Abstract
Objective
Non-alcoholic fatty liver disease (NAFLD) is a silent pandemic associated with obesity and the metabolic syndrome, and also increases cardiovascular- and cirrhosis-related morbidity and mortality. A complete understanding of adaptive compensatory metabolic programmes that modulate non-alcoholic steatohepatitis (NASH) progression is lacking.
Methods and results
Transcriptomic analysis of liver biopsies in patients with NASH revealed that NASH progression is associated with rewiring of metabolic pathways, including upregulation of de novo lipid/cholesterol synthesis and fatty acid remodelling. The modulation of these metabolic programmes was achieved by activating sterol regulatory element-binding protein (SREBP) transcriptional networks; however, it is still debated whether, in the context of NASH, activation of SREBPs acts as a pathogenic driver of lipotoxicity, or rather promotes the biosynthesis of protective lipids that buffer excessive lipid accumulation, preventing inflammation and fibrosis. To elucidate the pathophysiological role of SCAP/SREBP in NASH and wound-healing response, we used an Insig1 deficient (with hyper-efficient SREBPs) murine model challenged with a NASH-inducing diet. Despite enhanced lipid and cholesterol biosynthesis, Insig1 KO mice had similar systemic metabolism and insulin sensitivity to Het/WT littermates. Moreover, activating SREBPs resulted in remodelling the lipidome, decreased hepatocellular damage, and improved wound-healing responses.
Conclusions
Our study provides actionable knowledge about the pathways and mechanisms involved in NAFLD pathogenesis, which may prove useful for developing new therapeutic strategies. Our results also suggest that the SCAP/SREBP/INSIG1 trio governs transcriptional programmes aimed at protecting the liver from lipotoxic insults in NASH.
Keywords: Non-alcoholic fatty liver disease (NAFLD), De novo lipogenesis (DNL), Lipid remodelling, Western diet, Carbon tetrachloride (CCl4), Liver regeneration
Highlights
-
•
Human NASH biopsies’ transcriptomics analysis features metabolic pathway rewiring.
-
•
SCAP/SREBP/INSIG1 modulation promotes lipid/cholesterol synthesis/remodelling in NASH.
-
•
Loss of Insig1 promotes lipid remodelling, preventing hepatic lipotoxicity in NASH.
-
•
Loss of Insig1 improves liver damage and wound healing and restrains NASH progression.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) encompasses a spectrum of liver disease affecting up to one-third of Western populations [1], characterised by hepatic steatosis, leading to lipotoxicity, inflammation (non-alcoholic steatohepatitis, NASH), and fibrosis, potentially culminating in advanced liver disease [2] and conferring increased cardiovascular risk/mortality [3]. NAFLD is associated with obesity and/or adipose tissue dysfunction, insulin resistance (IR), and multiple features of the metabolic syndrome [4]. Moreover, the exponential increase in obesity and NAFLD means this condition is presently responsible for an enormous economic and health burden [5].
Despite recent advances in understanding NASH pathophysiology, important information is lacking regarding specific factors driving NAFL development and the mechanistic links between systemic metabolic stress and disease progression [6]. An increase in de novo lipogenesis (DNL) and lipid remodelling has been reported in multiple studies of subjects with NAFLD [[7], [8], [9]] and shown to be a significant factor in NASH pathophysiology [10]. Dysregulation of DNL and cholesterol synthesis in liver metabolism is a feature of NASH [11,12] and fibrogenesis [13].
In our cohort of biopsy-proven NASH patients, transcriptomic analysis suggested that the transcriptional complex of sterol regulatory element-binding protein 1 and 2 (SREBP1/SREBF1 and SREBP2/SREBF2) and SREBP cleavage-activating protein (SCAP) were strongly activated; this was associated with enhanced metabolic programmes governing cholesterol/lipid synthesis and remodelling. However, observational “omics” were unable to explain 1) whether these transcriptional regulators induced a maladaptive response or their activation might represent a protective mechanism aimed at maintaining the liver's functionality when metabolically challenged by an excess nutrient/calorie load, and 2) the consequences, in terms of compensatory mechanisms, of activating these pathways on connected metabolic programmes (for example, synthesis of complex lipids, lipid catabolism, and lipoprotein handling) [14].
To study the biological consequences of these processes, we used a murine model characterised by unrestrained SREBP1 and SREBP2 activity resulting from the absence of insulin-induced gene 1 (INSIG1) along with a NASH-inducing diet. INSIG1 is part of negative regulatory feedback, acting as a brake on SREBP transcriptional function [[15], [16], [17]]. INSIG1 also has a secondary function mediating sterol-accelerated proteolytic degradation of HMG CoA reductase, thus impacting the mevalonate pathway [18]. In this report, we show that, in a NASH-like metabolic context with acute oxidative damage and hyper-efficient SREBP activation, despite the collateral cost of increased hepatic lipid deposition, INSIG1 deficiency leads to a beneficial outcome in NASH driven by an increase in desaturase activity and lipid remodelling as well as a multifaceted regulation of multiple metabolic pathways preventing hepatic lipotoxicity.
2. Methods
2.1. Study population (patients with NAFLD)
Our BioNASH cohort consisted of 58 consecutive patients recruited by the NAFLD service at Cambridge University Hospital's NHS Foundation Trust. This study was approved by the Research Ethics Committee (ref: 06/Q0106/70). All of the patients provided informed consent for the use of data (biochemistry and clinical history) and samples for research purposes; the principles of the Declaration of Helsinki were followed. All of the patients had a clinical diagnosis of NAFLD (with alternate diagnoses and aetiologies excluded), histology scored by an expert pathologist according to the NASH Clinical Research Network scoring system [19,20], and snap-frozen tissue for research purposes. For this study, histology was grouped into NAFL (fatty liver without inflammation and/or ballooning) and NASH (fatty liver with inflammation and ballooning) [21] with fibrosis scores of 1–2 (moderate) or 3–4 (advanced). The study population is described in Table S1.
2.2. Animals
All of the data were from male mice. Insig1 KO mice were generated by Cre-mediated deletion of three Insig1 exons (exons 2–4) as previously described [15] on a C57Bl6/J background and compared to wild-type and heterozygous littermates. The mice were maintained in a room with a controlled temperature (21 °C; 12 h light/dark cycle) with free access to food and water (ad libitum); the pathogen-free facilities were compliant with FELASA guidelines [22]. The UK Home Office and USC Bioethics Committee of the Universities of Cambridge and Newcastle approved all of the animal procedures. The mice were fed a chow diet (Safe Diets, Code ds-105) until they were enrolled in specific procedures.
2.3. Western diet and sugar water studies
At the age of 8 weeks, the mice were fed either low-fat control (Teklad, TD08485) or Western diets (Teklad, TD88137) plus drinking water supplemented with fructose and glucose (SW, 23.1 g/L of d-fructose +18.9 g/L of d-glucose) ad libitum starting from week 8; the total duration of the experimental challenge was 12 weeks. Fat/lean mass was calculated by time-domain nuclear magnetic resonance (TD-NMR) using a Minispec LF50 Live Mice Analyser (Bruker) the day before culling. Serum and tissue were collected in a fed condition the day of the culling.
2.4. Carbon tetrachloride studies
The 12-week-old Insig1 KO, Het, or WT mice underwent intraperitoneal (IP) injection of 2 μl/BW (g) of CCl4: virgin olive oil [1:1 (v/v)] mix or virgin olive oil alone. Fed mice were humanely culled under terminal isoflurane anaesthesia four days after the IP injection by cutting the heart and inferior vena cava. Blood was collected from the chest cavity. These experiments were approved by the Animal Welfare and Ethical Review Board and carried out at Newcastle University Comparative Biology Centre under a UK Home office license.
2.5. Data availability
All of the raw data (FASTQ files) and relevant metadata are available in the Array Express database (www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-9815 and E-MTAB-9864.
2.5.1. Additional methods provided in supplementary files
Biochemical serum analyses, murine tissue collection and processing, histology, immunohistochemistry, tissue imaging, quantification, and scoring, RNA extraction and RNA integrity, reverse transcription (RT) polymerase chain reaction (PCR), whole-transcriptome amplification and RNA sequencing, hepatic lipid extraction and lipidomics, quantification and statistical analysis, NGS data processing, and bioinformatics functional analyses.
3. Results
3.1. Progressive human NASH was characterised by increasing lipid and cholesterol biosynthesis transcriptional programmes
We first recruited 58 patients with biopsy-proven NAFLD clustered into three groups (NAFL, moderate NASH, and advanced NASH) according to the NASH Clinical Research Network's scoring system [[19], [20], [21]]. In addition to the expected differences in the histology, we also found a progressive increase in body mass index (BMI) and worsening systemic insulin resistance (HOMA-IR) in NASH (Table S1). No other significant changes among the groups were recorded with regards to metabolic biochemistry and transaminases.
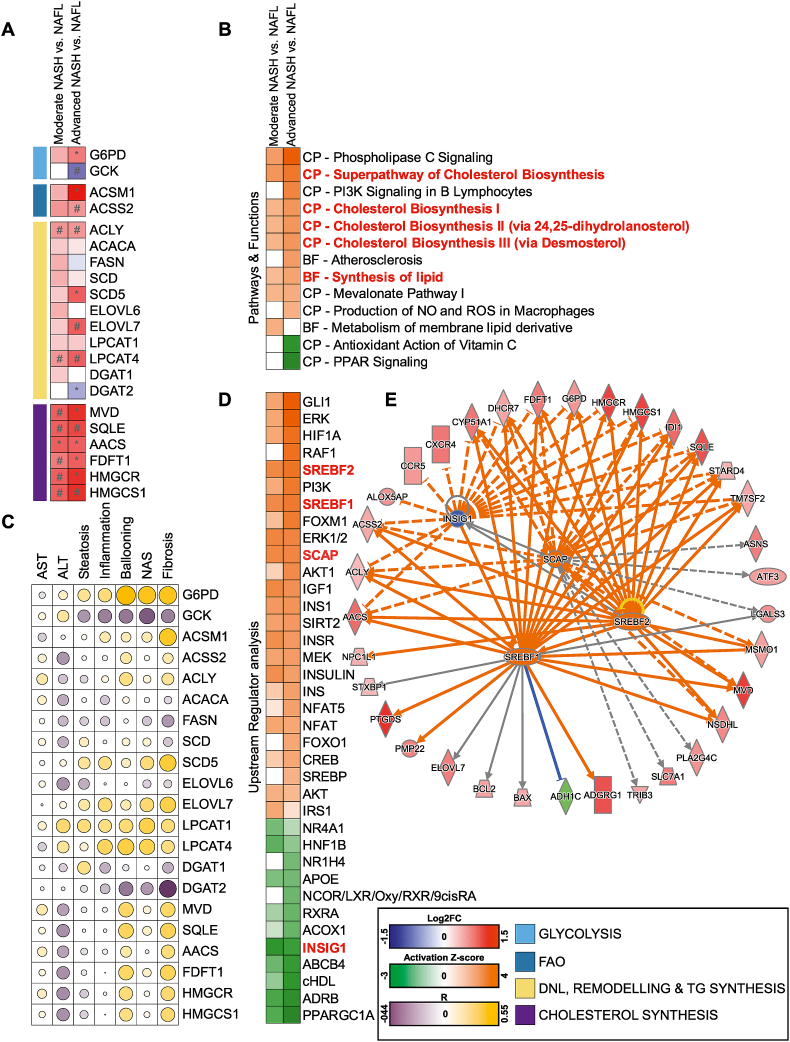
Using next-generation sequencing (NGS), we studied transcriptomic changes associated with NASH progression. We first focused on changes in the transcripts of individual genes involved in metabolic programmes and specific mechanisms of disease. We validated the upregulation of known drivers of disease progression involved in apoptosis, inflammation, hepatic stellate cell (HSC) activation and fibrosis, and compensatory hepatocyte proliferation (Figure S1A). Of relevance, we also found that the progression of NASH was associated with a coordinated and robust rewiring of metabolic pathways. Specifically, we found the upregulation of many transcripts involved in DNL, fatty acid remodelling, and triglyceride and cholesterol biosynthesis (Figure 1A) following the same trend of modulation observed for hyperinsulinaemia. The activation of these lipid metabolic transcriptional programmes with disease progression was further confirmed by the canonical pathway (CP) and biological functions (BF) of the Ingenuity Pathway Analysis (IPA; Figure 1B; a detailed list of significant hits is in Table S2). Many of these lipid-related genes correlated with hallmarks of metabolic impairment (Figure S1B), with the expression of critical drivers of disease progression (Figure S1C), elevated transaminases, and NASH histological features including steatosis, ballooning degeneration (cell death), inflammation, and fibrosis (Figure 1C).
Figure 1.
NGS analysis predicts a strong regulation of metabolic pathways in biopsy-proven NASH clustered against the disease stage. (A) Gene expression profiled by NGS and analysed with the Wald test (sample size defined in Table S1; #p < 0.05 and ∗p BH < 0.05) of key metabolic enzymes involved in glucose and lipid metabolism. (B) Selection of canonical pathways (CP) and bio-function (BF) predicted to be significantly (p < 0.05) enriched and activated (orange) or inhibited (green) according to the Ingenuity Pathway Analysis (IPA; −2 < Z score >2 in at least one comparison); expanded list in Table S2. (C) Correlation of genes with hallmarks of NASH progression (histology and transaminases). (D) Selection of upstream regulators predicted to be significantly (p < 0.05) enriched and activated (orange) or inhibited (green) according to the IPA (−2 < Z score >2 in at least one comparison); expanded list in Table S2. (E) Graphical representation in network format of SCAP/SREBF1 and 2 and INSIG1 transcriptional activity based on the regulation of their targets was differentially modulated (green: downregulated; red: upregulated), leading to the predicted inhibition (blue)/activation (orange). Data are shown in a heat map matrix format; details are in the graphical legend. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
To better understand the network of transcriptional regulators that might govern the metabolic adaptation to disease progression, we further refined the NGS analysis with an IPA “upstream regulator” analysis (URA), which predicts the degree of activation of transcription factors and signalling regulators accounting for changes in their downstream targets (Figure 1D; details in Table S2). The URA confirmed a strong activation of multiple downstream effectors of insulin signalling. Activation of insulin signalling was coupled with suppression of multiple lipid-sensing nuclear receptors and their coactivators, including farnesoid X receptor (FXR/NR1H4), liver X receptor (LXR), retinoid X receptor alpha (RXRα), and peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC1α/PPARGC1A) (Figure 1D). Of note, some of these hits are currently considered candidate targets for the pharmacological treatment of NASH [14].
Among the differentially activated upstream regulators, IPA predicted the activation of the two master transcription factors controlling DNL and cholesterol synthesis namely SREBP1/SREBF1, SREBP2/SREBF2, and their interacting partner SCAP; as well as suppressed activity of INSIG1 (Figure 1D,E), a target gene of SREBP1 and an inhibitor of SREBP1/2, functioning as a crucial negative feedback loop controlling SREBP1/2 cleavage and activity [15,17,23,24].
Altogether, these data suggested that the co-regulation of INSIG/SREBPs is tightly associated with the pathophysiology of NASH progression. The pathobiological consequences of DNL activation in a NASH context are under debate and yet to be fully understood. One view is that activation of SREBPs drives lipotoxicity [13,[25], [26], [27]]; an alternative view is that SREBPs also co-regulate desaturases and other lipid remodelling enzymes, potentially protecting from insulin resistance and lipotoxicity [28,29]. To determine whether SREBPs are protective or deleterious, we used an Insig1 KO mouse characterised by hyper-functional SREBP transcriptional activity that was subjected to a dietary challenge to model NAFLD.
3.2. Absence of Insig1 resulted in a more benign hepatic lipidome and decreased hepatic damage induced by a NASH-inducing challenge
To study the contribution of decreased INSIG1 in the early stages of NASH, we exposed Insig1 KO mice and their wild-type (WT) and heterozygous (Het) littermates to a Western diet (high in saturated fat, cholesterol, and refined carbohydrates) supplemented with sugar water (WDSW) ad libitum, thus promoting activation of SREBP1 and ChREBP transcriptional programmes [30].
In line with our previous observations where we defined that Insig1 KO mice were smaller/shorter (but not leaner) than their WT littermates [15], Insig1 KO mice body weights (Figure S2A) were reduced compared to the other genotypes without genotype-associated differences in fat mass (Figure S2B), liver weight to body weight ratio (Figure S2C), metabolic biochemistry (Figsure S2D–G: glucose, TG, cholesterol, and free fatty acid), or markers of systemic and peripheral insulin resistance (Figures. S2H–K). Insig1 KO mice were therefore indistinguishable from their WT/Het littermates with regards to the systemic metabolic phenotype as previously described by our group and others [15,17].
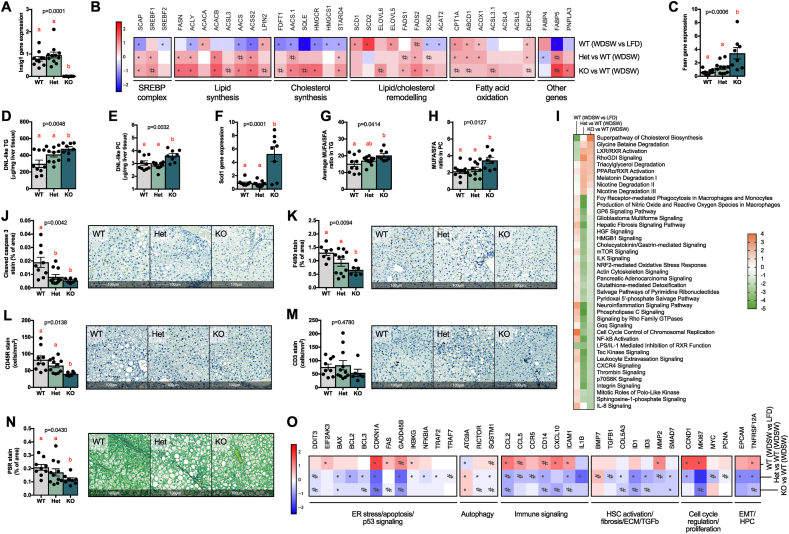
We previously described that INSIG1 in adipose tissue promotes a negative feedback loop on SREBP1 activation, ensuring SREBP1 function in the context of IR (when the transcriptional drive of both is reduced) [15]. Absence of Insig1 in the liver (Figure 2A) resulted in a marked upregulation of SREBP1 and SREBP2 target genes, leading to enhanced lipid/cholesterol synthesis and remodelling (Figure 2B). The total hepatic lipid and triglyceride (TG) composition was only marginally affected, with a mild upward trend in the KO mice without reaching statistical significance (Figures. S2L–M) despite an increase in Fasn (qPCR; Figure 2C) and “DNL-like” [8] lipids (Figure 2D–E); this was probably the consequence of the upregulation of genes encoding key beta-oxidation enzymes (Figure 2B) [31]. More broadly, the upregulation of lipid remodelling genes in the KO mice was reflected by an overall change in the triglyceride composition (elongation and desaturation) in the liver (Figure S2N). Furthermore, the upregulation of SREBP1 target Scd1 and of other desaturases (Figure 2B,F) led to qualitative alterations in the repertoire of fatty acid (FA) desaturation in TGs and phosphatidylcholines (PCs) as shown by the increased ratio of monounsaturated fatty acid (MUFA) to saturated fatty acid (SFA; Figure 2G–H). We did not detect differences in cholesterol ester (CE) species between the genotypes (Figure S2O), possibly due to feedback inhibition of cholesterol synthesis from the diet's cholesterol. Overall, an enhanced remodelling of the lipidome toward desaturated lipids is expected to reduce lipotoxicity in murine models of NASH [28]; accordingly, the Insig1 KO mice showed reduced hepatocyte damage with ALT values below the upper limit of normal (Figure S2P) which, although “clinically” significant, did not reach statistical significance. We therefore evaluated by NGS (Figure 2I) and histology (Figure 2J–N and Figure S2Q) whether the livers of the Insig1 KO mice exhibited a milder NASH phenotype compared to the WT and Het littermates with regards to hepatocellular damage, inflammation and fibrosis. Figure 2I shows than in Insig1 KO livers, there was downregulation of oxidative pathways and a concordant decrease in oxidative stress response. Consequently, we observed reduced apoptosis (gene expression in Figure 2I; cleaved caspase 3 IHC in Figure 2J), inflammation (gene expression in Figure 2I; inflammation markers' IHC in Figure 2K–M and Figure S2Q) and, despite this being a model of very mild NASH, extracellular matrix (ECM) deposition (picrosirius red, Figure 2N). Our study also revealed that the KO animals had decreased liver infiltration of lymphocytes (CD3+ T cells and CD45R+ B cells, Figure 2L–M) and a trend toward decreased myeloid cells (Ly6C/G+; monocytes/neutrophils, Figure S2Q), in keeping with the extensive literature detailing the involvement of these cell types in NAFLD progression [[32], [33], [34]]. The analysis of the hepatic transcriptome (Figure 2I and O; details in Table S3) and IPA confirmed that the Insig1 KO mice were protected from ER stress (for example, Ddit3), oxidative damage, and apoptosis (for example, Bax and Fasl), featuring a substantial suppression of pathways involved in the wound-healing response (inflammation, compensatory hepatocyte proliferation, and ECM remodelling).
Figure 2.
Insig1 ablation improved the lipidome and reduced cellular damage, inflammation, and extracellular matrix deposition in a Western diet supplemented with sugar water. (A) Relative Insig1 mRNA expression measured by RTqPCR. (B) NGS showing upregulation of key enzymes in lipid/cholesterol synthesis, remodelling, and oxidation. (C) Relative Fasn mRNA expression measured by RTqPCR. DNL-like triglycerides (D) and phosphatidylcholine (E) as measured by LC-MS. (F) Relative Scd1 mRNA expression as measured by RTqPCR. Monounsaturated fatty acid-containing triglycerides (G) and phosphatidylcholines (H). (I) Prediction of canonical pathways significantly enriched and predicted as activated (orange) or inhibited (green) according to IPA; data are shown as a heat map matrix format representing the prediction of activation (Z score). HALO imaging software analysis on scanned slides of whole tissue of: (J) caspase 3 IHC, (K) F4/80 IHC, (L) CD45R IHC, (M) CD3 IHC, and (N) Picrosirius Red. Representative tissues are shown. (O) Gene expression of selected genes involved in apoptosis, autophagy, compensatory hepatocyte proliferation, inflammation, hepatic stellate cell (HSC) activation and fibrosis, and epithelial–mesenchymal transition (EMT) as profiled by NGS. Data were analysed by ANOVA (p values < 0.05 are considered significant) with Tukey's post hoc test (a, reference group; groups with different letters are statistically different per post hoc comparison; differences between groups with the same letter are statistically not significant per post hoc comparison). NGS data were analysed with the Wald test (#p < 0.05 and ∗pBH < 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
These results indicated that INSIG1 inactivation in the context of NAFLD facilitated the hyper-efficient function of SREBP transcriptional programmes that prevented WDSW-induced hepatic lipotoxicity, leading to a milder NASH phenotype.
3.3. Absence of Insig1 protected the liver from carbon tetrachloride-induced acute hepatocellular damage
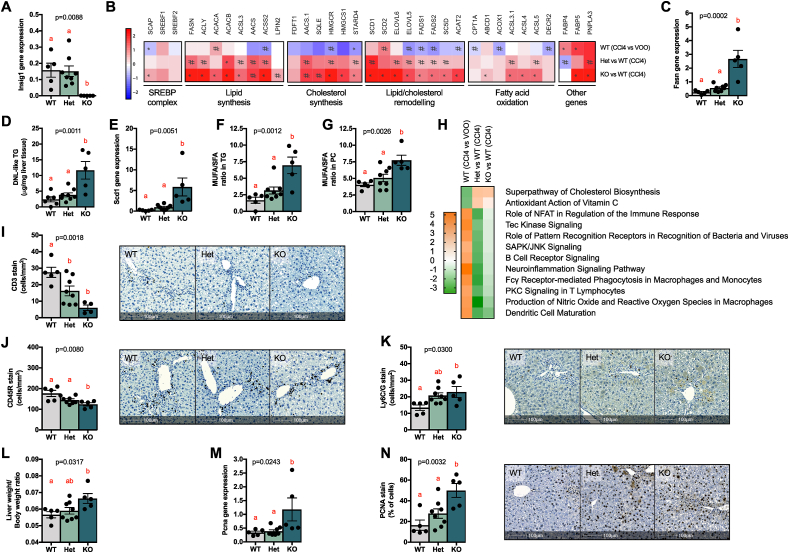
We next posited that the less harmful composition of the hepatic lipidome in the Insig1 KO mice, leading to reduced hepatocellular damage in the obesity-specific WDSW challenge, might also be protective in the context of other forms of liver damage. Following the transcriptomic changes in the WDSW model pointing to protection from oxidative damage, we investigated this paradigm in the acute carbon tetrachloride (CCl4) model that induces substantial reactive oxygen species-mediated hepatotoxicity. One day after CCl4 injection, the Insig1 Het and KO mice showed a trend of reduced hepatotoxicity (ALT, Figure S3A) compared to their WT littermates and this trend flattened on day four after the challenge (ALT, Figure S3B). We chose day four post-CCl4 injection as a time point to study the hepatic phenotype in the context of the wound-healing response (which peaks at 3–5 days) [35]. In line with the WDSW model and our previous report [15], the KO mice were smaller than their WT and Het littermates (BW, Figure S3C), but there were no genotype-associated differences in the systemic metabolism (TG, cholesterol, glucose, insulin, and HOMA-IR, Figures. S3D–H). However, as a result of the genetic deletion of Insig1 (Figure 3A), we found a statistically significant upregulation of SREBP1 and SREBP2 targets involved in lipid synthesis and remodelling (Figure 3B), reflected by an overall change in the triglyceride composition (elongation and desaturation) in the liver (Figure S3I). As in the WDSW model, Fasn upregulation (Figure 3C) led to the accumulation of “DNL-like” TG (Figure 3D) and preferential Scd1 upregulation (Fig. 3E) led to the accumulation of MUFA over SFA species in TGs and PCs (Figure 3F–G). We also observed increased cholesterol ester species in the KO animals (Figure S3J) in line with activation of cholesterol synthesis pathways (Figure 3B). Total hepatic lipid content remained unchanged (Figure S3K-L), possibly due to a mild upregulation in fatty acid oxidation pathways (Figure 3B). The NGS pathway analysis (Figure 3H; details in Table S4) confirmed that the KO livers exhibited increased antioxidant defences in the face of the CCl4 challenge (ascorbate pathway, G6pd upregulation in the KO), reduced reactive oxygen species production, and with regards to the inflammatory component of the wound-healing response, reduced signalling associated with lymphocyte infiltration, which was confirmed by IHC (reduced CD3+ T cells in Figure 3I and Cd45r+ B cells in Figure 3J). In this model, we did not detect any difference in F4/80 macrophages (Figure S3M) or αSMA-positive cells (that is, activated stellate cells, Figure S3N). Intriguingly, Ly6C/G+ cells (monocytes and neutrophils) increased in the livers of the KO mice (Figure 3K). We are tempted to speculate that this may be reflective of a more appropriate inflammatory response [36] to cytotoxic CCl4-induced damage and will be the focus of future studies aimed at ascertaining the biological mechanisms with cell-specific approaches.
Figure 3.
Insig1 ablation improved the lipidome and reduced cellular damage in animals treated with CCl4. (A) Relative Insig1 mRNA expression measured by RTqPCR. (B) NGS showing upregulation of key enzymes in lipid/cholesterol synthesis, remodelling, and oxidation. C) Relative Fasn mRNA expression measured by RTqPCR. (D) DNL-like triglycerides measured by LC-MS. (E) Relative Scd1 mRNA expression measured by RTqPCR, monounsaturated fatty acid-containing triglycerides (F), and phosphatidylcholines (G). (H) Prediction of canonical pathways was significantly enriched and predicted as activated (orange) or inhibited (green) according to the IPA; data are shown as a heat map matrix format representing the activation Z score prediction by the IPA. HALO imaging software analysis on scanned slides of whole tissue of (I) CD3 IHC, (J) CD45R IHC, and (K) Ly6C/G IHC. Representative tissues are shown. (L) Liver weight/body weight ratio. (M) Relative Pcna mRNA expression measured by RTqPCR. (N) PCNA IHC analysed by HALO software. Data were analysed by ANOVA (p values < 0.05 are considered significant) with Tukey's post hoc test (a, reference group; groups with different letters are statistically different per post hoc comparison; differences between groups with the same letter are statistically not significant per post hoc comparison). NGS data were analysed with the Wald test (#p < 0.05 and ∗pBH < 0.05). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Another critical aspect of the CCl4 model was that it allowed us to study the hepatocytes' compensatory regenerative programmes as part of the wound-healing responses activated by acute liver injury. This was an essential component of the Insig1 KO mice phenotype to characterise since 1) the Insig1 KO mice had increased LW to BW ratios (Figure 3L) in the CCl4 model, 2) hepatocyte proliferation was primed by pro-inflammatory pathways led by myeloid cells (increased in the Insig1 KO mice) [37,38], 3) SREBP1 has been proposed as a direct regulator of cell cycle and proliferation [39,40], and 4) we and others have shown that cholesterol synthesis and MUFA PC production are key metabolic processes for hepatocyte proliferation [35,38]. Analysis of proliferating cell nuclear antigen (Pcna mRNA by qPCR in Figure 3M and PCNA protein by IHC in Figure 3N) showed that Insig1 KO livers featured enhanced hepatocyte compensatory proliferation compared to the other genotypes. This phenotype was also confirmed by the IPA upstream regulator analysis of the KO vs WT mice, suggesting a strong activation of the proliferative programmes (CMYC, CCNE1, and CCDD1) supported by a dense and coherent activation of WNT and Hippo signalling pathways (β-CATENIN/CTNNBIP, FOXM1, JNK, YAP1, MTOR/MTORC1, CMYC, CCND1, TBX2, and TP73), growth factor signalling cascades (EGF, HGF, VEGF, and CSF2), STAT3 and STAT6 activation, and many other mediators of hepatocyte proliferation/survival and de-differentiation. Coherent tumour suppressors and cell cycle checkpoints (TP53 and PTEN) were suppressed (Figure S4; details in Table S5).
Overall, these data indicated that protection from hepatocellular damage in the Insig1 KO mice was replicated in a model of acute hepatic damage (CCl4), whereby the KO mice featured reduced adaptive immune cell infiltration paralleled by enhanced compensatory hepatocyte proliferation, resulting in hepatoprotection from CCl4. Therefore, we speculate that inhibiting INSIG1 function might not only be an efficient strategy to reduce hepatic cytotoxicity, but also to favour more efficient liver regeneration in the context of acute hepatocellular damage.
3.4. NGS analysis confirmed that Insig1 absence and increased DNL/lipid remodelling resulted in a favourable metabolic and inflammatory milieu in WDSW and CCl4 models
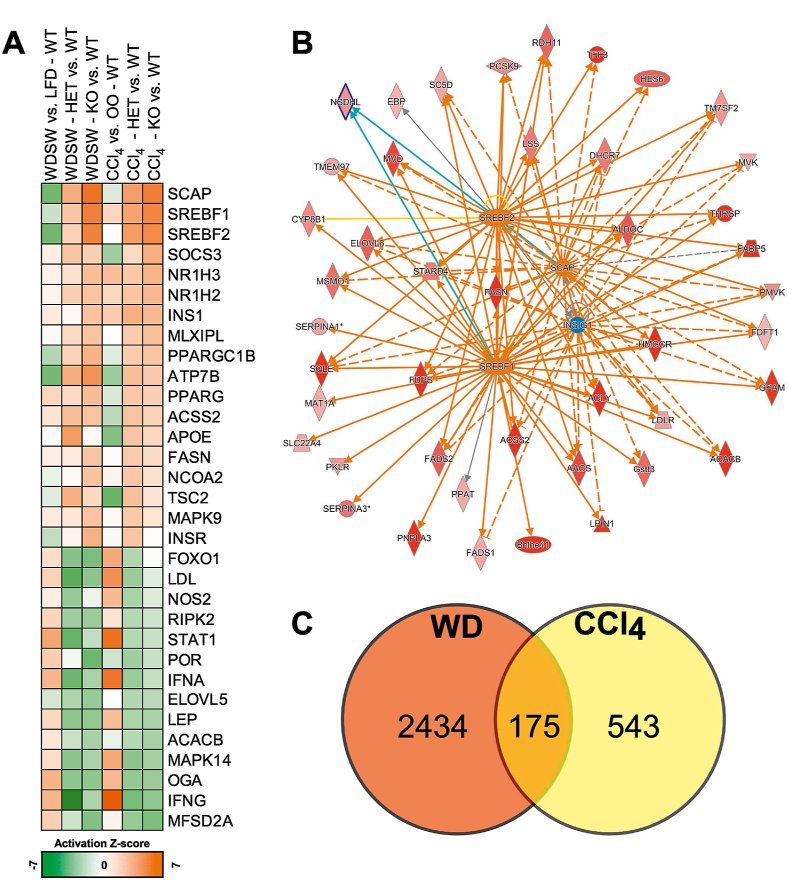
To interpret the complex mechanisms that characterise the deletion of Insig1 in liver damage, we performed the upstream regulator analysis (URA) in the IPA to highlight those adaptive changes that were associated with the absence of Insig1 independently from the challenge. In addition to the expected activation of SREBF1/2 (Figure 4A–B; details in Table S6), the Insig1 KO mice exhibited a pleiotropic activation of multiple regulators of hepatic metabolism that might have contributed to the more benign response to liver damage. Specifically, we found strong activation of lipid-sensing transcriptional programmes such as the liver X receptors (NR1H2/3), the peroxisome proliferator-activated receptor gamma (PPARG) and PPARG coactivator 1 beta (PGC1β/PPARGC1B). These transcriptional regulators contribute to lipid and cholesterol metabolism, fatty acid oxidation, lipoprotein secretion, and negative regulation of inflammation and fibrogenesis [14,39,[41], [42], [43]] potentially contributing to protecting the Insig1 KO mice from liver damage. IPA also predicted an improvement of other metabolic pathways and insulin sensitivity (activation of INS, INSR, and MAPK9), reduced ER stress (MAPK14 and FOXO1), and suppression of inflammatory modulators (NOS2, RIPK2, STAT1, IFNA, and IFNG). All of these upstream regulators have been shown to be involved in NASH progression, lending weight to the hypothesis that Insig1 downregulation might directly/indirectly lead to a pleiotropic protective set of mechanisms that attenuate inflammation and liver damage in NAFLD [41,42].
Figure 4.
NGS analysis confirmed Insig1 ablation impacted metabolic and inflammatory pathways in WDSW and CCl4models. (A) Prediction of upstream regulators significantly enriched and predicted (Z score) activated (orange) or inhibited (green) according to the IPA/URA in the Het and KO (vs WT) in the WDSW and CCl4 models (full list in Table S5). (B) Graphical representation in networks of differentially modulated genes (green, downregulated; red, upregulated) leading to the predicted activation (orange)/inhibition (blue) of the upstream regulators according to the IPA; data from the WDSW model. (C) Comparison of the genes differentially modulated in the treated Insig1 KO mice (vs WT), a subset of 175 genes were differentially modulated the WDSW and CCl4 models. (p < 0.05; −0.378 ≤ log2 [fold change] ≥ 0.378; full list in Table S6). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We also retrieved a list of 175 genes that are significantly modulated in the same direction in dietary and chemical models as a result of Insig1 deletion (Figure 4C; details in Table S6). These genes are not only limited to SREBP-mediated pathways (for example, Acacb, Fasn, Elovl6, Hmgcr, and Pnpla3), but also carbohydrate metabolism (Pfkfb1 and Pklr), anti-oxidant pathways (Thrsp, Slc23a1, and Cyb5r3), inflammation (Cd14, Il22ra, and Plin5), ECM remodelling (Col5a3 and Itga5), and proliferation (Blm), further confirming the predicted regulation of the previously described pathways and upstream regulators.
4. Discussion and conclusion
Increased hepatic DNL is associated with NAFLD development, especially the development of steatosis [[7], [8], [9], [10],43,44]. Although multiple reports have associated DNL with NASH progression [45,46], these anecdotal associations in observational studies have not been substantiated by translational studies demonstrating that DNL is unequivocally a direct causal event in NASH progression and fibrosis. Metabolic programmes governing DNL also substantially reshape lipid remodelling: DNL and lipid remodelling co-regulation could also be a biomarker of allostatic protective responses aimed at producing specific types of lipids required for membranes/organelle homeostasis and safe storage of lipids in the form of triglycerides, maintaining the liver's functionality [47,48]. In this study, we used a transcriptomic analysis of liver biopsy samples from 58 patients with biopsy-proven NAFLD to show that progression to NASH and fibrosis is associated with a rewiring of metabolic pathways, most notably those associated with DNL, lipid desaturation, fatty acid remodelling, and cholesterol synthesis (Figure 1). These findings confirm previous reports investigating circulating lipid species, suggesting that complex lipids enriched in DNL products could act as biomarkers of the disease [8,49,50]. The IPA upstream regulator analysis predicted the activation of the SREBF1/SREBF2/SCAP network and the suppression of INSIG1 as critical integrated events associated with increasing NASH/fibrosis stage. Our group previously suggested that INSIG1, a direct target of SREBP1, acts as a brake, tuning SCAP/SREBP functional activation. Thus, INSIG1 should be considered the keystone protein in a negative feedback loop, ensuring the maintenance of qualitative aspects of lipid desaturation and cholesterol composition. This allostatic adaptation's proposed goal is to maintain required biophysical properties and functionality of cellular membranes when confronted with the challenges of obesity, nutritional and metabolic changes [14,15], and cell proliferation [38], even at the expense of accumulating more lipids.
The literature focussing on INSIG1 function in metabolic homeostasis has been inconclusive, probably due to the pathway's complexity. The direct involvement of INSIG1 polymorphisms in regulating circulating TG and glucose levels is under debate [[51], [52], [53]], and previous studies suggested that the hepatocyte-specific deletion of Insig1 or Insig2 alone does not result in noticeable metabolic changes in the liver of healthy mice, requiring a double KO hit to observe metabolic dysfunction in the healthy liver [17]. Therefore, the specific role of INSIG1 in regulating hepatocellular metabolism and damage in the context of obesity, lipotoxic insults, and NASH was yet to be clearly determined. To mechanistically investigate whether deleting Insig1 promotes liver damage or protection as well as its effect on other metabolic parameters, we used a whole-body Insig1 deficient animal model. Our lab previously showed that in murine obesity, Insig1 is downregulated in adipose tissue. We posited that this response was part of an adaptive response promoting SREBP1 maturation, providing appropriate levels of fatty acid unsaturation and partially compensating the lipolytic and catabolic effect associated with insulin resistance [15] at the expense of facilitating the production and accumulation of lipids.
We challenged the Insig1 KO mice with a high cholesterol/fat/refined sugar (Western) diet supplemented with sugar water for 12 weeks to maximally drive the hepatic lipogenic programme by activating both SREBP1C and ChREBP [54]. In the context of overnutrition, the Insig1 KO mice showed a hyper-efficient activation of DNL and lipid remodelling programmes (Figure 2, Figure 4A). Despite this and in keeping with previous work from our laboratory and others [15,17], the Insig1 KO mice had no differences in systemic metabolism or insulin resistance compared to their Het/WT littermates. However, the Insig1 KO livers were protected from ER stress, oxidative damage and inflammation and exhibited reduced fibrogenesis (Figure 2I–O). Based on the transcriptional and lipidomic changes that we observed, we speculate that Insig1 KO-mediated “lipo-protection” occurs via optimisation of lipid composition via enhanced production of MUFAs [55,56], other pleiotropic mechanisms that may include improved signalling of PPARs and their co-activator PGC1β [39,57], which were predicted as being upregulated in the Insig1 KO mice (Figure 2, Figure 3, Figure 4). In our experimental setting, a significant role seemed to be played by the SREBP1-mediated upregulation of Scd1 and other remodelling enzymes. Hepatic SCD1 plays a crucial role in preventing steatohepatitis by partitioning excess lipid into MUFA that can be safely stored in TGs and lipid droplets [56]. Furthermore, Scd1 induction by ChREBP overexpression in the murine liver in conjunction with high-fat diet results in elevated MUFA, improved glucose tolerance, and insulin sensitivity despite the paradoxical increased hepatic steatosis; of relevance, this effect disappears upon SCD1 inhibition [28]. In contrast, Scd1 deletion results in lowering of MUFA, induction of ER stress, acute inflammation, and macrophage recruitment [55]. Our data confirm that SREBP1 in the liver preferentially induces Scd1, thus resulting in accumulation of unsaturated fatty acids and providing similar protection from hepatic damage in the context of Western diet-like feeding. Intriguingly, we observed similar results when the Insig1 WT/Het/KO mice were acutely challenged with CCl4, suggesting that the hepato-protection associated with INSIG1 suppression might not be limited to NASH and applicable to other acute and chronic liver diseases. To translate these findings back to human disease, we speculate that SREBPs activation in progressive human NASH likely reflects an attempt to metabolically adapt to nutritional surplus. The negative feedback loop induced by INSIG1 physiologically acts as a rheostat of the pathway; when excessively induced in the context of caloric excess, this feedback loop might restrain SREBPs and contribute to the failure of these equilibrium mechanisms [58].
This study has some limitations, such as the inability to conduct more comprehensive metabolically meaningful studies featuring advanced NASH: mice have a growth defect that becomes more prominent with age until adulthood, and as a result, in KO mice, length and BW progressively diverge with age. We also used a whole-body Insig1 KO mouse that does not restrict the phenotype mechanistically to the role of INSIG1 in hepatocytes or other cell types. Thus, the observed phenotype may also be influenced by the effects of Insig1 deletion in adipose tissue [15], immune cells [[59], [60], [61]], and hepatic stellate cells [62]. However, recapitulating a whole-body defect of DNL related to metabolic stress is a valid initial strategy to identify the potential relevance of this regulatory loop. It justifies and de-risks future studies focussed on the dissection of cell-specific mechanistic specificities. In this respect, these results represent a proof of concept that compensatory regulatory loops aiming to maintain lipid homeostasis provide a reasonable intellectual frame to devise combinatorial therapeutic strategies to inhibit effector mechanisms and promote resilience through potentiation of protective allostatic responses.
In conclusion, our study shows that activation and remodelling of lipid pathways in humans and animals in metabolic liver disease could be interpreted as a protective mechanism to limit liver damage and fibrosis. Our study provides actionable knowledge about the pathways and mechanisms involved in NAFLD pathogenesis, which may prove useful for developing new therapeutic strategies.
Author contributions
VA, MV, AVP, and MEDA conceived and designed the experiments, analysed and interpreted the results, prepared the figures, and wrote the manuscript. MEDA, MV, and SED provided human clinical and histologic data and tissues. VA, MV, SV, SC, JL, and FO conducted the animal experiments. MV, SV, MD, IK, AA, and EP processed and analysed the human/murine NGS data. MV carried out the IPA analyses. ZH and JG processed and analysed the lipidomics data. VA, MV, and AL conducted the histology analyses with HALO. All of the authors provided useful criticism and revised and approved the manuscript.
Acknowledgements
The authors are indebted to the following colleagues and institutions: MRC Metabolic Diseases Unit (MC_UU_00014/5); the Disease Model Core (DMC; Hannah Webber, Daniel Hart, Sarah Grocott, Charley Beresford, Darren Jessop, Eerika Rasijeff, and Amy Warner); the Biochemistry Assay Lab (Keith Burling and collaborators); the Genomics and Transcriptomics Core (Marcella Ma and Brian Y. H. Lam); the Histology Core (James Warner) and Imaging Core (Gregory Strachan); MRC Human Nutrition Research and Department of Biochemistry, University of Cambridge; and the staff at Translational Genomics, Discovery Sciences, Bio-Pharmaceuticals R&D, AstraZeneca, Gothenburg, Sweden, who contributed to the development of the Insig1 KO mouse model. The human NASH histological samples were from the Human Research Tissue Bank of Cambridge University Hospitals, which the NIHR Cambridge Biomedical Research Centre supports. MV, JLG, and AVP are supported by MRC programmes (MRC MDU Programme Grant, PO 4050281695, Lipotoxicity and the Metabolic Syndrome, and MRC DMC MC_UU_00014/2 to AVP; Lipid Profiling and Signalling, MC UP A90 1006 to JLG) and MRC receives adjunct funding as part of the Cambridge Initiative in Metabolic Diseases (Lipid Dynamics and Regulation: MC_PC_13030). VA, MV, MA, and AVP are supported by the Cambridge NIHR Biomedical Research Centre (Gastroenterology Theme). FO is supported by MRC programme grants MR/K0019494/1 and MR/R023026/1. JL is supported by a Medical Research Council PhD studentship and a CRUK programme grant (C18342/A23390). VA, MV, AVP, MA, and FO are contributing members of the European NAFLD Registry. MA, AVP, and JLG received funding from the Evelyn Trust. VA, MV, MA, FO, and AVP are members of the Elucidating Pathways of Steatohepatitis consortium, which is funded by the Horizon 2020 Framework Programme of the European Union under Grant Agreement 634413. SV and AJV are supported by the BHF (BHF Programme Grant RG/18/7/33636). The views expressed are those of the author(s) and not necessarily those of the relevant funding or supporting institutions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101210.
Contributor Information
Michael E.D. Allison, Email: michael.allison@addenbrookes.nhs.uk.
Antonio Vidal-Puig, Email: ajv22@medschl.cam.ac.uk.
Conflict of interest
JL owns stock in Fibrofind. FO is employed by and owns stock in Fibrofind. MB-Y and DL are employees of AstraZeneca. All of the other authors declare no conflicts of interest.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Bellentani S. The epidemiology of non-alcoholic fatty liver disease. Liver International. 2017;37(Suppl 1):81–84. doi: 10.1111/liv.13299. [DOI] [PubMed] [Google Scholar]
- 2.Argo C.K., Northup P.G., Al-Osaimi A.M., Caldwell S.H. Systematic review of risk factors for fibrosis progression in non-alcoholic steatohepatitis. Journal of Hepatology. 2009;51(2):371–379. doi: 10.1016/j.jhep.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 3.Ong J.P., Pitts A., Younossi Z.M. Increased overall mortality and liver-related mortality in non-alcoholic fatty liver disease. Journal of Hepatology. 2008;49(4):608–612. doi: 10.1016/j.jhep.2008.06.018. [DOI] [PubMed] [Google Scholar]
- 4.Anstee Q.M., Targher G., Day C.P. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nature Reviews Gastroenterology & Hepatology. 2013;10(6):330–344. doi: 10.1038/nrgastro.2013.41. [DOI] [PubMed] [Google Scholar]
- 5.Younossi Z.M., Tampi R., Priyadarshini M., Nader F., Younossi I.M., Racila A. Burden of illness and economic model for patients with nonalcoholic steatohepatitis in the United States. Hepatology. 2019;69(2):564–572. doi: 10.1002/hep.30254. [DOI] [PubMed] [Google Scholar]
- 6.Azzu V., Vacca M., Virtue S., Allison M., Vidal-Puig A. Adipose tissue-liver cross talk in the control of whole-body metabolism: implications in nonalcoholic fatty liver disease. Gastroenterology. 2020;158(7):1899–1912. doi: 10.1053/j.gastro.2019.12.054. [DOI] [PubMed] [Google Scholar]
- 7.Puri P., Wiest M.M., Cheung O., Mirshahi F., Sargeant C., Min H.K. The plasma lipidomic signature of nonalcoholic steatohepatitis. Hepatology. 2009;50(6):1827–1838. doi: 10.1002/hep.23229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanders F.W.B., Acharjee A., Walker C., Marney L., Roberts L.D., Imamura F. Hepatic steatosis risk is partly driven by increased de novo lipogenesis following carbohydrate consumption. Genome Biology. 2018;19(1):79. doi: 10.1186/s13059-018-1439-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luukkonen P.K., Sadevirta S., Zhou Y., Kayser B., Ali A., Ahonen L. Saturated fat is more metabolically harmful for the human liver than unsaturated fat or simple sugars. Diabetes Care. 2018;41(8):1732–1739. doi: 10.2337/dc18-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnelly K.L., Smith C.I., Schwarzenberg S.J., Jessurun J., Boldt M.D., Parks E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. Journal of Clinical Investigation. 2005;115(5):1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiappini F., Coilly A., Kadar H., Gual P., Tran A., Desterke C. Metabolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients. Scientific Reports. 2017;7:46658. doi: 10.1038/srep46658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall Z., Bond N.J., Ashmore T., Sanders F., Ament Z., Wang X. Lipid zonation and phospholipid remodeling in nonalcoholic fatty liver disease. Hepatology. 2017;65(4):1165–1180. doi: 10.1002/hep.28953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomita K., Teratani T., Suzuki T., Shimizu M., Sato H., Narimatsu K. Free cholesterol accumulation in hepatic stellate cells: mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology. 2014;59(1):154–169. doi: 10.1002/hep.26604. [DOI] [PubMed] [Google Scholar]
- 14.Vacca M., Allison M., Griffin J.L., Vidal-Puig A. Fatty acid and glucose sensors in hepatic lipid metabolism: implications in NAFLD. Seminars in Liver Disease. 2015;35(3):250–261. doi: 10.1055/s-0035-1562945. [DOI] [PubMed] [Google Scholar]
- 15.Carobbio S., Hagen R.M., Lelliott C.J., Slawik M., Medina-Gomez G., Tan C.Y. Adaptive changes of the Insig1/SREBP1/SCD1 set point help adipose tissue to cope with increased storage demands of obesity. Diabetes. 2013;62(11):3697–3708. doi: 10.2337/db12-1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takaishi K., Duplomb L., Wang M.Y., Li J., Unger R.H. Hepatic insig-1 or -2 overexpression reduces lipogenesis in obese Zucker diabetic fatty rats and in fasted/refed normal rats. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(18):7106–7111. doi: 10.1073/pnas.0401715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelking L.J., Liang G., Hammer R.E., Takaishi K., Kuriyama H., Evers B.M. Schoenheimer effect explained--feedback regulation of cholesterol synthesis in mice mediated by Insig proteins. Journal of Clinical Investigation. 2005;115(9):2489–2498. doi: 10.1172/JCI25614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sever N., Yang T., Brown M.S., Goldstein J.L., DeBose-Boyd R.A. Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Molecular Cell. 2003;11(1):25–33. doi: 10.1016/s1097-2765(02)00822-5. [DOI] [PubMed] [Google Scholar]
- 19.Brunt E.M., Janney C.G., Di Bisceglie A.M., Neuschwander-Tetri B.A., Bacon B.R. Nonalcoholic steatohepatitis: a proposal for grading and staging the histological lesions. American Journal of Gastroenterology. 1999;94(9):2467–2474. doi: 10.1111/j.1572-0241.1999.01377.x. [DOI] [PubMed] [Google Scholar]
- 20.Kleiner D.E., Brunt E.M., Van Natta M., Behling C., Contos M.J., Cummings O.W. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41(6):1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 21.Bedossa P., Poitou C., Veyrie N., Bouillot J.L., Basdevant A., Paradis V. Histopathological algorithm and scoring system for evaluation of liver lesions in morbidly obese patients. Hepatology. 2012;56(5):1751–1759. doi: 10.1002/hep.25889. [DOI] [PubMed] [Google Scholar]
- 22.Felasa working group on revision of guidelines for health monitoring of rodents, rabbits, Mahler Convenor M., Berard M., Feinstein R., Gallagher A. FELASA recommendations for the health monitoring of mouse, rat, hamster, Guinea pig and rabbit colonies in breeding and experimental units. Lab Animal. 2014;48(3):178–192. doi: 10.1177/0023677213516312. [DOI] [PubMed] [Google Scholar]
- 23.Engelking L.J., Kuriyama H., Hammer R.E., Horton J.D., Brown M.S., Goldstein J.L. Overexpression of Insig-1 in the livers of transgenic mice inhibits SREBP processing and reduces insulin-stimulated lipogenesis. Journal of Clinical Investigation. 2004;113(8):1168–1175. doi: 10.1172/JCI20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korn B.S., Shimomura I., Bashmakov Y., Hammer R.E., Horton J.D., Goldstein J.L. Blunted feedback suppression of SREBP processing by dietary cholesterol in transgenic mice expressing sterol-resistant SCAP(D443N) Journal of Clinical Investigation. 1998;102(12):2050–2060. doi: 10.1172/JCI5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimomura I., Bashmakov Y., Horton J.D. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. Journal of Biological Chemistry. 1999;274(42):30028–30032. doi: 10.1074/jbc.274.42.30028. [DOI] [PubMed] [Google Scholar]
- 26.Caballero F., Fernandez A., De Lacy A.M., Fernandez-Checa J.C., Caballeria J., Garcia-Ruiz C. Enhanced free cholesterol, SREBP-2 and StAR expression in human NASH. Journal of Hepatology. 2009;50(4):789–796. doi: 10.1016/j.jhep.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 27.Van Rooyen D.M., Farrell G.C. SREBP-2: a link between insulin resistance, hepatic cholesterol, and inflammation in NASH. Journal of Gastroenterology and Hepatology. 2011;26(5):789–792. doi: 10.1111/j.1440-1746.2011.06704.x. [DOI] [PubMed] [Google Scholar]
- 28.Benhamed F., Denechaud P.D., Lemoine M., Robichon C., Moldes M., Bertrand-Michel J. The lipogenic transcription factor ChREBP dissociates hepatic steatosis from insulin resistance in mice and humans. Journal of Clinical Investigation. 2012;122(6):2176–2194. doi: 10.1172/JCI41636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piccolis M., Bond L.M., Kampmann M., Pulimeno P., Chitraju C., Jayson C.B.K. Probing the global cellular responses to lipotoxicity caused by saturated fatty acids. Molecular Cell. 2019;74(1):32–44. doi: 10.1016/j.molcel.2019.01.036. e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asgharpour A., Cazanave S.C., Pacana T., Seneshaw M., Vincent R., Banini B.A. A diet-induced animal model of non-alcoholic fatty liver disease and hepatocellular cancer. Journal of Hepatology. 2016;65(3):579–588. doi: 10.1016/j.jhep.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schulz H. Fatty acid oxidation. In: Lennarz W.J., Lane M.D., editors. Encyclopedia of biological chemistry. Second ed. Academic Press; 2013. pp. 281–284. [Google Scholar]
- 32.Sutti S., Albano E. Adaptive immunity: an emerging player in the progression of NAFLD. Nature Reviews Gastroenterology & Hepatology. 2020;17(2):81–92. doi: 10.1038/s41575-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krenkel O., Puengel T., Govaere O., Abdallah A.T., Mossanen J.C., Kohlhepp M. Therapeutic inhibition of inflammatory monocyte recruitment reduces steatohepatitis and liver fibrosis. Hepatology. 2018;67(4):1270–1283. doi: 10.1002/hep.29544. [DOI] [PubMed] [Google Scholar]
- 34.Baeck C., Wei X., Bartneck M., Fech V., Heymann F., Gassler N. Pharmacological inhibition of the chemokine C-C motif chemokine ligand 2 (monocyte chemoattractant protein 1) accelerates liver fibrosis regression by suppressing Ly-6C(+) macrophage infiltration in mice. Hepatology. 2014;59(3):1060–1072. doi: 10.1002/hep.26783. [DOI] [PubMed] [Google Scholar]
- 35.Hall Z., Chiarugi D., Charidemou E., Leslie J., Scott E., Pellegrinet L. Lipid remodelling in hepatocyte proliferation and hepatocellular carcinoma. Hepatology. 2020;73(3):1028–1044. doi: 10.1002/hep.31391. [DOI] [PubMed] [Google Scholar]
- 36.Ramachandran P., Pellicoro A., Vernon M.A., Boulter L., Aucott R.L., Ali A. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(46):E3186–E3195. doi: 10.1073/pnas.1119964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fausto N., Campbell J.S., Riehle K.J. Liver regeneration. Hepatology. 2006;43(2 Suppl 1):S45–S53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 38.Vacca M., Degirolamo C., Massafra V., Polimeno L., Mariani-Costantini R., Palasciano G. Nuclear receptors in regenerating liver and hepatocellular carcinoma. Molecular and Cellular Endocrinology. 2013;368(1–2):108–119. doi: 10.1016/j.mce.2012.06.025. [DOI] [PubMed] [Google Scholar]
- 39.Bellafante E., Murzilli S., Salvatore L., Latorre D., Villani G., Moschetta A. Hepatic-specific activation of peroxisome proliferator-activated receptor gamma coactivator-1beta protects against steatohepatitis. Hepatology. 2013;57(4):1343–1356. doi: 10.1002/hep.26222. [DOI] [PubMed] [Google Scholar]
- 40.Seo Y.K., Chong H.K., Infante A.M., Im S.S., Xie X., Osborne T.F. Genome-wide analysis of SREBP-1 binding in mouse liver chromatin reveals a preference for promoter proximal binding to a new motif. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(33):13765–13769. doi: 10.1073/pnas.0904246106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardy T., Oakley F., Anstee Q.M., Day C.P. Nonalcoholic fatty liver disease: pathogenesis and disease spectrum. Annual Review of Pathology: Mechanisms of Disease. 2016;11:451–496. doi: 10.1146/annurev-pathol-012615-044224. [DOI] [PubMed] [Google Scholar]
- 42.Schuster S., Cabrera D., Arrese M., Feldstein A.E. Triggering and resolution of inflammation in NASH. Nature Reviews Gastroenterology & Hepatology. 2018;15(6):349–364. doi: 10.1038/s41575-018-0009-6. [DOI] [PubMed] [Google Scholar]
- 43.Lambert J.E., Ramos-Roman M.A., Browning J.D., Parks E.J. Increased de novo lipogenesis is a distinct characteristic of individuals with nonalcoholic fatty liver disease. Gastroenterology. 2014;146(3):726–735. doi: 10.1053/j.gastro.2013.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Puri P., Baillie R.A., Wiest M.M., Mirshahi F., Choudhury J., Cheung O. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46(4):1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 45.Lawitz E.J., Coste A., Poordad F., Alkhouri N., Loo N., McColgan B.J. Acetyl-CoA carboxylase inhibitor GS-0976 for 12 Weeks reduces hepatic de novo lipogenesis and steatosis in patients with nonalcoholic steatohepatitis. Clinical Gastroenterology and Hepatology. 2018;16(12):1983–1991. doi: 10.1016/j.cgh.2018.04.042. e1983. [DOI] [PubMed] [Google Scholar]
- 46.Gapp B., Jourdain M., Bringer P., Kueng B., Weber D., Osmont A. Farnesoid X receptor agonism, acetyl-coenzyme A carboxylase inhibition, and back translation of clinically observed endpoints of de novo lipogenesis in a murine NASH model. Hepatology Communications. 2020;4(1):109–125. doi: 10.1002/hep4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagen R.M., Rodriguez-Cuenca S., Vidal-Puig A. An allostatic control of membrane lipid composition by SREBP1. FEBS Letters. 2010;584(12):2689–2698. doi: 10.1016/j.febslet.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 48.Casares D., Escriba P.V., Rossello C.A. Membrane lipid composition: effect on membrane and organelle structure, function and compartmentalization and therapeutic avenues. International Journal of Molecular Sciences. 2019;20(9) doi: 10.3390/ijms20092167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorden D.L., Myers D.S., Ivanova P.T., Fahy E., Maurya M.R., Gupta S. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. The Journal of Lipid Research. 2015;56(3):722–736. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luukkonen P.K., Zhou Y., Sadevirta S., Leivonen M., Arola J., Oresic M. Hepatic ceramides dissociate steatosis and insulin resistance in patients with non-alcoholic fatty liver disease. Journal of Hepatology. 2016;64(5):1167–1175. doi: 10.1016/j.jhep.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 51.Smith E.M., Zhang Y., Baye T.M., Gawrieh S., Cole R., Blangero J. INSIG1 influences obesity-related hypertriglyceridemia in humans. The Journal of Lipid Research. 2010;51(4):701–708. doi: 10.1194/jlr.M001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krapivner S., Chernogubova E., Ericsson M., Ahlbeck-Glader C., Hamsten A., van 't Hooft F.M. Human evidence for the involvement of insulin-induced gene 1 in the regulation of plasma glucose concentration. Diabetologia. 2007;50(1):94–102. doi: 10.1007/s00125-006-0479-x. [DOI] [PubMed] [Google Scholar]
- 53.Szopa M., Meirhaeghe A., Luan J., Moreno L.A., Gonzalez-Gross M., Vidal-Puig A. No association between polymorphisms in the INSIG1 gene and the risk of type 2 diabetes and related traits. American Journal of Clinical Nutrition. 2010;92(1):252–257. doi: 10.3945/ajcn.2010.29422. [DOI] [PubMed] [Google Scholar]
- 54.Linden A.G., Li S., Choi H.Y., Fang F., Fukasawa M., Uyeda K. Interplay between ChREBP and SREBP-1c coordinates postprandial glycolysis and lipogenesis in livers of mice. The Journal of Lipid Research. 2018;59(3):475–487. doi: 10.1194/jlr.M081836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flowers M.T., Keller M.P., Choi Y., Lan H., Kendziorski C., Ntambi J.M. Liver gene expression analysis reveals endoplasmic reticulum stress and metabolic dysfunction in SCD1-deficient mice fed a very low-fat diet. Physiological Genomics. 2008;33(3):361–372. doi: 10.1152/physiolgenomics.00139.2007. [DOI] [PubMed] [Google Scholar]
- 56.Li Z.Z., Berk M., McIntyre T.M., Feldstein A.E. Hepatic lipid partitioning and liver damage in nonalcoholic fatty liver disease: role of stearoyl-CoA desaturase. Journal of Biological Chemistry. 2009;284(9):5637–5644. doi: 10.1074/jbc.M807616200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Regnier M., Polizzi A., Smati S., Lukowicz C., Fougerat A., Lippi Y. Hepatocyte-specific deletion of Pparalpha promotes NAFLD in the context of obesity. Scientific Reports. 2020;10(1):6489. doi: 10.1038/s41598-020-63579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kitano H., Oda K., Kimura T., Matsuoka Y., Csete M., Doyle J. Metabolic syndrome and robustness tradeoffs. Diabetes. 2004;53(Suppl 3):S6–S15. doi: 10.2337/diabetes.53.suppl_3.s6. [DOI] [PubMed] [Google Scholar]
- 59.Remmerie A., Scott C.L. Macrophages and lipid metabolism. Cellular Immunology. 2018;330:27–42. doi: 10.1016/j.cellimm.2018.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Howie D., Ten Bokum A., Necula A.S., Cobbold S.P., Waldmann H. The role of lipid metabolism in T lymphocyte differentiation and survival. Frontiers in Immunology. 2017;8:1949. doi: 10.3389/fimmu.2017.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hubler M.J., Kennedy A.J. Role of lipids in the metabolism and activation of immune cells. The Journal of Nutritional Biochemistry. 2016;34:1–7. doi: 10.1016/j.jnutbio.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.She H., Xiong S., Hazra S., Tsukamoto H. Adipogenic transcriptional regulation of hepatic stellate cells. Journal of Biological Chemistry. 2005;280(6):4959–4967. doi: 10.1074/jbc.M410078200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the raw data (FASTQ files) and relevant metadata are available in the Array Express database (www.ebi.ac.uk/arrayexpress) under accession numbers E-MTAB-9815 and E-MTAB-9864.
2.5.1. Additional methods provided in supplementary files
Biochemical serum analyses, murine tissue collection and processing, histology, immunohistochemistry, tissue imaging, quantification, and scoring, RNA extraction and RNA integrity, reverse transcription (RT) polymerase chain reaction (PCR), whole-transcriptome amplification and RNA sequencing, hepatic lipid extraction and lipidomics, quantification and statistical analysis, NGS data processing, and bioinformatics functional analyses.