Abstract
In this study we investigated the locomotor activity and non‐selective attention in spontaneously hypertensive rats (SHR) with control Wistar–Kyoto (WKY) rats, which were employed as an attention deficit hyperactivity disorder (ADHD) model. In open‐field test and làt maze, SHR rats were found to be much more spontaneously active than WKY rats. As compared with WKY rats, a lower level of galectin‐3 was observed in SHR brain prefrontal cortex (PFC), which was the major affected brain area of ADHD. Through miRNA microarray screening, rno‐let‐7d was noted to be solely upregulated in SHR PFC. Interestingly, rno‐let‐7d had a binding site at galectin‐3 mRNA and was shown to regulate galectin‐3 3′ untranslated region (UTR) directly. Mutation of galectin‐3 3′UTR by one nucleotide of the seed sequence prevented rno‐let‐7d regulation of the 3′ UTR completely. Although rno‐let‐7d did not directly regulate tyrosine hydroxylase (TH) 3′UTR, the level of galectin‐3 was important for cAMP response element binding protein, the major transcript factor for TH gene. Either overexpression or downexpression of galectin‐3 could result in modulation of TH expression in both PC12H and PC12L cells. In conclusion, our data suggested a novel function of rno‐let‐7d in regulation of galectin‐3 and in ADHD development. Rno‐let‐7d, which is increased in the PFC of SHR brain, negatively regulated galectin‐3, which is coupled with TH expression regulation.
Keywords: ADHD, galectin‐3, let‐7d
INTRODUCTION
Attention deficit hyperactivity disorder (ADHD) is a common children's disorder characterized by symptoms of inattention and/or impulsivity and hyperactivity 4, 17. To date, the pathogenesis and etiology of ADHD are still unclear. Several theories about ADHD development have been proposed and many researchers believe that it is caused by both genetic and environmental factors and relates to the structural and functional abnormalities of the brain (25). Spontaneously hypertensive rat (SHR) is a valid and currently accepted model for studying ADHD, and has received extensive genetic investigations. SHR is also the only animal model that has demonstrated all the behavioral characteristics of ADHD 12, 24.
MicroRNAs (miRNAs) are evolutionally conserved small non‐coding RNAs that regulate approximately 30% of human protein‐coding gene expressions at the post‐transcriptional level and play important roles in a wide variety of biological functions 1, 9, 32. In the central nervous system, miRNAs are particularly abundant and involved in neuronal development, differentiation, neurodegeneration and apoptosis. They are also involved in memory, mental retardation [reviewed by Corbin Rachel et al (11), Fiore et al (18), Kosik(29) and Bushati et al (5)]. miRNA let‐7 has recently been implicated as an important regulator for the development and morphogenesis of the brain 8, 51, although its role in the development and pathogenesis of ADHD is still completely unknown.
Among the alterations that have been identified in the central nervous system and that reflect possible neural disturbances of ADHD, dopamine (DA)‐deficit theory is the most important in current research (45). DA constitutes about 80% of the catecholamine content in the brain (48). Synthesis of DA occurs in two steps. First, tyrosine is catalyzed by the enzyme tyrosine hydroxylase (TH) and converted into l‐3,4‐dihydroxyphenylalanine (l‐DOPA). TH is thus considered the rate‐limiting enzyme in this pathway. Then DOPA is decarboxylated and produces DA, which is catalyzed by aromatic l‐amino acid decarboxylase. Therefore, TH is a cornerstone enzyme in the biosynthesis of DA, which acts as an important signaling molecule in the central nervous system. Previous studies have observed that TH was downregulated in the prefrontal cortex (PFC) of SHR brain, linking TH to ADHD 27, 31. The expression of TH gene is regulated by several transcription factors, 21, 28 among which cAMP response element binding protein (CREB) is the most important (33).
Galectin‐3, a member of the galectin family of carbohydrate‐binding mammalian lectins, exhibits pleiotropic biological functions in many physiological and pathological processes (30). The molecule is widely expressed in neural and nonneural cells and involved in neural cell adhesion, proliferation and neurite growth 20, 41. Galectin‐3 is able to modulate specific gene expression by interacting with particular transcription factors (15), such as SP1 and CREB, which is stabilized in binding through galectin‐3 (35). On the other hand, galectin‐3 gene (Lgals3) was also downregulated in SHR PFC determined by previous cDNA microarray (42). Therefore, in this study we searched and screened the possible miRNAs that were responsible for the regulation of galectin‐3 and TH in the brain of SHR.
MATERIALS AND METHODS
Animals
Six‐week old juvenile male SHR and age‐ and gender‐matched genetic control WKY (Wistar–Kyoto) rats weighing from 125 g to 150 g were obtained from Shanghai Slac Laboratory Animal CO. LTD. SHR at this age did not develop hypertension as in previous reports 39, 43, 47. All the rats were housed under standard laboratory conditions with a 12‐h light–dark cycle (lights on from 06:00 to 18:00) and with temperature and humidity maintained at 22 ± 1°C and 45%–55%, respectively, and the animals were allowed free access to standard laboratory rodent chow and water. The experiments were approved by the local Animal Ethics Committee.
Main reagents
Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS) and donor horse serum were from Gibco BRL (Carlsbad, CA, USA). Anti‐rat galectin‐3 polyclonal antibody against N‐terminal 1–160 amino acids was from Santa Cruz Biotechnology (sc‐20157), Inc. (Santa Cruz, CA, USA). Anti‐rat TH polyclonal antibody against N‐terminus was from Boster Biological Technology (BA1454), LTD (Wuhan, China). Anti‐rat α‐Tubulin monoclonal antibody was from Beyotime Institute of Biotechnology (AT819) (Haimen, China). The enhanced chemiluminescent (ECL) horseradish peroxidase (HRP) substrate was from Millipore (Billerica, CA, USA). Nerve growth factor (NGF) was from Takara (Tokyo, Japan). Retinoic acid (RA) was from Sigma‐Aldrich (St. Louis, MO, USA). miRNA array was from Exiqon (Skelstedet 16, 2950 Vedbaek, Denmark).
Behavioral assessments
Open‐field test
The procedure of open‐field test was performed according to the report by Ueno et al (47). Briefly, the open field box was a 90 cm × 90 cm × 50 cm cube constructed from black Plexiglas and the field was divided into 81 squares of equal size by stripes of black paint. The apparatus was located in a sound‐attenuated room. Rats were placed in the central square and allowed to move freely. The movements were tracked over 60 minutes from 11:00 pm to 12:00 pm. The procedure was recorded by a video camera.
Làt maze
The test of làt maze was done according to Aspideet al.'s report (2). The apparatus was a 60 × 60 × 40 cm3 black Plexiglas box with a 30 × 30 × 40 cm3 Plexiglas transparent smaller box inserted in the middle to result a corridor which was 60 cm long, 15 cm wide and 40 cm high. The apparatus was located in a sound attenuated room. Rats were placed in the corner of the corridor and allowed to explore the corridor freely. The movements were tracked over 30 minutes from 4:30 am to 5:00 am. The procedure was recorded by a video camera.
Vector construction
Plasmid pcDNA3.1B (Invitrogen, Carlsbad, CA, USA) was used an expressional vector for galectin‐3. Rat galectin‐3 full length complementary DNA (cDNA, 41‐948 from the start codon (ATG), version: NM_031832.1, GI:13929189) was obtained by reverse transcription–polymerase chain reaction (RT–PCR) from total RNA extracted from rat brain tissue. The primers consisted of 5′CCCAAGCTTATGGCAGACGGCTTCTCACTT3′ (sense) and 5′GCTCTAGATCTCTCAGCACGAGACTTTAT3′ (antisense). The amplified cDNA was cloned into pCDNA3.1B at the sites of Hind III and Xba I.
Plasmid pSilencer 4.1 (Ambion, Austin, TX, USA) was used to construct interference plasmids that produce specific shRNA for rat galectin‐3. Two sequences were used for shRNA in this study. For shRNA‐1, 5′GATCCGAGCGGCAAACCATTCAAATTCAAGAGATTTGAATGGTTTGCCGCTCAGA3′ was used as top strand and 5′AGCTTTCGAGCGGCAAACCATTCAAATCTCTTGAATTTGAATGGTTTGCCGCTCG3′ was used as bottom strand. For shRNA‐2, 5′GATCCAATGGCAGACGGCTTCTCATTCAAGAGATGAGAAGCCGTCTGCCATTAGA3′ was used as top strand and 5′AGCTTCTAATGGCAGACGGCTTCTCATCTCTTGAATGAGAAGCCGTCTGCCATTG3′ was used as bottom strand. Rno‐let‐7d expression plasmid construction was according to Jadhav et al (23) and Ghosh et al (19). Briefly, the precursor of rno‐let‐7d (accession ID MI0000601) was obtained by PCR from rat brain tissue and cloned into Hind III and Bam H I sites of pSilencer 4.1. These sites have documented validity for miRNA expression 19, 23. Luciferase report plasmid construction was based on the report by De Martino et al (13). Briefly, the 3′ untranslated region (UTR) sequences of galectin‐3 and TH mRNA were amplified by PCR and cloned into pGL3‐Control firefly luciferase report plasmid (Promega, Madison, WI, USA) at the Xba I site immediately downstream from the stop codon of luciferase. All constructed plasmids were confirmed by DNA sequence analysis. Plasmids with the random sequence were used as control.
Cell culture and transient transfection
Rat kidney low differentiated pheochromocytoma PC12 (PC12L) cells and rat kidney high differentiated pheochromocytoma PC12 (PC12H) cells (American Type Culture Collection, Rockville, MD, USA) were maintained in DMEM (High Glucose), supplemented with heat‐inactivated fetal calf serum (10%, v/v) and donor horse serum (10%, v/v). Rat hepatoma cell CBRH‐7919 cells (from the Institute of Cell and Biochemistry Research of Chinese Academy of Science, Shanghai, China) were maintained in DMEM supplemented with newborn bovine serum (10%, v/v). The culture was maintained at 37°C with 5% CO2 and humidified atmosphere. The medium was refreshed every 2 or 3 days and cells were divided once a week. For NGF induction, PC12H cells were incubated in a low concentration of serum (1% serum) for 48 h prior to treatment with 100 ng/mL NGF as described by Li et al (34). For RA induction, PC12L cells were incubated in DMEM with 1 µmol/L RA for 6 days as described in our previous report (50).
The day before transfection, cells were plated in 60 mm dishes (BD Falcon, Becton Dickinson Labwares, Franklin Lakes, NJ, USA) at 60%–80% confluence without antibiotics and incubated for 24 h. Transfections of plasmids were performed with SunBioTM Trans‐EZ (SunBio Medical Biotechnology CO., LTD, Shanghai, China), according to the manufacturer instruction. The transfection efficiency was evaluated with enhanced green fluorescent protein (EGFP) expressional pEGFP plasmid.
Western blots
To prepare protein samples, cultured cells were washed twice with phosphate‐buffered saline (PBS) and lysed in buffer consisting of 50 mmol/L Tris‐HCl (pH 8.0), 0.5% (v/v) Tween 20, 1 mmol/L ethylene diamine tetraacetic acid (EDTA), 2 mmol/L phenylmethylsulfonyl fluoride and 1 µg/mL aprotinin. For preparation of samples from brain tissue, rats were killed after being anesthetized by 10% chloral hydrate (4 mL/kg) solution through abdominal cavity and the brains were removed immediately on ice. The PFC, striatum (STR), midbrain (MB) and cerebellum (CB) were dissected according to the atlas of Paxinos and Watson (40) as described by Siuciak et al (44). The brain tissue samples were homogenized in the lysis buffer on ice. The protein concentrations of the extracts were determined by Lorry's method. The proteins were resolved by 10% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and were then electrotransferred onto a polyvinylidene difluoride membrane (Millipore, Bedford, MA, USA). After blocking with 5% non‐fat milk in PBST (PBS containing 0.1% Tween 20) for 2 h at room temperature and pre‐immune absorption, the membranes were incubated with specific primary antibodies (galectin‐3 and TH antibodies in 1:200 dilution) overnight at 4°C. Then the blots were washed three times with PBST and incubated for 2 h at room temperature with HRP‐conjugated secondary antibodies. Immunolabeling was detected by ECL after washing with PBST. All experiments were repeated at least three times independently.
Immunohistochemistry and immunofluorescence
PC12 cells were fixed with 4% paraformaldehyde after culturing on coverslips for 48 h and permeabilized with 0.2% Triton X‐100/PBS as our previous study (49). Then cells were blocked with 5% goat serum. Anti‐TH or anti‐galectin‐3 antibody was diluted at 1:100 as the primary antibody and HRP‐conjugated goat anti‐rabbit immunoglobulin was used as the secondary antibody. The samples were visualized by the HRP substrate diaminobenzidine tetrahydrochloride (DAB), and then nuclei were counterstained with 0.1% methyl green. To prepare slices of brain sections, rats were anesthetized by 10% Chloral Hydrate (4 mL/kg) solution and fixed by intracardiac perfusion with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4). After 10 minutes of perfusion, the brain was quickly removed and post‐fixed for 6 h in the same fixative solution. Coronal brain sections in 16 µm‐thickness were cut in a cryotome. Brain sections were permeabilized with 20% methanol at −20°C for 10 minutes and blocked with 5% goat serum. After sequential incubation with anti‐galectin‐3 antibody (in 1:100 dilution) and HRP‐conjugated goat anti‐rabbit immunoglobulin (1:1000), the samples were visualized by DAB and the nuclei were counterstained with 0.1% methyl green. Slices were then observed under an Olympus light microscope (Tokyo, Japan).
Immunofluorescence was performed based on our previous report (22). In brief, PC12 cells were fixed with 4% paraformaldehyde after culturing on coverslips for 48 h and permeabilized with 0.2% Triton X‐100/PBS. The cells were blocked with 5% goat serum and stained with anti‐galectin‐3 antibody, followed by incubation with Fluorescein isothiocyanate (FITC)‐conjugated anti‐Rabbit antibody. The nuclei were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI). Cells were then observed under an Olympus fluorescence microscope.
miRNA microarray
Microarray analysis was performed by Kangcheng Bio‐tech Inc. (Shanghai, China). Briefly, total RNAs were extracted using Trizol (Dingguo, China) and RNeasy mini kit (QIAGEN, Strasse, Hilden, Germany) according to manufacturer's instructions. After RNA measurement on the Nanodrop instrument, the samples were labeled using the miRCURY™ Hy3™ Power labeling kit (Exiqon) and hybridized on the miRCURY™ LNA Array (v.11.0, Exiqon). After hybridization, scanning was performed with the Axon GenePix 4000B microarray scanner (Molecular Devices, Downingtown, PA, USA). GenePix pro V6.0 (Molecular Devices) was used to read the raw intensity of the image. The intensity of green signal was calculated after background subtraction and four replicated spots of each probe on the same slide had been averaged. Median Normalization Method was used to obtain “Normalized Data”: Normalized Data = (Foreground − Background) / median, where the median was the 50% quantile of miRNA intensity, which was larger than 50 in all samples after background correction.
Reverse transcriptase–polymerase chain reaction (RT–PCR)
Complementary DNA (cDNA) was obtained by using RT (100 U of ReverTra Ace, Toyobo, Japan) and 1 µg of total RNA from PC12 cells, CBRH‐7919 cells and brain tissues. The RT reaction of miRNAs were performed according to the report (10).
PCR amplification of rat glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) was employed as sample load control: Its primers consisted of 5′ACAGCAACAGGGTGGTGGAC3′ (sense) and 5′ TTTGAGGGTGCAGCGAACTT3′ (antisense). The reaction was performed by a thermal cycler (Tianglong, Xian, China) with 28 cycles of 94°C for 4 minutes, 94°C for 30 s, 59°C for 30 s and 72°C for 30 s and the product size was 252 base pairs (bp). The primers of rat galectin‐3 included 5′GCTGATTTCCCTGAGGTTCT3′ (sense), and 5′CGACATCGCCTTCCACTTT3′ (antisense). The reaction steps of 30 cycles consisted of 94°C for 4 minutes, 94°C for 30 s, 57°C for 20 s and 72°C for 25 s. The expected product size was 238 bp. Rat TH primers included 5′GCTCTAGAATGCATAGGGTACCACCCACA3′ (sense), and 5′GCTCTAGAGGTTAGATTCTTTCCTTCCTTT3′ (antisense). The reaction was performed at 94°C for 4 minutes, 94°C for 30 s, 55°C for 30 s and 72°C for 30 s with 40 cycles and the product size was 278 bp.
U6 was used as the interior control for miRNA amplification. Its primers included reverse transcriptional primer (AAAATATGGAACGCT), sense PCR primer (5′CTCGCTTCGGCAGCACA3′) and antisense primer (5′AACGCTTCACGAATTTGCGT3′). PCR reaction (10) was carried out at 94°C for 2 minutes, 94°C for 30 s, 60°C for 15 s and 72°C for 15 s with 30 cycles and the product size was 97 bp. Rno‐let‐7d was detected by stem‐loop PCR with the reverse primer (GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACTAT), sense PCR primer (5′CGGGCAGAGGTAGTAGGTT3′) and antisense primer 5′GTGCAGGGTCCGAGGT3′. The PCR reaction consisted of 50 cycles of 94°C for 2 minutes, 94°C for 30 s, 60°C for 15 s and 72°C for 15 s and the product size was 61 bp. The amplified products were electrophoresed in 2% Agarose to confirm the expected size. Real‐time PCR was carried out to quantify the PCR product of microRNA by the Bio‐Rad cycler. The reaction was performed in a system containing samples, primers, and PCR Master Mix with SYBR green (DBI Bioscience, Shanghai, China) in 96‐well PCR plates (Axygen, Union City, CA, USA). Cycle threshold more than 35 was excluded.
Cotransfections and luciferase assay
The procedure of cotransfections for luciferase assay was based on Ghosh et al.'s report (19). The activity of luciferase was analyzed using the Luciferase Reporter Assay kit with the provided technical bulletin (Promega). The data of relative luciferase units were normalized to the control. Three independent repeats were performed.
Statistical analysis
All experiments were conducted at least three times. The results are expressed as the means ± standard error Statistical analysis was done by Student's t‐test, and differences with P‐value less than 0.05 were considered statistical significance.
RESULTS
Behavioral tasks
Behavioral differences between SHR and WKY rats were analyzed by the open‐field test and làt‐maze test. During the 1‐h open‐field test, SHR were more active than WKY rats. The total number of square crossings and the number of rearings in a 15‐minute test in SHR were significantly higher than those of WKY rats (Figure 1A,B). During the 30‐minute làt‐maze test, the numbers of square crossings and rearings in 10 minutes in SHR were also significantly higher than WKY rats (Figure 1C,D). These results supported that SHR was a valid ADHD animal model, which was then used as ADHD model in the following experiments.
Figure 1.
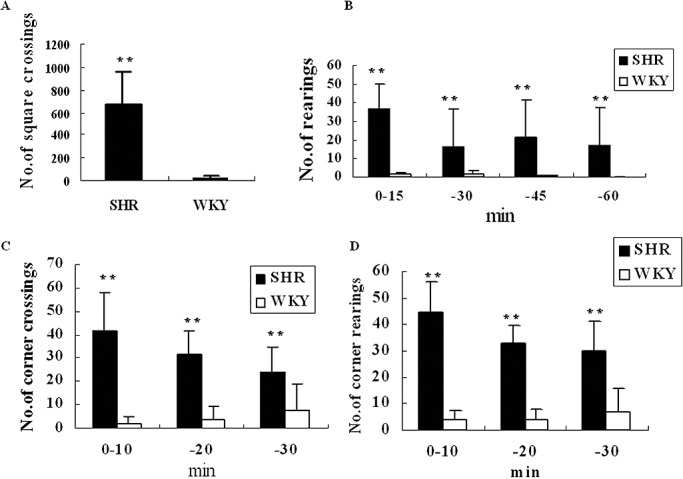
Behavioral task comparison between spontaneously hypertensive rats (SHR) and Wistar–Kyoto (WKY) rats. A. Comparison of the total number of square crossings in 1 h open‐field test; B. Comparison of the number of rearings among 15 minutes groups in 1‐h open‐field test; C. Number comparison of corner crossings among 10‐minute groups in 30‐minute làt‐maze test; D. The number comparison of corner rearings among 10‐minute groups in 30‐minute làt‐maze test. Data were given as an average of the number of crossings or rearings with standard errors, where n = 5. **P < 0.01.
Galectin‐3 expression in the brain of SHR
Galectin‐3 expression level was detected in PFC and other brain regions of SHR. In comparison with WKY rats, galectin‐3 was lowly expressed in PFC, STR, and MB of SHR in either protein level (Figure 2A–C) or mRNA level (Figure 2D), but similarly in the CB. Although SHR is a universally accepted ADHD animal model, it becomes hypertensive only after about 12 weeks. Therefore, we also detected the galectin‐3 protein level in aorta (AO), but did not observe any marked change in 6‐week‐old SHR (Figure 2A). There was also no marked change of galectin‐3 in the liver (Figure 2A). Therefore the downregulation of galectin‐3 expression in SHRs was not systemic, but only in PFC, STR and MD of SHR brain. The region of PFC is the site of integration of widespread glutamatergic inputs essential for executive functions and goal‐directed behaviors (36).
Figure 2.
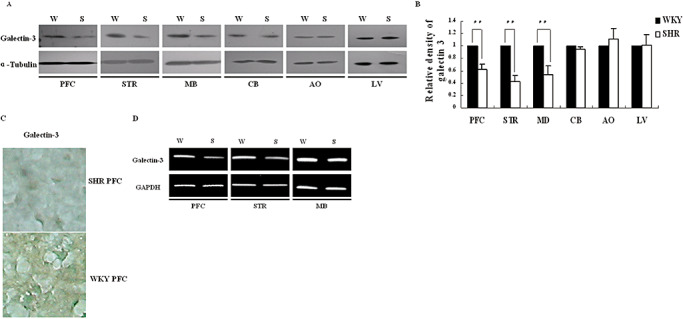
Comparison of galectin‐3 expression between ADHD and WKY rats. A. Galectin‐3 protein was analyzed by Western blot in prefrontal cortex (PFC), striatum (STR), midbrain (MB), cerebellum (CB), aorta (AO), liver (LV); B. Gray density analysis of the bands in A. The bands on X‐ray films was scanned and analyzed by the software Image J (Wayne Rasband, National Institute of Health, USA). Data were given as an average of relative density with standard errors, where n = 5. **P < 0.01; C. Immunostaining of the PFC slice was with galectin‐3 antibody, and cells were viewed in 400× magnifications. PFC slices of the brain were incubated with rabbit anti‐galectin‐3 antibody, HRP‐conjugated goat anti‐rabbit immunoglobulin and DAB sequentially, and counterstained with methyl green. The image was the representative of three independent experiments. D. Analysis of galectin‐3 mRNA level in PFC, STR and MB by RT–PCR. Abbreviations: ADHD = attention deficit hyperactivity disorder; DAB = diaminobenzidine tetrahydrochloride; HRP = horseradish peroxidase; RT–PCR = reverse transcriptase–polymerase chain reaction; SHR(S) = spontaneously hypertensive rats; WKY(W) = Wistar–Kyoto rats.
Galectin‐3 was regulated by rno‐let‐7d
To examine possible regulation of galectin‐3 by miRNAs, we analyzed the miRNA expression profile by miRNA microarray in PFC (Figure 3A) and the result showed that rno‐let‐7d was solely upregulated by twofold in SHR PFC. Then we searched for potential rno‐let‐7d binding sites in the galectin‐3 mRNA (Lgals3) 3′UTR by computational analysis. One predicted binding site (Figure 3B) of rno‐let‐7d in the Lgals3 3′UTR was found by rna22 (http://cbcsrv.watson.ibm.com/rna22_targets.html).
Figure 3.
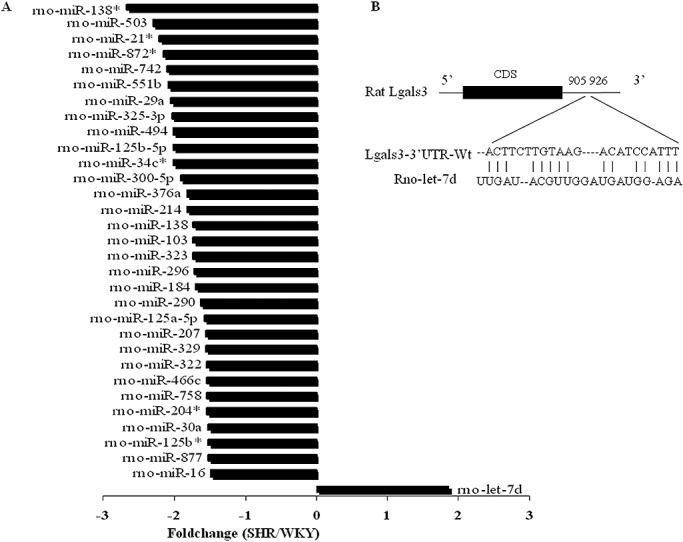
Rno‐let‐7d was upregulated in SHR prefrontal cortex. A. Foldchanges of all differentially expressed micro RNAs in the microarray results of the prefrontal cortex in SHR and WKY rats. B. Schematic representation of the interaction between rno‐let‐7d seeding sequence and the binding site on the wild‐type Lgals3‐3′UTR. Abbreviations: CDS = Coding Sequence; SHR = spontaneously hypertensive rat; UTR = untranslated region; Wt = wild type; WKY = Wistar–Kyoto rats.
To observe the direct effect of rno‐let‐7d on Lgals3‐3′UTR, rno‐let‐7d expressional plasmid was constructed and the validity was observed in CBRH‐7919 cells (used as non‐neuronal cell control) and PC12L cells (used as neuronal cells) (Figure 4A). Then a luciferase reporter assay was performed to verify direct rno‐let‐7d regulation of Lgals3‐3′UTR because miRNAs were thought to control gene expression by base pairing with specific recognizing elements in their target sequence. The luciferase reporter plasmid (pGL3‐control), which was constructed with 119 bp of Lgals3‐3′UTR, was cotransfected in CBRH‐7919 and PC12L cells with either rno‐let‐7d precursor or a random oligonucleotide control. The result indicated downregulation of luciferase expression by ∼45% in CBRH‐7919 cells (Figure 4B) and by ∼60% in PC12L cells. The downregulation of luciferase expression was prevented by mutating one nucleotide in the Lgals3 3′UTR (C→G, Figure 4B,C) to disrupt its seed sequence, supporting that the mode of effect of miRNAs is direct interaction with the mRNA targets, and that the “seed site” at the 5′ end of the miRNAs is necessary for the regulation.
Figure 4.
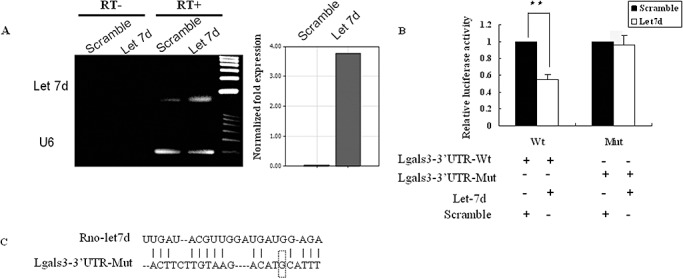
Rno‐let‐7d regulation of galectin‐3 expression. A. Validity of rno‐let‐7d expression plasmid was confirmed by PCR in both CBRH‐7919 and PC12L cells 48 h after transfection. The result was similar in two cells and the figure was the representative of three independent experiments. Reaction without reverse transcriptase (RT‐) was used as the negative control. The expression rno‐let‐7d was also confirmed by real‐time PCR (right panel). B. Luciferase activity analysis in CBRH‐7919 cells after co‐transfection of rno‐let‐7d precursor with luciferase report plasmid. The plasmid containing scramble sequence was as a control. Data were shown as an average of relative luciferase units which were normalized to the control, with standard errors (**P < 0.01), where n = 6. C. Schematic representation of the interaction between rno‐let‐7d seeding sequence and the binding site on the mutant‐type Lgals3‐3′UTR (C→G). Abbreviations: Mut = mutant type, the report plasmid included the mutant‐type Lgals3‐3′UTR; PCR = polymerase chain reaction; UTR = untranslated region; Wt = wild type, the report plasmid included the wild‐type Lgals3‐3′UTR.
It was also noted that galectin‐3 expression was higher in PC12L cells than that in PC12H cells (Figure 5A–C), while lower expression of rno‐let‐7d was observed in PC12L cells than that in PC12H cells (Figure 5D). After PC12L cells were treated with 1 µmol/L RA, galectin‐3 expression was downregulated (Figure 5A,B) while rno‐let‐7d was upregulated (Figure 5E). It seemed that there existed an inverse correlation between galectin‐3 and rno‐let‐7d expression, further suggesting rno‐let‐7d regulation of galectin‐3. Transfection of rno‐let‐7d in PC12 cells did not result in expression regulation of the transfection factors RUNX1,2 (data not shown), which could activate galectin‐3 promoter activity (49).
Figure 5.
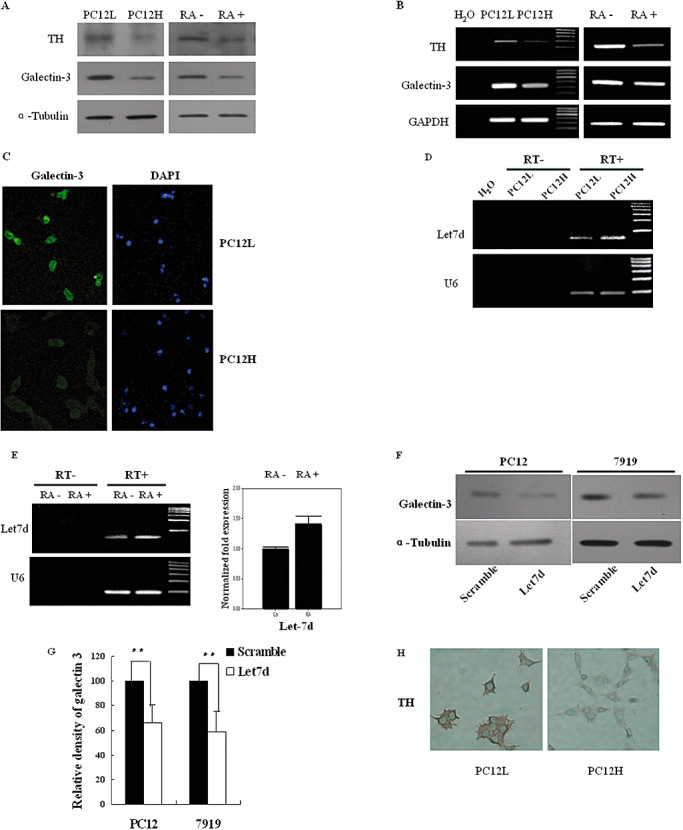
Galectin‐3, TH and rno‐let‐7d expressions in PC12 cells. A. Galectin‐3 and TH protein levels were detected by Western blot. Galectin‐3 and TH expressions were analyzed in both PC12L and PC12H cells at 48 h after passage, which showed that the expressions were higher in PC12L cells than in PC12H cells. Galectin‐3 and TH expressions in PC12L cells were then investigated at 6 days after incubation with 1 µmol/L RA. B. Galectin‐3 and TH mRNA levels were simultaneously detected by RT–PCR. C. Immunofluorescence staining of galectin‐3 in PC12L and PC12H cells, in 200× magnifications. D. Mature rno‐let‐7d was detected in PC12L and PC12H cells. Reactions without reverse transcriptase (RT‐) were as a negative control. E. Mature rno‐let‐7d was detected by either stem‐loop RT‐PCR (left panel) or real‐time PCR (right panel) in PC12L cells before and after 48 h incubation with 1 µmol/L RA. Reaction without reverse transcriptase (RT‐) was used as a negative control. The results were the representative of three independent experiments. F. In PC12L cells and CBRH‐7919 cells, galectin‐3 protein expression was reduced after transient transfection with rno‐let‐7d precursor. G. Density analysis of Western blot results in F. Density of the bands on X‐ray films was scanned, quantified by Image J (Wayne Rasband, National Institute of Health, USA). Data were shown as an average of relative density with standard errors (**P < 0.01), where n = 5. H. Immunohistochemistry observation of TH in PC12L and PC12H cells. Magnification: 200×. Abbreviations: DAPI = 4′,6‐diamidino‐2‐phenylindole; PC12L = low differentiated pheochromocytoma cells; PC12H = high differentiated pheochromocytoma PC12 cells; RA = retinoic acid; RT–PCR = reverse transcriptase–polymerase chain reaction; TH = tyrosine hydroxylase.
To further verify that rno‐let‐7d was an important regulator of galectin‐3, we observed the modulation of galectin‐3 protein levels in both CBRH‐7919 cells and PC12L cells. After transient transfection with rno‐let‐7d precursor, results showed that galectin‐3 protein expression was suppressed by ∼40% and 34% in CBRH‐7919 cells and PC12L cells, respectively (Figure 5F,G) although modulation of galectin‐3 mRNA levels was not so obvious.
TH expression downregulated
DA in the PFC is known to play an essential role in mediating executive functions, including working memory and attention. As the PFC is the major affected brain area in ADHD, and previous studies suggested that TH is downregulated in SHR PFC 27, 31, we investigated whether rno‐let‐7d or galectin‐3 could affect TH expression, which is important for the DA pathway. To test this, we transfected PC12L cells, which highly express TH (Figure 5A,B,H), with rno‐let‐7d precursor. In results, overexpression of rno‐let‐7d reduced TH expression by 38% approximately as evaluated by densitometric analysis (Figure 6A–C). To further investigate whether the effect was direct or indirect, TH 3′UTR (262 bp) was inserted into Xba I site, immediate downstream of the luciferase open reading frame, in PGL3 control plasmid. The reporter vector was then cotransfected into CBRH‐7919 and PC12 cells with rno‐let‐7d precursor. However, no altered luciferase activity was noted (Figure 6D) in the results, indicating no direct effect of rno‐let‐7d on TH 3′UTR.
Figure 6.
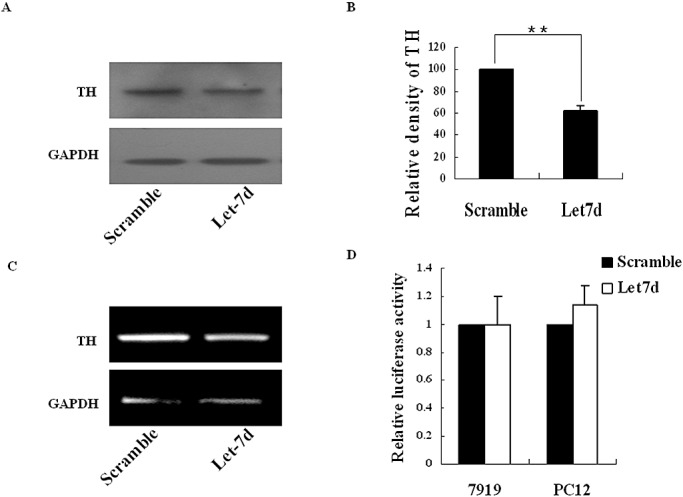
Observation of TH expression after rno‐let‐7d transfection. A. In PC12L cells, TH expression was detected by Western blot 72 h after transient transfection with rno‐let‐7d precursor. B. The band density in A was analyzed by the software Image J (Wayne Rasband, National Institute of Health, USA). Data were shown as an average of relative density with standard errors, where n = 3. (**P < 0.01). C. TH mRNA levels were detected by RT‐PCR in PC12L cells transfected with rno‐let‐7d precursor. D. Relative luciferase activity in CBRH‐7919 and PC12L cells was shown 48 h after co‐transfection with rno‐let‐7d precursor and report plasmids. The results were representative of three independent experiments. Abbreviations: PC12L = low differentiated pheochromocytoma cells; RA = retinoic acid; RT–PCR = reverse transcriptase–polymerase chain reaction; TH = tyrosine hydroxylase.
Relation between galectin‐3 and TH expressions
Previous reports suggested the role of nuclear galectin‐3 in regulation of gene transcription 15, 16. Through enhancement or stabilization of transcription factor binding to the cAMP response element (CRE) sites in the promoter site of target genes, galectin‐3 was shown to promote trans‐activation of transcription factors such as CREB and to induce the promoter activity.
Then we asked whether there was a relationship between galectin‐3 and TH expressions. PC12L and PC12H cells were used as a model for dopaminergic neurons because these cells synthesize DA. In the basal expressional level, PC12L cells were found to express higher galectin‐3 and TH than PC12H cells (Figure 5A–H). After PC12L cells were exposed to 1 µmol/L RA, both galectin‐3 and TH expression were downregulated simultaneously (Figure 5A,B). The PC12 cell line is an established model for NGF‐induced neurite formation (34). Therefore, PC12H cells were treated with 100 ng/mL NGF to induce neurite formation and differentiation, and the change of galectin‐3 and TH was investigated. In results, both of them were upregulated following NGF induction (Figure 7A,B). This seemed that there was a positive correlation between TH and galectin‐3 expressions.
Figure 7.
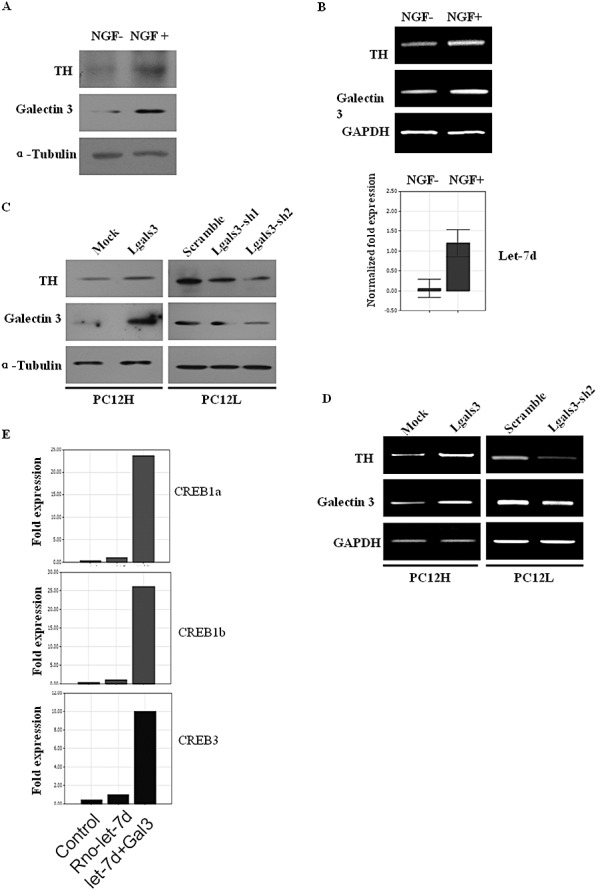
Association of galectin‐3 expression with TH regulation. A. Galectin‐3 and TH protein levels in PC12H cells were detected by Western blot 48 h after incubation with 100 ng/mL NGF. B. Galectin‐3 and TH mRNA levels were observed at the same time, indicating that their change was in a same tendency as the protein level. C. Galectin‐3 and TH protein levels were compared after 48 h transfection with galectin‐3 expression plasmid or interference plasmid. D. Galectin‐3 and TH mRNA levels were detected at the same time with (C). E. CREB transcription factors were detected by real‐time PCR after galectin‐3 and rno‐let‐7d transfection. Abbreviations: CREB = cAMP response element binding; NGF = nerve growth factor; PCR = polymerase chain reaction; TH = tyrosine hydroxylase.
Then we tested whether galectin‐3 level affected TH expression. PC12H cells were then transfected with galectin‐3 expression plasmid transiently. Interestingly, TH protein expression increased at 48 h after the transfection (Figure 7C). Then we transfected PC12L cells with galectin‐3 interference constructs and TH protein expression was interestingly found to be downregulated after the transfection (Figure 7C). TH mRNA decrease was also observed at the same time and was consistent with the protein expression (Figure 7D). After galectin‐3 transfection, the transcription factors CREB1a, 1b and 3, which are involved in TH promoter trans‐activation, were significantly upregulated (Figure 7E), while rno‐let‐7d transfection alone failed to induce CREB expression. After galectin‐3 knockdown by RNAi, CREB1a, b level also reduced although it was not so obvious because the basal level was low.
DISCUSSION
ADHD is characterized by symptoms of inattention, hyperactivity, and impulsivity 4, 17 and is an area of intense research due to its high incidence among children and severe hindrance of children's learning and education. Although many studies have made great contributions to understanding the mechanism of this disorder by using modern technologies or methods including molecular genetics to explore possible factors about this disorder, its pathogenesis and etiology remain unknown. Experiments in animal models have been the main venues of uncovering and revealing ADHD‐related molecules. Among the ADHD animal models, SHR has been the most extensively investigated genetic model and the only animal model that has demonstrated all the ADHD behavioral characteristics including hyperactivity, impulsivity and problems with sustained attention 12, 24. In the present study, we also assessed the behaviors of SHR with age and gender‐matched WKY rats using established open‐field and làt‐maze tests and observed that SHR was clearly more spontaneously active than WKY rats in the open‐field test, and SHR also had a higher level of spontaneous activity and non‐selective attention problem in the làt maze. Our results are consistent with the previous reports 2, 47 and support SHR as a valid ADHD model.
In this ADHD model a significantly lower expression level of galectin‐3 was noted in PFC, STR and MD of SHR brain than that in WKY rats. The PFC is the major affected brain area in ADHD (7). Thus our result suggested that galectin‐3 was downregulated in the major affected brain area of ADHD. A profile of miRNA expression was then investigated in SHR PFC and the results showed that rno‐let‐7d was upregulated. miRNA s are 21–23 nt long regulatory RNAs that can bind to target sites with partial complementation at 3′UTRs of mRNA and interfere with its stability or translational efficiency (19). A binding site of rno‐let‐7 was found in 3′UTR of galectin‐3 mRNA through computer analysis. Then we further investigated if rno‐let‐7d was responsible for galectin‐3 downregulation. Interestingly, overexpression of rno‐let‐7d resulted in downregulation of the protein level and 3′ UTR of galectin‐3, not only in PC12 cells but also in non‐neuron cells, which demonstrated a role of rno‐let‐7d in galectin‐3 post‐transcriptional regulation. Let‐7 is one of the first miRNAs discovered in Caenorhabditis elegans. In oncology, let‐7 expression is seen in various tumors and tumor cell lines with well‐documented tumor suppressor activity (37). Members of the let‐7 family were downregulated in both embryonic lung tissue and lung tumors (38). We observed that rno‐let‐7d expression was higher in PC12H cells than that in PC12L cells. After PC12 cell differentiation and growth inhibition induced by retinoic acid, rno‐let‐7d was increased. These results suggested rno‐let‐7d association with the differentiation of neurons. Galectin‐3 is also involved in regulation of development, immune reactions, wound repairing and differentiation 3, 52. Let‐7 controls multiple targets to regulate cell proliferation and differentiation through galectin‐3, as required for proper development (6). Abnormal expression of rno‐let‐7d in SHR PFC suggests that it might play an important role in galectin‐3 regulation, which might be associated with the neuron development or differentiation in this area. Galectin‐3 can serve as a substratum for neurite outgrowth of different neural cells. It is reasonable to suppose that let‐7 regulates neuron development through galectin‐3.
We also observed that TH expression was downregulated in PFC of SHR, which agreed with the previous work (31). In PC12 cells, rno‐let‐7d indeed resulted in lower expression of TH in both protein and mRNA levels. However, no luciferase activity alteration was detected after cotransfecting luciferase report vector with rno‐let‐7d precursor. This indicated that the lower expression of TH after rno‐let‐7d transfection might not be through the interaction between let‐7d and TH gene 3′UTR. However, we further observed that TH and galectin‐3 expressions were closely correlated and our data suggested that galectin‐3 level in the cells was important for the regulation of TH expression. Either overexpression or knockdown of galectin‐3 had subsequently effects on TH expression. The regulation of TH gene expression involves complicated signal pathways such as extracellular signal‐regulated kinase (ERK), c‐Jun NH2‐terminal kinase (JNK), Ras and Raf and the main cis‐elements of the gene are CRE, SP1 and AP‐1 (21). Galectin‐3 is a selective binding‐partner of activated K‐Ras (K‐Ras‐GTP) and seems to increase K‐Ras signaling to phosphatidylinositol‐3 kinase (PI3‐K) and to Raf‐1 as well as to a third, still unknown effector pathway that attenuates ERK activation 15, 16. It is also suggested that galectin‐3 can activate JNK pathways (46). Furthermore, galectin‐3 was found to be able to induce promoter activity of some genes like cyclin D1 14, 21, 26 in human breast epithelial cells independent of cell adhesion through multiple cis‐elements. Binding to the SP1 and CREB sites, galectin‐3 induction of the cyclin D1 promoter results from enhancement/stabilization of nuclear protein‐DNA complex formation at the CRE site of the cyclin D 1 promoter (35). Since CRE element is also present in TH gene promoter regions, it is possible for galactin‐3 to regulate TH gene promoter by trans‐activation. Our results also indicated the transcription factors CREB1a, 1b and 3 all were significantly upregulated by galectin‐3 transfection. CREB is considered as one of the major transcription factors responsible for TH gene expression. Thus galactin‐3 was able to enhance the expression of TH gene through CREB. In SHR PFC area, miRNA let‐7d targeted and downregulated galectin‐3 which eventually attenuated TH expression that might be involved in ADHD development.
In summary, abnormally lower level of galectin‐3 expression and elevated level of rno‐let‐7d expression were observed in PFC, STR and MB of SHR brain than those in WKY rat brain. There was a negative relationship between these two compounds, and rno‐let‐7d was found to interact with 3′UTR of galectin‐3 mRNA and downregulated galectin‐3 protein level. It was further observed that in both high and low levels, galectin‐3 was able to regulate CREB and affect TH gene expression. Our data demonstrate that a novel function of rno‐let‐7 in the regulation of galectin‐3 and the important roles in ADHD development.
ACKNOWLEDGMENTS
This work was supported by Wen Zhou Scientific and Technological Bureau (H2006023), the National Natural Science Foundation of China (30970641, 30570414), and Shanghai Leading Academic Discipline Project (B110).
REFERENCES
- 1. Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355. [DOI] [PubMed] [Google Scholar]
- 2. Aspide R, Fresiello A, De Filippis G, Gironi Carnevale UA, Sadile AG (2000) Non‐selective attention in a rat model of hyperactivity and attention deficit: subchronic methylphenydate and nitric oxide synthesis inhibitor treatment. Neurosci Biobehav Rev 24:59–71. [DOI] [PubMed] [Google Scholar]
- 3. Bellac CL, Coimbra RS, Simon F, Imboden H, Leib SL (2007) Gene and protein expression of galectin‐3 and galectin‐9 in experimental pneumococcal meningitis. Neurobiol Dis 28:175–183. [DOI] [PubMed] [Google Scholar]
- 4. Brown RT, Freeman WS, Perrin JM, Stein MT, Amler RW, Feldman HM et al (2001) Prevalence and assessment of attention‐deficit/hyperactivity disorder in primary care settings. Pediatrics 107:E43. [DOI] [PubMed] [Google Scholar]
- 5. Bushati N, Cohen SM (2008) MicroRNAs in neurodegeneration. Curr Opin Neurobiol 18:292–296. [DOI] [PubMed] [Google Scholar]
- 6. Bussing I, Slack FJ, Grosshans H (2008) let‐7 microRNAs in development, stem cells and cancer. Trends Mol Med 14:400–409. [DOI] [PubMed] [Google Scholar]
- 7. Casey BJ, Epstein JN, Buhle J, Liston C, Davidson MC, Tonev ST et al (2007) Frontostriatal connectivity and its role in cognitive control in parent‐child dyads with ADHD. Am J Psychiatry 164:1729–1736. [DOI] [PubMed] [Google Scholar]
- 8. Chandrasekar V, Dreyer JL (2009) microRNAs miR‐124, let‐7d and miR‐181a regulate cocaine‐induced plasticity. Mol Cell Neurosci 42:350–362. [DOI] [PubMed] [Google Scholar]
- 9. Chekulaeva M, Filipowicz W (2009) Mechanisms of miRNA‐mediated post‐transcriptional regulation in animal cells. Curr Opin Cell Biol 21:452–460. [DOI] [PubMed] [Google Scholar]
- 10. Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT et al (2005) Real‐time quantification of microRNAs by stem‐loop RT‐PCR. Nucleic Acids Res 33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Corbin R, Olsson‐Carter K, Slack F (2009) The role of microRNAs in synaptic development and function. BMB Rep 42:131–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. David G, Dennis LM (2005) Attention Deficit Hyperactivity Disorder: From Genes to Patients. Humana Press Inc: Totowa, NJ. [Google Scholar]
- 13. De Martino I, Visone R, Fedele M, Petrocca F, Palmieri D, Martinez Hoyos J et al (2009) Regulation of microRNA expression by HMGA1 proteins. Oncogene 28:1432–1442. [DOI] [PubMed] [Google Scholar]
- 14. DeCastro M, Nankova BB, Shah P, Patel P, Mally PV, Mishra R, La Gamma EF (2005) Short chain fatty acids regulate tyrosine hydroxylase gene expression through a cAMP‐dependent signaling pathway. Brain Res 142:28–38. [DOI] [PubMed] [Google Scholar]
- 15. Dumic J, Dabelic S, Flogel M (2006) Galectin‐3: an open‐ended story. Biochim Biophys Acta 1760:616–635. [DOI] [PubMed] [Google Scholar]
- 16. Elad‐Sfadia G, Haklai R, Balan E, Kloog Y (2004) Galectin‐3 augments K‐Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3‐kinase activity. J Biol Chem 279:34922–34930. [DOI] [PubMed] [Google Scholar]
- 17. Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, Sklar P (2005) Molecular genetics of attention‐deficit/hyperactivity disorder. Biol Psychiatry 57:1313–1323. [DOI] [PubMed] [Google Scholar]
- 18. Fiore R, Siegel G, Schratt G (2008) MicroRNA function in neuronal development, plasticity and disease. Biochim Biophys Acta 1779:471–478. [DOI] [PubMed] [Google Scholar]
- 19. Ghosh T, Soni K, Scaria V, Halimani M, Bhattacharjee C, Pillai B (2008) MicroRNA‐mediated up‐regulation of an alternatively polyadenylated variant of the mouse cytoplasmic {beta}‐actin gene. Nucleic Acids Res 36:6318–6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gustavsson P, Linsmeier CE, Leffler H, Kanje M (2007) Galectin‐3 inhibits Schwann cell proliferation in cultured sciatic nerve. Neuroreport 18:669–673. [DOI] [PubMed] [Google Scholar]
- 21. Hong SJ, Huh Y, Chae H, Hong S, Lardaro T, Kim KS (2006) GATA‐3 regulates the transcriptional activity of tyrosine hydroxylase by interacting with CREB. J Neurochem 98:773–781. [DOI] [PubMed] [Google Scholar]
- 22. Hu P, Shi B, Geng F, Zhang C, Wu W, Wu XZ (2008) E‐cadherin core fucosylation regulates nuclear beta‐catenin accumulation in lung cancer cells. Glycoconj J 25:843–850. [DOI] [PubMed] [Google Scholar]
- 23. Jadhav VM, Scaria V, Maiti S (2009) Antagomirzymes: oligonucleotide enzymes that specifically silence microRNA function. Angew Chem Int Ed 48:2557–2560. [DOI] [PubMed] [Google Scholar]
- 24. Kantak KM, Singh T, Kerstetter KA, Dembro KA, Mutebi MM, Harvey RC et al (2008) Advancing the spontaneous hypertensive rat model of attention deficit/hyperactivity disorder. Behav Neurosci 122:340–357. [DOI] [PubMed] [Google Scholar]
- 25. Kieling C, Goncalves RR, Tannock R, Castellanos FX (2008) Neurobiology of attention deficit hyperactivity disorder. Child Adolesc Psychiatr Clin N Am 17:285–307. [DOI] [PubMed] [Google Scholar]
- 26. Kim HS, Park JS, Hong SJ, Woo MS, Kim SY, Kim KS (2003) Regulation of the tyrosine hydroxylase gene promoter by histone deacetylase inhibitors. Biochem Biophys Res Commun 312:950–957. [DOI] [PubMed] [Google Scholar]
- 27. King JA, Barkley RA, Delville Y, Ferris CF (2000) Early androgen treatment decreases cognitive function and catecholamine innervation in an animal model of ADHD. Behav Brain Res 107:35–43. [DOI] [PubMed] [Google Scholar]
- 28. Kojima M, Suzuki T, Maekawa T, Ishii S, Sumi‐Ichinose C, Nomura T, Ichinose H (2008) Increased expression of tyrosine hydroxylase and anomalous neurites in catecholaminergic neurons of ATF‐2 null mice. J Neurosci Res 86:544–552. [DOI] [PubMed] [Google Scholar]
- 29. Kosik KS (2006) The neuronal microRNA system. Nat Rev 7:911–920. [DOI] [PubMed] [Google Scholar]
- 30. Krzeslak A, Lipinska A (2004) Galectin‐3 as a multifunctional protein. Cell Mol Biol Lett 9:305–328. [PubMed] [Google Scholar]
- 31. Leo D, Sorrentino E, Volpicelli F, Eyman M, Greco D, Viggiano D et al (2003) Altered midbrain dopaminergic neurotransmission during development in an animal model of ADHD. Neurosci Biobehav Rev 27:661–669. [DOI] [PubMed] [Google Scholar]
- 32. Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120:15–20. [DOI] [PubMed] [Google Scholar]
- 33. Lewis‐Tuffin LJ, Quinn PG, Chikaraishi DM (2004) Tyrosine hydroxylase transcription depends primarily on cAMP response element activity, regardless of the type of inducing stimulus. Mol Cell Neurosci 25:536–547. [DOI] [PubMed] [Google Scholar]
- 34. Li R, Kong Y, Ladisch S (1998) Nerve growth factor‐induced neurite formation in PC12 cells is independent of endogenous cellular gangliosides. Glycobiology 8:597–603. [DOI] [PubMed] [Google Scholar]
- 35. Lin HM, Pestell RG, Raz A, Kim HR (2002) Galectin‐3 enhances cyclin D(1) promoter activity through SP1 and a cAMP‐responsive element in human breast epithelial cells. Oncogene 21:8001–8010. [DOI] [PubMed] [Google Scholar]
- 36. Livingstone PD, Srinivasan J, Kew JN, Dawson LA, Gotti C, Moretti M et al (2009) alpha7 and non‐alpha7 nicotinic acetylcholine receptors modulate dopamine release in vitro and in vivo in the rat prefrontal cortex. Eur J Neurosci 29:539–550. [DOI] [PubMed] [Google Scholar]
- 37. Mendell JT (2009) Tumors line up for a letdown. Nat Genet 41:768–769. [DOI] [PubMed] [Google Scholar]
- 38. Navarro A, Marrades RM, Vinolas N, Quera A, Agusti C, Huerta A et al (2009) MicroRNAs expressed during lung cancer development are expressed in human pseudoglandular lung embryogenesis. Oncology 76:162–169. [DOI] [PubMed] [Google Scholar]
- 39. Pamplona FA, Pandolfo P, Savoldi R, Prediger RD, Takahashi RN (2009) Environmental enrichment improves cognitive deficits in Spontaneously Hypertensive Rats (SHR): relevance for Attention Deficit/Hyperactivity Disorder (ADHD). Prog Neuropsychopharmacol Biol Psychiatry 33:1153–1160. [DOI] [PubMed] [Google Scholar]
- 40. Paxinos G, Watson C (1997) The Rat Brain Sterotaxic Coordinates. New York Academic Press: New York. [Google Scholar]
- 41. Pesheva P, Kuklinski S, Schmitz B, Probstmeier R (1998) Galectin‐3 promotes neural cell adhesion and neurite growth. J Neurosci Res 54:639–654. [DOI] [PubMed] [Google Scholar]
- 42. Qiu J, Hong Q, Chen RH, Tong ML, Zhang M, Fei L et al (2010) Gene expression profiles in the prefrontal cortex of SHR rats by cDNA microarrays. Mol Biol Rep 37:1733–1740. [DOI] [PubMed] [Google Scholar]
- 43. Sagvolden T, Metzger MA, Schiorbeck HK, Rugland AL, Spinnangr I, Sagvolden G (1992) The spontaneously hypertensive rat (SHR) as an animal model of childhood hyperactivity (ADHD): changed reactivity to reinforcers and to psychomotor stimulants. Behav Neural Biol 58:103–112. [DOI] [PubMed] [Google Scholar]
- 44. Siuciak JA, Clark MS, Rind HB, Whittemore SR, Russo AF (1998) BDNF induction of tryptophan hydroxylase mRNA levels in the rat brain. J Neurosci Res 52:149–158. [DOI] [PubMed] [Google Scholar]
- 45. Swanson JM, Kinsbourne M, Nigg J, Lanphear B, Stefanatos GA, Volkow N et al (2007) Etiologic subtypes of attention‐deficit/hyperactivity disorder: brain imaging, molecular genetic and environmental factors and the dopamine hypothesis. Neuropsychol Rev 17:39–59. [DOI] [PubMed] [Google Scholar]
- 46. Takenaka Y, Fukumori T, Yoshii T, Oka N, Inohara H, Kim HR et al (2004) Nuclear export of phosphorylated galectin‐3 regulates its antiapoptotic activity in response to chemotherapeutic drugs. Mol Cell Biol 24:4395–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ueno KI, Togashi H, Mori K, Matsumoto M, Ohashi S, Hoshino A et al (2002) Behavioural and pharmacological relevance of stroke‐prone spontaneously hypertensive rats as an animal model of a developmental disorder. Behav Pharmacol 13:1–13. [DOI] [PubMed] [Google Scholar]
- 48. Vallone D, Picetti R, Borrelli E (2000) Structure and function of dopamine receptors. Neurosci Biobehav Rev 24:125–132. [DOI] [PubMed] [Google Scholar]
- 49. Wu XZ, Lu H, Zhou L, Huang Y, Chen H (1997) Changes of phosphatidylcholine‐specific phospholipase C in hepatocarcinogenesis and in the proliferation and differentiation of rat liver cancer cells. Cell Biol Int 21:375–381. [DOI] [PubMed] [Google Scholar]
- 50. Wu XZ, Shi PC, Hu P, Chen Y, Ding SS (2006) N‐all‐trans‐retinoyl‐L‐proline inhibits metastatic potential of hepatocellular carcinoma cells. Cell Biol Int 30:672–680. [DOI] [PubMed] [Google Scholar]
- 51. Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O et al (2007) Post‐transcriptional regulation of the let‐7 microRNA during neural cell specification. FASEB J 21:415–426. [DOI] [PubMed] [Google Scholar]
- 52. Yan YP, Lang BT, Vemuganti R, Dempsey RJ (2009) Galectin‐3 mediates post‐ischemic tissue remodeling. Brain Res 1288:116–124. [DOI] [PubMed] [Google Scholar]


