Abstract
Glioblastomas are morphologically and genetically heterogeneous, but little is known about the regional patterns of genomic imbalance within glioblastomas. We recently established a reliable whole genome amplification (WGA) method to randomly amplify DNA from paraffin‐embedded histological sections with minimum amplification bias [Huang et al (J Mol Diagn 11: 109–116, 2009)]. In this study, chromosomal imbalance was assessed by array comparative genomic hybridization (CGH; Agilent 105K, Agilent Technologies, Santa Clara, CA, USA), using WGA‐DNA from two to five separate tumor areas of 14 primary glioblastomas (total, 41 tumor areas). Chromosomal imbalances significantly differed among glioblastomas; the only alterations that were observed in ≥6 cases were loss of chromosome 10q, gain at 7p and loss of 10p. Genetic alterations common to all areas analyzed within a single tumor included gains at 1q32.1 (PIK3C2B, MDM4), 4q11–q12 (KIT, PDGFRA), 7p12.1–11.2 (EGFR), 12q13.3–12q14.1 (GLI1, CDK4) and 12q15 (MDM2), and loss at 9p21.1–24.3 (p16INK4a/p14ARF), 10p15.3–q26.3 (PTEN, etc.) and 13q12.11–q34 (SPRY2, RB1). These are likely to be causative in the pathogenesis of glioblastomas (driver mutations). In addition, there were numerous tumor area‐specific genomic imbalances, which may be either nonfunctional (passenger mutations) or functional, but constitute secondary events reflecting progressive genomic instability, a hallmark of glioblastomas.
Keywords: array CGH, glioblastoma, intratumoral heterogeneity, whole genome amplification
INTRODUCTION
Glioblastomas harbor a great variety of genetic alterations 2, 19, 22 and are also morphologically highly heterogeneous 9, 15. Histological criteria for the diagnosis of glioblastoma include marked mitotic activity and the presence of focal necrosis and/or microvascular proliferation (15). Some additional histological features tend to correlate with specific genetic alterations. Burger et al (1) reported that small cell glioblastomas are usually primary (de novo) glioblastomas and are typically associated with EGFR amplification. Similarly, Perry et al (23) observed that small cell glioblastomas frequently contain EGFR amplification (72%) and loss of heterozygosity (LOH) at 10q (100%), while none showed loss of 1p/19q. In our previous study of 420 cases, small cell glioblastomas had frequent EGFR amplification (46%) and p16INK4a homozygous deletion (39%), but infrequent PTEN mutations (19%), while the presence of multinucleated giant cells was associated with frequent TP53 mutations (45%) but infrequent EGFR amplification (24%) (9). The histological features of a glioblastoma often vary in different areas of the tumor. This has been interpreted as a reflection of genetic heterogeneity, but little is known about the extent of intratumoral heterogeneity at the genomic level.
We have recently developed a method for whole genome amplification (WGA) of DNA from small tumor areas on paraffin sections with minimum amplification bias, to generate sufficient quantity of DNA for array comparative genomic hybridization (CGH) (10). We found that a ligation step before WGA is important as it allows a short reaction time with Phi29 DNA polymerase to generate WGA‐DNA with significantly decreased amplification bias (10). Using this method, we assessed intratumoral genome‐wide chromosomal imbalance in WGA‐DNA of two to five small tumor areas from 14 glioblastomas on the same histological slide (total, 41 tumor areas), using array CGH.
MATERIALS AND METHODS
Tumor samples
We screened histological sections of 420 glioblastomas collected in our previous population‐based study 9, 20. We first selected a total of 20 cases that contained at least two relatively large (2–5 mm in diameter) tumor areas with distinct histological features. Of these, high‐quality DNA was available for array CGH in 14 cases. In seven cases, all tumor areas appeared to be similar, being either of the small cell type (monomorphic cells characterized by small, round to slightly elongated, densely packed cells with mildly hyperchromatic nuclei, high nuclear : cytoplastic ratio and modest atypia) or pleomorphic cell type (varying in size and shape of nuclei and cytoplasm) 9, 15. In the remaining seven cases, tumor areas with different predominant histological features (eg, cells resembling gemistocytes, multinucleated giant cells, oligodendroglial components) could clearly be recognized. Tumors were fixed in buffered formalin and embedded in paraffin. All cases were clinically diagnosed as primary glioblastomas, but one case (case PB574) contained an IDH1 mutation (18).
DNA extraction
For each glioblastoma, DNA was extracted as previously reported (10) from two to five separate tumor areas (approximately 2–5 mm in diameter; separated by at least 5 mm; Table 1), avoiding necrosis, vascular proliferation and inflammatory cells. For the histologically homogeneous glioblastomas, the tumor areas were randomly selected, while for the histologically heterogeneous glioblastomas, samples were chosen from the tumor areas displaying distinct histological features.
Table 1.
Regional patterns of genomic imbalance in glioblastomas
Briefly, tumor areas were scraped off from the histological slide and were deparaffinized in xylene for 15 minutes and then in 100% ethanol for 10 minutes. The pellets were dried in acetone and washed with 0.4% TWEEN 20® (Sigma, St Louis, MO, USA) solution and then with PBS (pH 7.4), before being dissolved in 400 µL of 1 M NaSCN solution. After overnight incubation at 37°C, the samples were suspended in 400 µL of DNA extraction buffer, composed of 360 µL of ATL buffer and 40 µL of proteinase K (DNeasy® Mini kit, Qiagen, Valencia, CA, USA), and were incubated overnight at 55°C. Additional proteinase K (40 µL) was added 24 h and 12 h later, with a total incubation time of 60 h. After incubation with 8 µL RNase (100 mg/mL) for 2 minutes at room temperature (RT), 420 µL ATL buffer was added, and the samples were separated into two parts (each 450 µL). Each part was mixed with 450 µL AL buffer and 450 µL 100% ethanol, and incubated at RT for 5 minutes. Samples were loaded into a DNeasy® Mini spin columns (Qiagen). After washing the column with AW1 buffer and drying the column membrane with 80% ethanol, the purified genomic DNA was eluted with 25 µL nuclease‐free H2O. The DNA concentration and quality was determined using an ND‐8000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Phi29‐based WGA of genomic DNA
Genomic DNA was ligated and amplified using REPLI‐g® FFPE Kit (Qiagen) as previously described (10). Briefly, purified DNA (300 ng) in a total volume of 10 µL was denatured at 95°C for 5 minutes. After cooling the samples on ice for 5 minutes, 8 µL FFPE buffer, 1 µL ligation enzyme and 1 µL FFPE enzyme were added, and the samples were incubated at 24°C for 30 minutes, followed by heat inactivation at 95°C for 5 minutes. The samples (20 µL) were then mixed with 30 µL of reaction mix (29 µL of reaction buffer and 1 µL Midi Phi29 DNA polymerase) and were incubated at 30°C for 1 h. The Phi29 enzyme was inactivated by heating at 95°C for 10 minutes.
Reproducibility of genetic alterations in DNA samples after WGA
To ensure that the DNA produced by WGA (WGA‐DNA) genetically represented the original DNA, the presence of previously reported genetic alterations 18, 20, 30 in each glioblastoma sample was determined. Briefly, direct DNA sequencing was carried out to screen for mutations of IDH1, TP53 and PTEN, as previously described 20, 29, 30. EGFR amplification was detected by differential PCR using the cystic fibrosis (CF) gene sequence as a reference, as previously described (29). We confirmed that all the known genetic alterations assessed were present in the WGA‐DNA.
Array CGH
The products of WGA were purified with NucleoTraP®CR kit (Macherey‐Nagel, Düren, Germany) before DNA labeling, as previously reported (10). Briefly, after the volume of the reaction mix was adjusted to 100 µL using PBS (pH 7.4), and adding buffer NT (400 µL), the samples were mixed thoroughly by vortex, 10 µL of the NucleoTraP®CR kit suspension containing silica matrix was added and the samples were incubated at RT for 10 minutes. After centrifugation at 10 000 × g for 30 s, the pellets were washed with buffers NT2, NT3 and NT3 (400 µL each), then dried at RT for 15 minutes. Nuclease‐free water (30 µL) was used to dissolve the pellets, and the samples were then incubated at 55°C for 15 minutes. After centrifuging at 10 000 × g for 30 s, the supernatants were transferred to new tubes. The yield of WGA‐DNA after purification was determined using the ND‐8000 spectrophotometer.
The genomic profile changes of paired DNA samples were compared using a 2× 105K CGH oligonucleotide microarrray (Agilent Technologies, Santa Clara, CA, USA; 15.0 Kb average probe resolution) according to the manufacturer's instructions. Briefly, the sample (1 µg) and the sex‐matched reference DNA were chemically labeled with ULS‐Cy5 and ULS‐Cy3, respectively, at 85°C for 30 minutes using an oligo aCGH labeling kit for FFPE samples (Agilent). The labeled samples were purified with the genomic DNA purification module (Agilent), combined, mixed with human Cot‐1 DNA, denatured at 95°C (oligo aCGH hybridization kit) and were applied to microarrays. After hybridization at 65°C for 40 h, the microarrays were washed in oligo aCGH wash buffer 1 at RT for 5 minutes and in wash buffer 2 at 37°C for 1 minute. After drying, the microarrays were scanned with a DNA microarray scanner G2565BA (Agilent), and data (log2) were extracted from the raw microarray‐image files using Feature Extraction software (version 9; Agilent). Data were analyzed by DNA Analytics software (version 3.5; Agilent) with default filter settings. The aberration detection method 2 algorithm with fuzzy zero correction was used to define aberrant intervals.
Validation of array CGH data
Amplification of KIT and PDGFRA
To validate amplification of the KIT and PDGFRA genes located at 4q12, differential PCR was performed as previously reported (26), using the CF sequence as a reference (29). The primer sequences were as follows: 5′‐TCC TGG ATG AAA CGA ATG AGA‐3′ (sense) and 5′‐TTG CTT AAG CCG TGT TTG TTG‐3′ (antisense) for KIT (PCR product, 106 bp), 5′‐GCT GTT TCT GTT GAC TTT TGA C‐3′ (sense) and 5′‐AAA CAA GGA ACT CAG AGA GGA‐3′ (antisense) for PDGFRA (product, 125 bp), and 5′‐GGC ACC ATT AAA GAA AAT ATC ATC TT‐3′ (sense) and 5′‐GTT GGC ATG CTT TGA TGA CGC TTC‐3′ (antisense) for the CF (product, 79 bp). PCR was carried out with 28 cycles, with an annealing temperature of 55°C. The PCR products were loaded on 8% acrylamide gels and stained with ethidium bromide. Quantitative analysis of the signal intensity was performed using the Molecular Imager and Quantity One Analysis Software (Bio‐Rad, Hercules, CA, USA). The mean KIT : CF and PDGFRA : CF ratios using DNA from peripheral blood of 14 healthy individuals were 0.78 and 0.98, with standard variations of 0.18 and 0.14, respectively. Threshold values of 2.12 and 2.34 were regarded as evidence of KIT and PDGFRA amplification, respectively, as previously reported (26).
AKT1 amplification
To validate amplification of the AKT1 genes (at 14q32.33), differential PCR was performed as previously described (26), using the CF sequence as a reference (29). The primer sequences were as follows: 5′‐CAC GCT ACT TCC TCC TCA AGA A‐3′ (sense) and 5′‐TAC GCG CCA CAG AGA AGT TG‐3′ (antisense; PCR product, 108 bp). PCR was carried out with 28 cycles, with an annealing temperature of 55°C. The mean AKT1/CF ratio using DNA from peripheral blood of 14 healthy individuals was 0.94, with standard variation of 0.06. Threshold values of 2.06 were regarded as evidence of AKT1 amplification, as previously reported (26).
LOC157740 amplification
To validate amplification of the LOC157740 gene (at 8p23.1), differential PCR was performed as previously described (26), using the CF sequence as a reference (29). The primer sequences were as follows: 5′‐ACT CCT TTG CTT GTT CCT TTC C‐3′ (sense) and 5′‐ACA GCA GAC CAG CAC TCA AAC A‐3′ (antisense; PCR product, 96 bp). PCR was carried out with 28 cycles, with an annealing temperature of 55°C. The mean LOC157740/CF ratio using DNA from peripheral blood of 12 healthy individuals was 0.83, with standard variation of 0.05. Threshold values of 1.81 were regarded as evidence of amplification, as previously reported (26).
LOH at 10q and 19q
LOH at 10q and 19q was assessed by quantitative microsatellite analysis as previously described (17), using microsatellite markers D10S536 at 10q23.31 and D10S1683 at 10q25.3, and three markers (D19S49, D19S931 and D10S932) at 19q12. PCR reactions were performed in a total volume of 20 µL with 10 µL of iQ Supermix (Bio‐Rad), 6.4 µL of primer sets (1.25 µmol/L of each primer), 2 µL of 1.5 µmol/L probe [21‐bp oligomer complementary to the microsatellite CA repeat: 5′,6‐carboxyfluorescein (FAM) TGTGTGTGTGTGTGTGTGTGT‐3′, black hole quencher‐1] and 20 ng of DNA, with cycling parameters as reported previously (17). PCR was performed in triplicate on a 96‐well optical plate with an iCycler iQ5 Detection System (Bio‐Rad). Amplification of a pool of six reference loci served to normalize for differences in the amount of total input DNA, as described previously. To calculate the average δCt [δCt (normal)], DNA was isolated from 14 FFPE normal tissues. The Ct, δCt [Ct (microsatellite) – Ct (reference pool)] and δδCt [δCt (tumor) – δCt (normal)] values, the relative copy number (2−δδCt) and the tolerance interval with confidence of 95% determined from the pooled standard deviation of normal DNA for the loci were calculated as reported previously (6). On the basis of this tolerance interval, copy numbers <1.37 were considered to represent losses.
RESULTS
Array CGH
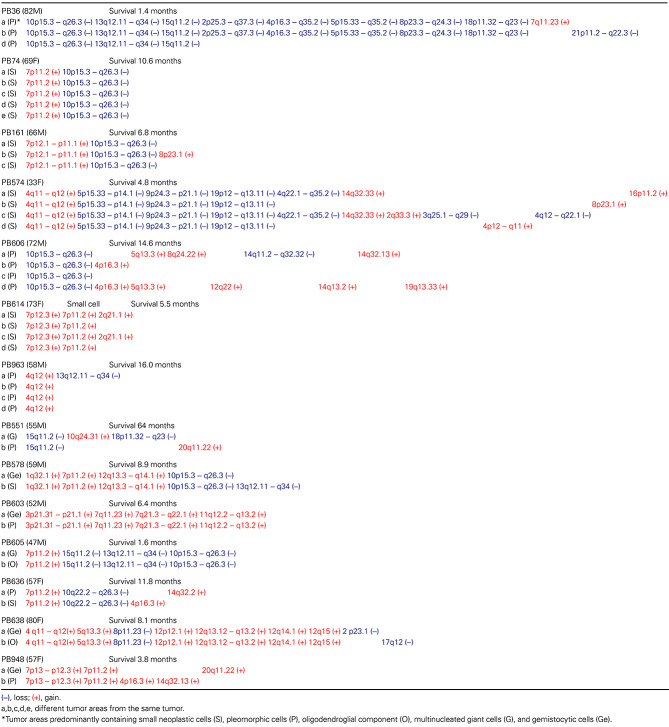
Chromosomal imbalances differed significantly between glioblastomas (Table 1); the only alterations that were observed in ≥2 cases were loss of chromosome 10q (seven cases; 50%), gain at 7p containing the EGFR locus (seven cases; 50%), loss of 10p (six cases, 43%), gain at 4q12 containing the KIT/PDGFRA locus (three cases; 21%), loss of 15q11.2 (three cases; 21%), gain at 12q14.1 containing the CDK4 locus (two cases, 14%) and loss at 13q12.11–q34 containing the SPRY2 and RB1 loci (two cases, 14%), while the other alterations were observed in only one case.
Comparison of different areas of the same tumor revealed that all glioblastomas had genetic alterations that were common to all areas analyzed and other alterations that were area specific (Table 1).
Genetic alterations that were common to all tumor areas analyzed included gains at 1q32.1 (PIK3C2B, MDM4), 3p21.31–p21.1, 4q11–q12 (KIT, PDGFRA), 5q13.3, 7p13–12.3, 7p12.3, 7p12.1–11.2 (EGFR), 7q11.23, 7q21.3–q22.1, 11q12.2–q13.2, 12p12.1, 12q13.12–12q13.2, 12q13.3–12q14.1 (GLI1, CDK4) and 12q15 (MDM2), and loss at 5p15.33–p14.1, 8p11.23, 9p21.1–24.3 (p16INK4a/p14ARF), 10p15.3–q26.3 (PTEN, etc.), 13q12.11–q34 (SPRY2, RB1), 15q11.2 and 19p12–q13.11 (Table 1; Figure 1).
Figure 1.

Graphic display of chromosomal imbalance detected by array CGH in representative cases. Case PB36 shows loss of 10p, 10q, 13q and 15q11.2 in all three tumor areas analyzed, while loss at 2p, 2q, 4p, 4q, 5p, 5q, 8p, 8q, 18p and 18q were observed in only two tumor areas. Gain at 7q11.23 and loss at 21p11.2–q22.3 were detected in only one tumor area. In case PB574, alterations common to all four tumor areas were gain at 4q11–q12 and loss at 5p15.33–p14.1, 9p24.3–p21.1 and 19p12–q13.11. In addition, tumor area‐specific alterations included gain at 14q32.33, 2q33.3, 4p12–q11, 8p23.1 and 16p11.2, and loss at 3q25.1–q29, 4q12–q22.1 and 4q22.1–q35.2.
Tumor area‐specific genetic alterations included gains at 2q21.1, 2q33.3, 4p12–q11 and 4p16.3 (three cases); 5q13.3, 7q11.23 and 8p23.1 (two cases); 8q24.22, 10q24.31, 12q22, 14q32.33 (AKT1), 14q13.2 and 14q32.13 (two cases); and 14q32.2, 16p11.2, 19q13.33 and 20q11.22 (two cases), and loss at 2p23.1, 2p25.3–q37.3, 3q25.1–q29, 4p16.3–q35.2, 4q22.1–q35.2, 4q12–q22.1, 5p15.33–q35.2, 8p23.3–q24.3 and 13q12.11–q34 (two cases); 14q11.2–q32.32, 17q12 and 18p11.32–q23 (two cases); and 21p11.2–q22.3 (Table 1; Figure 2).
Figure 2.

Array CGH analysis showing chromosomal imbalance at 14q32.33 in tumor areas a and c (case PB574) (A). Amplification of the AKT1 gene* (at 14q32.33) in the original DNA without whole genome amplification detected by differential PCR (B). M = molecular size marker.
There was no significant difference in the extent of chromosomal imbalance between tumors showing homogeneous histological features and those containing histologically distinct tumor areas.
Validation of array CGH data
Differential PCR was carried out to assess amplification of the PDGFRA and KIT genes in 10 tumor areas from three glioblastomas (cases PB574, PB638 and PB963), in which array CGH results showed gain at 4q12 (containing the PDGFRA and KIT loci). In all 10 tumor areas, amplification of the PDGFRA and KIT genes was confirmed in the WGA‐DNA and in the original DNA before WGA.
LOH analysis was carried out using two microsatellite markers for 10q23.31 and 10q25.3 in 21 tumor areas from seven glioblastomas (cases PB36, PB74, PB161, PB578, PB605, PB606, PB636), in which array CGH results showed LOH at 10q. In all 21 tumor areas, LOH 10q23.31 and 10q25.3 were confirmed in the WGA‐DNA and in the original DNA before WGA.
LOH analysis was carried out using three microsatellite markers for 19q12 in four tumor areas from one case (case PB574), in which array CGH results showed LOH 19q12. In all four tumor areas, LOH at 19q12 was confirmed in the WGA‐DNA and in the original DNA before WGA.
Differential PCR was carried out to assess amplification of the AKT1 gene in two tumor areas (a and c) from one case (case PB574), for which array CGH had revealed gain at 14q32.33 (containing the AKT1 locus). AKT1 amplification was detected in tumor areas a and c in the WGA‐DNA and in the original DNA before WGA. In contrast, AKT amplification was not detected in DNA from two other tumor areas (b and d) either before or after WGA (Figure 2).
Differential PCR was performed to assess amplification of the LOC157740 gene in one tumor area (b) from one case (case PB574), for which array CGH had shown gain at 8p23.1 containing the LOC157740 locus. LOC157740 amplification was detected in the tumor area b in the WGA‐DNA and in the original DNA before WGA. LOC157740 amplification was not detected in DNA from three other tumor areas (a, c and d) either before or after WGA.
DISCUSSION
Tumors develop through the sequential acquisition of genetic alterations. Recent genome‐wide sequencing has revealed the complexity of cancer genomes, showing that a typical solid tumor contains 40–80 miscoding mutations 5, 12, 28, 31. However, not all genetic alterations are considered to play a role in the development and progression of tumors. Some somatic mutations are involved in tumor initiation by conveying a growth advantage and positive selection (driver mutations), whereas others may not have functional consequences, do not promote growth and do not contribute to tumor initiation (passenger mutations) (7). A third type of genetic alteration results from the genomic instability of cancer cells and may have functional consequences, but does not play a role in the early stages of malignant transformation; such mutations can be a contributing factor in progression to a more malignant phenotype.
Little is known about the regional (tumor area‐specific) patterns of genomic imbalance in glioblastomas. Misra et al (16) assessed genetic alterations in two to three tumor areas from seven glioblastomas (total, 16 tumor areas), using a DNA fingerprinting technique involving randomly amplified polymorphic DNA (RAPD). In all cases except one, different areas of the tumor had a different fingerprint, indicating a remarkable extent of intratumoral genetic heterogeneity (16). Using conventional CGH, Harada et al (8) assessed chromosomal imbalance in two to three tumor areas from 11 glioblastomas. They showed that some chromosome imbalances were region independent, that is, common to all tumor areas analyzed, while in eight cases, there were also region‐dependent cytogenetic changes, suggesting intratumoral heterogeneity (8). Jung et al (13) carried out conventional CGH on WGA‐DNA produced by degenerate oligonucleotide‐primed PCR (DOP‐PCR) for a total of 20 different tumor areas from eight glioblastomas and observed that some genetic alterations were shared by all tumor areas examined (13). However, studies using RAPD or conventional CGH do not allow the precise chromosomal localization of such alterations.
The amount of DNA required for array CGH makes it difficult to apply this method to DNA samples extracted from small tumor areas on FFPE sections. To overcome this problem, several laboratories have attempted to develop a WGA protocol to generate a sufficient amount of DNA for aCGH, but most WGA methods (including DOP‐PCR) are associated with significant sequence representation bias or artifacts 3, 4, 24.
We have recently established an optimal protocol for WGA of DNA extracted from small tumor areas on formalin‐fixed, paraffin embedded histological sections. The use of Phi29 DNA polymerase, a short incubation time and a ligation step before amplification results in low amplification bias and high reproducibility (10). In the present study, we used this WGA method to obtain DNA for array CGH analysis of 41 small tumor areas from 14 glioblastomas. Before carrying out array CGH, we confirmed that the genetic alterations previously identified in each glioblastoma sample 18, 20, 30 were present in the WGA‐DNA (see Materials and Methods). The results obtained using array CGH were also validated by additional methods, such as differential PCR and LOH analysis. The results were identical for DNA samples before or after WGA, indicating that our method yields probes that reflect the genome of the original tumor sample.
In the present study, we observed remarkably different patterns of chromosomal imbalance in different glioblastomas. The only alterations found in >20% of the cases analyzed were loss of chromosome 10q and 10p, gain at 7p (containing EGFR), gain at 4q12 (containing KIT/PDGFRA) and loss of 15q11.2.
Comparison of different areas of the same tumor always revealed genetic alterations common to all tumor areas analyzed, while other imbalances were observed only in specific tumor areas. Genetic alterations that were common to all tumor areas are likely to convey a growth advantage and are considered to represent early genetic events that are involved in the pathogenesis of glioblastomas (driver mutations). These included loss of 10p and 10q, gain at 7p11.2 (EGFR), gain at 1q32.1 (PIK3C2B, MDM4), gain at 4q12 (KIT, PDGFRA), gain at 12q13.3–14.1 (CDK4, GLI1), gain at 12q15 (MDM2), loss at 13q12.11–q34 (SPRY2, RB1) and loss at 9p21.1–24.3 (p16INK4a, p14ARF). These loci contain well characterized oncogenes and tumor suppressor genes that play important roles in several signaling pathways and that have been reported to be commonly altered in glioblastomas 2, 11, 19, 20. Other chromosomal loci commonly altered in all tumor areas analyzed include gain at 3p21.31–p21.1, 5q13.3, 7p13–12.3, 7p12.3, 7q11.23, 7q21.3–q22.1, 11q12.2–q13.2, 12p12.1 and 12q13.12–12q13.2, and loss at 5p15.33–p14.1, 8p11.23, 15q11.2 and 19p12–q13.11, suggesting that, although rare, there may be additional driver mutations involved in the pathogenesis of glioblastomas. It is noted that gains at 1q32.1, 3p21.31–p21.1, 5q13.3 and 11q12.2–q13.2, and loss at 5p15.33–p14.1 and 19p12–q13.11 have not been detected as common alterations in previous reports on genetic heterogeneity in glioblastomas 8, 13.
We also observed genetic alterations in only one or two, but not in all tumor areas analyzed (tumor area‐specific genetic alterations). Some of these genetic alterations were reported previously (gains at 2q21.1, 4p12–q11, 4p16.3, 5q13.3, 7q11.23, 8p23.1, 8q24.22, 12q22, 19q13.33 and 20q11.22, and loss at 4p16.3–q35.2) 8, 13, but others that we detected in the present study have not been reported previously as tumor area‐specific genetic alterations (gains at 2q33.3, 10q24.31, 14q32.33, 14q13.2, 14q32.13, 14q32.2 and 16p11.2, and loss at 2p23.1, 2p25.3–q37.3, 3q25.1–q29, 5p15.33–q35.2, 8p23.3–q24.3, 13q12.11–q34, 14q11.2–q32.32, 17q12, 18p11.32–q23 and 21p11.2–q22.3). These genetic alterations may have been acquired as late genetic events, reflecting the genomic instability of neoplastic cells, and most are likely to constitute nonfunctional passenger mutations.
However, we also observed area‐specific but functional genetic alterations that may affect the biological behavior of the tumor. One example is gain at 14q32.33 and amplification of the AKT1 (PKBα) gene detected in two out of four tumor areas analyzed in one glioblastoma (case PB574). AKT1 is involved in the PI3K–Akt signaling pathway, which plays important roles in fundamental cellular functions such as cell proliferation and survival (21). Using U87MG glioblastoma cells, Pore et al (25) reported that AKT1 can augment hypoxia inducible factor (HIF)‐1α expression by increasing its translation under both normoxic and hypoxic conditions. AKT1 amplification has been reported in 1 gliosarcoma out of 103 glioblastomas analyzed using duplex PCR (14) and in 1 out of 9 glioblastomas using array CGH (27).
If regional genomic alterations convey a growth advantage, they are likely to become common genetic lesions in all tumor areas. These alterations may be also associated with resistance to radiotherapy or chemotherapy, and may be selected for during treatment, leading to new clones that are responsible for tumor recurrence and progression to a more malignant phenotype.
We analyzed seven cases of histologically homogeneous glioblastoma and seven cases in which different histological features were clearly recognized (eg, small cells, gemistocytes and multinucleated giant cells). There was no significant difference in terms of the overall numbers of genetic alterations. Thus, the morphological phenotype of glioblastoma does not always reflect the extent of genetic heterogeneity. In some tumors, DNA from areas with different histology showed the same pattern of genomic imbalance (cases PB603 and PB605). In other glioblastomas, the imbalance differed in histologically distinct tumor areas (cases PB551, PB578, PB636, PB638 and PB948). This may be because of the fact that in addition to chromosomal imbalance, other genetic/epigenetic alterations including point mutations, small deletions/insertions and promoter methylation that is not detectable by array CGH may also be responsible for genotype/phenotype correlations.
ACKNOWLEDGMENTS
We thank Ms Julie Vendrell and Ms Delphine Demangel for technical assistance.
REFERENCES
- 1. Burger PC, Pearl DK, Aldape K, Yates AJ, Scheithauer BW, Passe SM et al (2001) Small cell architecture—a histological equivalent of EGFR amplification in glioblastoma multiforme? J Neuropathol Exp Neurol 60:1099–1104. [DOI] [PubMed] [Google Scholar]
- 2. Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheung VG, Nelson SF (1996) Whole genome amplification using a degenerate oligonucleotide primer allows hundreds of genotypes to be performed on less than one nanogram of genomic DNA. Proc Natl Acad Sci U S A 93:14676–14679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daigo Y, Chin SF, Gorringe KL, Bobrow LG, Ponder BA, Pharoah PD, Caldas C (2001) Degenerate oligonucleotide primed‐polymerase chain reaction‐based array comparative genomic hybridization for extensive amplicon profiling of breast cancers: a new approach for the molecular analysis of paraffin‐embedded cancer tissue. Am J Pathol 158:1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garzon R, Calin GA, Croce CM (2009) MicroRNAs in Cancer. Annu Rev Med 60:167–179. [DOI] [PubMed] [Google Scholar]
- 6. Ginzinger DG, Godfrey TE, Nigro J, Moore DH, Suzuki S, Pallavicini MG et al (2000) Measurement of DNA copy number at microsatellite loci using quantitative PCR analysis. Cancer Res 60:5405–5409. [PubMed] [Google Scholar]
- 7. Greenman C, Wooster R, Futreal PA, Stratton MR, Easton DF (2006) Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics 173:2187–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Harada K, Nishizaki T, Ozaki S, Kubota H, Ito H, Sasaki K (1998) Intratumoral cytogenetic heterogeneity detected by comparative genomic hybridization and laser scanning cytometry in human gliomas. Cancer Res 58:4694–4700. [PubMed] [Google Scholar]
- 9. Homma T, Fukushima T, Vaccarella S, Yonekawa Y, Di Patre PL, Franceschi S, Ohgaki H (2006) Correlation among pathology, genotype, and patient outcomes in glioblastoma. J Neuropathol Exp Neurol 65:846–854. [DOI] [PubMed] [Google Scholar]
- 10. Huang J, Pang J, Watanabe T, Ng HK, Ohgaki H (2009) Whole genome amplification for array comparative genomic hybridization using DNA extracted from formalin‐fixed, paraffin‐embedded histological sections. J Mol Diagn 11:109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joensuu H, Puputti M, Sihto H, Tynninen O, Nupponen NN (2005) Amplification of genes encoding KIT, PDGFRalpha and VEGFR2 receptor tyrosine kinases is frequent in glioblastoma multiforme. J Pathol 207:224–231. [DOI] [PubMed] [Google Scholar]
- 12. Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P et al (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321:1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jung V, Romeike BF, Henn W, Feiden W, Moringlane JR, Zang KD, Urbschat S (1999) Evidence of focal genetic microheterogeneity in glioblastoma multiforme by area‐specific CGH on microdissected tumor cells. J Neuropathol Exp Neurol 58:993–999. [DOI] [PubMed] [Google Scholar]
- 14. Lee HG, Petersen RB, Zhu X, Honda K, Aliev G, Smith MA, Perry G (2003) Will preventing protein aggregates live up to its promise as prophylaxis against neurodegenerative diseases? Brain Pathol 13:630–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (2007) WHO Classification of Tumours of the Central Nervous System. IARC: Lyon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Misra A, Chattopadhyay P, Dinda AK, Sarkar C, Mahapatra AK, Hasnain SE, Sinha S (2000) Extensive intra‐tumor heterogeneity in primary human glial tumors as a result of locus non‐specific genomic alterations. J Neurooncol 48:1–12. [DOI] [PubMed] [Google Scholar]
- 17. Nigro JM, Takahashi MA, Ginzinger DG, Law M, Passe S, Jenkins RB, Aldape K (2001) Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real‐time quantitative polymerase chain reaction assay. Am J Pathol 158:1253–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nobusawa S, Watanabe T, Kleihues P, Ohgaki H (2009) IDH1 mutations as molecular signature and predictive factor of secondary glioblastomas. Clin Cancer Res 15:6002–6007. [DOI] [PubMed] [Google Scholar]
- 19. Ohgaki H, Kleihues P (2009) Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci 100:2235–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ohgaki H, Dessen P, Jourde B, Horstmann S, Nishikawa T, Di Patre PL et al (2004) Genetic pathways to glioblastoma: a population‐based study. Cancer Res 64:6892–6899. [DOI] [PubMed] [Google Scholar]
- 21. Osaki M, Oshimura M, Ito H (2004) PI3K‐Akt pathway: its functions and alterations in human cancer. Apoptosis 9:667–676. [DOI] [PubMed] [Google Scholar]
- 22. Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P et al (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perry A, Aldape KD, George DH, Burger PC (2004) Small cell astrocytoma: an aggressive variant that is clinicopathologically and genetically distinct from anaplastic oligodendroglioma. Cancer 101:2318–2326. [DOI] [PubMed] [Google Scholar]
- 24. Pinard R, De Winter A, Sarkis GJ, Gerstein MB, Tartaro KR, Plant RN et al (2006) Assessment of whole genome amplification‐induced bias through high‐throughput, massively parallel whole genome sequencing. BMC Genomics 7:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pore N, Jiang Z, Shu HK, Bernhard E, Kao GD, Maity A (2006) Akt1 activation can augment hypoxia‐inducible factor‐1alpha expression by increasing protein translation through a mammalian target of rapamycin‐independent pathway. Mol Cancer Res 4:471–479. [DOI] [PubMed] [Google Scholar]
- 26. Rollbrocker B, Waha A, Louis DN, Wiestler OD, Von Deimling A (1996) Amplification of the cyclin‐dependent kinase 4 (CDK4) gene is associated with high cdk4 protein levels in glioblastoma multiforme. Acta Neuropathol 92:70–74. [DOI] [PubMed] [Google Scholar]
- 27. Sasaki T, Arai H, Beppu T, Ogasawara K (2003) Detection of gene amplification and deletion in high‐grade gliomas using a genome DNA microarray (GenoSensor Array 300). Brain Tumor Pathol 20:59–63. [DOI] [PubMed] [Google Scholar]
- 28. Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD et al (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314:268–274. [DOI] [PubMed] [Google Scholar]
- 29. Tohma Y, Gratas C, Biernat W, Peraud A, Fukuda M, Yonekawa Y et al (1998) PTEN (MMAC1) mutations are frequent in primary glioblastomas (de novo) but not in secondary glioblastomas. J Neuropathol Exp Neurol 57:684–689. [DOI] [PubMed] [Google Scholar]
- 30. Watanabe T, Nobusawa S, Kleihues P, Ohgaki H (2009) IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol 174:1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ et al (2007) The genomic landscapes of human breast and colorectal cancers. Science 318:1108–1113. [DOI] [PubMed] [Google Scholar]