So far, few studies have mentioned the distinct function of guanylate-binding protein 7 (GBP7) on virus infection. Here, we reported that GBP7 expression was significantly upregulated in the lungs of mice, human PBMCs, and A549 cells during IAV infection.
KEYWORDS: guanylate-binding protein 7 (GBP7), influenza A virus, antiviral response, innate immunity, IFN, NF-κB
ABSTRACT
Guanylate-binding protein 7 (GBP7) belongs to the GBP family, which plays key roles in mediating innate immune responses to intracellular pathogens. Thus far, GBP7 has been reported to be a critical cellular factor against bacterial infection. However, the relationship between GBP7 and influenza A virus (IAV) replication is unknown. Here, we showed that GBP7 expression was significantly upregulated in the lungs of mice, human peripheral blood mononuclear cells (PBMCs), and A549 cells during IAV infection. Using the CRISPR-Cas9 system and overexpression approaches, it was found that GBP7 knockout inhibited IAV replication by enhancing the expression of IAV-induced type I interferon (IFN), type III IFN, and proinflammatory cytokines. Conversely, overexpression of GBP7 facilitated IAV replication by suppressing the expression of those factors. Furthermore, GBP7 knockout enhanced IAV-induced nuclear factor-κB (NF-κB) activation and phosphorylation of stat1 and stat2; overexpression of GBP7 had the opposite effect. Our data indicated that GBP7 suppresses innate immune responses to IAV infection via NF-κB and JAK-STAT signaling pathways. Taken together, upon IAV infection, the induced GBP7 facilitated IAV replication by suppressing innate immune responses to IAV infection, which suggested that GBP7 serves as a therapeutic target for controlling IAV infection.
IMPORTANCE So far, few studies have mentioned the distinct function of guanylate-binding protein 7 (GBP7) on virus infection. Here, we reported that GBP7 expression was significantly upregulated in the lungs of mice, human PBMCs, and A549 cells during IAV infection. GBP7 facilitated IAV replication by suppressing the expression of type I interferon (IFN), type III IFN, and proinflammatory cytokines. Furthermore, it was indicated that GBP7 suppresses innate immune responses to IAV infection via NF-κB and JAK-STAT signaling pathways. Taken together, our results elucidate a critical role of GBP7 in the host immune system during IAV infection.
INTRODUCTION
Influenza A virus (IAV) is a single-stranded, negative-sense RNA virus, with a genome consisting of eight RNA segments, that belongs to the Orthomyxoviridae family and poses a serious threat to global public and human health (1, 2). In the last hundred years, several serious worldwide pandemics have occurred. The Spanish influenza pandemic of 1918 to 1919 was caused by a founder H1N1 virus and resulted in approximately 50 million fatalities (3). The pandemics of 1957, 1968, and 2009 were caused by offspring of the 1918 virus, which acquired one or more genes by means of mutation or reassortment (4). The innate immune system, which typically involves a highly conserved host-cell signaling mechanism, is the first line of defense to protect the host against viral infections, and the complex interaction between the innate immunity of host cells and the influenza virus determines viral replication and deadly virus infections (5). Host factors serve as ideal drug targets, and a detailed understanding of host-virus interactions offers a worthy strategy to develop therapeutics for preventing or treating potentially fatal IAV infections (6).
During IAV infection, the host’s innate immune system effectively recognizes microbial components, called pathogen-associated molecular patterns (PAMPs), through a limited amount of pattern recognition receptors (PRRs) (7), and ultimately type I interferon (IFN-α/β), type III IFN (IFN-λ), and proinflammatory cytokines are produced, which play a crucial role in the antiviral response by inhibiting viral infection (8, 9). The nuclear factor-κB (NF-κB) signaling pathway performs several roles throughout the innate immune system. The NF-κB family consists of five members, including p65 (RelA), p50, p52, RelB, and c-Rel, which activate the expression of IFNs and proinflammatory cytokines (10–13). The NF-κB pathway can be activated by many extracellular signals, such as various viruses, bacteria, and cytokines secreted from the host, and is mediated by phosphorylation and degradation of the NF-κB inhibitor IκBα protein and p65 dimer translocation from the cytoplasm into the nucleus (12, 14, 15). The IFN-induced JAK-STAT signaling pathway gives rise to hundreds of IFN-stimulated genes (ISGs) whose expression inhibits viral infection (16). The signal transducer and activator of transcription 1 (stat1) and stat2 phosphorylation result in the transcriptional activation of ISGs (8).
Guanylate-binding proteins (GBPs) are a large family of 65- to 67-kDa GTPases induced by IFN that play a key role in innate immunity against intracellular pathogens (17–20). GBPs bind to GTP and hydrolyze GTP into GMP and GDP and are composed of a spherical N-terminal GTP binding domain, a C-terminal helix domain, and a middle domain (18, 20–22). Thus far, seven human GBPs (GBP1 to GBP7) have been confirmed, displaying a high degree of sequence homology. GBP7 reportedly protects the host against listeria or mycobacterial infection (17) and exhibits particular resistance to Shigella flexneri E3 ligase IpaH9.8 (23); however, few reports have mentioned the distinct function of GBP7, and the relationship between GBP7 and virus infection is unknown.
For the first time, our reports showed that GBP7 facilitates IAV replication by negatively modulating innate immune responses to IAV infection. Importantly, GBP7 could serve as a therapeutic target for controlling viral infection.
RESULTS
GBP7 expression is upregulated during IAV infection.
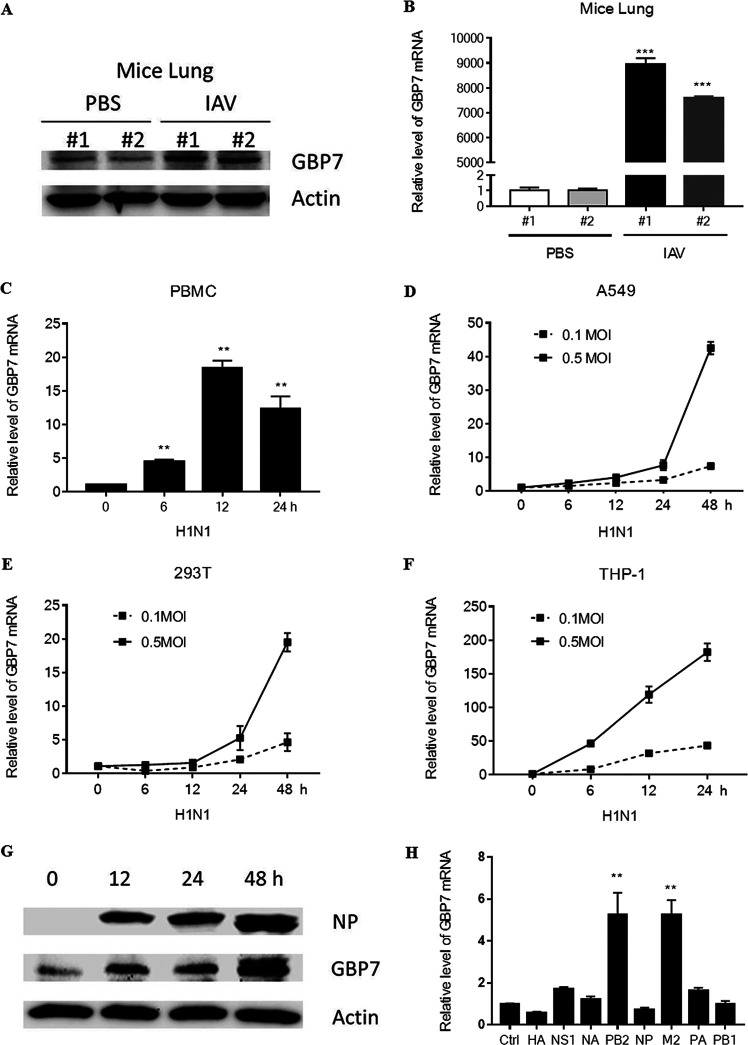
First, we determined whether GBP7 expression was induced during IAV infection. C57BL/6 mice were infected intranasally with the influenza A/Puerto Rico/8/34 H1N1(PR8) strain or inoculated with phosphate-buffered saline (PBS) for 48 h, and the protein level of GBP7 in the lungs of mice was assessed using Western blotting. We showed that GBP7 protein level in the lungs of mice infected with PR8 was significantly upregulated (Fig. 1A). Furthermore, GPB7 mRNA expression in the lungs of mice was measured using quantitative real-time PCR (qRT-PCR), and the results showed that GPB7 mRNA expression was also upregulated after IAV infection (Fig. 1B). Next, freshly isolated human peripheral blood mononuclear cells (PBMCs) were infected with PR8 for the specified times, and GBP7 mRNA was measured using qRT-PCR. The results suggested that GBP7 mRNA was induced during IAV infection (Fig. 1C).
FIG 1.
IAV infection induces GBP7 upregulation. (A and B) Mice were infected with PR8 (100 PFU; mouse 1 and mouse 2) or inoculated with PBS (mouse 1 and mouse 2) intranasally for 48 h. (A) The GBP7 protein levels in the lungs of mice were quantified using Western blotting. (B) The GBP7 mRNA levels in the lungs of mice were quantified using qRT-PCR. (C) Human PBMCs were infected with PR8 (MOI of 0.1) for the specified times. The GBP7 mRNA levels were quantified using qRT-PCR. (D to F) A549 cells, HEK293T cells, and THP-1 cells were infected with PR8 (MOI of 0.1 or 0.5) for the specified times. The levels of GBP7 mRNA were quantified using qRT-PCR. (G) A549 cells were infected with PR8 (MOI of 1) for the specified times. The levels of GBP7 protein were quantified using Western blotting. (H) Eight H1N1 plasmids, including HA, NS1, NA, PB2, NP, M2, PA, and PB1, were transfected into 293T cells for 24 h. The GBP7 mRNA levels were quantified using qRT-PCR. Ctrl, control. All experiments were independently performed at least three times. The results are expressed as means ± standard deviations (SD). **, P < 0.01.
Additionally, we infected A549 cells with PR8 at a multiplicity of infection (MOI) of 0.1 or 0.5 for the specified times and assessed the expression level of GBP7 mRNA using qRT-PCR. The results showed that GBP7 mRNA expression was upregulated in A549 cells during IAV infection in a time- and dose-dependent manner (Fig. 1D). The same experiments were performed in HEK293T cells and THP-1 cells, and mRNA expression of GBP7 agreed with previous results, indicating time- and dose-dependent upregulation during IAV infection (Fig. 1E and F). GBP7 protein levels in A549 cells infected with IAV were measured using Western blotting, exhibiting obvious upregulation during IAV infection and reaching a maximum peak at 48 h postinfection (Fig. 1G). We transfected 293T cells with H1N1 plasmids, including HA, NS1, NA, PB2, NP, M2, PA, and PB1, and the results suggested that PB2 and M2 proteins induced GBP7 mRNA expression (Fig. 1H). Taken together, these data suggested that GBP7 expression was upregulated during IAV infection.
GBP7 knockout inhibits IAV replication.
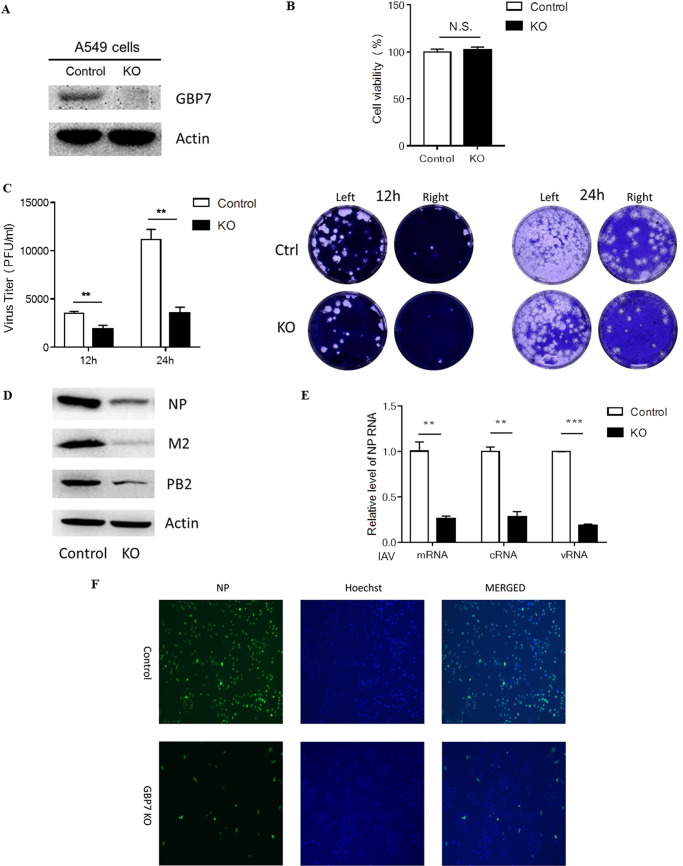
The finding that GBP7 was significantly upregulated in response to IAV infection prompted us to explore whether GBP7 influences IAV replication. We generated A549 cell lines with GBP7 knockout (KO) using CRISPR/Cas9 genome technology (24, 25). The GBP7 knockout cell lines were confirmed using Western blotting (Fig. 2A). GBP7 knockout exerted no effect on viability of A549 cells (Fig. 2B).
FIG 2.
GBP7 knockout in A549 cells inhibits IAV replication. (A) Western blotting showing GBP7 protein expression in control A549 cells and GBP7 knockout cells. (B) GBP7 KO and control cells were cultured for 48 h, and cell viability was measured using the MTT assay. (C) GBP7 KO and control cells infected with PR8 (MOI of 0.1) for 12 or 24 h. The supernatants of each group were collected, and the viral titers were determined using the plaque assay. Typical original images of plaque assays are shown above, and the image on the right is the plaque assay of the 10-fold-diluted supernatant from the left. (D and E) GBP7 KO and control cells infected with PR8 (MOI of 0.1) and harvested 12 h postinfection. (D) The protein levels of NP, M2, and PB2 were measured using Western blotting. (E) The relative levels of the IAV NP gene, including mRNA, cRNA, and vRNA, were measured using qRT-PCR. (F) GBP7 KO and control cells infected with PR8 (MOI of 0.1) and harvested 24 h postinfection. The viral NP proteins were measured using immunofluorescence microscopy assays. All experiments were independently performed at least three times. The results are expressed as means ± SD. **, P < 0.01; ***, P < 0.001; N.S., not significant.
A549 control cells and GBP7 KO cells were infected with PR8 at an MOI of 0.1 and collected 12 and 24 h after infection. The viral titer was assessed using the plaque assay. Results indicated that GBP7 knockout decreased the viral titer (Fig. 2C). Furthermore, compared with control cells, Western blotting indicated that GBP7 KO cells exhibited notably reduced IAV NP, PB2, and M2 protein expression (Fig. 2D) as well as decreased relative levels of IAV NP gene, including mRNA, cRNA, and vRNA, using qRT-PCR in the IAV replication experiment (Fig. 2E). Finally, immunofluorescence microscopy indicated that GBP7 knockout reduced the percentage of viral NP protein (Fig. 2F). In conclusion, GBP7 knockout inhibited IAV replication.
Overexpression of GBP7 facilitates IAV replication.
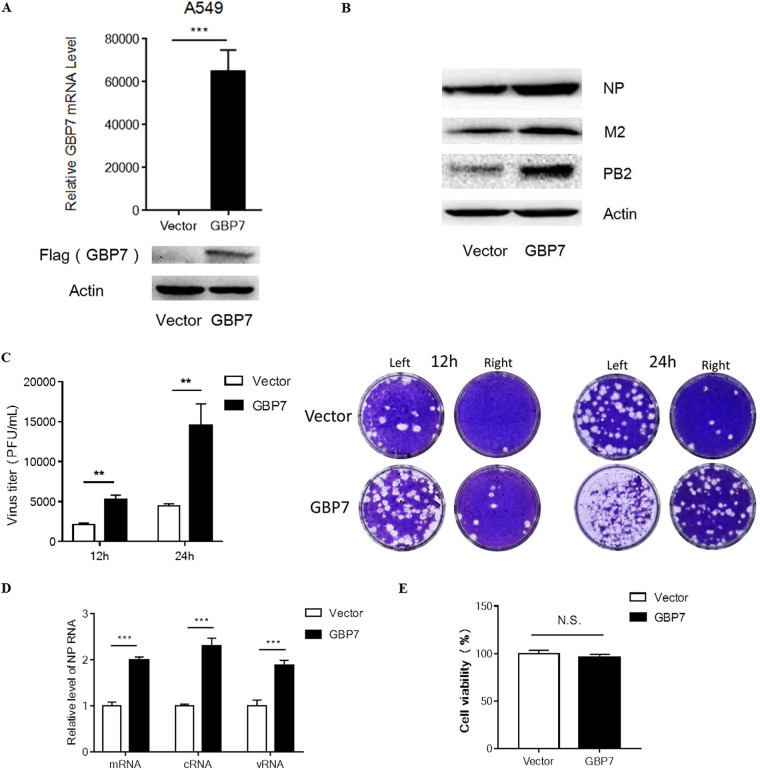
To further confirm the hypothesis that GBP7 facilitates IAV infection, we transfected A549 cells with the GBP7-expressing plasmid or empty vector plasmid. After 24 h, we infected the GBP7-overexpressing or vector cells with H1N1 at an MOI of 0.1 for 12 h. Thereafter, the infected cells were harvested. Increased GBP7 mRNA and Flag-GBP7 protein expression (Fig. 3A) in cells transfected with the GBP7 plasmid confirmed GBP7 overexpression. Western blotting results indicated that viral protein NP, PB2, and M2 expression levels were increased in GBP7-overexpressed cells (Fig. 3B). Furthermore, we observed that GBP7-overexpressed A549 cells exhibited higher virus titers than empty vector cells using the plaque assay (Fig. 3C). Using qRT-PCR, we observed that the relative levels of the IAV NP gene, including mRNA, cRNA, and vRNA, were enhanced following GBP7 overexpression (Fig. 3D). GBP7 overexpression exerted no effect on A549 cell viability (Fig. 3E). Thus, our results further confirmed that GBP7 facilitates IAV replication.
FIG 3.
Overexpression of GBP7 facilitates IAV replication. (A) A549 cells were transfected with the plasmid GBP7-expressing or empty vector for 24 h, the level of GBP7 mRNA was measured using qRT-PCR, and the protein expression of Flag-GBP7 was measured using Western blotting. (B) A549 cells were transfected with the plasmid GBP7-expressing or empty vector and, after 24 h, infected with PR8 (MOI of 0.1) and harvested 12 h postinfection. Protein levels of NP, M2, and PB2 were measured using Western blotting. (C) A549 cells were transfected with the plasmid GBP7-expressing or empty vector and, after 24 h, infected with PR8 (MOI of 0.1) and harvested 12 or 24 h postinfection. The supernatant of each group was collected and the viral titer was assessed using the plaque assay. Typical original images of plaque assays are shown above, and the image on the right is the plaque assay of the 10-fold-diluted supernatant from the left. (D) A549 cells were transfected with the plasmid GBP7-expressing or empty vector and, after 24 h, infected with PR8 (MOI of 0.1) and harvested 12 h postinfection. Relative levels of the IAV NP gene, including mRNA, cRNA, and vRNA, were measured using qRT-PCR. (E) A549 cells were transfected with the plasmid GBP7-expressing or empty vector. After 24 h, cell viability was measured using the MTT assay. All experiments were independently performed at least three times. The results are expressed as means ± SD. **, P < 0.01; ***, P < 0.001.
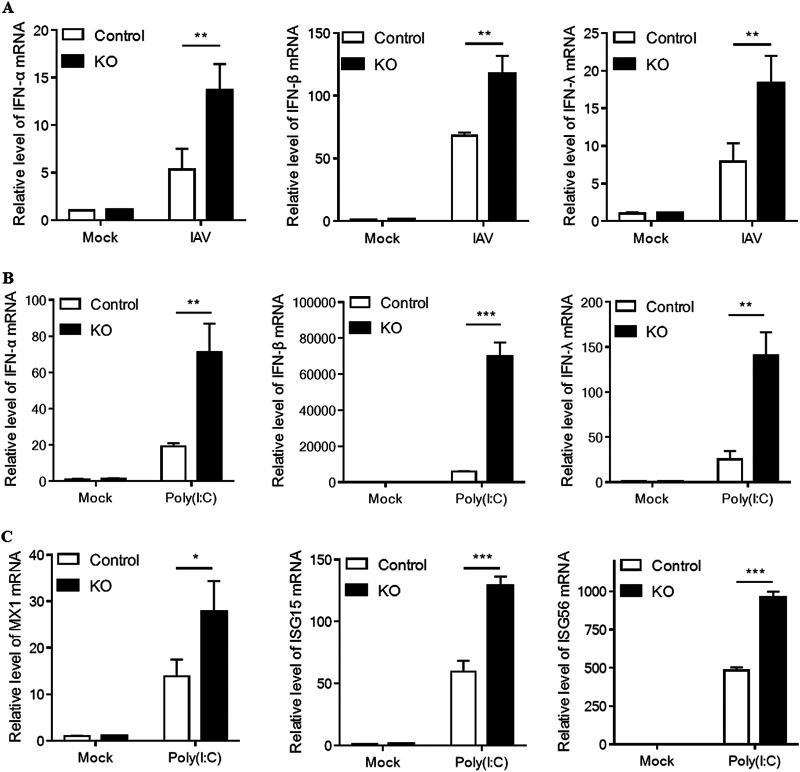
GBP7 knockout enhances IAV- or poly(I·C)-triggered expression of IFNs.
As our data demonstrated that GBP7 facilitates IAV replication, we investigated whether GBP7 influenced the antiviral response to a viral infection. Therefore, we aimed to explore the effect of GBP7 on IAV-induced expression of IFNs and ISGs. We found that after IAV infection, IFN-α, IFN-β, and IFN-λ mRNA levels were increased in GBP7 KO cells compared with control cells (Fig. 4A). Polyribocytidylic acid [poly(I·C)] is a synthetic analog of viral double-stranded RNA (dsRNA) that can be detected by cytoplasmic PRRs, including RIG-I, TLR3, and MDA-5 (26). It is widely accepted that the replicative intermediate dsRNA produced by IAV is critical for influenza infection (27). We treated GBP7 KO cells and control cells with poly(I·C); after 6 h of treatment, compared with control cells, type I and III IFN mRNA levels in GBP7 KO cells were significantly increased; in particular, a >65,000-fold increase in the IFN-β level was observed (Fig. 4B). Next, we determined ISG expression, including MX1, ISG15, and ISG56; the results showed that after 6 h of treatment with poly(I·C), compared with control cells, the mRNA levels of MX1, ISG15, and ISG56 in GBP7 KO cells increased notably (Fig. 4C). Together, the data indicated that endogenous GBP7 negatively modulates the IAV-induced expression of IFNs, which is associated with IAV replication.
FIG 4.
GBP7 knockout enhances IAV- or poly(I·C)-triggered expression of IFNs. (A) GBP7 KO and control cells were infected with PR8 (MOI of 0.1) or mock infected and harvested 6 h postinfection. The relative mRNA levels of IFN-α, IFN-β, and IFN-λ were measured using qRT-PCR. (B and C) GBP7 KO and control cells were treated with poly(I·C) (2 μg/ml) or mock infected and harvested 6 h postinfection. (B) The relative mRNA levels of IFN-α, IFN-β, and IFN-λ were measured using qRT-PCR. (C) The relative mRNA levels of MX1, ISG15, and ISG56 were measured using qRT-PCR. The data shown are one representative from at least three independent experiments. The results are expressed as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; N.S., not significant.
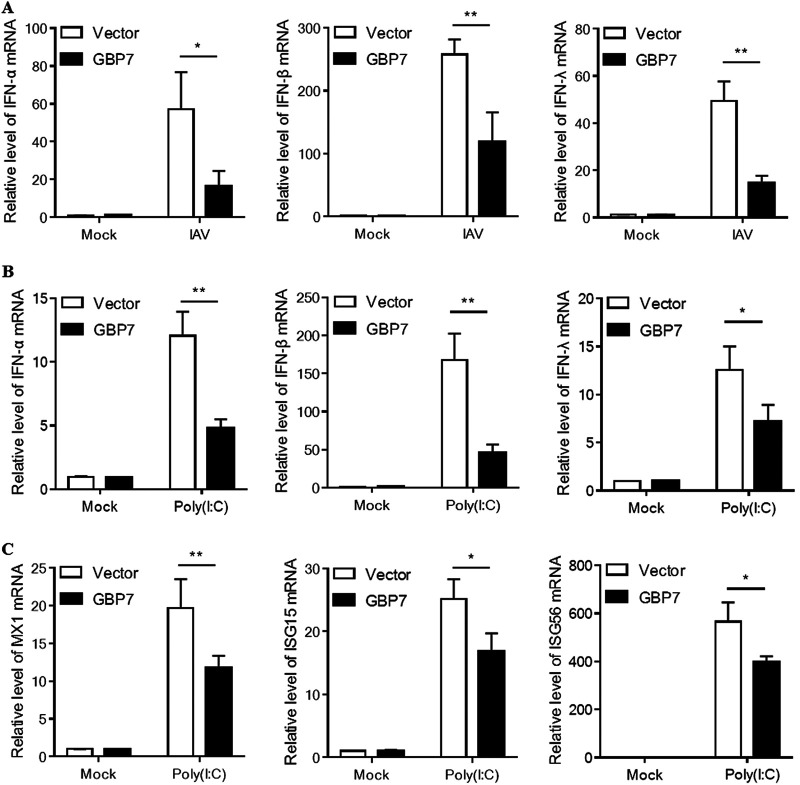
GBP7 suppresses IAV- and poly(I·C)-triggered IFNs.
Considering that GBP7 overexpression facilitated IAV replication, we hypothesized that GBP7 would negatively affect the innate immune responses to IAV infection. To evaluate this hypothesis, we transfected A549 cells with the GBP7-expressing plasmids or empty vector plasmids and infected the cells with PR8 after 24 h. The results showed that overexpression of GBP7 efficiently suppressed IAV-induced IFN-α, IFN-β, and IFN-λ mRNA levels in A549 cells (Fig. 5A). Furthermore, we treated GBP7-overexpressing or vector cells with poly(I·C) for 6 h, and the data showed GBP7 suppressed poly(I·C)-induced IFNs as well (Fig. 5B). We detected the expression of the ISGs, including MX1, ISG15, and ISG56, using qRT-PCR. The results indicated that after poly(I·C) treatment, GBP7 suppressed MX1, ISG15, and ISG56 mRNA expression (Fig. 5C). Collectively, GBP7 suppressed IAV- and poly(I·C)-triggered IFNs.
FIG 5.
GBP7 overexpression suppresses IAV- and poly(I·C)-triggered IFNs. (A) A549 cells were transfected with the plasmid GBP7-expressing or empty vector for 24 h, infected with PR8 (MOI of 0.1) or mock infected, and harvested 6 h postinfection. The relative mRNA levels of IFN-α, IFN-β, and IFN-λ were measured using qRT-PCR. (B and C) A549 cells were transfected with the plasmid GBP7-expressing or empty vector for 24 h and then treated with poly(I·C) (2 μg/ml) or mock treated and harvested 6 h postinfection. (B) The relative mRNA levels of IFN-α, IFN-β, and IFN-λ were measured using qRT-PCR. (C) The relative mRNA levels of MX1, ISG15, and ISG56 were measured using qRT-PCR. The data shown are one representative from at least three independent experiments. The results are expressed as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
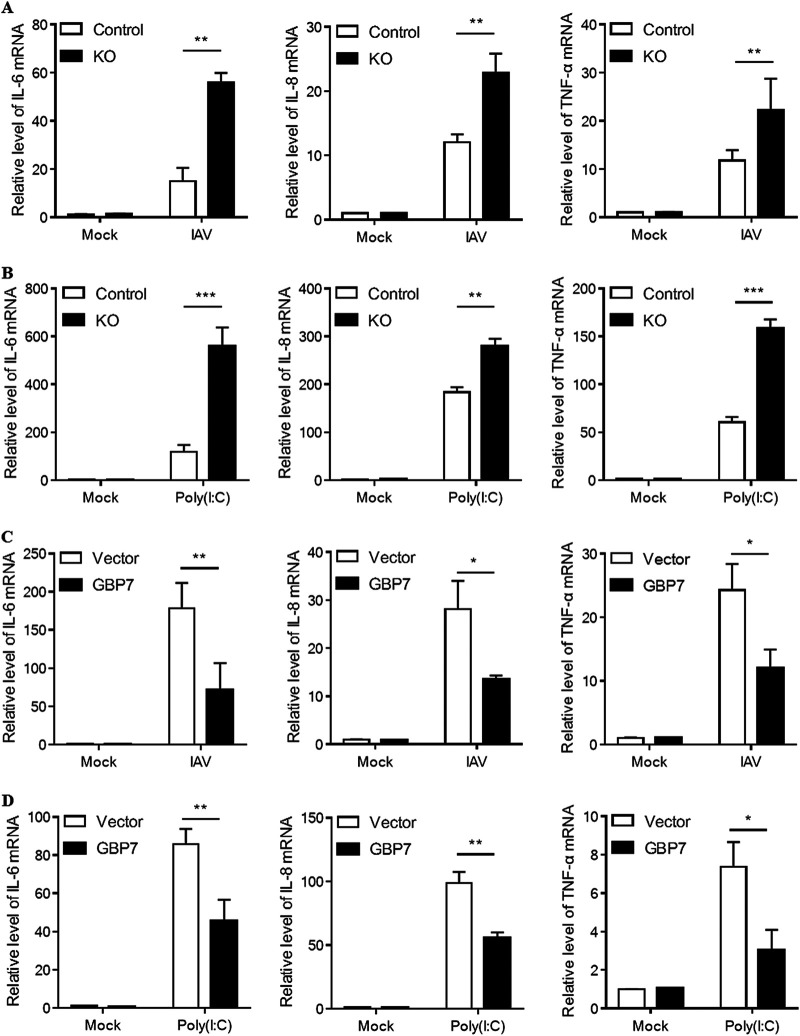
GBP7 is a negative regulator of IAV- and poly(I·C)-induced proinflammatory cytokines.
As our data suggested that GBP7 suppressed IAV-induced IFN production, we further investigated the role of GBP7 in the expression of IAV-induced proinflammatory cytokines, such as interleukin-6 (IL-6), IL-8, and tumor necrosis factor alpha (TNF-α). After IAV infection in control cells and GBP7 KO cells, results indicated that GBP7 knockout increased IL-6, IL-8, and TNF-α mRNA levels in response to IAV infection compared to controls (Fig. 6A). Furthermore, we treated GBP7 KO cells and control cells with poly(I·C), and a similar trend was observed in GBP7 KO cells (Fig. 6B). To confirm that GBP7 negatively regulates the antiviral response to IAV infection, we conducted GBP7 overexpression experiments. The data suggested that overexpression of GBP7 suppressed IAV-induced IL-6, IL-8, and TNF-α mRNA levels (Fig. 6C). Subsequently, we observed that overexpression of GBP7 inhibited IL-6, IL-8, and TNF-α mRNA expression after poly(I·C) treatment (Fig. 6D). Taken together, the results showed that GBP7 is a negative regulator of IAV- and poly(I·C)-induced proinflammatory cytokines. These results may provide a new target for treating cytokine storms.
FIG 6.
Exogenous GBP7 suppresses IAV- and poly(I·C)-triggered proinflammatory cytokines. (A) GBP7 KO and control cells were infected with PR8 (MOI of 0.1) or mock infected and harvested 6 h postinfection. (B) GBP7 KO and control cells were treated with poly(I·C) (2 μg/ml) or mock treated and harvested 6 h postinfection. (C) A549 cells were transfected with the plasmid GBP7-expressing or empty vector for 24 h, infected with PR8 (MOI of 0.1) or mock infected, and harvested 6 h postinfection. (D) A549 cells were transfected with the plasmid GBP7-expressing or empty vector for 24 h, treated with poly(I·C) (2 μg/ml) or mock treated, and harvested 6 h postinfection. (A to D) The relative mRNA levels of IL-6, IL-8, and TNF-α were measured using qRT-PCR. The data shown are one representative from at least three independent experiments. The results are expressed as means ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
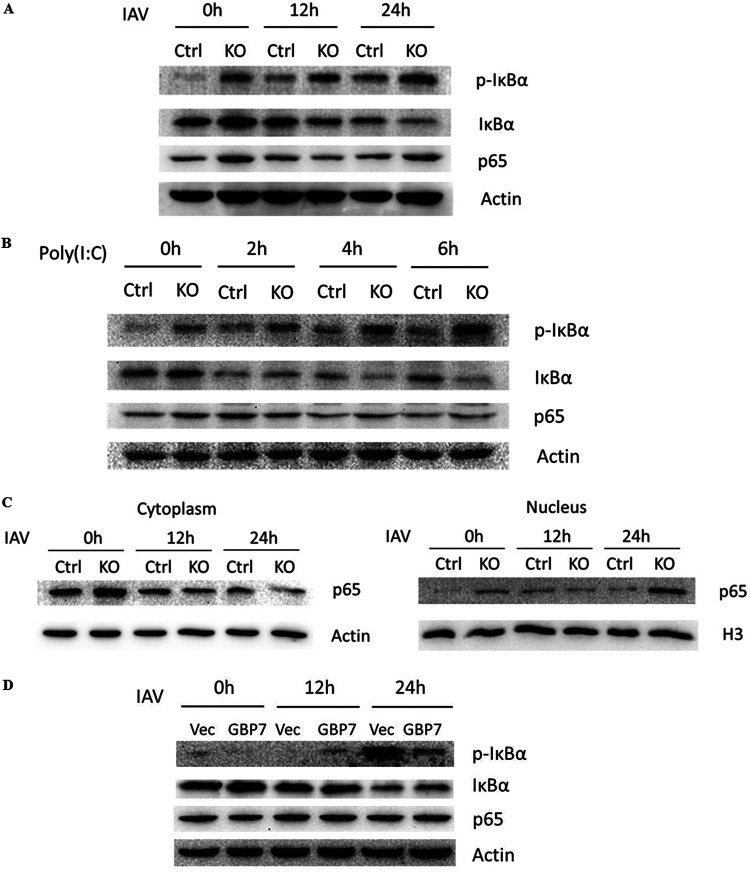
GBP7 negatively regulates NF-κB pathway induced by IAV infection and poly(I·C) treatment.
As the production of proinflammatory cytokines was related to the NF-κB signaling pathway, we next explored the role of GBP7 in modulating NF-κB signaling induced after IAV infection. We infected GBP7 KO cells and control cells with PR8 and assessed the protein expression of IκBα, p-IκBα, and p65 proteins in the nucleus and cytoplasm using Western blotting. Compared with control cells, GBP7 KO cells exhibited increased p-IκBα and decreased IκBα post-IAV infection (Fig. 7A). We treated GBP7 KO cells and control cells with poly(I·C) for the specified times; poly(I·C) treatment increased p-IκBα and reduced IκBα in GBP7 KO cells relative to control cells (Fig. 7B). Our data revealed that IAV infection increased p65 protein within the nucleus but reduced p65 in the cytoplasm in GBP7 KO cells relative to control cells (Fig. 7C). Next, we transfected A549 cells with the GBP7-expressing plasmids or empty vector plasmids and infected the cells with PR8 for the specified times. The results showed that overexpression of GBP7 efficiently suppressed IAV-induced p-IκBα expression (Fig. 7D). Together, the results suggested that endogenous GBP7 suppresses NF-κB pathway activation after IAV infection and poly(I·C) treatment.
FIG 7.
Endogenous GBP7 inhibits NF-κB activation induced by IAV infection and poly(I·C) treatment. (A) GBP7 KO and control cells were infected with PR8 (MOI of 0.1) for the specified times. Cells lysates were collected and p-IκBα, IκBα, and p65 proteins were measured using Western blotting. (B) GBP7 KO and control cells were treated with poly(I·C) (2 μg/ml) for the specified times. Cell lysates were collected and p-IκBα, IκBα, and p65 proteins were measured using Western blotting. (C) GBP7 KO and control cells were infected with PR8 (MOI of 0.1) for the specified times. Thereafter, cytoplasm and nuclear extraction was performed and p65, actin, and H3 proteins were measured using Western blotting. Actin was used as a cytoplasm marker, and H3 was used as a nuclear marker. (D) A549 cells were transfected with the plasmid GBP7-expressing or empty vector and, after 24 h, were infected with PR8 (MOI of 0.1) for the specified times. Cells lysates were collected, and p-IκBα, IκBα, and p65 proteins were measured using Western blotting. The results shown are one representative from at least three independent experiments.
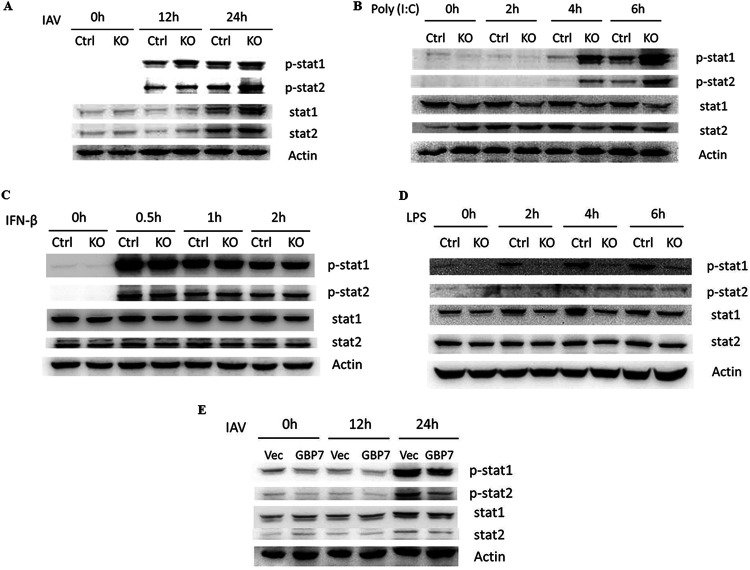
GBP7 suppresses stat1 and stat2 phosphorylation induced by IAV infection and poly(I·C) treatment.
As our results showed that GBP7 suppressed IFN production, we investigated whether GBP7 negatively mediated the JAK-STAT signaling pathway. We infected GBP7 KO cells and control cells with PR8 for the specified times, and protein expression of p-stat1, p-stat2, stat1, and stat2 was determined using Western blotting. Results indicated that protein levels of p-stat1 and p-stat2 gradually increased in GBP7 KO cells compared with control cells (Fig. 8A). We treated GBP7 KO cells and control cells with poly(I·C), IFN-β, or lipopolysaccharide (LPS) for the specified times. We found that poly(I·C) treatment increased p-stat1 and p-stat2 in GBP7 KO cells relative to control cells (Fig. 8B), and IFN-β treatment exerted no effect on the protein expression of p-stat1 and p-stat2 between GBP7 KO cells and control cells (Fig. 8C). The results suggested that GBP7 is not involved in IFN-β-induced p-stat1 and p-stat2, and the upregulation of p-stat1 and p-stat2 in GBP7 KO cells during IAV infection is caused by the upregulation of IFN-β after GBP7 deletion. Surprisingly, we found that LPS treatment inhibited p-stat1 in GBP7 KO cells compared with control cells (Fig. 8D). Next, we transfected A549 cells with the GBP7-expressing plasmids or empty vector plasmids and infected the cells with PR8 for the specified times. The results showed that overexpression of GBP7 efficiently suppressed IAV-induced stat1 and stat2 phosphorylation (Fig. 8E). Taken together, GBP7 suppressed JAK-STAT pathway activation by IAV infection.
FIG 8.
GBP7 suppresses IAV and poly(I·C)-triggered stat1 and stat2 phosphorylation. (A) GBP7 KO and control cells were infected with PR8 (MOI of 0.1) for the specified times. Cell lysates were collected and p-stat1, p-stat2, stat1, and stat2 proteins were measured using Western blotting. (B to D) GBP7 KO and control cells were treated with poly(I·C) (2 μg/ml) (B), IFN-β (20 ng/ml) (C), or LPS (100 ng/ml) (D) for the specified times. Cell lysates were collected and p-stat1, p-stat2, stat1, and stat2 proteins were measured using Western blotting. (E) A549 cells were transfected with the plasmid GBP7-expressing or empty vector (Vec) and, after 24 h, were infected with PR8 (MOI of 0.1) for the specified times. Cells lysates were collected and p-stat1, p-stat2, stat1, and stat2 proteins were measured using Western blotting. The results shown are one representative from at least three independent experiments. The results are presented as means ± SD. *, P < 0.05; **, P < 0.01.
DISCUSSION
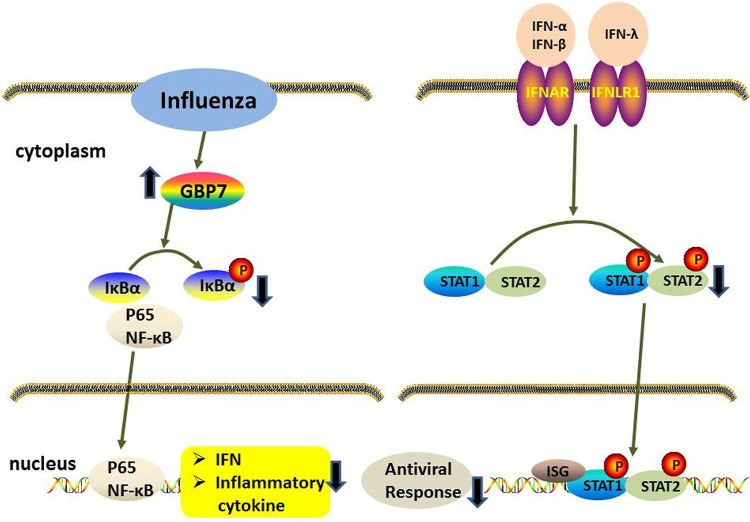
In this study, we demonstrated that GBP7 is significantly upregulated during IAV infection and facilitates IAV replication by suppressing the expression of IAV-induced IFNs and proinflammatory cytokines. GBP7 negatively modulates the innate immune responses to IAV infection by inhibiting NF-κB and JAK-STAT signaling pathways (Fig. 9).
FIG 9.
Hypothetical molecular model explains the role of GBP7 in the antiviral response. IAV infection induced GBP7 expression, and endogenous GBP7 protein suppressed the innate immunity to IAV infection by inhibiting the activation of NF-κB and JAK-STAT pathways. Mechanistically, GBP7 suppressed NF-κB activation by reducing phosphorylation of IκBα and p65 translocation from cytoplasm to nucleus. GBP7 inhibited IFN production, leading to reduction of stat1 and stat2 phosphorylation. Ultimately, induction of IFNs, proinflammatory cytokines, and ISGs decreased, which resulted in enhanced viral replication. Our research highlights the important role of GBP7 in facilitating IAV replication by negatively regulating innate immunity, which suggests that GBP7 acts as a therapeutic target in controlling IAV infection.
We found that GBP7 expression was markedly increased in the lungs of mice, human PBMCs, A549, THP-1, and HEK293T cells during H1N1 infection. Human PBMCs infected with IAV activate several genes involved in the antiviral response (28). These cells were used to detect the expression of GBP7, and we observed a similar trend. GBPs were previously reported as IFN-γ-induced proteins (18, 20, 29), but few studies mentioned GBP7 expression related to viral infection. Our data suggest that GBP7 can be upregulated significantly after infection with IAV, such as the H1N1 virus.
As GBP7 expression is related to IAV infection, we next examine the role of GBP7 in IAV replication. We found that GBP7 knockout not only restricted the H1N1 viral titer in supernatants of infected cells but also inhibited H1N1 viral gene and protein expression. Correspondingly, GBP7 overexpression facilitated the IAV viral titer, gene, and protein expression. GBPs play an important role in mediating viral replication, and GBP1, -2, -5 are well-characterized members of the GBP family that reduce replication of various viruses, including IAV (30–37). Here, we concluded that GBP7 facilitates IAV replication, which indicates a function inverse to that of some other GBP members, which inhibit IAV infection. GBP7 reportedly protects host cells from listeria or mycobacterial infection (17); in contrast, GBP7 promotes IAV replication.
We investigated whether GBP7-facilitated IAV replication was a consequence of the negative impact of GBP7 exerted on the innate immune system response to IAV infection. We measured the expression levels of innate immune genes in A549 cells exposed to IAV infection or poly(I·C) treatment. GBP7 knockout enhanced the expression of IFNs and proinflammatory cytokines, including IL-6, IL-8, and TNF-α, which promote antiviral responses. Overexpression of GBP7 exerted the opposite effect. However, excessive expression of IFNs and inflammatory cytokines are severely deleterious to the human host. Severe influenza virus infections, leading to human death, are usually associated with the excessive induction of proinflammatory cytokines, which is defined as a cytokine storm and can cause acute respiratory distress syndrome (ARDS), acute lung injury, or other organ damage (38–40). GBP5 reportedly enhances IFNs and proinflammatory cytokines, including IL-8, IL-6, and TNF-α expression (37). We showed that the effect of GBP7 in the innate immunity response to IAV was likely different from that of other GBP members. During a cytokine storm, a large amount of proinflammatory cytokines was produced; we guess that inducing GBP7 expression leads to the suppression of excessive inflammation. Finally, our study demonstrated that GBP7 facilitates influenza infection by suppressing the innate immune system response to viral infections.
The NF-κB pathway plays a critical role in the antiviral response through induction of IFNs and proinflammatory cytokines, such as IL-8 and TNF-α (10, 11, 37, 41, 42). Our results demonstrated that GBP7 inhibited phosphorylation of IκBα and suppressed NF-κB translocation from the cytoplasm to the nucleus (Fig. 9). It is plausible that GBP7 affects other signaling pathways to control IFN induction. Due to several reasons, we failed to further explore whether GBP7 phosphorylated IRF3 and IRF7, leading to IFN production (43). In our study, we revealed that GBP7 suppresses the innate immune responses to IAV infection and poly(I·C) stimuli by inhibiting the NF-κB signaling pathway.
Type I and III IFNs bind to IFN-α and IFN-β receptors (IFNAR) and the IFN-λ receptor (IFNLR1) (8), respectively, leading to stat1 and stat2 phosphorylation, as well as activation and induction of ISGs with an antiviral response (16). In our study, we showed that GBP7 knockout upregulated IAV-induced stat1 and stat2 phosphorylation and expression of IFN-α, IFN-β, IFN-λ, and poly(I·C)-induced ISGs, for instance, MX1, ISG15, and ISG56. It is known that IAV infection is sensitive to the antiviral response and could be restricted by ISGs (44, 45). We demonstrated the inhibitory effect of GBP7 on IFN expression and the JAK-STAT signaling pathway upon IAV infection. GBP7 was reported to protect the host from listeria or mycobacterial infection (17). Unexpectedly, we found that GBP7 knockout inhibited phosphorylation of stat1 in A549 cells treated with LPS, which perhaps clarified the mechanism of GBP7 against bacterial infections.
In conclusion, our results revealed, for the first time, that GBP7 facilitates IAV replication. We further highlighted the negative role of GBP7 in virus-induced innate immune responses by inhibiting NF-κB and JAK-STAT signaling pathways, which leads to reduced expression of IFNs and proinflammatory cytokines. Our research indicates that inhibition of GBP7 expression leads to inhibited IAV infection and suggests that GBP7 serves as a therapeutic target for controlling virus infection.
MATERIALS AND METHODS
Cells, virus infection, and poly(I·C) transfection.
PBMCs, A549, and THP-1 cells were cultured in RPMI 1640 basic (Gibco, Grand Island, NY, USA) medium. HEK-293T and MDCK cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (Gibco). All culture media were supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. All cells were cultured at 37°C with 5% CO2. Influenza virus strain A/Puerto Rico/8/34 H1N1 (PR8), used in this research, was purchased from South China Agricultural University, Guangzhou, China. Cells were washed with phosphate-buffered saline (PBS) twice, followed by incubation with PR8 for 1 h at 37°C with 5% CO2. Virus diluent was removed and replaced with medium containing 1% penicillin-streptomycin and no FBS. To treat A549 cells with poly(I·C) (Sigma, USA), poly(I·C) was transfected with PolyJet (Signagen, USA) into cells.
Antibodies.
The antibody used for GBP7 was purchased from Biorbyt (Cambridge, UK). The antibodies against influenza A virus nucleoprotein and M2 protein were purchased from Abcam (Cambridge, MA, USA). The antibody against PB2 was purchased from GeneTex (Alton Pkwy Irvine, CA, USA). Antibodies against Flag tag were purchased from Sigma (St. Louis, MO, USA). Antibodies against β-actin, H3, p-stat1, p-stat2, stat1, stat2, p-IκBα, IκBα, and p65, used in this research, were purchased from Cell Signaling Technology (Beverly, MA, USA).
Isolation of human PBMCs.
PBMCs were provided by healthy volunteers. The Histopaque-1077 kit (Sigma-Aldrich) was used to separate PBMCs by density gradient centrifugation. PBMCs were immediately infected with PR8 (MOI of 0.1) at 37°C supplied with 5% CO2 for the times indicated, and GBP7 mRNA was extracted and assessed using qRT-PCR.
Mice.
Female C57BL/6 mice, aged 6 to 8 weeks, used in this study were purchased from the Guangdong Medical Laboratory Animal Center. The Ethics Committee of Southern Medical University, Guangzhou, China, approved the animal experiments in this research. Mice were infected with PR8 (100 PFU) or PBS by intranasal inoculation and were euthanized 48 h postinfection. The lungs of mice were collected to detect GBP7 protein and mRNA expression following IAV infection using Western blotting and qRT-PCR.
MTT assay.
3-[4,5-Dimethylthiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT; Sigma, USA) was used to measure cell viability. The cells were cultured in 96-well plates with MTT solutions at 37°C supplied with 5% CO2 for 4 h. Before detection, we removed supernatants and added dimethyl sulfoxide to each well. Subsequently, the absorbance was assessed using a microplate reader at 570 nm (GENios Pro; TECAN, USA).
Plaque assay.
MDCK cells (1.0 × 106/ml) were inoculated in 6-well plates. When the cells covered 100% of the well, cells were infected with 10-fold-diluted cell supernatants as described above for 1 h. After aspirating the diluted virus, cells were cultured in 3 ml 2× MEM containing 2% agar (Sigma-Aldrich) and 1 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone (Sigma-Aldrich) at 37°C with 5% CO2 for 48 to 72 h. Finally, plaque number was obtained by counting after crystal violet staining (Sigma-Aldrich).
Measurement of IAV replication.
A549 cells were infected with PR8 at an MOI of 0.1. The viral titers were measured using plaque assay. NP RNA was reverse transcribed with the following primers: NP-cRNA, 5′-AGTAGAAACAAGG-3′; NP-vRNA, 5′-AGCGAAAGCAGG-3′; and NP-mRNA, oligo(dT). Relative levels of IAV NP gene, including mRNA, cRNA, and vRNA, were measured using qRT-PCR.
GBP7 knockout cells produced by the CRISPR-Cas9 system.
The Cas9 target design tool (https://www.genscript.com/gRNA-database.html) was used to design guide RNA sequences targeting the GBP7 gene. The guide RNA target sequence was 5′-TATACACTAGTGCCCAGAAA-3′. Primer sequences used for PCR in this research were the following: forward, 5′-CACCGTATACACTAGTGCCCAGAAA-3′; reverse, 5′-AAACTTTCTGGGCACTAGTGTATAC-3′. To package lentivirus, 1 μg GBP7 lentiCRISPR v2/empty lentiCRISPR v2 with the packaging plasmids, 500 ng pVSVG and 500 ng psPAX2, were cotransfected into HEK293T cells using PolyJet (Signagen, USA) according to the manufacturer’s instructions. After 24 h, fresh medium replaced the culture medium. Following another 24 h, lentiviruses were collected and stored at −80°C. A549 cells (2.0 × 105/ml) were seeded in 6-well plates 1 day in advance, and the cells reached 80% confluence before lentivirus was added to the well. The virus-infected cells were cultured for 24 h and were screened using purinomycin at 2 μg/ml for 3 days. Surviving single cells were added into 96-well plates containing RPMI medium 1640 basic, supplemented with 20% FBS, and were cultured for 2 weeks. Finally, the GBP7 knockout cells were verified using Western blotting.
Western blotting.
A549 cells were lysed in radioimmunoprecipitation assay buffer (Keygen, China) after two PBS washes. Proteins were quantified by standardization. Each protein sample (20 μl) was separated using 10% to 12% SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane. Subsequently, the membrane was immersed in 5% milk for 1 h at ambient temperatures and incubated with primary and secondary antibodies successively. Finally, the FluorChem E imaging system (ProteinSimple, USA) and Lumiglo reagent kit (Cell Signaling Technology, USA) were used to evaluate the chemiluminescent signal.
Immunofluorescence microscopy.
Cells were washed twice with prechilled PBS, and then the PBS was aspirated and cells were immobilized with 4% paraformaldehyde for 25 min. Paraformaldehyde was then aspirated and cells were permeated with 0.1% Triton X-100 for 5 min at room temperature. Subsequently, cells were blocked with 3% bovine serum albumin (BSA) for 1 h at room temperature, the blocking solution was then discarded, and cells were incubated with influenza NP protein antibody at 4°C overnight. Following 24 h of incubation, the subsequent experimental steps were performed in the dark. Cells were washed with PBS-Tween 20 (PBS-T) three times and incubated with the secondary antibody (Alexa Fluor 488-conjugated antibody; Cell Signaling Technology, USA) for 1 h. After washing with PBS-T buffer three times, cells were incubated with Hoechst for 5 min. Finally, cellular fluorescence was assessed using an inverted fluorescence microscope (Axio Observer; Zeiss, Germany).
Reverse transcription and quantitative real-time PCR.
RNA was isolated using the Cell Total RNA isolation kit (Foregene). RNA samples were reverse transcribed using the PrimeScript RT reagent kit (TaKaRa, China). qRT-PCR was performed using a Light Cycler 480 (Roche, Switzerland). The primary custom-designed primers used are shown in Table 1. The relative expression levels of target genes were calculated by the threshold cycle (2-ΔΔCT) method.
TABLE 1.
Primers for qRT-PCR
| Primer | Forward primer (5′–3′) |
|---|---|
| GBP7-human (F) | GTGGAGCGACTCCTTGTCTG |
| GBP7-human (R) | GTGGGGAATCTCACTTGCTGG |
| GBP7-mice (F) | AACTGAGGGTGAACTCCAAAGC |
| GBP7-mice (R) | GTTTCAGACCTAACTGTGGTGC |
| NP (F) | GGAGACTGATGGAGAACGCC |
| NP (R) | TCTTAGGATCCTTCCCCGCA |
| IFN-α (F) | GCCTCGCCCTTTGCTTTACT |
| IFN-α (R) | CTGTGGGTCTCAGGGAGATCA |
| IFN-β (F) | GGCAATTGAATGGGAGGCT |
| IFN-β (R) | GGCGTCCTCCTTCTGGAACT |
| IFN-λ (F) | CACATTGGCAGGTTCAAATCTCT |
| IFN-λ (R) | CCAGCGGACTCCTTTTTGG |
| IL-6 (F) | GTAGCCGCCCCACACAGA |
| IL-6 (R) | CATGTCTCCTTTCTCAGGGCTG |
| TNF-α (F) | CCCAGGGACCTCTCTCTAATCA |
| TNF-α (R) | GCTTGAGGGTTTGCTACAACATG |
| IL-8 (F) | ATAAAGACATACTCCAAACCTTTCCAC |
| IL-8 (R) | AAGCTTTACAATAATTTCTGTGTTGGC |
| MX1 (F) | GAGGTGGACCCCGAAGGA |
| MX1 (R) | CACCAGATCCGGCTTCGT |
| ISG15 (F) | CACCGTGTTCATGAATCTGC |
| ISG15 (R) | CTTTATTTCCGGCCCTTGAT |
| ISG56 (F) | CAGCCTAGAGGGCAGAACAG |
| ISG56 (R) | CACCTCAAATGTGGGCTTTT |
| GAPDH-human (F) | AGGGCAATGCCAGCCCCAGCG |
| GAPDH-human (R) | AGGCGTCGGAGGGCCCCCTC |
| GAPDH-mice (F) | AGGTCGGTGTGAACGGATTTG |
| GAPDH-mice (R) | GGGGTCGTTGATGGCAACA |
Overexpression of GBP7 in A549 cells.
GBP7 tagged with Flag-expressing plasmid was purchased from GeneCopoeia (EX-H5117-Lv242). According to the manufacturer’s instructions, the GBP7 overexpression plasmid or vector plasmid was transfected into A549 cells using the Lipofectamine 3000 reagent (Invitrogen, USA). Subsequently, A549 cells were infected with the virus 24 h after plasmid transfection. Finally, GBP7 overexpression cells were verified using Western blotting and qRT-PCR.
Statistical analyses.
Data processing and analysis were performed using GraphPad Prism 7. An unpaired Student's t test was used for statistical analysis between the two groups. One-way analysis of variance was performed for multiple comparisons between different groups. In all situations, a P value of <0.05 was considered statistically significant.
ACKNOWLEDGMENTS
This study was financially supported by grants from the National Natural Science Foundation of China (81773787), the National Major Science and Technology Project of China (2018ZX10301101), the Major Scientific and Technological Projects of Guangdong Province (2019B020202002), and the Chinese Academy of Traditional Chinese Medicine (ZZ13-035-02 and 2019XZZX-LG04) to S.L.
REFERENCES
- 1.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. 2012. Organization of the influenza virus replication machinery. Science 338:1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arranz R, Coloma R, Chichón FJ, Conesa JJ, Carrascosa JL, Valpuesta JM, Ortín J, Martín-Benito J. 2012. The structure of native influenza virion ribonucleoproteins. Science 338:1634–1637. doi: 10.1126/science.1228172. [DOI] [PubMed] [Google Scholar]
- 3.Taubenberger JK, Morens DM. 2006. 1918 influenza: the mother of all pandemics. Emerg Infect Dis 12:15–22. doi: 10.3201/eid1201.050979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morens DM, Taubenberger JK, Fauci AS. 2009. The persistent legacy of the 1918 influenza virus. N Engl J Med 361:225–229. doi: 10.1056/NEJMp0904819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Allen IC, Scull MA, Moore CB, Holl EK, McElvania-TeKippe E, Taxman DJ, Guthrie EH, Pickles RJ, Ting JP. 2009. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30:556–565. doi: 10.1016/j.immuni.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe T, Kawakami E, Shoemaker JE, Lopes TJ, Matsuoka Y, Tomita Y, Kozuka-Hata H, Gorai T, Kuwahara T, Takeda E, Nagata A, Takano R, Kiso M, Yamashita M, Sakai-Tagawa Y, Katsura H, Nonaka N, Fujii H, Fujii K, Sugita Y, Noda T, Goto H, Fukuyama S, Watanabe S, Neumann G, Oyama M, Kitano H, Kawaoka Y. 2014. Influenza virus-host interactome screen as a platform for antiviral drug development. Cell Host Microbe 16:795–805. doi: 10.1016/j.chom.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann HH, Schneider WM, Rice CM. 2015. Interferons and viruses: an evolutionary arms race of molecular interactions. Trends Immunol 36:124–138. doi: 10.1016/j.it.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mordstein M, Kochs G, Dumoutier L, Renauld J-C, Paludan SR, Klucher K, Staeheli P. 2008. Interferon-λ contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog 4:e1000151. doi: 10.1371/journal.ppat.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karin M, Ben-Neriah Y. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol 18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 11.Lawrence T. 2009. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb Perspect Biol 1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oeckinghaus A, Hayden MS, Ghosh S. 2011. Crosstalk in NF-κB signaling pathways. Nat Immunol 12:695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Lenardo MJ, Baltimore D. 2017. 30 Years of NF-kappaB: a blossoming of relevance to human pathobiology. Cell 168:37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao S, Song L, Li J, Zhang Z, Peng H, Jiang W, Wang Q, Kang T, Chen S, Huang W. 2012. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell Microbiol 14:1849–1866. doi: 10.1111/cmi.12005. [DOI] [PubMed] [Google Scholar]
- 15.Dai Y, Song J, Li W, Yang T, Yue X, Lin X, Yang X, Luo W, Guo J, Wang X, Lai S, Andrade KC, Chang J. 2019. RhoE fine-tunes inflammatory response in myocardial infarction. Circulation 139:1185–1198. doi: 10.1161/CIRCULATIONAHA.118.033700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schindler C, Levy DE, Decker T. 2007. JAK-STAT signaling: from interferons to cytokines. J Biol Chem 282:20059–20063. doi: 10.1074/jbc.R700016200. [DOI] [PubMed] [Google Scholar]
- 17.Kim BH, Shenoy AR, Kumar P, Das R, Tiwari S, MacMicking JD. 2011. A family of IFN-gamma-inducible 65-kD GTPases protects against bacterial infection. Science 332:717–721. doi: 10.1126/science.1201711. [DOI] [PubMed] [Google Scholar]
- 18.Kim BH, Shenoy AR, Kumar P, Bradfield CJ, MacMicking JD. 2012. IFN-inducible GTPases in host cell defense. Cell Host Microbe 12:432–444. doi: 10.1016/j.chom.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P, Jiang W, Yu Q, Liu W, Zhou P, Li J, Xu J, Xu B, Wang F, Shao F. 2017. Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 551:378–383. doi: 10.1038/nature24467. [DOI] [PubMed] [Google Scholar]
- 20.Cheng YS, Colonno RJ, Yin FH. 1983. Interferon induction of fibroblast proteins with guanylate binding activity. J Biol Chem 258:7746–7750. [PubMed] [Google Scholar]
- 21.Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. 2000. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature 403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- 22.Olszewski MA, Gray J, Vestal DJ. 2006. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J Interferon Cytokine Res 26:328–352. doi: 10.1089/jir.2006.26.328. [DOI] [PubMed] [Google Scholar]
- 23.Ji C, Du S, Li P, Zhu Q, Yang X, Long C, Yu J, Shao F, Xiao J. 2019. Structural mechanism for guanylate-binding proteins (GBPs) targeting by the Shigella E3 ligase IpaH9.8. PLoS Pathog 15:e1007876. doi: 10.1371/journal.ppat.1007876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. 2013. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A 103:8459–8464. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Goffic R, Pothlichet J, Vitour D, Fujita T, Meurs E, Chignard M, Si-Tahar M. 2007. Cutting Edge: influenza A virus activates TLR3-dependent inflammatory and RIG-I-dependent antiviral responses in human lung epithelial cells. J Immunol 178:3368–3372. doi: 10.4049/jimmunol.178.6.3368. [DOI] [PubMed] [Google Scholar]
- 28.Ronni T, Sareneva T, Pirhonen J, Julkunen I. 1995. Activation of IFN-alpha, IFN-gamma, MxA, and IFN regulatory factor 1 genes in influenza A virus-infected human peripheral blood mononuclear cells. J Immunol 154:2764–2774. [PubMed] [Google Scholar]
- 29.MacMicking JD. 2004. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol 25:601–609. doi: 10.1016/j.it.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Z, Shi Z, Yan W, Wei J, Shao D, Deng X, Wang S, Li B, Tong G, Ma Z. 2013. Nonstructural protein 1 of influenza A virus interacts with human guanylate-binding protein 1 to antagonize antiviral activity. PLoS One 8:e55920. doi: 10.1371/journal.pone.0055920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L-F, Yu J, Li Y, Wang J, Li S, Zhang L, Xia S-L, Yang Q, Wang X, Yu S, Luo Y, Sun Y, Zhu Y, Munir M, Qiu H-J. 2016. Guanylate-binding protein 1, an interferon-induced GTPase, exerts an antiviral activity against classical swine fever virus depending on its GTPase activity. J Virol 90:4412–4426. doi: 10.1128/JVI.02718-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zou Z, Meng Z, Ma C, Liang D, Sun R, Lan K, Sandri-Goldin RM. 2017. Guanylate-binding protein 1 inhibits nuclear delivery of Kaposi's sarcoma-associated herpesvirus virions by disrupting formation of actin filament. J Virol 91:e00632-17. doi: 10.1128/JVI.00632-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun E, Hotter D, Koepke L, Zech F, Groß R, Sparrer KMJ, Müller JA, Pfaller CK, Heusinger E, Wombacher R, Sutter K, Dittmer U, Winkler M, Simmons G, Jakobsen MR, Conzelmann KK, Pöhlmann S, Münch J, Fackler OT, Kirchhoff F, Sauter D. 2019. Guanylate-binding proteins 2 and 5 exert broad antiviral activity by inhibiting furin-mediated processing of viral envelope proteins. Cell Rep 27:2092–2104. doi: 10.1016/j.celrep.2019.04.063. [DOI] [PubMed] [Google Scholar]
- 34.Carter CC, Gorbacheva VY, Vestal DJ. 2005. Inhibition of VSV and EMCV replication by the interferon-induced GTPase, mGBP-2: differential requirement for wild-type GTP binding domain. Arch Virol 150:1213–1220. doi: 10.1007/s00705-004-0489-2. [DOI] [PubMed] [Google Scholar]
- 35.Krapp C, Hotter D, Gawanbacht A, McLaren PJ, Kluge SF, Sturzel CM, Mack K, Reith E, Engelhart S, Ciuffi A, Hornung V, Sauter D, Telenti A, Kirchhoff F. 2016. Guanylate binding protein (GBP) 5 is an interferon-inducible inhibitor of HIV-1 infectivity. Cell Host Microbe 19:504–514. doi: 10.1016/j.chom.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 36.Itsui Y, Sakamoto N, Kakinuma S, Nakagawa M, Sekine-Osajima Y, Tasaka-Fujita M, Nishimura-Sakurai Y, Suda G, Karakama Y, Mishima K, Yamamoto M, Watanabe T, Ueyama M, Funaoka Y, Azuma S, Watanabe M. 2009. Antiviral effects of the interferon-induced protein guanylate binding protein 1 and its interaction with the hepatitis C virus NS5B protein. Hepatology 50:1727–1737. doi: 10.1002/hep.23195. [DOI] [PubMed] [Google Scholar]
- 37.Feng J, Cao Z, Wang L, Wan Y, Peng N, Wang Q, Chen X, Zhou Y, Zhu Y. 2017. Inducible GBP5 mediates the antiviral response via interferon-related pathways during influenza A virus infection. J Innate Immun 9:419–435. doi: 10.1159/000460294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tisoncik JR, Korth MJ, Simmons CP, Farrar J, Martin TR, Katze MG. 2012. Into the eye of the cytokine storm. Microbiol Mol Biol Rev 76:16–32. doi: 10.1128/MMBR.05015-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Q, Zhou Y-H, Yang Z-Q. 2016. The cytokine storm of severe influenza and development of immunomodulatory therapy. Cell Mol Immunol 13:3–10. doi: 10.1038/cmi.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Y, Li H, Li J, Liu Y, Li Y, Zhou J, Zhou J, Lu X, Zhao W, Hou J, Wang XY, Chen Z, Zuo D. 2018. Macrophage scavenger receptor 1 contributes to pathogenesis of fulminant hepatitis via neutrophil-mediated complement activation. J Hepatol 68:733–743. doi: 10.1016/j.jhep.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia Z, Xu G, Yang X, Peng N, Zuo Q, Zhu S, Hao H, Liu S, Zhu Y. 2017. Inducible TAP1 negatively regulates the antiviral innate immune response by targeting the TAK1 complex. J Immunol 198:3690–3704. doi: 10.4049/jimmunol.1601588. [DOI] [PubMed] [Google Scholar]
- 42.Li ZW, Sun B, Gong T, Guo S, Zhang J, Wang J, Sugawara A, Jiang M, Yan J, Gurary A, Zheng X, Gao B, Xiao SY, Chen W, Ma C, Farrar C, Zhu C, Chan OTM, Xin C, Winnicki A, Winnicki J, Tang M, Park R, Winnicki M, Diener K, Wang Z, Liu Q, Chu CH, Arter ZL, Yue P, Alpert L, Hui GS, Fei P, Turkson J, Yang W, Wu G, Tao A, Ramos JW, Moisyadi S, Holcombe RF, Jia W, Birnbaumer L, Zhou X, Chu WM. 2019. GNAI1 and GNAI3 reduce colitis-associated tumorigenesis in mice by blocking IL6 signaling and down-regulating expression of GNAI2. Gastroenterology 156:2297–2312. doi: 10.1053/j.gastro.2019.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levy DE, Marié IJ, Durbin JE. 2011. Induction and function of type I and III interferon in response to viral infection. Curr Opin Virol 1:476–486. doi: 10.1016/j.coviro.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iwasaki A, Pillai PS. 2014. Innate immunity to influenza virus infection. Nat Rev Immunol 14:315–328. doi: 10.1038/nri3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller U, Steinhoff U, Reis L, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]