Abstract
Inflammasomes are macromolecular complexes involved in the host response to external and endogenous danger signals. Inflammasome-mediated sterile inflammation plays a central role in several human conditions such as autoimmune diseases, type-2 diabetes, and neurodegenerative disorders, indicating inflammasomes could be appealing therapeutic targets. Previous work has demonstrated that inhibiting the ATPase activity of the nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3 (NLRP3), disrupts inflammasome assembly and function. However, there is a necessity to find new potent compounds with therapeutic potential. Here we combine computational modeling of the target and virtual screening to discover a group of novel compounds predicted to inhibit NLRP3. We characterized the best compounds and determined their potency, specificity, and ability to inhibit processes downstream from NLRP3 activation. Moreover, we analyzed in mice the competence of a lead candidate to reduce lipopolysaccharide-induced inflammation. We also validated the active pharmacophore shared among all the NLRP3 inhibitors, and through computational docking, we clarify key structural features for compound positioning within the inflammasome ATP-binding site. Our study sets the basis for rational design and optimization of inflammasome-targeting probes and drugs.
Keywords: NLRP3, drug discovery, inflammasome, high-throughput screening (HTS), inflammation, NLRC4
Abbreviations: AD, Alzheimer’s disease; AIM-2, absent in melanoma 2; ASC, apoptosis-associated speck-like protein containing a CARD; CAPSs, cryopyrin-associated autoinflammatory syndromes; DAMPs, danger-associated molecular patterns; DMSO, dimethylsulfoxide; DRAQ5, deep red anthraquinone 5; FBS, fetal bovine serum; FC, frontal cortex; HMGB1, high mobility group box 1; hNLRP3, human NLRP3; IL-6, interleukin 6; IL-18, interleukin 18; IL-1β, interleukin 1β; LPS, lipopolysaccharide; LRR, leucine-rich repeat; NACHT, nucleotide-binding and oligomerization; NLRC4, NLR family CARD domain–containing 4; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3; NLRP7, NLR family pyrin domain containing 7; NLRs, nucleotide-binding domain leucine-rich repeat containing receptors; NOD2, nucleotide-binding oligomerization domain–containing protein 2; TNF-α, tumor necrosis factor alpha
The nucleotide-binding domain leucine-rich repeat (LRR)–containing receptors (NLRs) are a group of cytoplasmic pattern-recognition receptors that mediate the initial innate immune response to microbial infections, stress, and host-derived factors (1, 2). Upon activation, some NLRs form inflammasomes, which through recruitment and activation of caspase-1, promote the proteolytic cleavage, maturation, and secretion of the proinflammatory cytokines interleukin 1β (IL-1β) and interleukin 18 (IL-18) as schematized in Figures 1 and S1. In addition, activated caspase-1 cleaves Gasdermin-D, triggering a nonapoptotic type of cell death called pyroptosis, which mediates the secretion of the mature cytokines to the cytoplasm (3, 4, 5).
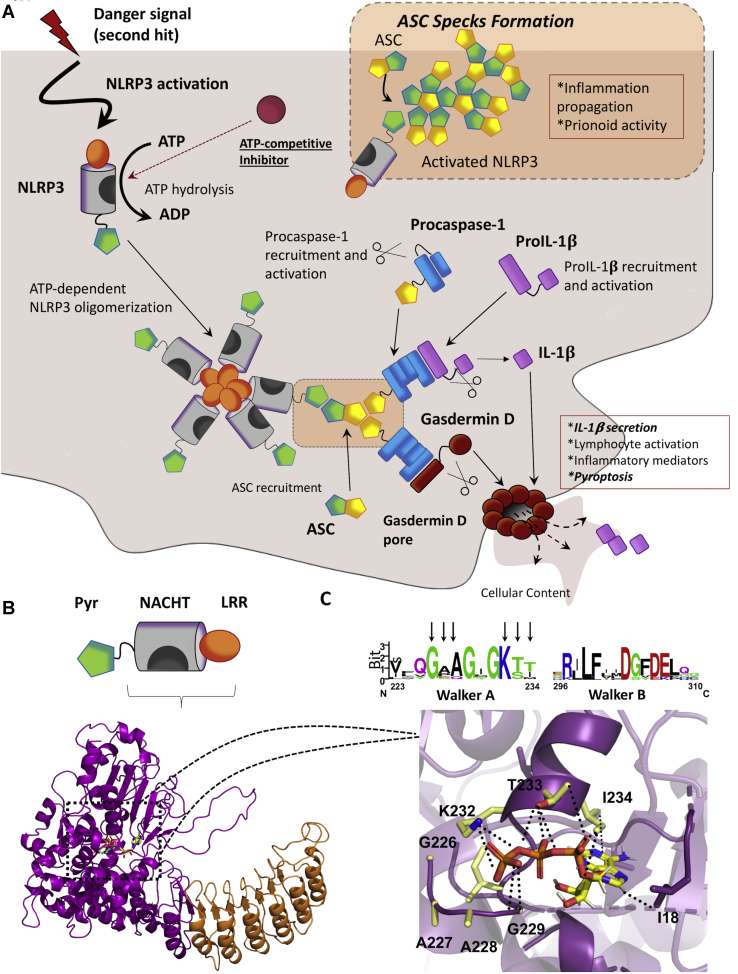
Figure 1.
The NLRP3 inflammasome.A, the NLRP3 inflammasome activation pathway. Generally, the NLRP3 inflammasome might be activated by a two-signal mechanism; the first signal (not shown in this scheme) increases the expression of the different components of the inflammasome. The second signal might trigger the activation and the assembly of the inflammasome. First, ATP binds to the nucleotide-binding and oligomerization (NACHT) domain, being subsequently hydrolyzed. Activated NLRP3 oligomerizes and recruits the apoptosis-associated speck-like protein containing a CARD (ASC). ASC is a scaffolding protein and recruits procaspase-1, allowing its autoproteolytic activation. Finally, activated caspase-1 cleaves pro-IL-1β into IL-1β, which is secreted and exerts inflammatory effects. Active caspase-1 also recruits and induces the activation of Gasdermin-D, which forms pores in the cell membrane, leading to the release of the cellular content and, subsequently, pyroptosis. In addition, activated ASC nucleates into large specks that have prionoid properties and contribute to inflammation propagation. Processes representing readouts for inflammasome activation are showed in italics. B, NLRP3 is organized in three different domains; a C-terminal leucine-rich repeat (LRR), a central NACHT, and N-terminal pyrin domain (PYD). The 3D model for NLRP3 is shown in the cartoon, with the bound ATP as green sticks. The model was built by homology modeling, using as a template the crystal structure of the nucleotide-binding oligomerization domain-containing protein 2 (NOD2). C, top, the sequence logo for the multiple sequence alignment of the highly conserved Walker A and Walker B motifs. NLRP3 numeration is used. Bottom, detail of the nucleotide-binding cavity. Residues from Walker A motive close to ATP (shown as sticks) are highlighted in sticks and colored in CPK code with carbons in yellow. Electrostatic interactions between K232 and ATP phosphate groups are displayed as black dots. NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3.
Among the NLRs, the nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3 (NLRP3) is so far the best studied and characterized one (6, 7, 8). In addition, another inflammasome, the NLR family CARD domain–containing 4 (NLRC4), is progressively collecting more attention (9, 10, 11). As displayed in Figures 1 and S1, structurally, both inflammasomes present an LRR domain and a nucleotide-binding and oligomerization (NACHT) domain (12). Also, NLRP3 has a pyrin domain that interacts with the apoptosis-associated speck-like protein containing a CARD (ASC) adaptor protein, which recruits caspase-1 (13). By its part, NLRC4 directly interacts with caspase-1 through a CARD domain (14). However, upon NLRP3 and NLRC4 activation, ASC homo-oligomerizes forming large specks that propagate inflammation (15, 16, 17). Thus, ASC oligomerization constitutes a readout for inflammasome activation. Interestingly, the NACHT domain of both proteins holds an ATPase activity that is essential for inflammasome activation (6), which makes this domain an appealing therapeutic target for the treatment of inflammatory conditions where either of the inflammasomes are involved. Indeed, it has been previously probed that the blockade of the ATPase activity with small-molecule inhibitors blockades the activation and assembly of the NLRP3 inflammasome (18, 19).
NLRP3 and NLRC4 have a primary role in mediating the host defense against microbial pathogens (20, 21), but their prolonged activation leads to a broad spectrum of diverse diseases. For instance, gain of function mutations in the NLRP3 NACHT domain cause different forms of cryopyrin-associated autoinflammatory syndromes (CAPSs) (22, 23, 24), whereas genetic alterations in the NLRC4 gene produce autoinflammatory syndromes (25). Moreover, NLRP3 and NLRC4 are indirectly involved in several conditions, where the prolonged activation of these inflammasomes have deleterious effects on disease progression (9, 26). Under these situations, danger-associated molecular patterns (DAMPs)—such as the alarmin high mobility group box 1 (HMGB1) or reactive oxygen species—trigger, respectively, inflammasome priming and activation, leading to a process known as sterile inflammation (27, 28). Sterile inflammation has an especially negative impact in a batch of neurological disorders such as Alzheimer’s disease (AD), Parkinson’s disease, amyotrophic lateral sclerosis, or multiple sclerosis (9, 26, 29, 30, 31), where inflammatory cytokines produced by microglia and astrocytes (32) establish a feedback loop that recruits more inflammatory cells, enhancing the inflammation process (33). Moreover, in AD, the role of the ASC adaptor protein deserves special mention. Inflammasome activation triggers the aggregation of the ASC into large fibrils forming specks, which are released to the extracellular medium, where they recruit more inflammatory cells, amplifying inflammation (16). Also, ASC specks recruit amyloid-β, promoting its oligomerization and aggregation, acting as an inflammation-driven cross-seed for amyloid-β pathology, and worsen the pathology of AD (16, 34). Thus, the optimization of inflammasome-targeting drugs would provide novel therapeutic in multiple immune-inflammatory disorders.
Currently, most of the therapeutic efforts to treat inflammasome-related pathologies focus on the specific inhibition of NLRP3. Among others, glyburide presents in vitro anti-NLRP3 activity, but a limited effect in vivo (35); β-hydroxybutyrate inhibits NLRP3 activation by disrupting NLRP3–ASC oligomerization (36); while MCC950 and CY-09 are very potent and specific NLRP3 inhibitors (37, 38). Both of these promising compounds directly bind to the ATP-binding site of the NLRP3 NACHT domain, inhibiting its ATPase activity and blocking the activation and assembly of the inflammasome (18, 19, 38). Using mouse models, MCC950 has been demonstrated to be effective in a broad spectrum of diseases, such as spontaneous colitis (39), diabetic nephropathy (40), and traumatic brain injury (41), among others. However, the drug was tested for rheumatoid arthritis, but the clinical trials were interrupted in phase II after it showed liver toxicity (42).
Although the selective inhibition of NLRP3 has been demonstrated as an effective treatment for inflammatory syndromes in a CAPS murine model (37), for the treatment of neurological disorders in which both NLRP3 and NLRC4 inflammasomes are involved—such as AD (9, 26, 43, 44), Parkinson’s disease (31, 44, 45, 46), or multiple sclerosis (29)—a less specific strategy appears as a better tactic. Following this latter approach, a few antagonists of the IL-1β receptor have been successfully used in patients with CAPS and rheumatoid arthritis (47, 48). Still, these drugs fail to block IL-18-mediated inflammation or ASC oligomerization. Consequently, there is a compelling need to develop new NLRP3 and NLRC4 inflammasome inhibitors.
Here we show a new therapeutic approach to treat inflammatory disorders, consisting of targeting the ATPase activity of the NACHT domain of NLR proteins. We develop a methodology to find inflammasome inhibitors, starting from the most fundamental computational level, and continuing with the in vitro validation and characterization of the hits. We apply this methodology to identify a family of compounds with anti-inflammasome properties, and we select and characterize a lead compound, that in addition to inhibiting the activation of the NLRP3 and NLRC4 inflammasomes in vitro, also present in vivo promising effects. Thus, the main scientific contributions of this work are (i) the identification and characterization of a new family of chemical analogs with proven anti-inflammatory properties and (ii) the detection of the structural features driving the allocation of the compounds within the ATP-binding pocket. Taken together, these findings pave the way for the future development of improved inflammasome inhibitors.
Results
A virtual screening targeting the ATP-binding site of the NLRP3 model allows the identification of 100 potential inhibitors
To find ATP-competitive inhibitors of the NLRP3 inflammasome, we targeted the ATPase activity of its NACHT domain (Fig. 1, A and B), which is essential for the activation and the activity of the inflammasome (49). Although a cryo-EM structure for human NLRP3 (hNLRP3) is currently available, it was not at the beginning of this project; thus, we modeled it. We built a homology model based on the crystal structure of nucleotide-binding oligomerization domain–containing protein 2 (NOD2) (50) (PDB entry: 5IRM). Despite the relatively low sequence homology between both proteins (28%), residues conforming the active site are highly conserved among NLR proteins (Fig. 1C), especially the Walker A and B motives, which increases the probability of getting an accurate model for the ATP-binding site. Starting from a virtual library of more than 17 million compounds, we identified 100 potential inhibitors, which we then screened and characterized in vitro, until selecting a lead candidate (Fig. 2A).
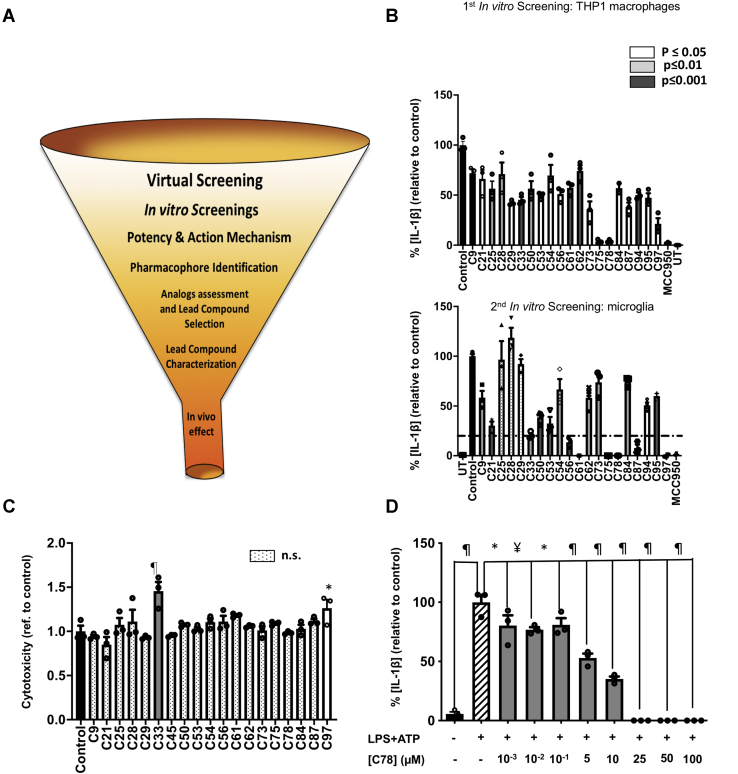
Figure 2.
In vitro screening.A, the diagram for the drug development process. The funnel represents the reduction in the number of compounds, starting from 17 million molecules in the virtual screening and finishing with a single compound to be tested in vivo. B, in vitro screenings for the 100 compounds from the in silico screening. The first screening tested the ability of the 100 molecules to inhibit NLRP3-mediated production of IL-1β by THP1 macrophages, primed, and activated with LPS and ATP, respectively. The compounds that inhibit IL-1β production with any level of statistical significance were tested in a secondary screening using murine primary microglia, primed, and activated with LPS (400 ng/ml, 3 h) and ATP (4.5 mM, 45 min). The compounds that inhibit IL-1β release below a 20% threshold were selected for further characterization. C, cytotoxicity exerted by compounds originating from the first screening, as measured by the lactate dehydrogenase released in cell supernatants, after treatment with 50 μM of the compounds. D, the dose-dependent inhibitory effect of one of the three compounds (C78), chosen according to potency criteria, to be deeper characterized. The data shown correspond to average values of biological triplicates (n = 3), ran in technical duplicates ± SEM. Statistical significances are referred to control samples; LPS + ATP in panels B and D and untreated cells in panel C. Samples and controls contained the same DMSO%. Positive controls for NLRP3 inhibition with the NLRP3 inhibitor MCC950 are included in panel B. Significance levels, as calculated by one-way ANOVA, are indicated as white (p ≤ 0.05), light gray (p ≤ 0.01), and dark gray (p ≤ 0.001) bars in panels B and C and as ∗p ≤ 0.05, ¥p ≤ 0.01, and ¶p ≤ 0.0001 in panel D. IL-1β, interleukin 1β; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3.
Two sequential in vitro screenings progressively reduced the number of compounds
To identify the compounds from the virtual screening that have an inhibitory effect on the NLRP3 inflammasome, we carried out two different in vitro screenings. Owing to the complexity of using primary cultures to screen such a high volume of molecules, we conducted a first in vitro screening using human immortalized THP1 monocytes. We found that 20 compounds among the 100 initial hits reduced NLRP3-mediated IL-1β production with statistical significance (Fig. 2B, Tables S1, S2, and S3). We then validated the 20 identified hits in a secondary screening, using primary murine microglia cultures, which is our cell target. To continue the study, we selected the most potent compounds. We established a threshold of 20% of residual activity, and we choose to characterize further the compounds C56, C61, C75, C78, C87, and C97, which we showed to inhibit IL-1β production below this threshold (Fig. 2B, Table 1, Fig. S2). The NLRP3 inhibitor MCC950 was also used as a positive control for inflammasome inhibition (37).
Table 1.
Inhibitory parameters obtained for the six compounds selected during the secondary in vitro screening and C77
| Compound number | % Residual activitya | [Cx] to reach 100% of inhibition (μM) | IC50 (μM) |
|---|---|---|---|
| C56 | 1.4 | >100 | 40.0 ± 2.4 |
| C61 | 0 | ≤50 | 15.4 ± 1.18 |
| C75 | 0 | ≈25 | 9.24 ± 1.06 |
| C77 | 0 | <25 | 4.10 ± 2.10 |
| C78 | 0 | ≤25 | 8.26 ± 1.16 |
| C87 | 14.3 | >100 | 57.8 ± 50 |
| C97 | 0 | ≈25 | 11.27 ± 1.09 |
IL-1β, interleukin 1β; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3.
Compounds selected for further characterization are highlighted in bold.
The percentage of IL-1β, referred to the control, released by microglia cultures pretreated with 100 μM of the corresponding compound before NLRP3 activation with LPS and ATP.
In addition, we analyzed the cytotoxicity of the 20 positive hits to eliminate toxic compounds from further studies. At 100 μM, only C33 and C97 exert some toxicity (Fig. 2C). However, because C97 at this concentration completely inhibited IL-1β concentration in culture supernatants, we did not eliminate it, and we continued to explore the cytotoxicity in a dose-dependent manner.
In NLRP3 target engagement studies, we treated primary microglia isolated from NLRP3 KO mice. We found that IL-1β concentration in supernatants from microglial cultures treated with LPS and ATP was not significantly higher than what was found in control cultures (vehicle-treated microglia) or than in microglia treated only with LPS (Fig. S3). This evidence suggests that challenging microglia with LPS + ATP specifically promotes the activation of the NLRP3 inflammasome, and not of other IL-1β-producing inflammasomes, revealing the specificity of the assay for NLRP3 activation. Consequently, the mitigation of the IL-1β release mediated by the compounds during the in vitro screenings was derived from NLRP3 inhibition.
The selected compounds inhibit NLRP3 in a dose-dependent manner, with three compounds possessing IC50 values in the low μM range
We next studied the dose-dependent inhibitory effect of the selected compounds to identify the most potent NLRP3 inhibitors. The six hits reduced the IL-1β concentration in the supernatant of activated microglia cultures in a dose-dependent manner (Figs. 2D, and S4A). The pretreatment with 100 μM of C61, C75, C78, and C97 completely prevented IL-1β secretion, whereas the concentration required to reach the 100% of inhibition was lower than 25 μM for C75, C78, and C97 (Table 1). Furthermore, the nonlinear fit of the data allowed the calculation of the IC50 for the studied molecules (Fig. S4B). According to these data, C75, C78, and C97 (Fig. 3A) were the most potent NLRP3 inhibitors, with IC50 values around 10 μM (Table 1, Fig. 3B). Then, we analyzed the toxicity of these three compounds (at concentrations ranging from 0.01 to 50 μM) on microglia cultures, finding that at 50 μM, none of the compounds were toxic (Fig. S5). Based on these results, we continued to study the specificity of the three compounds for NLRP3.
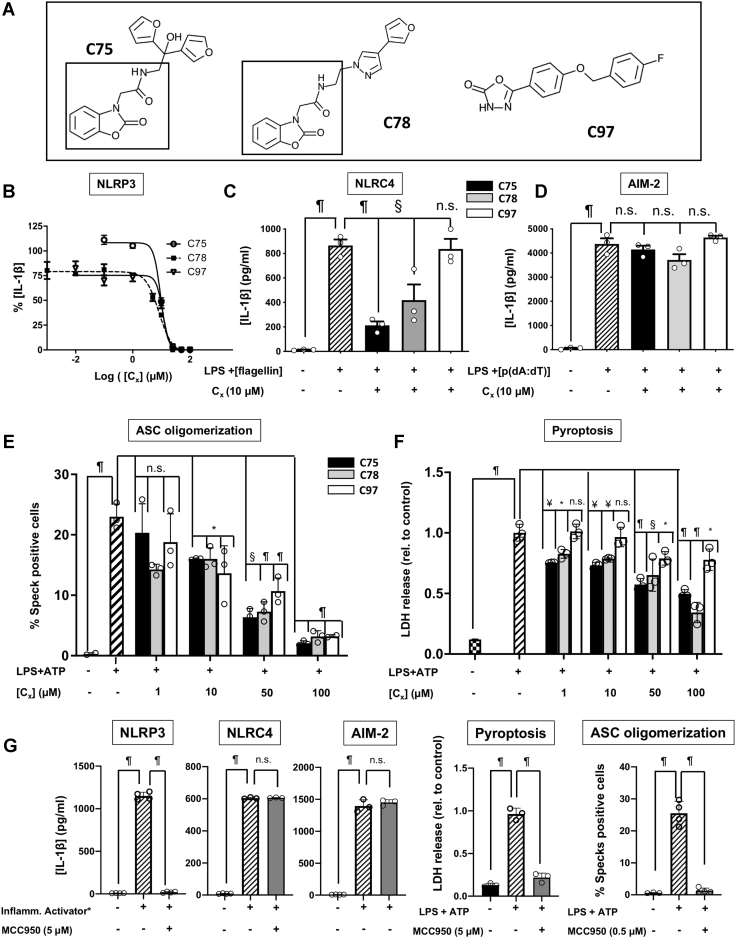
Figure 3.
Functional characterization of C75, C78, and C97.A, the structure of the three most potent compounds from the in vitro screening, C75, C78, and C97. The benzoxazolone acetamidyl group shared by C75 and C78 is squared. B, dose–response curves for the three compounds. C, the inhibitory effect of the three compounds on NLRC4-mediated IL-1β production. NLRC4 was primed and specifically activated in microglia from NLRP3 KO mice, using LPS (150 ng/ml, 2 h) and flagellin transfected with Lipofectamine (1 μg, 2 h). D, the effect of C75, C78, and C97 on the activity of the AIM-2 inflammasome. Microglia cultures from NLRP3 KO mice were primed with LPS (100 ng/ml, 2 h), and AIM-2 activation was induced by transfection of poly(dA:dT) (1 μg, 2 h). E, inhibition of ASC specks formation by different concentrations of the three compounds. Murine macrophages expressing m-cerulean-tagged ASC were primed and activated with LPS (250 ng/ml, 2.5 h) and ATP (4.5 mM, 45 min). F, the antipyroptotic effect of C75, C78, and C97. Primary microglia cultures pretreated with DMSO or different concentrations of the compounds were activated with LPS (400 ng/ml, 3 h) and ATP (4.5 mM, 45 min) to induce pyroptosis, or remained untreated. Pyroptosis was determined by measuring cell death using an lactate dehydrogenase assay. In all panels, C75, C78, and C97 are displayed as black, gray, and white bars, respectively. G, control experiments with the NLRP3-specific inhibitor MCC950. Reference controls are shown as crosswise bars. Data shown in panels B, C, D, and F correspond to average values of biological triplicates (n = 3), ran in technical duplicates ± SEM. Experiments displayed in panel E were performed by biological quadruplicates. Statistical significances are referred to control samples and consist of cells treated with the same DMSO% and the same LPS and ATP concentrations as the compound-treated samples. Significance levels, as calculated by one-way ANOVA, are indicated as: n.s. p > 0.05, ∗p ≤ 0.05, ¥p ≤ 0.01, §p ≤ 0.001, and ¶p ≤ 0.0001. ∗Inflamm. activator refers to the specific combination of stimuli for NLRP3, NLRC4, and AIM-2 inflammasome activation, as indicated in panels B, C, and D, and in the Experimental procedures section. ASC, apoptosis-associated speck-like protein containing a CARD; NLRC4, NLR family CARD domain–containing 4; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3.
C97 is specific for NLRP3, while C75 and C78 also inhibit the NLRC4 activity
To determine whether the three selected compounds are specific for NLRP3, or if they also inhibit other IL-1β-producing inflammasomes, we studied the effect of each compound on the IL-1β released upon activation of the absent in melanoma 2 (AIM-2) and the NLRC4 inflammasomes. We first primed murine microglia cultures with LPS, followed by the use of specific activators (double-stranded DNA and flagellin for AIM-2 and NLRC4, respectively) to activate the corresponding inflammasomes. As shown in Figure 3, C and D, C97 was specific for NLRP3, while C75 and C78 exhibited inhibitory activities against the NLRC4 inflammasome at 10 μM. As expected, none of the three compounds inhibited the activity of the AIM-2 inflammasome because the AIM-2 inflammasome lacks the NACHT domain the compounds are designed to target.
C75, C78, and C97 inhibit downstream processes of NLRP3 activation
We next evaluated whether C75, C78, and C97 block downstream processes of NLRP3 activation, such as ASC oligomerization or pyroptosis. As indicated in Figure 1A, ASC oligomerization is necessary for the assembly of the NLRP3 inflammasome, propagates inflammation, and has prionoid properties (16). Therefore, monitoring ASC oligomerization can be used as an endpoint to determine NLRP3 inflammasome activation (15). To evaluate the concentration-dependent ability of each compound to reduce the ratio of ASC-speck positive cells, we used murine macrophages that express the ASC protein with a fluorescent tag. Pretreatment with 10 μM of C75, C78, or C97 significantly reduced the rate of speck-positive cells compared with dimethylsulfoxide (DMSO)-treated cells. Furthermore, the inhibition of the ASC speck formation proportionally depended on the compound concentration (Fig. 3E). We also examined whether the three compounds inhibit pyroptosis, an inflammatory type of programmed cell death. Pyroptosis occurs downstream of NLRP3 activation and requires the action of caspase-1. C75 and C78 dose-dependently inhibited pyroptosis at concentrations starting from 1 μM, while higher concentrations of C97 were necessary to observe a similar effect (Fig. 3F). We used MCC950 as a positive control for pyroptosis and ASC oligomerization inhibition (37) (Fig. 3G).
The identification of the active pharmacophore allows the selection of chemical analogs with NLRP3-inhibitory activity
C75 and C78 possess a benzoxazolone acetamide pharmacophore (Fig. 3A), which could be crucial for their binding at the ATP pocket of the NLRP3 protein. After identifying the potential pharmacophore, we performed a homology search to find structurally related analogs of C75 and C78. Thus, using the MolPort web server, we found commercially available compounds with the same benzoxazolone acetamide scaffold and different R substituents (Fig. 4A).
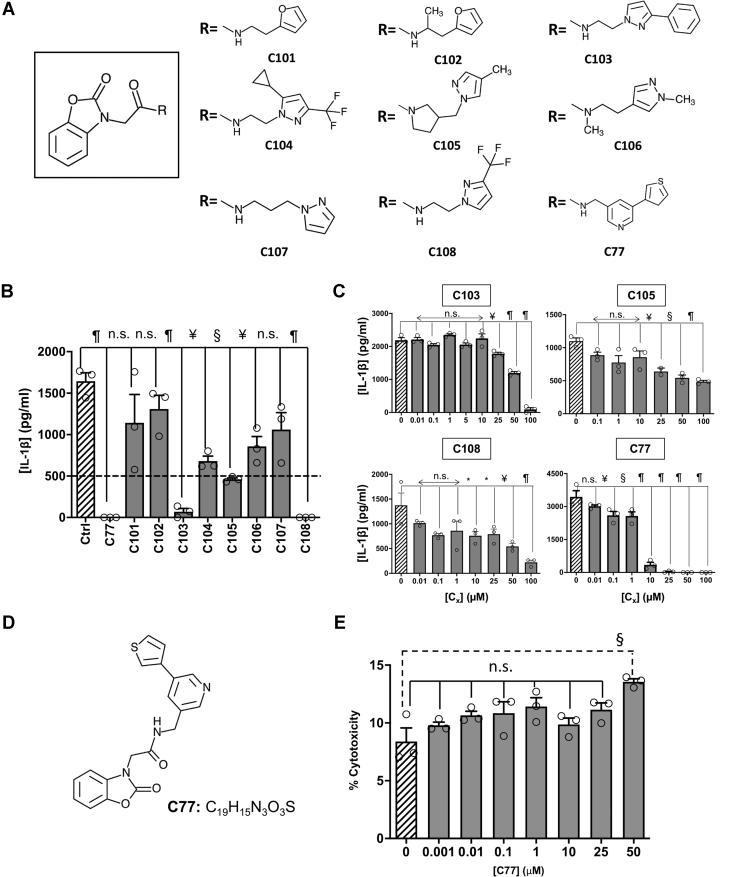
Figure 4.
Benzoxazolone acetamide analogs searching and assessment.A, the structure of the common core identified as the pharmacophore (squared). Chemical structure for the substituents of the selected benzoxazolone acetamide analogs. B, the inhibitory effect of 100 μM of each analog in NLRP3-mediated IL-1β production by primary murine microglia activated with LPS (400 ng/ml, 3 h) and ATP (4.5 mM, 45 min). The dashed line indicates 30% of activity regarding to controls. C, inhibition of NLRP3 activation by different concentrations of the selected analogs, as determined by measuring the IL-1β produced by microglia cultures. D, the structure and molecular formula of C77. E, cytotoxicity exerted by different concentrations of C77 in microglial murine cultures. The data shown correspond to average values of biological triplicates (n = 3), ran in technical duplicates ± SEM. Statistical significances are referred to control samples; LPS + ATP in panels B, C, and D and untreated cells in E. Samples and controls contained the same DMSO%. Significance levels, as calculated by one-way ANOVA, are indicated as ∗p ≤ 0.05, ¥p ≤ 0.01, §p ≤ 0.001, and ¶p ≤ 0.0001. IL-1β, interleukin 1β; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3.
Next, we assayed the ability of the analogs to inhibit IL-1β release, in primary microglia cultures, upon NLRP3 activation with LPS and ATP. Six of the analogs, C77, C103, C104, C105, C106, and C108, inhibited NLRP3 with statistical significance (Fig. 4B), which validates the nature of the benzoxazolone acetamide core as the active pharmacophore. We then selected those compounds that inhibited the IL-1β production below a 30% arbitrary threshold, and we performed dose–response curves to determine their potency (Fig. 4C). Through nonlinear regression, we obtained IC50 values of 87.2 ± 2.0, 45.4 ± 9.1, 35.7 ± 5, and 4.10 ± 2.1 μM for C103, C105, C108, and C77, respectively. Among the evaluated compounds, C77 completely inhibited NLRP3-mediated IL-1β production at concentrations lower than 25 μM, and it also presented the lowest IC50 value among all the evaluated compounds.
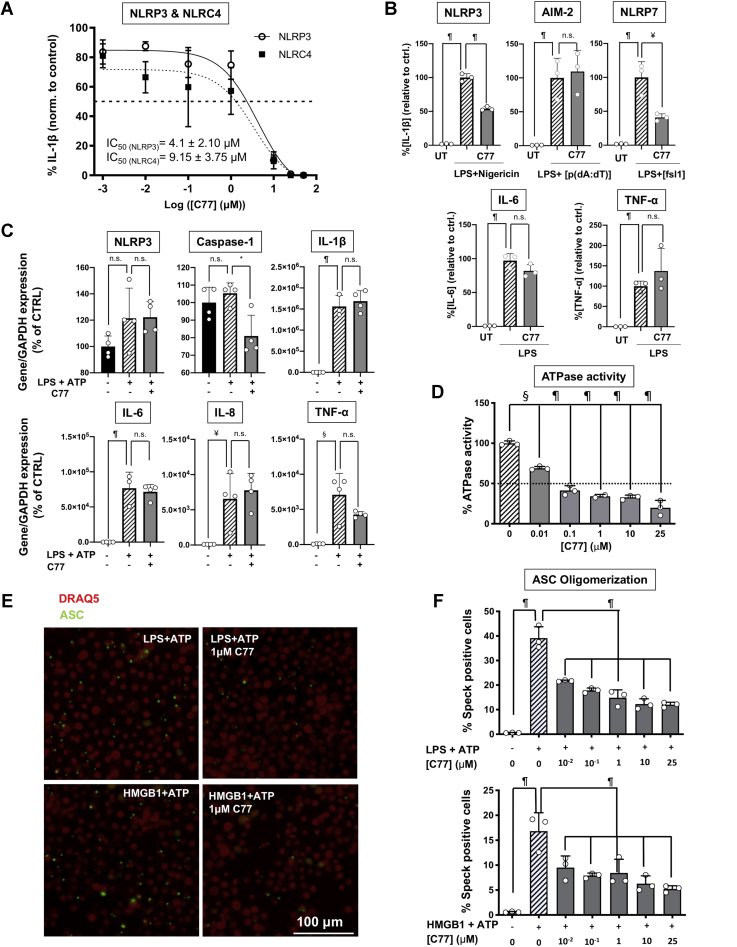
C77 potently inhibits NLRP3 and NLRC4 inflammasomes but does not affect AIM-2, IL-6, and TNF-α production, or the transcriptional priming of the inflammasome
As mentioned previously, we found that C77 was the most promising compound, and we thus characterized it further. C77 contains the benzoxazolone acetamide pharmacophore attached to a thiophenyl-pyridinyl substituent through a methylene linker (Fig. 4D). We first evaluated the toxicity of C77 using murine microglia cultures (Fig. 4E). The compound did not show significant toxicity levels at concentrations lower than 50 μM, although at this concentration, C77 significantly reduced the viability of the cultures. However, C77 inhibited NLRP3 activation at doses starting from 100 nM, while at 10 μM, the released IL-1β was reduced to less than 10%. Hence, from this point forward, and to avoid the toxicity of higher doses, we used C77 concentrations lower than 25 μM.
Then, we studied the effect of C77 on the IL-1β-producing inflammasomes, NLRC4 and NLR family pyrin domain containing 7 (NLRP7) (both of them presenting NACHT domains) and AIM-2 (which lacks this domain), and on the production of interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-α). C77 potently inhibited NLRC4, presenting an IC50 value similar to that for NLRP3 (Fig. 5A). This observation suggests the potential development of our novel compound to treat inflammasome-mediated immune-inflammatory responses, where both NLRP3 and NLRC4 inflammasomes are involved. We also assayed the ability of C77 to inhibit NLRP3 activation in response to the antibiotic nigericin. The treatment of LPS-primed microglia promoted the release of IL-1β, which was significantly reduced by treatment with C77 (Fig. 5B). In addition, 10-μM C77 also reduced NLRP7-mediated IL-1β production (Figs. 5B, S6). It is worthy to notice that C77 did not inhibit IL-6, TNF-α production, or AIM-2-mediated IL-1β release (Fig. 5B). As expected, these results suggest that C77 carries out its inhibitory activity by competitively binding to the NACHT domain and presumably to the ATP pocket, of NLRP3, NLRC4, and NLRP7 because AIM-2 does not present this NACHT domain. We also investigated the effect of 10-μM C77 on the expression level of the different components of the NLRP3 inflammasome and other inflammatory cytokines. C77 did not modify the mRNA levels of IL-6, L-8, or TNF-α and did not affect the transcriptional priming of NLRP3 or IL-1β, although it slightly reduced the expression of caspase-1 (Fig. 5C).
Figure 5.
Functional characterization of C77.A, dose–response inhibition of NLRP3 and NLRC4 by C77 and IC50 values for both inflammasomes in microglia cultures. NLRP3 activation was induced by treatment with LPS (400 ng/ml, 3 h) followed by ATP addition (4.5 mM, 45 min). For NLRC4-specific activation, primary microglia from NLRP3 KO mice were primed with LPS (150 ng/ml, 2 h) followed by transfection with flagellin (1 μg, 2 h). The dashed line indicates the 50% of inhibition, regarding the control average. For IC50 calculations, three independent experiments were conducted. A representative dose–response curve for NLRP3 and NLRC4 is displayed, while the IC50 values showed correspond to the average ± SEM of the IC50 obtained in the three independent experiments. B, the effect of 10-μM C77 on different inflammasomes and on IL-6 and TNF-α secretion, in primary microglia cultures. NLRP3 was primed and activated with LPS (400 ng/ml, 3 h) and nigericin (2 μM, 1 h). Activation of the AIM-2 inflammasome was promoted with LPS (100 ng/ml, 2 h) and transfection of p(dA:dT) (1 μg, 2 h). NLRP7 activation was induced with LPS (150 ng/ml, 1 h) and fsl1 transfection (400 ng/ml, 7 h). For IL-6 and TNF-α assessment, microglia cultures were treated with LPS (350 ng/ml, 6 h). The data shown correspond to average values ± SEM of three independent experiments, normalized to the average value of controls for each independent experiment. C, the effect of C77 on the RNA expression levels of the components of the NLRP3 inflammasome and other proinflammatory cytokines. THP1 human monocytes were incubated with 10-μM C77 (1 h) before treatment with LPS (400 ng/ml, 3 h). D, inhibition of the ATPase activity of NLRP3 at the protein level by C77. The ATPase activity of purified NLRP3 protein was measured by following ATP transformation into ADP, at different C77 concentrations. E and F, inhibition of ASC oligomerization by C77 in response to DAMPs and pathogen-associated molecular patterns. E, fluorescence microscope images for ASC speck formation after NLRP3 activation. Activation of the NLRP3 inflammasome of murine macrophages, expressing m-cerulean-tagged ASC, was induced through treatment with LPS (250 ng/ml, 2.5 h) and ATP (4.5 mM, 45 min) (upper panels) or HMGB1 (1 μg/ml, 3 h) and ATP (4.5 mM, 45 min) (lower panels). In the left panels, cells were pretreated with DMSO, while cells in the right panels were incubated with 1-μM C77 1 h before NLRP3 activation. F, the percentage of cells presenting ASC specks, at different concentrations of C77. NLRP3 priming was induced by treatment with LPS (upper panel) or HMGB1 (lower panel) (same conditions as in panel E). The data shown correspond to average values of biological triplicates (n = 3), ran in technical duplicates ± SEM (panels C, D, and F). In panel E, a representative image of each treatment group is displayed. Statistical significances are referred to control samples. Significance levels, as calculated by one-way ANOVA, are indicated as ∗p ≤ 0.05, ¥p ≤ 0.01, §p ≤ 0.001, and ¶p ≤ 0.0001. ASC, apoptosis-associated speck-like protein containing a CARD; DAMPs, danger-associated molecular patterns; IL-6, interleukin 6; NLRC4, NLR family CARD domain–containing 4; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3.
C77 dose-dependently inhibits NLRP3 ATPase activity and downstream processes
We analyzed the ability of C77 to reduce the NLRP3 activity at the protein level as described previously (49, 51). We incubated the NLRP3 protein with C77 concentrations ranging from 0.01 to 25 μM before ATP addition. C77 dose-dependently reduced the NLRP3 ATPase activity, showing statistically significant inhibition at concentrations as low as 10 nM (Fig. 5D). The nonlinear fit of the data allowed the estimation of an IC50 value for the ATPase activity of 40 nM. It is worth noticing that the IC50 value at the protein level is considerably lower than the IC50 value obtained when measuring the activation of NLRP3 in microglial cell cultures. This fact is not odd if we consider that the compound's actual intracellular concentration is lower than the concentration in the culture media because the concentration of C77 inside the cell depends on the cell membrane permeability to this compound.
Based on our demonstration that C77 directly inhibited the NLRP3 ATPase activity, we next analyzed the impact of the compound on ASC speck formation. In this case, we studied the response of ASC m-cerulean murine macrophages to two different activation stimuli. We induced NLRP3 activation with LPS and ATP, as well as with HMGB1 and ATP. HMGB1 is an alarmin released under stress situations, and it acts as a DAMP (52, 53), which promotes the priming of different inflammatory pathways (such as the NLRP3 inflammasome). Therefore, we demonstrated that C77 inhibited NLRP3 activation after inflammasome priming by a pathogen-associated molecular pattern and a DAMP (Fig. 5E). The reduction of the speck-positive cells depended on the compound concentration, with the inhibitory effect being observable at concentrations starting from 10 nM (Fig. 5F).
C77 reduces IL-1β in brains of LPS-treated mice, but it does not act on the inflammasome priming stage
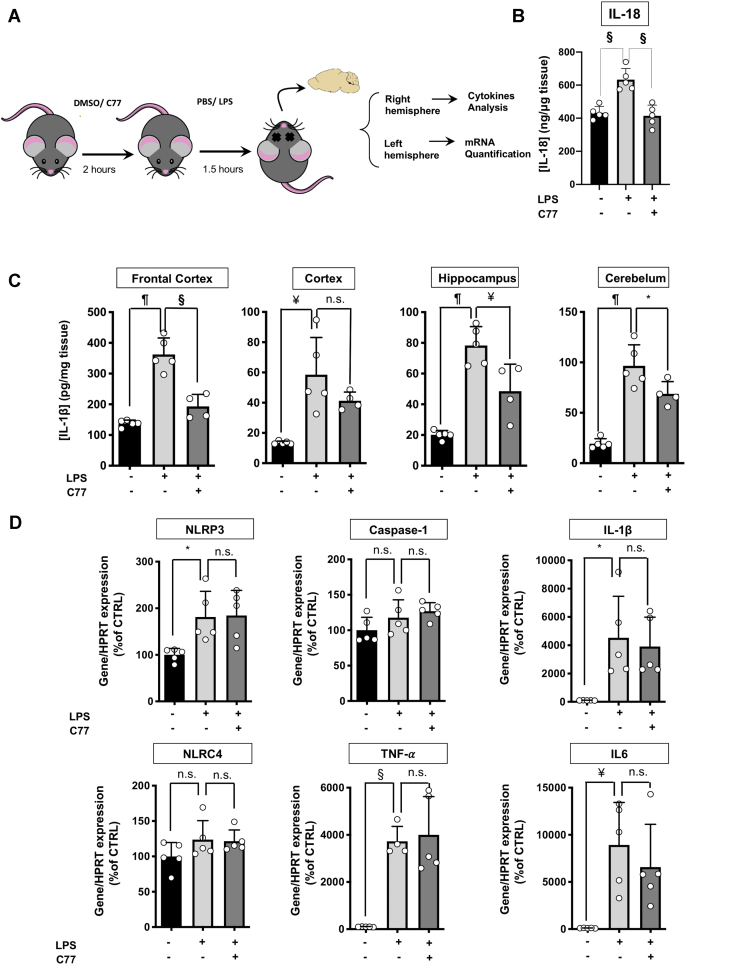
Finally, we evaluated the performance of C77 in vivo. We intraperitoneally administered mice with 50 mg/kg of C77 or vehicle (DMSO) followed by intraperitoneal injection of 15 mg/kg LPS. We quantified IL-1β in various brain regions and IL-18 in frontal cortex (FC) (Fig. 6A). LPS administration significantly increased IL-18 in the FC, while C77 reduced this effect (Fig. 6B). As shown in Figure 6C, LPS increased IL-1β in all brain regions, while pretreatment with C77 significantly reduced the IL-1β protein levels in the FC, hippocampus, and cerebellum of LPS-treated mice. In addition, we confirmed by Western blot that C77 reduced the cleaved form of IL-β and not pro IL-1β (Fig. S7). Next, we examined whether C77 might act on the inflammasome priming step. For that purpose, we analyzed the mRNA levels of inflammasome-related genes, such as NLRP3, caspase-1, IL-1β, and TNF-α, using the FC of the left hemisphere of each mouse, which was the region that presented the most significant differences at the protein level. In addition, we examined the C77 effect on IL-6 and NLRC4 expression. LPS treatment significantly increased the NLRP3, IL-1β, TNF-α, and IL-6 mRNA levels in mouse brains. Pretreatment with C77 for 2 h before LPS administration did not reduce the overexpression of these genes (Fig. 6C), indicating that C77 inhibits in vivo the inflammasome activation and not the priming stage.
Figure 6.
In vivo effect of C77.A, the scheme of the experimental procedure. B, IL-18 concentration in the frontal cortex of mice intraperitoneally administrated 50 mg/kg of C77 or vehicle (DMSO) 2 h before IP administration of 15 mg/kg of LPS or vehicle (PBS). C, IL-1β quantification in different brain regions of mice treated with vehicle, LPS, or C77+LPS. (Animals per group: vehicle, n = 5; LPS, n = 5; C77-LPS n = 4). D, RNA expression levels of inflammatory genes in the brain of mice treated with vehicle (DMSO), or C77 before PBS, or LPS IP injection. All treatment groups, n = 5. The data shown correspond to average values of biological replicates (ran in technical duplicates) ± SEM. Statistical significances are referred to control samples. Significance levels, as calculated by one-way ANOVA, are indicated as ∗p ≤ 0.05, ¥p ≤ 0.01, §p ≤ 0.001, and ¶p ≤ 0.0001. IP, intraperitoneal.
Discussion
NLRP3 is currently the best-studied member of the NLR family because of its direct implication in different CAPSs (22, 54) and to its involvement in the neuroinflammation associated with the onset and progression of several neurodegenerative conditions (31, 55, 56, 57, 58, 59, 60), where processes downstream of NLRP3 activation, such as ASC oligomerization or IL-1β secretion, play a detrimental effect. Therefore, therapeutic strategies targeting IL-1β-producing inflammasomes and downstream processes of the innate immunity are receiving major attention for the mitigation of chronic, progressive, and irreversible degenerative conditions in the brain and peripheral organs. Based on this, the overarching goal of this study is to elaborate new strategies for the discovery of novel inflammasome inhibitors by developing a 3D in silico model of the protein, a virtual screening, the subsequent in vitro characterization of the hits, and eventually, the in vivo assessment of the most promising lead compounds.
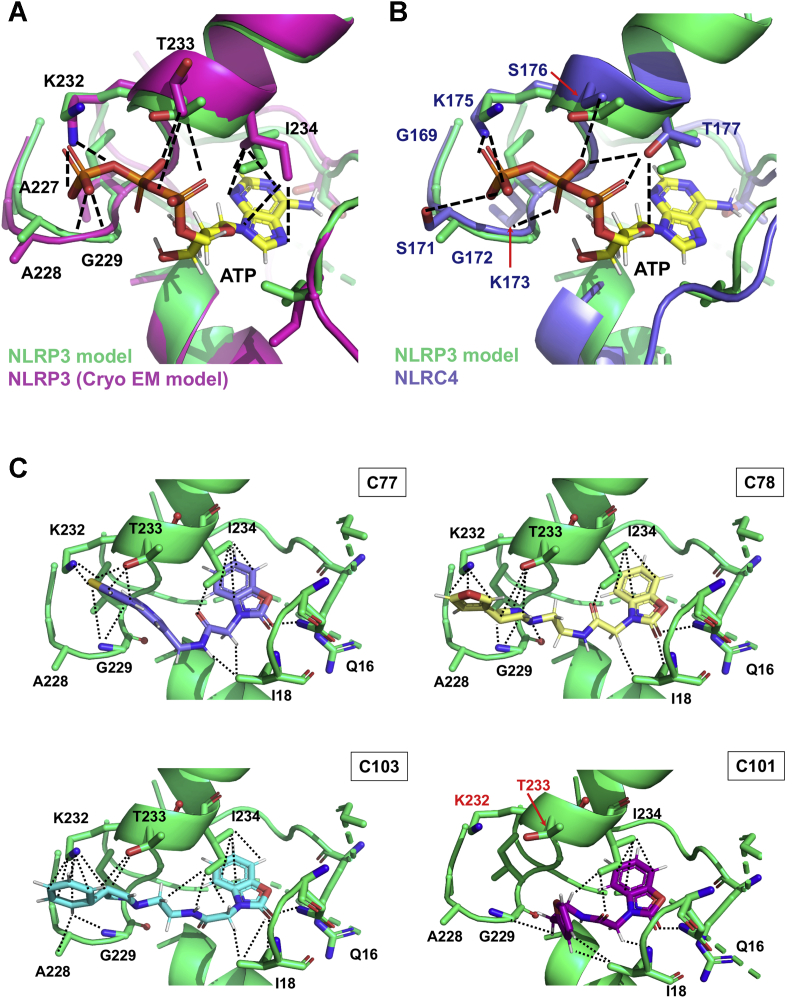
We started this project by building a homology model of NLRP3 based on the crystal structure of NOD2 (50). The alignment between our model and the later published NLRP3 cryo-EM structure (PDB entry: 6NPY (61)) exposes its accuracy to describe the NLRP3 active site. We find that critical residues of the catalytic pocket displayed resembling relative positions in both models, including G226, A227, A228, G229, and K232 from the Walker A motif (Figs. 1C and 7A). Interestingly, in both structures, K232 similarly interacts with the ATP γ-phosphate. This residue, together with T233, and G231, has already been described as crucial for ATP binding in a mutagenic study (6). The alignment between our NLRP3 model and NLRC4 reveals higher discrepancies between residues of the ATP pocket; however, the interaction between the lysine residue, K175 in the case of NLRC4, and the ATP γ-phosphate is conserved (Fig. 7B). Together, these structural insights with the fact that C75, C77, and C78 inhibit both NLRP3 and NLRC4 inflammasomes demonstrate the ability of our model to describe the ATP cavities of both proteins.
Figure 7.
Structural insights for the allocation of the adenine nucleotide ligands or the inhibitors within the ATP-binding site of the NACHT domain.A, structural alignment for the ATP-binding pockets of the NLRP3 model used for the virtual screening (green) and of the cryo-EM structure (magenta). B, structural alignment for the ATP-binding pockets of the NLRP3 model used for the virtual screening (green) and of the NLRC4 crystal structure. The ATP ligand is displayed in the CPK code with carbons in yellow. The lateral chains of the residues from the Walker A motif predicted to be involved in the ATP stabilization are highlighted in sticks and colored in CPK colors. Stabilizing interactions between residues from the NLRP3 cryo-EM structure (A) or NLRC4 (B) and ATP are shown as the dashed line. C, structural models for the allocation of C77 (light purple), C78 (pale yellow), C103 (cyan), and C101 (pink) within the ATP pocket of our NLRP3 model. Residues stabilizing the inhibitors are shown as sticks and colored in the CPK code. Interactions between side chains of these residues and the compounds are displayed as dotted lines. NACHT, nucleotide-binding and oligomerization; NLRC4, NLR family CARD domain–containing 4; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3.
In the next stage of the present work, we further screened, selected, and characterized the hits from the virtual screening. That way, we identified the compounds C75, C78, and C97, which dose-dependently inhibit NLRP3 activation, as well as downstream processes. Revealingly, C75 and C78 share the same chemical scaffold, which suggests that this structure might be particularly useful for the competent allocation of the compounds within the ATP-binding site, pointing to the benzoxazolone acetamidyl group as the potential key pharmacophore. To explore this hypothesis, we searched for structurally related analogs of the pharmacophore and checked their ability to inhibit NLRP3. Around 67% of the assayed compounds significantly reduced the inflammasome activity, which further supports the validity of the proposed pharmacophore.
To elucidate the structural basis behind the differences in the performance of the analogs, we carried out computational dockings to identify critical elements for NLRP3 compound interactions (Figs. 7C and S8). Our docking studies show that the benzoxazolone acetamidyl core interacts with the same residues that stabilize the ATP adenine ring (I18 and I234), suggesting that this group anchors the molecule in a beneficial position within the binding site. Interestingly, although all analogs maintain these interactions (Figs. 7C and S8), only those that also interact with K232 and T233 (residues that have been described to be critical for the ATPase activity (6)) exhibit in vitro activities (Figs. 7C and S8), especially the analogs that have an aromatic heterocycle with an electronegative heteroatom at an adequate distance from the pharmacophore; C75, C77, and C78. The positively charged lysine residue K232 may therefore interact with either the heteroatom or the heterocycle carbon atoms (which present negative partial charges established through resonance) to stabilize the compound in the ATP pocket. Thus, we can conclude that (i) the benzoxazolone acetamidyl is key for anchoring the compound within the ATP pocket and (ii) the length of the linker is crucial to allow the interaction of the pendant heterocycle with K232 and T233, with nine bonds being the optimal distance between the scaffold and the electronegative heteroatom.
Among the tested compounds, C77 was the most promising. C77 inhibits NLRP3, NLRC4, and NLRP7 but does not affect AIM-2, TNF-α, or IL-6 pathways (Fig. 5B). From a structural perspective, the three former inflammasomes present NACHT domains, while AIM-2 does not, and this motif is neither involved in IL-6 synthesis nor TNF-α secretion. Therefore, our data suggest that C77 exerts its inhibitory function by interacting with the inflammasome NACHT domain. We further demonstrate this concept by showing the concentration-dependent inhibition of the NLRP3 ATPase activity at the protein level (Fig. 5D). In addition, the mRNA levels of the different inflammasome components remain unaltered upon C77 treatment, both in the FC of LPS-administrated mice and activated THP1 cells, which reinforces the NACHT-C77 direct interaction. Considering the impact that ASC specks have on different neuroinflammatory disorders (9, 16, 34, 55, 56), our in vitro data suggest that C77 might have therapeutic effects against these conditions. Finally, the ability of C77 to reduce the IL-1β concentration in vivo reveals its therapeutic potential, suggesting that this compound can cross the blood–brain barrier.
In summary, starting from the most fundamental computational level, we identify and characterize small-molecule inhibitors of the NLRP3 and NLRC4 inflammasomes, and downstream processes, by using in silico, in vitro, and in vivo approaches. We start with a virtual screening to find NLRP3 inhibitors and use cell-free and cell-based assays to validate the hits, demonstrate the target engagement, and investigate their mechanism. We then prove in vivo the anti-inflammatory properties of the most promising candidate. In addition, we identify and validate the common benzoxazolone acetamidyl pharmacophore, as well as the critical residues involved in the stabilization of both the ATP substrate and the inhibitors within the protein active site. This study sheds light on the structural features for ligand’s allocation within the catalytic pocket of NLRs and paves the way for the future development of improved chemical analogs through medicinal chemistry structure–activity relationship studies.
Experimental procedures
NLRP3 modeling, structure refinement, and virtual screening
Sequence searches and alignment
The amino acid sequence of hNLRP3 (Q96P20 (NLRP3_HUMAN)) was obtained from the UniProtKB database. A SIB-BLAST search for the complete NLRP3 amino acid sequence (including NACHT, LRR, and PYRIN domains) against the mammalian taxonomic subset of the UniProtKB/SwissProt database was carried out at https://web.expasy.org/blast/. Multiple sequence alignment was produced using the multiple sequence alignment tool of EMBL-EBI, at https://www.ebi.ac.uk/Tools/msa/, with default parameters. Graphical representation of highly conserved motifs was produced with the WebLogo server http://weblogo.berkeley.edu/.
3D model for the NLRP3 protein
The structural model for hNLRP3 protein was built based on the crystal structure of NOD2 in the ADP-bound state (50) (PDB entry: 5IRM). The model was carried out via comparative modeling using the program MODELLER 9.15 (62). The model was further refined by using the loop modeling routine in MODELLER to rebuild the alignment insertions that are closer to the binding site. The model was then prepared for docking using the Protein Preparation Wizard in Maestro (Schrödinger) to define ionization states, optimize hydrogen bonds, and minimize the structure (heavy atom convergence to an RMSD of 0.3 Å).
Virtual screening
A virtual screening was carried out using the Glide program of the Schrödinger Small-Molecule Drug Discovery Suite (63, 64, 65). A library of more than 17 million commercially available drug-like compounds from the eMolecules collection was screened using the high-throughput virtual screening precision in Glide. In a second step, the top 100,000 hits from the high-throughput virtual screening screen were reranked using the standard precision, and finally the 10,000 hits from the standard precision screening were docked using XP precision. An additional simulation of membrane permeability was generated on the top 10,000 compounds from the virtual screening as an additional filter. Compounds with potential for chemical reactivity or instability were also excluded. Finally, candidate hits were further filtered to eliminate compounds containing assay interfering chemical groups and promiscuous scaffolds (66) and were visually inspected in the context of the binding site. Based on these structural and medicinal chemistry criteria, a final list of 100 compounds were selected for acquisition and experimental validation.
Benzoxazolone acetamide analog searching
To identify C75 and C78 chemical analogs containing benzoxazolone acetamide groups, the structure search tool of the MolPort server was utilized (www.molport.com). Compounds maintaining the benzoxazolone acetamide pharmacophore, but including different functional groups at the opposite part of the linker, were evaluated for acquisition. Pharmacokinetic properties and ADME parameters, as well as the drug-like properties of the compounds, were studied using the SwissADME server http://www.swissadme.ch/. Those compounds that presented the best-predicted properties, prioritizing solubility, and the ability to cross cell membranes, were acquired for their in vitro validation. In addition, C77, a compound included in the 100 initial hits from the virtual screening, was assessed at the same time as the analogs.
Computational docking
Computational docking of the benzoxazolone acetamide analogs was performed using the server SwissDock (http://www.swissdock.ch/). The ATP-binding pocket of our NLRP3 3D model was used as a target (coordinates: x = 6.1486, y = −7.406, z = 16.964, size: x = y = z = 20 Å). Dockings were analyzed using UCSF Chimera (https://www.cgl.ucsf.edu/chimera/), and the most favored poses for each molecule were represented with PyMOL (https://pymol.org/2/).
Cell culture and treatment
THP1 human macrophages
THP1 human monocytes were purchased from the American Type Culture Collection (TIB-202) and maintained in RPMI-1640 medium, supplemented with 10% fetal bovine serum (FBS) and 2-mercaptoethanol at a final concentration of 0.05 mM. Monocyte differentiation into macrophages was induced by treating with 100 ng/ml phorbol 12-myristate 13-acetate for 48 h. Then, cells were washed three times with culture medium without serum and let to rest for 24 h in the complete culture medium.
THP1 macrophages were preincubated for 1 h with the 100 hits from the primary screening, at a concentration of 100 μM—control samples were pretreated with the same DMSO volume. NLRP3 priming was then induced by the addition of 400 ng/ml LPS. After 3 h of incubation with LPS, 4.5-mM ATP was added for 45 min for inflammasome activation. Supernatants were collected for IL-1β determination and viability assays. 5 μM MCC950 was used as a positive control of NLRP3 inhibition. Experiments were run in biological triplicates.
Murine primary microglia
Mixed cortical cultures were prepared as described previously (67). Briefly, cortices from 2- to 3-day-old, WT, or NLRP3 KO pups were isolated, digested, and seeded at a density of eight cortices per 10-ml petri dish. Every 3 days, the medium was replaced by a fresh culture medium (Dulbecco's modified Eagle's medium [DMEM], 10% FBS, 1% penicillin–streptomycin). After 3 weeks, mixed glial cultures had reached confluence, and microglia cells were isolated by mild trypsinization as described previously (68). In brief, cells were washed with the culture medium without FBS and treated with a mixture of trypsin 0.25% (without EDTA) and DMEM-F12 medium in a 1:3 proportion. After an incubation period of approximately 40 min, an intact layer of mixed glial cells detached, leaving microglia attached firmly to the bottom of the petri dish. Pure microglia were then isolated by 15-min incubation with trypsin (0.05%)-EDTA at 37 °C, followed by gentle shaking. Cells were counted and seeded at the desired density, typically 7.5 × 104 cells/well in 24 well-plates.
NLRP3 inflammasome activation assay
Primary microglia cultures were incubated with the positive compounds from the primary in vitro screening (C9, C21, C25, C29, C33, C50, C53, C54, C56, C61, C62, C73, C75, C77, C78, C84, C87, C94, C95, and C97). Cells pretreated with 100 μM of the compounds (or with the same DMSO volume, in the case of control samples) were primed with 400 ng/ml LPS for 3 h and activated with 4.5-mM ATP for 45 min, or 2-μM nigericin for 1 h. Dose–response curves were conducted to determine the potency of the compounds that inhibit the IL-1β concentration below a 20% threshold. For that, microglia cultures were incubated with different concentrations of the selected compounds, ranging from 0.001 to 100 μM, 1 h before NLRP3 activation with LPS and ATP. Supernatants were collected for IL-1β and toxicity assessments. 5 μM MCC950 was used as a positive control of NLRP3 inhibition.
AIM-2, NLRP7, and NLRC4 inflammasome activation assays
Microglia from NLRP3 KO mice were pretreated with 100-μM C75, C78, C97, or C77, for 1 h. AIM-2 inflammasome activation was induced through the addition of 100 ng/ml LPS for 2 h, followed by transfection of 1-μg poly(dA:dT) using Lipofectamine 2000 during 2 h, following the manufacturer’s recommendations. NLRC4 was activated by treatment with 150 ng/ml LPS for 2 h, before transfection of 1-μg flagellin during 2 h, using Lipofectamine 2000 as a transfection reagent. Negative and positive controls were pretreated with the same DMSO percentage as cells treated with the inhibitors. Negative controls for inflammasome activation were kept untreated after DMSO addition, while LPS-poly(dA:dT) or LPS-flagellin was added to positive controls for AIM-2, and NLRC4 activation, respectively. For activation of the NLRP7 inflammasome, different conditions were assessed: treatment of WT microglia cultures with 400 ng/ml fsl-1 for 7 h, priming with 150 ng/ml LPS for 1 h and treatment with 400 ng/ml fsl-1 for 7 h, and priming with 150 ng/ml LPS and transfection of 400 ng/ml fsl-1 with Lipofectamine. Among the tried conditions, only microglia primed with LPS and transfected with fsl-1/Lipofectamine released IL-1β (Fig. S6B). In addition, the specific NLRP3 inhibitor MCC950 was used as a negative control for NLRC4 and AIM-2 inhibition. Cells were pretreated with 5-μM MCC950 before LPS addition, and 2 h later, the NLRC4 and AIM-2 activation was induced through addition of their specific activators. Cell supernatants were collected for IL-1β measurement. All conditions were assayed in triplicate.
m-Cerulean–ASC–labeled macrophages
NLRP3 KO immortalized murine macrophages, stably expressing m-Cerulean–ASC, and NLRP3–Flag were a kind donation of Prof. Eicke Latz, (Institute of Innate Immunity, University of Bonn, Germany). Murine macrophages were maintained in high-glucose DMEM supplemented with 10% FBS and 1% sodium pyruvate and passaged every 2 to 3 days, before reaching confluence.
For ASC speck formation evaluation, murine macrophages were seeded in 96-well SCREENSTAR black microplates with clear base, 24 h before stimulation. Then, cells were incubated for 1 h with different concentrations of the inhibitors or with the same DMSO volume for positive and negative controls for NLRP3 activation (DMSO–LPS–ATP or only DMSO, respectively). 0.5 μM MCC950 was used as a positive control for inhibition of NLRP3-mediated ASC oligomerization. NLRP3 activation was induced through treatment with 250 ng/ml LPS for 2.5 h, or 1 μg/ml HMGB1 for 3 h, followed by addition of 4.5-mM ATP for 45 min. Cells were then fixed and counterstained in a single step, as described previously (15). For that, a stock solution of deep red anthraquinone 5 (DRAQ5) in formaldehyde was used, the final concentrations being 1% formaldehyde and 2.5-μM DRAQ5.
Cell viability assays
The cytotoxic effect of the compounds identified by in vitro screening was assayed with the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega), following the recommendations of the manufacturer.
Pyroptosis measurement
The ability of C75, C78, and C97 to inhibit pyroptosis was determined by measuring the lactate dehydrogenase released by pyroptotic cells into the culture medium. 400 ng/ml LPS was added to primary microglial cultures pretreated with different concentrations of the inhibitors. After 3 h of treatment, pyroptosis was induced through the addition of 4.5-mM ATP, for 45 min. 5 μM MCC950 was used as a positive control of pyroptosis inhibition. Cell supernatants were collected for lactate dehydrogenase assessment, using the CytoTox 96 Non-Radioactive Cytotoxicity Assay (Promega), following the manufacturer recommendations. Experiments were run in triplicate.
ASC oligomerization determination
The inhibitory effect of the selected compounds on ASC oligomerization was tested using m-Cerulean–ASC–labeled macrophages. ASC specks in cells pretreated with C75, C77, C78, and C97, before NLRP3 activation (as indicated above), were visualized with a widefield Leica DMi8 fluorescence microscope, using the 20× objective. Cyanine5 (λexc 630 nm) and 4',6-diaminidino-2-phenylindole (λexc 405 nm) filters were used for m-cerulean and DRAQ5 visualization, respectively. Images were analyzed using ImageJ https://imagej.nih.gov/ij/index.html.
Speck quantification was performed as described previously (15). Nuclei were quantified using the automatic particle counting tool from ImageJ. The same threshold was applied to all samples. When required (overlapping nuclei), the watershed separation algorithm was used to separate the particles. All results were subject to manual validation. The rate of speck positive cells was calculated by normalizing the number of speck-positive cells to the number of nuclei. Three biological replicates were taken for each treatment group.
Cytokine quantification
IL-1β in cell culture supernatants and brain tissue samples—after the different treatment groups—was quantified using the mouse IL-1 beta/IL-1F2 DuoSet ELISA kit (R&D systems), IL-6 in the cell culture supernatant was measured through the mouse IL-6 ELISA kit (Thermo Fisher), TNF-α in the cell culture supernatants was determined using the mouse TNF-alpha DuoSet ELISA (R&D systems), and IL-18 in brain tissue was measured through the IL-18 mouse ELISA Kit (Invitrogen) following the manufacturers' recommended protocol. The IL-1β precursor and cleaved IL-1β in the FC of mice treated with vehicle, LPS, or C77-LPS were assessed by Western blot. All samples were run in technical duplicates.
Inhibition of the NLRP3 ATPase activity
The efficacy of C77 to inhibit ATP hydrolysis by NLRP3 was directly assayed using a cell-free assay. Human recombinant NLRP3 protein (BPS Bioscience) was incubated at 37 °C with different concentrations of C77 for 20 min, as described (49). 10 μM ultrapure ATP was then added, and reaction mixtures were further incubated for 45 min. ATP conversion into ADP was determined using the ADP-Glo Kinase Assay (Promega), following the recommendations of the manufacturer, in a SpectraMax i3x Multimode Detection Platform. Experiments were conducted in triplicate.
Gene expression analysis
RNA was isolated from the FC from C57BL/6J mice or from THP1 human cultures using the RNeasy Mini Kit (QIAGEN, Hilden, Germany), using the manufacturers' recommended protocol. The RNA concentration was determined, and cDNA was synthesized using the High-Capacity cDNA Reverse Transcription Kit with RNase Inhibitor (applied biosystems). The expression levels of inflammatory genes —NLRP3, caspase-1, IL-1β, TNF-α, NLRC4, IL-8, and IL-6—were determined in four replicates through PowerUp SYBR Green Master Mix (Thermo Fisher) using an ABI PRISM 7900HT Sequence Detection System. Hypoxanthine phosphoribosyltransferase was used as an internal control in samples from mouse brains, and GAPDH in the case of THP1 lysates. Gene expression levels were normalized to control samples (mice or THP1 cells treated with PBS). Primer sequences are displayed in Table S1.
In vivo assessments
Animals
WT C57BL/6J mice (stock #000664) were obtained from The Jackson Laboratory. Mice were housed in groups of five animals per cage on a 12:12 h light/dark cycle. Water and food were provided ad libitum. Mice aged 8 to 10 weeks were used for experiments. All procedures were approved by the Institutional Animal Care and Use Committee of the Icahn School of Medicine at Mount Sinai.
In vivo assessment of C77
The ability of C77 to inhibit NLRP3 in vivo was evaluated using WT C57BL/6J mice. C77 (50 mg/kg), or the same volume of DMSO vehicle, was intraperitoneally injected 2 h before LPS (15 mg/kg) or vehicle (PBS) administration. 1.5 h later, the animals were sacrificed, and their brain was isolated. The cortex, FC, hippocampus, and cerebellum regions from the right hemisphere were obtained for IL-1β analysis. The left hemisphere was used for gene expression analysis.
Statistical analysis
Unless otherwise indicated, all the experiments included in this article were performed in triplicate. Results are expressed as the mean ± SD or as the mean ± standard error of the regression. A one-way ANOVA followed by Tukey’s posttest (confidence interval = 95%) was performed using GraphPad Prism 8 software (GraphPad Software, San Diego, CA) to determine statistical significance between treatment groups. Unless otherwise indicated, significance intervals are expressed as ∗ p ≤ 0.05, ¥ p ≤ 0.01, § p ≤ 0.001, and ¶ p ≤ 0.0001.
Data availability
All data obtained during this study are contained within the present article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
R. S. contributed by producing the hNLRP3 3D model and by conducting the virtual screening. M. S.-V. contributed by performing the in vitro and in vivo studies and the computational docking. H. W. carried out the Western blot analysis. M. A. R. performed in vitro studies. K. K. and R. J. D. V. determined the purity of the compounds and advised with the chemistry. M. S.-V. and G. M. P. designed the study concept. All authors discussed and commented on the article.
Funding and additional information
This research was supported by a grant (AT008661) from the NIH's Office of Dietary Supplements (ODS) and the National Center for Complementary and Integrative Health, awarded to G. M. P. The study was supported by the generous support of the Altschul Foundation to G. M. P. G. M. P. holds a Senior VA Career Scientist Award. We acknowledge that the contents of this study do not represent the views of the NCCIH, the ODS, the National Institutes of Health, the U.S. Department of Veterans Affairs, or the United States Government.
Edited by Dennis Voelker
Supporting information
References
- 1.Franchi L., McDonald C., Kanneganti T.-D., Amer A., Núñez G. Nucleotide-binding oligomerization domain-like receptors: Intracellular pattern recognition molecules for pathogen detection and host defense. J. Immunol. 2006;177:3507–3513. doi: 10.4049/jimmunol.177.6.3507. [DOI] [PubMed] [Google Scholar]
- 2.Inohara N., Chamaillard M., McDonald C., Nuñez G. NOD-LRR proteins: Role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 2005;74:355–383. doi: 10.1146/annurev.biochem.74.082803.133347. [DOI] [PubMed] [Google Scholar]
- 3.He W., Wan H., Hu L., Chen P., Wang X., Huang Z., Yang Z.-H., Zhong C.-Q., Han J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015;25:1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 5.Kayagaki N., Stowe I.B., Lee B.L., O’Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., Liu P.S., Lill J.R., Li H., Wu J., Kummerfeld S. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 6.Duncan J.A., Bergstralh D.T., Wang Y., Willingham S.B., Ye Z., Zimmermann A.G., Ting J.P.-Y. Cryopyrin/NALP3 binds ATP/dATP, is an ATPase, and requires ATP binding to mediate inflammatory signaling. Proc. Natl. Acad. Sci. U. S. A. 2007;104:8041–8046. doi: 10.1073/pnas.0611496104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson K.V., Deng M., Ting J.P.-Y. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019;19:477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Y., Wang H., Kouadir M., Song H., Shi F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019;10:128. doi: 10.1038/s41419-019-1413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saadi M., Karkhah A., Pourabdolhossein F., Ataie A., Monif M., Nouri H.R. Involvement of NLRC4 inflammasome through caspase-1 and IL-1β augments neuroinflammation and contributes to memory impairment in an experimental model of Alzheimer’s like disease. Brain Res. Bull. 2020;154:81–90. doi: 10.1016/j.brainresbull.2019.10.010. [DOI] [PubMed] [Google Scholar]
- 10.Zhang L., Chen S., Ruan J., Wu J., Tong A.B., Yin Q., Li Y., David L., Lu A., Wang W.L., Marks C., Ouyang Q., Zhang X., Mao Y., Wu H. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350:404–409. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chear C.T., Nallusamy R., Canna S.W., Chan K.C., Baharin M.F., Hishamshah M., Ghani H., Ripen A.M., Mohamad S.B. A novel de novo NLRC4 mutation reinforces the likely pathogenicity of specific LRR domain mutation. Clin. Immunol. 2020;211:108328. doi: 10.1016/j.clim.2019.108328. [DOI] [PubMed] [Google Scholar]
- 12.Strowig T., Henao-Mejia J., Elinav E., Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- 13.Platnich J.M., Muruve D.A. NOD-like receptors and inflammasomes: A review of their canonical and non-canonical signaling pathways. Arch. Biochem. Biophys. 2019;670:4–14. doi: 10.1016/j.abb.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 14.Duncan J.A., Canna S.W. The NLRC4 inflammasome. Immunol. Rev. 2018;281:115–123. doi: 10.1111/imr.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stutz A., Horvath G.L., Monks B.G., Latz E. ASC speck formation as a readout for inflammasome activation. In: De Nardo C.M., Latz E., editors. Vol. 1040. Humana Press; Totowa, NJ: 2013. pp. 91–101. (The Inflammasome). [DOI] [PubMed] [Google Scholar]
- 16.Franklin B.S., Bossaller L., De Nardo D., Ratter J.M., Stutz A., Engels G., Brenker C., Nordhoff M., Mirandola S.R., Al-Amoudi A., Mangan M.S., Zimmer S., Monks B.G., Fricke M., Schmidt R.E. The adaptor ASC has extracellular and “prionoid” activities that propagate inflammation. Nat. Immunol. 2014;15:727–737. doi: 10.1038/ni.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Satoh T., Kambe N., Matsue H. NLRP3 activation induces ASC-dependent programmed necrotic cell death, which leads to neutrophilic inflammation. Cell Death Dis. 2013;4:e644. doi: 10.1038/cddis.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coll R.C., Hill J.R., Day C.J., Zamoshnikova A., Boucher D., Massey N.L., Chitty J.L., Fraser J.A., Jennings M.P., Robertson A.A.B., Schroder K. MCC950 directly targets the NLRP3 ATP-hydrolysis motif for inflammasome inhibition. Nat. Chem. Biol. 2019;15:556–559. doi: 10.1038/s41589-019-0277-7. [DOI] [PubMed] [Google Scholar]
- 19.Tapia-Abellán A., Angosto-Bazarra D., Martínez-Banaclocha H., de Torre-Minguela C., Cerón-Carrasco J.P., Pérez-Sánchez H., Arostegui J.I., Pelegrin P. MCC950 closes the active conformation of NLRP3 to an inactive state. Nat. Chem. Biol. 2019;15:560–564. doi: 10.1038/s41589-019-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franchi L., Park J.-H., Shaw M.H., Marina-Garcia N., Chen G., Kim Y.-G., Núñez G. Intracellular NOD-like receptors in innate immunity, infection and disease. Cell. Microbiol. 2008;10:1–8. doi: 10.1111/j.1462-5822.2007.01059.x. [DOI] [PubMed] [Google Scholar]
- 21.Fritz J.H., Ferrero R.L., Philpott D.J., Girardin S.E. Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 22.Menu P., Vince J.E. The NLRP3 inflammasome in health and disease: The good, the bad and the ugly: Inflammasomes: The good, the bad and the ugly. Clin. Exp. Immunol. 2011;166:1–15. doi: 10.1111/j.1365-2249.2011.04440.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aganna E., Martinon F., Hawkins P.N., Ross J.B., Swan D.C., Booth D.R., Lachmann H.J., Gaudet R., Woo P., Feighery C., Cotter F.E., Thome M., Hitman G.A., Tschopp J., McDermott M.F. Association of mutations in theNALP3/CIAS1/PYPAF1 gene with a broad phenotype including recurrent fever, cold sensitivity, sensorineural deafness, and AA amyloidosis. Arthritis Rheum. 2002;46:2445–2452. doi: 10.1002/art.10509. [DOI] [PubMed] [Google Scholar]
- 24.Neven B., Prieur A.-M., dit Maire P.Q. Cryopyrinopathies: Update on pathogenesis and treatment. Nat. Clin. Pract. Rheumatol. 2008;4:481–489. doi: 10.1038/ncprheum0874. [DOI] [PubMed] [Google Scholar]
- 25.Romberg N., Vogel T.P., Canna S.W. NLRC4 inflammasomopathies. Curr. Opin. Allergy Clin. Immunol. 2017;17:398–404. doi: 10.1097/ACI.0000000000000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L., Chan C. The role of inflammasome in Alzheimer’s disease. Ageing Res. Rev. 2014;15:6–15. doi: 10.1016/j.arr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frank M.G., Weber M.D., Watkins L.R., Maier S.F. Stress sounds the alarmin: The role of the danger-associated molecular pattern HMGB1 in stress-induced neuroinflammatory priming. Brain Behav. Immun. 2015;48:1–7. doi: 10.1016/j.bbi.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gong T., Liu L., Jiang W., Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020;20:95–112. doi: 10.1038/s41577-019-0215-7. [DOI] [PubMed] [Google Scholar]
- 29.Soares J.L., Oliveira E.M., Pontillo A. Variants in NLRP3 and NLRC4 inflammasome associate with susceptibility and severity of multiple sclerosis. Mult. Scler. Relat. Disord. 2019;29:26–34. doi: 10.1016/j.msard.2019.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Lehmann S., Esch E., Hartmann P., Goswami A., Nikolin S., Weis J., Beyer C., Johann S. Expression profile of pattern recognition receptors in skeletal muscle of SOD1 (G93A) amyotrophic lateral sclerosis (ALS) mice and sporadic ALS patients. Neuropathol. Appl. Neurobiol. 2018;44:606–627. doi: 10.1111/nan.12483. [DOI] [PubMed] [Google Scholar]
- 31.Fan Z., Pan Y.-T., Zhang Z.-Y., Yang H., Yu S.-Y., Zheng Y., Ma J.-H., Wang X.-M. Systemic activation of NLRP3 inflammasome and plasma α-synuclein levels are correlated with motor severity and progression in Parkinson’s disease. J. Neuroinflammation. 2020;17:11. doi: 10.1186/s12974-019-1670-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu L., Chan C. IPAF inflammasome is involved in interleukin-1β production from astrocytes, induced by palmitate; implications for Alzheimer’s Disease. Neurobiol. Aging. 2014;35:309–321. doi: 10.1016/j.neurobiolaging.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das S., Basu A. Inflammation: A new candidate in modulating adult neurogenesis. J. Neurosci. Res. 2008;86:1199–1208. doi: 10.1002/jnr.21585. [DOI] [PubMed] [Google Scholar]
- 34.Venegas C., Kumar S., Franklin B.S., Dierkes T., Brinkschulte R., Tejera D., Vieira-Saecker A., Schwartz S., Santarelli F., Kummer M.P., Griep A., Gelpi E., Beilharz M., Riedel D., Golenbock D.T. Microglia-derived ASC specks cross-seed amyloid-β in Alzheimer’s disease. Nature. 2017;552:355–361. doi: 10.1038/nature25158. [DOI] [PubMed] [Google Scholar]
- 35.Lamkanfi M., Mueller J.L., Vitari A.C., Misaghi S., Fedorova A., Deshayes K., Lee W.P., Hoffman H.M., Dixit V.M. Glyburide inhibits the Cryopyrin/Nalp3 inflammasome. J. Cell Biol. 2009;187:61–70. doi: 10.1083/jcb.200903124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Youm Y.-H., Nguyen K.Y., Grant R.W., Goldberg E.L., Bodogai M., Kim D., D’Agostino D., Planavsky N., Lupfer C., Kanneganti T.D., Kang S., Horvath T.L., Fahmy T.M., Crawford P.A., Biragyn A. The ketone metabolite β-hydroxybutyrate blocks NLRP3 inflammasome–mediated inflammatory disease. Nat. Med. 2015;21:263–269. doi: 10.1038/nm.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coll R.C., Robertson A.A.B., Chae J.J., Higgins S.C., Muñoz-Planillo R., Inserra M.C., Vetter I., Dungan L.S., Monks B.G., Stutz A., Croker D.E., Butler M.S., Haneklaus M., Sutton C.E., Núñez G. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang H., He H., Chen Y., Huang W., Cheng J., Ye J., Wang A., Tao J., Wang C., Liu Q., Jin T., Jiang W., Deng X., Zhou R. Identification of a selective and direct NLRP3 inhibitor to treat inflammatory disorders. J. Exp. Med. 2017;214:3219–3238. doi: 10.1084/jem.20171419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perera A.P., Fernando R., Shinde T., Gundamaraju R., Southam B., Sohal S.S., Robertson A.A.B., Schroder K., Kunde D., Eri R. MCC950, a specific small molecule inhibitor of NLRP3 inflammasome attenuates colonic inflammation in spontaneous colitis mice. Sci. Rep. 2018;8:8618. doi: 10.1038/s41598-018-26775-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang C., Zhu X., Li L., Ma T., Shi M., Yang Y., Fan Q. A small molecule inhibitor MCC950 ameliorates kidney injury in diabetic nephropathy by inhibiting NLRP3 inflammasome activation. Diabetes Metab. Syndr. Obes. Targets Ther. Volume. 2019;12:1297–1309. doi: 10.2147/DMSO.S199802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ismael S., Nasoohi S., Ishrat T. MCC950, the selective inhibitor of nucleotide oligomerization domain-like receptor protein-3 inflammasome, protects mice against traumatic brain injury. J. Neurotrauma. 2018;35:1294–1303. doi: 10.1089/neu.2017.5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangan M.S.J., Olhava E.J., Roush W.R., Seidel H.M., Glick G.D., Latz E. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat. Rev. Drug Discov. 2018;17:588–606. doi: 10.1038/nrd.2018.97. [DOI] [PubMed] [Google Scholar]
- 43.Lemprière S. NLRP3 inflammasome activation implicated in tau pathology. Nat. Rev. Neurol. 2020;16:4. doi: 10.1038/s41582-019-0299-5. 4. [DOI] [PubMed] [Google Scholar]
- 44.Freeman L., Guo H., David C.N., Brickey W.J., Jha S., Ting J.P.-Y. NLR members NLRC4 and NLRP3 mediate sterile inflammasome activation in microglia and astrocytes. J. Exp. Med. 2017;214:1351–1370. doi: 10.1084/jem.20150237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gustot A., Gallea J.I., Sarroukh R., Celej M.S., Ruysschaert J.-M., Raussens V. Amyloid fibrils are the molecular trigger of inflammation in Parkinson’s disease. Biochem. J. 2015;471:323–333. doi: 10.1042/BJ20150617. [DOI] [PubMed] [Google Scholar]
- 46.Codolo G., Plotegher N., Pozzobon T., Brucale M., Tessari I., Bubacco L., de Bernard M. Triggering of inflammasome by aggregated α–synuclein, an inflammatory response in synucleinopathies. PLoS One. 2013;8 doi: 10.1371/journal.pone.0055375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuemmerle-Deschner J.B., Hachulla E., Cartwright R., Hawkins P.N., Tran T.A., Bader-Meunier B., Hoyer J., Gattorno M., Gul A., Smith J., Leslie K.S., Jimenez S., Morell-Dubois S., Davis N., Patel N. Two-year results from an open-label, multicentre, phase III study evaluating the safety and efficacy of canakinumab in patients with cryopyrin-associated periodic syndrome across different severity phenotypes. Ann. Rheum. Dis. 2011;70:2095–2102. doi: 10.1136/ard.2011.152728. [DOI] [PubMed] [Google Scholar]
- 48.Dinarello C.A., Simon A., van der Meer J.W.M. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat. Rev. Drug Discov. 2012;11:633–652. doi: 10.1038/nrd3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cocco M., Pellegrini C., Martínez-Banaclocha H., Giorgis M., Marini E., Costale A., Miglio G., Fornai M., Antonioli L., López-Castejón G., Tapia-Abellán A., Angosto D., Hafner-Bratkovič I., Regazzoni L., Blandizzi C. Development of an acrylate derivative targeting the NLRP3 inflammasome for the treatment of inflammatory bowel disease. J. Med. Chem. 2017;60:3656–3671. doi: 10.1021/acs.jmedchem.6b01624. [DOI] [PubMed] [Google Scholar]
- 50.Maekawa S., Ohto U., Shibata T., Miyake K., Shimizu T. Crystal structure of NOD2 and its implications in human disease. Nat. Commun. 2016;7:11813. doi: 10.1038/ncomms11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shim D.-W., Shin W.-Y., Yu S.-H., Kim B.-H., Ye S.-K., Koppula S., Won H.-S., Kang T.-B., Lee K.-H. BOT-4-one attenuates NLRP3 inflammasome activation: NLRP3 alkylation leading to the regulation of its ATPase activity and ubiquitination. Sci. Rep. 2017;7:15020. doi: 10.1038/s41598-017-15314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venereau E., Casalgrandi M., Schiraldi M., Antoine D.J., Cattaneo A., De Marchis F., Liu J., Antonelli A., Preti A., Raeli L., Shams S.S., Yang H., Varani L., Andersson U., Tracey K.J. Mutually exclusive redox forms of HMGB1 promote cell recruitment or proinflammatory cytokine release. J. Exp. Med. 2012;209:1519–1528. doi: 10.1084/jem.20120189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Golen R.F., Reiniers M.J., Marsman G., Alles L.K., van Rooyen D.M., Petri B., Van der Mark V.A., van Beek A.A., Meijer B., Maas M.A., Zeerleder S., Verheij J., Farrell G.C., Luken B.M., Teoh N.C. The damage-associated molecular pattern HMGB1 is released early after clinical hepatic ischemia/reperfusion. Biochim. Biophys. Acta Mol. Basis Dis. 2019;1865:1192–1200. doi: 10.1016/j.bbadis.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 54.Yu J.R., Leslie K.S. Cryopyrin-associated periodic syndrome: An update on diagnosis and treatment response. Curr. Allergy Asthma Rep. 2011;11:12–20. doi: 10.1007/s11882-010-0160-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ising C., Venegas C., Zhang S., Scheiblich H., Schmidt S.V., Vieira-Saecker A., Schwartz S., Albasset S., McManus R.M., Tejera D., Griep A., Santarelli F., Brosseron F., Opitz S., Stunden J. NLRP3 inflammasome activation drives tau pathology. Nature. 2019;575:669–673. doi: 10.1038/s41586-019-1769-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tejera D., Mercan D., Sanchez-Caro J.M., Hanan M., Greenberg D., Soreq H., Latz E., Golenbock D., Heneka M.T. Systemic inflammation impairs microglial Aβ clearance through NLRP 3 inflammasome. EMBO J. 2019;38 doi: 10.15252/embj.2018101064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stancu I.-C., Cremers N., Vanrusselt H., Couturier J., Vanoosthuyse A., Kessels S., Lodder C., Brône B., Huaux F., Octave J.-N., Terwel D., Dewachter I. Aggregated Tau activates NLRP3–ASC inflammasome exacerbating exogenously seeded and non-exogenously seeded Tau pathology in vivo. Acta Neuropathol. 2019;137:599–617. doi: 10.1007/s00401-018-01957-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deora V., Lee J.D., Albornoz E.A., McAlary L., Jagaraj C.J., Robertson A.A.B., Atkin J.D., Cooper M.A., Schroder K., Yerbury J.J., Gordon R., Woodruff T.M. The microglial NLRP3 inflammasome is activated by amyotrophic lateral sclerosis proteins. Glia. 2020;68:407–421. doi: 10.1002/glia.23728. [DOI] [PubMed] [Google Scholar]
- 59.Debye B., Schmülling L., Zhou L., Rune G., Beyer C., Johann S. Neurodegeneration and NLRP3 inflammasome expression in the anterior thalamus of SOD1(G93A) ALS mice: Neurodegeneration and NLRP3 inflammasome expression. Brain Pathol. 2018;28:14–27. doi: 10.1111/bpa.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johann S., Heitzer M., Kanagaratnam M., Goswami A., Rizo T., Weis J., Troost D., Beyer C. NLRP3 inflammasome is expressed by astrocytes in the SOD1 mouse model of ALS and in human sporadic ALS patients: NLRP3 inflammasome expression in ALS. Glia. 2015;63:2260–2273. doi: 10.1002/glia.22891. [DOI] [PubMed] [Google Scholar]
- 61.Sharif H., Wang L., Wang W.L., Magupalli V.G., Andreeva L., Qiao Q., Hauenstein A.V., Wu Z., Núñez G., Mao Y., Wu H. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature. 2019;570:338–343. doi: 10.1038/s41586-019-1295-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinforma. 2014;47:5.6.1–5.6.32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 63.Friesner R.A., Banks J.L., Murphy R.B., Halgren T.A., Klicic J.J., Mainz D.T., Repasky M.P., Knoll E.H., Shelley M., Perry J.K., Shaw D.E., Francis P., Shenkin P.S. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 64.Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T. Extra precision Glide: Docking and scoring incorporating a model of hydrophobic enclosure for Protein−Ligand complexes. J. Med. Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 65.Halgren T.A., Murphy R.B., Friesner R.A., Beard H.S., Frye L.L., Pollard W.T., Banks J.L. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 66.Dahlin J.L., Nissink J.W.M., Strasser J.M., Francis S., Higgins L., Zhou H., Zhang Z., Walters M.A. PAINS in the assay: Chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. J. Med. Chem. 2015;58:2091–2113. doi: 10.1021/jm5019093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bronstein R., Torres L., Nissen J.C., Tsirka S.E. Culturing microglia from the neonatal and adult central nervous system. J. Vis. Exp. 2013 doi: 10.3791/50647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Saura J., Tusell J.M., Serratosa J. High-yield isolation of murine microglia by mild trypsinization. Glia. 2003;44:183–189. doi: 10.1002/glia.10274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data obtained during this study are contained within the present article.