Abstract
The incidence and mortality of papillary thyroid cancer (PTC) have steadily increased. Although conventional therapies are very effective toward differentiated PTC patients, very limited therapeutic options are applicable to those patients with distant metastases. Therefore, better understanding of the molecular biology of metastatic PTC helps identify novel targets and facilitates the development of new therapies. In this study, we first found that testicular orphan receptor 4 (TR4) was significantly increased in PTC tumors spreading to lymph nodes compared to the paired primary tumors. Experimental evidence suggested that TR4 drove PTC progression via promoting its cell invasion and cell migration. Mechanistically, TR4 transcriptionally regulated the expression level of circ-filamin A (FLNA), which competed with matrix metalloproteinase 9 (MMP9) for microRNA (miR)-149-5p binding and led to an increased protein level of MMP9. Interruption assays with various gene manipulations verified that the TR4/circ-FLNA/miR-149-5p/MMP9 signaling axis played a central role in cell invasion and cell migration of PTC cells. Moreover, a xenografted mouse model also confirmed that the TR4/circ-FLNA signal promoted PTC tumor growth. Overall, our study pinpoints the oncogenic role of TR4 in PTC development, and the targeting of TR4/circ-FLNA/miR-149-5p/MMP9 signaling may be an alternative option for metastatic PTC patients.
Keywords: thyroid cancer, TR4, circ-FLNA, cell invasion
Graphical abstract

Very limited therapeutic options are applicable to papillary thyroid cancer (PTC) with distance metastasis. The study of Wang and co-workers defined the role of TR4 in PTC progression through circ-FLNA/miR-149-5p/MMP9 signaling and builds rationale to develop TR4-targeted therapies for better treatment of metastatic PTC.
Introduction
Thyroid cancer (TC) is a common endocrine malignancy worldwide.1,2 According to the global survey in 2012, approximately 300,000 new cases of TC were diagnosed, and 40,000 deaths were reported.3 TC is usually divided into differentiated TC (DTC), medullary TC (MTC), and anaplastic TC (ATC), and papillary TC (PTC) accounts for the majority of DTC. Current treatments such as surgery, radioactive iodide therapy, and hormone-suppressive therapy show promising therapeutic efficacy in patients with PTC. However, about 30% of PTC will develop distant metastases;4,5 lymph node, bone, lung, and liver are desirable organs PTC prefers to invade. For such patients with metastasis, the mortality is rather high due to the worse 5-year survival rate (about 50%) and limited treatment options. Therefore, better understanding of the molecular biology of metastatic PTC will accelerate the development of novel targeted therapies.
Testicular orphan receptor 4 (TR4 or NR2C2) was first cloned from testis and was a nuclear receptor without an identified ligand.6 Early studies have demonstrated that TR4 is a key factor to maintain fertility, bone development, and metabolic homeostasis.7,8 As a transcription factor, activated TR4 is translocated to the nucleus where it binds its recognized DNA element and regulates a bundle of genes. Now increasing evidence suggests that TR4 may also contribute to cancer development.9, 10, 11, 12, 13 For instance, TR4 played a tumor-promoting role in renal cell carcinoma (RCC) by promoting vasculogenic mimicry formation and tumor metastasis.9 In prostate cancer (PCa), evidence suggested that TR4 could suppress PCa initiation via regulating ATM serine/threonine kinase to reduce DNA damages.12 Indeed, in vitro and in vivo studies showed that TR4 could increase PCa invasion/metastasis via its regulation on CCL2, transforming growth factor β receptor 2 (TGFBR2)/Smad3, and stem cell populations.11 However, little is known about the role of TR4 in PTC carcinogenesis and progression.
Circular RNAs (circRNAs) are pre-mRNA back-splicing products of exons and increasingly receive interest in the field of cancer research.14,15 The function of one certain circRNA relies on its regulation on gene expression;16 circRNA can act as a sponge of microRNA (miRNA) or nuclear circRNA or impair the alternative splicing of such pre-mRNAs and cause altered gene expression. The contributions of circRNAs to PTC development have been documented recently. For instance, circ-FOXM1 could promote PTC growth by acting as a sponge of microRNA (miR)-1179 to increase HMGB1 expression.17 Evidence also demonstrated that circ-BACH2 increased PTC cell proliferation, invasion, and migration by preventing miR-13p-5p from binding to the 3′ UTR of LMO4.18 These findings imply that circRNAs indeed play critical roles in PTC development and deserve our further investigations.
In this study, we first found that TR4 was increased in metastatic PTC patients compared to non-metastatic controls (Ctrls). Overexpression of TR4 (oeTR4) increased cell invasion/migration of PTC cells. Mechanistic dissection revealed that the circ-filamin A (FLNA)/miR-149-5p/matrix metalloproteinase 9 (MMP9) signaling axis was involved in TR4-induced cell invasion/migration of PTC cells, which was also verified in a xenografted mouse model. Together, our study builds a strong rationale to develop TR4-targeted therapies toward metastatic PTC patients.
Results
TR4 promotes cell invasion/migration of PTC cells
The role of TR4 in PTC development has not yet been investigated. Therefore, we first examined TR4 expression in 20 pairs of primary PTC tumors and the paired lymph node metastatic PTC tumors. The data obtained from immunohistochemistry (IHC) staining demonstrated that the TR4 expression level was significantly increased in metastatic PTC tumors compared to the paired primary PTC controls (Figure 1A), which was consistent with the analysis from The Cancer Genome Atlas (TCGA) dataset showing that the level of TR4 was also remarkably increased in TC patients with nodal metastasis (Figure 1B). More importantly, TC patients with a high TR4 expression level tended to have shorter overall survival (OS) time than patients with a low TR4 expression level (Figure 1C). These findings suggest that TR4 may play an oncogenic role in PTC development. To end this, we manipulated TR4 expression both in K-1 and TPC-1 cells by knockdown or overexpression strategies and examined the invasive and migration capacities of these cells. As shown in Figure 1D, TR4 was successfully reduced by short hairpin (sh)RNAs in K-1 cells, and knockdown of TR4 in K-1 cells led to a reduced cell invasion and migration (Figures 1E and 1F). On the contrary, cell invasion and migration of K-1 were dramatically increased upon TR4 transfection (Figures 1G−1I). What’s more, the data of TPC-1 cells also confirmed that knocking down TR4 could decrease cell invasion and migration capacities (Figures 1J−1L), and oeTR4 could increase cell invasion and migration capacities (Figures 1M−1O). At the same time, we also checked the proliferation and clonogenicity after manipulated TR4 in the PTC cells. The results showed that TR4 could promote PTC cell proliferation and clonogenicity capacities too (Figures S1A−S1H). Taken together, all of these data suggest that TR4 drives PTC progression via promoting its cell invasion, migration, and proliferation.
Figure 1.
TR4 promotes cell invasion/migration of PTC cells
(A) TR4 expression level was examined in 20 pairs of primary PTC tumors and the paired lymph node metastatic PTC tumors using IHC staining strategy. Left, representative images of TR4 signal in PTC patients; top, image from primary PTC tumor; bottom, image from the paired PTC tumors metastasizing into lymph node; right, quantification of TR4 signal in PTC patients. Paired t test was used to make the statistical analysis. (B) TR4 expression level was significantly increased in nodal metastasis (N1) TC patients compared to non-metastasis (N0) TC patients. (C) OS analysis of TC patients classified by TR4 expression level. The blue line stands for low TR4 TPM; red line stands for high TR4 TPM. (D) Western blot assay was used to check the efficiency of TR4 knockdown in K-1 cells. Actin was internal control. (E) Knockdown of TR4 suppressed the cell invasion of K-1 cells. Left, representative images of invading K-1 cells; right, statistical analysis of invading K-1 cells. (F) Wound-healing assay showed that knockdown of TR4 inhibited cell migration of K-1 cells. Left, representative images of migrating K-1 cells; right, statistical analysis of migrating K-1 cells. (G) Western blot assay was used to check the efficiency of TR4 overexpression (oeTR4) in K-1 cells. GAPDH was loading control. (H) oeTR4 promoted cell invasion of K-1 cells. Left, representative images of invading K-1 cells; right, statistical analysis of invading K-1 cells. (I) Wound-healing assay revealed that oeTR4 enhanced the migrating ability of K-1 cells. Left, representative images of migrating K-1 cells; right, statistical analysis of migrating K-1 cells. (J) Western blot assay was used to check the efficiency of TR4 knockdown in TPC-1 cells. GAPDH was internal control. (K) Knockdown of TR4 suppressed cell invasion of TPC-1 cells. Left, representative images of invading TPC-1 cells; right, statistical analysis of invading TPC-1 cells. (L) Wound-healing assay showed that knockdown of TR4 inhibited cell migration of TPC-1 cells. Left, representative images of migrating TPC-1 cells; right, statistical analysis of migrating TPC-1 cells. (M) Western blot assay was used to check the efficiency of oeTR4 in TPC-1 cells. GAPDH was loading control. (N) oeTR4 promoted cell invasion of TPC-1 cells. Left, representative images of invading TPC-1 cells; right, statistical analysis of invading TPC-1 cells. (O) Wound-healing assay revealed that oeTR4 enhanced the migrating ability of TPC-1 cells. Left, representative images of migrating TPC-1 cells; right, statistical analysis of migrating TPC-1 cells. All quantifications are presented as mean ± SD and p values calculated by t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; NS, no significance.
circ-FLNA is one of the factors contributing to TR4-mediated cell invasion/migration of PTC cells
To find the underlying mechanisms by which TR4 promotes cell invasion/migration of PTC cells, we turned our focus on circRNAs due to their potential roles in gene regulation. We first examined the expression levels of 10 circRNAs, which were differentially expressed between PTC tissues and the corresponding normal tissues according to the literature,19 in PTC cells before and after the manipulation of TR4. Results exhibited that four circRNAs (circRNA-0021553, circRNA-0079891, circRNA-0091894, and circRNA-0004771) were consistently altered in K-1 and TPC-1 cells when we altered TR4 expression (Figures 2A and 2B). To determine which circRNA was responsible for TR4-mediated cell invasion/migration of PTC cells, we applied a rescue assay in TPC-1 cells. Interestingly, we found that TR4-induced cell invasion of TPC-1 cells was reversed by circRNA-0091894 shRNAs but not by others (Figure 2C). Wound-healing assay also confirmed that knockdown of circRNA-0091894 blocked TR4-induced cell migration of TPC-1 cells (Figure 2D). As a back-splicing product of the FLNA precursor (Figure 2E), circRNA-0091894 was named as circ-FLNA for convenience, and its circular property was confirmed by RNase R digestion using linear glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA as a negative control (Figure 2F). Next, we oecirc-FLNA in TPC-1 cells and found that induction of circ-FLNA could prevent shTR4-decreased cell invasion and migration (Figures 2G and 2H). Similarly, shTR4-reduced cell invasion and migration were diminished in K-1 cells in the presence of circ-FLNA transfection (Figures 2I−2K). We also oeTR4/shcirc-FLNA in K-1 cells; the results showed that shcirc-FLNA could partly reverse the function of TR4 to PTC cells, both in invasion and migration capacities (Figures 2L and 2M). What’s more, we also checked the proliferation and clonogenicity after shTR4/oecirc-FLNA or oeTR4/shcirc-FLNA in TPC-1 and K-1 cells. The data were also consistent with the invasion and migration data (Figures S2A−S2D). Of note, we failed to observe any altered cell invasion of K-1 cells when FLNA was depleted (Figure S2E), suggesting circ-FLNA but not its host gene contributed to cell invasion/migration of PTC cells. Taken together, we conclude that circ-FLNA is one of the contributing factors responsible for TR4-mediated cell invasion/migration in PTC cells.
Figure 2.
circ-FLNA is one of the factors contributing to TR4-mediated cell invasion/migration of PTC cells
(A) qRT-PCR assay was used to check the expression levels of circRNAs in K-1 cells before and after TR4 knockdown. GAPDH was used to normalize gene expression. (B) qRT-PCR assay was used to check the expression levels of circRNAs in TPC-1 cells before and after oeTR4. GAPDH was used to normalize gene expression. (C) Transwell invasion assay showed that circRNA-0091894 (circ-FLNA) knockdown could prevent TR4-induced cell invasion of TPC-1 cells, but circRNA-0021533, circRNA-0079891, and circRNA-0004771 could not reverse the function of TR4. Left, representative images of invading TPC-1 cells; right, statistical analysis of invading TPC-1 cells. (D) Wound-healing assay revealed that TR4-induced cell migration of TPC-1 cells was reversed by shcirc-0091894 (shcirc-FLNA). Left, representative images of migrating TPC-1 cells; right, statistical analysis of migrating TPC-1 cells. (E) Nucleotide sequence of circ-FLNA. (F) The circular property of circ-FLNA was confirmed by RNase R digestion. GAPDH served as negative control. (G) oecirc-FLNA could block shTR4-reduced cell invasion of TPC-1 cells. Transwell invasion assay was used to check the invasion capacity after shTR4/oecirc-FLNA in the TPC-1 cells. Left, representative images of invading TPC-1 cells; right, statistical analysis of invading TPC-1 cells. (H) oecirc-FLNA could also prevent shTR4-reduced cell migration of TPC-1 cells. Wound-healing migration assay was used to check the migration capacity after shTR4/oecirc-FLNA in the TPC-1 cells. Left, representative images of migrating TPC-1 cells; right, statistical analysis of migrating TPC-1 cells. (I) qRT-PCR assay was used to check the efficiency of oecirc-FLNA in K-1 cells. (J) oecirc-FLNA could block shTR4-reduced cell invasion of K-1 cells. Transwell invasion assay was used to check the invasion capacity after shTR4/oecirc-FLNA in the K-1 cells. Left, representative images of invading K-1 cells; right, statistical analysis of invading K-1 cells. (K) oecirc-FLNA could also prevent shTR4-reduced cell migration of K-1 cells. Wound-healing migration assay was used to check the migration capacity after shTR4/oecirc-FLNA in the K-1 cells. Left, representative images of migrating K-1 cells; right, statistical analysis of migrating TPC-1 cells. (L) Knocking down of circ-FLNA could block TR4-induced cell invasion of K-1 cells. Transwell invasion assay was used to check the invasion capacity after oeTR4/shcirc-FLNA in the K-1 cells. Left, representative images of invading K-1 cells; right, statistical analysis of invading K-1 cells. (M) Knocking down of circ-FLNA could also prevent TR4-induced cell migration of K-1 cells. Wound-healing migration assay was used to check the migration capacity after oeTR4/shcirc-FLNA in the K-1 cells. Left, representative images of migrating K-1 cells; right, statistical analysis of migrating K-1 cells. All quantifications are presented as mean ± SD and p values calculated by t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
TR4 transcriptionally regulates circ-FLNA
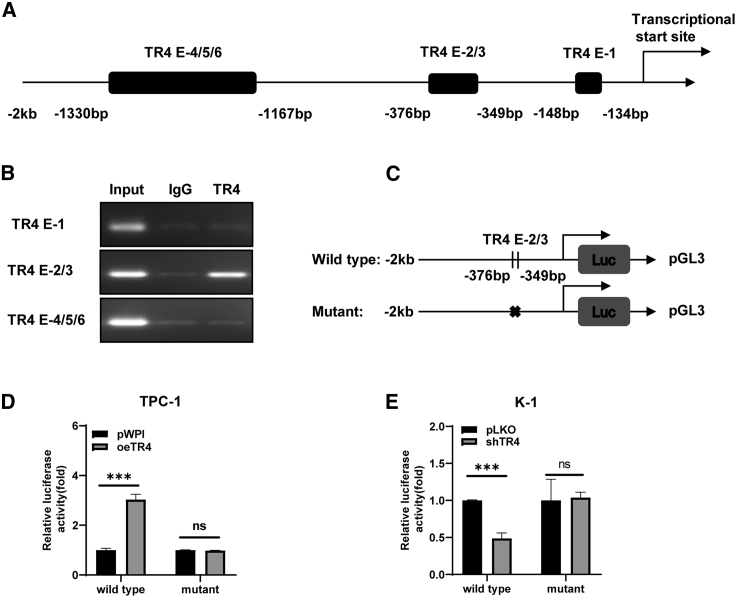
As a transcription factor, TR4 could enter the nucleus and regulate numerous genes. We first looked at the promoter region of the circ-FLNA host gene. As predicted in JASPAR, there were 6 TR4-binding elements (TR4Es) at the −2-kb promoter region of FLNA (Figure 3A). To check which binding site was associated with the regulation of TR4 on circ-FLNA, we performed a chromatin immunoprecipitation (ChIP) assay using TR4 antibody as a bait to pull down chromatin DNA fragments. The result revealed that TR4 could only bind TR4E-2/3 but not others (Figure 3B), implying that TR4E-2/3 at the promoter region of FLNA may be tightly linked with the regulation of circ-FLNA by TR4. To confirm whether TR4 could directly affect the promoter’s activity of FLNA and regulate the circ-FLNA expression level, we cloned the wild-type and mutated promoter region of FLNA, as depicted in Figure 3C, into pGL3-basic backbone and examined their activities. As shown in Figure 3D, oeTR4 significantly increased the wild-type promoter activity of FLNA in TPC-1 cells but had little effect on the mutated one. A significant decrease of wild-type promoter activity of FLNA was observed when TR4 was knocked down in K-1 cells, which was diminished when TR4-2/3 was mutated (Figure 3E). Together, these results support the notion that TR4 transcriptionally regulates circ-FLNA expression level via directly binding to TR4E-2/3.
Figure 3.
TR4 transcriptionally regulates circ-FLNA level
(A) Six TR4-binding sites were predicted in the upstream promoter of circ-FLNA. (B) ChIP assay exhibited that TR4 bonds to TR4E-2/3 in the upstream promoter of circ-FLNA. (C) Schematic depiction showed how wild-type and mutated promoter of circ-FLNA was constructed. (D) Luciferase activities of wild-type and mutated promoter of circ-FLNA before and after oeTR4 in TPC-1 cells. (E) Luciferase activities of wild-type and mutated promoter of circ-FLNA before and after TR4 depletion in K-1 cells. All quantifications are presented as mean ± SD and p values calculated by t test. ∗∗∗p < 0.001.
Mechanistic dissection shows that miR-149-5p is involved in TR4/circ-FLNA-mediated cell invasion/migration of PTC cells
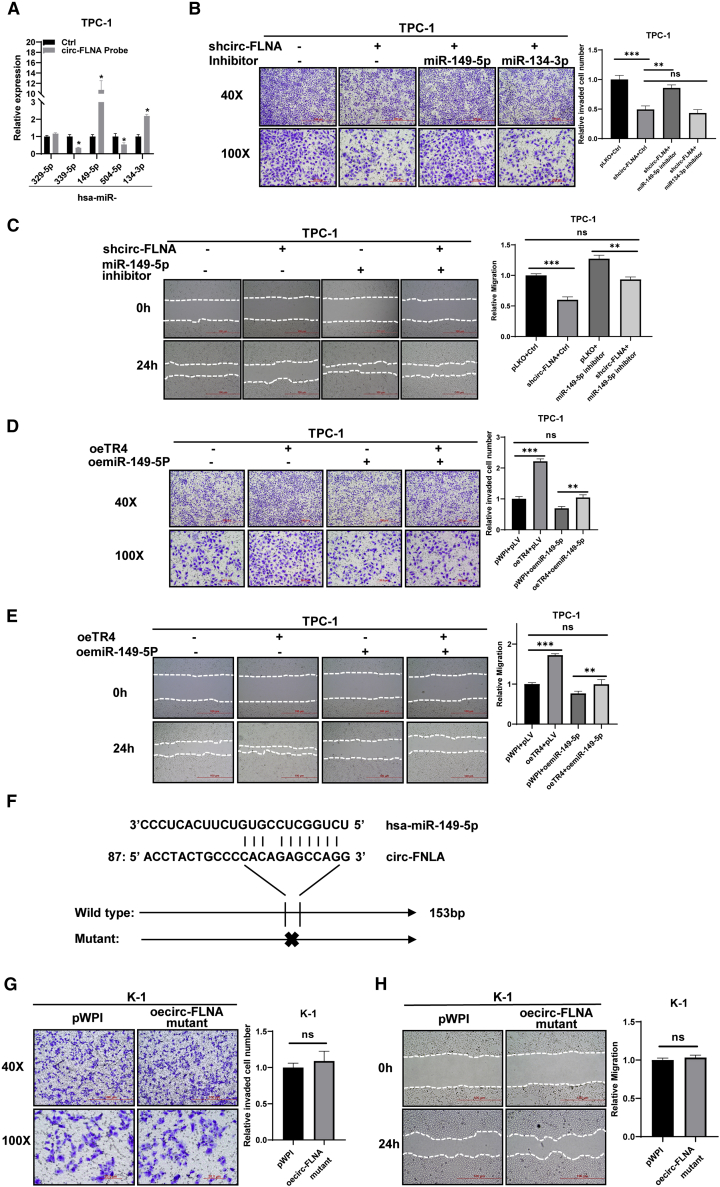
The sponge potential of circRNA in gene regulation has been wildly known. To find which miRNA was involved TR4/circ-FLNA-mediated progression of PTC cells, we pulled down circ-FLNA using the biotin probe and detected its associated miRNAs. The result exhibited that miR-149-5p and miR-134-3p were evidently enriched with circ-FLNA (Figure 4A). A rescue assay using miRNA inhibitor indicated that miR-149-5p but not miR-134-3p participated into circ-FLNA-mediated cell invasion of PTC cells, showing that a reduced cell invasion of TPC-1 cells by circ-FLNA knockdown was diminished in the presence of the miR-149-5p inhibitor (Figure 4B). Similarly, an inhibitory effect on cell migration of TPC-1 cells upon circ-FLNA knockdown was also lost when the miR-149-5p inhibitor was supplemented into these cells (Figure 4C). On the contrary, oemiR-149-5p (Figure S3A) could also block circ-FLNA-induced cell invasion and cell migration of K-1 cells (Figures S3B and S3C). Most importantly, TR4-induced cell invasion and cell migration of TPC-1 cells could also be attenuated when miR-149-5p was over-introduced into cells (Figures 4D and 4E), whereas shTR4 failed to reduce cell invasion and cell migration of K-1 cells when cells were supplemented with the miR-149-5p inhibitor (Figures S3D and S3E), implying the role of TR4 in PTC was also tightly connected with miR-149-5p. Furthermore, we also mutated the nucleotides of circ-FLNA responsible for miR-149-5p binding (Figure 4F) and examined whether it affected the biological functions of circ-FLNA. Our results showed that the mutated form of circ-FLNA lost its ability to influence cell invasion and cell migration of K-1 cells (Figures 4G and 4H), strengthening the point that the biological functions of circ-FLNA relied on its ability to interact with miR-149-5p. Together, all of these data suggest that miR-149-5p is involved in the TR4/circ-FLNA signal to regulate cell invasion/migration of PTC cells.
Figure 4.
Mechanistic dissection shows that miR-149-5p is involved in TR4/circ-FLNA-mediated cell invasion/migration of PTC cells
(A) RNA pull-down assay was used to choose the miRNAs that could potentially target by circ-FLNA. The result showed that miR-149-5p and miR-134-3p were associated with circ-FLNA. (B) Inhibition of miR-149-5p reversed shcirc-FLNA-reduced cell invasion of TPC-1 cells. Transwell invasion assay was used to check the invasion capacity after shcirc-FLNA/adding miR-149-5p inhibitor or miR134-3p inhibitor in the TPC-1 cells. Left, representative images of invasion capacities of TPC-1 cells; right, quantification of invading TPC-1 cells. (C) Inhibition of miR-149-5p reversed shcirc-FLNA-reduced cell migration of TPC-1 cells. Wound-healing migration assay was used to check the migration capacity after shcirc-FLNA/adding miR-149-5p inhibitor in the TPC-1 cells. Left, representative migration images of TPC-1 cells; right, quantification of migrating TPC-1 cells. (D and E) oemiR-149-5p inhibited cell invasion (D) and cell migration (E) of TPC-1 cells induced by TR4. Transwell invasion assay/wound-healing migration assay was used to check invasion/migration capacity after oeTR4/oemiR-149-5p in TPC-1 cells. Left, representative invasion/migration images of TPC-1 cells; right, quantification of invading or migrating TPC-1 cells. (F) The predicted binding nucleotides between miR-149-5p and circ-FLNA. (G and H) Mutated form of circ-FLNA failed to affect cell invasion (G) and cell migration (H) of K-1 cells. Left, representative images; right, quantification of invading or migrating cells. All quantifications are presented as mean ± SD and p values calculated by t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
MMP9 is the downstream effector controlling TR4/circ-FLNA/miR-149-5p-induced cell invasion/migration of PTC cells
To find the downstream targets contributing to TR4/circ-FLNA/miR-149-5p-mediated cell invasion/migration of PTC cells, we examined the expression levels of several metastasis-related genes (also predicated as miR-149-5p targeting genes) before and after the alteration of TR4 in TPC-1 and K-1 cells. Data showed that only MMP9 was consistently altered in TPC-1 and K-1 cells upon TR4 manipulation (Figure 5A), implying that MMP9 may be the candidate downstream effector responsible for TR4-induced cell invasion and migration of PTC cells. We also checked MMP9 expression after shTR4 in TPC-1 cells and oeTR4 in K-1 cells; the results were also consistent with the data in Figure 5A (Figure 5B). To test this postulation, we first examined the MMP9 expression level after the alteration of the TR4/circ-FLNA/miR-149-5p signaling axis. Our data demonstrated that either oecirc-FLNA or miR-149-5p inhibition with inhibitor could reverse the reduction of MMP9 expression level by TR4 depletion in K-1 cells (Figures 5C and 5D). In contrast, inhibition of circ-FLNA with shRNAs or oemiR-149-5p bore the capacity to prevent TR4-induced MMP9 expression level in TPC-1 cells (Figures 5E and 5F) and the reverse assay in K-1 cells also confirmed that inhibition of circ-FLNA with shRNAs could also prevent TR4 induced MMP9 expression (Figure 5G). Most importantly, knockdown of MMP9 had the capacity to impair TR4-induced cell invasion and cell migration of TPC-1 cells (Figures 5H and 5I), and shTR4/oeMMP9 in the same PTC cells also confirmed it (Figures 5J and 5K). What’s more, we also checked the proliferation and clonogenicity capacities of PTC cells, and the results were consistent with the invasion and migration data (Figures S4A−S4D), suggesting that MMP9 was involved in the TR4/circ-FLNA/miR-149-5p signal to control cell invasion/migration of PTC cells. In fact, analyses from TCGA dataset indicated that MMP9 expression level was correlated with TC development. MMP9 expression level was increased in TC tissues compared to normal tissues (Figure 5L). Disease-free survival (DFS) analysis also showed that MMP9 was a poor prognostic factor in TC progression (p = 0.017; Figure 5M), and TC patients with T3 + T4 stage or metastasis tended to express a higher expression level of MMP9 compared to the corresponding controls (Figures S4E−S4G). Interestingly, this trend only existed in TC patients with ages above 55 (Figures S4E−S4G). To further confirm that MMP9 was a direct target of miR-149-5p, we constructed wild-type and mutated 3′ UTR of MMP9 into the psiCHECK-2 enhancer and tested the luciferase activity upon the manipulation of miR-149-5p (Figure 5N). Our results demonstrated that miR-149-5p could significantly suppress the luciferase activity of wild-type but not the mutated 3′ UTR of MMP9 in TPC-1 cells (Figure 5O). A consistent result was gained from K-1 cells, revealing that the miR-149-5p inhibitor remarkably enhanced the wild-type luciferase activity of 3′ UTR of MMP9 (Figure 5P). However, the miR-149-5p inhibitor failed to affect the luciferase activity of mutated 3′ UTR of MMP9 (Figure 5P). Taken together, we conclude that MMP9 is a direct target of miR-149-5p and accounts for TR4/circ-FLNA/miR-149-5p-mediated cell invasion and migration of PTC cells.
Figure 5.
MMP9 is the downstream effector controlling TR4/circ-FLNA/miR-149-5p-induced cell invasion/migration of PTC cells
(A) Several genes as predicated as miR-149-5p were detected by western blotting before and after TR4 manipulation in K-1 cells and TPC-1 cells. GAPDH was served as internal control. (B) Western blot assay was used to check MMP9 expression after shTR4 in TPC-1 cells or oeTR4 in K-1 cells. α-tubulin was served as internal control. (C and D) oecirc-FLNA (C) or inhibition of miR-149-5p (D) could block shTR4-reduced MMP9 expression in K-1 cells by western blotting analysis. GAPDH was loading control. (E and F) Knockdown of circ-FLNA (E) or miR-149-5p (F) attenuated TR4-induced MMP9 expression in TPC-1 cells by western blotting analysis. GAPDH was internal control. (G) Western blot assay was used to check MMP9 expression after oeTR4/shcirc-FLNA in K-1 cells. GAPDH was internal control. (H and I) TR4-induced cell invasion (H) and cell migration (I) of TPC-1 cells were attenuated when MMP9 was knocked down. Transwell invasion assay/wound-healing migration assay was used to check invasion/migration capacity after oeTR4/shMMP9 in TPC-1 cells. Left, representative invasion/migration images of TPC-1 cells; right, quantification of invading or migrating TPC-1 cells. (J and K) shTR4-reduced cell invasion (J) and cell migration (K) of TPC-1 cells were attenuated when MMP9 was overexpressed. Transwell invasion assay/wound-healing migration assay was used to check invasion/migration capacity after shTR4/oeMMP9 in TPC-1 cells. Left, representative invasion/migration images of TPC-1 cells; right, quantification of invading or migrating TPC-1 cells. (L) MMP9 was increased in TC tissues compared to the corresponding controls. The data of the samples were collected from TCGA database. Left, normal versus tumor; right, TC tissues versus paired adjacent normal tissues. (M) DFS analysis of TC patients classified by MMP9 expression level. (N) Schematic depiction of how wild-type and mutated 3′ UTR of MMP9 was constructed. (O) The luciferase activity of wild-type but not mutated 3′ UTR of MMP9 was reduced by miR-149-5p in TPC-1 cells. (P) The luciferase activity of wild-type but not mutated 3′ UTR of MMP9 was enhanced by the miR-149-5p inhibitor in K-1 cells. All quantifications are presented as mean ± SD and p values calculated by t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
Xenografted mouse model confirms the role of TR4/circ-FLNA signal in PTC progression
Next, we applied a xenografted mouse model to verify the role of TR4/circ-FLNA in PTC progression. We subcutaneously implanted 1 × 106 K-1 cells with the following gene manipulations: (1) pWPI + pLKO, (2) TR4 + pLKO, (3) pWPI + shcirc-FLNA, and (4) TR4 + shcirc-FLNA, under the skin of 6-week male nude mice and monitored tumor growth. Data showed that TR4 evidently promoted K-1 tumor growth, whereas knockdown of circ-FLNA could suppress K-1 tumor growth (Figure 6A), and TR4-induced K-1 tumor growth was attenuated when circ-FLNA was depleted by shRNAs (Figure 6A), strengthening the role of TR4/circ-FLNA signal in papillary thyroid tumor growth. The curves of tumor volume also confirmed the data (Figure 6B). As the downstream effector of TR4/circ-FLNA/miR-149-5p, MMP9 expression level was examined by IHC staining in various K-1 tumors, showing that TR4 significantly promoted the MMP9 expression level, and circ-FLNA knockdown could prevent TR4-induced MMP9 expression (Figure 6C). We also constructed a metastatic mouse model by injecting K-1 cells through the tail vein to observe the lung metastasis, and the results showed it was consistent with our in vitro data and the mouse model of subcutaneous tumor formation. As it is shown in Figure 6D, the left picture showed the typical lung metastatic nodules in each group, the middle picture showed the hematoxylin-eosin (H&E) staining data of the lung metastasis, and the right picture showed the IVIS data of each group. From the data, we knew that oeTR4 could promote lung metastasis in the mice model, and shcirc-FLNA could decrease lung metastasis. What’s more, shcirc-FLNA could partly reverse the function of TR4 on the metastasis of PTC. Together, these in vivo results support the oncogenic role of the TR4/circ-FLNA/miR-149-5p/MMP9 signal in papillary thyroid tumor development.
Figure 6.
Xenografted mouse model confirms the role of TR4/circ-FLNA signal in PTC progression
(A) 2 × 106 K-1 cells were subcutaneously implanted into 6-week male nude mice with 1:1 Matrigel. Tumor growth of each group was shown after sacrificing the mice. (B) The growth curve of tumor volume of each group was shown. Tumor volume = length × (width × width). (C) IHC assay was used to check the MMP9 expression of the mice tumor in each group. (D) A metastatic mouse model by injecting K-1 cells through the tail vein to observe the lung metastasis was constructed, and the results showed it was consistent with our in vitro data and the mouse model of subcutaneous tumor formation. As it is shown in the figure, the left picture showed the typical lung metastatic nodules in each group, the middle picture showed the H&E data of the lung metastasis, and the right picture showed the IVIS data of each group. From the data, we knew that oeTR4 could promote lung metastasis in the mice model, and shcirc-FLNA could decrease lung metastasis. What’s more, shcirc-FLNA could partly reverse the function of TR4 on the metastasis of PTC. All quantifications are presented as mean ± SD and p values calculated by t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
To summarize, TR4 played a tumor-promoting role in PTC development by controlling the circ-FLNA/miR-149-5p/MMP9 signal to regulate cell proliferation, invasion, and migration.
Discussion
A small portion of PTC will progress to an advanced stage with distant metastasis, which has little response to either radioactive iodide treatment or hormone suppressive therapy.1 Therefore, understanding the molecular biology of metastatic PTC has a chance to develop novel targeted therapies toward advanced PTC patients. In this study, we first found that TR4 was significantly increased in PTC patients with distant metastasis compared to patients without metastasis, which was consistent with the analyses from TCGA dataset. A mechanistic study demonstrated that TR4 could transcriptionally regulate circ-FLNA expression level via directly binding to its upstream promoter, which served as a sponge of miR-149-5p to enhance MMP9 expression. Our identified TR4/circ-FLNA/miR-149-5p/MMP9 axis can promote cell invasion/migration of PTC cells. Furthermore, the oncogenic roles of TR4 and circ-FLNA were also verified in xenografted mouse model. Overall, our study defines the role of TR4 in PTC development, and targeting TR4 may be an alternative option for metastatic PTC patients.
The tumor-promoting role of TR4 has been documented in various cancers,9, 10, 11, 12, 13 promoting scientists to identify the TR4 inhibitor for better treatment of cancers. Therefore, it is of importance and interest to develop small molecules that can specifically inhibit TR4 activity. A previous study showed that metformin, a clinical drug used in patients with diabetes type II, can suppress TR4 transactivation via triggering AMP-activated protein kinase (AMPK) signaling.20 Given the fact that TR4 is overexpressed in metastatic PTC patients, it is reasonable to test the effect of metformin on PTC cell invasion and to prompt the clinical trial of metformin in metastatic PTC patients. In addition, a small molecule, tretinoin, was also reported to suppress RCC progression and affect sunitinib sensitivity via directly inhibiting the function of TR4.21 As fact, the potential anti-cancer role of tretinoin in TC has been recognized many decades ago. It is possible that the anti-cancer effect of tretinoin on TC relies on its inhibition on the function of TR4.
The pathological and physiological roles of noncoding RNA including circRNAs have been increasingly noticed recently.22, 23, 24, 25, 26 Some papers have reported that circ-FLNA played important roles in the regulation of many cancers. Wang et al.27 showed that upregulation of circ-FLNA contributes to laryngeal squamous cell carcinoma migration by the circ-FLNA-miR-486-3p-FLNA axis. Qu et al.28 reported that circ-FLNA acts as a sponge of miR-646 to facilitate the proliferation, metastasis, glycolysis, and apoptosis inhibition of gastric cancer by targeting PFKFB2. Our data showed that circ-FLNA acted as a sponge of miR-149-5p to increase MMP9 expression, leading to enhanced cell invasion of PTC cells, which was another example showing that circ-FLNA may influence cancer progression, and also provided a potential target for scientists to develop new therapies for better treatment of PTC. However, how to target circRNA remains a scientific concern because one circRNA may interact with several miRNAs, and one miRNA has hundreds of targets. Although we defined the role of circ-FLNA in TC cells, the translational value of circ-FLNA is currently limited.
Numerous studies have demonstrated that MMP proteins are tightly associated with cancer initiation and progression.29, 30, 31 As an important member of the MMP family, MMP9 was shown as the downstream effector of TR4/circ-FLNA/miR-149-5p to control PTC cell invasion/migration. Importantly, MMP9 was highly expressed in metastatic PTC patients and served as a poorly prognostic factor for DFS, according to TCGA dataset, suggesting that targeting MMP9 may be a promising therapeutic strategy toward metastatic PTC patients. In fact, clinical trials of MMP inhibitors have been examined in various cancers many decades ago. However, the results were disappointing due to the severe side effects of MMP inhibitors.32 Recently, a new generation of MMP inhibitors has been developed after the better understanding of MMP structure.33 Preclinical trials suggested that these new MMP inhibitors have improved toxicity and better efficacy. Therefore, it will be promising to see the therapeutic efficacy of a new generation of the MMP9 inhibitor toward metastatic PTC patients.
Conclusions
In summary, our results proved the oncogenic role of TR4 in PTC development. TR4 promoted PTC cell invasion/migration via increasing circ-FLNA level, which worked as a miR-149-5p sponge to increase MMP9 expression level. The TR4/circ-FLNA/miR-149-5p/MMP9 signal may be an ideal therapeutic target for metastatic PTC patients.
Materials and methods
Cell culture
K-1, TPC-1, and 293T were purchased from the Cell Bank in the Chinese Academy of Sciences (Shanghai, China). 10% fetal bovine serum (FBS) DMEM (100 units/mL penicillin, 100 μg/mL streptomycin) was used to culture cells in a humidified 5% CO2 environment at 37°C.
Lentivirus generation
Lentivirus was generated as previously described. Briefly, 20 μg plasmids with targeting sequence were co-transfected with psPAX2 (10 μg) and pMD2.G (10 μg) into 293T cells using the standard calcium phosphate transfection method. Lentivirus supernatant was collected with 0.45 filter after 48 h and infected TC cells. 5 μg/mL puromycin was used to select shRNA-infected TC cells.
The plasmids sequences used were as follows: pLKO.1-shTR4 sequence, forward: 5′-CCGGCCAGCACAAGCCAGATTGAAAGGATCCTTTCAATCTGGCTTGTGCTGGTTTTTG-3′, reverse: 5′-AATTCAAAAACCAGCACAAGCCAGATTGAAAGGATCCTTTCAATCTGGCTTGTGCTGG-3′; pWPI-oeTR4 sequence, forward: 5′-TTTCGACATTTAAATTTAATATGACCAGCCCCTCCCCACG-3′, reverse: 5′-ATTCCTGCAGCCCGTAGTTTCTATAGACTGGCTCCGGTGA-3′; pLKO.1-shFLNA sequence, forward: 5′-CCGGGACCGCCAATAACGACAAGAATTGGATCCGTTCTTGTCGTTATTGGCGGTCTTTTTG-3′, reverse: 5′-AATTCAAAAAGACCGCCAATAACGACAAGAACGGATCCAATTCTTGTCGTTATTGGCGGTC-3′; pLV-miR-149-5p (oemiR-149-5p) sequence, forward: 5′-CGCGTTCTGGCTCCGTGTCTTCACTCCCACCCGGTCGACGTGGGAGTGAAGACACGGAGCCAGATTTTTG-3′, reverse: 5′-CGCAAAAATCTGGCTCCGTGTCTTCACTCCCACGTCGACCGGGTGGGAGTGAAGACACGGAGCCAGAA-3′; pLKO.1-shMMP9 sequence, forward: 5′-CCGGCCACAACATCACCTATTGGATGGATCCATCCAATAGGTGATGTTGTGGTTTTTG-3′, reverse: 5′-AATTCAAAAACCACAACATCACCTATTGGATGGATCCATCCAATAGGTGATGTTGTGG-3′; pWPI-oeMMP9 sequence, forward: 5′-TTTCGACATTTAAATTTAATATGAGCCTCTGGCAGCCCCT-3′, reverse: 5′-ATTCCTGCAGCCCGTAGTTTCTAGTCCTCAGGGCACTGCA-3′; and pLKO.1-shcirc-FLNA sequence, forward: 5′-CCGGGACCAGCACGTGCCTGGCTATTTGGATCCGATAGCCAGGCACGTGCTGGTCTTTTTG-3′, reverse: 5′-AATTCAAAAAGACCAGCACGTGCCTGGCTATCGGATCCAAATAGCCAGGCACGTGCTGGTC-3′.
To construct pWPI-circ-FLNA (oecirc-FLNA), the specific DNA fragments, which were shown to promote circularization, were first engineered in the pBSK vector as pBSK (circArm). The exon of the FLNA gene was PCR amplified and then inserted into the pBSK (circArm) vector as the pBSK_circ-FLNA. Then we subcloned the fragments including the exon of FLNA and those two arms to the lentiviral pWPI vector for expressing circ-FLNA. The circ-FLNA mature sequence is 5′-GCTATGGTGGGCTCAGCCTGTCCATTGAGGGCCCCAGCAAGGTGGACATCAACACAGAGGACCTGGAGGACGGGACGTGCAGGGTCACCTACTGCCCCACAGAGCCAGGCAACTACATCATCAACATCAAGTTTGCCGACCAGCACGTGCCTG-3′.
Real-time qPCR
Isolation of total RNAs was done with TRIzol reagent following the standard RNA extraction protocol. 2 μg total RNAs was subjected to reverse transcription using SuperScript III transcriptase (Invitrogen). A reverse transcriptase (RT) product with 200 dilutions was used to perform quantitative real-time PCR in a Bio-Rad CFX96 machine with SYBR Green to determine the expression levels of interested genes. 18 s ribosome RNA was utilized to normalize gene expression.
RNase R digestion
RNase R digestion was done with 2 μg total RNAs in a 10-μL reaction at 37°C for 1 h: 0 unit or 20 units of RNase R (Epicentre), 1 × RNase R buffer, 1 unit murine ribonuclease inhibitor (NEB). Treated RNAs was used to perform RT to make cDNA. Then real-time PCR was performed as aforementioned.
Western blotting
After PBS wash, TC cells were lysed by radioimmunoprecipitation assay (RIPA) buffer, and proteins were collected. 20 μg of proteins was loaded for electrophoresis in 10%–12% SDS-PAGE gel. Then proteins were transferred to the polyvinylidene fluoride (PVDF) membrane and blocked with 10% BSA for 1 h. Incubation with specific primary antibody was performed at 4°C for at least 16 h. The next day, the membrane was washed and incubated with 1:2,500 horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h at room temperature. After an extensive TBST wash, blots were visualized using enhanced chemiluminescence (Thermo Fisher Scientific). The TR4 (MA5-26855, 1:500), actin (MA1-744, 1:500), IGF2BP1 (712138, 1:500), ELP5 (PA5-54745, 1:500), FOXC1 (PA1-807, 1:500), DAB2IP (PA5-85697, 1:500), MMP9 (MA5-15886, 1:500), MACC1 (PA5-20758, 1:500), and GAPDH (39-8600, 1:500) antibodies were purchased from Thermo Fisher Scientific.
Wound-healing assay
TC cells with gene manipulations were scratched using a pipette tip. Images were captured at 0 and 24 h. The experiment was done in triplicate, and quantification was gained using ImageJ software.
Transwell invasion assay
Transwell inserts (Corning) were pre-coated with 1:8 diluted Matrigel (BD Biosciences) before use. PTC cells with gene manipulations were collected and loaded into the inserts at the density of 1 × 105/well. 0% FBS medium in the lower chamber was used as a chemoattractant to attract cells. 36 h later, the invading cells in inserts were fixed with cold methanol and stained with 0.5% crystal violet. Images were taken by inverted microscope (Olympus), and quantifications of invading cells were done with ImageJ.
ChIP assay
TC cells were cross-linked with 4% formaldehyde for 10 min. Then cells were collected and sonicated to 300−500 bp DNA fragments. Cell lysates were pre-cleared with protein A-agarose and followed by the incubation with the TR4 primary antibody (immunoglobulin G [IgG] was used as a negative control). The next day, TR4-DNA fragments were intensively washed with high salt buffer. Proteins were digested with proteinase K, and DNA fragments were extracted for RT-PCR. Specific primer sets were designed to amplify a target sequence within the promoter of FLNA, and RT-PCR products were examined by agarose gel electrophoresis.
Xenografted mouse model
2 × 106 K-1 cells were subcutaneously implanted into 6-week male nude mice with 1:1 Matrigel (Corning). Tumor size was measured weekly by caliper. 8 weeks later, mice were sacrificed, and tumors were removed, washed, and fixed for IHC staining. The study was carried out under the approval of the Ethics Committee of Xiangya Hospital Central South University and followed the Interdisciplinary Principles and Guidelines for the Use of Animals in Research, Testing, and Education by the New York Academy of Sciences, Ad Hoc Animal Research Committee.
IHC staining
Tissues (humans and mice) were fixed in 10% (v/v) formaldehyde and embedded in paraffin. After hydration and antigen retrieval in boiling citrate buffer (pH 6.0), sections were treated with 3% peroxidase in methanol for 15 min and blocked with 10% goat serum PBS buffer for 30 min. The slides were incubated with 1:100 diluted TR4 or MMP9 primary antibody at 4°C overnight. The next day, 1:100 diluted biotin-labeled secondary antibody was used to incubate slides for 30 min and followed by 30 min incubation with streptavidin (PK-4000; Vectastain; Vector Laboratories, Burlingame, CA, USA). The protein signal was determined by DAB staining.
miRNA inhibitor transfection
The miR-149-5p inhibitor used in our study was purchased from Thermo Fisher Scientific (assay ID: MH12788, catalog #: 4464084). The transfection of miR-149-5p inhibitor was conducted as the “mirVana miRNA Inhibitors Protocol 2013” provided by Thermo Fisher Scientific.
Luciferase assay
The human 5′-promoter region of circ-FLNA was constructed into pGL3-basic luciferase reporter vector (Promega). Site-directed mutagenesis of the TR4 binding site in the circ-FLNA promoter was achieved with the QuickChange mutagenesis. The 193-bp wild-type or mutant 3′ UTR region of MMP9 was inserted into psiCHECK-2--basic vector (Promega) and transduced into control/miR-149-5p inhibitor and pLV/oemiR-149-5p stable cells using Lipofectamine 3000 (Invitrogen). Luciferase activities were measured 48 h after transfection by a Dual-Luciferase Assay (Promega).
Statistical analysis
All statistical analyses were performed using GraphPad Prism software. Data were presented as mean ± SE. Differences were analyzed with the one-way ANOVA test, and significance was set at p <0.05.
Acknowledgments
We thank Thelma for editing the manuscript for us. This work was supported by the National Natural Science Foundation of China (82073262 and 81903004).
Author contributions
G.L. and Z.W. designed and monitored the production of the experiments and helped to revise the manuscript. X.O. conducted the experiments and completed the manuscript. L.F., L.Y., and Y.X. helped to complete some of the experiments. X.H. and G.Z. helped to conduct the animal experiments.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.03.021.
Contributor Information
Guodong Liu, Email: guodongliu@csu.edu.cn.
Zhiming Wang, Email: wzm13808462382@126.com.
Supplemental information
References
- 1.Jin S., Borkhuu O., Bao W., Yang Y.T. Signaling Pathways in Thyroid Cancer and Their Therapeutic Implications. J. Clin. Med. Res. 2016;8:284–296. doi: 10.14740/jocmr2480w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2019. CA Cancer J. Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.La Vecchia C., Malvezzi M., Bosetti C., Garavello W., Bertuccio P., Levi F., Negri E. Thyroid cancer mortality and incidence: a global overview. Int. J. Cancer. 2015;136:2187–2195. doi: 10.1002/ijc.29251. [DOI] [PubMed] [Google Scholar]
- 4.Rivera M., Ghossein R.A., Schoder H., Gomez D., Larson S.M., Tuttle R.M. Histopathologic characterization of radioactive iodine-refractory fluorodeoxyglucose-positron emission tomography-positive thyroid carcinoma. Cancer. 2008;113:48–56. doi: 10.1002/cncr.23515. [DOI] [PubMed] [Google Scholar]
- 5.Sáez J.M. Treatment directed to signalling molecules in patients with advanced differentiated thyroid cancer. Anticancer. Agents Med. Chem. 2013;13:483–495. doi: 10.2174/1871520611313030011. [DOI] [PubMed] [Google Scholar]
- 6.Chang C., Da Silva S.L., Ideta R., Lee Y., Yeh S., Burbach J.P. Human and rat TR4 orphan receptors specify a subclass of the steroid receptor superfamily. Proc. Natl. Acad. Sci. USA. 1994;91:6040–6044. doi: 10.1073/pnas.91.13.6040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins L.L., Lee Y.F., Ting H.J., Lin W.J., Liu N.C., Meshul C.K., Uno H., Bao B.Y., Chen Y.T., Chang C. The roles of testicular nuclear receptor 4 (TR4) in male fertility-priapism and sexual behavior defects in TR4 knockout mice. Reprod. Biol. Endocrinol. 2011;9:138. doi: 10.1186/1477-7827-9-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin S.J., Ho H.C., Lee Y.F., Liu N.C., Liu S., Li G., Shyr C.R., Chang C. Reduced osteoblast activity in the mice lacking TR4 nuclear receptor leads to osteoporosis. Reprod. Biol. Endocrinol. 2012;10:43. doi: 10.1186/1477-7827-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai J., Yeh S., Qiu X., Hu L., Zeng J., Cai Y., Zuo L., Li G., Yang G., Chang C. TR4 nuclear receptor promotes clear cell renal cell carcinoma (ccRCC) vasculogenic mimicry (VM) formation and metastasis via altering the miR490-3p/vimentin signals. Oncogene. 2018;37:5901–5912. doi: 10.1038/s41388-018-0269-1. [DOI] [PubMed] [Google Scholar]
- 10.Wang M., Sun Y., Xu J., Lu J., Wang K., Yang D.R., Yang G., Li G., Chang C. Preclinical studies using miR-32-5p to suppress clear cell renal cell carcinoma metastasis via altering the miR-32-5p/TR4/HGF/Met signaling. Int. J. Cancer. 2018;143:100–112. doi: 10.1002/ijc.31289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin S.J., Yang D.R., Li G., Chang C. TR4 Nuclear Receptor Different Roles in Prostate Cancer Progression. Front. Endocrinol. (Lausanne) 2015;6:78. doi: 10.3389/fendo.2015.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin S.J., Lee S.O., Lee Y.F., Miyamoto H., Yang D.R., Li G., Chang C. TR4 nuclear receptor functions as a tumor suppressor for prostate tumorigenesis via modulation of DNA damage/repair system. Carcinogenesis. 2014;35:1399–1406. doi: 10.1093/carcin/bgu052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin R., Lin H., Li G., Xu J., Shi L., Chang C., Cai X. TR4 nuclear receptor suppresses HCC cell invasion via downregulating the EphA2 expression. Cell Death Dis. 2018;9:283. doi: 10.1038/s41419-018-0287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 15.Kristensen L.S., Hansen T.B., Venø M.T., Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37:555–565. doi: 10.1038/onc.2017.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holdt L.M., Kohlmaier A., Teupser D. Molecular functions and specific roles of circRNAs in the cardiovascular system. Noncoding RNA Res. 2018;3:75–98. doi: 10.1016/j.ncrna.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye M., Hou H., Shen M., Dong S., Zhang T. Circular RNA circFOXM1 Plays a Role in Papillary Thyroid Carcinoma by Sponging miR-1179 and Regulating HMGB1 Expression. Mol. Ther. Nucleic Acids. 2020;19:741–750. doi: 10.1016/j.omtn.2019.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Cai X., Zhao Z., Dong J., Lv Q., Yun B., Liu J., Shen Y., Kang J., Li J. Circular RNA circBACH2 plays a role in papillary thyroid carcinoma by sponging miR-139-5p and regulating LMO4 expression. Cell Death Dis. 2019;10:184. doi: 10.1038/s41419-019-1439-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng N., Shi L., Zhang Q., Hu Y., Wang N., Ye H. Microarray profiling of circular RNAs in human papillary thyroid carcinoma. PLoS ONE. 2017;12:e0170287. doi: 10.1371/journal.pone.0170287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim E., Liu N.C., Yu I.C., Lin H.Y., Lee Y.F., Sparks J.D., Chen L.M., Chang C. Metformin inhibits nuclear receptor TR4-mediated hepatic stearoyl-CoA desaturase 1 gene expression with altered insulin sensitivity. Diabetes. 2011;60:1493–1503. doi: 10.2337/db10-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H., Sun Y., He M., Yang X., Hamada M., Fukunaga T., Zhang X., Chang C. Targeting the TR4 nuclear receptor-mediated lncTASR/AXL signaling with tretinoin increases the sunitinib sensitivity to better suppress the RCC progression. Oncogene. 2020;39:530–545. doi: 10.1038/s41388-019-0962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong Y., Du Y., Yang X., Mo Y., Fan C., Xiong F., Ren D., Ye X., Li C., Wang Y. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer. 2018;17:79. doi: 10.1186/s12943-018-0827-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y., Mo Y., Gong Z., Yang X., Yang M., Zhang S., Xiong F., Xiang B., Zhou M., Liao Q. Circular RNAs in human cancer. Mol. Cancer. 2017;16:25. doi: 10.1186/s12943-017-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu P., Li X., Guo X., Chen J., Li C., Chen M., Liu L., Zhang X., Zu X. Circular RNA DOCK1 promotes bladder carcinoma progression via modulating circDOCK1/hsa-miR-132-3p/Sox5 signalling pathway. Cell Prolif. 2019;52:e12614. doi: 10.1111/cpr.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong Y., He D., Peng Z., Peng W., Shi W., Wang J., Li B., Zhang C., Duan C. Circular RNAs in cancer: an emerging key player. J. Hematol. Oncol. 2017;10:2. doi: 10.1186/s13045-016-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bai N., Peng E., Qiu X., Lyu N., Zhang Z., Tao Y., Li X., Wang Z. circFBLIM1 act as a ceRNA to promote hepatocellular cancer progression by sponging miR-346. Journal of experimental & clinical cancer research. J. Exp. Clin. Cancer Res. 2018;37:172. doi: 10.1186/s13046-018-0838-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J.X., Liu Y., Jia X.J., Liu S.X., Dong J.H., Ren X.M., Xu O., Zhang H.Z., Duan H.J., Shan C.G. Upregulation of circFLNA contributes to laryngeal squamous cell carcinoma migration by circFLNA-miR-486-3p-FLNA axis. Cancer Cell Int. 2019;19:196. doi: 10.1186/s12935-019-0924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu J., Yang J., Chen M., Wei R., Tian J. CircFLNA Acts as a Sponge of miR-646 to Facilitate the Proliferation, Metastasis, Glycolysis, and Apoptosis Inhibition of Gastric Cancer by Targeting PFKFB2. Cancer Manag. Res. 2020;12:8093–8103. doi: 10.2147/CMAR.S264674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kessenbrock K., Plaks V., Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liotta L.A., Tryggvason K., Garbisa S., Hart I., Foltz C.M., Shafie S. Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature. 1980;284:67–68. doi: 10.1038/284067a0. [DOI] [PubMed] [Google Scholar]
- 31.Winer A., Adams S., Mignatti P. Matrix Metalloproteinase Inhibitors in Cancer Therapy: Turning Past Failures Into Future Successes. Mol. Cancer Ther. 2018;17:1147–1155. doi: 10.1158/1535-7163.MCT-17-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coussens L.M., Fingleton B., Matrisian L.M. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 33.Fields G.B. The Rebirth of Matrix Metalloproteinase Inhibitors: Moving Beyond the Dogma. Cells. 2019;8:984. doi: 10.3390/cells8090984. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.