CHIKV nsP2 plays multiple roles in viral RNA replication and virus-host interactions. The helicase and protease regions of nsP2 are connected through a short linker.
KEYWORDS: alphavirus, nonstructural protein 2, interdomain linker, virus replication, chikungunya virus
ABSTRACT
Chikungunya virus (CHIKV) is a mosquito-borne alphavirus responsible for chikungunya fever. Nonstructural protein 2 (nsP2), a multifunctional protein essential for viral replication, has an N-terminal helicase region (nsP2h), which has both nucleotide triphosphatase and RNA triphosphatase activities, as well as a C-terminal cysteine protease region (nsP2p), which is responsible for nonstructural polyprotein processing. The two functional units are connected through a linker of 14 residues. Although crystal structures of the helicase and protease regions of CHIKV nsP2 have been solved separately, the conformational arrangement of the full-length nsP2 and the biological role of the linker remain elusive. Using the small-angle X-ray scattering (SAXS) method, we demonstrated that the full-length nsP2 is elongated and partially folded in solution. The reconstructed model of the structure of nsP2 contains a flexible interdomain linker, and there is no direct interaction between the two structured regions. To examine the function of the interdomain linker, we constructed and characterized a set of CHIKV mutants. The deletion of three or five amino acid residues in the linker region resulted in a modest defect in viral RNA replication and transcription but completely abolished viral infectivity. In contrast, increasing the flexibility of nsP2 by lengthening the interdomain linker increased both genomic RNA replication and viral infectivity. The enzymatic activities of the corresponding mutant proteins were largely unaffected. This work suggests that increasing the interdomain flexibility of nsP2 could facilitate the assembly of the replication complex (RC) with increased efficiency and promote virus production.
IMPORTANCE CHIKV nsP2 plays multiple roles in viral RNA replication and virus-host interactions. The helicase and protease regions of nsP2 are connected through a short linker. Here, we determined that the conformation of full-length CHIKV nsP2 is elongated and that the protein is flexible in solution. We also highlight the importance of the flexibility of the interdomain of nsP2 on viral RNA synthesis and infectivity. CHIKV mutants harboring shortened linkers fail to produce infectious virus particles despite showing only relatively mild defects in genomic and subgenomic RNA synthesis. Mutations increasing the length of the interdomain linker have only mild and generally beneficial impacts on virus replication. Thus, our findings link interdomain flexibility with the regulation of viral RNA replication and infectivity of the viral genome.
INTRODUCTION
Chikungunya virus (CHIKV, family Togaviridae) is an Old World alphavirus transmitted by Aedes aegypti and Aedes albopictus mosquitoes. CHIKV infection usually causes fever, rash, and polyarthritis in humans (1, 2). CHIKV has a 12-kb positive-sense RNA genome with a 5′ cap and a 3′ poly(A) tail. The genome contains two open reading frames (ORFs). The first ORF encodes precursors of nonstructural proteins (nsPs; polyproteins P1234 and P123) (3, 4). After proteolytic cleavage, four nsPs (nsP1, nsP2, nsP3, and nsP4), which have distinct roles, are formed. P1234, P123, their processing intermediates, and mature nsP1 to -4 together with host factors form a replication complex (RC) located at the plasma membrane. RCs, termed spherules, first carry out the synthesis of negative-sense and, subsequently, positive-sense genomic RNAs and subgenomic (SG) RNAs. The second ORF, which is expressed via SG RNA, encodes structural proteins for viral particle assembly (5).
nsP2 is the largest mature alphaviral protein that consists of an N-terminal helicase (nsP2h) region and a C-terminal protease (nsP2p) region connected through a short linker. nsP2h exhibits NTPase and RNA triphosphatase (RTPase) activities essential for viral RNA replication and 5′ cap synthesis. Interestingly, isolated nsP2h has no RNA unwinding and annealing activity (6–8). Studies have shown that only the full-length CHIKV nsP2 possesses 5′-3′ RNA helicase activity. It was hypothesized that because of the low processivity of nsP2 helicase, an additional region with a strong binding affinity to bind the double-stranded RNA (dsRNA) substrate is required for unwinding (9). nsP2p displays papain-like protease activity to sequentially process the junctions between nsP1/nsP2, nsP2/nsP3, and nsP3/nsP4 (10, 11). To date, crystal structures of both the N- and C-terminal regions of CHIKV nsP2 have been reported (8, 12), but the three-dimensional structure of full-length nsP2 remains unclear.
nsP2 is essential in viral genome replication and SG RNA transcription. Several studies have revealed that point mutations and the insertion of extra amino acid residues or large structured domains at certain positions of nsP2 could alter alphaviral replication and transcription (8, 13–16). For example, the F287A substitution disrupts the stacking interactions between the nsP2h core and RNA bases, resulting in the reduced synthesis of SG RNA and the attenuation of CHIKV infectivity. CHIKV harboring this substitution could restore infectivity through pseudoreversion or accumulation of adaptive mutations (8). The insertion of five extra residues after P647 and the substitution of P718G in the C-terminal region of nsP2 have been shown to reduce viral RNA replication and cytotoxicity (13). An enhanced green fluorescent protein (EGFP) inserted into the interdomain region exhibited a moderate effect on RNA replication and, as in the case of Sindbis virus (SINV), also affected polyprotein processing (14, 15, 17). During viral infection, the majority of nsP2 molecules are distributed in both the cytoplasm and nucleus of infected cells, and only a small fraction are associated with RCs (18). In the nucleus, nsP2 is responsible for inhibiting host cell transcription and blocking antiviral signaling; this is essential to promote viral replication without triggering the antiviral response by the host cell (19–21). Recent studies of the SINV particle proteome have revealed the presence of nsP2 in the virion, supporting suggestions that nsP2 might be involved in genome packaging (22–24). However, the biological role of the linker and its contribution to the full-length nsP2 structural arrangement and function remain elusive.
Using the small-angle X-ray scattering (SAXS) method, we determined that the full-length nsP2 adopts an elongated shape via a flexible interdomain linker. We subsequently showed that the viral replication process can tolerate a deletion of one residue or an insertion of 10 flexible residues at the linker region. Deletion of three or five residues of the linker became lethal to the virus, which could be due to defects in polyprotein processing and RC assembly. In summary, our study expands the understanding of the role of interdomain flexibility in the CHIKV RNA replication and virus production processes.
RESULTS
The solution structure of nsP2 is elongated and flexible.
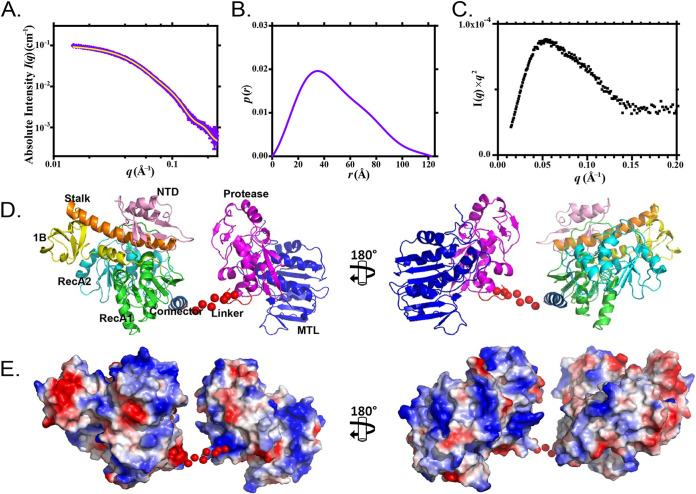
The SAXS data of nsP2 collected from a high-performance liquid chromatography (HPLC) system equipped with an online size exclusion column were analyzed by an in-house data reduction kit. The radius of gyration (Rg) of nsP2 was 38.2 ± 0.1 Å (Fig. 1A; Table 1), and the maximum particle dimension (Dmax) derived from the distance distribution function p(r) was 122.0 Å (Fig. 1B; Table 1). The p(r) profile of nsP2 reached the highest peak at approximately 32 Å, followed by a long-extended tail with a high r value, suggesting that nsP2 adopts an elongated conformation in solution. This finding was further supported by the Kratky plot of nsP2 (Fig. 1C), which exhibited a peak at a lower r value, followed by a slow descent to baseline, indicating that in solution, the full-length nsP2 is flexible. Next, we reconstructed the solution structure of nsP2 based on the crystal structures of nsP2h (residues 1 to 463; PDB ID 6JIM) and nsP2p (residues 471 to 791; PDB ID 4ZTB) using CORAL (25). The missing linker between residues 464 to 470 was represented as a relaxed coil of dummy residues. The calculated structural fitting from the CORAL model is in strong agreement with experimental data (Fig. 1A) and provides the possible positions of linker regions between the helicase and the protease regions. As shown in Fig. 1D, the CORAL model suggests that the interdomain linker of nsP2 is flexible and that there are no direct interactions between nsP2h and nsP2p. In addition, nsP2p is close to the N-terminal region of the helicase domain in this representative model. This arrangement shows the feasibility of nsP2 protease accessing the cleavage site between nsP1 and nsP2 in cis during P123 precursor processing (26).
FIG 1.
SAXS analysis of nsP2. (A) Scattering experimental data and (B) distance distribution function p(r) of nsP2 are shown in purple. The structural fitting of the CORAL model for nsP2 is shown in yellow. (C) Normalized Kratky plot of nsP2. (D) Solution architecture of nsP2 modeled by CORAL. (E) Electrostatic surface potential of nsP2 before (left) and after (right) 180° rotation. The CORAL model is displayed as ribbon diagrams. The random dummy residues are shown as red dots and were used to represent seven missing linker residues. The N-terminal domain (NTD) is in pink, the stalk domain is in orange, domain 1B is in yellow, the RecA1 domain is in green, the RecA2 domain is in cyan, the connector is in sky blue, the protease is in magenta, and the MTL domain is in blue.
TABLE 1.
Parameters of SAXS analysis of nsP2
| Parameter | Value for nsP2 |
|---|---|
| Beamline | BL23A, TLS, Taiwan |
| I(0) (cm−1) | 0.11 |
| Rg (Å) | 38.20 ± 0.10 Å |
| Dmax (Å) | 122.00 Å |
| Porod vol, Po (Å−3) | 132,000 |
| Molecular mass derived from Vc (kDa) | 94.52 |
| Molecular mass theory from ProtParam (kDa) | 89.63 |
| SASBDB accession code | SASDK82 |
Linker flexibility between the helicase and protease regulates CHIKV RNA replication and virus production.
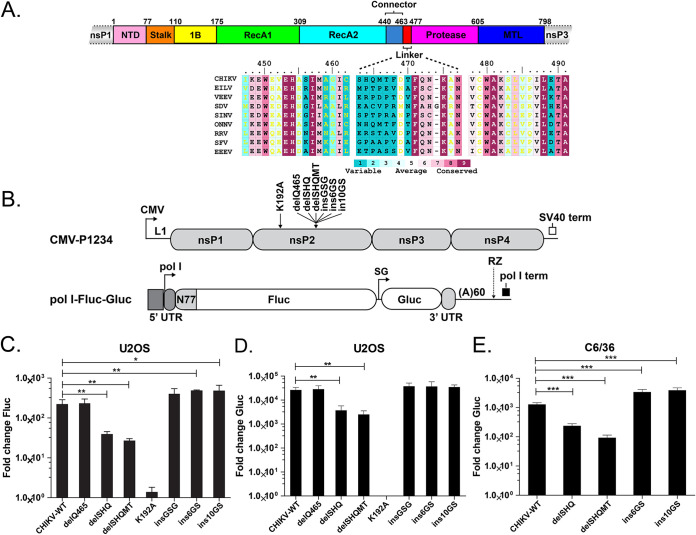
As shown in Fig. 2A, the N-terminal region of nsP2 (residues 1 to 462) consists of an N-terminal domain (NTD) and the superfamily 1 (SF1) RNA helicase core. The C-terminal region of nsP2 (residues 477 to 798) consists of a cysteine protease domain and a methyltransferase-like (MTL) domain. The N- and C-terminal regions are connected through a short linker (residues 463 to 476) (Fig. 2A). Alignment of nsP2 sequences from different alphaviruses revealed that the linker sequence, especially residues 463 to 468, is not well conserved (Fig. 2A). To characterize the biological roles of the nsP2 linker, a CHIKV trans-replicase system was used to investigate RNA synthesis (15, 27). Six mutations were introduced, with various linker lengths and thus various protein conformation flexibilities. Three nsP2 deletion mutations, the deletion of Q465 (delQ465), deletion of residues S463 to Q465 (delSHQ), and deletion of residues S463 to T467 (delSHQMT), were created. Three insertion mutations, consisting of three (insGSG), six (ins6GS), or 10 (ins10GS) flexible residues, were inserted after the Q465 position of nsP2. The previously described K192A substitution in the nsP2 nucleoside triphosphate (NTP) binding site and a mutation in the active site of nsP4 (8) were used as negative controls. As shown in Fig. 2C and D, an increase in neither the firefly luciferase (Fluc) (a marker of genomic RNA synthesis) or Gaussia luciferase (Gluc) (a marker of SG RNA synthesis) signal was observed for the construct harboring the K192A substitution. The mutant harboring one amino acid deletion (delQ465) had both replication and transcription activities similar to those of the wild-type (wt) trans-replicase. Significant reduction in both Fluc and Gluc expression levels was observed for mutants harboring larger deletions (delSHQ and delSHQMT); these decreases were approximately 10-fold, and the decreased expression levels were slightly more prominent in the case of the delSHQMT mutation. In contrast, replicases harboring insertions, especially the ins6GS and ins10GS mutants, had increased capacities to increase the Fluc expression. However, the Gluc expression of all three replicases harboring insertions was similar to that of the wt replicase (Fig. 2C and D). To determine if the observed effects were cell type specific, the two largest deletions (delSHQ and delSHQMT) and insertions (ins6GS and ins10GS) were also introduced into the Ubi-P1234 vector for the trans-replicase assay in Aedes albopictus cells (27). The results were similar to those obtained in human cells. Gluc expression in the deletion and insertion mutants was significantly reduced and significantly increased, respectively (Fig. 2E). These data suggest that the flexibility of the interdomain is critical for the synthesis of both genomic RNA and SG RNA. The effect is independent of the host cell type, as RNA synthesis in human and mosquito cells was affected similarly. In summary, our data indicate that linker flexibility is required for the efficient synthesis of viral RNA.
FIG 2.
Effects of nsP2 linker mutants on the activity of CHIKV trans-replicase. (A) Schematic representation of the domain arrangement of CHIKV nsP2. Domains of nsP2 are colored as in Fig. 1D. Sequence alignment of the nsP2 linker regions from alphaviruses, including CHIKV, Eilat virus (EILV), Venezuelan equine encephalitis virus (VEEV), sleeping disease virus (SDV), SINV, o’nyong’nyong virus (ONNV), Ross River virus (RRV), Semliki Forest virus (SFV) and Eastern equine encephalitis virus (EEEV). (B) Schematic representation of CMV-P1234 (Ubi-P1234 has the same design except that it uses the Aedes aegypti polyubiquitin promoter) and pol I-Fluc-Gluc constructs. The trans-replication system for human cells is composed of a cytomegalovirus promoter-based plasmid (CMV-P1234) for the expression of the CHIKV replicase and a plasmid for the production of replication-competent RNA containing firefly luciferase (Fluc) and Gaussia luciferase (Gluc) reporters. CMV, immediate early promoter of human cytomegalovirus; L1, leader region of herpes simplex virus thymidine kinase gene with an artificial intron; SV40 term, simian virus 40 late polyadenylation region; polI, truncated promoter for human or Aedes albopictus RNA polymerase I. The 5′ and 3′ untranslated regions (UTRs) are from CHIKV. N77, region encoding the 77 N-terminal amino acid residues of nsP1; SG, CHIKV SG promoter; RZ, antisense strand ribozyme of hepatitis delta virus; polI term, terminator for RNA polymerase I from mouse or from Aedes albopictus. Positions of linker mutations (delQ465, delSHQ, delSHQMT, insGSG, ins6GS, and ins10GS) and NTP binding site mutations (K192A) are indicated. In this assay, the increases in expression levels of Fluc and Gluc represent the production of full-length genomic RNA and SG RNA, respectively. (C and D) Effects of introduced mutations on CHIKV replicase activity in human cells. U2O2 cells were cotransfected with CMV-P1234 or its mutants and HSPolI-Fluc-Gluc plasmids; as a negative control, CMV-P1234GAA, which lacks polymerase activity, was used instead of CMV-P1234. Cells were incubated at 37°C and lysed 18 h posttransfection. Fluc (marker of replication) (C) and Gluc (marker of transcription) (D) activities produced by analyzed replicases were normalized to the P1234GAA control. The value obtained for the P1234GAA control was set as 1. The means and SD from three independent experiments are shown. *, P < 0.05; **, P < 0.01 (Student's unpaired t test). (E) Effects of introduced mutations on the transcriptional activity of CHIKV replicase in Aedes albopictus C6/36 cells. C6/36 cells were cotransfected with Ubi-P1234 or its mutants and the AlbPolI-Fluc-Gluc plasmid; as a negative control, Ubi-P1234GAA, which lacks polymerase activity, was used instead of Ubi-P1234. Cells were incubated at 28°C and lysed 48 h posttransfection. Gluc (marker of transcription) activities produced by analyzed replicases were normalized to the P1234GAA control. The value obtained for the P1234GAA control was set as 1. The means and SD from three independent experiments are shown; ***, P < 0.001 (Student's unpaired t test).
Deletion of three or five residues from the interdomain linker completely abolished the infectivity of CHIKV.
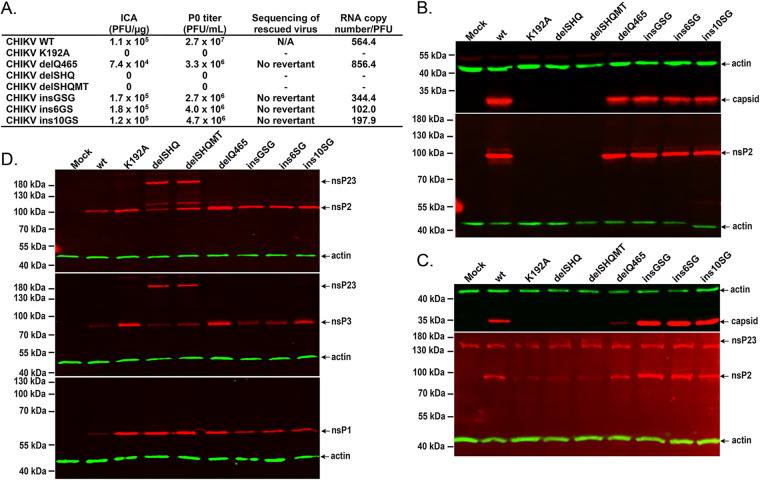
To analyze the effect of the mutations on viral infectivity, all the above mutations were introduced into an infectious cDNA (icDNA) clone of CHIKV. An infectious-center assay (ICA) performed using BHK-21 cells revealed that all three insertions slightly increased the infectivity of the corresponding icDNAs. A slight reduction was observed for the icDNA harboring the delQ465 mutation. Despite increased infectivity, the final titers of all mutant viruses harboring insertions in the linker region were lower than that of the wt CHIKV. Sequencing of mutated regions revealed that all introduced mutations were preserved in the genomes of the rescued viruses. Quantitative reverse transcription-PCR (qRT-PCR) analysis revealed a trend similar to that observed in the ICA. Virions harboring genomes with the delQ465 mutation had slightly reduced infectivity (i.e., the RNA copy number/PFU ratio was higher in the delQ465 mutant virus than in the wt virus). The opposite was observed for virions harboring the insertions, suggesting that the insGSG, ins6GS, and ins10GS mutations reduced particle production/release but had no negative impact on the viral infectivity (Fig. 3A).
FIG 3.
Effects of nsP2 linker mutants on CHIKV production. (A) Effects of introduced mutations on the efficiency of viral rescue and titers of viral progeny. The experiment was repeated three times with similar results, and data from one reproducible experiment are shown. Capsid protein (top) and CHIKV nsP2 (bottom) expression in (B) BHK-21 and (C) U2OS cells transfected with plasmids containing icDNA of wt CHIKV or its mutants. (D) CHIKV nsP2 (top), CHIKV nsP3 (middle), and CHIKV nsP1 (bottom) expression in U2OS cells transfected with plasmids expressing the indicated replicases. On the Western blots, nsP1 to -3 and capsid proteins were detected using corresponding polyclonal antibodies; antibodies against actin were used to detect the loading control.
Western blot analysis of nsP2 and capsid protein expression in the BHK-21 cells transfected with these icDNAs revealed that all these mutants expressed both of the proteins at a level very similar to that of wt CHIKV (Fig. 3B). Consistent with previously reported results (8), no plaques were obtained for the icDNA harboring the K192A substitution. Unexpectedly, the infectivity of icDNAs harboring the delSHQ and delSHQMT deletions was also below the detection limit (Fig. 3B). No release of infectious virus (Fig. 3A) or expression of viral proteins (Fig. 3B) was observed, confirming that these mutations were lethal for CHIKV. When the rescue experiment was performed using U2OS cells, identical data were obtained. Viruses from icDNAs harboring the wt CHIKV, delQ465 mutant, and insertion mutants were rescued, but viruses from icDNAs harboring the K192A, delSHQ, or delSHQMT mutation were unable to be rescued. Similarly, in BHK-21 cells, expression of the capsid protein was not detected in transfected cells with these three plasmids (Fig. 3B). Expression of nsP2 was detectable; however, as expected, it occurred at a lower level in BHK-21 cells than in cells transfected using icDNAs of the wt virus or viable mutants. In the lysates of cells transfected with icDNAs of delSHQ or delSHQMT, antibodies against nsP2 also recognized a protein with a higher molecular mass, indicating that P123 processing may be defective (Fig. 3C). Therefore, we further analyzed nsP expression in U2OS cells transfected using expression plasmids for replicases containing the above-mentioned mutations. As shown in Fig. 3D, the same additional band was detected for replicases carrying the delSHQ and delSHQMT mutations. Western blot analyses using CHIKV nsP1 and nsP3 antibodies revealed that this protein band corresponded to unprocessed nsP23.
Varying the length of the linker does not affect the intrinsic enzymatic activities of nsP2.
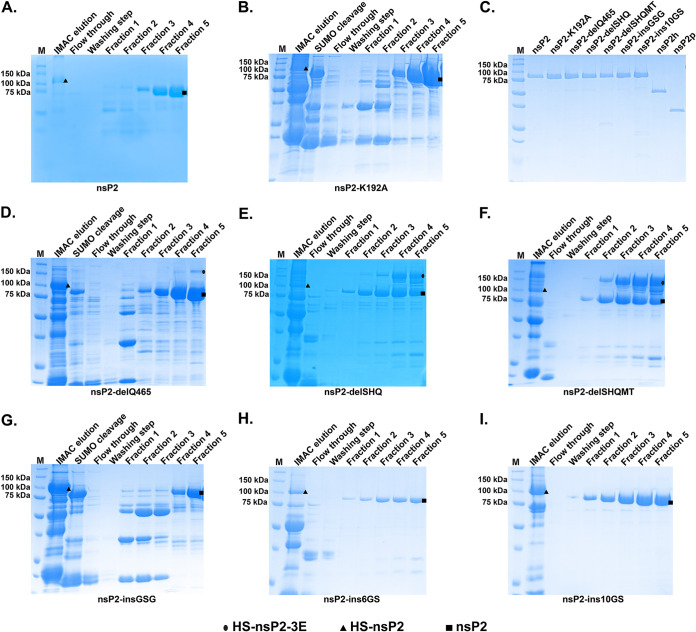
To determine whether the linker mutations affect the intrinsic enzymatic activities of nsP2, wt nsP2, linker mutants, nsP2h, and nsP2p were expressed and purified as recombinant proteins. The above-mentioned mutations were introduced into the His6-SUMO-nsP2-3-EGFP (HS-nsP2-3E) expression construct (15). The self-cleavage between nsP2 and nsP3-E was highly efficient in releasing HS-nsP2 during expression in Escherichia coli. Because nsP2 binds nucleic acids, heparin-Sepharose affinity chromatography was used to further purify the full-length nsP2 and mutants. During purification, the full-length P23E was not detected in any of the proteins except nsP23E-delSHQ (Fig. 4E) and nsP23E-delSHQMT (Fig. 4F). For these two recombinant proteins, the nsP2/nsP3 junction was not processed as efficiently as observed for wt nsP2 and other mutants (Fig. 4). This observation is in agreement with Western blot analysis of replicases containing delSHQ and delSHQMT in mammalian cells (Fig. 3D).
FIG 4.
SDS-PAGE of the purified recombinant proteins. After purification using immobilized metal affinity chromatography (IMAC), the purified proteins were incubated with SUMO protease to remove the N-terminal His6-SUMO tag before purification using heparin-Sepharose affinity chromatography. SDS-PAGE of (A) nsP2, (B) nsP2-K192A, (D) nsP2-delQ465, (E) nsP2-delSHQ, (F) nsP2-delSHQMT, (G) nsP2-insGSG, (H) nsP2-ins6GS, and (I) nsP2-ins10GS after heparin-Sepharose affinity chromatography. (C) Ten micromoles of purified recombinant proteins was separated using SDS-PAGE. Ovals, triangles, and squares indicate HS-nsP2-3E, HS-nsP2, and nsP2, respectively. HS, His6-SUMO tag; E, EGFP tag.
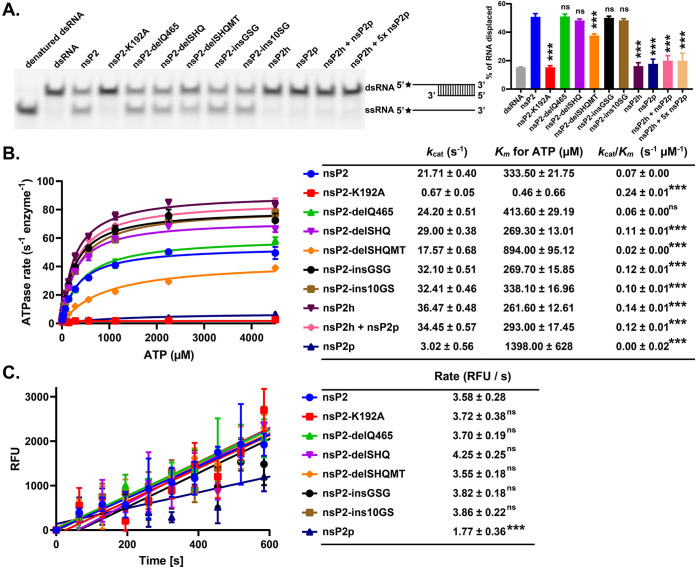
To further investigate how the flexibility of the nsP2 linker regulates the intramolecular communication between the helicase and protease, we compared the enzymatic activities of nsP2, nsP2 mutants, nsP2h, and nsP2p. As shown in Fig. 5A, the RNA unwinding activities of nsP2-delQ465, nsP2-delSHQ, nsP2-insGSG, and nsP2-ins10GS were similar to those of wt nsP2, while nsP2-delSHQMT possessed a slightly weaker RNA unwinding activity. Similar to what was reported previously (9), nsP2h alone did not possess any RNA unwinding activities above the background level. Furthermore, there was no detectable unwinding activity for nsP2h in the presence of an equimolar amount or 5-fold molar excess of nsP2p. Thus, to unwind dsRNA, nsP2h and nsP2p must be physically connected. Unlike in the previous report (9), the ATPase activity of nsP2 was found to be slightly lower than that of nsP2h. This discrepancy can be attributed to different assay conditions and/or different strategies used for obtaining these recombinant proteins. Importantly, the ATPase activity of nsP2h remained unchanged irrespective of the addition of the individual nsP2p, suggesting that nsP2p does not affect ATP hydrolysis of nsP2h when these domains are physically separated.
FIG 5.
Effects of nsP2 mutations on the RNA helicase, ATPase, and protease activities of the purified recombinant proteins. (A) RNA helicase activities of nsP2 and mutants were analyzed using the Alexa488-dsRNA28/16 substrate with a 12-base 5′ overhang. The band intensities were quantified by ImageJ. Alexa Fluor 488 is indicated by a black star. (B) ATP hydrolysis was carried out at various ATP concentrations. (C) Protease activities were determined by use of a FRET-based octapeptide substrate [DABCYL-RAGG↓YIFS-(Glu-EDANS)-NH2]. All enzymatic assays were performed in triplicate, and the data are means and SD. Significance was determined by one-way analysis of variance (ANOVA) with Dunnett’s posttest comparing mutants to nsP2 (vehicle control). ***, P < 0.001; ns, not significant.
While the ATPase activity of nsP2-delQ465 was similar to that of wt nsP2, the activities of nsP2-delSHQ, nsP2-insGSG, and nsP2-ins10GS were slightly increased (Fig. 5B). Notably, the ATPase activity of nsP2-delSHQMT was slightly decreased. For the FRET-based protease assay, the peptide (RAGG↓YIFS), corresponding to the nsP3/nsP4 junction of CHIKV P1234, was used as the substrate (28). nsP2p was found to be less active than full-length nsP2. As expected, the K192A mutation that completely inactivated the ATPase and RNA helicase activities of nsP2 had no negative impacts on the protease activity of nsP2 (Fig. 5C). All insertion/deletion mutants had a protease activity comparable to that of the wt nsP2. These data are different from the processing defect of the nsP2/nsP3 site of nsP23E-delSHQ and nsP23E-delSHQMT observed during purification (Fig. 4E and F) and in cells (Fig. 3C and D). This is likely because of the differences between the small peptide substrate and the junctions of large polyproteins and highlights the different requirements for nsP2/nsP3 cleavage from those for the other two cleavage sites (10, 13). Collectively, our findings suggest that small differences in the length of the linker have minor or no detectable impacts on the enzymatic activities of nsP2.
Varying the length of the linker affects the RNA binding and RNA-stimulated enzymatic activities of nsP2.
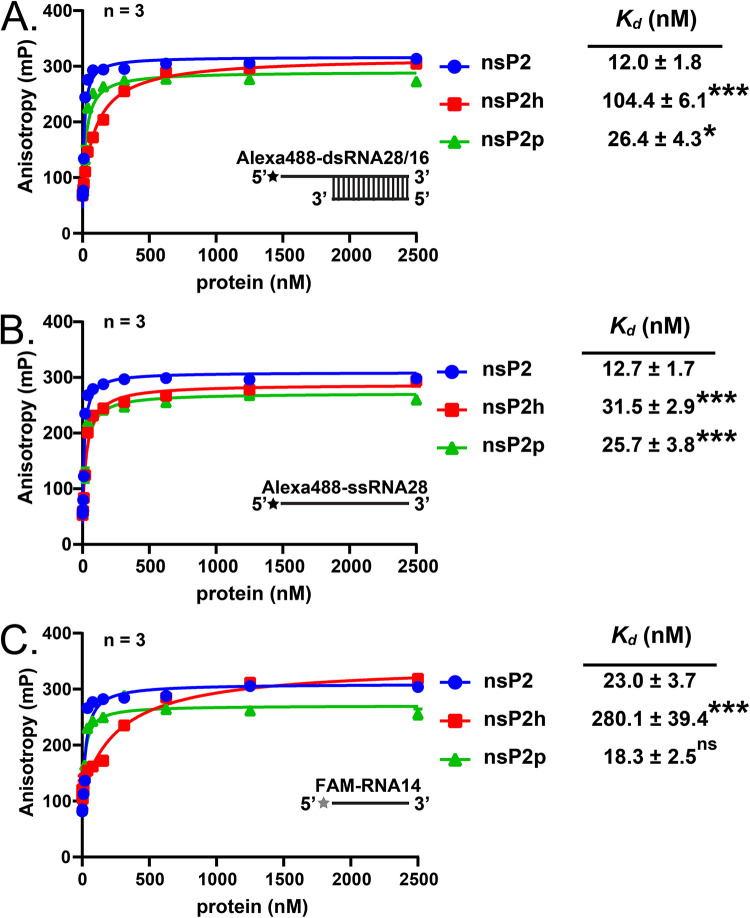
As the cleavage of the nsP2/nsP3 site of alphaviruses is driven by the macromolecular assembly (10), the observed defect (Fig. 3C and D and 4C and E) may indicate a possibility that a decrease in the length of the nsP2 linker has a specific impact on the processes requiring interactions between viral components. To probe the intermolecular communication, we measured the RNA-binding activities of nsP2, nsP2p, and nsP2h. An Alexa Fluor 488-labeled RNA helicase substrate on the 5′ end (Alexa488-dsRNA28/16 and Alexa488-ssRNA28) was used for RNA helicase assays (Fig. 5A), and the 5′ fluorescein (FAM)-labeled 14-mer single-stranded RNA (ssRNA) oligonucleotide (RNA14) described previously (8, 9) was used at various concentrations for RNA binding analyses. As shown in Fig. 6A to C, the RNA binding affinity of nsP2p was comparable to that of full-length nsP2. However, nsP2h had an ∼15-fold-lower RNA binding affinity against FAM-RNA14 than nsP2p (Fig. 6C). The RNA binding affinity of nsP2h was increased against the longer RNA substrates Alexa488-dsRNA28/16 and Alexa488-ssRNA28 (Fig. 6A and B). Nevertheless, the binding of these RNAs to nsP2h was still weaker than their binding to nsP2 and nsP2p. Thus, the protease region is an important contributor to RNA binding.
FIG 6.
Analysis of the relation of RNA binding with the purified recombinant proteins. RNA-binding analysis of the purified recombinant proteins using (A) Alexa488-dsRNA28/16, (B) Alexa488-ssRNA28, and (C) FAM-RNA14. The RNA-binding analysis was performed at various protein concentrations. Alexa Fluor 488 and FAM are indicated by black and gray stars, respectively. All enzymatic assays were performed in triplicate, and the data are means and SD. The data were analyzed by one-way ANOVA with Dunnett’s posttest for RNA-binding affinity between mutants and nsP2 (vehicle control). *, P < 0.05; ***, P < 0.001; ns, not significant.
Next, we evaluated the effect of RNA binding to the ATPase and protease activities of nsP2 and nsP2 mutants using RNA14 and a 28-mer ssRNA oligonucleotide (RNA28). ATPase activity of nsP2 and nsP2-delQ465 was stimulated only by RNA28 (Table 2). The abilities of nsP2, its mutants, and nsP2p to process the RAGG↓YIFS peptide (based on the nsP3/nsP4 junction) were upregulated in the presence of single-stranded RNA14 and RNA28 (Table 3). RNA14 increased the cleavage of this substrate by nsP2p, nsP2 and its mutant variants by 4- to 6-fold. The increased length of oligonucleotide RNA could further stimulate the proteolytic activity of all analyzed enzymes. In general, 8- to 10-fold stimulation was observed in the presence of RNA28; the exception was nsP2-delSHQMT activity, which had a 5-fold stimulation. However, the cleavage of other substrates, especially nsP2/nsP3 junctions, in the context of viral polyprotein may be affected differently. Overall, our findings suggest that the intramolecular flexibility and the dynamic interaction between nsP2 and RNA regulate viral polyprotein processing and may contribute to the formation and optimal function of viral RCs.
TABLE 2.
ATP hydrolysis studies of the purified recombinant proteins in the presence or absence of RNA28
| Protein |
kcat (s−1) |
Km for ATP (μM) |
kcat/Km (s−1 μM−1) |
|||
|---|---|---|---|---|---|---|
| Apo | RNA28 | Apo | RNA28 | Apo | RNA28 | |
| nsP2 | 21.71 ± 0.40 | 35.37 ± 0.55 | 333.50 ± 21.75 | 331.90 ± 18.23 | 0.07 ± 0.02 | 0.11 ± 0.02 |
| nsP2-delQ465 | 24.20 ± 0.51 | 46.07 ± 0.58 | 413.60 ± 29.19 | 257.70 ± 11.98 | 0.06 ± 0.01 | 0.18 ± 0.01 |
| nsP2-delSHQ | 29.00 ± 0.38 | 31.28 ± 0.49 | 269.30 ± 13.01 | 294.40 ± 16.48 | 0.11 ± 0.01 | 0.11 ± 0.01 |
| nsP2-delSHQMT | 17.57 ± 0.68 | 18.71 ± 0.84 | 894.00 ± 95.12 | 775.80 ± 99.60 | 0.02 ± 0.01 | 0.02 ± 0.01 |
| nsP2-insGSG | 32.10 ± 0.51 | 36.86 ± 0.60 | 269.70 ± 15.85 | 309.50 ± 18.00 | 0.12 ± 0.01 | 0.12 ± 0.01 |
| nsP2-ins10GS | 32.41 ± 0.46 | 34.88 ± 0.60 | 338.10 ± 16.96 | 339.20 ± 20.58 | 0.10 ± 0.00 | 0.10 ± 0.00 |
| nsP2h | 36.47 ± 0.48 | 37.70 ± 0.41 | 261.60 ± 12.61 | 278.30 ± 10.98 | 0.14 ± 0.02 | 0.14 ± 0.00 |
| nsP2h + nsP2p | 34.45 ± 0.57 | 37.22 ± 0.64 | 293.00 ± 17.45 | 337.30 ± 20.34 | 0.12 ± 0.06 | 0.11 ± 0.01 |
| nsP2-K192A | 0.67 ± 0.05 | 0.46 ± 0.66 | 0.24 ± 0.01 | |||
| nsP2p | 3.02 ± 0.56 | 1398.00 ± 628 | 0.00 ± 0.02 | |||
TABLE 3.
Fold change of proteolytic activity of recombinant proteins in the presence of RNA14 and RNA28
| Protein | Proteolysis rate (RFU/s)a | Fold change |
|
|---|---|---|---|
| RNA14 | RNA28 | ||
| nsP2 | 3.58 ± 0.28 | 4.95 ± 0.49 | 8.81 ± 0.40 |
| nsP2-K192A | 3.72 ± 0.38 | 4.96 ± 0.17 | 6.80 ± 0.61 |
| nsP2-delQ465 | 3.70 ± 0.19 | 5.94 ± 0.19 | 9.62 ± 0.39 |
| nsP2-delSHQ | 4.25 ± 0.25 | 4.58 ± 0.84 | 7.54 ± 0.44 |
| nsP2-delSHQMT | 3.55 ± 0.18 | 3.93 ± 0.04 | 4.83 ± 0.21 |
| nsP2-insGSG | 3.82 ± 0.18 | 5.74 ± 0.09 | 8.48 ± 0.95 |
| nsP2-ins10GS | 3.86 ± 0.22 | 5.43 ± 0.15 | 7.74 ± 0.51 |
| nsP2p | 1.77 ± 0.36 | 5.03 ± 0.79 | 6.53 ± 0.84 |
RFU, relative fluorescence units.
DISCUSSION
Alphavirus nsP2 is composed of an N-terminal helicase region and a C-terminal protease region and plays multiple roles in viral RNA replication and interactions with the host. The crystal structures of nsP2p from other alphaviruses, including Venezuelan equine encephalitis virus (VEEV) (PDB ID 2HWK), SINV (PDB ID 4GUA), and CHIKV (PDB ID 4ZTB), have been reported (12, 29, 30). In our recent study, we determined the crystal structure of a ternary complex of CHIKV nsP2h bound to a 14-mer ssRNA and the nucleotide analog ADP-AlF4 (8). However, there was no information concerning the structural and conformational arrangements of the full-length nsP2. Our SAXS model of full-length nsP2 suggests that the interdomain linker is flexible and loosely connected to the N-terminal helicase and C-terminal protease regions (Fig. 1B and C). We also observed that the CHIKV replicases harboring nsP2 linker insertion mutants possessed elevated viral RNA synthesis in both human and mosquito cells (Fig. 2C and E). In contrast, the mutant replicases with truncated linkers (delSHQ and delSHQMT) produced fewer genomic and SG RNAs in both cell types (Fig. 2D and E). Interestingly, though the negative effect of these deletion mutations on RNA synthesis was modest, virus rescue was not observed for icDNAs harboring the delSHQ and delSHQMT mutations (Fig. 3A).
The precise timing of nsP processing events has been determined to be critical in the infection cycle of alphaviruses, and these processing events are the key regulatory events in the trans-cleavage of nsP2/nsP3 (10). Studies have suggested that the presence of the conserved macrodomain of nsP3 and the N terminus of full-length nsP2 are needed for the precise positioning of the nsP2/nsP3 site by the nsP2 protease active-site residue (10). In this study, we observed that the flexibility of the interdomain linker of nsP2 influences the processing of the nsP2/nsP3 junction (Fig. 3C and D and 4E and F). These findings suggest that the decreased flexibility of the interdomain linker of nsP2 may restrict the accessibility of the N terminus of the full-length nsP2 to the nsP2/nsP3 site and subsequently affect its trans-cleavage (Fig. 4E and F), thereby contributing to the observed lack of infectivity (Fig. 2E). Compared to larger deletions, the removal of one residue from the interdomain did not reduce the linker flexibility, and the corresponding enzyme, replicase, and virus icDNAs had wt-like properties in all of the cell-free and cell-based assays used in this study. These data indicate that CHIKV can tolerate a slight shortening of the interdomain linker of nsP2. Additional studies are required to reveal the exact molecular mechanism(s) behind these observed defects.
Previous studies have shown that the RNA helicase activity of CHIKV nsP2 is dependent on the presence of the C-terminal protease region (9). Das et al. (9) hypothesized that a strong binding affinity is needed for nsP2 to bind the dsRNA substrate due to the low processivity of nsP2 helicase. Consistent with this hypothesis, we observed that nsP2p indeed possesses high RNA binding affinity against dsRNA that is slightly lower than that of the full-length nsP2 (Fig. 6A). Nevertheless, simply adding nsP2p to the reaction mixture did not facilitate the ability of nsP2h to unwind the dsRNA substrate (Fig. 6A). Thus, the RNA helicase activity of CHIKV requires nsP2h to be connected with nsP2p via a linker region. This finding further supports the idea that high RNA binding affinity is required for unwinding the dsRNA substrate. Our study shows that mildly increasing and decreasing the length of the flexible linker does not affect the functional cross talk between the two regions of nsP2 for unwinding the dsRNA substrate. On the whole, the presence of both the C-terminal protease region and the flexible linker is critical for maintaining the strong RNA binding affinity necessary to compensate for the low processivity of nsP2 in unwinding the dsRNA substrate.
The addition of flexible residues to the interdomain linker of nsP2 (insGSG, ins6GS, and ins10GS) slightly accelerated the synthesis of viral genomic RNA in human cells (Fig. 2C to E) and increased SG RNA synthesis in mosquito cells (Fig. 2F). These findings suggest that increased flexibility of the interdomain might provide a notable level of intramolecular movement freedom to facilitate the assembly of an efficient viral RC. The crystal structure of the uncleaved SINV P23 precursor suggests that the inaccessibility of the nsP2/nsP3 cleavage site by the nsP2 protease active site residue (C478) excludes the possibility of cis cleavage of the nsP2/nsP3 site (26, 30). Therefore, we hypothesized that the increase in flexibility of the nsP2 linker might accelerate the movement freedom of the nsP2 protease active site and subsequently affect the cis cleavage of the nsP1/nsP2 site and/or allow better coordination between multiple functions of nsP2. According to the solution structure of the full-length nsP2, the N terminus of nsP2 is 54 to 60 Å away from the nsP2 protease active site (Fig. 1D). The increase in linker flexibility might shorten the distance and reaction time between two domains, subsequently altering nonstructural-polyprotein processing and enhancing viral RNA replication. Early studies have shown that the insertion of EGFP into the interdomain linker of nsP2 of SINV slightly reduced polyprotein processing; the same insertion also slightly reduced RNA replication (14, 15, 17). EGFP insertion might restrict linker flexibility and the movement of freedom between the N- and C-terminal regions of nsP2. This observation is also in line with the above-described effects of the delSHQ and delSHQMT mutations.
It is a common observation that the intramolecular dynamics of multifunctional proteins play regulatory roles in viral replication, RC assembly, and infectivity. For example, nonstructural protein 3 (NS3) of flavivirus and hepacivirus (family Flaviviridae) also consists of protease and RNA helicase regions (31–34), verifying that the coexistence of the helicase-protease or protease-helicase domains in one molecule is common in the genomic organization of positive-strand ssRNA viruses. Furthermore, the functional interplay of both domains depends on the flexibility of the interdomain linker. Previous studies have indicated that the manipulation of the flexibility of the interdomain linker of NS3 exhibits a defect in the replication of dengue virus (DENV) (33) and hepatitis C virus (35), demonstrating the importance of linker flexibility in genomic RNA synthesis. Moreover, single-point mutations in the interdomain linker of DENV NS3 almost abolished infectious-particle formation but showed marginal inhibition of NS3 enzymatic activities (33, 36). Gebhard et al. (36) hypothesized that the multifunctional NS3 of DENV participates in the production of infectious particles (37) by interacting with host factors. These findings from different virus families further support the assertion that the flexibility of the helicase-protease interdomain region is crucial in infectious-particle production.
The interdomain linker of nsP2 does not contain any visible conserved sequences and, due to flexibility, may allow multiple conformations of nsP2 in solution. Interestingly, the reconstructed solution structure of nsP2 revealed a preferred orientation between the helicase and protease regions. The SAXS model showed that the N-terminal accessory domains of the helicase are arranged close to the N-terminal region of the protease domain (Fig. 1D). Based on our findings, we propose that the binding of dsRNA oligonucleotides harboring a 5′ overhang results in a conformational rearrangement of nsP2. Highly positive charges on the surface of the MTL domain (Fig. 1E) are responsible for the observed RNA binding affinity of nsP2p and nsP2 (Fig. 6A to C). The dsRNA substrate may bind to nsP2p to form an intermediate nsP2p-RNA complex. The flexible linker might arrange the 5′ overhang region of RNA of the complex toward nsP2h. Subsequently, the RNA binds to the central groove of nsP2h in a 5′-to-3′ direction (8). We also observed that complex formation between nsP2 and RNA could affect the protease activity. Importantly, we observed that the effect of RNA on processing varies; in the assay used in this study, the level of stimulation depended on the length of the RNA oligonucleotide used for nsP2 binding (Table 3). It is therefore plausible that in viral infection, the effect may depend on multiple factors, such as RNA (single or double stranded), nsP2 (fully or partially processed from the polyprotein precursor), the presentation of the cleavage site, and the presence of additional viral or host factors. Regardless, the flexible linker might play a role in coordinating RNA binding between the helicase and protease components for controlling the precise timing of processing events to regulate viral replication as well as to produce infectious particles in host cells.
In conclusion, the two functional regions of nsP2, helicase and protease, are connected with a flexible linker that gives the molecule a flexible and elongated structure in solution. The interdomain flexibility of nsP2 has evolved to be optimal “molecular gymnastics,” which enable nsP2 to perform multiple functions, such as processing polyproteins, assembling viral RCs, and interacting with the host.
MATERIALS AND METHODS
Expression constructs.
The CHIKV P123E4 plasmid, encoding a polyprotein containing the insertion of EGFP after residue 1,716 of P1234 (residue 383 of nsP3) (15), was used as a template for amplifying and cloning sequences encoding full-length nsP2. The region encoding CHIKV P23E (residues 536 to 1958 of P123E4) was cloned into a modified pSUMO-LIC expression vector downstream of a sequence encoding a cleavable N-terminal His6-SUMO tag. The full-length nsP2 (residues 536 to 1333 of P1234) was obtained from the P23E precursor after its self-cleavage. The construct used for nsP2h expression was described previously (8). The region encoding CHIKV nsP2p (residues 1008 to 1331 of P1234; GenBank accession no. KC149887.1) was cloned into a pNIC28-Bsa4 expression vector downstream of the region encoding a cleavable N-terminal His6 tag and tobacco etch virus (His6-TEV) protease cleavage site. Insertion of 3 to 10 flexible residues (Gly and Ser) after Q465, deletions in the region spanning from S463 to T467, and the K192A substitution were performed using site-directed mutagenesis. Fragments containing these mutations were introduced into the CHIKV P23E expression constructs, and CMV-ICRES1 (icDNA clone of the CHIKV LR2006OPY1 isolate, ECSA genotype) and the CHIKV trans-replicase system vectors CMV-P1234 and Ubi-P1234 were used for the subcloning procedures.
Recombinant-protein production.
For recombinant-protein production, a previously described method (8) was used. Briefly, all the constructs were transformed into the Escherichia coli Rosetta 2(DE3) strain. The bacterial culture containing the plasmid of interest was cultured at 37°C with shaking at 200 rpm in lysogeny broth (LB) medium supplemented with antibiotics. After the optical density at 600 nm (OD600) reached ∼0.8, isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added to a final concentration of 1 mM, and the culture was further cultured at 18°C with shaking at 200 rpm overnight. Subsequently, the bacterial cultures were harvested and lysed, and the proteins were purified from the supernatant using HisPur nickel-nitrilotriacetic acid (Ni-NTA) resin (Thermo Fisher Scientific). To remove the N-terminal His6-SUMO or His6-TEV tag, the purified proteins were incubated with the SUMO or TEV protease at an approximate molar ratio of 1:20 at 4°C overnight. The proteins were further purified through HiTrap heparin HP affinity columns (GE Healthcare Life Sciences) to remove the cleaved affinity tag and subsequently separated using either the HiLoad 16/600 Superdex 75 or 200 prep-grade column (GE Healthcare Life Sciences). The purified recombinant proteins were analyzed using 10% SDS-PAGE.
SAXS experiment and data analysis.
The purified recombinant nsP2 proteins were concentrated to 6.12 mg/ml. The samples were measured at 288 K at the 23A small/wide angle X-ray scattering (SWAXS) end station of the National Synchrotron Radiation Research Center (Hsinchu, Taiwan) (38). First, 0.1 ml of nsP2 was injected into an HPLC system (Agilent chromatographic system, 1260 series) equipped with an online size exclusion column at a flow rate of 0.035 ml/min for SAXS data collection. A 15-keV X-ray beam irradiated the sample flow within a thin-wall (30 μm) quartz capillary cell (2.0-mm diameter) for SAXS measurements (30 s/frame). The sample-to-detector distance (3.42 m) together with a Pilatus 1M-F area detector covers an X-ray momentum transfer q range of 0.01 Å−1 to 0.25 Å−1 (q = 4πλ−1sinθ, where 2θ is the scattering angle and λ [the wavelength of the X rays] is 0.8266 Å). The two-dimensional (2D) SAXS patterns were radially averaged, background subtracted, and scaled to the absolute scattering intensity of water at the same temperature. Primary SAXS data reduction was performed using the in-house data reduction kit. I(0) and Rg were evaluated using the Guinier approximation, with the limit qRg < 1.3 (33). Molecular modeling of nsP2 was conducted using the rigid body refinement implemented in CORAL (23), based on the crystal structures of nsP2h (PDB ID 6JIM) and nsP2p (PDB ID 4ZTB). Both nsP2h and nsP2p were treated as rigid bodies, leaving the linker as random loops in the fitting procedure.
Analysis of CHIKV replicase activity using a trans-replicase assay.
The CHIKV trans-replicase assay was performed as described previously (8, 15). One microgram of HSpol I-Fluc-Gluc and CMV-P1234 (or its mutant versions) plasmids was cotransfected into ∼4 × 105 U2OS cells growing on 12-well plates using the Lipofectamine LTX reagent (Thermo Fisher Scientific). After incubation at 37°C for 18 h, the transfected cells were lysed, and the Fluc and Gluc activities were measured using the dual-luciferase reporter assay and a GloMax SIS luminometer (Promega). For Aedes albopictus cells, 1 μg of Albpol I-Fluc-Gluc (27) and Ubi-P1234 (or its mutant versions) plasmids was cotransfected into ∼4 × 105 C6/36 cells growing on 12-well plates using the Lipofectamine LTX reagent (Thermo Fisher Scientific). After incubation at 28°C for 48 h, the transfected cells were lysed, and the Gluc activities were measured using the dual-luciferase reporter assay and a GloMax SIS luminometer (Promega). The constructs encoding inactive CHIKV nsP4 (CMV-P1234GAA and Ubi-P1234GAA) were used as negative controls in this study. All assays were repeated at least three times. For detection of nsP1, nsP2, and nsP3 expressed from CMV-P1234 and its mutant variants, U2OS cells were transfected as described above, except that HSpol I-Fluc-Gluc was not added. At 18 h posttransfection, the cells were collected, and samples were prepared and analyzed as described in the following section.
Virus analysis and Western blotting.
ICA was performed as previously described (13, 39). Briefly, 5 μg of endotoxin-free plasmids CMV-ICRES1 and its mutants containing the K192A substitution, delQ465, delSHQ, delSHQMT, insGSG, ins6GS, or ins10GS was transfected into BHK-21 cells using a Bio-Rad electroporator. Tenfold dilutions of cells were prepared using 10% transfected cells and seeded onto 6-well tissue culture plates containing 1.5 × 106 BHK-21 cells per well. After 2 h of incubation at 37°C, the cell culture medium was aspirated, and the cells were overlaid with 2 ml of growth medium supplemented with 0.8% carboxymethyl cellulose (Sigma Life Science). Plaques were stained with crystal violet after 2 to 3 days of incubation at 37°C. The rest of the transfected cells were plated into a 6-well plate, and viral stocks were harvested at 24 h posttransfection. The virus titers were determined using plaque titration on BHK-21 cells. All assays were repeated three times. Virus rescue was performed using U2OS cells. In this case, cell culture media were collected at 18 h posttransfection, and the presence of infectious virus was verified by infection of BHK-21 cells using collected media. The cells transfected using wt or mutant icDNA plasmids were harvested at the same time as the corresponding viral stocks. After being washed with 1 ml of PBS, the harvested cells were lysed, boiled, and subjected to 10% SDS-PAGE. The separated proteins were transferred to polyvinylidene difluoride membranes and detected using antibodies against CHIKV nsP1, nsP2, nsP3, capsid protein (all in-house), and β-actin (sc-47778; Santa Cruz Biotechnology). After incubation with appropriate secondary antibodies conjugated to fluorescent labels (Li-Cor), the proteins on the membrane were imaged using the Li-Cor Odyssey Fc imaging system.
RT-PCR and qRT-PCR.
RNA genomes of wt CHIKV and viruses containing the delQ465, insGSG, ins6GS, or ins10GS mutation were purified from 100 μl of P0 virus stock using a Quick-RNA miniprep kit (Zymo Research) according to the manufacturer’s instructions. RNA was eluted in 30 μl RNase-free water. Ten microliters of each obtained RNA was used for synthesis of cDNA using random primers and a First Strand cDNA synthesis kit (Thermo Fisher Scientific). Each reaction was carried out in a 20-μl volume. Each PCR was performed using DreamTaq DNA polymerase (Thermo Fisher Scientific). Eight microliters of each obtained cDNA and 300 nM primers (forward primer, 5′-CCCAAGGGTTAACCAGAAAAGGAGTTTACGC-3′; reverse primer, 5′-GGTTAAATCCGAACATTTTCCCTCCAGGCC-3′) was added to each mixture. PCR products were purified using a DNA Clean & Concentrator kit (Zymo Research) and further analyzed using Sanger sequencing. Genome copy numbers of wt CHIKV and mutant viruses were analyzed using qRT-PCR with a LightCycler 480 II instrument (Roche). Each reaction was performed in a 384-well white plate with a final volume of 10 μl containing 200 nM primers (forward primer, 5′-ATTCTGCTTACACACAGACGTCTCATG-3′; reverse primer, 5′-TTTGTCGAGTATGAGGGGTAGGCACC-3′) and 1× Hot FirePol EvaGreen qPCR Mix Plus (no ROX) (Solis BioDyne). A standard curve was obtained using 1.5 ng, 0.15 ng, 0.015 ng, 0.0015 ng, 0.00015 ng, and 0.000015 ng of CMV-ICRES1 plasmid. For accuracy, three different amounts of the above-described cDNAs (1 μl, 0.5 μl, and 0.25 μl) were used, and each sample was analyzed in triplicate. Nuclease-free water was used as a negative control. The qPCR program was as follows: 95°C for 12 min and 50 cycles of 95°C for 15 s, 63°C for 30 s and 72°C for 30 s. A melting curve analysis from 50°C to 95°C was performed after every qPCR run to exclude the formation of primer dimers and the amplification other nonspecific products.
Helicase assay.
For the helicase assay, a previously described method (9) was used with minor modifications. The 5′ Alexa Fluor 488-labeled 28-mer ssRNA oligonucleotide (Alexa488-ssRNA; 5′-AAAAAAAAAAAACCAGGCGACAUCAGCG-3′) and unlabeled 16-mer ssRNA oligonucleotide (5′-CGCUGAUGUCGCCUGG-3′) were used to generate dsRNA helicase substrate with a 12-base 5′ overhang. For the preparation of the dsRNA with a 12-base 5′ overhang, the labeled and unlabeled RNA oligonucleotides were mixed at a molar ratio of 1:1.1 in buffer containing 10 mM HEPES (pH 7.2) and 20 mM KCl. The mixture was incubated in a thermal cycler at 95°C for 1 min, followed by annealing using a decrease in temperature by 1°C per min until 22°C. The protein concentration and incubation time were optimized to study the RNA unwinding activity of nsP2 against 40 nM dsRNA substrate. Each RNA unwinding reaction mixture contained a 0.1 to 1 μM concentration of the protein of interest, a 40 nM concentration of dsRNA substrate with a 12-base 5′ overhang, an 800 nM concentration of unlabeled RNA trap (5′-CCAGGCGACAUCAGCG-3′), and 20 U of RNase-OUT inhibitor (Thermo Fisher Scientific) in the assay buffer (40 mM HEPES [pH 7.5], 2 mM dithiothreitol [DTT], and 12 mM NaCl). The reaction mixture was incubated for 20 min at 26°C in a thermal cycler before the addition of the 3.5 mM ATP-and-magnesium acetate (ATP-Mg2+) mixture. The reaction mixtures were incubated for 5 to 120 min and terminated by adding the stop solution (100 mM Tris-HCl [pH 7.5], 0.1% bromophenol blue, 1% SDS, 50 mM EDTA, and 50% glycerol). The ssRNA and dsRNA were separated by electrophoresis using a 15% nondenaturing polyacrylamide gel at 4°C and visualized using a Typhoon imager (GE Healthcare). The band intensity was quantified by ImageJ.
NADH coupled ATPase activity assay.
The NADH-coupled ATPase activity assay was carried out by a previously described method (8). Briefly, a total volume of 30 μl containing a 2.5 nM concentration of the protein of interest, 1 μM RNA, 1 mM NADH, 100 U/ml lactic dehydrogenase, 500 U/ml pyruvate kinase, and 2.5 mM phosphoenolpyruvic acid in the ATPase assay buffer (25 mM MOPS [pH 7.4], 150 mM KCl, 10 mM MgCl2, 2 mM DTT, and 0.01% Triton X-100) was transferred into a 96-well half-area clear flat-bottom microplate (Corning) and incubated for 2 h at room temperature before the addition of a series of 12 concentrations of ATP. The measurement was performed using a Spark 10M microplate reader (Tecan) at 340 nM.
Protease assay.
The protease assay was based on the use of a fluorescence resonance energy transfer (FRET)-based substrate as described previously (6, 28). The peptide containing 4 residues from both the P and P′ sides of the CHIKV nsP3/nsP4 cleavage site (RAGG↓YIFS) was labeled with a quencher, 4-[(4-(dimethylamino)phenyl-azo]benzoic acid (DABCYL), and a fluorescent molecule, 5-[(2-aminoethyl) amino] naphthalene-1-sulfonic acid (EDANS). The assay was performed in a 96-well half-area black clear-bottom microplate (Corning). The total volume of 30 μl contained 0.4 μM protein and 40 μM substrate [DABCYL-RAGG↓YIFS-(E-EDANS)-NH2; GL Biochem] in assay buffer (25 mM HEPES [pH 7.5], 2 mM DTT, and 5% DMSO). For the protease assay in the presence of RNA, the final concentration of RNA14 (5′-GUUUUUAAUAUUUC-3′) and RNA28 (5′-GUUUUUAAUAUUUCAAAAAAAAAAAAAA-3′) was 4.5 μM. The fluorescence readings of fluorophores released by protease activity were measured every 2 min for 1 h at 28°C using a Tecan Spark 10M microplate reader (Tecan) at excitation and emission wavelengths of 340 nm and 490 nm, respectively.
RNA-binding assay.
The RNA helicase substrate (Alexa488-dsRNA and Alexa488-ssRNA) and the 5′ fluorescein (FAM)-labeled 14-mer ssRNA oligonucleotide (FAM-RNA14; 5′-FAM-GUUUUUAAUAUUUC-3′; GenScript) described previously (8) were used for RNA binding analyses at various protein concentrations. Each reaction contained a 0 to 2,500 nM concentration of the protein of interest and 5 nM RNA in the assay buffer (25 mM HEPES [pH 7.5], 50 mM NaCl, 5 mM MgCl2, 50 mM TCEP [pH 7.5], and 0.01% Triton X-100). The reaction mixture was transferred into a 96-well half-area black flat-bottom microplate (Corning) and equilibrated at room temperature for 20 min. The measurement was performed by a Synergy H1 plate reader (BioTek) at an excitation wavelength of 480 nM and an emission wavelength of 510 nm. The data were analyzed using nonlinear regression with Prism 8 (GraphPad).
Data availability.
The SAXS data for nsP2 have been deposited in the SASBDB database (40) under accession code SASDK82.
ACKNOWLEDGMENTS
This research was supported by the Singapore Ministry of Education under its Singapore Ministry of Education Academic Research Fund Tier 2 (MOE2016T22097). Work in the A.M. laboratory was supported by the European Regional Development Fund through the Centre of Excellence in Molecular Cell Engineering, Estonia, 2014-2020.4.01.15-013, and by The Wellcome Trust (200171/Z/15/Z).
We acknowledge the technical services provided by the Synchrotron Radiation Protein Crystallography Facility of the National Core Facility Program for Biotechnology, Ministry of Science and Technology and the National Synchrotron Radiation Research Center, Taiwan, ROC. We thank beamline scientists from the Taiwan Light Source for their help with collecting diffraction data. We thank members of the D.L. and A.M. labs for their general support.
We declare that we have no conflicts of interest with the contents of this article.
A.M. and D.L. designed the research; Y.-S.L., S.W., Y.B.T., O.S., A.U., W.Y.G., B.-J.L., and M.W.C. performed the research; Y.-S.L., S.W., O.S., Y.B.T., A.U., M.W.C., U.-S.J., A.M., and D.L. analyzed the data; and Y.-S.L., O.S., A.M., and D.L. wrote the paper with input from all authors.
REFERENCES
- 1.Burt FJ, Rolph MS, Rulli NE, Mahalingam S, Heise MT. 2012. Chikungunya: a re-emerging virus. Lancet 379:662–671. doi: 10.1016/S0140-6736(11)60281-X. [DOI] [PubMed] [Google Scholar]
- 2.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58:491–562. doi: 10.1128/MR.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li G, Rice CM. 1993. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J Virol 67:5062–5067. doi: 10.1128/JVI.67.8.5062-5067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol 96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz O, Albert ML. 2010. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol 8:491–500. doi: 10.1038/nrmicro2368. [DOI] [PubMed] [Google Scholar]
- 6.Das PK, Puusepp L, Varghese FS, Utt A, Ahola T, Kananovich DG, Lopp M, Merits A, Karelson M. 2016. Design and validation of novel chikungunya virus protease inhibitors. Antimicrob Agents Chemother 60:7382–7395. doi: 10.1128/AAC.01421-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vasiljeva L, Merits A, Auvinen P, Kääriäinen L. 2000. Identification of a Novel Function of the alphavirus capping apparatus RNA 5′-triphosphatase activity of Nsp2. J Biol Chem 275:17281–17287. doi: 10.1074/jbc.M910340199. [DOI] [PubMed] [Google Scholar]
- 8.Law Y-S, Utt A, Tan YB, Zheng J, Wang S, Chen MW, Griffin PR, Merits A, Luo D. 2019. Structural insights into RNA recognition by the Chikungunya virus nsP2 helicase. Proc Natl Acad Sci U S A 116:9558–9567. doi: 10.1073/pnas.1900656116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das PK, Merits A, Lulla A. 2014. Functional cross-talk between distant domains of chikungunya virus non-structural protein 2 is decisive for its RNA-modulating activity. J Biol Chem 289:5635–5653. doi: 10.1074/jbc.M113.503433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lulla A, Lulla V, Merits A. 2012. Macromolecular assembly-driven processing of the 2/3 cleavage site in the alphavirus replicase polyprotein. J Virol 86:553–565. doi: 10.1128/JVI.05195-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kääriäinen L, Ahola T. 2002. Functions of alphavirus nonstructural proteins in RNA replication. Prog Nucleic Acid Res Mol Biol 71:187–222. doi: 10.1016/s0079-6603(02)71044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narwal M, Singh H, Pratap S, Malik A, Kuhn RJ, Kumar P, Tomar S. 2018. Crystal structure of chikungunya virus nsP2 cysteine protease reveals a putative flexible loop blocking its active site. Int J Biol Macromol 116:451–462. doi: 10.1016/j.ijbiomac.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Utt A, Das PK, Varjak M, Lulla V, Lulla A, Merits A. 2015. Mutations conferring a non-cytotoxic phenotype on chikungunya virus replicons compromise enzymatic properties of non-structural protein 2. J Virol 89:3145–3162. doi: 10.1128/JVI.03213-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atasheva S, Gorchakov R, English R, Frolov I, Frolova E. 2007. Development of Sindbis viruses encoding nsP2/GFP chimeric proteins and their application for studying nsP2 functioning. J Virol 81:5046–5057. doi: 10.1128/JVI.02746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Utt A, Quirin T, Saul S, Hellström K, Ahola T, Merits A. 2016. Versatile trans-replication systems for chikungunya virus allow functional analysis and tagging of every replicase protein. PLoS One 11:e0151616. doi: 10.1371/journal.pone.0151616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frolov I, Agapov E, Hoffman TA, Prágai BM, Lippa M, Schlesinger S, Rice CM. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J Virol 73:3854–3865. doi: 10.1128/JVI.73.5.3854-3865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartholomeeusen K, Utt A, Coppens S, Rausalu K, Vereecken K, Arien KK, Merits A. 2018. A chikungunya virus trans-replicase system reveals the importance of delayed nonstructural polyprotein processing for efficient replication complex formation in mosquito cells. J Virol 92:e00152-18. doi: 10.1128/JVI.00152-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. 2010. Functional Sindbis virus replicative complexes are rormed at the plasma membrane. J Virol 84:11679–11695. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fros JJ, Liu WJ, Prow NA, Geertsema C, Ligtenberg M, Vanlandingham DL, Schnettler E, Vlak JM, Suhrbier A, Khromykh AA, Pijlman GP. 2010. Chikungunya virus nonstructural protein 2 inhibits type I/II interferon-stimulated JAK-STAT signaling. J Virol 84:10877–10887. doi: 10.1128/JVI.00949-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akhrymuk I, Kulemzin SV, Frolova EI. 2012. Evasion of the innate immune response: the Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J Virol 86:7180–7191. doi: 10.1128/JVI.00541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akhrymuk I, Frolov I, Frolova EI. 2018. Sindbis virus infection causes cell death by nsP2-induced transcriptional shutoff or by nsP3-dependent translational shutoff. J Virol 92:e01388-18. doi: 10.1128/JVI.01388-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DY, Firth AE, Atasheva S, Frolova EI, Frolov I. 2011. Conservation of a packaging signal and the viral genome RNA packaging mechanism in alphavirus evolution. J Virol 85:8022–8036. doi: 10.1128/JVI.00644-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schuchman R, Kilianski A, Piper A, Vancini R, Ribeiro JMC, Sprague TR, Nasar F, Boyd G, Hernandez R, Glaros T. 2018. Comparative characterization of the Sindbis virus proteome from mammalian and invertebrate hosts identifies nsP2 as a component of the virion and sorting nexin 5 as a significant host factor for alphavirus replication. J Virol 92:e00694-18. doi: 10.1128/JVI.00694-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim DY, Atasheva S, Frolova EI, Frolov I. 2013. Venezuelan equine encephalitis virus nsP2 protein regulates packaging of the viral genome into infectious virions. J Virol 87:4202–4213. doi: 10.1128/JVI.03142-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HD, Konarev PV, Svergun DI. 2012. New developments in the ATSAS program package for small-angle scattering data analysis. J Appl Crystallogr 45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasiljeva L, Merits A, Golubtsov A, Sizemskaja V, Kääriäinen L, Ahola T. 2003. Regulation of the sequential processing of Semliki Forest virus replicase polyprotein. J Biol Chem 278:41636–41645. doi: 10.1074/jbc.M307481200. [DOI] [PubMed] [Google Scholar]
- 27.Utt A, Rausalu K, Jakobson M, Männik A, Alphey L, Fragkoudis R, Merits A. 2019. Design and use of chikungunya virus replication templates utilizing mammalian and mosquito RNA polymerase I-mediated transcription. J Virol 93:e00794-19. doi: 10.1128/JVI.00794-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saha A, Acharya BN, Priya R, Tripathi NK, Shrivastava A, Rao MK, Kesari P, Narwal M, Tomar S, Bhagyawant SS, Parida M, Dash PK. 2018. Development of nsP2 protease based cell free high throughput screening assay for evaluation of inhibitors against emerging Chikungunya virus. Sci Rep 8:10831. doi: 10.1038/s41598-018-29024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo AT, White MA, Watowich SJ. 2006. The crystal structure of the Venezuelan equine encephalitis alphavirus nsP2 protease. Structure 14:1449–1458. doi: 10.1016/j.str.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 30.Shin G, Yost SA, Miller MT, Elrod EJ, Grakoui A, Marcotrigiano J. 2012. Structural and functional insights into alphavirus polyprotein processing and pathogenesis. Proc Natl Acad Sci U S A 109:16534–16539. doi: 10.1073/pnas.1210418109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo D, Vasudevan SG, Lescar J. 2015. The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antiviral Res 118:148–158. doi: 10.1016/j.antiviral.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 32.Li K, Phoo WW, Luo D. 2014. Functional interplay among the flavivirus NS3 protease, helicase, and cofactors. Virol Sin 29:74–85. doi: 10.1007/s12250-014-3438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo D, Wei N, Doan DN, Paradkar PN, Chong Y, Davidson AD, Kotaka M, Lescar J, Vasudevan SG. 2010. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J Biol Chem 285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo D, Xu T, Hunke C, Gruber G, Vasudevan SG, Lescar J. 2008. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol 82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohlway A, Pirakitikulr N, Ding SC, Yang F, Luo D, Lindenbach BD, Pyle AM. 2014. The linker region of NS3 plays a critical role in the replication and infectivity of hepatitis C virus. J Virol 88:10970–10974. doi: 10.1128/JVI.00745-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gebhard LG, Iglesias NG, Byk LA, Filomatori CV, De Maio FA, Gamarnik AV. 2016. A proline-rich N-terminal region of the dengue virus NS3 is crucial for infectious particle production. J Virol 90:5451–5461. doi: 10.1128/JVI.00206-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patkar CG, Kuhn RJ. 2008. Yellow fever virus NS3 plays an essential role in virus assembly independent of its known enzymatic functions. J Virol 82:3342–3352. doi: 10.1128/JVI.02447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeng US, Su CH, Su C-J, Liao K-F, Chuang W-T, Lai Y-H, Chang J-W, Chen Y-J, Huang Y-S, Lee M-T, Yu K-L, Lin J-M, Liu D-G, Chang C-F, Liu C-Y, Chang C-H, Liang KS. 2010. A small/wide-angle X-ray scattering instrument for structural characterization of air–liquid interfaces, thin films and bulk specimens. J Appl Crystallogr 43:110–121. doi: 10.1107/S0021889809043271. [DOI] [Google Scholar]
- 39.Ülper L, Sarand I, Rausalu K, Merits A. 2008. Construction, properties, and potential application of infectious plasmids containing Semliki Forest virus full-length cDNA with an inserted intron. J Virological Methods 148:265–270. doi: 10.1016/j.jviromet.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valentini E, Kikhney AG, Previtali G, Jeffries CM, Svergun DI. 2015. SASBDB, a repository for biological small-angle scattering data. Nucleic Acids Res 43:D357–D363. doi: 10.1093/nar/gku1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The SAXS data for nsP2 have been deposited in the SASBDB database (40) under accession code SASDK82.