Abstract
Hirsutellones are fungal natural products containing a macrocyclic para-cyclophane connected to a decahydrofluorene ring system. We have elucidated the biosynthetic pathway for pyrrocidine B (3) and GKK1032 A2 (4). Two small hypothetical proteins, an oxidoreductase and a lipocalin-like protein, function cooperatively in the oxidative cyclization of the cyclophane; while an additional hypothetical protein in the pyrrocidine pathway catalyzes the exo-specific cycloaddition to form the cis-fused decahydrofluorene.
Graphical Abstract
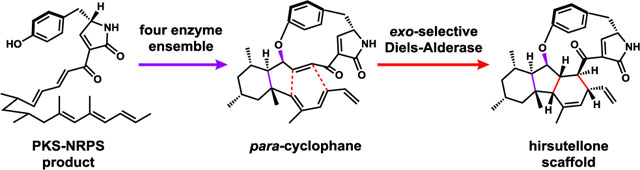
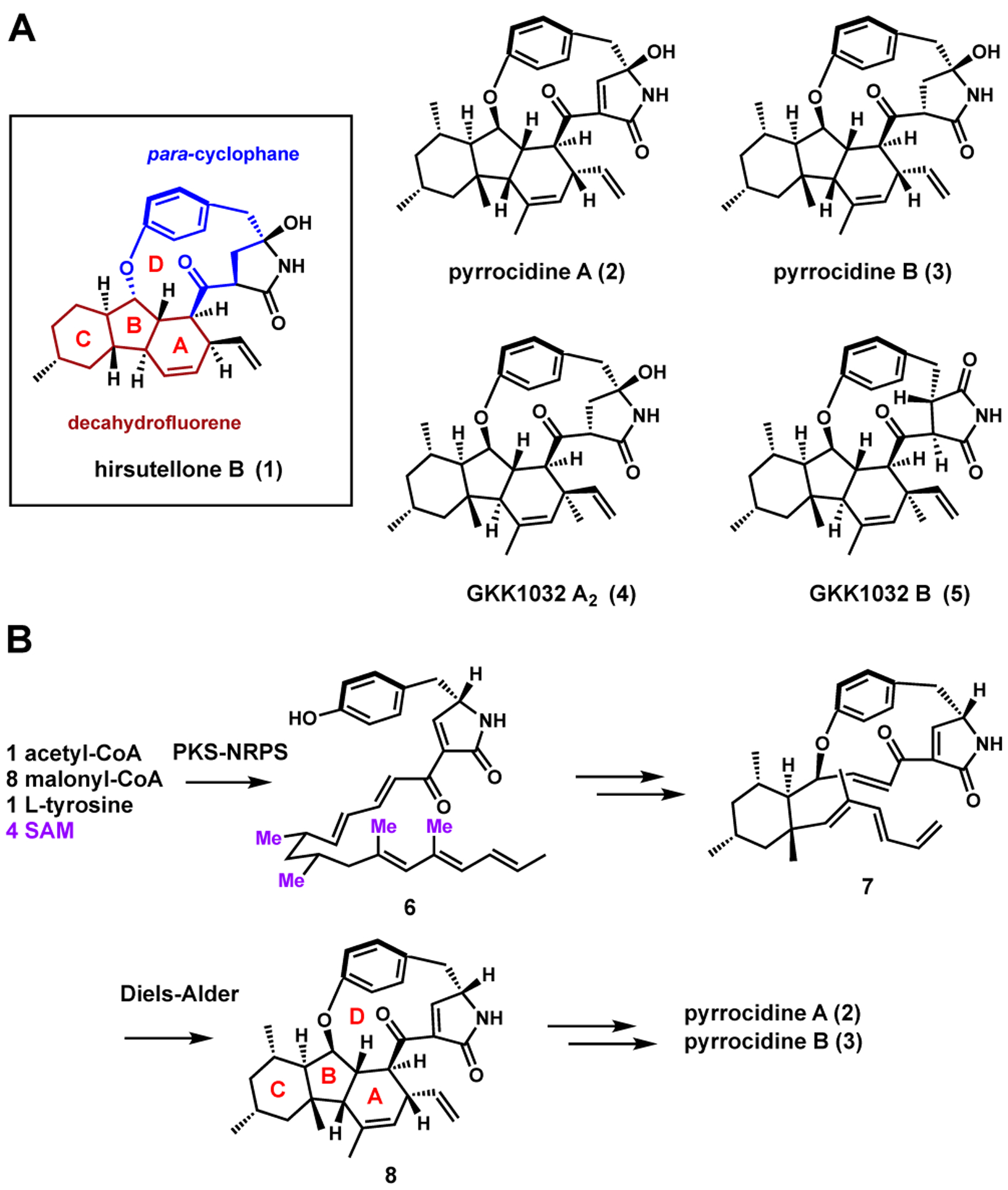
Members of the hirsutellone family of natural products are structurally complex and possess diverse biological activities.1,2 Hirsutellone B (1) from Hirsutella sp. displays submicromolar anti-tubercular activities.1,3 Pyrrocidine A (2) and B (3) isolated from Acremonium zeae showed potent antibiotic activities towards gram-negative bacteria.4,5 GKK1032A2 (4) and GKK1032B (5) from Penicillium sp. have antitumor activities.6–8 The unique structural feature of this family is the highly strained 12 or 13-membered para-cyclophane ring D (Figure 1), which is connected via an aryl-ether linkage to a decahydrofluorene core (rings A, B and C). Because of this feature, hirsutellones have attracted significant interest from synthetic chemists. Total syntheses of hirsutellone B were reported by Nicolaou9 and Uchiro.10,11 Synthesis of the bioactive decahydrofluorene core has been accomplished by many approaches, including a biomimetic electrophilic cyclization strategy used by Nay.12–15
Figure 1.
Hirsutellone-type natural products. (A) Representative compounds; (B) proposed biosynthesis of pyrrocidine.10,20
Biosynthetically, 2 and 3 are derived from the 3-pyrrolin-2-one intermediate 6 (Figure 1B), which is synthesized by a polyketide synthase-nonribosomal peptide synthetase (PKS-NRPS) from the producing host as suggested by Oikawa.16 Compound 6 is proposed to cyclize into the para-cyclophane 7 and form ring C through either electrophilic cyclization or a radical mechanism, connecting the tyrosine phenol and the acyclic polyketide chain, as shown by Nay using18O-labeled tyrosine and characterization of labeled pyrrocidine products.17 A subsequent Diels-Alder cycloaddition is proposed to form the A and B rings in 8.9 Based on the Nicolaou synthesis, formation of the cis-fused ring system in 2 and 3 from the Diels-Alder reaction is highly unfavorable, which implicates the involvement of an exo-specific pericyclase in this pathway. Fungal biosynthetic gene clusters containing PKS-NRPSs have been shown to produce natural products of diverse structural features.18,19 However, no biosynthetic gene cluster (BGC) responsible for biosynthesis of any hirsutellone has been reported. As a result, the enzymatic bases of both cyclophane and decahydrofluorene formation have remained obscure.
We sequenced the genome of 3-producing A. zeae and found three PKS-NRPS encoding BGCs (Figure S1). One of the BGC (pyd) encodes a lipocalin-like protein (LLP, pydB), with ~40% sequence similarity to Diels-Alderases that catalyze cycloaddition reactions in biosynthesis of decalin natural products.21 We reasoned that PydB may be the pericyclase needed in formation of cis-fused decahydrofluorene ring system. The pyd BGC also contains genes encoding PKS-NRPS (pydA) and its enoylreductase partner (ER, pydC), medium-chain dehydrogenase/reductase (MDR, pydE) and an α/β hydrolase (pydG) (Figure 2A). While A. zeae was verified to produce 3 (Figure 2B, NMRs in Figures S41–S48), attempts to perform genetic manipulations in A. zeae were unsuccessful. This prom¥pted us to perform heterologous reconstitution in A. nidulans A1145 ΔEMΔST, of which the emericellamide and sterigmatocystin pathways are inactivated (Figure 2B).22
Figure 2.
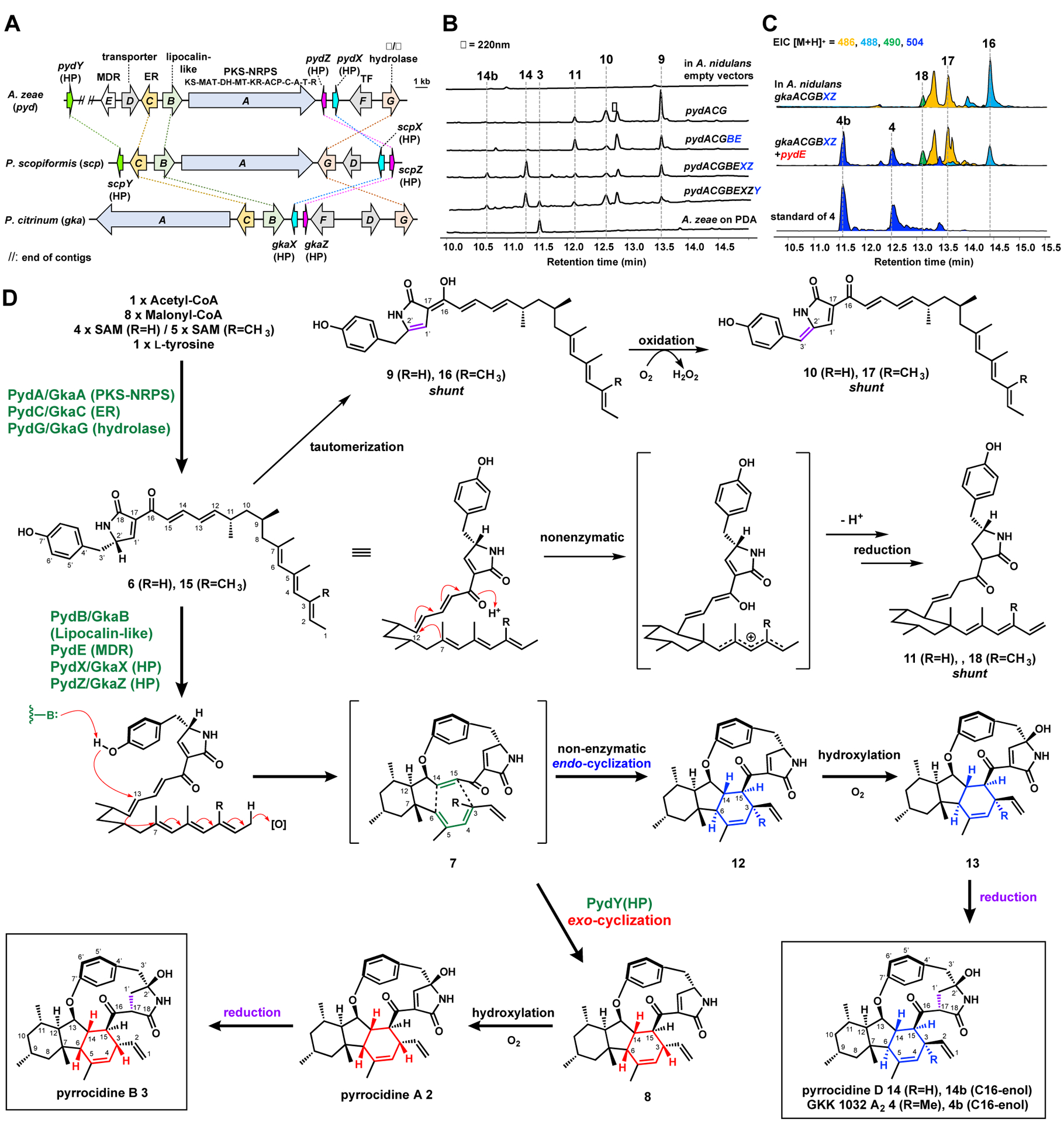
Biosynthesis of hirsutellones. (A) Three homologous clusters discussed in this work. Note: pydY and pydE are at ends of separate contigs. PKS-NRPS domains: KS, ketosynthase; MAT, malonyl-CoA:ACP transacylase; DH, dehydratase; MT, methyltransferase; KR, ketoreductase; ACP, acyl-carrier protein; C, condensation; A, adenylation; T, thiolation; R, reductase. ER, enoylreductase; MDR, medium-chain dehydrogenases/reductases; HP, hypothetical protein. (B) Metabolic profiling upon expressions of pyd genes in A. nidulans. The peak labeled with * is linoleic acid. (C) Reconstitution of gka pathway in A. nidulans led to biosynthesis of 4 and 4b. (D) Proposed biosynthetic pathway of 3, 4 and 14.
We first introduced the PKS-NRPS, ER and α/β hydrolase into A. nidulans to produce the proposed 6. PydA and PydB would synthesize the polyketide-tyrosyl acyl thioester product, which can be reductively offloaded by the terminal reductase (R) domain in PydA. The hydrolase PydG is required to catalyze the subsequent Knoevenagel condensation that affords the 3-pyrrolin-2-one ring.23 Three major products 9–11 with extended π-conjugation were observed from the extract (Figure 2B), with 9 (5 mg/L) having the same MWT 473 as 6, while 10 (5 mg/L) and 11 (1 mg/L) with −2 mu and +2 mu in MWT, respectively. Structure elucidation (Tables S2–S4 and Figures S13–S29) confirmed their relations to 6. 9 is an acyclic polyolefinic compound that contains a 4-pyrrolin-2-one moiety, an enamide that is the enolized and more stable form of 6, and is likely a shunt product in the absence of downstream enzymes (Figure S6).24 The red-shifted 10 contains an additional conjugation at C2’ and C3’, and is the air-oxidized product of 9, as seen in the myceliothermophin pathway.21
The minor product 11 contains a cyclohexane ring that corresponds to ring C in 3. However, examination of the remaining structural features suggests this is also a shunt product. While the triene moiety migrated as required for the cycloaddition, the C14-C15 conjugated double bond that serves as the dienophile is isomerized to C13-C14. This shift also precludes oxidative cyclization to form the cyclophane. We propose that cycloisomerization of the polyolefinic portion of 6 can take place nonenzymatically to form the C7-C12 bond, with the C18 carbonyl serving as the electron sink. Deprotonation of the resulting cation forms the migrated triene, followed by ketonization and C17-C1’ enoylreduction by an endogenous enzyme in A. nidulans, as observed in biosynthesis of other 3-pyrrolin-2-one compounds such as UCS-1025A.25
To advance the biosynthesis from 6 to 3, we expressed additional enzymes in the A. nidulans host. However, coexpression with the MDR PydE and the putative pericyclase PydB did not yield any new compounds, while 9–11 remained (Figure 2B). With no other gene candidates as leads, we searched for homologous gene clusters in other fungal genomes for clues. The two top hit clusters from Phialocephala scopiformis and Penicillium citrinum DSM 1997 shared similar genes except for the MDR (Figure 2A). Although these organisms have not been reported to produce hirsutellones, the PKS-NRPSs in these clusters share 60% identity to PydA.
On the fringes of the P. scopiformis scp cluster are two small predicted genes scpX and scpZ coding for hypothetical proteins (HPs) of 124 and 159 residues, respectively. These two genes had not been annotated by gene prediction algorithms in the other clusters. Using ScpX and ScpZ as queries, we identified conserved sequences in pyd and gka BGCs with > 70% similarity between ScpX, PydX and GkaX; and also >72% similarity between ScpZ, PydZ and GkaZ. Both PydX and PydZ proteins are predicted to be membrane-bound with multiple transmembrane regions (Figure S8). Furthermore, in the scp cluster, a third, small gene scpY encoding a 186-residue HP was found immediately upstream of the ER-encoding scpC. Using scpY as a lead, we found a highly similar pydY at one end of a different contig in the A. zeae genome (Figure 2A).
We next tested whether coexpression of these HPs can form 3. Starting with the strain that expresses PydABCDE, individual expression of any of the HPs did not lead to new products (Figure S4). However, when PydX and PydZ were coexpressed, we observed decreases in levels of shunt products, with the concomitant appearance of a new pair of interconverting compounds 14 and 14b (6 mg/L) with the same HRMS m/z 490.2965 (C31H40NO4) as 3 (Figure 2B). However, the difference in retention times suggests they are structurally distinct from 3. Purified 14 and 14b were left in deuterated chloroform overnight, which led to the disappearance of 14b and convergence of NMR signals that correspond to 14 (Table S5, Figures S30–S35). A key HMBC signal is that between H13 and the phenyl ring C7’, which established the C-O-C link forming the para-cyclophane. The carbon connectivity gleaned from the COSY and HMBC spectra supported the planar structure of 14 to be identical to that of 3. However, NOESY signals from the A and B rings established the configurations of 14 at C6 and C14 to be trans, compared to the cis found in 3. 14 is therefore a stereoisomer of 3 and is named pyrrocidine D, while 14b is the C16-enol tautomer as determined from NMR of the 14/14b mixture in DMSO-d6 (Table S5 and Figures S36–S40).
Different combinations of PydB (LLP), PydE (MDR), PydX (HP) and PydZ (HP) were introduced into A. nidulans, and only the coexpression of all four proteins can lead to formation of 14. Removing any single gene led to loss of 14, with only 9–11 detected (Figure S5). This suggests the four proteins may work as a complex to transform 6. The proposed mechanism is shown in Figure 2D. First, the ensemble of four proteins catalyzes the oxidative cyclization to form the cycloaddition precursor 7. In this cascade reaction, the acyclic chain of 6 is configured in an inverse S-shape, in which the phenol is positioned near C13, while the fully saturated portion of the chain forms a chair-like conformation aided by the two equatorially substituted methyl groups at C9 and C11. Upon deprotonation, the phenolate attacks the C13 olefin, which triggers the conjugated addition of C12 into the triene at C7 to form ring C. A hydride acceptor, possibly the NAD(P)+ cofactor present in the MDR, may position near C1 to complete the reaction. In the absence of a dedicated pericyclase, 7 can undergo nonenzymatic cycloaddition through the more favored endo transition state to give the trans-fused 12, as demonstrated in synthetic approaches to the decahydrofluorene fragment.9 From 12, hydroxylation of C2’ to 13 is nonenzymatic, while ene-reduction to 14 can be catalyzed by endogenous reductases in the host.25
The formation of 14 suggests the LLP PydB is not responsible for the exo selective cycloaddition required to form 3. To test whether the third HP PydY is the pericyclase that catalyzes this unfavorable cycloaddition, we coexpressed pydY in the strain that produced 14. A new compound with the same retention time and HRMS as the authentic 3 obtained from A. zeae emerged from the strain (Figure 2B). This compound was purified (0.4 mg/L), characterized by NMR (Table S6, Figures S41–S48), and confirmed to be the cis-fused decahydrofluorene pyrrocidine B (3). The NOSEY correlations from 7-Me to H-6 and H-14, and from H-1 to H-12 were clearly observed as in the original isolation of 3.4 Therefore, this result directly implicates the involvement of PydY in formation of 8, which is then nonenzymatically oxidized to pyrrocidine A (2), and reduced by endogenous reductase to 3.25
Reconstitution of 3 demonstrates the unexpected role of four small proteins (B, X, Y and Z), as well as the MDR PydE, in transforming the acyclic 6 into 8 or 14. Attempts to reconstitute the activities of these enzymes were not successful, hampered by the membrane-bound nature of PydX and PydZ, and inaccessible substrate 6. We propose one possible model in which the four proteins function synergistically to form the cyclophane. In this model, the LLP PydB and the membrane-bound PydX and PydZ are lipid binding proteins that can sequester and mold 6 into the inverse S-shape. Binding of the MDR PydE to the complex triggers the cascade oxidative cyclization and formation of 7, which can be subsequently captured by PydY to form 8. Although coexpression of PydY is not required for activity of the other four proteins, we cannot exclude PydY is part of such proposed multiprotein complex in formation of 8.
The colocalization of homologs of these HPs with PKS-NRPS containing BGCs indicates the potential to biosynthesize other members of the hirsutellone-family. To test this proposal, we reconstituted the gka cluster found in P. citrinum DSM 1997 (Figure 2A). While this strain has not been reported to produce hirsutellones, there is a report of an endophytic P. citrinum strain from which 4 had been isolated.8 The gka cluster lacks a homolog to PydY, which is consistent with the trans-fused decahydrofluorene portion of 4. The gka cluster also lacks a homolog to the MDR PydE, which is intriguing given our proposed role of PydE as a hydride acceptor. When the gka genes, which include gkaA (PKS-NRPS), gkaC (ER), gkaG (hydrolase), gkaB (LLP), gkaX and gkaZ (HPs), were introduced in A. nidulans, we observed the formation of compounds with mass and UV features consistent with shunt products 16–18 (Figures 2C, 2D and S12). We attribute the failed reconstitution of 4 to the absence of an MDR, which may be unclustered or shared with other pathways in the P. citrinum host (Figure S10). To test if the pyrrocidine MDR can complete the GKK1032 pathway, we coexpressed PydE with the gka genes. Gratifyingly, a new pair of interconverting compounds 4 and 4b emerged from strain (2.0 mg/L), which upon structural characterization (Table S7, Figures S49–S54), were confirmed to be GKK1032 A2 and its enol tautomer, respectively.
In conclusion, we identified and reconstituted the biosynthetic pathways of pyrrocidine and GKK1032, two biologically active hirsutellone cyclophanes. Our worked showed that a set of small proteins are essential to facilitate the oxidative cyclization to form the para-cyclophane, and the subsequent cycloaddition to form the decahydrofluorene. The establishment of the elusive hirsutellone biosynthetic pathways enables genome mining of other members of this family, as well as combinatorial biosynthesis of new compounds.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the NIH 1R35GM118056 to Y.T. Chemical characterization studies were supported by shared instrumentation grants from the NSF (CHE-1048804) and the NIH NCRR (S10RR025631). We thank Shushan Gao for assistance with NMR analysis.
Footnotes
Supporting Information
Experimental details, spectroscopic data. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- (1).Li X-W; Ear A; Nay B Hirsutellones and beyond: Figuring out the Biological and Synthetic Logics toward Chemical Complexity in Fungal PKS-NRPS Compounds. Nat. Prod. Rep 2013, 30 (6), 765. [DOI] [PubMed] [Google Scholar]
- (2).Gulder T; Baran SP Strained Cyclophane Natural Products: Macrocyclization at Its Limits. Nat. Prod. Rep 2012, 29 (8), 899–934. [DOI] [PubMed] [Google Scholar]
- (3).Isaka M; Rugseree N; Maithip P; Kongsaeree P; Prabpai S; Thebtaranonth Y Hirsutellones A–E, Antimycobacterial Alkaloids from the Insect Pathogenic Fungus Hirsutella Nivea BCC 2594. Tetrahedron 2005, 61 (23), 5577–5583. [Google Scholar]
- (4).He H; Yang HY; Bigelis R; Solum EH; Greenstein M; Carter GT Pyrrocidines A and B, New Antibiotics Produced by a Filamentous Fungus. Tetrahedron Lett. 2002, 43 (9), 1633–1636. [Google Scholar]
- (5).Wicklow DT; Poling SM Antimicrobial Activity of Pyrrocidines from Acremonium Zeae Against Endophytes and Pathogens of Maize. Phytopathology 2008, 99 (1), 109–115.. [DOI] [PubMed] [Google Scholar]
- (6).Koizumi F; Hasegawa K; Ando K; Ogawa T; Hara A Jpn. Kokai Tokkyo Koho, JP 2001147574 A2 200109, 2001. [Google Scholar]
- (7).Becker J; Liermann JC; Opatz T; Anke H; Thines E GKK1032A 2, a Secondary Metabolite from Penicillium Sp. IBWF-029–96, Inhibits Conidial Germination in the Rice Blast Fungus Magnaporthe Oryzae. J. Antibiot 2012, 65 (2), 99–102. [DOI] [PubMed] [Google Scholar]
- (8).Qader M; Kumar NS; Jayasinghe L; Fujimoto Y Production of antitumor antibiotic GKK1032B by Penicillium citrinum, an endophytic fungus isolated from Garcinia mangostana Fruits. Med. Aromat. Plants 2015, 5, 225. [Google Scholar]
- (9).Nicolaou KC; Sarlah D; Wu TR; Zhan W Total Synthesis of Hirsutellone B. Angew. Chem. Int. Ed 2009, 48 (37), 6870–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Uchiro H; Kato R; Arai Y; Hasegawa M; Kobayakawa Y Total Synthesis of Hirsutellone B via Ullmann-Type Direct 13-Membered Macrocyclization. Org. Lett 2011, 13 (23), 6268–6271. [DOI] [PubMed] [Google Scholar]
- (11).Sugata H; Inagaki K; Ode T; Hayakawa T; Karoji Y; Baba M; Kato R; Hasegawa D; Tsubogo T; Uchiro H Total Synthesis of GKK1032A2 via Direct 13-Membered Macrocyclization Using a Nucleophilic Aromatic Substitution of an (η6-Arene)Chromium Complex. Chem. Asian J 2017, 12 (6), 628–632. [DOI] [PubMed] [Google Scholar]
- (12).Li X-W; Ear A; Roger L; Riache N; Deville A; Nay B Bio-Inspired Formal Synthesis of Hirsutellones A–C Featuring an Electrophilic Cyclization Triggered by Remote Lewis Acid-Activation. Chem. Eur. J 2013, 19 (48), 16389–16393. [DOI] [PubMed] [Google Scholar]
- (13).Tanaka R; Ohishi K; Takanashi N; Nagano T; Suizu H; Suzuki T; Kobayashi S Synthetic Study of Pyrrocidines: First Entry to the Decahydrofluorene Core of Pyrrocidines. Org. Lett 2012, 14 (18), 4886–4889. [DOI] [PubMed] [Google Scholar]
- (14).Tilley SD; Reber KP; Sorensen EJ A Rapid, Asymmetric Synthesis of the Decahydrofluorene Core of the Hirsutellones. Org. Lett 2009, 11 (3), 701–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Uchiro H; Kato R; Sakuma Y; Takagi Y; Arai Y; Hasegawa D Synthetic Studies of GKK1032s: The Asymmetric Synthesis of the Decahydrofluorene Skeleton via a Novel Cyclization of Silyl Enol Ether and Sequential Retro Diels–Alder and Intramolecular Diels–Alder Reactions. Tetrahedron Lett. 2011, 52 (47), 6242–6245. [Google Scholar]
- (16).Oikawa H Biosynthesis of Structurally Unique Fungal Metabolite GKK1032A2: Indication of Novel Carbocyclic Formation Mechanism in Polyketide Biosynthesis. J. Org. Chem 2003, 68 (9), 3552–3557. [DOI] [PubMed] [Google Scholar]
- (17).Ear A; Amand S; Blanchard F; Blond A; Dubost L; Buisson D; Nay B Direct Biosynthetic Cyclization of a Distorted Paracyclophane Highlighted by Double Isotopic Labelling of L-Tyrosine. Org. Biomol. Chem 2015, 13 (12), 3662–3666. [DOI] [PubMed] [Google Scholar]
- (18).Boettger D; Hertweck C Molecular Diversity Sculpted by Fungal PKS–NRPS Hybrids. ChemBioChem 2013, 14 (1), 28–42. [DOI] [PubMed] [Google Scholar]
- (19).Minami A; Ugai T; Ozaki T; Oikawa H Predicting the Chemical Space of Fungal Polyketides by Phylogeny-Based Bioinformatics Analysis of Polyketide Synthase-Nonribosomal Peptide Synthetase and Its Modification Enzymes. Sci. Rep 2020, 10 (1), 13556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Tang M-C; Zou Y; Watanabe K; Walsh CT; Tang Y Oxidative Cyclization in Natural Product Biosynthesis. Chem. Rev 2017, 117 (8), 5226–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Li L; Yu P; Tang M-C; Zou Y; Gao S-S; Hung Y-S; Zhao M; Watanabe K; Houk KN; Tang Y Biochemical Characterization of a Eukaryotic Decalin-Forming Diels–Alderase. J. Am. Chem. Soc 2016, 138 (49), 15837–15840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Liu N; Hung Y-S; Gao S-S; Hang L; Zou Y; Chooi Y-H; Tang Y Identification and Heterologous Production of a Benzoyl-Primed Tricarboxylic Acid Polyketide Intermediate from the Zaragozic Acid A Biosynthetic Pathway. Org. Lett 2017, 19 (13), 3560–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sato M; Dander JE; Sato C; Hung Y-S; Gao S-S; Tang M-C; Hang L; Winter JM; Garg NK; Watanabe K; Tang Y Collaborative Biosynthesis of Maleimide- and Succinimide-Containing Natural Products by Fungal Polyketide Megasynthases. J. Am. Chem. Soc 2017, 139 (15), 5317–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Li H; Wei H; Hu J; Lacey E; Sobolev AN; Stubbs KA; Solomon PS; Chooi Y-H Genomics-Driven Discovery of Phytotoxic Cytochalasans Involved in the Virulence of the Wheat Pathogen Parastagonospora Nodorum. ACS Chem. Biol 2020, 15 (1), 226–233. [DOI] [PubMed] [Google Scholar]
- (25).Li L; Tang M-C; Tang S; Gao S; Soliman S; Hang L; Xu W; Ye T; Watanabe K; Tang Y Genome Mining and Assembly-Line Biosynthesis of the UCS1025A Pyrrolizidinone Family of Fungal Alkaloids. J. Am. Chem. Soc 2018, 140 (6), 2067–2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


