Abstract
Rationale: The computed tomography (CT) pattern of definite or probable usual interstitial pneumonia (UIP) can be diagnostic of idiopathic pulmonary fibrosis and may obviate the need for invasive surgical biopsy. Few machine-learning studies have investigated the classification of interstitial lung disease (ILD) on CT imaging, but none have used histopathology as a reference standard.
Objectives: To predict histopathologic UIP using deep learning of high-resolution computed tomography (HRCT).
Methods: Institutional databases were retrospectively searched for consecutive patients with ILD, HRCT, and diagnostic histopathology from 2011 to 2014 (training cohort) and from 2016 to 2017 (testing cohort). A blinded expert radiologist and pulmonologist reviewed all training HRCT scans in consensus and classified HRCT scans based on the 2018 American Thoracic Society/European Respriatory Society/Japanese Respiratory Society/Latin American Thoracic Association diagnostic criteria for idiopathic pulmonary fibrosis. A convolutional neural network (CNN) was built accepting 4 × 4 × 2 cm virtual wedges of peripheral lung on HRCT as input and outputting the UIP histopathologic pattern. The CNN was trained and evaluated on the training cohort using fivefold cross validation and was then tested on the hold-out testing cohort. CNN and human performance were compared in the training cohort. Logistic regression and survival analyses were performed.
Results: The CNN was trained on 221 patients (median age 60 yr; interquartile range [IQR], 53–66), including 71 patients (32%) with UIP or probable UIP histopathologic patterns. The CNN was tested on a separate hold-out cohort of 80 patients (median age 66 yr; IQR, 58–69), including 22 patients (27%) with UIP or probable UIP histopathologic patterns. An average of 516 wedges were generated per patient. The percentage of wedges with CNN-predicted UIP yielded a cross validation area under the curve of 74% for histopathological UIP pattern per patient. The optimal cutoff point for classifying patients on the training cohort was 16.5% of virtual lung wedges with CNN-predicted UIP and resulted in sensitivity and specificity of 74% and 58%, respectively, in the testing cohort. CNN-predicted UIP was associated with an increased risk of death or lung transplantation during cross validation (hazard ratio, 1.5; 95% confidence interval, 1.1–2.2; P = 0.03).
Conclusions: Virtual lung wedge resection in patients with ILD can be used as an input to a CNN for predicting the histopathologic UIP pattern and transplant-free survival.
Keywords: lung diseases, interstitial, idiopathic pulmonary fibrosis, deep learning
Interstitial lung disease (ILD) is a heterogenous group of fibrotic lung diseases with various etiologies, natural histories, and prognoses (1, 2). Idiopathic pulmonary fibrosis (IPF) is a progressive fibrotic ILD with a high mortality rate and few available therapies (1, 3–8). Increasing attention has been given to the accuracy of diagnosing IPF. The 2018 Fleischner Society white paper update for the diagnostic criteria for IPF offers guidance regarding the current role of surgical biopsy, high-resolution computed tomography (HRCT) of the chest, and clinical context in making the diagnosis of IPF (9). In the correct clinical context, a computed tomography (CT) pattern of definite or probable usual interstitial pneumonia (UIP) is diagnostic of IPF and may obviate the need for invasive surgical biopsy, which carries a significant risk of morbidity and mortality (10).
Advancements in computer vision and machine learning have led to new opportunities in the diagnosis and prognostication of various diseases (11). Deep learning using convolutional neural networks (CNNs) is a relatively new branch of machine learning that excels in classifying images (12). Few prior studies have attempted to classify ILD patterns using machine learning of CT scans (13–16). Previous studies are limited by small sample sizes, older machine-learning techniques, and/or lack of a histopathologic reference standard. The purpose of this study was to build a deep-learning network capable of predicting definite or probable UIP on HRCT scans using histopathology as the reference standard and to explore the association of the model with mortality.
Methods
This was a retrospective institutional review board–approved study with waiver of informed consent. An institutional radiology database was searched from January 2010 to December 2014 for all CT scans of the chest in patients with ILD using the search terms “fibrosis” or “interstitial” or “interstitial lung disease” or “fibrotic lung disease” or “UIP” or “usual interstitial pneumonia.” The search returned 7,005 CT scans. All CT scan reports and respective patient charts were reviewed to evaluate for a diagnosis of ILD and for history of surgical biopsy and/or lung transplant with explant yielding a histopathologic diagnosis. In total, 944 patients had a confirmed diagnosis of ILD. Of these, 676 were excluded because of a lack of histopathology, yielding 268 patients. Of these 268 patients, 14 were excluded because of an indeterminate biopsy, one with fibrosis due to ciliary dyskinesia, one with fibrosis attributed to cardiac disease, three because of no evidence of ILD on biopsy, and 28 because of poor CT quality. This resulted in 221 patients with diagnostic ILD protocol CT scans of the chest and diagnostic histopathology to be used as the training cohort in this study.
Subsequently, a search of a pathology database at the same institution was used to identify all patients who had a lung biopsy performed in 2016–2017 with the term “fibrosis” in the pathology report. This search yielded 386 biopsies. These reports were reviewed to include only those with surgical lung biopsies in the setting of ILD, resulting in 80 patients. All 80 patients had a diagnostic ILD protocol CT scan of the chest. These patients were held out and used as the final testing cohort. None of these patients were part of the training cohort. Figure 1 depicts a flowchart for the inclusion of the study patients.
Figure 1.

Flow chart for selection of patients in the training and testing cohorts. CT = computed tomography; ILD = interstitial lung disease.
CT Technique
CT images were acquired on one of three following GE scanners (GE Healthcare): Lightspeed VCT 64, Discovery CT750HD, or Revolution scanner. Scans were performed during end-inspiration phase using the breath-hold technique with patients in the supine position, from the apex of the lung to the costodiagphragmatic recesses. Tube voltage was 120 kvp, and milliampere-seconds (mAs) was modulated. Detector coverage was 64 × 0.625 mm, pitch speed was 1.375, and rotation time was 0.5 seconds. Slice thickness varied from 0.625 to 5.0 mm. Images were reconstructed using a high–spatial frequency lung kernel.
Image Analysis
One expert thoracic radiologist with 20 years of experience in ILD and one expert pulmonologist with 13 years of experience in ILD (blinded to pathology and the underlying diagnosis) reviewed all CT scans of the training cohort in consensus and classified patients into one of four categories as per the 2018 ATS/ERS/JRS/ALAT IPF, “typical UIP CT pattern,” “probable UIP CT pattern,” “CT pattern indeterminate for UIP” and “CT features most consistent with non-IPF diagnosis” (9). For purposes of comparing reader performance with the CNN performance “typical UIP CT pattern” and “probable UIP CT pattern” were grouped together as UIP and “CT pattern indeterminate for UIP” and “CT features most consistent with non-IPF diagnosis” were grouped together as not UIP.
Lung Segmentation and Deep Learning
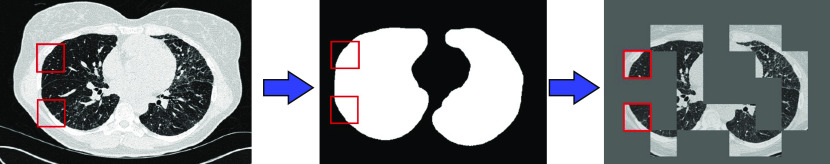
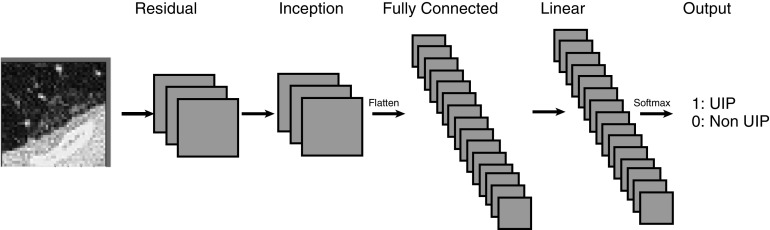
Each patient’s lung volumes on CT were automatically segmented. Hundreds of 4 × 4 × 2 cm peripheral wedges were created for each patient (Figure 2). A CNN was built (Figure 3) which accepted individual virtual wedges and output a binary result of UIP or not UIP using that patient’s surgical biopsy as reference standard. The CNN was trained and evaluated on the 221 patients in the training data set using fivefold cross validation, with subject levels splits of 44 patients per group with one group of 45 patients, and subsequently tested on the hold-out set of 80 patients from the testing set. The details of the segmentation process, CNN architecture and training process are explained in Appendix 1.
Figure 2.
Inputs to the network were generated by obtaining a 4 × 4 × 2 cm three-dimensional wedge of the subpleural regions of the bottom half of the lungs for each patient. Left: an example input slice with two example virtual wedge regions highlighted. Wedges were discarded if they contained no pleural space or no lung as defined by an automatically generated lung mask (middle). On average, 516 wedges were generated for each patient, totaling more than 114,000 wedges. Right: 18 example wedges from a patient. The red squares are sample wedges.
Figure 3.
Convolutional neural network architecture used. A custom 20-layer three-dimensional convolutional neural network was designed with random initializations. UIP = usual interstitial pneumonia.
Reference Standard
The original diagnostic histopathology reports from surgical resection, pneumonectomy or autopsy for each patient were reviewed by a pathologist and pulmonologist with experience in ILD and classified using the 2018 American Thoracic Society/European Respriatory Society/Japanese Respiratory Society/Latin American Thoracic Association (ATS/ERS/JRS/ALAT) IPF diagnosis statement, which describes the following four categories: UIP, probable UIP, indeterminate for UIP, and alternative diagnosis (9, 17). UIP and probable UIP were grouped together as UIP, and indeterminate for UIP and alternative diagnosis were grouped together as not UIP.
Statistics
The median percentage of CNN-predicted UIP wedges across patients in the training and testing cohorts with and without pathology-proven UIP was compared using the Wilcoxon rank-sum test. We built a univariable logistic regression model to assess the association between the percentage of CNN-predicted wedges of UIP per patient in the training cohort with pathology-proven UIP; the same logistic regression model was used to generate a receiver operating characteristic curve and compute the Youden Index needed to find the optimal binary cutoff of the percentage of CNN-predicted wedges for UIP in classifying patients as having UIP or not. The association of this binary per patient CNN-predicted UIP variable and pathology-proven UIP was then explored before and after controlling for age, sex, and human-predicted UIP using univariable and multivariable logistic regression models. In addition, the association of human CT–predicted UIP with pathology-proven UIP using univariable logistic regression model was explored. The Hosmer-Lemeshow test was used to assess the goodness of fit. We created a calibration plot for the multivariate model to demonstrate the goodness of fit (18). Cohen’s κ was used to evaluate the agreement of CNN-predicted UIP with histopathology-proven UIP as well as the agreement of human-predicted UIP with histopathology-proven UIP.
We also evaluated the predictive validity of the per-patient CNN-predicted UIP as well as the percentage of CNN-predicted UIP wedges against the time to death or lung transplantation (transplant-free survival). Kaplan-Meier curves of transplant-free survival were generated and compared using a log-rank test. We used Cox regression analysis to assess the associations of human- and CNN-predicted UIP with transplant-free survival.
We tested the cutoff of the percentage of CNN-predicted wedges for UIP derived from cross validation of the training cohort in the testing cohort. Figure 4 demonstrates a flow chart starting with the virtual wedge generation and ending in the per-patient classification.
Figure 4.
Flow chart depicting process from virtual wedge creation to per patient prediction of usual interstitial pneumonia. CNN = convolutional neural network.
Results
Patient Characteristics
A total of 221 patients in the training cohort (115 male and 106 female patients; median age 60 yr; interquartile range [IQR], 53–66 yr) had the following multidisciplinary team discussions (MDDs): 52 patients with IPF, 41 with hypersensitivity pneumonitis, 30 with unclassifiable ILD, 11 with sarcoidosis, 38 with connective tissue disease–related ILD, 33 with idiopathic nonspecific interstitial pneumonia, four with idiopathic desquamative interstitial pneumonia, one with smoking-related interstitial fibrosis, one with amyloidosis, two with cryptogenic organizing pneumonia, one with primary biliary cirrhosis–related ILD, one with respiratory bronchiolitis–associated ILD, one with Hermnasky-Pudlak Syndrome–related ILD, three with antineutrophil cytoplasmic antibodies vasculitis, one with pulmonary alveolar proteinosis, and one with bronchiolitis obliterans.
There were 57 patients (26%) with UIP, 14 patients (6%) with probable UIP, and 150 patients (68%) with alternative diagnosis pathology patterns on histopathology.
Reference standard histopathology was derived from surgical biopsy in 177 patients (80%), lung transplant and pneumonectomy/explant in 42 patients (19%), and autopsy in two patients (1%). During a median follow-up of 37 months (IQR, 10–77), 98 patients (44%) underwent lung transplantation and 29 patients (13%) died without undergoing lung transplantation.
Eighty patients in the testing cohort (45 male and 35 female patients; median age 66 yr; IQR, 58–69 yr) had the following MDDs: 17 patients with IPF, 40 with hypersensitivity pneumonitis, seven with unclassifiable ILD, one with sarcoidosis, four with connective tissue disease–related ILD, nine with idiopathic nonspecific interstitial pneumonia, one with smoking-related interstitial fibrosis, and one with ANCA vasculitis.
There were 13 patients (16%) with UIP, nine patients (11%) with probable UIP, nine patients (11%) with indeterminate for UIP, and 49 patients (61%) with alternative diagnosis pathology patterns on histopathology.
Reference standard histopathology was derived from surgical biopsy in all 80 patients (100%). During a median follow-up of 27 months (IQR, 13–36), eight patients (10%) underwent lung transplantation and six patients (8%) died without undergoing lung transplantation. Patient characteristics of both the training and testing cohorts are summarized in Table 1.
Table 1.
Patient characteristics
| Training Cohort | Testing Cohort | P Value | |
|---|---|---|---|
| Patients, n | 221 | 80 | — |
| Age, yr, median (IQR) | 60 (53–66) | 66 (58–69) | <0.001 |
| Sex, M:F | 115:106 | 45:35 | 0.42 |
| Pathologic UIP*, % | 71 (32) | 21 (26) | 0.44 |
| Transplants, n (%) | 98 (44) | 8 (10) | <0.001 |
| Deaths, n (%) | 29 (13) | 6 (8) | 0.18 |
Definition of abbreviations: IQR = interquartile range; UIP = usual interstitial pneumonia.
UIP or probable UIP.
UIP Prediction
The median percentage of CNN-predicted UIP virtual lung wedges was significantly different across patients in both the training and testing cohorts with and without pathology-proven UIP (Wilcoxon rank-sum test P <0.001), as shown in Figure 5.
Figure 5.
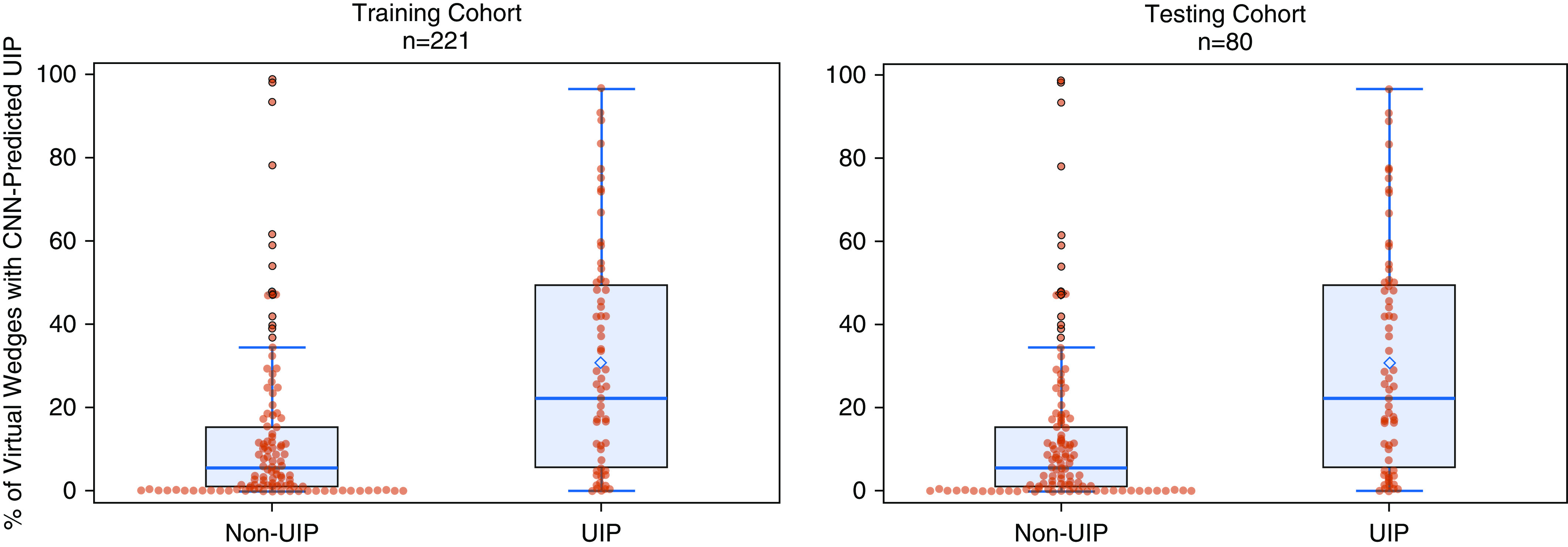
Difference in percentage convolutional neural network–predicted usual interstitial pneumonia wedges across patients in the training and testing cohorts with and without a histopathological diagnosis of usual interstitial pneumonia. CNN = convolutional neural network; UIP = usual interstitial pneumonia.
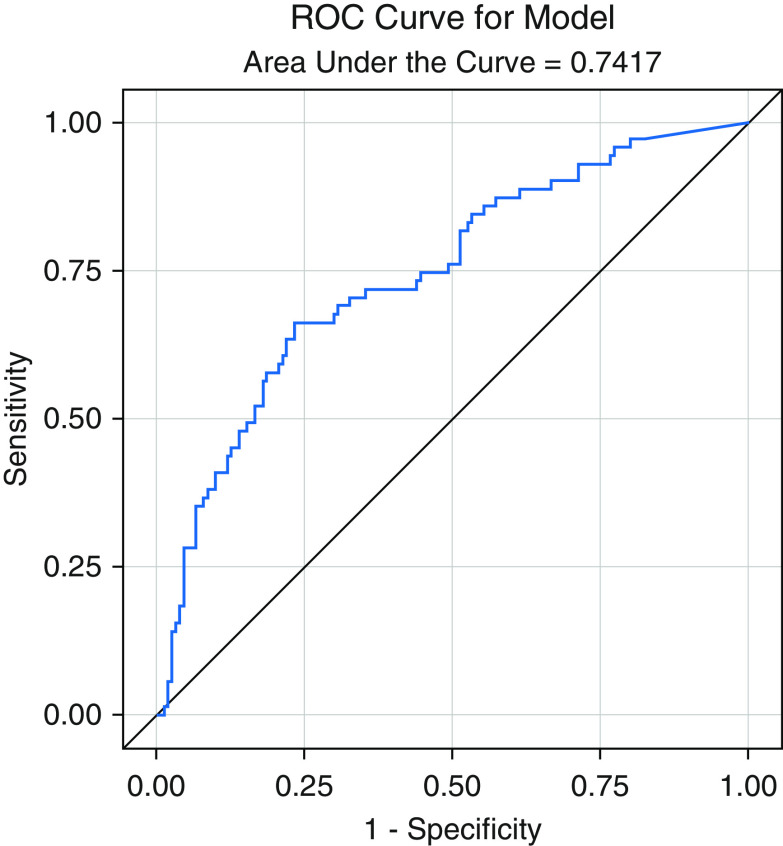
Univariable logistic regression analysis showed a significant association of the percentage of CNN-predicted UIP wedges per patient with pathology-proven UIP (odds ratio [OR], 6.43; 95% confidence interval [CI], 3.46–12.00; P <0.001). The area under the curve (AUC) of this logistic regression model was 0.74 (Figure 6) with sensitivity of 77%, specificity of 66%, and a maximal Youden Index of 0.43, which was ultimately used to compute the optimal cutoff of 16.5% of CNN-positive wedges to dichotomize the training cohort into per-patient CNN prediction of UIP. Multivariable logistic regression analysis showed that the association between CNN-predicted UIP and pathology-proven UIP was independent of other factors, including age, sex, and the human CT–predicted UIP (OR, 4.4; 95% CI, 2.3–8.6; P <0.001). These results are summarized in Table 2.
Figure 6.
Receiver operating characteristic curve from a univariable logistic regression model of convolutional neural network–predicted usual interstitial pneumonia wedges with histopathology on cross validation results from the training cohort. ROC = receiver operating characteristic.
Table 2.
Univariable and multivariable regression analysis in the training cohort for predicting histopathologic UIP
| Univariable Model |
Multivariable Model |
|||||
|---|---|---|---|---|---|---|
| OR | 95% CI | P Value | OR | 95% CI | P Value | |
| Age | 1.054 | 1.022–1.087 | <0.001 | 1.038 | 1.003–1.073 | 0.03 |
| Sex | 0.601 | 0.339–1.067 | 0.08 | 0.874 | 0.446–1.714 | 0.70 |
| Human UIP | 6.685 | 3.471–12.874 | <0.001 | 3.847 | 1.868–7.921 | <0.001 |
| CNN UIP | 6.434 | 3.460–11.967 | <0.001 | 4.422 | 2.266–8.628 | <0.0001 |
Definition of abbreviations: CI = confidence interval; CNN = convolutional neural network; OR = odds ratio; UIP = usual interstitial pneumonia.
The Hosmer-Lemeshow test results were not significant (P = 0.56), indicating goodness of fit. The test results as well as a calibration plot are depicted in Appendix 2.
Both the human CT– and CNN-predicted UIP showed moderate agreement with pathology-proven UIP in the training and testing cohorts, with κ of 0.40 and 0.41, respectively (Tables 3 and 4).
Table 3.
Agreement between human-predicted UIP on CT scan with histopathology in the training cohort
| Training Pathology Dx of UIP | Human CT–predicted UIP |
||
|---|---|---|---|
| No [n (%)] | Yes [n (%)] | Total [n (%)] | |
| No | 129 (58.37) | 21 (9.50) | 150 (67.87) |
| Yes | 34 (15.38) | 37 (16.74) | 71 (32.13) |
| Total | 163 (73.76) | 58 (26.24) | 221 (100.00) |
Definition of abbreviations: CT = computed tomography; Dx = diagnosis; UIP = usual interstitial pneumonia.
κ = 0.40 (0.27–0.53).
Table 4.
Agreement between CNN-predicted UIP on CT scan with histopathology in the training cohort
| Training Pathology Dx of UIP | CNN-predicted UIP |
||
|---|---|---|---|
| No [n (%)] | Yes [n (%)] | Total [n (%)] | |
| No | 115 (52.04) | 35 (15.84) | 150 (67.87) |
| Yes | 24 (10.86) | 47 (21.27) | 71 (32.13) |
| Total | 139 (62.90) | 82 (37.10) | 221 (100.100) |
Definition of abbreviations: CNN = convolutional neural network; CT = computed tomography; Dx = diagnosis; UIP = usual interstitial pneumonia.
κ = 0.41 (0.29–0.54).
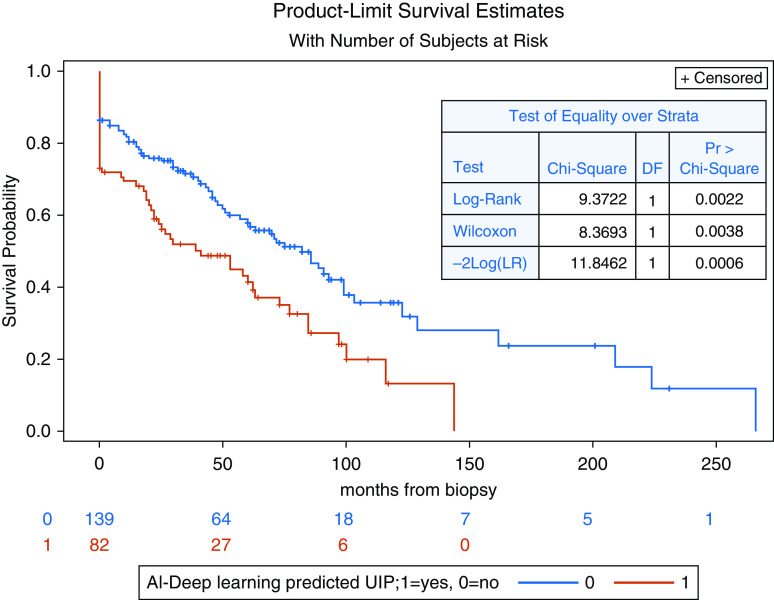
Multivariable Cox regression analysis from cross validation of the training cohort showed that CNN prediction of UIP was significantly associated with transplant-free survival from the time of pathology while controlling for age, sex, and human CT prediction of UIP (hazard ratio, 1.5; 95% CI, 1.1–2.2; P = 0.03). Human CT–predicted UIP was not significantly associated with transplant-free survival (P = 0.45). The Kaplan-Meier transplant-free survival curves for CNN-predicted UIP per patient and for the percentage of CNN-predicted UIP wedges by tertiles are shown in Figure 7 and Appendix 2, respectively.
Figure 7.
Kaplan-Meier transplant-free survival curves for convolutional neural network–predicted usual interstitial pneumonia per patient on cross validation results from the training cohort. AI = artificial intelligence; DF = degrees of freedom; LR = likelihood ratio; Pr = associated P value; UIP = usual interstitial pneumonia.
Applying the same training cohort cutoff of 16.5% of CNN-positive wedges to predict UIP per patient achieved an accuracy of 68%, sensitivity of 74%, and specificity of 58% in the testing cohort. CNN-predicted UIP showed moderate agreement with pathology-proven UIP in the testing cohort, with a κ of 0.39 (Table 5). Multivariable Cox regression analysis failed to show that either CNN prediction of UIP or histopathology of UIP were associated with transplant-free survival (P = 0.88 and 0.32, respectively). Figure 8 depicts four sample cases from the testing cohort.
Table 5.
Agreement between CNN-predicted UIP on CT scan with histopathology in the testing cohort
| Testing Pathology Dx of UIP | CNN-predicted UIP |
||
|---|---|---|---|
| No [n (%)] | Yes [n (%)] | Total [n (%)] | |
| No | 35 (44.87) | 21 (26.92) | 56 (71.79) |
| Yes | 4 (5.13) | 18 (23.08) | 22 (28.21) |
| Total | 39 (50.00) | 39 (50.00) | 78 (100.00) |
Definition of abbreviations: CNN = convolutional neural network; CT = computed tomography; Dx = diagnosis; UIP = usual interstitial pneumonia.
κ = 0.36 (0.17–0.55).
Figure 8.
Example network predictions with overlaid predictions from the virtual wedges evaluated by the network on the testing cohort. Top left: an example in which the network disagreed with histopathologic diagnosis of usual interstitial pneumonia (UIP). Top right: an example in which the network disagreed with histopathologic diagnosis of not UIP. Bottom left: an example in which the network agreed with histopathologic diagnosis of UIP. Bottom right: an example in which the network agreed with histopathologic diagnosis of not UIP.
Discussion
The results of this proof-of-concept study show that a deep-learning network can perform moderately well in predicting a patient’s histopathologic UIP pattern using HRCT images. Based on cross validation results, we suggest that this prediction is also associated with transplant-free survival. The potential applications of this include its use as an adjunctive tool during the initial work-up of a patient with ILD to be used in combination with expert radiologist interpretation or in lieu of it when not available, thereby helping the pulmonologist decide whether a surgical biopsy is necessary.
The role of surgical lung resection in the diagnosis of IPF has evolved over time. This evolution is partly due to the understanding that when a CT scan shows probable or definite UIP and the clinical scenario is consistent, a diagnosis of IPF can be made without tissue diagnosis (9). In cases with radiological and/or clinical uncertainty, wedge resection is recommended. Despite this recommendation, there is the acknowledgment that histology alone is not a perfect reference standard and must still be integrated into the MDD, which has been shown to increase the interobserver agreement and diagnostic confidence and to change the clinical diagnosis in 20–50% of patients (19, 20). Biopsies should be taken from different lobes and target abnormal but not end-stage lung. In addition, wedges should measure 2–3 cm wide and 1–2 cm deep from the pleura (9, 21). We attempted to simulate this process through virtual wedge resections of patients’ lungs on CT scans as the input to the CNN.
There has been prior work in machine learning of CT scans for ILD classification. Depeursinge and colleagues evaluated 33 patients with classic versus atypical UIP (13). The authors used Riesz filterbanks to characterize three-dimensional segments of the lung parenchyma through a support vector machine model and cross validation–supervised machine learning. The reference standard was two expert radiologists. They report an AUC of 0.81 for classifying classic UIP. Limitations include a very small sample size, use of the “leave one out” cross validation technique, which is prone to overfitting, lack of a histologic reference standard, and the use of an older form of machine learning.
Christe and colleagues built a computer-aided diagnosis system for classifying CT images of patients with ILD into the four CT UIP patterns. Two external databases of CT scans were used to train a CNN to classify two-dimensional regions of parenchyma into different morphologic patterns (normal, ground glass, honeycombing, etc.) Together with limited clinical data, the CNN output was fed into a random forest classifier to classify the CT pattern into a CT UIP pattern or not. The model was validated on 105 internal cases of ILD, including 51 cases of UIP and 54 cases of NSIP, as determined at a multidisciplinary ILD conference. The ground truth of the CT pattern for these 105 patients was consensus reads between two expert radiologists. The network’s performance was compared with two additional radiologists. Accuracies for the network, reader 1 and reader 2 in classifying “typical” or “probable” UIP versus the other 2 CT patterns were 0.81, 0.81, and 0.70. The F scores were similar at 0.80 (14). The major strength of this work is the separate training and testing cohorts. Nevertheless, there are weaknesses, such as using the CNN to classify morphologic patterns of the lungs rather than directly classify CT UIP patterns, using two-dimensional image input rather than three-dimensional input, using radiologist interpretation as the ground truth rather than histopathology, including only two types of ILD in the testing cohort, and finally, reporting accuracies and F scores without receiver operating characteristic AUCs and sensitivity/specificities.
Walsh and colleagues (16) published the largest machine-learning study on UIP diagnosis. The authors conducted a case-cohort study using 1,157 HRCT scans of patients with ILD. They trained, tested, and validated a CNN capable of predicting UIP, using expert radiologists’ consensus reads as the reference standard. Survival analysis showed that both human- and CNN-predicted UIP were similarly significant in predicting overall survival. Despite the large size and testing set, it is noteworthy that the reference standard was human consensus and not histopathology. Therefore, it is likely that Walsh and colleagues included many more straightforward cases of ILD given that surgical biopsy is more often obtained when there is discordance between different factors, including clinical history, serology, and HRCT. In addition, there is no mention of the start time for the overall survival analysis nor the handling of lung transplants, if relevant.
Our work builds on previous work through the following novelties and strengths. First, to our knowledge, this is the first study that has used histopathology as the reference standard rather than radiologist interpretation. The consequence of this inclusion criteria is the bias toward less straightforward HRCT scans, given the need for biopsy. Our approach of using peripheral three-dimensional wedges of the lung is meant to simulate the surgical biopsy itself and can be termed a “virtual surgical biopsy.” This allows for a three-dimensional map to be constructed that can be used for guiding the biopsy if one is deemed necessary. In addition, our cohort included a wide range of HRCT ILD patterns and MDD diagnoses, which was greater than those of the previously mentioned works. Finally, we conducted multivariable logistic regression analysis controlling for age, sex, and human prediction of UIP.
There are limitations to our study. We used data from a single institution for both training and testing, which limits generalizability. We did not perform direct radiologic-pathologic correlation because the surgically resected wedge was not identified on individual patient’s scans; in some cases, the CT scans used by the network were post resection. In fact, some patients’ resections were performed at an outside institution years before the in-house CT. This raises the issue of the interval between surgical resection and CT. To include as many patients as possible, we did not exclude patients with long intervals. To reproduce the surgical biopsy, we focused on virtual wedge resections. The added benefit of this approach was many wedges per patient, which resulted in a large overall number of wedges, a necessity when dealing with deep learning. On the other hand, a more intuitive approach would have been to use the entire three-dimensional lung volumes as a single input to a network. This would have allowed the network access to more data, including zonal distribution, airway-centric disease and other characteristics that undoubtably are diagnostically important. Unfortunately, this approach would have required a much larger number of patients. Although we used the histopathology report as the reference standard, there is the major caveat that histology is not the gold standard for diagnosing IPF, hence the importance of the MDD. In this regard, it is important to reiterate that the aim of our study was not to predict or replace the MDD. The next logical step will be to evaluate the utility of this tool in the setting of the MDD itself. There was heterogeneity in obtaining the histopathology in the training cohort, with 20% of patients in the training cohort having the pathologic diagnosis made through pneumonectomy during lung transplant or autopsy. On this note, it is important to stress that patients with histopathology derived from lung transplant pneumonectomy and autopsy did not contribute to the survival curves, as their follow-up time was 0. Although the survival analysis in the training cohort was significant, we could not reproduce these results with the testing cohort. The fact that neither CNN-predicted UIP nor histopathologic UIP were associated with transplant survival in the testing cohort suggests that the small sample size may be a limiting factor.
We used the 2018 ATS/ERS/JRS/ALAT diagnostic criteria for IPF rather than the Fleischner Society guidelines. The major difference between the two criteria is in the management of these patients and whether to biopsy or not rather than the radiologic and pathologic criteria. The diagnostic criteria were applied retrospectively by review of the original pathology reports. A more rigorous approach would have included blindly reexamining the original pathology slides themselves. There were significant differences between the training and testing cohorts. First and foremost, the histopathology was interpreted by different pathologists, which may explain the different distribution of the UIP categories. For example, there were no retrospective assignments of the indeterminate category in the training cohort. In addition, a larger percentage of patients in the training cohort underwent lung transplantation. This may be due to underlying changes for which patients proceed to biopsy over the years. Finally, our results are likely not adequate to be used in clinical practice and serve more as a proof of concept. On this note, it is important to reiterate that patients who undergo a surgical biopsy likely have a more ambiguous CT scan and/or clinical presentation, which is why the biopsy was necessary to begin with. Therefore, these cases are a challenging subset of all patients with ILD.
In conclusion, virtual wedge lung resection of HRCTs in patients with ILD can be used as input to a deep-learning model for predicting the probability of histologic UIP pattern with moderate accuracy, comparable to that of human prediction. This prediction may be associated with transplant-free survival. Additional research is needed to evaluate the utility of this approach in the setting of the MDD.
Acknowledgments
Acknowledgment
The authors thank Golan Pundak.
Footnotes
Author Contributions: H.S. was the principle investigator involved in all aspects. F.S.A. conducted the statistical analysis and contributed to the editing of the manuscript. D.L. and B.D’S. participated in data collection and writing of the manuscript. P.A. and S.H. collected the data. M.S. and A.S. analyzed the images and pathology reports. S.J. and S.M. conducted the machine learning.
This article has a data supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Travis WD, Costabel U, Hansell DM, King TE, Jr, Lynch DA, Nicholson AG, et al. ATS/ERS Committee on Idiopathic Interstitial Pneumonias. An official American Thoracic Society/European Respiratory Society statement: update of the international multidisciplinary classification of the idiopathic interstitial pneumonias. Am J Respir Crit Care Med. 2013;188:733–748. doi: 10.1164/rccm.201308-1483ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lederer DJ, Martinez FJ. Idiopathic pulmonary fibrosis. N Engl J Med. 2018;378:1811–1823. doi: 10.1056/NEJMra1705751. [DOI] [PubMed] [Google Scholar]
- 3.Alhamad EH, Al-Kassimi FA, Alboukai AA, Raddaoui E, Al-Hajjaj MS, Hajjar W, et al. Comparison of three groups of patients with usual interstitial pneumonia. Respir Med. 2012;106:1575–1585. doi: 10.1016/j.rmed.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 4.King TE, Jr, Bradford WZ, Castro-Bernardini S, Fagan EA, Glaspole I, Glassberg MK, et al. ASCEND Study Group. A phase 3 trial of pirfenidone in patients with idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2083–2092. doi: 10.1056/NEJMoa1402582. [DOI] [PubMed] [Google Scholar]
- 5.Noble PW, Albera C, Bradford WZ, Costabel U, du Bois RM, Fagan EA, et al. Pirfenidone for idiopathic pulmonary fibrosis: analysis of pooled data from three multinational phase 3 trials. Eur Respir J. 2016;47:243–253. doi: 10.1183/13993003.00026-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noble PW, Albera C, Bradford WZ, Costabel U, Glassberg MK, Kardatzke D, et al. CAPACITY Study Group. Pirfenidone in patients with idiopathic pulmonary fibrosis (CAPACITY): two randomised trials. Lancet. 2011;377:1760–1769. doi: 10.1016/S0140-6736(11)60405-4. [DOI] [PubMed] [Google Scholar]
- 7.Richeldi L, Costabel U, Selman M, Kim DS, Hansell DM, Nicholson AG, et al. Efficacy of a tyrosine kinase inhibitor in idiopathic pulmonary fibrosis. N Engl J Med. 2011;365:1079–1087. doi: 10.1056/NEJMoa1103690. [DOI] [PubMed] [Google Scholar]
- 8.Richeldi L, du Bois RM, Raghu G, Azuma A, Brown KK, Costabel U, et al. INPULSIS Trial Investigators. Efficacy and safety of nintedanib in idiopathic pulmonary fibrosis. N Engl J Med. 2014;370:2071–2082. doi: 10.1056/NEJMoa1402584. [DOI] [PubMed] [Google Scholar]
- 9.Lynch DA, Sverzellati N, Travis WD, Brown KK, Colby TV, Galvin JR, et al. Diagnostic criteria for idiopathic pulmonary fibrosis: a Fleischner society white paper. Lancet Respir Med. 2018;6:138–153. doi: 10.1016/S2213-2600(17)30433-2. [DOI] [PubMed] [Google Scholar]
- 10.Hutchinson JP, McKeever TM, Fogarty AW, Navaratnam V, Hubbard RB. Surgical lung biopsy for the diagnosis of interstitial lung disease in England: 1997-2008. Eur Respir J. 2016;48:1453–1461. doi: 10.1183/13993003.00378-2016. [DOI] [PubMed] [Google Scholar]
- 11.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saba L, Biswas M, Kuppili V, Cuadrado Godia E, Suri HS, Edla DR, et al. The present and future of deep learning in radiology. Eur J Radiol. 2019;114:14–24. doi: 10.1016/j.ejrad.2019.02.038. [DOI] [PubMed] [Google Scholar]
- 13.Depeursinge A, Chin AS, Leung AN, Terrone D, Bristow M, Rosen G, et al. Automated classification of usual interstitial pneumonia using regional volumetric texture analysis in high-resolution computed tomography. Invest Radiol. 2015;50:261–267. doi: 10.1097/RLI.0000000000000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christe A, Peters AA, Drakopoulos D, Heverhagen JT, Geiser T, Stathopoulou T, et al. Computer-aided diagnosis of pulmonary fibrosis using deep learning and CT images. Invest Radiol. 2019;54:627–632. doi: 10.1097/RLI.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milanese G, Mannil M, Martini K, Maurer B, Alkadhi H, Frauenfelder T. Quantitative CT texture analysis for diagnosing systemic sclerosis: effect of iterative reconstructions and radiation doses. Medicine (Baltimore) 2019;98:e16423. doi: 10.1097/MD.0000000000016423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh SLF, Calandriello L, Silva M, Sverzellati N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med. 2018;6:837–845. doi: 10.1016/S2213-2600(18)30286-8. [DOI] [PubMed] [Google Scholar]
- 17.Raghu G, Collard HR, Egan JJ, Martinez FJ, Behr J, Brown KK, et al. ATS/ERS/JRS/ALAT Committee on Idiopathic Pulmonary Fibrosis. An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am J Respir Crit Care Med. 2011;183:788–824. doi: 10.1164/rccm.2009-040GL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park SH, Han K. Methodologic guide for evaluating clinical performance and effect of artificial intelligence technology for medical diagnosis and prediction. Radiology. 2018;286:800–809. doi: 10.1148/radiol.2017171920. [DOI] [PubMed] [Google Scholar]
- 19.Flaherty KR, King TE, Jr, Raghu G, Lynch JP, III, Colby TV, Travis WD, et al. Idiopathic interstitial pneumonia: what is the effect of a multidisciplinary approach to diagnosis? Am J Respir Crit Care Med. 2004;170:904–910. doi: 10.1164/rccm.200402-147OC. [DOI] [PubMed] [Google Scholar]
- 20.Jo HE, Glaspole IN, Levin KC, McCormack SR, Mahar AM, Cooper WA, et al. Clinical impact of the interstitial lung disease multidisciplinary service. Respirology. 2016;21:1438–1444. doi: 10.1111/resp.12850. [DOI] [PubMed] [Google Scholar]
- 21.Monaghan H, Wells AU, Colby TV, du Bois RM, Hansell DM, Nicholson AG. Prognostic implications of histologic patterns in multiple surgical lung biopsies from patients with idiopathic interstitial pneumonias. Chest. 2004;125:522–526. doi: 10.1378/chest.125.2.522. [DOI] [PubMed] [Google Scholar]