Abstract
Purpose of review:
Artificial intelligence (AI) has already provided multiple clinically relevant applications in ophthalmology. Yet, the explosion of non-standardized reporting of high-performing algorithms are rendered useless without robust and streamlined implementation guidelines. The development of protocols and checklists will accelerate the translation of research publications to impact on patient care.
Recent findings:
Beyond technological scepticism, we lack uniformity in analyzing algorithmic performance generalizability, and benchmarking impacts across clinical settings. No regulatory guardrails have been set to minimize bias or optimize interpretability; no consensus clinical acceptability thresholds or systematized post-deployment monitoring has been set. Moreover, stakeholders with misaligned incentives deepen the landscape complexity especially when it comes to the requisite data integration and harmonization to advance the field. Therefore, despite increasing algorithmic accuracy and commoditization, the infamous ‘implementation gap’ persists.
Open clinical data repositories have been shown to rapidly accelerate research, minimize redundancies, and disseminate the expertise and knowledge required to overcome existing barriers. Drawing upon the longstanding success of existing governance frameworks and robust data use and sharing agreements, the ophthalmic community has tremendous opportunity in ushering AI into medicine. By collaboratively building a powerful resource of open, anonymized multimodal ophthalmic data, the next generation of clinicians can advance data-driven eye care in unprecedented ways.
Keywords: Artificial intelligence, open-source, translational big data
Summary:
This piece demonstrates that with readily accessible data, immense progress can be achieved clinically and methodologically to realize AI’s impact on clinical care. Exponentially progressive network effects can be seen by consolidating, curating, and distributing data amongst both clinicians and data scientists.
Introduction
“Information rules, information is costly to produce, but cheap to reproduce. So if you create information that can help solve a problem and contribute that information to a platform where it can be shared (or help create the platform), you will enable many others to use that valuable information at low or no cost”
- Eric Schmidt - former CEO, Google [1]
Profound mortality, morbidity, and economic downturn all characterize COVID-19’s global devastation. The pandemic’s reverberating effects drive disruptive social transformation and a race to find a cure to restore normality. In healthcare, for instance, beyond the urgent treatment necessity and supply shortfall, many unprecedented barriers are impeding routine clinical care provision. Consequently, the swiftly instinctive shift to increased telemedicine and remote monitoring services has accelerated the scaling up of the long-awaited digital technology adoption and movement towards patient-focused, decentralized care paradigms. Though not an absolute crisis solution, this transformation will be one of lasting consequences. Another subtle but significant transition is the provider community mindset; our notoriously analogue, disconnected and legacy health systems are welcoming disruptive innovations with new eyes – yesterday’s threats are today’s praiseworthy data-driven, technology-enabled breakthroughs.
Other converging forces ascertain that digital health will not only remain an entrenched norm, but continually develop beyond what we hope is a soon-to-be post-COVID-19 era. Recent years have seen a rapidly modernizing Food and Drug Administration (FDA) [2–4], maturation of the real-world-evidence landscape [5,6], the advent of superhuman artificial intelligence (AI) and the US HITECH Act’s impact on digital documentation for more than a decade[7]. March 2020 signified another momentous step forward; HHS (dept. health & human services) approved the Cures Act Final Rule, which enhances patient data access rights and legislatively prohibits information blocking by electronic medical record (EMR) companies. The interoperability vision now has a Senate-approved timeboxed roadmap towards seamless Application Programming Interface or API-driven data exchange and use of electronic health information [8–11].
Amidst these changes, AI has surreptitiously entered the medical arena and unsurprisingly, as a technological “pacesetter over time” [12], ophthalmology is leading the way. With almost 5000 Google Scholar results in searching “ophthalmology artificial intelligence” since 2019 alone, the specialty has evolved from ‘high potential’ to tangible, translational potency [13]. The paramount example is diabetic retinopathy (DR)-screening platform IDx-DR ®, which pioneered the first-ever algorithmic FDA-approved software in 2018 [14].
This piece summarizes a few current AI research feats in common ophthalmic conditions and outlines the accompanying persisting hurdles – the “AI-chasm” – why most of the abundant algorithms do not translate into meaningful applications[15,16]. Against this backdrop, we contend that a collaboratively curated, multisource and multimodal data-sharing platform will tremendously accelerate the realization of AI’s potential.
Text of review
Current trends in AI ophthalmic applications
As a specialty, ophthalmologists amass highly quantitative, structured clinical data, accompanied by a recent multi-modal imaging revolution, which are captured in high fidelity on routine clinical encounters. Moreover, the chronic nature of common ophthalmic disorders requires screening, monitoring and frequent therapeutic encounters, constructing a rich digitally-captured longitudinal patient-disease journey. Indeed, we produce copious troves of data ingredients for AI. Beyond academic analytics, AI holds promise to materially impact an unmet need: blindness. Globally, sight-loss costs $3B annually, its incidence is projected to triple by 2050, 80% of which is preventable [17]. The shortfall in specialist supply scrambling to address this unsustainably rising demand is felt unfairly disproportionately in the developing world [18]. However, even in the USA, one quarter of the 30 million Americans with diabetes mellitus are still unaware they have the disease [19] and adherence to recommended screening guidelines, a critical component in preventing irreversible changes, is less than 50% [20]. Severe vision loss is needlessly experienced by too many patients; regarding DR alone, appropriate treatment can reduce the risk of blindness or moderate vision loss by more than 90% [21].
Beyond screening, AI holds promise to augment accurate and efficient decision making, enhance triage and workflow automation and enrich care delivery through biomarker-driven prognostication and therapeutic personalization. Though shared-care models do shoulder some of the supply-demand mismatch burden, AI is increasingly powering many emerging solutions, such as decentralized screening programs, home-monitoring devices and a new wave of imaging hardware showcasing not only increasing resolution of information capture and modality sophistication, but also a proliferation of smartphone applications.
The compounding limitation of availability of retina specialists and trained human graders is also a major problem in many countries. Consequently, given the current population growth trends, it is inevitable that human surveilled automated applications are only going to expand [22].
Table 1 summarizes recent AI publications with translational potential.
Table 1:
Sample of significant publications exhibiting recent breakthrough trends in AI with translational impact, segmented by disease* (ophthalmic / non-ophthalmic) and methodological.
| Novelty & significance | Key new findings and implications |
|---|---|
| Diabetic retinopathy | |
| Screening; scale and generalizability [54] (2019) |
Study novelty - Numbers: > 100k patients; > 800k images; >400 clinics. - Generalizability: (“authors believe” so because of (i) sheer volume of patients and locations, and (ii) a previous study showing the EyeArt system was not affected by gender, ethnicity or camera type) [19]. Implication - Validated on large numbers in a real world setting against real world trained human graders; however, there was no gold standard comparing graders vs algorithm. - Although heterogenous in provider setting, the study would benefit from an additional external source of validation. |
| Severity prediction [55] (2020) |
Study novelty - Demonstrate feasibility of predicting future DR progression by leveraging CFPs of a patient acquired at a single visit from a 7-field color fundus photos (CFP). - The model predicted a 2-step or more ETDRS DR severity score worsening at 6, 12, and 24 months with an area under the curve (AUC) of 0.68 ± 0.13, 0.79 ± 0.05 and 0.77 ± 0.04 respectively. - Detected a predictive signal located in the peripheral retinal fields, which is not routinely collected for DR assessments, and the importance of microvascular abnormalities. Implication - May help stratify and identify high risk / rapidly progressive patients earlier that could prompt either intervention or for clinical trial recruitment assessing nuanced personalization |
| Treatment personalization [56] (2020) |
Study novelty - This algorithm achieved an average AUC of 0.866 in discriminating responsive from non-responsive patients with DME who are treated with various anti-VEGF agents. - Classification precision was significantly higher when differentiating between very responsive and very unresponsive patients. - A new feature-learning and classification framework. At the heart of this framework is a novel convolutional neural network (CNN) model called ‘CADNet’. - As compared to most previous studies that use longitudinal ocular coherence tomography (OCT) series, this model required the pre-treatment ocular coherence tomography (OCT) only to predict treatment outcomes. Implication - Critical step toward using non-invasive imaging and automated analysis to select the most effective therapy to select for a patient’s specific biomarker profile. - May help identify poor responders a priori, who could be more suitable for clinic trials. - However, need for larger studies (only 127 patients), further validation studies and better explainability (no specific predictive imaging features were identified). - Could not comparatively assess efficacy of different agents due to small sample. |
| Smartphones [57] (2019) |
Study novelty - First study assessing an offline AI algorithm on a smart phone for detection of referable DR (RDR = moderate DR or worse, or clinically significant macular edema (CSME)) in real-time in a real-world setting using local standards as benchmark for comparison (single ophthalmologist). - High sensitivity for detection of RDR (100% in both the eye-wise and patient-wise analysis). -Non-mydriatic patients; retained 100% sensitivity even without excluding low quality images (lower specificity; 88.4% vs 81.9%). Implication - Does not require internet, nor mydriasis; if well validated, ideal for deployment in developing world setting for screening (given high sensitivity). - Given low numbers (255 patients) and the gold standard of a single ophthalmologist (producing sensitivity of 100%), this algorithm requires both external validation and larger validation on both low and high quality images. |
| Glaucoma | |
| Remote / home monitoring [58] (2020) |
Study novelty - Demonstrates early reliability of remote intraocular pressure (IOP) measurement devices and visual field monitoring devices. - Both ancillary tests were easy-to-use. Implication - New reliable home devices can improve access, augment telemedicine, reduce burden on health systems and capture more data than infrequent office visits. - May improve patient engagement and compliance. - Wider validation required. |
| Surrogate device inference: (CFP from OCT) [59] (2019) |
Study novelty - Infer glaucomatous changes on a CFP from a corresponding SD-OCT with strong correlation between predicted and observed RNFL thickness values (Pearson r = 0.832; R2 . 69.3%; P < 0.001), with mean absolute error of the predictions of 7.39 mm. - Provided a quantitative assessment of the amount of neural tissue lost from a CFP alone. Implication - Avoided the need for human grading by leveraging quantitative data generated by OCT analysis; not only does this eliminate the burdensome workforce need, but human determinations of glaucoma are known to be highly variable. |
| Age-related Macular Degeneration (AMD) | |
| Biomarker-driven prognostication [60] (2020) |
Study novelty - A review of the phenotypic (clinical and imaging), demographic, environmental, genetic and molecular biomarkers that have been studied in prognostication of AMD progression. Implication - Risk factors can be combined in prediction models to predict disease progression, but the selection of the proper risk factors for personalised risk prediction will differ among individuals and is dependent on their current disease stage. |
| Predicting progression [61] (2019) |
Study novelty - Deep learning (DL) CNN predicted conversion to neovascular AMD with greater sensitivity than gradings from the AREDS dataset using both 4-step and 9-step grades for intermediate AMD or better. Implication - Promising, as this could further stratify high and low risk patients; needs external validation. |
| Genomic-based personalized risk [62] (2020) |
Study novelty - Fundus images coupled with genotypes could predict late AMD progression with an averaged AUC value of 0.85 from the AREDS dataset (31 652 fundus photos) and 52 known AMD - associated genetic variants - Fundus images alone showed an averaged AUC of 0.81 (95%CI: 0.80–0.83). Implication - Adding genetic information can improve prediction of AMD progression; currently routine generic testing not recommended [63]. |
| Non-ophthalmic diseases | |
| Cardiovascular (CV) risk factor identification [64] (2019) |
Study novelty - The prevalence and systemic risk factors for DR in multi-ethnic population could be determined accurately using a DL system, in significantly less time than human assessors. - Generalized across 5 races and 8 different datasets from 5 countries. Implication - This study highlights the potential use of AI for future epidemiology or clinical trials for DR grading in the global communities. - Notably, much larger and more diverse study than previous external validation of CV risk factor identification in a small Asian population from the original algorithm developed on images from the UK biobank [65]. |
| Anemia Detection [66] (2019) |
Study novelty -Using fundus imaging as a non-invasive way to detect anemia. -Achieving AUC 0.88 on a validation dataset of > 10k patients from the UK biobank, the DL algorithm could predict mean absolute error (MAE) of 0.63 g/dl in quantifying haemoglobin concentration. Implication -Ophthalmic imaging AI can be applied to systemic disease. -In this particular case, for diabetic patients who undergo regular screening retinal imaging are at increased risk of further morbidity and mortality from anemia. |
| Methods: OCT analysis | |
| Improving device agnostic analysis [67] (2020) |
Study novelty - New approach - unsupervised unpaired generative adversarial networks “cycleGANs” – translating OCT images from one vendor to another effectively reduces the covariate shift (the difference between the target and source domain), thereby demonstrating potential to overcome existing limitations is cross-device generalizability. - Applicability of existing methods is generally limited to samples that match training data. - Larger image patch sizes (256×256, 460×460) performed better in identifying and segmenting fluid, whereas for segmenting the thin photoreceptor layer, smaller image patch sizes (64×64, 128×128) achieved better performance. This suggests this image-context preservation is beneficial. Implication - Automated segmentation methods are expected to be part of routine diagnostic workflows, regardless of device; this method advances performance of cross-vendor translation and presents a new methodological basis for further investigation. |
(Publication year in brackets)
A detailed review of many ophthalmic subspecialties is contained elsewhere in this Journal’s edition.
Barriers to progress
“We always overestimate the change that will occur in the next 2 years and underestimate the change that will occur in the next ten. Don’t let yourself become lulled into inaction.”
– Bill Gates, co-founder of Microsoft ® and the Bill and Melinda Gates Foundation
One of many recent high-profile articles hinting at the potentially impending MD redundancy is the New Yorker piece “A.I. vs MD”, capturing both public attention and professional fears: “what will happen when diagnosis is automated?” [23]. This anticipatory anxiety mounts at a time of an unprecedented rapid convergence of factors intersecting health and technology. For example, just three years lay between AI reaching supra-human image-recognition performance in Stanford’s 2015 ImageNet competition and the first ever FDA-approved product-grade algorithm for autonomous consultation in ophthalmology [24]. The volume and variety of approved AI devices has since wildly proliferated and amidst this booming AI-device excitement, a concurrent tsunami of publications in medical journals tantalises readers with immense healthcare-AI clinical potential. But publication numbers are egregiously and disproportionately higher than clinical adoption. Very few medical algorithms (just 4 in Jan 2019 out of tens of thousands of papers) have undergone prospective real-world validation [25]. Peer-review publications of FDA-approved AI products are almost non-existent, inciting memories of Theranos. Long-term impact on clinical outcomes are also not yet known [25]. Indeed, this persisting ‘implementation gap’ both explains and results from the numerous non-trivial challenges in converting near-perfect ‘area under the curves’ (AUCs) into workflow-integrated solutions meeting genuine unmet patient needs (Table 2) [15,16]. Increasingly commoditized and automatically optimized algorithmic development is paradoxically the easy part. Overcoming the inertia against cultural and philosophical change in care paradigms is much more difficult.
Table 2:
AI implementation issue to overcome
| “Implementation-gap” | Description & underpinning issues | Current best practice / efforts to address the issue | Ultimate goal |
|---|---|---|---|
| Reporting standardization for (i) evaluation metrics and (ii) study variables | ● Recent meta-analysis review identified more than 20,000 studies in the field of DL; <1% of these studies had sufficiently high-quality design and reporting to be included [68]; notable finding variation in transparent reporting of: ○ Performance metrics ○ Code utilized ○ Algorithm details (eg – hyperparameters) ● Traditional reporting guidelines are not equipped to address many AI issues, particularly as AI studies are more complex, posing difficulty in interpretation of results, how relevant they are to a particular context and how to assess generalizability and bias [27]. |
● Consort-AI & SPIRIT-AI steering group: extending the work of CONSORT and SPIRIT groups (recommended reporting guidelines for clinical trials and protocol) to trials related to AI, using the EQUATOR framework (promoting transparent and accurate reporting and wider use of high-quality, robust reporting guidelines)[69]. ● STARD-AI: an AI-specific extension of the traditional STARD (Standards for Reporting Diagnostic Accuracy) recommendations, set to be published in 2020 [70,71] ● CLAIM: proposed checklist for AI manuscripts in imaging, based on STARD principles [72]. ● Tripod-ML: updated version of the proposed Tripod reporting structure for multivariate diagnostic probability functions; Tripod’s original focus on regression was not well placed to address AI / DL [73,74]. |
● Solve the “reproducibility crisis” ● Shift towards: ○ Journals mandating uniform and transparent reporting of methods (algorithm details), data, code, grading methodology results and predefining analysis variables (eg – operating thresholds / clinical definitions) ○ Improved documentation standardization in supplementary materials or online archives such as GitHub (https://github.com) ○ Reshaping academic goals to incentivize and foster a culture of sharing and collaboration for the greater good ● Algorithmic evaluation vs existing diagnostic pathways (clinical outcomes and using setting-dependant process for comparison against a gold standard; see examples in table 3 [75].) Note: as outlined in this table, many confounding variables exist and precise causal inference may be difficult to determine for each; increasing transparency will nonetheless, at least, improve capacity for study reproducibility and methodological evaluation. |
| Performance benchmarking |
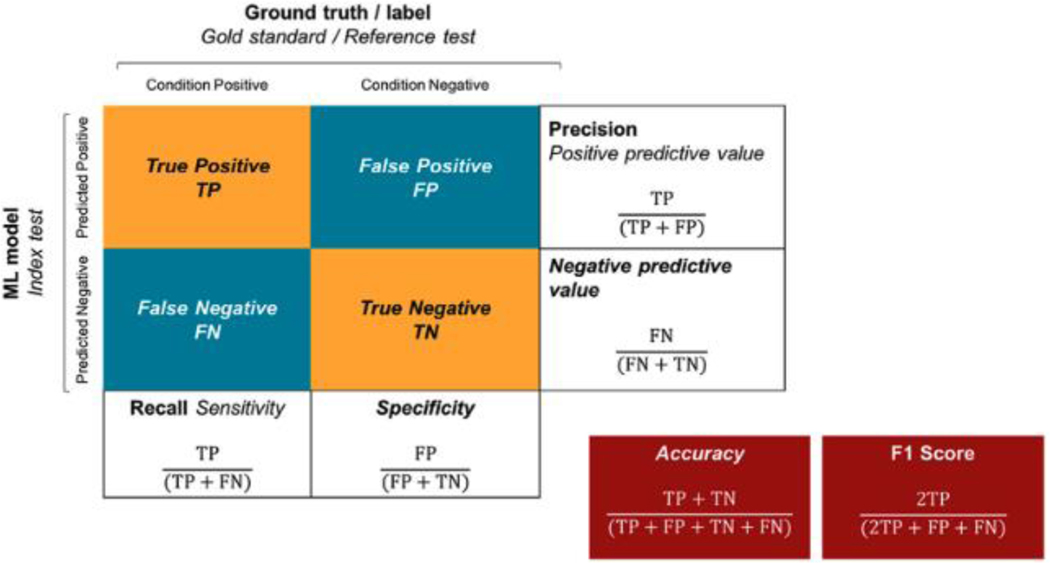
The gold standard: ● What constitutes an acceptable threshold for success? ● How to determine the clinically and ethically appropriate thresholds of sensitivity and specificity for a specific context to optimize for (see: Figure 1): ○ Low false negative (eg – in screening setting, particularly in the developing world, such as that described by Gulshan et al in their Indian validation study) ○ High true positive to minimize unnecessary referrals (eg – in a highly developed setting) ● To what degree should these factors be universal; are context, resourcing and incumbent process ethical determinants of influence (eg: developed vs developing world)? Bias/variance trade-off (generalizability): ● Balance between overfitting to the variables on which models train, vs losing accuracy to meet the requirements of all settings (‘1 size fits all’) ● Variables include: ○ Populations / ethnicities ○ Devices ○ Modalities ○ Disease states |
● No consensus approach; arbitrarily determined by authors +/− local specialist consultation ● Diagnostic ‘gold standards’ are highly heterogeneously defined, eg: ○ Physical examination ○ Image grader; proficiency varies (subspecialist / specialist / trained grader / technicial). Interobserver variability exists particularly between, but also within these categories. [47–49] These differences are amplified with increased disease classification granularity and severity. ○ Single vs multiple graders ○ Round robin / overlapping system to assess highest quality graders ○ Sophisticated adjudication methods for disagreement and edge cases[40] ○ Different imaging modality (eg – quantitative OCT outputs)[59] ● No clear regulatory benchmarks exist for (i) performance, (ii) grading schemes (comparison groups or gold standard determination) or (iii) validation process either pre or post deployment. ● Other variables potentially impacting performance also lack consensus guidelines (see below; eg – disease classification grading scheme; image quality and pre-processing methods etc.) Note: models that are not validated externally are more likely to overestimate performance [68] |
● Transparent reporting will iteratively inform the optimal recommendations and targets ● Consensus determination of: ○ Gold standards against which algorithms +/− graders will be compared ○ Validation requirements (external; prospective; post deployment etc) ○ Guidelines for post deployment performance validation and reporting (ensure no incurred bias or model drift etc.) ● Setting-dependant performance threshold acceptability (i.e. should it be assumed that ‘exceeding local grader performance’ is sufficient’?) ● Contingency table (Figure 1) should be provided at a clinically justified predetermined threshold that serves the algorithms intended purpose[52] ● Mandatory periodic reporting of clinical outcomes over time to maintain regulatory approval ● Reduce bias against publishing negative results to enhance ability to assess claims that a result is robust or generalizable. Note: Although evaluation of diagnostic accuracy is critical, we must evaluate the algorithm performance in (i) clinical trials and (ii) patient outcomes in the context of the whole patient pathway. |
| Variables in algorithmic development |
Diagnostic criteria variation (within a disease; eg -DR): ● ICDR [29,76] ● ETDRS (Early Treatment of Diabetic Retinopathy Study)[14,77] ● NHS DESP protocol [78] ● EURODIAB [79,80] Definition of clinical outcome (eg – RDR) ● Binary ● Mild DR (+/− DME ) vs worse [29] ● Vision-threatening DR (VTDR) [28,56] ● Graded ● 5-scale ICDR [81] Imaging modality & protocol (eg: CFP for DR screening) Modality ● Standard 45–60° fundus camera [14] ● Ultra-wide-field [82] ● Smartphones [83] Imaging Study Protocol ● Wide field stereoscopic retinal imaging protocol (4W-D); 4x covering 45–60° (equiv. ETDRS 7-field) ● ETDRS fields 1–5 ● Posterior pole only (40–45° macular centred) NB: each of the above can have +/− prephotographic pharmacological mydriasis Ground truth establishment: ● Manual grading vs exported from EMR; coded vs free-text? ● Image capture method variation [68] Model construction ● Input data format (eg image quality filtering / compression / pre-processing / augmentation / utilization of synthetic images[84,85]) ● Architecture and hyperparameter selection (transfer learning [86] / ensembling [87,88] etc.) ● Ground truth labels (accuracy and consistency) ● Data split (train / test / validate) +/− external validation |
Labelling and annotation methods ● Multiple open and paid software options ● Multiple human-skill levels to perform labelling ● Crowdsourcing, eg: Amazon Mechanical Turk[89,90], Labelme ®[91] ● Use image-generated measurement quantities as the ground truth (eg – OCT for RNFL thinning prediction in glaucoma) Evaluating annotations / labels ● Interobserver reliability, generally designated with a kappa score |
● Transparent reporting of all variables ● Ideally, gold standard should be used to compare algorithm vs real world setting annotators. This would ideally take place within the context of where that test would occur in the diagnostic pathway. ● Every method has its own strengths and weaknesses. The optimal approach may never be definitively discovered and require evolving inquiry Therefore, it is important to address a research question from multiple and hopefully orthogonal directions. This strategy combines the strengths of different methods to overcome their individual weaknesses |
|
Clinical Integration (patients, doctors and products) |
Cultural acceptance & fears ● Black box fears ● Data breaches ● Accuracy ● Workforce ● Trust ● Adversarial attacks Productization / workflow integration ● Building robust production-grade algorithmic software (large step from an experimental research code repository producing and AUC) ● Human operators / instructors + user manuals (out of the box ✇ ready) ● Referral and downstream systemic mapping ● HIPPA compliance ● Insurance ● Hardware considerations |
● Visualization and explainability methods (saliency mapping, class activation, eg – GradCam [92] ● Patient education and improved data control ● Physician engagement and explainability [52,53] |
Cultural acceptance ● Visualization integration ● Education about how individual data contributes towards the benefit of others ● AI products should be designed and used in ways and settings that respect and protect the privacy, rights and choices of patients and the public. ● Those determining the purpose and uses should include patients and the public as active partners ● Clinicians and health systems should demonstrate their continued trustworthiness by ensuring responsible and effective stewardship of patient data and data-driven technologies ● AI should be evaluated and regulated in ways that build understanding, confidence and trust, and guide their use ● Robust security regulation ● Increasing clinician engagement with AI development, data science and implementation oversight and monitoring Productization / workflow integration ● Post deployment validation to eliminate bias development ● Regular clinical review to assess for label leakage ● Security regulation |
It is hard enough to establish consensus guidelines and benchmarks for universal alignment and acceptance around standardization of reporting structures. It is hard enough to construct convention that caters to both a global gold standard and highly variable local realities across healthcare settings. It is hard enough to determine and regulate performance and validation requirements, particularly for software that iteratively learns (and that updates with increasing intelligence). This is exemplified by the FDA’s recent call for community feedback regarding their proposed regulatory framework, suggesting the organization’s quagmire in understanding the deeply sophisticated technical submissions [26]. It is hard enough to shift academic incentives away from the primitively absolute and competitive “publish or perish” maxim. Perhaps instead, we should adopt a balanced approach to sharing data, such as for unique surgical interventions, where no single centre could ever amass sufficient case numbers to conduct meaningful analysis (AI or otherwise), a healthy shift away from exceptionalism and towards collaboration would bring benefit to all.
It is also hard enough to reshape this academic culture to one that fosters transparency and collectively and earnestly demands the sharing required for translational progress underpinned by reproducibility and replicability [27]. Today’s jungle of confusion is a symptom of the pace of change, highlighted in a recent DR-screening review that demonstrates the difficulty in comparing different algorithms [28]. The authors outline the heterogeneity and arbitrary selection of many key variables: the validation datasets (internal; external; public; private; retrospective; prospective; size), the reference standards (grading methodology; performance metrics; disease classification scale; operating point/threshold), image processing methods and device variability (smartphones; modalities; field of view). This sentiment is echoed by thought leaders, noting remarkable and unjustifiable variation in choice of image quality (include vs exclude), publication of algorithm architecture and hyperparameters and the minimal external or prospective validation or comparison to healthcare experts, provision of heatmaps or other forms of explainability.
Harder still is striking a balance in building and implementing models that are both universally generalizable, but also purpose built for a specific setting or disease; that is to say, knowing that there will always be a trade-off, is ‘one-size fits all’ really the answer? Illustrating the nuances within this challenging issue is Gulshan et al.’s 2016 sentinel algorithm [29] and their subsequent validation study in India [30] (Table 3). The authors report performance superiority over the developing-world healthcare setting’s existing human grader infrastructure in screening patients for DR; however, the absolute performance was in fact weaker than in the original report. The papers were heralded as a victory for generalizability, but by what metric? One critical metric, the model’s predetermined operating threshold (the setting which can optimize for sensitivity or specificity) was selected without explicit justification and only published in the supplementary addendum in the validation paper (0.3; optimized for high sensitivity); while there may well be good reason for this choice, an explanation seems warranted. Threshold quantification was not provided beyond “high sensitivity (/specificity) operating point” in the original 2016 paper. Further, how should a reader evaluate the algorithmic ‘deployment’ performance when the original paper’s comparative reference was an army of US-board certified ophthalmologists, whereas the validation paper’s reference was the local grader and retinal specialist, which were then both compared to the ‘gold standard’ adjudication method outlined by Krause et al [31]. Moreover, the original abstract refers to RDR as moderate DR or worse (International Council of DR (ICDR) grade)[32], or DME or both, whereas the validation abstract references DR without DME. Notwithstanding, the validation study deployed would still de-burden an overrun Indian system and bring help to those who need it most. However, interrogating these papers also underscores the need for specialist clinical input to, amongst a myriad of other things, quantify the algorithm’s operating threshold and optimize its sensitivity whilst balancing the need for specificity for this unique screening setting, where per capita there are 80% less ophthalmologists than the USA [33]. A first mover in any field undergoes critical analysis, especially in disruptive medical innovation. However this group’s pioneering feats deserve the high accolades as frontier contributors to the advancement of AI, its safe and ethical investment in deep methodological groundwork and ultimately the impact on patients. Indeed, the aforementioned lessons gleaned should not be overlooked, but rather inspire and propel further cycles of inquiry, trial and subsequent appraisal.
Table 3:
varied recent innovative publications that have accelerated AI’s ophthalmology capability either clinically or methodologically; a repository (as described) would profoundly advance the volume of similar advancements
| Summary | Novel content & connection to dataset access | Implications / problems solved |
|---|---|---|
| “Retisort” automated image classification algorithm for AI research workflow optimization [93] (2020) | ● Constructed an automated sorting tool for preparing images for AI research: quality and laterality for both macular and disc photos. For disc centred photos, saliency mapping visually explains the model’s reliable predictions ● Externally validated on 3x public datasets: DIRETDBO, HEI – MED and Drishti-GS |
● Automated imaging dataset procurement is becoming increasingly important as their size and multi-modality increases, particularly as AI research continues to expand to address clinical and methodological problems ● Datasets are also becoming increasingly fused or compared for validation ● Development of validated algorithms (such as Retisort) |
| Real-world validation of a high performing DR screening algorithm assessing its generalizability in India [30] (2019) | ● Supra-human algorithm was originally developed and validated against retrospective datasets (Mesidoor and Eyepacs); this prospective validation was undertaken in a developing world setting to detect RDR (moderate of worse, or DME) ● Comparatively, the algorithm outperformed human graders within the developing-world healthcare setting’s existing infrastructure across 2 sites; gold standard was determined by adjudication methods previously described involving 3 retinal specialists [31] ● The model generalized to a new ethnicity and fundus camera: Eyepacs development – Centervue DRS, Optovue iCam, Canon CR1/DGi/CR2, and Topcon NW, Mesidoor validation on Topcon TRC NW6 nonmydriatic camera (both 45° fields of view, macula centred with ~ 40–45% pupil mydriasis) vs Topcon ® Forus 3nethra (45° fields of view, macula centred, no mydriasis). ● The study also evaluated the performance of an improved model optimized for 5-point ICDR grading published during its course, showing minimal performance improvements. Although, this analysis was performed retrospectively. |
● Absolute model performance was overall lower than the originally published algorithm. The Original study showed 96.1–97.5% sensitivity; AUC 0.99 for RDR (mod. DR or worse or DME at the sensitivity optimized operating point (OP; not quantified) vs prospective study, showing 88.9–92.1% sensitivity; AUC 0.963–0.98 for mod. DR or worse and 93.6–97.4% sensitivity; AUC 0.983 for referable DME (at 0.3 predefined OP, published in supplement). ● However, in the prospective study this still exceeded manual grading where sensitivity ranged 73.4%−89.8% for detection of mod. or worse DR and 57.5–79.5% sensitivity for DME. ● Note: original study did not report model performance separately for DME and mod. DR or worse at the sensitivity optimized OP (only at the ophthalmologist OP), whereas the prospective only reported them separately. ● DME; defined by hard exudate within 1 disc diameter; this would be improved using OCT (see below [94]) |
| Real world comparison of human graders in existing workflow vs automated algorithm [75] (2019) | ● Implementation of a robust algorithm [29] into an existing large-scale screening program for DR (vs human graders; nurses / technicians / ophthalmologists) ● Reference standard with an international panel of retinal specialists with an adjudication methodology for disagreement in referable cases only ● Model performance performed manual graders with sensitivity 0.968 (0.893–0.993) vs 0.734 (0.407–0.914); p value < 0.001. Of note, using mod. DR or worse as referral threshold, regional graders did not refer 12.1% and 14.1% of severe NPDR or PDR respectively, whereas the algorithm had a false negative rate for these classes of 3.3% and 4.5% respectively. ● Generalized to a new ethnic population (Thai), using 6 different camera manufacturers across 13 regions. |
● 5-grade model with gold standard adjudication methodology validated in a real world setting within an existing screening program, generalizing with excellent performance. ● As many countries have existing screening infrastructure, this study exemplifies the comparative evaluation should be assessed. ● Looking ahead, prospective validation should either precede or accompany integration of such an algorithm; moreover, it is integral to embed an iterative post deployment validation protocol into the implementation. |
| Reproduction study: DR screening using public data [95] (2019) | ● Exemplary publication of a negative finding; in attempting to replicate methods and results on 2 different openly available datasets (different to the originals, as no longer available), this reveals inferior algorithmic performance to the 2016 Gulshan et al publication. ● AUCs of 0.95 and 0.85 on Eyepacs (from Kaggle) and Messidoor 2 (different distribution) respectively do not compare to the AUC of 0.99 reported on both original internal (Eyepacs) and external (Messidor) validation datasets. ● Explores the confounding impact of different grading methodologies on a given algorithm; notes difficult to quantify the impact of multiple graders (this paper only had one). |
● Underscores the need for more replication and reproduction studies to validate DL performance across multiple datasets ● Highlights the need standards to mandate publication of source code and methodology to avoid the “reproducibility crisis.” (The code from this study was released). |
| Transfer learning detects geographic atrophy on CPF from multiple clinical trial datasets [96] (2020) | ● 3 clinical trial datasets were used to develop an automated segmentation and growth rate prediction model for GA using CPF ● Identified specific lesion features predictive of progression, and the rate of projected lesion growth |
● Progress towards automating clinical trial endpoints[97] ● While traditionally FAF and OCT would be used to characterize GA lesions, this may offer an adjunctive, additive or alternative method of automating GA lesion detection and predicting progression |
| Multiple OCT problems addressed through ‘RETOUCH’ public competition [98] (2019) | ● Dataset is substantially larger and more heterogenous (manufacturers) than anything previously available (112 OCTs with 11,334B-scans) ● At scale, addressed key clinical issues (eg cross-device volumetric measurement of all 3 fluid types) and technical questions (eg newer semantic seg-mentation algorithms, exploiting image context without losing the spatial resolution, assessing effects of motion artefacts, signal-to-noise ratio, spacing between and number of B scan slices and |
● When big datasets are released to multiple research groups, different methodological approaches are applied, allowing for direct comparison between them and findings to have more impactful outcomes. ● The combination of competition and experimental sandbox environment encourages creativity and incentive to confront challenging frontier questions. |
| Demonstration of potential of access to multimodal data [94] (2020) | ● Screening for DME is generally done through CPF review alone and lacks reliability and interobserver consistency ● This DL model draws OCT central macular thickness inference from CFP, outperforming retinal specialist grading across all metrics; ground truth for centre-involving DME (CI-DME) comparison was derived from OCT central subfield thickness maps > 250-um ● Model subsequently generalized to a second dataset (restricted access; EyePacs), outperforming the trained EyePacs graders (ground truth for CI-DME comparison was derived from OCT central subfield thickness maps > 300-um) ● Made possible through cross-modality dataset linkage of imaging, metadata and clinical data ● Visualizations reveal that DL-predictions are driven by the clinically appropriate presumption that the region of interest is in the central macula (model explainability) |
● Improved triaging, as patients with CI-DME and reduced VA more urgently require review and treatment ● CPF are significantly cheaper and more accessible than OCT; improving ● Future studies can improve model performance, utilize larger and more varied validation datasets to show generalizability across devices and settings ● Additionally, can link different and increasing modalities to refine performance of predictive disease detection |
| Meta-analysis using multiple open datasets to validate vessel segmentation [99] (2020) | ● Retinal vessel (RV) segmentation is a challenging task due to anatomic variability, inter-grader variability and other factors; previous automation methodologies (eg - vector geometry, low-level feature extraction from other ML techniques) have not shown superiority to human graders. ● Using multiple (seven) open-source datasets with accompanying manually segmented ground truths, the meta-analysis showed DL algorithmic performance was consistently comparable to human experts, largely owing to their capability to extract high dimensional features. |
● RV segmentation has an important role in screening, diagnosing and prognosticating CV, diabetes and RV disease; reliable DL automation techniques may play a significant role in improving assessment speed, reducing cost in an increasingly personalized manner ● Future studies can assess algorithmic. performance in RW settings, their reliability in various disease detection and evaluate impact on clinical systems and referral pathways. |
We know that the model architecture and training parameters, the quality of reference standard, the selection of images and the choice of operating point can significantly influence algorithmic performance and interpretability. In the absence of guidelines, methodological decisions are arbitrary, highly varied and therefore near-impractical to evaluate. But one must not be disheartened; although it may indeed take as long as Gates’ proverbial decade to achieve the idealized, streamlined and transparent data-driven future, we certainly should not underestimate or undervalue the progress made in this short time. That said, it is time we ask how to most effectively channel resources to support AI in achieving its promise in healthcare. Successful transition will bring an accompanying evolved and advanced system – cost and resource efficient, biomarker rich and globally democratized access.
Why open source:
“The goal of education is the advancement of knowledge and the dissemination of truth.”
- John F. Kennedy, 35th President of the USA
Underpinned by decentralized collaboration, the open source movement in organizational science is “any system of innovation or production that relies on goal-oriented yet loosely coordinated participants who interact to create a product (or service) of economic value, which they make available to contributors and noncontributors alike” [34]. This principle has propagated the network effects of information spread and progress through modern media such as Wikipedia ®, Ted Talks ® and internet forums. The communities founded around the Creative Common licence ® leverage peer-produced idea-sharing, build upon others’ endeavours yet respect tiers of self-declared privacy, where, instead of ‘all,’ only ‘some rights are reserved.’
This approach is the quantum leap required to release the aforementioned AI-implementation bottlenecks; in fact, the foundation of AI’s acceleration is literally built upon this very code (of conduct). Software libraries and algorithms and wrap-around support communities such as Python (open source programming language), TensorFlow (open-source machine learning library) and Imagenet-V3 (open source ImageNet-winning CNN-algorithm) are but a few examples of the free, core drivers of the AI-ophthalmology revolution [35–37].
Though patient privacy rightly constrains equivalent freedom of healthcare data, its sharing is more often curbed by variably incentivized stakeholders claiming ownership. Privacy is often ironically and naively the excuse provided to prevent secure, de-identified access to patients’ own information, which may help others through medical breakthrough. Despite institutional roadblocks, a recent proliferation of data repositories, with reasonable varied restrictions, has galvanized inspirational researcher engagement, productivity and discovery; a few diverse examples include the UK biobank (clinical, demographic, imaging and genomic), the NYU/Facebook “FastMRI” collaboration, the Medical Information Mart for Intensive Care or MIMIC and the Rotterdam study [38–41]. One particularly relevant and exceptional database is open-source sharing platform GISAID that pools and harmonizes genetic data from different laboratories: a global collaboration of experts in their respective disciplines work together to facilitate rapid access to data pertaining to viruses [42]. Accompanied by novel research tools, empowerment and capacity building through educational programs, the platform has traced hCoV-19 12,041 genome sequences, enabling Google Deepmind’s ® recently published “alpha-fold” protein structure computational-prediction model to study COVID-19’s structure, and subsequently opening their findings to the world [43]. Such molecular structures are often difficult to ascertain experimentally, but are critical to developing therapeutics, especially prudent in the fight against today’s pandemic. Not only does this collaborative effort exemplify the powerful possibilities of today, but also crystalizes a path forward for other fields.
Ophthalmic AI-implementation will momentously benefit from the promotion, coordination, and reuse of data through a globally representative data-sharing platform based on established academic and industry standards and best practices in data custodianship with strict but not exclusive user access [44,45]. Every challenge listed in Table 2 would be communally addressed more rapidly with such a resource; in keeping, all resulting insights, publications and code will be released under an open license.
Data sources may arise from:
Merging of the extensive, existing, yet paradoxically siloed open online datasets that have served as the primary source of algorithmic validation to date.
Clinical trial data: The National Eye Institute Age-Related Eye Disease Studies (AREDS) [46] have 134,500 high-quality fundus photographs and longitudinal clinical data of 4613 individuals, 595 of whom have dbGaP accessible genotype data. The AREDS dataset is available with authorized access via application. Additionally, DRCR.net recently announced its policy on data sharing (images & clinical data) with academia and industry alike. Vivli ®, a clinical research data-sharing platform provides access to anonymized individual patient data to enable secondary analyses, reproducibility assessment or systematic review [47]. A similar consortium of clinical study sponsors has emerged in the data sharing community, facilitating access to patient-level data from clinical studies on a research-friendly platform [48]. Patient privacy is tightly protected through controls preventing any data being exported. Similarly, albeit translucently, datasets are available in the ophthalmic world and have been outcome-modelled using AI with great efficacy, as they are often accompanied by reliably protocol-driven clinical data and consistent image grading.
Private contributors: The Moorfield’s Eye hospital has opened its AMD database [49,50]. But other hospitals have been slow to follow because of entrenched fears of data breach and a highly competitive academic culture. The concerns and issues may be addressed by an incentive-neutral platform and supported by robust but flexible data use agreements, accounting for variable preference of academic credit, commercial exclusivity and user access (e.g. in arrangement with other partners). Increasingly, federated learning systems and encryption services are facilitating privacy-preserving data sharing across sites without parting with data ownership [51]. It should be noted that democratizing access does not axiomatically prevent industry, government or any institution from intellectual property rights. In fact, those with significant resources may be well placed to benefit from rapid access to a vast experimental repository of varied datasets; in turn, this truly levelled playing field for data access is likely to hasten industry’s global impact and scientific contribution.
Key points
The volume and heterogeneity of studies evaluating AI applications in ophthalmology is expanding, demonstrating increasingly high performance.
The AI-chasm persists; many researchers lack the resources, data-access, networks and context-dependent gold standards for comparison of models. Reasons for this are complex and multi-faceted (Table 2).
A consolidated platform of multi-sourced, multimodal ophthalmic data would rapidly accelerate the rate of clinical validation, establishment of consensus for benchmarking image grading standards and model performance and create an experimental sandbox to drive data-science methodological advancements.
Conclusion
As stewards of patient care, clinicians and provider systems remain the gatekeepers in evaluating safety and efficacy of new technology and therapies. Modern trends suggest that iterative post-deployment validation of learning systems will increasingly become a service in demand. Barriers to clinicians engaging with AI are diminishing [52] and risk communication and management is improving for clinical end users to make more informed data-driven decisions [53].
Ophthalmologists stand only to benefit from encouraging investigation, fostering deep collaborations between clinical and data science communities and facilitating greater understanding of model development, validation and interrogation. In democratizing access to pooled, multi-modal and heterogeneous datasets, we transform dormant silos of pixels, numerics and texts into living data, promote organic scientific discovery and play an active role in ensuring safe oversight for our patients.
Figure 1: Overview of the confusion matrix / contingency table (previously published) [100].
Layout of a confusion matrix/contingency table.
Differences in nomenclature for machine learning (boldface type) and classical statistics (italic type) and where overlapping (boldface and italic) are highlighted.
Acknowledgements
1. Each author contributed to conception, writing and review of manuscript
2. Financial support and sponsorship:
• LAC is funded by the National Institute of Health through NIBIB R01 EB017205.
Footnotes
Conflict of interests:
None
Bibliography
- [1].Schmidt E. How Google Works. Hachette, UK; 2014. [Google Scholar]
- [2].Commissioner O of the. FDA’s Technology Modernization Action Plan. 2019.
- [3].Fda US. Digital Health Innovation Action Plan 2017.
- [4].Fda. Framework for FDA’s Real-World Evidence Program 2018.
- [5].Kern C, Fu DJ, Huemer J, Faes L, Wagner SK, Kortuem K, et al. AN OPEN-SOURCE DATASET OF ANTI-VEGF THERAPY IN DIABETIC MACULAR OEDEMA PATIENTS OVER FOUR YEARS & THEIR VISUAL OUTCOMES. Medrxiv 2019:19009332. doi: 10.1101/19009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Klonoff DC. The New FDA Real-World Evidence Program to Support Development of Drugs and Biologics. J Diabetes Sci Technology 2020;14:345–9. doi: 10.1177/1932296819832661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Blumenthal D. Launching HITECH. New Engl J Medicine 2009;362:382–5. doi: 10.1056/nejmp0912825. [DOI] [PubMed] [Google Scholar]
- [8].ONC’s Cures Act Final Rule n.d.
- [9].Lehne M, Luijten S, Imbusch P, Thun S. The Use of FHIR in Digital Health - A Review of the Scientific Literature. Stud Health Technol 2019;267:52–8. doi: 10.3233/shti190805. [DOI] [PubMed] [Google Scholar]
- [10].Hong N, Wen A, Shen F, Sohn S, Wang C, Liu H, et al. Developing a scalable FHIR-based clinical data normalization pipeline for standardizing and integrating unstructured and structured electronic health record data. Jamia Open 2019;2:570–9. doi: 10.1093/jamiaopen/ooz056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sayeed R, Gottlieb D, Mandl K. SMART Markers: collecting patient-generated health data as a standardized property of health information technology. Npj Digital Medicine 2020;3:9. doi: 10.1038/s41746-020-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Topol E. Deep Medicine: How Artificial Intelligence Can Make Healthcare Human Again. Basic Books; 2019. [Google Scholar]
- [13].ophthalmology artificial intelligence n.d https://scholar.google.com/scholar?as_ylo=2019&q=ophthalmology+artificial+intelligence&hl=en&as_sdt=0,5 (accessed April 18, 2020).
- [14].Abràmoff MD, Lavin PT, Birch M, Shah N, Folk JC. Pivotal trial of an autonomous AI-based diagnostic system for detection of diabetic retinopathy in primary care offices. Npj Digital Medicine 2018;1:39. doi: 10.1038/s41746-018-0040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Keane PA, Topol EJ. With an eye to AI and autonomous diagnosis. Npj Digital Medicine 2018;1:10–2. doi: 10.1038/s41746-018-0048-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Panch T, Mattie H, Celi LA. The “inconvenient truth” about AI in healthcare. Npj Digital Medicine 2019;2:4–6. doi: 10.1038/s41746-019-0155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Organization WH. Universal eye health: a global action plan 2014–2019. 2013. [Google Scholar]
- [18].Resnikoff S, Lansingh VC, Washburn L, Felch W, Gauthier T-M, Taylor HR, et al. Estimated number of ophthalmologists worldwide (International Council of Ophthalmology update): will we meet the needs? Brit J Ophthalmol 2019;104:bjophthalmol-2019. doi: 10.1136/bjophthalmol-2019-314336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].(CDC) C for DC and P. National diabetes statistics report 2020. n.d.
- [20].Lu Y, Serpas L, Genter P, Anderson B, Campa D, Ipp E. Divergent Perceptions of Barriers to Diabetic Retinopathy Screening Among Patients and Care Providers, Los Angeles, California, 2014–2015. Prev Chronic Dis 2016;13:E140. doi: 10.5888/pcd13.160193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Flaxel CJ, Adelman RA, Bailey ST, Fawzi A, Lim JI, Vemulakonda GA, et al. Diabetic Retinopathy Preferred Practice Pattern®. Ophthalmology 2019;127:P66–145. doi: 10.1016/j.ophtha.2019.09.025. [DOI] [PubMed] [Google Scholar]
- [22].Balyen L, Peto T. Promising Artificial Intelligence-Machine Learning-Deep Learning Algorithms in Ophthalmology. Asia-Pacific J Ophthalmol 2019;8:264–72. doi: 10.22608/apo.2018479. [DOI] [PubMed] [Google Scholar]
- [23].Mukherjee S. A.I. Versus M.D 2017.
- [24].Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, et al. ImageNet Large Scale Visual Recognition Challenge. Int J Comput Vision 2015;115:211–52. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- [25].Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019;25:44–56. doi: 10.1038/s41591-018-0300-7. [DOI] [PubMed] [Google Scholar]
- [26].O.v. Proposed Regulatory Framework for Modifications to Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD) - Discussion Paper and Request for Feedback 2019:1–20.
- [27].Schloss P. Identifying and Overcoming Threats to Reproducibility, Replicability, Robustness, and Generalizability in Microbiome Research. Mbio 2018;9:e00525–18. doi: 10.1128/mbio.00525-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Raman R, Srinivasan S, Virmani S, Sivaprasad S, Rao C, Rajalakshmi R. Fundus photograph-based deep learning algorithms in detecting diabetic retinopathy. Eye 2019;33:97–109. doi: 10.1038/s41433-018-0269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama 2016;316:2402–10. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- [30].Gulshan V, Rajan RP, Widner K, Wu D, Wubbels P, Rhodes T, et al. Performance of a Deep-Learning Algorithm vs Manual Grading for Detecting Diabetic Retinopathy in India. Jama Ophthalmol 2019;137:987–93. doi: 10.1001/jamaophthalmol.2019.2004.(**) (**)This paper demonstrated a pioneering milestone of real world algorithmic deployment, revealing its accompanying challenges.
- [31].Krause J, Gulshan V, Rahimy E, Karth P, Widner K, Corrado GS, et al. Grader Variability and the Importance of Reference Standards for Evaluating Machine Learning Models for Diabetic Retinopathy. Ophthalmology 2018;125:1264–72. doi: 10.1016/j.ophtha.2018.01.034. [DOI] [PubMed] [Google Scholar]
- [32].Wong TY, Sun J, Kawasaki R, Ruamviboonsuk P, Gupta N, Lansingh VC, et al. Guidelines on Diabetic Eye Care: The International Council of Ophthalmology Recommendations for Screening, Follow-up, Referral, and Treatment Based on Resource Settings. Ophthalmology 2018;125:1608–22. doi: 10.1016/j.ophtha.2018.04.007. [DOI] [PubMed] [Google Scholar]
- [33].Abrams C. Google’s Effort to Prevent Blindness Shows AI Challenges 2026. [Google Scholar]
- [34].Levine SS, Prietula MJ. Open Collaboration for Innovation: Principles and Performance. Organ Sci 2014;25:1414–33. doi: 10.1287/orsc.2013.0872. [DOI] [Google Scholar]
- [35].Python n.d.
- [36].Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. TensorFlow: A System for Large-Scale Machine Learning 2016:265–83. [Google Scholar]
- [37].Krizhevsky A, Sutskever I, Hinton GE, Pereira F, Burges CJC, Bottou L, et al. ImageNet Classification with Deep Convolutional Neural Networks, 2012, p. 1097–105. [Google Scholar]
- [38].Johnson AEW, Pollard TJ, Shen L, Lehman L-WH, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zbontar J, Knoll F, Sriram A, Muckley MJ, Bruno M, Defazio A, et al. fastMRI: An Open Dataset and Benchmarks for Accelerated MRI 2018. [Google Scholar]
- [40].Ikram MA, Brusselle GGO, Murad SD, van Duijn CM, Franco OH, Goedegebure A, et al. The Rotterdam Study: 2018 update on objectives, design and main results. Eur J Epidemiol 2017;32:807–50. doi: 10.1007/s10654-017-0321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. Plos Med 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Elbe S, Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Global Challenges 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, et al. Improved protein structure prediction using potentials from deep learning. Nature 2020;577:706–10. doi: 10.1038/s41586-019-1923-7. [DOI] [PubMed] [Google Scholar]
- [44].Emam I, Elyasigomari V, Matthews A, Pavlidis S, Rocca-Serra P, Guitton F, et al. PlatformTM, a standards-based data custodianship platform for translational medicine research. Sci Data 2019;6:149. doi: 10.1038/s41597-019-0156-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Wilkinson MD, Dumontier M, Aalbersberg IJJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bird AC, Bressler NM, Bressler SB, Chisholm IH, Coscas G, Davis MD, et al. An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol 1995;39:367–74. doi: 10.1016/s0039-6257(05)80092-x. [DOI] [PubMed] [Google Scholar]
- [47].Vivli n.d.
- [48].ClinicalStudyDataRequest.com n.d.
- [49].Fasler K, Fu DJ, Moraes G, Wagner S, Gokhale E, Kortuem K, et al. Moorfields AMD database report 2: fellow eye involvement with neovascular age-related macular degeneration. Brit J Ophthalmol 2019:bjophthalmol-2019–314446. doi: 10.1136/bjophthalmol-2019-314446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Fasler K, Moraes G, Wagner S, Kortuem KU, Chopra R, Faes L, et al. One- and two-year visual outcomes from the Moorfields age-related macular degeneration database: a retrospective cohort study and an open science resource. Bmj Open 2019;9:e027441. doi: 10.1136/bmjopen-2018-027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tong J, Duan R, Li R, Scheuemie MJ, Moore JH, Chen Y. Robust-ODAL: Learning from heterogeneous health systems without sharing patient-level data 2020;25:695–706. [PMC free article] [PubMed] [Google Scholar]
- [52].Faes L, Wagner SK, Fu DJ, Liu X, Korot E, Ledsam JR, et al. Automated deep learning design for medical image classification by health-care professionals with no coding experience: a feasibility study. Lancet Digital Heal 2019;1:e232–42. doi: 10.1016/s2589-7500(19)30108-6. [DOI] [PubMed] [Google Scholar]
- [53].Sendak MP, Gao M, Brajer N, Balu S. Presenting machine learning model information to clinical end users with model facts labels. Npj Digital Medicine 2020;3:41. doi: 10.1038/s41746-020-0253-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bhaskaranand M, Ramachandra C, Bhat S, Cuadros J, Nittala MG, Sadda SR, et al. The Value of Automated Diabetic Retinopathy Screening with the EyeArt System: A Study of More Than 100,000 Consecutive Encounters from People with Diabetes. Diabetes Technol The 2019;21:635–43. doi: 10.1089/dia.2019.0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Arcadu F, Benmansour F, Maunz A, Willis J, Haskova Z, Prunotto M. Deep learning algorithm predicts diabetic retinopathy progression in individual patients. Npj Digital Medicine 2019;2:92. doi: 10.1038/s41746-019-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Rasti R, Allingham MJ, Mettu PS, Kavusi S, Govind K, Cousins SW, et al. Deep learning-based single-shot prediction of differential effects of anti-VEGF treatment in patients with diabetic macular edema. Biomed Opt Express 2020;11:1139–52. doi: 10.1364/boe.379150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Natarajan S, Jain A, Krishnan R, Rogye A, Sivaprasad S. Diagnostic Accuracy of Community-Based Diabetic Retinopathy Screening With an Offline Artificial Intelligence System on a Smartphone. Jama Ophthalmol 2019;137:1182. doi: 10.1001/jamaophthalmol.2019.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hamzah JC, Daka Q, Azuara-Blanco A. Home monitoring for glaucoma. Eye 2020;34:155–60. doi: 10.1038/s41433-019-0669-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Medeiros FA, Jammal AA, Thompson AC. From Machine to Machine: An OCT-Trained Deep Learning Algorithm for Objective Quantification of Glaucomatous Damage in Fundus Photographs. Ophthalmology 2019;126:513–21. doi: 10.1016/j.ophtha.2018.12.033.(**) (**)This paper’s innovative use of objective, algorithmically generated measurements as its source of ground truth minimizes the limitations of human-derived ground truths and the well-known intergrader variability.
- [60].Heesterbeek TJ, Lorés-Motta L, Hoyng CB, Lechanteur YTE, Hollander AI den. Risk factors for progression of age-related macular degeneration. Ophthal Physl Opt 2020;40:opo.12675. doi: 10.1111/opo.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Babenko B, Balasubramanian S, Blumer KE, Corrado GS, Peng L, Webster DR, et al. Predicting Progression of Age-related Macular Degeneration from Fundus Images using Deep Learning 2019. [Google Scholar]
- [62].Yan Q, Weeks DE, Xin H, Swaroop A, Chew EY, Huang H, et al. Deep-learning-based prediction of late age-related macular degeneration progression. Nat Mach Intell 2020;2:141–50. doi: 10.1038/s42256-020-0154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Flaxel CJ, Adelman RA, Bailey ST, Fawzi A, Lim JI, Vemulakonda GA, et al. Age-Related Macular Degeneration Preferred Practice Pattern®. Ophthalmology 2020;127:P1–65. doi: 10.1016/j.ophtha.2019.09.024. [DOI] [PubMed] [Google Scholar]
- [64].Ting DSW, Cheung CY, Nguyen Q, Sabanayagam C, Lim G, Lim ZW, et al. Deep learning in estimating prevalence and systemic risk factors for diabetic retinopathy: a multi-ethnic study. Npj Digital Medicine 2019;2:24. doi: 10.1038/s41746-019-0097-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Poplin R, Varadarajan AV, Blumer K, Liu Y, McConnell MV, Corrado GS, et al. Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2018;2:158–64. doi: 10.1038/s41551-018-0195-0. [DOI] [PubMed] [Google Scholar]
- [66].Mitani A, Liu Y, Huang A, Corrado GS, Peng L, Webster DR, et al. Detecting Anemia from Retinal Fundus Images 2019. [Google Scholar]
- [67].Romo-Bucheli D, Seeböck P, Orlando JI, Gerendas BS, Waldstein SM, Schmidt-Erfurth U, et al. Reducing image variability across OCT devices with unsupervised unpaired learning for improved segmentation of retina. Biomed Opt Express 2020;11:346–63. doi: 10.1364/boe.379978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Liu X, Faes L, Kale AU, Wagner SK, Fu DJ, Bruynseels A, et al. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digital Heal 2019;1:undefined-undefined. doi: 10.1016/s2589-7500(19)30123-2. [DOI] [PubMed] [Google Scholar]
- [69].Group C-A and S-AS. Reporting guidelines for clinical trials evaluating artificial intelligence interventions are needed. Nat Med 2019;25:1467–8. doi: 10.1038/s41591-019-0603-3. [DOI] [PubMed] [Google Scholar]
- [70].Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: Explanation and elaboration. Bmj Open 2016;6:e012799. doi: 10.1136/bmjopen-2016-012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Network E. Reporting guidelines under development for other study designs n.d.
- [72].Mongan J, Moy L, Kahn CE. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiology Artif Intell 2020;2:e200029. doi: 10.1148/ryai.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Collins GS, Reitsma JB, Altman DG, Moons KGM. Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD). Ann Intern Med 2015;162:735. doi: 10.7326/l15-5093-2. [DOI] [PubMed] [Google Scholar]
- [74].Collins GS, Moons KGM. Reporting of artificial intelligence prediction models. Lancet 2019;393:1577–9. doi: 10.1016/s0140-6736(19)30037-6. [DOI] [PubMed] [Google Scholar]
- [75].Ruamviboonsuk P, Krause J, Chotcomwongse P, Sayres R, Raman R, Widner K, et al. Deep learning versus human graders for classifying diabetic retinopathy severity in a nationwide screening program. Npj Digital Medicine 2019;2:25. doi: 10.1038/s41746-019-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Association EMD, Others. International Clinical Diabetic Retinopathy Disease Severity Scale, Detailed Table 2002. [Google Scholar]
- [77].CANTRILL H. The Diabetic Retinopathy Study and the Early Treatment Diabetic Retinopathy Study. Int Ophthalmol Clin 1984;24:13–29. doi: 10.1097/00004397-198402440-00004. [DOI] [PubMed] [Google Scholar]
- [78].Core National Diabetic Eye Screening Programme team. Feature Based Grading Forms, Version 1.4. 2012. n.d. [Google Scholar]
- [79].van der Heijden AA, Abramoff MD, Verbraak F, van Hecke MV, Liem A, Nijpels G. Validation of automated screening for referable diabetic retinopathy with the IDx-DR device in the Hoorn Diabetes Care System. Acta Ophthalmol 2018;96:63–8. doi: 10.1111/aos.13613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Aldington SJ, Kohner EM, Meuer S, et al. Group TEICS. Methodology for retinal photography and assessment of diabetic retinopathy: the EURODIAB IDDM Complications Study. Diabetologia 1995;38:437–44. doi: 10.1007/bf00410281. [DOI] [PubMed] [Google Scholar]
- [81].International Council of Ophthalmology : Ophthalmologists Worldwide. n.d.
- [82].Nagasawa T, Tabuchi H, Masumoto H, Enno H, Niki M, Ohara Z, et al. Accuracy of ultrawide-field fundus ophthalmoscopy-assisted deep learning for detecting treatment-naïve proliferative diabetic retinopathy. Int Ophthalmol 2019;39:2153–9. doi: 10.1007/s10792-019-01074-z. [DOI] [PubMed] [Google Scholar]
- [83].Rajalakshmi R, Subashini R, Anjana RM, Mohan V. Automated diabetic retinopathy detection in smartphone-based fundus photography using artificial intelligence. Eye 2018;32:1138–44. doi: 10.1038/s41433-018-0064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Odaibo SG, D M, S M, S M. Generative Adversarial Networks Synthesize Realistic OCT Images of the Retina 2019. [Google Scholar]
- [85].Burlina PM, Joshi N, Pacheco KD, Liu TYA, Bressler NM. Assessment of deep generative models for high-resolution synthetic retinal image generation of age-related macular degeneration. Jama Ophthalmol 2018;9227:258. doi: 10.1001/jamaophthalmol.2018.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Mesnil G, Dauphin Y, Glorot X, Rifai S, Bengio Y, Goodfellow I, et al. Unsupervised and transfer learning challenge: a deep learning approach 2011:97–111. [Google Scholar]
- [87].Fraz MM, Remagnino P, Hoppe A, Uyyanonvara B, Rudnicka AR, Owen CG, et al. An ensemble classification-based approach applied to retinal blood vessel segmentation. Ieee T Bio-Med Eng 2012;59:2538–48. doi: 10.1109/tbme.2012.2205687. [DOI] [PubMed] [Google Scholar]
- [88].Hassan B, Hassan T, Li B, Ahmed R, Hassan O. Deep Ensemble Learning Based Objective Grading of Macular Edema by Extracting Clinically Significant Findings from Fused Retinal Imaging Modalities. Sensors 2019;19:2970. doi: 10.3390/s19132970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Mitry D, Zutis K, Dhillon B, Peto T, Hayat S, Khaw K-T, et al. The Accuracy and Reliability of Crowdsource Annotations of Digital Retinal Images. Transl Vis Sci Technology 2016;5:6. doi: 10.1167/tvst.5.5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Brady CJ, Villanti AC, Pearson JL, Kirchner TR, Gupta OP, Shah CP. Rapid grading of fundus photographs for diabetic retinopathy using crowdsourcing. J Med Internet Res 2014;16:e233. doi: 10.2196/jmir.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Russell BC, Torralba A, Murphy KP, Freeman WT. LabelMe: A Database and Web-Based Tool for Image Annotation. Int J Comput Vision 2008;77:157–73. doi: 10.1007/s11263-007-0090-8. [DOI] [Google Scholar]
- [92].Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. Int J Comput Vision 2016;128:336–59. doi: 10.1007/s11263-019-01228-7. [DOI] [Google Scholar]
- [93].Rim TH, Soh ZD, Tham Y-C, Yang HHS, Lee G, Kim Y, et al. Deep learning for automated sorting of retinal photographs. Ophthalmol Retin 2020. doi: 10.1016/j.oret.2020.03.007. [DOI] [PubMed] [Google Scholar]
- [94].Varadarajan AV, Bavishi P, Ruamviboonsuk P, Chotcomwongse P, Venugopalan S, Narayanaswamy A, et al. Predicting optical coherence tomography-derived diabetic macular edema grades from fundus photographs using deep learning. Nat Commun 2020;11:130. doi: 10.1038/s41467-019-13922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Voets M, Møllersen K, Bongo LA. Reproduction study using public data of: Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Plos One 2019;14:e0217541. doi: 10.1371/journal.pone.0217541.(*) (*) In publishing on inferior performance metrics, this paper highlights the need for increasing publication of both data and code to facilitate scientific reproducibility.
- [96].Liefers B, Colijn JM, González-Gonzalo C, Verzijden T, Wang JJ, Joachim N, et al. A Deep Learning Model for Segmentation of Geographic Atrophy to Study Its Long-Term Natural History. Ophthalmology 2020. doi: 10.1016/j.ophtha.2020.02.009. [DOI] [PubMed] [Google Scholar]
- [97].Fda/cder. Clinical Trial Imaging Endpoint Process Standards Guidance for Industry 2018.
- [98].Bogunovic H, Ciller C, Gopinath K, Gostar AK, Jeon K, Ji Z, et al. RETOUCH: The Retinal OCT Fluid Detection and Segmentation Benchmark and Challenge. Ieee T Med Imaging 2019;38:1858–74. doi: 10.1109/tmi.2019.2901398. [DOI] [PubMed] [Google Scholar]
- [99].Islam and Y-C (Jack) LHCYBAWTNPMdM. Artificial Intelligence in Ophthalmology: A Meta-Analysis of Deep Learning Models for Retinal Vessels Segmentation. J Clin Medicine 2020;9:1018. doi: 10.3390/jcm9041018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Faes L, Liu X, Wagner SK, Fu DJ, Balaskas K, Sim DA, et al. A Clinician’s Guide to Artificial Intelligence: How to Critically Appraise Machine Learning Studies. Transl Vis Sci Technology 2020;9:7. doi: 10.1167/tvst.9.2.7. [DOI] [PMC free article] [PubMed] [Google Scholar]