HBV biosynthesis begins at birth and increases during early postnatal liver development in the HBV transgenic mouse model of chronic infection. The levels of viral RNA and DNA synthesis correlate with pioneer transcription factor FoxA transcript plus Tet methylcytosine dioxygenase-generated 5hmC abundance but inversely with Dnmt transcript levels and HBV DNA methylation.
KEYWORDS: 5-hydroxymethylcytosine (5hmC), 5-methylcytosine (5mC), DNA demethylation, DNA methylation, DNA methyltransferase (Dnmt), Forkhead box protein A (FoxA), liver development, ten-eleven translocation (Tet) methylcytosine dioxygenase, hepatitis B virus (HBV)
ABSTRACT
Hepatitis B virus (HBV) transcription and replication increase progressively throughout postnatal liver development with maximal viral biosynthesis occurring at around 4 weeks of age in the HBV transgenic mouse model of chronic infection. Increasing viral biosynthesis is associated with a corresponding progressive loss of DNA methylation. The loss of DNA methylation is associated with increasing levels of 5-hydroxymethylcytosine (5hmC) residues which correlate with increased liver-enriched pioneer transcription factor Forkhead box protein A (FoxA) RNA levels, a rapid decline in postnatal liver DNA methyltransferase (Dnmt) transcripts, and a very modest reduction in ten-eleven translocation (Tet) methylcytosine dioxygenase expression. These observations are consistent with the suggestion that the balance between active HBV DNA methylation and demethylation is regulated by FoxA recruitment of Tet in the presence of declining Dnmt activity. These changes lead to demethylation of the viral genome during hepatocyte maturation with associated increases in viral biosynthesis. Consequently, manipulation of the relative activities of these two counterbalancing processes might permit the specific silencing of HBV gene expression with the loss of viral biosynthesis and the resolution of chronic HBV infections.
IMPORTANCE HBV biosynthesis begins at birth and increases during early postnatal liver development in the HBV transgenic mouse model of chronic infection. The levels of viral RNA and DNA synthesis correlate with pioneer transcription factor FoxA transcript plus Tet methylcytosine dioxygenase-generated 5hmC abundance but inversely with Dnmt transcript levels and HBV DNA methylation. Together, these findings suggest that HBV DNA methylation during neonatal liver development is actively modulated by the relative contributions of FoxA-recruited Tet-mediated DNA demethylation and Dnmt-mediated DNA methylation activities. This mode of gene regulation, mediated by the loss of DNA methylation at hepatocyte-specific viral and cellular promoters, likely contributes to hepatocyte maturation during liver development in addition to the postnatal activation of HBV transcription and replication.
INTRODUCTION
Hepatitis B virus (HBV) is a small enveloped human hepatotropic DNA virus that replicates by reverse transcription of the viral pregenomic 3.5-kb transcript (1–3). The HBV pregenomic 3.5-kb RNA is transcribed from viral covalently closed circular (CCC) DNA in the nucleus of the infected hepatocyte by cellular RNA polymerase II (4, 5). As the HBV pregenomic 3.5-kb RNA encodes both the core antigen (HBcAg) and the reverse transcriptase/DNA polymerase, the regulation of pregenomic RNA expression is a primary determinant of viral biosynthesis which occurs within cytoplasmic capsids (3, 5–8). Pregenomic RNA expression is regulated by the activity of the viral enhancers and the nucleocapsid promoter which are governed by liver-enriched transcription factors, including the nuclear receptors, hepatocyte nuclear factor 4 (HNF4), retinoid X receptor (RXR), peroxisome proliferator-activated receptor (PPAR), farnesoid X receptor (FXR), liver receptor homolog 1 (LRH1), the basic-leucine zipper factors, CCAAT-enhancer-binding proteins (C/EBP), and the winged-helix transcription factors Forkhead box protein A/hepatocyte nuclear factor 3 (FoxA/HNF3) (4, 9, 10).
FoxA can serve as a liver-enriched transcription factor governing RNA synthesis from the viral nucleocapsid promoter (4, 11). In addition, FoxA proteins are pioneer transcription factors which are essential for the establishment of competence within the foregut endoderm and the onset of hepatogenesis (12–15). The changes in gene expression associated with FoxA binding have also been linked to epigenetic changes, including genomic DNA demethylation (16–18). Furthermore, the FoxA pioneer transcription factors have been specifically shown to regulate the developmental demethylation and expression of the HBV genomic DNA in the liver of HBV transgenic mice (19). Specifically, liver-specific FoxA-deficient HBV transgenic mice expressing a single allele of FoxA3 in hepatocytes fail to demethylate viral genomic DNA and hence fail to display viral biosynthesis within the liver. Additionally, wild-type neonatal HBV transgenic mice do not transcribe the viral genome due to its silencing by DNA methylation at CpG sequences flanking the CpG island region, which spans the viral enhancer and nucleocapsid sequences located between nucleotide coordinates 1000 to 2000 (19). In contrast, adult wild-type HBV transgenic mice completely lack any DNA methylation of the viral genome in a major fraction of hepatocytes, and this is associated with high levels of HBV transcription and replication (19).
In the current study, the developmental relationships between HBV biosynthesis and DNA methylation are investigated. Viral RNA and DNA synthesis increase progressively throughout neonatal development, reaching maximal viral biosynthesis at approximately 4 weeks of age. HBV genomic DNA methylation decreased in parallel with increasing HBV biosynthesis. In contrast, the levels of HBV DNA-associated 5-hydroxymethylcytosine (5hmC) increased throughout neonatal development, suggesting the active ten-eleven translocation (Tet) methylcytosine dioxygenase-mediated demethylation of viral DNA during liver maturation. Given the previously identified roles of FoxA in recruitment of Tet to chromatin (14, 20–22), control of liver development (12, 23), and HBV DNA methylation (19), FoxA1, FoxA2, and FoxA3 expression levels throughout liver maturation were determined and shown to increase modestly during neonatal hepatocyte development. In contrast, postnatal liver DNA methyltransferase (Dnmt) expression decreased approximately 10-fold, whereas a very modest reduction in Tet expression was observed. These findings support a model where FoxA-dependent Tet recruitment mediates efficient postnatal HBV DNA demethylation as Dnmt activity declines during hepatocyte maturation.
RESULTS
FoxA expression during postnatal liver development in HBV transgenic mice.
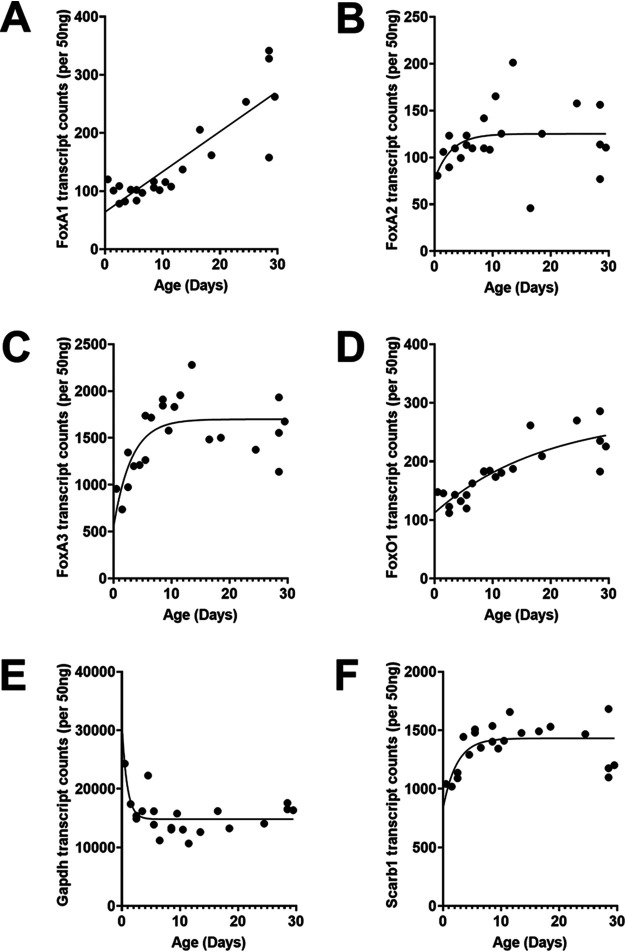
FoxA, a pioneer transcription factor, regulates HBV RNA and DNA synthesis in cell culture (11, 20, 24–29) and viral biosynthesis in the liver of HBV transgenic mice (30, 31), in part by modulating viral genomic DNA methylation during postnatal liver development (19). A detailed analysis of FoxA1, FoxA2, and FoxA3 RNA levels throughout postnatal liver development in HBV transgenic mice indicated that these transcripts increased about 2- to 3-fold from birth to weaning at 4 weeks of age (Fig. 1). FoxO1 RNA also showed a similar modest increase in abundance (Fig. 1D), similar to the FoxA RNAs (Fig. 1A to C), indicating the expression of these genes was not greatly changing throughout postnatal liver development despite their critical role in controlling the hepatocyte-specific gene expression profiles (12, 32, 33). FoxA3 appears to be the most highly expressed member of the FoxA family and displayed increased expression levels, primarily during the first week of postnatal liver development (Fig. 1C).
FIG 1.
Developmental expression of Fox transcript levels in livers of HBV transgenic mice. Quantitative analysis of the liver FoxA1 (A), FoxA2 (B), FoxA3 (C), FoxO1 (D), Gapdh (Northern analysis control) (E), and Scarb1 (a NanoString control) (F) transcripts by NanoString gene expression analysis in the HBV transgenic mice. One-phase decay analysis was used to estimate the changes in gene expression throughout postnatal liver development in the HBV transgenic mice (GraphPad Prism 8.4).
HBV biosynthesis during postnatal liver development in HBV transgenic mice.
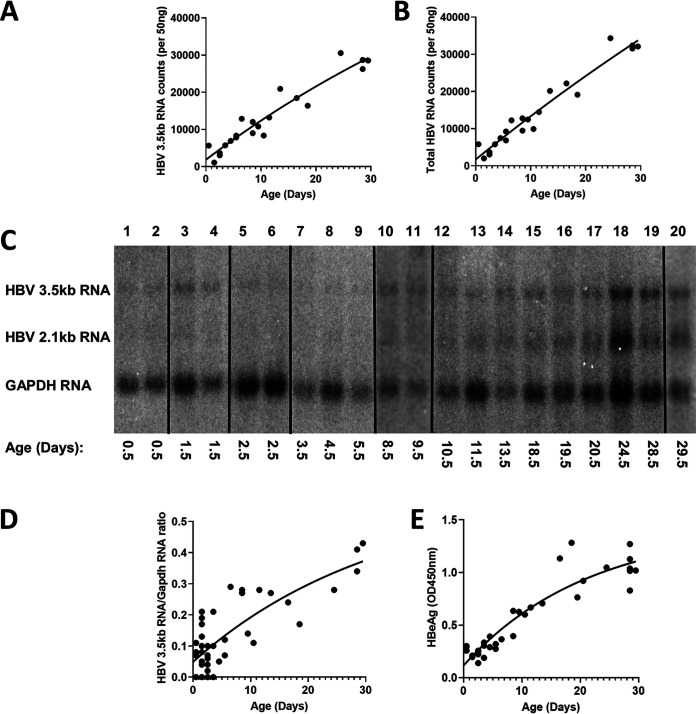
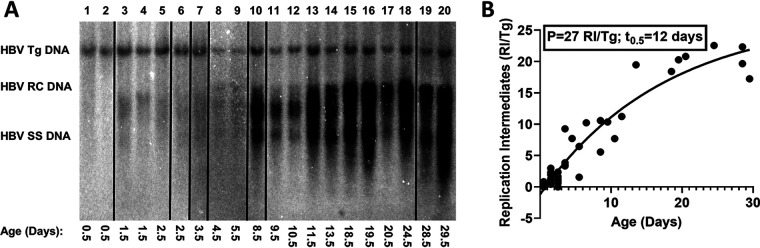
Increasing levels of HBV transcripts during postnatal liver development were estimated by RNA counts (NanoString) and RNA filter hybridization analysis (Fig. 2). At birth, the liver expresses very limited amounts of viral transcripts in the HBV transgenic mouse model of chronic infection. Viral RNA levels increased progressively throughout postnatal liver development, reaching adult transcript abundance around 4 weeks of age. Consistent with these observations, serum HBeAg translated from the HBV precore 3.5-kb transcript displays a similar expression profile throughout neonatal development of the HBV transgenic mouse (Fig. 2E). Furthermore, intracellular HBV DNA replication intermediates displayed a similar increase in abundance throughout neonatal development (Fig. 3). As noted for HBV transcripts, viral DNA replication intermediates were not detectable at birth but increased progressively toward maximal levels by 4 weeks of age (Fig. 3). The rate of increase in observed DNA replication intermediates decreases with time, with half-maximal viral biosynthesis occurring after approximately 12 days with an estimated upper limit equal to about 30 genome equivalents per hepatocyte (plateau, 27 HBV DNA replication intermediates [RI]/HBV transgene [Tg]; Fig. 3B), in agreement with previous analyses of adult HBV transgenic mice (19, 30, 31, 34–38).
FIG 2.
Liver HBV transcripts and serum HBeAg levels throughout postnatal developmental of HBV transgenic mice. (A and B) Quantitative analysis of liver viral 3.5-kb RNA and total HBV RNA (3.5-kb plus 2.1-kb RNA), respectively, by NanoString gene expression analysis in the HBV transgenic mice. One-phase decay analysis was used to estimate the changes in gene expression throughout postnatal liver development in the HBV transgenic mice (GraphPad Prism 8.4). (C) RNA (Northern) filter hybridization analyses of liver transcripts isolated from HBV transgenic mice of various postnatal developmental ages are shown. Noncontiguous lanes from a single analysis are presented. The probes used were HBVayw genomic DNA plus glyceraldehyde-3-phosphate dehydrogenase (Gapdh) cDNA. The Gapdh transcript was used as an internal control for the quantitation of the HBV 3.5-kb RNA. (D and E) Quantitative analysis of RNA (Northern) filter hybridization of the HBV 3.5-kb transcript (D) and serum HBeAg (E) in the HBV transgenic mice. One-phase decay analysis was used to estimate the changes in HBV RNA gene expression and serum HBeAg throughout postnatal liver development in the HBV transgenic mice (GraphPad Prism 8.4).
FIG 3.
DNA (Southern) filter hybridization analysis of HBV DNA replication intermediates throughout postnatal liver development in HBV transgenic mice. (A) DNA (Southern) filter hybridization analysis of liver replication intermediates isolated from HBV transgenic mice of various postnatal developmental ages are shown. Noncontiguous lanes from a single analysis are presented. The probe used was HBVayw genomic DNA. The HBV transgene (Tg) was used as an internal control for the quantitation of the HBV replication intermediates. RC, HBV relaxed circular replication intermediates; SS, HBV single-stranded replication intermediates. (B) Quantitative analysis of DNA (Southern) filter hybridization of the HBV replication intermediates in the HBV transgenic mice. One-phase decay analysis was used to estimate the changes in HBV DNA replication intermediates throughout postnatal liver development in the HBV transgenic mice (P, plateau, maximum calculated number of HBV DNA replication intermediates per cell in adult mouse liver; t0.5, days required to reach half-maximal levels of HBV replication intermediates; GraphPad Prism 8.4).
Effect of postnatal development on HBV DNA methylation in the livers of HBV transgenic mice.
Postnatal liver development in HBV transgenic mice is associated with a progressive increase in HBV RNA and DNA synthesis (Fig. 2 and 3). Furthermore, viral biosynthesis in neonatal mice is very limited with essentially complete HBV DNA methylation outside the CpG island region, which spans the HBV enhancers and nucleocapsid promoter regions (19). In contrast, HBV biosynthesis in adult mice is robust, while viral DNA methylation is essentially absent from viral sequences in the majority of hepatocytes (19). Therefore, it is important to determine the relationships between viral biosynthesis and HBV DNA methylation throughout postnatal liver development in the HBV transgenic mouse model of chronic viral infection.
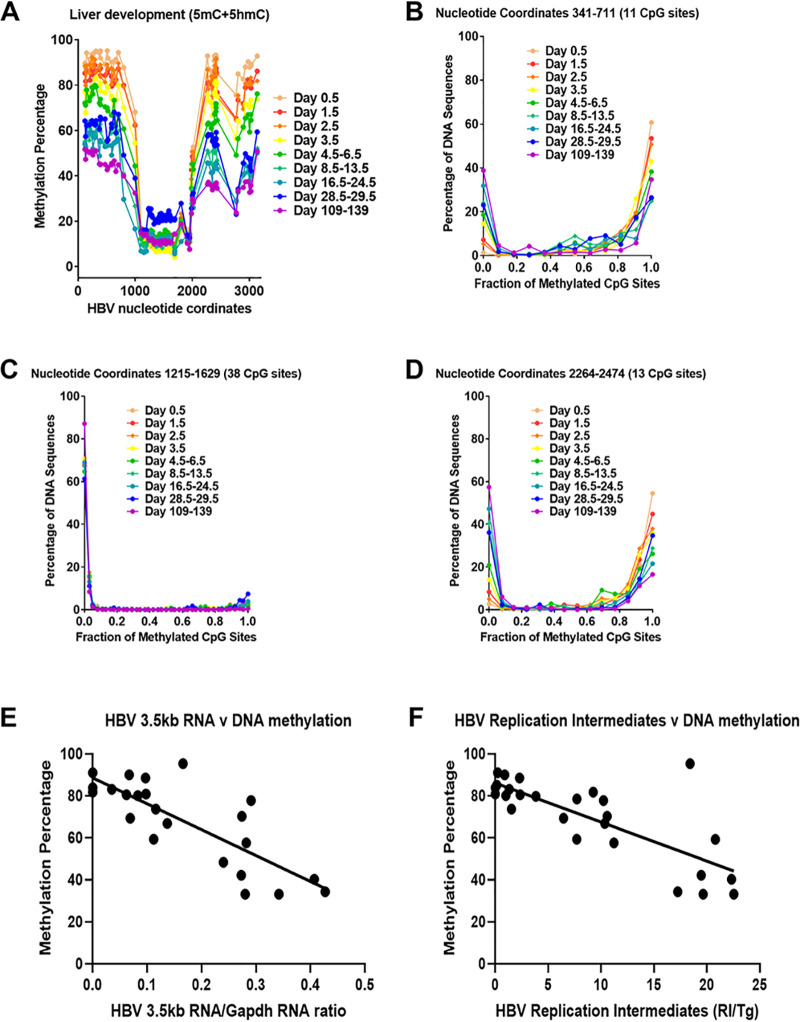
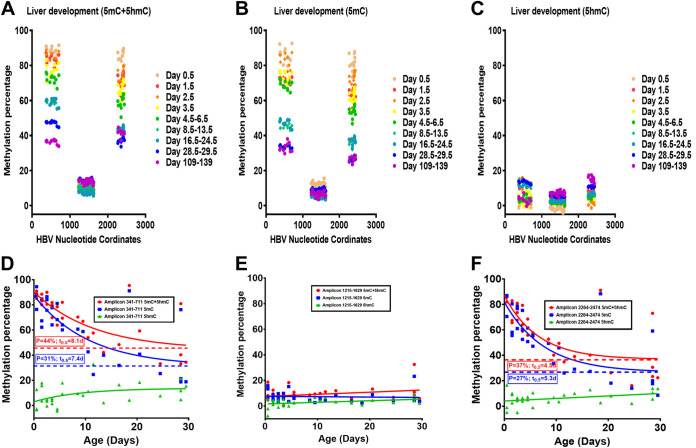
Bisulfite genomic sequencing of the HBV transgene DNA from wild-type HBV transgenic mice was performed throughout postnatal liver development to determine the timing of the loss of viral DNA methylation (Fig. 4). Regardless of the postnatal stage of liver development, the CpG island located between approximately HBV nucleotide coordinates 1000 to 2000 was hypomethylated (Fig. 4A and C) (19, 39). This region encompasses the enhancer 1/X gene promoter and enhancer 2/core gene promoter regions (4, 9), and the limited DNA methylation within this transcriptional regulatory region did not change in a consistent manner throughout postnatal liver development (Fig. 4). In contrast, the percentage of DNA methylation occurring from approximate nucleotide coordinates 2000 to 3182 (Fig. 4A and D), which includes the large and major surface antigen promoter (24, 25), and nucleotide coordinates 1 to 1000 decreases progressively throughout postnatal liver development (Fig. 4). Furthermore, the loss of DNA methylation occurs most rapidly within the first 2 weeks of postnatal liver development (Fig. 4) when the increases in HBV biosynthesis are highest (Fig. 3).
FIG 4.
Effect of postnatal liver development on HBV DNA methylation in HBV transgenic mice. (A) Percentage of CpG DNA methylation (5mC plus 5hmC; bisulfite treatment) at each position within the HBV genome from the livers of HBV transgenic mice throughout postnatal development is shown. The numbers of mice per group are 10 on day 0.5, 11 on day 1.5, 8 on day 2.5, 6 on day 3.5, 5 on days 4.5 to 6.5, 6 on days 8.5 to 13.5, 5 on days 16.5 to 24.5, 5 on days 28.5 to 29.5, and 2 on days 109 to 139. The viral transcription initiation sites for the X gene, the precore/pregenomic, the large surface antigen, and the middle/major surface antigen transcripts plus the CpG island are located at nucleotide coordinates 1310, 1785/1821, 2809, and 3159/3178 plus 1000 to 2000, respectively. (B to D) Effect of postnatal liver development on HBV DNA methylation distribution. The CpG DNA methylation (5mC plus 5hmC; bisulfite treatment) frequency distributions across the 11, 38, and 13 sites within HBV nucleotide coordinates 341 to 711, 1215 to 1629, and 2264 to 2474, respectively, for the viral genome from the livers of HBV transgenic mice throughout postnatal development are shown. The numbers of mice per group are 10 on day 0.5, 11 on day 1.5, 8 on day 2.5, 6 on day 3.5, 5 on days 4.5 to 6.5, 6 on days 8.5 to 13.5, 5 on days 16.5 to 24.5, 5 on days 28.5 to 29.5, and 2 on days 109 to 139. (E) Linear regression analysis was used to correlate HBV 3.5-kb RNA levels with the average viral DNA methylation percentage across HBV nucleotide coordinates 341 to 711 throughout postnatal liver development in the HBV transgenic mice (GraphPad Prism 8.4; R2, 0.64; P < 0.001; y-intercept, 89%). (F) Linear regression analysis was used to correlate HBV DNA replication intermediate levels with the average viral DNA methylation percentage across HBV nucleotide coordinates 341 to 711 throughout postnatal liver development in the HBV transgenic mice (GraphPad Prism 8.4; R2, 0.59; P < 0.001; y-intercept, 86%).
Previously, the distribution of DNA methylation sites within the three HBV DNA amplicons that contained the largest number of CpG sites was evaluated in adult wild-type HBV transgenic mice (19). This analysis indicated that HBV DNA was either completely (or almost completely) methylated or completely (or almost completely) unmethylated with virtually none of the sequences displaying an intermediate level of DNA methylation. To determine how HBV DNA demethylation occurs, the distribution of DNA methylation on individual amplicons was evaluated throughout postnatal liver development (Fig. 4). The HBV DNA amplicon spanning nucleotide coordinates 1215 to 1629 contains 38 CpG sites (Fig. 4C) and is located within the CpG island and, as such, is hypomethylated (19, 40). Methylation of any CpG sequence within this region was very limited as expected for a CpG island, so almost all these sequences were unmethylated, with only a few singly methylated CpG sequences (Fig. 4C).
The HBV DNA amplicons spanning nucleotide coordinates 341 to 711 and 2164 to 2474 contain 11 and 13 CpG sites, respectively (Fig. 4B and D). The HBV transgene DNA was either hypo- or hypermethylated, with up to two CpG sites being methylated or unmethylated, respectively, within any single amplicon sequence. These observations are consistent with the suggestion that the HBV transgene DNA is either almost completely methylated or completely unmethylated within these regions of the genome throughout postnatal liver development (Fig. 4). Partial methylation of the HBV transgene DNA is essentially absent, suggesting that the lack of methylation of one CpG site enhances the probability that the adjacent CpG sites will also be unmethylated. Together, these observations suggest that the loss of HBV DNA methylation occurs by the simultaneous elimination of all (or most) methylated CpG sites across whole individual viral genome sequences by a processive mechanism. This could occur by either DNA replication in the absence of de novo CpG methylation or active demethylation involving Tet-mediated oxidation of 5mC to 5hmC, 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) with subsequent nucleotide excision repair (18, 41). Regardless of the precise mechanism, it is apparent that HBV DNA demethylation occurs progressively throughout postnatal liver development, resulting in increasing numbers of hepatocytes expressing viral transcripts from viral genomes completely (or almost completely) devoid of CpG methylation (Fig. 2 and 4). The extent of HBV DNA demethylation through development is essentially complete by 4 weeks of age, as the level of viral methylation in these mice is very similar to that observed in 15- to 20-week-old mice. Additionally, the levels of HBV 3.5-kb RNA and replication intermediates correlated with the extent of viral CpG demethylation, with biosynthesis being undetectable when HBV DNA methylation was greater than 85% as measured across viral nucleotide coordinates 341 to 711 (Fig. 4E and F). These findings are consistent with extensive viral DNA methylation outside the HBV CpG island region preventing HBV transcription, and hence replication, throughout postnatal liver development.
Tet-mediated oxidation of 5mC contributes to the developmental demethylation of HBV transgene DNA in the liver.
Bisulfite genomic DNA sequencing measures the levels of 5mC plus 5hmC, whereas oxidative-bisulfite (oxBS) genomic DNA sequencing measures only 5mC content (Fig. 5). The difference in cytosine modifications detected by these two methods permits an estimate of 5hmC in genomic DNA sequences. The levels of 5mC and 5hmC present throughout liver development were estimated for the three major viral amplicons covering 62 of the 99 CpG sequences present within the genomic HBV transgene DNA to determine the levels of these modifications associated with viral DNA demethylation (Fig. 5). At birth, the HBV DNA amplicons spanning nucleotide coordinates 341 to 711, 1215 to 1629, and 2264 to 2474 (Fig. 5) demonstrate very low to undetectable levels of 5hmC, indicating that the HBV transgene DNA is neonatally hypermethylated with 5mC residues at nucleotide coordinates 341 to 711 and 2264 to 2474. Throughout postnatal liver development, the HBV DNA spanning nucleotide coordinates 341 to 711 and 2264 to 2474 is progressively demethylated, while the level of 5hmC simultaneously increases to approximately 10 to 15% of CpG sequences. Despite increasing 5hmC levels throughout postnatal liver development, the rate of HBV DNA demethylation decreased with time, with half of the total demethylation occurring within approximately the first 5 to 8 days, which correlates with the increase in viral biosynthesis (Fig. 3 and 5). Furthermore, the changes in the 5mC and 5hmC levels associated with HBV DNA throughout liver development are essentially complete by 4 weeks of age, as approximately equivalent levels of these modifications are observed in 4-week-old and adult mice (Fig. 5A to C). Interestingly, the estimated level of HBV DNA 5mC modification present in 4-week-old and adult mice is approximately 30% (Fig. 5B; Fig. 5D and F; P = 31% and 27%, respectively), which is modestly lower than the estimated fraction of nonparenchymal cells in the adult mouse liver (42, 43). This observation suggests that HBV DNA in the majority of adult hepatocytes may be completely unmethylated, whereas it is fully methylated in the other liver-resident cells, including Kupffer, biliary epithelial, stellate, and endothelial cells.
FIG 5.
Effect of postnatal liver development on HBV DNA 5mC and 5hmC levels in HBV transgenic mice. (A to C) Percentages of CpG DNA methylation (5mC plus 5hmC; bisulfite treatment) at each of the 11, 38, and 13 sites located within HBV nucleotide coordinates 341 to 711, 1215 to 1629, and 2264 to 2474, respectively, for the viral genome from the livers of HBV transgenic mice throughout postnatal development are shown. (A) 5mC plus 5hmC; bisulfite treatment; (B) 5mC; oxidative bisulfite treatment; (C) 5hmC; the difference between panels A and B. The numbers of mice per group are 3 on day 0.5, 3 on day 1.5, 3 on day 2.5, 3 on day 3.5, 4 on days 4.5 to 6.5, 5 on days 8.5 to 13.5, 3 on days 16.5 to 24.5, 3 on days 28.5 to 29.5, and 2 on days 109 to 139. The viral transcription initiation sites for the X gene, the precore/pregenomic, the large surface antigen, and the middle/major surface antigen transcripts plus the CpG island are located at nucleotide coordinates 1310, 1785/1821, 2809, and 3159/3178 plus 1000 to 2000, respectively. (D to F) Quantification of 5mC and 5hmC levels present in HBV DNA nucleotide coordinates 341 to 711, 1215 to 1629, and 2264 to 2474, respectively, throughout postnatal liver development in HBV transgenic mice. One-phase decay analysis was used to estimate the changes in HBV DNA 5mC and 5hmC levels throughout postnatal liver development in the HBV transgenic mice (P, plateau, minimal calculated percent HBV DNA methylation in adult mice; t0.5, days required to reduce DNA methylation by half; GraphPad Prism 8.4).
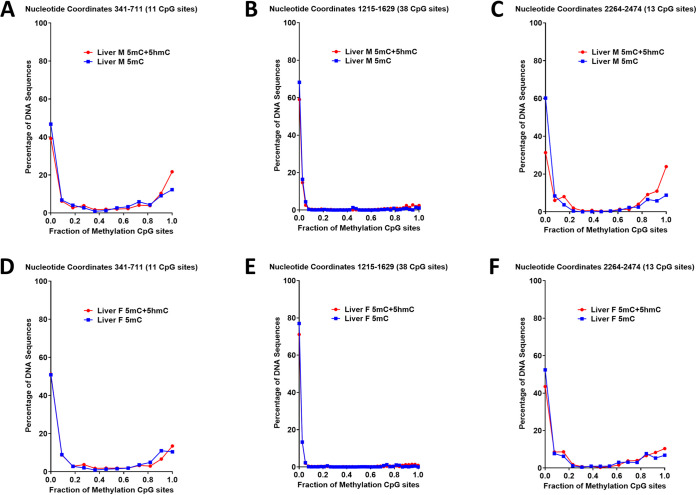
Examination of the distribution of 5mC plus 5hmC (determined from bisulfite sequencing) and 5mC alone (determined from oxidative bisulfite sequencing) within individual amplicon sequences are very similar, with either all (or almost all) or none (or very few) of the CpG sequences being methylated (Fig. 6). However, amplicons that were completely methylated occasionally displayed one or two 5hmC residues. Similarly, a limited number of amplicons that possess one or two methylated CpG sequences were solely 5hmC modified, accounting for the greater number of unmethylated amplicon sequences when DNA was modified with oxidative bisulfite compared to bisulfite treatment (Fig. 6). These results indicate that Tet-mediated oxidation of 5mC residues contributes to the active demethylation of HBV DNA. In addition, it appears that all the methylated CpG sequences present outside the CpG island region are generally demethylated in a rapid and synchronized manner to generate transcriptionally active unmethylated viral genomic DNA.
FIG 6.
Distribution of HBV DNA 5mC and 5hmC in adult transgenic mouse liver. The CpG DNA methylation (5mC plus 5hmC, bisulfite treatment; 5mC, oxidative bisulfite treatment) frequency distributions across the 11, 38, and 13 sites within HBV nucleotide coordinates 341 to 711, 1215 to 1629, and 2264 to 2474, respectively, for the viral genome from the livers of a male (A to C) and female (D to F) adult HBV transgenic mouse is shown.
Developmental reduction in DNA methyltransferase activity may contribute to active HBV transgene DNA demethylation by FoxA-dependent, Tet-mediated oxidation of 5mC in the liver.
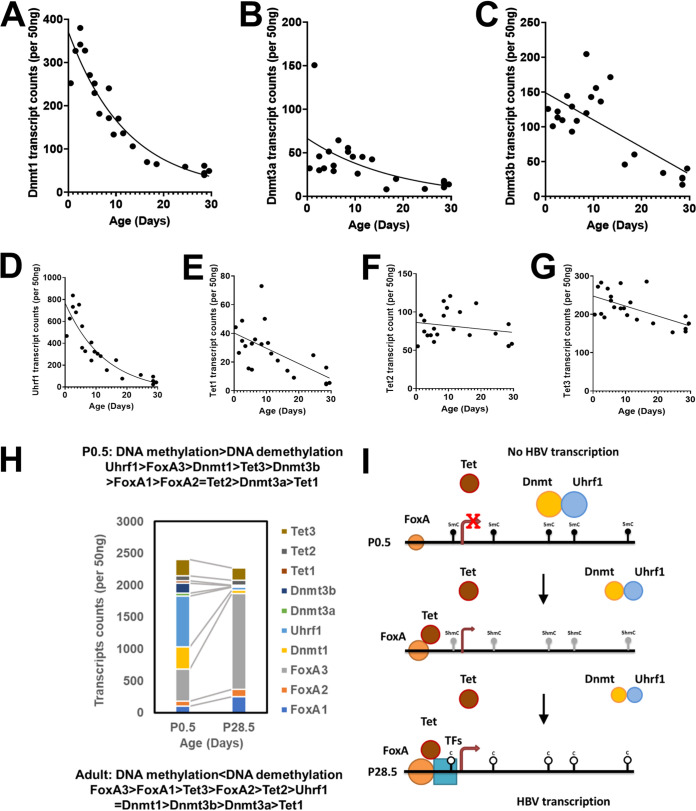
The pioneer transcription factor FoxA regulates HBV biosynthesis in the liver of HBV transgenic mice by modulating viral genomic DNA methylation during postnatal liver development (19). However, FoxA RNA levels only increase approximately 3-fold during postnatal liver development (Fig. 1). Interestingly, for the maintenance Dnmt1 and Uhrf1, the associated DNA methylation regulator (44–46), RNA levels decrease dramatically throughout postnatal liver development (Fig. 7A and D) whereas the de novo Dnmt3a and Dnmt3b RNA levels also decrease but to a much lesser extent (Fig. 7B and C). In contrast, the Tet2 and Tet3 RNA levels encoding the major liver isoforms are largely unaffected during postnatal liver development, whereas Tet1 RNA levels are limited at birth and decrease to very low levels by 4 weeks of age in HBV transgenic mice (Fig. 7E to G). These observations suggest that FoxA recruitment of Tet1, Tet2, and/or Tet3 during postnatal liver development in the context of decreasing levels of Dnmt activity may represent a major factor accounting for the progressive HBV DNA demethylation and associated increases in viral biosynthesis observed in the HBV transgenic mouse model of chronic infection (Fig. 7H and I) (16, 17, 22, 23, 47, 48).
FIG 7.
Developmental expression of DNA methyltransferase and ten-eleven translocation methylcytosine dioxygenase transcript levels in livers of HBV transgenic mice. Quantitative analysis of the liver Dnmt1 (A), Dnmt3a (B), Dnmt3b (C), Uhrf1 (D), Tet1 (E), Tet2 (F), and Tet3 (G) transcripts by NanoString gene expression analysis in the HBV transgenic mice. One-phase decay analysis was used to estimate the changes in gene expression throughout postnatal liver development in the HBV transgenic mice (GraphPad Prism 8.4). (H) Stacked column chart indicating the relative abundances of FoxA1, FoxA2, FoxA3, Dnmt1, Dnmt3a, Dnmt3b, Uhrf1, Tet1, Tet2, and Tet3 in neonatal (postnatal day 0.5) and adult (postnatal day 28.5) HBV transgenic mice (Fig. 1 and 7). (I) Model for the developmental regulation of HBV DNA methylation and transcription. In the neonatal (P0.5) wild-type HBV transgenic mice, the HBVtransgene DNA is maintained in an extensively methylated state by Dnmt1 and Uhrf1 with possible additional contributions from Dnmt3a and Dnmt3b. FoxA is expressed at birth and marks the HBV genome for later developmental expression upon recruitment of additional epigenetic-modifying activities and transcription factors to the viral promoters. During postnatal development, FoxA expression levels increase modestly while Dnmt levels rapidly decline, permitting the recruitment of additional transcriptional factors to the viral enhancer and promoter sequences. FoxA alone, or in combination with the additional transcription factors, leads to Tet recruitment and the oxidative conversion of 5mC to 5hmC. The conversion of 5mC to 5hmC, in combination with declining Dnmt activity, in the presence of hepatocyte proliferation leads to both active and passive demethylation of HBV DNA throughout postnatal liver development. In adult HBV transgenic mice, viral DNA is maintained in an unmethylation state due to the balance between Tet-mediated demethylation and Dnmt-dependent methylation.
DISCUSSION
The majority of HBV infections worldwide result from vertical transmission between chronic carrier mothers and their children due to the transfer of maternal blood to the neonate at birth (49, 50). Interestingly in the HBV transgenic mouse model of chronic viral infection, viral biosynthesis does not occur in the liver of neonates but progressively increases throughout postnatal maturation (38, 51). Increases in HBV replication parallel the changes in viral transcription, which reflects, in part, the increases in liver-enriched transcription factor abundances associated with hepatocyte terminal differentiation (52). However, an additional layer of regulation mediated by FoxA-dependent demethylation of viral DNA is a critical determinant of HBV gene expression and biosynthesis (19). Interestingly, HBV DNA is always hypomethylated across the CpG island region spanning the enhancer I/X gene and the enhancer II/core promoters (nucleotide coordinates 1000 to 2000; Fig. 4A and Fig. 5A to C), suggesting hypermethylation across the rest of the genome leads to a chromatin structure, possibly heterochromatin, preventing the transcriptional machinery from accessing the viral genome. FoxA is a pioneer transcription factor capable of binding to nucleosomal DNA, responsible for marking genes for subsequent tissue-specific gene expression, and is critical for the initiation of liver development (12, 53). In the specific case of HBV transgenic DNA, viral DNA is transcriptionally inactivated and extensively methylated in neonatal liver but essentially completely devoid of methylated CpG sequences in hepatocytes, supporting active transcription and replication in adult mice (19). The developmental demethylation of HBV DNA is dependent upon FoxA expression (19). However, the developmental timing and molecular determinants of HBV DNA demethylation are unknown.
As FoxA deficiency prevents the developmental demethylation of HBV DNA and viral biosynthesis in hepatocytes (19), it was critical to examine FoxA expression throughout postnatal liver development (Fig. 1). FoxA1, FoxA2, and FoxA3 transcript levels increase modestly throughout liver development, although the kinetics appear to be somewhat different. FoxA3 transcripts appear to represent the most abundantly expressed isoform, increasing approximately 3-fold during the first week of postnatal liver development (Fig. 1C). These observations are similar to those previously reported for the postnatal expression of FoxA proteins in mouse liver (33). It is unclear if the limited changes in FoxA transcription and protein levels could be responsible for the observed changes in developmental demethylation of HBV DNA with corresponding increases in viral biosynthesis (Fig. 2 and 5). Liver-specific developmental analysis of HBV RNA and DNA synthesis plus associated loss of viral DNA methylation at CpG sequences suggested that these processes occur with similar kinetics (Fig. 2 and 5), with approximately half the maximal effects being observed within the range of 1 to 2 weeks. Given the postnatal growth rate of mice and the associated increasing liver size, HBV DNA demethylation could be mediated solely by inhibition of Dnmt1 activity acting upon hemi-methylated DNA generated as a consequence of hepatocyte proliferation (54). Consistent with this possibility is the observation that HBV DNA demethylation is progressive, producing either completely (or almost completely) demethylated viral genomes which could result from cellular DNA replication in the absence of subsequent maintenance methylation of the generated hemi-methylated DNA. However, the developmental analysis of 5hmC residues within HBV DNA demonstrated that oxidation of 5mC residues by Tet increases throughout postnatal hepatocyte differentiation (Fig. 5). These findings suggest that Tet-mediated oxidation of 5mC to 5hmC either prevents the maintenance methylation of hemi-methylated HBV DNA post-cellular DNA replication or supports the active demethylation of 5hmC modified HBV DNA by base-excision repair (BER) (41, 55, 56). If BER is involved, it must remove tracks of 5hmC extending across the whole HBV genome to account for the observed all-or-none distribution of 5mC and 5hmC residues seen in the hepatocytes of the postnatal livers (Fig. 4 and 6). The observation that Dnmt1 and Uhrf1 transcript abundances decline rapidly, whereas Tet RNAs, particularly the more abundant liver Tet2 and Tet3 isoforms, are quite stable suggests that the relative contribution of the opposing DNA-methylating and -demethylating activities might determine the overall degree of HBV DNA methylation throughout postnatal hepatocyte development (Fig. 7).
Overall, it appears that changes in the relative abundances of factors involved in controlling gene-specific DNA methylation contribute to the developmental timing of HBV transcription and likely also contribute to the differential activation of gene expression responsible for hepatocyte differentiation and liver development (23). Specifically, it suggests that increasing levels of FoxA binding activity associated with enhanced expression levels (Fig. 1) mediate the progressive postnatal recruitment of Tet1, Tet2, and/or Tet3, either directly or indirectly, to the HBV enhancer and promoter sequences. HBV DNA in the liver of neonatal transgenic mice is essentially completely methylated and transcriptionally silent (Fig. 2 and 4) (19). However, within 1 to 2 days, HBV RNA and DNA synthesis are apparent in the liver (Fig. 2 and 3). Viral biosynthesis is associated with HBV DNA demethylation and an increasing abundance of 5hmC residues within viral CpG sequences (Fig. 4 and 5). The presence of 5hmC indicates that Tet1, Tet2, and/or Tet3 are actively oxidizing viral 5mC, leading to HBV DNA demethylation. In the context of decreasing maintenance of Dnmt activity, reflected as reduced Dnmt1 and Uhrf1 RNA levels throughout liver postnatal development (Fig. 7), HBV DNA is demethylated in the majority of hepatocytes (Fig. 5), which leads to robust HBV biosynthesis within the liver of adult HBV transgenic mice (Fig. 3).
HBV nuclear covalently closed circular DNA (HBV cccDNA), the molecular replication intermediate, which serves as the template for viral transcription, is resistant to current treatments (57). This prevents the resolution of chronic HBV infections which afflict over 250 million individuals worldwide and are responsible for approximately 600,000 deaths annually (58, 59). The current observations suggest that modulating the balance between HBV DNA methylation and demethylation by therapeutically targeting activities involved in these processes may represent an approach to inactivating HBV cccDNA, blocking viral transcription, and thus resolving chronic HBV infection.
MATERIALS AND METHODS
Transgenic mice.
The production and characterization of the HBV transgenic mouse lineage 1.3.32 have been described (51). These HBV transgenic mice contain a single copy of the terminally redundant, 1.3-genome-length copy of the HBVayw genome integrated into the mouse chromosomal DNA. High levels of HBV replication occur in the livers of adult mice. The mice used were heterozygous for the HBV transgene and were maintained on the SV129 genetic background (60). HBV transgenic mice were fed normal rodent chow, and water was available ad libitum. Mice were sacrificed after the indicated number of days of postnatal development. Liver tissue was frozen in liquid nitrogen and stored at −70°C prior to DNA and RNA extraction. All animal experiments were Institutional Animal Care and Use Committee (IACUC) approved and performed according to institutional guidelines with the University of Illinois at Chicago (UIC) Institutional Biosafety and Animal Care Committee approval (ACC number 19-190). All animal procedures were performed in the College of Medicine Research Building at the UIC and adhere to the policies of the NIH Office of Laboratory Animal Welfare (OLAW), the standards of the Animal Welfare Act, the Public Health Service Policy, and the Guide for the Care and Use of Laboratory Animals.
HBV DNA and RNA analysis.
Total DNA and RNA were isolated from liver of HBV transgenic mice as described (61, 62). Protein-free DNA was isolated in an identical manner to the total DNA except the proteinase K digestion was omitted (36). DNA (Southern) filter hybridization analyses were performed using 20 μg of HindIII-digested DNA (62). Filters were probed with 32P-labeled HBVayw genomic DNA (63) to detect HBV sequences. RNA (Northern) filter hybridization analyses were performed using 10 μg of total cellular RNA as described (62). Filters were probed with 32P-labeled HBVayw genomic DNA to detect HBV sequences and mouse glyceraldehyde-3-phosphate dehydrogenase (Gapdh) cDNA to detect the Gapdh transcript used as an internal control (64). Filter hybridization analyses were quantified by phosphorimaging using a Molecular Dynamics Typhoon 8600 phosphor imager system.
NanoString nCounter gene expression transcript counting was used to quantify the levels of FoxA1, FoxA2, FoxA3, FoxO1, Gapdh, Dnmt1, Dnmt3a, Dnmt3b, Uhrf1, Tet1, Tet2, and Tet3 and HBV 3.5-kb and total HBV 3.5-kb plus 2.1-kb transcript levels in 50 ng mouse liver RNA using a specifically designed code set. Data were quality controlled and normalized using the nSolver Analysis software 4.0 (NanoString Technologies). RNA expression levels throughout postnatal liver development were normalized to mouse Cnot1, Rps29, Slc9a8, Mrps5, and Scarb1 RNA controls.
Serum HBV antigen.
HBeAg analysis was performed using 2 μl of mouse serum and the HBe enzyme-linked immunosorbent assay as described by the manufacturer (Epitope Diagnostics). The level of antigen was determined in the linear range of the assay.
DNA methylation analysis.
Bisulfite (BS-seq; identifies 5mC plus 5hmC) and oxidative-bisulfite sequencing (oxBS-seq; identifies 5mC) of protein-free genomic DNA for methylation analysis was performed using the EZ DNA Methylation-Lightning kit (catalog no. D5030; Zymo Research, Inc., Irvine, CA, USA) and TrueMethyl oxBS module kit (catalog no. 0414-32; NuGen Technologies, Inc., Redwood City, CA, USA), respectively, according to the manufacturer’s instructions (65, 66). The 99 CpG sequences in the HBV DNA genome were targeted using PCR amplification of the bisulfite- and oxidative bisulfite-treated DNA, followed by sequencing of the amplicons on an Illumina MiSeq instrument. Preparation of DNA for high-throughput amplicon sequencing was performed in two PCR steps as described previously (19). Twelve HBV primer pairs targeting 94 of the 99 CpG sites were used. The primer pairs targeting the bisulfite converted HBV DNA were (i) 5′-ACACTGACGACATGGTTCTACACAATACCTAAACCTTTACCC-3′ (oligonucleotide CS1FP1; HBV nucleotide coordinates 1131 to 1150) and 5′-TACGGTAGCAGAGACTTGGTCTGTTTTAGTTAGTGGGGGT-3′ (oligonucleotide CS2RP1; HBV coordinates 1214 to 1197), (ii) 5′-ACACTGACGACATGGTTCTACACAAACTTTCACTTTCTC-3′ (oligonucleotide CS1FP1a; HBV coordinates 1086 to 1102) and 5′-TACGGTAGCAGAGACTTGGTCTGTTGATGGTTTATGATTAA (oligonucleotide CS2RP1a; HBV coordinates 1233 to 1215), (iii) 5′-ACACTGACGACATGGTTCTACACCCCCACTAACTAAAAC-3′ (oligonucleotide CS1FP2; HBV nucleotide coordinates 1198 to 1214) and 5′- TACGGTAGCAGAGACTTGGTCTGGGTAATATTTGGTGG-3′ (oligonucleotide CS2RP2; HBV nucleotide coordinates 1645 to 1630), (iv) 5′-ACACTGACGACATGGTTCTACATTAAACTCTCAACAATATCA-3′ (oligonucleotide CS1FP3; HBV nucleotide coordinates 1668 to 1687) and 5′-TACGGTAGCAGAGACTTGGTCTAAGTTATTTAAGGTATAGTTTG-3′ (oligonucleotide CS2RP3; HBV nucleotide coordinates 1896 to 1875), (v) 5′-ACACTGACGACATGGTTCTACAGTGGTTTTGGGGTATGG-3′ (oligonucleotide CS1FP4; HBV nucleotide coordinates 1890 to 1906) and 5′- TACGGTAGCAGAGACTTGGTCTCAAATTAACACCCACCC-3′ (oligonucleotide CS2RP4; HBV nucleotide coordinates 2130 to 2114), (vi) 5′-ACACTGACGACATGGTTCTACAAGTTATAGAGTATTTGGTGT-3′ (oligonucleotide CS1FP5a; HBV nucleotide coordinates 2244 to 2263) and 5′-TACGGTAGCAGAGACTTGGTCTCCCAATAAAATTCCCCA-3′ (oligonucleotide CS2RP5a; HBV nucleotide coordinates 2491 to 2475), (vii) 5′- ACACTGACGACATGGTTCTACAGATTGTAATTGATTATGTTTG -3′ (oligonucleotide CS1FP6; HBV nucleotide coordinates 2625 to 2645) and 5′- TACGGTAGCAGAGACTTGGTCTCCATACTATAAATCTTATTCCC-3′ (oligonucleotide CS2RP6; HBV nucleotide coordinates 2832 to 2853), (viii) 5′-ACACTGACGACATGGTTCTACAGGGAATAAGATTTATAGTATGG (oligonucleotideCS1FP7; HBV nucleotide coordinates 2832 to 2853) and 5′-TACGGTAGCAGAGACTTGGTCTTAAACCTAAAAACTCCACC-3′ (oligonucleotide CS2RP7; HBV nucleotide coordinates 3061 to 3043, (ix) 5′-ACACTGACGACATGGTTCTACAAATCAAAAAAACAACCTACC-3′ (oligonucleotide CS1FP8; HBV nucleotide coordinates 3115 to 3134) and 5′-TACGGTAGCAGAGACTTGGTCTGTGAGTGATTGGAGGT-3′ (oligonucleotide CS2RP8; HBV nucleotide coordinates 340 to 325), (x) 5′-ACACTGACGACATGGTTCTACAACCTCCAATCACTCAC-3′ (oligonucleotide CS1FP9; HBV nucleotide coordinates 325 to 340) and 5′-TACGGTAGCAGAGACTTGGTCTGTTAAATAGTGGGGGAAAG-3′ (oligonucleotide CS2RP9; HBV nucleotide coordinates 730 to 712), (xi) 5′-ACACTGACGACATGGTTCTACAGGATGATGTGGTATTGG-3′ (oligonucleotide CS1FP10; HBV nucleotide coordinates 743 to 759) and 5′-TACGGTAGCAGAGACTTGGTCTCAAAACCCAAAAAACCCAC-3′ (oligonucleotide CS2RP10; HBV nucleotide coordinates 1020 to 1002), and (xii) 5′-ACACTGACGACATGGTTCTACATTGATGTTTTTGTATGTA-3′ (oligonucleotide CS1FP11; HBV nucleotide coordinates 1053 to 1070) and 5′-TACGGTAGCAGAGACTTGGTCTATAACCAAACCCCAACC-3′ (oligonucleotide CS2RP11; HBV nucleotide coordinates 1222 to 1206). These primers contained linker sequences (underlined) at the 5′ ends of the oligonucleotides, termed common sequences (CS1 and CS2). These PCRs were performed using ZymoTaq PreMix according to the manufacturer’s instructions (Zymo Research). PCR amplification involved 10 min denaturation at 95°C, followed by 40 cycles of 95°C for 30 s, 50°C for 40 s (ramp rate of 95 to 50 was set to 0.3 C/s), and 72°C for 60 s. Finally, a 7-min incubation at 72°C was performed. The HBV amplicons were subsequently subjected to a second-stage PCR (MyTaq HS mix, 2 times; Bioline USA Inc., Taunton, MA, USA), used to incorporate unique barcodes and sequencing adapters. The PCR primers used were 5′-AATGATACGGCGACCACCGAGATCTACACTGACGACATGGTTCTACA-3′ (oligonucleotide PE1CS1) and 5′-CAAGCAGAAGACGGCATACGAGATNNNNNNNNNNTACGGTAGCAGAGACTTGGTCT-3′ (oligonucleotide PE2BCCS2). The 10-base sample-specific barcodes are indicated with bold Ns and represent barcode sequences from a set of 384 unique primer pairs (Fluidigm Access Array barcode library for Illumina sequencers). PCR amplification involved 5 min denaturation at 95°C, followed by 8 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s. Finally, a 7-min incubation at 72°C was performed. The resulting final PCR products (libraries) contain Illumina sequencing adapters and a sample-specific barcode.
Pooled libraries were sequenced using an Illumina MiSeq instrument, and data were analyzed using the Casava 1.8 pipeline. Sequencing was performed using MiSeq V3 chemistry, with paired-end 2 × 300 base reads, employing Access Array custom sequencing primers. Raw paired-end sequence data were merged without trimming using the software package PEAR (67). Merged reads were mapped to the HBV reference DNA sequence (63) using Bismark (68) in nondirectional mode to account for amplicon design targeting both strands. Methylation status per read was obtained with the Bismark_methylation_extractor. The percent methylation levels plus counts of methylated and unmethylated bases were then summarized per CpG. In addition, the methylation status of each read was assessed individually by counting the number of methylated CpG sites observed within that read. The distribution of methylation levels per read allows for the characterization of the cellular heterogeneity in the methylation status at each amplicon.
Data availability.
Data for this study, both demultiplexed raw FASTQ files and methylation statistics per read, are accessible on the Gene Expression Omnibus (GEO) under GEO IDs GSE89832 and GSE157451.
ACKNOWLEDGMENTS
We thank Luca G. Guidotti and Francis V. Chisari (The Scripps Research Institute, La Jolla, CA) for providing the HBV transgenic mice.
This work was supported by National Institutes of Health grants R01 AI125401 and R01 CA238328.
REFERENCES
- 1.Seeger C, Mason WS. 2015. Molecular biology of hepatitis B virus infection. Virology 479-480:672–686. doi: 10.1016/j.virol.2015.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tuttleman JS, Pourcel C, Summers J. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47:451–460. doi: 10.1016/0092-8674(86)90602-1. [DOI] [PubMed] [Google Scholar]
- 3.Summers J, Mason WS. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 4.Oropeza CE, Tarnow G, Sridhar A, Taha TY, Shalaby RE, McLachlan A. 2020. The regulation of HBV transcription and replication. Adv Exp Med Biol 1179:39–69. doi: 10.1007/978-981-13-9151-4_3. [DOI] [PubMed] [Google Scholar]
- 5.Will H, Reiser W, Weimer T, Pfaff E, Büscher M, Sprengel R, Cattaneo R, Schaller H. 1987. Replication strategy of human hepatitis B virus. J Virol 61:904–911. doi: 10.1128/JVI.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartenschlager R, Schaller H. 1992. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J 11:3413–3420. doi: 10.1002/j.1460-2075.1992.tb05420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirsch RC, Loeb DD, Pollack JR, Ganem D. 1991. cis-acting sequences required for encapsidation of duck hepatitis B virus pregenomic RNA. J Virol 65:3309–3316. doi: 10.1128/JVI.65.6.3309-3316.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ou J-H, Bao H, Shih C, Tahara SM. 1990. Preferred translation of human hepatitis B virus polymerase from core protein- but not from precore protein-specific transcript. J Virol 64:4578–4581. doi: 10.1128/JVI.64.9.4578-4581.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang H, Banks KE, Anderson AL, McLachlan A. 2001. Hepatitis B virus transcription and replication. Drug News Perspect 14:325–334. [PubMed] [Google Scholar]
- 10.Kosovsky MJ, Qadri I, Siddiqui A. 1998. The regulation of hepatitis B virus gene expression: an overview of the cis- and trans-acting components, p 21–50. In Koshy R, Caselmann WH (ed), Hepatitis B virus: molecular mechanisms in disease and novel strategies for therapy. Imperial College Press, London, UK. [Google Scholar]
- 11.Johnson JL, Raney AK, McLachlan A. 1995. Characterization of a functional hepatocyte nuclear factor 3 binding site in the hepatitis B virus nucleocapsid promoter. Virology 208:147–158. doi: 10.1006/viro.1995.1138. [DOI] [PubMed] [Google Scholar]
- 12.Lee CS, Friedman JR, Fulmer JT, Kaestner KH. 2005. The initiation of liver development is dependent on Foxa transcription factors. Nature 435:944–947. doi: 10.1038/nature03649. [DOI] [PubMed] [Google Scholar]
- 13.Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25:2227–2241. doi: 10.1101/gad.176826.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. 2016. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell 62:79–91. doi: 10.1016/j.molcel.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev 10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- 16.Sérandour AA, Avner S, Percevault F, Demay F, Bizot M, Lucchetti-Miganeh C, Barloy-Hubler F, Brown M, Lupien M, Métivier R, Salbert G, Eeckhoute J. 2011. Epigenetic switch involved in activation of pioneer factor FOXA1-dependent enhancers. Genome Res 21:555–565. doi: 10.1101/gr.111534.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donaghey J, Thakurela S, Charlton J, Chen JS, Smith ZD, Gu H, Pop R, Clement K, Stamenova EK, Karnik R, Kelley DR, Gifford CA, Cacchiarelli D, Rinn JL, Gnirke A, Ziller MJ, Meissner A. 2018. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat Genet 50:250–258. doi: 10.1038/s41588-017-0034-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Zhang D, Li Q, Liang J, Sun L, Yi X, Chen Z, Yan R, Xie G, Li W, Liu S, Xu B, Li L, Yang J, He L, Shang Y. 2016. Nucleation of DNA repair factors by FOXA1 links DNA demethylation to transcriptional pioneering. Nat Genet 48:1003–1013. doi: 10.1038/ng.3635. [DOI] [PubMed] [Google Scholar]
- 19.McFadden VC, Shalaby RE, Iram S, Oropeza CE, Landolfi JA, Lyubimov AV, Maienschein-Cline M, Green SJ, Kaestner KH, McLachlan A. 2017. Hepatic deficiency of the pioneer transcription factor FoxA restricts hepatitis B virus biosynthesis by the developmental regulation of viral DNA methylation. PLoS Pathog 13:e1006239. doi: 10.1371/journal.ppat.1006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 21.Kohler S, Cirillo LA. 2010. Stable chromatin binding prevents FoxA acetylation, preserving FoxA chromatin remodeling. J Biol Chem 285:464–472. doi: 10.1074/jbc.M109.063149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang YA, Zhao JC, Fong K-w, Kim J, Li S, Song C, Song B, Zheng B, He C, Yu J. 2016. FOXA1 potentiates lineage-specific enhancer activation through modulating TET1 expression and function. Nucleic Acids Res 44:8153–8164. doi: 10.1093/nar/gkw498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reizel Y, Sabag O, Skversky Y, Spiro A, Steinberg B, Bernstein D, Wang A, Kieckhaefer J, Li C, Pikarsky E, Levin-Klein R, Goren A, Rajewsky K, Kaestner KH, Cedar H. 2018. Postnatal DNA demethylation and its role in tissue maturation. Nat Commun 9:2040. doi: 10.1038/s41467-018-04456-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raney AK, Zhang P, McLachlan A. 1995. Regulation of transcription from the hepatitis B virus large surface antigen promoter by hepatocyte nuclear factor 3. J Virol 69:3265–3272. doi: 10.1128/JVI.69.6.3265-3272.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raney AK, McLachlan A. 1997. Characterization of the hepatitis B virus major surface antigen promoter hepatocyte nuclear factor 3 binding site. J Gen Virol 78:3029–3038. doi: 10.1099/0022-1317-78-11-3029. [DOI] [PubMed] [Google Scholar]
- 26.Tang H, McLachlan A. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci U S A 98:1841–1846. doi: 10.1073/pnas.041479698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang H, McLachlan A. 2002. Mechanisms of inhibition of nuclear hormone receptor-dependent hepatitis B virus replication by hepatocyte nuclear factor 3 beta. J Virol 76:8572–8581. doi: 10.1128/jvi.76.17.8572-8581.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen M, Hieng S, Qian X, Costa R, Ou JH. 1994. Regulation of hepatitis B virus ENI activity by hepatocyte-enriched transcription factor HNF3. Virology 205:127–132. doi: 10.1006/viro.1994.1627. [DOI] [PubMed] [Google Scholar]
- 29.Ori A, Shaul Y. 1995. Hepatitis B virus enhancer binds and is activated by the hepatocyte nuclear factor 3. Virology 207:98–106. doi: 10.1006/viro.1995.1055. [DOI] [PubMed] [Google Scholar]
- 30.Banks KE, Anderson AL, Tang H, Hughes DE, Costa RH, McLachlan A. 2002. Hepatocyte nuclear factor 3 beta inhibits hepatitis B virus replication in vivo. J Virol 76:12974–12980. doi: 10.1128/JVI.76.24.12974-12980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Oropeza CE, Kaestner KH, McLachlan A. 2009. Limited effects of fasting on hepatitis B virus (HBV) biosynthesis in HBV transgenic mice. J Virol 83:1682–1688. doi: 10.1128/JVI.02208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaestner KH. 2010. The FoxA factors in organogenesis and differentiation. Curr Opin Genet Dev 20:527–532. doi: 10.1016/j.gde.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy P, Bisgaard HC, Thorgeirsson SS. 1994. Expression of hepatic transcription factors during liver development and oval cell differentiation. J Cell Biol 126:223–233. doi: 10.1083/jcb.126.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guidotti LG, Eggers CM, Raney AK, Chi SY, Peters JM, Gonzalez FJ, McLachlan A. 1999. In vivo regulation of hepatitis B virus replication by peroxisome proliferators. J Virol 73:10377–10386. doi: 10.1128/JVI.73.12.10377-10386.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson AL, Banks KE, Pontoglio M, Yaniv M, McLachlan A. 2005. Alpha/beta interferon differentially modulates the clearance of cytoplasmic encapsidated replication intermediates and nuclear covalently closed circular hepatitis B virus (HBV) DNA from the livers of hepatocyte nuclear factor 1 alpha-null HBV transgenic mice. J Virol 79:11045–11052. doi: 10.1128/JVI.79.17.11045-11052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raney AK, Eggers CM, Kline EF, Guidotti LG, Pontoglio M, Yaniv M, McLachlan A. 2001. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1 alpha-null hepatitis B virus transgenic mice. J Virol 75:2900–2911. doi: 10.1128/JVI.75.6.2900-2911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reese VC, Moore DD, McLachlan A. 2012. Limited effects of bile acids and small heterodimer partner on hepatitis B virus biosynthesis in vivo. J Virol 86:2760–2768. doi: 10.1128/JVI.06742-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Oropeza CE, Sainz B, Uprichard SL, Gonzalez FJ, McLachlan A. 2009. Developmental regulation of hepatitis B virus biosynthesis by hepatocyte nuclear factor 4 alpha. PLoS One 4:e5489. doi: 10.1371/journal.pone.0005489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Li C, Zhang Y, Zhu H, Kang Y, Liu H, Wang J, Qin Y, Mao R, Xie Y, Huang Y, Zhang J. 2013. Comparative analysis of CpG islands among HBV genotypes. PLoS One 8:e56711. doi: 10.1371/journal.pone.0056711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Straussman R, Nejman D, Roberts D, Steinfeld I, Blum B, Benvenisty N, Simon I, Yakhini Z, Cedar H. 2009. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol 16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 41.Shen L, Wu H, Diep D, Yamaguchi S, D'Alessio AC, Fung H-L, Zhang K, Zhang Y. 2013. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell 153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baratta JL, Ngo A, Lopez B, Kasabwalla N, Longmuir KJ, Robertson RT. 2009. Cellular organization of normal mouse liver: a histological, quantitative immunocytochemical, and fine structural analysis. Histochem Cell Biol 131:713–726. doi: 10.1007/s00418-009-0577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kmieć Z. 2001. Cooperation of liver cells in health and disease. Adv Anat Embryol Cell Biol 161:1–151. doi: 10.1007/978-3-642-56553-3. [DOI] [PubMed] [Google Scholar]
- 44.Sharif J, Muto M, Takebayashi S-i, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 45.Bostick M, Kim JK, Estève P-O, Clark A, Pradhan S, Jacobsen SE. 2007. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. 2013. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun 4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- 47.Li J, Wu X, Zhou Y, Lee M, Guo L, Han W, Mo W, Cao WM, Sun D, Xie R, Huang Y. 2018. Decoding the dynamic DNA methylation and hydroxymethylation landscapes in endodermal lineage intermediates during pancreatic differentiation of hESC. Nucleic Acids Res 46:2883–2900. doi: 10.1093/nar/gky063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ancey PB, Ecsedi S, Lambert MP, Talukdar FR, Cros MP, Glaise D, Narvaez DM, Chauvet V, Herceg Z, Corlu A, Hernandez-Vargas H. 2017. TET-catalyzed 5-hydroxymethylation precedes HNF4A promoter choice during differentiation of bipotent liver progenitors. Stem Cell Rep 9:264–278. doi: 10.1016/j.stemcr.2017.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lavanchy D. 2004. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat 11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 50.Dienstag JL. 2008. Hepatitis B virus infection. N Engl J Med 359:1486–1500. doi: 10.1056/NEJMra0801644. [DOI] [PubMed] [Google Scholar]
- 51.Guidotti LG, Matzke B, Schaller H, Chisari FV. 1995. High-level hepatitis B virus replication in transgenic mice. J Virol 69:6158–6169. doi: 10.1128/JVI.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. 2006. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev 20:2293–2305. doi: 10.1101/gad.390906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iwafuchi-Doi M, Zaret KS. 2014. Pioneer transcription factors in cell reprogramming. Genes Dev 28:2679–2692. doi: 10.1101/gad.253443.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moreno-Carranza B, Bravo-Manríquez M, Baez A, Ledesma-Colunga MG, Ruiz-Herrera X, Reyes-Ortega P, de Los Ríos EA, Macotela Y, Martínez de la Escalera G, Clapp C. 2018. Prolactin regulates liver growth during postnatal development in mice. Am J Physiol Regul Integr Comp Physiol 314:R902–R908. doi: 10.1152/ajpregu.00003.2018. [DOI] [PubMed] [Google Scholar]
- 55.de Mendoza A, Lister R, Bogdanovic O. 2020. Evolution of DNA methylome diversity in eukaryotes. J Molecular Biology 432:1687–1705. doi: 10.1016/j.jmb.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 56.Wu X, Zhang Y. 2017. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 18:517–534. doi: 10.1038/nrg.2017.33. [DOI] [PubMed] [Google Scholar]
- 57.Lucifora J, Protzer U. 2016. Attacking hepatitis B virus cccDNA - the holy grail to hepatitis B cure. J Hepatol 64:S41–S48. doi: 10.1016/j.jhep.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 58.Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ. 2015. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet 386:1546–1555. doi: 10.1016/S0140-6736(15)61412-X. [DOI] [PubMed] [Google Scholar]
- 59.Ott JJ, Stevens GA, Groeger J, Wiersma ST. 2012. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 60.Lee SS-T, Pineau T, Drago J, Lee EJ, Owens JW, Kroetz DL, Fernandez-Salguero PM, Westphal H, Gonzalez FJ. 1995. Targeted disruption of the alpha isoform of the peroxisome proliferator-activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol Cell Biol 15:3012–3022. doi: 10.1128/mcb.15.6.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 62.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, New York, NY. [Google Scholar]
- 63.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. 1979. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature 281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 64.Sabath DE, Broome HE, Prystowsky MB. 1990. Glyceraldehyde-3-phosphate dehydrogenase mRNA is a major interleukin 2-induced transcript in a cloned T-helper lymphocyte. Gene 91:185–191. doi: 10.1016/0378-1119(90)90087-8. [DOI] [PubMed] [Google Scholar]
- 65.Booth MJ, Ost TW, Beraldi D, Bell NM, Branco MR, Reik W, Balasubramanian S. 2013. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc 8:1841–1851. doi: 10.1038/nprot.2013.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Booth MJ, Branco MR, Ficz G, Oxley D, Krueger F, Reik W, Balasubramanian S. 2012. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science 336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 67.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data for this study, both demultiplexed raw FASTQ files and methylation statistics per read, are accessible on the Gene Expression Omnibus (GEO) under GEO IDs GSE89832 and GSE157451.