Infectious bursal disease virus (IBDV) infects chicks and is the causative agent of Gumboro disease. During IBDV outbreaks in recent decades, the emergence of very virulent variants and the lack of effective prevention/treatment strategies to fight this disease have had devastating consequences for the poultry industry.
KEYWORDS: birnavirus, double-stranded RNA virus, endosomes, phosphoinositides, viral replication
ABSTRACT
Infectious bursal disease virus (IBDV) is the archetypal member of the family Birnaviridae and the etiological agent of Gumboro disease, a highly contagious immunosuppressive infection of concern to the global poultry sector for its adverse health effects in chicks. Unlike most double-stranded RNA (dsRNA) viruses, which enclose their genomes within specialized cores throughout their viral replication cycle, birnaviruses organize their bisegmented dsRNA genome in ribonucleoprotein (RNP) structures. Recently, we demonstrated that IBDV exploits endosomal membranes for replication. The establishment of IBDV replication machinery on the cytosolic leaflet of endosomal compartments is mediated by the viral protein VP3 and its intrinsic ability to target endosomes. In this study, we identified the early endosomal phosphatidylinositol 3-phosphate [PtdIns(3)P] as a key host factor of VP3 association with endosomal membranes and consequent establishment of IBDV replication complexes in early endosomes. Indeed, our data reveal a crucial role for PtdIns(3)P in IBDV replication. Overall, our findings provide new insights into the replicative strategy of birnaviruses and strongly suggest that it resembles those of positive-strand RNA (+ssRNA) viruses, which replicate in association with host membranes. Furthermore, our findings support the role of birnaviruses as evolutionary intermediaries between +ssRNA and dsRNA viruses and, importantly, demonstrate a novel role for PtdIns(3)P in the replication of a dsRNA virus.
IMPORTANCE Infectious bursal disease virus (IBDV) infects chicks and is the causative agent of Gumboro disease. During IBDV outbreaks in recent decades, the emergence of very virulent variants and the lack of effective prevention/treatment strategies to fight this disease have had devastating consequences for the poultry industry. IBDV belongs to the peculiar family Birnaviridae. Unlike most dsRNA viruses, birnaviruses organize their genomes in ribonucleoprotein complexes and replicate in a core-independent manner. We recently demonstrated that IBDV exploits host cell endosomes as platforms for viral replication, a process that depends on the VP3 viral protein. In this study, we delved deeper into the molecular characterization of IBDV-endosome association and investigated the role of host cell phosphatidylinositide lipids in VP3 protein localization and IBDV infection. Together, our findings demonstrate that PtdIns(3)P serves as a scaffold for the association of VP3 to endosomes and reveal its essential role for IBDV replication.
INTRODUCTION
Gumboro disease, also known as infectious bursal disease (IBD), is a highly contagious immunosuppressive disease in young chickens (Gallus gallus) caused by infectious bursal disease virus (IBDV) (1). Despite the designation of IBD as a reportable disease by the World Organization for Animal Health and the implementation of worldwide vaccination programs, IBD outbreaks remain difficult to control and a burden on the poultry industry (2).
IBDV is the archetypal and best-characterized member of the family Birnaviridae, a family integrated by nonenveloped icosahedral double-stranded RNA (dsRNA) viruses that infect a wide range of vertebrate and invertebrate hosts (3). Except for birnaviruses, virtually all other dsRNA viruses have their genomes enclosed within specialized icosahedral capsids, internal layers known as T=1 cores (4–6). T=1 cores are essential for viral replication, as they protect the viral genome and its replicative intermediates from the RNA surveillance and degradation mechanisms of host cells (4, 7). Birnaviruses present unique structural and functional features among dsRNA viruses (8–11), as they lack T=1 cores and instead organize their genomes in ribonucleoprotein complexes (RNPs), which are filamentous structures composed of the multifunctional protein VP3, the RNA-dependent RNA polymerase VP1, and the dsRNA genome segments (10, 12, 13). RNPs are transcriptionally active and have been proposed to be functionally equivalent to T=1-based inner cores (10, 14).
The infectious cycle of IBDV begins with viral attachment to cell receptors, entry into the host cell via macropinocytosis, and subsequent trafficking to endosomes (15–17). In the endosome, the viral capsid dismantles exposing pore-forming peptides that may promote the escape of RNPs from the endosomes into the cytosol (18–21). In the cytoplasm, RNPs associate with the exofacial leaflet of endosomal membranes to establish IBDV replication complexes (21, 22). This association is mediated by the IBDV multifunctional protein VP3, which has an intrinsic ability to bind exofacially to endosomal membranes (21, 22). Crystallographic analysis of the central region of VP3 revealed that this protein exists as a dimer in solution (23) and bears two positively charged conformational domains, Patch 1 and Patch 2 (24). It is evident that the binding of VP3 to endosomes depends on Patch 2, since the obliteration of this domain prevented its binding to endosomes and reduced IBDV progeny (21). These IBDV-co-opted endosomal membranes may act as hideouts where the dsRNA molecules generated during viral genome replication are protected from host cell antiviral defenses. Interestingly, the mechanism involving the utilization of host membranes to create replicative niches resembles those previously described for positive-sense single-stranded RNA (+ssRNA) viruses (25–27). In the present study, we delved deeper into analysis of the birnavirus replication strategy by investigating the role of host phosphatidylinositides (PIs) in the association of IBDV VP3 protein with endosomal membranes and its significance for IBDV infection.
PIs are negatively charged signaling lipids found in eukaryotic cytoplasmic and organellar membranes, which control membrane trafficking and cytoskeleton remodeling, among other important cellular functions (28–30). Different PI species show their inositol head groups phosphorylated at positions 3, 4, and/or 5 (31), and because the composition of PIs varies among different cellular membranes, these signaling lipids contribute to determining the identity of plasma membrane domains and intracellular compartments (29).
Several intracellular pathogens, including many viruses, hijack membrane PIs to control host cells’ pathways. In doing so, pathogens secure metabolic resources and an intracellular membranous niche for replication (32–38). The co-opting of PI metabolism for the establishment of replicative niches is well documented for many +ssRNA virus families (33, 38). For example, several species of picornaviruses and hepatitis C virus (HCV) generate unique replicative organelles enriched in phosphatidylinositol 4-phosphate [PtdIns(4)P] (39, 40), while members of the family Tombusviridae build their replicative organelles on PtdIns(3)P-enriched compartments (41).
Here, we show that PtdIns(3)P, a major regulator of early endosomal function and membrane trafficking (42), mediates the association of IBDV VP3 protein with endosomal membranes, a pivotal step for the establishment of IBDV replication complexes on the exofacial surfaces of these organelles. More importantly, we demonstrate that PtdIns(3)P is a host factor required for the efficient completion of the IBDV replication cycle. By depending on host cell PIs for replication, birnaviruses follow a unique replicative strategy among dsRNA viruses that is often found in +ssRNA viruses. Therefore, our findings highlight the atypical features of this viral family among dsRNA viruses and provide further evidence supporting the hypothesis that birnaviruses are an evolutionary link between +ssRNA and dsRNA viruses (8, 43).
RESULTS
VP3 protein and the IBDV replication complexes localize on the surfaces of PtdIns(3)P-enriched endosomes.
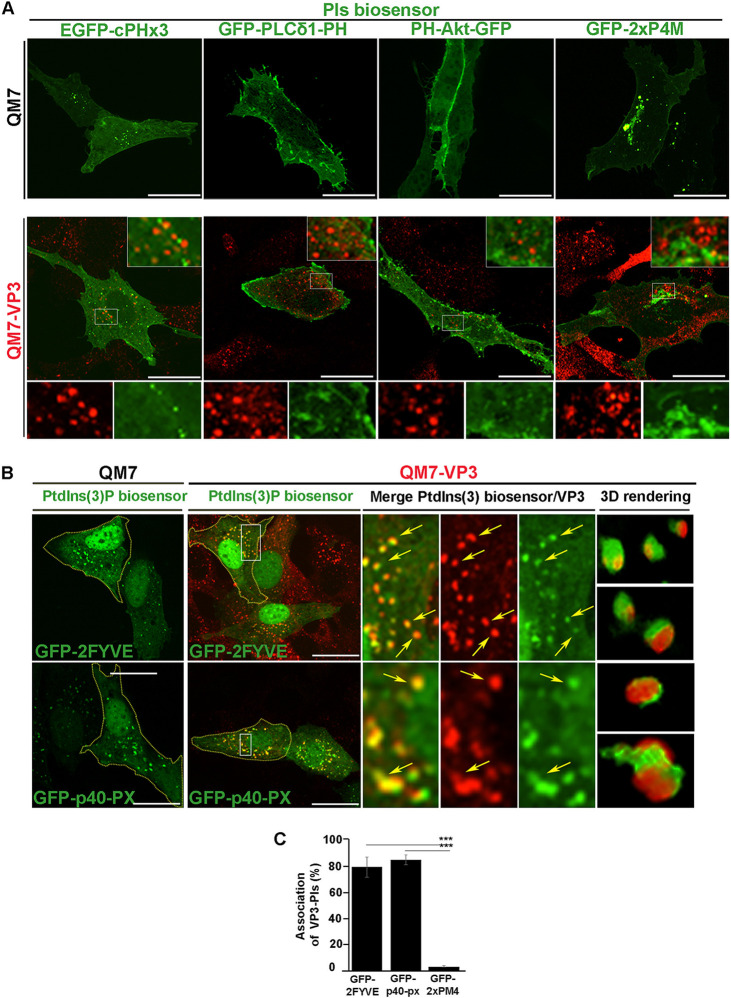
The IBDV VP3 protein (referred to here as VP3) is a moonlighting protein (44) that participates in multiple stages of the IBDV infectious cycle (23, 24, 45, 46). By associating with endosomes, VP3 facilitates the establishment of IBDV replication machinery on the cytoplasm-facing surfaces of these compartments, as we showed previously (21, 22). VP3 binding to endosomes depends on its electropositive conformational domain Patch 2 (21). Because VP3 has the ability to bind negatively charged PIs in vitro (21), we next sought to investigate whether the location of VP3 in host cells could be driven by its interaction with these lipids. To this end, we transiently expressed a panel of well-known green fluorescent protein (GFP)-tagged PI biosensors in QM7 cells and in QM7 cells stably expressing VP3 (QM7-VP3) (21). As shown in Fig. 1A (top) and Fig. 1B (left, QM7 cells), the subcellular localization of the PIs biosensors in avian cells resembled those previously reported for mammalian cells. Specifically, GFP-tagged phospholipase Cδ1 with the pleckstrin homology domain (GFP-PLCδ1-PH) and PH-Akt-GFP, biosensors of PtdIns(4,5)P2 (47) and PtdIns(3,4,5)P3/PtdIns(3,4)P2 (48), respectively, localize at the plasma membrane; pGFP-2xP4M, a biosensor of PtdIns(4)P, predominantly labels the Golgi complex (49–52); enhanced GFP (EGFP)-cPHx3, a biosensor of PtdIns(3,4)P2, decorates plasma membrane microdomains and nascent endosomes (53); and the PtdIns(3)P biosensors GFP-2FYVE and GFP-p40-PX label early endosomes (EEs) (54–59).
FIG 1.
IBDV VP3 localizes on PtdIns(3)P-enriched compartments. (A and B) Subcellular distribution of PIs biosensors and VP3 viral protein. QM7 or QM7-VP3 cells were transfected with several PIs biosensors (green): GFP-PLCδ-PH, which binds to PtdIns(4,5)P2 (47); PH-Akt-GFP, which binds both PtdIns(3,4,5)P3 and PtdIns(3,4)P2 (91); GFP-2FYVE and GFP-p40-PX biosensors, which bind to PtdIns(3)P (54, 55); EGFP-cPHx3, which binds to PtdIns(3,4)P2 (53), and the probe for PtdIns(4)P, GFP-2xP4M (49–52). After 12 h, cells were fixed, permeabilized, stained with antibodies against VP3 (red), and analyzed by spinning-disc confocal microscopy. Main panels and insets show representative images of merged z-stacks. The images are representative of three independent experiments. Bars, 10 μm. Arrows (B) indicate VP3-PtdIns(3)P-positive compartments, and 3D renderings of those compartments are shown on the right. (C) Quantitative analysis of VP3-PI association. QM7-VP3 cells were treated as below in panels A and B, and the percent association between VP3 and PIs was calculated after analysis of cell images with Volocity software. Data are the percentage of VP3 puncta associated with PtdIns(3)P or PtdIns(4)P biosensor-derived signals, calculated as described in Materials and Methods. Data are means and SD. ***, P < 0.01.
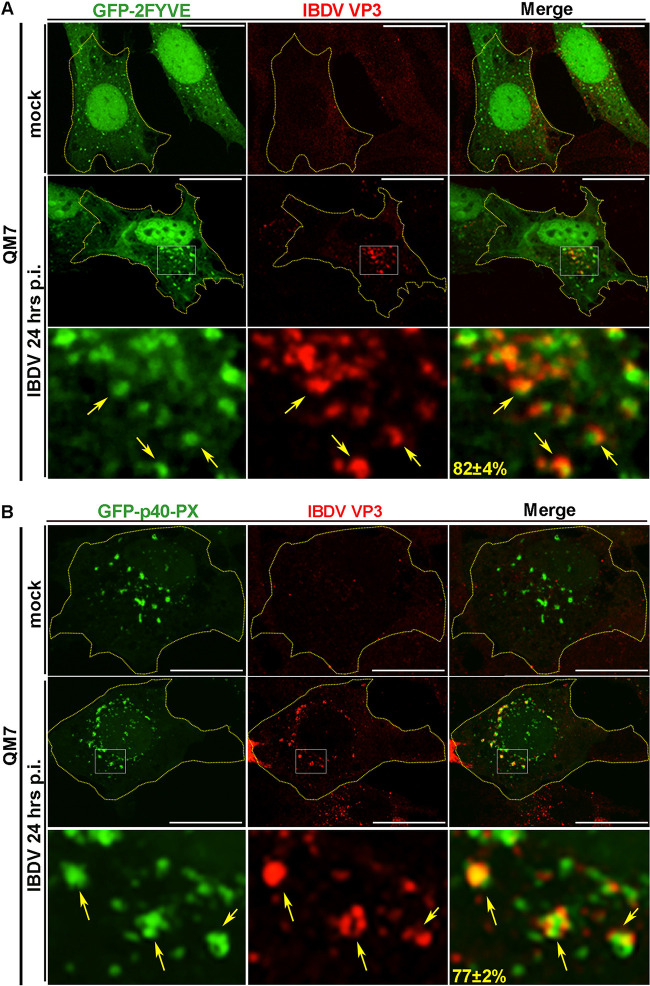
When expressed in cells, VP3 forms puncta in the cytoplasm (21, 22). The distribution of the VP3 puncta with respect to the PI biosensors in QM7-VP3 cells showed that VP3 localized exclusively with the biosensors for PtdIns(3)P, GFP-2FYVE, and GFP-p40-PX (Fig. 1A, bottom, and Fig. 1B and C). Importantly, this was not a phenomenon solely attributable to the heterologous expression of VP3, because when we labeled IBDV RNPs in infected cells using anti-VP3 antibodies, as previously described (22), it also showed a strong association with both PtdIns(3)P biosensors (Fig. 2A and B). Thus, collectively, our results indicate that VP3 puncta and the viral replication complexes associate with PtdIns(3)P-enriched EEs, which aligns with our previous findings showing that VP3 colocalizes with the early endosomal small GTPase Rab5 (21).
FIG 2.
IBDV replication complexes localize on PtdIns(3)P-enriched compartments. (A and B) Subcellular distribution of VP3 protein and PtdIns(3)P biosensors. QM7 cells were transiently transfected with GFP-2FYVE or GFP-p40-PX constructs for 12 h and either infected with IBDV at an MOI of 1 PFU/cell or incubated with virus-free culture medium (mock condition). After 1 h of adsorption at 37°C, cells were infected for 24 h and processed for IIF against VP3 protein as described in Materials and Methods. Arrows indicate IBDV replication complexes on PtdIns(3)P-positive compartments. The z-stack images are representative of three independent experiments. Bars, 10 μm. Numbers show the percentages of VP3 puncta associated with PtdIns(3)P biosensor-derived signals, determined with Volocity software and calculated as described in Materials and Methods. Data are means and SD.
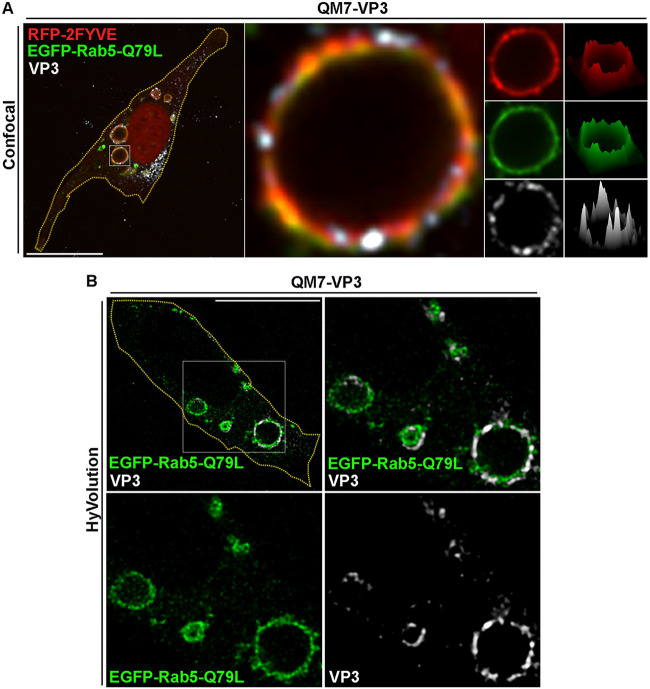
A more detailed inspection of the VP3 puncta in PtdIns(3)P-enriched EEs revealed that the viral protein does not completely overlap PtdIns(3)P biosensors, but instead, it lies in close apposition to PtdIns(3)P-enriched endosomes (Fig. 1B, three-dimensional [3D] renderings; Fig. 2A and B, arrows) and GFP-Rab5-positive compartments (data not shown). To investigate this phenomenon in more detail, we resorted to the constitutively active mutant of the small GTPase Rab5 (EGFP-Rab5-Q79L). The expression of EGFP-Rab5-Q79L in cells causes the formation of enlarged EEs that are enriched in PtdIns(3)P; therefore, we took advantage of the large surface area of these compartments to better visualize the distribution of VP3 protein in EEs. As shown in Fig. 3A, in VP3-QM7 cells, VP3 was recruited to large GFP-Rab5-Q79L-positive EEs formed clusters. Meanwhile, however, RFP (red fluorescent protein)-2FYVE and GFP-Rab5-Q79L were distributed continuously along endosomal membranes. These protein clusters were clearly distinguishable as fluorescence peaks in the surface intensity plot shown in Fig. 3A, and we further confirmed their occurrence by Hyvolution enhanced resolution imaging (Fig. 3B). VP3 clustering might respond to the proclivity of this protein to oligomerize (60), and this may be critical for the establishment of IBDV replication complexes, which we previously showed to also form clusters associated with endosomal membranes (21).
FIG 3.
VP3 displays a discontinuous distribution along the membrane of PtdIns(3)P-enriched compartments. (A) Spinning-disc confocal microscopy analysis of VP3 protein distribution phenotype along the membrane of enlarged Rab5Q79L/PtdIns(3)P compartments. QM7-VP3 cells were cotransfected with RFP-2FYVE and EGFP-Rab5-Q79L for 12 h, and the cells were fixed, permeabilized, and stained with antibodies against VP3 (white) prior to analysis by spinning-disc confocal microscopy. (Left) Merged image showing a single confocal plane. (Middle) Magnified single plane from the framed region showing a giant endosome positive for VP3-, RFP-2FYVE-, and EGFP-Rab5-Q79L-derived signals. (Right) Single channels and surface intensity plots corresponding to the endosome in the middle. The images are representative of three independent experiments. Bars, 10 μm. (B) Improved-resolution analysis of VP3 distribution in enlarged EEs. QM7-VP3 cells where transfected with EGFP-Rab5-Q79L and processed as described for panel A, and the cells were analyzed using enhanced resolution imaging, acquired with a Hyvolution microscopy system as described in Materials and Methods. (Top left) Merged image showing a single confocal plane; (top right) Merged image of a magnified single plane from the framed region on the left showing enlarged endosomes positive for VP3- and EGFP-Rab5-Q79L-derived signals. (Bottom) Single planes of the framed region above. Bars, 10 μm. The images are representative of three independent experiments.
PtdIns(3)P is necessary for the endosomal localization of VP3.
PtdIns(3)P is rapidly synthetized at the membrane of EEs and converted into other PI species as these compartments mature (61, 62). Most PtdIns(3)P is produced by vacuolar protein sorting 34 (Vps34), the catalytic subunit of the PI3K-III enzymatic complex (63, 64), which phosphorylates phosphatidylinositol to produce PtdIns(3)P (65). Other minor synthetic pathways of PtdIns(3)P depend on class II and class I phosphatidylinositol 3-kinases (PI3Ks) (66, 67).
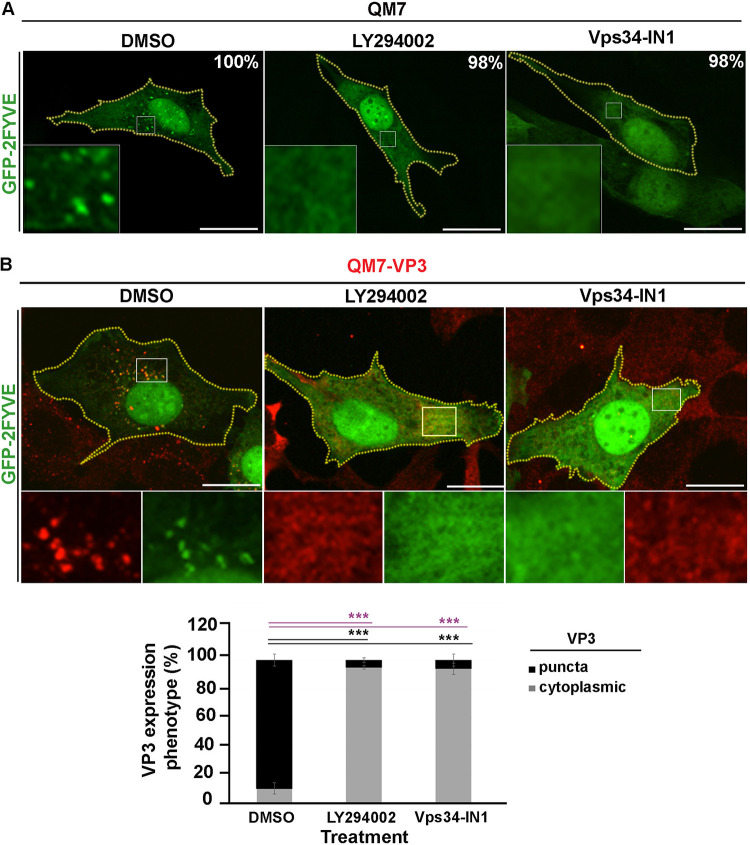
To determine whether PtdIns(3)P mediates the binding of VP3 to endosomal membranes, we blocked its synthesis in QM7 and QM7-VP3 with either the pan-PI3K inhibitor LY294002, which inhibits class I, II, and III PI3K isoforms (68, 69), or the specific class III PI3K Vps34 inhibitor Vps34-IN1 (70). As shown in Fig. 4A, in 98% of the cells treated with these inhibitors GFP-2FYVE dissociated from EEs, bona fide evidence for the depletion of PtdIns(3)P in EEs (71). The depletion of PtdIns(3)P by either inhibitor caused the dissociation of VP3 from EEs and the disaggregation of VP3 puncta. Importantly, however, Vps34-IN1 treatment sufficed to redistribute the VP3 protein, thus indicating that PtdIns3(P) is required for the targeting of VP3, and consequently IBDV replication complexes, to EE membranes (Fig. 4B).
FIG 4.
The chemical depletion of PtdIns(3)P prevents VP3 localization on EEs. (A) Analysis of the subcellular distribution of GFP-2FYVE in avian cells treated with PI3K inhibitors. QM7 cells were transfected with GFP-2FYVE for 12 h and then treated with DMSO (control vehicle), 100 μM LY294002, or 1 μM Vps34-IN1 for 2 h. Subsequently, cells were fixed and visualized by spinning-disc confocal microscopy. Main panels show representative images of merged z-stacks. Framed regions show amplified images that depict the intracellular localization of GFP-2FYVE under the different conditions. The images are representative of three independent experiments. Bars, 10 μm. The percentages were calculated from 100 cells per condition. (B) Analysis of GFP-2FYVE and VP3 protein subcellular distributions in avian cells treated with PI3K inhibitors. QM7-VP3 cells were transfected with GFP-2FYVE and for 12 h and treated as described for panel A. Subsequently, cells were fixed, permeabilized, stained using antibodies against VP3 (red), and analyzed using spinning-disc confocal microscopy. Main panels show representative images of merged single confocal planes. The smaller boxes are amplified images that depict the intracellular localization of GFP-2FYVE and VP3. Bars, 10 μm. VP3 expression phenotypes (punctate or cytoplasmic) were determined for QM7-VP3 cells with Volocity as described in Materials and Methods. Fifty cells per condition were scored for each experiment. The images are representative of three independent experiments. Data are means and SD. ***, P < 0.01.
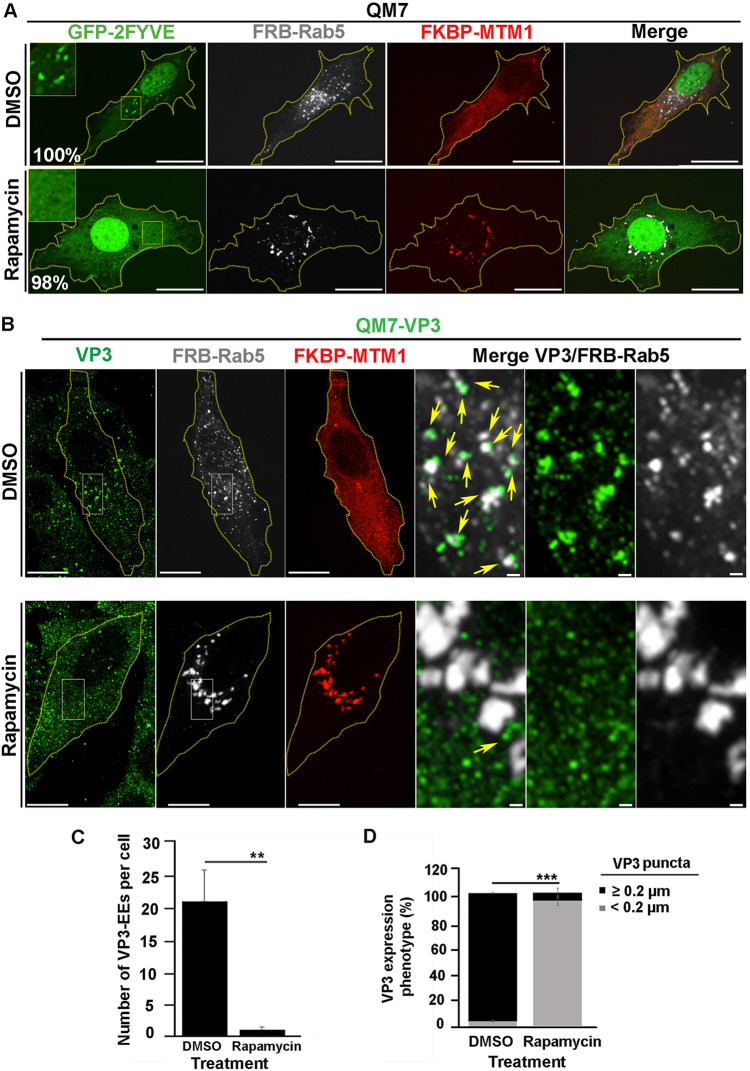
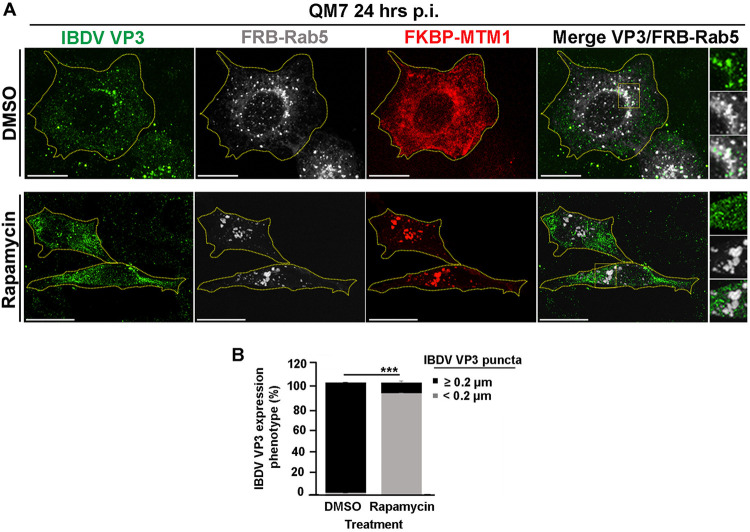
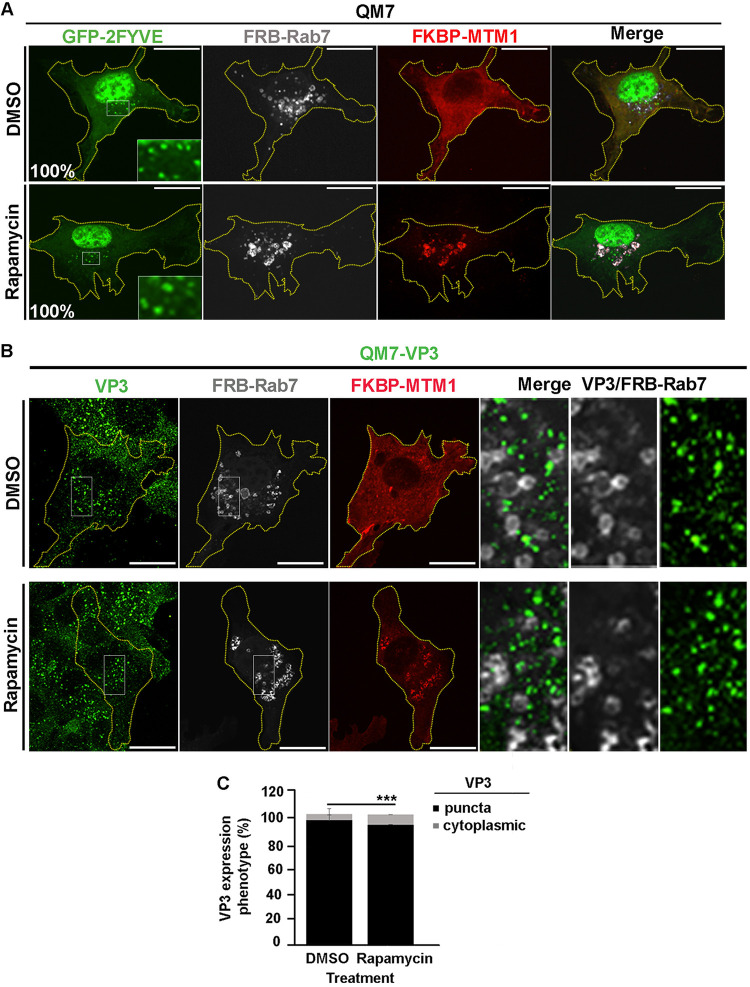
We further confirmed these findings utilizing an alternative strategy to deplete PtdIns(3)P in EEs. To this end, we used the fluorescent proteins mCherry-FKBP-MTM1 and iRFP-FRB-Rab5, which bind to each other in the presence of rapamycin, as described elsewhere (49, 72, 73). Briefly, in resting cells, mCherry-FKBP-MTM1, which encodes the enzyme 3-phosphatase myotubularin phosphatase 1 (MTM1) fused to the rapamycin-binding protein FKBP, is expressed in the cytoplasm; meanwhile iRFP-FRB-Rab5, consisting of Rab5 GTPase fused to the rapamycin binding domain FRB, mainly localizes at EEs. Upon brief incubation with rapamycin, mCherry-FKBP-MTM1 is recruited to EEs via dimerization of the FKBP-FRB rapamycin-binding moieties, causing the localized dephosphorylation of PtdIns(3)P. As shown in Fig. 5A, the recruitment of mCherry-FKBP-MTM1 to iRFP-FRB-Rab5-positive endosomes resulted in the detachment of GFP-2FYVE from these compartments (Fig. 5A, bottom) and substantially changed the distribution pattern of VP3 protein in QM7-VP3 cells (Fig. 5B, top). Indeed, we found that EEs, in this case seen as Rab5-positive compartments, lost their association with VP3 upon the induction of MTM1 recruitment to EEs (Fig. 5B and C). Intriguingly, and differently from what we described above for the treatment with PI3K inhibitors, VP3 puncta remained detectable in the cytoplasm of QM7-VP3 cells, although their numbers and size were noticeably reduced in 93% of cells assessed, as per the average puncta size determined by the imaging analysis algorithm used for punctum identification and quantification (see Materials and Methods) (Fig. 5B and D). In agreement with these observations, targeting mCherry-FKBP-MTM1 to EEs in IBDV-infected QM7 cells also caused a clear dissociation of IBDV RNPs (labeled with anti-VP3 antibodies [22]) from these compartments, resulting in a drastic change in VP3 punctate distribution (Fig. 6A and B). Importantly, the effect exerted by MTM1 3-phosphatase activity on VP3 and IBDV RNPs was attributed to mCherry-FKBP-MTM1 acting on PtdIns(3)P in EEs, as targeting the chimeric 3-phosphatase to Rab7 positive endosomal compartments by coexpressing it with iRFP-FRB-Rab7 altered neither PtdIns(3)P nor VP3 distribution in EEs of QM7 and QM7-VP3 cells (Fig. 7A and B). Thus, collectively, our results demonstrated that PtdIns(3)P mediates and is necessary for the association of IBDV VP3 protein and RNPs with EEs.
FIG 5.
The localized depletion in endosomal PtdIns(3)P prevents VP3 localization on EEs. (A) Analysis of GFP-2FYVE subcellular distribution in QM7 cells after rapamycin-induced localized depletion of PtdIns(3)P in EEs. QM7 cells were transiently cotransfected with mCherry-FKBP-MTM1, iRFP-FRB-Rab5, and GFP-2FYVE for 12 h and subsequently treated for 15 min with DMSO or 1 μM rapamycin to induce the recruitment of FKBP-MTM1 to the FRB-Rab5-decorated membranes to trigger PtdIns(3)P dephosphorylation in EEs. Cells were fixed and visualized by spinning-disc confocal microscopy. Main panels show representative images of merged z-stacks. Framed regions show amplified images that depict the intracellular localization of GFP-2FYVE. The images are representative of three independent experiments. Bars, 10 μm. Percentages were calculated by analyzing 50 cells per condition. (B to D) Analysis of VP3 distribution in cells locally depleted of PtdIns(3)P. QM7-VP3 cells were transiently cotransfected with mCherry-FKBP-MTM1 and iRFP-FRB-Rab5 for 12 h and subsequently treated for 15 min with DMSO or 1 μM rapamycin. The cells were fixed, permeabilized, VP3 (green) stained as described in Materials and Methods, and analyzed by spinning-disc confocal microscopy. (B) Representative images of merged z-stacks. The images are representative of three independent experiments. Bars, 10 μm (left panels) or 0.5 μm (right panels). (C) The number of VP3-Rab5-positive EEs per cell were calculated as described in Materials and Methods. The images are representative of three independent experiments. Data are means and SD. **, P < 0.05. (D) VP3 expression phenotypes were determined employing Volocity software as described in Materials and Methods. Twenty-five cells per condition were scored for each experiment. Data are means and SD. ***, P < 0.01.
FIG 6.
IBDV replication complexes detach from EEs after rapamycin-induced localized depletion of PtdIns(3)P. (A) Subcellular distribution of IBDV RNPs in infected QM7 cells after the localized depletion of PtdIns(3)P from EEs. QM7 cells were transiently cotransfected with mCherry-FKBP-MTM1 and iRFP-FRB-Rab5 for 12 h and subsequently infected with IBDV at an MOI of 1 PFU/cell or incubated with virus-free culture medium. After 1 h of adsorption at 37°C, the cells were infected for 24 h. Before the end of the infection period, the cells were treated for 30 min with DMSO or 1 μM rapamycin to induce the recruitment of FKBP-MTM1 to the FRB-Rab5-decorated membranes in order to trigger the dephosphorylation of PtdIns(3)P in EEs. The cells were fixed and permeabilized, and IBDV replication complexes were stained with antibodies against VP3 (red) as described in Materials and Methods. Cells were analyzed by spinning-disc confocal microscopy. Representative images of merged z-stacks are shown. The images are representative of three independent experiments. Bars, 10 μm. (B) VP3 expression phenotypes were determined with Volocity software as described in Materials and Methods. Twenty-five cells per condition were scored for each experiment. The images are representative of three independent experiments. Data are means and SD. ***, P < 0.01.
FIG 7.
The rapamycin-induced recruitment of MTM1 to Rab7-positive compartments does not prevent VP3-EE association. (A) Analysis of GFP-2FYVE subcellular distribution in QM7 cells after rapamycin-induced recruitment of MTM1 to late endosomes. QM7 cells were transiently cotransfected with mCherry-FKBP-MTM1, iRFP-FRB-Rab7, and GFP-2FYVE for 12 h and subsequently treated for 15 min with DMSO or 1 μM rapamycin, to induce the recruitment of FKBP-MTM1 to the FRB-Rab7-decorated membranes. Cells were fixed and visualized by spinning-disc confocal microscopy. Main panels show representative images of merged z-stacks. Insets show amplified images that depict the intracellular localization of GFP-2FYVE. The images are representative of three independent experiments. Bars, 10 μm. Percentages were calculated by analyzing 50 cells per condition. (B and C) Analysis of VP3 subcellular distribution after recruitment of MTM1 to late endosomes. (B) QM7-VP3 cells were transiently cotransfected with mCherry-FKBP-MTM1 and iRFP-FRB-Rab7 for 12 h and subsequently treated for 15 min with DMSO or 1 μM rapamycin. The cells were fixed, permeabilized, VP3 (green) stained as described in Materials and Methods, and analyzed by spinning-disc confocal microscopy. Representative images of merged z-stacks are shown. The images are representative of three independent experiments. Bars, 10 μm. (C) VP3 expression phenotypes (punctate or cytoplasmic) were determined employing Volocity software and following the criteria described in Materials and Methods. Twenty-five cells per condition were scored for each experiment. The images are representative of three independent experiments. Data are means and SD. ***, P < 0.01.
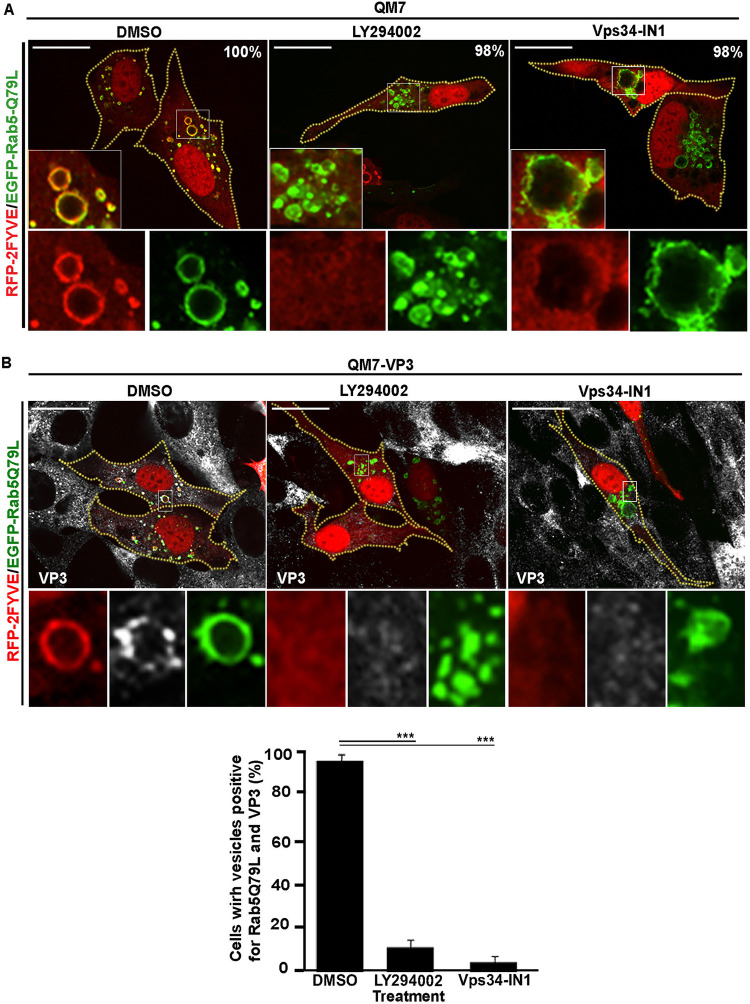
Because Rab5 GTPase recruits multiple proteins to EEs that are involved in the trafficking and function of these compartments (74), we next investigated if this small GTPase could also be involved in the recruitment of VP3 to EEs. To this end, we used PI3K inhibitors to deplete PtdIns(3)P in QM7-VP3 cells coexpressing EGFP-Rab5-Q79L and RFP-2FYVE. As shown in Fig. 8A, the binding of Rab5-Q79L to the EEs was not affected by the pharmacological treatments, whereas RFP-2FYVE and VP3 clearly dissociated from the enlarged compartments (Fig. 8B). Thus, together, our results demonstrated that Rab5 is not involved in the targeting of VP3 protein to EE membranes, which solely requires PtdIns(3)P.
FIG 8.
The constitutive active mutant of Rab5 does not prevent VP3 dissociation from PtdIns(3)P-depleted endosomes. (A) Analysis of GFP-2FYVE subcellular distribution in avian cells overexpressing the constitutive active mutant of Rab5 and treated with PI3K inhibitors. QM7 cells were cotransfected with EGFP-Rab5-Q79L and RFP-2FYVE for 12 h and then treated with DMSO, 100 μM LY294002, or 1 μM Vps34-IN1 for 2 h. Subsequently, cells were fixed and analyzed by spinning-disc confocal microscopy. Main panels show representative images of merged single confocal planes. Insets show amplified images that depict the intracellular localization of EGFP-Rab5-Q79L and RFP-2FYVE. The images are representative of three independent experiments. Bars, 10 μm. The percentages depicted in main panels were calculated from 100 cotransfected cells per condition. (B) Analysis of RFP-2FYVE and VP3 protein subcellular distribution in avian cells expressing the constitutive active mutant of Rab5 and treated with PI3K inhibitors. QM7-VP3 cells were cotransfected with EGFP-Rab5-Q79L and RFP-2FYVE for 12 h, treated, processed, and analyzed as for panel A. The larger panels show representative images of merged single confocal planes. The smaller panels depict the subcellular distribution of VP3, RFP-2FYVE, and EGFP-Rab5-Q79L. The images are representative of three independent experiments. Bars, 10 μm. The percentages of cells positive for EGFP-Rab5-Q79L and VP3 vesicles were analyzed with Volocity software, and results are presented in the graph. Fifty cells per condition were scored for each experiment. The images are representative of three independent experiments. Data are means and SD. ***, P < 0.01.
PtdIns(3)P is required for efficient IBDV replication.
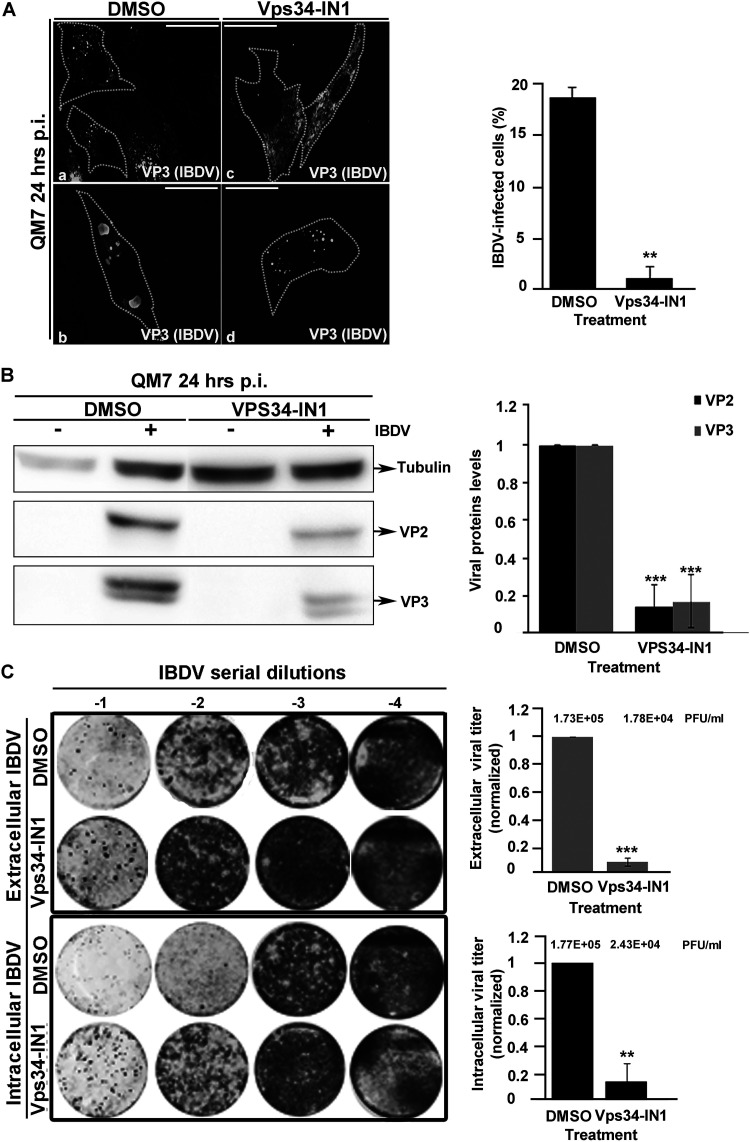
Considering that PtdIns(3)P mediates VP3 association with EEs, we next investigated whether PtdIns(3)P is required for IBDV replication and the generation of viral progeny. To this end, we treated IBDV-infected QM7 cells with the inhibitor Vps34-IN1. To prevent any potential effect of the drug in IBDV internalization, we added the inhibitor 2 h after the onset of the infection for a total of 22 h. For untreated IBDV-infected cells, the viral replication complexes depicted a punctate distribution, mainly at the juxtanuclear region, shown in Fig. 9A and previously described (22) (Fig. 9A, panel a) while in cells at the late phase of the infection, VP3 protein formed large cytoplasmic aggregates (14) (Fig. 9A, panel b). These phenotypes changed radically after Vps34-IN1 treatment. Indeed, IBDV replication complexes completely lost their perinuclear punctate distribution (Fig. 9A, panel c), whereas the VP3 cytoplasmic aggregates shrank in size and increased in number (Fig. 9A, panel d). These changes correlated with a reduction in the number of infected cells (Fig. 9A, right) despite the Vps34-IN1 treatment not affecting cell viability as per an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assay (data not shown).
FIG 9.
The chemical depletion of PtdIns(3)P in avian cells inhibits IBDV replication and reduces its infectivity. (A) Treatment of infected avian cells with Vps34-IN1 impairs IBDV infection. QM7 cells were either mock infected or infected with IBDV at an MOI of 1 PFU/cell and at 2 h p.i. were treated with DMSO or 1 μM Vps34-IN1. At 24 h p.i., cells were fixed, permeabilized, stained with antibodies against VP3 (white) and analyzed by spinning-disc confocal microscopy. Panels a to d show representative images of infected cells. Images are merged z-stacks. The images are representative of three independent experiments. Bars, 10 μm. The percentages of IBDV-infected cells were calculated by scoring 100 cells per condition, and the results are presented in the graph. Data are means and SD. **, P < 0.05. (B) Intracellular accumulation of VP2 and VP3 proteins in QM7 cells depleted of PtdIns(3)P. QM7 cells were either mock infected or infected and treated as for panel A and at 24 h p.i. were processed by Western blotting using the corresponding anti-VP2 and VP3 antibodies described in Materials and Methods. The Western blots and the data shown in the normalized bar graph correspond to an experiment representing three independent trials. Data are means and SD. ***, P < 0.01. (C) QM7 cells were either mock infected or infected and treated as for panel A. At 24 h p.i., the supernatants were collected for extracellular virus titration, and the cellular pellets were processed for intracellular viral titration as described in Materials and Methods. The image shows a representative plaque assay results from three independent trials. Normalized extra- and intracellular viral titers are shown in the graphs. The crude viral titers are shown above the bars. Error bars show SD. ***, P < 0.01; **, P < 0.05.
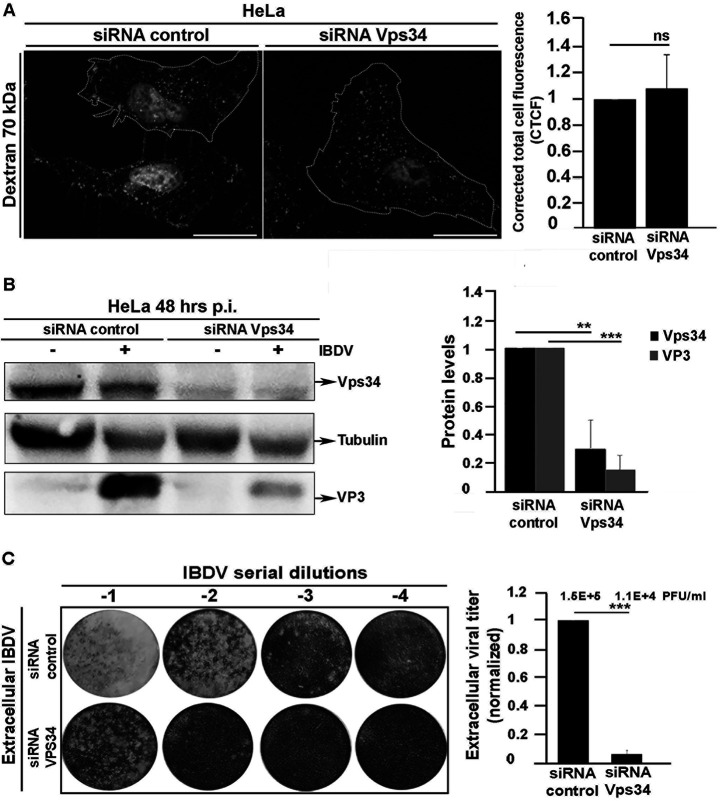
Considering these results, we next sought to investigate whether the depletion of PtdIns(3)P might cause a reduction in viral infectivity. To test this possibility, we first evaluated the accumulation of the IBDV proteins VP2 and VP3 produced in QM7-infected cells treated with Vps34-IN1. As shown in Fig. 9B, Vps34-IN1 treatment caused a drastic decrease in the intracellular accumulation of the assessed IBDV structural proteins, strongly suggesting that PtdIns(3)P is required for IBDV replication. To confirm this assumption, we next investigated whether PtdIns(3)P depletion has an effect on the production of IBDV infectious progeny. As shown in Fig. 9C, the production of both intra- and extracellular IBDV in QM7 cells, determined by plaque assay, was significantly reduced due to the treatment of infected cells with Vps34-IN1. To further confirm these results, we assessed the effect of siRNA-mediated knockdown of Vps34 on IBDV replication and infectivity. For practical reasons, we chose HeLa cells (which have been shown to be able to support IBDV replication [15, 22, 24, 75]) as an alternative infection model for this experiment, since neither the Vps34 gene sequence nor Vps34 antibodies were available for our avian model. As shown in Fig. 10A, the treatment with Vps34 siRNA did not interfere with macropinocytosis, which is the entry route of IBDV (15–17). However, knocking down Vps34 drastically reduced the levels of VP3 protein expressed in infected cells (Fig. 10B); also, in line with the results obtained with Vps34NI-1, the siRNA treatment significantly reduced the production of IBDV progeny (Fig. 10C). Collectively, our results demonstrate that PtdIns(3)P is an essential host cell factor required for the establishment of IBDV replication complexes on endosomes and therefore a crucial mediator of a pivotal step in the viral replication cycle.
FIG 10.
siRNA-mediated depletion of Vps34 inhibits IBDV replication and decreases its infectivity. (A) Evaluation of macropinocytosis of Vps34 knockdown cells. HeLa cells were transfected with control siRNA or a Vps34 siRNA duplex for 72 h and then incubated with the fluid-phase reagent 70-kDa dextran-tetramethylrhodamine, as described in Materials and Methods. Cells were fixed and analyzed by spinning-disc confocal microscopy. Images are merged z-stacks and representative of three independent trials. Bars, 10 μm. The corrected total cell fluorescence (CTCF) was determined with ImageJ software, as described in Materials and Methods. Data are means and SD. ns, not significant. (B) Intracellular accumulation of VP3 protein in Vps34 knockdown cells infected with IBDV. HeLa cells were transfected with control siRNA or a Vps34 siRNA duplex for 24 h and either mock infected or infected with IBDV at an MOI of 1 PFU/cell. After 1 h of adsorption at 37°C, cells were infected for 48 h. At 72 h, they were processed by Western blotting with anti-VP3 and anti-Vps34 antibodies as described in Materials and Methods. The Western blots and the data shown in the normalized bar graph correspond to an experiment representing three independent trials. Data are means and SD. **, P < 0.05; ***, P < 0.01. (C) HeLa cells were transfected and infected as described above, and at 72 h, the cell supernatants were collected for extracellular virus titration, as described in Materials and Methods. The image shows representative plaque assays from three independent trials. Normalized extracellular viral titers are shown in the graph. The crude viral titers are shown above the bars. Error bars show SD. ***, P < 0.01.
DISCUSSION
PIs were recently identified as a prime target of several viruses that co-opt PIs’ signaling functions for their replication in host cells (25, 27, 76, 77). However, only a few cases of viruses that utilize PtdIns(3)P have been described. These include African swine fever virus (ASFV), a large DNA virus of the family Asfarviridae, and the +ssRNA tombusviruses tomato bushy stunt virus (TBSV) and carnation Italian ringspot virus (CIRV) (35, 41). ASFV hijacks the host FYVE finger-containing phosphoinositide kinase (PIKfyve) to control the conversion of PtdIns(3)P to PtdIns(3,5)P2. This allows the virus to reorganize late endosomal membranes around the cellular perinuclear region in order to build its replication factories (35). On the other hand, TBSV and CIRV hijack hosts’ Vps34 to produce PtdIns(3)P at their replicative organelles (41). Here, we demonstrated that IBDV behaves like this small group of viruses known to depend on host PtdIns(3)P to complete their intracellular cycles. Our results indicate that PtdIns(3)P mediates the recruitment of the viral protein VP3 and IBDV replication complexes to EEs. Therefore, PtdIns(3)P is an essential host factor for IBDV replication.
PI-binding structural domains are commonly found in eukaryotic and pathogenic bacterial effector proteins, but intriguingly, this is not the case for viruses, despite their being obligately intracellular parasites (32, 78). The binding of viral proteins to PIs is instead mediated by small polycationic conformational domains. Examples of this are the IBDV protein VP5 (32, 79), the vaccinia virus protein H7, the poliovirus protein 3CD (80, 81), and the hepatitis C virus protein NS5A(82). In the case of VP3, which binds to different PIs in protein-lipid overlay assays (21), a basic conformational domain, Patch 2, mediates VP3 association to endosomes. However, we show here that in the host cell, VP3 binds exclusively to PtdIns(3)P-containing endosomes. Factors intrinsic to the organellar membrane (28, 83, 84) which modulate the interaction of the negatively charged phosphorylated inositol ring in PIs with the Patch 2 domain in the VP3 protein may account for the discrepancies between the in vitro affinity assay (21) and our results in the context of the host cell. Furthermore, the interactions of VP3 with host membranes could be either positively or negatively modulated by its interactions with yet-to-be-identified cellular proteins (78).
The clustering of VP3 we observed on the surface of EEs might be the consequence of VP3 oligomerization at the endosomal membrane (60). However, VP3 oligomerization alone does not completely explain the formation of VP3 clusters, since our observations indicate that they occurred only in association with PtdIns(3)P-enriched EE membranes. In fact, the depletion of PtdIns(3)P led to the dissociation of VP3 clusters from the endosomes and their disaggregation in the cytoplasm. Considering our evidence, we propose a mechanism by which PtdIns(3)P facilitates the docking of VP3 oligomers to the surface of the EE. The interaction of VP3 oligomers with PtdIns(3)P may induce their aggregation and stabilization into micrometric clusters on the surface of EEs. Similarly, Ebola virus protein VP40 undergoes extensive oligomerization at the host cell plasma membrane upon interacting with PtdIns(4,5)P2 (36). Furthermore, VP3 protein clusters may induce membrane curvatures in the surface of EEs, leading to infolding and the formation of hideouts for IBDV replication complexes and viral genome synthesis. Such a mechanism has been described for several viral proteins that can enhance membrane curvature to form distinguishable functional subdomains in host membranes (40, 77, 85–87). Further research to elucidate the nature of VP3 interaction with PtdIns(3)P in EE membranes will be required to validate our assumptions.
We have shown in previous studies that IBDV replication complexes localize not only to EEs but also to compartments that were positive for lysosome-associated protein 1 and 2 (Lamp-1/2) but that they lack the Rab7 small GTPase and acidification, both distinctive features of mature endolysosomal compartments (21, 22). Therefore, IBDV may interfere with the endosomal maturation pathway, stalling endosomes in an intermediate maturation stage functional for IBDV replication. By targeting RNPs to EEs, VP3 could be pivotal in capturing these compartments and preventing their maturation into degradative organelles, to instead hide IBDV replication complexes from the intracellular antiviral surveillance. In addition, the persistence of PtdIns(3)P at the VP3-containing endosomes may be the consequence of VP3 altering PtdIns(3)P metabolism and therefore facilitating the recruitment of the retrograde trafficking machinery for the transport of IBDV replication complexes toward the Golgi complex, which is involved in the late stage of the IBDV replication cycle (22, 88, 89). Collectively our results indicate that IBDV replication requires PtdIns(3)P to co-opt endosomal membranes, possibly to secure viral replication niches from host surveillance. This replication strategy is unique among dsRNA viruses, which may respond to the singularity of birnaviruses’ genome organization in RNPs instead of T=1 cores (12). Whereas T=1 cores shield the dsRNA genomes from host cell surveillance and degradation, IBDV must associate with endosomal membranes to replicate. Collectively, our findings further indicate that the replication strategy of IBDV resembles those of +ssRNA viruses, which invariably induce the formation of organelle-like replicative compartments (27, 77, 86), and therefore highlights the uniqueness of birnaviruses among dsRNA viruses.
MATERIALS AND METHODS
Cell lines and culture conditions.
Quail muscle clone 7 cells (QM7; ATCC CRL-1962) were cultured in a tissue culture incubator at 37°C in Dulbecco’s modified Eagle medium (DMEM; catalogue number 12-800058; Thermo Fisher Scientific, Canada) containing 10% fetal bovine serum (Gibco FBS: catalogue number 10270-106; Thermo Fisher Scientific, Canada) and 25 mM HEPES buffer (catalogue number 15630080; Thermo Fisher Scientific, Canada). QM7-VP3 cells, described elsewhere (21), were cultured in DMEM containing 10% FBS, 25 mM HEPES buffer and 0.5 mg/ml of Geneticin (G418 sulfate; catalogue number 10131035; Thermo Fisher Scientific, Canada). HeLa cells (ATCC CCL-2) were cultured in DMEM containing 10% FBS.
Viral stocks production.
Serotype I IBDV (Soroa strain) was propagated in QM7 cells as described elsewhere (90). Briefly, 70% confluent QM7 cells were infected with IBDV at a multiplicity of infection (MOI) of 0.05 PFU/cell. Upon completion of the cytopathic effect, at 72 to 96 h postinfection (p.i.), supernatants were exposed to 3 freeze-thaw cycles (−80°C for 15 min and 37°C for 5 min) and then centrifuged at 800 × g at 4°C for 15 min. The supernatant was mixed 1:5 with 20% polyethylene glycol 8000 (PEG 8000; catalog number 89510; Sigma-Aldrich, Canada) in a 3 M sodium chloride solution and incubated at 4°C with gentle shaking. After 12 h of incubation, the viruses were pelleted at 4°C and 12,000 × g for 30 min and resuspended in PES buffer [25 mM piperazine-N,N-bis(2-ethanesulfonic acid) (pH 6.2), 150 mM NaCl, and 20 mM CaCl2], and viral suspensions were aliquoted and stored at −80°C.
Plasmids and transfection methods.
Plasmids encoding GFP-2FYVE and RFP-2FYVE were kindly provided by Stenmark Harald, Norwegian Radium Hospital (Oslo, Norway). GFP-p40-PX was a gift from Christine M. Pacold, University of Illinois at Chicago (Chicago, IL, USA). GFP-PLCδ1-PH was a gift from Tobias Meyer, Stanford University (Stanford, CA). Plasmids encoding PH-Akt-GFP, GFP-2xP4M, and GFP-Lac-C2 were generously provided by Sergio Grinstein, Sick Kids Hospital (Toronto, Canada). EGFP-cPHx3 was a gift from Gerald Hammond, University of Pittsburgh School of Medicine (Pittsburgh, PA, USA). Philip D. Stahl, Washington University (St. Louis, MO), generously supplied the EGFP-Rab5wt- and EGFP-Rab5-Q79L-encoding plasmids. mCherry-FKBP-MTM1, iRFP-FRB-Rab5, and iRFP-FRB-Rab7 were developed by Tamas Balla and supplied by Addgene (plasmids 51614, 51612, and 51613, respectively). For transient transfections, QM7 cells were cultured to reach 70 to 80% confluence and transfected or cotransfected using FuGENE HD (catalogue number E2311; Promega, Canada) per the supplier’s instructions. At 12 h posttransfection, the cells were either fixed with paraformaldehyde (4% PFA) for 15 min at room temperature (RT) or infected with IBDV at an MOI of 1 PFU/cell.
Antibodies.
Antibodies and dilutions utilized for Western blotting and indirect immunofluorescence (IIF) include mouse antitubulin (1:1,000 for Western blotting; catalogue number T5201; Sigma-Aldrich), rabbit polyclonal anti-VP3 (1:1,000 for Western blotting and 1:500 for IIF; kindly provided by José F. Rodriguez, CNS-CSIC, Madrid, Spain), rabbit polyclonal anti-VP2 (1:1,000 for Western blotting; kindly provided by José F. Rodriguez), rabbit polyclonal anti-PI3 kinase class III (D9A5) (anti-Vps34 [1:500] for Western blotting; catalogue number 4263; Cell Signaling Technology), donkey anti-rabbit Alexa Fluor 555 (1:500 for IIF; catalogue number A32732; Thermo Fisher Scientific), goat anti-rabbit Alexa Fluor 647 (1:500 for IIF; catalogue number A32733; Thermo Fisher Scientific), goat anti-rabbit Alexa 488 (1:500 for IIF; catalogue number A3273; Thermo Fisher Scientific), horseradish peroxidase (HRP)-conjugated goat anti-rabbit immunoglobulin (1:5,000 for Western blotting; catalogue number A0545; Jackson Immunoresearch), and goat anti-mouse IgG–HRP (1:1,000 for Western blotting; catalogue number A9044; Jackson Immunoresearch).
Indirect immunofluorescence.
QM7 or QM7-VP3 cells on 12-mm glass coverslips were washed twice with phosphate-buffered saline (PBS) (pH 7.4) and fixed in 4% PFA solution for 15 min at room temperature (RT). Cells were permeabilized with a solution of 0.05% saponin in PBS, containing 0.2% bovine serum albumin (BSA) for 20 min at RT. Permeabilized cells were incubated with anti-VP3 antibodies for 90 min at RT, extensively washed with PBS, and incubated with secondary antibodies for 90 min at RT. Coverslips were mounted with Dako fluorescent mounting medium (catalogue number S3023; Aligent, Canada) or, in the case of Hyvolution microscopy, with ProLong Gold antifade mountant (catalogue number P10144; Thermo Fisher Scientific, Canada).
Microscopy and image analysis.
Images were acquired with a Leica DMI6000B spinning-disc confocal microscope (SDCM) by Quorum Technologies, Inc. (Guelph, Ontario, Canada), equipped with an ORCA-R2 camera (Hamamatsu, Japan) driven by MetaMorph software. Images analyzed and processed by Volocity 6.1 software (PerkinElmer) and Adobe Photoshop and Illustrator (Adobe Systems, Inc.) were deconvolved using calculated point spread function in Volocity. Hyvolution microscopy was performed using an inverted Leica DMi8 scanning confocal microscope, driven by LASX software (Leica Microsystems, Mannheim, Germany). Association analysis of VP3 and PI biosensors or GFP-Rab5 were performed on spinning-disc confocal images using Volocity software. Images were deconvolved with a 100% confidence interval and 30 iterations. Fluorescent objects were identified using Volocity’s measurement tool, and Pearson’s correlation coefficient (PCC) was calculated. PCC values above 0 were considered positive for the association of VP3 with PIs or GFP-Rab5. The percentage of association per cell was calculated by dividing associated VP3 objects by total VP3 objects in the cell.
PtdIns(3)P depletion assays.
The pharmacological inhibitor LY294002 (catalogue number L9908; Sigma-Aldrich) was used at a final concentration of 100 μM, while the Vps34-IN1 inhibitor (catalogue number S7980; Selleckchem) was utilized at a final concentration of 0.5 or 1 μM (70) in culture medium in the presence of FBS.
The rapamycin-dependent endosomal targeting of MTM1 utilizing mCherry-FKBP-MTM1/iRFP-FRB-Rab5 and mCherry-FKBP-MTM1/iRFP-FRB-Rab7 was performed as described elsewhere (49). Briefly, QM7 or QM7-VP3 cells at 80% confluence were cotransfected with these constructs for 12 h and subsequently treated with 1 μM rapamycin for 15 min (catalogue number 73362; Stem Cell Technologies) to target MTM1 to Rab5- and Rab7-positive endosomes.
Analysis of VP3 distribution phenotypes.
VP3 distribution phenotypes were assessed in confocal images using Volocity. To this end, we defined puncta as VP3 aggregates equal to or larger than 0.2 μm (corresponding to the limit of resolution of optical microscopes). To classify VP3 cellular phenotypes into puncta or cytoplasmic, the average number of VP3 puncta was calculated per cell. Cells with fewer than 10 VP3 puncta were considered to reflect the VP3 cytoplasmic distribution phenotype, whereas cells with more than 10 puncta were considered to reflect the VP3 punctate distribution phenotype.
siRNA-mediated depletion of Vps34 protein and IBDV infections in HeLa cells.
HeLa cells grown in a 6-well plate at 80% confluence were transfected either with 30 pmol of control small interfering RNA (siRNA) (catalogue number AM4611; Thermo Fisher Scientific) or siRNA against human Vps34 (siRNA ID, s10517; Thermo Fisher Scientific) using Lipofectamine RNAiMAX (catalogue number 13778075; Thermo Fisher Scientific), following the manufacturer’s recommendations. At 24 h, cells were infected with IBDV at an MOI of 1 PFU/cell for 1 h. The infection medium was discarded, and the cells were washed with PBS and incubated in the same transfection medium for an additional 48 h.
MTT viability assay.
QM7 cells were seeded in a 96-well plate and either mock infected or infected with IBDV at an MOI of 1 PFU/cell. At 2 h p.i., the cells were treated with dimethyl sulfoxide (DMSO) and 0.5 or 1 μM Vps34-IN1 for 22 h. At 24 h p.i., the medium was discarded, serum-free medium and MTT reagent were added, and cells were incubated at 37°C for 3 h. After the addition of MTT solvent, the plate was shaken for 15 min at room temperature, and the absorbance was measured at an optical density (OD) of 590 nm using a BioTek Synergy H4 hybrid microplate reader. The percentage of cell viability under each condition was determined following the manufacturer’s instructions (MTT viability assay kit; catalogue number ab211091; Abcam).
Macropinocytosis assay.
HeLa cells were grown to 80% confluence, transfected with control siRNA or Vps34 siRNA for 72 h, and then incubated with 1 mg/ml 70-kDa dextran-tetramethylrhodamine (catalog number D1818; Thermo Fisher Scientific) for 1 h at 37°C in culture medium. Next, cells were extensively washed with PBS and fixed for fluorescence microscopy visualization. For the measurement of dextran uptake efficiency, ImageJ software was employed to calculate the corrected total cell fluorescence (CTCF) as follows: integrated density − (area of selected cell × mean fluorescence of background readings).
Western blotting.
Cells were lysed using Laemmli sample buffer consisting of 0.5 M Tris (pH 6.8), glycerol, 10% SDS, 2 mM dithiothreitol (DTT), and 5% bromophenol blue and heated to 95°C for 10 min. Electrophoresis in 12% acrylamide gels was performed to resolve proteins, which were then transferred to nitrocellulose membranes. Subsequently, membranes were blocked in 10% nonfat milk prepared in PBS for 2 h at RT or overnight (ON) at 4°C and then incubated with primary antibodies against VP3, VP2, tubulin, and Vps34 proteins ON at 4°C. Following three washes of 15 min each with a PBS–0.05% Tween 20 solution, membranes were incubated in HRP-conjugated secondary antibodies for 90 min at RT. Following washes in PBS–0.05% Tween 20, immunoreactive bands were observed by employing a chemiluminescent detection kit (WBKLS0100 [Millipore] or WBLUR0500 [Merck-Millipore, Canada]), and the data were collected using a ChemiDoc XRS+ imaging system (Bio-Rad, Canada). Adobe Photoshop CS5 was used to quantify the intensity of protein bands.
Viral titration employing a standard plaque assay.
QM7 cells were cultured until 70% confluence and infected with serial dilutions of IBDV. Following 1 h of viral absorption at 37°C, cells were washed twice with PBS and incubated with a 1:1 mix of 2× DMEM and 1.4% of low-melting-point agarose aqueous solution, plus 2% (final concentration) FBS. After 5 days of incubation, cells were fixed in a 10% formaldehyde aqueous solution for 2 h at RT, the semisolid medium was removed, and the monolayers were stained using a 1% crystal violet aqueous solution for lysis plaque enumeration and estimation of viral titers (PFU per milliliter). For titration of extracellular IBDV, the supernatants of infected cells were collected and used to perform the plaque assays. For intracellular IBDV titration, IBDV-infected monolayers were harvested in 1 ml of DMEM, and exposed to three freeze-thaw cycles (–80°C for 15 min and 37°C for 5 min). The cell lysates were centrifuged to remove the cellular debris, and the supernatants, containing intracellular viral particles, were then employed for plaque assays.
Statistical analysis.
Ky-Plot software version 2.0, beta 15, was used to conduct Student’s t test. All results are means and standard deviations (SD) from at least three independent trials.
ACKNOWLEDGMENTS
We sincerely appreciate José F. Rodríguez for generously providing us with IBDV Soroa strain and antibodies. We thank the Centre for Neurobiology of Stress at the University of Toronto at Scarborough for the use of the imaging facilities and Paul Paroutis from the Imaging Facility of The Hospital for Sick Children for his technical support during the acquisition and deconvolution processes of high-resolution images. We thank Roberto J Botelho, Ryerson University, for his helpful feedback and scientific discussion and Cynthia Bongard for polishing the last version of our manuscript.
We declare no competing interests.
This work was supported by a Discovery grant from the Natural Sciences and Engineering Research Council to M.R.T. and by funds from the Agencia Nacional de Promoción Científica y Tecnológica (MINCYT, PICT 2016-0528, to L.R.D), SeCTyP 2013-2015 M006 and SeCTyP 2016-2018 M029 (Universidad Nacional de Cuyo) to L.R.D., and PIP-CONICET 2015-2017 (112 20150100114 CO) to L.R.D. M.C.G. was supported by funds of a NSERC Discovery grant to M.R.T. and a fellowship from the Consejo Nacional de Investigaciones Científicas y Tecnológicas (CONICET).
REFERENCES
- 1.Cosgrove AS. 1962. An apparently new disease of chickens: avian nephrosis. Avian Dis 6:385–389. doi: 10.2307/1587909. [DOI] [Google Scholar]
- 2.Müller H, Islam MR, Raue R. 2003. Research on infectious bursal disease–the past, the present and the future. Vet Microbiol 97:153–165. doi: 10.1016/j.vetmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Delmas B, Attoui H, Ghosh S, Malik YS, Mundt E, Vakharia VN, Ictv Report Consortium. 2019. ICTV virus taxonomy profile: Birnaviridae. J Gen Virol 100:5–6. doi: 10.1099/jgv.0.001185. [DOI] [PubMed] [Google Scholar]
- 4.Mertens P. 2004. The dsRNA viruses. Virus Res 101:3–13. doi: 10.1016/j.virusres.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Luque D, Mata CP, González-Camacho F, González JM, Gómez-Blanco J, Alfonso C, Rivas G, Havens WM, Kanematsu S, Suzuki N, Ghabrial SA, Trus BL, Castón JR. 2016. Heterodimers as the structural unit of the T=1 capsid of the fungal double-stranded RNA Rosellinia necatrix quadrivirus 1. J Virol 90:11220–11230. doi: 10.1128/JVI.01013-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luque D, González JM, Garriga D, Ghabrial SA, Havens WM, Trus B, Verdaguer N, Carrascosa JL, Castón JR. 2010. The T=1 capsid protein of Penicillium chrysogenum virus is formed by a repeated helix-rich core indicative of gene duplication. J Virol 84:7256–7266. doi: 10.1128/JVI.00432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tyler KL. 2008. Segmented double-stranded RNA viruses: structure and molecular biology. Lancet Infect Dis 8:224. doi: 10.1016/S1473-3099(08)70063-X. [DOI] [Google Scholar]
- 8.Coulibaly F, Chevalier C, Gutsche I, Pous J, Navaza J, Bressanelli S, Delmas B, Rey FA. 2005. The birnavirus crystal structure reveals structural relationships among icosahedral viruses. Cell 120:761–772. doi: 10.1016/j.cell.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 9.Coulibaly F, Chevalier C, Delmas B, Rey FA. 2010. Crystal structure of an Aquabirnavirus particle: insights into antigenic diversity and virulence determinism. J Virol 84:1792–1799. doi: 10.1128/JVI.01536-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luque D, Saugar I, Rejas MT, Carrascosa JL, Rodríguez JF, Castón JR. 2009. Infectious bursal disease virus: ribonucleoprotein complexes of a double-stranded RNA virus. J Mol Biol 386:891–901. doi: 10.1016/j.jmb.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan J, Vakharia VN, Tao YJ. 2007. The structure of a birnavirus polymerase reveals a distinct active site topology. Proc Natl Acad Sci U S A 104:7385–7390. doi: 10.1073/pnas.0611599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luque D, Rivas G, Alfonso C, Carrascosa JL, Rodríguez JF, Castón JR. 2009. Infectious bursal disease virus is an icosahedral polyploid dsRNA virus. Proc Natl Acad Sci U S A 106:2148–2152. doi: 10.1073/pnas.0808498106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hjalmarsson A, Carlemalm E, Everitt E. 1999. Infectious pancreatic necrosis virus: identification of a VP3-containing ribonucleoprotein core structure and evidence for O-linked glycosylation of the capsid protein VP2. J Virol 73:3484–3490. doi: 10.1128/JVI.73.4.3484-3490.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalton RM, Rodríguez JF. 2014. Rescue of infectious birnavirus from recombinant ribonucleoprotein complexes. PLoS One 9:e87790. doi: 10.1371/journal.pone.0087790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gimenez MC, Rodríguez Aguirre JF, Colombo MI, Delgui LR. 2015. Infectious bursal disease virus uptake involves macropinocytosis and trafficking to early endosomes in a Rab5-dependent manner. Cell Microbiol 17:988–1007. doi: 10.1111/cmi.12415. [DOI] [PubMed] [Google Scholar]
- 16.Yip CW, Hon CC, Zeng F, Leung FCC. 2012. Cell culture-adapted IBDV uses endocytosis for entry in DF-1 chicken embryonic fibroblasts. Virus Res 165:9–16. doi: 10.1016/j.virusres.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 17.Ye C, Han X, Yu Z, Zhang E, Wang L, Liu H. 2017. Infectious bursal disease virus activates c-Src to promote α4β1 integrin-dependent viral entry by modulating the downstream Akt-RhoA GTPase-actin rearrangement cascade. J Virol 91:e01891-16. doi: 10.1128/JVI.01891-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chevalier C, Galloux M, Pous J, Henry C, Denis J, Da Costa B, Navaza J, Lepault J, Delmas B. 2005. Structural peptides of a nonenveloped virus are involved in assembly and membrane translocation. J Virol 79:12253–12263. doi: 10.1128/JVI.79.19.12253-12263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galloux M, Libersou S, Morellet N, Bouaziz S, Da Costa B, Ouldali M, Lepault J, Delmas B. 2007. Infectious bursal disease virus, a non-enveloped virus, possesses a capsid-associated peptide that deforms and perforates biological membranes. J Biol Chem 282:20774–20784. doi: 10.1074/jbc.M701048200. [DOI] [PubMed] [Google Scholar]
- 20.Galloux M, Libersou S, Alves ID, Marquant R, Salgado GF, Rezaei H, Lepault J, Delmas B, Bouaziz S, Morellet N. 2010. NMR structure of a viral peptide inserted in artificial membranes. J Biol Chem 285:19409–19421. doi: 10.1074/jbc.M109.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gimenez MC, Zanetti FA, Terebiznik MR, Colombo MI, Delgui LR. 2018. Infectious bursal disease virus hijacks endosomal membranes as the scaffolding structure for viral replication. J Virol 92:e01964-17. doi: 10.1128/JVI.01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delgui LR, Rodriguez JF, Colombo MI. 2013. The endosomal pathway and the Golgi complex are involved in the infectious bursal disease virus life cycle. J Virol 87:8993–9007. doi: 10.1128/JVI.03152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casañas A, Navarro A, Ferrer-Orta C, González D, Rodríguez JF, Verdaguer N. 2008. Structural insights into the multifunctional protein VP3 of birnaviruses. Structure 16:29–37. doi: 10.1016/j.str.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Valli A, Busnadiego I, Maliogka V, Ferrero D, Castón JR, Rodríguez JF, García JA. 2012. The VP3 factor from viruses of Birnaviridae family suppresses RNA silencing by binding both long and small RNA duplexes. PLoS One 7:e45957. doi: 10.1371/journal.pone.0045957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackenzie J. 2005. Wrapping things up about virus RNA replication. Traffic 6:967–977. doi: 10.1111/j.1600-0854.2005.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol Cell 97:147–172. doi: 10.1042/BC20040058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Den den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu Rev Microbiol 64:241–256. doi: 10.1146/annurev.micro.112408.134012. [DOI] [PubMed] [Google Scholar]
- 28.Di Paolo G, De Camilli P. 2006. Phosphoinositides in cell regulation and membrane dynamics. Nature 443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 29.Payrastre B, Missy K, Giuriato S, Bodin S, Plantavid M, Gratacap M-P. 2001. Phosphoinositides: key players in cell signalling, in time and space. Cell Signal 13:377–387. doi: 10.1016/S0898-6568(01)00158-9. [DOI] [PubMed] [Google Scholar]
- 30.Mayinger P. 2012. Phosphoinositides and vesicular membrane traffic. Biochim Biophys Acta 1821:1104–1113. doi: 10.1016/j.bbalip.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Billcliff PG, Lowe M. 2014. Inositol lipid phosphatases in membrane trafficking and human disease. Biochem J 461:159–175. doi: 10.1042/BJ2014036. [DOI] [PubMed] [Google Scholar]
- 32.Qiu S, Côté M. 2019. From hitchhiker to hijacker: pathogen exploitation of endosomal phosphoinositides. Biochem Cell Biol 97:1–9. doi: 10.1139/bcb-2017-0317. [DOI] [PubMed] [Google Scholar]
- 33.Bishé B, Syed G, Siddiqui A. 2012. Phosphoinositides in the hepatitis C virus life cycle. Viruses 4:2340–2358. doi: 10.3390/v4102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh HS, Banerjee S, Aponte-Diaz D, Sharma SD, Aligo J, Lodeiro MF, Ning G, Sharma R, Arnold JJ, Cameron CE. 2018. Multiple poliovirus-induced organelles suggested by comparison of spatiotemporal dynamics of membranous structures and phosphoinositides. PLoS Pathog 14:e1007036. doi: 10.1371/journal.ppat.1007036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cuesta-Geijo M, Barrado-Gil L, Galindo I, Muñoz-Moreno R, Alonso C. 2017. Redistribution of endosomal membranes to the African swine fever virus replication site. Viruses 9:133. doi: 10.3390/v9060133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson KA, Taghon GJF, Scott JL, Stahelin RV. 2016. The Ebola virus matrix protein, VP40, requires phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) for extensive oligomerization at the plasma membrane and viral egress. Sci Rep 6:19125. doi: 10.1038/srep19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fernandes F, Chen K, Ehrlich LS, Jin J, Chen MH, Medina GN, Symons M, Montelaro R, Donaldson J, Tjandra N, Carter CA. 2011. Phosphoinositides direct equine infectious anemia virus Gag trafficking and release. Traffic 12:438–451. doi: 10.1111/j.1600-0854.2010.01153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altan-Bonnet N. 2017. Lipid tales of viral replication and transmission. Trends Cell Biol 27:201–213. doi: 10.1016/j.tcb.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee C. 2012. Roles of phosphoinositides and phosphoinositides kinases in hepatitis C virus RNA replication. Arch Pharm Res 35:1701–1711. doi: 10.1007/s12272-012-1001-2. [DOI] [PubMed] [Google Scholar]
- 40.Banerjee S, Aponte-Diaz D, Yeager C, Sharma SD, Ning G, Oh HS, Han Q, Umeda M, Hara Y, Wang RYL, Cameron CE. 2018. Hijacking of multiple phospholipid biosynthetic pathways and induction of membrane biogenesis by a picornaviral 3CD protein. PLoS Pathog 14:e1007086. doi: 10.1371/journal.ppat.1007086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng Z, Xu K, Kovalev N, Nagy PD. 2019. Recruitment of Vps34 PI3K and enrichment of PI3P phosphoinositide in the viral replication compartment is crucial for replication of a positive-strand RNA virus. PLoS Pathog 15:e1007530. doi: 10.1371/journal.ppat.1007530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kutateladze T. 2006. Phosphatidylinositol 3-phosphate recognition and membrane docking by the FYVE domain. Biochim Biophys Acta 1761:868–877. doi: 10.1016/j.bbalip.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ahlquist P. 2005. Virus evolution: fitting lifestyles to a T. Curr Biol 15:R465–R467. doi: 10.1016/j.cub.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 44.Mertens J, Casado S, Mata CP, Hernando-Pérez M, de Pablo PJ, Carrascosa JL, Castón JR. 2015. A protein with simultaneous capsid scaffolding and dsRNA-binding activities enhances the birnavirus capsid mechanical stability. Sci Rep 5:13486. doi: 10.1038/srep13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye C, Jia L, Sun Y, Hu B, Wang L, Lu X, Zhou J. 2014. Inhibition of antiviral innate immunity by birnavirus VP3 protein via blockage of viral double-stranded RNA binding to the host cytoplasmic RNA detector MDA5. J Virol 88:11154–11165. doi: 10.1128/JVI.01115-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferrero D, Garriga D, Navarro A, Rodríguez JF, Verdaguer N. 2015. Infectious bursal disease virus VP3 upregulates VP1-mediated RNA-dependent RNA replication. J Virol 89:11165–11168. doi: 10.1128/JVI.00218-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lemmon MA, Ferguson KM, O’Brien R, Sigler PB, Schlessinger J. 1995. Specific and high-affinity binding of inositol phosphates to an isolated pleckstrin homology domain. Proc Natl Acad Sci U S A 92:10472–10476. doi: 10.1073/pnas.92.23.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Várnai P, Balla T. 1998. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol 143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammond GRV, Machner MP, Balla T. 2014. A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol 205:113–126. doi: 10.1083/jcb.201312072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida A, Hayashi H, Tanabe K, Fujita A. 2017. Segregation of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate into distinct microdomains on the endosome membrane. Biochim Biophys Acta Biomembr 1859:1880–1890. doi: 10.1016/j.bbamem.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 51.Levin R, Hammond GRV, Balla T, De Camilli P, Fairn GD, Grinstein S. 2017. Multiphasic dynamics of phosphatidylinositol 4-phosphate during phagocytosis. Mol Biol Cell 28:128–140. doi: 10.1091/mbc.E16-06-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Harrison RE, Bucci C, Vieira OV, Schroer TA, Grinstein S. 2003. Phagosomes fuse with late endosomes and/or lysosomes by extension of membrane protrusions along microtubules: role of Rab7 and RILP. Mol Cell Biol 23:6494–6506. doi: 10.1128/mcb.23.18.6494-6506.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goulden BD, Pacheco J, Dull A, Zewe JP, Deiters A, Hammond GRV. 2019. A high-avidity biosensor reveals plasma membrane PI(3,4)P2 is predominantly a class I PI3K signaling product. J Cell Biol 218:1066–1079. doi: 10.1083/jcb.201809026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ellson CD, Gobert-Gosse S, Anderson KE, Davidson K, Erdjument-Bromage H, Tempst P, Thuring JW, Cooper MA, Lim ZY, Holmes AB, Gaffney PRJ, Coadwell J, Chilvers ER, Hawkins PT, Stephens LR. 2001. Ptdlns(3)P regulates the neutrophil oxidase complex by binding to the PX domain of p40phox. Nat Cell Biol 3:679–682. doi: 10.1038/35083076. [DOI] [PubMed] [Google Scholar]
- 55.Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wills RC, Goulden BD, Hammond GV. 2018. Genetically encoded lipid biosensors. Mol Biol Cell 29:1526–1532. doi: 10.1091/mbc.E17-12-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJG, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL. 2001. The crystal structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate. Mol Cell 8:829–839. doi: 10.1016/S1097-2765(01)00372-0. [DOI] [PubMed] [Google Scholar]
- 58.Burd CG, Emr SD. 1998. Phosphatidylinositol(3)-phosphate signaling mediated by specific binding to RING FYVE domains. Mol Cell 2:157–162. doi: 10.1016/s1097-2765(00)80125-2. [DOI] [PubMed] [Google Scholar]
- 59.Huotari J, Helenius A. 2011. Endosome maturation. EMBO J 30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maraver A, Oña A, Abaitua F, González D, Clemente R, Ruiz-Díaz JA, Castón JR, Pazos F, Rodriguez JF. 2003. The oligomerization domain of VP3, the scaffolding protein of infectious bursal disease virus, plays a critical role in capsid assembly. J Virol 77:6438–6449. doi: 10.1128/jvi.77.11.6438-6449.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takenawa T, Itoh T. 2006. Membrane targeting and remodeling through phosphoinositide-binding domains. IUBMB Life 58:296–303. doi: 10.1080/15216540600732039. [DOI] [PubMed] [Google Scholar]
- 62.Liu K, Jian Y, Sun X, Yang C, Gao Z, Zhang Z, Liu X, Li Y, Xu J, Jing Y, Mitani S, He S, Yang C. 2016. Negative regulation of phosphatidylinositol 3-phosphate levels in early-to-late endosome conversion. J Cell Biol 212:181–198. doi: 10.1083/jcb.201506081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shin H-W, Hayashi M, Christoforidis S, Lacas-Gervais S, Hoepfner S, Wenk MR, Modregger J, Uttenweiler-Joseph S, Wilm M, Nystuen A, Frankel WN, Solimena M, De Camilli P, Zerial M. 2005. An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J Cell Biol 170:607–618. doi: 10.1083/jcb.200505128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simonsen A, Lippe R, Christoforidis S, Gaullier J-M, Brech A, Callaghan J, Toh B-H, Murphy C, Zerial M, Stenmark H. 1998. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 65.Raiborg C, Schink KO, Stenmark H. 2013. Class III phosphatidylinositol 3-kinase and its catalytic product PtdIns3P in regulation of endocytic membrane traffic. FEBS J 280:2730–2742. doi: 10.1111/febs.12116. [DOI] [PubMed] [Google Scholar]
- 66.Lindmo K, Stenmark H. 2006. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci 119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 67.Wallroth A, Haucke V. 2018. Phosphoinositide conversion in endocytosis and the endolysosomal system. J Biol Chem 293:1526–1535. doi: 10.1074/jbc.R117.000629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gharbi SI, Zvelebil MJ, Shuttleworth SJ, Hancox T, Saghir N, Timms JF, Waterfield MD. 2007. Exploring the specificity of the PI3K family inhibitor LY294002. Biochem J 404:15–21. doi: 10.1042/BJ20061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vlahos CJ, Matter WF, Hui KY, Brown RF. 1994. A specific inhibitor of phosphatidylinositol 3-kinase, 2–(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem 269:5241–5248. [PubMed] [Google Scholar]
- 70.Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, Shpiro N, Ward R, Cross D, Ganley IG, Alessi DR. 2014. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J 463:413–427. doi: 10.1042/BJ20140889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ikonomov OC, Sbrissa D, Venkatareddy M, Tisdale E, Garg P, Shisheva A. 2015. Class III PI 3-kinase is the main source of PtdIns3P substrate and membrane recruitment signal for PIKfyve constitutive function in podocyte endomembrane homeostasis. Biochim Biophys Acta 1853:1240–1250. doi: 10.1016/j.bbamcr.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fili N, Calleja V, Woscholski R, Parker PJ, Larijani B. 2006. Compartmental signal modulation: endosomal phosphatidylinositol 3-phosphate controls endosome morphology and selective cargo sorting. Proc Natl Acad Sci U S A 103:15473–15478. doi: 10.1073/pnas.0607040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schaletzky J, Dove SK, Short B, Lorenzo O, Clague MJ, Barr FA. 2003. Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr Biol 13:504–509. doi: 10.1016/s0960-9822(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 74.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. 2005. Rab conversion as a mechanism of progression from early to late endosomes. Cell 122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 75.Cubas-Gaona LL, Diaz-Beneitez E, Ciscar M, Rodríguez JF, Rodríguez D. 2018. Exacerbated apoptosis of cells infected with infectious bursal disease virus upon exposure to interferon alpha. J Virol 92:e00364-18. doi: 10.1128/JVI.00364-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tao YJ, Ye Q. 2010. RNA virus replication complexes. PLoS Pathog 6:e1000943. doi: 10.1371/journal.ppat.1000943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Romero-Brey I, Bartenschlager R. 2014. Membranous replication factories induced by plus-strand RNA viruses. Viruses 6:2826–2857. doi: 10.3390/v6072826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hammond GRV, Balla T. 2015. Polyphosphoinositide binding domains: key to inositol lipid biology. Biochim Biophys Acta 1851:746–758. doi: 10.1016/j.bbalip.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Méndez F, Romero N, Cubas LL, Delgui LR, Rodríguez D, Rodríguez JF. 2017. Non-lytic egression of infectious bursal disease virus (IBDV) particles from infected cells. PLoS One 12:e0170080. doi: 10.1371/journal.pone.0170080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kolli S, Meng X, Wu X, Shengjuler D, Cameron CE, Xiang Y, Deng J. 2015. Structure-function analysis of vaccinia virus H7 protein reveals a novel phosphoinositide binding fold essential for poxvirus replication. J Virol 89:2209–2219. doi: 10.1128/JVI.03073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shengjuler D, Chan YM, Sun S, Moustafa IM, Li Z-L, Gohara DW, Buck M, Cremer PS, Boehr DD, Cameron CE. 2017. The RNA-binding site of poliovirus 3C protein doubles as a phosphoinositide-binding domain. Structure 25:1875–1886.E7. doi: 10.1016/j.str.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cho N-J, Lee C, Pang PS, Pham EA, Fram B, Nguyen K, Xiong A, Sklan EH, Elazar M, Koytak ES, Kersten C, Kanazawa KK, Frank CW, Glenn JS. 2015. Phosphatidylinositol 4,5-bisphosphate is an HCV NS5A ligand and mediates replication of the viral genome. Gastroenterology 148:616–625. doi: 10.1053/j.gastro.2014.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Teasdale RD, Collins BM. 2012. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: structures, functions and roles in disease. Biochem J 441:39–59. doi: 10.1042/BJ20111226. [DOI] [PubMed] [Google Scholar]
- 84.Balla T. 2005. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. J Cell Sci 118:2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- 85.Paul D, Bartenschlager R. 2015. Flaviviridae replication organelles: oh, what a tangled web we weave. Annu Rev Virol 2:289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- 86.Paul D, Bartenschlager R. 2013. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2:32–48. doi: 10.5501/wjv.v2.i2.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kujala P, Ikäheimonen A, Ehsani N, Vihinen H, Auvinen P, Kääriäinen L. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J Virol 75:3873–3884. doi: 10.1128/JVI.75.8.3873-3884.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burda P, Padilla SM, Sarkar S, Emr SD. 2002. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J Cell Sci 115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- 89.Neefjes J, Jongsma MML, Berlin I. 2017. Stop or go? endosome positioning in the establishment of compartment architecture, dynamics, and function. Trends Cell Biol 27:580–594. doi: 10.1016/j.tcb.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 90.Lombardo E, Maraver A, Castón JR, Rivera J, Fernández-Arias A, Serrano A, Carrascosa JL, Rodriguez JF. 1999. VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J Virol 73:6973–6983. doi: 10.1128/JVI.73.8.6973-6983.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Franke TF, Kaplan DR, Cantley LC, Toker A. 1997. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]