Abstract
Background
Antimicrobial resistance (AMR) poses a complex threat to global health security and universal health coverage. Recently, nosocomial infections with carbapenemase-producing Gram-negative bacilli (GNB) is increasing worldwide. We report the molecular characterization and detection of genes associated with carbapenemase producing Gram negative bacteria isolated from hospitalized patients at Soba University Hospital (SUH) in Khartoum State, Sudan.
Results
Between October 2016 and February 2017, a total of 206 GNB clinical specimens were collected from hospitalized patients in SUH. Of 206 carbapenem resistance isolates, 171 (83 %) were confirmed as phenotypically resistant and 121 (58.7 %) isolates harboured one or more carbapenemase genes. New Delhi metallo-β-lactamase (NDM) types were the most predominant genes, blaNDM 107(52 %), followed by blaIMP 7 (3.4 %), blaOXA-48 5(2.4 %) and blaVIM 2 (0.9 %). Co-resistance genes with NDM producing GNB were detected in 87 (81.3 %) of all blaNDM producing isolates. NDM-1 was the most frequent subtype observed in 75 (70 %) blaNDM producing isolates. The highest percentage of resistance was recorded in ampicillin (98 %), cephalexin (93.5 %) amoxicillin clavulanic acid (90 %), cefotaxime (89.7 %), ceftriaxone (88.4 %), ceftazidime (84.2 %), sulfamethoxazole-trimethoprim (78.4 %) and nitrofurantoin (75.2 %), aztreonam (66 %) and temocillin (64 %). A close correlation between phenotypic and carbapenemase genes detection in all GNB was observed.
Conclusions
The frequency of carbapenemase producing bacilli was found to be high in SUH. NDM was found to be the most prevalent carbapenemase gene among clinical isolates. Close surveillance across all hospitals in Sudan is required. The relative distribution of carbapenemase genes among GNB in nosocomial infections in Africa needs to be defined.
Keywords: Hospitalized patients, Carbapenemase resistant genes, Multidrug‐resistant, Gram negative bacteria, Sudan
Background
Antimicrobial resistance (AMR) poses a complex threat to global health security and universal health coverage. The prevalence and distribution of antimicrobial resistant bacterial infections in the nosocomial settings in Africa is poorly defined [1, 2]. Carbapenems have been considered as a robust group of antibiotics to treat extended spectrum β-lactamase (ESBL)- producing bacteria in the past ten years and are widely prescribed for treatment of multidrug-resistant Gram-negative bacilli in systemic infections [3]. Overuse of these drugs can favor the selection and spread of multidrug resistant bacteria as well as Carbapenem resistant Enterobacteriales (CRE) [4] CRE, carbapenem-resistant Pseudomonas aeruginosa, and carbapenem-resistant Acinetobacter baumannii were ranked as ‘critical’ and ‘high’ priority pathogens by the World Health Organization (WHO) in 2017 [5].
Carbapenem resistance is associated with the production of carbapenemase, encoded by what is termed carbapenemase encoding genes, and categorized as four classes of ß-lactamase, including class A carbapenemases such as Klebsiella pneumoniae carbapenemases (blaKPC), imipenem- hydrolyzing β-lactamase (blaIMI) and Serratia marcescens enzyme (blaSME). Class B Metallo-beta-lactamases such as New Delhi Metallo-beta-lactamases (blaNDM), verona integron metallo-beta-lactamases (blaVIM), imipenemase (blaIMP), German imipenemase (bla GIM-1) and Sao Paulo Metallo-beta-lactamases (blaSPM). While class D carbapenemases including oxacillinase-group (blaOXA-48, OXA- 181, OXA-204, OXA-162, OXA-23, OXA-24) and a rare class C ß-lactamase, including cephamycin-hydrolyzing β-lactamase (blaCMY-10) [6, 7]. Resistance to carbapenems can occur by other mechanisms including overproduction of ESBLs or AmpC enzyme in combination with porin mutations, which reduces outer membrane permeability, and activation of multidrug efflux pumps in response to antibiotic exposure [8]. Plasmids encoding carbapenemases may also carry co-resistance genes for resistance to other β-lactam and non β-lactam antibiotics [7]. There is a significant challenge in controlling the spread of carbapenemases. Surveys of the molecular epidemiology of carbapenemases revealed that the dissemination of carbapenemases, including NDM, VIM, IMP, OXA-48 and KPC producers, are rapid and widespread among healthcare facilities [9, 10].
Detection of carbapenemase producing isolates by clinical microbiology laboratories is essential to provide targeted therapy, antimicrobial stewardship and update local antibiotic guidelines for clinicians. Furthermore, screening of resistance mechanisms using of antimicrobial susceptibility test in addition to detection of genes associated by phenotype and molecular analysis provides confirmation of clinically observed treatment failure. Whilst over the past decade, there has been an increase in reports globally of nosocomial infections caused by carbapenem resistant Gram-negative bacilli (GNB), data from Africa have been scanty and antimicrobial stewardship is not optimally practiced. This study aimed to detect and characterize carbapenem resistance GNB isolated from patients treated at Soba University Hospital in Khartoum state, Sudan.
Results
Demographic distribution
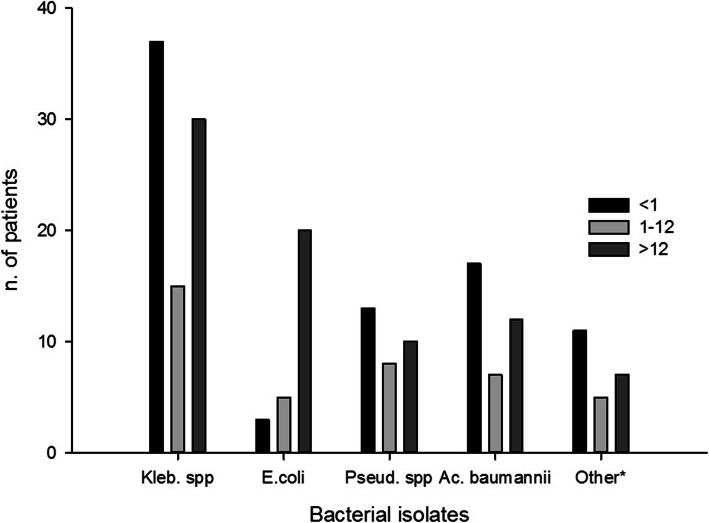
The demographic characteristics of the inpatients and the frequency of GNB isolates according to age groups are shown in Fig. 1. Most of the isolates were from pediatric patients less than one year old (42.5 %), followed by age group ranged from 13 to 80 years (38 %) and the rest of pediatric patients age group ranged from 1 to 12 years (19.5 %). Males 53.4 % (110/206) were predominant among inpatients, with females at 46.6 % (96/206). With regard to the distribution of carbapenemase producers by hospital location, the most carbapenemase producing isolates were found in the Neonatal Intensive Care Unit 32(26 %), followed by Medicine wards 26(22 %), Pediatric wards 22 (18 %), Surgery 18(15 %), ICU 15(12 %) and the Renal Unit 8(7 %). Carbapenemase producing isolates were most frequently distributed among the following clinical specimens; blood (36 %) followed by wound samples (24 %), urine (21 %), body fluids (7 %), catheter tips (6 %) and sputum samples (6 %).
Fig. 1.
Distribution of bacterial species isolates among different age group
Antimicrobial susceptibility and phenotypic screening
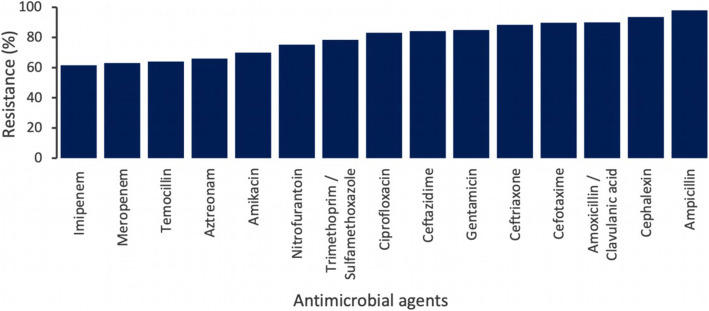
The antibiotic resistance pattern is shown in Fig. 2. Out of 206 isolates tested, the highest percentage of resistance was recorded in ampicillin (98 %) and cephalexin (93.5 %), amoxicillin clavulanic acid (90 %), cefotaxime (89.7 %), ceftriaxone (88.4 %), ceftazidime (84.2 %), sulfamethoxazole-trimethoprim (78.4 %) and nitrofurantoin (75.2 %), aztreonam (66 %) and temocillin (64 %). The resistance rate was also high in ciprofloxacin (83.1 %), gentamicin (85 %) and amikacin (70 %). The resistance rate for meropenem and imipenem was 63.1 % and 61.6 %, respectively.
Fig. 2.
Antimicrobial Resistance pattern among different Gram-negative bacilli isolated from patients treated at Khartoum state hospitals
Phenotypic carbapenemase activity was detected in 171 (83 %) of the 206 clinical isolates. These isolates were positive for the production of one or more carbapenemase enzymes. Details of the carbapenemase activity among different isolates and phenotypic tests are given in Table 1.
Table 1.
Frequency of carbapenemase producing among Gram-negative bacilli by phenotypic tests
| Positive isolates for a particular phenotypic test | Total | ||||
|---|---|---|---|---|---|
| Bacterial isolates | EDTA | MHT + BA | EDTA + BA | EDTA + AB + MHT | |
| n (%) | n (%) | n (%) | n (%) | n (%) | |
| E.coli (n = 28) | 17 (16.4) | 2 (8.3) | 3 (11.1) | 4 (26.6) | 26 (15.2) |
| Klebsiella spp. (n = 82) | 41 (39) | 10 (41.6) | 12 (44.4) | 8 (53.3) | 71 (41.5) |
| Pseudomonas spp. (n = 45) | 19 (18) | 9 (37.5)a | 4 (14.9) | 1 (6.6) | 33(19.2) |
| Acin. baumannii (n = 36) | 19 (18) | 3 (12.6) a | 6 (22.2) | 2 (13.3) | 30 (17.6) |
| Burkholderia cepacia (n = 2) | 2 (1.9) | 0 (0) | 0 (0) | 0 (0) | 2 (1.2) |
| Enterobacter spp. (2) | 2 (1.9) | 0 (0) | 0 (0) | 0 (0) | 2 (1.2) |
| Other GNBb (n = 11) | 5 (4.8) | 0 (0) | 2 (7.4) | 0(0) | 7 (4) |
| Total (n = 206) | 105(50.9) | 24 (11.7) | 27 (13.1) | 15 (7.2) | 171(83) |
-MHT positive = KPC + OXA48 - Boronic acid positive = KPC
-MHT + Boronic acid positive = KPC -MHT positive + boronic acid negative = OXA 48
Note: MHT applied only for Enterobacterceae. -EDTA positive = Metallo-β-lactamase.aThese isolates just positive by BA and not MHT.bOther Gram-negative bacilli include: Citrobacter species (3), Serratia species (1), Proteus spp. (2), Stenotrophomonas maltophilia (3), Vibrio vurneficus (1) and Morganella morganii (1).
Prevalence and distribution of carbapenemase genes among Gram negative bacilli
One or more carbapenemase genes were detected in 121 (58.7 %) of the 206 study isolates using PCR. blaNDM was the most commonly detected among the isolates, predominantly in K. pneumonia. blaNDM was also detected more often in A. baumannii, P. aeruginosa and E. coli. The most prevalent gene was blaNDM 107(52 %), followed by blaIMP 7 (3.4 %), blaOXA-48 5(2.4 %), blaVIM 2 (0.9 %) and blaKPC 0 (0 %). ESBLs were detected among these isolates with high prevalence in 183 isolates (88.8 %), with the following genes: blaCTXM 126(61.6 %), blaSHV 84(40.7 %) and blaTEM 78(37.8). The genes were unevenly distributed among the different study isolates and more details are given in Table 2.
Table 2.
Distributions of carbapenemase and ESBL genes among GNB isolates
| Bacterial isolates | Carbapenemase genes | ESBL genes | ||||||
|---|---|---|---|---|---|---|---|---|
| NDM | OXA-48 | IMP | VIM | KPC | CTXM | SHV | TEM | |
| n(%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | n (%) | |
| K. pneumoniae (n = 82) | 58 (54.3) | 1 (20) | 3 (43) | 1 (50) | 0 (0) | 53 (41.7) | 28 (33.4) | 32 (40) |
| E.coli (n = 28) | 9 (8.4) | 1 (20) | 2 (28.5) | 0 (0) | 0 (0) | 14 (11.6) | 6 (7.2) | 4 (5) |
| Pseudomonas Spp.(n = 45) | 14 (13.1) | 2 (40) | 2 (28.5) | 1 (50) | 0 (0) | 28 (22.3) | 18 (21.5) | 16 (20) |
| A. baumannii (n = 36) | 17 (15.8) | 1 (20) | 0 (0) | 0 (0) | 0 (0) | 20 (15.7) | 24 (28.6) | 18 (22.5) |
| Burkholderia (n = 2) | 0 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (1.6) | 2 (2.4) | 4 (5) |
| Enterobacter Spp.(n = 2) | 2 (1.8) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (1.6) | 1 (1.2) | 1 (1.25) |
|
Other GNBa (n = 11) |
7 (6.5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 7 (5.5) | 4 (5.7) | 5 (6.25) |
| Total (n = 206) | 107 (52) | 5 (2.4) | 7 (3.4) | 2 (0.9) | 0 (0) | 126 (61.1) | 84 (40.7) | 78 (37.8) |
aOther Gram-negative bacilli include: Citrobacter species (3), Serratia species (1), Proteus spp. (2), Stenotrophomonas maltophilia (3), Vibrio vurneficus (1) and Morganella morganii (1)
Co-resistance carbapenemase genes were observed in a small number of isolates; blaNDM + blaOXA-48 were detected in three isolates, while blaNDM + blaVIM and blaNDM + blaIMP were detected in two different isolates.
Out of 107 blaNDM genes detected, 75 (70 %) were blaNDM-1. Other subtypes of blaNDM were identified by sequencing, including blaNDM- 5, and blaNDM-6. The sequence of the 14 NDM genes was analyzed to confirm the presumed most prevalent (NDM) gene type and all showed 97–100 % similarity with blaNDM genes from the NCBI database with accession number MF379688 and MG764089. Thereafter, all sequences have been deposited in the GenBank database under the following accession numbers: MK033562, MK033563, MK033564, MK363705, MK363706, MK363707, MK363708, MK363709, MK363710, MK371542, MK371543, MK371544, MK37154 5, and MK371546.
Our analysis revealed that phenotypic test versus Carbapenemase or ESBL gene detection were strongly correlated (P = 0.0000001; P = 0.01, respectively). The correlation between phenotypic and carbapenemase genes detection was highly significant for K. pneumonia, E. coli, P. aerugnosa and A. baumannii (P = 0.0000031; P = 0.00079; P = 0.015; P = 0.02, respectively) while for ESBL genes correlation was only significant for P. aerugnosa (P = 0.038) Table 3.
Table 3.
Comparison between phenotypic and genotypic results of carbapenem resistance among different GNB strains
| Bacterial species | Resistant isolates n |
Phenotypic testa n (%) |
Carbapenemase genesb n (%) |
Correlation with carbapenemase | ESBL genesc n (%) |
Correlation with ESBL | |||
|---|---|---|---|---|---|---|---|---|---|
| positive | negative | positive | negative | p-value | positive | negative | p-value | ||
| K. pneumoniae | 82 | 71 (86.6) | 11(13.4) | 63 (76.8) | 19 (23.2) | 0.0000031 | 72 (87.8) | 10 (12.2) | 0.017 |
| E.coli | 28 | 26 (92.8) | 2 (7.2) | 12 (42.8) | 16 (57.2) | 0.00079 | 19 (67.8) | 9 (32.2) | 0.123 |
| P. aeruginosa | 45 | 33 (73) | 12 (27) | 19 (42) | 26 (58) | 0.0015 | 39 (86.6) | 6 (13.4) | 0.038 |
| A. baumannii | 36 | 30 (83.3) | 6 (16.7) | 18 (50) | 18 (50) | 0.022 | 31 (86) | 5 (14) | 0.14 |
| Other GNBd | 15 | 11 (73) | 4 (27) | 9 (60) | 6 (40) | 0.055 | 15 (100) | 0 (0) | 0.08 |
| Total | 206 | 171 (83) | 35 (17) | 121(58.7) | 85 (41.3) | 0.0000001 | 176 (85.4) | 30 (14.6) | 0.01 |
aPhenotypic test include: EDTA, Borinic Acid and Modified Hodge Test
bCarbapenemase genes include: NDM, VIM, IMP and OXA-48
cESBL genes include: CTX-M, SHV, and TEM
dOther GNB include: Citrobacter species (3), Burkholderia spp. (2), Enterobacter Spp.(2), Serratia species (1), Proteus spp. (2), Stenotrophomonas maltophilia (3), Vibrio vurneficus (1), and Morganella morganii (1)
Co‐resistance genes carried with NDM producing gram‐negative bacilli
Several isolates carried more than one type of gene including blaNDM. ESBL were often observed together with blaNDM in 87 (81.3 %) of blaNDM producing isolates (107). Most of the isolates carried blaNDM with one ESBL gene in 38(43.5 %) as the following: blaNDM + blaCTXM in (24 isolates, 27.6 %), blaNDM + blaTEM (8 isolates, 9.1 %), and blaNDM + blaSHV (6 isolates, 6.8 %). Isolates carried blaNDM with two ESBL genes in (39.2 %) as the following: blaNDM + blaCTXM + blaSHV (10 isolates, 11.5 %), blaNDM + blaCTXM + blaTEM (10 isolates, 11.5 %), blaNDM + blaSHV + blaTEM (14 isolates, 16.2 %). Isolates carried blaNDM with three ESBL genes, blaNDM + blaCTXM + blaSHV + blaTEM in 15 isolates (17.3 %). The distribution of co-resistance genes among different Gram-negative bacilli is shown in Table 4.
Table 4.
Co resistance genes with blaNDM among Gram-negative bacilli
| Bla genes | K. pn | E.coli | P. aer | A. bau | Enter | Total (%) |
|---|---|---|---|---|---|---|
| NDM + CTXM | 15 | 2 | 5 | 2 | 0 | 24 (27.6 %) |
| NDM + SHV | 2 | 0 | 2 | 2 | 0 | 6 (6.8 %) |
| NDM + TEM | 4 | 1 | 1 | 1 | 1 | 8 (9.1 %) |
| NDM + CTXM + SHV | 7 | 0 | 1 | 1 | 1 | 10 (11.5 %) |
| NDM + CTXM + TEM | 6 | 2 | 0 | 2 | 0 | 10 (11.5 %) |
| NDM + SHV + TEM | 7 | 0 | 1 | 6 | 0 | 14 (16.2 %) |
| NDM + All | 9 | 1 | 3 | 2 | 2 | 15 (17.3 %) |
| Total | 50 | 6 | 13 | 16 | 2 | 87 (100 %) |
- K. pn = Klebsiella pneumoniae, P. aer = Pseudomonasa aeruginosa, A. bau = Acinetobacter baumannii, Enter = Enterobacter spp
Discussion
Carbapenems have become the drug of choice for the treatment of severe nosocomial infections caused by Gram-negative bacilli. Carbapenem resistant Enterobacteriales (CRE) is a considerable health problem globally and are associated with increased mortality [3], and thus rapid detection of carbapenem resistance and adequate treatment of such cases is mandatory in health care settings. This study was undertaken to determine the prevalence of different types of carbapenemase producing bacteria among Gram-negative bacilli isolated from various hospitalised patients at Soba University Hospital. The accurate detection of carbapenemase producing microorganisms is challenging for laboratories and requires phenotypic and genotypic analysis. Of 206 isolates, 171(83 %) were positive by phenotypic analysis, including isolates with resistance to carbapenems. The genotypic analysis detected 121 (58.7 %) positive isolates.
Our results revealed that the prevalence of carbapenemase production among different Gram-negative isolates is increasing (up to 83 %). Of note, this finding is higher than the incidence observed in a previous study conducted in Khartoum State in 2017, which showed a prevalence of 56 % by phenotypic tests (unpublished data). The high frequency of MBL in Khartoum State might be attributed to the excessive use of meropenem in the treatment of patients associated with ESBL infections. Our results are in agreement with a study in Egypt, which reported that carbapenem resistance was 62.7 % among Enterobacterial [12]. High rates of carbapenem resistance in 28.6 % of isolates have also been observed in Uganda by Okoche in 2015 [13]. In Tanzania, the prevalence of carbapenemase producing isolates was 35 % [14]. In South Africa, it was found to be 68 % [15], and in Nigeria, 11.9 % [16]. Carbapenem resistance in low and middle-income countries (LMICs) in Africa is likely to increase as a result of unrestricted usage of antibiotics, as the majority of the population consumes antibiotics without a clinical prescription [17].
Carbapenemase genes have been recently recognized, and these genes are associated with mobile genetic elements that allow their rapid circulation among bacterial isolates. For instance, blaNDM has the potential for rapid spread, as evidenced in Turkey and other countries across the globe [18]. In this study, carbapenemase genes were detected using PCR in 121 (58.7 %) of the sampled isolates. The most prevalent gene among the isolates was blaNDM (88.4 %), mainly in K. pneumonia and other Gram-negative bacilli, including A. baumannii, P. aeruginosa and E. coli. Our results are comparable with those reported in India, South Africa, Saudi Arabia and other Middle Eastern countries [15, 19–21].
Carbapenemase genes are reported to be more frequent in some regions. For example, blaKPC genes are dominant in some countries such as Greece, Israel, and USA, while blaNDM genes are prevalent in isolates reported from the Far East, India, and Pakistan [11]. OXA-48 was first reported from Turkey, followed by reports from Central Asia and Europe [22]. In the current study, the genes were unevenly distributed among the different bacterial isolates. The blaNDM gene was found in high prevalence (52 %) compared to other genes. Our findings, however, differ with several studies (for instance, in the Okoche study, the most common gene was blaVIM (10.7 %), and blaNDM-1 (2.6 %) was the lowest gene [13], while Mushi reported IMP types were the most predominant at 21.6 % in their study [14]. Other studies reported that blaOXA-48 was the most prevalent gene [23, 24]. In contrast to global reports of a high prevalence of blaKPC genes [11, 25], we have not detected blaKPC among the tested isolates. NDM variants have been described differing by several amino acid changes include, blaNDM-2, blaNDM-4, blaNDM-5, blaNDM-6 and blaNDM-7 [26]. In this study, 107 blaNDM producer isolates had been identified using PCR, the most common subtype 75 (70 %) was blaNDM-1. Other subtypes of blaNDM were detected by sequencing including blaNDM-5, and blaNDM-6 among different Gram-negative bacilli, including K. pneumoniae, E. coli, A. baumannii, P. aeruginosa and Enterobacter spp.
Carbapenemase producers are becoming highly distributed among Enterobacterales, A. baumannii, P. aeruginosa and other Gram-negative bacilli. The highest prevalence of carbapenemase production in this study was observed in K. pneumoniae, followed by P. aeruginosa, A. baumannii and E. coli Table 1. Similar studies in which K. pneumonia, A. baumannii and Pseudomonas spp. as the most common carbapenemase producing isolates were reported [27–29]. The prevalence of carbapenemase producing isolates varies across hospital settings. This variation could be attributed to a wide range of factors including differences in collection time of isolates, study designs and target populations. Common carbapenemase encoding genes have been associated with bacteria isolated from blood, urine, wounds and sputum, as reported in many studies in Africa and India [13, 14, 16, 30]. In this finding, carbapenem producers were more frequently isolated from blood followed by wounds and urine as these results are compatible with a study in South Africa in which blood was the most common specimen type (25 %), followed by urine (22 %) [15].
Young patient age has long been considered as a risk factor for Carbapenem resistant Enterobacteriales (CRE) infection, which agrees with this study’s finding that carbapenemase-producing Gram negative bacilli were most frequent in children less than one year of age, located in the neonatal and pediatric wards. High rates of carbapenem resistant infections were observed among elderly patients from Medicine wards (22 %) and ICU (12 %), which agrees with another study that found CRE to be more frequently isolated in the elderly [31].
Carbapenem resistant Gram-negative bacilli are usually resistant to other routinely used antimicrobial agents [32–34]. The plasmids carrying carbapenemase genes like blaNDM-1 are diverse and can harbor a high number of additional resistance genes (e.g., ESBL-alleles) as well as other carbapenemase genes like blaOXA-48, blaVIM. These plasmids were considered as the source of multidrug resistance in one single bacterium [20, 35]. Moreover, mechanisms of resistance to β-lactam antibiotics via the production of ESBL, AmpC and carbapenemase were also noticed among the isolates that produce different combinations of the enzymes. In the present study, co-resistance of blaNDM with blaOXA-48, blaVIM and blaIMP were reported in few isolates. In connection to co-resistance with ESBL, blaCTXM, blaSHV and blaTEM were detected in a high prevalence of blaNDM positive isolates. Most of the isolates carried blaNDM with one or more ESBL genes. This is in agreement with various studies that have reported co-resistance among clinical isolates [36, 37]. These multiple resistance genes found in some isolates, as observed in this study, are indicative of the existence of multidrug resistant pathogens, which are responsible for treatment failure, outbreaks of infections and higher treatment costs [38].
Sudan is a large country that shares its borders with seven other countries. People move freely between these borders with the potential passage of antibiotic resistant strains. The dynamic movements of people will make it challenging to monitor AMR in these countries, especially at the borders. These challenges may also represent an opportunity for wider continental monitoring and collaboration between countries rather than country-specific. Such an approach will aid in universal and intergovernmental initiatives to control and limit the AMR spread.
Conclusions
The frequency of carbapenemase producing bacilli was found to be high in Soba University Hospital (SUH). blaNDM was found to be the most prevalent carbapenemase gene among clinical isolates. Improved antibiotic stewardship and infection control measures and close surveillance across all hospitals in Sudan is required. The relative distribution of carbapenemase genes among Gram-negative bacilli (GNB) in nosocomial infections in Africa needs to be investigated.
Methods
Study design and clinical isolates
A cross-sectional laboratory-based study was conducted at the microbiology department in Soba University Hospital and Unit of Molecular Biology, Institute of Endemic Diseases (IEND), University of Khartoum; involving Gram negative clinical bacterial isolates, suspected as carbapenemase producing based on breakpoints zone diameter of carbapenems (CLSI, 2017) [39]. These were isolated from cultures of various clinical specimens; blood, urine, wound swabs, sputum, tips of catheters and other body fluids, between 1st October 2016 and 25th February 2017 from inpatients at Soba University Hospital. Quality control strains [E. coli (ATCC #25,922) and P. aeruginosa (ATCC #27,853)] were used in antimicrobial susceptibility testing. Standard biochemical tests were used for primary identification [40] and molecular identification using PCR [8] was used for all study isolates with universal primer (16SrRNA). For species-specific isolates identified on biochemical testing, species-specific primers for Klebsiella pneumoniae, Escherichia coli, Pseudomonas aeruginosa and Acinetobacter baumannii were used for confirmation [41–52]. All isolates were stored in 20 % glycerol at -20°C until use.
Subculture, susceptibility testing, phenotypic screening and confirmatory tests for carbapenemase resistance
Selection of antimicrobial panels and interpretation of disk diffusion for each bacteria was completed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [39].
Isolates were subcultured on blood agar (BA) and then subjected to susceptibility testing with the following antimicrobials (Mast Diagnostic, UK): amoxicillin-clavulanate (AMC) (30µg); cefuroxime (CXM) (30µg); cephalexin (CL) (30µg); ceftriaxone (CRO) (30µg); ceftazidime (CAZ) (30µg); cefotaxime (CTX) (30µg); meropenem (MEM) (10µg); imipenem (IPM) (10µg); amikacin (AK)(30µg); gentamicin (CN)(10µg); ciprofloxacin (CIP)(5µg); trimethoprim-sulfamethoxazole (SXT)(25µg); temocillin (TEM)(30µg); aztreonam (AZT)(30µg). The Kirby Bauer(disk diffusion) was performed; each isolate was swabbed onto Muller-Hinton agar and the antibiotic discs were placed on top, incubated at 37 °C for 18–24 hours [39, 40].
Bacterial isolates that showed intermediate or resistance to imipenem or meropenem were considered as suspected carbapenemase producers and were screened. Phenotypic confirmatory tests for carbapenemase production were conducted using the boronic acid synergy test for class A β-lactamases, the EDTA synergy test for metallo-β-lactamase and the Modified Hodge Test (MHT) for Enterobacterales to detect KPC and OXA-48 producers in addition to temocillin sensitivity [46].
Molecular identification of carbapenemase encoding genes
PCR was carried out using a thermal cycler and the following primers (Macrogen, Korea), blaVIM, blaIMP, blaNDM, blaNDM-1, blaKPC, blaOXA-48, blaTEM, blaSHV and blaCTX-M genes were used [47–51]. The reaction was carried out in a total reaction volume of 25µl (5µl master mix, Maxime RT premix kit) [8]. The purity and integrity of each PCR product was evaluated, and the amplified product was confirmed with reference to the standard DNA ladder.
DNA sequencing and genes analysis
The PCR product of blaNDM genes and 16SrRNA were purified and Sanger sequencing was performed by Macrogen Company (Seoul, Korea). Fourteen blaNDM gene products have been selected randomly to represent the study isolates.
Bioinformatics analysis
Firstly, DNA sequences were clarified and determined the overall quality of the sequences by reviewing nucleotide chromatogram by using Finch TV software version 1.4.0 () to ensure the ambiguous sites. Thereafter, nucleotide sequences of the NDM genes identified were searched for sequence similarity using nucleotide BLAST [52] (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical analysis
Data were analysed using SPSS software version 20.0. Cross tabulation was used to present the relationships between data of antimicrobial sensitivity, phenotypic tests and resistant gene detection among the study isolates, qualitative data were performed through χ2 test and significance was set at p ≤ 0.05.
Acknowledgements
We would like to thank the medical and technical staff of Medical Microbiology Department in Soba University Hospital, University of Khartoum for their help in data collection. We thank Dr. Mohamed Osman for English corrections. FN; LE; TDM; AA; GI; AZ; AYO; HE; MMAH are members of the Pan African Network for Rapid Research, Response, and Preparedness for Infectious Diseases Epidemics consortium (PANDORA-ID-NET) supported by EDCTP2.
Abbreviations
- AK
Amikacin
- AMC
Amoxicillin clavulanate
- Amp C
Class C β-lactamases
- AMR
Antimicrobial resistance
- ATCC
American Type Culture Collection
- AZT
Azteroname
- BLAST
Basic local alignment search tool
- bp
Base pair
- CAZ
Ceftazidime
- CIP
Ciprofloxacin
- CL
Cephalexin
- CLSI
Clinical and Laboratory Standards Institute
- CN
Gentamicin
- CPR
Carbapenemase producer Enterobacteriales
- CRE
Carbapenem resistance Enterobacteriales
- CRO
Ceftriaxone
- CTX
Cefotaxime
- CTX-M
CTX for cefotaximase and M for Munich
- CXM
Cefuroxime
- DNA
Deoxyribonucleic acid
- E. coli
Escherichia coli
- EDTA
Ethylene diamine tetraacetic acid
- ESBL
Extended-Spectrum β-Lactamase
- GNB
Gram-negative bacilli
- ICU
Intensive care unit
- IEND
Institute of Endemic Diseases
- IMP
Imipenemase Metallo-β-lactamase
- IPM
Imipenem
- K. pneumonia
Klebsiella pneumonia
- KPC
Klebsiella pneumoniae carbapenemase-producer
- LMICs
Low and middle income countries
- MBL
Metallo-β-lactamas
- MDR
Multi drug resistance
- MEM
Meropenem
- MHT
Modified Hodge test
- NDM
New Delhi Metallo-β-lactamase
- OXA
oxacillinases
- P. aeruginosa
Pseudomonas aeruginosa
- PCR
Polymerase Chain Reaction
- S. maltophilia
Stenotrophomonas maltophilia
- SHV
Sulphydryl Reagent Variable
- SPSS
Statistical Package for the Social Sciences
- SUH
Soba University Hospital
- SXT
Trimethoprim-sulfamethoxazole
- TEM
Named after patient Temoniera
- TEM
Temocillin
- VIM
Verona integron-encoded Metallo-β-lactamase
- WHO
World Health Organisation
- °C
Celsius degree
- 16SrRNA
16 small ribosomal RNA
- μg
Microgram
Authors’ contributions
HE, KE, AA and MMAH designed the study. HE carried out the microbiological analysis. HE, MMAH and HA analysed the data. HE and MMAH wrote the first draft. MMAH, KE, EM, HA, FN, LE, TDM, JT, AYO, GI and AZ were major contributors in revising the manuscript critically for important intellectual content. MMAH, EM, and KE supervised the work. MMAH, and HA secured funding for the practical work. AZ and AYO revised the final draft. All authors read and approved the final manuscript.
Funding
This research received partial research funding from the Ministry of Higher Education and Scientific Research, Sudan, grant number: SUD/MOH/MMAH/08/2016. FN, LE, TDM, MMAH, JT, GI, AYO and AZ are part of PANDORA-ID-NET Consortium supported by the European and Developing Countries Clinical Trials Partnership (EDCTP2) programme (RIA2016E-1609). The funders had no role in the study design, data collection, analysis, interpretation or writing of the manuscript.
Availability of data and materials
The antimicrobial data datasets used and/or analyzed during the current study are available at . blaNDM gene sequences are deposited at NCBI under the accession numbers: MK033562- MK033564, MK363705-MK363710 and MK371542- MK371546 for. The 16S rRNA sequences generated in this study are available from GenBank under sequential accession numbers MW034199–MW034227.
Ethics approval and consent to participate
This study was approved by the Institutional Research Ethics Committee of the Institute of Endemic Diseases (IEND), University of Khartoum and Soba University Hospital (Ref: 12/2017). Written informed consent was obtained from all participants.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hana S. Elbadawi and Muzamil M. Abdel Hamid contributed equally to this work.
Contributor Information
Hana S. Elbadawi, Email: Hanasalah200@gmail.com
Muzamil M. Abdel Hamid, Email: mahdi@iend.org, Email: mahdi@uofk.edu.
References
- 1.Tadesse BT, Ashley EA, Ongarello S, Havumaki J, Wijegoonewardena M, González IJDS. Antimicrobial resistance in Africa: a systematic review. BMC Infect Dis. 17(1):616. [DOI] [PMC free article] [PubMed]
- 2.Kumwenda GP, Sugawara Y, Abe R, Akeda Y, Kasambara W, Chizani K, et al. First identification and genomic characterization of multidrug-resistant carbapenemase-producing Enterobacteriaceae clinical isolates in Malawi, Africa. J Med Microbiol. 10.1099/jmm.0.001087. [DOI] [PubMed]
- 3.Hall BG, Barlow M. Evolution of the serine β-lactamases: past, present and future. Drug Resist Updat. 2007;7:111–123. doi: 10.1016/j.drup.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Harris P, Paterson D, Rogers B. Facing the challenge of multidrug- resistant gramnegative bacilli in Australia. Clin Focus. 2015;202:243–247. doi: 10.5694/mja14.01257. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization . Global priority list of antibiotic-resistant bacteria to guide research, discovery and development of new antibiotics. 2017. [Google Scholar]
- 6.Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase- producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Queenan AM, Bush K. Carbapenemases : the versatile B-lactamases. Clin Microbiol Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nordmann P, Gniadkowski M, Giske CG, Poirel L, Woodford N, Miriagou V. Identification and screening of carbapenemase-producing Enterobacteriaceae. Clin Microbiol Infect. 2012;18:432–438. doi: 10.1111/j.1469-0691.2012.03815.x. [DOI] [PubMed] [Google Scholar]
- 9.Fournier S, Lepainteur M, Kassis-Chikhani N, Huang M, BrunBuisson C, Jarlier V, AP-HP Outbreaks Control Group Link between carbapenemase-producing Enterobacteria carriage and crossborder exchanges: eight-year surveillance in a large French multihospitals institution. J Travel Med. 2012;19:320–323. doi: 10.1111/j.1708-8305.2012.00641.x. [DOI] [PubMed] [Google Scholar]
- 10.Huang TD, Bogaerts P, Berhin C, Hoebeke M, Bauraing C, Glupczynski Y. Increasing proportion of carbapenemase-producing Enterobacteriaceae and emergence of a MCR-1 producer through a multicentric study among hospital-based and private laboratories in Belgium from September to November 2015. Eurosurveillance. 2017:1–9. [DOI] [PMC free article] [PubMed]
- 11.Pitout JDD, Nordmann P. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Anti Microb Agents Chemother. 2015;59(10):5873–5884. doi: 10.1128/AAC.01019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amer WH, Khalil HS, Abd ELWahab MAA. Characterization of carpabenem resistant Enterobacteriaceae in risk factors , phenotypic and genotypic characterization of carbapenem resistant Enterobacteriaceae in Tanta University Hospitals , Egypt. Int J Infect Control. 2016;12:1–11. doi: 10.3396/IJIC.v12i2.012.16. [DOI] [Google Scholar]
- 13.Okoche D, Asiimwe BB, Katabazi FA, Kato L. Prevalence and characterization of Carbapenem-resistant Enterobacteriaceae isolated from Mulago National Referral. PLOS. 2015;48:1–11. doi: 10.1371/journal.pone.0135745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mushi MF, Mshana SE, Imirzalioglu C, Bwanga F. Carbapenemase genes among multidrug resistant gram negative clinical isolates from a tertiary Hospital in Mwanza , Tanzania. Biomed Res Int. 2014:3–8. [DOI] [PMC free article] [PubMed]
- 15.Perovic O, Micro FSA, Micro M, Britz E, Chb MB, Chetty V. IN PRACTICE molecular detection of carbapenemase-producing genes in referral Enterobacteriaceae in South Africa : a short report. SAMJ. 2016;106(10):975–977. doi: 10.7196/SAMJ.2016.v106i10.11300. [DOI] [PubMed] [Google Scholar]
- 16.Yusuf I, Arzai AH, Haruna M, Sharif AA, Getso MI. Detection of multi drug resistant bacteria in major hospitals in Kano. Braz J Microbiol. 2014;45(3):791–798. doi: 10.1590/S1517-83822014000300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukonzo JK, Namuwenge PM, Okure G, Mwesige B, Namusisi OK, Mukanga D. Over-the-counter suboptimal dispensing of antibiotics in Uganda. J Multidiscip Healthc. 2013;6:303–310. doi: 10.2147/JMDH.S49075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karabay O, Altindis M, Koroglu M, Karatuna O, Aydemir ÖA. The carbapenem - resistant Enterobacteriaceae threat is growing : NDM - 1 epidemic at a training hospital in Turkey. Ann Clin Microbiol Antimicrob. 2016;15:1–6. doi: 10.1186/s12941-016-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deshpande P, Rodrigues C, Shetty A, Kapadia F, Hedge A, Soman R. New Delhi Metallo-beta lactamase (NDM-1) in Enterobacteriaceae: treatment options with carbapenems compromised. J Assoc Physicians India. 2010;58:147–149. [PubMed] [Google Scholar]
- 20.Kumarasamy K, Toleman M, Walsh T, Al. E. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 2010;10:597–602. [DOI] [PMC free article] [PubMed]
- 21.Sonnevend Á, Ghazawi A, Hashmey R, Jamal W, Rotimi V, Shibl A, et al. Characterization of carbapenem-resistant Enterobacteriaceae with high rate of autochtho- nous transmission in the arabian peninsula. PLoS One. 2015;10:e0131372. doi: 10.1371/journal.pone.0131372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gülmez D, Woodford N, Palepou M-F, Mushtaq S, Metan G, Yakupogullari Y, et al. Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from Turkey with OXA-48-like carbapenemases and outer membrane protein loss. Int J Antimicrob Agents. 2008;31:523–526. doi: 10.1016/j.ijantimicag.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 23.Dahab RA, Ibrahim AM, Altayb HN. Phenotypic and genotypic detection of carbapenemase enzymes producing gram-negative bacilli isolated from patients in Khartoum state [version 1; peer review: awaiting peer review] F1000Research. 2017;6:1656. doi: 10.12688/f1000research.12432.1. [DOI] [Google Scholar]
- 24.Memish Z, Assiri A, Almasri M, Al. E Molecular characterization of carbapenemase production among gram-negative bacteria in Saudi Arabia. Microb Drug Resist. 2015;21(3):307–314. doi: 10.1089/mdr.2014.0121. [DOI] [PubMed] [Google Scholar]
- 25.Tängdén T, Giske CG. Global dissemination of extensively drug-resistant carbapenemase-producing Enterobacteriaceae: clinical perspectives on detection, treatment and infection control. J Intern Med. 2015;277(5):501–512. doi: 10.1111/joim.12342. [DOI] [PubMed] [Google Scholar]
- 26.Bonnin A, Nordmann P. First identification of novel NDM Carbapenemase , NDM- 7 , in Escherichia coli in France. 2013;8(4):1–5. [DOI] [PMC free article] [PubMed]
- 27.Doi Y, Paterson DL. Carbapenemase-producing Enterobacteriaceae. Semin Respir Crit Care Med. 2015;36:74–84. doi: 10.1055/s-0035-1544208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zowawi HM, Sartor AL, Sidjabat HE, Balkhy HH, Walsh TR, Al Johani M. Molecular epidemiology of Carbapenem-resistant Acinetobacter baumannii isolates in the Gulf cooperation council States : dominance of OXA-23-type producers. J Clin Microbiol. 2015;53(3):896–903. doi: 10.1128/JCM.02784-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karaaslan A, Soysal A, Gelmez GA, Kadayifci EK. Molecular characterization and risk factors for carbapenem-resistant gram-negative bacilli colonization in children : emergence of NDM-producing Acinetobacter baumannii in a newborn intensive care unit in Turkey. J Hosp Infect. 2016;92:67–72. doi: 10.1016/j.jhin.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 30.Bhaskar MM, Anand R, Harish BN. Prevalence of blaNDM-1 producing blood isolates of Escherichia coli, Klebsiella species and Enterobacter species in a tertiary care centre in South India. J Microbiol Res Rev. 2013;1(6):61–68. [Google Scholar]
- 31.Lin MY, Lyles-Banks RD, Lolans K, et al. The importance of long-term acute care hospitals in the regional epidemiology of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clin Infect Dis. 2013;57:1246–1252. doi: 10.1093/cid/cit500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yusuf I, Arzai AH, Haruna M, Sharif AA, Getso MI. Detection of multi drug resistant bacteria in major hospitals in Kano, North-West, Nigeria. Braz J Microbiol. 2014;45(3):791–798. doi: 10.1590/S1517-83822014000300005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy HG, Gould I, Endimiani A, Al E. Detection, treatment, and prevention of carbapenemase-producing Enterobacteriaceae: recom- mendations from an international working group. J Chemother. 2013;25(3):129–140. doi: 10.1179/1973947812Y.0000000062. [DOI] [PubMed] [Google Scholar]
- 34.Legese MH, Weldearegay GM, Asrat D. Extended-spectrum beta-lactamase- and among Ethiopian children. Infect Drug Resist. 2017;10:27–34. doi: 10.2147/IDR.S127177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nordmann P, Poirel L, Toleman MA, Walsh T. Does broad-spectrum B-lactam resistance due to NDM-1 herald the end of the antibiotic era for treatment of infections caused by gram-negative bacteria? J Antimicrob Chemother. 2011;66(4):689–692. doi: 10.1093/jac/dkq520. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Chen D, Xu G, Huang W, Wang X. Molecular epidemiology and drug resistant mechanism in carbapenem-resistant Klebsiella pneumoniae isolated from pediatric patients in Shanghai, China. PLoS One. 2018;20:1–10. doi: 10.1371/journal.pone.0194000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nithya N, Remitha R, Jayasree PR, Faisal M, Manish Kumar PR. Analysis of beta-lactamases, blaNDM-1phylogeny & plasmid replicons in multidrug-resistant Klebsiella spp. from a tertiary care centre in south India. Indian J Med Res. 2017;146(Supplement):S38–S45. doi: 10.4103/ijmr.IJMR_31_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen S, Leverstein-Van J, Hall M. Dutch working party on the detection of highly resistant microorganisms. Guideline for pheno- typic screening and confirmation of carbapenemases in Enterobacte- riaceae. Int J Antimicrob Agents. 2010;36(3):205–210. doi: 10.1016/j.ijantimicag.2010.05.014. [DOI] [PubMed] [Google Scholar]
- 39.CLSI . M100 performance standards for antimicrobial. 27. 2017. pp. 106–143. [Google Scholar]
- 40.Wauters G, Vaneechoutte M. Approaches to the identification of aerobic gram-negative Bacteria. In: Manual of clinical microbiology. 11th ed. Washington, DC; 2015. p. 613–34.
- 41.Srinivasan R, Karaoz U, Volegova M, MacKichan J, Kato-Maeda M, Miller S, et al. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS One. 2015;10(2):e0117617. doi: 10.1371/journal.pone.0117617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Liu C, Zheng W, Zhang X, Yu J, Gao Q, et al. PCR detection of Klebsiella pneumoniae in infant formula based on 16S-23S internal transcribed spacer. Int J Food Microbiol. 2008;125(3):230–235. doi: 10.1016/j.ijfoodmicro.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Sabat G, Rose P, Hickey WJ, Harkin JM. Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl Environ Microbiol. 2000;66(2):844–849. doi: 10.1128/AEM.66.2.844-849.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42(5):2074–2079. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Higgins PG, Wisplinghoff H, Krut O, Seifert H. A PCR-based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. CMI. 2007;13:1199–1201. doi: 10.1111/j.1469-0691.2007.01819.x. [DOI] [PubMed] [Google Scholar]
- 46.Aguirre-Quiñonero A, Martínez-Martínez L. Non-molecular detection of carbapenemases in Enterobacteriaceae clinical isolates. J Infect Chemother. 2017;23(1):1–11. doi: 10.1016/j.jiac.2016.09.008. [DOI] [PubMed] [Google Scholar]
- 47.Hong D, Bae I, Jang I, Jeong S, Kang H, Lee K. Epidemiology and characteristics of Metallo-β-lactamase-producing Pseudomonas aeruginosa. Infect Chemother. 2015;42(2):81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suwantarat N, Carroll KC. Epidemiology and molecular characterization of multidrug-resistant gram-negative bacteria in Southeast Asia. Antimicrob Resist Infect Control. 2016:1–8. [DOI] [PMC free article] [PubMed]
- 49.Pasteran F, Mendez T, Guerriero L, Rapoport M, Corso A. Sensitive screening tests for suspected class A Carbapenemase production in species of Enterobacteriaceae. J Clin Microbiol. 2009;47(6):1631–1639. doi: 10.1128/JCM.00130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lartigue M, Poirel L, Decousser J, Nordmann P. Multidrug-resistant Shigella sonnei and salmonella enterica serotype Typhimurium isolates producing CTXMbeta- lactamases as causes of community-acquired infection in France. Clin Infect Dis. 2005;40:1069–1070. doi: 10.1086/428667. [DOI] [PubMed] [Google Scholar]
- 51.Isturiz RECC. Antibiotic use in developing countries. Infect Control Hosp Epidemiol. 2000;21(6):394–397. doi: 10.1086/501780. [DOI] [PubMed] [Google Scholar]
- 52.Atschul S, Madden T, Schaffer A, et al. Gapped BLAST and PSI-BLAST. A new generation of protein database search programmes. NucleicAcid Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The antimicrobial data datasets used and/or analyzed during the current study are available at . blaNDM gene sequences are deposited at NCBI under the accession numbers: MK033562- MK033564, MK363705-MK363710 and MK371542- MK371546 for. The 16S rRNA sequences generated in this study are available from GenBank under sequential accession numbers MW034199–MW034227.