Abstract
Background
Hypertension-related microRNA(miR)-1283 and its target gene, activating transcription factor-4 (ATF4), can regulate vascular endothelial dysfunction. This study aimed to explore whether miR-1283 prevents hypertension through targeting ATF4.
Material/Methods
Transcriptome sequencing was performed after overexpression or inhibition of miR-1283 in human amniotic epithelial cells (HAECs). After miR-1283 was overexpressed or inhibited in HAECs, ATF4+/− and wild-type mice were induced with a high-salt diet. We detected the expression of ATF4, C/EBP-homologous protein (CHOP), BH3-interacting domain death agonist (BID), Bcl-2, Bcl-2-like protein 11 (BIM), Bcl-2-like protein 1 (BCL-X), and caspase-3 by PCR and western blotting. We detected the changes of vasoactive substances including nitric oxide (NO), endothelin 1 (ET-1), endothelial protein C receptor (EPCR), thrombin (TM), and von Willebrand factor (vWF) by ELISA.
Results
Compared with that of the miR-1283- inhibited group, NO was higher in the miR-1283 overexpression group, while the expression of ET-1, EPCR, TM, and vWF were lower. Similarly, compared with that of the miR-1283 inhibited group, the expression of ATF4, CHOP, BID, BIM, and caspase-3 in the miR-1283 overexpression group was downregulated, while the expression of BCL-2 and BCL-X was upregulated (P<0.05). In vivo experiments showed the lack of ATF4 gene could prevent hypertension in mice induced by high-salt diet and protect endothelial function.
Conclusions
The mechanism of regulating blood pressure and endothelial function of the miR-1283/ATF4 axis was related to inhibiting endoplasmic reticulum stress and cell apoptosis through the ATF4/CHOP signaling pathway. Therefore, the miR-1283/ATF4 axis may be a target for the prevention and treatment of hypertension.
Keywords: Activating Transcription Factor 4, Endoplasmic Reticulum Stress, Endothelial Cells, Hypertension
Background
Hypertension is a common chronic disease characterized by elevated systemic arterial pressure, which can damage target organs such as the heart, brain, kidney, and blood vessels [1,2]. Cardiovascular disease is responsible for causing 45% of deaths from noncommunicable diseases worldwide, according to the World Health Organization. Epidemiological statistics predict that by 2025, the number of patients with hypertension will reach 1.56 billion, with a prevalence of 29.2% [3]. Currently, of the people who have high blood pressure, less than one-third are receiving treatment, and less than 1 in 12 attain good blood pressure control. Further, 2 million people die each year from hypertension and its complications, according to a recent report from China [4–6]. The etiology of hypertension is not completely clear; however, current research shows that a diet high in salt intake is the main cause of hypertension [4–6]. Hypertension is often associated with vascular dysfunction, endoplasmic reticulum (ER) stress, and cell apoptosis. Studies found that long-term treatment with an ER chemical chaperone can improve arterial blood pressure and restore vascular function in spontaneous hypertension rat models [7–10]. Cheng et al found that metformin prevents vascular damage in hypertension through the AMPK/ER stress pathway [11]. The reasons for the occurrence and development of hypertension is a hot topic, which researchers are urgently trying to solve [12].
In previous studies in patients with hypertension, we used serum intervention in human umbilical vein endothelial cells (HUVECs) and found that microRNA (miR)-1283 expression in patients with high blood pressure was reduced and the expression of activating transcription factor 4 (ATF4) was increased [13,14]. ATF4 is the target gene of miR-1283, as confirmed by TargetScan Soft and luciferase activity assays [13,14]. The results also indicated that the pathogenesis of vascular endothelial cell injury may be related to ER stress. Other studies also showed that miR-1283 is associated with essential hypertension and gestational hypertension [15,16]. ATF4 is a key mRNA for miR-1283 regulating the ER stress pathway and it might be a hypertension-specific biomarker [14]. Therefore, the study of the pathogenesis of hypertension from the perspective of the miR-1283/ATF4 axis is significant.
Vascular endothelial cells provide a physical barrier between circulating blood and the blood vessel wall and they secrete a variety of vasoactive substances, which regulate blood viscosity, vasoconstriction, and the relaxation function [17]. All of these processes are involved in maintaining vascular endothelial function and are important for maintaining normal blood pressure [18]. For example, our previous study found that ER stress and the apoptosis of vascular endothelial cells play an important role in hypertension [14].
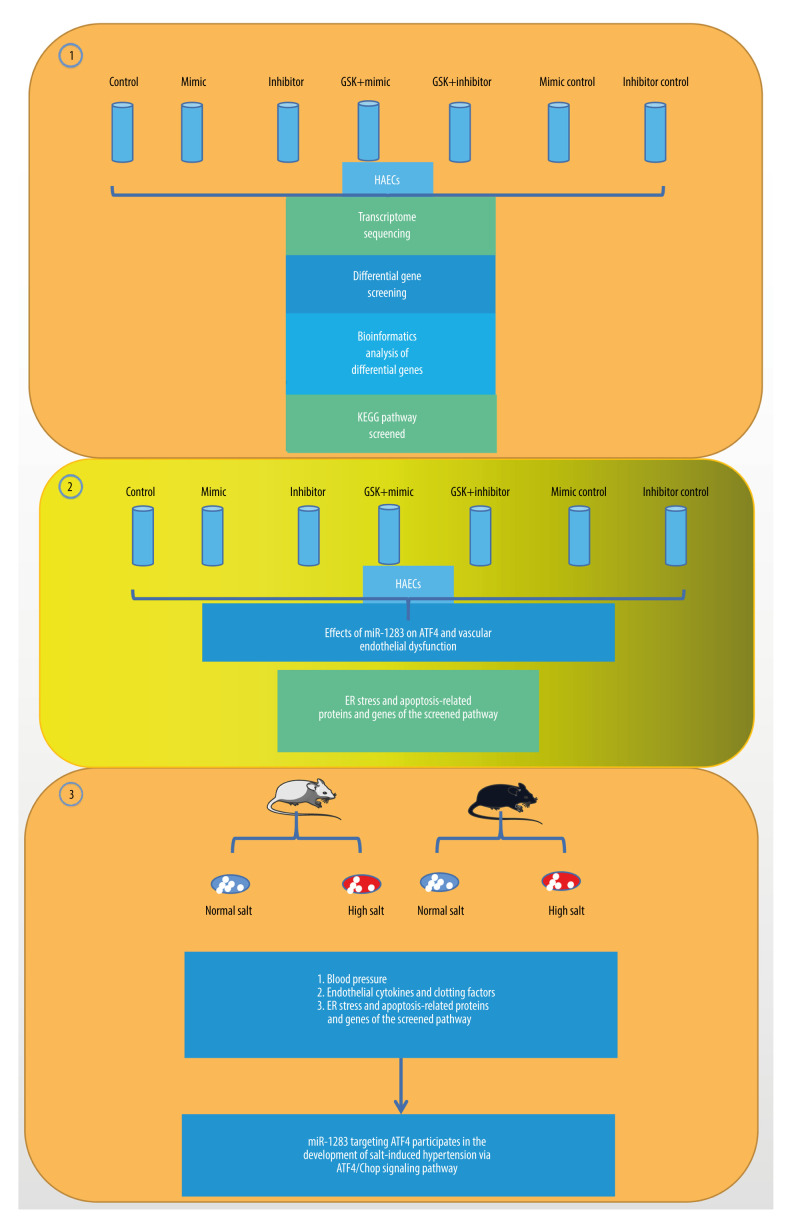
The present study used a human amniotic epithelial cell (HAEC) and ATF4 gene knockout mouse model and the study was conducted in 3 parts (Figure 1). First, we investigated the overexpression and inhibition of miR-1283 in HAECs, conducted transcriptome analysis of the target relationship between miR-1283 and ATF4, and analyzed biological information. Second, we investigated the regulatory effects of ATF4, the target gene of miR-1283, on salt-induced hypertension and vascular endothelial dysfunction. Third, combinations of in vivo and in vitro experiments were conducted to further study the mechanism of the miR-1283 and ATF4 axis participating in the formation of hypertension.
Figure 1.
The illustration of miR-1283 targeting of activating transcription factor 4 (ATF4) participating in the development of salt-induced hypertension via the ATF4/CHOP signaling pathway. Control – control group; mimic – miR-1283 mimic group; inhibitor – miR-1283 inhibitor group; GSK – GSK2656157; mimic control – miR-1283 mimic negative control group; inhibitor control – miR-1283 inhibitor negative control group.
Material and Methods
Cell Culture Experimental Groups
When HAECs (American Type Culture Collection) reached a fusion degree of more than 90% in a T25 culture flask, the supernatant was discarded, washed with phosphate-buffered saline for 3 times, and then digested with 0.25% trypsin (Gibco, USA) for 1 min. The cells were then observed under an inverted microscope. When most of the cells became round, 1 mL of Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) containing 10% fetal bovine serum (FBS) was added immediately to terminate digestion. Then, the cells were gently blown with a pipette for peeling and collecting. The cells were centrifuged at 4°C and 1000 rpm for 5 min. Finally, we added 4 mL of DMEM containing 10% FBS and inoculated the mixture in a ratio of 1: 2 into a new T25 culture flask. After mixing, the flask was put into a cell incubator at 37.0°C and cultured in 5% CO2. A total of 7 groups were created with 3 repetitions per group: (1) GSK2656157 (1 μM)+miR-1283 mimic group; (2) GSK2656157 (1 μM)+miR-1283 inhibitor group; (3) miR-1283 mimic group; (4) miR-1283 inhibitor group; (5) miR-1283 mimic negative control group; (6) miR-1283 inhibitor negative control group; and (7) a normal control group. GSK2656157 (CAS no: 1337532-29-2) is an ATP-competitor and highly selective PERK inhibitor. Pretreating cells with GSK2656157 can inhibit the activation of PERK and reduce the expression of downstream substrates including eIF2a, ATF4, and the C/EBP-homologous protein (CHOP). For processing, cells in the normal control group did not undergo any treatment. Cells in the miR-1283 mimic group were transfected with 50 nmol/L of miR-1283 mimic for 24 h. Cells in the miR-1283 inhibitor group were treated with 50 nmol/L of inhibitor for 24 h. The GSK2656157 + miR-1283 mimic group cells were pretreated with 1 μM of GSK2656157 for 1 h and transfected with a 50 nmol/L mimic for 24 h. The GSK2656157+miR-1283 inhibitor group cells were pretreated with 1μM of GSK2656157 for 1 h, followed by treatment with 50 nmol/L inhibitor for 24 h. Cells in the miR-1283 mimic negative control group were transfected with 50 nmol/L of mimic negative control for 24 h. Cells in the miR-1283 inhibitor negative control group were negatively controlled by 50 nmol/L of inhibitor for 24 h.
Animals and Animal Experimental Protocol
Male ATF4+/− heterozygous and ATF4+/+ wild-type mice with a C57BL/6J background were obtained from GemPharmatech Co., Ltd. The temperature in the feeding room was 21±3°C, humidity was 50%, and a there was a 12-h light/dark cycle. The animals were able to freely access water and food. All animal experiments were supervised by the Medical Ethics Committee of Jinan University (approval No. 20190221-02). Before the experiments, the mice acclimated to the environment for at least 1 week. There were a total of 18 ATF4+/− mice and 18 wild-type mice. They mice were divided into 4 groups: (1) ATF4 knockout mice with normal salt group (KO-NS, n=6); (2) ATF4 knockout mice with 8% high-salt diet groups (KO-HS, n=12); (3) wild-type mice with normal diet group (WT-NS, n=6); and (4) wild-type with 8% high-salt diet group (WT-HS, n=12). For processing, the WT-NS and KO-NS groups were fed with normal mouse feed, while the WT-HS and KO-HS groups were fed with 8% high-salt feed customized by the Nantong Trofield Co. Food and water were made freely available to the mice for 28 days. At the end of the experiment, the mice were euthanized with sodium pentobarbital (60 mg/kg IP), blood was collected from the eyeball, and thoracic aorta tissue was collected for further research.
Transcriptome Sequencing
RNA Extraction
The cells in the control group, miR-1283 mimic group, and miR-1283 inhibitor group were used for transcriptome sequencing; each group was set up with 3 repetitions. Total RNA was extracted from the tissue using TRIzol reagent, according the manufacturer’s instructions (Invitrogen), and genomic DNA was removed using DNase I (TaKaRa). Next, RNA quality was determined using a 2100 Bioanalyser (Agilent) and was quantified with an ND-2000 spectrophotometer (NanoDrop Technologies). The high-quality RNA sample, which met the standard of OD 260/280=1.8~2.2, OD 260/230 ≥2.0, 28S: 18S ≥1.0, RIN ≥6.5, >2 μg, was included in the sequencing library.
Sequencing and Read Mapping
After the RNA-seq transcriptome library was completed using 1 μg of total RNA according to TruSeqTM RNA sample preparation kit, the RNA-seq sequencing library was then sequenced using the Illumina HiSeq X Ten/NovaSeq 6000 sequencer (2×150 bp read length). The raw paired-end reads were trimmed and quality controlled by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) with default parameters. Next, the clean reads were separately aligned to the reference genome with an orientation mode using TopHat (http://tophat.cbcb.umd.edu/, version2.0.0) software [18].
Differential Gene Expression Analysis and Functional Enrichment
The expression level of each transcript was calculated based on the fragments per kilobase of exon per million mapped reads method to estimate the differentially expression genes between 2 different groups. The differential expression analysis was performed using R statistical package software EdgeR (http://www.bioconductor.org/packages/2.12/bioc/html/edgeR.html) [19,20]. In addition, functional enrichment analysis, including Gene Ontology (GO), was performed to identify which differentially expression genes were significantly enriched in GO terms at a Bonferroni-corrected P value ≤0.05, compared with the whole transcriptome background. GO functional enrichment was carried out by DAVID bioinformatics resources 6.8 (https://david.ncifcrf.gov/).
Blood Pressure and Weight Measurements
The mice adapted to the detection environment for 2 weeks before the experiment, and then the blood pressure of each group was measured by the non-invasive tail sleeve method with a BP-2000 Blood Pressure Analysis System (Visitech, USA) before the intervention. The arterial systolic pressure was measured with multiple readings, and the mean value was obtained after obtaining 6 stable measurements. Blood pressure was measured every 4 days after intervention for a total of 7 times, within a total period of 28 days. Body weight was also measured. Each step was performed by the same operator to maintain the consistency of readings.
Measurement of Vasoactive Substances by Enzyme-Linked Immunoassay
The cell suspension was centrifuged at 1000 rpm for 5 min to remove the residual cells, and the supernatant was collected and stored at −80°C. After 28 days of feeding, the mice were euthanized with sodium pentobarbital. Blood samples were drawn from the eyeball, naturally solidified at room temperature for 20 min, centrifuged at 3000 rpm for 20 min at 2°C to 8°C, and the supernatant was collected and stored at −80°C. The concentrations of nitric oxide (NO), endothelin 1 (ET-1), endothelial protein C receptor (EPCR), thrombin (TM), von Willebrand factor (vWF), tissue plasminogen activator (tPA), and plasminogen activator inhibitor (PAI) in the supernatant were detected, according to the instructions of the enzyme-linked immunoassay kit (Huyu, China).
Quantitative Reverse Transcription-Polymerase Chain Reaction
Total RNA was extracted from tissues and cells by TRIzol reagent (Invitrogen, USA), and the synthesis of complementary DNA (cDNA) and quantitative reverse transcription-polymerase chain reaction were done using a reverse transcriptase kit (Takara, China) and SYBR Green PCR kit (Takara, China), respectively. cDNA samples of target genes ATF4, CHOP, BCL-2, BCL-X, BH3-interacting domain death agonist (BID), Bcl-2-like protein 11 (BIM), caspase-3, and GAPDH were detected using the Applied Biosystems 7500 Real-Time PCR System (Applied Biosystems, CA, USA). Lastly, the mRNA expression levels were normalized by GAPDH and the gene expression levels were calculated using the 2−ΔΔCT method. The primer sequences of mouse and human aortic cells are shown in Table 1.
Table 1.
Primer sequences of mouse and human aortic cells.
| Gene | Sample | Primer |
|---|---|---|
| ATF4 | Mouse | F: 5′-CAAAACAAGACAGCAGCCACTA-3′ |
| R: 5′-CTTCTTCCCCCTTGCCTTAC-3′ | ||
| HAEC | F: 5′-TGCCCGTCCCAAACCTTAC-3′ | |
| R: 5′-TGCTCCGCCCTCTTCTTCT-3′ | ||
| BCL-X | Mouse | F: 5′-GGAGAGCGTTCAGTGATC-3′ |
| R: 5′-CAATGGTGGCTGAAGAGA-3′ | ||
| HAEC | F: 5′-GGGCATTCAGTGACCTGACA-3′ | |
| R: 5′-ATGCGACCCCAGTTTACCC-3′ | ||
| BCL-2 | Mouse | F: 5′-AGGGGGAAACACCAGAATC-3′ |
| R: 5′-GGTAGCGACGAGAGAAGTCA-3′ | ||
| HAEC | F: 5′-AACATCGCCCTGTGGATGAC-3′ | |
| R: 5′-AGAGTCTTCAGAGACAGCCAGGAG-3′ | ||
| BID | Mouse | F: 5′-TGGAGAAGGACCTGATAACC-3′ |
| R: 5′-CTTGTAGGCAGTGACGATGA-3′ | ||
| HAEC | F: 5′-AATGGCCTTCATATCATCCACACA-3′ | |
| R: 5′-GGGAACCTGCACAGTGGAAATAA-3′ | ||
| Caspase-3 | Mouse | F: 5′-GTCCCACTGTCTGTCTCA-3′ |
| R: 5′-GTCTCGCTCTACTGTAAG-3′ | ||
| HAEC | F: 5′-GACTCTGGAATATCCCTGGACAACA-3′ | |
| R: 5′-AGGTTTGCTGCATCGACATCTG-3′ | ||
| CHOP | Mouse | F: 5′-CTCGCTCTCCAGATTCCAGT-3′ |
| R: 5′-CTGCTCCTTCTCCTTCATGC-3′ | ||
| HAEC | F: 5′-TCTAAGGCACTGAGCGTATCATGT-3′ | |
| R: 5′-TTTCAGGTGTGGTGATGTATGAAG-3′ | ||
| BIM | Mouse | F: 5′-GGCCAAGCAACCTTCTGACT-3′ |
| R: 5′-TGCCTTCTCCATACCAGACG-3′ | ||
| HAEC | F: 5′-CATCATCGCGGTATTCGGTTC-3′ | |
| R: 5′-AAGGTTGCTTTGCCATTTGGTC-3′ | ||
| GAPDH | Mouse | F: 5′-tggccgtggggctgcccag-3′ |
| R: 5′-ggaaggccatgccagtgagc-3′ | ||
| HAEC | F: 5′-GCACCGTCAAGGCTGAGAAC-3′ | |
| R: 5′-TGGTGAAGACGCCAGTGGA-3′ |
Western Blot Assay
An amount of 5×105 HAEC cells or 100 mg mouse thoracic aorta were extracted, and a 600 uL radioimmunoprecipitation assay buffer containing phenylmethanesulfonyl fluoride was added, and the mice were lysed on ice for 30 min. The pyrolysis products were centrifuged at 4°C and 12000 rpm for 5 min. The supernatant was collected, and the total protein concentration was determined by the bicinchoninic acid method with bovine serum albumin as the standard reference. The 30 ug of protein extracted from each sample was separated with 10% to 15% sodium dodecyl sulfate polyacrylamide gel electrophoresis. The separated protein was transferred to a polyvinylidene fluoride membrane and incubated at room temperature with 5% dried skimmed milk powder sealant for 1 h. Next, the diluted primary antibodies anti-ATF4 (11815S), anti-CHOP (2895S), anti-BCL-2 (D038-3), anti-BCL-XL (2764S), anti-BID (AB1735), anti-BIM (2933S), anti-caspase-3 (9662S), and anti-GAPDH (ab181602) were prepared and stored at 4°C overnight. Next, the primary antibodies were removed and washed with TBST. They were incubated with a 1: 1000 horseradish peroxidase-conjugated goat anti-rabbit IgG (Beyotime, China) for 1 h, and then rinsed with TBST and developer (Millipore, USA). Finally, the optical signal was observed by enhanced chemiluminescence, and the optical density of the band was determined by the ImageJ program and normalized to a GAPDH signal.
Statistical Analysis
SPSS version 13.0 was used for data analysis, and the measurement data were expressed as mean±standard deviation. One-way analysis of variance was used for between group mean comparisons. The Student-Newman-Keuls method was used for between group comparison when the variances were homogeneous, and Tamhane’s T2 method was used for between group comparison when the variances were heterogeneous. A value of P<0.05 indicated statistical significance.
Results
Comparison of Differences Between Groups
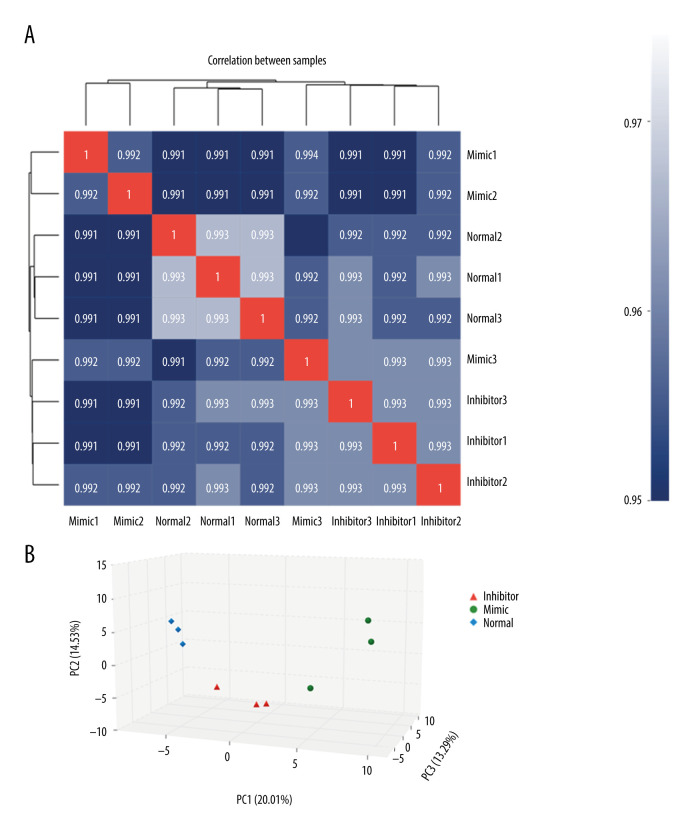
The results of the correlation and clustering analyses between samples showed that the correlation coefficients of samples in the control group, miR-1283 mimic group, and miR-1283 inhibitor group were all greater than 0.99, and there was no specificity in each sample directly (Figure 2A). Principal component analysis results showed that the control group, miR-1283 mimic group, and miR-1283 inhibitor group had similar sample composition within the groups; however, the differences between the groups were large, indicating there was consistency within the groups and differences between the groups (Figure 2B).
Figure 2.
Comparison of differences between groups. (A) The correlation analysis; (B) principal component analysis (PCA). Mimic – miR-1283 mimic group; inhibitor – miR-1283 inhibitor group; normal – control group.
Differential Gene Screening
The raw counts of the differentially expressed genes were analyzed by DESeq2 software, based on a negative binomial distribution, and genes with a different expression between the comparison groups were obtained based on certain screening conditions (parameter: P value <0.05). There were 4840 differentially expressed genes in the miR-1283 mimic group, of which 2419 were upregulated and 2421 were downregulated. There were 4410 differentially expressed genes in miR-1283 inhibitor group, of which 2144 were upregulated and 2266 were downregulated. There was an intersection of 2144 upregulated differentially expressed genes in the miR-1283 inhibitor group and control group. Taken together, there were 23 common differentially expressed genes related to miR-1283 (Table 2).
Table 2.
Common differentially expressed genes.
| Gene ID | Gene name |
|---|---|
| ENSG00000177606 | JUN |
| ENSG00000115808 | STRN |
| ENSG00000115758 | ODC1 |
| ENSG00000067057 | PFKP |
| ENSG00000100664 | EIF5 |
| ENSG00000144959 | NCEH1 |
| ENSG00000184575 | XPOT |
| ENSG00000100422 | CERK |
| ENSG00000033867 | SLC4A7 |
| ENSG00000146648 | EGFR |
| ENSG00000136244 | IL6 |
| ENSG00000179295 | PTPN11 |
| ENSG00000127314 | RAP1B |
| ENSG00000128272 | ATF4 |
| ENSG00000105993 | DNAJB6 |
| ENSG00000025772 | TOMM34 |
| ENSG00000175197 | DDIT3 |
| ENSG00000151012 | SLC7A11 |
| ENSG00000026025 | VIM |
| ENSG00000198959 | TGM2 |
| ENSG00000139318 | DUSP6 |
| ENSG00000116016 | EPAS1 |
| ENSG00000067082 | KLF6 |
Bioinformatics Analysis of Differential Genes
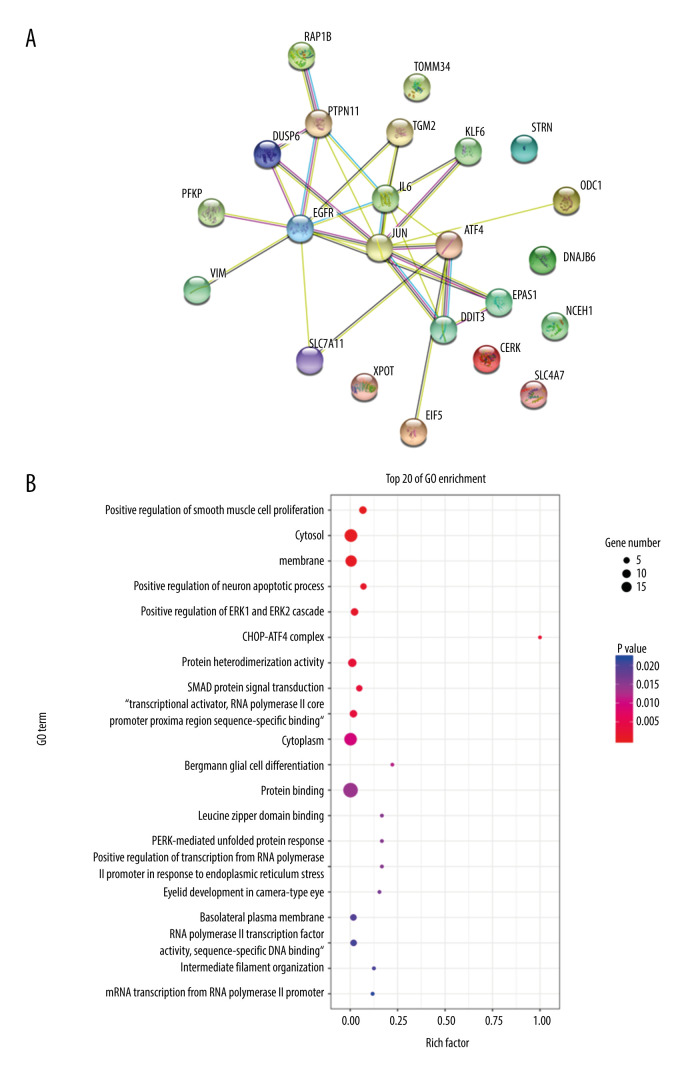
The protein-protein interaction correlation analysis of these 23 genes showed that JUN, ATF4, EGFR, and IL6 played a core role (Figure 3A). GO enrichment analysis was performed on the 23 genes, including PERK-mediated unfolded protein response, intrinsic apoptotic signaling pathway in response to ER stress, CHOP-ATF4 complex, and 66 other functions, and according to the number of vasoactive substances and clotting factors gene enrichment, the top 20 GO functions were plotted into a histogram (Figure 3B).
Figure 3.
Bioinformatics analysis of differential genes. (A) Protein-protein interaction (PPI) correlation analysis of these 23 genes. (B) Gene Ontology enrichment analysis.
miR-1283 Regulated Vasoactive Substances and Clotting Factors
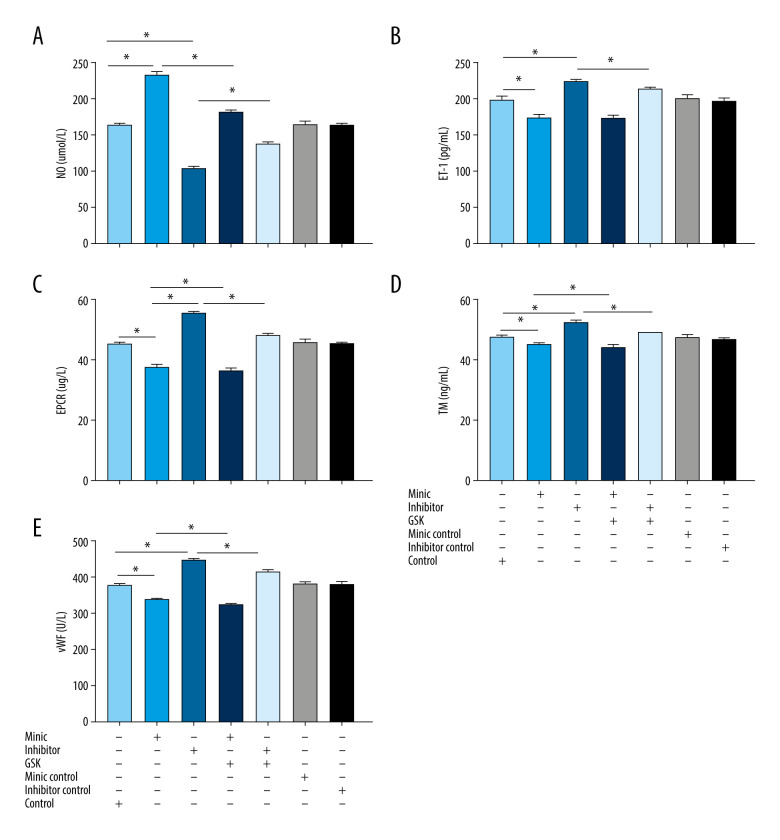
The results showed that NO was higher and ET-1, EPCR, TM, and vWF were lower in the miR-1283 mimic group than in the control group (P<0.05). By contrast, the NO level was lower and ET-1, EPCR, TM, and vWF levels were higher in the miR-1283 inhibitor group than in the control group (P<0.05). Compared with that of the miR-1283 mimic group, NO, EPCR, TM, and vWF levels were lower in the GSK2656157+miR-1283 mimic group (P<0.05), and there was no significant change in ET-1 (P>0.05). Compared with that of the miR-1283 inhibitor group, NO in the GSK2656157+miR-1283 inhibitor group was higher, and ET-1, EPCR, TM and vWF were lower (P<0.05). There was no significant change in NO, ET-1, EPCR, TM, and vWF in the control group, miR-1283 mimic negative control group, and miR-1283 inhibitor negative control group (P>0.05). These results indicated that the inhibition of miR-1283 may lead to a decrease in the expression of HAEC vasodilators and an increase in coagulation factors, which further leads to cell endothelial dysfunction, while overexpression of miR-1283 can increase the expression of HAEC vasodilators and reduce the release of clotting factors (Figure 4). All findings indicated that miR-1283 regulated vasoactive substances and coagulation factors.
Figure 4.
The content of vasoactive substances and clotting factors in the cell supernatant intervened by miR-1283. (A–E) nitric oxide (NO), endothelin 1 (ET-1), endothelial protein C receptor (EPCR), thrombin (TM), von Willebrand factor (vWF). Control – control group; mimic – miR-1283 mimic group; inhibitor – miR-1283 inhibitor group; GSK – GSK2656157; mimic control – miR-1283 mimic negative control; inhibitor control – miR-1283 inhibitor negative control group. * P<0.05.
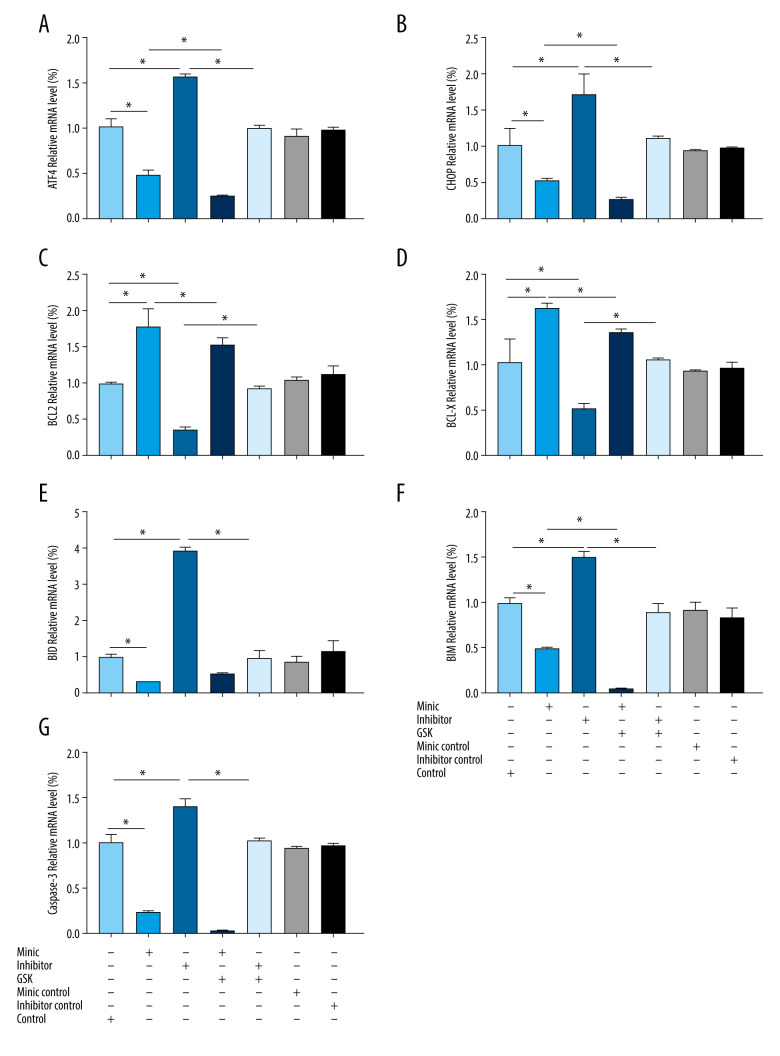
miR-1283 Regulated mRNA of ER Stress and Apoptosis-Related Genes
Compared with that of the control group, the expression of ATF4, CHOP, BID, BIM, and caspase-3 mRNA in the miR-1283 mimic group were lower, while the expression of BCL-X and BCL-2 mRNA were higher (P<0.05). However, the miR-1283 inhibitor group showed a higher expression of ATF4, CHOP, BID, BIM, and caspase-3 mRNA, while the expression of BCL-X and BCL-2 mRNA were lower than that of control (P<0.05). There were no significant changes in the mRNA expression of ATF4, CHOP, BCL-X, BCL-2, BID, BIM, and caspase-3 in the control group, miR-1283 mimic negative control group, and miR-1283 inhibitor negative control group (P>0.05). The results revealed that the inhibition of miR-1283 could lead to the upregulation of ATF4 and its downstream genes and pro-apoptotic factors on the ER stress pathway and the downregulation of anti-apoptotic factors, while the overexpression of miR-1283 could improve these results and reduce ER stress (Figure 5). All the findings showed that miR-1283 regulated mRNA of ER stress- and apoptosis-related genes.
Figure 5.
Expression of endoplasmic reticulum stress-related mRNA in human amniotic epithelial cells intervened by miR-1283. (A–G) of activating transcription factor 4 (ATF4), C/EBP-homologous protein (CHOP), BCL-2, BCL-X, BH3-interacting domain death agonist (BID), Bcl-2-like protein 11 (BIM), caspase-3 mRNA expression. Control – control group; mimic – miR-1283 mimic group; inhibitor – miR-1283 inhibitor group; GSK – GSK2656157; mimic control – miR-1283 mimic negative control; inhibitor control – miR-1283 inhibitor negative control group. * P<0.05.
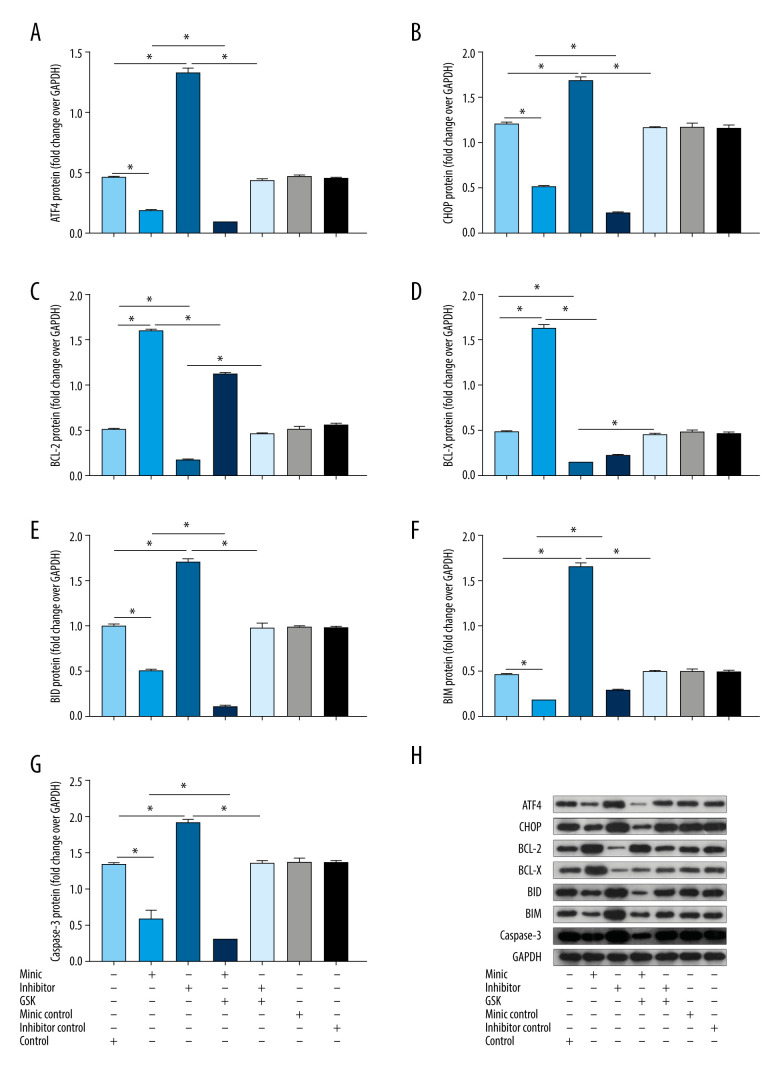
miR-1283 Regulated ER Stress and Apoptosis-Related Proteins
Compared with that of the control group, the expression of ATF4, CHOP, BID, BIM, and caspase-3 protein in the miR-1283 mimic group was lower, while the expression of BCL-X and BCL-2 was higher (P<0.05). The miR-1283 inhibitor group had the opposite result, which mainly showed that the expression of ATF4, CHOP, BID, BIM, and caspase-3 protein was higher, while the expression of BCL-X and BCL-2 was lower than that of control (P<0.05). The expressions of ATF4, CHOP, BCL-X, BCL-2, BID, BIM, and caspase-3 did not change significantly among the control group, miR-1283 mimic negative control group, and miR-1283 inhibitor negative control group (P>0.05). The results showed that the inhibition of miR-1283 could lead to the upregulation of ATF4 and its downstream proteins and pro-apoptotic factors on the ER stress pathway and the downregulation of anti-apoptotic factors, while the overexpression of miR-1283 could improve these results and reduce ER stress, consistent with the results of mRNA levels (Figure 6). All the results showed that miR-1283 regulated ER stress and apoptosis-related proteins.
Figure 6.
The protein expression levels of endoplasmic reticulum stress and apoptosis-related genes in human amniotic epithelial cells intervened by miR-1283. (A–G) of activating transcription factor 4 (ATF4), C/EBP-homologous protein (CHOP), BCL-2, BCL-X, BH3-interacting domain death agonist (BID), Bcl-2-like protein 11 (BIM), caspase-3 protein expression. (H) Western blotting of indicated proteins. Control – control group; mimic – miR-1283 mimic group; inhibitor – miR-1283 inhibitor group; GSK – GSK2656157; mimic control – miR-1283 mimic negative control; inhibitor control – miR-1283 inhibitor negative control group. * P<0.05.
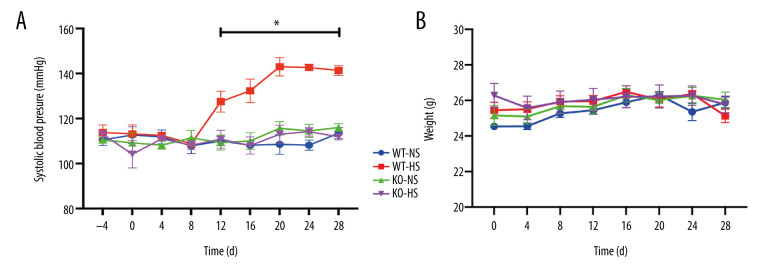
Lack of Target Gene ATF4 Blocking Salt-Induced Blood Pressure Rise in Mice
The non-invasive tail sleeve method was used to measure the arterial systolic blood pressure of mice. We found that the blood pressure of mice in the WT-HS group gradually increased from day 8 to 20 after the high-salt diet, compared with no increase in the WT-NS group. The blood pressure of mice in the WT-HS group reached a plateau (P<0.05) on days 20 to 28, while the blood pressure of mice in the KO-HS group was lower compared with that of the WT-NS group (P<0.05). There was no significant difference in body weight in the WT-NS, WT-HS, KO-NS, and KO-HS groups (P>0.05). The results showed that the high-salt diet did not affect the body weight of the mice but could induce the occurrence of hypertension, while the lack of the target gene ATF4 could inhibit the increase of blood pressure caused by the high-salt diet (Figure 7).
Figure 7.
The blood pressure and weight of of activating transcription factor 4 (ATF4) knockout mice induced by hyper-salt. (A) Blood pressure, (B) weight. WT-NS – wild-type+normal salt group; WT-HS – wild-type+high salt group; KO-NS – ATF4+/−+normal salt group; KO-HS – ATF4+/−+high salt group. * P<0.05.
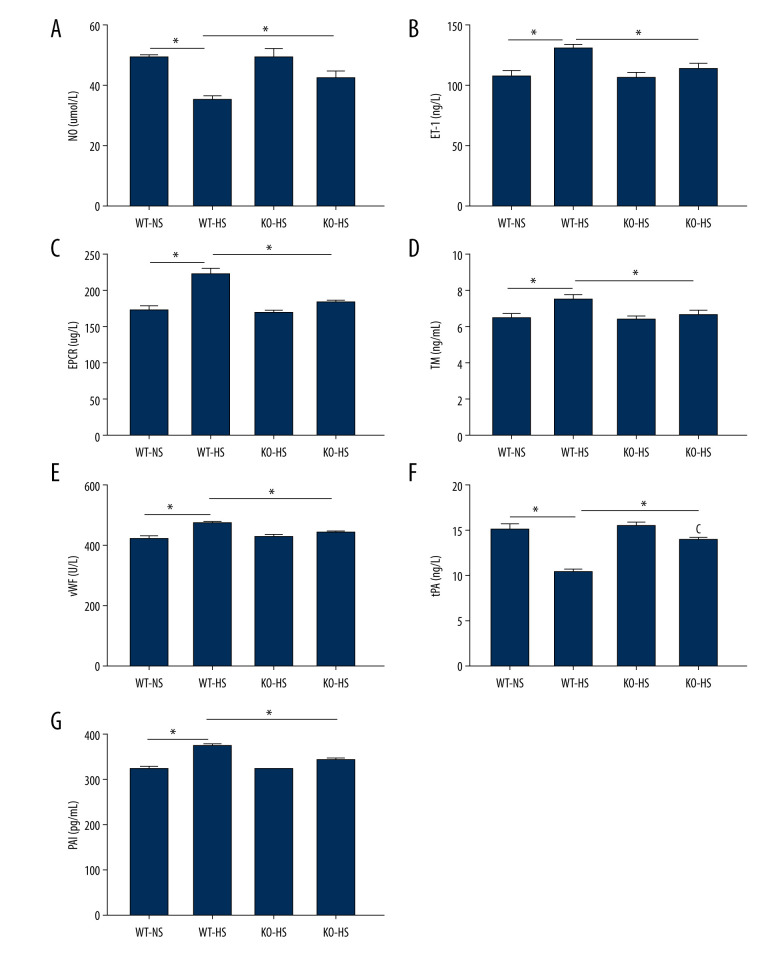
Lack of Target Gene ATF4 Protected High-Salt-Induced Vasoactive Substances and Clotting Factors in Mice
The results showed that, compared with the WT-NS and KO-HS groups, the WT-HS group had significantly higher ET-1, EPCR, TM, vWF and PAI, and lower NO and tPA (P<0.05). There were no significant changes in NO, ET-1, EPCR, TM, vWF, tPA, and PAI in groups WT-NS, KO-NS, and KO-HS (P>0.05). The results showed that the high-salt diet could lead to vascular endothelial dysfunction, while the lack of the target gene could improve vascular endothelial dysfunction in mice (Figure 8).
Figure 8.
The content of vasoactive substances and clotting factors in serum of of activating transcription factor 4 (ATF4) knockout mice induced by high salt. (A–G) nitric oxide (NO), endothelin 1 (ET-1), endothelial protein C receptor (EPCR), thrombin (TM), von Willebrand factor (vWF), tissue plasminogen activator (tPA), and plasminogen activator inhibitor (PAI). WT-NS – wild-type+normal salt group; WT-HS – wild-type+high salt group; KO-NS – ATF4+/−+normal salt group; KO-HS – ATF4+/−+high salt group. * P<0.05.
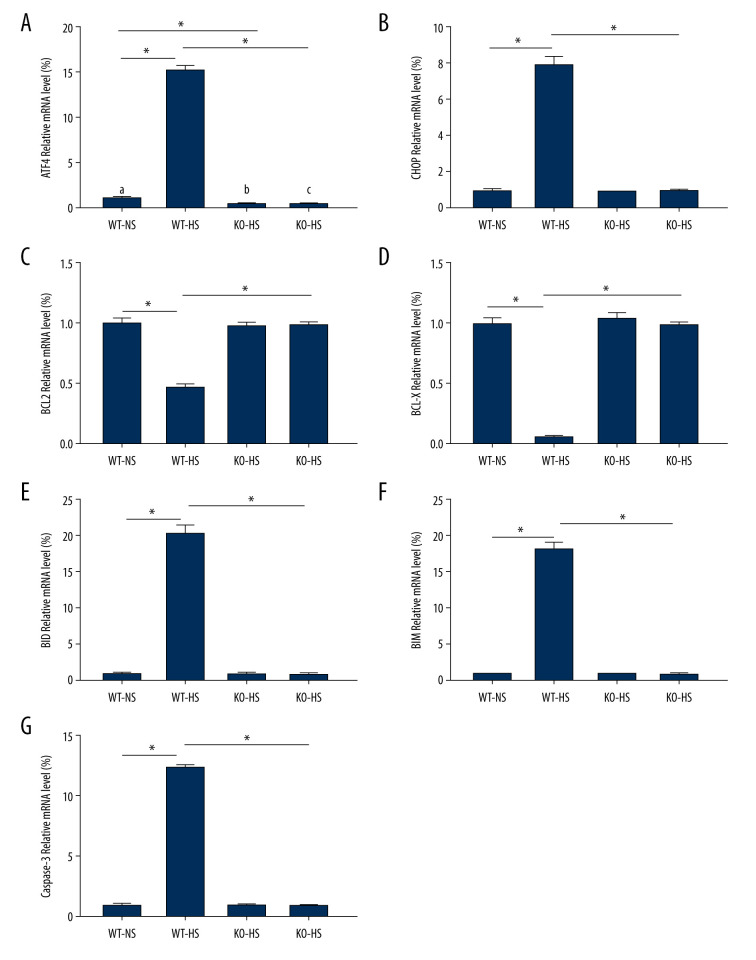
ATF4 Deficiency Blocked mRNA of Genes Related to ER Stress and Apoptosis in Mice Induced by High Salt
Compared with that of the WT-NS group, ATF4 mRNA expression was lower in the KO-NS group (P<0.05). Compared with that of the WT-NS and KO-HS groups, the expressions of ATF4, CHOP, BID, BIM, and caspase-3 mMRAs in the WT-HS group were significantly higher, and the expressions of BCL-X and BCL-2 mRNA were significantly lower (P<0.05). The mRNA expressions of ATF4, CHOP, BCL-X, BCL-2, BID, BIM, and caspase-3 did not change significantly in groups WT-NS, KO-NS, and KO-HS (P>0.05). These results suggested that a high-salt diet could lead to the upregulation of the ER pathway-related mRNAs (ATF4 and CHOP) and pro-apoptosis-related mRNAs (BID, BIM, and caspase-3) and the downregulation of anti-apoptosis-related mRNAs (BCL-X, BCL-2), while the lack of the target gene ATF4 can improve cell apoptosis and reduce ER stress (Figure 9).
Figure 9.
The levels of mRNA related to endoplasmic reticulum stress and apoptosis in of activating transcription factor 4 (ATF4) knockout mice induced by high salt. (A–G) ATF4, C/EBP-homologous protein (CHOP), BCL-2, BCL-X, BH3-interacting domain death agonist (BID), Bcl-2-like protein 11 (BIM), caspase-3 mRNA expression. WT-NS – wild-type+normal salt group; WT-HS – wild-type+high salt group; KO-NS – ATF4+/−+normal salt group; KO-HS – ATF4+/−+high salt group. * P<0.05.
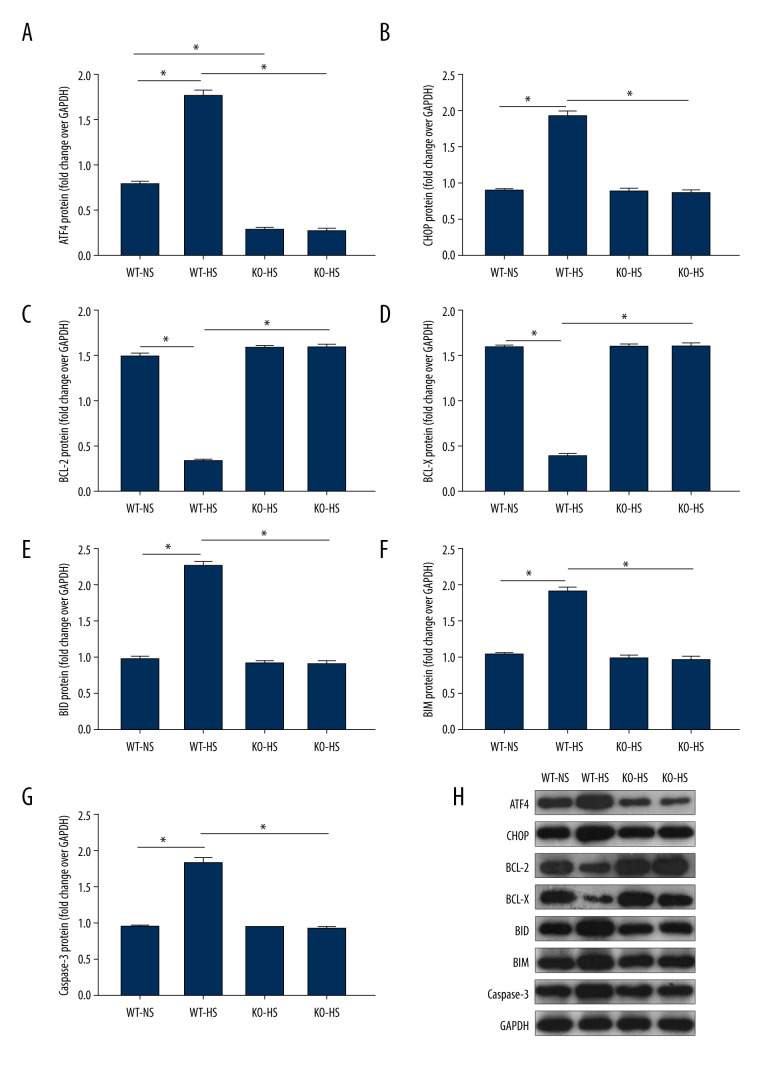
ATF4 Deficiency Improved ER Stress and Apoptosis-Related Proteins in Mice Induced by High Salt
Compared with that of the WT-NS group, ATF4 protein expression was lower in the KO-NS group (P<0.05). Compared with that of the WT-NS and KO-HS groups, the protein expressions of ATF4, CHOP, BID, BIM, and caspase-3 in the WT-HS group were significantly higher, while the protein expressions of BCL-X and BCL-2 were lower (P<0.05). There were no significant changes in ATF4, CHOP, BCL-X, BCL-2, BID, BIM, and caspase-3 protein expressions in groups WT-NS, KO-NS, and KO-HS (P>0.05). The results showed that ATF4+/− mice were successfully knocked out, and a high-salt diet could lead to an increased expression of ER stress pathway-related proteins ATF4 and CHOP and apoptosis-related proteins BID, BIM, and caspase-3. Also, the expression of anti-apoptotic proteins BCL-X and BCL-2 decreased. However, the lack of the target gene ATF4 could improve the ER stress disorder and the degree of apoptosis, which was consistent with the results of the mRNA level (Figure 10).
Figure 10.
The proteins expression of endoplasmic reticulum stress and apoptosis-related genes in of activating transcription factor 4 (ATF4) knockout mice induced by high salt. (A–G) ATF4, C/EBP-homologous protein (CHOP), BCL-2, BCL-X, BH3-interacting domain death agonist (BID), Bcl-2-like protein 11 (BIM), caspase-3 protein expression. (H) Western blotting of indicated proteins. WT-NS – wild-type+normal salt group; WT-HS – wild-type+high salt group; KO-NS – ATF4+/−+normal salt group; KO-HS – ATF4+/−+high salt group. * P<0.05.
Discussion
The protein encoded by the ATF4 gene belongs to a family of DNA-binding proteins that is involved in protein-protein interactions. Also, ATF4 is involved in cell damage repair, differentiation, proliferation, and apoptosis via the ER stress pathway [21,22]. Moreover, miR-1283 could directly target ATF4, which has been demonstrated by bioinformatics analysis and luciferase reporter assay [23]. In our previous study, we found that the expression of miR-1283 was decreased and the expression of ATF4 was increased in patients with hypertension [13]. In the present study, the overexpression or inhibition of miR-1283 also reduced or increased the expression of ATF4. Researchers modulated ER stress, and the miR-1283 inhibitor increased apoptosis in HUVECs and mouse cardiac myocytes [14]. This was confirmed in the HAECs in our present study. Compared with that of the miR-1283 mimic group, the protein and mRNA expression levels of ATF4, CHOP, BID, BIM, and caspase-3 were significantly higher and the levels of Bcl-X and Bcl-2 were lower in the miR-1283 inhibitor group in HAECs. Among these genes, CHOP can increase the expression of pro-apoptotic genes such as Bax/Bak, BIM, and BID and inhibit the expression of anti-apoptotic genes such as BCL-X and BCL-2, leading to apoptosis [24–26]. We found the same results in our present animal experiments. Compared with that of the WT-HS group, the protein and mRNA expression levels of ATF4, CHOP, BID, BIM, and caspase-3 were significantly higher and the levels of Bcl-X and Bcl-2 were lower in the KO-HS group (ATF4 knockout mice induced by high salt). Moreover, we overexpressed and inhibited miR-1283 in HAECs, followed by transcriptome analysis. Comparing the overexpressed group with the inhibited group, the differentially expression genes were involved with the ER stress pathway. Also, JUN, ATF4, EGFR, and IL6, the related genes of this pathway, played a key role, according to protein-protein interaction correlation analysis. Through the GO function enrichment analysis, the 23 identified differentially expression genes were involved in the PERK-mediated unfolded protein response, intrinsic apoptotic signaling pathway in response to ER stress, CHOP-ATF4 complex, and 66 other functions. These results showed that miR-1283 causes ER stress and participates in apoptosis of vascular endothelial cells by targeting ATF4.
The apoptosis of vascular endothelial cells also brings about changes in serum components. In the HAEC supernatant, compared with that of the miR-1283 inhibitor group, the miR-1283 mimic group showed a significantly higher NO level, and lower ET-1, EPCR, TM, and vWF levels. This result also appeared in a previous study of HUVECs [14]. With the induction of high salt, the wild-type mice were detected to have the highest ET-1, EPCR, TM, and vWF levels and the lowest NO level of the groups. Also, their blood pressure increased to cause hypertension. However, compared with the levels of the WT-HS mice, the levels of NO, ET-1, EPCR, TM, and vWF in the KO-HS mice were all significantly lower, and the blood pressure was stable and similar to that of the WT-NS group. It is known that NO is an in vivo signaling molecule and a protective factor for HUVECs, while ET-1, EPCR, TM, and vWF are markers of vascular injury or vascular functional disturbances [27–30]. Therefore, we believe that miR-1283/ATF4 is involved in the formation of hypertension through ER stress and apoptosis.
ER stress mediates 2 different stress responses in cells, namely a protective response and apoptotic response [31]. ER stress is related to excessive activation of the PERK-eIF2α-ATF4-CHOP pathway, triggering the apoptotic pathway [32,33], in which ER stress mediates the apoptotic response within the cell. Related studies have reported that vascular endothelial cell dysfunction caused by ER stress is involved in various pathophysiological processes caused by vascular injury and related cardiovascular diseases such as hypertension, atherosclerosis, diabetic heart disease, myocardial hypertrophy, and heart failure [34,35].
A high-salt diet can activate the renin angiotensin aldosterone system (RAAS), and the expression of the angiotensin II receptor in the activated RAAS is increased, leading to ER stress. Sun et al found that inhibiting the expression of angiotensin converting enzymes in renal tubules of diabetic rats can block RAAS and relieve ER stress [35]. Studies have reported that the activation of PERK, ATF6, and IRE-1 in an ER stress reaction induced the synthesis of CHOP proteins [36,37], and CHOP promoted apoptosis by downregulating the expression of BCL-2 anti-apoptotic factors [38]. The above results suggest that the ATF4/CHOP pathway induced by ER stress may be an important mechanism for the occurrence and development of myocardial hypertrophy caused by various factors.
Vascular endothelial cells not only constitute a mechanical barrier between circulating blood and the vascular wall, but they also maintain vascular intima smooth muscle and vascular homeostasis, preventing platelet and white blood cell adhesion and harmful substances from invading the vascular wall. Vascular endothelial cells secrete a variety of vasoactive substances, including NO, ET-1, and TM, which can regulate blood viscosity and vasoconstriction, thus maintaining normal blood flow and vasomotor state. Vascular endothelial cells secreting NO can inhibit platelet aggregation, leukocyte adhesion, and proliferation of vascular smooth muscle, the action of regulating vascular tension [39]. ET-1 is the strongest known endogenous vasoconstriction active peptide for the constriction of blood vessels [40]. TM is an important multifunctional protease in the blood coagulation system, in which the degree of viscosity can often react in blood vessels [41]. EPCR and vWF are considered specific markers of endothelial cell injury and dysfunction [42,43], tPA can better reflect the degree of damage to vascular endothelial function, and PAI is a fast and efficient inhibitor of tPA [44,45]. Under pathological conditions, vascular endothelial cells are damaged, the secretion of NO and tPA decreased, the secretion of ET-1, TM, EPCR, vWF, and PAI increased, endocrine function was disrupted, the balance of vasodilator and systolic factors was broken, resulting in abnormal vascular tone regulation, and the blood was in a highly condensed and viscous state.
Our transcriptome sequencing results showed that ATF4 was downregulated in the miR-1283 overexpression group and upregulated in the miR-1283 inhibition group, which further verified that there was also a clear target and regulatory relationship between miR-1283 and ATF4 in HAECs, consistent with previous research results [14]. The results of our animal experiments showed that the high-salt diet did not affect the body weight of mice but could induce the occurrence of hypertension, while the knockout of the mouse miR-1283 target gene ATF4 could inhibit the blood pressure increase caused by the high-salt diet. The results also showed that, compared with in the WT-HS group, ET-1, EPCR, TM, vWF, and PAI were lower, and NO and tPA were higher in the KO-HS group, indicating that the lack of ATF4, the target gene of miR-1283 in mice, could protect the function of endothelial cells. Compared with that of the WT-HS group, the pro-apoptotic genes and proteins of ATF4, CHOP, BID, BIM, and caspase-3 in the KO-HS group were downregulated, while the anti-apoptotic genes and proteins of BCL-X and BCL-2 were upregulated, indicating that the lack of ATF4 gene may inhibit ER stress and apoptosis by blocking the ATF4/CHOP signaling pathway. Moreover, we found in the cell experiments that the inhibition of miR-1283 could lead to increased expression of NO and decreased expression of ET-1, EPCR, TM, and vWF, indicating that the inhibition of miR-1283 could lead to cell endothelial dysfunction, while the overexpression of miR-1283 could improve endothelial function. In addition, miR-1283 downregulated ATF4, CHOP, BID, BIM, and caspase-3, upregulated BCL-X and BCL-2 anti-apoptosis-genes, and increased the cell apoptosis rate in the miR-1283 inhibition group, suggesting that miR-1283 may improve vascular endothelial function and inhibit ER stress and cell apoptosis through the ATF4/CHOP signaling pathway. This result was also consistent with the results of our animal experiments. Combining the results of our in vivo and in vitro experiments, we found that the miR-1283/ATF4 axis contributed to ER stress and apoptosis by regulating the ATF4/CHOP signaling pathway in the development of hypertension. Of course, additional clinical evidence is needed to further support these conclusions.
Conclusions
In general, our study showed that there was a clear regulatory relationship between hypertension-related miR-1283 and ATF4 in HAECs. The lack of ATF4 protected against high-salt diet-induced hypertension and endothelial function. In addition, the combined results of in vivo and in vitro experiments demonstrated that miR-1283 regulated its target gene ATF4 to inhibit ER stress and cell apoptosis through the ATF4/CHOP signaling pathway, which may be one of the pathways involved in hypertension, and protected endothelial function.
Footnotes
Data Availability
There are no restrictions on data availability. All relevant data are available upon request from the lead corresponding author.
Conflicts of Interest
None.
Source of support: This work was supported by the National Natural Sciences Foundation of China (No. 81673848 and 82074307), Natural Sciences Foundation of Guangdong Province (No. 2016A030310084 and 2017A030313658), Science and Technical Plan of Guangzhou, Guangdong, China (No. 201804010213 and 201707010100), Administration of Traditional Medicine of Guangdong Province (No. 20161063 and 20181068), Fundamental Research Funds for the Central Universities (No. 21616315), and the Medical Scientific Research Foundation of Guangdong Province (No. 2016111221315850)
References
- 1.Lorigo M, Mariana M, Oliveira N, et al. Vascular pathways of testosterone: Clinical implications. J Cardiovasc Transl. 2020;13(1):55–72. doi: 10.1007/s12265-019-09939-5. [DOI] [PubMed] [Google Scholar]
- 2.Oparil S, Acelajado MC, Bakris GL, et al. Hypertension. Nat Rev Dis Primers. 2018;4:18014. doi: 10.1038/nrdp.2018.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher N, Curfman G. Hypertension – a public health challenge of global proportions. JAMA. 2018;320(17):1757. doi: 10.1001/jama.2018.16760. [DOI] [PubMed] [Google Scholar]
- 4.Lewington S, Lacey B, Clarke R, et al. The burden of hypertension and associated risk for cardiovascular mortality in China. JAMA Intern Med. 2016;176:524–32. doi: 10.1001/jamainternmed.2016.0190. [DOI] [PubMed] [Google Scholar]
- 5.Tae-Hoon K, Pil-Sung Y, Tae YH, et al. Effect of hypertension duration and blood pressure level on ischaemic stroke risk in atrial fibrillation: Nationwide data covering the entire Korean population. Eur Heart J. 2019;(10):809–19. doi: 10.1093/eurheartj/ehy877. [DOI] [PubMed] [Google Scholar]
- 6.Carey RM, Muntner P, Bosworth HB, et al. Prevention and control of hypertension: JACC health promotion series. J Am Coll Cardiol. 2018;72(11):1278–93. doi: 10.1016/j.jacc.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spitler KM, Webb RC. Endoplasmic reticulum stress contributes to aortic stiffening via proapoptotic and fibrotic signaling mechanisms. Hypertension. 2014;63(3):e40–45. doi: 10.1161/HYPERTENSIONAHA.113.02558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L, Liu J, Huang Z, et al. Berberine improves endothelial function by inhibiting endoplasmic reticulum stress in the carotid arteries of spontaneously hypertensive rats. Biochem Biophys Res Commun. 2015;458(4):796–801. doi: 10.1016/j.bbrc.2015.02.028. [DOI] [PubMed] [Google Scholar]
- 9.Carlisle RE, Werner KE, Yum V, et al. Endoplasmic reticulum stress inhibition reduces hypertension through the preservation of resistance blood vessel structure and function. J Hypertens. 2016;34(8):1556–69. doi: 10.1097/HJH.0000000000000943. [DOI] [PubMed] [Google Scholar]
- 10.Choi SK, Lim M, Byeon SH, Lee YH. Inhibition of endoplasmic reticulum stress improves coronary artery function in the spontaneously hypertensive rats. Sci Rep. 2016;6:31925. doi: 10.1038/srep31925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C, Kassan A, Castañeda D, et al. Metformin prevents vascular damage in hypertension through the AMPK/ER stress pathway. Hypertens Res. 2019;42(7):960–69. doi: 10.1038/s41440-019-0212-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z, Chen Z, Zhang L, et al. Status of hypertension in China: Results from the China hypertension survey, 2012–2015. Circulation. 2018;137(22):2344–56. doi: 10.1161/CIRCULATIONAHA.117.032380. [DOI] [PubMed] [Google Scholar]
- 13.He L, Fang MX, Chen LG, et al. Transcriptome analysis of blood stasis syndrome in subjects with hypertension. J Tradit Chin Med. 2016;2:173–80. doi: 10.1016/s0254-6272(16)30024-3. [DOI] [PubMed] [Google Scholar]
- 14.He L, Xu QY, Chen RX, et al. miRNA-1283 regulates the PERK/ATF4 pathway in vascular injury by targeting ATF4. Plos One. 2016;118(11):e159171. doi: 10.1371/journal.pone.0159171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang SB, Gao YJ, Liu GH, et al. The human ATF1 rs11169571 polymorphism increases essential hypertension risk through modifying miRNA binding. FEBS Lett. 2015;589:2087–93. doi: 10.1016/j.febslet.2015.06.029. [DOI] [PubMed] [Google Scholar]
- 16.Wang D, Song W, Na Q. The emerging roles of placenta-specific microRNAs in regulating trophoblast proliferation during the first trimester. Aust NZJ Obstet Gynaecol. 2012;52(6):565–70. doi: 10.1111/j.1479-828X.2012.01481.x. [DOI] [PubMed] [Google Scholar]
- 17.Anne KG, Blocki A, Franke RP, et al. Vascular endothelial cell biology: An update. Int J Mol Sci. 2019;20(18):84411. doi: 10.3390/ijms20184411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chignalia AZ, Weinberg G, Dull RO. Norepinephrine induces lung microvascular endothelial cell death by NADPH oxidase-dependent activation of caspase-3. Oxid Med Cell Longev. 2020;2020(4):1–11. doi: 10.1155/2020/2563764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Patcher L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics (Oxford, England) 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dewey CN, Li B. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;112(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonsson WO, Margolies NS, Anthony TG. Dietary sulfur amino acid restriction and the integrated stress response: mechanistic insights. Nutrients. 2019;11(6):1349. doi: 10.3390/nu11061349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Das S, Joshi MB, Parashiva GK, et al. Stimulation of cytoprotective autophagy and components of mitochondrial biogenesis/proteostasis in response to ionizing radiation as a credible pro-survival strategy. Free Radic Biol Med. 2020;152:715–27. doi: 10.1016/j.freeradbiomed.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Chen H, Zhang Y, Su H, et al. Overexpression of miR-1283 inhibits cell proliferation and invasion of glioma cells by targeting ATF4. Oncol Res. 2019;27:325–34. doi: 10.3727/096504018X15251282086836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyce M, Yuan J. Cellular response to endoplasmic reticulum stress: A matter of life or death. Cell Death Differ. 2006;13(3):363–73. doi: 10.1038/sj.cdd.4401817. [DOI] [PubMed] [Google Scholar]
- 25.Fu HY, Minamino T, Tsukamoto O, et al. Ablation of C/EBP homologous protein attenuates endoplasmic reticulum-mediated apoptosis and cardiac dysfunction induced by pressure overload. J Card Fail. 2010;16(9):S145–46. doi: 10.1161/CIRCULATIONAHA.109.917914. [DOI] [PubMed] [Google Scholar]
- 26.Tabas I. The role of endoplasmic reticulum stress in the progression of atherosclerosis. Circ Res. 2010;107(7):839–50. doi: 10.1161/CIRCRESAHA.110.224766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamada H, Suzuki M, Yuasa S, et al. Dilated cardiomyopathy caused by aberrant endoplasmic reticulum quality control in mutant KDEL receptor transgenic mice. Mol Cell Biol. 2004;24(18):8007–17. doi: 10.1128/MCB.24.18.8007-8017.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao W, Iwai C, Liu J, et al. Darbepoetin alfa exerts a cardioprotective effect in autoimmune cardiomyopathy via reduction of ER stress and activation of the PI3K/Akt and STAT3 pathways. J Mol Cell Cardiol. 2008;45(2):250–60. doi: 10.1016/j.yjmcc.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer B, Hemmens B. Biosynthesis and action of nitric oxide in mammalian cells. Trends Biochem Sci. 1998;22:477–81. doi: 10.1016/s0968-0004(97)01147-x. [DOI] [PubMed] [Google Scholar]
- 30.Shilo NR, Morris CR. Pathways to pulmonary hypertension in sickle cell disease: The search for prevention and early intervention. Expert Review of Hematology. 2017;10(10):875–90. doi: 10.1080/17474086.2017.1364989. [DOI] [PubMed] [Google Scholar]
- 31.Hou Y, Xue P, Woods CG, et al. Association between arsenic suppression of adipogenesis and induction of CHOP10 via the endoplasmic reticulum stress response. Environ Health Perspect. 2013;121:237–43. doi: 10.1289/ehp.1205731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uehara Y, Hirose J, Yamabe S, et al. Reticulum stress-induced apoptosis contributes to articular cartilage degeneration via C/EBP homologous protein. Osteoarthr Cartil. 2014;22(7):1007–17. doi: 10.1016/j.joca.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 33.Matsumoto H, Miyazaki S, Matsuyama S, et al. Selection of autophagy or apoptosis in cells exposed to ER-stress depends on ATF4 expression pattern with or without CHOP expression. Biol Open. 2013;2(10):1084–90. doi: 10.1242/bio.20135033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.San CW, Yuen NC, Yen TY, et al. Black tea protects against hypertension-associated endothelial dysfunction through alleviation of endoplasmic reticulum stress. Sci Rep. 2015;5:10340. doi: 10.1038/srep10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun HL, Sun L, Li YY, et al. ACE-inhibitor suppresses the apoptosis induced by endoplasmic reticulum stress in renal tubular in experimental diabetic rats. Exp Clin Endocrinol Diabetes. 2009;117(7):336–44. doi: 10.1055/s-0028-1112148. [DOI] [PubMed] [Google Scholar]
- 36.Tsutsui H, Ishibashi Y, Imanaka YK, et al. Alterations in sarcoplasmic reticulum calcium-storing proteins in pressure-overload cardiac hypertrophy. Am J Physiol. 1997;272:H168–75. doi: 10.1152/ajpheart.1997.272.1.H168. [DOI] [PubMed] [Google Scholar]
- 37.Zwadlo C, Borlak J. Disease-associated changes in the expression of ion channels, ion receptors, ion exchangers and Ca(2+)-handling proteins in heart hypertrophy. Toxicol Appl Pharmacol. 2005;207(3):244–56. doi: 10.1016/j.taap.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 38.McCullough KD, Martindale JL, Klotz LO, et al. Gadd153 sensitizes cells to endoplasmic reticulum stress by down-regulating Bcl2 and perturbing the cellular redox state. Mol Cell Biol. 2001;21(4):1249–59. doi: 10.1128/MCB.21.4.1249-1259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Madamanchi NR, Vendrov A, Runge MS. Oxidative stress and vascular disease. Arterioscler Thromb Vasc Biol. 2005;25(1):29–38. doi: 10.1161/01.ATV.0000150649.39934.13. [DOI] [PubMed] [Google Scholar]
- 40.Archer CR, Robinson EL, Drawnel FM, et al. Endothelin-1 promotes hypertrophic remodelling of cardiac myocytes by activating sustained signalling and transcription downstream of endothelin type A receptors. Cell Signal. 2017;36:240–54. doi: 10.1016/j.cellsig.2017.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrano GR, Weiss EJ, Zheng YW, et al. Role of thrombin signalling in platelets in haemostasis and thrombosis. Nature. 2001;413(6851):74–78. doi: 10.1038/35092573. [DOI] [PubMed] [Google Scholar]
- 42.Wu MD, Atkinson TM, Lindner JR. Platelets and von Willebrand factor in atherogenesis. Blood. 2017;129:1415–19. doi: 10.1182/blood-2016-07-692673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Medina P, Navarro S, Estellés A, et al. Polymorphisms in the endothelial protein C receptor gene and thrombophilia. J Thromb Haemost. 2007;98:564–69. [PubMed] [Google Scholar]
- 44.Sakata K, Miura F, Sugino H, et al. Impaired fibrinolysis early after percutaneous transluminal coronary angioplasty is associated with restenosis. Am Heart J. 1996;131(1):1–6. doi: 10.1016/s0002-8703(96)90043-5. [DOI] [PubMed] [Google Scholar]
- 45.Keijer J, Linders M, Wegman JJ, et al. On the target specificity of plasminogen activator inhibitor 1: The role of heparin, vitronectin, and the reactive site. Blood. 1991;78(5):1254–61. [PubMed] [Google Scholar]