Abstract
The novel coronavirus infectious disease-2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has traumatized the whole world with the ongoing devastating pandemic. A plethora of microbial domains including viruses (other than SARS-CoV-2), bacteria, archaea and fungi have evolved together, and interact in complex molecular pathogenesis along with SARS-CoV-2. However, the involvement of other microbial co-pathogens and underlying molecular mechanisms leading to extortionate ailment in critically ill COVID-19 patients has yet not been extensively reviewed. Although, the incidence of co-infections could be up to 94.2% in laboratory-confirmed COVID-19 cases, the fate of co-infections among SARS-CoV-2 infected hosts often depends on the balance between the host's protective immunity and immunopathology. Predominantly identified co-pathogens of SARS-CoV-2 are bacteria such as Streptococcus pneumoniae, Staphylococcus aureus, Klebsiella pneumoniae, Haemophilus influenzae, Mycoplasma pneumoniae, Acinetobacter baumannii, Legionella pneumophila and Clamydia pneumoniae followed by viruses including influenza, coronavirus, rhinovirus/enterovirus, parainfluenza, metapneumovirus, influenza B virus, and human immunodeficiency virus. The cross-talk between co-pathogens (especially lung microbiomes), SARS-CoV-2 and host is an important factor that ultimately increases the difficulty of diagnosis, treatment, and prognosis of COVID-19. Simultaneously, co-infecting microbiotas may use new strategies to escape host defense mechanisms by altering both innate and adaptive immune responses to further aggravate SARS-CoV-2 pathogenesis. Better understanding of co-infections in COVID-19 is critical for the effective patient management, treatment and containment of SARS-CoV-2. This review therefore necessitates the comprehensive investigation of commonly reported microbial co-pathogens amid COVID-19, their transmission pattern along with the possible mechanism of co-infections and outcomes. Thus, identifying the possible co-pathogens and their underlying molecular mechanisms during SARS-CoV-2 pathogenesis may shed light in developing diagnostics, appropriate curative and preventive interventions for suspected SARS-CoV-2 respiratory infections in the current pandemic.
Keywords: COVID-19, SARS-CoV-2, Microbial co-infections, Molecular pathogenesis
1. Introduction
The novel coronavirus infectious disease-2019 (COVID-19) is a rapidly transmissible pneumonia-like disease caused by the SARS-CoV-2 which emerged in Wuhan, China in December 2019, and is currently circulating throughout the world [1,2]. The positive-sense enveloped RNA virus (SARS-CoV-2) is genetically different from the previously known coronaviruses such as SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV) [[1], [2], [3]]. Immediately after its first outbreak inChina, this fearsome virus has emerged as one of the deadliest human pathogens in the last hundred years after the Spanish Flu in 1918–1920 [4,5]. The SARS-CoV-2 infection has become a public health challenge worldwide, and thus, the World Health Organization (WHO) has declared this disease as a public health emergency of international concern [1,2]. The deadly outbreaks of SARS in 2003 and MERS in 2012, with case fatality rate of 9.6% and 34.4%, respectively were successfully contained within six months. The COVID-19 disease affected total 217 countries and territories until April 7, 2021, and more than 133,688,126 cases have been confirmed globally with 2,901,038 deaths (https://www.worldometers.info/coronavirus/?utm_campaign=homeAdvegas1?). Therefore, this quickly spreading COVID-19 pandemic highlights the critical need for rapid development of vaccines and antiviral treatments to reduce the number of hospitalizations and deaths by this worrisome pathogen [6].
Co-infections and superinfections are common in many respiratory viral infectious diseases [6,7]. Bacterial co-infections can significantly increase the mortality rate in patients infected with any viral infections [6,8,9]. Previously, bacterial co-infections were also reported in MERS-CoV patients receiving intensive care [10]. The co-infection of the SARS-CoV-2 with other microorganisms is a very important factor in COVID-19 pathogenesis that may complicate the accurate diagnosis, treatment, prognosis of COVID-19, and even increase the mortality rates [11]. Clinical trials and metagenomic investigations indicated the co-presence of other viruses, bacteria, archaea and fungi with SARS-CoV-2 in COVID-19 patients [[11], [12], [13]]. About 50% of the patients who died of COVID-19 had secondary bacterial infections [12,14] which further intensifies the pathophysiological progression of COVID-19. Better understanding of co-infections in COVID-19 is critical for the effective patient management, treatment and containment of SARS-CoV-2. It is therefore, necessary to strengthen the investigation of the co-infection in COVID-19 patients. Moreover, regarding COVID-19 several issues such as useful strategies to prevent disease spread, collection of appropriate clinical specimens, transmission route, viral dynamics and effective drug treatments are still largely unknown. However, the possibility of co-infections with other respiratory pathogens including bacteria, archaea, viruses (other than betacoronavirus) and fungi are not yet clearly understood. The association of these secondary pathogens to causing co-infections should be an important concern for the clinician in the management of COVID-19 cases. The emerging infectious diseases section of the Centers for Disease Control and Prevention (CDC, USA) endorsed testing for other respiratory pathogens, suggesting that evidence of another infection could aid the evaluation of patients with potential COVID-19 in the absence of widely available rapid testing for SARS-CoV-2 [15].
The two earlier coronaviruses (SARS-CoV-1 and MERS-CoV), influenza virus, and SARS-CoV-2 show highly similar respiratory symptoms, including high fever, cough, headache and even pneumonia [[16], [17], [18], [19]]. Recent clinical and in silico studies showed that viral co-infections mainly includes respiratory viruses such as entero/rhinovirus (hRV), human metapneumovirus (hMPV), Respiratory Syncytial Virus (RSV), Siphovirus, Alphapapillomavirus, Myovirus, Tombusvirus, Victorivirus, Partitivirus, Chrysovirus, Totivirus, and other coronaviruses (non-SARS-CoV-2) [11,20]. Concurrent co-infections in COVID-19 can also change the respiratory microbiome homeostasis, and thus triggers the infection and stimulates immune cells to produce more severe inflammation [11,20]. The gut bacterial diversity of the COVID-19 patients is also reduced with the increased relative abundance of opportunistic pathogens, and the lower relative abundance of the beneficial symbionts [21]. Recent metagenomic studies reveal the concurrent association of bacteria, archaea and non-COVID viruses in nasal swabs of COVID-19 patient [11]. During the earlier epidemics of SARS-CoV-1 and MERS-CoV, patients receiving invasive mechanical ventilation were easily developed bacterial co-infections, and had higher mortality rates [16,18]. Therefore, bacterial co-infections might be a key element that promotes severities of the COVID-19 and mortality rates [22]. Recent study on deceased patients showed that sepsis (100%) acted as one of the main complications [23], indicating that co-infection is of great importance to prognosis and subsequent treatment of COIVD-19 patients. Furthermore, co-infection has been associated with more severe outcomes in pandemic and seasonal influenza [24]. It has been suggested that influenza‐related bacterial infections overall may account for up to 30% of community-acquired pneumonia (CAP) cases [25]. Several studies of hospitalized patients with COVID-19 noted the empiric use of antibiotics in majority of patients [24,26,27]. However, there is an evidence of increased inflammatory serological markers associated with bacterial infections including procalcitonin and C-reactive protein in patients with COVID-19 without a corresponding bacterial co-infections [24,28]. Meanwhile, several descriptive studies showed that the ecosystem of commensal microbiota can both regulate and be regulated by invading viruses, facilitating either stimulatory or suppressive effects [23,29,30]. More importantly, the coinfected microorganisms may also be a new strategy for the development of new treatment of SARS-CoV-2 infection. Despite increasing evidence for its salience to COVID-19 outcomes, the effect of co-infection clearance on SARS-CoV-2 load has not yet been systematically reviewed or critically discussed. This systematic review updates our knowledge on the microbial co-infections associated with SARS-CoV-2 pandemic, and the possible molecular mechanisms of co-infections in COVID-19 to emphasize that microbial co-infections.
2. Rationale and review methodology
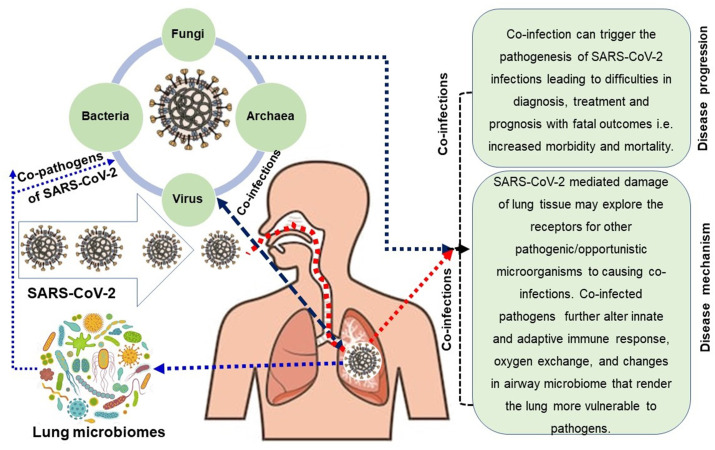
To date, thousands of reports on genomics, origin, genome evolution, molecular diagnosis and vaccine and/or therapeutics of SARS-CoV-2 have been published. However, a comprehensive review on microbial (virus, bacteria, fungus, archaea) co-infections associated with COVID-19 and the impact of co-infections on COVID-19 patients, characterization of co-pathogens, and their underlying molecular mechanisms in COVID-19 patients are lacking. Therefore, we conducted a rigorous literature survey on the co-infections, identifying of co-infecting microorganisms and their pathogenesis. The concept and evidence of co-infection with COVID-19 disease and a rationale of this comprehensive review are described in the introduction section. Later sections of this review were arranged coherently from the literature available in the PubMed central, Google Scholar, ResearchGate, bioRxiv, medRxiv, Preprints archives, World Health Organization (WHO) COVID-19 blog, National Institute of Health (NIH), Centers for Disease Control and Prevention (CDC, USA), Clinical Trials Registry databases, and COVID-19 vaccine and therapeutics tracker (https://biorender.com/covid-vaccine-tracker). The original research articles that discussed the evidence and significance of co-infections amid COVID-19, detection and possible molecular mechanisms of co-infections were considered for the content of this review. This literature survey also included case studies, case series and observational studies published from the very beginning of COVID-19 outbreak in Wuhan city, China in late December 2019 to April 7, 2021. The literature search was done through screening of titles, abstracts and full articles for eligibility. Proposed molecular mechanisms of co-infections concurrent in SARS-CoV-2 infections have been represented in Fig. 1 .
Fig. 1.
Inter-relationship between SARS-CoV-2 and respiratory microbiomes leading to co-infections in CoVID-19 patients. COVID-19 patients can be co-infected with different microbial domains including viruses (other than SARS-CoV-2), bacteria, archaea and fungi. These diverse microbial communities concurrently complicate the pathophysiology and disease progression of SARS-CoV-2 infections.
3. Microbial co-infections in hospitalized patients with COVID-19
Co-infection refers to the concurrent infection of a cell or host by two or multiple pathogen species and/or strains, whereas, superinfection is a scenario where one pathogen infects the host some time before infection by the second pathogen [31]. For both of these cases, the fate of the infected host often depends on a balance between the host's protective immunity and immunopathology [32]. The universal pervasiveness or incidence of co-infection among humans is unknown, but it is thought to be commonplace, sometimes more common than single infection [33]. Co-infecting pathogens can alter the population of the primary pathogen, as for example, Van der Hoek et al. (2004) reported that in respiratory co-infections, the human coronavirus (hCoV) load was much lower than for a single infection [34]. Co-infections may occur by multiple infectious agents of viral, bacterial, archaeal and fungal origin (Fig. 1), and appear to occur simultaneously with the initial onset of illness [35]. Co-infection morbidity has previously been studied within certain cohorts (e.g., age and sex), and is often reported to be worse than single infections [36]. Recently, several observational and cohort studies reported that pulmonary complications occurred in 51·2% COVID-19 patients, of which 82.6% accounted for deaths, and independent risk factors for mortality were male sex, age 65 years or older [37]. However, the occurrence of co-infection in death across age and sex cohorts of COVID-19 patients has not been studied yet. Co-infections in COVID-19 may not accounted for by age group, and unexplained heterogeneity in COVID-19 severity may be due to differences between studies in disease severity, patient comorbidities, treatment differences (such as corticosteroid administration), use of antibiotics prior to and during hospitalization, or other unidentified covariates. However, underestimation of secondary bacterial or fungal infections developing later in the course of COVID-19 is likely.
3.1. Viral co-infections in hospitalized patients with COVID-19
Co-infections with other viruses are very common in the viral infections of respiratory diseases. The prevalence of respiratory viral co-infection varies from 3.0% to 68.0% [20,38]. Several clinical studies indicated that viral co-infections of SARS-CoV-2 occurred with other virus from different countries [23,26]. Lin et al. (2020) reported that in Shenzhen Third People's Hospital, 3.2% SARS-CoV-2 patients suffered from viral co-infections [20]. Association of other viruses, bacteria, fungi, with SARS-CoV-2 infection has been reported [13]. Bacteriophages are naturally occurring viruses that use bacteria as hosts, and play an extremely important part in allowing relatively harmless bacteria to become pathogens [19,39]. The bacteriophages are overlooked human pathogens that imply in triggering and worsening of a number of human diseases [19]. The COVID-19 causing SARS-CoV-2 strains show neighboring relationship to human classic coronavirus, the SARS coronavirus isolate Tor2 (SARS-CoV Tor2) corroborating with the recent findings [40]. In a recent metagenomic study, Hoque et al. (2020) reported that COVID-19 samples have sole association with 16 viral genera (other than betacoronavirus), and of them, Tombusvirus, Victorivirus, Partitivirus, Chrysovirus and Totivirus were the most abundant genera associated with SARS-CoV-2 co-infections [11].
3.1.1. SARS-CoV-2 and influenza: the disarray
Influenza is an acute and highly contagious respiratory disease that is responsible for significant morbidity and mortality worldwide. Annually, influenza can affect approximately 9% of the world's population, with up to 1 billion infections, 3 to 5 million severe cases, and 0.3 to 0.65 million deaths [41]. The current COVID-19 pandemic caused by SARS-CoV-2 demonstrates similar symptoms of influenza such as fever, headache, sore throat and so on. Although, at the early stage of COVID-19 pandemic, the co-infection of SARS-CoV-2 and influenza did not draw any significant attention, the first report of influenza and SARS-CoV-2 co-infection [42] indicated that only 0.4% influenza positive patients were infected with SARS-CoV-2 in January 2020. The incidence of co-infection by influenza viruses in COVID-19 patients have been reported frequently from different countries other than China [27]. A case report of a 78-year old woman in Japan, showed the presence of Influenza A co-infection with SARS-CoV-2 [43] which intensified the necessity for specific and accurate detection of etiologic agents considering the travel history and medical condition of the patients. Another study showed 4.35% presence of influenza co-infection in hospitalized COVID-19 positive patients [44]. Although no anomalies in hematology screening were reported, all coinfected patients were clinically cured after treatment with oxygen inhalation, oseltamivir and antimicrobial agents without any invasive ventilator, intensive care unit (ICU) care and extracorporeal membrane oxygenation treatment [44]. In another retrospective study in Jiangsu Province in China demonstrated that 94.2% of laboratory-confirmed COVID-19 patients had co-infection, specifically 31% of them had viral co-infections with influenza virus [45]. In a recent retrospective cohort study on COVID-19 patients in a hospital in Barcelona showed that only 0.4% patients were co-infected with community-acquired influenza A virus, whereas 3.1% patients infected with community-acquired bacterial infections, 4.7% of which were hospital-acquired superinfections [46]. However, the concomitant outbreak of influenza and COVID-19 in the winter season confirmed the presence of co-infection in multiple cases and inevitably emphasized the simultaneous laboratory diagnosis facilities for both SARS-CoV-2 and influenza viruses. The amphipathic symptoms of both COVID-19 and typical influenza have necessitated the co-diagnosis of influenza in COVID-positive patients, specifically with recent travel in influenza-endemic areas.
3.1.2. Hepatitis-B co-infections in COVID-19 patients
Hepatic complications associated with COVID-19 are particularly concerning among people living with hepatitis B virus (HBV) co-infection with pre-existing liver complications (e.g., cirrhosis, liver failure, hepatocellular carcinoma) [47,48]. Co-infections among COVID-19 patients with hepatitis symptoms before developing the respiratory syndromes have been documented in several studies [49,50]. While our understanding of SARS-CoV-2's pathogenesis continues to grow, initial studies suggest that the virus could lead to liver injury mainly by binding to angiotensin-converting enzyme 2 (ACE2) receptors on hepatocytes or causing an immune-mediated hepatic injury through activation of cytokine storm [51]. It is now clear that COVID-19 could lead to liver injuries and elevate alanine aminotransferase (ALT), aspartate aminotransferase (AST), and total bilirubin, particularly among the severe COVID-19 cases who need ICU [[52], [53], [54]]. Abnormal liver functions in COVID-19 patients have also been associated with increased disease severity and risk of mortality. Interestingly, viral co-infections caused by non-respiratory infectious agents have been reported from the Wuhan city in China, which stated the co-incidence of hepatitis B virus infections in 12.2% patients with acute COVID-19 [23]. The significant rate of liver cirrhosis and abnormally higher liver functions in severe COVID-19 patients corroborated with the findings of co-infection with hepatitis B including elevated ALT, AST, gamma-glutamyl transferase (GGT) levels accompanied by moderately elevated prothrombin time (PT), and total bilirubin (TB) levels in COVID-19 patients [55,56]. The possibilities of liver impairment in severe COVID-19 patients have been suggested due to viral tropism to hepatic tissues, drug toxicity and systemic inflammation [54]. Therefore, extensive screening for hepatitis B infections in critical COVID-19 patients can be more useful for disease progression analysis, and effective treatment plan application in patients with remarkable diagnostic indications of abnormalities in liver functions.
3.1.3. SARS-CoV-2 and dengue: the deadly duo
The spread of SARS-CoV-2 in temperate countries such as Switzerland and France, where arboviral dengue fever is endemic, has been described with a contemporary travel history [57]. In France, the first diagnostic test of SARS-CoV-2 RT-PCR revealed negative with a flu-like syndrome in patients, which became positive a week later with severe clinical onsets of COVID-19 symptoms like fever, fatigue, loss of appetite and diarrhea [57]. The appearance of diffuse maculopapular exanthema made the clinicians to screen for Leptospira spp., Rift Valley fever virus, dengue virus and Chikungunya virus infections which indicated that RT-PCR for type-1 dengue virus was positive. Another report of two patients from Singapore revealed that rapid serological tests for dengue can generate false positive results accelerating the respiratory complications in patients who later became positive for SARS-CoV-2 [58]. The initially worsening fever, increasing thrombocytopenia and sero-positivity for dengue misguided the clinicians who lately found the absence of dengue specific immunoglobulins, but with positive results from RT-PCR test of nasopharyngeal swab for SARS-CoV-2. In several countries of South America, a significant numbers of dengue cases were reported along with a gradual increase of COVID-19 cases. A study from Brazil depicted the possibility of under-reporting of dengue cases due to extensive mobilization of epidemiological sero-surveillance response team for COVID-19 emergency response, which may indirectly affected the reporting and treatment of dengue during COVID-19 outbreak [59]. That study urged the robust integrated national strategy for combined surveillance, treatment and prevention plan for dengue and COVID-19 management in Brazil. Another study from Columbia analyzed the dual epidemiological features of dengue and SARS-CoV-2 in the first 20 weeks of COVID-19 pandemic [60]. The viral interference resulting in blocking entry and replication of dengue during SARS-CoV-2 infection may also have contributed to the decreased onset of clinical dengue in subclinically or, mildly COVID-19 infected population [61]. Contemporary reports from Ecuador revealed that the eco-epidemiological dynamics and high endemic-epidemic transmission of dengue in large coastal areas can drastically affect the mitigation campaign and COVID-19 containment measures through extensive loads of patients on public health facilities for diagnosis, treatment and preventive actions [62]. These reports indicated the urgency of sero-surveillance for dengue virus infections in dwellers and travelers from temperate endemic areas. It has also been stated that the physical distancing and reduced public mobility may contribute to the containment of dengue infections amid COVID-19 [63].
3.1.4. COVID-19 patients with chronic viral diseases: HIV and HCV
The triple burden of COVID-19, human immunodeficiency virus (HIV) and hepatitis C virus (HCV) is one of the major and persistent global health challenges of the twenty-first century. The HIV, HCV and newly emerging infectious diseases such as coronavirus epidemics are expected to overlap in high HIV and HCB or HCV burden countries [64]. How COVID-19 will manifest itself in persons co-infected with HIV/HCV is still unclear [64]. Populations infected with HIV and HCV may be at elevated risk for severe responses if they are infected with COVID-19. In the future, lung lesions associated with COVID-19 may increase the risk of HIV or HCV, which induces a truly vicious circle of HIV-HCV-COVID-19 co-infections [64,65]. While COVID-19 continues to spread across the world, many areas face the risk of infection with SARS-CoV-2 and the obstacles and challenges to sustaining the continuum of HIV and HCV treatment in high-burden HIV/HCV countries are increasing [64]. In fact, the pathogenicity of COVID-19 could be accelerated in people living with HIV, who have compromised immunity [65]. Recent evidence has indicated a substantial association between coronavirus-related lower respiratory tract infections (LRTIs) and increased risk of death in immuno-compromised individuals [64]. Co-infections of HIV and SARS-CoV-2 in five individuals—three male and two transgender patients in Spain has been reported [32]. Interestingly, the HIV/SARS-CoV-2 patients had similar clinical, laboratory and radiographical features to the HIV-negative patients infected with SARS-CoV-2. During the current SARS-CoV-2 pandemic, this lack of information is a concern in countries with high HIV cases, especially in Sub-Saharan Africa, where 70% of people living with HIV infection [32]. As the patient had the history of co-infection with HIV-1 and HCV before 4 years, the follow-up study of anti-SARS-CoV-2 immune response revealed the delayed antibody response but with repeatedly negative RT-PCR test for SARS-CoV-2 RNA [66]. However, the compromised immune status of the patient caused the delayed humoral response development against the SARS-CoV-2, but the anti-HIV therapeutics and elevated level of activated IFN-γ due to anti-HIV agents may be suppressed to SARS-CoV-2 infection leading to persistently undetectable RNA in RT-PCR tests [66]. Therefore, history of viral co-infection with immuno-compromised status and antiviral therapeutics may lead to delayed antibody response along with indistinct COVID-19 diagnosis.
3.1.5. SARS-CoV-2 with hCoV-HKU1
The endemic human coronaviruses (hCoVs) have been known to cause co-infections, sequential infections or can be co-detected with each other or with other respiratory viruses, including influenza A/B, Respiratory Syncytial Virus (RSV), metapneumovirus, enterovirus, and adenovirus [67]. A critical case of co-infection by human coronavirus HKU1 in a COVID-19 patient, reported from Indonesia, indicated the sequential infections by hCoV-HKU1 and SARS-CoV-2, which was confirmed by a FilmArray Respiratory Panel (RP) test [67]. Thus, clinicians need to be aware of hCoV co-infections among COVID-19 patients. A high degree of suspicion in this rapidly evolving outbreak is required to make the diagnosis, and thereby, to contain and control the spread of the COVID-19.
3.2. Bacterial, archaeal and fungal co-infections in hospitalized patients with COVID-19
Like other well studied respiratory viral infections including the 1918 influenza outbreak [4], and 2009 H1N1 pandemic [68], the current pandemic outbreak of SARS-CoV-2 is also reported to associate with secondary microbial infections [11,12,69]. Bacterial co-infections were previously reported for respiratory diseases including SARS-CoV, MERS-CoV and influenza patients receiving intensive care [10,70]. Several retrospective studies showed that during the 1918 Spanish flu pandemic, bacterial pneumonia was a major cause of morbidity and mortality [71]. In a previous study, the prevalence of bacterial co-infections during the pandemic of influenza A (H1N1) between 2009 and 2012 was 23.0% [68]. Recent study reported that about 65% of laboratory-confirmed cases of influenza infection are known to be complicated by bacterial co-infections [72].
Bacterial co-infections develop in patients amid or after the primary infection initiated by an infectious agent. Bacterial co-infections also play a significant role during COVID-19 and are associated with an increasing rate of disease severity and case fatality [35]. Nevertheless until now, the incidence of bacterial co-infections in patients admitted to the ICU for acute respiratory failure associated with SARS-CoV-2 pneumonia is poorly studied [73,74]. SARS-CoV-2 primarily enters human body through nasopharyngeal tract and then gradually move to lung to initiate infection. The pathophysiology of COVID-19 can be attributed to aberrant immune responses in clearing the virus [75,76]. Given the unequivocal association between viral and bacterial co-infection and respiratory disease severity, there is a pressing need to better understand how interactions of the SARS-CoV-2 with the host microbiome in the respiratory tracts correlate with viral infections that facilitate opportunistic co-infections [77]. However, little is known the outcome of the interactions of SARS-CoV-2 with nasal commensal bacteria which is thought to be critical for ultimate severity of COVID-19 disease development. The prevalence rate of bacterial co-infections in critically ill hospitalized COVID-19 patients was around 14% revealed by a meta-analysis [78]. A recent North American study reported 41% prevalence of bacterial co-infections among 17 COVID-19 patients admitted to ICU [79]. The incidence of co-infection associated with bacterial pneumonia ranged between 11% and 35% among the patients who had been infected with respiratory viruses [72]. Recently, Fu et al. (2020) reported that among ICU admitted COVID-19 patients, 13.9% were suffering from bacterial co-infections. Despite having a varying rate of bacterial co-infections among COVID-19 patients, the rate of prevalence could be as high as 50% among the non-survivors [80]. A series of retrospective case studies on SARS-COV-2 confirmed that severely and non-severely ill patients had 7.7% and 3.2%, bacterial and fungal co-infections, respectively [54]. In Italy, a study conducted among 16,654 patients with critical condition, who died of SARS-CoV-2 infection depicted that 11% of those cases were associated with bacterial and fungal co-infections [81].
Co-infections with Streptococcus pneumoniae, Staphylococcus aureus, or other colonizing bacteria during the pathophysiology of COVID-19 impairs both innate and adaptive antibacterial host defenses and temporarily compromise the physical and immunological barrier to cause secondary bacterial pneumonia, leading to severe and deadly disease in people with pre-existing comorbidities and previously healthy people [82]. Data regarding bacterial co-infections in COVID-19 pneumonia are still emerging, but an association has been made between the detection of bacterial pathogens in samples with disease severity in COVID-19 patients. The most commonly identified coinfected bacterial pathogens include, Acinetobacter baumannii, Klebsiella pneumoniae, Mycoplasma pneumoniae, Legionella pneumophila, Streptococcus pneumoniae, and Clamydia pneumoniae [11,70,83,84], while Aspergillus flavus, Candida glabrata, and Candida albicans are the most common coinfected fungi [78]. In addition, bacterial pathogens such as Staphylococcus aureus, Haemophilus influenzae, Klebsiella pneumoniae, Streptococcus pneumoniae, Neisseria meningitides [83,84] as well as some genera of Proteus, Enterobacter, and Citrobacter species have also been reported in hospitalized COVD-19 patients. In a recent microbiome study, Hoque et al. reported 527 and 306 bacterial genera in COVID-19 patients of Bangladesh and China, respectively [11]. Moreover, Pseudomonas aeruginosa and E. coli are the most frequently isolated multi-drug resistant (MDR) pathogens to be associated with hospital acquired superinfections [46]. Remarkably, SARS-CoV-2 RNA has also been detected in fecal samples of COVID-19 patients. It raises the question of gastrointestinal infection of SARS-CoV-2 and a possible fecal-oral route of disease transmission [85,86]. Moreover, high expression levels of ACE2 mRNA in the gastrointestinal system revealed a strong interaction of SARS-CoV-2 with the gastrointestinal system that has high microbiome diversity and possible chances of immune suppression and bacterial co-infections [85,86]. The role of SARS-CoV-2 in modulation of microbiome diversity in the gastrointestinal tracts is an important question for further research. The infection of SARS-CoV-2 of intestinal cells can lead to the change of intestinal microbiota. Gu et al. reported that the gut bacterial community of COVID-19 patients significantly reduced compared to those of healthy humans [87]. This dysbiosis (loss of beneficial commensals) was reflected by the significantly higher abundance of opportunistic pathogens such as Streptococcus, Rothia, Veillonella and Actinomyces in COVID-19 patients, while the relative of beneficial symbionts abundance, such as Blautia, Romboutsia, Collinsella, and Bifidobacterium remained much lower [87].
A study on hospitalized COVID-19 patients with oropharyngeal candidiasis (OPC) showed that C. albicans was found to be the most prevalent pathogen, which was counted for 70.7%, followed by other fungi including C. glabrata (10.7%), C. dubliniensis (9.2%), C. tropicalis (3%), and C. krusei (1.5%) [88]. On the other hand, Chen et al. reported 5% prevalence of fungal co-infections in 99 COVID-19 patients in China, including one case of Aspergillus flavus, one case of Candida glabrata and three cases of C. albicans [23]. Yang et al. found that 5.8% (3/52) of the critical COVID-19 patients had fungal co-infections with A. flavus, A. fumigatus and C. albicans [89]. In addition, 8–15% incidence of non-specific co-infections among COVID-19 patients were reported in different studies from China, but it is not clear whether it is bacterial or fungal infections [52,90]. A significant percentage of the SARS-CoV-2 infected patients developed co-infections associated with MDR typically from nosocomial pathogens [91]. An appreciable minority cases of superinfection, most commonly pneumonia and bacteremia can be developed due to MDR bacterial pathogens and fungus specially Aspergillus spp. Fungal originated co-infections including pulmonary aspergillosis and candidiasis were reported to complicate SARS-CoV-2 infection [46]. To date COVID-19-associated pulmonary aspergillosis (CAPA) has been documented in >30% of the cases [78].
Until now, most of the reported respiratory tract co-infections are limited to viral, bacterial, and fungal pathogens [5,78,90,92], while a plethora of other concomitant microbial components including archaea could also be found [11,93]. Unlike bacteria, the incidence, diversity and composition of these co-pathogens always remain much lower compared to the infectious agent of COVID-19. Metagenomic investigations confirmed presence of Methanosarcina, Methanocaldococcus, Thermococcus, Methanothermobacter, Haloarcula, Staphylothermus, Natronomonas, Ferroglobus, Caldivirga, Halobacterium, Natrialba, Methanosphaerula and Picrophilus as the archaeal genera in samples of COVID-19 patients [11,93].
4. Molecular mechanism of co-infection in COVID-19
Co-infections can augment the pathogenesis, morbidity and mortality in most of the respiratory viral diseases [35]. Table 1 discusses the commonly reported microbial co-pathogens amid COVID-19, their transmission pattern, possible mechanism of co-infections and outcomes. Co-infections in COVID-19 patients may also complicate the clinical outcomes of the disease. The SARS-CoV-2 enters human cells by binding to the ACE2 protein of the cells lining the upper and lower airways. Recently, Lee et al. reported that the ACE2 receptor protein robustly localizes within the motile cilia of airway epithelial cells, which likely represents the initial or early subcellular site of SARS-CoV-2 viral entry during host respiratory transmission [94]. However, the ciliary ACE2 expression in the upper airway is influenced by patient demographics (age, sex and smoking), clinical characteristics, comorbidities/co-infections or medication use [94]. Remarkably, specific molecular kinetics of these additional infections in COVID-19 patients are still remained unclear although a few studies proposed some models for the co/superinfections in COVID-19 patients in different countries [26]. Based on available literature, we propose a plausible mechanism of co-infection in COVID-19 patients (Fig. 1, Table 1).
Table 1.
Commonly reported microbial co-pathogens amid COVID-19, their transmission pattern along with the possible mechanism of co-infections and outcomes.
| Type of co-infection | Co-pathogens | Route of transmission | Person to person transmission | Possible mechanism of co-infection and pathogenesis | Possible outcomes |
|---|---|---|---|---|---|
| Viral | Influenza | Respiratory | Yes | IFN induced overexpression of ACE2 triggered by influenza virus aids SARS-CoV-2 infection [112]. | Influenza co-infection can provoke COVID-19 hyper-inflammatory states. Higher incidence of acute cardiac injury was reported [113] |
| HBV | Body fluid | Yes | Increased liver tissue damage and inflammatory responses due to COVID-19 may aid HBV co-infection by overexpressing host cell receptors [114]. It may also fuel the reactivation of pre-existing chronic HBV [115]. | Elevation of ALT, AST, TBIL, ALP, and γ-GT. [116] Higher risk of liver injury. [117] | |
| Dengue | Mosquito bite | No | NR | Increase the severity of symptoms [118]. Decrease in white blood cell, neutrophils, lymphocytes and platelets count and eventual higher mortality rate [119] | |
| HIV | Body fluid | Yes | Suppression of T lymphocyte mediated immunity (as observed in HIV patients) leads to the prognosis of increased disease severity and higher mortality rate during COVID-19 co-infection [120]. | HIV Patients under ART exhibits mild COVID-19 symptoms. But ART-naïve patients show acute COVID-19 clinical representation [121]. Higher maximum body temperatures, longer duration of fever and longer improvement time of chest CT image was reported due to co-infection [122] | |
| HCV | Body fluid | Yes | Both SARS-CoV-2 E and HCV p7 proteins can form similar ion channels which ensure their success in attacking their host and effective replication during co-infection [123]. | The actual outcome is not reported till date. It has been speculated that some investigational COVID‐19 drugs may adversely affect the HCV‐related decompensated cirrhosis patients [124]. | |
| Rhinovirus | Respiratory | Yes | Major disease-causing rhinovirus serotype HRV-A16 infection upregulates ACE2 and TMPRSS2 expression in epithelial cells by inducing IFNb1. This event facilitates SARS-CoV-2 transmission and further disease severity [125] | One case has been reported in a young patient expressing critical illness as the outcome of co-infection [126] | |
| Adenovirus | Respiratory | Yes | Similar ion channel forming capability of SARS-CovV-2 E and Adenovirus 6K proteins facilitates co-infection [123] | Unfavorable prognostic outcome including ARDS [127] | |
| Bacterial | Streptococcus pneumoniae | Respiratory | Yes | Opportunistic normal flora of human upper respiratory track | Severe respiratory distress followed by pleural effusion and necrotizing pneumonia [128], higher mortality rate [129] |
| Staphylococcus aureus | Respiratory/Digestive/Contact | Yes | Opportunistic normal flora of human upper respiratory track, gut mucosa and skin | Necrotizing pneumonia [130]. Bacteremia and higher mortality [131] |
|
| Pseudomonas aeruginosa | Contact | Yes | Opportunistic pathogen causing HAI mostly related with poor hygiene, mechanical ventilation and urinary catheterization. | NR | |
| Acinetobacter baumannii | Contact | Yes | Mechanical ventilation | NR | |
| Klebsiella pneumoniae | Respiratory/Contact | Yes | Opportunistic normal flora of human mouth, skin, and intestines | Fatal sepsis [132] | |
| Mycoplasma pneumoniae | Respiratory/contact | Yes | NR | Severe pneumonia [133]. Increased morbidity, mortality and disease severity [134] | |
| Clamydia pneumoniae | Respiratory/contact | Yes | NR | Severe pneumonia [133]. | |
| Legionella pneumophila | Digestive/Respiratory | Yes | NR | Elevated aspartate aminotransferase, blood urea nitrogen, creatinine, lactate dehydrogenase and C-reactive protein [135] | |
| Haemophilus influenzae | Respiratory/contact | Yes | Opportunistic normal flora of human upper respiratory track | NR | |
| Neisseria meningitides | Respiratory/contact | Yes | NR | Convulsion [136], elevated C-reactive protein, headache, neck stiffness, rigors, confusion, and a new purpuric rash over hands and feet [137] | |
| Mycobacterium tuberculosis | Respiratory | Yes | Cytokine storm produced by COVID-19 may reactivate latent TB or boost the development of active TB. Lung damages caused by TB may also escalate the disease severity caused by SARS-CoV-2 [138]. | Co-infection is associated with disease severity and disease progression rate [139]. 2.17 times higher risk-of-death and 25% lower risk-of-recovery was reported. Also shorter time-to-death and longer time-to-recovery was found [140]. | |
| Fungal | Aspergillus spp. | Respiratory | No | Pro-inflammatory cytokines (especially IL-6 and IL-10) released during COVID-19 results in tissue necrosis and ARDS, which eventually makes patient more vulnerable to Aspergillosis [141]. | Invasive pulmonary aspergillosis, higher case fatality rate (64.7% reported) [141] |
| Candida spp. | Perinatal/Contact | No | Opportunistic pathogen found in human skin. | Candidemia and increased mortality rate [142]. |
IFN: Interferon; ACE2: Angiotensin-converting enzyme 2; SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2; COVID-19: Coronavirus disease 2019; HBV: Hepatitis B Virus; HIV: Human Immunodeficiency Virus; HCV: Hepatitis C Virus; ALT: Alanine transaminase; AST: Aspartate transaminase; TBIL: Total bilirubin; ALP: Alkaline phosphatase; γ-GT: Gamma-glutamyl transferase; ART: Antiretroviral therapy; CT: Computed Tomography; HRV-A16: Human rhinovirus A16; TMPRSS2: Transmembrane protease, serine 2; IFNb1: Interferon Beta 1; ARDS: Acute respiratory distress syndrome; HAI: Hospital Acquired Infections TB: Tuberculosis; IL-6: Interleukin 6; IL-10: Interleukin 10; NR: Not Reported.
Respiratory viruses are frequently collaborated by secondary bacterial infections due to the outgrowth of opportunistic bacterial pathogens. Although the specific molecular mechanisms of co-infections in COVID-19 patients remain unclear, it may include virus-induced airway damage, cell loss, goblet cell hyperplasia, altered mucus secretion, reduced ciliary beat frequency, function and clearance, reduced oxygen exchange, and damage to the immune system [27,45,95]. Co-infection increases the levels of C-reactive protein (CRP) and procalcitonin (PCT) [5]. Viral infections damage the respiratory airway both histologically and functionally [69,96]. The co-infection mechanisms include virus-induced airway damage, cell loss, goblet cell hyperplasia, altered mucus secretion, reduced ciliary beat frequency, reduced mucociliary clearance, dis-coordinated mucociliary functions, reduced oxygen exchange, and damage to the immune system [69,96,97]. Viral co-infection can also facilitate bacterial adhesion, disrupt the tight junction and epithelial barrier integrity favoring paracellular transmigration of bacteria, and alter both innate and adaptive immune responses that render the lung more vulnerable to SARS-CoV-2 infections [20,38]. Many viruses can destroy the airway epithelium which facilitates other viral co-infection [98]. The COVID-19 patient having co-infected with HIV had a longer progression of the disease and slower generation of specific antibody because of the collapse of immune system [99]. SARS-CoV-2 infection may cause liver damage [5], and thus, drug-induced liver injury (DILI) is more likely to occur in patients who already have certain viral infections including HCV and HIV [23]. Therefore, the development and outcome of SARS-CoV-2 associated co-infections with other viruses are highly dependent on the host immune response, especially in the elderly [23]. The co-infection of viruses is associated with different molecular mechanisms by which the predisposition of the virus occurs in the respiratory tract that promotes simultaneous bacterial infection. The epithelial cells of the respiratory tract help bacterial adherence using different mechanisms during viral infection, while the disease severity varies upon virus, bacterial strain, other co-pathogens and hosts immunity. Respiratory viruses can up-regulate the expression of host cell membrane protein to facilitate their binding [69]. Respiratory Syncytial Virus (RSV) reported to bind directly with Haemophilus influenzae and Staphylococcus pneumonia, and thus, favoring bacterial proximity to the epithelial monolayer and supplementing attachment to the host cell receptors. Moreover, the expression and localization of the RSV glycoprotein on the host cell membrane during infection can further act as bacterial receptors for pneumococcal binding [100]. Previous studies demonstrated that influenza virus can make mice susceptible to pneumonia caused by S. aureus where both virus and bacterial load increased during co-infection [100,101]. Several respiratory viruses such as RSV, parainfluenzavirus-3, and influenza viruses reported to increase the bacterial adherence upon infection, in both primary and immortalized epithelial cells [69]. The surface glycoprotein adhesion molecule-1 (ICAM-1) expression is upregulated during RSV and adenovirus infection, and thereby, increases adherence for S. pneumoniae in human nasopharyngeal cells (HEp-2) and pneumocyte type II cells (A549). The enhanced pneumococcal adherence in epithelial cells results in bacterial accumulation which may facilitate other bacterial co-infections [69,102]. Methanogenic archaea coexist and interact closely with anaerobic bacteria [39]. Methanogenic archaea utilize low molecular weight compounds, such as H2 + CO2, formic acid, or acetate, and therefore, have symbiotic relationships with the producers of these substrates. It is reasonable to assume that the presence or increase in level of methanogenic archaea modulates the composition of the polymicrobial community and changes the virulence property of the microflora [39]. Although, several earlier studies stated that archaeal co-infection is frequently detected in viral and bacterial hosts [92,103], the systematic tests of the factors explaining variation in viral co-infection across different taxa and environments are still lacking.
Recent studies suggested that COVID-19 patients having microbial co-infections are characterized by lymphopenia and enhanced levels of proinflammatory cytokines including interleukin-6 (IL-6) and IL-1β as well as MCP-1, IP-10, and granulocyte colony-stimulating factor (G-CSF) in the plasma. It has been proposed that high levels of proinflammatory cytokines might lead to shock as well as respiratory failure or multiple organ failure, and several trials to assess inflammatory mediators are under way [104]. Cytokine storm, or hypercytokinemia, describes hyperactivation of the immune system that may be provoked or worsened by co-infections. This can lead to devastating and irreparable destruction of lung tissue as proinflammatory cytokines damage the alveoli, tiny sacs in the lungs responsible for gas exchange and oxygenation [51]. Damage to lung tissues caused by SARS-CoV-2 may explore the receptors for other pathogenic or opportunistic microorganisms facilitating secondary infections. Specific domains in which viruses play such facilitating role including enhancement of bacterial adhesion by unmasking cryptic receptors and upregulation of adhesion proteins, disruption of tight junction integrity favoring paracellular transmigration of bacteria and loss of epithelial barrier integrity, increased availability of nutrients, such as mucins and iron, alteration of innate and adaptive immune responses, and disabling defense against bacteria, and lastly, changes in airway microbiome that render the lung more vulnerable to pathogens [105]. Moreover, SARS-CoV-2 infection can damage lymphocytes, especially B cells, T cells, and NK cells, which will lead to the immune system's impairment during the period of disease [26]. The decrease of lymphocytes and host immune function may be the main reason for further super- or secondary infection [106]. The mortality is more significant in severe cases compared with the non-severe group [107] due to the higher co-infection rate in severe patients [106,107]. The mechanistic evidence of influenza and pneumococcal co-infections showed that influenza virus causes a depletion of alveolar macrophage which allows a smaller inoculum of bacteria to establish productive infection [108], and consequent severity of the bacterial co-infection is exacerbated by preexisting host factors such as obesity [109]. For more severely ill patients, they are more likely to receive treatment with invasive catheters, resulting in increased sensitivity to co-infections with multidrug-resistant pathogens such as Acinetobacter baumannii, Escherichia coli, Pseudomonas aeruginosa, and Enterococcus species [110]. In addition, during the pathophysiology of COVID-19, SARS-CoV-2 viruses can interact with a large number of cellular proteins (virus-host interactome) and protein-protein interactions between unrelated viruses, bacteria, archaea and fungi are also possible [110]. Co-infections may result in genetic exchange among heterologous viruses, and/or agents [111] leading to the generation of recombinant or chimeric pathogens, and this recombination effects can influence viral evolution, disease dynamics, sensitivity to antiviral therapy, and eventually the fate of the host [110] Though not studied yet, similar mechanistic events may be found in SARS-CoV-2 and associated co-infections aggravating the pathophysiology of COVID-19. In addition, four factors including host ecology, host taxonomy or phylogeny, host defense mechanisms, and the interactions of co-pathogens with SARS-CoV-2 are likely to play vital role in the pathophysiology and severity of COVID-19 disease. The relevance and importance of these molecular events of pathogenesis are likely to vary for cross-infectivity, culture co-infection, and single-cell co-infection [92]. Overall, due to some risk factors including the epithelial lung damage, immune system dysregulation, prolonged period of hospitalization etc., the possible development of superinfections is somewhat expected in severely ill COVID-19 patients [78]. However, the actual scenario of prevalence, incidence and characteristics of microbial co-infections in SARS-CoV-2 infected patients is yet to be elucidated and analyzed. This review highlights that understanding the immunological mechanisms of co-infections and underlying the diverse clinical presentations of COVID-19 is a crucial step in the design of rational therapeutic strategies. Therefore, the effect of SARS-CoV-2 replication and induction of innate immune response on the composition of the human or animal upper respiratory tract (URT) microbiome remains to be elucidated and analyzed in depth on a community wide scale. Further extensive investigation is warranted for a better understanding and evaluating the risk factors associated and the disease spectrum of co/secondary and superinfection in critically ill patients suffering from SARS-CoV-2 infection.
5. Conclusion and perspectives
Co-infections with various microorganisms are commonly found in SARS-CoV-2 infected patients that significantly influences the severity and mortality rates of COVID-19. However, our understanding about co-infecting organisms, their cross-talks and ultimate interactions with the hosts are poor. The COVID-19 co-infections are associated with multiple domains of microorganisms including viruses, bacteria, fungi and archaea. Although the specific molecular events of co-pathogenesis in SARS-CoV-2 pathophysiology is yet unknown, the co-infecting pathogens may participate to damage the respiratory airway, cell loss, goblet cell hyperplasia, alter mucus secretion, reduced ciliary beat frequency, function and clearance, reduced oxygen exchange, and damage the immune system. Furthermore, viral co-infection facilitates bacterial adhesion, disrupt the tight junction and epithelial barrier integrity favoring paracellular transmigration of bacteria, and alter both innate and adaptive immune responses that render the lung more vulnerable to SARS-CoV-2 infections. Although, this review provides a comprehensive scenario of co-infection in COVID-19 patients, further studies are needed to focus the epidemiology, clinical and laboratory characteristics of co-infecting pathogens among diverse group of COVID-19 patients from different geo-climatic conditions, and assess the effect of co-infecting microorganisms on the outcome of COVID-19 patients.
Data availability
This is a Literature Review article without any data or code.
Funding information
The authors received no funding for this review article.
Author contributions
MNH, SA, IDM, MRI, MSR, MA, II and MMH conceived and designed the structure of this review, and wrote the manuscript. MMR, MS, TI and MAH critically reviewed and edited the drafted manuscript.
Declaration of competing interest
The authors declare no conflict of interests.
Acknowledgements
The authors would like to thank ASM Rubayet-Ul-Alam for his kind support and constructive criticism on the review.
References
- 1.Hoque M.N., Chaudhury A., Akanda M.A.M., Hossain M.A., Islam M.T. Genomic diversity and evolution, diagnosis, prevention, and therapeutics of the pandemic COVID-19 disease. PeerJ. 2020;8:e9689. doi: 10.7717/peerj.9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahman M.S., Hoque M.N., Islam M.R., Akter S., Rubayet-Ul-Alam A., Siddique M.A., et al. Epitope-based chimeric peptide vaccine design against S, M and E proteins of SARS-CoV-2 etiologic agent of global pandemic COVID-19: an in silico approach. PeerJ. 2020;8 doi: 10.7717/peerj.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China. N. Engl. J. Med. 2019:2020. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reid A.H., Fanning T.G., Hultin J.V., Taubenberger J.K. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. Unit. States Am. 1999;96:1651–1656. doi: 10.1073/pnas.96.4.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mirzaei R., Goodarzi P., Asadi M., Soltani A., Aljanabi H.A.A., Jeda A.S., et al. Bacterial co‐infections with SARS‐CoV‐2. IUBMB Life. 2020;72:2097–2111. doi: 10.1002/iub.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paget C., Trottein F. Mechanisms of bacterial superinfection post-influenza: a role for unconventional T cells. Front. Immunol. 2019;10:336. doi: 10.3389/fimmu.2019.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia L., Xie J., Zhao J., Cao D., Liang Y., Hou X., et al. Mechanisms of severe mortality-associated bacterial co-infections following influenza virus infection. Frontiers in cellular and infection microbiology. 2017;7:338. doi: 10.3389/fcimb.2017.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quah J., Jiang B., Tan P.C., Siau C., Tan T.Y. Impact of microbial Aetiology on mortality in severe community-acquired pneumonia. BMC Infect. Dis. 2018;18:451. doi: 10.1186/s12879-018-3366-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Memish Z.A., Perlman S., Van Kerkhove M.D., Zumla A. Middle East respiratory syndrome. Lancet. 2020;395(10229):1063–1077. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoque M.N., Rahman M.S., Ahmed R., Hossain M.S., Islam M.S., Crandall K.A., et al. 2020. Diversity and Genomic Determinants of the Microbiomes Associated with COVID-19 and Non-COVID Respiratory Diseases. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., et al. Genomic diversity of SARS-CoV-2 in coronavirus disease 2019 patients. Clin. Infect. Dis. 2020;71(15):713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020 doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martines R.B., Ritter J.M., Matkovic E., Gary J., Bollweg B.C., Bullock H., et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:2005. doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Assiri A., Al-Tawfiq J.A., Al-Rabeeah A.A., Al-Rabiah F.A., Al-Hajjar S., Al-Barrak A., et al. Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis. 2013;13:752–761. doi: 10.1016/S1473-3099(13)70204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;395(10229):1054–1062. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zahariadis G., Gooley T.A., Ryall P., Hutchinson C., Latchford M.I., Fearon M.A., et al. Risk of ruling out severe acute respiratory syndrome by ruling in another diagnosis: variable incidence of atypical bacteria coinfection based on diagnostic assays. Can. Respir. J. J. Can. Thorac. Soc. 2006;13 doi: 10.1155/2006/862797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng T., Li J., Ni Y., Kang K., Misiakou M.-A., Imamovic L., et al. Mining, analyzing, and integrating viral signals from metagenomic data. Microbiome. 2019;7:1–15. doi: 10.1186/s40168-019-0657-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin X., Gong Z., Xiao Z., Xiong J., Fan B., Liu J. Novel coronavirus pneumonia outbreak in 2019: computed tomographic findings in two cases. Korean J. Radiol. 2020;21:365–368. doi: 10.3348/kjr.2020.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guan W-j, Ni Z-y, Hu Y., Liang W-h, Ou C-q, He J-x, et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCullers J.A. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat. Rev. Microbiol. 2014;12:252–262. doi: 10.1038/nrmicro3231. [DOI] [PubMed] [Google Scholar]
- 23.Chen X., Liao B., Cheng L., Peng X., Xu X., Li Y., et al. The microbial coinfection in COVID-19. Appl. Microbiol. Biotechnol. 2020:1–9. doi: 10.1007/s00253-020-10814-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lansbury L., Lim B., Baskaran V., Lim W.S. Co-infections in people with COVID-19: a systematic review and meta-analysis. J. Infect. 2020;81(2):266–275. doi: 10.1016/j.jinf.2020.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joseph C., Togawa Y., Shindo N. Bacterial and viral infections associated with influenza. Influenza and other Respiratory Viruses. 2013;7:105–113. doi: 10.1111/irv.12089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. J. Am. Med. Assoc. 2020;323:1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X., Cai Y., Huang X., Yu X., Zhao L., Wang F., et al. Co-infection with SARS-CoV-2 and influenza A virus in patient with pneumonia, China. Emerg. Infect. Dis. 2020;26:1324. doi: 10.3201/eid2606.200299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan S., Xiang Y., Fang W., Zheng Y., Li B., Hu Y., et al. Clinical features and treatment of COVID‐19 patients in northeast Chongqing. J. Med. Virol. 2020;92(7):797–806. doi: 10.1002/jmv.25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kalantar-Zadeh K., Ward S.A., Kalantar-Zadeh K., El-Omar E.M. Considering the effects of microbiome and diet on SARS-CoV-2 infection: nanotechnology roles. ACS Nano. 2020;14(5):5179–5182. doi: 10.1021/acsnano.0c03402. [DOI] [PubMed] [Google Scholar]
- 30.Netea M.G., Giamarellos-Bourboulis E.J., Domínguez-Andrés J., Curtis N., van Crevel R., van de Veerdonk F.L., et al. Trained immunity: a tool for reducing susceptibility to and the severity of SARS-CoV-2 infection. Cell. 2020;181(5):969–977. doi: 10.1016/j.cell.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salas-Benito J.S., Nova-Ocampo D. Viral interference and persistence in mosquito-borne flaviviruses. J. Immunol. Res. 2015;2015 doi: 10.1155/2015/873404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makoti P., Fielding B.C. HIV and human Coronavirus coinfections: a historical perspective. Viruses. 2020;12:937. doi: 10.3390/v12090937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths E.C., Pedersen A.B., Fenton A., Petchey O.L. The nature and consequences of coinfection in humans. J. Infect. 2011;63:200–206. doi: 10.1016/j.jinf.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Der Hoek L., Pyrc K., Berkhout B. Human coronavirus NL63, a new respiratory virus. FEMS (Fed. Eur. Microbiol. Soc.) Microbiol. Rev. 2006;30:760–773. doi: 10.1111/j.1574-6976.2006.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bengoechea J.A., Bamford C.G. SARS‐CoV‐2, bacterial co‐infections, and AMR: the deadly trio in COVID‐19? EMBO Mol. Med. 2020;12(7) doi: 10.15252/emmm.202012560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griffiths E., Pedersen A., Fenton A., Petchey O. Reported co-infection deaths are more common in early adulthood and among similar infections. BMC Infect. Dis. 2015;15:411. doi: 10.1186/s12879-015-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nepogodiev D., Glasbey J.C., Li E., Omar O.M., Simoes J.F., Abbott T.E., et al. Mortality and pulmonary complications in patients undergoing surgery with perioperative SARS-CoV-2 infection: an international cohort study. Lancet. 2020;396(10243):27–38. doi: 10.1016/S0140-6736(20)31182-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nickbakhsh S., Mair C., Matthews L., Reeve R., Johnson P.C., Thorburn F., et al. Virus–virus interactions impact the population dynamics of influenza and the common cold. Proc. Natl. Acad. Sci. Unit. States Am. 2019;116:27142–27150. doi: 10.1073/pnas.1911083116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoque M.N., Istiaq A., Clement R.A., Sultana M., Crandall K.A., Siddiki A.Z., et al. Metagenomic deep sequencing reveals association of microbiome signature with functional biases in bovine mastitis. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-49468-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32:108185. doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lambert L.C., Fauci A.S. Influenza vaccines for the future. N. Engl. J. Med. 2010;363:2036–2044. doi: 10.1056/NEJMra1002842. [DOI] [PubMed] [Google Scholar]
- 42.Zheng X., Wang H., Su Z., Li W., Yang D., Deng F., et al. Co-infection of SARS-CoV-2 and influenza virus in early stage of the COVID-19 epidemic in Wuhan, China. J. Infect. 2020;81(2):128–129. doi: 10.1016/j.jinf.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azekawa S., Namkoong H., Mitamura K., Kawaoka Y., Saito F. Co-infection with SARS-CoV-2 and influenza A virus. IDCases. 2020;20 doi: 10.1016/j.idcr.2020.e00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ding Q., Lu P., Fan Y., Xia Y., Liu M. The clinical characteristics of pneumonia patients coinfected with 2019 novel coronavirus and influenza virus in Wuhan, China. J. Med. Virol. 2020;92(9):1549–1555. doi: 10.1002/jmv.25781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu X., Ge Y., Wu T., Zhao K., Chen Y., Wu B., et al. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005. doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia-Vidal C., Sanjuan G., Moreno-García E., Puerta-Alcalde P., Garcia-Pouton N., Chumbita M., et al. Incidence of co-infections and superinfections in hospitalized patients with COVID-19: a retrospective cohort study. Clin. Microbiol. Infect. 2020;27(1):83–88. doi: 10.1016/j.cmi.2020.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunutsor S.K., Laukkanen J.A. Hepatic manifestations and complications of COVID-19: a systematic review and meta-analysis. J. Infect. 2020;81:e72–e74. doi: 10.1016/j.jinf.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang C., Shi L., Wang F.-S. Liver injury in COVID-19: management and challenges. The lancet Gastroenterology & Hepatology. 2020;5:428–430. doi: 10.1016/S2468-1253(20)30057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wander P., Epstein M., Bernstein D. COVID-19 Presenting as Acute Hepatitis. Am J Gastroenterol. 2020 doi: 10.14309/ajg.0000000000000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alqahtani S.A., Schattenberg J.M. Liver injury in COVID-19: the current evidence. United European Gastroenterology Journal. 2020;8:509–519. doi: 10.1177/2050640620924157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., et al. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet (London, England) 2020;395:1033. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khinda J., Janjua N.Z., Cheng S., van den Heuvel E.R., Bhatti P., Darvishian M. Association between markers of immune response at hospital admission and COVID‐19 disease severity and mortality: a meta‐analysis and meta‐regression. J. Med. Virol. 2020;93(2):1078–1098. doi: 10.1002/jmv.26411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang G., Hu C., Luo L., Fang F., Chen Y., Li J., et al. Clinical features and short-term outcomes of 221 patients with COVID-19 in Wuhan, China. J. Clin. Virol. 2020:104364. doi: 10.1016/j.jcv.2020.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ali N. Relationship between COVID-19 infection and liver injury: a review of recent data. Front. Med. 2020;7:458. doi: 10.3389/fmed.2020.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cai Q., Huang D., Yu H., Zhu Z., Xia Z., Su Y., et al. COVID-19: abnormal liver function tests. J. Hepatol. 2020;73(3):566–574. doi: 10.1016/j.jhep.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Epelboin L., Blondé R., Nacher M., Combe P., Collet L. COVID-19 and dengue co-infection in a returning traveller. J. Trav. Med. 2020;27 doi: 10.1093/jtm/taaa114. taaa114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yan G., Lee C.K., Lam L.T., Yan B., Chua Y.X., Lim A.Y., et al. Covert COVID-19 and false-positive dengue serology in Singapore. Lancet Infect. Dis. 2020;20:536. doi: 10.1016/S1473-3099(20)30158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lorenz C., Bocewicz A.C.D., de Azevedo Marques C.C., Santana L.M.R., Chiaravalloti-Neto F., Gomes A.H.A., et al. Have measures against COVID-19 helped to reduce dengue cases in Brazil? Trav. Med. Infect. Dis. 2020 doi: 10.1016/j.tmaid.2020.101827. arXiv:2103.08669, Submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cardona‐Ospina J.A., Arteaga‐Livias K., Villamil‐Gómez W.E., Pérez‐Díaz C.E., Katterine Bonilla‐Aldana D., Mondragon Cardona Á., et al. Dengue and COVID‐19, overlapping epidemics? An analysis from Colombia. J. Med. Virol. 2020;93(1):522–527. doi: 10.1002/jmv.26194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinky L., Dobrovolny H.M. Coinfections of the respiratory tract: viral competition for resources. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155589. e0155589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Navarro J.-C., Arrivillaga-Henríquez J., Salazar-Loor J., Rodriguez-Morales A.J. COVID-19 and dengue, co-epidemics in Ecuador and other countries in Latin America: pushing strained health care systems over the edge. Trav. Med. Infect. Dis. 2020;37 doi: 10.1016/j.tmaid.2020.101656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lorenz C., Azevedo T.S., Chiaravalloti-Neto F. COVID-19 and Dengue Fever: A Dangerous Combination for the Health System in Brazil. Trav. Med. Infect. Dis. 2020;35 doi: 10.1016/j.tmaid.2020.101659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tamuzi J.L., Ayele B.T., Shumba C.S., Adetokunboh O.O., Uwimana-Nicol J., Haile Z.T., et al. Implications of COVID-19 in high burden countries for HIV/TB: a systematic review of evidence. BMC Infect. Dis. 2020;20:1–18. doi: 10.1186/s12879-020-05450-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soriano V., Barreiro P. Impact of new coronavirus epidemics on HIV-infected patients. AIDS Rev. 2020;22 doi: 10.24875/AIDSRev.M20000031. [DOI] [PubMed] [Google Scholar]
- 66.Tang X., Zhang S., Peng Q., Ling L., Shi H., Liu Y., et al. Sustained IFN-I stimulation impairs MAIT cell responses to bacteria by inducing IL-10 during chronic HIV-1 infection. Science Advances. 2020;6:eaaz0374. doi: 10.1126/sciadv.aaz0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaung J., Chan D., Pada S., Tambyah P.A. Coinfection with COVID‐19 and Coronavirus HKU1–the critical need for repeat testing if clinically indicated. J. Med. Virol. 2020;92(10):1785–1786. doi: 10.1002/jmv.25890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.MacIntyre C.R., Chughtai A.A., Barnes M., Ridda I., Seale H., Toms R., et al. The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a (H1N1) pdm09. BMC Infect. Dis. 2018;18:637. doi: 10.1186/s12879-018-3548-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Manna S., Baindara P., Mandal S.M. Molecular pathogenesis of secondary bacterial infection associated to viral infections including SARS-CoV-2. Journal of Infection and Public Health. 2020;13(10):1397–1404. doi: 10.1016/j.jiph.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Langford B.J., So M., Raybardhan S., Leung V., Westwood D., MacFadden D.R., et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin. Microbiol. Infect. 2020;26(12):1622–1629. doi: 10.1016/j.cmi.2020.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morens D.M., Taubenberger J.K., Fauci A.S. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J. Infect. Dis. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Klein E.Y., Monteforte B., Gupta A., Jiang W., May L., Hsieh Y.H., et al. The frequency of influenza and bacterial coinfection: a systematic review and meta‐analysis. Influenza and other Respiratory Viruses. 2016;10:394–403. doi: 10.1111/irv.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huttner B., Catho G., Pano-Pardo J.R., Pulcini C., Schouten J. COVID-19: don't neglect antimicrobial stewardship principles! Clin. Microbiol. Infect. 2020;26:808–810. doi: 10.1016/j.cmi.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cox M.J., Loman N., Bogaert D., O'Grady J. Co-infections: potentially lethal and unexplored in COVID-19. The Lancet Microbe. 2020;1:e11. doi: 10.1016/S2666-5247(20)30009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yeoh Y.K., Zuo T., Lui G.C.-Y., Zhang F., Liu Q., Li A.Y., et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mostafa H.H., Fissel J.A., Fanelli B., Bergman Y., Gniazdowski V., Dadlani M., et al. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect covid-19 patients. mBio. 2020:11. doi: 10.1128/mBio.01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaul D., Rathnasinghe R., Ferres M., Tan G.S., Barrera A., Pickett B.E., et al. Microbiome disturbance and resilience dynamics of the upper respiratory tract during influenza A virus infection. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-16429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bassetti M., Kollef M.H., Timsit J.-F. Bacterial and fungal superinfections in critically ill patients with COVID-19. Intensive Care Med. 2020;46:2071–2074. doi: 10.1007/s00134-020-06219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lehmann C.J., Pho M.T., Pitrak D., Ridgway J.P., Pettit N.N. Community acquired co-infection in COVID-19: a retrospective observational experience. Clin. Infect. Dis. 2020;72(8):1450–1452. doi: 10.1093/cid/ciaa902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu Y., Yang Q., Xu M., Kong H., Chen H., Fu Y., et al. Secondary bacterial infections in critical ill patients of COVID-19. Open Forum Infectious Diseases. 2020;7(6) doi: 10.1093/ofid/ofaa220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lai C.-C., Wang C.-Y., Hsueh P.-R. Co-infections among patients with COVID-19: the need for combination therapy with non-anti-SARS-CoV-2 agents? J. Microbiol. Immunol. Infect. 2020;53(4):505–512. doi: 10.1016/j.jmii.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ginsburg A.S., Klugman K.P. COVID-19 pneumonia and the appropriate use of antibiotics. The Lancet Global Health. 2020;8:e1453–e1454. doi: 10.1016/S2214-109X(20)30444-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khatiwada S., Subedi A. Lung microbiome and coronavirus disease 2019 (COVID-19): possible link and implications. Human Microbiome J. 2020;17:100073. doi: 10.1016/j.humic.2020.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peddu V., Shean R.C., Xie H., Shrestha L., Perchetti G.A., Minot S.S., et al. Metagenomic analysis reveals clinical SARS-CoV-2 infection and bacterial or viral superinfection and colonization. Clin. Chem. 2020;66(7):966–972. doi: 10.1093/clinchem/hvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gao Q.Y., Chen Y.X., Fang J.Y. 2019 Novel coronavirus infection and gastrointestinal tract. J. Digestive Diseases. 2020;21:125–126. doi: 10.1111/1751-2980.12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gu S., Chen Y., Wu Z., Chen Y., Gao H., Lv L., et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020;71:2669–2678. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. Am. J. Roentgenol. 2020:1–7. doi: 10.2214/AJR.20.23034. [DOI] [PubMed] [Google Scholar]
- 89.Yang X., Yu Y., Xu J., Shu H., Liu H., Wu Y., et al. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. The Lancet Respiratory Medicine. 2020;8(5):475–481. doi: 10.1016/S2213-2600(20)30079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Song G., Liang G., Liu W. Fungal co-infections associated with global COVID-19 pandemic: a clinical and diagnostic perspective from China. Mycopathologia. 2020:1–8. doi: 10.1007/s11046-020-00462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Clancy C.J., Buehrle D.J., Nguyen M.H. PRO: the COVID-19 pandemic will result in increased antimicrobial resistance rates. JAC-Antimicrobial Resistance. 2020;2:dlaa049. doi: 10.1093/jacamr/dlaa049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Díaz-Muñoz S.L. Viral coinfection is shaped by host ecology and virus–virus interactions across diverse microbial taxa and environments. Virus Evolution. 2017;3 doi: 10.1093/ve/vex011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Contou D., Claudinon A., Pajot O., Micaëlo M., Flandre P.L., Dubert M., et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann. Intensive Care. 2020;10:1–9. doi: 10.1186/s13613-020-00736-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lee I.T., Nakayama T., Wu C.-T., Goltsev Y., Jiang S., Gall P.A., et al. ACE2 localizes to the respiratory cilia and is not increased by ACE inhibitors or ARBs. Nat. Commun. 2020;11:1–14. doi: 10.1038/s41467-020-19145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.You H.-L., Chang S.-J., Yu H.-R., Li C.-C., Chen C.-H., Liao W.-T. Simultaneous detection of respiratory syncytial virus and human metapneumovirus by one-step multiplex real-time RT-PCR in patients with respiratory symptoms. BMC Pediatr. 2017;17:89. doi: 10.1186/s12887-017-0843-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Avadhanula V., Rodriguez C.A., DeVincenzo J.P., Wang Y., Webby R.J., Ulett G.C., et al. Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species-and cell type-dependent manner. J. Virol. 2006;80:1629–1636. doi: 10.1128/JVI.80.4.1629-1636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vareille M., Kieninger E., Edwards M.R., Regamey N. The airway epithelium: soldier in the fight against respiratory viruses. Clin. Microbiol. Rev. 2011;24:210–229. doi: 10.1128/CMR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Denney L., Ho L.-P. The role of respiratory epithelium in host defence against influenza virus infection. Biomed. J. 2018;41:218–233. doi: 10.1016/j.bj.2018.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang M., Luo L., Bu H., Xia H. Case report: one case of coronavirus disease 2019 (COVID-19) in patient Co-infected by HIV with a low CD4+ T cell count. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Iverson A.R., Boyd K.L., McAuley J.L., Plano L.R., Hart M.E., McCullers J.A. Influenza virus primes mice for pneumonia from Staphylococcus aureus. JID (J. Infect. Dis.) 2011;203:880–888. doi: 10.1093/infdis/jiq113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smith A.M., Adler F.R., Ribeiro R.M., Gutenkunst R.N., McAuley J.L., McCullers J.A., et al. Kinetics of coinfection with influenza A virus and Streptococcus pneumoniae. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nguyen D.T., Louwen R., Elberse K., van Amerongen G., Yüksel S., Luijendijk A., et al. Streptococcus pneumoniae enhances human respiratory syncytial virus infection in vitro and in vivo. PLoS One. 2015;10 doi: 10.1371/journal.pone.0127098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Roux S., Hawley A.K., Beltran M.T., Scofield M., Schwientek P., Stepanauskas R., et al. Ecology and evolution of viruses infecting uncultivated SUP05 bacteria as revealed by single-cell-and meta-genomics. Elife. 2014;3 doi: 10.7554/eLife.03125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Arunachalam P.S., Wimmers F., Mok C.K.P., Perera R.A., Scott M., Hagan T., et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rossi G.A., Fanous H., Colin A.A. Viral strategies predisposing to respiratory bacterial superinfections. Pediatr. Pulmonol. 2020;55:1061–1073. doi: 10.1002/ppul.24699. [DOI] [PubMed] [Google Scholar]
- 106.Luo Y., Xie Y., Zhang W., Lin Q., Tang G., Wu S., et al. Combination of lymphocyte number and function in evaluating host immunity. Aging (Albany NY) 2019;11:12685. doi: 10.18632/aging.102595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., et al. Dysregulation of immune response in patients with COVID-19 in Wuhan, China. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa248. covidwho-7937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Smith A.M., Smith A.P. A critical, nonlinear threshold dictates bacterial invasion and initial kinetics during influenza. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep38703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.McArdle A.J., Turkova A., Cunnington A.J. When do co-infections matter? Curr. Opin. Infect. Dis. 2018;31:209. doi: 10.1097/QCO.0000000000000447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kumar N., Sharma S., Barua S., Tripathi B.N., Rouse B.T. Virological and immunological outcomes of coinfections. Clin. Microbiol. Rev. 2018;31 doi: 10.1128/CMR.00111-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roux S., Enault F., Bronner G., Vaulot D., Forterre P., Krupovic M. Chimeric viruses blur the borders between the major groups of eukaryotic single-stranded DNA viruses. Nat. Commun. 2013;4:1–10. doi: 10.1038/ncomms3700. [DOI] [PubMed] [Google Scholar]
- 112.Suwanwongse K., Shabarek N. Can coinfection with influenza worsen COVID-19 outcomes? Journal of Investigative Medicine High Impact Case Reports. 2020;8 doi: 10.1177/2324709620953282. 2324709620953282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ma S., Lai X., Chen Z., Tu S., Qin K. Clinical characteristics of critically ill patients Co-infected with SARS-CoV-2 and the influenza virus in Wuhan, China. Int. J. Infect. Dis. 2020 doi: 10.1016/j.ijid.2020.05.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wu J., Song S., Cao H.-C., Li L.-J. Liver diseases in COVID-19: etiology, treatment and prognosis. World J. Gastroenterol. 2020;26:2286. doi: 10.3748/wjg.v26.i19.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lozano-Sepulveda S.A., Galan-Huerta K., Martínez-Acuña N., Arellanos-Soto D., Rivas-Estilla A.M. SARS-CoV-2 another kind of liver aggressor, how does it do that? Ann. Hepatol. 2020;19:592–596. doi: 10.1016/j.aohep.2020.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zou X., Fang M., Li S., Wu L., Gao B., Gao H., et al. Characteristics of liver function in patients with SARS-CoV-2 and chronic HBV coinfection. Clin. Gastroenterol. Hepatol. 2020;19(3):597–603. doi: 10.1016/j.cgh.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lin Y., Yuan J., Long Q., Hu J., Deng H., Zhao Z., et al. Patients with SARS-CoV-2 and HBV co-infection are at risk of greater liver injury. Genes & Diseases. 2020 doi: 10.1016/j.gendis.2020.11.005. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Verduyn M., Allou N., Gazaille V., Andre M., Desroche T., Jaffar M.-C., et al. Public Library of Science; San Francisco, CA USA: 2020. Co-infection of Dengue and COVID-19: A Case Report. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Saddique A., Rana M.S., Alam M.M., Ikram A., Usman M., Salman M., et al. Emergence of co-infection of COVID-19 and dengue: a serious public health threat. J. Infect. 2020;81(6):e16–e18. doi: 10.1016/j.jinf.2020.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xu B., Fan C-y, Wang A-l, Zou Y-l, Yu Y-h, He C., et al. 2020. Suppressed T Cell-Mediated Immunity in Patients with COVID-19: a Clinical Retrospective Study in Wuhan, China. China. (3/16/2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu Y., Ma J., Huang H., Vermund S.H. Coinfection with HIV and SARS-CoV-2 in Wuhan, China: a 12-person case series. J. Acquir. Immune Defic. Syndr. 2020;85(1):1–5. doi: 10.1097/QAI.0000000000002424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang R., Gui X., Zhang Y., Xiong Y., Gao S., Ke H. Clinical characteristics of COVID-19 patients with HIV coinfection in Wuhan, China. Expet Rev. Respir. Med. 2020:1–7. doi: 10.1080/17476348.2021.1836965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alothaid H., Aldughaim M.S., El Bakkouri K., AlMashhadi S., Al-Qahtani A.A. Similarities between the effect of SARS-CoV-2 and HCV on the cellular level, and the possible role of ion channels in COVID19 progression: a review of potential targets for diagnosis and treatment. Channels. 2020;14:403–412. doi: 10.1080/19336950.2020.1837439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Reddy K.R. SARS‐CoV‐2 and the liver: considerations in hepatitis B and hepatitis C infections. Clinical Liver Disease. 2020;15:191. doi: 10.1002/cld.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Murphy R.C., Lai Y., Barrow K.A., Hamerman J.A., Lacy-Hulbert A., Piliponsky A.M., et al. Effects of asthma and human rhinovirus A16 on the expression of SARS-CoV-2 entry factors in human airway epithelium. Am. J. Respir. Cell Mol. Biol. 2020;63:859–863. doi: 10.1165/rcmb.2020-0394LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Orozco-Hernández J.P., Montoya-Martínez J.J., Pacheco-Gallego M.C., Céspedes-Roncancio M., Porras-Hurtado G.L. SARS-CoV-2 and rhinovirus/enterovirus co-infection in a critically ill young adult patient in Colombia. Biomedica. 2020;40:34–43. doi: 10.7705/biomedica.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Motta J.C., Gómez C.C. Adenovirus and novel coronavirus (SARS-Cov2) coinfection: a case report. IDCases. 2020;22 doi: 10.1016/j.idcr.2020.e00936. e00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nieto‐Moro M., Ecclesia F.G., Tomé‐Masa I., Caro‐Patón G.D.L., Leoz‐Gordillo I., Cabrero‐Hernández M., et al. SARS‐CoV‐2 and Streptococcus pneumoniae coinfection as a cause of severe pneumonia in an infant. Pediatr. Pulmonol. 2020;55(9):2198–2200. doi: 10.1002/ppul.24916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rodriguez-Nava G., Yanez-Bello M.A., Trelles-Garcia D.P., Chung C.W., Egoryan G., Friedman H.J. A retrospective study of coinfection of SARS-CoV-2 and Streptococcus pneumoniae in 11 hospitalized patients with severe COVID-19 pneumonia at a single center. Med. Sci. Mon. Int. Med. J. Exp. Clin. Res. 2020;26 doi: 10.12659/MSM.928754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Duployez C., Le Guern R., Tinez C., Lejeune A.-L., Robriquet L., Six S., et al. Panton-valentine leukocidin–secreting staphylococcus aureus pneumonia complicating COVID-19. Emerg. Infect. Dis. 2020;26:1939. doi: 10.3201/eid2608.201413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Cusumano J.A., Dupper A.C., Malik Y., Gavioli E.M., Banga J., Berbel Caban A., et al. Oxford University Press US; 2020. Staphylococcus aureus Bacteremia in Patients Infected with COVID-19: A Case Series. Open Forum Infectious Diseases; p. ofaa518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hosoda T., Harada S., Okamoto K., Ishino S., Kaneko M., Suzuki M., et al. COVID-19 and Fatal Sepsis Caused by Hypervirulent Klebsiella pneumoniae, Japan, 2020. Emerg. Infect. Dis. 2020:27. doi: 10.3201/eid2702.204662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Oliva A., Siccardi G., Migliarini A., Cancelli F., Carnevalini M., D'Andria M., et al. Co-infection of SARS-CoV-2 with Chlamydia or Mycoplasma pneumoniae: a case series and review of the literature. Infection. 2020;48:871–877. doi: 10.1007/s15010-020-01483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Amin D., McKitish K., Shah P.S. Association of mortality and recent Mycoplasma pneumoniae infection in COVID‐19 patients. J. Med. Virol. 2020;93(2):1180–1183. doi: 10.1002/jmv.26467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Arashiro T., Nakamura S., Asami T., Mikuni H., Fujiwara E., Sakamoto S., et al. SARS-CoV-2 and Legionella co-infection in a person returning from a Nile cruise. J. Trav. Med. 2020;27 doi: 10.1093/jtm/taaa053. taaa053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Moriguchi T., Harii N., Goto J., Harada D., Sugawara H., Takamino J., et al. A first case of meningitis/encephalitis associated with SARS-Coronavirus-2. Int. J. Infect. Dis. 2020;94:55–58. doi: 10.1016/j.ijid.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gallacher S.D., Seaton A. Meningococcal meningitis and COVID-19 co-infection. BMJ Case Reports CP. 2020;13 doi: 10.1136/bcr-2020-237366. e237366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Crisan-Dabija R., Grigorescu C., Pavel C.A., Artene B., Popa I.V., Cernomaz A., et al. medRxiv; 2020. Tuberculosis and COVID-19 in 2020: Lessons from the Past Viral Outbreaks and Possible Future Outcomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu Y., Bi L., Chen Y., Wang Y., Fleming J., Yu Y., et al. medRxiv; 2020. Active or Latent Tuberculosis Increases Susceptibility to COVID-19 and Disease Severity. [Google Scholar]
- 140.Sy K.T.L., Haw N.J.L., Uy J. Previous and active tuberculosis increases risk of death and prolongs recovery in patients with COVID-19. Infectious Diseases. 2020;52:902–907. doi: 10.1080/23744235.2020.1806353. [DOI] [PubMed] [Google Scholar]
- 141.Lai C.-C., Yu W.-L. COVID-19 associated with pulmonary aspergillosis: a literature review. J. Microbiol. Immunol. Infect. 2020;54(1):46–53. doi: 10.1016/j.jmii.2020.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Al-Hatmi A.M., Mohsin J., Al-Huraizi A., Khamis F. COVID-19 associated invasive candidiasis. J. Infect. 2020;82(2):e45–e46. doi: 10.1016/j.jinf.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is a Literature Review article without any data or code.