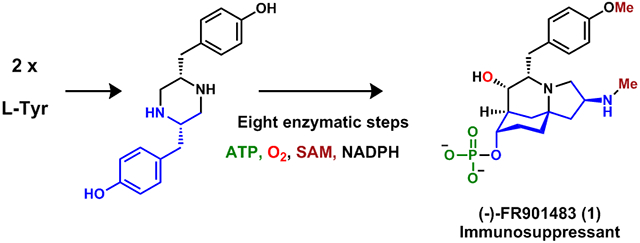
Abstract
We report characterization of the biosynthetic pathway of the potent immunosuppressant (−)-FR901483 (1) through heterologous expression and enzymatic assays. The biosynthetic logic to form the azatricyclic alkaloid is consistent with those proposed in biomimetic syntheses, and involves aza-spiro annulation of dityrosyl-piperazine to form a ketoaldehyde intermediate, followed by regioselective aldol condensation, stereoselective ketoreduction and phosphorylation. A possible target of 1 is proposed based on the biosynthetic studies.
Graphical Abstract
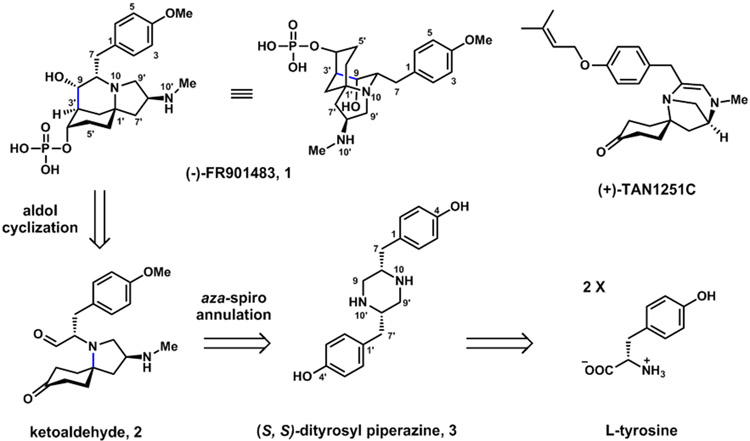
The fungal alkaloid (−)-FR901483 (1) (Figure 1), isolated from Cladobotryum sp. No 11231, is a potent immunosuppressant that shows efficacy in animal models.1 The mode of action of 1 is distinct from cyclosporine A and FK-506, and was proposed to function through the inhibition of purine nucleotide biosynthesis.1 Structurally, 1 has a rigid core that contains a 2-azabicyclo[3.3.1]nonane and a spiro-fused pyrrolidine ring. The core is substituted with a p-methoxybenzyl group at C8 and a phosphate ester at C4’, which are essential for the biological activity of 1.1 Because of the potent activity and the unique structure, 1 and the related (+)-TAN1251C have attracted significant interests as synthetic targets, with more than ten reports of total synthesis of 1.2,3 The first enantioselective synthesis reported by Snider was biomimetic,3a which suggested the potential biosynthetic origin of 1 from l-tyrosine (Figure 1) via the spirocyclic pyrrolidine 2. The most recent synthesis from Gaunt is also biomimetic, with a significantly shortened route to 2 using a photocatalytic olefin hydroaminoalkylation step.3r However, all biomimetic syntheses suffer from low overall yield (<10%), mainly due to steps from 2 to 1.2 The aldol condensation established by Snider and used by others to form the C9-C3’ bond in 2 is <40% yield due to the formation of undesirable regioisomers and diastereomers.3a,b,d,g,r In the next step, chemical reduction of C4’ ketone gives only the equatorial isomer, which must be inverted by the Mitsunobu reaction with concomitant phosphate addition with yield of ~ 30%, as demonstrated by Sorensen.3b,g,j,o,r The low yields of these synthetic steps prompted us to investigate the biosynthesis of 1 and uncover how nature catalyzes these challenging transformations.
Figure 1.
Structures of (−)-FR901483 1 and related (+)-TAN1251C.
Based on Snider’s proposal, we hypothesize biosynthesis of 1 starts with the condensation of two l-tyrosines by a nonribosomal peptide synthetase (NRPS) to yield (S, S)-dityrosyl-piperazine 3, a precursor that is also formed in the biosynthesis of herquline B.4 Oxidative aza-spiro annulation, which results in formation of the N10-C1’ bond and cleavage of the C9-N10’ bond, forms the ketoaldehyde 2. Regio- and stereoselective aldol condensation would form the C9-C3’ bond, followed by C4’ reduction to the axial alcohol and phosphorylation to give 1.
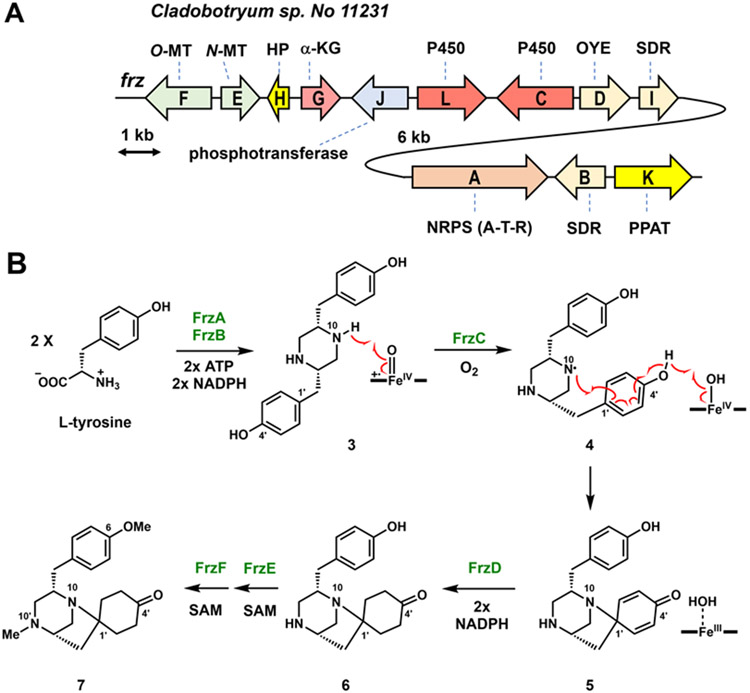
Given the requirement to phosphorylate C4’, the phosphotransferase PsiK5 from the psilocybin biosynthetic pathway was used as a query to search the sequenced genome of Cladobotryum sp. No 11231, and hits were refined by searching for a nearby NRPS-encoding gene. We identified the frz cluster shown in Figure 2A to be the best candidate. The cluster contains 12 genes (frzA-frzL) of which the encoded protein functions are consistent with the structure of 1, including a single-module NRPS with a terminal reductase domain (FrzA), two short-chain dehydrogenase/reductases (SDRs, FrzB and FrzI), two P450 monooxygenases (FrzC and FrzL), two methyltransferases (FrzE and FrzF), an old-yellow enzyme like ene-reductase (OYE, FrzD), a non-heme, iron and α-ketoglutarate dependent oxygenase (αKG, FrzG), a phosphotransferase (FrzJ), a hypothetic protein (HP, FrzH) and a phosphoribosylpyrophosphate amidotransferase (PPAT, FrzK).
Figure 2.
Characterization of FR901483 biosynthesis. A) The frz cluster encodes a NRPS (A-T-R, A, adenylation, T, thiolation; R, reductase) FrzA and tailoring enzymes; B) Biosynthesis of 7 from l-Tyr.
The heterologous host Aspergillus nidulans A1145 ΔEMΔST was chosen for stepwise reconstitution.6 The NRPS FrzA and SDR FrzB show 58% and 64% sequence identities to HqlA and HqlB, respectively, which produce the dityrosyl piperazine (3) from two l-tyrosines in herquline B biosynthesis.4 Coexpression of FrzA and FrzB produced 3 (MW 298) with a titer of 10 mg/L (Figure S2, Table S5, Figures S12-13). In the hql pathway, the P450 HqlC performs aryl coupling between C3 and C3’ to form the constrained 12-membered ring found in herquline.4 The P450 FrzC shows 45% sequence identity to HqlC (Figure S3) and is hence a candidate for oxidative transformation of 3. When coexpressed with FrzAB, a new metabolite 5 (MW 296) was produced (Figure S2). NMR and HRMS characterization revealed structure of 5, which contains a new N10-C1’ bond that morphs the piperazine 3 into a 1,4-diazabicyclo[3.2.1]octane spiro-fused to a 2,5-cyclohexadienone (Table S6, Figures S14-18). The newly installed N10-C1’ bond and the quaternary C1’ in 5 are preserved in the final product 1. Several mechanisms can be proposed for FrzC, depending on the site of the first hydrogen abstraction (Figure S4). One possible mechanism is shown in Figure 2B, and is based on the computationally predicted mechanism of GsfF that generates the oxa-spiro core of griseofulvin.7 We propose that FrzC first catalyzes homolysis of the N-H bond with the ferryl-oxo Compound I to generate the N10 radical 4. This is immediately followed by an O-H abstraction by Compound II to give the phenolic radical which can be delocalized to C1’. Radical coupling between N10• and C1’• then forms 5 (Figure 2). While both catalyze oxidative intramolecular cyclization of 3, the difference in regioselectivities between HqlC (C3-C3’ coupling) and FrzC (N10-C1’ coupling) showcases the versatility of P450 enzymes in generating structural complexity early in the respective biosynthetic pathways.
Additional frz enzymes were then coexpressed with FrzA-C to determine the subsequent biosynthetic steps. The dienone portion of 5 is reduced to cyclohexanone in the proposed intermediate 2. The mostly plausible candidate is FrzD, which encodes for flavin-dependent OYE that catalyzes reduction of α,β-unsaturated carbonyls. Coexpression of FrzA-D in A. nidulans led to formation of 6 (MW 300) (Figure S2, trace iv). However, the titer of 6 was unexpectedly low, which prevented isolation and structural characterization. To obtain a downstream product, we coexpressed FrzE and FrzF, which are predicted to be N-MT and O-MT, respectively. This led to the emergence of 7 (MW 328), which was structurally confirmed to contain the reduced cyclohexanone, and methylations at both the C4 phenolic oxygen and the C10’ amine (Figure 2, Table S7, Figures S19-23). We then expressed and purified recombinant versions of FrzD, FrzE and FrzF (Figure S5). When 1 μM of FrzD was added to 0.1 mM of 5 in the presence of 0.8 mM NADPH, near complete conversion of 5 to 6 was observed (Figure S6). Further addition of 1 μM of both MTs and 0.4 mM SAM resulted in the near complete conversion of 6 to 7 (Figure S6). The order of the two MTs appear to be interchangeable, as adding either MT alone in the above assay resulted in formation of singly methylated products (Figure S7). Collectively, the reconstitution studies so far established a biosynthetic pathway to the aza-spiro intermediate 7 from two l-Tyr residues. The only reported natural products with the same core structure as 7 are TAN-1251 series compounds (Figure 1) isolated from Penicillium thomii RA-89.8 In these compounds, the piperazine portion is further dehydrogenated, while O-methylation is replaced with O-prenylation.
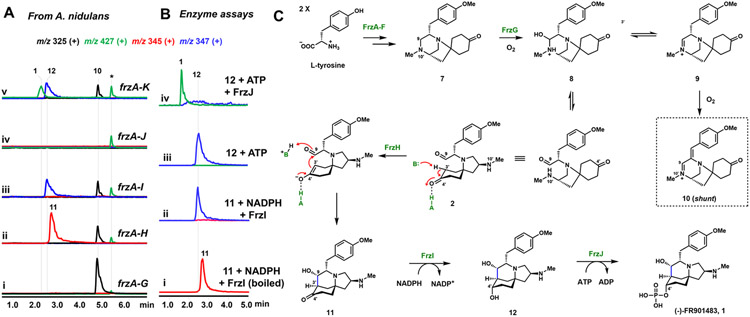
We next aimed to complete the pathway from 7 to 1. Based on the structure of the proposed biosynthetic intermediate 2, it is evident that cleavage of the C9-N10’ bond in 7 must take place, possibly through an hemiaminal intermediate such as 8 or the dehydrated imine 9 (Figure 3C). Two remaining enzyme candidates in the frz cluster, the P450 FrzL or the αKG FrzG, could catalyze hydroxylation of C9 in 7. FrzG shares 50% identity with AusE,9 which catalyzes multi-step oxidations in austinol biosynthesis; while FrzL shares 36% identity with FtmE,10 which performs oxidative C-N bond formation in fumitremorgin biosynthesis. Only coexpression of αKG FrzG with FrzA-F led to formation of a new product 10 (MW 325) (Figure 3A, trace i). Compared to 7 which does not have significant UV absorption above 280 nm, 10 has a strong λmax of 381 nm, indicating presence of extended π-conjugation. Compound 10 was very difficult to isolate in stable form, and 5 mg was recovered from 8 L of solid culture. HRMS and NMR characterization led to the determination of the structure to be a conjugated iminium (Figure 3C, Table S8, Figures S24-28). In addition to the expected oxidation of C9, an additional dehydrogenation between C7 and C8 occurred to give 10, which is likely a shunt product. The accumulation of 10 indicates the role of FrzG in hydroxylation of C9 in 7 to give the hemiaminal 8, which is in equilibrium with both the amino aldehyde 2 and the iminium 9. We propose that in the absence of a downstream enzyme that can advance 2 in the biosynthetic route, 10 is formed as the thermodynamic product through nonenzymatic oxidation of 9. The next biosynthetic step is most likely the intramolecular aldol condensation that is leveraged in the biomimetic syntheses.
Figure 3.
Reconstitution of the biosynthesis of 1. (A) LC-MS analysis of products profiles of heterologous expression of different combinations of frz genes; (B) LC-MS analysis of in vitro assay from 11 to 1; (C) Proposed pathway of 7 to 1. Note: Peak labeled with * is identified by selected ion monitoring but is unrelated to the frz pathway.
Of the remaining five enzymes FrzH-L, there is no candidate that shows sequence similarity to known aldolases. Analysis of hypothetical protein FrzH using I-TASSER server11 showed it has 21% sequence identity to Pseudomonas putida ketosteroid isomerase (KSI, Genbank accession AAA64437.1) (Figure S8), which catalyzes olefin isomerization of 3-oxo-Δ5 ketosteroids to the Δ4-enone isomers in steroid biosynthesis.12 The mechanism involves formation of C3 dienolate which is stabilized by nearby aspartic acid and tyrosine. Although the sequence identity of FrzH to KSI is low, it nevertheless hints that FrzH may catalyze the aldol reaction via the enolate intermediate (Figure 3C). To test this hypothesis, we coexpressed FrzH in the strain that produced 10. Metabolite analysis showed the emergence of 11 (MW 344) with decrease of 10 (Figure 3A, trace ii). Characterization of 11 showed it is the aza-tricyclic product with the newly formed C9-C3’ bond (Table S9, Figures S29-33), thereby establishing FrzH to be the aldolase. To complete the biosynthesis of dephospho-(−)-FR901483 12, we coexpressed the SDR FrzI in the cluster. The heterologous strain produced a compound with the same MW 346 as 12 (3 mg/L) (Figure 3A, trace iii). Isolation and characterization of the compound led to full confirmation of the structure (Table S10, Figures S34-38). The activity of FrzI was reconstituted in vitro using recombinant protein and NADPH (Figure 3B, trace i and ii). Together, FrzH and FrzI catalyze the two-step conversion of 2 to 12, which are the most challenging synthetic steps in the biomimetic syntheses.
The final step in biosynthesis of 1 is phosphorylation of C4’-OH of 12 with phosphate (Figure 3C). This step is critical for biological activity as 12 is completely inactive.1 The enzyme FrzJ shares 20% identity with PsiK, which catalyzes the C4-OH phosphorylation step in psilocybin biosynthesis.5 We directly performed an in vitro assay using purified FrzJ, ATP and MgCl2, and observed near complete transformation of 12 into 1 (Figure 3B, traces iii and iv). The identity of enzymatically prepared 1 was confirmed by comparison to authentic standard of (−)-FR901483, HRMS, and full NMR characterization (Table S11, Figures S39-43).
Lastly, we attempted to recapitulate heterologous biosynthesis of 1 by coexpression of FrzA-J in A. nidulans. Surprisingly, while FrzA-I produced 3 mg/L of 12, coexpression of FrzJ led to no detectable amounts of either 1 or 12 (Figure 3A, trace iv). This led us to propose that the potent inhibitory nature of 1 is toxic to the host, which leads to either degradation of 1, or transcriptional repression of the frz genes. 1 was proposed to inhibit purine nucleotide biosynthesis, although no biological target of 1 has been validated.1,13 The frzK gene encodes the only copy of PPAT in the producing host. PPAT catalyzes the first step of de novo purine biosynthesis in which 5-phosphoribosyl-1-pyrophosphate (PRPP) is converted to 5-phosphoribosyl-1-amine (PRA) (Figure S9).14 PPAT has been targeted for disruption of DNA synthesis, and synthetic small molecule inhibitors have been found.15 Based on the colocalization of self-resistant enzymes (SREs) with biosynthetic gene clusters,16 the presence of FrzK raises the intriguing possibility that PPAT could be the target of 1, with FrzK serving as a resistant housekeeping PPAT to protect the host. When FrzA-K were coexpressed, 12 reemerged along with the accumulation of 1 at a titer of 1 mg/L (Figure 3A, trace v). Our complete reconstitution of 1 only in the presence of FrzK reveals the possible biological target of 1 as PPAT, although this hypothesis requires full biochemical validation.
The only unassigned enzyme in the cluster is the second P450 FrzL. No clear change in metabolite profile was observed when this enzyme was coexpressed with the pathway that produced 1 (Figure S10). In conclusion, we reconstituted the biosynthesis of 1 using 11 enzymes from the frz BGC, and uncovered the biosynthetic logic to be consistent with those proposed in the biomimetic syntheses. Several enzymes that catalyze synthetic challenging steps are found, and can serve as useful biocatalytic tools to achieve most robust synthesis of 1 and derivatives.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NIH R35GM118056 to YT and JSPS 19KK0150.
Footnotes
Supporting Information
Experimental details, spectroscopic and computational data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no conflict of interest.
REFERENCES
- (1).Sakamoto K; Tsujii E; Abe F; Nakanishi T; Yamashita M; Shigematsu N; Izumi S; Okuhara M Fukuda T FR901483, a novel immunosuppressant isolated from Cladobotryum sp. No. 11231. Taxonomy of the producing organism, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot 1996, 49, 37–44. [DOI] [PubMed] [Google Scholar]
- (2).Ruan ZW; Li C; Shen DF; Huang S; Hong R FR901483: Synthetic efficiency remains a challenge. Synthesis. 2019, 51, 2237–2251. [Google Scholar]
- (3).(a) Snider BB; Lin H Total synthesis of (−)-FR901483. J. Am. Chem. Soc. 1999, 121, 7778–7786. [Google Scholar]; (b) Scheffler G; Seike H; Sorensen EJ An enantiospecific synthesis of the potent immunosuppressant FR901483. Angew. Chem. Int. Ed. Engl. 2000, 39, 4593–4596. [DOI] [PubMed] [Google Scholar]; (c) Suzuki H; Yamazaki N; Kibayashi C Synthesis of the azatricyclic core of FR901483 by bridgehead vinylation via an anti-Bredt iminium ion. Tetrahedron lett. 2001, 42, 3013–3015. [Google Scholar]; (d) Ousmer M; Braun NA; Ciufolini MA Total synthesis of FR901483. Org. Lett. 2001, 3, 765–767. [DOI] [PubMed] [Google Scholar]; (e) Maeng JH; Funk RL Total synthesis of the immunosuppressant FR901483 via an amidoacrolein cycloaddition. Org. Lett. 2001, 3, 1125–1128. [DOI] [PubMed] [Google Scholar]; (f) Brummond KM; Lu J Tandem cationic aza-Cope rearrangement–Mannich cyclization approach to the core structure of FR901483 via a bridgehead iminium ion. Org. Lett. 2001, 3, 1347–1349. [DOI] [PubMed] [Google Scholar]; (g) Ousmer M; Braun NA; Bavoux C; Perrin M; Ciufolini MA Total synthesis of tricyclic azaspirane derivatives of tyrosine: FR901483 and TAN1251C. J. Am. Chem. Soc. 2001, 123, 7534–7538. [DOI] [PubMed] [Google Scholar]; (h) Kan T; Fujimoto T; Ieda S; Asoh Y; Kitaoka H; Fukuyama T Stereocontrolled total synthesis of potent immunosuppressant FR901483. Org. Lett. 2004, 6, 2729–2731. [DOI] [PubMed] [Google Scholar]; (i) Brummond KM; Hong SP A formal total synthesis of (−)-FR901483, using a tandem cationic aza-Cope rearrangement/Mannich cyclization approach. J. Org. Chem. 2005, 70, 907–916. [DOI] [PubMed] [Google Scholar]; (j) Kropf JE; Meigh IC; Bebbington MW; Weinreb SM Studies on a total synthesis of the microbial immunosuppresive agent FR901483. J. Org. Chem. 2006, 71, 2046–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Diaba F; Ricou E; Solé D; Teixidó E; Valls N; Bonjoch J Studies in the FR901483 tricyclic skeleton synthesis and a new approach to the perhydropyrrolo[2,1-i]indole ring system. Arkivoc. 2007, iv, 320–330. [Google Scholar]; (l) Seike H; Sorensen EJ; A synthesis of the tricyclic core structure of FR901483 featuring an Ugi four-component coupling and a remarkably selective elimination reaction. Synlett. 2008. 5, 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]; (m) Carson C; Kerr MA Total synthesis of FR901483. Org. Lett. 2009, 8, 777–779. [DOI] [PubMed] [Google Scholar]; (n) Ma AJ; Tu YQ; Peng JB; Dou QY; Hou SH; Zhang FM; Wang SH Total synthesis of (−)-FR901483. Org. Lett. 2012, 12, 3604–3607. [DOI] [PubMed] [Google Scholar]; (o) Huo HH; Zhang HK; Xia XE; Huang PQ A formal enantioselective total synthesis of (−)-FR901483. Org. Lett. 2012, 12, 4834–4837. [DOI] [PubMed] [Google Scholar]; (p) Perreault S; Rovis T Enantioselective synthesis of the tricyclic core of FR901483 featuring a Rhodium-catalyzed [2+2+2] cycloaddition. Synthesis. 2013, 45, 0719–0728. [DOI] [PMC free article] [PubMed] [Google Scholar]; (q) Huo HH; Xia XE; Zhang HK; Huang PQ Enantioselective total syntheses of (−)-FR901483 and (+)-8-epi-FR901483. J. Org. Chem. 2013, 78, 455–465. [DOI] [PubMed] [Google Scholar]; (r) Reich D; Trowbridge A; Gaunt MJ Rapid Syntheses of (−)-FR901483 and (+)-TAN1251C Enabled by Complexity-Generating Photocatalytic Olefin Hydroaminoalkylation. Angew. Chem. Int. Ed. Engl. 2020, 59, 2256–2261. [DOI] [PubMed] [Google Scholar]
- (4).Yu X; Liu F; Zou Y; Tang MC; Hang L; Houk KN; Tang Y Biosynthesis of strained piperazine alkaloids: uncovering the concise pathway of herquline A. J. Am. Chem. Soc. 2016, 138, 13529–13532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Fricke J; Blei F; Hoffmeister D Enzymatic Synthesis of Psilocybin. Angew. Chem. Int. Ed. Engl. 2017, 56, 12352–12355. [DOI] [PubMed] [Google Scholar]
- (6).Zhang Z; Jamieson CS; Zhao YL; Li D; Ohashi M; Houk KN; Tang Y Enzyme-catalyzed inverse-electron demand Diels-Alder reaction in the biosynthesis of antifungal ilicicolin H. J. Am. Chem. Soc. 2019, 141, 5659–5663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Grandner JM; Cacho RA; Tang Y; Houk KN Mechanism of the P450-catalyzed oxidative cyclization in the biosynthesis of Griseofulvin. ACS Catal. 2016, 6, 4506–4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Shirafuji H; Tsubotani S; Ishimaru T; Harada S TAN-1251 compounds and their production from Penicillium thomii. U.S. Patent 5,310,741, 1994. [Google Scholar]
- (9).Matsuda Y; Awakawa T; Wakimoto T; Abe I Spiro-ring formation is catalyzed by a multifunctional dioxygenase in austinol biosynthesis. J. Am. Chem. Soc. 2013, 135, 10962–10965. [DOI] [PubMed] [Google Scholar]
- (10).Kato N; Suzuki H; Takagi H; Asami Y; Kakeya H; Uramoto M; Usui T; Takahashi S; Sugimoto Y; Osada H Identification of cytochrome P450s required for fumitremorgin biosynthesis in Aspergillus fumigatus. Chembiochem, 2009, 10, 920–928. [DOI] [PubMed] [Google Scholar]
- (11).Roy A; Kucukural A; Zhang Y I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc, 2010, 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).(a) Fried SD; Bagchi S; Boxer SG Extreme electric fields power catalysis in the active site of ketosteroid isomerase. Science, 2014, 346, 1510–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pollack RM Enzymatic mechanisms for catalysis of enolization: ketosteroid isomerase. Bioorg Chem, 2004, 32, 341–353. [DOI] [PubMed] [Google Scholar]
- (13).Atta-Ur-Rahman. Studies in natural products chemistry: Bioactive natural product (part L). Synthesis of immunosuppressant FR901483 and biogenetically related TAN 1251 alkaloids. Ohjoch J; Diaba F Elsevier, 2005; pp 3–60. [Google Scholar]
- (14).(a) Smith JL; Zaluzec EJ; Wery JP; Niu L; Switzer RL; Zalkin H; Satow Y Structure of the allosteric regulatory enzyme of purine biosynthesis. Science, 1994, 264, 1427–1433. [DOI] [PubMed] [Google Scholar]; (b) Smith JL Glutamine PRPP amidotransferase: snapshots of an enzyme in action. Curr. Opin. Struct. Biol. 1998, 8, 686–694. [DOI] [PubMed] [Google Scholar]
- (15).Kamal MA; Christopherson RI Accumulation of 5-phosphoribosyl-1-pyrophosphate in human CCRF-CEM leukaemia cells treated with antifolates. Int J. Biochem. Cell. Biol. 2004, 36, 545–551. [DOI] [PubMed] [Google Scholar]
- (16).Yan Y; Liu N; Tang Y Recent developments in self-resistance gene directed natural product discovery. Nat. Prod. Rep. 2020, doi: 10.1039/c9np00050j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.