614 | Study stopped earlier due to poor recruitment, no published data. |
| Ozgur 2019 | Inappropriate study design: only 1 blastocyst underwent PGT‐A; the rest were morphologically scored |
| Rubio 2017 | Incorrect unit of analysis, e.g. per started cycle. Multiple ovum pick‐ups were allowed to collect multiple oocytes before a transfer was attempted, and it was not possible to calculate the exact numbers per first treatment cycle. |
| Scott 2013 | Only inclusion of patients with an embryo transfer. Study in favour of PGT‐A. If randomisation occurred at cycle start, a percentage would not have had euploid embryos to biopsy or transfer, thus likely altering success rates, based on an intention‐to‐treat analysis. |
| Stevens 2004 | The participants included in this study were also included in another, larger study. |
| Yang 2012 | Only inclusion of patients with an embryo transfer. Study in favour of PGT‐A. If randomisation occurred at cycle start, some percentage would not have had euploid embryos to biopsy or transfer, thus likely altering success rates, based on an intention‐to‐treat analysis. |
DET: Double embryo transfer
PGS: Preimplantation genetic screening
PGT‐A: Preimplantation genetic testing for aneuploidies
SET: Single embryo transfer
Characteristics of studies awaiting classification [ordered by study ID]
NCT02223221.
| Methods | Randomised controlled trial |
| Participants | 189 participants, at least 2 failed pregnancies |
| Interventions | IVF cycles with PGT‐A. Select embryos by SNP‐array‐based PGS for the number of all chromosomes on day 5, only euploid embryos will be transferred. A maximum of 2 embryos will be transferred for each treatment cycle. Up to 3 treatment cycles will be offered. |
| Outcomes | Pregnancy outcome, ongoing pregnancy rate, clinical pregnancy, implantation of transferred embryo |
| Notes | Results information has been submitted to ClinicalTrials.gov by the sponsor or investigator, but is not yet publicly available (or 'posted') on ClinicalTrials.gov. The submitted information may not be available if it is pending quality control (QC) review by the National Library of Medicine (NLM) or if issues identified during QC review are being addressed or corrected by the sponsor or investigator. NLM's limited QC review assesses for apparent errors, deficiencies, or inconsistencies. NLM staff do not verify the scientific validity or relevance of the submitted information. |
IVF: in vitro fertilisation
PGT‐A: Preimplantation genetic testing for aneuploidies
PGS: Preimplantation genetic screening
SNP: single‐nucleotide polymorphism
Characteristics of ongoing studies [ordered by study ID]
NCT01946945.
| Study name | Comparison of Standard ART Practice vs. Trophectoderm Biopsy, Whole Chromosome Analysis by Next Generation Sequencing, and Replacement of a Single Euploid Embryo |
| Methods | Randomised controlled trial |
| Participants | Sample size: 240 Inclusion criteria: all patients medically cleared to do a fresh or frozen embryo transfer; age up to 42 years Exclusion criteria: microsurgical epididymal sperm aspiration (MESA) and testicular sperm extraction (TESE) patients; at least 1 partner carrier of a chromosomal or genetic disease; abnormal ovarian reserve, defined as follicle stimulating hormone (FSH) of > 10 IU/L on day 2 to 4 of the cycle and anti‐mullerian hormone (AMH) < 1 ng/mL (if only 1 of the 2 parameters altered, then patient is acceptable); egg donor cycle (sperm donor is acceptable); gender selection cycles, thaw cycles |
| Interventions |
Control group: all blastocyst embryos will be biopsied on day 5/6, but the biopsies will be frozen and will not be analysed before replacement. Blastocyst embryos will be vitrified for future frozen embryo transfer (FET) cycle. Participants will have a single hatching blastocyst thawed and transferred into the uterus in an FET cycle based on standard embryo quality assessment without NGS. After transfer, all biopsied samples will be analysed (the replaced embryo also, in order to do a non‐selection study). If participants in the control group do not have a pregnancy to term from that FET cycle, euploid frozen blastocysts will be thawed and transferred on the next FET transfer. Test group: all blastocyst embryos will be biopsied on day 5/6, and the biopsies will be analysed using NGS; biopsied blastocyst embryos will be vitrified for a future FET cycle. Participants will have a single hatching euploid blastocyst thawed and transferred into the uterus in an FET cycle. |
| Outcomes | Primary outcome: ongoing implantation rate |
| Starting date | September 2013 |
| Contact information | Allen Kung, BSc Telephone 973‐758‐7970 Email: akung@reprogenetics.com |
| Notes | All embryos will be biopsied, results can therefore not be taken into account in the review. First posted 20 September 2013, last updated 9 September 2014 |
NCT01977144.
| Study name | Screening of Low Responders for Aneuploidy to Improve Reproductive Efficiency (Solaire) |
| Methods | Randomised controlled trial |
| Participants | Sample size: 400 Inclusion criteria: age of female partner < 43 years; AMH < 1.1 or AFC < 8 (within previous year); male must have > 100,000 motile sperm; BMI < 32 Exclusion criteria: diagnosis of endometrial insufficiency; use of oocyte donor/gestational carriers; use of surgical sperm or DNA Banking Communicating hydrosalpinges; single‐gene disorders or sex selection; participation in another study |
| Interventions | The study group will have their embryo(s) biopsied for CCS on day 5, if appropriate, for a fresh day 6 embryo transfer. |
| Outcomes | Primary outcome: delivery rate |
| Starting date | September 2013 |
| Contact information | Christine V Reda, BSN, RN Reproductive Medicine Associates of New Jersey Telephone: 973‐656‐2841 Email: clinicalresearchteam@rmanj.com |
| Notes | First posted 6 November 2013, last updated 5 April 2019 |
NCT02032264.
| Study name | Next Generation Sequencing Screening for Embryonic Ploidy Status (Nexgen) |
| Methods | Randomised controlled trial |
| Participants | Sample size: 309 Inclusion criteria: patient undergoing IVF/CCS (no PGD banking); patient meets ASRM guidelines for double embryo transfer (DET); donor sperm OK; AMH ≥ 1.2; FSH ≤ 12; AFC ≥ 12; max 1 prior failed IVF cycle for patients 35 to 45 years old; patients < 35 years old MUST have 1 prior failed IVF cycle Exclusion criteria: chronic endometrial insufficiency; use of oocyte donor or gestational carriers; medical contraindications to DET; male factor (< 100,000 sperm or surgical sperm); communicating hydrosalpinx (on HSG); single‐gene disorders or sex selection |
| Interventions | Trophectoderm biopsy will be performed on all blastocysts and CCS via next‐generation sequencing screening performed on biopsy samples. Participants will proceed with a single or double embryo transfer of the 1 or 2 morphologically best euploid embryos. |
| Outcomes | Primary outcome: implantation rate |
| Starting date | December 2013 |
| Contact information | Richard T Scott, MD Reproductive Medicine Associates of New Jersey |
| Notes | First posted 10 January 2014, last updated 17 March 2017 |
NCT02353364.
| Study name | Blastocyst Euploidy Assessment and Conditioned embryO traNsfer (BEACON) |
| Methods | Randomised controlled trial |
| Participants | Sample size: 1000 Inclusion criteria: patient undergoing IVF; normal uterine function by ultrasound and absence of hysteromyoma; regular menstrual cycle of 25 to 35 days; normal hormone levels (WHO standard) for LH,E2, progesteron, testosterone, and TSH; FSH 1 to 12 IU/L and follicle number > 5 on day 2 to 3 of menstrual cycle; minimum of 3 blastocysts on day 5 of embryo development; signed consent form Exclusion criteria: known endometriosis; abnormal vaginal bleeding with no known cause; known genital organ system malformation, unsuitable to conceive; known currently active pelvic inflammation; abnormal liver, kidney lab results, with clinical implications; known endocrine or metabolic disorders (pituitary gland, adrenal glands, pancreas, liver, or kidney); known ovarian, breast, uterine, adrenal glands, pituitary gland, or hypothalamus tumour; known abnormal cervical cancer lesions, with clinical implications, within 1 year before PGS; history of chemo‐ or radiotherapy; seropositive for HIV, Hep B, Hep C, or syphilis; known ovarian poor response in previous cycles, i.e. after administration of GnRH for > 20 days; more than 2 implantation failures; more than 2 miscarriages; known altered parental karyotype such as Robertsonian or reciprocal translocation; use of sperm or oocyte donors; severe male factor (surgical retrieval of sperm); preimplantation genetic diagnosis cycles for single‐gene diseases or sex selection; participation in other IVF research studies; patient refusal or inability to follow the protocol for any good reason, including clinical visit or lab test |
| Interventions | Transfer of 1 or 2 biopsied euploid embryo of high morphological grade based on NGS testing using CNV‐Seq (PGS) |
| Outcomes | Primary outcome: ongoing pregnancy |
| Starting date | November 2014 |
| Contact information | Li Wang, PhD, Yao Yuanqing, MD Chinese PLA General Hospital, Beijing, China |
| Notes | First posted 2 February 2015, last updated 2 February 2015 |
NCT02868528.
| Study name | A Study of Preimplantation Genetic Screening With Next Generation Sequencing Technology on Advanced Age Women |
| Methods | Randomised controlled trial |
| Participants | Sample size: 238 Inclusion criteria: premenopausal females, age ≥ 37 years ≤ 44 years; have given birth to a healthy baby; bilateral ovaries; antral follicle count (AFC) ≥ 10 and anti‐mullerian hormone (AMH) ≥ 2.0 ng/mL Exclusion criteria: endometriosis disease; intrauterine adhesions history; intrauterine membrane polyp, tuberculosis and inflammation, uterine malformation, multiple uterine myoma, uterine intramural myoma > 3 cm, submucous myoma; unprocessed hydrosalpinx; adverse reproductive history; greater than or equal to 2 times history of unexplained abortion Chromosomal abnormalities or other genetic disease; infertility caused by male factors, such as puncture testicular without high‐quality embryos in past controlled ovarian hyperstimulation (COH) cycles; patients with poor ovarian response |
| Interventions | After blastocyst culture, blastocyst embryo trophoblast biopsy will be performed and chromosome screening with NGS technology, at the same time, the blastocysts will be frozen, then the blastocysts with normal chromosome will be thawed and transferred. |
| Outcomes | Primary outcome: live birth rate |
| Starting date | September 2016 |
| Contact information | Yi Tang, doctor Reproductive & Genetic Hospital of CITIC‐XIANGYA Telephone +86‐731‐82355100 ext 8608 Email: cstangyi@sina.com |
| Notes | First posted 16 August 2016, last updated 10 May 2017 |
NCT02941965.
| Study name | Preimplantation Genetic Screening in Patients With Male Factor Infertility |
| Methods | Randomised controlled trial |
| Participants | Sample size: 480 Inclusion criteria:
Exclusion criteria:
|
| Interventions | ICSI with PGS, PGS will be applied to select embryos on day 5, only euploid embryos will be transferred. A maximum of 2 embryos will be transferred for each treatment cycle. |
| Outcomes | Primary outcome: live birth rate |
| Starting date | May 2017 |
| Contact information | He‐Feng Huang, MD International Peace Maternity & Child Health Hospital, Shanghai Telephone +86‐21‐18017310186 Email: hefenghuang@126.com |
| Notes | First posted 21 October 2016, last updated 15 March 2017 |
NCT03118141.
| Study name | Cumulative Live Birth Rate With eSET After Preimplantation Genetic Screening Versus Conventional In‐vitro Fertilization (CESE‐PGS) |
| Methods | Randomised controlled trial |
| Participants | Sample size: 1208 Inclusion criteria: female with the first IVF cycle; 20 to 37 years old; women with 3 or more blastocysts on day 5 of embryo culture will be randomised |
| Interventions | Participants in the PGS group will have blastocyst biopsy and sequencing done with 3 good‐quality embryos on day 5. Principle of freeze‐all and single thawed blastocyst transfer will be applied. The transfer order of euploid embryos will be determined by blastocyst morphologic score. The outcome of all euploids transfers within 1 year after randomisation will be followed up. During study, every participant will have at most 1 live birth. |
| Outcomes | Primary outcome: cumulative live birth after transfers of up to 3 single blastocysts |
| Starting date | June 2017 |
| Contact information | Zi‐Jiang Chen, Professor Shandong University, China Telephone +0086 531 85651190 Emaill: chenzijiang@vip.163.com |
| Notes | First posted 18 April 2017, last updated 20 September 2019 |
NCT03173885.
| Study name | Investigating the Cryopreserved Blastocyst's ImplantatiOn Potential After Genetic Screening (BIOPS) |
| Methods | Randomised controlled trial |
| Participants | Sample size: 276 |
| Interventions | Intent to transfer single cryopreserved embryo, selection based on euploid status (after preimplantation genetic screening) and standard morphological assessment, with:
|
| Outcomes | Primary outcome: clinical pregnancy |
| Starting date | 18 May 2017 |
| Contact information | Sara Somers, MSc University Hospital of Ghent Email: arg.studies@uzgent.be |
| Notes | First posted June 2017, last updated 2 June 2017 |
NCT03214185.
| Study name | Effects of PGS 2.0 in Patients With Unexplained RPL |
| Methods | Randomised controlled trial |
| Participants | Sample size: 710 Inclusion criteria: couple has experienced 2 or more failed pregnancies; karyotypes of both husband and wife are normal (polymorphic chromosomes are considered normal either); women age ≥ 20 and < 38 years old Exclusion criteria:
|
| Interventions | Blastocysts are selected by PGS 2.0 (NGS based), and only euploid embryos will be transferred. |
| Outcomes | Primary outcome: cumulative live birth rate |
| Starting date | 6 February 2018 |
| Contact information | Caixia Lei, MD Shanghai Ji Ai Genetics & IVF Institute, Obstetrics and Gynecology Hospital, Fudan University Telephone: 86‐18917958213 Email: green3318@163.com |
| Notes | First posted 11 July 2017, last updated 6 February 2018 |
NCT03371745.
| Study name | The PrISICE Clinical Trial (Pre‐Implantation Screening and Investigation on the Cryopreservation of Embryos) (PrISICE) |
| Methods | Randomised controlled trial |
| Participants | Sample size: 1539 Inclusion criteria: 18 to 42 years |
| Interventions | Group 1: PGS‐FET group: deferred transfer of day 5/6/7 (blastocyst stage) embryos cryopreserved following trophectoderm biopsy Group 2: "freeze‐only" (FET) group: deferred transfer of frozen/thawed embryos without biopsy Group 3: "fresh" ET group: immediate transfer of "fresh" embryos in the stimulation cycle |
| Outcomes | Primary outcome: live birth |
| Starting date | August 2018 |
| Contact information | Rebecca Wong Yale University Telephone 415‐353‐4305 Email: rebecca.wong@ucsf.edu or Nik Lenhart Telephone 415‐885‐3598 Email: lenhartn@obgyn.ucsf.edu |
| Notes | First posted 13 December 2017, last updated 15 October 2019 |
AMH: Anti‐Mullerian Hormone
ART: Assisted reproductive technology
ASRM: American Society for Reproductive Medicine
BMI: Body mass index
CCS: Comprehensive Chromosome Screening
ESHRE: European Society of Human Reproduction and Embryology
E2: Estradiol
FET: Frozen embryo transfer
FSH: Follicle stimulating hormone
GnRH: gonadotropin‐releasing hormone
HSG: hysterosalpingogram
ICSI: intracytoplasmic sperm injection
IU: International United
IVF: in vitro fertilisation
LH: Luteinizing hormone
NGS: Next Generation Sequencing
PGD: Preimplantation genetic diagnosis
PGS: Preimplantation genetic screening
RPL: recurrent pregnancy loss
WHO; World Health Organisation
Differences between protocol and review
We substituted the previous term 'preimplantation genetic screening' (PGS) for 'preimplantation genetic testing for aneuploidies' (PGT‐A), following a revision of terminology used in infertility care.
In the title, we only used the term 'in vitro fertilisation' and did not use the term 'intracytoplasmic sperm injection' anymore.
We substituted the comparisons by indication for comparisons by genetic analysis techniques used: genome‐wide analysing versus fluorescence in situ hybridisation (FISH), as genome‐wide analysing techniques have begun to replace the methods of FISH over the last decade.
We substituted the previous term 'live birth rate' for 'live birth rate after the first embryo transfer, defined as the birth of a living child after 20 weeks of gestation'. We changed the outcome live birth rate after the first embryo transfer from a primary outcome to a secondary outcome. In turn, we added cumulative live birth rate as a primary outcome, as it is today generally considered to be the outcome of choice for patients and therefore an important primary outcome. A reason for this is the increasing efficiency of embryo cryopreservation and thawing protocols, and improved outcome after transfer of cryopreserved embryos.
We removed the outcome women whose child has a congential malformation from the outcome list.
In the previous version of the review, data were compared per type of participant (i.e. advanced maternal age, repeated IVF failure, and good prognosis). At that time only evidence with the genetic analysing technique FISH existed. In this new update, we compared data per type of genetic analysing technique used, with subgroups for the type of biopsy. However, it is still important to be aware of the type of participants included in the trial.
Contributions of authors
For this update SC and MZ conducted the literature searches for the review, selected relevant trials, produced data and information about studies, assessed the validity and checked the data extraction for each trial, entered all study information, data, and text into Review Manager 5, performed the analyses, wrote the Abstract, Background, Methods, Results, and Authors' conclusions sections of the review, and approved the final version of the review.
EK and SM took part in writing the Abstract, Background, Methods, Results, and Authors' conclusions sections of the review, and approved the final version of the review.
KF and MvW commented upon the review and agreed with its content.
Sources of support
Internal sources
There were no internal sources of support, Other
External sources
There were no external sources of support, Other
Declarations of interest
Simone Cornelisse: no conflicts to declare. Miriam Zagers: no conflicts to declare. Elena Kostova: no conflicts to declare. Kathrin Fleischer: no conflicts to declare. Madelon van Wely: no conflicts to declare. Sebastiaan Mastenbroek has performed a randomised controlled trial on the effect of PGT‐A in IVF in women aged 35 and over (Mastenbroek 2007). This was an independent trial funded by the Netherlands Organisation for Health Research and Development.
New search for studies and content updated (no change to conclusions)
References
References to studies included in this review
Blockeel 2008 {published data only}
- Blockeel C, Schutyser V, De Vos A, Verpoest W, De Vos M, Staessen C, et al. Prospectively randomized controlled trial of PGS in IVF/ICSI patients with poor implantation. Reproductive Biomedicine Online 2008;17(6):848-54. [DOI] [PubMed] [Google Scholar]
Debrock 2010 {published data only}
- Debrock S, Melotte C, Spiessens C, Peeraer K, Vanneste E, Meeuwis L, et al. Preimplantation genetic screening for aneuploidy of embryos after in vitro fertilization in women aged at least 35 years: a prospective randomized trial. Fertility and Sterility 2010;93(2):364-73. [DOI] [PubMed] [Google Scholar]
Hardarson 2008 {published data only}
- Hardarson T, Hanson C, Lundin K, Hillensjo T, Nilsson L, Stevic J, et al. Preimplantation genetic screening in women of advanced maternal age caused a decrease in clinical pregnancy rate: a randomized controlled trial. Human Reproduction 2008;23(12):2806-12. [DOI] [PubMed] [Google Scholar]
Jansen 2008 {published data only}
- Jansen RP, Bowman MC, De boer KA, Leigh DA, Lieberman DB, McArthur SJ. What next for preimplantation genetic screening (PGS)? Experience with blastocyst biopsy and testing for aneuploidy. Human Reproduction 2008;23(7):1476-8. [DOI] [PubMed] [Google Scholar]
Mastenbroek 2007 {published data only}
- Mastenbroek S, Twisk M, Van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, et al. In vitro fertilization with preimplantation genetic screening. New England Jounal of Medicine 2007;537(1):9-17. [DOI] [PubMed] [Google Scholar]
Meyer 2009 {published data only}
- Meyer LR, Klipstein S, Hazlett WD, Nasta T, Mangan P, Karande VC. A prospective randomized controlled trial of preimplantation genetic screening in the "good prognosis" patient. Fertility and Sterility 2009;91(5):1731-8. [DOI] [PubMed] [Google Scholar]
Munné 2019 {published data only}
- Munné S, Kaplan B, Frattarelli JL, Child T, Nakhuda G, Shamma FN, et al, on behalf of the STAR study group. Preimplantation genetic testing for aneuploidy versus morphology as selection criteria for single frozen-thawed embryo transfer in good-prognosis patients: a multicenter randomized clinical trial. Fertility and Sterility December 2019;112(6):1071-79. [DOI: 10.1016/j.fertnstert.2019.07.1346] [DOI] [PubMed] [Google Scholar]
Rubio 2013 {published data only}
- Rubio C, Bellver J, Rodrigo L, Bosch E, Mercader A, Vidal C, et al. Preimplantation genetic screening using fluorescence in situ hybridization in patients with repetitive implantation failure and advanced maternal age: two randomized trials. Fertility and sterility 2013;99(5):1400-7. [PMID: ] [DOI] [PubMed] [Google Scholar]
Schoolcraft 2009 {published data only}
- Schoolcraft WB, Katz-Jaffe MG, Stevens J, Rawlins M, Munne S. Preimplantation aneuploidy testing for infertile patients of advanced maternal age. Fertility and Sterility 2009;92(1):157-62. [DOI] [PubMed] [Google Scholar]
Staessen 2004 {published data only}
- Staessen C, Platteau P, Van Assche E, Michiels A, Tournaye H, Camus M, et al. Comparison of blastocyst transfer with or without preimplantation genetic diagnosis for aneuploidy screening in couples with advanced maternal age: a prospective randomised controlled trial. Human Reproduction 2004;19(12):2849-58. [DOI] [PubMed] [Google Scholar]
Staessen 2008 {published data only}
- Staessen C, Verpoest W, Donoso P, Haentjens P, Van der Elst J, Liebaers I, et al. Preimplantation genetic screening does not improve delivery rate in women under the age of 36 following single-embryo transfer. Human Reproduction 2008;23(12):2818-25. [DOI] [PubMed] [Google Scholar]
Verpoest 2018 {published data only}
- Verpoest W, Staessen C, Bossuyt PM, Goossens V, Altarescu G, Bonduelle M, et al. Preimplantation genetic testing for aneuploidy by microarray analysis of polar bodies in advanced maternal age: a randomized clinical trial. Human Reproduction 2018;33(9):1767-76. [PMID: ] [DOI] [PubMed] [Google Scholar]
Werlin 2003 {published data only}
- Werlin L, Rodi I, DeCherney A, Marello E, Hill D, Munné S. Preimplantation genetic diagnosis as both a therapeutic and diagnostic tool in assisted reproductive technology. Fertility and Sterility 2003;80(2):467-8. [DOI: 10.1016/S0015-0282(03)00605-8] [DOI] [PubMed] [Google Scholar]
References to studies excluded from this review
Forman 2013 {published data only}
- Forman EJ, Hong KH, Ferry KM, Tao X, Taylor D, Levy B, et al. In vitro fertilization with single euploid blastocyst transfer: a randomized controlled trial. Fertility and Sterility 2013;100(1):100-7. [DOI: 10.1016/j.fertnstert.2013.02.056] [DOI] [PubMed] [Google Scholar]
Forman 2014 {published data only}
- Forman EJ, Hong KH, Franasiak JM, Scott RT Jr. Obstetrical and neonatal outcomes from the BEST Trial: single embryo transfer with aneuploidy screening improves outcomes after in vitro fertilization without compromising delivery rate. American Journal of Obstetrics and Gynecology 2014;210(2):157-160. [DOI: 10.1016/j.ajog.2013.10.016] [DOI] [PubMed] [Google Scholar]
Gianaroli 1997 {published data only}
- Gianaroli L, Magli MC, Ferraretti AP, Fiorentine A, Garrisi J, Munne S. Preimplantation genetic diagnosis increases the preimplantation rate in human in vitro fertilization by avoiding the transfer of chromosomally abnormal embryos. Fertility and Sterility 1997;68(6):1128-31. [DOI] [PubMed] [Google Scholar]
Gianaroli 1999 {published data only}
- Gianaroli L, Magli MC, Ferraretti AP, Munne S. Preimplantation diagnosis for aneuploidies in patients undergoing in vitro fertilization with a poor prognosis: identification of the categories for which it should be proposed. Fertility and Sterility 1999;72(5):837-8. [DOI] [PubMed] [Google Scholar]
Mersereau 2008 {published data only}
- Mersereau JE, Pergament E, Zhang X, Milad MP. Preimplantation genetic screening to improve in vitro fertilization pregnancy rates: a prospective randomized controlled trial. Fertility and Sterility 2008;90(4):1287-9. [DOI] [PubMed] [Google Scholar]
Moayeri 2016 {published data only}
- Moayeri M, Saeidi H, Modarresi MH, Hashemi M. The effect of preimplantation genetic screening on implantation rate in women over 35 years of age. Cell Journal 2016;18(1):13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
NCT02265614 {published data only}
- NCT02265614. PGS Using Microarray in IVF Patients With Repeated Implantation Failure [Preimplantation Genetic Screening (PGS) Using Microarray Technique: Method to Select the Embryo With the Greatest Chance for Successful Implantation During in Vitro Fertilization in Couples With a History of Unsuccessful IVF Attempts]. https://clinicaltrials.gov/ct2/show/NCT02265614 16-10-2014.
Ozgur 2019 {published data only}
- Ozgur K, Berkkanoglu M, Bulut H, Yoruk GD, Candurmaz NN, Coetzee K. Single best euploid versus single best unknown-ploidy blastocyst frozen embryo transfers: a randomized controlled trial. Journal of Assisted Reproduction and Genetics 2019;36(4):629-36. [DOI: 10.1007/s10815-018-01399-1] [DOI] [PMC free article] [PubMed] [Google Scholar]
Rubio 2017 {published data only}
- Rubio C, Bellver J, Rodrigo L, Castillón G, Guillén A, Vidal C, et al. In vitro fertilization with preimplantation genetic diagnosis for aneuploidies in advanced maternal age: a randomized, controlled study. Fertility and Sterility 2017;107(5):1145-1146. [DOI: 10.1016/j.fertnstert.2017.03.011.] [DOI] [PubMed] [Google Scholar]
Scott 2013 {published data only}
- Scott RT, Upham KM, Forman EJ, Hong KH, Scott KL, Taylor D, et al. Blastocyst biopsy with comprehensive chromosome screening and fresh embryo transfer significantly increases in vitro fertilization implantation and delivery rates: a randomized controlled trial. Fertility and Sterility 2013;100(3):697-703. [DOI: 10.1016/j.fertnstert.2013.04.035] [DOI] [PubMed] [Google Scholar]
Stevens 2004 {published data only}
- Stevens J, Wale P, Surrey ES, Schoolcraft WB. Is aneuploidy screening for patients aged 35 or over beneficial? A prospective randomized trial. Fertility and Sterility 2004;82(Suppl 2):249. [DOI: ] [Google Scholar]
Yang 2012 {published data only}
- Yang Z, Liu J, Collins GS, Salem SA, Liu X, Lyle SS, et al. Selection of single blastocysts for fresh transfer via standard morphology assessment alone and with array CGH for good prognosis IVF patients: results from a randomized pilot study. Molecular Cytogenetics 2012;5(1):24. [DOI: 10.1186/1755-8166-5-24] [DOI] [PMC free article] [PubMed] [Google Scholar]
References to studies awaiting assessment
NCT02223221 {unpublished data only}
- NCT02223221. Effects of PGS in Infertile Female Patients With RPL [Effects of Preimplantation Genetic Screening for Aneuploidies in Infertile Female Patients With Recurrent Spontaneous Abortion History]. https://clinicaltrials.gov/ct2/show/NCT02223221 (first received August 2014).
References to ongoing studies
NCT01946945 {unpublished data only}
- NCT01946945. Comparison of Standard ART Practice vs. Trophectoderm Biopsy and Whole Chromosome Analysis [Comparison of Standard ART Practice vs. Trophectoderm Biopsy, Whole Chromosome Analysis by Next Generation Sequencing, and Replacement of a Single Euploid Embryo]. clinicaltrials.gov/ct2/show/NCT01946945 (first received 20 September 2013).
NCT01977144 {unpublished data only}
- NCT01977144. Solaire [Screening of Low Responders for Aneuploidy to Improve Reproductive Efficiency]. clinicaltrials.gov/ct2/show/NCT01977144 (first received 6 November 2013).
NCT02032264 {unpublished data only}
- NCT02032264. Nexgen [Evaluation of the Efficacy of Next Generation Sequencing in Predicting Embryonic Karyotype and Subsequent Pregnancy Outcomes in in Vitro Fertilization Cycles (IVF)]. clinicaltrials.gov/ct2/show/NCT02032264 (first received 10 January 2014).
NCT02353364 {unpublished data only}
- NCT02353364. Blastocyst Euploidy Assessment and Conditioned embryO traNsfer (BEACON) [Prospective Randomised Control Trial of Blastocyst Euploidy Assessment and Conditioned Embryo Transfer for Infertility Patients of Advanced Maternal Age]. clinicaltrials.gov/ct2/show/NCT02353364 (first received 2 February 2015).
NCT02868528 {unpublished data only}
- NCT02868528. Prospective Randomized Controlled Study of Preimplantation Genetic Screening With Next Generation Sequencing Technology on Advanced Age Women [A Study of Preimplantation Genetic Screening With Next Generation Sequencing Technology on Advanced Age Women]. clinicaltrials.gov/ct2/show/NCT02868528 (first received 16 August 2016).
NCT02941965 {unpublished data only}
- NCT02941965. Preimplantation Genetic Screening in Patients With Male Factor Infertility [Preimplantation Genetic Screening in Patients With Male Factor Infertility]. clinicaltrials.gov/ct2/show/NCT02941965 (first received 21 October 2016).
NCT03118141 {unpublished data only}
- NCT03118141. Cumulative Live Birth Rate With eSET After Preimplantation Genetic Screening Versus Conventional In-vitro Fertilization (CESE-PGS) [Cumulative Live Birth Rate With eSET After In-vitro Fertilization With Preim-plantation Genetic Screening by Next Generation Sequencing Versus Conventional In-vitro Fertilization: a Pragmatic Randomized Controlled Clinical Trial]. clinicaltrials.gov/ct2/show/NCT03118141 (first received 18 April 2017).
NCT03173885 {unpublished data only}
- NCT03173885. Investigating the Cryopreserved Blastocyst's ImplantatiOn Potential After Genetic Screening (BIOPS) [An RCT Evaluating the Implantation Potential of Vitrified Embryos Screened by Next Generation Sequencing Following Trophectoderm Biopsy, Versus Vitrified Unscreened Embryos in Good Prognosis Patients Undergoing IVF]. clinicaltrials.gov/ct2/show/NCT03173885 (first received 2 June 2017).
NCT03214185 {unpublished data only}
- NCT03214185. Effects of PGS2.0 in Patients With Unexplained RPL [Effects of Preimplantation Genetic Screening 2.0 on the Clinical Outcomes of Assisted Reproductive Treatment in Patients With Recurrent Pregnancy Loss: a Multi-center-based Prospective Randomized Clinical Trial]. clinicaltrials.gov/ct2/show/NCT03214185 (first received 11 July 2017).
NCT03371745 {unpublished data only}
- NCT03371745. The PrISICE Clinical Trial (Pre-Implantation Screening and Investigation on the Cryopreservation of Embryos) (PrISICE) [A Prospective, Randomized, Controlled Clinical Trial Evaluating the Superiority of Preimplantation Genetic Screening (PGS) and Deferred Transfer of Cryopreserved Embryos Over "Freeze-Only" Deferred Transfer Without PGS or Immediate Embryo Transfer During a "Fresh" In Vitro Fertilization Cycle]. clinicaltrials.gov/ct2/show/NCT03371745 (first received 13 December 2017).
Additional references
Angell 1983
- Angell RR, Aitken RJ, Van Look PF, Lumsden MA, Templeton AA. Chromosome abnormalities in human embryos after in vitro fertilization. Nature 1983;303(5915):336-8. [DOI: 10.1038/303336a0] [DOI] [PubMed] [Google Scholar]
ASRM 2018
- Practice Committees of the American Society for Reproductive Medicine and the Society for Assisted Reproductive Technology. The use of preimplantation genetic testing for aneuploidy (PGT-A): a committee opinion. Fertility and Sterility 2018;109(3):429-36. [DOI: 10.1016/j.fertnstert.2018.01.002] [DOI] [PubMed] [Google Scholar]
Baart 2006
- Baart EB, Martini E, Van den Berg I, Macklon NS, Galjaard RJ, Fauser BC, et al. Preimplantation genetic screening reveals a high incidence of aneuploidy and mosaicism in embryos from young women undergoing IVF. Human Reproduction 2006;21(1):223-33. [DOI: 10.1093/humrep/dei291] [DOI] [PubMed] [Google Scholar]
Belandres 2019
- Belandres D, Shamonki M, Nabil A. Current status of spent embryo media research for preimplantation genetic testing. Journal of Assisted Reproduction and Genetics 2019;36(5):819-26. [DOI: 10.1007/s10815-019-01437-6] [DOI] [PMC free article] [PubMed] [Google Scholar]
Benadiva 1996
- Benadiva CA, Kligman I, Munné S. Aneuploidy 16 in human embryos increases significantly with maternal age. Fertility and Sterility 1996;66:248-55. [PubMed] [Google Scholar]
Blockeel 2018 [pers comm]
- Blockeel C. Definition of live birth [personal communication]. Personal communication with the authors of the PGS Cochrane 2011 Unknown.
Bolton 2016
- Bolton H, Graham SJL, Van der Aa N, Kumar P, Theunis K, Fernandez E, et al. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nature Communications 2016;7:11165. [DOI: 10.1038/ncomms11165] [DOI] [PMC free article] [PubMed] [Google Scholar]
Capalbo 2018
- Capalbo A, Romanelli V, Patassini C, Poli M, Girardi L, Giancani A, et al. Diagnostic efficacy of blastocoel fluid and spent media as sources of DNA for preimplantation genetic testing in standard clinical conditions. Fertility and Sterility 2018;110(5):870-79. [DOI: 10.1016/j.fertnstert.2018.05.031] [DOI] [PubMed] [Google Scholar]
Chaimani 2013
- Chaimani A, Higgins JPT, Mavridis D, Spyridonos P, Salanti G. Graphical tools for network meta‐analysis in STATA. PLoS ONE 2013;8(10):e76654. [DOI] [PMC free article] [PubMed] [Google Scholar]
Cimadomo 2020
- Cimadomo D, Rienzi L, Capalbo A, Rubio C, Innocenti F, García-Pascual C, et al. The dawn of the future: 30 years from the first biopsy of a human embryo. The detailed history of an ongoing revolution. Human Reproduction Update 2020;26(4):453-473. [DOI: https://doi.org/10.1093/humupd/dmaa019 ] [DOI] [PubMed] [Google Scholar]
Covidence [Computer program]
- Veritas Health Innovation Covidence. Version accessed 14 September 2019. Melbourne, Australia: Veritas Health Innovation. Available at covidence.org.
Cram 2019
- Cram DS, Leigh S, Handyside A, Rechitsky L, Xu K, Harton G, et al. PGDIS Position Statement on the Transfer of Mosaic Embryos 2019. Reproductive BioMedicine Online 2019;39:e1-4. [DOI: 10.1016/j.rbmo.2019.06.012] [DOI] [PubMed] [Google Scholar]
De Geyter 2018
- De Geyter C, Calhaz-Jorge C, Kupka MS, Wyns C, Mocanu E, Mortenko T et al. ART in Europe, 2014: results generated from European registries by ESHRE: The European IVF-monitoring Consortium (EIM) for the European Society of Human Reproduction and Embryology (ESHRE). Human Reproduction 2018;33(9):1586-01. [DOI: 10.1093/humrep/dey242 ] [DOI] [PubMed] [Google Scholar]
De Rycke 2017
- De Rycke M, Goossens V, Kokkali G, Meijer-Hoogeveen M, Coonen E, Moutou C. ESHRE PGD Consortium data collection XIV-XV: cycles from January 2011 to December 2012 with pregnancy follow-up to October 2013. Human Reproduction 2017;32:1974-94. [DOI] [PubMed] [Google Scholar]
DeBrock 2010 [pers comm]
- Debrock S. Results after the first treatment cycle[personal communication]. Personal communication with the authors of the PGS Cochrane 2011 Unknown.
Delhanty 1997
- Delhanty JD, Harper JC, Ao A, Handyside AH, Winston RM. Multicolour FISH detects frequent chromosomal mosaicism and chaotic division in normal preimplantation embryos from fertile patients. Human Genetics 1997;99:755-60. [DOI: 10.1007/s004390050443 ] [DOI] [PubMed] [Google Scholar]
Dokras 1990
- Dokras A, Sargent IL, Ross C, Gardner RL, Barlow DH. Trophectoderm biopsy in human blastocysts. Human Reproduction 1990;5(7):821-5. [DOI: 10.1093/oxfordjournals.humrep.a137191 ] [DOI] [PubMed] [Google Scholar]
Dyer 2016
- Dyer S, Chambers GM, Mouzon J, Nygren KG, Zegers-Hochschild F, Mansour R, et al. International Committee for Monitoring Assisted Reproductive Technologies world report: Assisted Reproductive Technology 2008, 2009 and 2010. Human Reproduction 2016;31(7):1588-609. [DOI] [PubMed] [Google Scholar]
ESHRE PGT‐SR/PGT‐A Working Group 2020
- ESHRE PGT-SR/PGT-A Working Group, Coonen E, Rubio C, Christopikou D, Dimitriadou E, Gontar J, et al. ESHRE PGT Consortium good practice recommendations for the detection of structural and numerical chromosomal aberrations. Progress in Human Reproduction Open 2020;3:1-12. [DOI: 10.1093/hropen/hoaa017 ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Geraedts 2011
- Geraedts J, Montag M, Magli MC, Repping S, Handyside A, Staessen C, et al. Polar body array CGH for prediction of the status of the corresponding oocyte. Part I: clinical results. Human Reproduction 2011;26(3):3173-80. [DOI: 10.1093/humrep/der294 ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Geraedts 2016
- Geraedts J, Sermon K. Preimplantation genetic screening 2.0: the theory. Molecular Human Reproduction 2016;22(8):539-544. [DOI: 10.1093/molehr/gaw033. ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Gianaroli 2014
- Gianaroli L, Magli MC, Pomante A, Crivello AM, Cafueri G, Valerio M, et al. Blastocentesis: a source of DNA for pre implantation genetic testing. Results from a pilot study. Fertility and Sterility 2014;102:1692–99. [DOI] [PubMed] [Google Scholar]
Goossens 2009
- Goossens V, Harton G, Moutou C, Traeger-Synodinos J, Van Rij M, Harper JC. ESHRE PGD Consortium data collection IX: cycles from January to December 2006 with pregnancy follow-up to October 2007. Human Reproduction 2009;24(8):1786-810. [DOI: 10.1093/humrep/dep059] [DOI] [PubMed] [Google Scholar]
GRADEpro GDT [Computer program]
- McMaster University (developed by Evidence Prime) GRADEpro GDT. Version accessed 15 January 2020. Hamilton (ON): McMaster University (developed by Evidence Prime). Available at gradepro.org.
Greco 2015
- Greco E, Minasi MG, Fiorentino F. Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. New England Journal of Medicine 2015;373:2089-90. [DOI: 10.1056/NEJMc1500421] [DOI] [PubMed] [Google Scholar]
Griesinger 2016
- Griesinger G. Beware of the 'implantation rate'! Why the outcome parameter 'implantation rate' should be abandoned from infertility research. Human Reproduction 2016;31(2):249-51. [DOI: 10.1093/humrep/dev322] [DOI] [PubMed] [Google Scholar]
Hammond 2017
- Hammond ER, McGillivray BC, Wicker SM, Peek JC, Shelling AN, Stone P, et al. Characterizing nuclear and mitochondrial DNA in spent embryo culture media: genetic contamination identified. Fertility and Sterility 2017;107(1):220-8. [DOI: 10.1016/j.fertnstert.2016.10.015.] [DOI] [PubMed] [Google Scholar]
Handyside 1989
- Handyside AH, Pattinson JK, Penketh RJ, Delhanty JD, Winston RM, Tuddenham EG. Biopsy of human preimplantation embryos and sexing by DNA amplification. Lancet 1989;18(8634):347-9. [DOI: 10.1016/s0140-6736(89)91723-6 ] [DOI] [PubMed] [Google Scholar]
Harton 2011
- Harton GL, De Rycke M, Fiorentino F, Moutou C, SenGupta S, Traeger-Synodinos J, et al, on behalf of the European Society for Human Reproduction and Embryology (ESHRE) PGD Consortium. ESHRE PGD consortium best practice guidelines for amplification-based PGD. Human Reproduction 2011;26:33-40. [DOI: 10.1093/humrep/deq231] [20966462] [DOI] [PubMed] [Google Scholar]
Higgins 2011
- Higgins JP, Green S, editor(s). Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. Available from handbook.cochrane.org.
Kahraman 2000
- Kahraman S, Bahce M, Samli H, Imirzalioglu N, Yakisn K, Cengiz G, et al. Healthy births and ongoing pregnancies obtained by preimplantation genetic diagnosis in patients with advanced maternal age and recurrent implantation failure. Human Reproduction 2000;15(9):2003-7. [DOI: 10.1093/humrep/15.9.2003] [DOI] [PubMed] [Google Scholar]
Lawrenz 2019
- Lawrenz B, El Khatib I, Liñán A, Bayram A, Arnanz A, Chopra R, et al. The clinicians' dilemma with mosaicism - an insight from inner cell mass biopsies. Human Reproduction 2019;34(6):998-1010. [DOI: 10.1093/humrep/dez055] [DOI] [PubMed] [Google Scholar]
Leaver 2020
- Leaver M, Wells D. Non-invasive preimplantation genetic testing (niPGT): the next revolution in reproductive genetics? Human Reproduction Update 2020;26(1):16-42. [DOI: 10.1093/humupd/dmz033] [DOI] [PubMed] [Google Scholar]
Lefebvre 2011
- Lefebvre C, Manheimer E, Glanville J. Chapter 6: Searching for studies. In: Higgins JP, Green S, editor(s). Cochrane Handbook for Systematic Reviews of Interventions Version 5.1.0 (updated March 2011). The Cochrane Collaboration, 2011. In: Available from handbook.cochrane.org. [Google Scholar]
Maheshwari 2015
- Maheshwari A, McLernon D, Bhattacharya S. Cumulative live birth rate: time for a consensus? Human Reproduction 2015;30(12):2703-07. [DOI: 10.1093/humrep/dev263] [PMID: ] [DOI] [PubMed] [Google Scholar]
Mastenbroek 2007 [pers comm]
- Mastenbroek S. Data of the first treatment cycle [personal communication]. Email to: Cornelisse S 19-06-2020.
Mastenbroek 2011
- Mastenbroek S, Twisk M, Veen F, Repping S. Preimplantation genetic screening: a systematic review and meta-analysis of RCTs. Human Reproduction Update 2011;17(4):454-66. [DOI: 10.1093/humupd/dmr003] [DOI] [PubMed] [Google Scholar]
Mastenbroek 2014
- Mastenbroek S, Repping S. Preimplantation genetic screening: back to the future. Hum Reprod 2014;29(9):1846-50. [DOI: 10.1093/humrep/deu163] [DOI] [PubMed] [Google Scholar]
Mochizuki 2020
- Mochizuki L, Gleicher N. The PGS/PGT-A controversy in IVF addressed as a formal conflict resolution analysis. J Assist Reprod Genet 2020;37(3):677-678. [DOI: 10.1007/s10815-020-01688-8] [DOI] [PMC free article] [PubMed] [Google Scholar]
Montag 2004
- Montag M, Van der Ven K, Dorn C, Van der Ven H. Outcome of laser-assisted polar body biopsy and aneuploidy testing. Reproductive Biomedicine Online 2004;9(4):425-9. [DOI: 10.1016/s1472-6483(10)61278-3 ] [DOI] [PubMed] [Google Scholar]
Montag 2009
- Montag M, Van der Ven K, Rosing B, Van der Ven H. Polar body biopsy: a viable alternative to preimplantation genetic diagnosis and screening. Reproductive Biomedicine Online 2009;18(1 Suppl 1):6-11. [DOI: 10.1016/s1472-6483(10)60109-5 ] [DOI] [PubMed] [Google Scholar]
Munné 1993
- Munné S, Weier HU, Griffo J, Cohen J. A fast and efficient method for simultaneous X and Y in situ hybridization of human blastomeres. Journal of Assisted Reproduction & Genetics 1993;10(1):82-90. [DOI: 10.1007/bf01204446 ] [DOI] [PubMed] [Google Scholar]
Munné 1999
- Munné S, Magli MC, Cohen J, Morton P, Sadowy S, Gianaroli L, et al. Positive outcome after preimplantation diagnosis of aneuploidy in human embryos. Human Reproduction 1999;14(9):2191-9. [DOI: 10.1093/humrep/14.9.2191 ] [DOI] [PubMed] [Google Scholar]
Munné 2003
- Munné S, Sandalinas M, Escudero T, Velila E, Walmsley R, Sadowy S, et al. Improved implantation after preimplantation genetic diagnosis of aneuploidy. Reproductive Biomedicine Online 2003;7(1):91-7. [DOI: 10.1016/s1472-6483(10)61735-x ] [DOI] [PubMed] [Google Scholar]
Munné 2005
- Munné S, Chen S, Fischer J, Colls P, Zheng X, Stevens J, et al. Preimplantation genetic diagnosis reduces pregnancy loss in women aged 35 years and older with a history of recurrent miscarriages. Fertility and Sterility 2005;84(2):331-5. [DOI: 10.1016/j.fertnstert.2005.02.027 ] [DOI] [PubMed] [Google Scholar]
Obasaju 2001
- Obasaju M, Kadam A, Biancardi T, Sultan K, Fateh M, Munné S. Pregnancies from single normal embryo transfer in women older than 40 years. Reproductive Biomedicine Online 2001;2(2):98-101. [DOI: 10.1016/s1472-6483(10)62232-8 ] [DOI] [PubMed] [Google Scholar]
Patrizio 2019
- Patrizio P, Shoham G, Shoham Z, Leong M, Barad DH, Gleicher N. Worldwide live births following the transfer of chromosomally "Abnormal" embryos after PGT/A: results of a worldwide web-based survey. Journal Assisted Reproductive Genetics 2019;36(8):1599-1607. [DOI: 10.1007/s10815-019-01510-0] [DOI] [PMC free article] [PubMed] [Google Scholar]
Paulson 2017
- Paulson RJ. Preimplantation genetic screening: what is the clinical efficiency? Fertility and Sterility 2017;108(2):228-30. [DOI: 10.1016/j.fertnstert.2017.06.023] [DOI] [PubMed] [Google Scholar]
Pehlivan 2003
- Pehlivan T, Rubio C, Rodrigo L, Romero J, Remohi J, Simon C, et al. Impact of preimplantation genetic diagnosis on IVF outcome in implantation failure patients. Reproductive Biomedicine Online 2003;6(2):232-7. [DOI: 10.1016/s1472-6483(10)61715-4 ] [DOI] [PubMed] [Google Scholar]
Pellicer 1999
- Pellicer A, Rubio C, Vidal F, Minguez Y, Gimenez C, Egozcue J, et al. In vitro fertilization plus preimplantation genetic diagnosis in patients with recurrent miscarriage: an analysis of chromosome abnormalities in human preimplantation embryos. Fertility and Sterility 1999;71(6):1033-9. [DOI: 10.1016/s0015-0282(99)00143-0 ] [DOI] [PubMed] [Google Scholar]
Platteau 2004
- Platteau P, Staessen C, Michiels A, Tournaye H, Van Steirteghem A, Liebaers I, et al. Comparison of the aneuploidy frequency in embryos derived from testicular sperm extraction in obstructive azoospermic men. Human Reproduction 2004;19(7):1570-4. [DOI] [PubMed] [Google Scholar]
Platteau 2005
- Platteau P, Staessen C, Michiels A, Van Steirteghem A, Liebaers I, Devroey P. Preimplantation genetic diagnosis for aneuploidy screening in women older than 37 years. Fertility and Sterility 2005;84(2):319-24. [DOI: 10.1016/j.fertnstert.2005.02.019 ] [DOI] [PubMed] [Google Scholar]
Popovic 2018
- Popovic M, Dheedene A, Christodoulou C, Taelman J, Dhaenens L, Van Nieuwerburgh F, et al. Chromosomal mosaicism in human blastocysts: the ultimate challenge of preimplantation genetic testing? Humun Reproduction 2018;33(7):1342-54. [DOI: 10.1093/humrep/dey106] [DOI] [PubMed] [Google Scholar]
Popovic 2020
- Popovic M, Dhaenens L, Boel A, Menten B, Heindryckx B. Chromosomal mosaicism in human blastocysts: the ultimate diagnostic dilemma. Human Reproduction Update 2020;26(3):313-34. [DOI: 10.1093/humupd/dmz050] [DOI] [PubMed] [Google Scholar]
Review Manager 2014 [Computer program]
- Nordic Cochrane Centre, The Cochrane Collaboration Review Manager 5 (RevMan 5). Version 5.3. Copenhagen: Nordic Cochrane Centre, The Cochrane Collaboration, 2014.
Rosenwaks 2018
- Rosenwaks Z, Handyside AH, Fiorentino F, Gleicher N, Paulson RJ, Schattman GL, et al. The pros and cons of preimplantation genetic testing for aneuploidy: clinical and laboratory perspectives. Fertily and Sterility 2018;110(3):353-61. [DOI] [PubMed] [Google Scholar]
Rubio 2003
- Rubio C, Simon C, Vidal F, Rodrigo L, Pehlivan T, Remohi J, et al. Chromosomal abnormalities and embryo development in recurrent miscarriage couples. Human Reproduction 2003;18(1):192-8. [DOI: 10.1093/humrep/deg015 ] [DOI] [PubMed] [Google Scholar]
Rubio 2005
- Rubio C, Pehlivan T, Rodrigo L, Simon C, Remohi J, Pellicer A. Embryo aneuploidy screening for unexplained recurrent miscarriage: a minireview. American Journal of Reproductive Immunology 2005;53(4):159-65. [DOI: 10.1111/j.1600-0897.2005.00260.x ] [DOI] [PubMed] [Google Scholar]
Scriven 2010
- Scriven PN, Bossuyt PM. Diagnostic accuracy: theoretical models for preimplantation genetic testing of a single nucleus using the fluorescence in situ hybridization technique. Hum Reprod 2010;25(10):2622-2628. [DOI: 10.1093/humrep/deq196] [DOI] [PubMed] [Google Scholar]
Sermon 2016
- Sermon K, Capalbo A, Cohen J, Coonen E, De Rycke M, De Vos A, et al. The why, the how and the when of PGS 2.0: current practices and expert opinions of fertility specialists, molecular biologists, and embryologists. Molecular Human Reproduction 2016;22(8):845-57. [DOI: 10.1093/molehr/gaw034 ] [PMID: ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Silber 2003
- Silber S, Escudero T, Lenahan K, Abdelhadi I, Kilani A, Munné S. Chromosomal abnormalities in embryos derived from testicular sperm extraction. Fertility and Sterility 2003;79(1):30-8. [DOI: 10.1016/s0015-0282(02)04407-2] [DOI] [PubMed] [Google Scholar]
Singla 2020
- Singla S, Iwamoto-Stohl LK, Zhu M, Zernicka-Goetz M. Autophagy-mediated apoptosis eliminates aneuploid cells in a mouse model of chromosome mosaicism. Nature Communications 2020;11:2958. [DOI: 10.1038/s41467-020-16796-3] [DOI] [PMC free article] [PubMed] [Google Scholar]
Taylor 2014
- Taylor TH, Gitlin SA, Patrick JL, Crain JL, Wilson JM, Griffin DK. The origin, mechanisms, incidence and clinical consequences of chromosomal mosaicism in humans. Hum Reprod Update 2014;20(4):571-81. [DOI: 10.1093/humupd/dmu016] [DOI] [PubMed] [Google Scholar]
Treff 2013
- Treff NR, Fedick A, Tao X, Devkota B, Taylor D, Scott RT. Evaluation of targeted next-generation sequencing-based preimplantation genetic diagnosis of monogenic disease. Fertility and Sterility 2013;99(5):1377-84. [DOI: 10.1016/j.fertnstert.2012.12.018 ] [PMID: ] [DOI] [PubMed] [Google Scholar]
Twisk 2006
- Twisk M, Mastenbroek S, Wely M, Heineman MJ, Van der Veen F, Repping S. Preimplantation genetic screening for abnormal number of chromosomes (aneuploidies) in in vitro fertilisation or intracytoplasmic sperm injection. Cochrane Database of Systematic Reviews 2006, Issue 1. Art. No: CD005291. [DOI: 10.1002/14651858.CD005291.pub2] [DOI] [PubMed] [Google Scholar]
van Echten‐Arends 2011
- Echten-Arends J, Mastenbroek S, Sikkema-Raddatz B, Korevaar JC, Heineman MJ, Veen F, et al. Chromosomal mosaicism in human preimplantation embryos: a systematic review. Human Reproduction Update 2011;17(5):620-27. [DOI: 10.1093/humupd/dmr014] [DOI] [PubMed] [Google Scholar]
van Uum 2012
- Uum CM, Stevens SJ, Dreesen JC, Drüsedau M, Smeets HJ, Hollanders-Crombach B, et al. SNP array-based copy number and genotype analyses for preimplantation genetic diagnosis of human unbalanced translocations. European Journal of Human Genetics 2012;20:938-44. [DOI: 10.1038/ejhg.2012.27] [DOI] [PMC free article] [PubMed] [Google Scholar]
Vera‐Rodriquez 2018
- Vera-Rodriguez M, Diez-Juan A, Jimenez-Almazan J, Martinez S, Navarro R, Peinado V, et al. Origin and composition of cell-free DNA in spent medium from human embryo culture during preimplantation development. Human Reproduction 2018;33(4):745-56. [DOI: 10.1093/humrep/dey028] [PMID: ] [DOI] [PubMed] [Google Scholar]
Verlinsky 1995
- Verlinsky Y, Cieslak J, Freidine M, Ivakhnenko V, Wolf G, Kovalinskaya L, et al. Pregnancies following pre-conception diagnosis of common aneuploidies by fluorescent in-situ hybridization. Human Reproduction 1995;10(7):1923-27. [DOI: 10.1093/oxfordjournals.humrep.a136207 ] [PMID: 8583011 ] [DOI] [PubMed] [Google Scholar]
Verpoest 2018 [pers comm]
- Sermon K. Cochrane review update: preimplantation genetic screening. Cornelisse S 27-02-2019.
Wells 2008
- Wells D, Alfarawati S, Fragouli E. Use of comprehensive chromosomal screening for embryo assessment: microarrays and CGH. Molecular Human Reproduction 2008;14(12):703-10. [DOI: 10.1093/molehr/gan062 ] [PMID: 18957518 ] [DOI] [PMC free article] [PubMed] [Google Scholar]
Wilding 2004
- Wilding M, Forman R, Hogewind G, Di Matteo L, Zullo F, Cappiello F, et al. Preimplantation genetic diagnosis for the treatment of failed in vitro fertilization embryo transfer and habitual abortion. Fertility and Sterility 2004;891(5):1302-7. [DOI: 10.1016/j.fertnstert.2003.10.028 ] [DOI] [PubMed] [Google Scholar]
Wilton 2002
- Wilton L. Preimplantation genetic diagnosis for aneuploidy screening in early human embryos: a review. Prenat Diagn 2002;22(6):512-18. [DOI: 10.1002/pd.388] [DOI] [PubMed] [Google Scholar]
Wong 2014
- Wong KM, Mastenbroek S, Repping S. Cryopreservation of human embryos and its contribution to in vitro fertilization success rates. Fertility and Sterility 2014;102(1):19-26. [DOI: ] [PMID: 24890275 ] [DOI] [PubMed] [Google Scholar]
Zegers‐Hochschild 2017
- Zegers-Hochschild F, Adamson GD, Dyer S, Racowsky C, Mouzon J, Sokol R, et al. The International Glossary on Infertility and Fertility Care, 2017. Human Reproduction 2017;32:1786-1801. [DOI: 10.1093/humrep/dex234 ] [DOI] [PMC free article] [PubMed] [Google Scholar]
References to other published versions of this review
Twisk M 2006
- Twisk M, Mastenbroek S, Wely M, Heineman MJ, Van der Veen F, Repping S. Preimplantation genetic screening for abnormal number of chromosomes (aneuploidies) in in vitro fertilisation or intracytoplasmic sperm injection. Cochrane Database of Systematic Reviews 2006, Issue 1. [DOI: 10.1002/14651858] [DOI] [PubMed] [Google Scholar]


