Abstract
Co‐deletion of chromosomes 1p and 19q is a common event in oligodendroglial tumors (OTs), suggesting the presence of OT‐related genes. The aim of this study was to identify the target genes residing in the minimally deleted regions on chromosome 1p36.31–p36.32 that might be involved in OTs. A novel gene KIAA0495/p53‐dependent apoptosis modulator (PDAM) was found frequently deregulated, with 37 of 58 (63.8%) OTs examined showing reduced expression compared with normal brain. Chromosome 1p loss and epigenetic modifications were the major mechanisms contributing to PDAM downregulation. The role of PDAM in chemosensitivity was also evaluated. PDAM knockdown had no effect on sensitivity to vincristine, lomustine, temozolomide and paclitaxel, but could induce cisplatin resistance in glioma cells harboring wild‐type p53. B‐cell CCL/lymphoma 2 (BCL2)‐like 1 (BCL2L1) exhibited significant upregulation, while BCL2 showed partial derepression in PDAM‐silenced cells after cisplatin treatment, suggesting that alteration of anti‐apoptotic genes contributed in part to cisplatin resistance. Knockdown of BCL2L1 abrogated the induced cisplatin‐resistant phenotype. Moreover, our data suggested that PDAM might function as a non‐protein‐coding RNA. Collectively, these findings suggest that PDAM deregulation may play a role in OT development and that PDAM may possess the capacity to modulate apoptosis via regulation of p53‐dependent anti‐apoptotic genes.
Keywords: BCL2L1, chromosome 1p, cisplatin resistance, KIAA0495/PDAM, oligodendroglial tumors
INTRODUCTION
Oligodendroglial tumors (OTs) are primary brain neoplasms of adults that comprise the classic oligodendrogliomas and mixed oligoastrocytomas. Unlike other glioma types, OTs appear to have a more favorable prognosis and are likely sensitive to chemotherapy (36). The genetic hallmark of OTs is a combined deletion of chromosome arms 1p and 19q, which is detectable in 60%–70% of tumors 2, 4, 20, 39. Such frequent genomic deletions suggest that these chromosome arms carry critical OT‐related genes, and their alteration is involved in the early stage of OT development (2). 1p/19q co‐deletion is associated with oligodendrogliomas showing the “classic” histology 8, 30, 38, implicating its use as an adjunct marker in OT diagnosis. This genetic signature has also been correlated with chemosensitivity and better outcome in OTs 9, 17, 40, and recent randomized clinical trials have confirmed the prognostic significance of 1p/19q co‐deletion 6, 15, 22.
1p/19q co‐deletion frequently involves the entire chromosomal arms, which may be attributable to the centromeric or pericentromeric t(1;19)(q10;p10) translocation with loss of one copy each of 1p and 19q 16, 21. Several postulates have been put forward to explain the consequent effects of such rearrangement event on OT development: activation or inactivation of genes at the break points, haploinsufficiency of multiple genes and epigenetic modifications. Molecular analysis of Notch homolog 2 (Drosophila; NOTCH2), which is located at the juxta‐centromeric region of 1p and implicated in oligodendrocyte differentiation (18), revealed that loss of NOTCH2 was correlated with prognosis (5), but no gene rearrangement or somatic mutation was found in OTs (3). Profiling analysis has demonstrated that numerous genes located on 1p or 19q have concurrent reduced expressions in tumors with 1p/19q co‐deletion 13, 14, 19, 42, supporting the haploinsufficiency hypothesis. However, some genes appeared to have aberrant expression unaccountable by the gene dosage effect. Identification and biological characterization of deregulated genes mapped to 1p and 19q may help understand the molecular pathogenesis of OTs.
Our group had conducted a detailed deletion mapping in OTs and delineated two contiguous minimally deleted regions, bound by D1S468 and D1S1608, on chromosome 1p36.31–p36.32 (12). Current genome annotation (GRCh37 assembly) indicates 11 genes [TP73, KIAA0495, CCDC27, LOC388588, leucine‐rich repeat containing 47 (LRRC47), KIAA0562, DNA fragmentation factor, beta polypeptide (DFFB), chromosome 1 open reading frame 174 (C1orf174), LOC100133612, LOC284661 and adherens junctions‐associated protein 1 (AJAP1)] in these intervals, with seven of them being novel and of these, at least two are noncoding RNAs. We previously reported frequent downregulation of TP73 in OTs (10), but the discovery of numerous alternatively spliced TP73 isoforms, with some of them having anti‐apoptotic activity (7), has complicated the role of this gene in OT formation. The aim of this study was to investigate if any of the remaining 10 genes located in the minimally deleted regions might be involved in the etiology of OTs. Our results revealed frequent downregulation of a novel gene KIAA0495 in OTs and uncovered a novel mechanism of cisplatin resistance, in which KIAA0495 might function to modulate apoptosis through p53‐dependent regulation of anti‐apoptotic genes. We therefore named KIAA0495 p53‐dependent apoptosis modulator (PDAM).
MATERIALS AND METHODS
Clinical samples and cell lines
A cohort of 58 OTs, containing 27 oligodendrogliomas, 14 oligoastrocytomas, 15 anaplastic oligodendrogliomas and 2 anaplastic oligoastrocytomas, were collected from Prince of Wales Hospital (Hong Kong) and Huashan Hospital (Shanghai). All tumors were classified according to the current World Health Organization Classification of Tumors of the Central Nervous System (36). A portion of tumor tissues resected at the time of surgery was immersed immediately in the RNALater® solution (Ambion, Austin, TX, USA) to preserve RNA integrity and stored at −80°C until use. Histologic examination confirmed that the RNALater‐preserved samples had tumor cell content greater than 80%. The median age of patients was 43 years (range 10–77 years) and the male/female ratio was 1.46:1 (Table 1).
Table 1.
Clinicopathological and molecular data of 58 oligodendroglial tumors studied. Abbreviations: m = male; f = female; O = oligodendroglioma; AO = anaplastic oligodendroglioma; OA = oligoastrocytoma; AOA = anaplastic oligoastrocytoma; LOH = loss of heterozygosity; ROH = retention of heterozygosity; M = methylation; U = unmethylation; NA = not available; ND = not determined.
| Case number | Sex | Age (year) | Histologic subtype | Tumor location | PDAM expression (fold change relative to normal brain) | PDAM methylation | Allelic status | |
|---|---|---|---|---|---|---|---|---|
| Chr 1p | Chr 19q | |||||||
| 390 | m | 10 | O | Frontal | −50.3 | M | ROH | ROH |
| 383 | m | 37 | O | Frontal | −43.6 | M | LOH | LOH |
| 330 | m | 47 | O | Frontal | −25.7 | M | ROH | ROH |
| 379 | f | 53 | O | Parietal | −17.8 | U | LOH | ROH |
| 380 | m | 35 | O | Frontal | −17.4 | M | LOH | LOH |
| 332 | m | 53 | O | Parieto‐occipital | −15.1 | M | LOH | LOH |
| 304 | f | 43 | O | Frontal | −14.2 | M | LOH | LOH |
| 363 | f | 22 | O | Frontal | −12.7 | M | LOH | ROH |
| 303 | m | 48 | O | Frontal | −10.3 | U | LOH | LOH |
| 325 | f | 42 | O | Frontal | −9.9 | M | LOH | LOH |
| 331 | f | 46 | O | Frontal | −7.6 | M | LOH | LOH |
| 387 | m | 39 | O | Thalamic | −5.7 | M | ROH | ROH |
| 370 | m | 66 | O | Frontal | −5.4 | M | ROH | ROH |
| 391 | m | 30 | O | Frontal | −4.0 | M | LOH | LOH |
| 329 | f | 69 | O | Frontal | −3.2 | M | LOH | LOH |
| 367 | m | 48 | O | NA | −3.0 | M | ROH | ROH |
| 389 | f | 32 | O | Frontal | −2.7 | M | LOH | LOH |
| 326 | f | 60 | O | Frontal | −2.5 | M | ROH | ROH |
| 400 | f | 49 | O | Temporal | −2.3 | M | LOH | LOH |
| 316 | m | 45 | O | Frontal | −1.9 | M | ROH | ROH |
| 378 | f | 38 | O | Frontal | −1.6 | U | ROH | ROH |
| 371 | f | 56 | O | Temporal | −1.4 | U | ROH | ROH |
| 311 | m | 40 | O | Temporal | −1.3 | ND | LOH | LOH |
| 322 | m | 55 | O | Frontal | −1.1 | U | ROH | ROH |
| 369 | m | 42 | O | Frontal | 1.0 | U | ROH | ROH |
| 324 | m | 42 | O | Frontal | 1.7 | ND | ROH | ROH |
| 323 | m | 23 | O | Temporal | 2.5 | U | LOH | LOH |
| 388 | m | 29 | AO | Frontal | −17.4 | M | ROH | ROH |
| 333 | f | 43 | AO | Frontal | −15.5 | M | ROH | ROH |
| 313 | m | 46 | AO | Frontal | −15.1 | M | LOH | LOH |
| 315 | f | 31 | AO | Frontal | −13.3 | M | LOH | ROH |
| 312 | m | 53 | AO | Frontal | −11.9 | U | ROH | ROH |
| 360 | m | 51 | AO | Ventricular | −2.8 | U | ROH | ROH |
| 302 | m | 35 | AO | Occipital | −2.5 | U | ROH | ROH |
| 365 | f | 47 | AO | NA | −2.5 | M | LOH | LOH |
| 335 | f | 45 | AO | Frontal | −2.1 | U | LOH | LOH |
| 319 | m | 77 | AO | Parietal | −1.7 | U | LOH | ROH |
| 310 | m | 35 | AO | Parietal | −1.6 | M | ROH | ROH |
| 377 | m | 77 | AO | Parietal | −1.5 | M | LOH | LOH |
| 334 | m | 19 | AO | Frontal | −1.3 | M | ROH | LOH |
| 318 | f | 57 | AO | Frontal | 1.3 | M | ROH | ROH |
| 358 | f | 50 | AO | Cerebellar | 1.7 | U | ROH | ROH |
| 382 | f | 37 | OA | Frontal | −80.9 | M | LOH | LOH |
| 381 | f | 28 | OA | Fronto‐temporal | −68.6 | M | LOH | LOH |
| 384 | m | 30 | OA | Frontal | −27.0 | M | ROH | ROH |
| 394 | f | 31 | OA | Frontal | −20.6 | M | LOH | LOH |
| 397 | m | 57 | OA | Fronto‐temporal | −18.9 | M | LOH | LOH |
| 301 | m | 43 | OA | Frontal | −6.3 | U | LOH | LOH |
| 362 | m | 41 | OA | Frontal | −4.6 | M | ROH | ROH |
| 375 | m | 28 | OA | Frontal | −3.1 | M | LOH | ROH |
| 398 | m | 54 | OA | Frontal | −2.3 | M | LOH | LOH |
| 395 | f | 44 | OA | Fronto‐temporal | −1.6 | U | ROH | ROH |
| 393 | f | 49 | OA | Fronto‐temporal | −1.4 | U | ROH | ROH |
| 314 | f | 38 | OA | Temporal | 1.9 | M | ROH | ROH |
| 368 | m | 61 | OA | Temporo‐parietal | 2.1 | U | ROH | ROH |
| 399 | m | 34 | OA | Frontal | 2.5 | U | ROH | ROH |
| 366 | m | 35 | AOA | Parietal | −1.3 | U | ROH | ROH |
| 364 | f | 48 | AOA | NA | 1.7 | U | ROH | ROH |
Nine glioma cell lines [A172, GOS‐3, HOG (35), KG‐1‐C (33), LNZ308, SKMG‐3, TC620 (27), U373MG and U87MG], two gastric tumor lines (AGS and MKN450) and the human embryonal kidney cells 293FT were included in this study. Sources and culture conditions of these cells were described in the Supporting Information and Methods.
Quantitative reverse transcription–polymerase chain reaction
Quantitative TaqMan®‐based PCR (25) was conducted in a 20‐µL volume containing 30 ng of cDNA, 1X TaqMan Universal PCR master mix and the commercially available PDAM or glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) probes and primers (PDAM: Hs00390315_m1, spans exons 1 and 2 of NM_207306; GAPDH: Hs99999905_m1; Applied Biosystems, Foster City, CA, USA). Amplification was conducted under the conditions of 95°C for 10 minutes, and 40 cycles of 95°C for 15 s and 60°C for 1 minute in the iCycler™ thermocycler (Bio‐Rad Laboratories, Hercules, CA, USA). Standard curves were generated to determine copy numbers of PDAM and GAPDH. The expression level of PDAM was normalized to that of GAPDH and was considered downregulated when the level was at least twofold lower than the mean PDAM transcript level of eight normal brain samples. Total RNA of normal brains was obtained from Ambion, Clontech Laboratories (Mountain View, CA, USA) and Origene Technologies (Rockville, MD, USA).
SYBR® Green‐based PCR analysis was performed in a 20‐µL reaction mix containing 30 ng of cDNA, 1X Power SYBR Green PCR master mix (Applied Biosystems) and 125 nM of each primer pair. PCR was conducted under the conditions of 95°C for 10 minutes and 40 cycles of 95°C for 15 s, 58°C–62°C for 20 s and 72°C for 20 s. Results were analyzed using the comparative threshold method. All quantitative PCR was carried out in triplicate, and the experiments were performed at least three times. Primer sequences are listed in Table S1.
Bisulfite sequencing
One microgram of genomic DNA from each sample was treated with sodium bisulfite using Methylamp™ One‐step DNA modification kit (Epigentek, Brooklyn, NY, USA) according to the manufacturer's recommendation. Each PCR mix was prepared as described for conventional reverse transcription–polymerase chain reaction (RT‐PCR) except that one‐tenth of modified DNA was substituted as template. PCR products were cloned into the pCR®2.1‐TA vector (Invitrogen, Hong Kong, China). Five to 10 colonies of each sample were picked and subjected to DNA sequencing. Products were resolved on Genetic Analyzer 3130xl and analyzed by the DNA Sequencing Analysis software (Applied Biosystems). Samples were considered methylation positive when more than 50% of analyzed clones showed methylation in more than 60% of CpG sites examined.
Demethylaton treatment
Cells were treated with the demethylating agent 5‐azacytidine (AzaC; Sigma‐Aldrich, St. Louis, MO, USA) at 2, 5 or 10 µM continuously for 3 days or with the histone deacetylase inhibitor trichostatin A (TSA; Sigma‐Aldrich) at 0.33 µM for 24 h. For combinational treatment, cells were exposed to AzaC for 3 days and to TSA for 24 h. Cells were then harvested, and RNA was extracted. Untreated cells were used as a control.
Cloning of full‐length PDAM cDNA
The full‐length PDAM cDNA of ∼6.3 kb (NM_207306) was assembled from two DNA fragments generated using the KAPA HiFi DNA polymerase (KAPA Biosystems, Woburn, MA, USA). The 5′ fragment comprises exons 1–3 and part of exon 4, whereas the 3′ fragment contains exon 4 (Table S1). Both fragments were cloned into pCR®2.1, and the full‐length cDNA was created by ligating the fragments at an internal AatII restriction site. The full‐length cDNA was then subcloned at the NotI–HindIII sites of pBluescript vector (pBS‐PDAM). The full‐length green fluorescent protein (GFP) cDNA was also cloned into pBluescript (pBS‐GFP). Additionally, constructs with the PDAM and GFP cDNA fragments cloned in reverse orientation were prepared (pBS‐PDAMr and pBS‐GFPr). The authenticity of all clones was confirmed by DNA sequencing.
Transfection
Cells were seeded to yield 20%–30% confluency at the time of transfection. Five nM of siRNA was transfected into cells using Lipofectamine™ 2000 according to the manufacturer's recommendation (Invitrogen). The transfection condition was optimized using siRNA concentration that suppressed at least 70% of GAPDH transcript level. Two siRNAs were employed to suppress PDAM transcripts, with siPDAM‐1 targeting exon 1 and siPDAM‐2 targeting the 36‐basepair (bp) repeat region in exon 4 (Table S1). As control, the siRNA negative control #1 was used. All siRNAs were obtained from Applied Biosystems.
Drug sensitivity assay
Cells were seeded at a density of 1600–2200 cells/well on 96‐well dish a day prior to transfection. The next day, cells in each well were transfected with 5 nM of siRNA using Lipofectamine 2000 (Invitrogen). After incubation for 24 h, fresh media containing various concentrations of vincristine (Pharmachemie BV, Haarlem, the Netherlands), lomustine (Sigma‐Aldrich), temozolomide (Schering‐Plough, Kenilworth, NJ, USA), cisplatin (Pharmachemie BV) or paclitaxel (Hospira, VIC, Australia) were added. Two days after drug treatment, cell viability was assessed by reduction of 3‐(4,5‐dimethylthiazol‐2‐yl)‐2,5‐diphenyl tetrazolium bromide (MTT) by mitochrondrial succinate dehydrogenase. MTT (USB, Cleveland, OH, USA) was added to a final concentration of 450 µg/mL. After incubation at 37°C for 4 h, the formed formazan crystal was solubilized with dimethyl sulfoxide and measured spectrophotometrically at a wavelength of 570 nm (Victor™ 3 Multilabel Plate Reader, Perkin‐Elmer, Waltham, MA, USA). Each measurement was carried out in triplicate, and the experiment was repeated three times. The half maximal inhibitory concentration (IC50) was defined as the drug concentration yielding 50% nonsurviving cells compared with vehicle‐treated controls.
Western blot analysis
Cell lysates were prepared as described (25). Soluble proteins electroblotted onto polyvinylidene fluoride (PVDF) membrane were probed with rabbit anti‐PDAM polyclonal antibody (1:100 dilution; Supporting Information Materials and Methods), mouse antihuman p53 antibody (0.1 µg/mL; clone 1C12; Cell Signaling Technology, Danvers, MA, USA), mouse antihuman GAPDH antibody (0.1 µg/mL; Abonva, Taiwan) or rabbit anti‐GFP antibody (1:10 000 dilution; Clontech). Protein expression was detected by SuperSignal® West Pico Chemiluminescent substrate (Thermo Scientific, Rockford, IL, USA) with appropriate horseradish peroxidase‐conjugated secondary antibody.
In vitro transcription–translation
The in vitro transcription–translation reaction was carried out using the TNT® Quick Coupled Transcription/Translation system with T7 RNA polymerase (Promega, Madison, WI, USA) according to the manufacturer's recommendation. Briefly, the 10‐µL reaction mix containing 400 ng of plasmid DNA, 8 µL of TNT Quick master mix and 1 µL of L‐[35S]methionine (1000 Ci/mmol at 10 mCi/mL; Perkim‐Elmer) was incubated at 30°C for 90 minutes. The products were resolved on 15% sodium dodecyl sulphate–polyacrylamide gel and fixed in 50% methanol–10% acetic acid solution for 30 minutes. The gel was then treated with the fluorographic enhancer Amplify™ (GE Healthcare, Hong Kong) and dried onto Whatman 3MM filter paper (GE Healthcare), followed by exposure to Kodak X‐OMAT AR film (Sigma‐Aldrich).
Statistical analysis
Statistical analysis was performed using the software SPSS 16 (SPSS, Chicago, IL, USA). The correlation between molecular and clinicopathological parameters was evaluated by chi‐squared test or Fisher's exact test, whichever was appropriate. Statistical comparisons were based on Student's t‐test. An obtained P‐value less than 0.05 (two‐sided) was considered statistically significant.
RESULTS
PDAM expression was downregulated in OTs
To identify differentially expressed genes in the minimally deleted regions on 1p36.31–p36.32, we subjected 12 OT samples that had undergone microdissection to conventional RT‐PCR analysis (Supporting Information Materials and Methods). The transcript expression of 10 genes, including PDAM, CCDC27, LOC388588, LRRC47, KIAA0562, DFFB, C1orf174, LOC100133612, LOC284661 and AJAP1, was determined. PDAM, KIAA0562, C1orf174 and AJAP1 showed no detectable transcripts in seven (58.3%), three (25.0%), three (25.0%) and one case (8.3%), respectively (Figure 1). Concurrent absent expression of PDAM with KIAA0562, AJAP1 and/or C1orf174 was observed in four cases (#313, #331, #333 and #335). The oligodendroglioma cell line KG‐1‐C exhibited no AJAP1 expression. LOC388588, LRRC47 and DFFB expression were found in all OTs, glioma lines and normal brain tissues examined, whereas CCDC27, LOC100133612 and LOC284661 transcripts were not detectable in any samples.
Figure 1.
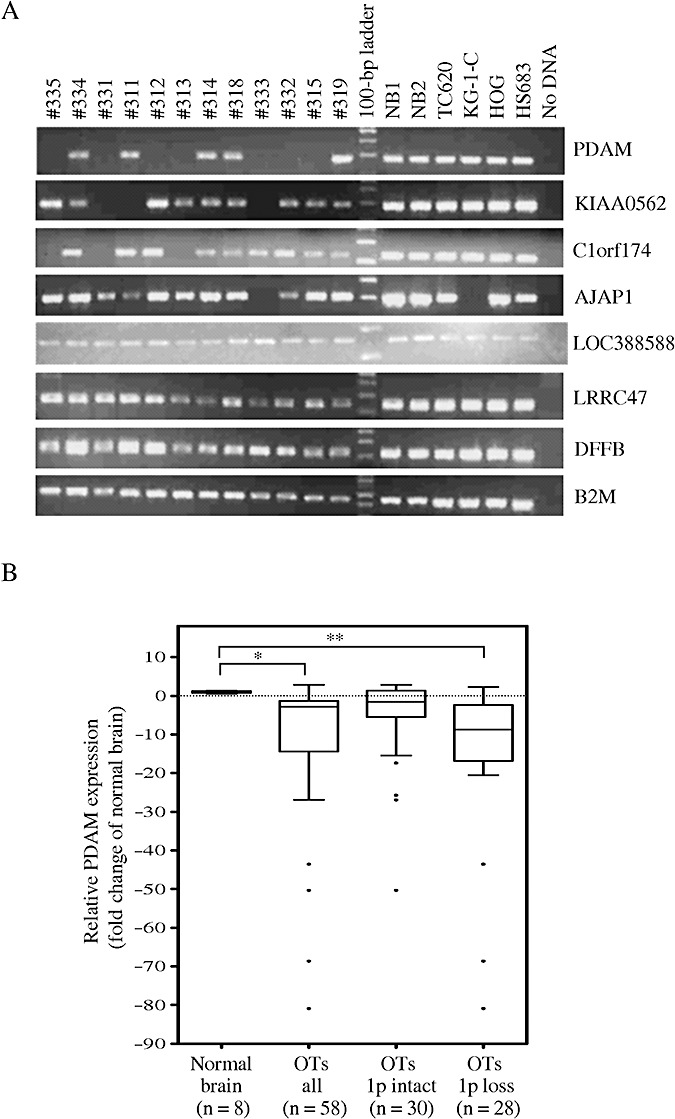
A. Reverse transcription–polymerase chain reaction (RT‐PCR) analysis of candidate target genes located in the minimally deleted regions on 1p36.31–p36.32 in OTs, glioma cell lines and normal brain tissues. Absent expressions of p53‐depdendent apoptosis modulator (PDAM), KIAA0562, C1orf174 and AJAP1 are found, respectively, in 58.3%, 25.0%, 25.0% and 8.3% of cases examined. β‐2 microglobulin (B2M) serves as the internal reference gene. B. Quantitative RT‐PCR analysis reveals significant PDAM expression differences between OTs and normal brain (*P = 0.002), and between 1p and deleted tumors and normal brain (**P = 0.0001).
Because PDAM displayed the highest frequency of absent expression in 12 OTs examined, we sought to determine its transcript level in a larger series of OTs by quantitative real‐time RT‐PCR. Our results revealed that PDAM mRNA level was significantly decreased in OTs compared with normal brain samples (P = 0.002). Thirty‐seven of 58 (63.8%) OTs showed downregulation of PDAM transcript by at least twofold, with 19 of them exhibiting >10‐fold reduced expression, relative to the mean expression level of eight normal brain samples (Figure 1 and Table 1). When correlated with clinicopathological parameters, reduced PDAM expression was significantly associated with 1p/19q co‐deletion (P = 0.005) and tumor location in frontal lobe (P = 0.009), but not with tumor grade, histologic subtype, gender or age (<40 years vs. ≥40 years; <50 years vs. ≥50 years).
Inactivating mechanisms for PDAM expression
To determine mechanisms for PDAM downregulation, we evaluated allelic change on chromosome 1p36 (Supporting Information Materials and Methods). Twenty‐eight of 58 (48.3%) tumors exhibited chromosome 1p loss. Of these cases, 24 (85.7%) showed decreased PDAM expression (P = 0.001), indicating a strong association for genomic loss in reduced gene expression. However, there were some discordant cases with diminished PDAM expression but no allelic alteration on chromosome 1p. We speculated that other inactivating mechanisms might exist for PDAM.
Using MethPrimer (26), two contiguous CpG islands were predicted on an interval spanning the putative promoter region and part of exon 1 of the PDAM gene (Figure 2). We questioned if promoter hypermethylation played a role in PDAM downregulation. A stretch of 30 CpG dinucleotides located in the putative promoter region was interrogated for aberrant methylation by bisulfite sequencing. A total of 56 OTs and four normal brain samples were assessed. Our results revealed rare hypermethylation at CpG island 1 in normal brain tissues but hypermethylation in 36 of 56 (64.3%) OTs examined (Table 1 and Figure 2). Promoter hypermethylation of PDAM was detected in 30 of 37 (81.1%) OTs with reduced PDAM expression, and the two parameters were significantly associated (P = 0.0004). To further investigate the role of promoter hypermethylation on PDAM expression, we treated glioma lines A172 and TC620 (exhibiting PDAM expression of twofold to threefold below normal brain level) with the demethylating agent AzaC, alone or combined with the histone deacetylase inhibitor TSA, followed by determination of PDAM transcript expression by quantitative RT‐PCR. Our results demonstrated that AzaC treatment induced significant enhancement of PDAM expression in glioma cells by 30%–50% compared with untreated cells (P < 0.05), whereas TSA alone showed no apparent effect on PDAM level (Figure 2). Notably, combined treatment with both AzaC and TSA showed a synergistic effect on PDAM expression in A172 and TC620 (2‐ to 2.6‐fold higher than that of untreated cells), suggesting that epigenetic change is an important mechanism to regulate PDAM expression.
Figure 2.
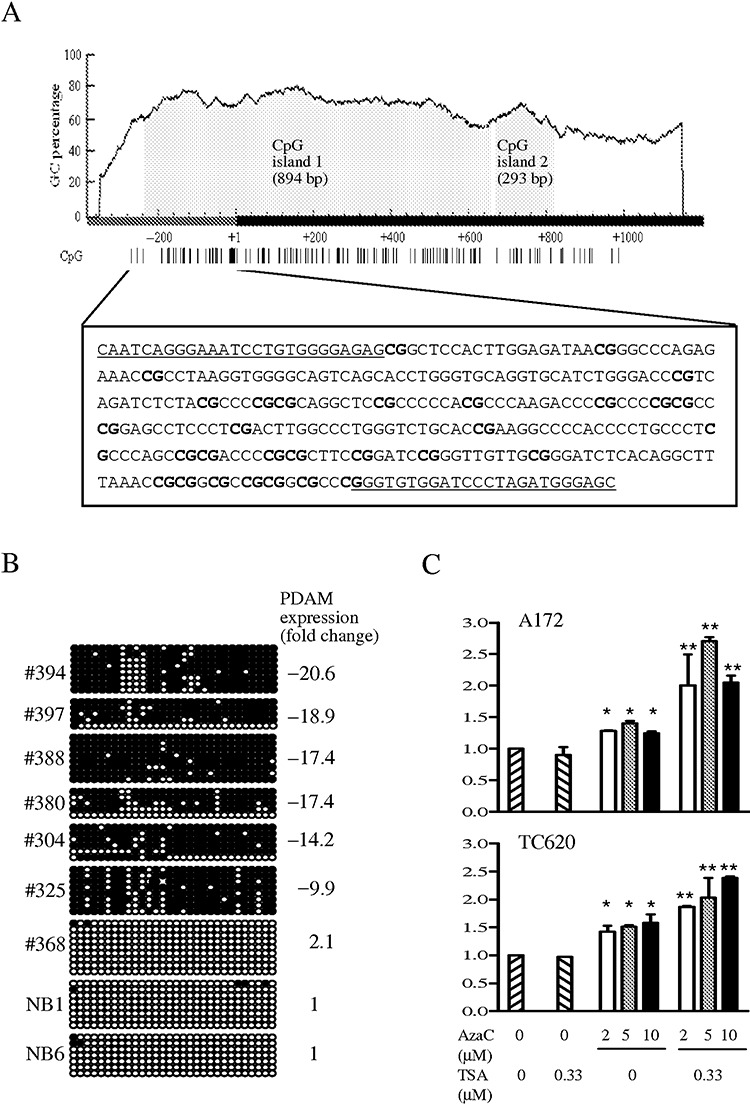
A. Two CpG islands are predicted on the p53‐depdendent apoptosis modulator (PDAM) gene by MethPrimer using prediction criteria of GC content >50% and a ratio of observed CpG to expected CpG >0.6 over a minimum length of 500 bp. CpG island 1 spans 242 bp of the putative promoter region and 652 bp of exon 1, whereas CpG island 2 lies in exon 1. The dark bar represents the 5′ end of PDAM transcript with the transcriptional start site designated as +1. The distribution of CpG sites (vertical lines) is shown below the graph. The boxed DNA sequence indicates the putative promoter region in which 30 CpG dinucleotides (bold) are interrogated for methylation by bisulfite sequencing. The underlined sequences represent regions where primers for amplification of modified DNA are designed. B. Representative results of bisulfite sequencing of 30 CpG sites examined. Each circle represents the methylation status of a CpG dinucleotide, with dark circle indicating methylation and open circle representing unmethylation. The CpG sites arranged in order from 5′ to 3′ are shown in left‐to‐right orientation. Case numbers and PDAM transcript level relative to normal brain (NB) expressed as fold change are also shown. C. Effects of azacytidine (AzaC) alone or in combination with trichostatin A (TSA) on PDAM expression monitored by quantitative reverse transcription–polymerase chain reaction. PDAM expression was significantly increased in AzaC‐treated A172 and TC620 cells (*P < 0.05), and was further enhanced upon combined treatment with AzaC and TSA (**P < 0.01).
Taken together, 20 of 37 (54.1%) OTs with decreased PDAM expression exhibited both promoter hypermethylation and chromosome 1p loss, whereas 10 cases (27.0%) with diminished PDAM level showed promoter hypermethylation and balanced chromosome 1p. These data indicate that chromosome 1p loss and epigenetic modifications are the major mechanisms contributing to PDAM downregulation. Additionally, hypermethylation of PDAM was significantly associated with chromosome 19q loss (P = 0.024), 1p/19q co‐deletion (P = 0.045) and tumor location in frontal lobe (P = 0.018).
Genomic structure of PDAM
The PDAM gene, located at chr1:3 652 550–3 663 886 (GRCh37 assembly), is 11 337 bp in length and contains four exons (Figure 3). Its full‐length transcript (NM_207306) is 6358 bp, and the predicted coding region, which spans exons 1 and 2, has a size of 606 bp, encoding a peptide of 201 amino acid residues (NP_997189). Of note is an unusual sequence feature found around the translational start region, at which an in‐frame stop codon (TGA) is positioned immediately upstream of the predicted translational start site. An AluSg sequence is also detected at 4484–4764 bp (Repbase). Additionally, a unique sequence of 36 bp is found to be repeated 27 times in exon 4 (2158–3125 bp). Base variations are observed in some repeats, and the consensus sequence of 27 repeats is 5′‐GGTTGATTGTG(C/T)GGGTTAGATAGAGGTCATCAGC(C/T)G‐3′ (Figure S1). Phylogenetic comparative analysis indicates that PDAM DNA sequence is conserved in the primate infraorder Simiiformes, which comprises the monkeys, apes and humans (Figure S1).
Figure 3.
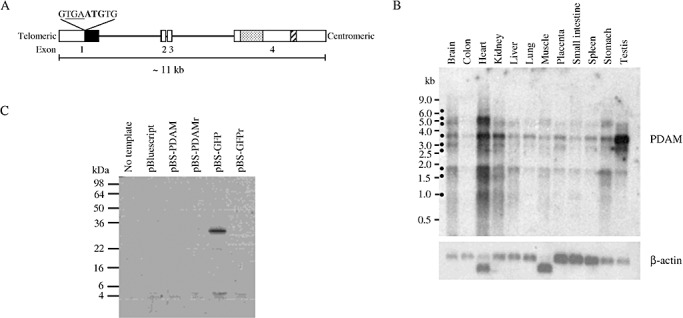
A. Genomic organization of the human p53‐depdendent apoptosis modulator (PDAM) gene. Open box represents exon and dark box indicates predicted coding region (NM_207306). The early cryptic stop codon (underlined) positions upstream of the translational start codon (bold) is also shown. The 36‐bp repeat region is represented by dotted box and the AluSg sequence is indicated by hatched box. B. Tissue distribution of PDAM transcripts. Blot containing polyA‐RNA of various human tissues was probed with the radioactively labeled PDAM‐predicted coding cDNA. At least nine isoforms (dark circle) are discernable. The 3.7‐kb isoform appears to be present in all tissues examined. C. Coupled in vitro transcription–translation assay shows no translatable proteins from the full‐length PDAM cDNA [pBluescript (pBS)‐PDAM]. A product of ∼27 kDa is observed with pBS green fluorescent protein (pBS‐GFP). As controls, cDNAs cloned in reverse orientation (pBS‐PDAMr and pBS‐GFPr) yield no detectable products.
Expression distribution
Northern blot analysis of 12 different human tissues was performed to determine the expression distribution of PDAM (Supporting Information Materials and Methods). Figure 3 reveals that at least nine PDAM transcript isoforms were detectable, and their sizes were approximated to be 6.4 kb, 5.2 kb, 4.8 kb, 3.7 kb, 3.1 kb, 2.7 kb, 1.8 kb, 1.6 kb and 1.0 kb. The 3.7‐kb transcript appeared to be the most prominent isoform and was detectable in all tissues examined, with the highest levels observed in heart and testis; moderate abundances in brain, kidney, lung, placenta, spleen and stomach; and low levels in colon, liver and small intestine.
Determination of PDAM gene product
To determine the PDAM product, an antibody specific to amino acid residues 66–79 of PDAM‐predicted coding sequence was generated (Supporting Information Materials and Methods) and used to probe against various glioma cell lines expressing PDAM transcripts of different levels. However, Western blot analysis revealed no specific protein band in cells expressing detectable transcripts (Figure S2; Supporting Information Results). Immunofluorescence staining also failed to detect any signals in cells expressing PDAM transcripts (data not shown). These findings suggest that the PDAM product may be present at a level undetectable by the methods employed. Alternatively, the PDAM coding sequence may have been predicted incorrectly.
To evaluate whether the PDAM transcript could be translatable into any proteins, we cloned the full‐length PDAM cDNA of 6.3 kb and subjected it to in vitro transcription and translation analysis. Figure 3 shows that no products, in the range of 4–98 kDa, could be detected using the full‐length PDAM cDNA as a template. As control, a product of ∼27 kDa was observed when GFP cDNA was used as a template. These results suggest that PDAM may not encode any protein product.
PDAM knockdown induces cisplatin resistance
Because chromosome 1p deletion alone or in combination with chromosome 19q loss has been shown to be a predictor of chemosensitivity in OT patients 9, 17, 40, we asked if downregulation of genes residing on chromosome 1p might render glioma cells sensitive to chemotherapeutic drugs. To address this issue, we knocked down PDAM in glioma cell lines and then evaluated drug sensitivity by the MTT assay. Because none of the available oligodendroglioma cell lines carry chromosome 1p deletion, we included glioma cells that show partial chromosome 1p loss (24) in this part of the study. Transfection of either siRNAs targeting PDAM into glioma cells led to ∼60% reduction of PDAM transcript, indicating comparable efficacy of these siRNAs in suppressing PDAM. Our results showed that knockdown of PDAM did not significantly alter the IC50 values of drugs commonly used in treatment of OTs (vincristine, lomustine and temozolomide) and of paclitaxel in all five glioma lines examined (Figure S3). Both PDAM‐targeting siRNAs generated similar IC50 values for each drug tested. These findings indicate that PDAM does not play a role in mediating sensitivity to the drugs examined. Surprisingly, knockdown of PDAM led to elevated IC50 values in two (A172 and U87MG) of five glioma lines treated with cisplatin (Figures 4A, S3). The cisplatin IC50 values for A172 increased significantly from 60.4 ± 5.2 µM in siRNA negative control‐transfected cells to 77.7 ± 2.2 µM and 82.5 ± 3.1 µM in siPDAM‐1‐ and siPDAM‐2‐transfected cells, respectively (P < 0.01). Similarly, the cisplatin IC50 values for U87MG rose from 14.1 ± 2.0 µM in control cells to 19.8 ± 2.3 and 19.4 ± 0.9 µM in siPDAM‐1‐ and siPDAM‐2‐transfected cells, respectively (P < 0.03). One major genetic difference between the cisplatin‐resistant and ‐sensitive lines is that the former cells harbor wild‐type p53, whereas the latter cells carry mutant p53. Because p53 plays a pivotal role in DNA damage response, we examined its levels after cisplatin treatment. Western blot analysis confirmed p53 accumulation and stabilization in cells following cisplatin treatment. Notably, p53 abundance was further enhanced when PDAM was suppressed (Figure 4C). Such data were supported by the observation that p53 transcript was also increased in PDAM‐suppressed and cisplatin‐treated cells (P < 0.02) (Figure 4B). In addition, the transcript level of CDKN1A/p21, a p53 target gene, showed concordant elevation with p53 (P < 0.002).
Figure 4.
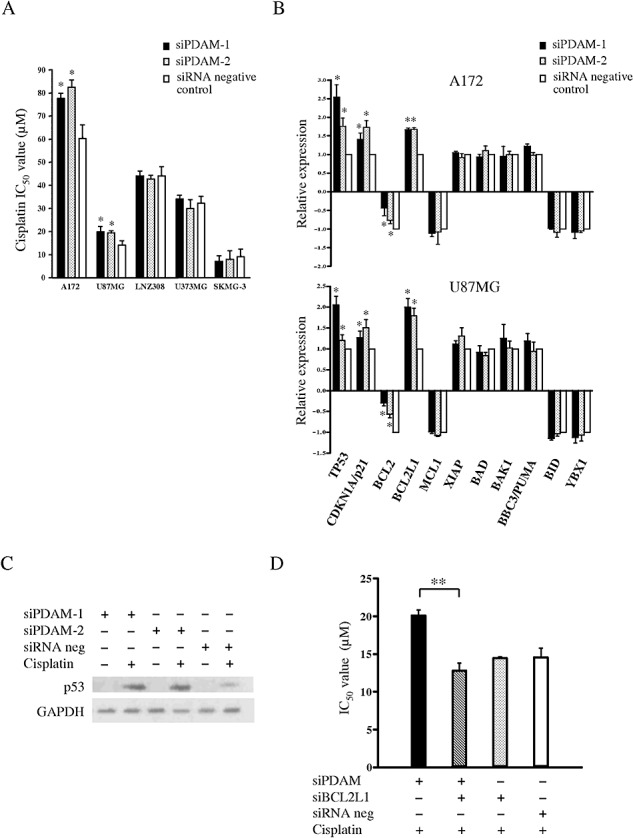
A. Drug sensitivity assay reveals that the cisplatin IC50 values are significantly elevated when p53‐depdendent apoptosis modulator (PDAM) is silenced in A172 and U87MG cells (*P < 0.05). Such cisplatin‐resistant phenotype is not observed in LNZ308, U373MG and SKMG‐3. B. Effects of PDAM knockdown on gene expression after cisplatin treatment as evaluated by quantitative reverse transcription–polymerase chain reaction analysis. Transcript levels of p53, CDNK1A/p21, BCL2 and BCL2L1 show significant changes after PDAM knockdown compared with those in cells without PDAM suppression (*P < 0.05). C. Western blot analysis reveals that p53 abundance is accumulated after cisplatin treatment and is further elevated when PDAM is suppressed. D. BCL2L1 knockdown reduces the cisplatin IC50 value from 20.1 ± 0.72 µM to 12.7 ± 1.1 µM in PDAM‐suppressed cells, thus abolishing the cisplatin‐resistant phenotype induced by PDAM suppression. **P = 0.005. Abbreviation: GAPDH = glyceraldehyde 3‐phosphate dehydrogenase.
To elucidate the mechanism of cisplatin resistance, we sought for expression changes in genes that are associated with apoptosis [B‐cell CLL/lymphoma 2 (BCL2)‐associated agonist of cell death (BAD), BCL2‐antagonist/killer 1 (BAK1), BCL2, BCL2‐like 1 (BCL2L1), BH3 interacting domain death agonist (BID), myeloid cell leukemia sequence 1 (MCL1), BCL2 binding component 3 (BBC3/PUMA) and X‐linked inhibitor of apoptosis (XIAP)] and with cisplatin resistance (Y box binding protein 1 (YBX1)) (34). Cells were transfected either with siPDAM or siRNA negative control, treated with cisplatin or vehicle, and then subjected to quantitative RT‐PCR. Of nine genes examined, BCL2 showed decreased expression upon cisplatin treatment compared with vehicle‐treated control in both A172 and U87MG cells (Figure 4B). However, when PDAM was suppressed, BCL2 level was found to be not reduced as much (25%–75% less) as that in cells without PDAM knockdown (P < 0.04). Additionally, BCL2L1 expression was increased slightly (∼10%) after cisplatin treatment, but its level was significantly elevated (70%–100% higher than that of control cells) when PDAM was suppressed (P < 0.01). The expressions of the remaining seven genes examined did not change appreciably following PDAM knockdown. Collectively, these findings suggest that BCL2 and BCL2L1, both possess anti‐apoptotic activity, may play a role in conferring p53‐dependent cisplatin resistance in cells with PDAM knockdown.
To substantiate the role of BCL2L1 in cisplatin resistance, cells were transfected with siPDAM and siBCL2L1, treated with cisplatin, and followed by IC50 determination. Our results showed that the cisplatin IC50 value was significantly reduced from 20.1 ± 0.72 µM to 12.7 ± 1.1 µM (P = 0.005) in PDAM‐suppressed cells after BCL2L1 knockdown (Figure 4D). This finding provides strong evidence that BCL2L1 is an important factor contributing to cisplatin resistance in PDAM‐silenced cells.
DISCUSSION
Recurrent deletion of chromosomes 1p in OTs strongly implies the presence of OT‐related genes on this chromosome arm. Our finer deletion mapping analysis had delineated two contiguous minimally deleted regions on chromosome 1p36.31–p36.32, and we hypothesized that OT‐related genes might reside in these intervals. One novel gene PDAM located in the minimally deleted regions was found to have significantly reduced expression in 63.8% of OTs examined compared with normal brain tissues. Further analyses demonstrated that PDAM expression status was associated with chromosome 1p allelic status and epigenetic modifications. Of 37 OTs with reduced PDAM expression, 20 (54.1%) of them exhibited both chromosome 1p36 loss and promoter hypermethylation of PDAM, and 10 (27.0%) cases displayed aberrant hypermethylation but balanced chromosome 1p. In the latter cases, biallelic hypermethylation may be attributable to PDAM downregulation. These findings strongly indicate that genomic loss and epigenetic modifications are the major mechanisms contributing to PDAM inactivation. The remaining seven cases with diminished PDAM expression showed no detectable epigenetic alteration and had intact chromosome 1p status; it is likely that other inactivation mechanisms may exist for PDAM. Mutation analysis of the PDAM‐predicted coding sequence was also performed, but no somatic base alterations were detected in the series (data not shown). Taken together, our data demonstrate that PDAM is frequently deregulated in OTs, suggesting a role of this gene in tumorigenesis.
Epigenetic regulation plays an important role in silencing transcription of cancer‐related genes. We and others have demonstrated that promoter hypermethylation of many tumor suppressor genes and DNA repair genes is frequent in OTs 11, 45, 46. However, despite the uncovering of numerous genes with decreased expression on chromosome 1p, only a limited number of 1p‐located genes are reported to be inactivated by epigenetic changes 37, 43. In this study, we have demonstrated that promoter hypermethylation is one of the key mechanisms for silencing PDAM, implicating specific inactivation of PDAM during OT formation.
In addition to PDAM, three other genes (AJAP1, KIAA0562 and C1orf174) mapped within the minimally deleted regions also showed no detectable transcripts in some OTs examined. Our data were in line with the study of McDonald et al in which AJAP1 was found to be downregulated in most OT examined compared with normal brain(28). Ectopic expression of AJAP1 in glioma cells did not inhibit cell growth but attenuated cell adhesion and migration ability (28). KIAA0562 was first isolated as a glycine‐, glutamate‐ and thienylcyclohexylpiperidine‐binding protein (23). Our data indicated that KIAA0562 expression was not detectable in 25% of tumors, suggesting that it might be involved in a subset of OTs. C1orf174 is a novel gene that has yet to be characterized. In addition, although our initial expression screen did not identify the apoptosis‐related DFFB as a candidate gene, McDonald et al showed that DFFB expression was reduced in OTs with deleted 1p compared with those with intact 1p (29). Taken together, multiple genes from the minimally deleted regions on chromosome 1p36.3 were shown to have aberrant expression, and their deregulation might contribute to the etiology of OTs.
Cisplatin is a widely used anticancer drug in treatment of various solid cancers. It is believed that cisplatin exerts its cytotoxic effects mainly through formation of DNA adducts and induction of DNA damaging signals, culminating in the activation of apoptosis (44). During such process, several signal transduction pathways and their associated transcriptional factors can be activated. One of the key factors that mediates cisplatin‐induced DNA‐damaging signals is the p53 tumor suppressor.
One major finding of this study was the revelation of a link between PDAM and p53‐dependent cisplatin resistance. When PDAM was knocked down, glioma cells harboring wild‐type p53, but not those carrying mutant p53, exhibited induced cisplatin resistance. Further investigation indicated that PDAM knockdown led to partial derepression of BCL2 with concurrent enhancement of BCL2L1 after cisplatin treatment, suggesting that BCL2 and BCL2L1 might be responsible for the observed induced cisplatin resistance. Moreover, silencing BCL2L1 in PDAM‐suppressed cells abrogated the induced cisplatin‐resistant phenotype, providing additional evidence that upregulation of BCL2L1 in part antagonizes apoptosis. Collectively, these findings implicate that PDAM may possess the capacity to modulate apoptosis via p53‐dependent regulation of the anti‐apoptotic genes. Based on these findings, KIAA0495 is named PDAM, for p53‐dependent apoptosis modulator.
Numerous mechanisms have been attributed to cisplatin resistance. These include decreased intracellular drug accumulation, increased detoxification, enhanced DNA repair, increased tolerance of DNA damage and altered signaling pathways (41). Because cisplatin induces apoptotic cell death through DNA adduct formation, genes involved in DNA damage, apoptosis and survival signaling may also contribute to resistance. The importance of the apoptosis pathway in chemoresistance is supported by the strong correlation between overexpression of BCL2 and BCL2L1, and decreased sensitivity to cisplatin 1, 32. Our glioma model confirms the pivotal role that BCL2 and BCL2L1 play in cisplatin resistance, and highlights PDAM as a potential modulator of p53‐dependent anti‐apoptotic genes.
What is the gene product of PDAM? We had attempted to verify PDAM expression using different techniques. First, an antibody against PDAM product based on the predicted coding sequence was generated. Western blotting and immunoflourescence staining using such antibody failed to detect any unique protein in all cell lines examined, even in cells with PDAM transcript level equivalent to that of normal brain level. Second, in vitro transcription and translation assay using the full‐length PDAM cDNA as a template did not reveal any protein product. These results suggest that PDAM may not encode a protein and raise the possibility that PDAM is a noncoding RNA.
In conclusion, our study reveals for the first time that aberrant PDAM expression is frequent in OTs, suggesting that PDAM deregulation may be involved in OT development. A novel mechanism of cisplatin resistance is also uncovered, in which PDAM may function to antagonize p53‐dependent DNA damage response. Additionally, our study suggests that PDAM is a noncoding RNA. Accumulative evidence has indicated that noncoding RNAs play an important role in brain development (31); the finding of PDAM deregulation in OTs suggests that noncoding RNA may contribute to oncogenesis of brain tumors.
Supporting information
Figure S1. Sequences of 36‐bp repeat region and phylogenetic sequence analysis. A. The unique 36‐bp repeat sequences observed in exon 4 of the PDAM gene are shown. Repeat 13, which is located in the center of the repeat region, shows loose base conservation after first 21 bp and has a 4‐bp deletion at the 3′ end. B. Phylogenetic comparative analysis reveals that human PDAM is conserved in the primate infraorder Simiiformes (UCSC Genome Browser, http://genome.ucsc.edu/).
Figure S2. Detection of endogenous PDAM by Western blot analysis. The specificity of the anti‐PDAM antibody is validated by its reactivity to PDAM based on the predicted coding sequence (pPDAM) and GFP‐tagged PDAM fusion proteins (pPDAM‐GFP and pGFP‐PDAM). The latter two proteins are also detected by the anti‐GFP antibody. The 293FT cells show PDAM mRNA of normal brain level, the glioma lines exhibit twofold to 10‐fold lower level relative to normal brain and gastric tumor lines AGS and MKN45 display no detectable PDAM transcript. None of the cell lines expressing detectable PDAM transcripts show any unique protein bands that are reactive to the anti‐PDAM antibody.
Figure S3. Drug sensitivity assays and cell survival curves. A. Drug sensitivity assay reveals that PDAM knockdown has no effects on sensitivity to vincristine, lomustine, temozolomide and paclitaxel in all five glioma lines examined. B Cell survival curves indicate increased resistance to cisplatin when PDAM is knocked down in A172 and U87MG cells.
Table S1. Oligonucleotides used in this study.
Supporting info item
ACKNOWLEDGMENTS
This study was supported by the Research Grants Council of Hong Kong (CUHK4407/06M).
REFERENCES
- 1. Bauer JA, Trask DK, Kumar B, Los G, Castro J, Lee JS et al (2005) Reversal of cisplatin resistance with a BH3 mimetic, (‐)‐gossypol, in head and neck cancer cells: role of wild‐type p53 and Bcl‐xL. Mol Cancer Ther 4:1096–1104. [DOI] [PubMed] [Google Scholar]
- 2. Bello MJ, Leone PE, Vaquero J, De Campos JM, Kusak ME, Sarasa JL et al (1995) Allelic loss at 1p and 19q frequently occurs in association and may represent early oncogenic events in oligodendroglial tumors. Int J Cancer 64:207–210. [DOI] [PubMed] [Google Scholar]
- 3. Benetkiewicz M, Idbaih A, Cousin PY, Boisselier B, Marie Y, Crinière E et al (2009) NOTCH2 is neither rearranged nor mutated in t(1;19) positive oligodendrogliomas. PLoS One 4:e4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bigner SH, Matthews MR, Rasheed BK, Wiltshire RN, Friedman HS, Friedman AH et al (1999) Molecular genetic aspects of oligodendrogliomas including analysis by comparative genomic hybridization. Am J Pathol 155:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boulay JL, Miserez AR, Zweifel C, Sivasankaran B, Kana V, Ghaffari A et al (2007) Loss of NOTCH2 positively predicts survival in subgroups of human glial brain tumors. PLoS One 2:e576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brandes AA, Tosoni A, Cavallo G, Reni M, Franceschi E, Bonaldi L et al (2006) Correlations between O6‐methylguanine DNA methyltransferase promoter methylation status, 1p and 19q deletions, and response to temozolomide in anaplastic and recurrent oligodendroglioma: a prospective GICNO study. J Clin Oncol 24:4746–4753. [DOI] [PubMed] [Google Scholar]
- 7. Buhlmann S, Pützer BM (2008) DNp73 a matter of cancer: mechanisms and clinical implications. Biochim Biophys Acta 1785:207–216. [DOI] [PubMed] [Google Scholar]
- 8. Burger PC, Minn AY, Smith JS, Borell TJ, Jedlicka AE, Huntley BK et al (2001) Losses of chromosomal arms 1p and 19q in the diagnosis of oligodendroglioma. A study of paraffin‐embedded sections. Mod Pathol 14:842–853. [DOI] [PubMed] [Google Scholar]
- 9. Cairncross JG, Ueki K, Zlatescu MC, Lisle DK, Finkelstein DM, Hammond RR et al (1998) Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst 90:1473–1479. [DOI] [PubMed] [Google Scholar]
- 10. Dong S, Pang JC, Hu J, Zhou LF, Ng HK (2002) Transcriptional inactivation of TP73 expression in oligodendroglial tumors. Int J Cancer 98:370–375. [DOI] [PubMed] [Google Scholar]
- 11. Dong SM, Pang JC, Poon WS, Hu J, To KF, Chang AR, Ng HK (2001) Concurrent hypermethylation of multiple genes is associated with grade of oligodendroglial tumors. J Neuropathol Exp Neurol 60:808–816. [DOI] [PubMed] [Google Scholar]
- 12. Dong Z, Pang JC, Ng MH, Poon WS, Zhou L, Ng HK (2004) Identification of two contiguous minimally deleted regions on chromosome 1p36.31‐p36.32 in oligodendroglial tumours. Br J Cancer 91:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ducray F, Idbaih A, De Reyniès A, Bièche I, Thillet J, Mokhtari K et al (2008) Anaplastic oligodendrogliomas with 1p19q codeletion have a proneural gene expression profile. Mol Cancer 7:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ferrer‐Luna R, Mata M, Núñez L, Calvar J, Dasí F, Arias E et al (2009) Loss of heterozygosity at 1p‐19q induces a global change in oligodendroglial tumor gene expression. J Neurooncol 95:343–354. [DOI] [PubMed] [Google Scholar]
- 15. Giannini C, Burger PC, Berkey BA, Cairncross JG, Jenkins RB, Mehta M et al (2008) Anaplastic oligodendroglial tumors: refining the correlation among histopathology, 1p 19q deletion and clinical outcome in Intergroup Radiation Therapy Oncology Group Trial 9402. Brain Pathol 18:360–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Griffin CA, Burger P, Morsberger L, Yonescu R, Swierczynski S, Weingart JD, Murphy KM (2006) Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol 65:988–994. [DOI] [PubMed] [Google Scholar]
- 17. Hoang‐Xuan K, Capelle L, Kujas M, Taillibert S, Duffau H, Lejeune J et al (2004) Temozolomide as initial treatment for adults with low‐grade oligodendrogliomas or oligoastrocytomas and correlation with chromosome 1p deletions. J Clin Oncol 22:3133–3138. [DOI] [PubMed] [Google Scholar]
- 18. Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T et al (2003) F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell 115:163–175. [DOI] [PubMed] [Google Scholar]
- 19. Huang H, Okamoto Y, Yokoo H, Heppner FL, Vital A, Fevre‐Montange M et al (2004) Gene expression profiling and subgroup identification of oligodendrogliomas. Oncogene 23:6012–6022. [DOI] [PubMed] [Google Scholar]
- 20. Husemann K, Wolter M, Büschges R, Boström J, Sabel M, Reifenberger G (1999) Identification of two distinct deleted regions on the short arm of chromosome 1 and rare mutation of the CDKN2C gene from 1p32 in oligodendroglial tumors. J Neuropathol Exp Neurol 58:1041–1050. [DOI] [PubMed] [Google Scholar]
- 21. Jenkins RB, Blair H, Ballman KV, Giannini C, Arusell RM, Law M et al (2006) A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res 66:9852–9861. [DOI] [PubMed] [Google Scholar]
- 22. Kros JM, Gorlia T, Kouwenhoven MC, Zheng PP, Collins VP, Figarella‐Branger D et al (2007) Panel review of anaplastic oligodendroglioma from European Organization For Research and Treatment of Cancer Trial 26951: assessment of consensus in diagnosis, influence of 1p/19q loss, and correlations with outcome. J Neuropathol Exp Neurol 66:545–551. [DOI] [PubMed] [Google Scholar]
- 23. Kumar KN, Babcock KK, Johnson PS, Chen X, Ahmad M, Michaelis EK (1995) Cloning of the cDNA for a brain glycine‐, glutamate‐ and thienylcyclohexylpiperidine‐binding protein. Biochem Biophys Res Commun 216:390–398. [DOI] [PubMed] [Google Scholar]
- 24. Law ME, Templeton KL, Kitange G, Smith J, Misra A, Feuerstein BG, Jenkins RB (2005) Molecular cytogenetic analysis of chromosomes 1 and 19 in glioma cell lines. Cancer Genet Cytogenet 160:1–14. [DOI] [PubMed] [Google Scholar]
- 25. Li KK, Pang JC, Ching AK, Wong CK, Kong X, Wang Y et al (2009) miR‐124 is frequently down‐regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum Pathol 40:1234–1243. [DOI] [PubMed] [Google Scholar]
- 26. Li LC, Dahiya R (2002) MethPrimer: designing primers for methylation PCRs. Bioinformatics 18:1427–1431. [DOI] [PubMed] [Google Scholar]
- 27. Manuelidis L, Yu RK, Manuelidis EE (1977) Ganglioside content and pattern in human gliomas in culture. Acta Neuropathol 38:129–135. [DOI] [PubMed] [Google Scholar]
- 28. McDonald JM, Dunlap S, Cogdell D, Dunmire V, Wei Q, Starzinski‐Powitz A et al (2006) The SHREW1 gene, frequently deleted in oligodendrogliomas, functions to inhibit cell adhesion and migration. Cancer Biol Ther 5:300–304. [DOI] [PubMed] [Google Scholar]
- 29. McDonald JM, Dunmire V, Taylor E, Sawaya R, Bruner J, Fuller GN et al (2005) Attenuated expression of DFFB is a hallmark of oligodendrogliomas with 1p‐allelic loss. Mol Cancer 4:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McDonald JM, See SJ, Tremont IW, Colman H, Gilbert MR, Groves M et al (2005) The prognostic impact of histology and 1p/19q status in anaplastic oligodendroglial tumors. Cancer 104:1468–1477. [DOI] [PubMed] [Google Scholar]
- 31. Mehler MF, Mattick JS (2007) Noncoding RNAs and RNA editing in brain development, functional diversification, and neurological disease. Physiol Rev 87:799–823. [DOI] [PubMed] [Google Scholar]
- 32. Michaud WA, Nichols AC, Mroz EA, Faquin WC, Clark JR, Begum S et al (2009) Bcl‐2 blocks cisplatin‐induced apoptosis and predicts poor outcome following chemoradiation treatment in advanced oropharyngeal squamous cell carcinoma. Clin Cancer Res 15:1645–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miyake E (1979) Establishment of a human oligodendroglial cell line. Acta Neuropathol 46:51–55. [DOI] [PubMed] [Google Scholar]
- 34. Ohga T, Koike K, Ono M, Makino Y, Itagaki Y, Tanimoto M et al (1996) Role of the human Y box‐binding protein YB‐1 in cellular sensitivity to the DNA‐damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res 56:4224–4228. [PubMed] [Google Scholar]
- 35. Post GR, Dawson G (1992) Characterization of a cell line derived from a human oligodendroglioma. Mol Chem Neuropathol 16:303–317. [DOI] [PubMed] [Google Scholar]
- 36. Reifenberger G, Kros JM, Louis DN, Collins VP (2007) Oligodendrogliomas. In: WHO Classification of Tumors of the Central Nervous System. Louis DN, Ohgaki H, Wiestler OD, Cavenee WK (eds), pp. 54–59. International Agency for Research on Cancer: Lyon, France. [Google Scholar]
- 37. Riemenschneider MJ, Reifenberger J, Reifenberger G (2008) Frequent biallelic inactivation and transcriptional silencing of the DIRAS3 gene at 1p31 in oligodendroglial tumors with 1p loss. Int J Cancer 122:2503–2510. [DOI] [PubMed] [Google Scholar]
- 38. Sasaki H, Zlatescu MC, Betensky RA, Johnk LB, Cutone AN, Cairncross JG, Louis DN (2002) Histopathological‐molecular genetic correlations in referral pathologist‐diagnosed low‐grade “oligodendroglioma.” J Neuropathol Exp Neurol 61:58–63. [DOI] [PubMed] [Google Scholar]
- 39. Smith JS, Alderete B, Minn Y, Borell TJ, Perry A, Mohapatra G et al (1999) Localization of common deletion regions on 1p and 19q in human gliomas and their association with histological subtype. Oncogene 18:4144–4152. [DOI] [PubMed] [Google Scholar]
- 40. Smith JS, Perry A, Borell TJ, Lee HK, O'Fallon J, Hosek SM et al (2000) Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol 18:636–645. [DOI] [PubMed] [Google Scholar]
- 41. Stewart DJ (2007) Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol 63:12–31. [DOI] [PubMed] [Google Scholar]
- 42. Tews B, Felsberg J, Hartmann C, Kunitz A, Hahn M, Toedt G et al (2006) Identification of novel oligodendroglioma‐associated candidate tumor suppressor genes in 1p36 and 19q13 using microarray‐based expression profiling. Int J Cancer 119:792–800. [DOI] [PubMed] [Google Scholar]
- 43. Tews B, Roerig P, Hartmann C, Hahn M, Felsberg J, Blaschke B et al (2007) Hypermethylation and transcriptional downregulation of the CITED4 gene at 1p34.2 in oligodendroglial tumours with allelic losses on 1p and 19q. Oncogene 26:5010–5016. [DOI] [PubMed] [Google Scholar]
- 44. Torigoe T, Izumi H, Ishiguchi H, Yoshida Y, Tanabe M, Yoshida T et al (2005) Cisplatin resistance and transcription factors. Curr Med Chem Anticancer Agents 5:15–27. [DOI] [PubMed] [Google Scholar]
- 45. Watanabe T, Nakamura M, Yonekawa Y, Kleihues P, Ohgaki H (2001) Promoter hypermethylation and homozygous deletion of the p14ARF and p16INK4a genes in oligodendrogliomas. Acta Neuropathol 101:185–189. [DOI] [PubMed] [Google Scholar]
- 46. Wolter M, Reifenberger J, Blaschke B, Ichimura K, Schmidt EE, Collins VP, Reifenberger G (2001) Oligodendroglial tumors frequently demonstrate hypermethylation of the CDKN2A (MTS1, p16INK4a), p14ARF, and CDKN2B (MTS2, p15INK4b) tumor suppressor genes. J Neuropathol Exp Neurol 60:1170–1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sequences of 36‐bp repeat region and phylogenetic sequence analysis. A. The unique 36‐bp repeat sequences observed in exon 4 of the PDAM gene are shown. Repeat 13, which is located in the center of the repeat region, shows loose base conservation after first 21 bp and has a 4‐bp deletion at the 3′ end. B. Phylogenetic comparative analysis reveals that human PDAM is conserved in the primate infraorder Simiiformes (UCSC Genome Browser, http://genome.ucsc.edu/).
Figure S2. Detection of endogenous PDAM by Western blot analysis. The specificity of the anti‐PDAM antibody is validated by its reactivity to PDAM based on the predicted coding sequence (pPDAM) and GFP‐tagged PDAM fusion proteins (pPDAM‐GFP and pGFP‐PDAM). The latter two proteins are also detected by the anti‐GFP antibody. The 293FT cells show PDAM mRNA of normal brain level, the glioma lines exhibit twofold to 10‐fold lower level relative to normal brain and gastric tumor lines AGS and MKN45 display no detectable PDAM transcript. None of the cell lines expressing detectable PDAM transcripts show any unique protein bands that are reactive to the anti‐PDAM antibody.
Figure S3. Drug sensitivity assays and cell survival curves. A. Drug sensitivity assay reveals that PDAM knockdown has no effects on sensitivity to vincristine, lomustine, temozolomide and paclitaxel in all five glioma lines examined. B Cell survival curves indicate increased resistance to cisplatin when PDAM is knocked down in A172 and U87MG cells.
Table S1. Oligonucleotides used in this study.
Supporting info item


