Extracellular vesicles mediate cell-to-cell communication and convey messages important for cell homeostasis. Pathways of EV biogenesis are often hijacked by pathogens to facilitate their dissemination and to establish a favorable microenvironment for the infection.
KEYWORDS: extracellular vesicles, tetraspanins, CD63, CD81, HSV-1, ESCRT, biogenesis of extracellular vesicles, L-particles, biogenesis of extracellular vesicles
ABSTRACT
Extracellular vesicles (EVs) are released by all types of cells as a means of intercellular communication. Their significance lies in the fact that they can alter recipient cell functions, despite their limited capacity for cargo. We have previously demonstrated that herpes simplex virus 1 (HSV-1) infection influences the cargo and functions of EVs released by infected cells and that these EVs negatively impact a subsequent HSV-1 infection. In the present study, we have implemented cutting-edge technologies to further characterize EVs released during HSV-1 infection. We identified distinct EV populations that were separable through a gradient approach. One population was positive for the tetraspanin CD63 and was distinct from EVs carrying components of the endosomal sorting complexes required for transport (ESCRT). Nanoparticle tracking analysis (NTA) combined with protein analysis indicated that the production of CD63+ EVs was selectively induced upon HSV-1 infection. The ExoView platform supported these data and suggested that the amount of CD63 per vesicle is larger upon infection. This platform also identified EV populations positive for other tetraspanins, including CD81 and CD9, whose abundance decreased upon HSV-1 infection. The stimulator of interferon genes (STING) was found in CD63+ EVs released during HSV-1 infection, while viral components were found in ESCRT+ EVs. Functional characterization of these EVs demonstrated that they have opposite effects on the infection, but the dominant effect was negative. Overall, we have identified the dominant population of EVs, and other EV populations produced during HSV-1 infection, and we have provided information about potential roles.
IMPORTANCE Extracellular vesicles mediate cell-to-cell communication and convey messages important for cell homeostasis. Pathways of EV biogenesis are often hijacked by pathogens to facilitate their dissemination and to establish a favorable microenvironment for the infection. We have previously shown that HSV-1 infection alters the cargo and functions of the released EVs, which negatively impact the infection. We have built upon our previous findings by developing procedures to separate EV populations from HSV-1-infected cells. We identified the major population of EVs released during infection, which carries the DNA sensor STING and has an antiviral effect. We also identified an EV population that carries selected viral proteins and has a proviral role. This is the first study to characterize EV populations during infection. These data indicate that the complex interactions between the virus and the host are extended to the extracellular environment and could impact HSV-1 dissemination and persistence in the host.
INTRODUCTION
In 1967, Peter Wolf observed microparticles that he termed “platelet dust” while performing blood coagulation experiments using human platelets (1). He proposed that these microparticles facilitate blood coagulation, which was supported by follow-up studies (1–4). In 1983, electron microscopy studies demonstrated extracellular release of the transferrin receptor within vesicles that were considered “trash bags” of cells, used to eliminate unwanted plasma membrane proteins (5). Today, it has been established that such vesicles are released by all types of cells, as they constitute a major mechanism of intercellular communication and are important for cellular homeostasis (6–11).
Extracellular vesicles (EVs) are highly heterogeneous in cargo, origin, and size, but most investigators divide them into two broad groups (12–15). The first group includes vesicles 30 to 150 nm in diameter that are formed as intraluminal vesicles (ILVs) within endosomal multivesicular bodies (MVBs). After fusion of the MVBs with the plasma membrane, ILVs are released to the extracellular space and are now referred to as exosomes. The other major group is comprised of larger vesicles (100 to 1,000 nm in diameter) that are generated by outward budding of the plasma membrane. These vesicles are referred to as microvesicles or ectosomes. A third group of vesicles (1,000 to 5,000 nm) that also bud from the plasma membrane are known as apoptotic bodies; these vesicles are considered end-death products and are not a subject of this study (12–15).
The cargo of EVs is a key determinant of their function; thus, it is of interest to identify mechanisms of cargo sorting. The endosomal sorting complexes required for transport (ESCRT) were first identified in Saccharomyces cerevisiae and studied for their role in sorting proteins into ILVs. This machinery is composed of four multisubunit complexes, ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III, that work in an orchestrated manner to sort ubiquitinated cargo into late endosomes (16–20). However, silencing of ESCRT components such as the tumor susceptibility gene 101 protein (TSG101), a core component of the ESCRT-I complex, VPS22/EAP30, which is an ESCRT-II factor, or Vps24/CHMP3, which is an ESCRT-III factor, does not abrogate MVB formation, suggesting that alternate mechanisms can regulate EV biogenesis (21). In support of this, ESCRT-independent pathways that involve tetraspanins, lipids, and certain Rab GTPases have been proposed. Tetraspanins belong to a family of proteins with diverse roles in cell adhesion, cell morphology and motility, protein trafficking, and plasma membrane dynamics (22). They carry four transmembrane domains, have a short and a long extracellular loop, and are clustered in tetraspanin-enriched microdomains (TEMs) in ceramide-enriched membranes (23–26). TEMs are distinct from lipid rafts, and the implication is that they constitute a new type of signaling platform. Tetraspanins interact with numerous proteins through a hypervariable domain located within the large extracellular loop that varies in sequence and length between family members and potentially facilitate cargo sorting to ILVs (22–26).
In addition to the above-mentioned mechanisms, other, less characterized mechanisms can contribute to cargo sorting (15). For example, proteins carrying late domains, termed late to reflect their function late in virus budding, can interact with proteins carrying WW domains (35 to 40 amino acids in length with two invariant tryptophan residues) that provide a platform for assembly of multiprotein networks, which can be sorted into MVBs (27, 28). Posttranslational modifications, including phosphorylation, glycosylation, and citrullination, can influence cargo sorting in the appropriate context (15, 29). Analysis of various membrane anchors demonstrated that myristoylation has a great impact in the formation of microvesicles (30). SUMOylation could also contribute to cargo sorting by exploiting the ESCRT complex (31). ISGylation has so far been linked to attenuation of EV secretion by promoting aggregation and degradation of ISGylated TSG101 (32). A bioinformatics analysis revealed an enrichment of EV cargo in proteins with coiled-coil domains (33). Finally, the RNA content of EVs is very distinct compared to the cytoplasm of the donor cells, which suggests active RNA sorting to EVs. The current mechanism for RNA cargo sorting involves RNA-binding proteins targeted to EVs by mechanisms similar to other proteins. Also, cis-acting elements appear to control RNA sorting to the EVs (21, 34).
Pathogens often hijack EV biogenesis pathways for their assembly (hepatitis A virus and HIV), to package their virions (e.g., noroviruses and rotaviruses), genome (hepatitis C virus), or selected factors that modulate the microenvironment of infection (herpes simplex virus 1 [HSV-1]) (35–43). Other viruses (human herpesvirus 6 [HHV-6]) are released via an exosomal release pathway (44). In support of this, we have previously demonstrated that HSV-1 infection stimulates production of EVs that carry viral factors (viral mRNAs, microRNAs [miRNAs], and proteins) and host factors such as the stimulator of interferon genes (STING) (38, 39, 45). These EVs can activate innate immune response in uninfected recipient cells and suppress a subsequent HSV-1 infection (45). Our laboratory has developed procedures to separate EVs from virions that enabled us to identify distinct EV populations. We found that during HSV-1 infection, the dominant EV population is produced through the CD63 tetraspanin pathway. These CD63+ EVs carry STING but no viral proteins or ESCRT components. We also identified a population of EVs that carries ESCRT components and viral factors, but not CD63, that we termed ESCRT+ EVs. Individually, these EV populations seem to have opposite effects on the infection, but the net effect is negative. This is the first study to provide insight into the nature of EVs populating the microenvironment of HSV-1 infection, with implications for virus dissemination and persistence in the host.
RESULTS
Separation of EV populations through an iodixanol/sucrose gradient during HSV-1 infection.
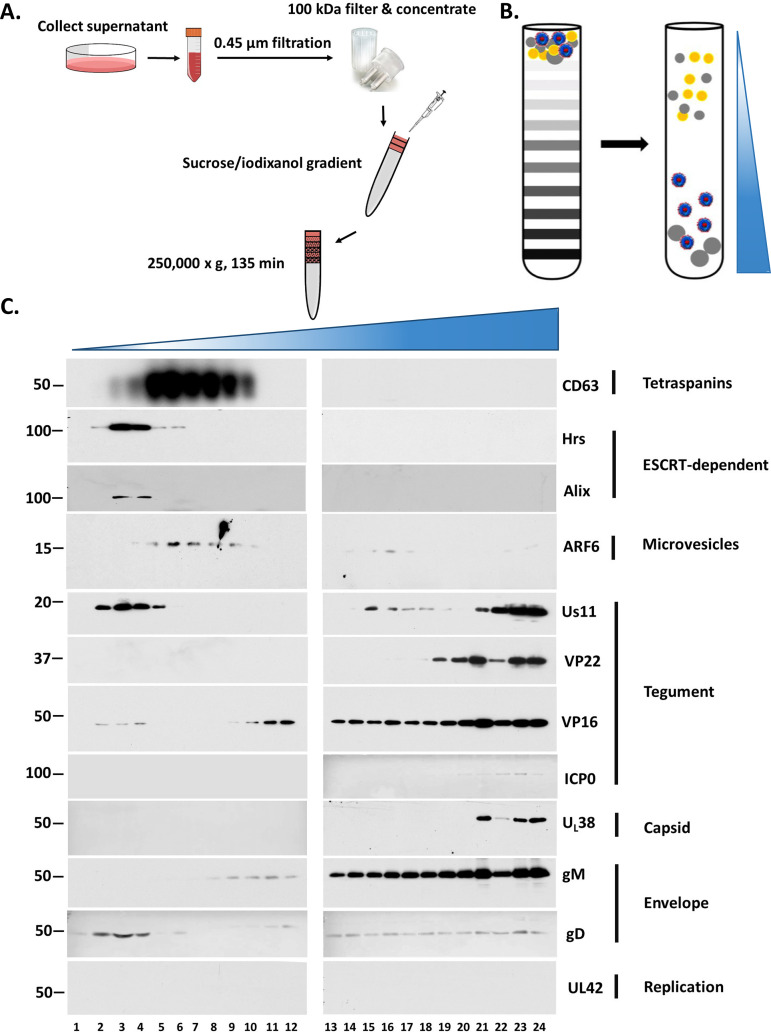
Previously we described a procedure to separate EVs from HSV-1 virions based on their density difference (45, 46). For this study, we adapted the same procedure but increased the volume of the sample loaded on the gradient (from ∼300 μl to 1.5 ml). HEL cells were infected with HSV-1(F) (0.1 PFU/cell), and the supernatant was collected at 48 h postinfection, subjected to two rounds of low-speed centrifugation to remove cell debris and nuclei, filtered through 0.45-μm-pore size filters, concentrated through 100-kDa filters, and loaded on top of an iodixanol/sucrose discontinuous gradient with the iodixanol concentration ranging between 6% and 18% (increments of 1.2%) (Fig. 1A and B). Fractions (500 μl) were collected from the top to the bottom of the gradient and analyzed by Western blotting using antibodies against viral components such as the capsid protein UL38, the envelope proteins gM and gD, the tegument proteins VP22, Us11, ICP0, and VP16, the component of the viral replication machinery, UL42, and different EV factors such as components of the ESCRT machinery Hrs and Alix, the tetraspanin CD63, and the small GTPase ARF6, which serves as a marker of microvesicles (13, 47). Consistent with our previous findings, the viral components concentrated in high-density fractions where infectious virus accumulates, whereas all EV markers were found in light-density fractions (Fig. 1C). UL42 served as a contamination control and was undetectable. Three major observations were made. (i) The ESCRT components Hrs and Alix concentrated in lighter-density fractions than CD63, suggesting that they belong to different EV populations. (ii) Selected viral proteins (gD, VP16, and Us11) cofractionated with the ESCRT components. Since infectious virus is absent from the light-density fractions, as we have previously described, it is likely that some viral factors are packaged in EVs (43). Notably, these viral factors are selectively packaged in ESCRT+ but not in CD63+ EVs (Fig. 1C). (iii) ARF6, a marker of microvesicles, overlapped with CD63+ EVs, suggesting that perhaps a small number of microvesicles cofractionate with CD63+ EVs.
FIG 1.
Separation of EVs from HSV-1 virions. (A and B) Schematic of the procedures used for EV isolation. HEL cells were infected with HSV-1(F) (0.1 PFU/cell). At 48 h postinfection, the supernatant was collected, centrifuged at 1,200 rpm for 5 min followed by a centrifugation at 3,500 rpm for 20 min, and filtered through a 0.45-μm filter. The supernatant was concentrated using Centricon Plus 70 (100-kDa cutoff) (Millipore). The sample was then loaded on top of an iodixanol/sucrose gradient ranging from 6 to 18%, with a 1.2% increment in the concentration of iodixanol for a total of 11 different concentrations. The 60% iodixanol was diluted in 10 mM Tris (pH 8) and 0.25 M sucrose. Samples were centrifuged in an SW41Ti rotor for 135 min at 250,000 × g and 4°C in a Beckman Coulter OPTIMA XPN-80 ultracentrifuge. Fractions of 500 μl were collected from the top to the bottom of the gradient. (C) Equal volumes from the fractions were separated in denaturing polyacrylamide gels and analyzed by immunoblot analysis using antibodies against EV components such as CD63, Hrs, Alix, and ARF6, viral tegument proteins such as Us11, VP22, VP16, and ICP0, viral capsid proteins such as UL38, viral envelope proteins such as glycoprotein M (gM) and gD, and a component of the viral replication machinery, UL42. Numbers on the left are molecular weight markers.
To confirm that ESCRT components are in separate vesicles from CD63, we increased the centrifugation time of our gradient from 135 min to 16 h. We observed that Hrs (an ESCRT component) migrated to lighter-density fractions than CD63 (Fig. 2A). These distinct populations were also observed in EVs from human epithelial cells (HEp-2). As shown in Fig. 2B, Alix, an ESCRT factor, separated from CD63+ EVs. Consistent with Fig. 1C, Us11 was enriched in ESCRT+ EVs (Fig. 2B). ARF6, a microvesicle marker, was enriched in CD63+ EVs as in HEL cells (47). However, we noticed that the distribution of ARF6 in the gradient when using EVs from HEp-2 cells, compared to EVs from HEL cells, was not as consistent as the other markers, perhaps because HEp-2 is a cancer cell line and cancer cells are known to alter EV biogenesis pathways to facilitate tumor growth (48, 49).
FIG 2.
Separation of EV populations from HSV-1-infected cells. (A) Samples from HSV-1(F)-infected HEL cells prepared as for Fig. 1 were centrifuged either for 135 min or 16 h in an SW41Ti rotor in a Beckman Coulter OPTIMA XPN-80 ultracentrifuge. Fractions of 500 μl were collected from the top to the bottom of the gradient, and equal volumes were electrophoretically separated in denaturing polyacrylamide gels, transferred to nitrocellulose sheets, and analyzed by immunoblot analysis using antibodies against CD63 or Hrs. (B) HEp-2 cells were infected with HSV-1(F) (0.1 PFU/cell), the supernatant was processed as for Fig. 1, the fractions containing the ESCRT+ EVs and the CD63+ EVs were isolated separately, and equal numbers of EVs were analyzed by immunoblot analysis using antibodies against Alix, CD63, ARF6, or Us11. (C) The supernatant of HSV-1(F)-infected cells (0.1 PFU/cell) was collected at 48 h postinfection. CD63+ EVs were separated from the ESCRT+ EVs using procedures as for panel A, and the fractions of the gradient containing either CD63+ EVs or ESCRT+ EVs were pooled. Protein analysis was performed on equal amounts of EVs. Total lysates served as a control. Differences in the mobility of glycoproteins between virions and EVs are most likely due to sucrose present in EV samples. (D) HEL cells seeded in a 6-well plate were lysed using the triple detergent buffer (see Materials and Methods), and equal amounts of cell lysates were mixed with different loading buffers (A, B, C, and D), electrophoretically separated on a 12% SDS-polyacrylamide gel, and transferred to a nitrocellulose sheet, and immunoblot analysis was performed using an anti-CD63 antibody. β-Actin served as a loading control. β-ME, β-mercaptoethanol.
Previous studies have reported production of noninfectious light particles (L-particles) from HSV-1-infected cells that carry viral tegument and envelope proteins but lack capsid proteins and viral genome (50). The L-particles appear to increase virus infectivity and provide immune evasion functions (50–52). Since the ESCRT+ EVs carry viral proteins like the L-particles, we wanted to determine if they are distinct or not. Thus, after separating the ESCRT+ EVs from the CD63+ EVs through the gradient described in Fig. 1, we analyzed them for viral proteins. In the ESCRT+ EVs we detected components of the viral entry and fusion machinery such as gD and gH, and the tegument protein Us11, but we did not detect ICP4 and VP22 proteins, which are abundantly present in L-particles (Fig. 2C) (50–52). As discussed above, the CD63+ EVs did not carry viral proteins. The differences observed in the mobility of the glycoproteins between EVs and cell lysates is most likely due to sucrose present in EV samples. These data suggest that the ESCRT+ EVs are distinct from the L-particles.
During these studies, consistent detection of CD63 by immunoblot analysis was found to be difficult, likely because CD63 is extensively and variably glycosylated. To circumvent this issue, we tested increasing concentrations of the denaturing agents sodium dodecyl sulfate (SDS) and β-mercaptoethanol in the sample loading buffer, or seminative conditions under which both agents were removed from the buffer. Consistent detection of glycosylated CD63 was observed under seminative conditions, whereas the presence of SDS in the loading buffer obstructed CD63 detection (Fig. 2D).
Overall, we have developed procedures to separate EVs from HSV-1 virions, to dissect EV populations, and to consistently detect CD63 tetraspanin.
HSV-1 infection stimulates production of CD63+ EVs.
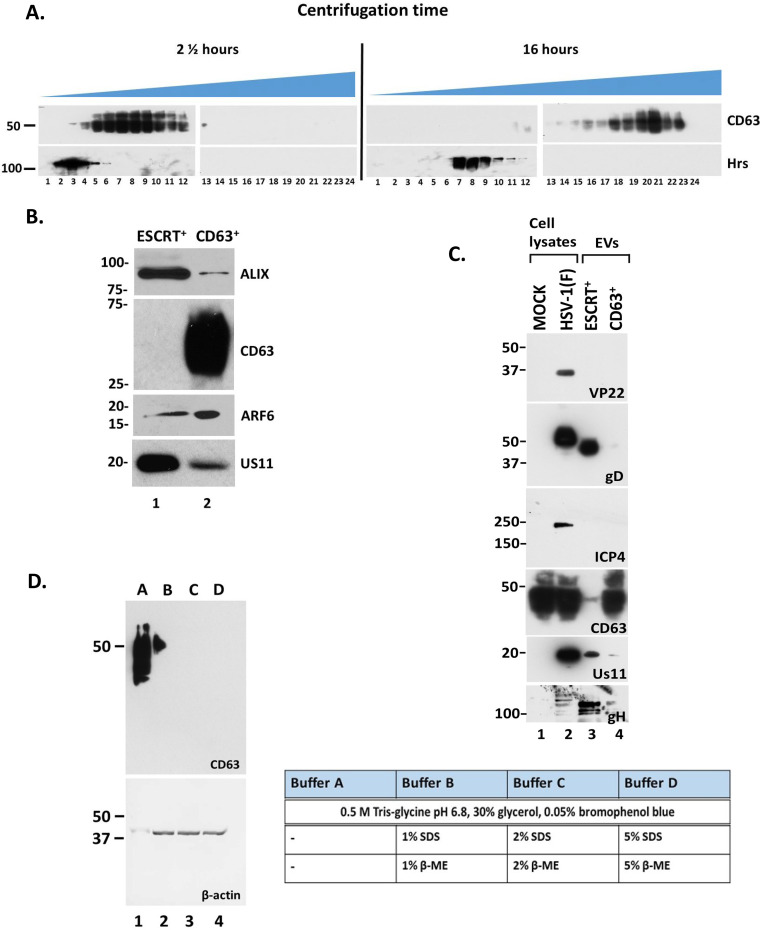
We performed qualitative and quantitative comparisons of EVs isolated from uninfected and HSV-1(F)-infected cells, as in Fig. 1C, focusing on gradient fractions enriched in EVs but devoid of infectious virus. These fractions were pooled, EVs were quantified by nanoparticle tracking analysis (NTA), and equal numbers of EVs were analyzed by Western blotting, probing for selected EV markers. Consistent with our previous findings, CD63 was increased in EVs from infected cells. However, no substantial difference was noted in the exocytosis of ARF6 and Alix between infected and uninfected cells (Fig. 3A), suggesting that microvesicle production (as indicated by ARF6) and EV production through the ESCRT pathway (as indicated by Alix) are not affected (stimulated or inhibited) during HSV-1 infection.
FIG 3.
Increased production of EVs upon HSV-1 infection. (A) Total EVs were isolated from the supernatant of infected or uninfected HEL cells as for Fig. 1. The first seven fractions containing the EVs were pooled, washed with PBS, and quantified by NTA. Equal numbers of EVs were analyzed by immunoblot analysis using antibodies against CD63, ARF6, Alix, and STING. The STING dimers are prevalent in EVs released from HSV-1-infected cells. (B) EVs were isolated as for Fig. 1 and quantified using NTA. The quantity of the EVs and their size distribution are depicted. Results represent the averages from three independent EV isolations. The color-shaded regions are error bars that are used to show the variation in the concentration and size of EVs between different EV isolations. (C) Chips coated with separate capture spots for anti-CD63, anti-CD81, anti-CD9, or mouse isotype control IgG1 were placed in the center of separate wells in a 24-well plate. Diluted sample (40 μl) prepared as for Fig. 1 was applied to each chip. Sterile water was added to the void space between wells to form a humidity chamber, and the plate was sealed and allowed to incubate for 16 h at room temperature. Subsequently, wells containing chips were washed three times with buffer supplied by the manufacturer (NanoView Biosciences, USA). Anti-CD9-Alexa Fluor 488, anti-CD81-Alexa Fluor 555, or anti-CD63-Alexa Fluor 647 was applied to the chips for 1 h at room temperature. After washing, the chips were dried and imaged using an ExoView R100 reader on nScan2 2.7.6 software and analyzed using NanoViewer 2.8.9. (D) Interferometric detection of EVs from panel C, which were captured using different anti-tetraspanin antibodies, was performed using NanoViewer 2.8.9 supplied by the manufacturer (NanoView Biosciences). Total captured EVs on each chip were quantified. (E) Mean fluorescence intensity (MFI) of CD63 in single EVs from uninfected (mock) and infected (HSV-1) samples was quantified using NanoViewer 2.8.9 supplied by the manufacturer (NanoView Biosciences). MFI values were divided in 3 arbitrary groups; the percentage of EVs from mock or infected samples in each group is depicted. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Previously, we demonstrated that HSV-1-infected cells produce at least two times more EVs than uninfected cells (46). These findings were recapitulated after quantifying the EVs isolated as described above using NTA (Fig. 3B). A qualitative analysis of EVs released by HSV-1-infected cells was performed using the ExoView platform (NanoView Biosciences, USA). The ExoView tetraspanin kit features chips with immobilized antibodies against the tetraspanins CD63, CD81, and CD9, which were used to capture EVs from infected and uninfected cells followed by interferometric analysis and detection of EVs with fluorophore-conjugated antibodies against the same tetraspanins. Representative images of captured EVs stained for the different tetraspanins are depicted in Fig. 3C, and interferometric analysis is shown in Fig. 3D. The results of this analysis can be summarized as follows. (i) After incubation with equal numbers of EVs from infected and uninfected cells, the anti-CD63 capture spots captured more EVs from infected than from uninfected cells (Fig. 3C, CD63 capture, compare panel f to panel e), suggesting that HSV-1 infection stimulates release of CD63+ EVs. Interferometric analysis confirmed that infected cells produce almost two times more CD63+ EVs than do uninfected cells (Fig. 3D). (ii) After incubation with equal numbers of EVs from infected and uninfected cells, the anti-CD81 capture spots captured fewer EVs from infected than from uninfected cells (Fig. 3C, CD81 capture, compare panel d to panel c). This corroborates our previous findings showing that the CD81 protein is degraded late during HSV-1 infection (44). (iii) anti-CD81 and anti-CD9 capture spots captured fewer EVs from infected cells than the anti-CD63 capture spots. These data support the notion that CD63+ EVs are the dominant population released during HSV-1 infection. Also, most likely CD63+ EVs are distinct from EVs carrying other tetraspanins. Indeed, interferometric analysis showed that at least half of CD63+ EVs are devoid of CD81 or CD9 (Fig. 3D), indicating that CD81 and CD9 colocalize on the same EVs more often, even though CD63 is the dominant tetraspanin, further supporting distinct pathways of biogenesis. Also, a chip coated with the CD9 antibody captured more CD81+ EVs than CD63+ EVs (Fig. 3C, CD9 capture). (iv) Analysis of mean fluorescence intensity per EV of CD63+ EVs from infected compared to uninfected cells revealed that approximately 80% of CD63+ EVs from infected cells display high fluorescent intensity, indicating that they most likely carry more CD63 molecules per vesicle (Fig. 3E). In contrast, about 75% of CD63+ EVs from uninfected cells display low fluorescent intensity (Fig. 3E). (v) The CD9 captured EVs displayed weaker staining with the CD9 antibody than with CD81 (Fig. 3C). Consistently, the CD81 captured EVs displayed weaker staining with CD9 antibody than with CD81 (Fig. 3C). These data suggest that CD9 is the least abundant of the three tetraspanins in EVs released from infected and uninfected fibroblasts. CD9+ EVs are highly positive for CD81 but not for CD63.
Overall, HSV-1 infection stimulates biogenesis of EVs through the CD63 pathway, and these EVs are enriched in CD63 tetraspanin.
Features of CD63+ versus ESCRT+ EVs during HSV-1 infection.
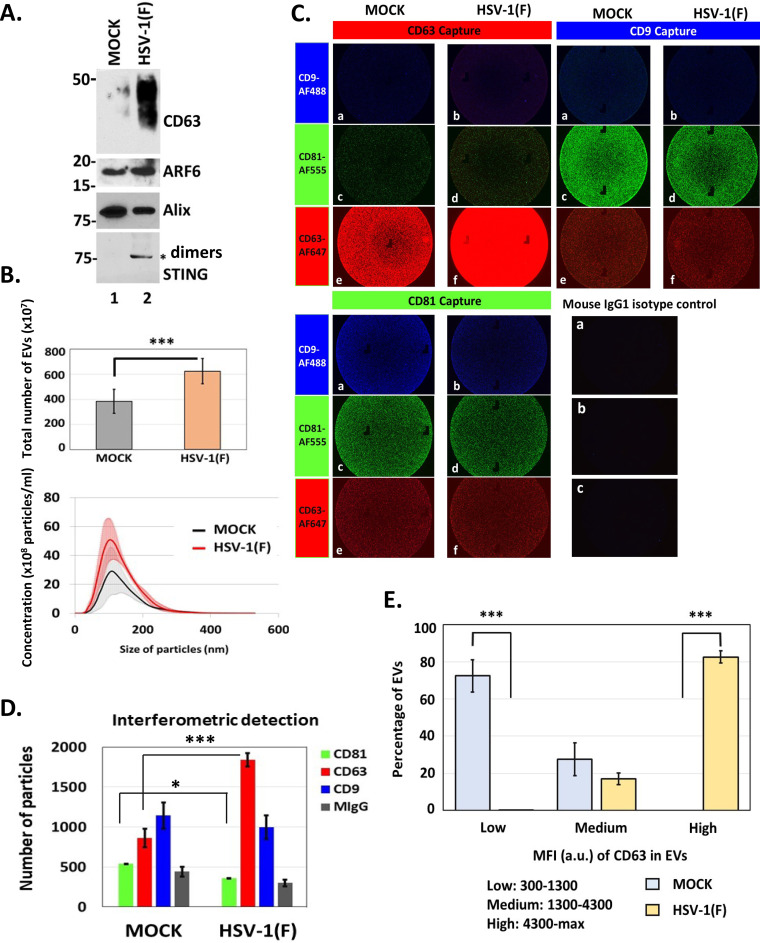
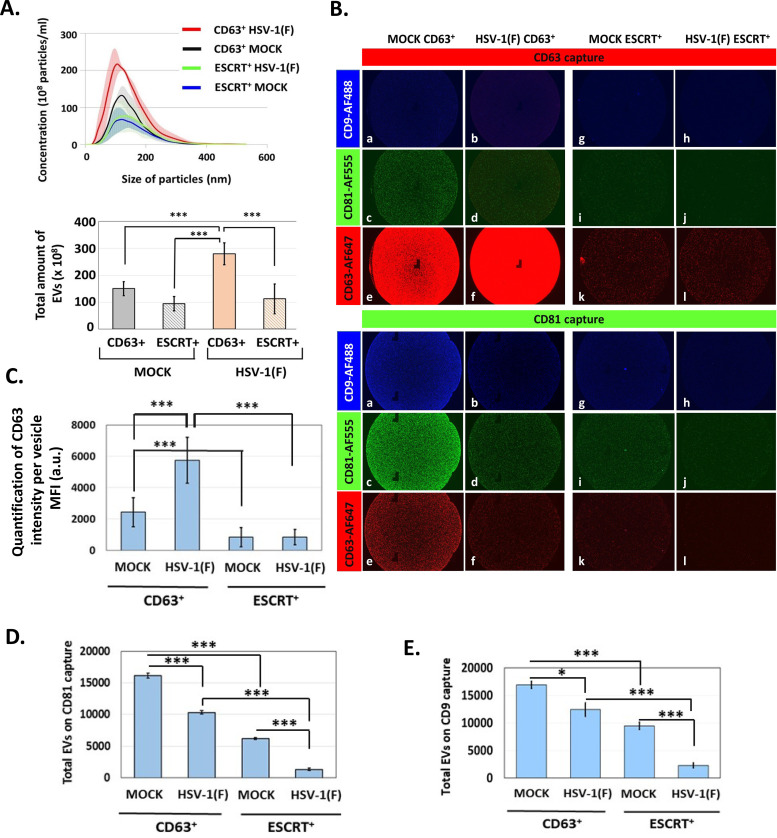
EVs from infected and uninfected HEL cells were isolated through the gradient described earlier, and CD63+ EVs were separated from ESCRT+ EVs (Fig. 1). Quantification of these EVs was done by NTA. As shown in Fig. 4A and B, CD63+ EV production was induced in infected cells, but production of ESCRT+ EVs was not. ExoView analysis of CD63+ and ESCRT+ EVs from infected and uninfected cells demonstrated the following. (i) After incubation with equal numbers of EVs from infected and uninfected cells, anti-CD63 capture spots captured more CD63+ EVs from infected than from uninfected cells (Fig. 4B, CD63 capture, compare panel f versus panel e). Comparison of the mean fluorescence intensity per EV of CD63+ EVs from infected versus uninfected cells suggested that CD63+ EVs from infected cells carry more CD63 molecules than CD63+ EVs from uninfected cells (Fig. 4C). (ii) ESCRT+ EVs are devoid of CD63 protein, further supporting the notion that this population is distinct from the CD63+ population (Fig. 4B and C). These results also highlight the efficiency of separation of ESCRT+ EVs from CD63+ EVs using our gradient. (iii) a chip coated with CD81 antibody captured fewer CD81+ EVs when using the CD63+ population from infected than from uninfected cells (Fig. 4B, CD81 capture, compare panel c to panel d, and Fig. 4D) supporting the idea that CD81+ EVs are down-modulated during infection. The same pattern was observed using ESCRT+ EVs (Fig. 4B and D), although CD81 was less prevalent in ESCRT+ EVs. (iv) Only small amounts of CD63, CD81, and CD9 were detected in ESCRT+ EVs, suggesting that at least in HEL fibroblasts the ESCRT biogenesis pathway does not involve these tetraspanins (Fig. 4B, D, and E). (v) CD9 is more prevalent in CD63+ EVs from uninfected cells than from infected cells (Fig. 4B, CD81 capture, compare panel a to panel b, and Fig. 4E). The pattern of CD9 in CD63+ EVs and ESCRT+ EVs from infected and uninfected cells resembles that of CD81 (Fig. 4, compare panel E to panel D).
FIG 4.
Increased production of CD63+ EVs during HSV-1 infection. A. EVs were isolated from the supernatant of infected or uninfected HEL cells as for Fig. 1, and the ESCRT+ EVs were collected separately from the CD63+ EVs. Quantification of the two EV populations derived from either infected or uninfected cells was done by NTA. The color-shaded regions are error bars that are used to show the variation in the concentration and size of EVs between different EV isolations. (B) Chips coated with separate capture spots for anti-CD63, anti-CD81, anti-CD9, or mouse isotype control IgG1 (shown in Fig. 3) were incubated with either ESCRT+ EVs or CD63+ EVs, as for Fig. 3. Anti-CD9-Alexa Fluor 488, anti-CD81-Alexa Fluor 555, or anti-CD63-Alexa Fluor 647 was applied to the chips for 1 h at room temperature. After washing, the chips were dried and imaged using an ExoView R100 reader on nScan2 2.7.6 software and analyzed using NanoViewer 2.8.9. (C) MFI of CD63 in single EVs from ESCRT+ and CD63+ fractions from mock or infected cells was quantified using NanoViewer 2.8.9 supplied by the manufacturer (NanoView Biosciences). (D and E) The number of EVs on each chip from panel C was quantified using NanoViewer 2.8.9. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Overall, we have developed a procedure to isolate the dominant EV populations during HSV-1 infection. Furthermore, we have established that HSV-1 infection timulates production of EVs through the CD63 pathway. Production of EVs through the ESCRT pathway is not altered during HSV-1 infection, although ESCRT components may be utilized in virus-induced cargo sorting processes and for virion assembly.
STING is present in CD63+ EVs released by HSV-1-infected cells.
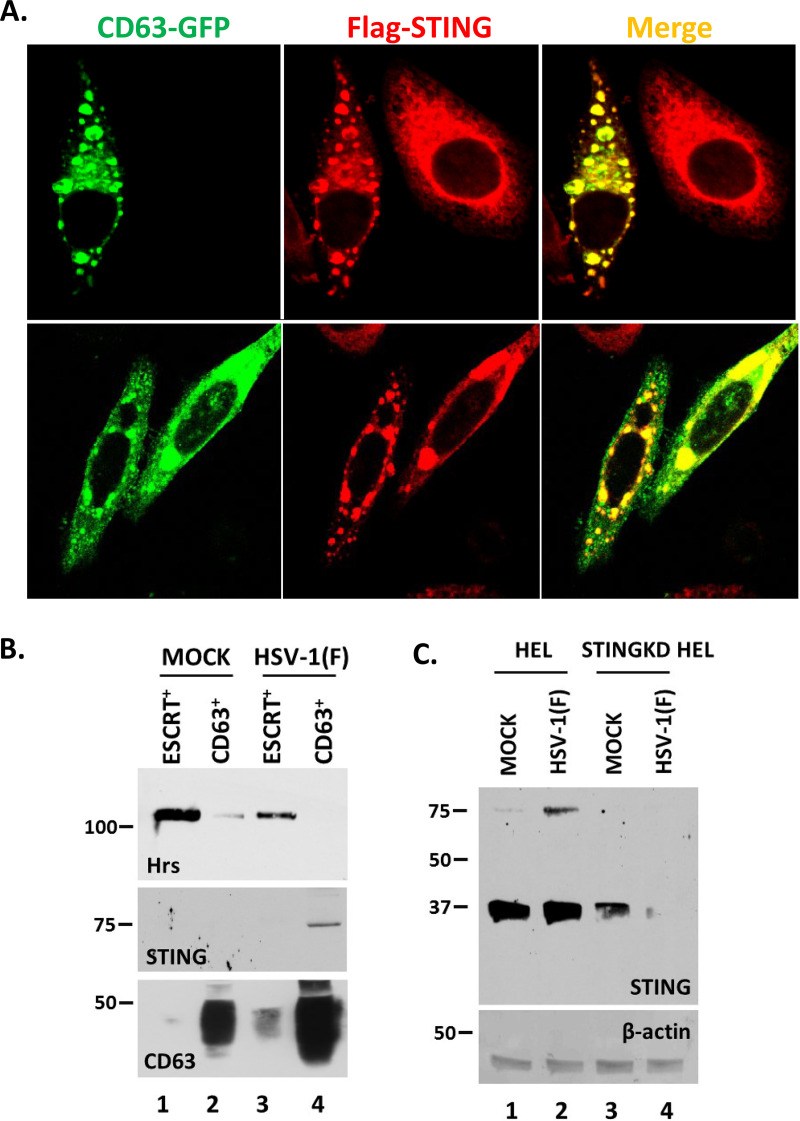
Earlier, we published that STING is present in EVs released by HSV-1-infected cells (38, 39, 45). Thus, we sought to determine the population of EVs in which STING is packaged. First, we examined potential colocalization of STING with CD63. For this, we established a HEp-2 cell line expressing Flag-STING by using a lentiviral vector carrying the STING open reading frame. The Flag-STING HEp-2 cells were transfected with a green fluorescent protein (GFP)-CD63-expressing plasmid for 24 h, followed by HSV-1(F) infection (10 PFU/cell). The cells were fixed at 10 h postinfection, stained with an anti-Flag antibody, and examined under a confocal microscope. STING and CD63 form globular structures in the cytoplasm and appear to colocalize (Fig. 5A). Second, we isolated CD63+ and ESCRT+ EVs produced by infected and uninfected cells as shown in Fig. 1 and analyzed them for Hrs, CD63, and STING. EVs carrying either Hrs or CD63 were efficiently separated, and STING was found exclusively in CD63+ EVs derived from infected cells (Fig. 5B). Notably, it is the STING dimers that are predominantly detected in the CD63+ EVs. STING dimerization is a hallmark of STING translocation that occurs upon HSV-1 infection (Fig. 5C, lane 2 compare to lane 1). Antibody specificity was demonstrated using STING knockdown (STING-KD) HEL cells (Fig. 5C, lanes 3 and 4).
FIG 5.
STING protein associates with the CD63+ EVs released from HSV-1-infected cells. (A) The Flag-STING-expressing HEp-2 cell line was transfected with a CD63-GFP-expressing plasmid. At 24 h posttransfection, the cells were infected with HSV-1(F) (10 PFU/cell). The cells were fixed using 4% paraformaldehyde at 10 h postinfection and stained with an anti-Flag antibody. Images were obtained using a Leica confocal microscope. (B) HEL cells either left uninfected or infected with HSV-1(F) (0.1 PFU/cell) were harvested at 48 h postinfection, and CD63+ EVs and ESCRT+ EVs were isolated as for Fig. 1 and 2. Equal numbers of EVs were analyzed in denaturing polyacrylamide gels, and immunoblot analysis was performed using antibodies against Hrs, STING, and CD63. (C) HEL or STING-KD HEL cells were either left uninfected or infected with HSV-1(F) (0.1 PFU/cell). The cells were harvested at 24 h postinfection, and equal amounts of proteins were analyzed by immunoblot analysis using a mouse monoclonal antibody against STING. β-Actin was used as a loading control.
We conclude that STING is exocytosed in CD63+ EVs during HSV-1 infection.
Antiviral effect of CD63+ EVs and proviral role of ESCRT+ EVs during HSV-1 infection.
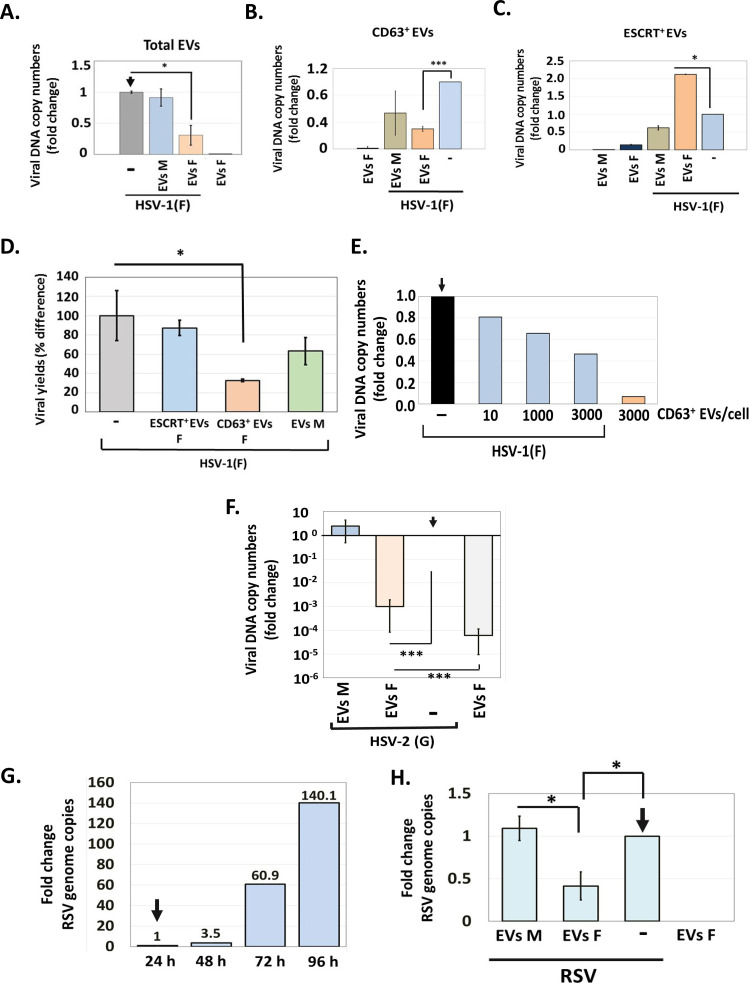
Using the entire population of EVs, we have demonstrated that EVs from infected cells, but not from uninfected cells, negatively impact HSV-1 infection (45). The goal of these studies was to determine the functions of the distinct EV populations described above. First, we isolated the entire EV population from infected and uninfected cells as in Fig. 1C, quantified EVs by NTA, and used them to expose replicate cultures of HEL cells (1,000 EVs/cell). At 2 h post-EV exposure, the cells were infected with HSV-1(F) (0.01 PFU/cell). The cells were harvested at 24 h postinfection, and viral DNA copy numbers were quantified by quantitative PCR (qPCR) analysis. Cells infected with HSV-1(F) or uninfected cells treated with EVs served as controls. EVs derived from infected cells caused a 70% reduction in viral DNA copy numbers in recipient cells compared to EVs derived from uninfected cells (Fig. 6A). Treatment with EVs alone served as a contamination control and showed that the viral genome is not detectable in cells exposed to EVs derived from HSV-1-infected cells.
FIG 6.
Effect of EVs on HSV-1 infection. (A) EVs from infected or uninfected HEL cells present in the first seven fractions of the gradient described in the legend for Fig. 1 were quantified by NTA and used to expose uninfected HEL cells (1,000 EVs/cell). At 2 h post-EV exposure, the cells were infected with HSV-1(F) (0.01 PFU/cell). The cells were harvested at 24 h postinfection, and quantification of the viral genome was done by qPCR analysis as detailed in Materials and Methods. Cells exposed only to EVs or to virus served as controls. (B and C) CD63+ EVs (B) and ESCRT+ EVs (C) were isolated from infected and uninfected HEL cells at 24 h postinfection (0.01 PFU/cell), as for Fig. 1. Equal volumes of EVs were used to expose HEL cells for 2 h, followed by HSV-1 infection (0.01 PFU/cell). The cells were harvested at 48 h postinfection, and viral genome copy numbers were quantified as described above. Relative fold gene expression was calculated using the delta-delta threshold cycle (CT) method. (D) CD63+ and ESCRT+ EVs were isolated from infected and uninfected HEL cells at 24 h postinfection (0.01 PFU/cell), as for Fig. 2. Equal volumes of EVs were used to expose HEL cells for 2 h, followed by HSV-1 infection (0.01 PFU/cell). The cells were harvested at 48 h postinfection, and quantification of the infectious virus was done by plaque assay in Vero cells. (E) CD63+ EVs were isolated from HSV-1(F)-infected HEL cells as described above, and different doses were used to expose HEL cells for 2 h prior to HSV-1(F) infection (0.01 PFU/cell). The cells were harvested at 48 h postinfection, and viral genome copy numbers were quantified as described above. Exposure to CD63+ EVs from infected cells served as a contamination control. Fold change is relative to infected but non-EV-treated cells. (F) HEL cells were exposed to CD63+ EVs isolated from HSV-1(F)-infected or uninfected cells as for panel B, but infection was done with HSV-2(G) (0.01 PFU/cell). The cells were harvested at 48 h postinfection, and viral genome copy numbers were quantified as described above using primer pairs targeting the gG region of HSV-2 genome. (G) HEL cells were infected with RSV (0.01 PFU/cell). The cells were harvested every 24 h up to 96 h postinfection. Total RNA was extracted, reversed transcribed, and used to quantify the RSV genome. (H) HEL cells were infected with RSV (0.01 PFU/cell) for 48 h, followed by exposure to CD63+ EVs derived from HSV-1(F)-infected or uninfected cells as for panel B for another 48 h. The cells were harvested at 96 h postinfection, and viral genome copy numbers were quantified as detailed in Materials and Methods. All values are derived from triplicate samples. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Second, we sought to determine the role of the different EV populations released during HSV-1 infection. For this, EVs were isolated from HSV-1-infected or uninfected HEL cells and ESCRT+ EVs were separated from CD63+ EVs by collecting separately the corresponding fractions of the gradient. Quantification of each EV population was done by NTA. Subsequently, those EVs were used to expose HEL cells. At 2 h after exposure to EVs, the cells were infected with HSV-1(F) (0.01 PFU/cell). The cells were harvested at 48 h postinfection, and quantification of the viral genome was done by qPCR analysis. As shown in Fig. 6B, CD63+ EVs from infected cells downregulated HSV-1 replication, whereas CD63+ EVs from uninfected cells did not. Treatment with CD63+ EVs from infected cells for 48 h showed negligible viral genome. In contrast, ESCRT+ EVs from HSV-1-infected cells enhanced viral replication, as opposed to ESCRT+ EVs from uninfected cells (Fig. 6C). Exposure to ESCRT+ EVs without subsequent infection served as a contamination control. Virus growth was determined under the same conditions using a plaque assay (Fig. 6D). Consistent with the data above, CD63+ EVs from infected cells caused a decrease in virus yields by 70%, which was found to be significant. The ESCRT+ EVs did not interfere with the virus growth. Also, EVs from uninfected cells did not have a significant effect on virus yields. The effect of CD63+ EVs is dose dependent (Fig. 6E).
We conclude that CD63+ EVs released from HSV-1-infected cells have an antiviral effect on a subsequent infection, whereas ESCRT+ EVs do not interfere with a subsequent infection and occasionally display a modest proviral effect. The total EV population has an overall negative impact on HSV-1 infection.
Considering that innate immunity factors (i.e., STING) are involved in the antiviral function of CD63+ EVs, we sought to determine if EVs from HSV-1(F)-infected cells exert broad antiviral activity and can impact infection by other viruses. Thus, CD63+ EVs isolated from uninfected and HSV-1(F)-infected HEL cells were used to pretreat HEL cells (1,000 EVs/cell) for 2 h, followed by HSV-2(G) infection (0.01 PFU/cell). The cells were harvested at 48 h post-EV exposure, and HSV-2(G) genome copy numbers were determined using primer pairs against the HSV-2 gG sequence. The primers we chose specifically amplify the HSV-2 genome, but not HSV-1. Exposure to EVs from HSV-1-infected cells served as a control. We found that CD63+ EVs from HSV-1(F)-infected cells caused an approximately 1,000-fold decrease in HSV-2 genome copy numbers compared to the case with EVs from uninfected cells (Fig. 6G). Thus, EVs derived from HSV-1-infected cells exert stronger antiviral activity against HSV-2.
Next, we asked if EVs from HSV-1-infected cells can affect RNA viruses, and for this assay, we chose respiratory syncytial virus (RSV). We determined that RSV replicates in HEL cells and that at 0.01 PFU/cell, onset of virus replication is observed after 48 h of infection (Fig. 6G). To assess the impact of EVs from HSV-1 on RSV infection, we first exposed HEL to RSV (0.01 PFU/cell), and at 48 h postinfection, the cells were exposed to CD63+ EVs from HSV-1-infected or uninfected cells. The cells were collected at 48 h post-EV exposure and RSV genome copy numbers were determined by quantitative PCR analysis using primer pairs targeting the NS1 sequence of RSV. We observed that EVs from HSV-1-infected but not from uninfected cells caused a decrease in RSV genome copy numbers by almost 60% (Fig. 6H).
We conclude that CD63+ EVs released from HSV-1-infected cells have a broader antiviral activity and can downregulate both HSV-2 and RSV infection.
DISCUSSION
HSV-1 infection increases production of EVs that negatively impact the infection (38, 39, 45, 46). This increased production of EVs occurs postreplication and results in accumulation of CD63+ EVs in the milieu of the infection (46). One factor found in EVs released from HSV-1-infected cells is STING. In our previous studies, we demonstrated that EVs released by HSV-1-infected cells suppress virus replication in a STING-dependent manner (38, 45).
The salient findings of this study can be summarized as follows.
Two major EV populations were found during HSV-1 infection that were separable through our gradient approach. The first carries ESCRT components, such as Alix and Hrs, and was found in lighter-density fractions than the second, which is enriched in CD63. These two EV populations were present in uninfected cells too, but the production of the CD63+ EVs was substantially increased upon HSV-1 infection, whereas the amount of ESCRT+ EVs remained unaltered. The ExoView platform highlighted not only that CD63+ EVs are the dominant population during HSV-1 infection but also that each vesicle carries multiple CD63 molecules.
In addition to this major EV population, other, minor populations were discovered. These included CD81+ CD9+ EVs, which cofractionate with CD63+ EVs but appear to constitute a separate population since they did not stain positive for CD63. CD81 was reduced in EVs released by infected cells. This is consistent with our previous findings showing that CD81 is degraded during the late stages of HSV-1 infection (46). The levels of extracellular CD9 were also reduced during HSV-1 infection. EV populations carrying other combinations of the three tetraspanins cannot be excluded, but they appear to be minor populations since at least half of the EVs released by infected cells carry only CD63. Although the CD63+ EVs produced during HSV-1 infection most likely represent exosomes, the origin of the CD63− EVs carrying CD81 and CD9 is not clear yet. All three tetraspanins can be found in exosomes, but CD81 and CD9 are frequently found in microvesicles produced from the plasma membrane (13). Microvesicle production was not altered upon HSV-1 infection, as determined by the levels of ARF6 in EVs. In the gradient, ARF6 had a distribution similar to that of CD63, but these markers likely belong to separate EV populations since only CD63, and not ARF6, exocytosis was induced upon infection.
A striking observation was that selected viral proteins were present in the ESCRT+ EVs released by HSV-1-infected cells. Among these proteins were VP16, a major viral transactivator, Us11, a viral RNA-binding protein, and components of the HSV-1 entry and fusion machinery such as glycoprotein D (gD), gH, and gB. Other viral proteins such as the tegument protein VP22, the viral gM, and the ICP4 transactivator were absent. ESCRT+ EVs are free of infectious virus, as evidenced by the absence of viral genome or through a plaque assay using all the fractions of the gradient as previously published (45). While ESCRT+ fractions carry viral proteins, CD63+ fractions do not, suggesting that viral proteins are preferentially packaged in ESCRT+ EVs. This example shows selectivity during EV cargo packaging. One mechanism by which these viral proteins could be packaged into ESCRT+ EVs is through their interaction with ESCRT accessory proteins. Sequencing analysis of VP16 and gD identified putative late domains through which they could interact with the ESCRT accessory protein Alix and sorted into EVs. The Us11 protein does not carry late domains but it interacts with host proteins known to be packaged into EVs, such as Hsp90 (53, 54).
The similarities of ESCRT+ EVs released during HSV-1 infection with previously described light particles (L-particles) released from infected cells prompted us to further analyze the ESCRT+ EVs. We discovered that ESCRT+ EVs are noninfectious like L-particles, carry viral proteins, and play a proviral role. A substantial difference between ESCRT+ EVs and L-particles is that proteins abundantly present in L-particles, such as VP22, gM, UL46, and ICP4, were absent from ESCRT+ EVs. Both types of particles carry components of the viral entry and fusion machinery and Us11. Considering that viral proteins that distinguish L-particles from ESCRT+ EVs (VP22, gM, UL46, and ICP4) accumulated only in high-density fractions, this suggests that L-particles overlap infectious virions in our gradient. Also, since ESCRT components (Alix, Hrs, and TSG101) were detected only in light-density fractions, this suggests that ESCRT+ EVs and L-particles are distinct particles and separable through our gradient. The host proteins present in L-particles are unknown. An intriguing possibility is that L-particles represent a population of extracellular vesicles generated by the virus egress pathway. Based on this scenario, L-particles could be heterogeneous in composition and lighter L-particles (lacking VP22, gM, UL46, and ICP4) may comigrate with ESCRT components in our gradient. Thus, further work is required to determine the nature, properties, and roles of these particles. STING, one of the first host proteins that we discovered in EVs released by HSV-1-infected cells, was found in CD63+ EVs, but not in ESCRT+ EVs (38, 39). STING localizes to the membranes of the endoplasmic reticulum, and following ligand binding, a conformational change facilitates STING oligomerization and translocation to the trans-Golgi network and perinuclear compartments (55–57). During HSV-1 infection, a fraction of STING appears to enter the CD63 pathway of exocytosis and is packaged in CD63+ EVs. We noticed that the dimeric form of STING is preferentially detected in CD63+ EVs. The mechanism of STING exocytosis currently remains unknown, but it is unlikely to involve STING downstream signaling since the virus has evolved multiple mechanisms to obstruct this pathway (58, 59).
Previously, we found that EVs released by HSV-1-infected cells impact the infection negatively in a STING-dependent manner (45). Since STING was found in the dominant population of EVs produced by infected cells and viral proteins were found in a separate, less abundant population, we speculated that these EVs may have different roles during infection. Indeed, CD63+ EVs were found to suppress virus replication, whereas EVs carrying viral proteins were found to either enhance it or not affect it. The variability observed with the ESCRT+ EVs could be due to variability in the amount of proviral cargo present in these EVs. Total EVs derived from HSV-1-infected cells had a suppressive role in virus replication, suggesting that the dominant effect of these EVs is negative. This is perhaps because CD63+ EVs are at least two times more in number than ESCRT+ EVs in the entire EV population. An interesting observation is that the CD63+ EVs released by HSV-1-infected cells have an antiviral effect against other DNA and RNA viruses e.g., HSV-2 and RSV. The extent of the effect may depend on the ability of these viruses to counteract antiviral responses. Notably, RNA viruses can be restricted by the DNA sensor STING (60).
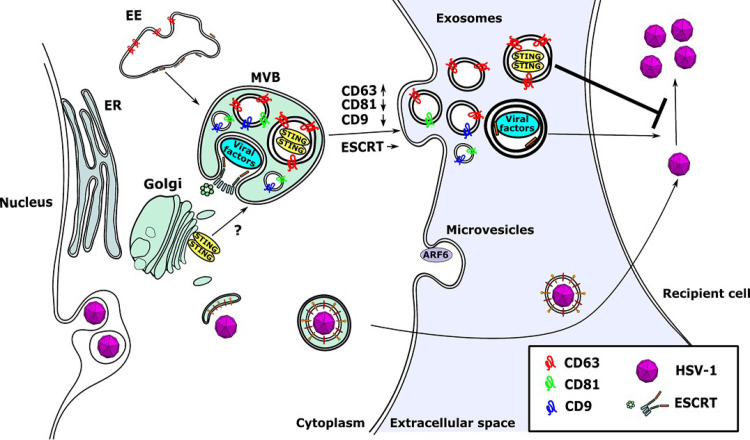
Overall, it appears that EVs with proviral and antiviral roles are released during HSV-1 infection (Fig. 7). In the environment of the infection the outcome will depend on multiple factors, including the time of release of each population, their efficiency of uptake by recipient cells, whether both populations are taken up by the same type of cells, and the relative stability of the cargo responsible for these effects. The EVs with proviral roles (ESCRT+ EVs) appear to lack tetraspanins, and our previous studies demonstrated that their production does not depend on ceramide (45). These data suggest differences in targeting and uptake of these EVs. In addition, increased production of CD63+ EVs appears to require late gene expression, whereas viral factors could be packaged in EVs as they are expressed. Nevertheless, our data indicate that intercellular communication could be a key determinant of HSV-1 persistence and pathogenesis.
FIG 7.
Model of EV exocytosis during HSV-1 infection. STING is sorted into CD63+ ILVs, whereas viral factors are sorted into ESCRT+ ILVs. The ILVs are released to the extracellular space after fusion of the MVBs to the plasma membrane and are referred to as exosomes. Production of CD63+ EVs is selectively enhanced during HSV-1 infection, but the quantity of ESCRT+ EVs remains unaltered. Also, the production of the microvesicles released from the plasma membrane is unaffected. CD81 is degraded during the late stages of HSV-1 infection and therefore is underrepresented in EVs. A small population of EVs carrying CD81 and CD9 appears to be distinct from the CD63+ EVs. The CD63+ EVs appear to have an antiviral role and suppress subsequent HSV-1 infection, but the ESCRT+ EVs seem to have an opposite effect.
MATERIALS AND METHODS
Cell lines and virus.
HEL cells (immortalized human embryonic lung fibroblasts, human telomerase reverse transcriptase [hTERT] transformed) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (FBS). Vero, HEp-2, and HEK-293 cells (ATCC) were cultured according to the manufacturer’s instructions. HSV-1(F) and HSV-2(G) are limited-passage isolates that have been describe before (61). Human respiratory syncytial virus (RSV) strain A2 was obtained through the ATCC.
Extracellular vesicle purification.
Isolation of EVs was done as detailed in the legend for Fig. 1 (45). For functional assays, isolated EVs were washed with phosphate-buffered saline (PBS) through 100-kDa-cutoff filters and used to expose cells for 2 h prior infecting them with HSV-1(F) or HSV-2(G) at 0.01 PFU/cell or RSV (0.01 PFU/cell). For protein analysis, isolated EVs were mixed with either buffer A (Fig. 2) for CD63 detection or buffer B (Fig. 2) for detection of other proteins.
NTA.
Nanoparticle tracking analysis (NTA) was performed using the NanoSight LM10 instrument (NanoSight, Salisbury, United Kingdom). For each sample, nine different acquisitions were obtained, of 60 s each. NTA software version 2.3 was used to analyze 60-s videos of data collection to obtain the mean, median, and mode of vesicle size and concentration. Results represent the averages of three independent EV isolations.
Single-particle interferometric reflectance imaging sensing (NanoView) analysis.
Samples were diluted according to the manufacturer’s protocol for the ExoView tetraspanin kit (NanoView Biosciences; EV-TETRA) and incubated for 16 h on ExoView tetraspanin chips coated with antibodies against CD81, CD63, or CD9 or the mouse IgG1 isotype control in triplicate. The chips were then washed four times in incubation buffer on an orbital shaker for 3 min and incubated with conjugated antibodies for fluorescent labeling of the captured EVs (anti-CD9-Alexa Fluor 488 [AF488], anti-CD81-AF555, and anti-CD63-AF647) for 1 h. After labeling, the chips were washed once with incubation solution, three times with wash solution, and once with rinse solution. The chips were pulled dry from the rinse solution and placed in the reader for analysis. All data were gathered using an ExoView R100 reader equipped with nScan2 2.7.6 software and analyzed using NanoViewer 2.8.9.
Development of STING-expressing or STING-KD cells with the aid of lentiviral vectors.
The pLKO.1 plasmid expressing Flag-STING was developed by inserting the Flag-STING open reading frame into the BamHI/SalI/Klenow site of pLKO.1 GFP CMV Puro (658-5; Addgene). A HEp-2 cell line expressing Flag-STING was developed as described before (46). The STING-KD HEL cell line has been described before (62).
Immunoblot and immunofluorescence analyses.
Cells were solubilized in triple detergent buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% Nonidet P-40, 0.5% sodium deoxycholate, 100 μg ml−1 of phenylmethylsulfonyl fluoride) supplemented with phosphatase inhibitors (10 mM NaF, 10 mM β-glycerophosphate, 0.1 mM sodium vanadate) and protease inhibitor cocktail (Sigma) and briefly sonicated. Protein concentration was determined with the Bradford method (Bio-Rad Laboratories). Purified EVs were mixed with either buffer A (CD63 detection) or buffer B (all other proteins) (Fig. 2C). The mouse monoclonal antibodies to ICP0, UL42, CD63, VP16, gD, Hrs (Santa Cruz), β-actin, Flag epitope (Sigma), Alix (Cell Signaling Technologies), ARF6 (Thermo Fisher Scientific), and STING (R&D systems) were used in a 1:1,000 dilution. The rabbit polyclonal antibodies against UL38, VP22, and gM and the mouse monoclonal antibody against Us11 were gifts from B. Roizman (University of Chicago) and were used in a 1:1,000 dilution. The mouse monoclonal antibody against gH was a gift from R. M. Longnecker (Northwestern University). Proteins were visualized with 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (VWR) or with ECL Western blotting detection reagents (Amersham Biosciences). Procedures for immunofluorescence have been described before (63, 64).
Viral DNA or RNA quantification.
Total DNA was extracted from the cells using the NucleoSpin DNA RapidLyse kit (Macherey-Nagel). To detect HSV-1 DNA, we used primer pairs targeting the area of the viral genome encoding gI. To detect HSV-2 DNA, we used primer pairs targeting the area of the viral genome encoding glycoprotein G (forward, 5′-TAC GCT CTC GTA AAT GCT TC-3′, and reverse, 5′-GCC CAC CTC TAC CCA CAA-3′) (65). For normalization, we used primers targeting β-actin DNA. Both primer pairs have been described before (45, 46). To detect RSV RNA, cells were lysed in 1 ml of TRIzol (Life Technologies) and total RNA was extracted using phenol-chloroform procedures. DNase treatment was performed using Turbo DNase enzyme (Ambion) according to the manufacturer’s instructions. cDNA synthesis was performed using the Superscript IV first-strand synthesis system (Invitrogen). Semiquantitative PCR analysis was performed using SYBR green master mix (Life Technologies) and cDNA that was generated from 1 μg of total RNA per sample. Primers against the NS1 sequence of RSV were used for quantification of the RSV genome. Primer sequences were as follows: NS1 forward, 5′-GCTTTGGCTAAGGCAGTGAT-3′, and NS1 reverse, 5′-TGGCATTGTTGTGAAATTGG-3′. 18S rRNA (Ambion) was used for normalization.
Statistical analysis.
Prism 7 software (GraphPad) was used for statistical analysis of the NanoSight and qPCR data. P values were calculated using either a standard unpaired Student t test or analysis of variance (ANOVA). P values of ≤0.05 were considered significant. In figures, asterisks represent P values as follows: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. Statistical analyses were performed using biological replicates.
ACKNOWLEDGMENTS
B. Roizman (University of Chicago) kindly provided the antibodies against UL38, VP22, gM, and Us11. R. M. Longnecker (Northwestern University) kindly provided the gH antibody. We thank Hope Waisner for editing the manuscript.
M. Kalamvoki has been funded through KUMC startup funds, grants CBID COBRE P20 GM113117, R21AI144883, and R21AI158229, and KUMC Research Institute enhancement award Y6K00081.
REFERENCES
- 1.Wolf P. 1967. The nature and significance of platelet products in human plasma. Br J Haematol 13:269–288. doi: 10.1111/j.1365-2141.1967.tb08741.x. [DOI] [PubMed] [Google Scholar]
- 2.Edelstein LC. 2017. The role of platelet microvesicles in intercellular communication. Platelets 28:222–227. doi: 10.1080/09537104.2016.1257114. [DOI] [PubMed] [Google Scholar]
- 3.Hargett LA, Bauer NN. 2013. On the origin of microparticles: from “platelet dust” to mediators of intercellular communication. Pulm Circ 3:329–340. doi: 10.4103/2045-8932.114760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Meijden PEJ, Heemskerk JWM. 2019. Platelet biology and functions: new concepts and clinical perspectives. Nat Rev Cardiol 16:166–179. doi: 10.1038/s41569-018-0110-0. [DOI] [PubMed] [Google Scholar]
- 5.Harding C, Heuser J, Stahl P. 1983. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J Cell Biol 97:329–339. doi: 10.1083/jcb.97.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieu M, Martin-Jaular L, Lavieu G, Thery C. 2019. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 21:9–17. doi: 10.1038/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- 7.Raposo G, Stoorvogel W. 2013. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sampey GC, Meyering SS, Zadeh MA, Saifuddin M, Hakami RM, Kashanchi F. 2014. Exosomes and their role in CNS viral infections. J Neurovirol 20:199–208. doi: 10.1007/s13365-014-0238-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoorvogel W, Kleijmeer MJ, Geuze HJ, Raposo G. 2002. The biogenesis and functions of exosomes. Traffic 3:321–330. doi: 10.1034/j.1600-0854.2002.30502.x. [DOI] [PubMed] [Google Scholar]
- 10.van NG, D’Angelo G, Raposo G. 2018. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 11.Veerman RE, Gucluler AG, Eldh M, Gabrielsson S. 2019. Immune cell-derived extracellular vesicles—functions and therapeutic applications. Trends Mol Med 25:382–394. doi: 10.1016/j.molmed.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Colombo M, Raposo G, Thery C. 2014. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 13.Jeppesen DK, Fenix AM, Franklin JL, Higginbotham JN, Zhang Q, Zimmerman LJ, Liebler DC, Ping J, Liu Q, Evans R, Fissell WH, Patton JG, Rome LH, Burnette DT, Coffey RJ. 2019. Reassessment of exosome composition. Cell 177:428–445. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kowal J, Tkach M, Thery C. 2014. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 29:116–125. doi: 10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Anand S, Samuel M, Kumar S, Mathivanan S. 2019. Ticket to a bubble ride: cargo sorting into exosomes and extracellular vesicles. Biochim Biophys Acta Proteins Proteom 1867:140203. doi: 10.1016/j.bbapap.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 16.Raiborg C, Stenmark H. 2009. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 17.Frankel EB, Audhya A. 2018. ESCRT-dependent cargo sorting at multivesicular endosomes. Semin Cell Dev Biol 74:4–10. doi: 10.1016/j.semcdb.2017.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juan T, Furthauer M. 2018. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin Cell Dev Biol 74:66–77. doi: 10.1016/j.semcdb.2017.08.022. [DOI] [PubMed] [Google Scholar]
- 19.Saksena S, Sun J, Chu T, Emr SD. 2007. ESCRTing proteins in the endocytic pathway. Trends Biochem Sci 32:561–573. doi: 10.1016/j.tibs.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 20.Schuh AL, Audhya A. 2014. The ESCRT machinery: from the plasma membrane to endosomes and back again. Crit Rev Biochem Mol Biol 49:242–261. doi: 10.3109/10409238.2014.881777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villarroya-Beltri C, Baixauli F, Gutierrez-Vazquez C, Sanchez-Madrid F, Mittelbrunn M. 2014. Sorting it out: regulation of exosome loading. Semin Cancer Biol 28:3–13. doi: 10.1016/j.semcancer.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Termini CM, Gillette JM. 2017. Tetraspanins function as regulators of cellular signaling. Front Cell Dev Biol 5:34. doi: 10.3389/fcell.2017.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charrin S, Jouannet S, Boucheix C, Rubinstein E. 2014. Tetraspanins at a glance. J Cell Sci 127:3641–3648. doi: 10.1242/jcs.154906. [DOI] [PubMed] [Google Scholar]
- 24.Hemler ME. 2005. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol 6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 25.Hemler ME. 2014. Tetraspanin proteins promote multiple cancer stages. Nat Rev Cancer 14:49–60. doi: 10.1038/nrc3640. [DOI] [PubMed] [Google Scholar]
- 26.Maecker HT, Todd SC, Levy S. 1997. The tetraspanin superfamily: molecular facilitators. FASEB J 11:428–442. doi: 10.1096/fasebj.11.6.9194523. [DOI] [PubMed] [Google Scholar]
- 27.Freed EO. 2002. Viral late domains. J Virol 76:4679–4687. doi: 10.1128/jvi.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingham RJ, Colwill K, Howard C, Dettwiler S, Lim CS, Yu J, Hersi K, Raaijmakers J, Gish G, Mbamalu G, Taylor L, Yeung B, Vassilovski G, Amin M, Chen F, Matskova L, Winberg G, Ernberg I, Linding R, O’Donnell P, Starostine A, Keller W, Metalnikov P, Stark C, Pawson T. 2005. WW domains provide a platform for the assembly of multiprotein networks. Mol Cell Biol 25:7092–7106. doi: 10.1128/MCB.25.16.7092-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ageta H, Tsuchida K. 2019. Post-translational modification and protein sorting to small extracellular vesicles including exosomes by ubiquitin and UBLs. Cell Mol Life Sci 76:4829–4848. doi: 10.1007/s00018-019-03246-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen B, Wu N, Yang JM, Gould SJ. 2011. Protein targeting to exosomes/microvesicles by plasma membrane anchors. J Biol Chem 286:14383–14395. doi: 10.1074/jbc.M110.208660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunadt M, Eckermann K, Stuendl A, Gong J, Russo B, Strauss K, Rai S, Kugler S, Falomir LL, Schwalbe M, Krumova P, Oliveira LM, Bahr M, Mobius W, Levin J, Giese A, Kruse N, Mollenhauer B, Geiss-Friedlander R, Ludolph AC, Freischmidt A, Feiler MS, Danzer KM, Zweckstetter M, Jovin TM, Simons M, Weishaupt JH, Schneider A. 2015. Extracellular vesicle sorting of alpha-Synuclein is regulated by sumoylation. Acta Neuropathol 129:695–713. doi: 10.1007/s00401-015-1408-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Villarroya-Beltri C, Baixauli F, Mittelbrunn M, Fernandez-Delgado I, Torralba D, Moreno-Gonzalo O, Baldanta S, Enrich C, Guerra S, Sanchez-Madrid F. 2016. ISGylation controls exosome secretion by promoting lysosomal degradation of MVB proteins. Nat Commun 7:13588. doi: 10.1038/ncomms13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mathivanan S, Lim JW, Tauro BJ, Ji H, Moritz RL, Simpson RJ. 2010. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteomics 9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Li S, Li L, Li M, Guo C, Yao J, Mi S. 2015. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics 13:17–24. doi: 10.1016/j.gpb.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng Z, Hirai-Yuki A, McKnight KL, Lemon SM. 2014. Naked viruses that aren’t always naked: quasi-enveloped agents of acute hepatitis. Annu Rev Virol 1:539–560. doi: 10.1146/annurev-virology-031413-085359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKnight KL, Xie L, Gonzalez-Lopez O, Rivera-Serrano EE, Chen X, Lemon SM. 2017. Protein composition of the hepatitis A virus quasi-envelope. Proc Natl Acad Sci U S A 114:6587–6592. doi: 10.1073/pnas.1619519114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santiana M, Ghosh S, Ho BA, Rajasekaran V, Du WL, Mutsafi Y, De Jesus-Diaz DA, Sosnovtsev SV, Levenson EA, Parra GI, Takvorian PM, Cali A, Bleck C, Vlasova AN, Saif LJ, Patton JT, Lopalco P, Corcelli A, Green KY, Altan-Bonnet N. 2018. Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell Host Microbe 24:208–220. doi: 10.1016/j.chom.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kalamvoki M, Du T, Roizman B. 2014. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs. Proc Natl Acad Sci U S A 111:E4991–E4996. doi: 10.1073/pnas.1419338111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kalamvoki M, Deschamps T. 2016. Extracellular vesicles during herpes simplex virus type 1 infection: an inquire. Virol J 13:63. doi: 10.1186/s12985-016-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lippincott-Schwartz J, Freed EO, van Engelenburg SB. 2017. A consensus view of ESCRT-mediated human immunodeficiency virus type 1 abscission. Annu Rev Virol 4:309–325. doi: 10.1146/annurev-virology-101416-041840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Votteler J, Sundquist WI. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe 14:232–241. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patman G. 2014. Hepatitis: exosomal route of HCV transmission exists in patients. Nat Rev Gastroenterol Hepatol 11:704. doi: 10.1038/nrgastro.2014.179. [DOI] [PubMed] [Google Scholar]
- 43.Dogrammatzis C, Waisner H, Kalamvoki M. 2020. Cloaked viruses and viral factors in cutting edge exosome-based therapies. Front Cell Dev Biol 8:376. doi: 10.3389/fcell.2020.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mori Y, Koike M, Moriishi E, Kawabata A, Tang H, Oyaizu H, Uchiyama Y, Yamanishi K. 2008. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9:1728–1742. doi: 10.1111/j.1600-0854.2008.00796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deschamps T, Kalamvoki M. 2018. Extracellular vesicles released by herpes simplex virus 1-infected cells block virus replication in recipient cells in a STING-dependent manner. J Virol 92:e01102-18. doi: 10.1128/JVI.01102-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dogrammatzis C, Deschamps T, Kalamvoki M. 2019. Biogenesis of extracellular vesicles during herpes simplex virus type 1 infection: the role of the CD63 tetraspanin. J Virol 93:e01850-18. doi: 10.1128/JVI.01850-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muralidharan-Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G, D’Souza-Schorey C. 2009. ARF6-regulated shedding of tumor cell-derived plasma membrane microvesicles. Curr Biol 19:1875–1885. doi: 10.1016/j.cub.2009.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fonseca P, Vardaki I, Occhionero A, Panaretakis T. 2016. Metabolic and signaling functions of cancer cell-derived extracellular vesicles. Int Rev Cell Mol Biol 326:175–199. doi: 10.1016/bs.ircmb.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 49.Yang C, Robbins PD. 2011. The roles of tumor-derived exosomes in cancer pathogenesis. Clin Dev Immunol 2011:842849. doi: 10.1155/2011/842849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szilágyi JF, Cunningham C. 1991. Identification and characterization of a novel non-infectious herpes simplex virus-related particle. J Gen Virol 72:661–668. doi: 10.1099/0022-1317-72-3-661. [DOI] [PubMed] [Google Scholar]
- 51.McLauchlan J, Rixon FJ. 1992. Characterization of enveloped tegument structures (L particles) produced by alphaherpesviruses: integrity of the tegument does not depend on the presence of capsid or envelope. J Gen Virol 73:269–276. doi: 10.1099/0022-1317-73-2-269. [DOI] [PubMed] [Google Scholar]
- 52.McLauchlan J, Addison C, Craigie MC, Rixon FJ. 1992. Noninfectious L-particles supply functions which can facilitate infection by HSV-1. Virology 190:682–688. doi: 10.1016/0042-6822(92)90906-6. [DOI] [PubMed] [Google Scholar]
- 53.Liu X, Main D, Ma Y, He B. 2018. Herpes simplex virus 1 inhibits TANK-binding kinase 1 through formation of the Us11-Hsp90 complex. J Virol 92:e00402-18. doi: 10.1128/JVI.00402-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Graner MW. 2016. HSP90 and immune modulation in cancer. Adv Cancer Res 129:191–224. doi: 10.1016/bs.acr.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Ergun SL, Fernandez D, Weiss TM, Li L. 2019. STING polymer structure reveals mechanisms for activation, hyperactivation, and inhibition. Cell 178:290–301. doi: 10.1016/j.cell.2019.05.036. [DOI] [PubMed] [Google Scholar]
- 56.Shang G, Zhang C, Chen ZJ, Bai XC, Zhang X. 2019. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature 567:389–393. doi: 10.1038/s41586-019-0998-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang C, Shang G, Gui X, Zhang X, Bai XC, Chen ZJ. 2019. Structural basis of STING binding with and phosphorylation by TBK1. Nature 567:394–398. doi: 10.1038/s41586-019-1000-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deschamps T, Kalamvoki M. 2017. Evasion of the STING DNA-sensing pathway by VP11/12 of herpes simplex virus 1. J Virol 91:e00535-17. doi: 10.1128/JVI.00535-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng C. 2018. Evasion of cytosolic DNA-stimulated innate immune responses by herpes simplex virus 1. J Virol 92:e00099-17. doi: 10.1128/JVI.00099-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ni G, Ma Z, Damania B. 2018. cGAS and STING: at the intersection of DNA and RNA virus-sensing networks. PLoS Pathog 14:e1007148. doi: 10.1371/journal.ppat.1007148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ejercito PM, Kieff ED, Roizman B. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J Gen Virol 2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 62.Kalamvoki M, Roizman B. 2014. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc Natl Acad Sci U S A 111:E611–E617. doi: 10.1073/pnas.1323414111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kalamvoki M, Qu J, Roizman B. 2008. Translocation and colocalization of ICP4 and ICP0 in cells infected with herpes simplex virus 1 mutants lacking glycoprotein E, glycoprotein I, or the virion host shutoff product of the UL41 gene. J Virol 82:1701–1713. doi: 10.1128/JVI.02157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalamvoki M, Roizman B. 2008. Nuclear retention of ICP0 in cells exposed to HDAC inhibitor or transfected with DNA before infection with herpes simplex virus 1. Proc Natl Acad Sci U S A 105:20488–20493. doi: 10.1073/pnas.0810879105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hong YJ, Lim MS, Hwang SM, Kim TS, Park KU, Song J, and, Kim EC. 2014. Detection of herpes simplex and varicella-zoster virus in clinical specimens by multiplex real-time PCR and melting curve analysis. Biomed Res Int 2014:261947. doi: 10.1155/2014/261947. [DOI] [PMC free article] [PubMed] [Google Scholar]