Abstract
Objective:
To develop 3D T1ρ and T2 imaging based on the same sequence structure on MR systems from multiple vendors, and to evaluate intra-site repeatability and inter-site inter-vendor reproducibility of T1ρ and T2 measurements of knee cartilage.
Methods:
3D magnetization-prepared angle-modulated partitioned k-space spoiled gradient echo snapshots (3D MAPSS) were implemented on MR systems from Siemens, GE and Philips. Phantom and human subject data were collected at four sites using 3T MR systems from the three vendors with harmonized protocols. Phantom data were collected by means of different positioning of the coil. Volunteers were scanned and rescanned after repositioning. Two traveling volunteers were scanned at all sites. Data were transferred to one site for centralized processing.
Results:
Intra-site average coefficient of variations (CVs) ranged from 1.09% to 3.05% for T1ρ and 1.78% to 3.30% for T2 in phantoms, and 1.60% to 3.93% for T1ρ and 1.44% to 4.08% for T2 in volunteers. Inter-site average CVs were 5.23% and 6.45% for MAPSS T1ρ and T2, respectively in phantoms, and 8.14% and 10.06% for MAPSS T1ρ and T2, respectively, In volunteers.
Conclusion:
This study showed promising results of multi-site, multi-vendor reproducibility of T1ρ and T2 values in knee cartilage. These quantitative measures may be applied in large-scale multi-site, multi-vendor trials with controlled sequence structure and scan parameters and centralized data processing.
Keywords: Magnetic resonance imaging, Cartilage imaging, T1ρ, T2, Reproducibility, Cartilage, Quantitative MRI, Multi-site, Multi-vendor
INTRODUCTION
Osteoarthritis (OA) is a degenerative disease affecting multiple joint tissues and characterized by pain, stiffness and loss of function.1 The disease is the most common form of synovial joint disorder, affecting 30.8 million adults in the USA.2 Despite its significant impact, early diagnosis and prognosis of OA remain challenging. As yet, despite extensive efforts, there are no clinically approved disease-modifying OA drugs (DMOADs) available. One of the hurdles for DMOAD development is the lack of sensitive and reliable non-invasive biomarkers that can detect treatment effects over a short-time window.3, 4
Radiographic imaging is the current clinical imaging tool to evaluate structural abnormalities in OA. Joint space narrowing measured by radiographs is the only FDA-approved imaging marker for measuring clinical efficacy in DMOAD development. Radiographs, however, have limited contrast for soft tissues such as cartilage and meniscus, and thus fail to detect early joint degeneration.
Magnetic resonance imaging (MRI), however, provides high-resolution 3D images of the whole joint with superior soft tissue contrast and without ionizing radiation. Conventional MRI techniques focus on morphological information, advanced quantitative cartilage MRI techniques, such as T1ρ, T2, delayed gadolinium-enhanced MRI of cartilage (dGEMRIC), diffusion MRI, chemical exchange saturation transfer (gagCEST), and sodium MRI, provide in-depth information about the compositional changes in cartilage proteoglycan-collagen matrix known to occur in the early stage of OA.5, 6 Numerous studies have suggested that T1ρ and T2 imaging can not only distinguish patients with early OA or those who are at risk of developing OA from healthy controls, as summarized by two recent systematic reviews7, 8, but can also predict disease progression in OA or acutely injured knees9, 10. Moreover, T1ρ and T2 relaxometry do not require contrast agent injection or special equipment, thus enabling these techniques to be applied to large multi-center, multi-vendor trials and translated into clinical practice.
T1ρ and T2 values have been evaluated in terms of repeatability in single-site studies11–16 and reproducibility in multi-site studies with a single vendor (average CV of 4.9% for T1ρ and 3.3–6.5% for T2)17–19, but few studies have examined the reliability in a multi-site, multi-vendor context. Only one study has reported inter-vendor reliability of T2 with mean differences of 5.4–10.0ms (10–25%).19 Understanding and documenting these variations is critical for large-scale multi-vendor multi-site clinical trials using T1ρ and T2 imaging.
It is well known that measured T1ρ and T2 values are dependent on MRI pulse sequences (the term “sequence” will be used throughout the text).20–22 Although T2 imaging based on multi-echo spin-echo acquisition is available as product sequences on most MR systems, differences in implementation details between vendors are unknown, and the T2 values derived from this sequence are known to be susceptible to B1 inhomogeneity-invoked stimulated echoes.23 Since T1ρ imaging is only available as research prototypes on MR systems, published T1ρ studies have used sequences with different structures (balanced or non-balanced gradient-echo vs spin-echo readout, variable flip angle vs constant flip angle, with and without RF cycling, etc.) on different MR systems24–26. To date, no T1ρ imaging acquisition with similar sequence structure is available across the different vendors’ MR systems. Acquisition of T1ρ and T2 using the exact same readout will facilitate comparisons of these two measures, as well as exploration of interesting new markers such as R2 - R1ρ (a composite relaxation rate created to describe extracellular matrix using a single metric).27
The goals of this study were to: 1) implement T1ρ and T2 mapping using the same sequence structure at 3T on three vendor platforms (Siemens, GE, Philips) located at four geographically different sites; 2) standardize imaging protocols and data processing procedures; and 3) measure intra-site repeatability and inter-site reproducibility of T1ρ and T2 measures in phantoms and human volunteers.
METHODS
Study Design
Phantoms and volunteers were scanned at four sites using 3T MRI systems from three vendors to evaluate intra- and inter-site variability (Table 1a). The phantoms consisted of six tubes with three different agarose concentrations (2, 3, and 4%, weight/volume) fixed in a cylinder holder (manufactured by the Phantom Lab), each scanned three times in three different positions. Five knees from volunteers were scanned in feet-first supine position at each site after informed consent, including three knees from two traveling volunteers who were scanned at all four sites. All procedures were approved by the Institutional Review Board (IRB) at all participating sites. All volunteers were required to sit for at least 30 minutes before scans to equilibrate loading status to knee cartilage.
Table 1.
Hardware information and imaging parameters for data collection
| a) MR systems and RF coils | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Site 1 | Site 2 | Site 3 | Site 4 | |||||||
| MR system | Siemens Prisma | Siemens Prisma | GE Discovery™ MR750 wide bore | Philips Ingenia wide bore | ||||||
| Software | VE11C | VE11C | DV25R2 | 5.3.1 | ||||||
| RF coil | QED 1 Tx/15Rx knee coil | QED 1Tx/15Rx knee coil | Invivo 1Tx/8Rx knee coil | Invivo 1Tx/16Rx coil | ||||||
| b) T1ρ and T2 imaging | ||||||||||
| Phantom | Volunteer | |||||||||
| FOV | 140×140×80 mm3 | 140×140×96 mm3 | ||||||||
| Imaging matrix | 256×128×20 | 320×160×24 | ||||||||
| Image orientation | Transversal | Sagittal | ||||||||
| Time of recovery | 1.5 seconds | |||||||||
| Number of echoes | 8 | |||||||||
| Bandwidth per pixel (Hz) | 400 Hz | |||||||||
| Views per segment | 76 (Site 1, 3, 4) 128 (Site 2) |
92 (Site 1, 3, 4) 160 (Site 2) |
||||||||
| Parallel imaging | Factor 2 (Site 1, 3, 4) None (Site 2) |
Factor 2 (Site 1, 3) Factor 1.6×1.6 (Site 4) None (Site 2) |
||||||||
| Time of Spin-Lock (TSL) | 0, 10, 20, 30, 40, 50, 60, 70 ms | |||||||||
| Spin-Lock Frequency | 500 Hz | |||||||||
| T2 preparation TE | 0, 9.7, 21.3, 32.9, 44.5, 56.1, 67.6, 79.2 ms (Site 1) 5, 10, 20, 30, 40, 50, 60, 70 ms (Site 2) 0, 8.8, 17.7, 26.5, 35.4, 44.2, 53.1, 61.9 ms (Site 3) 0, 6.83, 15.3, 23.7, 32.1, 40.6, 49.0, 57.4 ms (Site 4) |
|||||||||
| Acquisition time | 10:40 (Site 1) 6:24 (Site 2) 10:41 (Site 3) 9:04 (Site 4) |
13:43 (Site 1) 7:48 (Site 2) 14:42(Site 3) 9:11 (Site 4) |
||||||||
| c) List of scan parameters used to evaluate scan parameter dependency | ||||||||||
| T1ρ imaging | Combinations of time of recovery (0.8, 1.5, and 3 seconds), views per segment (64 and 128), with and without parallel imaging | |||||||||
| T2 imaging | TE_set1 = [1.86 8.59 17.18 25.78 34.37 42.96 51.55 60.14] ms TE_set2 = [1.86 9.99 19.98 29.98 39.97 49.96 59.95 69.94] ms TE_set3 = [1.86 11.59 23.18 34.78 46.37 57.96 69.55 81.14] ms | |||||||||
| d) High-resolution gradient echo imaging | ||||||||||
| Site 1 (DESS) | Site 2 (DESS) | Site 3 (SPGR) | Site 4 (FFE) | |||||||
| FOV (mm3) | 140×140×112 | 140×140×112 | 140×140×114 | 140×140×112 | ||||||
| Imaging matrix | 384×307×160 | 256×238×160 | 384×300×114 | 388×310×160 | ||||||
| TR (ms) | 17.55 | 17.55 | 13.036 | 18 | ||||||
| TE (ms) | 6.02 | 6.02 | 2.716 | 6.5 | ||||||
| Flip angle (°) | 25 | 25 | 25 | 25 | ||||||
| Bandwidth per pixel (Hz) | 185 | 240 | 139 | 185 | ||||||
To measure T1ρ and T2, three sites (1, 3, and 4) used sequences based on 3D magnetization-prepared angle-modulated partitioned k-space spoiled gradient-echo snapshots (3D MAPSS)11, 24. Site 2 used a different strategy employing 3D T1ρ and T2 imaging based on segmented gradient echo acquisition with a constant flip angle25 (termed as “segmented GRE acquisition” in this study). After data collection, all data were transferred to one site (Site 1) for centralized image processing and data analysis.
3D MAPSS Sequence Implementation
The 3D MAPSS sequence for combined T1ρ and T2 imaging was implemented on MRI systems from all three vendors following the general description in previous papers.11, 24 Briefly, T1ρ preparation used continuous spin-lock pulses with 180° phase shift in the second half28, and T2 preparation used an MLEV 16 (Malcolm Levitt’s composite pulse decoupling sequence) refocusing pulse train. Echo time (TE) correction was applied as follows.29
| e.q. [1] |
where TE is the original TE, k is the number of refocusing pulses for the TE, pw is the width of the refocusing pulse, and T1 and T2 are the relaxation times of the target tissue (assumed T1 over T2 of 40 for cartilage and 50 for phantom). Composite pulses as proposed by Dixon et al30 were applied for the tip-down and tip-up pulses before and after the T1ρ or T2 preparation in order to reduce artifacts caused by B0 and B1 inhomogeneity.
Multiple k-space lines (views per segmentation [VPS]) were acquired immediately after each magnetization preparation. RF cycling was applied to eliminate the adverse impact of longitudinal relaxation on quantitative accuracy. This RF cycling scheme also yields a transient signal evolution that is independent of the prepared magnetization; consequently, a same variable flip angle train can be applied to provide a flat signal response to eliminate the filtering effect in k-space caused by transient signal evolution after each magnetization preparation. As a result of RF cycling, the scan time was doubled compared to the sequence used in Site 2. The ordering of the collected k-space lines after a magnetization preparation differed from site to site. Sites 1 and 2 collected entire phase-encoding lines in centric order for a partition encode after a magnetization preparation, which led to the number of preparations for an image being equal to the number of partitions. Sites 3 and 4 used segmented centric view ordering.11
Phantom Evaluation
The T1ρ and T2 sequence protocols were imaged at three different coil locations (left 70mm, center, and right 70mm) to evaluate the effect of knee laterality. At each site, phantoms were imaged three times on different days within a week. Phantoms were kept in the scanner room at least overnight before each scan and the room temperature was recorded. Phantom orientation inside the coil was the same for all sites. Table 1b lists the parameters used for phantom imaging. Each site used the minimum allowed TR and TE. The acquisition parameter dependence of 3D MAPSS was investigated at Site 1 by evaluating T1ρ and T2 reproducibility with sequence parameters changes (Table 1c).
Volunteer Evaluation
In addition to T1ρ and T2 imaging, a high-resolution gradient echo (GRE) imaging (spoiled gradient-echo [SPGR] for GE, T1 fast field echo [T1-FFE] for Philips, and dual-echo steady-state [DESS] for Siemens) was performed for cartilage segmentation. All scans were performed with fat suppression using frequency selective excitation, followed by crusher gradient. The parameters for each site are listed in Table 1d. All volunteers were repositioned in the magnet between scan and rescan acquisitions.
Image Processing
Phantom
An automatic segmentation method was used to select the region of interest (ROI) for each tube by removing the background, finding six connected components, and eroding 10 pixels in the periphery to avoid edge artifact.
Volunteers
A standardized workflow was established for robust image registration to ensure efficient and objective intra- and inter-site comparison of images as diagrammed in Figure 1a, b, c. Detailed descriptions are listed below.
Figure 1.
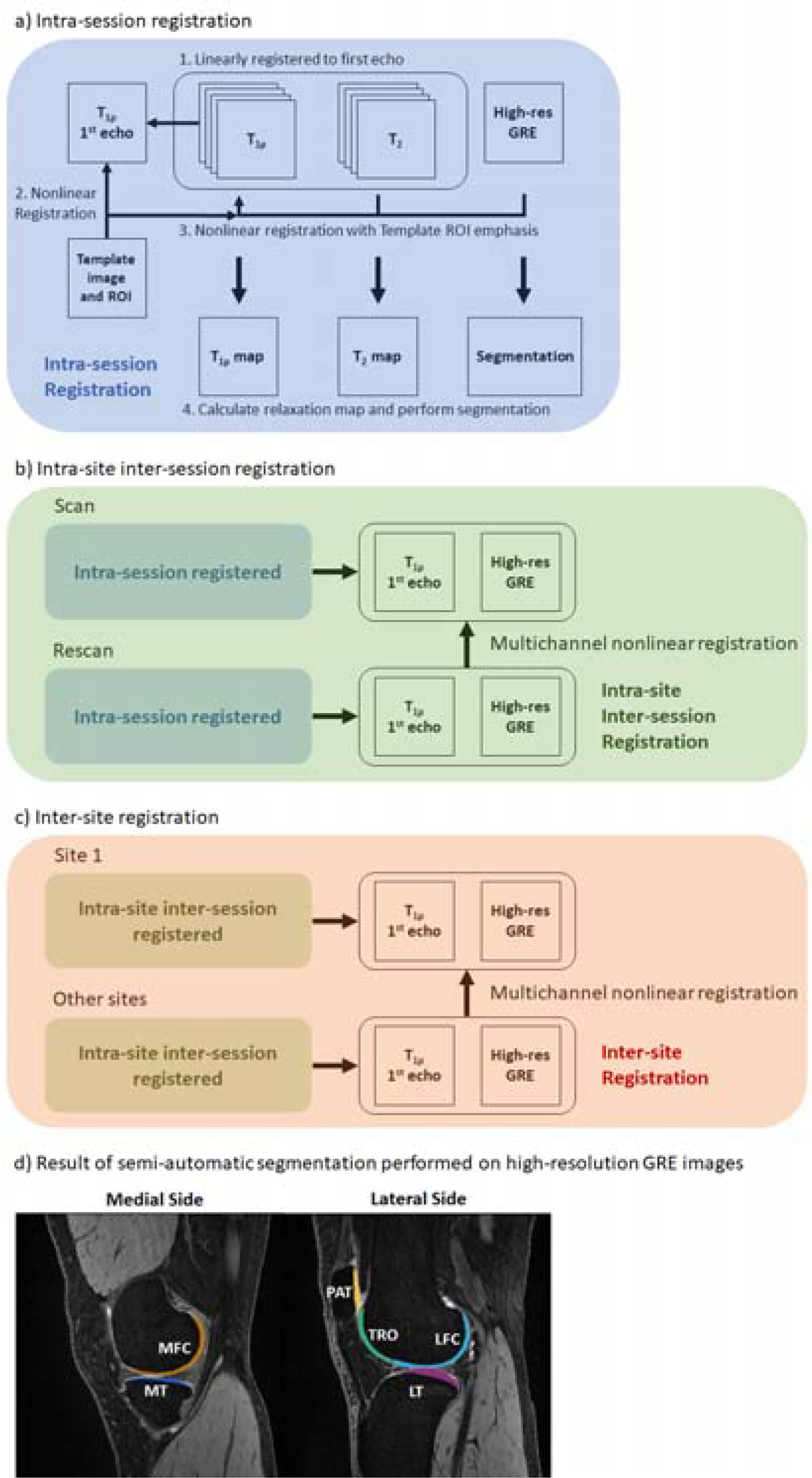
a, b, c) Flowchart of image registrations performed on volunteer data. For intra-site evaluation, segmentation was performed on the high-resolution GRE image of the first scan. For inter-site evaluation, segmentation was performed only on the first scan high-resolution GRE from Site 1. d) Example segmentation map on registered high-resolution GRE image.
Each volunteer dataset consisted of high-resolution GRE, T1ρ, and T2 images. All images within a dataset were registered to the first echo of the T1ρ images, using Elastix’s rigid registration.31 A mask covering the areas surrounding the patella, tibia, fibula, and femur, generated by warping a predefined mask on a template image, was used in Elastix to guide the registration. The template image was generated by averaging images from the Osteoarthritis Initiative (OAI) dataset.
The intra-site rescan images were nonlinearly registered to the initial scan images using ANTS32 by registering the first echo of T1ρ and high-resolution GRE images simultaneously. Registration was evaluated for subtle shifts visually by switching between different images. The resulting nonlinear transformation was then applied to the other rescan images for the subject. The cartilage ROIs were nonlinearly warped to the rescan images.
For inter-site comparison, each traveling subject’s images across the four sites were registered to the scan of that subject acquired at Site 1, using the same method as in b).
Cartilage compartments, including medial/lateral femur (MFC/LFC), medial/lateral tibial (MT/LT), trochlear (TRO), and patella (PAT), were segmented using a custom graphical image interface and adjusting control points on b-spline curves (Figure 1d). The segmentation was based on the registered high-resolution GRE image, which was guided by gradient image as the reader selected control points around the cartilage region, making the process semi-automatic. To confirm the reproducibility of segmentation, the primary and another trained segmenter independently segmented a total of 24 compartments from four knees. The inter-segmenter CVs for T1ρ and T2 were 1.17% and 1.38%, respectively, indicating small inter-observer variation. For traveling volunteers, the cartilage segmentation was performed once on the registered high-resolution GRE image from Site 1. Following registration and segmentation, statistical measures for each cartilage compartment were computed after a mono-exponential fitting of the registered T1ρ, and T2 images.
Relaxation time fitting
A voxel-wise two parameter mono-exponential fitting was performed to calculate T2 and T1ρ, based on the Levenberg-Marquardt algorithm using MATLAB (MathWorks, Natick, MA) with linear regression solution using logarithmic transformation for the initial guess. After relaxation time maps were calculated, the mean and standard deviation (SD) of relaxation times of ROIs, each tube for phantom data and cartilage compartment for volunteer data were calculated. To evaluate the feasibility of using fewer echoes, maps fitted using 4 of the 8 echoes in phantom and volunteer data from Site 1, were created using two different sets of echoes representing strategies with equal/unequal spaces between echoes (Echo_set1 = [1,3,5,7], Echo_set2 = [1,2,4,8]).
SNR Efficiency Estimation
SNR efficiency was calculated as follows:
The ratio of the acquisition time at each site with respect to the acquisition time of Site 1 was used for calculation. For volunteer images, a low signal area in the fat-suppressed bone marrow was used, because background signal was filtered out by one vendor.
Statistical Analysis
CVs were calculated to evaluate variability in phantoms from changing scan parameters. CVs were combined by averaging in both phantoms and volunteers. CVs and intraclass correlation coefficients (ICC) were calculated to investigate intra- and inter-site correlation between sites. ICC(2,1) was calculated for intra-site repeatability and ICC(4,1) was calculated for inter-site reproducibility. The inter-site ICCs and CVs were calculated using data from all sites, and only data from three sites that used 3D MAPSS (Site 1, 3, and 4). Bland-Altman analysis was conducted for volunteer data, and the concordance correlation coefficient (CCC) was calculated between sites for inter-site reproducibility.
RESULTS
Scan Parameter Dependency of T1ρ and T2 measures using MAPSS sequences
The MAPSS T1ρ sequence yielded consistent relaxation time measures from the experiments, regardless of differences in time of recovery or views per segment, with and without parallel imaging reconstruction (Figure 2, average CV 0.4%). The differences in T2 measures with different T2 preparation TEs were subtle (average CV 1.02%).
Figure 2.
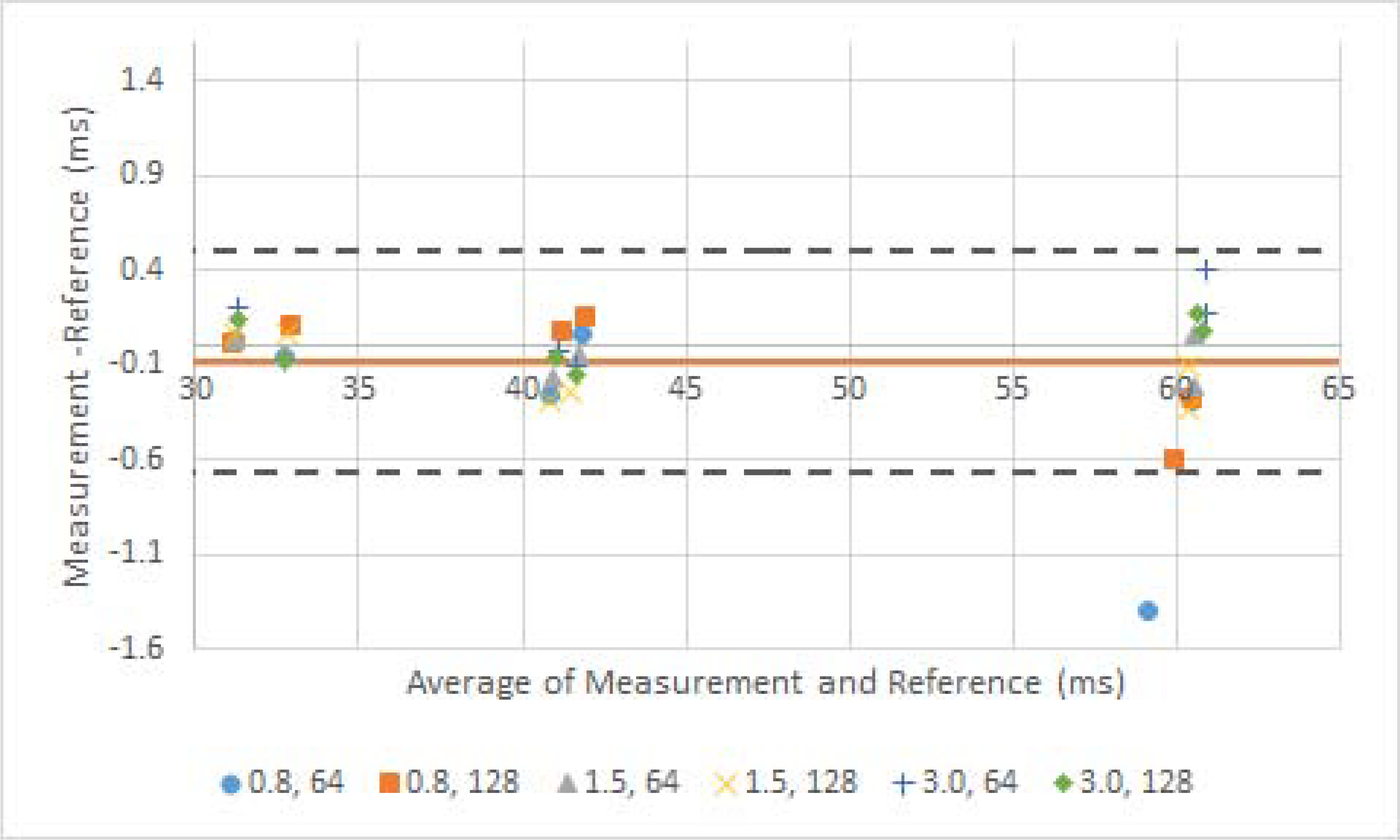
Bland-Altman plots of sequence evaluation with different scan parameters. a) T1ρ measurement with different pairs of time of recovery (seconds) and views per segments. Reference value was measured with the phantom scan protocols which used 1.5 seconds time of recovery and 76 views per segments with GRAPPA 2 reconstruction. b) T2 measurement with different sequences of T2 preparation TEs. The sequences of preparation TEs are listed in the methods.
Phantom Evaluation
Figure 3a shows the photo of the phantom. Figure 3b demonstrates phantom measures from each site. T1ρ values were higher than T2 values, as expected, using MAPSS acquisitions (Site 1, 3 and 4), while T2 values were higher than T1ρ values, using segmented GRE acquisitions (Site 2). Figure 3c shows the scatter plot of the relaxation time values of all sites with respect to values from Site 1. While no position-related systematic differences were found (T1ρ/T2 CV (%) = 0.50/0.92 for Site 1, 2.59/2.11 for Site 2, 1.98/2.98 for Site 4), T1ρ for Site 4 showed higher value when positioned in the center. Data from Site 3 had to be excluded due to incomplete data.
Figure 3.
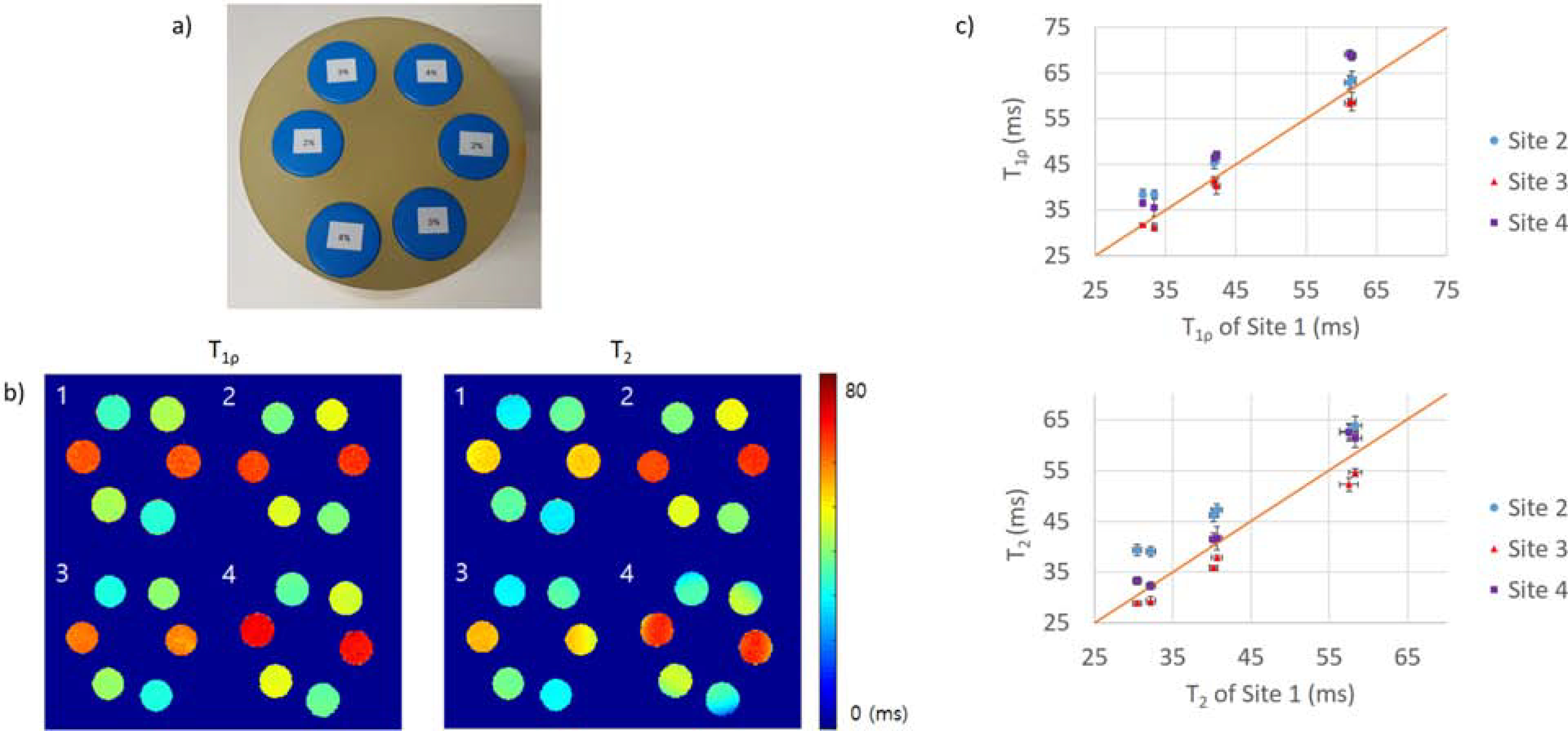
a) Photo of phantom used in this study. b) Example phantom T1ρ and b) T2 relaxation time maps from all sites. The number in the top left corners of the maps indicates the site. c) Graph of T1ρ and T2 of each sites with respect to the value from Site 1. The diagonal red line shows the reference line of X = Y.
Excellent intra-site repeatability was observed for all sites, with average CVs ranging from 1.09 to 3.05% for T1ρ and from 1.78 to 3.30% for T2 (Table 2a; measurements from different coil locations were considered separately when calculating the CVs). Average inter-site CVs for all four sites were 6.54% and 8.44% for T1ρ and T2, respectively (Table 2b). The average inter-site CVs fell to 6.45% and 5.23% for T1ρ, and T2 respectively when the CV was calculated for data collected by 3D MAPSS only (Site 1, 3, and 4). Between sites, MAPSS relaxation times were highly correlated: ICC of 0.925 (95% CI [0.438, 0.990]) for T1ρ and 0.939 (95% CI [0.492, 0.992]) for T2. The temperature of the scanner rooms were 20.7°C (Site 1), 17.8°C (Site 2), 22.2°C (Site 3), and 20°C (Site 4).
Table 2.
Results for phantom T1ρ and T2 measurements.
| a) Average relaxation time ± SD (ms) (above) and intra-site CV (%) (below) calculated for phantom T1ρ and T2 measurements | |||||||
|---|---|---|---|---|---|---|---|
| T1ρ | 2%-1 | 2%-2 | 3%-1 | 3%-2 | 4%-1 | 4%-2 | Average |
| Site 1 | 61.23±0.74 | 61.53±0.55 | 41.94±0.42 | 42.26±0.44 | 33.41±0.37 | 31.76±0.39 | |
| 1.22 | 0.90 | 1.01 | 1.04 | 1.12 | 1.23 | 1.09 | |
| Site 2 | 62.96±1.52 | 63.57±1.84 | 45.44±1.39 | 46.69±1.22 | 38.39±0.98 | 38.41±1.17 | |
| 2.18 | 2.60 | 2.80 | 2.41 | 2.36 | 2.82 | 2.53 | |
| Site 3 | 58.48±1.02 | 58.8±2.43 | 41.34±1.25 | 40.32±2.18 | 31.3±1.04 | 31.76±0.21 | |
| 1.75 | 4.14 | 3.03 | 5.42 | 3.31 | 0.67 | 3.05 | |
| Site 4 | 69.11±1.03 | 68.74±0.95 | 46.31±0.77 | 47.05±0.59 | 35.59±1.9 | 36.49±0.45 | |
| 1.50 | 1.39 | 1.66 | 1.25 | 5.34 | 1.23 | 2.06 | |
| T2 | 2%-1 | 2%-2 | 3%-1 | 3%-2 | 4%-1 | 4%-2 | Average |
| Site 1 | 57.55±1.24 | 58.33±0.87 | 40.22±0.56 | 40.65±0.72 | 32.21±0.66 | 30.47±0.56 | |
| 2.15 | 1.48 | 1.40 | 1.77 | 2.03 | 1.83 | 1.78 | |
| Site 2 | 62.78±1.63 | 63.81±1.9 | 46.2±1.38 | 47.24±1.24 | 39.09±0.99 | 39.36±1.23 | |
| 2.59 | 2.97 | 3.00 | 2.63 | 2.54 | 3.12 | 2.81 | |
| Site 3 | 52.26±1.61 | 54.62±0.95 | 35.88±0.47 | 37.87±0.69 | 29.37±1 | 28.88±0.6 | |
| 3.08 | 1.75 | 1.31 | 1.81 | 3.40 | 2.08 | 2.24 | |
| Site 4 | 62.46±1.7 | 61.33±1.96 | 41.48±1.35 | 41.67±2.38 | 32.31±0.77 | 33.34±0.85 | |
| 2.72 | 3.20 | 3.25 | 5.72 | 2.39 | 2.55 | 3.30 | |
| b) Inter-site CV (%) calculated for phantom T1ρ and T2 measurements | |||||||
| T1ρ | 2%-1 | 2%-2 | 3%-1 | 3%-2 | 4%-1 | 4%-2 | Average |
| All | 6.09 | 5.71 | 5.13 | 6.17 | 7.55 | 8.57 | 6.54 |
| MAPSS | 7.05 | 6.59 | 5.49 | 6.53 | 5.87 | 7.19 | 6.45 |
| T2 | 2%-1 | 2%-2 | 3%-1 | 3%-2 | 4%-1 | 4%-2 | Average |
| All | 6.33 | 5.47 | 8.10 | 8.32 | 10.50 | 11.94 | 8.44 |
| MAPSS | 6.61 | 4.62 | 5.23 | 5.01 | 3.96 | 5.93 | 5.23 |
| c) SNR efficiency of first echo of phantom measurements | |||||||
| Site 1 | Site 2 | Site 3 | Site 4 | ||||
| SNR efficiency | 149.8±19.2 | 133.4±10.56 | 196.6±23.5 | 226.9±10.4 | |||
All: using data from all four sites. MAPSS: using data from sites 1,3,4 that were collected with the MAPSS sequence.
SNR efficiency varied, ranging from 150 to 227 (Table 2c). All CVs between the phantom relaxation times fitted with 8 echoes and two sets of 4 echoes were smaller than 0.5%.
Volunteer Evaluation
Excellent intra-site repeatability for cartilage relaxation times for volunteers was observed at all sites, with average CVs ranging from 1.60 to 3.93% for T1ρ and from 1.44 to 4.08% for T2 (Table 3a). However, the average inter-site CVs were 13.61% and 20.34% for T1ρ and T2, respectively (Table 3b). The inter-site CVs were reduced to 8.14% and 10.06% for T1ρ and T2, respectively, when the data compared were restricted to MAPSS sites only. Figure 4 shows example relaxation maps for one of the traveling volunteers. Both T1ρ and T2 values were higher at Site 2 than those from other sites. The first traveling volunteer was not scanned at Site 2.
Table 3.
Results for volunteer T1ρ and T2 measurements.
| a) Average relaxation time ± SD (ms) (above) and intra-site CV (%) (below) calculated for volunteer T1ρ and T2 measurements | |||||||
|---|---|---|---|---|---|---|---|
| T1ρ | LFC | MFC | LT | MT | TRO | PAT | Average |
| Site 1 | 49.02±4.65 | 43.81±4.79 | 46.86±2.55 | 42.04±6.79 | 47.06±4.59 | 44.78±2.45 | |
| 1.27 | 2.78 | 2.70 | 5.09 | 1.56 | 4.15 | 2.93 | |
| Site 2 | 57.75±2.31 | 52.35±5 | 59.35±1.02 | 53.67±3.04 | 63.08±4.05 | 62.7±5.22 | |
| 1.13 | 0.92 | 0.82 | 1.73 | 2.59 | 2.40 | 1.60 | |
| Site 3 | 44.92±4.33 | 41.74±3.21 | 43.32±3.98 | 38.45±5.18 | 45.48±4.72 | 42.81±5 | |
| 2.44 | 2.42 | 2.62 | 0.63 | 3.45 | 0.91 | 2.08 | |
| Site 4 | 41.88±2.04 | 38.5±3.44 | 40.5±2.44 | 37.14±4.3 | 45.07±3.75 | 44.47±4.83 | |
| 2.32 | 4.87 | 2.87 | 2.75 | 4.47 | 6.32 | 3.93 | |
| T2 | LFC | MFC | LT | MT | TRO | PAT | Average |
| Site 1 | 38.88±4.85 | 35.04±6.43 | 34.77±4.11 | 32.59±6.65 | 32.7±6.95 | 28.95±5.95 | |
| 1.14 | 3.74 | 2.36 | 6.13 | 4.03 | 7.06 | 4.08 | |
| Site 2 | 52.3±2.95 | 47.35±4.81 | 54.91±4.17 | 47.66±4.06 | 54.06±3.1 | 51.36±6.25 | |
| 0.93 | 1.26 | 1.48 | 0.77 | 2.21 | 1.99 | 1.44 | |
| Site 3 | 35.14±3.87 | 32.49±3.81 | 33.13±3.69 | 29.44±5.19 | 32.38±2.92 | 30.5±3.3 | |
| 3.01 | 4.13 | 2.35 | 1.87 | 2.04 | 2.03 | 2.57 | |
| Site 4 | 36.67±3.34 | 31.93±4.02 | 34.18±3.06 | 30.37±5.75 | 37.3±1.86 | 34.84±4.36 | |
| 2.50 | 4.27 | 0.83 | 1.59 | 2.38 | 1.59 | 2.20 | |
| b) Inter-site CV (%) calculated for volunteer T1ρ and T2 measurements | |||||||
| T1ρ | LFC | MFC | LT | MT | TRO | PAT | Average |
| All | 9.87 | 12.35 | 12.74 | 11.82 | 17.12 | 17.78 | 13.61 |
| MAPSS | 6.36 | 8.79 | 7.33 | 7.33 | 9.17 | 9.85 | 8.14 |
| T2 | LFC | MFC | LT | MT | TRO | PAT | Average |
| All | 15.49 | 18.35 | 19.68 | 18.54 | 23.15 | 26.84 | 20.34 |
| MAPSS | 5.96 | 8.37 | 9.03 | 9.54 | 10.56 | 16.87 | 10.06 |
| c) SNR efficiency of first and last echo of volunteer measurements | |||||||
| Site 1 | Site 2 | Site 3 | Site 4 | ||||
| First echo | 33.82±4.74 | 31.92±3.23 | 23.81 ±4.99 | 33.83±5.55 | |||
| Last echo (T1ρ) | 7.25±0.81 | 10.24±1.29 | 4.88±0.64 | 5.16±0.87 | |||
| Last echo (T2) | 6.53±1.52 | 10.51 ±0.87 | 6.46±0.91 | 5.89±0.84 | |||
All: using data from all four sites. MAPSS: using data from sites 1,3,4 that were collected with the MAPSS sequence.
Figure 4.
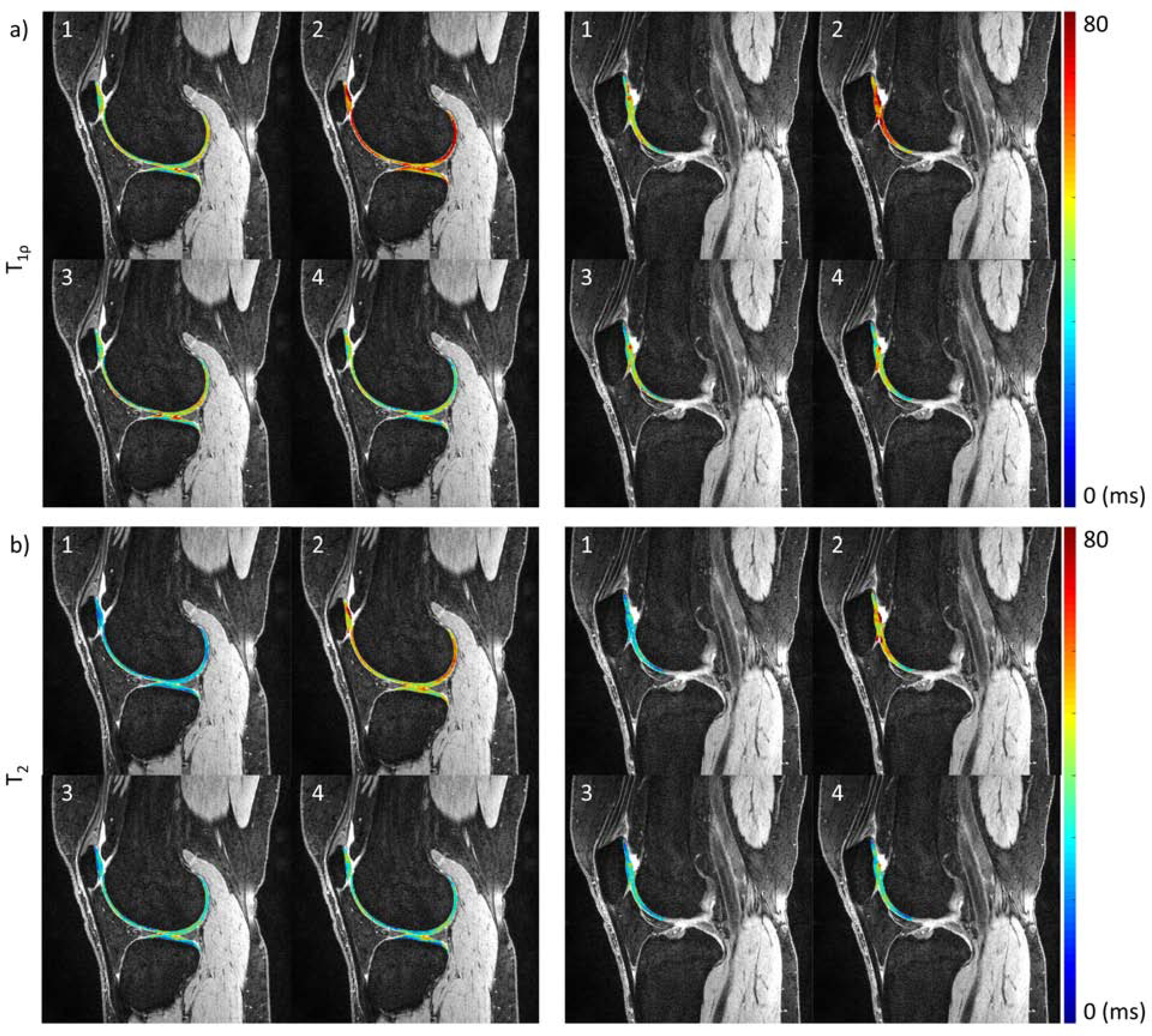
Example volunteer a) T1ρ and b) T2 relaxation time maps from all sites, overlaid on DESS image from Site 1. The columns show different slice positions. The number on the top left of the images indicates the site.
shows the Bland-Altman analysis, ICCs for intra-site repeatability, and CCCs for inter-site reproducibility. All intra-site ICCs showed high correlation, except for T1ρ at Site 4, where two subjects showed more than 10ms difference between scan/rescan for the patellar and trochlear cartilages. The relaxation times had low correlation between sites. Relaxation times from Site 2 were higher than at other sites: 14.88ms (95% CI [3.48, 26.27]) for T1ρ and 19.02ms (95% CI [8.16, 29.88]) for T2.
The SNR efficiency for volunteers ranged across sites from 24 to 34 (Table 3c). The CVs between the volunteer relaxation times fitted with 8 echoes and the two sets of 4 echoes were less than 3%.
DISCUSSION
To successfully use cartilage of T1ρ and T2 values in large-scale studies and clinical trials, accuracy and stability of these measures across sites and equipment needs to be ensured. In this study, we implemented 3D T1ρ and T2 imaging based on the same sequence structure (MAPSS) on MR platforms from three vendors (Siemens, GE, Philips), and analyzed the repeatability and reproducibility of the relaxation measurements in a multi-site, multi-vendor approach.
In our single-site phantom analysis, 3D MAPSS showed robust changes in parameters, including with or without parallel imaging, time of recovery, and views-per-segment. Compared with using the constant flip angle, MAPSS sequence design (magnetization reset, RF cycling and variable flip angle train) helped mitigate the effect of T1 recovery and provided stability to views-per-segment.24 The choice of preparation TEs did not influence the T2 measures, showing minimal variation (CV=1.02%), thus ensuring that the different corrected TEs on different MR systems would have no significant effects on T2 measurement.
The acquisition time for 8-echo T1ρ or T2 imaging for the whole knee with 8-echo was longer than 10 minutes, which increased the risk of motion during the scan and thus was not ideal for clinical implementation. Since our fitting results from 8 echoes and 4 echoes for T1ρ and T2 in both phantoms and volunteers showed little differences (<1% in phantoms and <3% in volunteers), clinical applications using mono-exponential fitting of only 4 echoes may provide reliable measures with shorter imaging time. Using 4 echoes, the acquisition time would be reduced to 7 minutes. Furthermore, this may be combined with fast T1ρ and T2 imaging based on novel techniques such as compressed sensing or deep-learning reconstruction that are being developed,33–35 which will greatly facilitate clinical translation of T1ρ and T2 imaging.
Our intra-site phantom and volunteer measurements showed good repeatability at all four sites with low intra-site CVs (CV<3.5% for phantoms and <5% for volunteers), with results comparable to previous single-site studies11, 18, 19, indicating successful implementation on all MR platforms.
Our inter-site CVs were higher than single-site repeatability as summarized by Mackay et al8, and higher than previous single-vendor multi-site studies reports (average CV of 4.9% for T1ρ and 4.4% for T218 and 3.3–6.5% for T217). The factors to be addressed for reduction of inter-vendor inter-site variability can be categorized by their sources: hardware (MRI systems, RF coils), software (acquisition sequences and reconstruction and post-processing software), and environmental factors.
The most significant inter-site differences observed during this study were caused by different sequence structures. When we limited our analysis to sites using the same sequence structure (MAPPS), i.e, we excluded data from the one site using a different pulse sequence structure (Site 2), the overall inter-site CVs decreased from 14% to 8% and from 20% to 10% for T1ρ and T2, respectively, confirming the dependency of relaxation times on sequences reported in previous studies.20–22 The difference between the two sequences was more pronounced for T2 since the acquisitions had greater variation in the T2 preparation scheme as compared to the T1ρ, preparation, in addition to the different read-out structures. 3D MAPSS utilized a train of refocusing pulses with phase modulation (MLEV) for T2 preparation and TE correction.29 On the other hand, the segmented GRE acquisition used T2 preparation with single refocusing pulse. Another factor for slightly lower CVs for T1ρ compared to T2 could be less magic angle effect in T1ρ imaging, due to spin-lock pulses.36, 37
Another potential source of variability is inter-site scanner difference (including B0 inhomogeneity profiles) due to different loading inside the scanner and different shimming algorithms employed by each vendor. The RF coils used in this study also had different structures, which will introduce different B1 profiles. Although the MAPSS sequence is designed to be robust to B0 and B1 inhomogeneity with composite tip-down/tip-up pulses, phase shift in the middle of spin-lock pulses, and RF cycling,11, 24, 38 residual artifacts could have caused higher inter-site variability, especially for volunteer imaging where inhomogeneity issues are greater.
In addition, different coils may result in different SNR, which can potentially introduce variations in relaxation time fitting, as consistent with previous reports,17, 18 especially for the volunteer imaging with relatively low SNR of last echo images.
To prevent variations that can be introduced by different fitting algorithms, centralized processing was performed at a single site.39, 40 However, the differences in DICOM image reconstruction between vendors (different filtering and regularization during reconstruction) should be noted. Although the retrieval, storage, and transmission of raw data, including complex data, is challenging, these inter-vendor variations in image reconstruction indicate a potential need to collect the raw k-space data for centralized, uniform, single-algorithm image reconstruction across MR systems and sites in future studies.
Environmental factors, such as phantom temperature at the time of measurement, may explain some of the variability in our phantom measurement results, as the site with the lowest temperature had longer relaxation times. Potential differences in knee positioning of traveling volunteers at each site may have added to inter-site variation. Also, even though all images of traveling volunteers were visually checked for registration success, high inter-site CVs were observed for patellar and trochlear cartilage, the areas that are most challenging to register given differences in knee rotation. These regions are also highly susceptible to magic angle artefact due to their structural orientations.
The phantom measurements across sites were highly correlated, and with the high correlation of T1ρ and T2 values between sites, a calibration model could be built for pool analysis between sites. However, large data sets for both phantoms and traveling volunteers need to be collected before a calibration model can be developed. Our current human cartilage relaxation times were uncorrelated, but this could be due to the limited number of volunteer data points and the smaller range of relaxation times compared with the phantom (since the data was collected with healthy volunteers only).
When measuring treatment effect during clinical trials, it is important to consider the measurement imprecision that is due to both between- and within-subject (i.e., test-retest measurement error) variability. Obuchowski et al41 determined how to estimate the necessary sample size for a clinical trial using a quantitative imaging biomarker with known measurement error. For example, cross-sectional studies comparing controls and osteoarthritis patients report mean T1p and T2 values for 20 subjects.16 Using this methodology, an intra-site measurement error of 4% would increase the sample size of such a clinical trial by 1–2 subjects, whereas an inter-site measurement error of 10% would increase the sample size by 9 subjects. For a longitudinal study of ACL-injured knees reporting one-year changes in injured and control knees of 40 patients and 15 controls,27 factoring in these intra-site and inter-site measurement errors would increase the sample size by 5 and 12 subjects, respectively. Thus, the results from our study suggest that T1ρ and T2 imaging with harmonized sequences and protocols and centralized post-processing are promising quantitative imaging biomarkers for future OA clinical trials.
Despite the promising results, there are several limitations to this study. The phantoms were not scanned before they were distributed to different sites. However, the four phantoms used in this study were manufactured from one batch. Furthermore, we analyzed T1p and T2 of three other identical phantoms that were manufactured from the same batch, but not used in this study at Site 1. The CVs of T1p and T2 values among these three phantoms and the one used in this study at Site 1 were approximately 1.2% and 1.44%, respectively, suggesting minimal inter-phantom variations. The pre-scan activities of traveling volunteers were not standardized and the scans were not performed at the same time of day, which may introduce diurnal variations.11 However, all volunteers sat for 30 minutes before the scan to minimize potential effects from loading status differences. B0 and B1 corrections were not performed, which may have helped mitigate the inter-site variation. Lastly, the study was limited by the small sample size, and no patients with diagnosed cartilage degeneration were studied.
In conclusion, 3D T1ρ and T2 imaging at 3T with the same sequence structure (MAPSS) has been developed on three major MR platforms, showing promising reproducibility with controlled sequence structure and parameters, and centralized post-processing methods. Additionally, our results indicate that the required scan times can be reduced when using four echoes, rather than eight, without loss of data reliability, making T1ρ and T2 mapping more feasible and cost-effective for clinical practice. Larger-scale studies with controls and OA patients are warranted to further develop ways to mitigate inter-site and inter-vendor variation of cartilage relaxation time. Strategies for measurement calibration between sites and vendors are required in order to facilitate the application of the quantitative measures for multi-site multi-vendor clinical trials.
Figure 5.
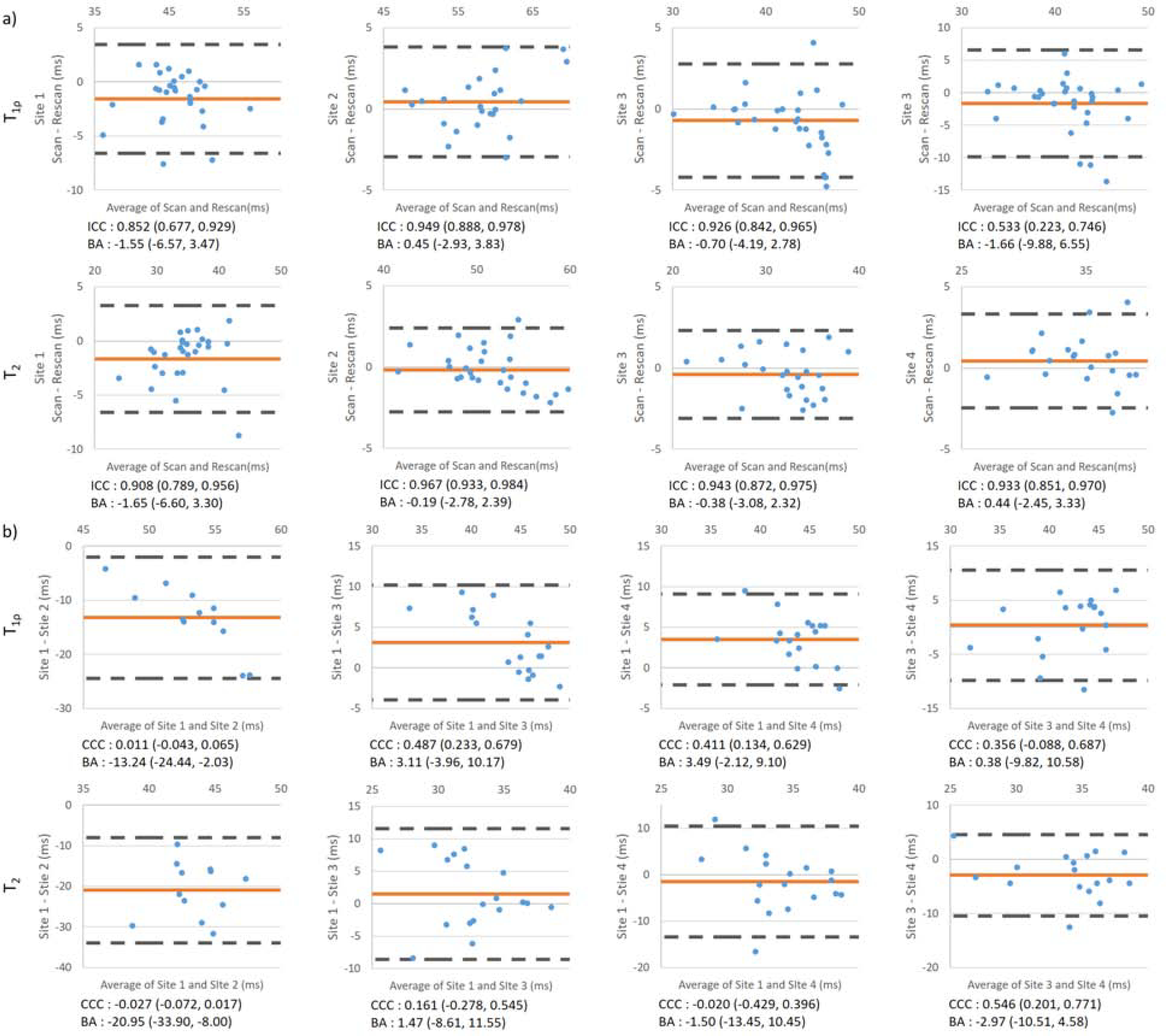
Bland-Altman analysis for (a) intra-site and (b) inter-site T1ρ and T2. Red line indicates the average of the difference value; dashed black line indicates the upper and lower limit of agreement (LOA). ICCs for intra-site repeatability and CCCs for inter-site reproducibility are listed below each plot. Values in the brackets are 95% confidence intervals. Mean difference and 95% limit of agreement were also listed.
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Ari Borthakur for sharing Segmented GRE T1ρ and T2 imaging sequences.
ROLE OF THE FUNDING SOURCE
The study was supported by the Arthritis Foundation. Study sponsors had no role in the design and conduct of the study; collection, management, analysis and interpretation of data; and preparation, review or approval of manuscript.
Footnotes
COMPETING INTERESTS
The authors declare no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kraus VB, Blanco FJ, Englund M, Karsdal MA, Lohmander LS. Call for standardized definitions of osteoarthritis and risk stratification for clinical trials and clinical use. Osteoarthritis and cartilage. 2015;23(8):1233–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cisternas MG, Murphy L, Sacks JJ, Solomon DH, Pasta DJ, Helmick CG. Alternative Methods for Defining Osteoarthritis and the Impact on Estimating Prevalence in a US Population-Based Survey. Arthritis care & research. 2016;68(5):574–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karsdal MA, Michaelis M, Ladel C, Siebuhr AS, Bihlet AR, Andersen JR, et al. Disease-modifying treatments for osteoarthritis (DMOADs) of the knee and hip: lessons learned from failures and opportunities for the future. Osteoarthritis and cartilage. 2016;24(12):2013–21 [DOI] [PubMed] [Google Scholar]
- 4.Oo WM, Yu SP, Daniel MS, Hunter DJ. Disease-modifying drugs in osteoarthritis: current understanding and future therapeutics. Expert opinion on emerging drugs. 2018;23(4):331–47 [DOI] [PubMed] [Google Scholar]
- 5.Guermazi A, Alizai H, Crema MD, Trattnig S, Regatte RR, Roemer FW. Compositional MRI techniques for evaluation of cartilage degeneration in osteoarthritis. Osteoarthritis and cartilage. 2015;23(10):1639–53 [DOI] [PubMed] [Google Scholar]
- 6.Link TM, Neumann J, Li X. Prestructural cartilage assessment using MRI. Journal of magnetic resonance imaging : JMRI. 2017;45(4):949–65 [DOI] [PubMed] [Google Scholar]
- 7.Atkinson HF, Birmingham TB, Moyer RF, Yacoub D, Kanko LE, Bryant DM, et al. MRI T2 and T1rho relaxation in patients at risk for knee osteoarthritis: a systematic review and meta-analysis. BMC musculoskeletal disorders. 2019;20(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.MacKay JW, Low SBL, Smith TO, Toms AP, McCaskie AW, Gilbert FJ. Systematic review and meta-analysis of the reliability and discriminative validity of cartilage compositional MRI in knee osteoarthritis. Osteoarthritis and cartilage. 2018;26(9): 1140–52 [DOI] [PubMed] [Google Scholar]
- 9.Joseph GB, Baum T, Alizai H, Carballido-Gamio J, Nardo L, Virayavanich W, et al. Baseline mean and heterogeneity of MR cartilage T2 are associated with morphologic degeneration of cartilage, meniscus, and bone marrow over 3 years--data from the Osteoarthritis Initiative. Osteoarthritis and cartilage. 2012;20(7):727–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prasad AP, Nardo L, Schooler J, Joseph GB, Link TM. T(1)rho and T(2) relaxation times predict progression of knee osteoarthritis. Osteoarthritis and cartilage. 2013;21(1):69–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Wyatt C, Rivoire J, Han E, Chen W, Schooler J, et al. Simultaneous acquisition of T1rho and T2 quantification in knee cartilage: repeatability and diurnal variation. Journal of magnetic resonance imaging : JMRI. 2014;39(5):1287–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carballido-Gamio J, Link TM, Majumdar S. New techniques for cartilage magnetic resonance imaging relaxation time analysis: texture analysis of flattened cartilage and localized intra- and inter-subject comparisons. Magnetic resonance in medicine. 2008;59(6):1472–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jordan CD, McWalter EJ, Monu UD, Watkins RD, Chen W, Bangerter NK, et al. Variability of CubeQuant T1rho, quantitative DESS T2, and cones sodium MRI in knee cartilage. Osteoarthritis and cartilage. 2014;22(10):1559–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedoia V, Li X, Su F, Calixto N, Majumdar S. Fully automatic analysis of the knee articular cartilage T1rho relaxation time using voxel-based relaxometry. Journal of magnetic resonance imaging : JMRI. 2016;43(4):970–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welsch GH, Apprich S, Zbyn S, Mamisch TC, Mlynarik V, Scheffler K, et al. Biochemical (T2, T2* and magnetisation transfer ratio) MRI of knee cartilage: feasibility at ultra-high field (7T) compared with high field (3T) strength. European radiology. 2011;21(6):1136–43 [DOI] [PubMed] [Google Scholar]
- 16.Zuo J, Li X, Banerjee S, Han E, Majumdar S. Parallel imaging of knee cartilage at 3 Tesla. Journal of magnetic resonance imaging : JMRI. 2007;26(4):1001–9 [DOI] [PubMed] [Google Scholar]
- 17.Dardzinski BJ, Schneider E. Radiofrequency (RF) coil impacts the value and reproducibility of cartilage spin-spin (T2) relaxation time measurements. Osteoarthritis and cartilage. 2013;21(5):710–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Pedoia V, Kumar D, Rivoire J, Wyatt C, Lansdown D, et al. Cartilage T1rho and T2 relaxation times: longitudinal reproducibility and variations using different coils, MR systems and sites. Osteoarthritis and cartilage. 2015;23(12):2214–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balamoody S, Williams TG, Wolstenholme C, Waterton JC, Bowes M, Hodgson R, et al. Magnetic resonance transverse relaxation time T2 of knee cartilage in osteoarthritis at 3-T: a cross-sectional multicentre, multivendor reproducibility study. Skeletal Radiol. 2013;42(4):511–20 [DOI] [PubMed] [Google Scholar]
- 20.Pai A, Li X, Majumdar S. A comparative study at 3 T of sequence dependence of T2 quantitation in the knee. Magnetic resonance imaging. 2008;26(9):1215–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matzat SJ, McWalter EJ, Kogan F, Chen W, Gold GE. T2 Relaxation time quantitation differs between pulse sequences in articular cartilage. Journal of magnetic resonance imaging : JMRI. 2015;42(1):105–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka MS, Pedoia V, Chandramohan D, Chen W, Li X. Comparison of T1ρ quantification, SNR, and reproducibility in 3D Magnetization-Prepared Angle-Modulated Partitioned k-space Spoiled Gradient Echo Snapshots (3D MAPSS) and 3D Fast Spin Echo (CUBEQuant) techniques. Annual conference of International Society of Magnetic Resonance in Medicine (ISMRM); Apr 22–27; Honolulu, USA 2017. [Google Scholar]
- 23.Hennig J, Weigel M, Scheffler K. Multiecho sequences with variable refocusing flip angles: optimization of signal behavior using smooth transitions between pseudo steady states (TRAPS). Magnetic resonance in medicine. 2003;49(3):527–35 [DOI] [PubMed] [Google Scholar]
- 24.Li X, Han ET, Busse RF, Majumdar S. In vivo T(1rho) mapping in cartilage using 3D magnetization-prepared angle-modulated partitioned k-space spoiled gradient echo snapshots (3D MAPSS). Magnetic resonance in medicine. 2008;59(2):298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saxena V, D’Aquilla K, Marcoon S, Krishnamoorthy G, Gordon JA, Carey JL, et al. T1rho Magnetic Resonance Imaging to Assess Cartilage Damage After Primary Shoulder Dislocation. Am J Sports Med. 2016;44(11):2800–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goto H, Iwama Y, Fujii M, Aoyama N, Kubo S, Kuroda R, et al. A preliminary study of the T1rho values of normal knee cartilage using 3T-MRI. European journal of radiology. 2012;81(7):e796–803 [DOI] [PubMed] [Google Scholar]
- 27.Russell C, Pedoia V, Majumdar S, Consortium A-A. Composite metric R2 - R1rho (1/T2 – 1/T1rho ) as a potential MR imaging biomarker associated with changes in pain after ACL reconstruction: A six-month follow-up. J Orthop Res. 2017;35(3):718–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charagundla SR, Borthakur A, Leigh JS, Reddy R. Artifacts in T(1rho)-weighted imaging: correction with a self-compensating spin-locking pulse. Journal of magnetic resonance (San Diego, Calif : 1997). 2003;162(1):113–21 [DOI] [PubMed] [Google Scholar]
- 29.Foltz WD, Stainsby JA, Wright GA. T2 accuracy on a whole-body imager. Magnetic resonance in medicine. 1997;38(5):759–68 [DOI] [PubMed] [Google Scholar]
- 30.Dixon WT, Oshinski JN, Trudeau JD, Arnold BC, Pettigrew RI. Myocardial suppression in vivo by spin locking with composite pulses. Magnetic resonance in medicine. 1996;36(1):90–4 [DOI] [PubMed] [Google Scholar]
- 31.Klein S, Staring M, Murphy K, Viergever MA, Pluim JP elastix: a toolbox for intensity-based medical image registration. IEEE transactions on medical imaging. 2010;29(1):196–205 [DOI] [PubMed] [Google Scholar]
- 32.Avants BB, Tustison N, Song G. Advanced normalization tools (ANTS). Insight j. 2009;2:1–35 [Google Scholar]
- 33.Zibetti MVW, Sharafi A, Otazo R, Regatte RR. Compressed sensing acceleration of biexponential 3D-T1rho relaxation mapping of knee cartilage. Magnetic resonance in medicine. 2019;81(2):863–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y, Liu Y, Ying L, Peng X, Wang YJ, Yuan J, et al. SCOPE: signal compensation for low-rank plus sparse matrix decomposition for fast parameter mapping. Physics in medicine and biology. 2018;63(18):185009. [DOI] [PubMed] [Google Scholar]
- 35.Cai C, Wang C, Zeng Y, Cai S, Liang D, Wu Y, et al. Single-shot T2 mapping using overlapping-echo detachment planar imaging and a deep convolutional neural network. Magnetic resonance in medicine. 2018;80(5):2202–14 [DOI] [PubMed] [Google Scholar]
- 36.Akella SV, Regatte RR, Wheaton AJ, Borthakur A, Reddy R. Reduction of residual dipolar interaction in cartilage by spin-lock technique. Magnetic resonance in medicine. 2004;52(5):1103–9 [DOI] [PubMed] [Google Scholar]
- 37.Wang N, Xia Y Experimental issues in the measurement of multi-component relaxation times in articular cartilage by microscopic MRI. Journal of magnetic resonance (San Diego, Calif : 1997). 2013;235:15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W Errors in quantitative T1rho imaging and the correction methods. Quantitative imaging in medicine and surgery. 2015;5(4):583–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raya JG, Dietrich O, Horng A, Weber J, Reiser MF, Glaser C. T2 measurement in articular cartilage: impact of the fitting method on accuracy and precision at low SNR. Magnetic resonance in medicine. 2010;63(1):181–93 [DOI] [PubMed] [Google Scholar]
- 40.Koff MF, Amrami KK, Felmlee JP, Kaufman KR. Bias of cartilage T2 values related to method of calculation. Magnetic resonance imaging. 2008;26(9):1236–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Obuchowski NA, Mozley PD, Matthews D, Buckler A, Bullen J, Jackson E. Statistical Considerations for Planning Clinical Trials with Quantitative Imaging Biomarkers. Journal of the National Cancer Institute. 2019;111(1):19–26 [DOI] [PubMed] [Google Scholar]


