Abstract
How modification of gene expression generates novel traits is key to understanding the evolutionary process. We investigated the genetic basis for the origin of the piscine gas bladder from lungs of ancestral bony vertebrates. Distinguishing these homologous organs is the direction of budding from the foregut during development; lungs bud ventrally and the gas bladder buds dorsally.
We investigated whether this morphological inversion is associated with molecular inversion of conserved genes regulating lung and gasbladder development. Using laser-capture microdissection and RNA-seq, we assayed transcript abundance and compared expression patterns between dorsal and ventral foregut tissues at three developmental stages spanning gasbladder development. Our focal taxon, bowfin (Amia calva), representing the sistergroup to teleosts, is an early diverging ray-finned fish with a gas bladder. We discovered a number of genes with unknown function during lung development that are differentially expressed during gas bladder development and annotated to functions relevant for organ budding. We also identified several known lung-regulatory genes exhibiting inverted dorsoventral expression during gasbladder relative to lung development. Specifically, we found Tbx5 is strongly expressed in the dorsal mesoderm surrounding the gas bladder during bowfin development, and several interacting genes are co-expressed dorsally with Tbx5. In contrast, in mouse and bichir (Polypterus senegalus), the only ray-finned fish that have lungs, Tbx5 is expressed in the ventral lung mesoderm during development. Our data demonstrating dorsoventral inversion of conserved genes suggests that these genes may have contributed to the evolutionary transition between ventral lungs and a dorsal gas bladder in ray-finned fishes.
Keywords: gas bladder, Tbx5, dorsoventral, lungs
Graphical Abstract

INTRODUCTION
Phenotypic evolution depends on the generation of variation through the modification of development. How that variation is generated is fundamental to understanding the evolutionary process. Increasing evidence points towards the existence of conserved genes across distantly related taxa regulating major aspects of the body plan (Averof and Patel, 1997; Carroll et al., 2013). Modification to the temporal and spatial expression patterns of those conserved genes during development produces novel traits and body plans (Carroll et al., 2013). For example, recent molecular studies have supported the controversial 19th-century hypothesis that the dorsoventral body axis in chordates is inverted relative to that in arthropods (Geoffroy St-Hilaire, E., 1822; Arendt and Nubler-Jung, 1994; Holley, et al., 1995; Ferguson, 1996; De Robertis and Sasai, 2000). The expression patterns and function of orthologous genes important for dorsoventral axis development in both arthropods and chordates parallel the morphological inversion of the body axis.
Here we investigate another example of dorsal-ventral patterning, leading either to the development of ventral lungs in tetrapods or a dorsal gas bladder in ray-finned fishes. These two air-filled organs are homologous (Darwin, 1859; Romer and Parsons, 1970; Cass et al., 2013; Longo et al., 2013) with the common ancestor of tetrapods and ray-finned fishes characterized by ventral paired lungs (Fig. 1B). The gas bladder is an important innovation originating in the Actinopteri, a clade of more than 30,000 species of ray-finned fishes, including all teleosts, sturgeon, paddlefish, gar, and bowfin (Nelson et al., 2016; Betancur-R et al., 2017). The gas bladder lies dorsal to the gut and ventral to the spine (Fig. 1A) and in most fishes, functions to control buoyancy, but in some groups, it serves as a respiratory organ or aids in hearing and sound production (Alexander, 1966; Helfman et al., 2009). The evolution of the gas bladder from lungs involved a shift in the direction of budding from ventral (lungs) to dorsal (gas bladder). Thus, we investigate whether this inverted morphology is associated with the molecular inversion of conserved gene regulatory networks specific to both lungs and gas bladders.
Figure 1:
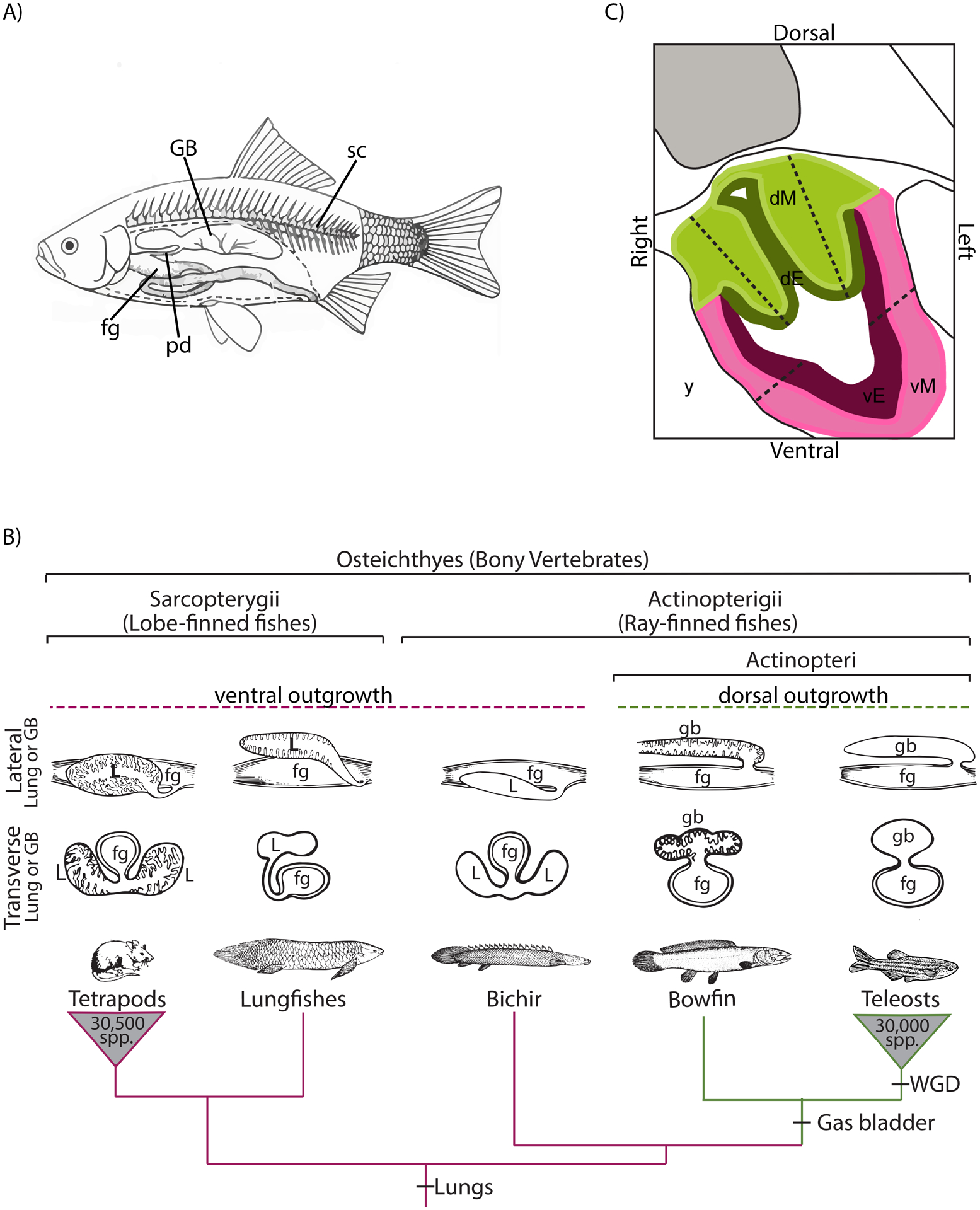
A) Lateral view of a cutaway of a schematic teleost fish showing the location of the gas bladder (GB), which is located dorsal to the foregut (fg) and ventral to the spinal column (sc). The gas bladder is an outgrowth of the foregut, and in many species, it remains connected to the gut via the pneumatic duct (pd). B) Highly-pruned bony vertebrate phylogeny showing the relationship of bichir, bowfin, and teleosts (phylogeny after Betancur et al., 2017), the clade containing the majority of extant ray-finned fishes (at least 30,000 extant species; Nelson et al., 2016). Lobed-finned fishes, represented here by tetrapods and lungfishes, develop ventral lungs (magenta lineages). The dorsal gas bladder arose after the divergence of bichirs (green lineages), which have lungs. The gas bladder is characteristic of the Actinopteri, including bowfin and teleosts. That bichirs, sister to Actinopteri, have lungs as do tetrapods and lobed-finned fishes, indicating that lungs are the ancestral state for bony vertebrates. Transverse and lateral sectional drawings of either lungs or gas bladders (modified from Romer 1970) are shown above each taxon. Note that members of the Actionpteri have dorsal gasbladders (gb) and this clade is nested within the bony vertebrates, the common ancestor of which had ventral lungs (L). The teleost whole-genome duplication (WGD) is also shown on the phylogeny. C) Diagram of a transverse section of a bowfin embryo showing the tissue regions of the foregut dissected using laser capture microscopy (LCM). Dorsal mesoderm, dM, pictured in light green; dorsal endoderm, dE, in dark green; ventral mesoderm, vM, in pink; and ventral endoderm, vE, in purple. Tissue was sampled between the dotted lines, which were intentionally placed well within the actual boundaries of dorsal and ventral regions to avoid sample contamination from adjacent tissue. Three individuals and 4 tissues were sampled at three developmental stages.
Reflecting their homology, lungs and gas bladders are similar in many ways. Morphologically, both develop from the anterior foregut endoderm, and both are supplied by the pulmonary artery. At a molecular level, both lungs and gas bladders express a suite of orthologous genes known to have lung-specific interactions, and both produce pulmonary surfactant proteins (Goodrich, 1958; Daniels et al., 2004; Cass et al., 2013; Longo et al., 2013). Despite these commonalities, several differences exist between the lungs and the gas bladder. For example, the lungs are usually paired and branching (Kardong, 2015), while the gas bladder is usually single and non-branching (Helfman et al., 2009). However, exceptions include snakes and caecilians, which can have a single lung (Wallach, 1998), and toadfishes, which can have bilaterally paired gas bladders (Rice and Bass, 2009). The defining difference between the lungs and the gas bladder is the direction of budding from the anterior foregut during development. As illustrated in Figure 1B, lungs bud ventrally and gas bladders bud dorsally (Wilder, 1877; Graham, 1997; Cass et al., 2013). Thus, similar to the arthropod versus chordate body plans, the evolution of the gas bladder from lungs represents an instance of dorsoventral inversion.
Development of the tetrapod lungs, based on mouse and chick, is well characterized with the earliest stages beginning as a ventral evagination from the foregut endoderm. This ventral budding is known to depend on the expression of a gene regulatory network (GRN) that includes Nkx2.1, Sox2, and Bmp4. We previously investigated the expression of these candidate genes during gasbladder development in bowfin and zebrafish (Funk et al., in review), hypothesizing that the expression of Nkx2.1 and Sox2 would show an inverted expression pattern relative to the pattern during tetrapod lung development. We found that dorsal and ventral spatial expression of these two genes was similar during lung and gasbladder development. In both tetrapod lungs and bowfin gas bladder, Nkx2.1 is expressed ventrally. In tetrapod lungs, Sox2 is expressed dorsally, while in bowfin gas bladder, Sox2 expression is expanded to the entire foregut. However, we made the unexpected discovery that paralogous genes Bmp16 and Bmp4 show an inverted expression pattern during later gas bladder development in bowfin compared to lung development in mouse, respectively (Funk et al., in review), suggesting that Bmp-signaling played a role in the lung-to-gas bladder evolutionary transition. Known core genes patterning the dorsal-ventral axis during lung budding appear not to exhibit an inverted expression pattern associated with the evolution of a dorsally budding gas bladder. To discover additional genes that may be driving the evolutionary transition from ventrally budding lungs to a dorsally budding gas bladder in ray-finned fishes, we took a genome-wide approach. We sequenced the transcriptome at three developmental stages to screen for the suite of genes differentially expressed between dorsal and ventral foregut tissue during gasbladder development. We found many genes with unknown function in lung development that show differential expression dorsoventrally during gas bladder development. A number of these novel genes are annotated to functions (GO terms) relevant to the process of organ budding, such as extracellular matrix, cell polarity, membrane trafficking, growth factors and patterning, and angiogenesis. Additionally, we identified several lung-development genes that are also differentially expression during gas bladder development. In particular, we found the expression pattern of Tbx5 during gas bladder development is inverted dorsoventrally compared to the that during lung development.
MATERIALS & METHODS
Taxon selection
To identify genes potentially involved in the ventral-to-dorsal shift in budding location underlying the lung-to-gas bladder transition in ray-finned fishes, we chose bowfin as our focal species. Bowfin are an early-diverging ray-finned fish within the sister group to teleosts, the clade containing the majority of extant fishes (over 30,000 species; Fig. 1B). Bowfin develop a gas bladder that exhibits ancestral characteristics, including maintaining a connection to the foregut, the pneumatic duct, and being highly vascularized for respiration (Liem, 1989; Graham, 1997). Conveniently, bowfin diverged prior to the whole genome duplication that occurred at the base of the teleost clade (Fig. 1B; Hoegg et al., 2004). After identifying specific genes-of-interest from the tissue expression profiles in bowfin, we investigated those expression patterns in the bichir, Polypterus senegalus. Bichirs are the sister group to all other ray-finned fishes, and they are the only living ray-finned fishes which develop ventral lungs, making them the most pertinent species to study lung development in comparison to gas bladder development in bowfin.
Tissue sampling
Recently fertilized bowfin eggs were collected from nests in Oneida Lake NY during May of 2018 (for details, see Funk et al., in review). Eggs were reared at 12.8 degrees Celsius in water collected from Oneida Lake in the animal care facility at Cornell University. Bowfin embryos were sampled just before gas bladder budding, at budding, and during outgrowth at stages 24, 25, and 27 respectively (Ballard, 1986). Sampled embryos were euthanized with a lethal dose of MS-222, rinsed in deionized water, and equilibrated in TissueTek optimal cutting temperature (O.C.T.) embedding medium for 2 minutes (IACUC Protocol 2006–0013). Following equilibration, the embryos were transferred to individual cryomolds with fresh O.C.T. and flash frozen in a 2-Methylbutane bath with liquid nitrogen. Additionally, a number of samples were fixed in 4% paraformaldehyde overnight and flash frozen in 2:1 O.C.T:30%-sucrose embedding medium for RNAscope in situ hybridization assays. All embedded samples were stored at −80°C until further processing.
Bichir eggs were obtained from a private aquarist. Eggs were shipped overnight in water treated with methylene blue to prevent fungal growth. We reared the embryos in freshwater at 24 degrees Celsius. Bichir embryos were sampled between 2 and 10 days post hatching to capture a developmental series spanning lung budding (IACUC Protocol 2006–0013; Budgett, 1902). Sampled embryos were euthanized with a lethal dose of MS-222, rinsed in PBS, and fixed overnight in 4% paraformaldehyde (PFA). After fixing, the samples were flash frozen in 2:1 O.C.T:30%-sucrose embedding medium. Embedded samples were stored at −80°C until further processing.
Laser capture microdissection and RNA-seq
Using a cryostat (Leica CM 1950), embedded bowfin embryos were sectioned transversely at 15 μm from the anterior to posterior ends of the gas bladder bud and mounted on polyethylene naphthalate (PEN) membrane slides. For stage 24 embryos, we sectioned the entire foregut along the anterior-posterior axis. For stage 25, we sectioned the entire gas bladder bud and corresponding foregut tissue along the anterior-posterior axis. For stage 27, we ended sectioning of the gas bladder and foregut just anterior to the stomach to avoid contamination. From each transverse section, the dorsal and ventral regions of the foregut endoderm and mesoderm were excised using the Zeiss Palm Microbeam LCM System at the Cornell University Biotechnology Resource Center. When dissecting the dorsal and ventral regions of the foregut, we sampled well within the actual dorsal-ventral boundary to avoid contamination from adjacent tissue regions (Fig. 1C). LCM was completed in 2 hours or less for each slide to minimize RNA degradation in the samples. Immediately following LCM, we added buffer RLT with β-Mercaptoethanol (Qiagen RNeasy Micro) to the tube containing dissected tissue to stabilize the RNA and homogenized the sample by vortexing for 2 minutes and then storing at −20°C. We sampled 3 developmental stages × 4 tissue regions × 3 replicates for 36 samples total.
RNA extraction was performed using the Qiagen RNeasy Micro Kit following the manufacturer’s instructions. RNA amplification and cDNA reverse transcription was completed using the NuGEN Ovation RNA-seq System V2. We used NEBNext Ultra II DNA Library Prep Kit to prepare cDNA libraries for sequencing. All libraries were quality checked on a Tape Station by the Cornell BRC Genomics Facility and showed a single peak of between 170 and 240 base pairs. The individually barcoded libraries (36 total) were pooled and sequenced in one lane of the Illumina NextSeq500 (single end, 150 bp) by the Cornell BRC Genomics Facility.
Bioinformatics
Trimmomatic (version 0.38; Bolger et al., 2014) was used to remove adaptor sequences and low quality regions from the reads. Reads were aligned to the bowfin genome (Braasch pers. comm.) using STAR aligner (version 2.6; Dobin et al., 2013), and read counts per gene were quantified using STAR quant mode. We generated PCA plots in R (version 3.6.0) with the gene expression profiles of all tissue regions for each stage separately. For principle component axes 1–3 at each stage, we ranked the loadings from positive to negative. Using the loadings as pre-ranked lists, we performed Gene Set Enrichment Analysis (GSEA, version 4.0.3) as implemented in the Broad Institute Java script software (Mootha et al., 2003; Subramanian et al., 2005). We tested for enrichment in the GSEA Hallmarks v7.0 gene sets and performed 1000 permutations to establish significance.
To determine differential expression between dorsal and ventral tissue regions, we analyzed each developmental stage separately (12 samples per stage) by building generalized linear models (GLM) in edgeR (Robinson et al., 2010) carried out in R (version 3.6.0). We filtered low expressed genes and included only those genes expressed at minimum of 1 counts per million (cpm) in at least half of the samples or genes expressed at 0.5 cpm in at least three quarters of the samples. We manually curated the functional categories related to tissue budding for our differentially expressed genes based on biological process GO (gene ontology) terms and gene descriptions from UniProt (Anon, 2008).
Tbx5 RNAscope
Fixed and embedded bowfin and bichir embryos were cryosectioned transversely at 15 μm and mounted onto Superfrost Plus slides (Fisher Scientific). We performed RNAscope Multiplex Fluorescent Assays to validate Tbx5 expression patterns during gas bladder development in bowfin and to determine the expression patterns these two genes during lung development in bichir. Species-specific RNA probes were designed for both species and genes by Advanced Cell Diagnostics. We used the RNAscope Multiplex Fluorescent v2 Kit (Advanced Cell Diagnostics, Newark, CA) and Perkin Elmer Cy3 fluorophore following the manufacturer’s protocol. To visualize the fluorescently labeled expression patterns, we used a Zeiss Observer.Z1/ApoTome.2 inverted microscope with AxiocamHRc camera.
Results
Functional categories of genes expressed during bowfin gasbladder development
At each stage of development, gene expression patterns distinctly separated foregut-tissue samples by tissue type, endoderm or mesoderm, along the first two principle component (PC) axes (Fig. 2). At pre-budding (stage 24), gene expression patterns show no separation between dorsal and ventral regions of the foregut and gas bladder (Fig. 2A). Some separation between dorsal and ventral regions of the mesoderm is evident during budding (stage 25) along PC 3 (Fig. 2B) and during outgrowth along PC 1 (Fig. 2C). Dorsal versus ventral identity does not explain much of the variation in gene expression patterns between foregut/gas bladder tissue regions implying few genes are differentially expressed dorsoventrally. This is consistent with our results from the differential expression analysis between dorsal and ventral tissue regions (see below).
Figure 2: Principle Component Analysis (PCA) plots.

showing the main axes of variation in gene expression patterns among foregut/gas bladder tissue samples at each stage. Top row of plots depicts PC1 and PC2, and bottom row depicts PC2 and PC3. Principal components reflect the expression level of all genes in the sampled tissue regions. Endoderm samples are colored in orange, and mesoderm samples are colored in blue. Dorsal samples are labeled D, and ventral samples are labeled V. A) At pre-budding (stage 24), gene expression patterns separate samples by tissue type, endoderm and mesoderm, along PC2. B) During budding (stage 25), gene expression levels also separate by tissue type along PC2. Along PC3, gene expression in the mesoderm shows some separation between dorsal and ventral tissue regions. C) During outgrowth (stage 27), gene expression patterns distinguish endoderm and mesoderm samples along PC1. Gene expression patterns in mesoderm samples show separation along PC1 as well.
To identify conserved gene functions enriched along the PC axes, we ranked the genes contributing to each PC based on their loadings (Table 1; Table S1). The PC axes that discriminate endoderm and mesoderm samples at each stage show similar functional enrichment. At pre-budding and outgrowth stages, genes with positive loadings along these PC axes showed highest enrichment, based on the normalized enrichment score (NES), in epithelial to mesenchymal transition (Table 1). Epithelial to mesenchymal transition was also one of the top enriched functions for genes with negative loadings along PC 2 at budding stage. Additionally, several processes related to cell cycle are enriched along the PC axes at each stage including G2M checkpoint, E2F targets, mitotic spindle, and DNA repair (Table 1, Table S1). Several signaling pathways, including Kras signaling, Notch signaling, Wnt/Beta-catenin signaling, Hedgehog signaling, and Il6/Jak/Stat3 signaling, are enriched as well. (Table 1, Table S1)
Table 1:
Top 10 Hallmark v7.0 gene sets enriched along the principle component axis that distinguishes endoderm (endo) and mesoderm (meso) at each stage of gas bladder development.
| Pre-budding (stage 24) | Budding (stage 25) | Outgrowth (stage 27) | |||
|---|---|---|---|---|---|
| PC2: Separates endo and meso | PC2: Separates endo and meso | PC1: Separates endo and meso | |||
| Positive loading | Positive loading | Positive loading | |||
| NAME | NES | NAME | NES | NAME | NES |
| Epithelial mesenchymal transition | 1.81 | E2F targets | 1.50 | Epithelial mesenchymal transition | 2.05 |
| G2M checkpoint | 1.35 | G2M checkpoint | 1.38 | UV response Dn | 1.74 |
| Spermatogenesis | 1.34 | Androgen response | 1.32 | Myogenesis | 1.61 |
| Apical junction | 1.25 | Xenobiotic metabolism | 1.30 | Apical junction | 1.56 |
| E2F targets | 1.24 | Complement | 1.29 | Coagulation | 1.49 |
| Allograft rejection | 1.19 | Estrogen response late | 1.26 | Angiogenesis | 1.49 |
| Peroxisome | 1.19 | Unfolding protein response | 1.26 | Hypoxia | 1.42 |
| Tnfa signaling via Nfkb | 1.13 | P53 pathway | 1.24 | Complement | 1.40 |
| Glycolosis | 1.10 | DNA repair | 1.24 | Hedgehog signaling | 1.37 |
| Myc targets V1 | 1.09 | Mitotic spindle | 1.23 | Cholesterol homeostasis | 1.30 |
| Negative loading | Negative loading | Negative loading | |||
| NAME | NES | NAME | NES | NAME | NES |
| Interferon alpha response | −1.48 | Il6 Jak Stat3 signaling | −1.35 | Oxidative phosphorylation | −1.95 |
| Interferon gamma response | −1.42 | Wnt Beta-catenin signaling | −1.29 | Myc targets V2 | −1.78 |
| P53 pathway | −1.34 | Apical junction | −1.27 | Myc targets V1 | −1.51 |
| Estrogen response late | −1.33 | Hypoxia | −1.23 | Interferon alpha response | −1.19 |
| Complement | −1.30 | Allograft rejection | −1.10 | Uv response Up | −1.18 |
| Apical surface | −1.27 | Epithelial mesenchymal transition | −0.97 | Fatty acid metabolism | −1.16 |
| Cholesterol homeostasis | −1.17 | Notch signaling | −0.83 | G2M checkpoint | −1.15 |
| Kras signaling Dn | −1.16 | Tnfa signaling via Nfkb | −0.81 | Kras signaling Dn | −1.13 |
| Estrogen response early | −1.11 | Bile acid metabolism | −0.72 | Androgen response | −1.11 |
| Xenobiotic metabolism | −1.06 | Protein secretion | −0.65 | Wnt Beta-catenin signaling | −1.06 |
As development progresses and the gas bladder becomes more differentiated, more genes become differentially expressed between dorsal and ventral foregut tissue. Twelve, 45, and 160 genes are differentially expressed during pre-budding (stage 24), budding (stage 25), and outgrowth (stage 27) stages, respectively, at a false discovery rate (FDR) of 0.1 (Fig. 3). By manual curation, we found many of the genes differentially expressed between dorsal and ventral tissues are annotated to functions relevant to the process of gas bladder budding, including extracellular matrix and cell adhesion, cell polarity and actin cytoskeleton, cell migration, membrane trafficking, growth factors and patterning, and angiogenesis (Table 2).
Figure 3: Differentially expressed genes between dorsal and ventral tissues of the foregut during gas bladder development.

Heat maps showing differentially expressed genes between dorsal and ventral foregut tissue at 3 stages of gasbladder development in bowfin. Each column is a sample and each row is a gene; gene names are listed along the right hand side. Below each heatmap, samples (columns) are labelled dorsal (D) or ventral (V) and endoderm or mesoderm. Warmer colors indicate higher relative gene expression, and cooler colors indicate lower relative expression. A) Twelve genes are differentially expressed at pre-budding (stage 24). B) At budding (stage 25), 45 genes are differentially expressed bewteen dorsal and ventral tissues. C) During outgrowth, 160 genes show differential expression dorsoventrally. Note that as gas bladder development proceeds, more genes are differentially expressed between dorsal and ventral foregut tissues.
Table 2:
Dorsoventral differentially expressed (DE) genes manually curated by function based on GO terms and gene descriptions from UniProt (Anon, 2008).
| FUNCTION | Stage 24 DE Genes | Stage 25 DE Genes | Stage 27 DE Genes |
|---|---|---|---|
| Extracellular Matrix Cell Adhesion |
st3gal5 | col6a3, dcn, pcdhgc5 | postn, itgb5, tnc, mmp11, matn4, itga3, ddr2, col3a1, adam19, rgcc, pkp3, loxla, cdh31, epcam, mcam, pkp1, megf10, nectin1, sorbs3 |
| Angiogenesis | N/A | dcn, map3k3 | thbs2a, loxla, gata6, mcam, ptgis, plcg1, prox1, rgcc, notch3 |
| Cell polarity Actin |
N/A | actr2, was, fnbp1l, fryl, arhgef25 | limch1, prex1, rab11fip2, actc1a, cotl1, ttn, acta2, tpm3, ripor1 |
| Cell migration Cancer |
insr, brd9 | dcn, map3k3 | hgfb, limch1, itgb5, loxla, spata13, epcam, igfbp5, itga3, plcg1, ddr2, megf10, ripor1, prox1, rgcc, arid2, tbx5, tead3, kiaa1549, vwce |
| Growth factors Patterning |
arl6 | tead3, esrp2, foxa2, tbx5, rab7 | hgfb, igfbp5, gata6, mid1, fzd7a, plcg1, fibl3, c3, igf2, pitx3, rgcc, olfml, irx2 |
| Membrane Trafficking Membrane Organization | arl6 | fnbp1l, tmem111, actr2, stx16, rab7, aftph | pkp3, krt18, itga3, tlcd1, gpd1l, mrap, sfxn2, abcc8 |
Expression of lung development genes during bowfin gasbladder development
We manually curated the differentially expressed genes to identify those known to be involved in lung development in mice. During gasbladder budding (stage 25), these include Tbx5 and FoxA2 (Fig. 4; Weidenfeld et al., 2002; Arora et al., 2012), and during outgrowth (stage 27), these include Fzd7a, Gata4, Gata5, Gata6, Irx2, and Notch3 (Fig. 5; Becker et al., 2001; Weidenfeld et al., 2002; Wang et al., 2005; Ackerman et al., 2007; Hussain et al., 2017), all of which are significantly expressed dorsoventrally in our datasets. Of these 8 genes, Tbx5 is particularly interesting because its early expression in the gas bladder is inverted relative to its spatial expression during mouse lung development. In the gas bladder during pre-budding, Tbx5 is expressed at similar levels in both dorsal and ventral mesoderm. However, at budding and outgrowth, Tbx5 expression increases in the dorsal mesoderm while decreasing in the ventral mesoderm (Fig. 6A). During gasbladder development, Tbx5 is expressed in a gradient across the dorsoventral axis of the foregut with strong expression dorsally and weak expression ventrally. This is in contrast to mouse lung development, during which Tbx5 is expressed strongly in the ventral mesoderm.
Figure 4: Tissue-specific expression of known lung-development genes, differentially expressed at budding, during gas bladder development in bowfin.

Average level of expression of lung-development genes (FoxA2 and Tbx5) differentially expressed during gasbladder budding (stage 25) in the A) dorsal mesoderm, B) dorsal endoderm, C) ventral mesoderm, and D) ventral endoderm, at all stages of gasbladder development in bowfin. From these line plots, the pattern of Tbx5 expression is particularly interesting because expression is stronger in the dorsal mesoderm than the ventral, which is opposite of the expression pattern during lung development. Sample size of 3 for each stage and tissue region. Error bars show the standard error of the mean.
Figure 5: Tissue-specific expression of known lung-development genes, differentially expressed at outgrowth, during gas bladder development in bowfin.
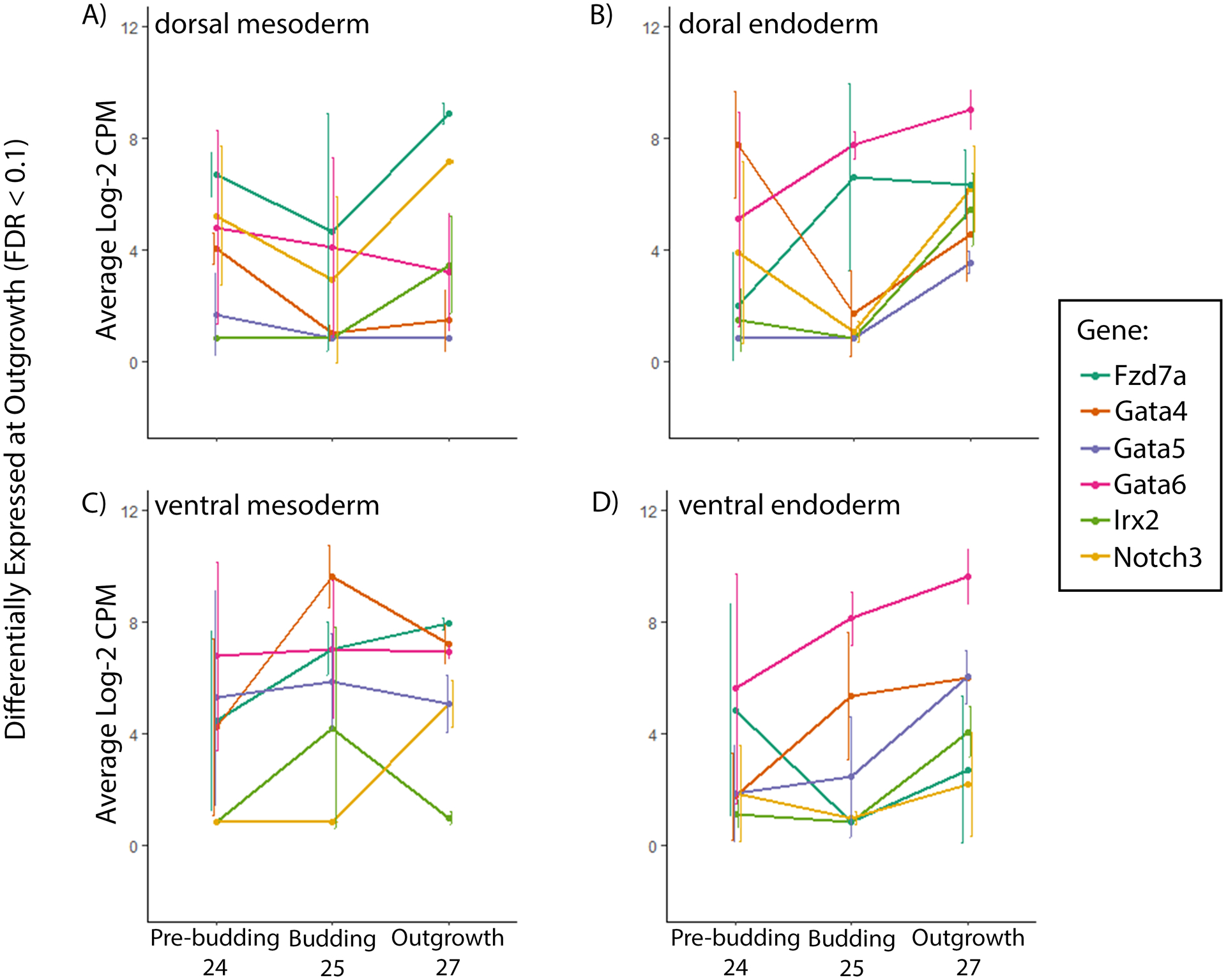
Average level of expression of lung-development genes (Fzd7a, Gata4, Gata5, Gata6, Irx2, and notch3) differentially expressed during outgrowth (stage 27) in the A) dorsal mesoderm, B) dorsal endoderm, C) ventral mesoderm, and D) ventral endoderm at all stages of gasbladder development in bowfin. These genes, all with known roles in lung development, are differentially expressed across the dorsoventral axis during gasbladder development in bowfin. Sample size of 3 for each stage and tissue region. Error bars show the standard error of the mean.
Figure 6: Dorsoventral expression of Tbx5 during gas bladder development in bowfin.
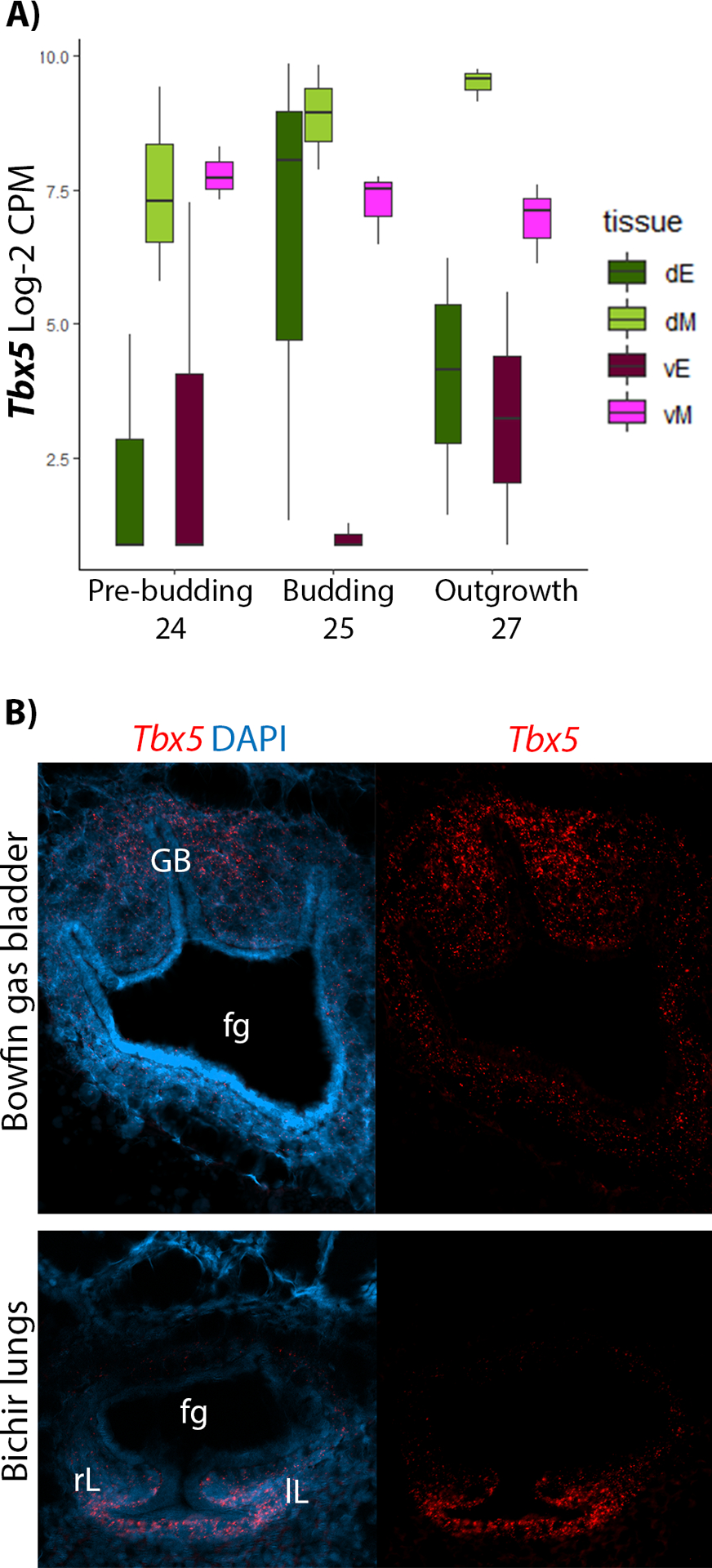
A) Boxplot of Tbx5 expression at pre-budding, budding, and outgrowth stages of gasbladder development. Tbx5 expression in the dorsal and ventral mesoderm is equally high during pre-budding (pink and light green), but as development proceeds through budding and outgrowth, dorsal expression increases while ventral expression decreases. B) RNAscope in situ hybridization of Tbx5 during gas bladder development in bowfin compared with lung development in bichir. During gas bladder outgrowth (stage 27) in bowfin, Tbx5 is expressed most strongly in the dorsal mesoderm surrounding the gas bladder (top panels). During lung outgrowth (stage 35) in bichir, Tbx5 is expressed in the ventral mesoderm surrounding the developing lungs (bottom panels). The left side of panels show Tbx5 expression in bright red overlaid with DAPI (bright blue), which stains nuclei, and the right panels show Tbx5 expression alone in bright red. Abbreviations: fg, foregut; gb, gas bladder; lL, left lung; rL, right lung.
We used RNAscope in situ hybridization to confirm the dorsoventral expression patterns of Tbx5 during bowfin gas bladder development. In addition, we also investigated the expression patterns of Tbx5 during bichir lung development to better rule out the possibility that the change in expression pattern is associated with the evolution of ray-finned fishes (Fig. 1B), and determine whether changes to Tbx5 expression are associated with the clade within ray-finned fishes that has a dorsally budding gas bladder. Bichir are ray-finned fish more closely related to bowfin than to tetrapods; however, importantly, bichir retain ventrally budding lungs (Fig. 1B). During the outgrowth stage of gasbladder development, Tbx5 shows a clear dorsoventral expression gradient across the foregut, in which Tbx5 is expressed strongly in the dorsal mesoderm surrounding the gasbladder bud (Fig. 6B). In contrast, during lung outgrowth in mouse and bichir (Fig. 6B), Tbx5 is expressed strongly in the ventral mesoderm surrounding the lung buds and is absent from the dorsal mesoderm.
In mouse, Tbx5 is known to interact with a number of genes during lung development including Tbx4, Fgf10, and Wnt2/Wnt2b (Morrisey and Hogan, 2010; Arora et al., 2012; Hines and Sun, 2014). None of these genes were found to be significantly differentially expressed across the dorsoventral axis during gasbladder development in bowfin. However, all were expressed in foregut and gasbladder tissue and showed supporting trends in expression pattern differences dorsoventrally. Tbx4 is strongly expressed in the dorsal and ventral mesoderm during gasbladder budding (stage 25). During gasbladder outgrowth (stage 27), Tbx4 expression increases in the dorsal mesoderm while it remains the same in the ventral mesoderm creating a dorsoventral gradient similar to Tbx5 (Fig. 7A). Wnt2ba, a paralogue of Wnt2b, shows increasing expression in the dorsal mesoderm between pre-budding (stage 24) and outgrowth (stage 27) during gasbladder development. Expression of Wnt2ba also increases in the ventral mesoderm from budding (stage 25) to outgrowth (stage 27); however, expression is weaker than that observed in the dorsal mesoderm (Fig. 7B). Fgf10 expression is weak or absent during gasbladder pre-budding (stage 24) and budding (stage 25). But, during gasbladder outgrowth (stage 27), Fgf10 is strongly expressed in the ventral and dorsal mesoderm (Fig. 7C).
Figure 7: Expression of Tbx4, Wnt2ba, and Fgf10 during gas bladder development in bowfin.
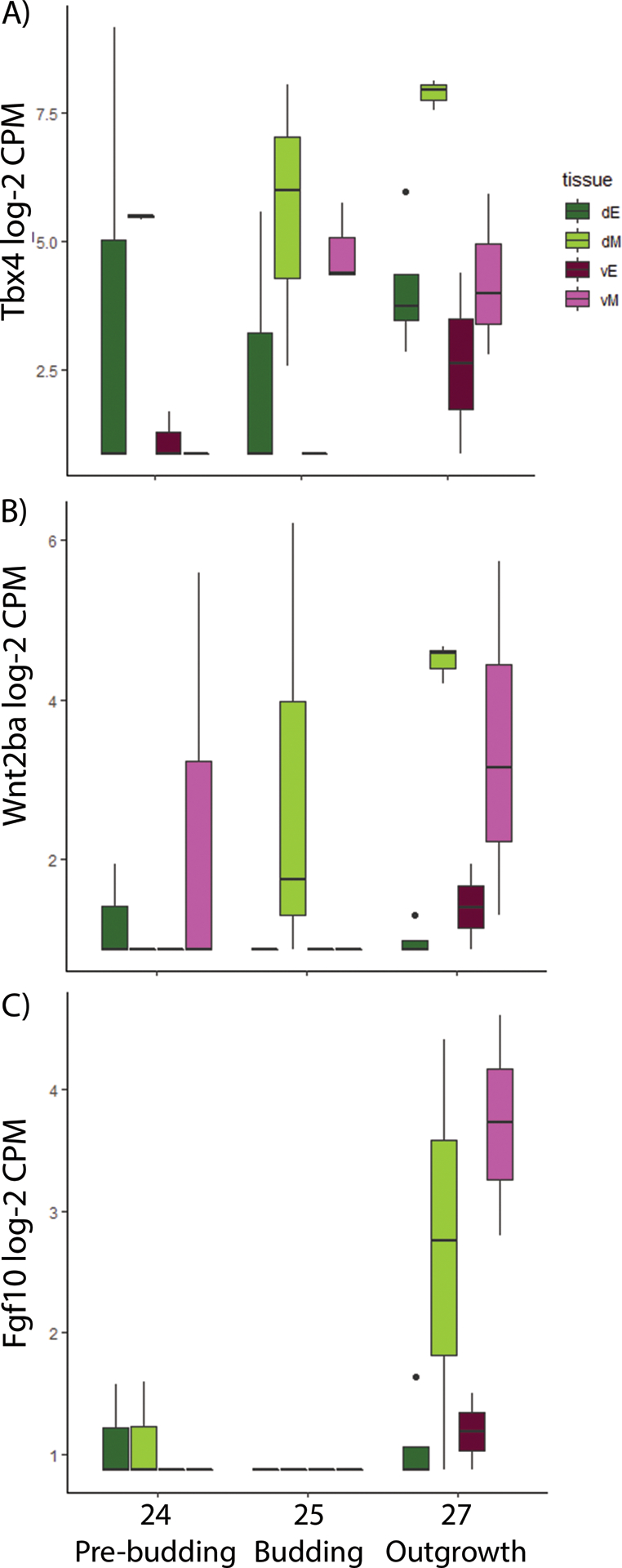
Tbx4, Wnt2ab, and Fgf10 are all expressed during gas bladder development; however, none were found to be significantly differentially expressed between dorsal and ventral tissue regions. A) Tbx4 is expressed in the dorsal mesoderm (dM) during pre-budding and budding stages. During outgrowth, expression of Tbx4 increases in the dorsal mesoderm. Though Tbx4 is also expressed in the ventral mesoderm (vM) at budding and outgrowth stages, expression is weaker than in the dorsal mesoderm. B) Wnt2ba is expressed in the dorsal mesoderm during gas bladder budding and shows an increase in expression in the dorsal mesoderm as development proceeds to the outgrowth stage. C) Fgf10 shows weak or no expression in any foregut tissue regions during pre-budding and budding; however, during gas bladder outgrowth, expression in both the dorsal and ventral mesoderm increases. Abbreviations: dE, dorsal endoderm; dM, dorsal mesoderm; vE, ventral endoderm; vM, ventral mesoderm.
Discussion
RNA-seq identifies novel genes associated with gas bladder budding in bowfin
Using RNA-seq, we aimed to discover genes that are differentially expressed across the dorsoventral axis of the foregut during gas bladder development and thus may play a role in regulating the dorsal direction of budding. By manual curation, we discovered many of the differentially expressed dorsoventral genes are annotated to GO terms that are related to organ development, e.g. gas bladder budding (Table 2). The extracellular matrix (ECM) is an interface between the epithelium and the mesenchyme such that during organ budding, the ECM is continuously remodeled through both chemical and mechanical signals (Kim and Nelson, 2012). Thus, the ECM plays a role in regulating cell proliferation, cell differentiation, tissue shape change, and the establishment of cell polarity, all of which are happening during the budding process. Specifically, during lung branching morphogenesis, the thinning of the ECM adjacent to the tip of nascent lung buds in mice occurs simultaneously with high rates of epithelial cell proliferation (Kim and Nelson, 2012). Both ECM-cell and cell-cell adhesion can be regulated by the actin cytoskeleton producing contractile and tensile forces to cause shape changes to tissue and organs, such as budding (Kim and Nelson, 2012). During lung budding, the epithelial cells begin migrating out into the surrounding mesenchyme before cell proliferation occurs (Nogawa et al., 1998; Lü et al., 2005), and Fgf10, a growth factor expressed in the mesenchyme, regulates the direction of this cell migration (Park et al., 1998). The epithelial cells of the lungs or gas bladder form a monolayer over the surface of the organ serving as a barrier between extracellular environments (Drubin and Nelson, 1996; Mostov et al., 2000). To function properly as a barrier and regulate membrane trafficking of proteins, these epithelial cells are polarized, meaning the distribution of cellular components inside a single cell is asymmetrical (Drubin and Nelson, 1996; El‐Hashash and Warburton, 2011). As cells proliferate and differentiate during budding, the polarity of new epithelial cells must be established. The key players stimulating cell proliferation, differentiation, and migration are growth factors.
Lung-gas bladder inversion
Of the genes differentially expressed between the dorsal and ventral foregut during gas bladder development, we identified those known to regulate lung development and asked whether they show an inverted expression pattern during gas bladder development relative to lungs. Tbx5 is an early marker of a dorsally budding gas bladder and is strongly expressed in the dorsal mesenchyme surrounding the gas bladder both at budding and outgrowth stages in bowfin. In addition, the paralogous gene, Tbx4, is also strongly expressed in the dorsal mesenchyme during gas bladder outgrowth. The dorsal expression of these genes during gasbladder development contrasts with the ventral expression patterns observed during bichir and tetrapod (Sakiyama et al., 2003; Arora et al., 2012; Tatsumi et al., 2016) lung development. We confirmed the early expression of Tbx5 in the ventral mesenchyme surrounding the bichir lung at budding and outgrowth stages of development using RNAscope in situ hybridization assays. Notably, in mouse and chicken, Tbx5 and Tbx4 are known to contribute to the budding and elongation of the lung. Specifically, Tbx5 alone is important for bilateral specification during initial lung budding in mouse. When Tbx5 expression is reduced in mouse foregut tissue cultures, Nkx2.1 expression, the first genetic marker of tetrapod lung development, is lost in one lung bud (Arora et al., 2012). Later in development, during tetrapod lung elongation, both Tbx5 and Tbx4 are important for proper branching morphogenesis (Sakiyama et al., 2003; Arora et al., 2012). Thus, the expression pattern of Tbx5 and Tbx4 during development of the dorsal gas bladder in bowfin is inverted relative to that during development of the ventral lungs in bichir and tetrapods. These data suggest Tbx5 may be involved in regulating the direction of budding of both gas bladders and lungs.
Resulting from the teleost whole genome duplication (WGD), zebrafish have two copies of Tbx5: Tbx5a and Tbx5b (Albalat et al., 2010; Boyle Anderson and Ho, 2018). Although spatial expression of either Tbx5a or Tbx5b in the gas bladder has not yet been examined in zebrafish, in Tbx5b-knockdown embryos, the gas bladder does not inflate (Boyle Anderson and Ho, 2018) providing evidence supporting a role for Tbx5 in gas bladder development. However, non-inflation of the gas bladder is common and known to be the result of many distinct genetic mutations (McCune and Carlson, 2004). Both the dorsal expression pattern of Tbx5 during gas bladder development in bowfin and the functional role that Tbx5b plays in zebrafish gas bladder development are consistent with the hypothesis that inverted spatial expression of Tbx5 contributed to the morphological inversion of budding location from ventral lungs to dorsal gas bladder in ray-finned fishes.
Genes interacting with Tbx5
After revealing the inverted expression pattern of Tbx5 mirroring the inverted budding direction during lung versus gasbladder development, we examined whether genes known to interact with Tbx5 exhibit similar expression patterns to Tbx5 during development of the gas bladder. Tbx5 and Tbx4 expression overlaps with Fgf10 expression in the dorsal mesoderm during gas bladder development in bowfin suggesting that these genes may interact to regulate outgrowth as they do during tetrapod lung development. T-box genes have a known role as transcriptional regulators, and Tbx5 and Tbx4 have been shown to regulate Fgf10 expression during mouse and chick lung development, respectively (Ng et al., 2002; Cebra‐Thomas et al., 2003; Sakiyama et al., 2003; Arora et al., 2012). Fgf10 plays an important role in lung budding as evidenced by the absence of lungs in Fgf10-knockdown mice (Min et al., 1998; Sekine et al., 1999). Later during lung development, Fgf10 is also critical for regulating branching morphogenesis (Morrisey and Hogan, 2010). During lung development in mice, Tbx4 and Tbx5 show overlapping expression in the lung mesenchyme and may be functionally redundant (Chapman et al., 1996; Naiche and Papaioannou, 2003; Cardoso and Lü, 2006). In fact, when Tbx4 and Tbx5 expression is reduced, the mutant mice show decreased Fgf10 expression during lung branching and accordingly, the lungs were smaller than wild type (Cebra‐Thomas et al., 2003; Arora et al., 2012). In chick, Tbx4 is is co-expressed with Fgf10 in the ventral lung mesenchyme (Sakiyama et al., 2003; Morrisey and Hogan, 2010). When Tbx4 is ectopically expressed in the visceral mesoderm of the foregut, Fgf10 expression is induced and ectopic lung buds form on the esophagus, whereas when Tbx4 function is disrupted in chick, Fgf10 expression is repressed and lung buds fail to form (Sakiyama et al., 2003). During bowfin gasbladder development, both Tbx4 and Tbx5 are expressed most strongly in the dorsal mesenchyme surrounding the gasbladder bud at all stages. Fgf10 expression also overlaps with Tbx4 and Tbx5 in the dorsal mesenchyme but not until the outgrowth stage. Compared to lung development, the delayed onset of Fgf10 expression suggests it is not involved in initiating gasbladder budding. However, the overlapping expression patterns between Tbx4, Tbx5, and Fgf10 during outgrowth make apparent the possibility of gene interactions regulating gasbladder development.
During bowfin gas bladder development, Wnt2ba, whose ortholog is known to act upstream of Tbx4 and Tbx5 during lung development, is co-expressed with Tbx4, Tbx5, and Fgf10 in the dorsal mesenchyme. In mouse, Wnt2 and Wnt2b are expressed ventrally in the lung mesenchyme during lung specification and outgrowth (Goss et al., 2009), and when both are lost in mice, the lung buds fail to develop. Additionally, the loss of Wnt2 leads to reduced expression of Fgf10 (Goss et al., 2009). Though the exact interactions between Tbx5, Fgf10, and Wnt2/2b during lung development have not yet been determined, the interaction of these genes is established in other contexts, such as fin bud initiation and outgrowth in zebrafish (Ng et al., 2002). These comparative data suggest that Wnt2ba could be regulating Tbx5 and Tbx4 expression, which in turn, are interacting with Fgf10 to promote gasbladder development.
Interactions between genes expressed in the mesenchyme and those expressed the in epithelium are also critical. Specifically, Fgf10 expression in the mesenchyme and Bmp4 expression in the epithelium interact to regulate lung outgrowth and branching (Weaver et al., 2000; Morrisey and Hogan, 2010; Hines and Sun, 2014). In this study, all tissues, dorsal and ventral endoderm and mesoderm, show equal RNA expression of Bmp4 at all stages of gas bladder development in bowfin (Fig. S1A). However, previously, we showed that Bmp4 protein expression appears absent from the foregut and gas bladder during both bowfin and zebrafish development (Funk et al., in review). Instead, Bmp16, a paralogue of Bmp4 that has been lost from the mammalian genome but retained in ray-finned fishes, is expressed strongly in the gas bladder epithelium during outgrowth (Funk et al., in review; Feiner et al., 2009). Bmp16 RNA expression, on the other hand, is weak or absent at all developmental stages (Fig. S1B). The discrepancy between RNA and protein expression is not unusual for signaling molecules like Bmps, which are not necessarily active in the same location that they are transcribed. Because the protein, translated from RNA, is the molecule that carries out biological processes, protein expression is more reliable for studying Bmp patterns. Bmp16 protein expression during outgrowth (stage 27; Funk et al., in review), the same stage at which Fgf10 expression increases, suggests an interaction between Bmp16 and Fgf10 may contribute to the regulation of gas bladder expansion.
Conclusion
Here we show that Tbx5, known to be important for lung development, is expressed in the dorsal mesenchyme surrounding the gas bladder. Spatial expression of Tbx5 is inverted in the bowfin gas bladder relative to its expression during mouse lung development. Additionally, we identified Tbx4, Wnt2, and Fgf10, all of which interact with Tbx5 during mouse lung development, as potential players in gas bladder development based on their overlapping expression patterns with Tbx5. Functional experiments are not yet possible to perform on bowfin because we cannot collect single-cell stage embryos from the wild and we are unable to breed them in the lab. However, zebrafish could provide the opportunity to test the function of these candidate genes during gasbladder development.
Supplementary Material
Figure S1: Bmp4 and Bmp16 expression during gasbladder development in bowfin. A) Boxplot of average Bmp4 expression at pre-budding (24), budding (25), and outgrowth (27) stages of gas bladder development. Bmp4 is expressed at all stages. During outgrowth, Bmp4 is expressed strongest dorsally, in both endoderm and mesoderm. B) Boxplot of average Bmp16 expression at all stages. No or weak expression of Bmp16 is detected at all stages in all tissue regions. Abbreviations: dE, dorsal endoderm; dM, dorsal mesoderm; vE, ventral endoderm; vM, ventral mesoderm.
Acknowledgements
E. Funk was supported by the National Science Foundation Graduate Research Fellowship Program and the Cornell Presidential Life Sciences Fellowship. E. Lencer is supported by the National Institutes of Health Ruth L. Kirschstein National Research Service Award (T32CA17468). Funding for this project was provided by the American Society for Ichthyology and Herpetology Edward C. Raney Fund, the Cornell Department of Ecology and Evolutionary Biology, and McCune Lab Funds. Many thanks to Ken Zeedyk for breeding his bichirs and providing many eggs, allowing us to study bichir lung development. This study could not have been done without his generosity and interest. We also thank Joe Fetcho and Nikki McGuire for generously providing zebrafish embryos from the Fetcho Lab zebrafish colony. Thank you to Francis Feng for allowing us to use his boat ramp on Oneida Lake easing our access to bowfin spawning sites. In addition, we are pleased to thank Bhargav Sanketi for sharing his technical expertise and experience with RNAscope. We are very grateful to Natasza Kurpios for access to her lab. Bob Reed provided invaluable advice on methodology and feedback on the manuscript. Bioinformatic analyses were performed on the BioHPC cloud managed by the Cornell Biotechnology Resource Center Bioinformatics Facility.
Footnotes
Data Availability
The data supporting the findings of this publication are openly available on NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession number GSE152992 (https://www.ncbi.nim.nih.gov/geo/query/acc.cgi?acc=GSE152992).
References
- Ackerman KG, Wang J, Luo L, Fujiwara Y, Orkin SH, Beier DR. 2007. Gata4 Is Necessary for Normal Pulmonary Lobar Development. Am J Respir Cell Mol Biol 36:391–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albalat R, Baquero M, Minguillón C. 2010. Identification and characterisation of the developmental expression pattern of tbx5b, a novel tbx5 gene in zebrafish. Gene Expr Patterns 10:24–30. [DOI] [PubMed] [Google Scholar]
- Alexander RMcN. 1966. Physical Aspects of Swimbladder Function. Biol Rev 41:141–176. [DOI] [PubMed] [Google Scholar]
- Anon. 2008. The Universal Protein Resource (UniProt). Nucleic Acids Res 36:D190–D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D, Nubler-Jung K. 1994. Inversion of dorsoventral axis? | Nature. Nature 371:26. [DOI] [PubMed] [Google Scholar]
- Arora R, Metzger RJ, Papaioannou VE. 2012. Multiple Roles and Interactions of Tbx4 and Tbx5 in Development of the Respiratory System. PLOS Genet 8:e1002866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Averof M, Patel NH. 1997. Crustacean appendage evolution associated with changes in Hox gene expression. Nature 388:682–686. [DOI] [PubMed] [Google Scholar]
- Ballard WW. 1986. Stages and rates of normal development in the holostean fish, Amia calva. J Exp Zool 238:337–354. [Google Scholar]
- Becker M-B, Zülch A, Bosse A, Gruss P. 2001. Irx1 and Irx2 expression in early lung development. Mech Dev 106:155–158. [DOI] [PubMed] [Google Scholar]
- Betancur-R R, Wiley EO, Arratia G, Acero A, Bailly N, Miya M, Lecointre G, Ortí G. 2017. Phylogenetic classification of bony fishes. BMC Evol Biol 17:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinforma Btu 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle Anderson EAT, Ho RK. 2018. A transcriptomics analysis of the Tbx5 paralogues in zebrafish. PLoS ONE 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budgett JS. 1902. On the Structure of the Larval Polypterus. Trans Zool Soc Lond 16:315–346. [Google Scholar]
- Cardoso WV, Lü J. 2006. Regulation of early lung morphogenesis: questions, facts and controversies. Development 133:1611–1624. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK, Weatherbee SD. 2013. From DNA to diversity: molecular genetics and the evolution of animal design. John Wiley & Sons. Available from: https://books.google.com/books?hl=en&lr=&id=VF6gomATy1AC&oi=fnd&pg=PT291&dq=carroll+grenier+weatherbee+2013&ots=C3q6og0ts6&sig=nlEWIUBwctxWe2NrTNwZ-cRLxWc [Google Scholar]
- Cass AN, Servetnick MD, McCune AR. 2013. Expression of a lung developmental cassette in the adult and developing zebrafish swimbladder. Evol Dev 15:119–132. [DOI] [PubMed] [Google Scholar]
- Cebra‐Thomas JA, Bromer J, Gardner R, Lam GK, Sheipe H, Gilbert SF. 2003. T-box gene products are required for mesenchymal induction of epithelial branching in the embryonic mouse lung. Dev Dyn 226:82–90. [DOI] [PubMed] [Google Scholar]
- Chapman DL, Garvey N, Hancock S, Alexiou M, Agulnik SI, Gibson‐Brown JJ, Cebra‐Thomas J, Bollag RJ, Silver LM, Papaioannou VE. 1996. Expression of the T-box family genes, Tbx1–Tbx5, during early mouse development. Dev Dyn 206:379–390. [DOI] [PubMed] [Google Scholar]
- Daniels CB, Orgeig S, Sullivan LC, Ling N, Bennett MB, Schürch S, Val AL, Brauner CJ. 2004. The Origin and Evolution of the Surfactant System in Fish: Insights into the Evolution of Lungs and Swim Bladders. Physiol Biochem Zool Ecol Evol Approaches 77:732–749. [DOI] [PubMed] [Google Scholar]
- Darwin C 1859. On the origins of species by means of natural selection. Lond Murray [Internet] 247. Available from: http://sciencestudies.pbworks.com/f/OoS.pdf [Google Scholar]
- De Robertis EM, Sasai Y. 2000. A common plan for dorsoventral patterning in Bilateria. Shaking Tree Read Nat Hist Life:89. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. 1996. Origins of Cell Polarity. Cell 84:335–344. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El‐Hashash AH, Warburton D. 2011. Cell polarity and spindle orientation in the distal epithelium of embryonic lung. Dev Dyn 240:441–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner N, Begemann G, Renz AJ, Meyer A, Kuraku S. 2009. The origin of bmp16, a novel Bmp2/4relative, retained in teleost fish genomes. BMC Evol Biol 9:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson EL. 1996. Conservation of dorsal-ventral patterning in arthropods and chordates - ScienceDirect. Curr Opin Genet Dev 6:424–431. [DOI] [PubMed] [Google Scholar]
- Funk EC, Breen C, Sanketi BD, Kurpios NA, McCune AR. in review. Changes in Nkx2.1, Sox2, Bmp4, and Bmp16 expression underlying the lung-to-gas bladder evolutionary transition in ray-finned fishes. Evol Dev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoffroy St-Hilaire E 1822. Considerations gererales sur la vertebre. Mem Mus Hist Nat 9:89–119. [Google Scholar]
- Goodrich ES. 1958. Studies on the structure and development of vertebrates. 2nd ed. New York: Dover Publications Inc. [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, Morrisey EE. 2009. Wnt2/2b and β-Catenin Signaling Are Necessary and Sufficient to Specify Lung Progenitors in the Foregut. Dev Cell 17:290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham JB. 1997. Air-Breathing Fishes: Evolution, Diversity, and Adaptation. Academic Press. [Google Scholar]
- Helfman G, Collette BB, Facey DE, Bowen BW. 2009. The Diversity of Fishes: Biology, Evolution, and Ecology, 2nd Edition. 2nd ed. Wiley-Blackwell. [Google Scholar]
- Hines EA, Sun X. 2014. Tissue Crosstalk in Lung Development. J Cell Biochem 115:1469–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. 2004. Phylogenetic Timing of the Fish-Specific Genome Duplication Correlates with the Diversification of Teleost Fish. J Mol Evol 59:190–203. [DOI] [PubMed] [Google Scholar]
- Holley SA, Jackson PD, Sasai Y, Lu B, DeRobertis EM, Hoffman FM, Ferguson EL. 1995. A conserved system for dorsal-ventral patterning in insects and vertebrates involving sog and chordin | Nature. Nature 376. [DOI] [PubMed] [Google Scholar]
- Hussain M, Xu C, Ahmad M, Yang Y, Lu M, Wu X, Tang L, Wu X. 2017. Notch Signaling: Linking Embryonic Lung Development and Asthmatic Airway Remodeling. Mol Pharmacol 92:676–693. [DOI] [PubMed] [Google Scholar]
- Kardong KV. 2015. The Respiratory System. In: Vertebrates: comparative anatomy, function, evolution. 7th ed. New York: McGraw-Hill. p 413–450. [Google Scholar]
- Kim HY, Nelson CM. 2012. Extracellular matrix and cytoskeletal dynamics during branching morphogenesis. Organogenesis 8:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem KF. 1989. Respiratory Gas Bladders in Teleosts: Functional Conservatism and Morphological Diversity. Am Zool 29:333–352. [Google Scholar]
- Longo S, Riccio M, McCune AR. 2013. Homology of lungs and gas bladders: Insights from arterial vasculature. J Morphol 274:687–703. [DOI] [PubMed] [Google Scholar]
- Lü J, Izvolsky KI, Qian J, Cardoso WV. 2005. Identification of FGF10 Targets in the Embryonic Lung Epithelium during Bud Morphogenesis. J Biol Chem 280:4834–4841. [DOI] [PubMed] [Google Scholar]
- McCune AR, Carlson RL. 2004. Twenty ways to lose your bladder: common natural mutants in zebrafish and widespread convergence of swim bladder loss among teleost fishes. Evol Dev 6:246–259. [DOI] [PubMed] [Google Scholar]
- Min H, Danilenko DM, Scully SA, Bolon B, Ring BD, Tarpley JE, DeRose M, Simonet WS. 1998. Fgf-10 is required for both limb and lung development and exhibits striking functional similarity to Drosophila branchless. Genes Dev 12:3156–3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273. [DOI] [PubMed] [Google Scholar]
- Morrisey EE, Hogan B. 2010. Preparing for the First Breath: Genetic and Cellular Mechanisms in Lung Development. Dev Cell 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostov KE, Verges M, Altschuler Y. 2000. Membrane traffic in polarized epithelial cells. Curr Opin Cell Biol 12:483–490. [DOI] [PubMed] [Google Scholar]
- Naiche LA, Papaioannou VE. 2003. Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois | Development. Development 130:2681–2693. [DOI] [PubMed] [Google Scholar]
- Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the World. 5th ed. John Wiley & Sons. [Google Scholar]
- Ng JK, Kawakami Y, Büscher D, Raya Á, Itoh T, Koth CM, Esteban CR, Rodríguez-León J, Garrity DM, Fishman MC, Belmonte JCI. 2002. The limb identity gene Tbx5 promotes limb initiation by interacting with Wnt2b and Fgf10. Development 129:5161–5170. [DOI] [PubMed] [Google Scholar]
- Nogawa H, Morita K, Cardoso WV. 1998. Bud formation precedes the appearance of differential cell proliferation during branching morphogenesis of mouse lung epithelium in vitro. Dev Dyn 213:228–235. [DOI] [PubMed] [Google Scholar]
- Park WY, Miranda B, Lebeche D, Hashimoto G, Cardoso WV. 1998. FGF-10 Is a Chemotactic Factor for Distal Epithelial Buds during Lung Development. Dev Biol 201:125–134. [DOI] [PubMed] [Google Scholar]
- Rice AN, Bass AH. 2009. Novel vocal repertoire and paired swimbladders of the three-spined toadfish, Batrachomoeus trispinosus: insights into the diversity of the Batrachoididae. J Exp Biol 212:1377–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, McCarthy D, Smyth G. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:138–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer AS, Parsons TS. 1970. Vertebrate body. 4th ed. Philadelphia: Saunders. [Google Scholar]
- Sakiyama J, Yamagishi A, Kuroiwa A. 2003. Tbx4-Fgf10 system controls lung bud formation during chicken embryonic development. Development 130:1225–1234. [DOI] [PubMed] [Google Scholar]
- Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, Yagishita N, Matsui D, Koga Y, Itoh N, Kato S. 1999. Fgf10 is essential for limb and lung formation. Nat Genet 21:138–141. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumi N, Kobayashi R, Yano T, Noda M, Fujimura K, Okada N, Okabe M. 2016. Molecular developmental mechanism in polypterid fish provides insight into the origin of vertebrate lungs. Sci Rep 6:30580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallach V 1998. The Pulmonary System: The Lungs of Snakes. In: Biology of the Reptilia: Morphology G. Visceral Organs. Vol. 19. Ithaca, New York: Society for the Study of Amphibians and Reptiles. p 93–295. [Google Scholar]
- Wang Z, Shu W, Lu MM, Morrisey EE. 2005. Wnt7b Activates Canonical Signaling in Epithelial and Vascular Smooth Muscle Cells through Interactions with Fzd1, Fzd10, and LRP5. Mol Cell Biol 25:5022–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M, Dunn NR, Hogan BL. 2000. Bmp4 and Fgf10 play opposing roles during lung bud morphogenesis. Development 127:2695–2704. [DOI] [PubMed] [Google Scholar]
- Weidenfeld J, Shu W, Zhang L, Millar SE, Morrisey EE. 2002. The WNT7b Promoter Is Regulated by TTF-1, GATA6, and Foxa2 in Lung Epithelium. J Biol Chem 277:21061–21070. [DOI] [PubMed] [Google Scholar]
- Wilder BG. 1877. Gar-pikes, old and young. Pop Sci Mon:1–12. [Google Scholar]
- [dataset]Funk EC, Lencer ES, McCune AR; 2020; Dorso-ventral inversion in the air-filled organ (lungs, gas bladder) of vertebrates: RNA-sequencing of laser capture microdissected embryonic tissue; NCBI Gene Expression Omnibus; Accession #GSE152992; https://www.ncbi.nim.nih.gov/geo/query/acc.cgi?acc=GSE152992 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Bmp4 and Bmp16 expression during gasbladder development in bowfin. A) Boxplot of average Bmp4 expression at pre-budding (24), budding (25), and outgrowth (27) stages of gas bladder development. Bmp4 is expressed at all stages. During outgrowth, Bmp4 is expressed strongest dorsally, in both endoderm and mesoderm. B) Boxplot of average Bmp16 expression at all stages. No or weak expression of Bmp16 is detected at all stages in all tissue regions. Abbreviations: dE, dorsal endoderm; dM, dorsal mesoderm; vE, ventral endoderm; vM, ventral mesoderm.


