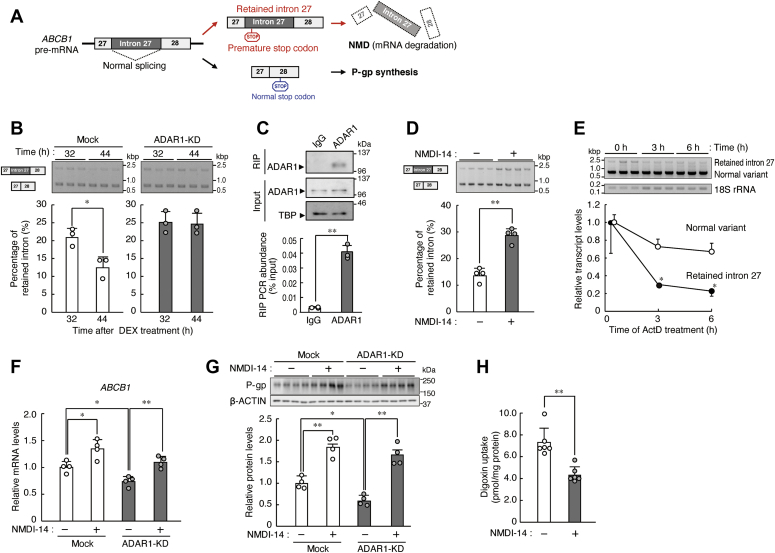
Abstract
The expression and function of some xenobiotic transporters vary according to the time of the day, causing the dosing time-dependent changes in drug disposition and toxicity. P-glycoprotein (P-gp), encoded by the ABCB1 gene, is highly expressed in the kidneys and functions in the renal elimination of various drugs. The elimination of several P-gp substrates was demonstrated to vary depending on administration time, but the underlying mechanism remains unclear. We found that adenosine deaminase acting on RNA (ADAR1) was involved in the circadian regulation of P-gp expression in human renal proximal tubular epithelial cells (RPTECs). After synchronization of the cellular circadian clock by dexamethasone treatment, the expression of P-gp exhibited a significant 24-h oscillation in RPTECs, but this oscillation was disrupted by the downregulation of ADAR1. Although ADAR1 catalyzes adenosine-to-inosine (A-to-I) RNA editing in double-stranded RNA substrates, no significant ADAR1-regulated editing sites were detected in the human ABCB1 transcripts in RPTECs. On the other hand, downregulation of ADAR1 induced alternative splicing in intron 27 of the human ABCB1 gene, resulting in the production of retained intron transcripts. The aberrant spliced transcript was sensitive to nonsense-mediated mRNA decay, leading to the decreased stability of ABCB1 mRNA and prevention of the 24-h oscillation of P-gp expression. These findings support the notion that ADAR1-mediated regulation of alternative splicing of the ABCB1 gene is a key mechanism of circadian expression of P-gp in RPTECs, and the regulatory mechanism may underlie the dosing time-dependent variations in the renal elimination of P-gp substrates.
Keywords: RNA editing, circadian rhythm, ABC transporter, posttranscriptional regulation, alternative splicing, pharmacokinetics
Abbreviations: ADAR, adenosine deaminase acting on RNA; AhR, aryl hydrocarbon receptor; A-to-I, adenosine-to-inosine; ATF4, activating transcription factor 4; DEX, dexamethasone; dsRNA, double-stranded RNA; HLF, hepatic leukemia factor; KD, knockdown; NMD, nonsense-mediated mRNA decay; P-gp, P-glycoprotein; RIP, RNA immunoprecipitation; RPTEC, renal proximal tubular epithelial cell; SCN, suprachiasmatic nucleus; shRNA, small hairpin RNA; SLC, solute carrier; snRNP, small nuclear ribonucleoprotein
The daily variations in biological functions are thought to be important factors affecting the efficacy and/or toxicity of drugs; a large number of drugs cannot be expected to have the same potency at different administration times (1, 2). Dosing time-dependent differences in the therapeutic effects of drugs are, at least in part, due to diurnal changes in drug disposition such as absorption, distribution, metabolism, and elimination (3). ABC transporters are widely known as energy-dependent efflux pumps that extrude cytotoxic substances from cells. They are expressed in epithelial cells of several organs, such as the brain, liver, intestine, and kidneys (4) and function in the biliary, intestinal, and renal elimination of drugs (5).
The expression and function of several ABC transporters exhibit diurnal variation, resulting in dosing time-dependent differences in drug disposition (6, 7, 8). We previously demonstrated that the expression and function of P-glycoprotein (P-gp), encoded by the ABCB1 gene, exhibit diurnal oscillation in the small intestine of mice and cynomolgus monkeys (9, 10). In both rodents and monkeys, the oscillation of P-gp expression was associated with the dosing time-dependent differences in the intestinal absorption of its substrate drugs. The efflux pump activity of P-gp was also suggested to exhibit diurnal oscillation in humans because of the dosing time-dependent differences in renal clearance of substrate drugs (11, 12), but diurnal variations in the expression and function of P-gp in human renal cells remain unclear.
In general, protein levels are regulated at nearly all stages of the gene expression process. In addition to their synthesis, mRNA and proteins are both also subject to degradation, which influences the abundance of membrane proteins. The circadian expression of clock and clock-controlled proteins occurs at the transcriptional, posttranscriptional, translational, and posttranslational levels (13, 14, 15). Among posttranscriptional regulation mechanisms, adenosine-to-inosine (A-to-I) RNA editing is the most prevalent nucleotide conversion in mammals (16). Inosine, an analog of guanosine, forms a base pair with cytidine, altering the amino acid sequence, microRNA targeting, and splicing of RNA (16). A-to-I RNA editing is catalyzed by adenosine deaminase acting on RNA (ADAR) enzymes. They convert adenosines in the double-stranded RNA (dsRNA) structure to inosines by hydrolytic deamination. There are two functional members of the ADAR family in humans: ADAR1 and ADAR2. ADAR1 has two isoforms: ADAR1-p110 (110-kDa protein) and ADAR1-p150 (150-kDa protein). ADAR1-p110 is constitutively expressed in the nucleus, whereas ADAR1-p150 has an interferon-inducible promoter and is predominantly localized in the cytoplasm (16, 17). Recently, A-to-I RNA editing is demonstrated to exhibit diurnal oscillation in the liver of mice, suggesting that A-to-I RNA editing generates diurnal rhythmicity of a wide range of mRNAs (18).
ADAR1-mediated RNA editing is also reported to regulate the expression of drug metabolism enzymes (19). For example, ADAR1 creates microRNA targeting sites of aryl hydrocarbon receptor (AhR) mRNA, thereby inducing the downregulation of AhR in human liver cells (20). The downregulation of AhR interferes with drug-inducible expression of Cytochrome 1A1. During the analysis of the role of ADAR1 in the regulation of renal expression of xenobiotic transporters, we found that the levels of P-gp were reduced by the downregulation of ADAR1 in human renal proximal tubular epithelial cells (RPTECs). The expression of P-gp exhibited significant 24-h oscillation in circadian clock-synchronized cultured RPTECs, but this oscillation was disrupted in ADAR1-knockdown (KD) RPTECs. Therefore, we investigated the mechanism by which ADAR1 regulates the circadian expression of P-gp in human RPTECs.
Results
Effects of downregulation of ADAR1 on xenobiotic transporters in human renal cells
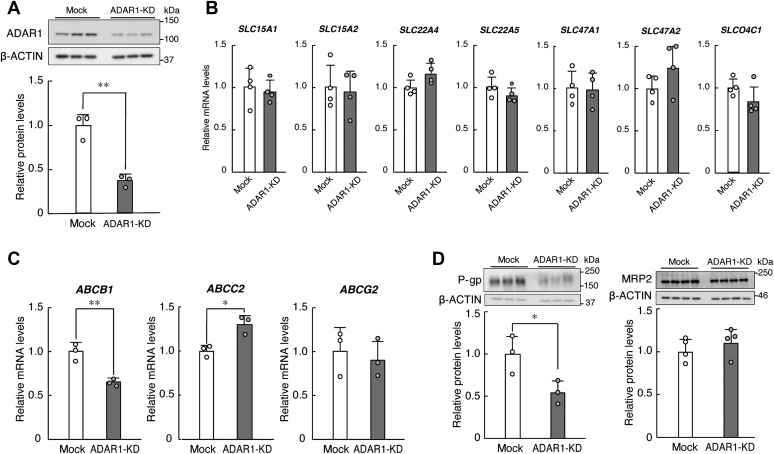
In the first set of experiments, we used nonsynchronized human RPTECs to investigate which renal transporters are under the control of ADAR1. To achieve this, we prepared ADAR1-KD human RPTECs by stable expression of small hairpin RNA (shRNA). Although no notable bands derived from ADAR1-p150 were detectable in RPTECs, the expression levels of the ADAR1-p110 isoform were significantly reduced to 40% in anti-ADAR1 shRNA-transduced cells (p < 0.01, Fig. 1A). Therefore, we used the ADAR1-KD RPTECs to assess the mRNA levels of xenobiotic transporters that are known to regulate renal drug elimination (21). As results of quantitative real-time RT-PCR analysis, the mRNA levels of the solute carrier (SLC) group of membrane transport proteins, SLC15A1, SLC15A2, SLC22A4, SLC22A5, SLC47A1, SLC47A2, and SLCO4C1, were not significantly different between mock-transduced and ADAR1-KD RPTECs (Fig. 1B). The transport proteins encoded by SLC22A2, SLC22A6, and SLC22A8 genes also regulate tubular secretion of various drugs, but their mRNA expression was undetectable in RPTECs. On the other hand, the mRNA levels of ABCB1 and ABCC2 in ADAR1-KD RPTECs decreased and increased, respectively, whereas ABCG2 mRNA levels in ADAR1-KD RPTECs were not significantly different from those in mock-transduced cells (Fig. 1C). Furthermore, downregulation of ADAR1 reduced the expression of P-gp encoded by the ABCB1 gene, but not of MRP2 encoded by the ABCC2 gene, in RPTECs (Fig. 1D). This suggests that ADAR1 positively regulates the expression of ABCB1 mRNA and P-gp in tubular cells of the human kidney. Due to the stability of MRP2 protein (22), ADAR1-induced expression of ABCC2 mRNA might not be reflected to its protein levels.
Figure 1.
Downregulation of ADAR1 reduces the expression of P-gp in RPTECs.A, construction of ADAR1-knockdown (KD) RPTECs with stable expression of shRNA. The expression levels of ADAR1-p110 protein were normalized to that of β-ACTIN. Values are the mean with S.D. (n = 3). The value of ADAR1-p110 in mock-transduced RPTECs was set at 1.0. ∗∗p < 0.01; significant difference between the two groups (t4 = 6.988, p = 0.002; unpaired t-test, two-sided). B, the mRNA levels of solute carrier (SLC) transporters in mock-transduced and ADAR1-KD RPTECs. The mRNA levels were assessed by quantitative real-time RT-PCR analysis and their expression levels were normalized to that of 18S rRNA. Values are the mean with S.D. (n = 4). C, the mRNA levels of ABC transporters in mock-transduced and ADAR1-KD RPTECs. The mRNA levels were normalized to that of 18S rRNA. Values are the mean with S.D. (n = 3). ∗∗p < 0.01, ∗p < 0.05; significant difference between the two groups (t4 = 5.656, p = 0.005 for ABCB1; t4 = 4.175, p = 0.014 for ABCC2; unpaired t-test, two-sided). D, The protein levels of P-gp and MRP2 in mock-transduced and ADAR1-KD RPTECs. The protein levels were normalized to that of β-ACTIN. Values are the mean with S.D. (n = 3–4). ∗p < 0.05; significant difference between the two groups (t4 = 2.938, p = 0.043 for P-gp; unpaired t-test, two-sided).
ADAR1 is involved in circadian regulation of P-gp expression in human RPTECs
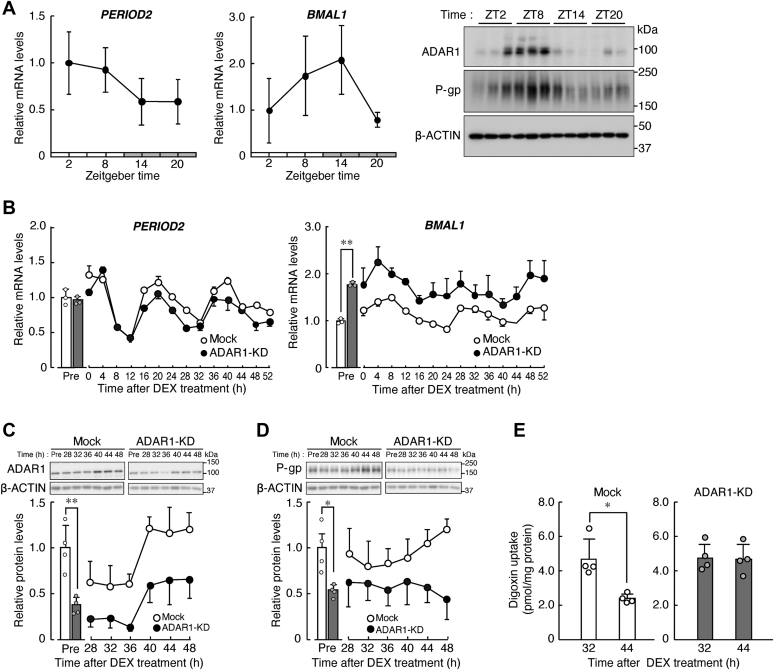
The elimination rate of several P-gp substrates is changed depending on their administration time of the day (11, 12), suggesting that efflux pump function of P-gp varies during the day due to changes in its expression level. Indeed, we observed diurnal oscillations in the protein levels of ADAR1 and P-gp in the kidney of cynomolgus monkeys (Fig. 2A) kept under the light/dark cycle (ZT, zeitgeber time; ZT0, lights on; ZT12, lights off). This prompted us to explore the possibility that ADAR1 is also involved in circadian regulation of P-gp expression in the human kidney. We therefore investigated the temporal expression profiles of ADAR1 and P-gp in cultured human RPTECs after synchronization of their circadian clock. Treatment of mock-transduced RPTECs with 100 nM dexamethasone (DEX) for 2 h induced significant 24-h oscillations in the mRNA expression of the major circadian clock genes PERIOD2 and BMAL1 (p < 0.01, respectively, Fig. 2B). Similar 24-h oscillations of circadian gene expression were also observed in DEX-treated ADAR1-KD RPTECs (p < 0.01, respectively, Fig. 2B), although BMAL1 mRNA levels increased in ADAR1-KD RPTECs. Synchronization of the circadian clock persisted for up to 52 h after DEX treatment. In this in vitro culture model, the oscillation in the expression of clock genes after 24 h following synchronization is thought to be generated by cell-autonomous circadian machinery (23, 24), which constitute fundamental properties that characterize circadian rhythms. Therefore, we investigated the expression of ADAR1 and P-gp from 28 h to 48 h after DEX treatment. A significant 24-h oscillation in the protein levels of ADAR1 was observed in mock-transduced RPTECs (p < 0.01, Fig. 2C), but the oscillation of ADAR1 protein levels was dampened in ADAR1-KD cells. The expression of P-gp in circadian clock-synchronized mock-transduced RPTECs also exhibited significant 24-h oscillation (p < 0.05, Fig. 2D), accompanying significant time-dependent differences in the intracellular accumulation of digoxin, a typical substrate of P-gp (p < 0.05, Fig. 2E). Low intracellular accumulation of digoxin was observed around the peak time of P-gp expression, demonstrating that the levels of P-gp and its efflux pump activity increased at the time after the elevation of ADAR1 levels. In contrast, the expression levels of P-gp in ADAR1-KD RPTECs decreased at all examined time points, and the amplitude of P-gp oscillation was reduced by the downregulation of ADAR1 (Fig. 2D). Consequently, the time dependency of digoxin accumulation was not observed in ADAR1-KD RPTECs (Fig. 2E) due to the increased digoxin accumulation at both examined time points. This suggests that ADAR1 is involved in circadian regulation of P-gp expression in human RPTECs.
Figure 2.
ADAR1 is involved in circadian regulation of P-gp expression in human RPTECs.A, circadian oscillation of PERIOD2 and BMAL1 mRNA (left), and ADAR1 and P-gp protein (right) in the kidney of cynomolgus monkeys. The mRNA levels were assessed by quantitative real-time RT-PCR analysis and their expression levels were normalized to that of β-ACTIN. Values are the mean with S.D. (n = 3). The protein levels of β-ACTIN are shown as loading controls. B, temporal expression profiles of PERIOD2 and BMAL1 mRNA in mock-transduced and ADAR1-KD RPTECs after treatment with 100 nM DEX for 2 h. The mRNA levels were assessed by quantitative real-time RT-PCR analysis and their expression levels were normalized to that of 18S rRNA. Values are the mean with S.D. (n = 3). The value of mock-transduced RPTECs before DEX treatment (Pre) was set at 1.0. There were significant time-dependent variations in PERIOD2 and BMAL1 mRNA expression (F13,28 = 56.917, p < 0.001 and F13,28 = 46.243, p < 0.001 for PERIOD2 in mock-transduced and ADAR1-KD RPTECs, respectively; F13,28 = 14.565, p < 0.001 and F13,28 = 5.157, p < 0.001 for BMAL1 in mock-transduced and ADAR1-KD RPTECs, respectively; ANOVA). ∗∗p < 0.01; significant difference between the two groups (t4 = 38.454, p < 0.001; unpaired t-test, two-sided). C, temporal protein expression profiles of ADAR1 in mock-transduced and ADAR1-KD RPTECs after treatment with 100 nM DEX. The protein levels were normalized to that of β-ACTIN. Values are the mean with S.D. (n = 4). The value of mock-transduced RPTECs before DEX treatment (Pre) was set at 1.0. There were significant time-dependent variations (F5,18 = 10.414, p < 0.001 for mock-transduced; F5,18 = 8.214, p < 0.001 for ADAR1-KD; ANOVA). ∗∗p < 0.01; significant difference between the two groups (t6 = 4.641, p = 0.004; unpaired t-test, two-sided). D, temporal expression profiles of P-gp in mock-transduced and ADAR1-KD RPTECs after treatment with 100 nM DEX. The protein levels were normalized to that of β-ACTIN. Values are the mean with S.D. (n = 4). The value of mock-transduced RPTECs before DEX treatment (Pre) was set at 1.0. There was a significant time-dependent variation in mock-transduced RPTECs (F5,18 = 2.899, p = 0.043 for mock-transduced; ANOVA). ∗p < 0.05; significant difference between the two groups (t6 = 2.617, p = 0.040; unpaired t-test, two-sided). In panel C and D, β-ACTIN was reused as loading control because the same protein samples were used for Western blotting for detection of ADAR1 and P-gp. E, intracellular accumulation of digoxin in mock-transduced and ADAR1-KD RPTECs 32 and 44 h after DEX treatment. Concentrations of digoxin were measured by LC-MS/MS analysis 1 h after incubation with the drug. Values are the mean with S.D. (n = 4). ∗p < 0.05; significant difference between the two groups (F3,12 = 7.487, p = 0.004; ANOVA with Tukey–Kramer’s post hoc test).
Circadian time-dependent differences in the stability of ABCB1 mRNA
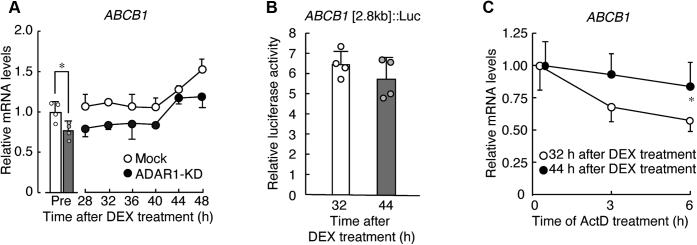
To investigate the underlying mechanism of disruption of the circadian expression of P-gp in ADAR1-KD RPTECs, the temporal expression profiles of ABCB1 mRNA were assessed in circadian clock-synchronized RPTECs. A significant 24-h oscillation in the mRNA levels of ABCB1 was observed in mock-transduced RPTECs after DEX treatment (p < 0.01, Fig. 3A). The mRNA levels of ABCB1 decreased in DEX-treated ADAR1-KD RPTECs, although still exhibited significant 24-h oscillation (p < 0.01, Fig. 3A). This suggests that downregulation of ADAR1 alters the circadian expression of ABCB1 mRNA in human RPTECs, but disruption of the circadian expression of P-gp in ADAR1-KD RPTECs is not completely dependent on the alternation of the ABCB1 mRNA rhythm.
Figure 3.
The time dependency of ABCB1 mRNA expression in human RPTECs.A, temporal expression profiles of ABCB1 mRNA in mock-transduced and ADAR1-KD RPTECs after treatment with 100 nM DEX. The mRNA levels were assessed by quantitative real-time RT-PCR analysis and their expression levels were normalized to that of 18S rRNA. Values are the mean with S.D. (n = 4). The value of mock-transduced RPTECs before DEX treatment was set at 1.0. There were significant time-dependent variations in ABCB1 mRNA levels in mock-transduced and ADAR1-KD RPTECs (F5,18 = 12.280, p < 0.001 for mock-transduced, F5,18 = 9.835, p < 0.001 for ADAR1-KD; ANOVA). ∗p < 0.05; significant difference between the two groups (t6 = 2.994, p = 0.024; unpaired t-test, two-sided). B, the luciferase activity of ABCB1 [2.8kb]::Luc in RPTECs 32 and 44 h after DEX treatment. Values are the mean with S.D. (n = 4). C, the stability of ABCB1 mRNA in RPTECs 32 or 44 h after DEX treatment. Cells were treated with 5 μM ActD 32 or 44 h after DEX treatment, and then RNA was extracted at the indicated time points after ActD treatment. The mRNA levels were assessed by semiquantitative RT-PCR analysis and their expression levels were normalized to that of 18S rRNA. Values are the mean with S.D. (n = 6). The value at 0 h (the time of the initiation of ActD) was set at 1.0. ∗p < 0.05; significant difference between the two groups at the corresponding time point (F7,40 = 16.020, p < 0.001; ANOVA with Tukey–Kramer’s post hoc test).
Next, we investigated the mechanism by which ABCB1 mRNA expression exhibits 24-h oscillation in circadian clock-synchronized RPTECs. For this purpose, we constructed a reporter vector in which the human ABCB1 gene 5′-flanking region around the transcription start site spanning from −2091 to +701 (the number is the distance in base pairs from the putative transcription start site, +1) was inserted upstream of the luciferase gene (ABCB1 [2.8kb]::Luc) because the 5′-flanking region contains clock gene response elements (10). Luciferase activity from ABCB1 [2.8kb]::Luc-transfected RPTECs was assessed at 32 and 44 h after DEX treatment, which were decreasing and increasing times of ABCB1 mRNA expression in circadian clock-synchronized cells, respectively (Fig. 3A). There was no significant time-dependent variation in luciferase activity driven by ABCB1 [2.8kb]::Luc in DEX-treated RPTECs (Fig. 3B), suggesting that the 24-h oscillation in ABCB1 mRNA expression is not dependent on its transcriptional level.
Circadian oscillation of gene expression is not only caused by the transcription process, but also induced by posttranscriptional regulation (13, 18). Thus, we also investigated the time dependence of the stability of ABCB1 mRNA in circadian clock-synchronized RPTECs. Cells were treated with 5 μM actinomycin D (ActD) at 32 or 44 h after synchronization of their circadian clock, and ABCB1 mRNA levels were then assessed until 6 h after the initiation of ActD treatment. The stability of ABCB1 mRNA at 32 h after synchronization of the circadian clock was significantly lower than that at 44 h (p < 0.05, Fig. 3C). This suggests that the 24-h oscillation of ABCB1 mRNA levels in circadian clock-synchronized RPTECs is caused by the time-dependent differences in their stability, but not due to the time-dependent variation in transcriptional activity.
ADAR1 regulates the stability of ABCB1 mRNA
In the circadian clock-synchronized RPTECs, ABCB1 mRNA was stabilized around the time when ADAR1 protein levels increased (Figs. 2C and 3C). Therefore, we investigated whether ADAR1 is involved in the regulation of ABCB1 mRNA expression. As described above, mRNA levels of ABCB1 in mock-transduced and ADAR1-KD RPTECs were assessed after treatment with ActD. The reduction of mRNA levels of ABCB1 in ActD-treated ADAR1-KD RPTECs was significantly greater than that in mock-transduced cells (Fig. 4A). ADAR1 had negligible effects on the transcriptional activity of the human ABCB1 gene because the luciferase activity driven by ABCB1 [2.8kb]::Luc was not significantly different between mock-transduced and ADAR1-KD RPTECs (Fig. 4B).
Figure 4.
ADAR1 regulates the stability of ABCB1 mRNA.A, the stability of ABCB1 mRNA in mock and ADAR1-KD RPTECs. Cells were treated with 5 μM ActD and then RNA was extracted at the indicated time points. ABCB1 mRNA levels were measured by semiquantitative RT-PCR. The mRNA levels were normalized to that of 18S rRNA. A schematic diagram of the PCR-amplified product is shown in the upper panel. Values are the mean with S.D. (n = 5–6). The basal value at 0 h (the time of the initiation of ActD) was set at 1.0. ∗∗p < 0.01, ∗p < 0.05; significant difference between the two groups at the corresponding time point (F5,30 = 6.837, p < 0.001; ANOVA with Tukey–Kramer’s post hoc test). B, the luciferase activity of ABCB1 [2.8kb]::Luc in mock-transduced and ADAR1-KD RPTECs. Schematic diagrams of luciferase reporter constructs containing the 5′-flanking region of ABCB1 gene are shown in the upper panel. The numbers in the upper panel indicate the distance in base pairs from the putative transcription start site (TSS, +1). Values are the mean with S.D. (n = 4). C, the luciferase activity of ABCB1 3′ UTR::Luc in mock-transduced and ADAR1-KD RPTECs. Schematic diagrams of luciferase reporter constructs containing ABCB1 3′ UTR are shown in the upper panel. The numbers in the upper panel indicate the distance in base pairs from the stop codon (+1). Values are the mean with S.D. (n = 4).
The stability of mRNA is often regulated via its 3′ untranslated region (3′ UTR) (25). Thus, we also constructed reporter vector in which the human ABCB1 mRNA 3′ UTR was inserted downstream of the luciferase gene (ABCB1 3′ UTR::Luc) and then transfected it into cells. The luciferase activity of reporter vector was reduced by insertion of the 3′ UTR of the ABCB1 gene, but the reporter activity of ABCB1 3′ UTR::Luc in ADAR1-KD RPTECs was not significantly different from that in mock-transduced cells (Fig. 4C). This suggests that the decreased stability of ABCB1 mRNA in ADAR1-KD RPTECs is not due to 3′ UTR-mediated regulation by ADAR1.
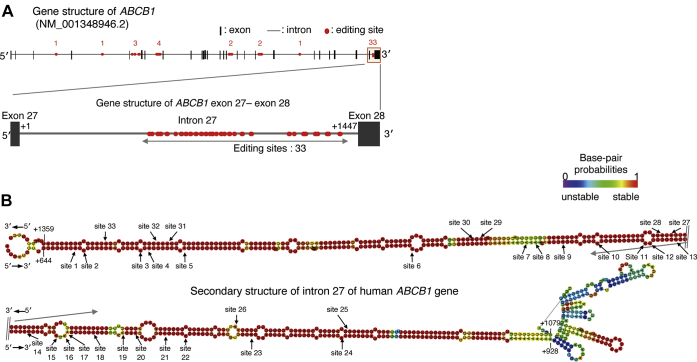
Searching for A-to-I RNA editing sites and structure analysis of the ABCB1 gene transcripts
To investigate the underlying mechanism of ADAR1-mediated regulation of ABCB1 gene expression, we conducted a comprehensive analysis of RNA editing events in the human ABCB1 gene transcript using REDIPortal, a searching database of A-to-I RNA editing sites (26). A total of 47 A-to-I RNA editing sites were registered in the human ABCB1 gene pre-mRNA in the database. Among them, 33 were identified in intron 27 (Fig. 5A). As A-to-I RNA editing by ADAR1 is often observed in the long regions of dsRNA (27, 28), we also performed in silico prediction analysis for the secondary structure of pre-mRNA of ABCB1 gene (29). The results indicated that intron 27 forms stable dsRNA of approximately 280 bp in length (Fig. 5B), whereas the other introns involving putative A-to-I editing sites failed to show stable form of dsRNA structure (Fig. S1). However, the results of direct sequence analysis revealed that no significant editing sites were detected in the intron 27 region of the ABCB1 gene pre-mRNA prepared from mock-traduced, ADAR1-KD, and ADAR1-overexpressing RPTECs (Figs. S2 and S3 and Table S1).
Figure 5.
Estimation of the secondary structure of intron 27 of the human ABCB1 gene.A, schematic image of human ABCB1 mRNA (NM_001348946.2). Red dots indicate A-to-I RNA editing sites registered in REDIPortal. The nucleotide numbering refers to the 5′ end of intron 27 as +1. B, in silico prediction of RNA secondary structure of intron 27 of the human ABCB1 gene by RNAfold. The minimum free energy structure with base-pair probabilities calculated to have the lowest value of free energy. Base-pair probabilities are shown by a color spectrum. The RNA editing sites registered in REDIportal are indicated by arrows.
ADAR1 is involved in the regulation of nonsense-mediated mRNA decay of ABCB1 gene transcripts
During the analysis of transcript variants of the human ABCB1 gene, we noted that ABCB1 transcripts are subjected to alternative splicing that generates the transcripts of retained intron 27 (registered as ENST00000491360.1). Retention of intron 27 results in a premature stop codon, suggesting the cause of nonsense-mediated mRNA decay (NMD) (Fig. 6A). To address this possibility, we constructed minigene plasmid vectors in which the DNA fragment of the human ABCB1 gene from exon 27 to exon 28 was inserted into the multicloning site of pcDNA3.1 (ex27–ex28 minigene) and then transfected them into mock-transduced and ADAR1-KD RPTECs. The amount of splicing variants from the ex27–ex28 minigene were assessed at 32 and 44 h after synchronization of their circadian clock by DEX treatment. The percentage of retained intron 27 transcripts among the sum of all transcripts and normal splicing variant (intron 27 removal transcript) in mock-transduced cells was significantly higher at 32 h than at 44 h (p < 0.05, Fig. 6B). The generation of retained-intron 27 transcripts from the ex27–ex28 minigene in ADAR1-KD cells increased at both time points.
Figure 6.
ADAR1 regulates the time-dependent differences in the alternative splicing of the human ABCB1 gene.A, schematic diagram of the retention of intron 27 of the ABCB1 gene. Intron-retaining ABCB1 mRNA may be degraded through nonsense-mediated mRNA decay (NMD). B, ADAR1 is involved in the time-dependent differences in the production of retained-intron 27 ABCB1 transcripts. Cells were transfected with the ex27–ex28 minigene and treated with DEX. Minigene-derived normal splicing transcripts and retained-intron transcripts were assessed by semiquantitative RT-PCR at the indicated time points after DEX treatment. The percentage of retained intron was calculated by dividing the signal intensity of retained intron by the summed signal intensity of retained intron and normal splicing transcripts. Values are the mean with S.D. (n = 3). ∗p < 0.05; significant difference between the two groups (F3,8 = 11.373, p = 0.003; ANOVA with Tukey–Kramer’s post hoc test). C, ADAR1 binds to intron 27 of ABCB1 pre-mRNA. RPTECs were transfected with the ABCB1 ex27–ex28 minigene, and then RIP assay was conducted at 24 h posttransfection. Upper panel shows western blotting analysis of ADAR1-RIP immunoprecipitates. Lower panel shows quantitative real-time RT-PCR analysis of ADAR1-RIP immunoprecipitates using intron 27 specific primers listed in Table 1. Values are the mean with S.D. (n = 3). ∗∗p < 0.01; significant difference between the two groups (t4 = 12.964, unpaired t-test, two-sided). D, retained-intron 27 transcripts of the ABCB1 gene are degraded by NMD. Cells were transfected with the ex27–ex28 minigene and treated with 1 μM NMDI-14 for 6 h. Minigene-derived normal splicing transcripts and retained-intron transcripts were assessed by semiquantitative RT-PCR. Values are the mean with S.D. (n = 4). ∗∗p < 0.01; significant difference between the two groups (t6 = 8.022, unpaired t-test, two-sided). E, the stability of retained-intron 27 transcript of the ABCB1 gene. Cells were transfected with the ex27–ex28 minigene. Minigene-derived normal splicing transcripts and retained-intron transcripts were assessed by semiquantitative RT-PCR. The mRNA levels were normalized to that of 18S rRNA. Values are the mean with S.D. (n = 3). The value at 0 h (the time of the initiation of ActD) was set at 1.0. ∗p < 0.05; significant difference between the two groups at the corresponding time point (F5,12 = 13.641, p < 0.001; ANOVA with Tukey–Kramer’s post hoc test). F and G, NMD inhibition restores levels of ABCB1 mRNA and P-gp in ADAR1-KD RPTECs. Mock-transduced and ADAR1-KD cells were treated with 1 μM NMDI-14 for 24 h. The mRNA levels were assessed by quantitative real-time RT-PCR, and their levels were normalized to that of 18S rRNA. The levels of P-gp were normalized that of β-ACTIN. Values are the mean with S.D. (n = 4). The value of vehicle (0.01% DMSO)-treated mock cells was set at 1.0. ∗∗p < 0.01, ∗p < 0.05; significant difference between the indicated groups (F3,12 = 15.727, p < 0.001 for ABCB1 mRNA; F3,12 = 37.583, p < 0.001 for P-gp; ANOVA with Tukey–Kramer’s post hoc test). H, intracellular accumulation of digoxin in NMDI-14-treated ADAR1-KD RPTECs. Concentrations of digoxin were measured by LC-MS/MS analysis 1 h after incubation with the drug. Values are the mean with S.D. (n = 6). ∗∗p < 0.01; significant difference between the two groups (t10 = 4.868, unpaired t-test, two-sided).
The regulation of gene function by ADAR1 depends on both its editing activity and binding capacity to dsRNA (30). Although no significant editing sites were found in the intron 27 region of the ABCB1 gene pre-mRNA (Figs. S2 and S3), the results of RNA immunoprecipitation (RIP) analysis using RNA extracted from the minigene-transfected RPTECs indicated that ADAR1 can bind to intron 27 region of pre-mRNA of the ABCB1 gene (p < 0.01, Fig. 6C). The NMD inhibitor NMDI-14 (31) significantly increased the percentage of retained-intron 27 transcripts in ex27–ex28 minigene-transfected cells (p < 0.01, Fig. 6D), suggesting that the retention of intron 27 produces NMD-sensitive transcripts. The stability of retained-intron 27 transcripts was significantly lower than that of normal splicing variants (Fig. 6E). Lower expression levels of ABCB1 mRNA and P-gp in ADAR1-KD RPTECs were increased by treatment with NMDI-14 (Fig. 6, F and G). Furthermore, the NMD inhibition in RPTECs by NMDI-14 also decreased intracellular accumulation of digoxin (Fig. 6H). These findings suggest that ADAR1 prevents the generation of retained-intron 27 transcripts from the ABCB1 gene and increases the stability of ABCB1 mRNA. The ADAR1-dependent maturation of ABCB1 mRNA may also function in the production of P-gp in RPTECs.
Discussion
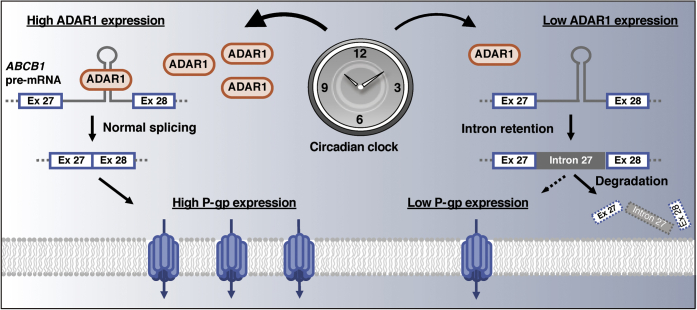
The circadian clock machinery controls many downstream events through transcription, translation, or degradation processes. In this study, we demonstrated that the 24-h oscillation of P-gp expression in circadian clock-synchronized RPTECs is generated through a posttranscriptional mechanism, by which ADAR1-mediated regulation of alternative splicing is associated with the maturation and stabilization of ABCB1 mRNA. ADAR1 time-dependently prevented the production of aberrant spliced transcripts of the ABCB1 gene, resulting in circadian expression of P-gp (Fig. 7).
Figure 7.
Schematic diagram of the ADAR1-mediated regulation of diurnal expression of P-gp in human renal cells. ADAR1 time-dependently prevents the production of intron 27-retaining ABCB1 transcripts, resulting in the diurnal changes in the maturation and stability of ABCB1 mRNA. The time-dependent differences in the ABCB1 mRNA levels lead to the diurnal expression of P-gp in human renal cells.
To evaluate the circadian profiles of gene expression, it is necessary to collect tissue samples every few hours. Therefore, experimental animals are often used for chronopharmacological and chronopharmacokinetic studies. Diurnal expression of drug metabolism enzymes and xenobiotic transporters has been identified in mice and rats (6, 32). In contrast to experimental animals, our understanding about circadian characteristics of human pharmacokinetics regulators is limited because of restrictions on frequent sampling from tissues. In mammals, nearly all cells contain circadian oscillators organized in a hierarchical fashion, with a master pacemaker located in the suprachiasmatic nucleus (SCN) of the anterior hypothalamus (33). Clock genes and clock-controlled output genes are expressed rhythmically not only in the SCN, but also in other brain regions and peripheral tissues (34). Such rhythmic expression of genes is also observed in cultured cells after brief treatment with certain compounds, such as high-concentration serum, forskolin, phorbol-12-myristate-13-acetate, calcimycin, or dexamethasone (23, 24, 35), because these compounds synchronized rhythmic phase of clock gene expression in each individual cell (36). Therefore, the peripheral oscillator in cultured human cells constitutes an in vitro model for the molecular oscillator in human tissues. The exposure of RPTECs to 100 nM DEX for 2 h resulted in the circadian expression of P-gp and ADAR1, suggesting that the expression of these proteins time-dependently varies in the human kidney. This notion was also supported by the expression of P-gp and ADAR1 exhibiting 24-h oscillation in the kidneys of cynomolgus monkeys, which are diurnal active animals similar to humans.
The function of ADAR1 is highly conserved in mammals, with emerging evidence that A-to-I RNA editing is not rare and represents a widespread phenomenon regulating a large fraction of the human genome (16). The rhythmic phase of P-gp expression in circadian clock-synchronized RPTECs and kidney of monkeys was similar to the ADAR1 protein rhythm. The most parsimonious explanation for our results is that a temporal increase in ADAR1 levels prevents the production of aberrant spliced transcripts of the ABCB1 gene, thereby promoting the maturation of its mRNA. The time-dependent difference in the ADAR1-mediated maturation of ABCB1 mRNA may cause the circadian expression of P-gp (Fig. 7). RNA editing and splicing are two major RNA processes that diversify transcriptomes in eukaryotes. These processes interact with each other. RNA editing occurs cotranscriptionally on nascent RNA preceding its splicing (37). Recently, it was reported that ADAR1 regulates cassette exons of CCDC15 (coiled-coil domain containing 15) in both RNA editing-dependent and -independent manners (38). In addition, active-site-mutated ADAR2 causes aberrant splicing of its own pre-mRNA (39). This suggests that ADARs can regulate the splicing of pre-mRNA through binding to double-stranded regions without A-to-I RNA editing. In this study, computer-based analysis revealed that the intron 27 region of ABCB1 pre-mRNA forms a double-stranded structure, although no significant A-to-I RNA editing sites were detected. The length of the double-stranded structure formed by intron 27 of ABCB1 pre-mRNA was sufficient for ADAR1 binding because ADAR1 can bind to dsRNA that is longer than approximately 50 bp (27, 28). In fact, the results of RIP analysis indicated the ability of ADAR1 to bind to intron 27 region of ABCB1 gene pre-mRNA. This suggested that ADAR1 regulates the splicing of the intron 27 region of ABCB1 pre-mRNA in an editing-independent manner.
Splicing is widely known as RNA processing to remove introns from pre-mRNA and ligate exons to form mature mRNA. To define exons and introns, there are conserved sequences at the 5′ splice site, 3′ splice site, branch point site, and polypyrimidine tract (40). Splicing is catalyzed by the spliceosome, which is an RNA–protein complex containing five small nuclear ribonucleoproteins (snRNPs) and an assembly of auxiliary proteins. For example, the U1 and U2 snRNPs recognize the 5′ splice site and branch point site, respectively, specifying the intron–exon structure. Intron retention is a common variant of alternative splicing in which selected introns are specifically retained in otherwise spliced and polyadenylated transcripts (41). The weaker splice sites that impede identification by the spliceosome are often located in the intron-retaining region of transcripts (42). In addition, the intron-retaining region also often has weaker polypyrimidine tracts. As a consequence, intron-retaining mRNA transcripts are susceptible to degradation via NMD, thus intron retention reduces gene expression through posttranscriptional processing (41). In the present study, we demonstrated that downregulation of ADAR1 increased intron 27-retaining ABCB1 mRNA transcripts, suggesting that ADAR1 protects against intron retention. Indeed, the weak polypyrimidine tract is also located in the intron 27 region of the ABCB1 gene. Considering the function of ADAR1 as a splicing regulator, its downregulation affects the action of the spliceosome on the ABCB1 intron 27 region, causing intron retention.
Although the rhythmicity in the expression of P-gp was disrupted in circadian clock-synchronized ADAR1-KD cells, the mRNA levels of ABCB1 still exhibited time-dependent oscillation. These findings suggest that disruption of the circadian expression of P-gp in ADAR1-KD RPTECs is not completely dependent on the alternation of the ABCB1 mRNA rhythm. As retention of intron 27 in ABCB1 mRNA results in a premature stop codon, ADAR1 may also be involved in the translation process from ABCB1 mRNA to P-gp. Furthermore, the expression and sequence of microRNA are also altered depending on the A-to-I RNA editing activity of ADAR1 (43, 44). Several types of microRNA were found to regulate the expression of P-gp (45, 46, 47). ADAR1-mediated regulation of microRNA function may also underlie the disruption of the rhythmicity in P-gp expression in circadian clock-synchronized ADAR1-KD cells. Further studies are required to clarify the role of ADAR1 in the regulation of the translation process from ABCB1 mRNA to P-gp.
We previously demonstrated that the expression and activity of P-gp exhibited diurnal oscillation in the small intestine of mice and cynomolgus monkeys (9, 10). The diurnal oscillation was regulated at the transcriptional level. Hepatic leukemia factor (HLF) and E4 promoter-binding protein-4 (E4BP4) inversely regulated transcription of the Abcb1 gene through the same PAR bZip response element (PARRE). Although ABCB1 [2.8kb]::Luc contains PARRE, the luciferase activity did not exhibit time-dependent variation in circadian-clock synchronized RPTECs. Indeed, the amplitude of the HLF mRNA rhythm was different between the small intestine and kidney of monkeys; HLF mRNA levels in the small intestine at the peak time were approximately 3.5-fold higher than those at the trough time, whereas the peak levels of the HLF mRNA rhythm in the kidney were approximately 1.5-fold higher than those at the trough time (10). Although HLF and E4BP4 may also be involved in the circadian regulation of P-gp expression in renal cells, their role in the generation of P-gp rhythm in the kidney is different from that in the small intestine.
The expression of ADAR1 protein exhibited circadian oscillation in DEX-treated human RPTECs and kidney of monkeys. However, the underlying mechanism remains unclear. Indeed, the nucleotide sequence having homology with cAMP-response element (CRE) is located near the transcription start site of the human and monkey ADAR1 gene. Activating transcription factor 4 (ATF4) was reported to act as an output component of the circadian clock via CRE-mediated transcription (6, 48). ATF4 expression also exhibited circadian oscillation in the mouse kidney and DEX-treated embryonic fibroblasts (49). Collectively, CRE-mediated transcription by ATF4 may mediate the action of the circadian clock to generate the diurnal expression of ADAR1 in RPTECs and kidney of monkeys.
In this study, we demonstrated that ADAR1 was a regulator of 24-h oscillation of P-gp expression in circadian clock-synchronized human renal cells. Downregulation of ADAR1 induced alternative splicing in intron 27 of the human ABCB1 gene, resulting in the production of retained intron transcripts. The aberrant spliced transcript was sensitive to NMD, leading to the decreased stability of ABCB1 mRNA. Although no significant editing sites were found in the intron 27 region of the ABCB1 gene pre-mRNA (Figs. S2 and S3), ADAR1 binds to intron 27 region of ABCB1 gene pre-mRNA. Therefore, the temporal increase in ADAR1 levels seemed to prevent the production of aberrant spliced transcripts of the ABCB1 gene through binding to intron 27 region, thereby promoting the maturation of its mRNA. The time-dependent difference in the ADAR1-mediated maturation of ABCB1 mRNA may cause the circadian expression of P-gp (Fig. 7). The pharmacokinetics of many drugs vary depending on the rhythm of absorption, distribution, metabolism, and elimination. Our present study suggests a mechanism underlying the dosing time-dependent differences in drug elimination and provides a molecular link between the circadian clock and xenobiotic detoxification.
Experimental procedures
Cell culture and treatment
Human renal proximal tubular epithelial cells (RPTECs; SA7K clone) were commercially obtained from Sigma-Aldrich. Cells were cultured under 5% CO2 at 37 °C in MEMα supplemented with 5.5% RPTEC Complete Supplement (Sigma-Aldrich), 2.33 mM l-glutamine (Sigma-Aldrich), 28 μM gentamicin (Sigma-Aldrich), and 14 nM amphotericin B (Sigma-Aldrich). For synchronization of the circadian clock in RPTECs, cells were treated with 100 nM DEX (FUJIFILM Wako Pure Chemical Corporation) for 2 h. The medium was replaced with fresh medium, and cells were collected to extract RNA and protein at the indicated time points. For the mRNA stability assay, RPTECs were treated with 5 μM actinomycin D (ActD; FUJIFILM Wako Pure Chemical Corporation), a transcription inhibitor, and cells were collected for RNA extraction at the indicated time points. For splicing analysis, RPTECs were treated with 1 μM NMDI-14 (Merck Millipore). Six or twenty-four hours later, cells were collected to extract RNA or protein, respectively.
Animals and treatment
Cynomolgus monkeys (4–7 years old, male) were purchased from Shin Nippon Biomedical Laboratories, Ltd. They were kept separately one per cage and maintained on 90 to 100 g of solid food (Oriental Yeast Co, Ltd) and approximately 30 g of fresh fruit once a day (fed from ZT1.5 to ZT3), with free access to water, under a stable temperature (24 °C ± 2 deg. C), stable humidity (50 ± 20%), and a 12-h light/dark cycle. Under the light/dark cycle, ZT0 was designated as “lights on” and ZT12 as “lights off.” Animals were painlessly sacrificed under anesthesia at ZT2, ZT8, ZT14, or ZT20. The kidneys were quickly removed and frozen in liquid N2 and stored at −80 °C. This study was approved by the Committee on the Care and Use of Laboratory Animals of TransGenic Inc and Daiichi Sankyo Co, Ltd.
Construction of ADAR1-KD RPTECs
RPTECs were infected with lentiviral particles expressing small hairpin RNA (shRNA) against the human ADAR1 gene (sc-37657-V; Santa Cruz Biotechnology), which contained 3 target-specific constructs encoding 19- to 25-nucleotide shRNA designed to KD ADAR1. To select clones stably expressing shRNA, cells were maintained in a medium containing 5 μg/ml of puromycin (FUJIFILM Wako Pure Chemical Corporation). KD of ADAR1 was confirmed by western blotting.
Western blottings
Total protein of RPTECs and kidney of monkey was prepared using CelLytic MT (Sigma-Aldrich) according to the manufacturer’s instructions. Denatured samples containing 10 μg of protein were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto polyvinylidene difluoride membranes. The membranes were incubated with primary antibodies against ADAR1 (1:1000, sc-73408; Santa Cruz Biotechnology), P-gp (1:1000, C494, Thermo Fisher Scientific), MRP2 (1:1500, sc-5770; Santa Cruz Biotechnology), TBP (1:1000, ab51841; abcam), and β-ACTIN (1:10,000, sc-1616; Santa Cruz Biotechnology). Specific antigen–antibody complexes were visualized using horseradish-peroxidase-conjugated anti-mouse antibodies (1:10,000, sc-2005; Santa Cruz Biotechnology) against ADAR1, P-gp and TBP, or anti-goat antibodies (1:10,000, sc-2020; Santa Cruz Biotechnology) against MRP2 and ImmunoStar LD (FUJIFILM Wako Pure Chemical Corporation). Visualized images were scanned using an ImageQuant LAS3000 (FUJIFILM).
Quantitative and semiquantitative RT-PCR analysis
Total RNA was extracted from RPTECs and kidney of monkeys using RNAiso Plus (Takara Bio Inc) according to the manufacturer’s instructions. RNA was treated with DNase I to prevent contamination of genomic DNA and then reversed transcribed using the ReverTra Ace qPCR RT Kit (TOYOBO Co, Ltd). For quantitative real-time RT-PCR, the cDNA equivalent of 10 ng of RNA was amplified by PCR using the LightCycler 96 system (Roche Diagnostics) with THUNDERBIRD SYBR qPCR Mix (TOYOBO Co, Ltd). Sequences of primers are listed in Table 1. For semiquantitative RT-PCR, the cDNA equivalent of 10 ng of RNA was amplified by the GoTaq Green Master Mix (Promega) with the gene-specific primers listed in Table 1. We confirmed no significant amplification of PCR products without reverse transcription. The PCR-amplified products were separated by electrophoresis using 1% agarose gel containing ethidium bromide. Signals from the agarose gel were detected using LAS3000 (FUJIFILM). PCR-amplified products were identified by gel purification and Sanger sequencing.
Table 1.
Primer sets for quantitative and semiquantitative RT-PCR analysis of gene expression
| Gene | Primers |
|---|---|
| Human SLC15A1 | |
| Forward | 5′-TCACCTGTGGCGAAGTGGTC-3′ |
| Reverse | 5′-AGCACCGACTTCATGTTGGAAG-3′ |
| Human SLC15A2 | |
| Forward | 5′-TCGTCTGGTCTCCAAGTGTGG-3′ |
| Reverse | 5′-TGGGCTGGGGCCATTTCATTT-3′ |
| Human SLC22A4 | |
| Forward | 5′-TGCCCAGGCGTTATATCATAGC-3′ |
| Reverse | 5′-AGCATGACCAGACCAATGGATAAG-3′ |
| Human SLC22A5 | |
| Forward | 5′-ATGGGTGTGGGAGTCAGCTC-3′ |
| Reverse | 5′-GAATGTAGGGCAGGAAGCGG-3′ |
| Human SLC47A1 | |
| Forward | 5′-ATGTGTCCAGGCTTACCCAGAC-3′ |
| Reverse | 5′-GACAAGGTTGGCTGCAACTCC-3′ |
| Human SLC47A2 | |
| Forward | 5′-CTCCGCCTATGCCAACATCATC-3′ |
| Reverse | 5′-GGGACAGCCAGGGAGAAGAAG-3′ |
| Human SLCO4C1 | |
| Forward | 5′-ACTCATTGTGCGAAACTGCCC-3′ |
| Reverse | 5′-AGGCTAGGGACCGTTGTCTG-3′ |
| Human SLC22A2 | |
| Forward | 5′-CATCGTCACCGAGTTTAACCTG-3′ |
| Reverse | 5′-AGCCGATACTCATAGAGCCAAT-3′ |
| Human SLC22A6 | |
| Forward | 5′-AGGAGCCAAATTGAGTATGGAGG-3′ |
| Reverse | 5′-GTATGCAAAGCTAGTGGCAAACC-3′ |
| Human SLC22A8 | |
| Forward | 5′-AGCACCGTCATCTTGAATGTGG-3′ |
| Reverse | 5′-AGGTGTAGCAGTACCCGAGTG-3′ |
| Human ABCB1 | |
| Forward | 5′-CCATGCTCAGACAGGATGTGA-3′ |
| Reverse | 5′-ATCATTGGCGAGCCTGGTAG-3′ |
| Human ABCC2 | |
| Forward | 5′-TGCAACTCTACTTTTTGGAA-3′ |
| Reverse | 5′-TCTCTTGGTCCTGGATTTAT-3′ |
| Human ABCG2 | |
| Forward | 5′-ATGTAATTCAGGTTACGTGG-3′ |
| Reverse | 5′-CTAACTCTTGAATGACCCTG-3′ |
| Human PERIOD2 | |
| Forward | 5′-GACATGAGACCAACGAAAACTGC-3′ |
| Reverse | 5′-AGGCTAAAGGTATCTGGACTCTG-3′ |
| Human BMAL1 | |
| Forward | 5′-AAGGGAAGCTCACAGTCAGAT-3′ |
| Reverse | 5′-GGACATTGCGTTGCATGTTGG-3′ |
| Human ABCB1 [1.2 kb] | |
| Forward | 5′-TTCGCAACCCCAAGATCCTC-3′ |
| Reverse | 5′-ATGTGCCACCAAGTAGGCTC-3′ |
| Human ABCB1 [RIP assay] | |
| Forward | 5′-ATGGAGTCTCGCTCTGTCG-3′ |
| Reverse | 5′-GTGCCTGTAGTCCCAGCTAG-3′ |
| Human 18S rRNA | |
| Forward | 5′-AGAAGCCCCTGGCACTCTAT-3′ |
| Reverse | 5′-GCAAAGTGGGCACAGTGATG-3′ |
| Monkey PERIOD2 | |
| Forward | 5′-GGGCCCTTTGAATGAGGACG-3′ |
| Reverse | 5′-CTGGAGGAGGTCTGGCTCAT-3′ |
| Monkey BMAL1 | |
| Forward | 5′-CGGCGCCGGGATAAAATGAA-3′ |
| Reverse | 5′-GCTGCCCTGAGAATGAGGTG-3′ |
| Monkey β-ACTIN | |
| Forward | 5′-CCTTCAACACCCCAGCCATG-3′ |
| Reverse | 5′-TAGGGCGACATAGCACAGCT-3′ |
Measurement of intracellular concentrations of digoxin
Mock-transduced and ADAR1-KD RPTECs were treated with 100 nM DEX (FUJIFILM Wako Pure Chemical Corporation) for 2 h to synchronize their circadian clock. The medium was replaced with fresh medium and incubated for 32 or 44 h at 37 °C. Then, the medium was replaced with Krebs–Ringer buffer (117 mM NaCl, 4.7 mM KCl, 1.2 mM MgCl2, 1.2 mM NaH2PO4·2H2O, 10 mM HEPES, 2.5 mM CaCl2, and 11 mM d-glucose) containing 1 μM digoxin (Chugai Pharmaceutical Co, Ltd) for 1 h at 37 °C. To measure the intracellular content of digoxin, cells were washed with saline and homogenized with 150 μl of methanol containing an internal standard (3.3 μM digitonin; Sigma-Aldrich). The homogenates were centrifuged at 12,000g for 5 min at 4 °C, and the supernatants were filtered using 0.22-μm hydrophilic polyvinylidene difluoride membrane (Merck Millipore). Samples were then evaporated and dissolved into 50-μl aliquots of the mobile phase. The liquid chromatograph tandem mass spectrometer system was ACQUITY (Waters). Liquid chromatography was performed using a CORTECS C18+ column (2.7 μm, 2.1 mm × 100 mm; Waters). The mobile phases consisted of 10 mM ammonium formate–methanol (1:4, v/v). The flow rate was set at 0.2 ml/min. The total run time was 6 min. The mass spectrometer was operated in the multiple reaction monitoring (MRM) mode using positive ESI. The MRM was set at 798.8-651.5 and 1247-781.6 mass-to-charge ratio (m/z) for digoxin and digitonin, respectively. The amount of digoxin was normalized to protein concentrations measured with the Pierce BCA Assay Kit (Thermo Fisher Scientific).
Plasmid constructions
The upstream region (from bp −2091 to +701, where +1 indicates the transcription start site) and 3′ UTR (from bp +1 to +391, where +1 indicates the stop codon) of the human ABCB1 gene were amplified using PrimeSTAR MAX DNA polymerase (Takara Bio Inc). These sequences were subcloned into the pGL4.12 luciferase-reporter vector (Promega) using KpnI and NheI restriction sites and into the pGL4.13 luciferase-reporter vector (Promega) using XbaI and FseI restriction sites. The minigene sequence was amplified using PrimeSTAR MAX DNA polymerase (Takara Bio Inc). The sequence was subcloned into the pcDNA3.1 (Invitrogen; Life Technologies) vector using NheI and KpnI restriction sites. Sequences of primers are listed in Table 2.
Table 2.
Primer sets for plasmid construction and minigene assay
| Gene | Primers |
|---|---|
| Human ABCB1 promoter | |
| Forward for −2091 bp | 5′-GCTGGTACCTCAACTTGCAAGGGGACCAG-3′ |
| Reverse for +703 bp | 5′-ATAGCTAGCCGACCTGAAGAGAAACCGCA-3′ |
| Human ABCB1 3′ UTR | |
| Forward for +1 bp | 5′-GCGCTCTAGAAACTCTGACTGTATGAGATG-3′ |
| Reverse for +391 bp | 5′-TAAGGCCGGCCAGTCACATGAAAGTTTAG-3′ |
| Human ABCB1 minigene | |
| Forward for exon 27 | 5′-CGTGCTAGCGCAAGCTGTTAGAACTTTACTTTCA-3′ |
| Reverse for exon 28 | 5′-CGCGGTACCACAGGCAGTTTGGACAAGATGA-3′ |
The numbers indicate the distance from the transcription site (+1) for the promoter vector or from the stop codon (+1) for the 3′ UTR vector.
Luciferase reporter assay
RPTECs were seeded at a density of 1.5 × 105/well in 24-well culture plates. Lipofectamine LTX regent (Invitrogen; Life Technologies) was used for reverse transfection of cells with 500 ng of the pGL4.12 reporter construct or with 200 ng of the pGL4.13 reporter constructs. A total of 10 ng of the pRL-TK vector (Promega) was also transfected as an internal control reporter. Cells were harvested 24 h after transfection and lysates were analyzed using the Dual-Luciferase reporter assay system (Promega). The ratio of firefly (expressed from pGL4.12 reporter constructs) to Renilla (expressed from pRL-TK) luciferase activity in each sample served as a measure of normalized luciferase activity.
In silico prediction of RNA secondary structure
The secondary structure of the human ABCB1 RNA was predicted using RNAfold (29), which computes the minimum free energy and predicts an optional secondary structure (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The range from +634 to +1370 of ABCB1 intron 27 was input as data with all default options.
RNA immunoprecipitation (RIP)
A 10-cm dish of RPTECs was transfected with 50 μg of the minigene plasmid. At 24 h post transfection, cells were collected and lysed in 1 ml of nuclear isolation buffer (1.28 M sucrose, 40 mM Tris-HCl pH 7.5, 20 mM MgCl2, and 4% Triton X-100). After addition of 1 ml of PBS and 3 ml to lysates, they were centrifuged at 2500g for 15 min at 4 °C and resuspended in RIP buffer (150 mM KCl, 25 mM Tris-HCl pH 7.4, 5 mM EDTA, 0.5 mM dithiothreitol, 0.5% Nonidet P-40, 2 μg/ml leupeptin, 2 μg/ml aprotinin, and 100 U/ml recombinant RNase inhibitor) (TOYOBO Co, Ltd). The resuspended nuclei were homogenized and centrifuged at 13,000 rpm for 10 min, and the supernatant was then incubated with anti-ADAR1 antibody (sc-73408; Santa Cruz Biotechnology) or mouse IgG (Fujifilm Wako Pure Chemical) for 2 h at 4 °C with gentle rotation, followed by incubation with protein G beads (Thermo Fisher Scientific) for 1 h at 4 °C. Then, the beads were washed with RIP buffer for three times. A total of 10% of beads was used for protein elution while the rest was subjected to RNA extraction using RNAiso Plus (Takara Bio Inc) and quantitative real-time RT-PCR analysis was performed as mentioned above. %Input = 2−ΔCt × 100: ΔCt = CtRIP − [Ctinput − Log2(input dilution factor)]. Sequences of primers for amplification of intron 27 region are listed in Table 1.
Splicing analysis by semiquantitative PCR
RPTECs were seeded at a density of 1.5 × 105/well in 24-well culture plates and incubated for 24 h to semiconfluency. Cells were reverse transfected with 1 μg of the minigene plasmid using Lipofectamine LTX reagent (Invitrogen; Life Technologies). RNA was extracted using the ReliaPrep RNA Cell Miniprep System (Promega) and treated with DNase I on a column. A total of 500 ng of RNA was used for cDNA synthesis with the ReverTra Ace qPCR RT Kit (TOYOBO Co, Ltd). Exogenous cDNA from the minigene was amplified by the GoTaq Green Master Mix (Promega) with the minigene-specific primers used in vector construction. PCR cycle conditions were as follows: initial denaturation at 95 °C for 2 min followed by 28 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 2 min. PCR-amplified products were separated by electrophoresis using 1% agarose gel containing ethidium bromide. Signals from the agarose gel were detected using LAS3000 (FUJIFILM). PCR-amplified products were identified by gel purification and Sanger sequencing. Sequences of primers are listed in Table 2.
Statistical analysis
All statistical analyses were carried out using JMP pro 14 (SAS institute Japan). The significance of the 24-h variations was tested by ANOVA. The significance of differences among groups was analyzed by ANOVA, followed by Tukey–Kramer’s post hoc test. The unpaired t-test was used for the comparison of data between two groups. Equal variances were not formally tested. A 5% level of probability was considered to be significant.
Data availability
All data supporting the results of the present study are included in the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are grateful for the technical support provided by the Research Support Center, Graduate School of Medical Sciences, Kyushu University. We are indebted to M. Iwasaki (Daiichi Sankyo RD Novare Co, Ltd, Tokyo, Japan) for treatment of monkeys.
Author contributions
Y. O., S. K., and S. O. designed the study and wrote the paper. Y. O. and T. Y. performed and analyzed the experiments shown in figures and contributed to the preparation of the figures. A. T. and N. M. provided technical assistance. All the authors reviewed the results and approved the final version of the article.
Funding and additional information
This study was supported in part by a Grant-in-Aid for Scientific Research A (18H04019 to S. K.), the Naito Foundation (S. K.), and the Platform Project for Supporting Drug Discovery, and Life Science Research [Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)] from AMED (Grant Number JP20am0101091).
Edited by Ronald Wek
Contributor Information
Satoru Koyanagi, Email: koyanagi@phar.kyushu-u.ac.jp.
Shigehiro Ohdo, Email: ohdo@phar.kyushu-u.ac.jp.
Supporting information
References
- 1.Ohdo S., Koyanagi S., Matsunaga N., Hamdan A. Molecular basis of chronopharmaceutics. J. Pharm. Sci. 2011;100:3560–3576. doi: 10.1002/jps.22656. [DOI] [PubMed] [Google Scholar]
- 2.Dallmann R., Okyar A., Lévi F. Dosing-time makes the poison: Circadian regulation and pharmacotherapy. Trends Mol. Med. 2016;22:430–445. doi: 10.1016/j.molmed.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 3.Ohdo S., Koyanagi S., Matsunaga N. Chronopharmacological strategies focused on chrono-drug discovery. Pharmacol. Ther. 2019;202:72–90. doi: 10.1016/j.pharmthera.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 4.Sissung T.M., Goey A.K.L., Ley A.M., Strope J.D., Figg W.D. Pharmacogenetics of membrane transporters: A review of current approaches. Methods Mol. Biol. 2014;1175:91–120. doi: 10.1007/978-1-4939-0956-8_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shitara Y., Horie T., Sugiyama Y. Transporters as a determinant of drug clearance and tissue distribution. Eur. J. Pharm. Sci. 2006;27:425–446. doi: 10.1016/j.ejps.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Hamdan A.M., Koyanagi S., Wada E., Kusunose N., Murakami Y., Matsunaga N., Ohdo S. Intestinal expression of mouse Abcg2/breast cancer resistance protein (BCRP) gene is under control of circadian clock-activating transcription factor-4 pathway. J. Biol. Chem. 2012;287:17224–17231. doi: 10.1074/jbc.M111.333377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu F., Zhang T., Zhou C., Xu H., Guo L., Chen M., Wu B. The circadian clock gene Bmal1 controls intestinal exporter MRP2 and drug disposition. Theranostics. 2019;9:2754–2767. doi: 10.7150/thno.33395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato M., Tsurudome Y., Kanemitsu T., Yasukochi S., Kanado Y., Ogino T., Matsunaga N., Koyanagi S., Ohdo S. Diurnal expression of MRP4 in bone marrow cells underlies the dosing-time dependent changes in the oxaliplatin-induced myelotoxicity. Sci. Rep. 2020;10:13484. doi: 10.1038/s41598-020-70321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami Y., Higashi Y., Matsunaga N., Koyanagi S., Ohdo S. Circadian clock-controlled intestinal expression of the multidrug-resistance gene mdr1a in mice. Gastroenterology. 2008;135:1636–1644. doi: 10.1053/j.gastro.2008.07.073. [DOI] [PubMed] [Google Scholar]
- 10.Iwasaki M., Koyanagi S., Suzuki N., Katamune C., Matsunaga N., Watanabe N., Takahashi M., Izumi T., Ohdo S. Circadian modulation in the intestinal absorption of P-glycoprotein substrates in monkeys. Mol. Pharmacol. 2015;88:29–37. doi: 10.1124/mol.114.096735. [DOI] [PubMed] [Google Scholar]
- 11.Choi J., Park H. Circadian changes in pharmacokinetics of cyclosporin in healthy volunteers. Res. Commun. Mol. Pathol. Pharmacol. 1999;130:109–112. [PubMed] [Google Scholar]
- 12.Ferrazzini G., Sohl H., Robieux I., Johnson D., Giesbrecht E., Koren G. Diurnal variation of methotrexate disposition in children with acute leukaemia. Eur. J. Clin. Pharmacol. 1991;41:425–427. doi: 10.1007/BF00626363. [DOI] [PubMed] [Google Scholar]
- 13.Fustin J.M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., Isagawa T., Morioka M.S., Kakeya H., Manabe I., Okamura H. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 14.Tsurudome Y., Koyanagi S., Kanemitsu T., Katamune C., Oda M., Kanado Y., Kato M., Morita A., Tahara Y., Matsunaga N., Shibata S., Ohdo S. Circadian clock component PERIOD2 regulates diurnal expression of Na+/H+ exchanger regulatory factor-1 and its scaffolding function. Sci. Rep. 2018;8:9072. doi: 10.1038/s41598-018-27280-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okazaki H., Matsunaga N., Fujioka T., Okazaki F., Akagawa Y., Tsurudome Y., Ono M., Kuwano M., Koyanagi S., Ohdo S. Circadian regulation of mTOR by the ubiquitin pathway in renal cell carcinoma. Cancer Res. 2014;74:543–551. doi: 10.1158/0008-5472.CAN-12-3241. [DOI] [PubMed] [Google Scholar]
- 16.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patterson J.B., Samuel C.E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: Evidence for two forms of the deaminase. Mol. Cell. Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terajima H., Yoshitane H., Ozaki H., Suzuki Y., Shimba S., Kuroda S., Iwasaki W., Fukada Y. ADARB1 catalyzes circadian A-to-I editing and regulates RNA rhythm. Nat. Genet. 2017;49:146–151. doi: 10.1038/ng.3731. [DOI] [PubMed] [Google Scholar]
- 19.Nakano M., Nakajima M. Significance of A-to-I RNA editing of transcripts modulating pharmacokinetics and pharmacodynamics. Pharmacol. Ther. 2018;181:13–21. doi: 10.1016/j.pharmthera.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 20.Nakano M., Fukami T., Gotoh S., Takamiya M., Aoki Y., Nakajima M. RNA editing modulates human hepatic aryl hydrocarbon receptor expression by creating MicroRNA recognition sequence. J. Biol. Chem. 2016;291:894–903. doi: 10.1074/jbc.M115.699363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giacomini K.M., Huang S.M., Tweedie D.J., Benet L.Z., Brouwer K.L.R., Chu X., Dahlin A., Evers R., Fischer V., Hillgren K.M., Hoffmaster K.A., Ishikawa T., Keppler D., Kim R.B., Lee C.A. Membrane transporters in drug development. Nat. Rev. Drug Discov. 2010;9:215–236. doi: 10.1038/nrd3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones B.R., Li W., Cao J., Hoffman T.A., Gerk P.M., Vore M. The role of protein synthesis and degradation in the post-transcriptional regulation of rat multidrug resistance-associated protein 2 (Mrp2, Abcc2) Mol. Pharmacol. 2005;68:701–710. doi: 10.1124/mol.105.013144. [DOI] [PubMed] [Google Scholar]
- 23.Balsalobre A., Damiola F., Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 24.Balsalobre A., Brown S.A., Marcacci L., Tronche F., Kellendonk C., Reichardt H.M., Schütz G., Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 25.Fabian M.R., Sonenberg N., Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 26.Picardi E., D’Erchia A.M., Lo Giudice C., Pesole G. REDIportal: A comprehensive database of A-to-I rna editing events in humans. Nucleic Acids Res. 2017;45:D750–D757. doi: 10.1093/nar/gkw767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eggington J.M., Greene T., Bass B.L. Predicting sites of ADAR editing in double-stranded RNA. Nat. Commun. 2011;2:319. doi: 10.1038/ncomms1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishikura K., Yoo C., Kim U., Murray J.M., Estes P.A., Cash F.E., Liebhaber S.A. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991;10:3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz R., Bernhart S.H., Höner zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L. ViennaRNA package 2.0. Algorithms Mol. Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakurai M., Shiromoto Y., Ota H., Song C., Kossenkov A.V., Wickramasinghe J., Showe L.C., Skordalakes E., Tang H.-Y., Speicher D.W., Nishikura K. ADAR1 controls apoptosis of stressed cells by inhibiting Staufen1-mediated mRNA decay. Nat. Struct. Mol. Biol. 2017;24:534–543. doi: 10.1038/nsmb.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin L., Grigoryan A., Wang D., Wang J., Breda L., Rivella S., Cardozo T., Gardner L.B. Identification and characterization of small molecules that inhibit nonsense-mediated rna decay and suppress nonsense p53 mutations. Cancer Res. 2014;74:3104–3113. doi: 10.1158/0008-5472.CAN-13-2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsunaga N., Ikeda M., Takiguchi T., Koyanagi S., Ohdo S. The molecular mechanism regulating 24-hour rhythm of CYP2E1 expression in the mouse liver. Hepatology. 2008;48:240–251. doi: 10.1002/hep.22304. [DOI] [PubMed] [Google Scholar]
- 33.Hastings M.H., Reddy A.B., Maywood E.S. A clockwork web: Circadian timing in brain and periphery, in health and disease. Nat. Rev. Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 34.Akhtar R.A., Reddy A.B., Maywood E.S., Clayton J.D., King V.M., Smith A.G., Gant T.W., Hastings M.H., Kyriacou C.P. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 35.Balsalobre A., Marcacci L., Schibler U. Multiple signaling pathways elicit circadian gene expression in cultured rat-1 fibroblasts. Curr. Biol. 2000;10:1291–1294. doi: 10.1016/s0960-9822(00)00758-2. [DOI] [PubMed] [Google Scholar]
- 36.Nagoshi E., Saini C., Bauer C., Laroche T., Naef F., Schibler U. Circadian gene expression in individual fibroblasts: Cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 37.Hsiao Y.H.E., Bahn J.H., Yang Y., Lin X., Tran S., Yang E.W., Quinones-Valdez G., Xiao X. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res. 2018;28:812–823. doi: 10.1101/gr.231209.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang S.J., Shen H., An O., Hong H., Li J., Song Y., Han J., Tay D.J.T., Ng V.H.E., Bellido Molias F., Leong K.W., Pitcheshwar P., Yang H., Chen L. Cis- and trans-regulations of pre-mRNA splicing by RNA editing enzymes influence cancer development. Nat. Commun. 2020;11:799. doi: 10.1038/s41467-020-14621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Agranat L., Sperling J., Sperling R. A novel tissue-specific alternatively spliced form of the A-to-I RNA editing enzyme ADAR2. RNA Biol. 2010;7:253–262. doi: 10.4161/rna.7.2.11568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Will C.L., Luhrmann R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong J.J.L., Au A.Y.M., Ritchie W., Rasko J.E.J. Intron retention in mRNA: No longer nonsense. Bioessays. 2016;38:41–49. doi: 10.1002/bies.201500117. [DOI] [PubMed] [Google Scholar]
- 42.Sakabe N., de Souza S. Sequence features responsible for intron retention in human. BMC Genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawahara Y., Zinshteyn B., Chendrimada T.P., Shiekhattar R., Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the dicer–TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawahara Y., Zinshteyn B., Sethupathy P., Iizasa H., Hatzigeorgiou A.G., Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng D.D., Zhang H., Zhang P., Zheng Y.S., Zhang X.J., Han B.W., Luo X.Q., Xu L., Zhou H., Qu L.H., Chen Y.Q. Down-regulated miR-331-5p and miR-27a are associated with chemotherapy resistance and relapse in leukaemia. J. Cell. Mol. Med. 2011;15:2164–2175. doi: 10.1111/j.1582-4934.2010.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bao L., Hazari S., Mehra S., Kaushal D., Moroz K., Dash S. Increased expression of P-glycoprotein and doxorubicin chemoresistance of metastatic breast cancer is regulated by miR-298. Am. J. Pathol. 2012;180:2490–2503. doi: 10.1016/j.ajpath.2012.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shang Y., Zhang Z., Liu Z., Feng B., Ren G., Li K., Zhou L., Sun Y., Li M., Zhou J., An Y., Wu K., Nie Y., Fan D. miR-508-5p regulates multidrug resistance of gastric cancer by targeting ABCB1 and ZNRD1. Oncogene. 2014;33:3267–3276. doi: 10.1038/onc.2013.297. [DOI] [PubMed] [Google Scholar]
- 48.Ushijima K., Koyanagi S., Sato Y., Ogata T., Matsunaga N., Fujimura A., Ohdo S. Role of activating transcription factor-4 in 24-hour rhythm of serotonin transporter expression in the mouse midbrain. Mol. Pharmacol. 2012;82:264–270. doi: 10.1124/mol.112.079079. [DOI] [PubMed] [Google Scholar]
- 49.Koyanagi S., Hamdan A.M., Horiguchi M., Kusunose N., Okamoto A., Matsunaga N., Ohdo S. cAMP-response element (CRE)-mediated transcription by activating transcription factor-4 (ATF4) is essential for circadian expression of the Period2 gene. J. Biol. Chem. 2011;286:32416–32423. doi: 10.1074/jbc.M111.258970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the results of the present study are included in the article.