Abstract
Coronavirus disease 2019 (COVID-19) is an ongoing pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Although viral infection is known to trigger inflammatory processes contributing to tissue injury and organ failure, it is unclear whether direct viral damage is needed to sustain cellular injury. An understanding of pathogenic mechanisms has been handicapped by the absence of optimized methods to visualize the presence and distribution of SARS-CoV-2 in damaged tissues. We first developed a positive control cell line (Vero E6) to validate SARS-CoV-2 detection assays. We then evaluated multiple organs (lungs, kidneys, heart, liver, brain, intestines, lymph nodes, and spleen) from fourteen COVID-19 autopsy cases using immunohistochemistry (IHC) for the spike and the nucleoprotein proteins, and RNA in situ hybridization (RNA ISH) for the spike protein mRNA. Tissue detection assays were compared with quantitative polymerase chain reaction (qPCR)-based detection. SARS-CoV-2 was histologically detected in the Vero E6 positive cell line control, 1 of 14 (7%) lungs, and none (0%) of the other 59 organs. There was perfect concordance between the IHC and RNA ISH results. qPCR confirmed high viral load in the SARS-CoV-2 ISH-positive lung tissue, and absent or low viral load in all ISH-negative tissues. In patients who die of COVID-19-related organ failure, SARS-CoV-2 is largely not detectable using tissue-based assays. Even in lungs showing widespread injury, SARS-CoV-2 viral RNA or proteins were detected in only a small minority of cases. This observation supports the concept that viral infection is primarily a trigger for multiple-organ pathogenic proinflammatory responses. Direct viral tissue damage is a transient phenomenon that is generally not sustained throughout disease progression.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Diffuse alveolar damage, RNA in situ hybridization, Spike protein, Nucleoprotein
1. Introduction
COVID-19 is an ongoing pandemic caused by the SARS-CoV-2 virus. To date, almost 73 million cases have been reported worldwide resulting in more than 1.62 million deaths (coronavirus.jhu.edu; checked in December 2020). Clearly, there is an urgent need to better understand the pathogenesis of the disease in a way that will inform preventive and therapeutic strategies. Progress could be accelerated by detection assays that define the tissue distribution of the virus in infected patients. Instead, the capriciousness of tissue-based detection assays has thwarted insight into the role and timing of SARS-CoV-2 in disease progression. While some studies have consistently visualized the virus in organs such as the lungs, brain, heart, and gastrointestinal tract of infected patients [[1], [10], [11], [2], [3], [4], [5], [6], [7], [8], [9]], and have used these findings as direct evidence of tissue specific tropism and viral-mediated tissue injury, others have been unable to detect virus widely in these same tissues, suggesting alternative mechanisms of organ damage that do not require persistent active viral infection [[12], [13], [14], [15]].
Access to tissues for investigative purposes has largely come through the postmortem examinations of fatal cases. The value of these samples has been restricted by two factors that have mired tissue-based research. First, postmortem samples are highly susceptible to tissue degradation, limiting the effectiveness of detection assays that rely on the integrity of viral protein, DNA, and RNA. Second, there has been a conspicuous absence of a positive control to guide interpretation of detection assays rendering detection assays vulnerable to bias and erroneous interpretation. In the absence of positive controls to establish standards of interpretation, even electron microcopy—the historic benchmark for confirming cellular viral infections—has led to erroneous affirmation of SARS-CoV-2 due to misinterpretation of nonspecific ultrastructural findings [[16], [17], [18], [19]]. For tissue-based platforms, discordant detection rates across studies may underscores the absence of well-defined and universally applied standards for probe validity and test interpretation [10]. These two factors, taken together, have obscured the ability to understand fundamental aspects of cell injury and organ dysfunction such as the distribution of SARS-CoV-2 in human tissues. It remains unclear, for example, whether the consistent inability to detect virus using conventional immunohistochemical and in situ hybridization assays in damaged tissues reflects postmortem viral degradation, ineffectiveness of detection assays, absence of tropism toward a particular tissue, low viral load, or effective viral clearance. Conversely, it is not always clear whether the ready identification of SARS-CoV-2 in various tissues reflects the true presence of virus, or the misinterpretation of nonspecific changes. Resolution of key mechanisms of viral pathogenesis and, more specifically, the relative importance of direct viral injury versus subsequent inflammatory damage, cannot move forward until a positive control is established to guide the interpretation of detection assays. To more definitively define the tissue distribution of SARS-CoV-2 in fatal cases of COVID-19, we 1) developed a positive cell line control to guide interpretation of various detection assays for detection of viral protein and RNA, 2) used these detection assays to look for SARS-CoV-2 RNA or proteins in tissues from multiple organs, and then 3) compared histologic detection with a highly sensitive polymerase chain reaction (PCR)-based detection assay.
2. Cases and methods
2.1. Generation of SARS-CoV-2–infected Vero E6 cells
A SARS-CoV-2–infected cell line was generated as previously described in detail [20]. Briefly, 1,000,000 Vero E6 cells (ATCC CRL-1586) per well were seeded in a 6-well cell culture plate and maintained in Dulbecco's modified Eagle medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS), 10 ml of Pen-Strep (Gibco; catalog #15140122) and 10 ml of HEPES buffer (Gibco, catalog #15630080). After a day, the medium was removed, and SARS-CoV-2 isolate USA–WA1/2020 (obtained from BEI Resources NR-52281) was added to the cells at a multiplicity of infection of 0.01. The SARS-CoV-2 stock was diluted and added to the cells in 1X MEM that was supplemented with 2% FBS. The virus-containing cell media was left on the cells and cells were scraped off and collected in PBS at three intervals: 24 h, 48 h, and 72 h. Cells were fixed with 10% formaldehyde for 24 h before use.
Ten ml of fixed Vero cell culture fluid suspension was centrifuged at 1600 rpm for 10 min resulting in a well-formed pellet. The tube was decanted and 5 drops of Histogel was added to the pellet. This mixture was vortexed for 3 s and refrigerated for 5 min to harden the pellet. The hardened pellet was then wrapped in biopsy tissue paper, placed in a tissue cassette, and fixed in formalin. Transmission electron microscopy was used to confirm the presence of SARS-CoV-2 viral particles within the infected cell line.
Uninfected Vero E6 cells were used as a control to compare day 3 morphology for the presence of cytopathic effect and as a negative control for the immunohistochemistry (IHC), in situ hybridization (ISH), and immunofluorescence (IF).
2.2. Cases
Fourteen COVID-19 autopsy cases were included: nine full autopsies and five additional lung-only cases. All patients had been admitted to the Mount Sinai for progressing COVID-related signs and symptoms, and a positive nasopharyngeal swap PCR SARS-CoV-2 test. Autopsies were preferentially selected on the basis of short intervals from time of death to autopsy and/or short time intervals from onset of symptoms to death. To assess tissue-specific tropism, formalin-fixed and paraffin-embedded tissue blocks were selected from multiple organs including the lung (multiple samples from each lung), heart, liver, kidney, small intestine, brain (frontal lobe), spleen, and lymph nodes. Paired fresh frozen specimens were collected at the time of autopsy from seven of the cases for quantitative PCR (qPCR) analysis. Archived autopsy blocks from 2019 (ie, pre-COVID cases) were used as negative tissue controls.
2.3. Immunohistochemistry
Four μm–thick sections were cut from the FFPE autopsy blocks and the SARS-CoV-2–infected cell pellets. Immunostaining was performed using antibodies against the SARS-CoV-2 spike protein (ProSci, Poway, CA; catalog #3525; polyclonal; 1:10000) and the SARS-CoV-2 nucleoprotein (ProSci, Poway, CA; catalog #35-579; monoclonal; 1:24000). A rabbit peroxidase conjugated with 3,3′-diaminobenzidine (DAB) as brown chromogen was used as the secondary antibody. Titration was optimized using the SARS-CoV-2–infected cell line pellet as the positive control. The H1 (citrate buffer) and H2 (EDTA buffer) retrieval methods were used at two different time points. The staining results were reviewed by two study pathologists (SEJ and WHW). The patterns of IHC staining in the SARS-CoV-2–infected Vero cell line was used to guide interpretation of IHC staining in the autopsy blocks. CD68 (Leica, Buffalo Grove, IL; 514H12) was used to highlight macrophage distribution, and CAM5.2 (Leica, Buffalo Grove, IL; 5D3) to highlight pneumocytes distribution in SARS-CoV-2–positive cases.
2.4. RNA in-situ hybridization
RNA ISH was performed using the RNAscope® kit (Advanced Cell Diagnostics, Inc., Hayward, CA) in accordance with the manufacturer's instructions. The RNAscope® has been shown to be highly reliable in detecting viral RNA. Briefly, 4 μm sections of the autopsy blocks and the infected Vero cell pellet were pretreated with heat and protease before hybridization. Sections were hybridized separately with probes specific for 1) the sense (catalog# 848561) strand detecting viral genomic RNA for the spike protein; and 2) the anti-sense strand (catalog# 845701) detecting the complementary RNA generated during active viral replication. The preamplifier, amplifier, and horseradish in situ peroxidase–labeled probes were then hybridized sequentially, followed by color development with DAB brown chromogen. UBC ISHRNA probe was used as to confirm the RNA integrity in the tissue blocks (Advanced Cell Diagnostics, Inc., Hayward, CA, reference RS7760). The staining results were reviewed by two study pathologists (SEJ and WHW). The patterns of hybridization signals in the SARS-CoV-2–infected Vero cell pellet were used to guide recognition of positive hybridization signals in the autopsy blocks.
2.5. Immunofluorescence
Immunofluorescence studies were performed on formalin-fixed paraffin-embedded tissue blocks and cell line pellet after 1-h deparaffinization with subsequent rehydration in decreasing ethanol gradient, and antigen retrieval in boiling citrate buffer at pH 6 for 30min (Vector Labs, H-3300). Cells were blocked in 10% NDS/0.5% TX for 30min, incubated with SARS-CoV-2 nucleoprotein (ProSci Inc, 35-579, 1:500) and ACE2 (Abcam, ab15348, 1:500) primary antibodies in 1% NDS/0.25% Triton-X overnight at 4C, and incubated with Cy3 (SARS/NEUN) and AF-488 (ACE2) secondary antibodies in 1% NDS/0.25% TX at RT. Nuclear counterstain was with DAPI (1:1000). Secondary-only and nonspecific IgG isotype controls (NEUN, which is not expressed by Vero cells) (MAB377, 1:100) were similarly performed. Cells were imaged on a Zeiss LSM 780 confocal microscope. Immunofluorescence studies on formalin-fixed and paraffin-embedded lung tissue were performed identically, with the exception of incubation in primary SARS-CoV-2 nucleoprotein at 1:100.
2.6. Quantitative polymerase chain reaction
Fresh tissue samples were systematically collected from multiple organs at the time of autopsy in seven cases. RNA from fresh tissue was stored in RNAlater and then isolated using the Direct-zol RNA Miniprep Plus (Zymo) kit. RNA from frozen tissue was isolated. 1 μg of RNA was retrotranscribed using the enzyme AffinityScript Multiple Temperature Reverse Transcriptase (Agilent Technologies), and PCR reaction was performed using the TaqMan™ Universal PCR Master Mix (Thermo Fisher). Previously reported primers and probes against E and RdRp genes (IP2) genes [[21], [22], [23]] were used for SARS-CoV-2 detection:
E-SARS-CoV2-F: ACAGGTACGTTAATAGTTAATAGCGT.
E-SARS-CoV2-R: ATATTGCAGCAGTACGCACACA.
E-SARS-CoV2-P: TET - ACACTAGCCATCCTTACTGCGCTTCG - BHQ-1.
IP2-SARS-CoV2-F: ATGAGCTTAGTCCTGTTG.
IP2-SARS-CoV2-R: CTCCCTTTGTTGTGTTGT.
IP2-SARS-CoV2-P: TET - AGATGTCTTGTGCTGCCGGTA - BHQ-1.
Primers and probes for housekeeping genes GAPDH (assay ID Hs02758991_g1 FAM) and 18S (assay ID Hs03928990_g1 FAM) were obtained from Thermo Fisher Scientific. RNA-positive control was obtained from Biodefense and Emerging Infections Research Resources Repository (BEI Resources, NR-52358). Reactions were run in 384 plates in the thermocycler (ABI7900HT; Applied Biosystems). All samples were run in triplicates. Thermal Cycler protocol used for amplification was 50 °C for 2 min; 95 °C for 10 min; 40 cycles of 95 C for 15 s and 60° C for 1 min SDSv2.4 (Applied Biosystems) software was used for analysis.
To minimize the problem of nonspecific amplification, two independent primers (ie, E gene and IP2) were used with positive detection requiring the amplification of both primers within 40 amplification cycles. The limit of detection of this assay for both primers was determined to be 1–5 RNA copies/reaction. Virus RNA was considered “detected” in a specific tissue when RNA amplification was found with both E and IP2 primer sets at a cycle threshold [Ct] value lower than 40 amplification cycles. Cases where viral RNA was detected by only one of the primers sets were considered inconclusive.
3. Results
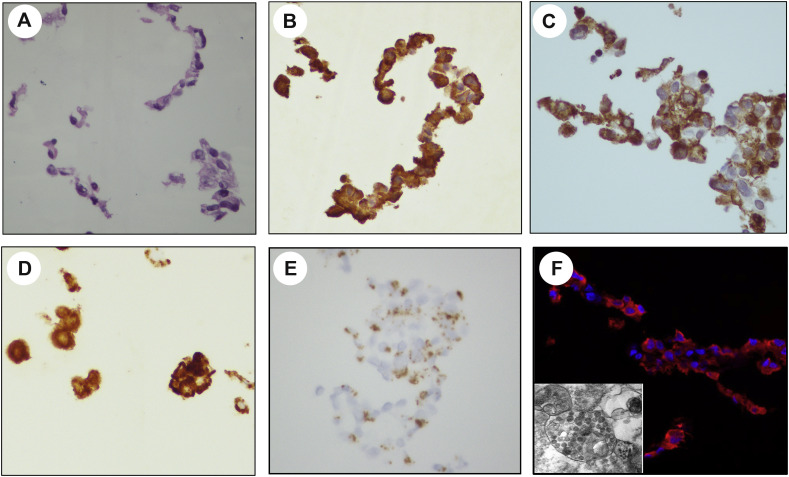
SARS-CoV-2 detection in the Vero E6 cells is shown in Fig. 1 . Viral immunostaining of the nucleoprotein (Fig. 1B) and spike protein (Fig. 1C) was equally intense at 24, 48, and 72 h preparations. In these suspensions of cultured cells, the intense staining together with cytoplasmic overlay of the nucleus made it difficult to confirm cytoplasmic restriction of the signals. RNA ISH with the sense strand RNA probe (S) showed abundant dot-like hybridization signals in the cytoplasm that coalesced into brown globules that effaced cellular and nuclear details (Fig. 1D). The antisense RNA ISH probe (SS), specific for the SARS-CoV-2 spike protein negative RNA strand, showed dot-like hybridization signals confined to the cytoplasm, a pattern indicative of active viral replication within the infected cells (Fig. 1E). Immunofluorescence using the identical SARS-CoV-2 nucleoprotein primary antibody used for IHC but at 1:500 concentration showed strong cytoplasmic staining within most SARS-CoV-2–infected Vero cells at 24 and 48 h of infection (Fig. 1F). Transmission electron microscopy confirmed the presence of densely packed viral particles within the infected cells (Fig. 1F, insert). The presence of dot-like signals in infected cells is consistent with the formation of replication–transcription complexes during infection by coronaviruses and other positive sense RNA viruses [[24], [25], [26]].
Fig. 1.
SARS-CoV-2 detection in infected Vero cell line. Cell pellet stained with hematoxylin and eosin (A). Immunohistochemical stain for the SARS-COV-2 nucleoprotein protein (B) and spike (C). RNA in situ hybridization for the sense RNA strand –S (D) and antisense RNA strand –SS (E) show cytoplasmic singles indicative of infected cells with active viral replication, although the density and confluence of signals was prone to obscure the nucleus in some cells. Immunofluorescence shows strong cytoplasmic staining within the majority of SARS-CoV-2–infected Vero cells (red) (F, inset of electron microscopy showing vesicles containing multiple viral particles).
Uninfected Vero cells showed no recognizable nucleoprotein immunoreactivity and minimal to no background staining was detected using secondary only and nonspecific monoclonal IgG isotype controls. All of the detection assays were negative in the uninfected Vero E6 cells used as a negative control. The general RNA ISH probe confirmed the presence of intact RNA in the Vero cell lines.
The autopsy cases were from eight (57%) females and six (43%) males (Table 1 ). Their ages ranged from 32 to 94 years (median, 66.5 years; mean, 67 years). Eleven (79%) of the autopsies were performed within 24 h of death (median postmortem interval, 7.5 h). The duration of disease from the onset symptoms to death ranged from 1 to 34 days (median, 7.5 days; mean, 10 days). All patients had chronic medical conditions known to increase the risk of severe COVID-19 illness including chronic hypertension (n = 10), chronic kidney disease (n = 4), asthma (n = 4), diabetes (n = 4), cirrhosis (n = 2), dementia (n = 2), and/or severe obesity.
Table 1.
Summary of demographic and clinical information.
| Case | Age | Sex | Disease duration (days) | Postmortem interval (hours) |
Past medical history |
|---|---|---|---|---|---|
| 1 | 94 | f | 11 | 7 |
|
| 2 | 66 | m | 25 | 7 |
|
| 3 | 88 | f | 17 | 11 |
|
| 4 | 67 | f | 3 | 72 |
|
| 5 | 85 | f | 13 | 7 |
|
| 6 | 68 | m | 34 | 6 |
|
| 7 | 64 | m | 3 | 6 |
|
| 8 | 57 | f | 8 | 4 |
|
| 9 | 68 | m | 7 | 8 |
|
| 10 | 86 | f | 1 | 189 |
|
| 11 | 60 | f | 1 | 24 |
|
| 12 | 67 | f | 3 | 72 |
|
| 13 | 32 | m | 6 | 19 |
|
| 14 | 48 | m | 5 | 48 |
|
A summary of the major pathological findings at the time of autopsy are tabulated in Table 2 . The histopathologic findings have been previously reported as part of a larger Mount Sinai COVID-19 autopsy cohort [27]. Most of the injury was in the lungs. Diffuse alveolar damage (DAD) was present in all cases, predominantly in the acute phase, and judged to be the major factor contributing to patient death. All cases showed some degree of hypertensive changes in the heart manifesting as myocyte hypertrophy and interstitial fibrosis. Two cases showed end-stage renal disease. Two cases showed liver cirrhosis. Five cases showed thrombi in the brain with evidence of acute ischemic injury. Hematophagocytosis was a consistent finding in the lymph nodes and spleen.
Table 2.
Summary of major pathologic findings at autopsy.
| Case | Cause of death | Major pathologic findings |
||||||
|---|---|---|---|---|---|---|---|---|
| Lung | Heart | Kidneys | Liver | Brain | LN | Spleen | ||
| 1 | COVID-19 pneumonia with AP | DAD AP |
HTN | ATN ANS |
none | HTN Infarcts Microthrombi |
HP | TNA |
| 2 | COVID-19 pneumonia with AP | DAD AP |
HTN | none | none | Acute ischemia Microthrombi |
none | TNA |
| 3 | COVID-19 pneumonia with AP | DAD AP |
HTN Ischemia |
none | none | Acute ischemia | HP | HP |
| 4 | COVID-19 pneumonia | DAD | HTN | none | none | HTN | TNA | TNA |
| 5 | COVID-19 pneumonia | DAD | HTN | none | ischemia | TNA | TNA | HP |
| 6 | COVID-19 pneumonia | DAD | HTN | none | ischemia | Thrombi Infarction |
HP | HP |
| 7 | COVID-19 pneumonia | DAD | HTN | ANS | none | HTN | HP | HP |
| 8 | COVID-19 pneumonia | DAD | HTN | ESRD | none | HTN | HP | HP |
| 9 | COVID-19 pneumonia | DAD | HTN | ATN | cirrhosis | Microthrombi | none | HP |
| 10 | COVID-19 pneumonia | DAD | HTN Severe AS |
ESRD | none | TNA | TNA | TNA |
| 11 | COVID-19 pneumonia | DAD | HTN | none | none | Thrombi Infarcts |
none | none |
| 12 | COVID-19 pneumonia | DAD | HTN | none | none | TNA | none | TNA |
| 13 | COVID-19 pneumonia complicated by multiorgan failure | DAD | HTN | none | none | TNA | none | TNA |
| 14 | COVID-19 pneumonia | DAD | HTN | none | none | NTA | None | TNA |
Abbreviations: DAD, diffuse alveolar damage; AP, acute pneumonia; HTN, hypertensive changes; TNA, tissue not available; ANS, acute nephrotic syndrome; HP, hemophagocytosis; ESRD, end-stage renal disease; AS, atherosclerosis.
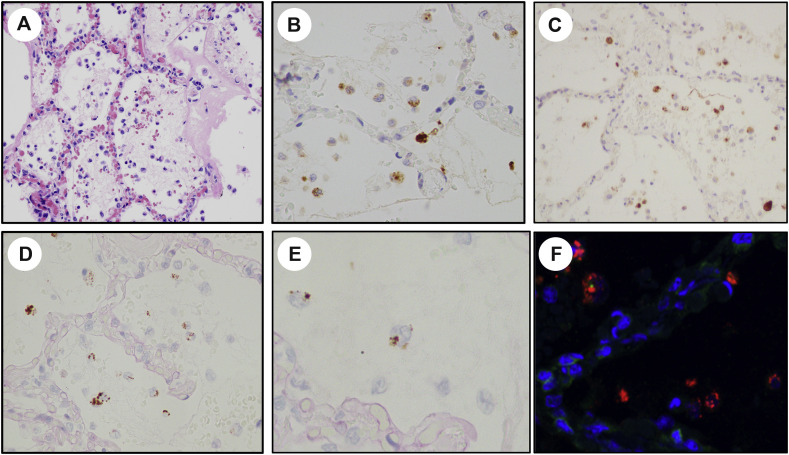
The general RNA ISH probe confirmed the presence of intact RNA in all of the tested autopsy tissue blocks. Detection of SARS-CoV-2 in the autopsy sections as a function of anatomic site and detection method is summarized in Table 2. Despite the constant presence of diffuse alveolar damage in the active stage in all of the lungs, SARS-CoV-2 was only detected in one (7%) of 14 cases (case 7). In this positive case, the SARS-CoV-2 nucleoprotein and spike proteins were detected using IHC and immunofluorescence (Fig. 2 B, C, and G). In addition, abundant signals were detected using the RNA ISH probes against the sense strands (S), but signals were not detected with probes against the antisense strand (SS) (Fig. 2D). The virus products were diffusely present throughout all lung fields, and localized to possibly both the macrophages present in the alveolar spaces, which could have acquired these viral products by phagocytosis, and sporadically in the pneumocytes that lined the alveolar septum and sloughed in the alveolar spaces, indicative of viral infection in these cells. The presence of SARS-CoV-2 in this case was confirmed by immunofluorescence. Immunofluorescence demonstrated strong cytoplasmic SARS-CoV-2 nucleoprotein immunoreactivity within the lung parenchyma of case 7 (Fig. 2F), similar to the staining pattern seen within the positive control SARS-CoV-2–infected Vero cells and showing the same distribution pattern as noted with the IHC and RNA ISH assays. The other organs in case 7 were negative for SARS-CoV-2. The tissue assays did not detect SARS-CoV-2 in any the other 59 organ sections taken in the other 13 patients including the lung sections from 5 patients selected on the basis of short time of disease progress. The overall concordance between IHC and RNA ISH for tissue detection of SARS-CoV-2 was 100%.
Fig. 2.
SARS-CoV-2 detection postmortem lung in case 7. The injured lung shows changes of diffuse alveolar damage including interstitial inflammation and hyaline membrane formation (A, hematoxylin and eosin stain). Virus is present in mostly within the alveolar spaces as detected by immunohistochemistry for the spike protein (B) and nucleoprotein (C), and in situ hybridization for the sense RNA strand (D). The viral infected cells show the same intra-alveolar distribution as the CD68 positive mononuclear phagocytic cells (E). The presence of SARS-CoV-2 was further confirmed by immunofluorescence for the nucleoprotein (F, red labeling).
Correlation between qPCR testing and the histologic detection assays is shown in Table 3 . Using qPCR, virus RNA was detected at a low amplification cycle threshold (Ct 24.07) in the lungs from case 7. In this same patient, virus RNA was also detected in a hilar lymph node, but at higher amplification cycle (34.06). For the other seven cases tested, virus RNA was detected in three lungs (cases 1, 5, and 6), 1 spleen (case 3), and 1 lymph node (case 5) with Ct values ranging from 31.06 to 36.75 (Table 3). Of note, only the lungs in case 7 showed positivity for SARS-CoV-2 by IHC (for spike and NP proteins) and by ISH for S strand. None of the other tissues in this case or the other cases in which the virus detected by PCR showed positivity by IHC or ISH.
Table 3.
Detection of SARS-CoV-2 using various detection platforms.
4. Discussion
In this study, we have investigated the tissue distribution of SARS-CoV-2 in fatal cases of COVID-19 using multiple detection platforms. Following construction of a SARS-CoV-2–infected Vero cell line to guide interpretation of assay results, we found that tissue damage in the lungs and other sites does not always directly correlate with the histologic presence of virus. This disconnect between the presence of virus and tissue damage points to other mechanisms of cellular injury not requiring the sustained presence of SARS-CoV-2.
Although direct viral cytotoxic effects may play some role in initiating a chain of events culminating in patient death, our findings suggest that the sustained presence of SARS-CoV-2 is not necessary to drive disease progression. Of the 73 organs we tested from 14 patients who died of COVID-19, we were able to histologically detect the presence of SARS-CoV-2 protein and RNA in the lungs of only a single patient. In this single positive case, the uncoupling of sense and antisense spike protein RNA expression suggests low levels of RNA replication at the time of staining. Admittedly, SARS-CoV-2 was identified by qPCR—albeit at relatively high cycle thresholds, likely indicative of low viral loads—in the lungs, spleen, and lymph nodes from four patients where virus products were not detected using tissue-based assays. This uncoupling of qPCR-based and tissue-based detection does not invalidate methods of histologic detection, but only underscores that virus may be present at levels below histologic detection thresholds. Low level viral RNA detection by qPCR align with emerging models of disease progression where SARS-CoV-2 RNA is shed into the blood [15,28]. In fact, PCR-based detection could simply reflect virus in transit and should not be taken as unequivocal evidence of organ-specific tropism.
The uncoupling of tissue injury SARS-CoV-2 with the ability to consistently visualize the presence of virus using histologic detection assays has been intimated in smaller studies. Schaefer et al. [1] could detect high numbers (ie, >5 cells per 4 mm2) of infected cells in just two out of seven post mortem lungs. Massoth et al. [13] were unable to detect SARS-CoV-2 in any extrapulmonary sites from seven postmortem cases, and Santoriello et al. [12] were unable to detect virus in ten postmortem kidneys. Although Dorward et al. [14] were able to histologically detect SARS-CoV-2 in various postmortem organs, virus distribution did not topographically correlate with pathologic responses. This difficulty in unequivocally identifying virus in injured tissues mirrors the experience with SARS-CoV-1: viral detection often eludes detection by IHC and ISH, and most of the extrapulmonary manifestations have been attributed to systemic effects of abnormal inflammatory or immune reactions to the virus [14,[29], [30], [31]]. This phenomenon is not unique to the coronavirus family but has also been reported influenza virus infections [[32], [33], [34]].
Even though sustained viral presence may not be required to drive disease progression, tissue infection with cytotoxic injury may be still be required to trigger the cascade of diffuse endothelial injury, hypercoagulability, and a hyperinflammatory state culminating in respiratory demise, multiorgan failure, and death [14,35,36]. Histologic detection of SARS-CoV-2 could help define the temporal sequence of these events. In the progression of COVID-19, the temporal role of SARS-CoV-2 as a triggering event in the lung infections appears to be very early and transitory as some have suggested that SARS-CoV-2 tissue infection occurs at an early acute phase of disease progression [1,37]. Some have suggested that SARS-CoV-2 tissue infection occurs at an early acute phase of disease progression [1,37]. With this in mind, we selectively evaluated postmortem lungs from five cases with short intervals disease intervals (mean 3 days). SARS-CoV-2 was not detected in any of these cases. In light of these findings, the temporal role of SARS-CoV-2 as a triggering event in the lungs lung infections appears to be very early and transitory.
In summary, what was initially conceptualized as a primarily respiratory viral disease, COVID-19 is now recognized as a heterogeneous illness with a diverse array of symptoms and complications that may not necessarily be linked to direct viral injury. Our findings support the view that this complex process is likely related to ongoing and progressive immune dysregulation rather than persistent viral replication within the lung and other tissues and may advocate to focus on attenuating the pathological host response rather than targeting the actual virus in managing and treating patients with COVID-19.
Acknowledgments
I“n loving memory of Dr. Mary Fowkes”.
This research was partly funded by CRIP (Center for Research for Influenza Pathogenesis), a NIAID supported Center of Excellence for Influenza Research and Surveillance (CEIRS, contract # HHSN272201400008C); by NIAID grant U19AI142733 and NCI grant U54CA260560; by the generous support of the JPB Foundation and the Open Philanthropy Project (research grant 2020-215611 (5384); and by anonymous donors to A.G-S.; partial funding by NIH grant R01NS106229-02S2, United States We thank Dr. Florian Kramer for providing the infected cell lines and Jason Reidy for providing the EM picture.
Footnotes
Competing interests: The AG-S laboratory has received research support from Pfizer, Pharmamar, Blade Therapeutics, Avimex, Dynavax, Kenall Manufacturing, ImmunityBio, Nanocomposix, Senhwa Biosciences, and 7Hills Pharma. AG-S has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Accurius, and Esperovax.
References
- 1.Schaefer I.M., Padera RF, Solomon IH, et al. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod Pathol. 2020 doi: 10.1038/s41379-020-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su H., Yang M, Wan C, et al. Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int. 2020;98:219–227. doi: 10.1016/j.kint.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tavazzi G., Pellegrini C, Maurelli M, et al. Myocardial localization of coronavirus in COVID-19 cardiogenic shock. Eur J Heart Fail. 2020;22:911–915. doi: 10.1002/ejhf.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiao F., Tang M, Zheng X, et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158 doi: 10.1053/j.gastro.2020.02.055. 1831-1833.e1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puelles V.G., Lütgehetmann M, Lindenmeyer MT, et al. Multiorgan and renal tropism of SARS-CoV-2. N Engl J Med. 2020 doi: 10.1056/NEJMc2011400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun F., Lütgehetmann M, Pfefferle S, et al. SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet. 2020;396:597–598. doi: 10.1016/S0140-6736(20)31759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bussani R., Schneider E, Zentilin L, et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine. 2020;61:103104. doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martines R.B., Ritter JM, Matkovic E, et al. Pathology and pathogenesis of SARS-CoV-2 associated with fatal coronavirus disease, United States. Emerg Infect Dis. 2020;26:2005–2015. doi: 10.3201/eid2609.202095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borczuk A.C., Salvatore SP, Seshan SV, et al. COVID-19 pulmonary pathology: a multi-institutional autopsy cohort from Italy and New York City. Mod Pathol. 2020 doi: 10.1038/s41379-020-00661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szabolcs M., Sauter JL, Frosina D, et al. Identification of immunohistochemical reagents for in situ protein expression analysis of coronavirus-associated changes in human tissues. Appl Immunohistochem Mol Morphol. 2020 doi: 10.1097/PAI.0000000000000878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauter J.L., Baine MK, Butnor KJ, et al. Insights into pathogenesis of fatal COVID-19 pneumonia from histopathology with immunohistochemical and viral RNA studies. Histopathology. 2020;77:915–925. doi: 10.1111/his.14201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santoriello D., Khairallah P, Bomback AS, et al. Postmortem kidney pathology findings in patients with COVID-19. J Am Soc Nephrol. 2020;31:2158–2167. doi: 10.1681/ASN.2020050744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massoth L.R., Desai N, Szabolcs A, et al. Comparison of RNA in situ hybridization and immunohistochemistry techniques for the detection and localization of SARS-CoV-2 in human tissues. Am J Surg Pathol. 2020 doi: 10.1097/PAS.0000000000001563. [DOI] [PubMed] [Google Scholar]
- 14.Dorward D.A., Russell CD, Um IH, et al. Tissue-specific immunopathology in fatal COVID-19. Am J Respir Crit Care Med. 2021;203:192–201. doi: 10.1164/rccm.202008-3265OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhatnagar J., Gary J, Reagan-Steiner S, et al. Evidence of SARS-CoV-2 replication and tropism in the lungs, airways and vascular endothelium of patients with fatal COVID-19: an autopsy case-series. J Infect Dis. 2021 doi: 10.1093/infdis/jiab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldsmith C.S., Miller S.E., Martines R.B., Bullock H.A., Zaki S.R. Electron microscopy of SARS-CoV-2: a challenging task. Lancet. 2020;395:e99. doi: 10.1016/S0140-6736(20)31188-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller S.E., Brealey J.K. Visualization of putative coronavirus in kidney. Kidney Int. 2020;98:231–232. doi: 10.1016/j.kint.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullock H.A., Goldsmith C.S., Zaki S.R., Martines R.B., Miller S.E. Difficulties in differentiating coronaviruses from subcellular structures in human tissues by electron microscopy. Emerg Infect Dis. 2021;27 doi: 10.3201/eid2704.204337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dittmayer C., Meinhardt J, Radbruch H, et al. Why misinterpretation of electron micrographs in SARS-CoV-2-infected tissue goes viral. Lancet. 2020;396:e64–e65. doi: 10.1016/S0140-6736(20)32079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amanat F., Stadlbauer D, Strohmeier S, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020 doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corman V.M., Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etievant S., Bal A, Escuret V, et al. Performance assessment of SARS-CoV-2 PCR assays developed by WHO referral laboratories. J Clin Med. 2020;9 doi: 10.3390/jcm9061871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.P. Institut Pasteur . 2020. Protocol: real-time RT-PCR assays for the detection of SARS-CoV-2. https://www.who.int/docs/default-source/coronaviruse/real-time-rt-pcr-assays-for-the-detection-of-sars-cov-2-institut-pasteur-paris.pdf?sfvrsn=3662fcb6_2b. [Google Scholar]
- 24.Knoops K., Kikkert M, van den Worm SHE, et al. SARS-Coronavirus Replication Is Supported by a Reticulovesicular Network of Modified Endoplasmic Reticulum, In: PloS Biol. Vol. 6. 2008 doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Hemert M.J., van den Worm SH, Knoops K, et al. SARS-coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller S., Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–374. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryce C., Grimes Z, Pujadas E, et al. Pathophysiology of SARS-CoV-2: the Mount Sinai COVID-19 autopsy experience. Mod Pathol. 2021 doi: 10.1038/s41379-021-00793-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Solomon I.H., Normandin E, Bhattacharyya S, et al. Neuropathological features of covid-19. N Engl J Med. 2020;383:989–992. doi: 10.1056/NEJMc2019373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.To K.F., Tong JH, Chan PK, et al. Tissue and cellular tropism of the coronavirus associated with severe acute respiratory syndrome: an in-situ hybridization study of fatal cases. J Pathol. 2004;202:157–163. doi: 10.1002/path.1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsiao C.H., Chang M.F., Hsueh P.R., Su I.J. Immunohistochemical study of severe acute respiratory syndrome-associated coronavirus in tissue sections of patients. J Formos Med Assoc. 2005;104:150–156. [PubMed] [Google Scholar]
- 31.Nicholls J.M., Butany J, Poon LL, et al. Time course and cellular localization of SARS-CoV nucleoprotein and RNA in lungs from fatal cases of SARS. PLoS Med. 2006;3:e27. doi: 10.1371/journal.pmed.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guarner J., Shieh WJ, Dawson J, et al. Immunohistochemical and in situ hybridization studies of influenza A virus infection in human lungs. Am J Clin Pathol. 2000;114:227–233. doi: 10.1309/HV74-N24T-2K2C-3E8Q. [DOI] [PubMed] [Google Scholar]
- 33.Guarner J., Paddock CD, Shieh WJ, et al. Histopathologic and immunohistochemical features of fatal influenza virus infection in children during the 2003-2004 season. Clin Infect Dis. 2006;43:132–140. doi: 10.1086/505122. [DOI] [PubMed] [Google Scholar]
- 34.Shieh W.J., Blau DM, Denison AM, et al. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am J Pathol. 2010;177:166–175. doi: 10.2353/ajpath.2010.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cordon-Cardo C., Pujadas E, Wajnberg A, et al. COVID-19: staging of a new disease. Canc Cell. 2020;38:594–597. doi: 10.1016/j.ccell.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta A., Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26:1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schurink B., Roos E, Radonic T, et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]