Abstract
The gene for the tissue inhibitor of metalloproteinase 3 (TIMP3) on 22q12.3 had been reported to be inactivated by promoter methylation in various types of cancers, with controversial findings in meningiomas. We performed direct sodium bisulfite sequencing in a series of 50 meningiomas, including 27 benign meningiomas [World Health Organization (WHO) grade I], 11 atypical meningiomas (WHO grade II) and 12 anaplastic meningiomas (WHO grade III), and found hypermethylation of TIMP3 in 67% of anaplastic meningiomas, but only 22% of atypical and 17% of benign meningiomas. Moreover, TIMP3 methylation scores were significantly inversely correlated with TIMP3 mRNA expression levels (P = 0.0123), and treatment of the meningioma cell line Ben‐Men‐1 with demethylating agents induced an increased TIMP3 mRNA expression. TIMP3 is located in the chromosomal band 22q12, the allelic loss of which occurs early in meningioma tumorigenesis and preferentially targets the NF2 tumor suppressor gene. In our tumor panel, all meningiomas with TIMP3 hypermethylation—except for a single case—exhibited allelic losses on 22q12.3. Thus, TIMP3 inactivation by methylation seems fairly exclusive to meningiomas with allelic losses on 22q12 but—in contrast to NF2 mutation—appears to be involved in meningioma progression as it is associated with a more aggressive, high‐grade meningioma phenotype.
Keywords: meningioma, methylation, NF2, TIMP3, tumor suppressor
INTRODUCTION
Tumor cell dissemination and angiogenesis require basement membrane and extracellular matrix degradation that can be carried out by enzymes such as matrix metalloproteinases (MMPs). The TIMP3 (tissue inhibitor of metalloproteinase 3) gene on 22q12.3 codes for a protein that can specifically inhibit MMPs by covalent binding to the active site of the enzymes and thus reduces the invasion and the metastatic potential of tumor cells 1, 15. In addition, TIMP3 appears to possess unique tumor suppressor‐like properties that are not related to its properties to abrogate MMPs. As such, it had been shown that overexpression of TIMP3 in vitro induces apoptosis and suppresses tumor growth and angiogenesis in a spectrum of different cancer cell lines 1, 2, 3, 8, 10, 12, 13.
In human tumors, decreased expression of TIMP3 had been linked to poor prognosis in a range of different neoplasms, including esophageal squamous cell carcinoma, secondary glioblastoma and breast, colorectal and prostate cancers 5, 16, 17, 24, 26. Moreover, hypermethylation of the TIMP3 promoter had been identified as a common cause explaining the decreased TIMP3 expression levels in many of the affected tumors 4, 11, 23, 30, 32, 33.
In meningiomas, TIMP3, among other genes, had been investigated for hypermethylation in two preceding studies with controversial results. While Bello and colleagues reported on the hypermethylation of the TIMP3‐associated CpG island in 24% of meningiomas, Liu et al, in a subsequent study, investigated the identical chromosomal region and could not denote TIMP3 hypermethylation in meningiomas 7, 19. To clarify the role of TIMP3 in the pathogenesis of these tumors, we examined a series of 50 human meningiomas representing the different World Health Organization (WHO) grades for TIMP3 methylation, mRNA and protein expression. As TIMP3 maps to the chromosomal region 22q12, which contains the NF2 tumor suppressor gene and is lost in about 50% of sporadic meningiomas early in tumorigenesis (18), we also assessed the allelic status on 22q12.3 in our tumor panel. Our results indicate hypermethylation and transcriptional downregulation of TIMP3 as a mechanism that fairly exclusively occurs in meningiomas with allelic losses on 22q12 but—in contrast to NF2 mutation—appears to be involved in meningioma progression as it is significantly associated with a more aggressive, high‐grade meningioma phenotype.
MATERIALS AND METHODS
Patients and materials
Tumors were selected from the tumor tissue collection of the Department of Neuropathology, Heinrich‐Heine‐University, Düsseldorf, Germany, and investigated according to protocols approved by the institutional review board. All tumors were classified according to the WHO classification of tumors of the nervous system of 2007 (21). The tumor series comprised 50 human meningiomas, including 27 benign meningiomas (WHO grade I, MN I; 8 meningotheliomatous, 8 transitional, 7 fibromatous, 2 secretory, 1 psammomatous and 1 angiomatous meningioma), 11 atypical meningiomas (WHO grade II, MN II) and 12 anaplastic meningiomas (WHO grade III, MN III). Parts of each tumor were snap‐frozen immediately after operation and stored at −80°C. To ensure that the tumor fragments taken for molecular analysis contained a sufficient proportion of tumor cells, each tissue specimen used for nucleic acid extraction was histologically evaluated. Only tissue samples with an estimated tumor cell content of 80% or higher were used for molecular analyses. DNA obtained at autopsy from non‐neoplastic leptomeningeal tissue of a 60‐year‐old male donor and peripheral blood leukocytes of a 57‐year‐old male individual were used as reference for the methylation analyses. As a positive control, we in vitro methylated both non‐neoplastic reference DNA samples by using SssI (CpG) methylase (New England Biolabs, Beverly, MA, USA). Leptomeningeal RNA obtained from autopsy specimens of a 74‐ and an 86‐year‐old male donor served as reference for the expression studies at the mRNA level. From all 39 meningioma patients that underwent microsatellite analysis, we extracted leukocyte DNA from peripheral blood as constitutional reference.
DNA and RNA extraction
Extraction of DNA and RNA from frozen tumor tissue was performed by ultracentrifugation as previously described (9). DNA extraction from peripheral blood leukocytes was performed according to a standard protocol.
Microsatellite analysis
Peripheral blood samples were available from 39 meningioma patients (25 MN I, 10 MN II and 4 MN III patients). To investigate the respective tumors for 22q allelic losses, we employed loss of heterozygosity (LOH) analysis at the following three microsatellite markers [nucleotide (nt) numbering according to the University of California Santa Cruz (UCSC) genome browser at http://www.genome.ucsc.edu, primer sequences supplied in supplementary Table S1]: D22S430 (nt 28 941 777–28 941 860) located proximal, as well as D22S304 (nt 33 700 683–33 700 793) and D22S929 (nt 55 782 709–55 783 071) located distal to the TIMP3 locus (nt 31 562 802–31 589 028).
TIMP3 methylation analyses
Sodium bisulfite treatment of 1 µg DNA was performed overnight (16 h) according to a standard protocol (22). Two different regions from the TIMP3 5′‐CpG island located between nt 31 527 381 and 31 528 267 at 22q12.3 (nt numbering according to the UCSC genome browser at http://www.genome.ucsc.edu; Genbank accession no. NM_011520) were investigated for TIMP3 methylation. Region 1 was investigated by methylation‐specific PCR analysis (CpG sites −42 to −40 and −62 to −59 relative to the TIMP3 translation start site, TLS; 1, 2) and direct bisulfite sequencing (CpG sites −72 to −50, 1, 2). This region overlapped with a smaller fragment of the TIMP3 5′‐CpG island that in two preceding studies had been investigated by methylation‐specific PCR analysis with divergent results (CpG sites −55 to −46, Figure 1) 7, 19. Another region of the TIMP3 5′‐CpG island located telomerically and directly covering the translation start site of TIMP3 (region 2, CpG site −18 to +2; 1, 3, 4) was additionally investigated by direct sodium bisulfite sequencing.
Figure 1.
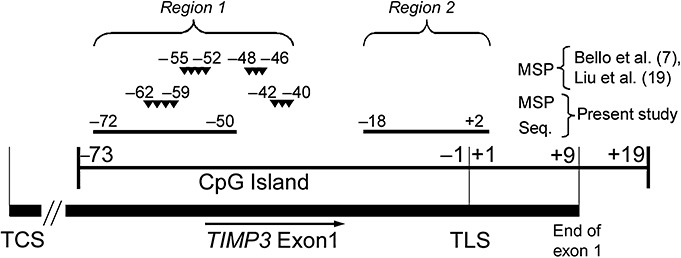
Overview of the TIMP3 5′‐CpG island. Two different regions were investigated for TIMP3 methylation. Region 1 was investigated by methylation‐specific PCR analysis (CpG sites −42 to −40 and −62 to −59 relative to the TIMP3 translation start site, TLS) and direct sodium bisulfite sequencing (CpG sites −72 to −50). This region overlapped with a smaller fragment of the TIMP3 5′‐CpG island that in two preceding studies had been investigated by methylation‐specific PCR analysis with divergent results (CpG sites −55 to −46) 7, 19. Another region of the TIMP3 5′‐CpG island located telomerically and directly covering the translation start site of TIMP3 (region 2, CpG site −18 to +2) was additionally investigated by direct sodium bisulfite sequencing. TCS = transcription start site.
Figure 2.
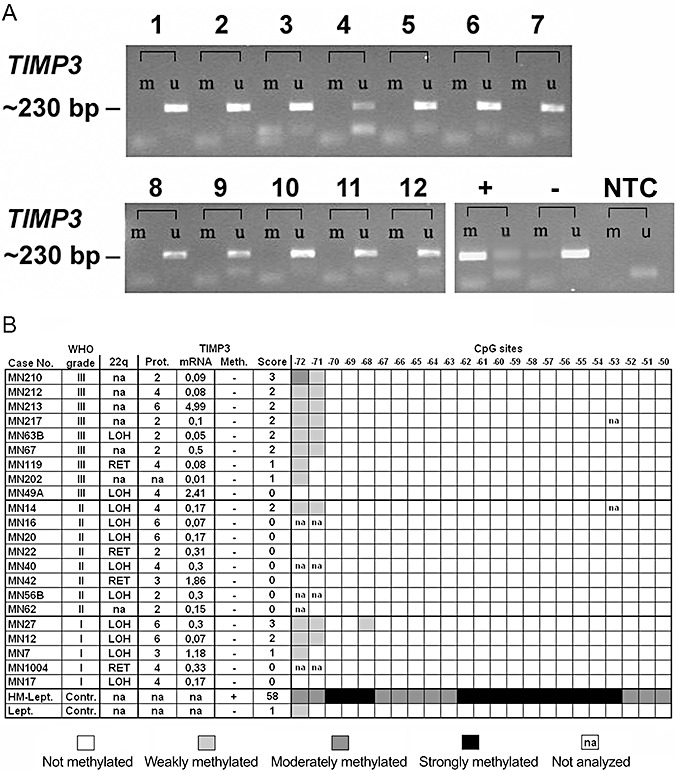
Results of the methylation analysis of region 1. A. Methylation‐specific PCR analysis. 1, MN49A; 2, MN210; 3, MN63B; 4, MN64; 5, MN213; 6, MN119; 7, MN217; 8, MN53; 9, MN58; 10, MN212; 11, MN1038; 12, MN202; +, in vitro hypermethylated DNA control; −, non‐methylated DNA control; NTC, no tissue PCR negative control; m, signal resulting from DNA amplified with primers for methylated DNA, u, signal resulting from DNA amplified with primers for unmethylated DNA. B. TIMP3 region 1 direct bisulfite sequencing analysis (CpG sites −72 to −50) in synopsis with mRNA expression levels relative to normal leptomeningeal tissue, protein (Prot.) expression scores and allelic status on 22q in selected meningiomas. Meth., TIMP3 methylation; Score, TIMP3 methylation score; Lept., non‐neoplastic leptomeningeal tissue; HM‐Lept.; in vitro hypermethylated non‐neoplastic leptomeningeal DNA; LOH, loss of heterozygosity; RET, retention of heterozygosity. Note that neither methylation‐specific PCR (A) nor direct bisulfite sequencing analysis (B) reveals significant hypermethylation in this specific region of the TIMP3 5′‐CpG island.
Figure 3.
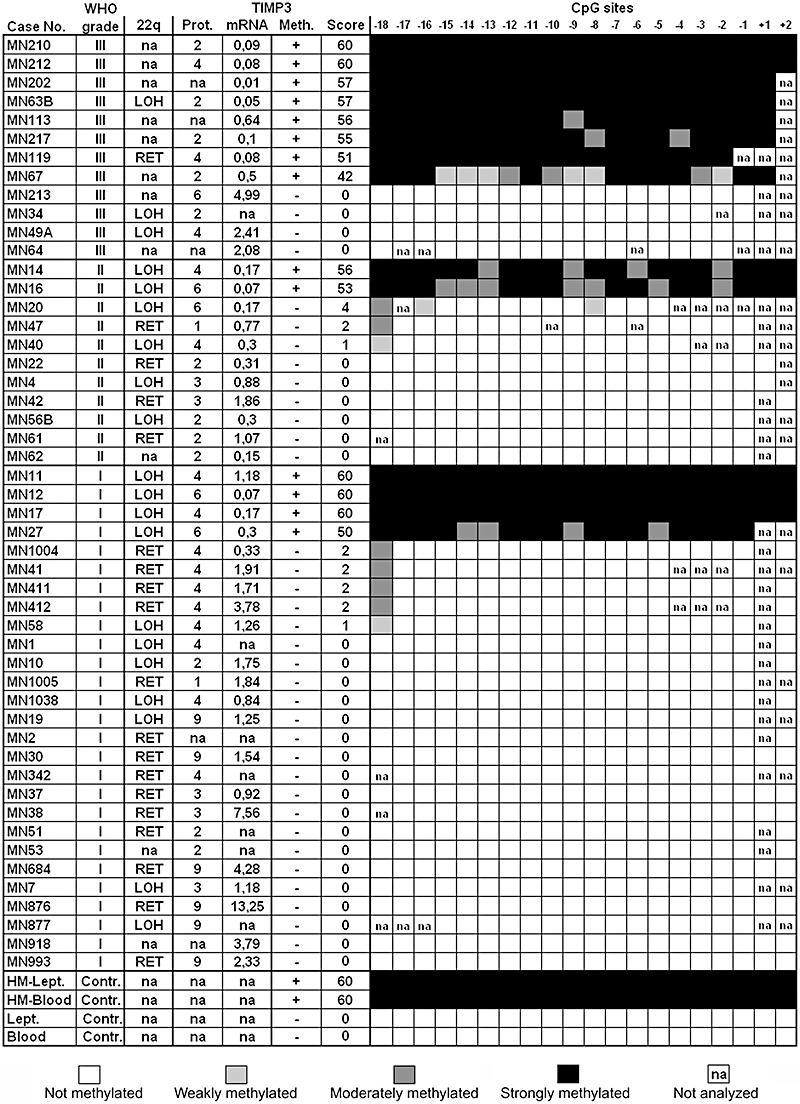
Methylation status (Meth.), methylation score (Score) and methylation pattern in region 2 of the 5′‐CpG island of TIMP3 (CpG sites −18 to +2 relative to the transcription start site). The results are visualized in synopsis with the protein (Prot.) and mRNA expression levels as well as the allelic status on 22q in all investigated meningiomas. Lept., non‐neoplastic leptomeningeal tissue; Blood, non‐neoplastic blood control sample; HM‐Lept./HM‐Blood; non‐neoplastic control samples after in vitro hypermethylation; LOH, loss of heterozygosity; RET, retention of heterozygosity. Note that anaplastic meningiomas (8 out of 12 cases, 67%) show the highest frequency of TIMP3 hypermethylation (methylation score ≥42) associated with marked transcriptional downregulation in the majority of cases. Except for a single anaplastic meningioma (MN119), TIMP3 hypermethylation is exclusively observed in tumors that carry allelic losses on 22q.
Figure 4.
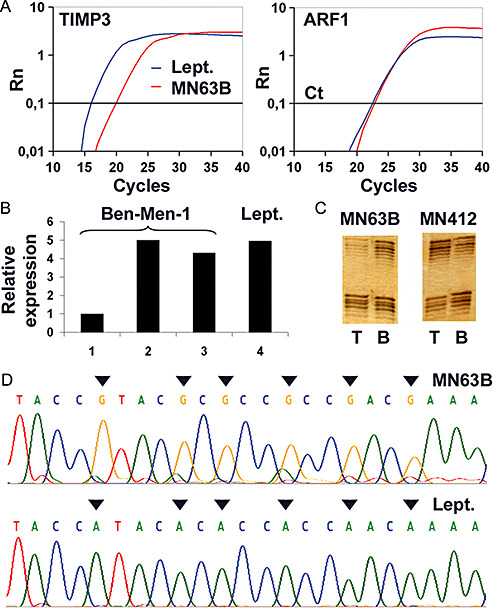
A. Real‐time reverse transcription (RT)‐PCR analysis of TIMP3 expression in a selected case of anaplastic meningioma (MN63B) with TIMP3 5′‐CpG island methylation (compare D) and allelic loss on 22q (compare C). Abscissa, cycle number; ordinate, relative amount of PCR product. While the reference mRNA curves (ARF1) for non‐neoplastic leptomenigeal tissue (Lept.) and MN63B pass the threshold (Ct) at an approximately equal cycle number, the TIMP3 mRNA curve for MN63B is shifted to the right relative to the NB curve, indicating a markedly decreased TIMP3 expression in MN63B. B. Re‐expression of TIMP3 transcripts as assessed by real‐time RT‐PCR analysis in the meningioma cell line Ben‐Men‐1 after treatment with the demethylating agent 5‐aza‐2′‐deoxycytidine and the histone deacetylase inhibitor trichostatin A. 1, cells grown under standard conditions; 2, 500 nM 5‐aza‐2′‐deoxycytidine for 48 h plus 1 µM trichostatin A for 24 h; 3, 1 µM 5‐aza‐2′‐deoxycytidine for 72 h plus 1 µM trichostatin A for 24 h; 4, relative TIMP3 expression level in non‐neoplastic leptomeningeal tissue, Lept. C. Selected examples of the microsatellite analyses for allelic losses on 22q in meningiomas. Note the loss of heterozygosity of the polymorphic microsatellite marker D22S304 in MN63B and retention of D22S304 in MN412 in the tumor tissue (T), while in corresponding blood samples of the patients both alleles have been retained. D. Example of the methylation pattern of TIMP3 (region 2, CpG sites −6 to −11, reverse strand) in the anaplastic meningioma MN63B and non‐neoplastic leptomeningeal tissue (Lept.). Sodium bisulfite sequencing analysis reveals methylation at all CpG sites (marked with arrows) in the tumor, while the non‐neoplastic leptomeningeal tissue lacks CpG site methylation.
Methylation‐specific PCR (MSP) was carried out for 43 cycles using primers specific for the methylated or unmethylated DNA sequence (for primer sequences, see online supplementary Table S1). PCR products were separated on 2% agarose gels, and ethidium bromide‐stained bands were recorded with the GelDoc™ 1000 system (Bio‐Rad, Hercules, CA, USA). For direct bisulfite sequencing, PCR fragments were amplified from sodium bisulfite‐modified DNA (online supplementary Table S1 for the respective primer sequences) for 43 cycles, and the PCR products were then purified using the Jetquick PCR Product Purification Kit (Genomed, Löhne, Germany). Sequencing was performed with the BigDye® Cycle Sequencing Kit and an ABI PRISM 377 semi‐automated DNA sequencer (Applied Biosystems, Foster City, CA, USA). To calculate the degree of CpG site methylation in the investigated DNA segment, the methylation status at each analyzed CpG sites was semi‐quantitatively rated using the following scale: 0, completely unmethylated; 1, a weak methylated signal detectable in the sequence; 2, methylated signal approximately equal to unmethylated signal; and 3, methylated signal markedly stronger than unmethylated signal. Based on this rating, a regional methylation score was calculated for each tumor by adding the figures determined at the individual CpG sites. Methylation scores in region 2 clearly segregated tumors in two groups with either low or absent methylation (methylation score ≤4) or strong methylation (methylation score ≥42).
Real‐time reverse transcription (RT)‐PCR analysis
Three micrograms of total RNA from each tumor were reverse‐transcribed into cDNA using random hexanucleotide primer sequences and SuperScript®reverse transcriptase according to the manufacturer's protocol (Invitrogen Corporation, Carlsbad, CA, USA). Primers for TIMP3 expression analysis are supplied in supplementary Table S1. TIMP3 mRNA expression levels were determined by real‐time reverse transciption PCR analysis using the ABI PRISM 5700 sequence detection system (Applied Biosystems), which allows the continuous measurement of the PCR product amount by means of incorporation of SybrGreen fluorescent dye. Fluorescent data were converted into cycle threshold measurements using the SDS system software (Version 2.0, Applied Biosystems) and exported to Microsoft Excel. Fold expression changes relative to non‐neoplastic leptomeningeal tissue were calculated with the ΔΔCT method (20) using ARF1 (ADP‐ribosylation factor 1) as the reference transcript (27).
Induction of re‐expression of TIMP3 in vitro by 5‐Aza‐2′‐deoxycytidine and trichostatin A treatment
To assess for a causal relationship between TIMP3 hypermethylation and decreased TIMP3 mRNA expression, we treated the benign meningioma cell line Ben‐Men‐1 (kindly provided by Prof. W. Paulus, Münster, Germany) with the demethylating agent 5‐aza‐2′‐deoxycytidine and the histone deacetylase inhibitor trichostatin A. Cells were grown under standard conditions or under two different treatment conditions with either 500 nM 5‐aza‐2′‐deoxycytidine for 48 h plus 1 µM trichostatin A for 24 h or 1 µM 5‐aza‐2′‐deoxycytidine for 72 h plus 1 µM trichostatin A for 24 h. (Figure 4B). After harvesting of the cells and extraction of the mRNA, re‐expression of TIMP3 mRNA was assessed by real‐time RT‐PCR analysis as described above.
Immunohistochemistry
Immunohistochemistry was performed on formalin‐fixed paraffin‐embedded tissue sections using a commercially available TIMP3‐specific rabbit polyclonal antibody (Millipore, Billerica, MA, USA; 1:50 dilution). Immunoreactivity was detected with the EnVision™ Detection System (Dako, Hamburg, Germany) using 3,3′‐diaminobenzidine tetrahydrochloride (DAB) as a chromogen. TIMP3 protein expression was evaluated in 45 meningiomas (25 benign, 11 atypical and 9 anaplastic meningiomas) of the tumor panel. Negative controls without primary antibodies were performed for all reactions. As positive controls for TIMP3 staining, we employed selected breast, colon and pancreatic cancer tissues. Protein expression levels were semi‐quantitatively assessed by a composite numerical score, based on the percentage of positive stained tumor cells multiplied by staining intensity, potentially ranging from 0 to 12. The percentage of labeled cells was scored as follows: 0 (no or minimal reactivity, similar to non‐neoplastic brain tissue), 1 (<10%), 2 (10–50%), 3 (50–90%) and 4 (>90%). Staining intensity was graded as 0 (negative), 1 (weak), 2 (moderate) or 3 (strong).
Statistical analyses
Two‐sided Student's t‐test analyses were employed to compare TIMP3 mRNA expression between meningiomas of the different WHO grades (Figure 5A) and to assess the significance of TIMP3 mRNA expression differences between the group of tumors with low/absent and strong TIMP3 methylation (Figure 5B). The non‐parametric Mann–Whitney U‐test was used to compare protein expression scores between the different WHO grades. Fisher's exact test was utilized to further assess frequency differences of TIMP3 methylation between meningiomas of the different WHO grades (Figure 5C) and the association between TIMP3 hypermethylation and the allelic status on 22q12.3 (Figure 5D). A P‐value of <0.05 was considered as significant for all analyses.
Figure 5.
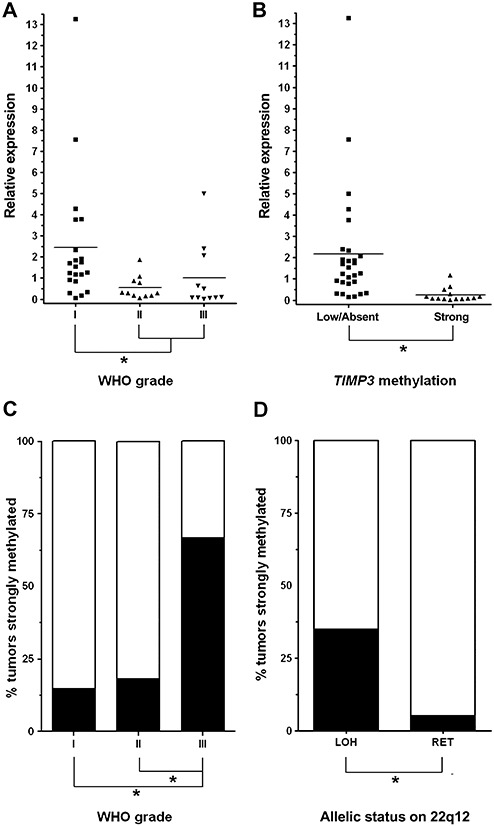
Associations between TIMP3 methylation (region 2), mRNA expression, WHO grade and allelic status on 22q. A. Mean TIMP3 transcript levels are significantly lower in atypical and anaplastic meningiomas than in benign meningiomas of WHO grade I. B. Meningiomas with strong TIMP3 5′‐CpG island methylation (methylation score ≥42) exhibit significantly lower TIMP3 mRNA expression levels as compared to meningiomas with low or absent TIMP3 methylation (methylation scores ≤4). C. Distribution of the TIMP3 methylation frequency over the different meningioma grades. Note that TIMP3 5′‐CpG island methylation is significantly more frequent in anaplastic (8 out of 12 cases, 67%) than in atypical (2 out of 11, 18%) and benign meningiomas (4 out of 27, 15%). D. TIMP3 methylation is significantly more frequent in meningiomas with allelic losses on 22q than in tumors with retention of this chromosome arm.
RESULTS
We first assessed TIMP3 5′‐CpG island methylation in a region that had been investigated in two preceding studies with controversial results (region 1, Figure 1). Both methylation‐specific PCR analysis and direct sodium bisulfite sequencing in our hands did not reveal tumor‐specific TIMP3 hypermethylation in a subset of 22 meningiomas representing the different WHO grades (Figure 2). Using direct sodium bisulfite sequencing, we then investigated the entire panel of 50 meningiomas for TIMP3 5′‐CpG island methylation in a region (region 2) located telomerically to region 1 and overlapping with the translation start site of TIMP3 (Figure 1). Tumors could be segregated into two clearly different groups with either strong methylation involving virtually the entire sequenced region (methylation score ≥42) or low or absent methylation corresponding to methylation scores ≤4 (3, 4). Of note, strong TIMP3 5′‐CpG island methylation was most frequent in anaplastic meningiomas (8 out of 12 cases, 67%), while atypical (2 out of 11 cases, 18%) and benign meningiomas (4 out of 27 cases, 15%) exhibited significantly lower TIMP3 methylation frequencies (Fisher's exact test; MN III vs. MN II, P = 0.0361; MN III vs. MN I, P = 0.0024; Figure 5C).
Using real‐time RT‐PCR analysis, we then assessed TIMP3 mRNA expression in 43 of the 50 meningiomas of our panel for which sufficient and high‐quality RNA was available. Relative to non‐neoplastic leptomeningeal tissue, TIMP3 expression showed a more than threefold reduction most frequently in atypical (7 out of 11, 64%) and anaplastic meningiomas (6 out of 11, 55%), while in benign meningiomas of WHO grade I, a respective decrease in TIMP3 mRNA expression was only observed in 3 out of 21 cases (14%; 3, 4). Moreover, comparing the mean TIMP3 mRNA expression between the different meningioma grades, we found that atpical (MN II; mean: 0.6; standard deviation, SD: 0.5) and anaplastic meningiomas (MN III; mean: 1.0; SD: 1.6) exhibited lower mean TIMP3 mRNA expression levels than benign meningiomas of WHO grade I (MN I; mean: 2.4; SD: 3.0), and these findings proved significant in Student's t‐test analyses (MN III and II vs. MN I, P = 0.0211; Figure 5A).
We next investigated whether the decrease in TIMP3 mRNA expression in higher grade meningiomas was also reflected at the protein level by performing immunhistochemistry in 45 of the 50 meningiomas of our tumor panel (3, 6). Benign meningiomas of WHO grade I had the highest median labeling scores (median labeling index: 4; interquartile range, IQR: 3). Intermediate TIMP3 immunoreactivity scores were denoted in atypical meningiomas (median: 3; IQR: 2), while anaplastic meningiomas in their majority exhibited only weak TIMP3 immunoreactivity in a fraction of tumor cells (median: 2; IQR: 2; Figure 6). Consistent with the mRNA data, TIMP3 protein expression was significantly lower in higher grade (atypical and anaplastic) meningiomas when compared with benign meningiomas (Mann–Whitney U‐test; MN III and II vs. MN I; P = 0.02).
Figure 6.
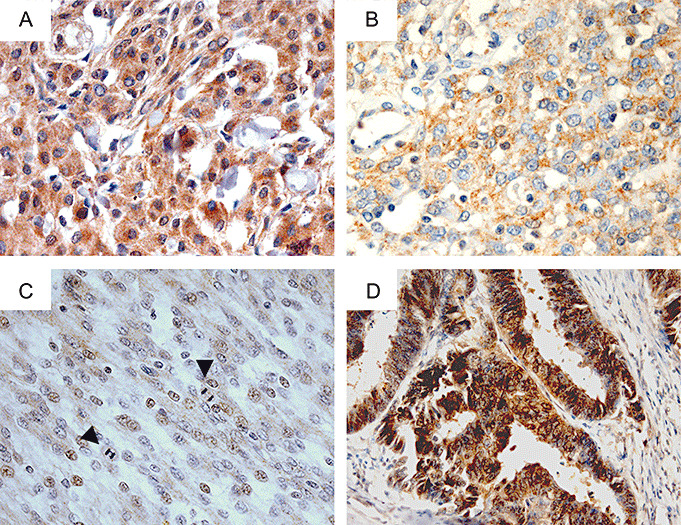
Immunohistochemical analysis of TIMP3 protein expression in meningiomas. A. Example of a benign meningioma of WHO grade I (MN30) with strong and diffuse immunoreactivity for TIMP3, consistent with retained expression. TIMP3 protein expression significantly decreases with malignant progression with intermediate levels in atypical (B, MN14) and lowest mean expression levels in anaplastic meningiomas (C, MN210). Note that both MN14 and MN210 exhibit concomitant TIMP3 hypermethylation. Magnification of all meningioma images, ×400. Arrowheads, mitoses. D. Colorectal carcinoma tissue served as a positive control for TIMP3 immunostaining. Magnification, ×200. All sections are counterstained with hematoxylin.
To assess the potential causal link between TIMP3 5′‐CpG island methylation and reduced TIMP3 expression, we treated the meningioma cell line Ben‐Men‐1 with the demethylating agent 5‐aza‐2′‐deoxycytidine and the histone deacetylase inhibitor trichostatin A. After harvesting of the cells and extraction of the mRNA, expression of TIMP3 transcripts increased in the range of four‐ to fivefold in the treated cells as compared with the untreated cells. Expression levels after treatment reflected those observed in non‐neoplastic leptomeningeal tissue (Figure 4B). Moreover, the finding of an association between TIMP3 5′‐CpG island methylation and decreased TIMP3 mRNA expression in meningiomas was reinforced by statistical associations in our tumor panel. When segregating tumors into two groups for strong (methylation score ≥42) and low/absent (methylation score ≤4) TIMP3 5′‐CpG island methylation, the 14 meningiomas with strong TIMP3 methylation exhibited significantly lower TIMP3 transcript levels (mean: 0.3; SD: 0.3) than the 29 tumors with low or absent TIMP3 methylation (mean: 2.2; SD: 2.7; Student's t‐test, P = 0.0123).
As TIMP3 is located close to the NF2 tumor suppressor gene on 22q12.3, a region that is often lost in meningiomas, we assessed the allelic status on 22q by means of microsatellite analyses in those 39 cases of our tumor panel for which corresponding patient blood samples were available (25 MN I, 10 MN II, 4 MN III). Allelic losses on 22q spanning the TIMP3 locus were detected in 20 of the 39 investigated meningiomas (51%; 3, 4). Of note, TIMP3 5′‐CpG island methylation was significantly more frequent in meningiomas with allelic losses on 22q (7 out of 20 cases, 35%) than in tumors with retention in this chromosomal region (1 out of 19, 5%; Fisher's exact test, P = 0.0436; Figure 5D). Thus, TIMP3 methylation—except for a single case (MN119)—was exclusive to meningiomas with 22q loss, while in the reverse comparison only about one‐third of tumors with 22q loss carried a TIMP3 methylation.
DISCUSSION
TIMP3 at 22q12.3 has been described as a gene with unique tumor suppressor‐like properties that when inactivated as a result of hypermethylation is associated with poor prognosis in a range of different cancers 4, 11, 23, 30, 32, 33. Here, we addressed the role of TIMP3 aberrations in the molecular pathogenesis of meningiomas and provide evidence that TIMP3 hypermethylation and transcriptional downregulation is associated with allelic losses on 22q12 and a more aggressive, high‐grade meningioma phenotype.
In a panel of 50 meningiomas covering the different WHO grades, we assessed methylation of the TIMP3 5′‐CpG island in two distinct regions. Region 1 had been investigated by methylation‐specific PCR analysis in two preceding studies with divergent results: While Bello and colleagues detected hypermethylation of this region of the TIMP3‐associated CpG islands in about 24% of meningiomas, Liu et al could not validate these findings 7, 19. We assessed TIMP3 methylation in region 1 by methylation‐specific PCR analysis and additional direct sodium bisulfite sequencing. Both methods did not indicate TIMP3 promoter hypermethylation in this region, thus corroborating the negative findings in the latter of the two preceding studies (19). The divergent results reported in the paper by Bello and colleagues might be a result of the methylation‐specific PCR analysis being restricted to detection of the methylation status of the few selected CG sites within the primer sequence that in individual tumors might not necessarily prove representative for the methylation status of the entire amplified sequence (7). We thus feel comfortable that the additional direct bisulfite sequence of region 1 performed in our study provides a more comprehensive picture of the methylation status of TIMP3 in region 1 as each of the 23 covered CG sites can be individually assessed. We then continued to investigate the TIMP3 5′‐CpG island in a region located telomerically to region 1 and directly covering the translation start site of TIMP3 (region 2). Of note, direct bisulfite sequencing of 21 CG sites revealed strong and explicit TIMP3 methylation in the majority (67%) of anaplastic meningiomas, while in atypical or benign meningiomas TIMP3 methylation was detected at significantly lower frequencies.
Assessing TIMP3 mRNA expression in our tumor panel, we found a significant reduction of the mean TIMP3 transcript levels in the clinically more aggressive, higher‐grade meningioma subtypes (WHO grades II and III), and a similar observation was made for the median TIMP3 protein expression levels as assessed by immunohistochemistry. These results are in line with reports in other tumor entities that describe a downregulation of TIMP3 expression in conjunction with high‐grade tumor phenotype and poor prognosis 5, 16, 17, 24, 26. Furthermore, we could provide in vitro and in vivo evidence for a causal relationship between TIMP3 methylation and the downregulation of TIMP3 transcripts in meningiomas. Treatment of the meningioma cell line Ben‐Men‐1 with the demethylating agent 5‐aza‐2′‐deoxycytidine and the histone deacetylase inhibitor trichostatin A resulted in increased expression of TIMP3 mRNA similar to the expression observed in non‐neoplastic leptomeningeal tissue. Moreover, in the tumor samples, meningiomas with strong TIMP3 methylation had significantly lower mean TIMP3 mRNA expression levels than the group of tumors that did not bear this epigenetic alteration. Nevertheless, it has to be mentioned that five atypical meningiomas in our series exhibited a more than threefold reduction of TIMP3 mRNA expression in the absence of TIMP3 methylation (Figure 3), which was also reflected by a mean TIMP3 mRNA expression decrease in the presence of an only negligible increase of methylation frequency in this tumor group (Figure 5A, C). A possible explanation for this discrepancy could be that TIMP3 hypermethylation is an important but not the sole cause of TIMP3 transcriptional downregulation. Indeed, recent reports indicate that other alterations might contribute to the transcriptional silencing of the TIMP3 gene product. As such, ERBB2 overexpression and colon‐carcinoma‐derived specific p53 mutant proteins have both been demonstrated to suppress the transcription of TIMP3 in murine fibroblasts 6, 29. Also miR‐21 has been indicated to downregulate TIMP3 expression in glioma cells in vitro and in a glioma model in nude mice (14). It requires further investigation if these mechanisms add to the inactivation of TIMP3 in meningiomas and may contribute to the more aggressive clinical behavior, particularly in atypical meningiomas, by substituting for the lower TIMP3 methylation frequency in these tumors in comparison with the anaplastic variants.
Unfortunately, we do not have clinical follow‐up data on our meningioma patients to assess whether patients with WHO grade III tumors that lacked TIMP3 inactivation showed a longer survival or whether patients with WHO grade I tumors and concomitant TIMP3 inactivation had a worse prognosis than the other patients within the respective grades. Nonetheless, our tumor panel partly overlapped with a panel of meningiomas for which CGH data had been generated in a previous publication (31). Indeed, the three WHO grade I meningiomas included in both studies that had a reduction in TIMP3 expression (MN12, MN17, MN27) showed a number of allelic gains and losses in addition to 22q deletions, as for example, 1p deletions in all three tumors. These molecular alterations may represent a precondition for a more aggressive clinical course in these selected tumors. However, the two WHO grade III lesions included in both studies that did not have TIMP3 inactivation (MN49A, MN64) showed an accumulation of multiple chromosomal gains and deletions as it is typical for anaplastic meningiomas, thus underlining the need for further investigation of the prognostic effect of TIMP3 inactivation in clinically well‐documented patient series.
TIMP3 maps to 22q12, a chromosomal band that is lost in about 50% of sporadic meningiomas and contains the tumor suppressor gene NF2 25, 28. Mutations of the NF2 gene—in addition to chromosomal deletions on 22q12—serve as a second hit to completely inactivate the NF2 gene product Merlin. NF2 mutation frequency does neither significantly differ between WHO grades nor does it increase in meningioma progression, suggesting that NF2 mutations arise early in meningiomagenesis and are relevant to meningioma initation 25, 28. In our meningioma panel, we detected loss of heterozygosity on 22q in 20 of the 39 investigated meningiomas (51%), which matches the frequencies reported in the literature. Notably, TIMP3 methylation—except for a single case—occurred exclusively in meningiomas with deletions on 22q. Thus, TIMP3 5′‐CpG island methylation seems selected for in a genetic background of 22q loss. However, in contrast to NF2 mutations, TIMP3 methylation was preferentially observed in high‐grade anaplastic meningiomas, thus appearing as a late event in meningiomagenesis and contributing to meningioma progression. As such, TIMP3 silencing by methylation may induce a more aggressive, high‐grade phenotype, preferentially in the subgroup of meningiomas that carry preexisting 22q deletions. Nevertheless, the high frequency of cases with 22q loss that do not bear TIMP3 methylation (about one‐third of tumors) underlines the major role of other tumor suppressor genes (NF2) in this region and confines the role of TIMP3 to a (more aggressive) subset of meningiomas.
Taken together, we here present a comprehensive study investigating TIMP3 methylation, mRNA and protein expression in meningiomas and correlating the results to the allelic status on 22q. The results indicate hypermethylation and transcriptional downregulation of TIMP3 as a mechanism that fairly exclusively occurs in meningiomas with allelic losses on 22q12 but—in contrast to NF2 mutation—appears to be involved in meningioma progression as it is significantly associated with a more aggressive, high‐grade meningioma phenotype.
Supporting information
Table S1. Primers used for TIMP3 methylation, expression and 22q12.3 microsatellite analyses (according to GenBank Accession no. NM_011520).
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
ACKNOWLEDGMENTS
The authors would like to thank Peter Roerig for helpful suggestions in the initiation of the study, and Britta Friedensdorf for excellent technical assistance. This work was supported by the Max‐Eder Junior Research Group Program of the German Cancer Aid (Deutsche Krebshilfe, grant no. 107709 to Markus J. Riemenschneider). Prof. W. Paulus, Münster, is kindly acknowledged for providing the Ben‐Men‐1 cell line.
REFERENCES
- 1. Ahonen M, Baker AH, Kahari VM (1998) Adenovirus‐mediated gene delivery of tissue inhibitor of metalloproteinases‐3 inhibits invasion and induces apoptosis in melanoma cells. Cancer Res 58:2310–2315. [PubMed] [Google Scholar]
- 2. Anand‐Apte B, Bao L, Smith R, Iwata K, Olsen BR, Zetter B et al (1996) A review of tissue inhibitor of metalloproteinases‐3 (TIMP‐3) and experimental analysis of its effect on primary tumor growth. Biochem Cell Biol 74:853–862. [DOI] [PubMed] [Google Scholar]
- 3. Anand‐Apte B, Pepper MS, Voest E, Montesano R, Olsen B, Murphy G et al (1997) Inhibition of angiogenesis by tissue inhibitor of metalloproteinase‐3. Invest Ophthalmol Vis Sci 38:817–823. [PubMed] [Google Scholar]
- 4. Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK et al (1999) Methylation‐associated silencing of the tissue inhibitor of metalloproteinase‐3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res 59:798–802. [PubMed] [Google Scholar]
- 5. Bai YX, Yi JL, Li JF, Sui H (2007) Clinicopathologic significance of BAG1 and TIMP3 expression in colon carcinoma. World J Gastroenterol 13:3883–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beckers J, Herrmann F, Rieger S, Drobyshev AL, Horsch M, Hrabe de Angelis M et al (2005) Identification and validation of novel ERBB2 (HER2, NEU) targets including genes involved in angiogenesis. Int J Cancer 114:590–597. [DOI] [PubMed] [Google Scholar]
- 7. Bello MJ, Aminoso C, Lopez‐Marin I, Arjona D, Gonzalez‐Gomez P, Alonso ME et al (2004) DNA methylation of multiple promoter‐associated CpG islands in meningiomas: relationship with the allelic status at 1p and 22q. Acta Neuropathol 108:413–421. [DOI] [PubMed] [Google Scholar]
- 8. Bian J, Wang Y, Smith MR, Kim H, Jacobs C, Jackman J et al (1996) Suppression of in vivo tumor growth and induction of suspension cell death by tissue inhibitor of metalloproteinases (TIMP)‐3. Carcinogenesis 17:1805–1811. [DOI] [PubMed] [Google Scholar]
- 9. Van Den Boom J, Wolter M, Kuick R, Misek DE, Youkilis AS, Wechsler DS et al (2003) Characterization of gene expression profiles associated with glioma progression using oligonucleotide‐based microarray analysis and real‐time reverse transcription‐polymerase chain reaction. Am J Pathol 163:1033–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Deng X, Bhagat S, Dong Z, Mullins C, Chinni SR, Cher M (2006) Tissue inhibitor of metalloproteinase‐3 induces apoptosis in prostate cancer cells and confers increased sensitivity to paclitaxel. Eur J Cancer 42:3267–3273. [DOI] [PubMed] [Google Scholar]
- 11. Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L et al (2001) Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res 61:3410–3418. [PubMed] [Google Scholar]
- 12. Edwards DR (2004) TIMP‐3 and endocrine therapy of breast cancer: an apoptosis connection emerges. J Pathol 202:391–394. [DOI] [PubMed] [Google Scholar]
- 13. Finan KM, Hodge G, Reynolds AM, Hodge S, Holmes MD, Baker AH et al (2006) In vitro susceptibility to the pro‐apoptotic effects of TIMP‐3 gene delivery translates to greater in vivo efficacy versus gene delivery for TIMPs‐1 or ‐2. Lung Cancer 53:273–284. [DOI] [PubMed] [Google Scholar]
- 14. Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS et al (2008) MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 28:5369–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han X, Zhang H, Jia M, Han G, Jiang W (2004) Expression of TIMP‐3 gene by construction of a eukaryotic cell expression vector and its role in reduction of metastasis in a human breast cancer cell line. Cell Mol Immunol 1:308–310. [PubMed] [Google Scholar]
- 16. Helleman J, Jansen MP, Ruigrok‐Ritstier K, Van Staveren IL, Look MP, Meijer‐van Gelder ME et al (2008) Association of an extracellular matrix gene cluster with breast cancer prognosis and endocrine therapy response. Clin Cancer Res 14:5555–5564. [DOI] [PubMed] [Google Scholar]
- 17. Hilska M, Roberts PJ, Collan YU, Laine VJ, Kossi J, Hirsimaki P et al (2007) Prognostic significance of matrix metalloproteinases‐1, ‐2, ‐7 and ‐13 and tissue inhibitors of metalloproteinases‐1, ‐2, ‐3 and ‐4 in colorectal cancer. Int J Cancer 121:714–723. [DOI] [PubMed] [Google Scholar]
- 18. Leone PE, Bello MJ, De Campos JM, Vaquero J, Sarasa JL, Pestana A et al (1999) NF2 gene mutations and allelic status of 1p, 14q and 22q in sporadic meningiomas. Oncogene 18:2231–2239. [DOI] [PubMed] [Google Scholar]
- 19. Liu Y, Pang JC, Dong S, Mao B, Poon WS, Ng HK (2005) Aberrant CpG island hypermethylation profile is associated with atypical and anaplastic meningiomas. Hum Pathol 36:416–425. [DOI] [PubMed] [Google Scholar]
- 20. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- 21. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (eds) (2007) WHO Classification of Tumours of the Central Nervous System, 3rd edn. IARC Press: Lyon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mueller W, Nutt CL, Ehrich M, Riemenschneider MJ, Von Deimling A, Van Den Boom D et al (2007) Downregulation of RUNX3 and TES by hypermethylation in glioblastoma. Oncogene 26:583–593. [DOI] [PubMed] [Google Scholar]
- 23. Nakamura M, Ishida E, Shimada K, Kishi M, Nakase H, Sakaki T et al (2005) Frequent LOH on 22q12.3 and TIMP‐3 inactivation occur in the progression to secondary glioblastomas. Lab Invest 85:165–175. [DOI] [PubMed] [Google Scholar]
- 24. Ninomiya I, Kawakami K, Fushida S, Fujimura T, Funaki H, Takamura H et al (2008) Quantitative detection of TIMP‐3 promoter hypermethylation and its prognostic significance in esophageal squamous cell carcinoma. Oncol Rep 20:1489–1495. [PubMed] [Google Scholar]
- 25. Perry A, Gutmann DH, Reifenberger G (2004) Molecular pathogenesis of meningiomas. J Neurooncol 70:183–202. [DOI] [PubMed] [Google Scholar]
- 26. Riddick AC, Shukla CJ, Pennington CJ, Bass R, Nuttall RK, Hogan A et al (2005) Identification of degradome components associated with prostate cancer progression by expression analysis of human prostatic tissues. Br J Cancer 92:2171–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Riemenschneider MJ, Koy TH, Reifenberger G (2004) Expression of oligodendrocyte lineage genes in oligodendroglial and astrocytic gliomas. Acta Neuropathol 107:277–282. [DOI] [PubMed] [Google Scholar]
- 28. Riemenschneider MJ, Perry A, Reifenberger G (2006) Histological classification and molecular genetics of meningiomas. Lancet Neurol 5:1045–1054. [DOI] [PubMed] [Google Scholar]
- 29. Thomas S, Reisman D (2006) Localization of a mutant p53 response element on the tissue inhibitor of metalloproteinase‐3 promoter: mutant p53 activities are distinct from wild‐type. Cancer Lett 240:48–59. [DOI] [PubMed] [Google Scholar]
- 30. Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP, Hruban RH et al (2000) Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res 60:1835–1839. [PubMed] [Google Scholar]
- 31. Weber RG, Bostrom J, Wolter M, Baudis M, Collins VP, Reifenberger G et al (1997) Analysis of genomic alterations in benign, atypical, and anaplastic meningiomas: toward a genetic model of meningioma progression. Proc Natl Acad Sci U S A 94:14719–14724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yuan Y, Wang J, Li J, Wang L, Li M, Yang Z et al (2006) Frequent epigenetic inactivation of spleen tyrosine kinase gene in human hepatocellular carcinoma. Clin Cancer Res 12:6687–6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zochbauer‐Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD (2001) Aberrant promoter methylation of multiple genes in non‐small cell lung cancers. Cancer Res 61:249–255. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used for TIMP3 methylation, expression and 22q12.3 microsatellite analyses (according to GenBank Accession no. NM_011520).
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


