Abstract
In Ngsk prion protein (PrP)‐deficient mice (NP0/0), ectopic expression of PrP‐like protein Doppel (Dpl) in central neurons induces significant Purkinje cell (PC) death resulting in late‐onset ataxia. NP0/0 PC death is partly prevented by either knocking‐out the apoptotic factor BAX or overexpressing the anti‐apoptotic factor BCL‐2 suggesting that apoptosis is involved in Dpl‐induced death. In this study, Western blotting and immunohistofluorescence show that both before and during significant PC loss, the scrapie‐responsive gene 1 (Scrg1)—potentially associated with autophagy—and the autophagic markers LC3B and p62 increased in the NP0/0 PCs whereas RT‐PCR shows stable mRNA expression, suggesting that the degradation of autophagic products is impaired in NP0/0 PCs. At the ultrastructural level, autophagic‐like profiles accumulated in somatodendritic and axonal compartments of NP0/0, but not wild‐type PCs. The most robust autophagy was observed in NP0/0 PC axon compartments in the deep cerebellar nuclei suggesting that it is initiated in these axons. Our previous and present data indicate that Dpl triggers autophagy and apoptosis in NP0/0 PCs. As observed in amyloid neurodegenerative diseases, upregulation of autophagic markers as well as extensive accumulation of autophagosomes in NP0/0 PCs are likely to reflect a progressive dysfunction of autophagy that could trigger apoptotic cascades.
Keywords: autophagy, doppel, neuronal death, prion protein, Purkinje cell
INTRODUCTION
Doppel (Dpl) was the first identified homologue of the cellular prion protein (PrPc) that is implicated in the pathogenesis of transmissible spongiform encephalopathies (48). Large deletions in the PrPc gene Prnp, for example in the Ngsk Prnp 0/0 (NP0/0) mouse line, result in the ectopic expression of Dpl in brain neurons (51) that induces significant levels of cerebellar Purkinje cell (PC) death (53) as early as 6 months after birth (26). PrP‐deficient cells have been shown to undergo Dpl‐induced apoptosis in a dose‐dependent, cell‐autonomous manner 40, 51, 54. Expression of the N‐terminal truncated form of PrP (ΔPrP) in Prnp‐ablated mouse lines (21) has neurodegenerative effects in PCs, similar to those observed when Dpl is overexpressed 1, 64. Moreover, the neurotoxic effects of Dpl and ΔPrP are both antagonized by PrPc 1, 11, 21, 64, suggesting that Dpl and ΔPrP may cause cell death by similar mechanisms, perhaps by interfering with a cellular signalling pathway essential for cell survival and normally controlled by a full‐length PrPc 1, 56, 61. To date, only a few studies have investigated the mechanism by which Dpl kills neurons. Oxidative stress linked with glial activation may play a role in the death of neurons because NOS activity is induced by Dpl both in vitro and in vivo 2, 11, 62.
In a recent study that investigated the involvement of the mitochondrial proapoptotic factor BAX in the Dpl‐induced apoptosis of PCs, we have shown that deletion of Bax expression rescues many PCs from Dpl‐induced cell death in NP0/0:Bax −/− double knockout mutant mice (26). As PrPc has the ability to counteract Dpl neurotoxicity and has BCL‐2‐like properties that promote neuronal survival, we also examined the capacity of the antiapoptotic factor BCL‐2 to prevent Dpl neurotoxicity in vivo. Hu‐bcl‐2 overexpression in NP0/0‐Hu‐bcl‐2 double mutants was found to rescue PCs in similar proportions to that observed when Bax was deleted in NP0/0:Bax −/− double knockout mutants (27). The capacity of BCL‐2 to compensate PrPc deficiency and rescue PCs from Dpl‐induced death suggests that this BCL‐2‐like property of PrPc impairs Dpl‐like neurotoxic pathways in wild‐type (WT) neurons. These studies indicate that the proapoptotic factor BAX contributes to the neurotoxic mechanisms triggered by Dpl in the NP0/0 PCs and that the antiapoptotic factor BCL‐2 can counteract this neurotoxic mechanism. However, NP0/0 PC numbers are not fully restored to WT levels, suggesting that the ectopic expression of Dpl induces both BAX‐dependent and BAX‐independent cell death pathways.
A Dpl‐activated, BAX‐independent cell death mechanism may involve neuronal autophagy as NP0/0 PCs have been recently shown to express Scrg1, a novel protein having a potential link with autophagy (18). Indeed, neuronal Scrg1 mRNA and protein levels are increased in prion‐diseased brains 12, 13, 17. At the ultrastructural level, Scrg1 is associated with dictyosomes of the Golgi apparatus and autophagic vacuoles in degenerating neurons of scrapie‐infected Scrg1‐overexpressing transgenic and WT mice (18). Indeed, autophagy was first reported in experimentally induced scrapie a long time ago (6), and recently has been implicated in an alternative cell death program in prion diseases (35).
For this reason, we investigated the expression of Scrg1 and two characteristic markers of autophagy, LC3B and p62, 4, 5, 37, 63 prior to a significant PC loss in the 3‐ to 4‐month old NP0/0 cerebellum and when PC degeneration peaks in the 6‐ to 8‐month‐old NP0/0 cerebellum (26). We also looked for ultrastructural signs of autophagy in the dendrites, axons and somata of PCs in the cerebellar cortex and deep cerebellar nuclei (DCN) of NP0/0 mice.
MATERIALS AND METHODS
Animals and genotyping
As previously reported, NP 0/0 mice were generated by deleting the entire open reading frame (ORF) of the Prnp gene, located in exon 3, as well as 5′ and 3′ non‐coding flanking regions (52). The deleted sequences were replaced by a Neo cassette. The Prnp ORF was identified using the following primers: forward 5′CCGCTACCCTAACCAAGTGT3′ and reverse 5′CCTAGACCACGAGAATGCGA3′, both located within the Prnp ORF. The following primers were used to identify the NP0/0 mutants: forward 5′TGCCGCACTTCTTTGTGAAT3′ and reverse 5′CGGTGGATGTGGAATGTGT3′ (within the Neo domain).
For this study, founding mice (NP0/0 gift from S. Katamine) were first backcrossed with C57BL/6 mice for at least 10 generations. These mice were then intercrossed, and NP0/0 and WT pups were selected and bred at the animal facility of the Neurosciences IFR37 in Strasbourg, according to the National Institutes of Health (NIH) guidelines (NIH Publication 80–23, revised 1996) and the European Communities Council Directive of November 24, 1986 (86/609/EEC).
Western blot analysis of LC3B, p62 and Lamp1
Three‐ to four‐ and 6‐ to 8‐month‐old WT (n = 3/age) and NP0/0 (n = 3/age) littermates were anesthetized with a mixture of ketamine 5% and xylazine 2% (0.1 mL of the mix per 30 g by intraperitoneally) and then killed by decapitation. Cerebella were dissected and first homogenized in cold extraction TX‐DOC buffer [50 mM Tris‐HCl pH 7.4, 150 mM NaCl, 2 mM ethylenediaminetetraacetic acid (EDTA), 0.5% sodium deoxycholate, 0.5% Triton X‐100, 1/500 Sigma protease inhibitor cocktail). After centrifugation, the cleared extracts were taken up in Laemmli sample buffer [10 mM Tris pH 7.0, 1 mM EDTA, 3% sodium dodecyl sulfate (SDS), 10% glycerol, 20 mM dithiotreithol, 10% bromophenol blue] before heat denaturation. The remaining protein pellets were washed with phosphate‐buffered saline (PBS) before extraction with 2% SDS‐containing sample buffer (insoluble fraction). An equivalent of 100 µg of brain tissue per well was run on 4%–20% (LC3B and p62) and 4%–12% (p62 and Lamp1) Nupage gels (Invitrogen, Carlsbad, CA, USA). Proteins were transferred onto nitrocellulose membrane following the manufacturer's recommendations and routinely monitored with Ponceau S staining. Blots were pre‐incubated for 1 h in a blocking solution [5% milk powder (Sigma, St. Louis, MO, USA), 0.1% Tween‐20 in 0.1 M PBS pH 7.3]. Antibodies were diluted in blocking solution and blots were incubated overnight with anti‐actin mouse monoclonal antibody (1/10 000; Sigma‐Aldrich), anti‐Lamp1 rabbit polyclonal antibody (1/1000; Abcam Ltd., Cambridge, UK), anti‐LC3B mouse monoclonal antibody (1/200; Nanotools, Munich, Germany) and anti‐p62 mouse monoclonal antibody (1/1000; BD Transduction Labs, San Jose, CA, USA). The anti‐LC3B antibody was directed against the N‐terminal end of the LC3B molecule and reacts with both LC3B‐I and LC3B‐II proteins. The anti‐p62 antibody was directed against amino acids 257–437 sequence of the human p62 molecule. The anti‐Lamp1 antibodies were directed against a synthetic peptide within residues 350 to the C‐terminus of human Lamp1 (100% identity with mouse Lamp1). Immunoreactivity was revealed using the SuperSignal West Dura Extended Duration Substrate reagent kit (Pierce, Rockford, IL, USA) and images were obtained with a Chemi‐Smart 5000 camera using the Chemi‐capt software (Vilber‐Lourmat, Cedex 1, France). Quantification of protein bands was done using the Bio1D software (Vilber‐Lourmat). Values have been corrected for variation in actin values and are expressed as a percentage of the values obtained for control animals.
RT‐PCR analysis of LC3B and p62
Total RNA was extracted from isolated cerebellum from 3‐ to 6‐month‐old control WT mice and age‐matched NP0/0 mice with the GenElute Kit (Sigma‐Aldrich), and mRNA was transcribed into cDNA using oligo(dT) and Superscript RNase H‐ Moloney murine leukemia virus reverse transcriptase (Invitrogen). PCR amplification was performed using specific primers for p62: forward—5′GATGTGGAACATGGAGGGAAGAG3′ and reverse—5′AGTCATCGTCTCCTCCTGAGCA3′; PCR product 246 bp (GI: 118130186), for LC3: forward—5′ATGCCGTCCGAGAAGACCTTC3′ and reverse—5′TTACACAGCCATTGCTGTC3′; PCR product 377 bp (GI: 141601716) and for actin: forward—5′GTGGGCCGCTCTAGGCACCAA3′ and reverse—5′CTCTTTGATGTCACGCACGATTTC3′; PCR product 540 bp (GI: 141601716). PCR cycles consisting of 94°C for 45 s, 55°C, (except for actin 60°C) for 30 s and 70°C for 30 s were varied between 18 and 27 cycles for β‐actin and 21 and 33 cycles for p62 and LC3. Aliquots of PCR products were then migrated on 2% agarose gels and stained with ethidium bromide. The relative expression of the autophagic markers produced in the exponential phase of the PCR amplifications was calculated by a semi‐quantitative analysis using the Image J program (National Center for Biotechnology Information, Bethesda, MD, USA). Similar measurements on the endogenous reference gene β‐actin served as a control for the quantity of RNA input.
Immunohistofluorescence
WT (n = 2) and NP0/0 (n = 3) mice aged 6–8 months were anesthetized as described earlier and transcardiacally perfused with 4% paraformaldehyde (PAF) in 0.1 M phosphate buffer (PB), pH 7.3. The cerebellum was dissected and immersed in the same fixative at 4°C. After immersion in isobutanol for 4 days, the cerebella were embedded in paraffin and sagittal sections (10‐µm thick) were cut with a microtome (Leica, Heerbrugg, St. Gallen, Switzerland). The sections were de‐paraffinized in toluene and rehydrated in ethanol. After rinsing in PBS, the sections were pre‐incubated for 45 minutes in a blocking solution [3% normal goat serum (NGS) in 0.5% Triton X‐100 PBS (PBST)]. The sections were incubated overnight at 4°C in PBST containing 0.3% NGS and mouse monoclonal antibodies against LC3B (Nanotools) diluted 1/10 and rabbit polyclonal antibodies against 28 kDa calcium‐binding protein (CaBP; gift from Dr Thomasset) diluted 1/1000, or rabbit polyclonal antibodies against Lamp1 (Abcam) diluted 1/100 for double immunohistofluorescence, or with mouse monoclonal antibodies against p62 (BD Transduction Labs) diluted 1/100. The sections were then rinsed in PBS and incubated with Alexa 488‐coupled goat anti‐mouse and Alexa 546‐coupled goat anti‐rabbit immunoglobulins (Molecular Probes, Eugene, OR, USA) diluted 1/1000 in PBS containing 0.3% NGS. After rinsing in PBS, the sections were mounted in Mowiol and examined by epifluorescence (Axioskop‐II, Zeiss, Jena, Germany).
Scrg1 immunocytochemistry
Tissue preparation and sampling
WT (8 months, n = 2) and NP0/0 (3, 6, 8 months, n = 2/age) mice were anesthetized as described earlier and transcardiacally perfused with PAF 1% in PB. The cerebellum was dissected and immersed in the same fixative at 4°C for 4 h.
Immunohistofluorescence
The cerebellum was cryoprotected in sucrose 0.44 M in PB overnight at 4°C before freezing in liquid nitrogen. Sagittal sections (10‐µm thick) were cut with a cryostat (Leica), rinsed in PBS and pre‐incubated for 45 minutes in a blocking solution (3% NGS in PBST). After overnight incubation at 4°C in PBST containing 0.3% NGS and rabbit polyclonal antibody against Scrg1 diluted 1/60 (gift from M. Dron) (28), the sections were rinsed in PBS and incubated with goat anti‐rabbit immunoglobulins coupled to Alexa 488 (Molecular Probes) diluted 1/500 in PBS containing 0.3% NGS. The sections were then rinsed in PBS and processed for fluorescence microscopy as described earlier.
Control sections processed either without the anti‐LC3 and anti‐Scrg‐1 primary antibodies or the fluorescent secondary antibodies were devoid of any specific fluorescent signal.
Quantitative analysis of Scrg1‐immunofluorescent PCs in the NP0/0 cerebellum
Scrg1 immunofluorescent PCs were counted in seven sagittal sections separated from each other by 400 µm (total sampling distance = 2470 µm) in the cerebellar vermis of the 8‐month‐old WT (n = 2) and NP0/0 mice (n = 3). Data are given as mean ± standard deviation.
Pre‐embedding immunogold for electron microscopy
Transversal vibratome sections (60‐µm thick) of 1% PAF‐fixed cerebellum were cut and processed, as previously described (16). Briefly, sections were pre‐incubated in a blocking solution [0.5% bovine serum albumin (BSA) 0.1% cold water, fish skin gelatine and 0.5% NGS in PB) before being rinsed in PBS containing 0.15% acetylated BSA (BSAc; Aurion, Wageningen, the Netherlands) and incubated overnight at 4°C with anti‐Scrg1 antibodies (1/50 in PBS‐BSAc). After washing in PBS‐BSAc, the sections were incubated in Ultra small nanogold F(ab′) fragments of goat anti‐rabbit immunoglobulin G (IgG) (H and L chains; Aurion) diluted 1/100 in PBS‐BSAc. After several rinses in PBS‐BSAc and in PB, sections were postfixed in glutaraldehyde 2% in PB before washing in PB and distilled water. Gold particles were then silver enhanced using the R‐Gent SE‐EM kit (Aurion) before being washed in distilled water and PB. Finally, the sections were postfixed in 0.5% OsO4 in PB before classical processing for Araldite embedding (Sigma, St. Louis, MO, USA) and ultramicrotomy. The ultrathin sections were counterstained with uranyl acetate and observed with a Hitachi 7500 transmission electron microscope (Hitachi High Technologies Corporation, Tokyo, Japan) equipped with an AMT Hamamatsu digital camera (Hamamatsu Photonics, Hamamatsu City, Japan). In control sections processed without anti‐Scrg‐1 primary antibodies or gold‐labeled secondary antibodies, no gold particles were observed.
Post‐embedding immunogold for detection of gamma‐aminobutyric acid (GABA)
DCN were sampled from transverse cerebellar vibratome sections of 10‐month‐old NP0/0 mice (n = 2) perfused with a mixture of 2% PAF and 2% glutaraldehyde in 0.1 M PB. After embedding in Araldite, ultrathin sections were submitted to a standard post‐embedding immunogold protocol for the detection of GABA (15). The sections were immersed in 1% sodium borohydride, and after rinsing in distilled water and Tris‐buffered saline (TBS) they were incubated with 10% NGS in TBS and then with the GABA‐specific primary antiserum (0.5 mg/mL; overnight at 4°C) in TBS containing 1% NGS. The sections were rinsed in TBS and incubated in goat anti‐rabbit IgG coupled to 15 nm gold particles (Aurion), diluted 1/200 in TBS containing 1% NGS. After rinsing in TBS and distilled water, sections were counterstained with 1% uranyl acetate. Substitution of the primary antiserum with pre‐immune serum or TBS abolished labeling, as did omission of the gold‐conjugated secondary antibodies. Axonal varicosities establishing asymmetric synapses were always devoid of labeling. Non‐specific or background labeling was very low in these preparations.
Semi‐quantitative ultrastructural analysis of PC autophagy
Tissue preparation and sampling
Cerebellar cortex and DCN were sampled from transverse vibratome sections of 3 and 6 month old NP0/0 and WT mice (n = 2/genotype and age) perfused with a mixture of 2% PAF and 2% glutaraldehyde in 0.1 M PB. Regions of the cerebellar cortex were randomly sampled in the cerebellar lobules either in the vermis or in the right or left hemispheres in one section of the anterior cerebellum (1 block) and in one section of the posterior cerebellum (1 block). Regions in the fastigial, interposed and dentate nuclei was randomly sampled in sections from the anterior, median and posterior levels (1 block/level) of each right or left nucleus. After embedding in Araldite, ultrathin sections were obtained from each block. In the cerebellar cortex, 100 PCs in the anterior cerebellum and 100 PCs in the posterior cerebellum were examined. In the DCN, 7000 µm2 were examined in the synaptic neuropile between the deep cerebellar neurons and an equivalent area in the synaptic neuropile close to the DCN. This was done at the anterior, median and posterior levels of the three DCN. The deep cerebellar neurons were identified by their large size (mean maximum diameter: 23.07 ± 0.36 µm) and by absence of GABA immunogold labeling. Autophagy was considered to be activated in the neuronal profiles which displayed recognizable structural features of the macroautophagic process. These included isolation membranes or phagophores surrounding neuroplasm with or without organelles and autophagic vacuoles such as complete double‐membraned autophagosomes and autolysosomes or autophagolysosomes (47).
Semi quantitative analysis of autophagic profiles in the NP0/0 cerebella
In the cerebellar cortex, activation of autophagy was estimated by the number of PC soma exhibiting abnormal numbers of autophagic features (at least three autophagic vacuoles per soma) as a percentage of the total number of PCs examined in each animal. In the DCN, activation of autophagy was estimated by the number of PC presynaptic and preterminal axonal profiles exhibiting abnormal numbers of autophagic features (at least two autophagic vacuoles per profile) as a percentage of the total number of PC axonal profiles examined in each nucleus.
RESULTS
Scrg1, LC3B, p62 and Lamp1 are upregulated in NP0/0 PCs
As expression of Scrg1 has been previously reported to be a potential marker of autophagy and to increase in central neurons of scrapie‐infected mice and in PCs of 12‐month‐old NP0/0 mice (18), we first investigated the immunohistochemical distribution of Scrg1 in the cerebellar cortex of young, 6‐ to 8‐month‐old NP0/0 mice. In these mutants, intense Scrg1 immunofluorescence accumulated in large dots in the apical pole of the soma of a few PCs, reminiscent of Golgi apparatus staining (Figure 1A). No or very weak labeling was seen in the PC dendrites. Scrg1 labeling was observed in less than 10 PCs per sagittal cerebellar section and was absent in PCs from WT age‐matched littermates (Figure 1B and C).
Figure 1.

Immunohistofluorescence of Scrg1 in the 8 month‐old cerebellar cortex of NP0/0 and wild‐type mice. A few PCs (arrowheads) display Scrg1 fluorescence in the cerebellar cortex of a NP0/0 mouse (A). PCs remain devoid of Scrg1 fluorescence in the cerebellar cortex of the wild‐type mouse (B). A and B, bar = 50 μm. Quantitative analysis of Scrg1‐immunofluorescent PCs. The number of Scrg1‐immunoreactive PCs per 7 sagittal cerebellar sections sampled in each of 3 wild‐type (WT) and 3 NP0/0 mice is plotted on a y axis (C). ML, molecular layer; IGL, internal granular layer.
To further substantiate the occurrence of autophagy in the NP0/0 PCs, the cerebellar expression of LC3B, a newly discovered autophagy‐related variant of LC3, the classical marker of autophagy (63), and in parallel, the expression of p62—a functional autophagic ligand of LC3 5, 45—were analyzed (Figure 2). First, RT‐PCR analysis of mRNA levels did not reveal any differences in the expression of either LC3B or p62 in the cerebellum of 3‐ and 6‐month‐old NP0/0 mice compared with WT mice (Figure 2A). Next, changes in the LC3 and p62 protein levels were analyzed by Western blot. The results indicated an increase in the levels of both the autophagy‐specific LC3B‐II fragment (4.6‐fold) and its ligand p62 (threefold) in the Triton X‐soluble fraction (Figure 2B) but not in the insoluble fraction (data not shown) of the cerebellar extracts from the 3‐ to 4‐ and 6‐ to 8‐month‐old NP0/0 mice compared with the age‐matched WT littermates. This increased expression of LC3B‐II in the cerebellar cortex of the NP0/0 mutants was confirmed in histological sections (3, 4). LC3B (Figure 3D–I) and p62 (Figure 4B) staining was observed in the soma of NP0/0 PCs, but was barely detected in WT PCs (3, 4). In addition, CaBP‐positive axon terminals of PCs displayed LC3B (Figure 3M–O) and p62 (Figure 4D) immunohistofluorescence in the DCN of the NP0/0 mice whereas LC3B (Figure 3J–L) and p62 (Figure 4C) could not be immunodetected in WT DCN. In agreement, the level of expression of the lysosomal marker Lamp1 was increased (4.1‐fold) in the cerebellar extract of all NP0/0 mice (Figure 2B), implicating an upregulation of lysosome recruitment. This was confirmed using Lamp1 and LC3B double immunohistofluorescence that showed colocalization of LC3B‐ and Lamp1‐positive granules in the NP0/0 PCs somata in the the cerebellar cortex (Figure 5C) and in the NP0/0 PC axonal profiles in the DCN (Figure 5L).
Figure 2.
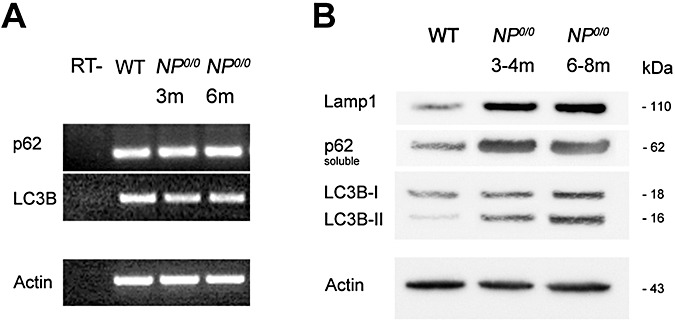
RT‐PCR analysis of mRNA expression of p62 and LC3 in the NP0/0 cerebellum. mRNA levels of LC3B and p62 in both 3 month‐old (lane 3) and 6 month‐old mice (lane 4) are equivalent to the wild‐type (WT) (lane 2). The expression of b‐actin was analyzed as an internal control. Negative control samples which were not reverse transcribed (RT‐) are shown (lane 1) (A). Western blot analysis of the LC3B, p62 and Lamp1 proteins in the NP0/0 cerebellum. The 16‐kDa LC3B‐II protein, the 62 kDa p62 protein and the 110 kDa Lamp1 protein are increased in both 3–4 month‐old (lane 2) and 6–8 month‐old (lane 3) extracts of cerebellum from NP0/0 mice compared with adult wild‐type (lane 1) (B). As a control for equivalent protein loading, 42‐kDa actin was revealed.
Figure 3.
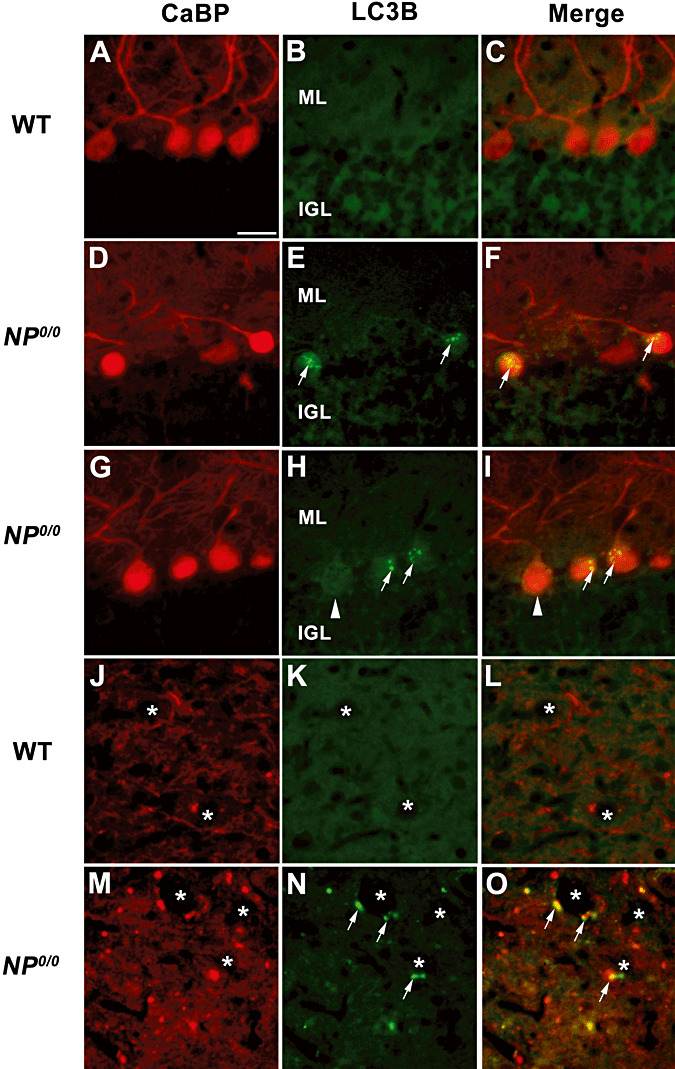
Immunohistofluorescence for LC3B and calcium binding protein (CaBP) in the cerebellum of the 6–8 month‐old NP0/0 and wild‐type (WT) mice. In the cerebellar cortex, double immunohistofluorescence for CaBP (A, D, G, J, M, red) and LC3B (B, E, H, K, N, green) shows that the NP0/0 PC somata contain a punctuate LC3B staining (arrows in E, F, H, I). Some NP0/0 PCs do not display LC3B fluorescence (arrowheads) like wild‐type PCs (B, C). In a deep cerebellar nucleus, some NP0/0 PC axon terminals close to deep cerebellar neurons (asterisks) display red CaBP (M, O) and green LC3B (N, O) immunohistofluorescence. Some other CaBP‐positive PC terminals do not display LC3B labeling in the merged image (O). All CaBP‐labelled PC axon terminals (J, L) are LC3B‐negative (K, L) in the wild‐type. Bar = 20 μm in A to O. ML, molecular layer; IGL, internal granular layer.
Figure 4.
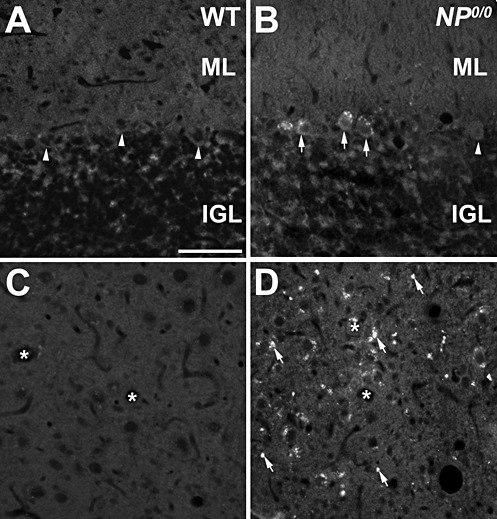
Immunohistofluorescence for p62 in the cerebellum of 6–8 month‐old NP0/0 and wild‐type (WT) mice. A, B. In the cerebellar cortex, p62 appears as cytoplasmic dots in the soma of NP0/0 PCs (arrows) displays. Some NP0/0 PCs as well as wild‐type PCs (A) do not display p62 fluorescence (arrowheads). C, D. P62 immunofluorescence of presumptive PC axon terminals is observed in the deep cerebellar nucleus of NP0/0 mouse (arrows, D) but not wild‐type mouse (C). Asterisks indicate deep cerebellar neurons soma. Bar = 20 μm in A to D. ML, molecular layer; IGL, internal granular layer
Figure 5.
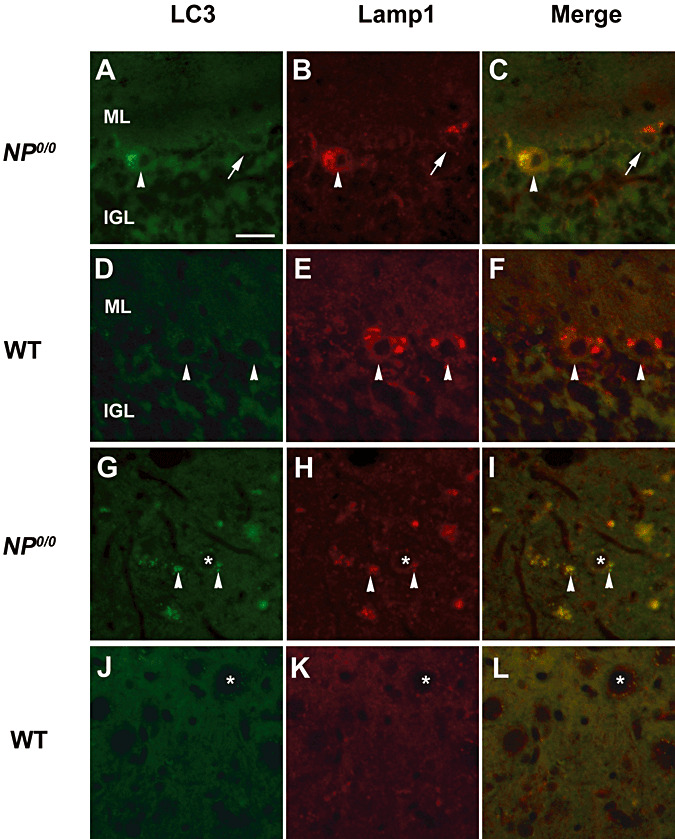
Immunohistofluorescence for Lamp1 and LC3B in the cerebellum of 6 month‐old NP0/0 and wild‐type (WT) mice. Double immunohistofluorescence for LC3B (A, D, G, J, green) and Lamp1 (B, E, H, K, green) shows colocalization of Lamp1 and LC3B staining in a NP0/0 PC soma (arrowhead in C). Some NP0/0 PCs do not display LC3B fluorescence (arrows in A, C) like wild‐type PCs (D, F). In a deep cerebellar nucleus, some NP0/0 PC axon terminals close to deep cerebellar neurons (asterisks) display red Lamp1 (arrowheads in H, I) and green LC3B (arrowheads in G, I) immunohistofluorescence. All PC axon terminals in wild‐type mice are LC3B‐ (J, L) and Lamp1‐negative (K, L). Bar = 20 μm in A to L. ML, molecular layer; IGL, internal granular layer.
Scrg1 immunogold labels autophagic NP0/0 PC somata but not axon terminals
Ultrastructural immunogold labeling of PCs from the 6‐ to 8‐month old NP0/ 0 mice revealed the association of Scrg1 with vesicles and saccules of dictyosomes in the somatic Golgi apparatus. The labeled dictyosomes sometimes had a normal ultrastructural appearance (Figure 6A), but in most cases, double‐membrane saccules and intertwined membrane shapes were evident (Figure 6B–F). Unlabeled autophagic vacuoles, membrane whorls and concentric arrays of membranes (phagophores) also seemed to be derived from other organelles, such as multivesicular bodies, ergastoplasmic saccules (Figure 7G) and mitochondria (Figure 6H). Profiles of neurodegeneration were rarely observed in the molecular layer, although autophagosomes and autophagolysosomes were found in some PC dendrites (Figure 6I). At the nuclear level, no ultrastructural features of apoptotic cell death, such as chromatin condensation and nuclear fragmentation, were observed. Presumptive PC myelinated axons exhibiting GABA immunogold labeling and dystrophic shapes contained phagophores, autophagosomes and autophagolysosomes in the deep molecular layer (Figure 6J), as well as in the granule cell layer and the cerebellar white matter (Figure 7C–E). None of these dendritic and axonal autophagic profiles displayed Scrg1 immunogold. Although most PCs containing autophagic profiles did not display any other ultrastructural signs of neurodegeneration, some soma clearly displayed features of advanced neurodegeneration, including shrunken soma and a condensed neuroplasm filled with unlabeled phagophores, autophagosomes and autophagolysosomes (Figure 7A–B).
Figure 6.
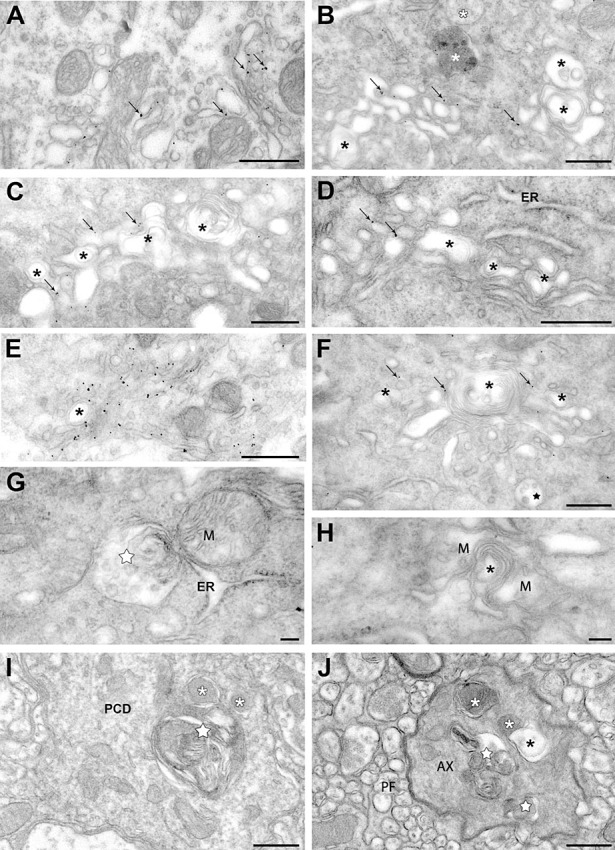
Autophagy in the somato‐dendritic and axonal compartments of Scrg1‐immunogold labelled Purkinje cells of the NP0/0 mouse. A–H. Purkinje cell soma. Immunogold particles (arrows) label saccules and vesicles of an apparently intact dictyosome of the Golgi apparatus (A). B–F. Golgi apparatus dictyosomes with Scrg1‐immunogold labelling (arrows) and autophagic‐like double membrane vacuoles and membrane whorls (asterisks). White asterisks in B, lysosomes. ER in D, rough endoplasmic reticulum. Star in F, multivesicular body. A multivesicular body (white star) is engaged in autophagic‐like membrane wrapping with a rough ER saccule (G). Mitochondria (M) around autophagic‐like membrane whorls (asterisk) (H). None of the organelles in G and H are labelled with Scrg1 immunogold. I, J. Molecular layer. Fusion of an autophagosome (white star) with lysosomes (white asterisks) in a Scrg1‐negative Purkinje cell primary dendrite (PCD) (I). Autophagosomes (white stars) and autophagolysosomes (white asterisks) and in a Scrg1‐negative myelinated presumptive Purkinje cell recurrent axon collateral (AX) (J). Note the fusion of a lysosome with a double membrane autophagosome (black asterisk). PF, parallel fibres. Bars: 500 nm in A–F, I, J; 100 nm in G, H.
Figure 7.
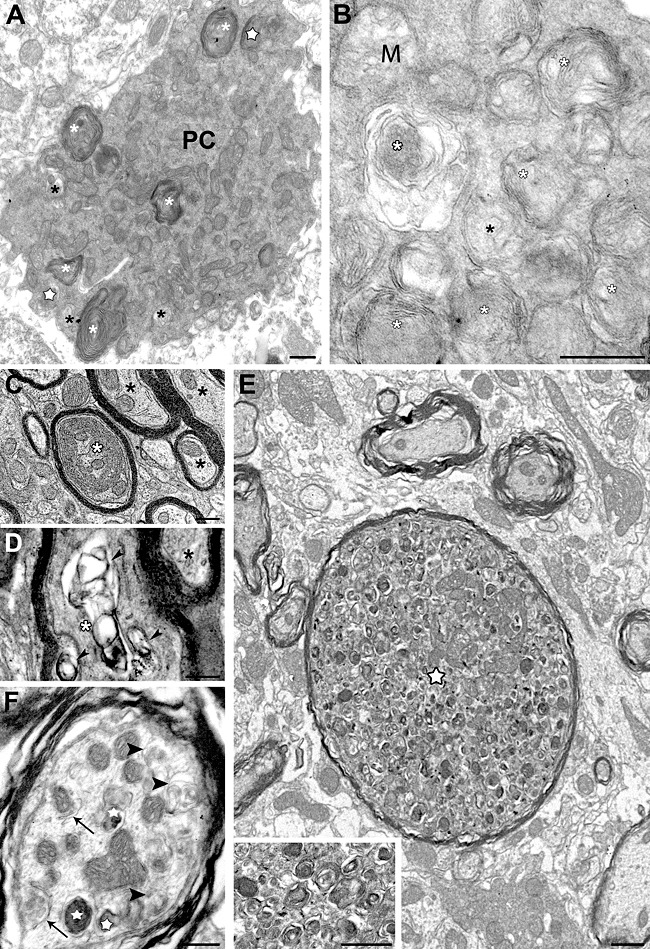
Autophagy in the soma and axons of NP0/0 Purkinje cells. Immunogold electron microscopy did not reveal Scrg1 in any of the areas shown. A degenerating autophagic Purkinje cell (PC) filled with autophagosomes, autophagolysosomes (white asterisks) and lysosomes (white stars) and has a shrunken shape (A). High magnification of neuroplasmic content of an autophagic Purkinje cell soma (B). M, mitochondria; white asterisks, autophagic vacuoles; asterisk, phagophore. C–D. Degenerating myelinated Purkinje cell axons in the white mater surrounding the fastigial deep cerebellar nucleus. A phagophore made of concentric wrapping of membrane around a core of axoplasm (white asterisk) fills a myelinated axon profile (C). Collapsed autophagic‐like vacuoles (arrowheads) in a myelinated axon (asterisk) (D). Black asterisks show intact myelinated axons in C and D. A dystrophic axon (star) with features of acute autophagy: thinned myelin sheath, swollen shape and numerous autophagic vacuoles at different stages of maturation and electron‐dense lysosomes (see higher magnification in inset). Compare with surrounding intact myelinated axons (E). Numerous reticular phagophores (white arrowhead) encircling axoplasmic material, autophagosomes (black arrowheads) and autophagolysosomes (white stars) reflect advanced stages of axonal autophagy (F). Bars: 500 nm in A–D, inset E and F; 2 μm in E.
In all of the DCN, PC axon terminals make inhibitory synapses on the dendrites and soma of the deep cerebellar neurons. Many of these PC presynaptic varicosities displayed GABA immunogold labeling (Figure 8C) and contained autophagic vacuoles at all ages studied (3–8 months, Figure 8A–E), but other terminals displayed normal ultrastructural characteristics (Figure 8D and F), similar to those of PC terminals in the DCN of WT animals (Figure 8G). Scrg1 immunogold did not label autophagic vacuoles in NP0/0 PC axon terminals.
Figure 8.
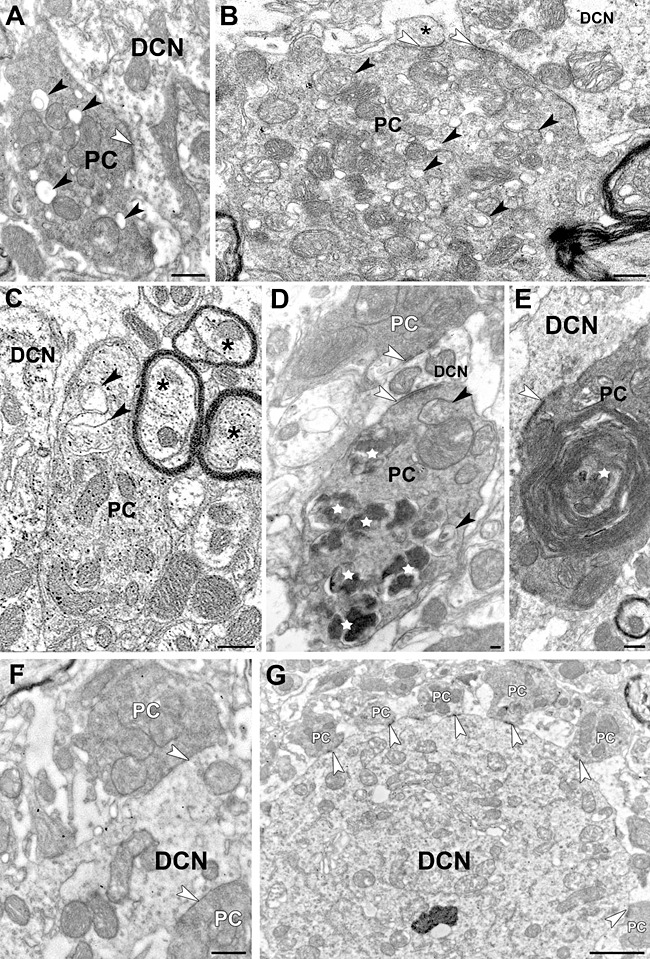
Purkinje cell axon terminals in the fastigial deep cerebellar nucleus of NP0/0 (A–F) and wild‐type (G) mice. A–F. NP0/0 mouse. Immunogold electron microscopy does not reveal Scrg1 in any of these areas. A–B. Early stages of autophagy in presynaptic PC terminals making symmetric axo‐somatic (A) and axo‐dendritic (B) synapses (white arrowheads) on fastigial neurons (DCN). Black arrowheads point to autophagic double‐membrane and small vacuoles within the PC terminals. Asterisk indicates a postsynaptic fastigial dendritic spine. Immunogold labelling for GABA. In a GABA‐immunolabelled preterminal PC axon terminal close to a DCN, cytoplasmic material is isolated by an endoplasmic saccule (arrowhead) in an autophagic‐like manner featuring an early stage of autophagy. See the surrounding double membrane autophagic‐like vacuole (arrowhead). Asterisks show intact myelinated axons (C). An autophagic PC axon terminal containing autophagosomes (arrowheads) and many autophagolysosomes (white stars). This PC terminal makes an symmetric synapse (white arrowhead) on a postsynaptic fastigial dendrite (DCN) similar to that made by an intact PC terminal (white PC) on the opposite side of the dendrite (D). Huge autophagosome featured by multiple membrane wrapping around cytoplasmic material (white star) in a PC terminal making a symmetric synapse (white arrowhead) on a fastigial dendrite (DCN) (E). Two intact presynaptic PC axon terminals make symmetric synapses (white arrowheads) on a fastigial dendrite (DCN) (F). Six intact presynaptic PC axon terminals make symmetric synapses (white arrowheads) on a large fastigial dendrite (DCN) (G). Bars: 500 nm.
Presynaptic autophagy in NP0/0 PCs
Semi‐quantitative estimation of the percentage of autophagic PCs somata and axon terminals clearly indicated that autophagy was more prevalent in axons and terminal varicosities (14%, Figure 9A) than in soma and dendrites (2%, Figure 9A) of the NP0/0 PCs at both ages studied (3 and 6 months). No autophagic PC soma and axons were observed in the WT cerebella. These results indicate that autophagy is already activated in 3‐month‐old NP0/0 PCs prior to significant PC loss (Figure 9B) (26). Furthermore, the semi‐quantitative data suggest that autophagy is largely restricted to the axonal compartment of the 3‐ to 6‐month‐old NP0/0 PCs, whereas the somatodendritic compartment displays a quasinormal ultrastructure with only discrete alterations including Golgi‐derived autophagic‐like shapes.
Figure 9.
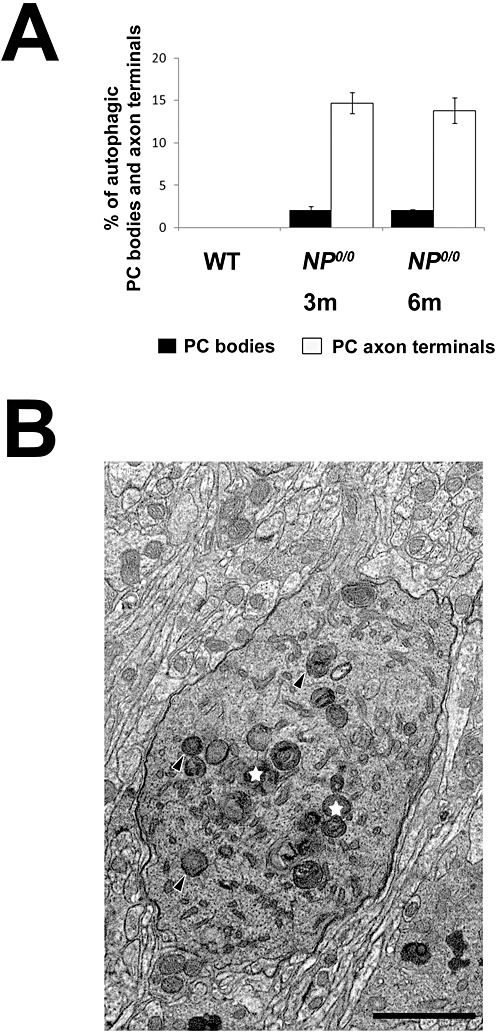
A Semi‐quantitative analysis of autophagic PC somata and axon terminals in the cerebellar cortex and deep cerebellar nuclei of the 3 and 6 month‐old wild‐type (WT) and NP0/0 mice. The percentage of autophagic somata (black) and axon terminals (white) in the wild‐type and NP0/0 mice is plotted on y axis (A). Autophagosomes (black arrowheads) and autophagolysosomes (white stars) in the somatic neuroplasm of a PC in the cerebellar cortex of a 3 month‐old mouse (B). Bar = 2 μm.
DISCUSSION
The mechanisms by which the prion protein‐like Dpl provokes premature PC death in the PrP‐deficient NP0/0 mice are still poorly understood. Neither Prnd knockout mice nor Prnp and Prnd double knockout mice exhibit overt neurological phenotypes indicating that PCs survive in these mutants and that Dpl is responsible for PC death in NP0/0 mice 23, 44. Here, we investigated whether neuronal autophagy is activated during Dpl‐induced PC death in NP0/0 mice.
Using immunochemistry at the biochemical, histological and ultrastructural levels to evaluate this possibility, we demonstrate that NP0/0 PCs undergo autophagy as indicated by the upregulation of Scrg1, a presumed autophagy‐associated protein, and two mediators of autophagic degradation LC3B‐II and p62, as well as the presence of ultrastructural autophagic profiles in all PC compartments.
Upregulation of Scrg1 and p62/LC3 reflects different phases of autophagy in degenerating NP0/0 PCs
The microtubule‐associated light chain 3 (LC3) is an essential component of autophagy (29) that acts as an adaptor protein between microtubules and autophagosomes through its N‐terminal domain (33). Three isoforms LC3, LC3A and LC3B, have been shown to be associated with autophagic membranes (63) and thus can be used as autophagosomal markers. The punctuate LC3B immunoreactivity observed in the soma of NP0/0—but not WT PCs—is likely to indicate the formation of autophagic organelles resulting from the activation of the autophagic machinery in the mutant PCs. In agreement with immunohistochemical data, the Western blot analysis clearly showed that the autophagic membrane‐bound marker LC3B‐II was increased in the NP0/0 cerebellum. Furthermore, the increased expression of p62 whose interaction with LC3 is instrumental in mediating autophagic degradation 5, 45 confirms that autophagy is abnormally activated in the NP0/0 cerebellum (see further discussion). The very early increase of these two markers at 3–4 months when PC loss is not yet significant (26) suggests that autophagy precedes apoptosis. In agreement, PCs with clear ultrastructural signs of advanced autophagy can be found in the cerebellar cortex of the 3‐month‐old NP0/0 mice. In the NP0/ 0 cerebellum, the increases of p62 and LC3B‐II protein levels observed with age are likely to reflect both an increased number of autophagic PCs as indicated by increased PC loss at this age (26), and a decreased cerebellar mass as previously shown by the reduced size of the NP0/0 cerebellum (26). In many NP0/0 PCs, this increase may correspond to the enhanced cleavage of LC3B‐I to form LC3B‐II. This would take place only in those PCs that have entered a stage of advanced neurodegeneration as indicated at the ultrastructural level by robust formation and accumulation of autophagic structures in a fraction of PC population at this time. However, LC3 has been shown to be incorporated into intracellular inclusion bodies induced in a number of pathological conditions, indicating that LC3‐labeled dots do not necessarily represent autophagic structures (34). In the NP0/0 PCs, this possibility is unlikely because inclusion bodies were not particularly abundant at the ultrastructural level in these neurons and the quantity of p62 protein in the soluble NP0/0 cerebellar fraction increases in parallel. The high levels of p62 and LC3B‐II in NP0/0 cerebellum may not only reflect an augmentation in the density of autophagic PCs, but may also be because of an impairment in the autophagic flux (10) (see following discussion). In agreement with this latter possibility, the absence of any detectable changes in the mRNA levels of LC3B and p62 in the NP0/0 cerebellum compared with WT cerebellum strongly argues for a deficient autophagic degradation.
Scrg1, a gene primarily expressed in brain neurons, has been recently shown to be upregulated in several conditions of brain injury, following prion infection of human and animal brains, as well as in the cerebellum of the NP0/0 mutant mouse (18). In scrapie‐infected central neurons of both WT and Scrg1‐overexpressing transgenic mice, Scrg1 was associated with dictyosomes of the Golgi apparatus and autophagic vacuoles 12, 14 that are hallmarks of prion‐induced degeneration 6, 28, 36. In the cerebellum of the 8‐ to 12‐month‐old NP0/0 mice (18), upregulation of Scrg1 protein was immunodetected in fewer PCs than p62/LC3B‐II, but not in any PCs of the WT cerebellum. Scrg1‐labeling and p62/LC3B‐II production was detected only in a few NP0/0 PCs at any given time, indicating that the proteins may be involved in restricted phases of Dpl‐induced stress. These two events may represent distinct steps in the neurodegenerative process because the ultrastructural data indicate that immunogold labeling for Scrg1 was mostly associated with Golgi complexes in NP0/0 PC somata that display aberrant structures suggesting inaugural signs of autophagy. Interestingly, the Golgi apparatus of both NP0/0 and prion‐infected PCs displayed similar ultrastructural alterations suggesting that Scrg1 labels Golgi apparatus of PCs that are likely to later undergo autophagy. In NP0/0 PCs, upregulation of Scrg1 at the dictyosomes may be an early sign of Golgian autophagy that precedes p62/LC3B‐II production as very little, if any, Scrg1 labeling of phagophores 25, 37, 41, autophagosomes and autophagolysosomes was observed in these neurons. Nevertheless, the chronology of the two events could not be more accurately determined because simultaneous immunohistochemical staining of Scrg1 and LC3B markers is not possible because of the incompatibility of the fixation protocols required to reveal Scrg1 (light 1% PAF tissue fixation) and LC3B (strong 4% PAF tissue fixation).
The specific Golgian localization confirms the spatial restriction of Scrg1 in the degenerative process of NP0/0 PCs and is distinct from the more generalized association of LC3 with autophagosomal membranes (29). Thus, upregulation of Scrg1 and LC3 are not only chronologically, but also spatially distinct events. To date, the relationship of Scrg1 with autophagy remains circumstantial and the functional significance to the autophagic process is unknown. Although a potential role in the autophagic machinery was not confirmed by Scrg1 overexpression in transgenic mice, further investigations in knockout mice are merited.
Autophagic stress is a hallmark of NP0/0 PC death
In addition to its known induction during starvation, differentiation and normal growth control 9, 30, 59, autophagy has been demonstrated in various neurodegenerative disorders including Alzheimer's disease and prion disease 31, 35, 37, 42. Here, autophagic vacuoles were revealed by electron microscopy in both somatodendritic and axonal compartments of GABA immunogold‐labeled NP0/0 PCs suggesting that autophagy is concomitant to Dpl‐induced cell death mechanisms. Nevertheless, the increased concentration of autophagic vacuoles is not necessarily correlated with increased autophagic activity. Instead the striking accumulation of autophagic profiles in degenerating neurons often reflects an imbalance between the rates of autophagic sequestration and completion of the lysosomal degradation process 20, 31. Increased or continued requirements for autophagy coupled with an impairment of the late phases of autophagic flux may lead to autophagic stress (10). This is likely to occur in known neurodegenerative conditions such as Alzheimer's disease (66) and in the NP0/0 cerebellum where the accumulation of cytoplasmic autophagic profiles in PCs, and increased p62 protein levels without apparent changes in mRNA expression are likely to reflect a deficit in autophagic flux (67). Robust autophagy may be detrimental, particularly when successful recycling of cellular components is impaired leading to the accumulation of cytoplasmic autophagic vacuoles and contributing to neuronal death (10). The electron dense content of many autophagic vacuoles and phagophores found in the NP0/0 PCs are features of autolysosomal stages that suggest that autophagolysosome formation occurs in NP0/0 PCs (16). Indeed, an increased lysosomal recruitment is signalled by the upregulation of Lamp1 in NP0/0 PCs. Furthermore, double immunohistofluorescence for LC3B and Lamp1 shows a co‐localization of both molecules in the same granules within NP0/0 PCs, suggesting that formation of autophagolysosomes does occur. Thus, the upregulation of autophagic and lysosomal markers as well as the extensive accumulation of autophagosomes and autophagolysosomes in the NP0/0 PCs probably reflect a subsequent blockade of autophagic proteolysis. As observed in amyloid neurodegenerative diseases and in other neurological disorders such as ceroid lipofuscinosis (7), leakage of hydrolases induced by membrane destabilization of autophagolysosomes and lysosomes could trigger apoptotic cascades in the NP0/0 PCs. Along this line, a genetic deficiency of the glycogen‐degrading lysosomal enzyme acid‐alpha glucosidase in Pompe‐diseased mice induces severe skeletal and cardiac myopathy involving a massive autophagic buildup in myofibers 22, 49. In this case, autophagy is induced, but the muscle pathology manifests as a functional autophagic deficit because of impaired autophagosomal–lysosomal fusion that leads to an accumulation of autophagic substrates (50).
In the cerebellum of the NP0/0 mutants, there are both intact PCs, as well as PCs exhibiting different degrees of macroautophagy. This ranges from PCs with Scrg1‐labeled Golgi dictyosomes and moderate amounts of all types of unlabeled autophagic profiles to PCs filled with phagophores, autophagosomes and autophagolysosomes, probably in a late step of cell death. This differential autophagic status of PCs suggests that PC death is asynchronous in NP0/0 cerebellum. Also, autophagic NP0/0 PCs displayed differential intracellular distribution of autophagic profiles that were semi‐quantitatively estimated to be more frequent in the axonal compartment (ie, PC axons and terminals in the deep cerebellar white matter and nuclei) than in the somatodendritic compartment. In NP0/0 PC, the higher frequency of autophagic presynaptic terminals forming synapses on the deep cerebellar neurons compared with relatively few autophagic PCs found in the cerebellar cortex suggests that at least some of the axon terminals displaying autophagic profiles have an intact somatodendritic compartment. This may indicate that in NP0/0 PCs, axonal autophagy precedes somatodendritic autophagy. Indeed, axonal autophagy has been previously reported in other cases of degenerating central neurons 31, 68, including the PCs of the Lurcher mutant mouse where autophagosomes form in axons before being transported to the soma 60, 67. As Scrg1 labeling was restricted to somatic dictyosomes in NP0/0 PCs, the chronology of axonal autophagy and somatic Scrg1 upregulation could not be determined (see earlier discussion). With our current state of knowledge, we cannot tell if autophagy in the axon terminals of the NP0/0 PCs is an initial step of a Dpl‐induced autophagic cell death mechanism or a protective reaction of neurons to maintain axonal homeostasis (32) in response to Dpl‐induced stress. Nevertheless, the early autophagic destruction of axons observed in the white matter and DCN of the 3‐month‐old NP0/0 mice, well before significant PC loss, argues for a contribution of an autophagy‐dependent mechanism (see earlier discussion) for Dpl‐induced PC death rather than for autophagy as an epiphenomenon. To date, the precise subcellular localization of Dpl is unknown, although its close structural homology with PrPc 38, 58 suggests that both proteins should have a similar subcellular expression pattern at the plasma membrane of axons and terminals (3). At these sites, Dpl may induce rapid physiological alterations within axons that locally trigger the autophagic machinery.
Dpl‐induced PC death: apoptosis and/or autophagy?
To date, little is known about how Dpl kills neurons. Our recent data show that Bax knockout or overexpression of Bcl‐2 partly rescues PCs from Dpl‐induced cell death in NP0/0:Bax −/− (26) and NP0/0‐Hu‐bcl‐2 (27) double mutant mice. This partial recovery suggests that Bcl‐2 family‐dependent apoptotic pathways are not the only death mechanism implicated in the Dpl neurotoxicity. According to the present results, another possibility may be related to non‐apoptotic autophagic cell death. Indeed, Dpl has been shown to induce the production of reactive oxygen species 11, 62, which are able to activate both apoptosis (43) and autophagy (55). However, the respective contribution of apoptosis and autophagy to Dpl‐induced cell death remains unclear. There are several explanations for the occurrence of both autophagy and Bax‐dependent apoptosis of PCs in the cerebellum of the NP0/0 mouse. First, intrinsic differences between PCs could make them susceptible to different cell death pathways in response to Dpl‐induced stress. Supporting this hypothesis, PCs exhibit different resistance levels to Dpl toxicity related to both their anterior versus posterior position and their aldolase C expression in the NP0/0 cerebellum (27). This may determine a differential specificity of Dpl‐triggered cell death program among PCs. Second, a defective autophagic flux in NP0/0 PCs may ultimately trigger apoptotic cell death. Indeed, recent findings obtained both in non‐neuronal cells 19, 24, 39, 57, 65 and neurons (8), argue against a clear‐cut distinction between autophagy and apoptotic cell death. Recognition of the extensive cross‐talk between different cell death pathways is beginning to provide insight into the complex patterns of neuronal cell death seen in nervous system diseases (41). Recent data highlight the ability of BCL‐2 to not only function as an anti‐apoptotic factor, but also as an anti‐autophagic factor via its inhibitory interaction with Beclin1 (46). This could contribute to NP0/0 PC survival in the NP0/0‐Hu‐bcl‐2 double mutants (27). In these mutants, the NP0/0 PCs that die in spite of Bcl‐2 overexpression are likely to undergo Bcl‐2‐independent cell death processes that need further investigations.
To conclude, our results reveal that the autophagic machinery is activated early in the process leading to Dpl‐induced neuronal death in NP0/0 PCs. This shows that autophagy is related to Dpl‐induced PC neurodegeneration. In this way, NP0/0 PCs display many signs of impaired autophagic flux that could convert an initial autophagic defense reaction to Dpl neurotoxicity into a neurodegenerative mechanism. Our present data support emerging concepts in the field of neuronal death such as axonal initiation of neuronal autophagy and cross‐talk between autophagic and apoptotic pathways. Designing specific drugs to modulate autophagy are challenging but promising avenues for future therapy of neurodegenerative diseases.
AKNOWLEDGMENTS
The authors are greatly indebted to Dr Patrice Codogno for the helpful discussion and critical reading of the manuscript; to Drs Françoise Dandoy‐Dron and Michel Dron for helpful assistance in the Scrg1 experiments; to Tamou Thahouly for helpful assistance in the RT‐PCR experiments; and to Dr Sophie Reibel‐Foisset and Nicolas Lethenet for excellent assistance in breeding the NP0/0 mice in the Plateforme d'Expérimentation Animale of the IFR37 des Neurosciences de Strasbourg. This study was supported by the G.I.S. Maladies à Prions. S Heitz was supported by a 1‐year salary provided by the Roche Research Foundation and the Fondation Novartis pour la Recherche.
REFERENCES
- 1. Anderson L, Rossi D, Linehan J, Brandner S, Weissmann C (2004) Transgene‐driven expression of the Doppel protein in Purkinje cells causes Purkinje cell degeneration and motor impairment. Proc Natl Acad Sci USA 101:3644–3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Atarashi R, Sakaguchi S, Shigematsu K, Arima K, Okimura N, Yamaguchi N et al (2001) Abnormal activation of glial cells in the brains of prion protein‐deficient mice ectopically expressing prion protein‐like protein, PrPLP/Dpl. Mol Med 7:803–809. [PMC free article] [PubMed] [Google Scholar]
- 3. Bailly Y, Haeberlé AM, Blanquet‐Grossard F, Chasserot‐Golaz S, Grant N, Schulze T et al (2004) Prion protein (PrPc) immunocytochemistry and expression of the green fluorescent protein reporter gene under control of the bovine PrP gene promoter in the mouse brain. J Comp Neurol 473:244–269. [DOI] [PubMed] [Google Scholar]
- 4. Bjørkøy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A et al (2005) p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin‐induced cell death. J Cell Biol 171:603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bjørkøy G, Lamark T, Johansen T (2006) p62/SQSTM1: a missing link between protein aggregates and the autophagy machinery. Autophagy 2:138–139. [DOI] [PubMed] [Google Scholar]
- 6. Boellaard JW, Kao M, Schlote W, Diringer H (1991) Neuronal autophagy in experimental scrapie. Acta Neuropathol 82:225– 228. [DOI] [PubMed] [Google Scholar]
- 7. Boland B, Nixon RA (2006) Neuronal macroautophagy: from development to degeneration. Mol Aspects Med 27:503–519. [DOI] [PubMed] [Google Scholar]
- 8. Canu N, Tufi R, Serafino AL, Amadoro G, Ciotti MT, Calissano P (2005) Role of the autophagic‐lysosomal system on low potassium‐induced apoptosis in cultured cerebellar granule cells. J Neurochem 92:1228–1242. [DOI] [PubMed] [Google Scholar]
- 9. Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, Cotman SL (2006) Autophagy is disrupted in a knock‐in mouse model of juvenile neuronal ceroid lipofuscinosis. J Biol Chem 281:20483–20493. [DOI] [PubMed] [Google Scholar]
- 10. Chu CT (2006) Autophagic stress in neuronal injury and disease. J Neuropathol Exp Neurol 65:423–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cui T, Holme A, Sassoon J, Brown DR (2003) Analysis of doppel protein toxicity. Mol Cell Neurosci 23:144–155. [DOI] [PubMed] [Google Scholar]
- 12. Dandoy‐Dron F, Guillo F, Benboudjema L, Deslys J‐P, Lasmézas C, Dormont D et al (1998) Gene expression in Scrapie. Cloning of a new scrapie‐responsive gene and the identification of increased levels of seven other mRNA transcripts. J Biol Chem 273:7691– 7697. [DOI] [PubMed] [Google Scholar]
- 13. Dandoy‐Dron F, Benboudjema L, Guillo F, Jaegly A, Jasmin C, Dormont D et al (2000) Enhanced levels of scrapie responsive gene mRNA in BSE‐infected mouse brain. Brain Res Mol Brain Res 76:173–179. [DOI] [PubMed] [Google Scholar]
- 14. Dandoy‐Dron F, Griffond B, Mishal Z, Tovey MG, Dron M (2003) Scrg1, a novel protein of the CNS is targeted to the large dense‐core vesicles in neuronal cells. Eur J Neurosci 18:2449–2459. [DOI] [PubMed] [Google Scholar]
- 15. De Biasi S, Vitellaro‐Zuccarello L, Bernardi P, Valtschanoff JG, Weinberg RJ (1994) Ultrastructural and immunocytochemical characterization of primary afferent terminals in the rat cuneate nucleus. J Comp Neurol 347:275–287. [DOI] [PubMed] [Google Scholar]
- 16. Ding WX, Yin XM (2008) Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 4:141–150. [DOI] [PubMed] [Google Scholar]
- 17. Dron M, Dandoy‐Dron F, Guillo F, Benboudjema L, Hauw J‐J, Lebon P et al (1998) Gene expression in scrapie. Cloning of a new scrapie‐responsive gene and the identification of increased levels of seven other mRNA transcripts. J Biol Chem 273:7691– 7697. [DOI] [PubMed] [Google Scholar]
- 18. Dron M, Bailly Y, Beringue V, Haeberlé A‐M, Griffond B, Risold P‐Y et al (2005) Scrg1 is induced in TSE and brain injuries, and associated with autophagy. Eur J Neurosci 22:133–146. [DOI] [PubMed] [Google Scholar]
- 19. Espert L, Denizot M, Grimaldi M, Robert‐Hebmann V, Gay B, Varbanov M et al (2006) Autophagy is involved in T cell death after binding of HIV‐1 envelope proteins to CXCR4. J Clin Invest 116:2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerød L, Fisher EM et al (2007) Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol 179:485–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flechsig E, Hegyi I, Leimeroth R, Zuniga A, Rossi D, Cozzio A et al (2003) Expression of truncated PrP targeted to Purkinje cells of PrP knockout mice causes Purkinje cell death and ataxia. EMBO J 22:3095–3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fukuda T, Roberts A, Ahearn M, Zaal K, Ralston E, Plotz PH, Raben N (2006) Autophagy and lysosomes in Pompe disease. Autophagy 2:318–320. [DOI] [PubMed] [Google Scholar]
- 23. Genoud N, Behrens A, Miele G, Robay D, Heppner FL, Freigang S, Aguzzi A (2004) Disruption of Doppel prevents neurodegeneration in mice with extensive Prnp deletions. Proc Natl Acad Sci USA 101:4198–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gonzalez‐Polo RA, Boya P, Pauleau AL, Jalil A, Larochette N, Souquère S et al (2005) The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci 118:3091–3102. [DOI] [PubMed] [Google Scholar]
- 25. Hariri M, Millane G, Guimond MP, Guay G, Dennis JW, Nabi IR (2000) Biogenesis of multilamellar bodies via autophagy. Mol Biol Cell 11:255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heitz S, Lutz Y, Rodeau J‐L, Zanjani H, Gautheron V, Bombarde G et al (2007) BAX contributes to Doppel‐induced apoptosis of prion protein‐deficient Purkinje cells. Dev Neurobiol 67:670–686. [DOI] [PubMed] [Google Scholar]
- 27. Heitz S, Gautheron V, Lutz Y, Rodeau J‐L, Zanjani H, Sugihara I et al (2008) BCL‐2 opposes Dpl‐induced apoptosis of prion protein‐deficient Purkinje cells in the Ngsk Prnp 0/0 mouse. Dev Neurobiol 68:332–348. [DOI] [PubMed] [Google Scholar]
- 28. Jeffrey M, Goodsir CM, Bruce ME, McBride PA, Scott JR, Halliday WG (1992) Infection specific prion protein (PrP) accumulates on neuronal plasmalemma in scrapie infected mice. Neurosci Lett 147:106–109. [DOI] [PubMed] [Google Scholar]
- 29. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T et al (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19:5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Klionsky DJ (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cel Biol 8:931–937. [DOI] [PubMed] [Google Scholar]
- 31. Koike M, Shibata M, Waguri S, Yoshimura K, Tanida I, Kominami E et al (2005) Participation of autophagy in storage of lysosomes in neurons from mouse models of neuronal ceroid‐lipofuscinoses (Batten disease). Am J Pathol 167:1713–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Komatsu M, Wang QJ, Holstein GR, Friedrich VL Jr, Iwata J, Kominami E et al (2007) Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc Natl Acad Sci U S A 104:14489–14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kouno T, Mizuguchi M, Tanida I, Ueno T, Kanematsu T, Mori Y et al (2005) Solution structure of microtubule‐associated protein light chain 3 and identification of its functional subdomains. J Biol Chem 280:24610–24617. [DOI] [PubMed] [Google Scholar]
- 34. Kuma A, Matsui M, Mizushima N (2007) LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: caution in the interpretation of LC3 localization. Autophagy 3:323–328. [DOI] [PubMed] [Google Scholar]
- 35. Larsen KE, Sulzer D (2002) Autophagy in neurons: a review. Histol Histopathol 17:897–908. [DOI] [PubMed] [Google Scholar]
- 36. Liberski PP, Sikorska B, Bratosiewicz‐Wasik J, Gajdusek DC, Brown P (2004) Neuronal cell death in transmissible spongiform encephalopathies (prion diseases) revisited: from apoptosis to autophagy. Int J Biochem Cell Biol 36:2473–2490. [DOI] [PubMed] [Google Scholar]
- 37. Liberski PP, Brown DR, Sikorska B, Caughey B, Brown P (2008) Cell death and autophagy in prion diseases (transmissible spongiform encephalopathies). Folia Neuropathol 46:1–25. [PubMed] [Google Scholar]
- 38. Lu K, Wang W, Xie Z, Wong BS, Li R, Petersen RB et al (2000) Expression and structural characterization of the recombinant human doppel protein. Biochemistry 39:13575–13583. [DOI] [PubMed] [Google Scholar]
- 39. Martin DN, Baehrecke EH (2004) Caspases function in autophagic programmed cell death in Drosophila. Development 131:275–284. [DOI] [PubMed] [Google Scholar]
- 40. Nishida N, Tremblay P, Sugimoto T, Shigematsu K, Shirabe S, Petromilli C et al (1999) A mouse prion protein transgene rescues mice deficient for the prion protein gene from Purkinje cell degeneration and demyelination. Lab Invest 79:689–697. [PubMed] [Google Scholar]
- 41. Nixon RA (2006) Autophagy in neurodegenerative disease: friend, foe or turncoat? Trends Neurosci 29:528–535. [DOI] [PubMed] [Google Scholar]
- 42. Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM (2005) Extensive involvement of autophagy in Alzheimer disease: an immuno‐electron microscopy study. J Neuropathol Exp Neurol 64:113–122. [DOI] [PubMed] [Google Scholar]
- 43. Orrenius S (2007) Reactive oxygen species in mitochondria‐mediated cell death. Drug Metab Rev 39:443–455. [DOI] [PubMed] [Google Scholar]
- 44. Paisley D, Banks S, Selfridge J, McLennan NF, Ritchie AM, McEwan C et al (2004) Male infertility and DNA damage in Doppel knockout and prion protein/Doppel double‐knockout mice. Am J Pathol 164:2279–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H et al (2007) p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 282:24131–24145. [DOI] [PubMed] [Google Scholar]
- 46. Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N et al (2005) Bcl‐2 antiapoptotic proteins inhibit Beclin 1‐dependent autophagy. Cell 122:927–939. [DOI] [PubMed] [Google Scholar]
- 47. Pattingre S, Espert L, Biard‐Piechaczyk M, Codogno P (2008) Regulation of macroautophagy by mTOR and Beclin 1 complexes. Biochimie 90:313–323. [DOI] [PubMed] [Google Scholar]
- 48. Prusiner SB (1998) Prions. Proc Natl Acad Sci U S A 95:13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Raben N, Takikita S, Pittis MG, Bembi B, Marie SKN, Roberts A et al (2007) Deconstructing Pompe disease by analyzing single muscle fibers. Autophagy 3:546–552. [DOI] [PubMed] [Google Scholar]
- 50. Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N et al. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet [Epub of print. [DOI] [PMC free article] [PubMed]
- 51. Rossi D, Cozzio A, Flechsig E, Klein MA, Rülicke T, Aguzzi A, Weissmann C (2001) Onset of ataxia and Purkinje cell loss in PrP null mice inversely correlated with Dpl level in brain. EMBO J 20:694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sakaguchi S, Katamine S, Shigematsu K, Nakatani A, Moriuchi R, Nishida N et al (1995) Accumulation of proteinase K‐resistant prion protein (PrP) is restricted by the expression level of normal PrP in mice inoculated with a mouse‐adapted strain of the Creutzfeldt‐Jakob disease agent. J Virol 69:7586–7592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sakaguchi S, Katamine S, Nishida N, Moriuchi R, Shigematsu K, Sugimoto T et al (1996) Loss of cerebellar Purkinje cells in aged mice homozygous for a disrupted PrP gene. Nature 380:528–531. [DOI] [PubMed] [Google Scholar]
- 54. Sakudo A, Lee DC, Nakamura I, Taniuchi Y, Saeki K, Matsumoto Y et al (2005) Cell‐autonomous PrP‐Doppel interaction regulates apoptosis in PrP gene‐deficient neuronal cells. Biochem Biophys Res Commun 333:448–454. [DOI] [PubMed] [Google Scholar]
- 55. Scherz‐Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J 26:1749–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shmerling D, Hegyi I, Fischer M, Blattler T, Brandner S, Gotz J et al (1998) Expression of amino‐terminally truncated PrP in the mouse leading to ataxia and specific cerebellar lesions. Cell 93:203–214. [DOI] [PubMed] [Google Scholar]
- 57. Shimizu S, Kanaseki T, Mizushima N, Mizuta T, Arakawa‐Kobayashi S, Thompson CB, Tsujimoto Y (2004) Role of Bcl‐2 family proteins in a non‐apoptotic programmed cell death dependent on autophagy genes. Nat Cell Biol 6:1221–1228. [DOI] [PubMed] [Google Scholar]
- 58. Silverman GL, Qin K, Moore RC, Yang Y, Mastrangelo P, Tremblay P et al (2000) Doppel is an N‐glycosylated, glycosylphosphatidylinositol‐anchored protein. Expression in testis and ectopic production in the brains of Prnp(0/0) mice predisposed to Purkinje cell loss. J Biol Chem 275:26834–26841. [DOI] [PubMed] [Google Scholar]
- 59. Vittorini S, Paradiso C, Donati A, Cavallini G, Masini M, Gori Z et al (1999) The age‐related accumulation of protein carbonyl in rat liver correlates with the age‐related decline in liver proteolytic activities. J Gerontol A Biol Sci Med Sci 54:B318–23. [DOI] [PubMed] [Google Scholar]
- 60. Wang QJ, Ding Y, Kohtz DS, Mizushima N, Cristea IM, Rout MP et al (2006) Induction of autophagy in axonal dystrophy and degeneration. J Neurosci 26:8057–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Watts JC, Westaway D (2007) The prion protein family: diversity, rivalry, and dysfunction. Biochim Biophys Acta 1772:654–672. [DOI] [PubMed] [Google Scholar]
- 62. Wong BS, Liu T, Paisley D, Li R, Pan T, Chen SG et al (2001) Induction of HO‐1 and NOS in doppel‐expressing mice devoid of PrP: implications for doppel function. Mol Cell Neurosci 17:768–775. [DOI] [PubMed] [Google Scholar]
- 63. Wu J, Dang Y, Su W, Liu C, Ma H, Shan Y et al (2006) Molecular cloning and characterization of rat LC3A and LC3B—two novel markers of autophagosome. Biochem Biophys Res Commun 339:437–442. [DOI] [PubMed] [Google Scholar]
- 64. Yamaguchi N, Sakaguchi S, Shigematsu K, Okimura N, Katamine S (2004) Doppel‐induced Purkinje cell death is stoichiometrically abrogated by prion protein. Biochem Biophys Res Commun 319:1247–1252. [DOI] [PubMed] [Google Scholar]
- 65. Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L et al (2006) Calpain‐mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol 8:1124–1132. [DOI] [PubMed] [Google Scholar]
- 66. Yu WH, Cuervo AM, Kumar A, Peterhoff CM, Schmidt SD, Lee JH et al (2005) Macroautophagy—a novel Beta‐amyloid peptide‐generating pathway activated in Alzheimer's disease. J Cell Biol 171:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Yue Z, Wang QJ, Komatsu M (2007) Neuronal autophagy: going the distance to the axon. Autophagy 4:94–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zanjani SH, Selimi F, Vogel MW, Haeberlé AM, Boeuf J, Mariani J, Bailly YJ (2006) Survival of interneurons and parallel fiber synapses in a cerebellar cortex deprived of Purkinje cells: studies in the double mutant mouse Grid2Lc/+;Bax(−/−). J Comp Neurol 497:622–635. [DOI] [PubMed] [Google Scholar]


