Several enteric pathogens, including enterohemorrhagic E. coli (EHEC) strains, require CvpA to robustly colonize the intestine. This inner membrane protein is also important for secretion of a colicin and for EHEC resistance to the bile salt deoxycholate (DOC), but its function is unknown.
KEYWORDS: E. coli, EHEC, bacterial genetics, bile stress, envelope stress, extracytoplasmic stress, inner membrane protein, sigma factor
ABSTRACT
The function of cvpA, a bacterial gene predicted to encode an inner membrane protein, is largely unknown. Early studies in Escherichia coli linked cvpA to colicin V secretion, and recent work revealed that it is required for robust intestinal colonization by diverse enteric pathogens. In enterohemorrhagic E. coli (EHEC) strains, cvpA is required for resistance to the bile salt deoxycholate (DOC). Here, we carried out genome scale transposon insertion (TIS) mutagenesis and spontaneous suppressor analysis to uncover the genetic interactions of cvpA and identify common pathways that rescue the sensitivity of a ΔcvpA EHEC mutant to DOC. These screens demonstrated that mutations predicted to activate the σE-mediated extracytoplasmic stress response bypass the ΔcvpA mutant’s susceptibility to DOC. Consistent with this idea, we found that deletions in rseA and msbB and direct overexpression of rpoE restored DOC resistance to the ΔcvpA mutant. Analysis of the distribution of CvpA homologs revealed that this inner membrane protein is conserved across diverse bacterial phyla in both enteric and nonenteric bacteria that are not exposed to bile. Together, our findings suggest that CvpA plays a role in cell envelope homeostasis in response to DOC and similar stress stimuli in diverse bacterial species.
IMPORTANCE Several enteric pathogens, including enterohemorrhagic E. coli (EHEC) strains, require CvpA to robustly colonize the intestine. This inner membrane protein is also important for secretion of a colicin and for EHEC resistance to the bile salt deoxycholate (DOC), but its function is unknown. Genetic analyses carried out here showed that activation of the σE-mediated extracytoplasmic stress response restored the resistance of a cvpA mutant to DOC, suggesting that CvpA plays a role in cell envelope homeostasis. The conservation of CvpA across diverse bacterial phyla suggests that this membrane protein facilitates cell envelope homeostasis in response to varied cell envelope perturbations.
INTRODUCTION
Enteric pathogens encounter a broad range of host-derived stressors during their transit through the gastrointestinal (GI) tract. These pathogens enter the GI tract through consumption of contaminated food or water, and the diverse challenges they face in the host environment include substantial fluctuations in osmolarity, oxygen concentration, pH, and nutrient availability, as well as mechanical shear force from peristalsis and the bactericidal activities of antimicrobial peptides and bile salts. Pathogens must also compete with the microbiota and face the threat of immune cells and effectors (1, 2). To survive and successfully colonize the host, enteric pathogens must rapidly sense and respond to these stressors.
Bile in particular poses a complex threat to enteric pathogens. Bile is an aqueous solution of bile salts, bilirubin, fats, and inorganic salts produced by the liver and secreted into the intestine, where it plays a critical role in the digestion of fats (3, 4). Bile salts are potent antimicrobial compounds that can damage diverse components of the bacterial cell. Bile salts are thought to enter the bacterial cell both by passive diffusion and through outer membrane proteins (OMPs) such as porins (5, 6). Enteric pathogens employ a variety of strategies to combat the perturbations caused by bile salts, including utilizing efflux pumps to remove bile salts from the cell cytosol and expressing chaperones and proteases to ameliorate protein folding stress (7–9).
To coordinate responses to bile, bacteria utilize a host of global stress response programs. For example, bile-induced DNA damage activates the expression of the SOS regulon, a large suite of genes that assist in repairing and responding to DNA damage (10–12). Bile also triggers the activation of alternate sigma factors, which enable RNA polymerase (RNAP) to selectively transcribe sets of genes in response to environmental conditions. During normal growth conditions, RNAP associates with the housekeeping sigma subunit, RpoD (σ70). The cellular stresses provoked by bile promote utilization of the alternative stress response sigma factor RpoS (σS), which governs the expression of a large number (>500) of stress response genes (13–15). RpoE (σE), a sigma factor which mediates the response to “extracytoplasmic” stress, such as misfolded proteins in the periplasmic space or disruptions in lipopolysaccharide (LPS) biosynthesis, may also be important for resistance to the membrane-damaging effects of bile (16, 17). σE function intersects with a variety of other cell envelope stress response pathways in Gram-negative bacteria, such as the two-component systems CpxRA and BaeSR; together, these pathways create a complex regulatory network mediating bacterial envelope homeostasis (16, 18).
The gene cvpA was identified in screens for genes required for intestinal colonization in several enteric pathogens, including Vibrio cholerae, Vibrio parahaemolyticus, and Salmonella enterica subsp. enterica serovar Typhimurium (19–21), but the mechanism(s) that accounts for the colonization defects has not been characterized. We recently found that CvpA is required for the foodborne intestinal pathogen enterohemorrhagic Escherichia coli (EHEC) O157:H7 to optimally colonize the colon of infant rabbits; moreover, EHEC cvpA deletion mutants were highly sensitive to bile and in particular to the bile salt deoxycholate (DOC) (22). EHEC causes hemorrhagic colitis and, in some cases, the severe complication hemolytic uremic syndrome, a condition in which damage to blood vessels of the kidney microvasculature leads to renal failure. Previous studies have led to the idea that EHEC strains use bile as a cue to induce expression of virulence genes, flagella, and genes promoting survival in vivo, including the AcrAB efflux pump, which enables removal of bile salts from the cell interior (12, 23–29), but the role of CvpA in contributing to bile resistance in EHEC strains is unknown.
cvpA was originally described as a gene required for the production of colicin V (ColV), a small peptide antibiotic produced by some strains of pathogenic E. coli (30). Although the ColV secretion system has been characterized (31), the specific role for CvpA in this mechanism was not identified. Here, we provide evidence that mutations in genes linked to activation of the σE stress response pathway can bolster the ΔcvpA mutant’s fitness in DOC. We also show that CvpA is widely conserved across diverse bacterial phyla and hypothesize that its function is linked to bacterial cell envelope homeostasis.
RESULTS
CvpA localizes to the EHEC cell periphery.
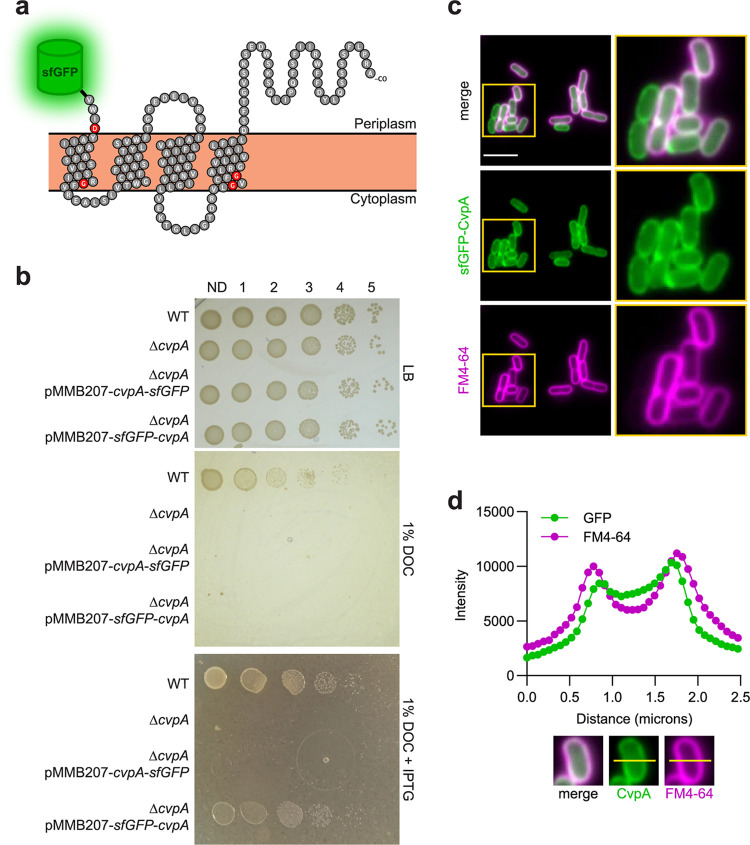
CvpA bears structural similarity to the inner membrane mechanosensitive ion channels MscS and MscL (HHPred and Phyre2 algorithms) (22), and whole-proteome localization analysis in E. coli assigned CvpA to the inner membrane (32). Furthermore, several protein modeling algorithms (PSLPred, HHPred, Phobius, Phyre2, and SPOCTOPUS) (22) predict that CvpA is a 4-pass inner membrane protein with a periplasmic C terminus (Fig. 1a). To experimentally verify these assignments, we generated two plasmids that inducibly express superfolder green fluorescent protein (sfGFP)-CvpA fusions, with sfGFP on either the N or C terminus, enabling visualization of the localization of CvpA in EHEC. Only the sfGFP fusion to the N terminus of CvpA yielded a construct that complemented the growth defect of an EHEC strain lacking the cvpA gene (ΔcvpA) on plates containing DOC, implying that this construct contains a functional CvpA protein (Fig. 1b). Induction of this fusion protein revealed a clear signal that outlined the EHEC cell membrane in a similar pattern to that shown by the membrane stain FM4-64 (Fig. 1c and d), supporting the predicted localization of CvpA.
FIG 1.
CvpA localizes to the EHEC cell periphery. (a) Schematic of the predicted CvpA topology (based on reference 22), with location of N-terminal superfolder green fluorescent protein (sfGFP) fusion shown. Red residues are nearly 100% conserved across bacterial phyla. (b) Dilution series of wild type (WT), ΔcvpA mutant, and ΔcvpA mutant with an isopropyl-β-d-1-thiogalactopyranoside (IPTG)-inducible sfGFP-cvpA fusion complementation plasmid plated on LB, LB 1% deoxycholate (DOC), and LB 1% DOC 1 mM IPTG. (c) Micrographs of an N-terminal sfGFP-CvpA fusion protein expressed in EHEC and stained with the FM4-64 membrane dye. Bar, 5 μm. (d) Horizontal line scan of GFP and FM4-64 signal.
EHEC cvpA mutants do not secrete functional ColV and are sensitive to the bile salt DOC.
A previous report suggested that E. coli CvpA is required for the export of the peptide antibiotic colicin V (ColV) (30). We sought to reproduce this observation in EHEC. The entire ColV production and immunity operon and endogenous promoters derived from the naturally occurring ColV plasmid, pColV-K30, were inserted into the cloning vector pBR322, yielding pBR322-ColV. Although both the wild-type (WT) and ΔcvpA EHEC strains were transformable with pBR322-ColV, only the WT strain grew robustly in LB broth while carrying pBR322-ColV. In contrast, the ΔcvpA mutant elongated and lysed (see Fig. S1A in the supplemental material). This phenotype was not observed in a ΔcvpA mutant carrying an empty vector plasmid (Fig. S1A). The abilities of these strains to secrete functional ColV were evaluated by testing whether they create a zone of growth inhibition on a sensitive indicator strain. The WT strain carrying pBR322-ColV killed the indicator strain, creating a distinct zone of clearing (Fig. S1B), whereas the ΔcvpA strain carrying the same plasmid failed to establish such a zone (Fig. S1B). These data are consistent with previous reports (30) and suggest that ΔcvpA mutants are unable to secrete functional ColV and may be sensitive to self-intoxication.
Our previous work revealed that CvpA is required for EHEC to robustly colonize the infant rabbit colon and that ΔcvpA mutants are sensitive to the bile salt DOC but not to the bile salt cholate (CHO) (22). Bile salts like DOC are thought to cause a wide range of deleterious effects in bacteria, including damage to the cell envelope and DNA, and widespread protein misfolding and redox stress (5, 6). The exact mechanisms through which bile salts mediate these diverse deleterious effects are not completely understood (5, 6). We attempted to determine which aspect of DOC toxicity was most damaging to ΔcvpA mutants by testing a variety of agents that cause individual aspects of bile stress, including the membrane-damaging detergent SDS, DNA-damaging antibiotics, cell wall-perturbing antibiotics, protein synthesis-disrupting antibiotics, and the redox stress-inducing compounds diamide and hydrogen peroxide (22) (Table 1). However, the ΔcvpA mutant was not more sensitive than the WT strain to any of these agents. These observations argue against the idea that the sensitivity of the ΔcvpA mutant to DOC results from a general cell envelope defect and instead suggest that DOC toxicity (and potentially the toxicity of ColV accumulation) reflects a more specific defect in cells lacking cvpA. Below, we moved forward with DOC as a tool to probe the function of CvpA.
TABLE 1.
MIC values for various compounds against WT and ΔcvpA EHEC
| Compound (concn) | Type of stress caused | MIC against: |
|
|---|---|---|---|
| WT | ΔcvpA EHEC | ||
| DOC (%) | Multiple | 2.5 | 0.08 |
| Diamide (mM) | Disulfide bond formation | 0.6 | 0.6 |
| H2O2 (mM) | Reactive oxygen species | 3.2 | 3.2 |
| Ampicillina (μg/ml) | Cell wall damage | 7.8 | 7.8 |
| Piperacillina (μg/ml) | Cell wall damage | 0.3 | 0.3 |
| SDSa (%) | Detergent | 5 | 5 |
| Polymyxin Ba (μg/ml) | Detergent | 1.2 | 1.2 |
| Nalidixic acida (μg/ml) | DNA damage | 2.5 | 2.5 |
Compound originally presented in reference 22 and reproduced here for comparison.
Genetic screens link CvpA to the σE response.
To further assess the function of CvpA, we carried out three genetic screens in EHEC strains to define the cvpA gene’s network of genetic interactions. Two of the screens relied on transposon insertion (TIS) mutagenesis, and one used spontaneous suppressor analysis. Similar genetic approaches have been fruitful for other membrane proteins (33–36), which are often difficult to probe using biochemical approaches to identify protein-protein interactions.
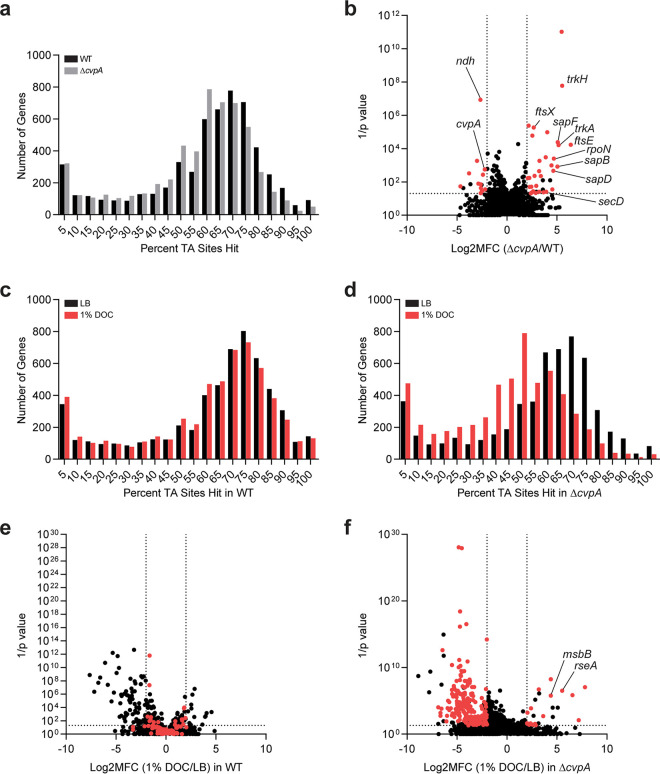
For the transposon screens, we began by generating high-density transposon insertion mutant libraries in the WT and ΔcvpA EHEC strains by mutagenizing the strains with the mariner transposon Himar1, which inserts randomly at the TA dinucleotide. The abundance of each transposon insertion mutant in the two libraries was quantified by deep sequencing the sites of transposon insertion. Genes were binned by the percentage of TA sites disrupted (Fig. 2a) and, as expected for libraries of high insertion density, this distribution was bimodal (37). The leftmost minor peak represents genes with few to no insertions, and the rightmost major peak represents genes with a majority of TA sites disrupted (37). The distributions of transposon insertions in the WT and ΔcvpA libraries were similar and displayed complexity, enabling high-resolution analysis of transposon insertion frequency (19) (Fig. 2a).
FIG 2.
Transposon insertion sequencing screens reveal CvpA’s genetic interactions. (a) Distribution of the percentage of TA sites disrupted for all genes in the library constructed in the WT and ΔcvpA backgrounds. (b) Volcano plot depicting the relative abundance of read counts mapped to individual genes in transposon libraries in WT and ΔcvpA mutant backgrounds. The mean log2 fold change (Log2MFC) and Mann-Whitney U P value are shown for each gene. Genes highlighted in red have more than 5 unique transposon mutants, a log2FC of >2 or <−2, and a P value of <0.05. A modified ΔcvpA locus can sustain insertions at the 1 TA dinucleotide in the stop codon. (c and d) Distribution of the percentage of TA sites disrupted for all genes in the library constructed in the WT (c) or ΔcvpA background (d) after growth in LB or 1% DOC. (e and f) Volcano plot depicting the relative abundance of read counts mapped to individual genes in transposon libraries in WT (e) or ΔcvpA background (f) in LB versus in 1% DOC. The log2 mean fold change and Mann-Whitney U P value are shown for each gene. Genes highlighted in red are those for which the mean log2FC is >2 or <−2 and the P value is <0.05 in the ΔcvpA mutant background but not in the WT background.
We first completed a synthetic screen in which we compared the distributions of the transposon insertions across the genome in the WT and ΔcvpA libraries. This comparison yields the following three categories of genes: (i) loci with similar insertion profiles in both backgrounds that are deemed “neutral,” (ii) loci with fewer insertions in the ΔcvpA versus the WT background that are considered “synthetic sick,” and (iii) loci with more insertions in the ΔcvpA compared to the WT background that are deemed “synthetic healthy.” This approach has been used to identify genetic interactions in several other organisms (38–41). We identified 32 candidate synthetic healthy genes for which transposon insertion mutants were overrepresented at least 4-fold in the ΔcvpA mutant background compared to the WT (Fig. 2b; see also Table S1 in the supplemental material). Several of these genes were linked to ion transport systems, including trkH and trkA, which are components of the cellular potassium uptake system, and three genes of the sap transporter complex (sapB, sapD, and sapF). SapD is also known to interact with the Trk potassium transport system (42). Several chaperones, including secB, the alternative sigma factor rpoN, and several cell shape/septal structure genes, including ftsEX, were also on the list of synthetic healthy genes. A total of 14 candidate synthetic sick genes, which were underrepresented at least 4-fold in the ΔcvpA mutant background compared to the WT (Fig. 2b and Table S1), were also identified. Most of these genes encode proteins of unknown function, but they include ndh, which encodes NADH dehydrogenase II (NDH-2), a redox protein that catalyzes electron transfer (43, 44). Collectively, these results suggest that disrupting ion or electron homeostasis can alter the fitness of the ΔcvpA strain.
A second transposon screen was designed to identify insertion mutations in genes that suppress the DOC sensitivity phenotype in the ΔcvpA mutant. The WT and ΔcvpA transposon insertion mutant libraries were screened on plates containing either LB or LB supplemented with 1% DOC. As above, after enumerating the abundance of transposon insertion mutants with sequencing, genes were binned according to the percentage of disrupted TA sites (Fig. 2c and d). The distribution of disrupted TA sites per gene in the WT library was not markedly different between the LB and DOC selection, as expected because a majority of mutants do not have a phenotype in DOC (Fig. 2c). However, the distributions in the ΔcvpA library were different between the two conditions; there was a leftward shift in the distribution of insertions when the ΔcvpA library was exposed to DOC versus that in LB (Fig. 2d). This leftward shift is indicative of a bottleneck, a stochastic, genotype-independent constriction of population size (37), and reflects the profound DOC sensitivity of the ΔcvpA mutant.
To identify mutants in both libraries that have markedly better or worse phenotypes than expected in DOC, we used an analytical pipeline that mitigates the effects of bottlenecks to identify differentially abundant mutants (19). For each library, the abundances of each mutant were compared between the LB and DOC conditions to identify the following three categories of genes: (i) loci with similar insertion profiles in both LB and DOC are “neutral,” (ii) loci with fewer insertions in the DOC condition than expected compared to those in LB are “depleted,” and (iii) loci with more insertions in the DOC condition than expected compared to those in LB are “enriched” (Fig. 2e and f). In the ΔcvpA background, we identified 233 “depleted” genes in which mutations are expected to increase baseline DOC sensitivity, and 26 “enriched” genes in which mutations are expected to suppress DOC sensitivity (Fig. 2f). Nineteen of these 26 genes did not show differential abundance in DOC versus LB comparison in the WT background and were classified as “neutral” (Fig. 2e and f). These genes are of particular interest as candidate suppressors, as they have cvpA-specific DOC phenotypes. Many of these genes were linked to stress response pathways, redox processes, and cell envelope integrity, and they include the anti-sigma factor gene rseA, the σE-regulated periplasmic chaperone gene skp, the two-component system genes basSR, the redox genes sodC and cyoC, the disulfide/redox bond formation genes dsbA and dsbB, and the cell membrane synthesis genes msbB, wcaM, and rfaC.
In addition to transposon-based screens, we also characterized spontaneous suppressors of DOC sensitivity in the ΔcvpA mutant. In this screen, a broth culture of the ΔcvpA mutant was plated on 1% DOC, and putative suppressor colonies were picked. After repatching on 1% DOC, which confirmed their resistance, whole-genome sequencing of the suppressors was carried out, and these genome sequences were compared to that of the ΔcvpA parent strain to identify putative suppressor mutations. Single-nucleotide polymorphisms (SNPs) or deletions that were not present in the ΔcvpA parent strain were found in 14 of the 16 sequenced strains. Notably, 12 of the 14 strains had mutations in genes linked to LPS biogenesis, including several genes in the lipopolysaccharide transport (LPT) system (lptB, lptE, lptF, and lptG) and the gene for an enzyme required for lipid A synthesis, msbB; genes related to redox, including nuoH and cyoDC, were also identified (Table 2). Interestingly, msbB was also identified as a candidate suppressor in the transposon DOC screen described above. LPT genes are essential and thus are not highly represented in our transposon libraries. Although it was not statistically robust, we analyzed the transposon DOC screen data for the LPT genes with at least 1 insertion mutant and found that lptCDFG had large, positive fold changes in 1% DOC versus LB in the ΔcvpA background but not in the WT (see Table S2 in the supplemental material), supporting the findings of the spontaneous suppressor screen.
TABLE 2.
Mutations in suppressors of ΔcvpA DOC sensitivity
| Suppressor | Locus | Gene | Genome position | Nucleotide change | Amino acid changea | Mutationb | Functionc |
|---|---|---|---|---|---|---|---|
| 1 | RS00350 | araC | 75253 | A 20 C | Asp 7 Ala | SNP | Arabinose operon transcriptional regulator |
| RS02475 | yajR | 511698 | G 1022 A | Gly 341 Asp | SNP | Putative transport protein | |
| RS13840 | msbB | 2635593 | G 894 A | Trp 298 * | SNP | Lipid A biosynthesis acyltransferase | |
| 2 | RS16885 | nuoH | 3209841 | T 778A | Trp 260 Arg | SNP | NADH-quinone oxidoreductase subunit |
| RS27795 | lptG | 5391979 | G 187 A | Asp 63 Asn | SNP | LPS export ABC transporter permease | |
| 3 | RS16885 | nuoH | 3209841 | T 778 A | Trp 260 Arg | SNP | NADH-quinone oxidoreductase subunit |
| RS21750 | lptB | 4174645 | T 722 G | Leu 241 Arg | SNP | LPS export ABC transporter ATP-binding protein | |
| 4 | RS13840 | msbB | 2635854 | T 633 G | Asp 211 Glu | SNP | Lipid A biosynthesis acyltransferase |
| 5 | RS21750 | lptB | 4174548 | A 625 C | Ile 209 Leu | SNP | LPS export ABC transporter ATP-binding protein |
| 6 | RS27795 | lptG | 5392372 | A 580 C | Thr 194 Pro | SNP | LPS export ABC transporter permease |
| 7 | RS27795 | lptG | 5391946 | G 154 C | Ala 52 Pro | SNP | LPS export ABC transporter permease |
| 8 | Intergenic | Upstream lptE | 753329 | C to T | SNP | LPS assembly lipoprotein | |
| RS02485 | cyoD | 513798 | T 302 del | Asn 101 fs | DEL | Cytochrome bo terminal oxidase complex subunit IV | |
| 9 | RS13840 | msbB | 2636269–2636276 | CAGAGCGA 211–218 del | Ser 71 fs | DEL | Lipid A biosynthesis acyltransferase |
| 10 | RS16885 | nuoH | 3209841 | T 778 A | Trp 260 Arg | SNP | NADH-quinone oxidoreductase subunit |
| RS27795 | lptG | 5391979 | G 187 A | Asp 63 Asn | SNP | LPS export ABC transporter permease | |
| 11 | RS27790 | lptF | 5390961 | C 269 A | Ala 90 Asp | SNP | LPS export ABC transporter permease |
| 14 | RS12645 | dtpA | 2407894 | G 938 C | Gly 313 Ala | SNP | Proton-dependent di/tripeptide symporter |
| RS02495 | cyoB | 516208 | G 487 A | Gly 163 Ser | SNP | Cytochrome bo3 ubiquinol oxidase subunit | |
| 15 | RS13840 | msbB | 2635801 | C 686 A | Thr 229 Lys | SNP | Lipid A biosynthesis acyltransferase |
| RS16885 | nuoH | 3209841 | T 778 A | Trp 260 Arg | SNP | NADH-quinone oxidoreductase subunit | |
| 16 | RS12645 | dtpA | 2407894 | G 938 C | Gly 313 Ala | SNP | Di-/tripeptide permease |
| RS02495 | cyoB | 516208 | G 487 A | Gly 163 Ser | SNP | Cytochrome bo3 ubiquinol oxidase subunit |
Asterisk, stop codon; fs, frameshift.
SNP, single-nucleotide polymorphism; DEL, deletion mutation.
LPS, lipopolysaccharide.
Mutations linked to σE activation and direct expression of rpoE rescue the sensitivity of the ΔcvpA mutant to DOC.
Several independent pieces of evidence suggesting that activation of the σE response could bypass the sensitivity of the ΔcvpA mutant to DOC were found in the 3 genetic screens described above. σE is normally sequestered at the inner membrane by association with an anti-sigma factor, RseA. When the appropriate stimuli are detected, proteases YaeL and DegS sequentially cleave RseA, liberating σE to activate expression of ∼100 genes in response (45, 46). A variety of perturbations to the cell envelope can trigger this pathway, including misfolded OMPs, disruptions in LPS biosynthesis leading to precursor accumulation, and changes in cellular redox state (47–52). Notably, the genetic screens revealed a variety of mutations, including in rseA, lptBCDEFG, and msbB, that likely trigger the σE response, which appeared to rescue the sensitivity of ΔcvpA to DOC. Inactivating mutations in rseA increase the basal activity of σE by preventing its membrane sequestration, and mutations in LPS biosynthesis genes msbB and the LPS transport system trigger σE through accumulation of LPS precursors (48, 49).
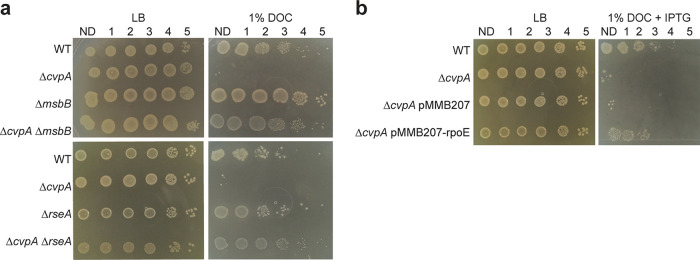
To directly test whether mutations known to activate σE rescue the ΔcvpA mutant’s sensitivity to DOC, deletion mutants of rseA and msbB in the WT and ΔcvpA backgrounds were created. These deletions both restored the resistance of the ΔcvpA mutant to 1% DOC to WT levels (Fig. 3a). Similarly, direct overexpression of rpoE from a plasmid rescued the sensitivity of the ΔcvpA mutant to DOC (Fig. 3b). Thus, activation of σE, and presumably of its downstream regulon of genes that mediate cell envelope repair, circumvents the deleterious effects that DOC causes to the ΔcvpA mutant.
FIG 3.
Activation of RpoE rescues the ΔcvpA mutant’s sensitivity to DOC. (a) Dilution series of WT, ΔcvpA, ΔcvpA ΔmsbB, and ΔcvpA ΔrseA strains plated on LB or LB–1% deoxycholate (DOC). (b) Dilution series of WT, ΔcvpA, ΔcvpA strains transformed with empty expression vector pMMB207 and the ΔcvpA strain transformed with pMMB207 carrying IPTG-inducible rpoE plated on LB and LB–1% DOC with 0.1 mM IPTG.
CvpA is highly conserved across bacterial phyla.
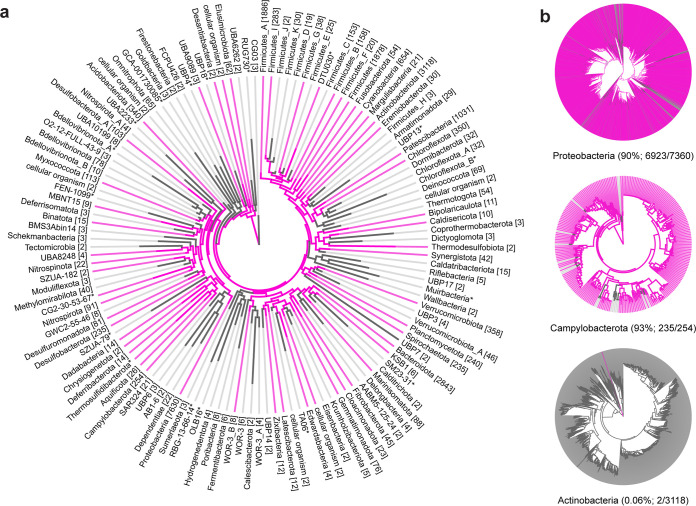
The phylogeny tool AnnoTree (53) was used to analyze the distribution of CvpA homologs across more than 27,000 bacterial genomes. CvpA homologs (≥30% amino acid identity and ≥70% alignment) were found in 9,830 genomes distributed across 55 phyla (Fig. 4a; see also Table S3 in the supplemental material). Homologs are present in the majority of represented genomes in the Campylobacterota (236/254, 93%), Deferribacterota (13/14, 93%), and Proteobacteria (6,923/7,630, 91%) phyla (Fig. 4b). In contrast, only 2 of 3,118 strains in Actinobacteria have a CvpA homolog (0.06%) (Fig. 4b).
FIG 4.
CvpA is highly conserved across many bacterial phyla. (a) Phylogeny of CvpA distribution across bacterial phyla. Pink lines represent phyla where at least one species has a CvpA homolog with at least 30% amino acid identity and at least 70% alignment to E. coli CvpA. The numbers shown in brackets denote the number of individual genomes in that phylum. (b) Phyla with various degrees of CvpA conservation at the species level. Pink lines represent species that contain a CvpA homolog. Numbers in parentheses after phylum names indicate the percentages of species in those phyla with a CvpA homolog.
Sequence alignment of the primary amino acid sequences of 20 CvpA homologs from bacterial species from diverse phyla revealed several highly conserved residues, including an aspartate and three glycines with nearly 100% conservation (see Fig. S2 in the supplemental material; highlighted in red in Fig. 1a). The motif GXXXG, which is common in transmembrane segments and indicative of helix-helix interactions (54), is also present. All 20 CvpA homologs are predicted by PSLPred to localize to the inner membrane. All homologs share greater similarity in alignments of their respective N-terminal regions and have more variable lengths in their final periplasmic segments, which could reflect different functions or binding partners at the C terminus.
This conservation across diverse phyla highlights that the function of CvpA is not strictly related to bile. While homologs are present in enteric pathogenic and commensal strains, such as Campylobacter jejuni, Yersinia enterocolitica, Fibrobacter succinogenes, and Bacteroides fragilis, many strains with a CvpA homolog inhabit nonintestinal niches. These strains include nonenteric pathogens such as Neisseria meningitidis, plant symbionts such as Rhizobium tropici, and nonpathogenic environmental isolates from diverse environments such as tundra soil (Granulicella tundricola), hydrothermal vents (Thermovibrio ammonificans), and marine environments (Spirochaeta cellobiosiphila).
DISCUSSION
Since the original report describing cvpA as a gene required for production of the plasmid-borne ColV peptide from E. coli more than 30 years ago (30), very little progress has been made determining the function of this widely conserved inner membrane protein. More recently, cvpA has been found to be important for intestinal colonization of Vibrio cholerae, V. parahaemolyticus, Salmonella Typhimurium, and EHEC in studies based on in vivo transposon screens (19–22). In EHEC, we found that a ΔcvpA mutant was highly sensitive to DOC (22), a bile component, thus presenting a possible explanation for the role of cvpA role in the colonization of distinct niches in the small and large intestine by diverse enteric pathogens. Here, we used genetic screens to further our understanding of cvpA function. These screens suggested that activation of the σE extracytoplasmic stress response pathway could restore the resistance of the ΔcvpA mutant to DOC, and this hypothesis was experimentally confirmed. Our findings link cvpA to cell envelope homeostasis. However, the relationship between cvpA and the σE pathway requires further elucidation. CvpA itself could be an upstream factor that can trigger the σE pathway; alternatively, expression of downstream components of the σE regulon may compensate for the absence of cvpA without a direct functional link between cvpA and rpoE.
A variety of cellular perturbations can activate the σE response. Misfolded outer membrane proteins, disruptions in LPS biogenesis, and disruptions in redox state have all been linked to increased σE activity (47–50). Mutations in the regulators of these pathways, such as inactivation of rseA, the anti-sigma factor which controls σE activity (47); msbB, an enzyme required for the biosynthesis of lipid A (48); or redox enzymes such as dsbAB or sodC (50–52), can also trigger the activation of σE in the absence of external stimuli. Mutations in genes involved in the β-barrel assembly machinery (Bam) also activate σE due to an accumulation of mislocalized (periplasmic) OMPs (16, 55). Many genes identified in our screens as candidate modulators of the fitness of ΔcvpA in DOC are linked to these σE-activating pathways (Fig. 2 and Table 2).
One potential model for these observations is that CvpA is required to activate the σE response when cells are exposed to DOC. In a ΔcvpA mutant, this pathway is unable to respond to deleterious stimuli, and additional σE -activating mutations are required to bypass this block (Table 2 and Fig. 3). Although we did not measure σE activity directly, we found that deletion of rseA, LPS biosynthetic gene mutations, and direct overexpression of rpoE could rescue the ΔcvpA mutant’s DOC sensitivity. Although redox mutants were identified in our screens (cyoBCD, nuoH, sodC, and dsbAB), we found that clean deletions in a subset of these genes did not suppress the ΔcvpA mutant’s DOC sensitivity. Notably, we did not identify any mutations related to the Bam machinery that suppressed the ΔcvpA mutant’s DOC sensitivity, raising the possibility that CvpA function is limited to cellular responses to LPS damage/defects.
Another possibility is that CvpA is required for envelope homeostasis during DOC stress but is functionally unrelated to the σE response. In this scenario, increased σE activity provides the ΔcvpA cell membrane with the “resiliency” required to survive DOC-induced stress. Activation of σE induces expression of several proteases and chaperones which could repair the DOC-damaged envelope, and other effector proteins, like PtsN, that reduce overall envelope stress (56). Other mechanisms that could promote envelope resiliency during DOC stress may include eliminating the assembly of large, energetically costly transmembrane complexes such as the electron transport chain (57, 58). The respiratory complex NDH-I is one of the largest protein complexes in the E. coli membrane, and downregulation of nuo genes and components of the partner cytochrome bo3 cyo genes have been shown to reduce envelope stress (16). Many of the spontaneous suppressors we identified had mutations in components of these structures (Table 2). Furthermore, mutations in respiratory complex protein NDH-2 identified in the synthetic lethal transposon screen were predicted to decrease the fitness of the ΔcvpA mutant, which would make the cell more reliant on envelope-destabilizing NDH-1 (57, 58). Interestingly, these redox mutations were often identified in suppressors that also carried mutations in LPS biosynthesis genes. This may explain why deletions in individual redox genes were not sufficient to rescue the ΔcvpA mutant’s DOC sensitivity and reflect that multiple mechanisms of buttressing envelope resiliency promote survival under these conditions.
The conservation of CvpA across diverse bacterial phyla, including in species which are never exposed to bile, suggests that CvpA function is not restricted to responding to bile stress (Fig. 4). Bile can impair several processes in the bacterial cell, including inflicting damage to the cell envelope and DNA and generating protein folding stress and alterations in redox state (5, 6). We were unable to isolate which individual aspect of DOC stress kills ΔcvpA mutants (Table 1), raising the possibility either that these deleterious effects need to occur together or that bile impairs additional processes that were not tested. One possibility is that bile, and DOC in particular, perturbs ion homeostasis, which could in turn disrupt membrane potential. The increased fitness of insertions in several genes associated with K+ transport (Fig. 2b) links cvpA and ion transport processes and supports this idea. Furthermore, in eukaryotic cells, DOC exposure leads to changes in mitochondrial membrane potential (59). Interestingly, ΔcvpA mutants are not sensitive to the very similar bile salt cholate (CHO) (22), which may reflect a difference in the ability of these salts to move through membranes (60) or the increased hydrophobicity of DOC (61).
Given the similarity of CvpA to the mechanosensitive solute transporters MscS and MscL, and the numerous genes related to ion transport that were identified in the synthetic lethal screen (Fig. 2b), it is tempting to speculate that CvpA is itself an ion channel that monitors and promotes membrane potential homeostasis. Notably, colicin V mediates its bactericidal action by disrupting the membrane potential of the inner membrane (62, 63). If ΔcvpA mutants have heightened sensitivity to perturbations of membrane potential, this could explain why they cannot produce functional colicin V. Maintenance of ion homeostasis and membrane potential is a critical function for all bacterial species. DOC may represent a stimulus that leads to perturbation of ion levels; other stimuli that lead to similar perturbations are likely encountered in the disparate niches inhabited by the diverse bacterial species encoding CvpA homologs. Clearly, additional biochemical studies to elucidate the mechanistic bases of CvpA function are warranted.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains were cultured in LB medium or on LB agar plates at 37°C. Antibiotics and supplements were used at the following concentrations: 20 μg/ml chloramphenicol (Cm), 50 μg/ml kanamycin (Km), 10 μg/ml gentamicin (Gm), 50 μg/ml carbenicillin (Cb), 0.3 mM diaminopimelic acid (DAP), 1% deoxycholate (DOC), 0.1 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG).
A gentamicin-resistant mutant of E. coli O157:H7 strain EDL933 (ΔlacI::aacC1) (22) was used in all experiments in this study as the wild type (WT). All mutants were constructed in this strain background using standard allelic exchange techniques (64) with the pTOX plasmid system (65) or lambda red recombineering (66).
Colicin V production and immunity operons and endogenous promoters were cloned from plasmid pColV-K30::Tn10 (a gift from Roberto Kolter) into pBR322 (yielding pBR322-ColV) using isothermal assembly. The resulting plasmids were transformed into WT or ΔcvpA strains.
To construct inducible N- or C-terminal sfGFP fusions to CvpA, the sfGFP sequence was amplified from pBad-sfGFP (Addgene plasmid no. 85482) and linked to the cvpA sequence amplified from EHEC genomic DNA with the linker sequence GCAGCGGCCGGCGGAGGG through isothermal assembly into pMMB207’s (67) (ATCC no. 37809) multiple cloning site (MCS). Similarly, to construct the rpoE expression plasmid, the rpoE sequence was amplified from genomic DNA and cloned into the pMMB207 MCS. The resulting plasmids were transformed into WT or ΔcvpA strains by electroporation.
ColV production assay.
pBR322-ColV was transformed into WT or ΔcvpA strains. Individual colonies of either strain were selected and grown overnight in LB-Carb and then normalized to an optical density at 600 nm (OD600) of 0.5. To prepare indicator plates, 100 μl of WT EHEC was added to 5 ml of 0.8% molten LB agar, vortexed to mix, and then poured quickly onto prewarmed LB plates, which were allowed to solidify undisturbed. Five microliters of OD600 0.5 ColV-producing strain (WT or ΔcvpA strain transformed with pBR322-ColV) was jabbed into soft agar plate and incubated upright overnight at 37°C. The next day, plates were examined for zones of clearing around the ColV-producing strain.
DOC sensitivity assay.
To determine sensitivity to DOC, each strain was grown at 37°C until the mid-exponential phase (OD600 = 0.5). For inducible expression, at the mid-exponential phase, IPTG was added to a final concentration of 1 mM, and cells were incubated at 37°C shaking for 1 h. Cultures were normalized to an OD600 of 0.5, serially diluted, and plated onto LB agar plates with and without 1% DOC and 0.1 mM IPTG.
Microscopy.
To prepare sfGFP-CvpA fusion cells for microscopy, strains were grown to the mid-exponential phase (OD600 = 0.5). IPTG was then added to a final concentration of 1 mM. After 1 h, cells were stained with the membrane dye FM4-64 as described previously (38, 68). Briefly, 1 μg/ml FM4-64 was added to 100 μl of culture and incubated at room temperature for 5 min. To prepare pColV-containing cells for microscopy, overnight culture was first normalized to an OD600 of 0.5. A 3-μl aliquot of culture was then immobilized on 0.8% agarose pads and imaged with a Nikon Ti Eclipse microscope equipped with a wide-field Andor NeoZyla camera and a 100× oil phase 3 1.4-numerical-aperture objective. Images were processed in ImageJ/FIJI (69).
MIC.
To determine MIC values of various compounds listed in Table 1, an assay was performed as described previously (22, 70). Briefly, compounds to be tested were prepared in serial 2-fold dilutions in LB (50 μl total volume) in a 96-well plate. Overnight cultures of bacterial strains were diluted 1:1,000 in LB, grown for 1 h at 37°C, and then diluted again 1:1,000 in LB. Fifty microliters of this diluted culture was added to each well. Plates were incubated statically for 24 h at 37°C.
Transposon insertion library construction.
TIS libraries were generated in EHEC EDL933 ΔlacI::aacC1 and ΔcvpA mutant backgrounds as described previously (22). Briefly, conjugation was performed to transfer the transposon-containing suicide vector pSC189 (71) from a DAP-auxotrophic donor strain (E. coli MFDλpir) into the recipient strain. Overnight culture of donor and recipient (600 μl each) was pelleted, washed with 1 ml LB, and resuspended in 60 μl LB (each). The cultures were mixed and spotted onto a 0.45-μm hemagglutinin filter (Millipore) on an LB-DAP agar plate and incubated at 37°C for 1 h. The filters were then washed in 24 ml LB and immediately spread across three 245-mm2 (Corning) LB-agar plates containing Gm and Km and incubated overnight at 37°C. Plates were scraped to collect colonies, which were resuspended in LB and stored as 1-ml aliquots in LB plus 20% glycerol (vol/vol) at −80°C. An aliquot was thawed, and genomic DNA (gDNA) was isolated for analysis.
DOC-selected TIS library.
Aliquots of WT and ΔcvpA library were thawed and diluted to an OD600 of 1 with LB. Five milliliters of each diluted culture was added to a flask with 75 ml of LB. The cultures were grown with shaking at 37°C for 1 h, at which their OD600 values were measured as ∼0.25. Each respective culture (5 ml) was plated on one of two 245-mm2 (Corning) LB-agar plates containing Gm and Km and on one of two plates containing Gm and Km and 1% deoxycholate (DOC). Plates were incubated overnight (∼16 h) at 37°C and immediately scraped to collect colonies. Colonies were resuspended in LB and stored as 1-ml aliquots in LB plus 20% glycerol (vol/vol) at −80°C.
Characterization of transposon insertion libraries.
Transposon insertion libraries were characterized as described previously (22). Briefly, for each library, gDNA was isolated using the Wizard genomic DNA extraction kit (Promega). gDNA was fragmented to 400 to 600 bp using a Covaris E200 sonicator and end repaired using the NEB Quick Blunting kit. PCR was used to amplify transposon junctions, and PCR products were gel purified to recover 200- to 500-bp fragments. To estimate library concentration, purified PCR products were subjected to quantitative PCR (qPCR) using primers designed for the Illumina P5 and P7 hybridization sequences. Libraries were mixed in an equimolar fashion and sequenced with a MiSeq instrument. Sequencing information is available at GEO (accession number GSE162346).
Reads were trimmed of transposon and adapter sequences using CLC Genomics Workbench (Qiagen) and mapped to E. coli O157:H7 EDL933 (NCBI accession numbers NZ_CP008957.1 [chromosome] and NZ_CP008958.1 [pO157 plasmid]) using Bowtie, allowing for no mismatches. Reads were discarded if they did not map to any sites in the genome, and reads mapping to multiple sites were randomly distributed. The data were normalized for chromosomal replication biases and differences in sequence depth using a LOESS correction of 100,000-bp (chromosome) and 10,000-bp (plasmid) windows. Reads at each TA site were tallied and binned by protein-coding gene.
For identification of loci with synthetic fitness between WT and the ΔcvpA mutant, the Con-ARTIST pipeline was used (19, 22). First, the WT library was normalized to simulate any bottlenecks or differences in sequencing depth as observed in the ΔcvpA library using multinomial distribution-based random sampling (n = 100). Next, a Mann-Whitney U test was applied to compare these 100 simulated data sets to the ΔcvpA mutant library. Genes for which there were more than 5 individual transposon insertion mutants were considered to have sufficient data for analysis (otherwise “insufficient data”). Genes for which mutant abundance in the ΔcvpA background was a mean log2 fold change (log2FC) of >2 compared to the WT with a P value of <0.05 were considered to be “synthetic healthy,” while genes with a mutant abundance log2FC of <−2 compared to the WT with a P value of <0.05 were considered to be “synthetic sick.” The remaining genes were classified as “neutral.”
A similar analysis was performed to identify mutants conditionally depleted or enriched in 1% DOC compared to LB for each strain. First, for each strain, the LB library was normalized to simulate any bottlenecks or differences in sequencing depth as observed in the corresponding 1% DOC library using multinomial distribution-based random sampling (n = 100). Next, a Mann-Whitney U test was applied to compare these 100 simulated data sets to the 1% DOC mutant library. Genes for which there were more than 5 individual transposon insertion mutants were considered to have sufficient data for analysis (otherwise insufficient data). Genes for which mutant abundance in the 1% DOC background has a mean log2FC of ≥2 compared to the WT with a P value of <0.05 were considered to be “enriched,” while genes with a mutant abundance log2FC of ≤−2 compared to the WT with a P value of <0.05 were considered to be “depleted.” The remaining genes were classified as “neutral.”
Spontaneous suppressor isolation and sequencing.
To isolate and identify spontaneous suppressor mutations, the ΔcvpA mutant was first grown to the mid-exponential phase (OD600 = 0.5). Two milliliters of this culture was plated on a 150- by 15-mm (Fisher) LB agar plate containing 1% DOC and incubated for 24 h at 37°C. The next day, suppressor colonies were picked and restreaked on LB agar plates containing 1% DOC. Fifteen suppressor colonies were picked, grown overnight, and subjected to gDNA isolation with the Wizard genomic DNA extraction kit (Promega). gDNA from the ΔcvpA mutant was also isolated as the parental control strain. gDNA was either submitted for sequencing at the Microbial Genome Sequencing Center at the University of Pittsburgh or prepared for whole-genome sequencing in-house using the Nextera XT DNA library preparation kit (Illumina). Libraries were sequenced using the NextSeq 550 or MiSeq platforms, respectively.
Reads were mapped to the E. coli O157:H7 EDL933 genome (NCBI accession numbers NZ_CP008957.1 [chromosome] and NZ_CP008958.1 [pO157 plasmid]) using CLC Genomics Workbench (Qiagen) with default parameters. Basic variant detection was performed, assuming a ploidy of one and filtering out variants with a frequency of less than 35%. Known variants were filtered against the ΔcvpA strain. Variant calls were then manually curated to identify SNPs with a frequency greater than 90%.
Bioinformatics/protein prediction.
Protein predictions about CvpA structure were inferred from several programs, namely, PSLPred, HHPred, Phobius, Phyre2, and SPOCTOPUS (72–76). Protter (77) was used to create a topological diagram, which originally appeared in Warr et al. (22).
The topology diagram for CvpA was made using Protter. To examine the conservation of the CvpA amino acid sequence across bacterial phyla, we used AnnoTree version 89 (53). The query term “CvpA” (Kyoto Encyclopedia of Genes and Genomes [KEGG] orthology K03558) was used to find bacterial proteins with at least 30% identity and 70% alignment across 27,000 genomes. A total of 20 CvpA sequences from genomes across bacterial phyla were selected for comparison and aligned using MUSCLE (78) within the MEGA X platform (79) using default parameters.
Data availability.
Sequencing reads and processed data for transposon insertion sequencing (Tn-Seq) experiments are available in the Gene Expression Omnibus (GEO) repository under accession number GSE162346.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to members of the Waldor lab, particularly Jacob Lazarus, for comments on this project and the manuscript.
M.K.W. is funded by NIH grant R01-AI-042347 and by the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Fang FC, Frawley ER, Tapscott T, Vazquez-Torres A. 2016. Bacterial stress responses during host infection. Cell Host Microbe 20:133–143. doi: 10.1016/j.chom.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Woodward SE, Krekhno Z, Finlay BB. 2019. Here, there, and everywhere: how pathogenic Escherichia coli sense and respond to gastrointestinal biogeography. Cell Microbiol 21:e13107. doi: 10.1111/cmi.13107. [DOI] [PubMed] [Google Scholar]
- 3.Boyer JL. 2013. Bile formation and secretion. Compr Physiol 3:1035–1078. doi: 10.1002/cphy.c120027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maldonado-Valderrama J, Wilde P, Macierzanka A, Mackie A. 2011. The role of bile salts in digestion. Adv Colloid Interface Sci 165:36–46. doi: 10.1016/j.cis.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Begley M, Gahan CGM, Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol Rev 29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Urdaneta V, Casadesús J. 2017. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front Med 4:163. doi: 10.3389/fmed.2017.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barnett Foster D. 2013. Modulation of the enterohemorrhagic E. coli virulence program through the human gastrointestinal tract. Virulence 4:315–323. doi: 10.4161/viru.24318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sistrunk JR, Nickerson KP, Chanin RB, Rasko DA, Faherty CS. 2016. Survival of the fittest: how bacterial pathogens utilize bile to enhance infection. Clin Microbiol Rev 29:819–836. doi: 10.1128/CMR.00031-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sengupta C, Ray S, Chowdhury R. 2014. Fine tuning of virulence regulatory pathways in enteric bacteria in response to varying bile and oxygen concentrations in the gastrointestinal tract. Gut Pathog 6:38. doi: 10.1186/s13099-014-0038-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prieto AI, Ramos-Morales F, Casadesús J. 2006. Repair of DNA damage induced by bile salts in Salmonella enterica. Genetics 174:575–584. doi: 10.1534/genetics.106.060889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodríguez-Beltrán J, Rodríguez-Rojas A, Guelfo JR, Couce A, Blázquez J. 2012. The Escherichia coli SOS gene dinF protects against oxidative stress and bile salts. PLoS One 7:e34791. doi: 10.1371/journal.pone.0034791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernstein C, Bernstein H, Payne CM, Beard SE, Schneider J. 1999. Bile salt activation of stress response promoters in Escherichia coli. Curr Microbiol 39:68–72. doi: 10.1007/s002849900420. [DOI] [PubMed] [Google Scholar]
- 13.Vijayakumar SRV, Kirchhof MG, Patten CL, Schellhorn HE. 2004. RpoS-regulated genes of Escherichia coli identified by random lacZ fusion mutagenesis. J Bacteriol 186:8499–8507. doi: 10.1128/JB.186.24.8499-8507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paget MS. 2015. Bacterial sigma factors and anti-sigma factors: structure, function and distribution. Biomolecules 5:1245–1265. doi: 10.3390/biom5031245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hews CL, Cho T, Rowley G, Raivio TL. 2019. Maintaining integrity under stress: envelope stress response regulation of pathogenesis in Gram-negative bacteria. Front Cell Infect Microbiol 9:313. doi: 10.3389/fcimb.2019.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ray S, Da Costa R, Das M, Nandi D. 2019. Interplay of cold shock protein E with an uncharacterized protein, YciF, lowers porin expression and enhances bile resistance in Salmonella Typhimurium. J Biol Chem 294:9084–9099. doi: 10.1074/jbc.RA119.008209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guest RL, Raivio TL. 2016. Role of the Gram-negative envelope stress response in the presence of antimicrobial agents. Trends Microbiol 24:377–390. doi: 10.1016/j.tim.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Pritchard JR, Chao MC, Abel S, Davis BM, Baranowski C, Zhang YJ, Rubin EJ, Waldor MK. 2014. ARTIST: high-resolution genome-wide assessment of fitness using transposon-insertion sequencing. PLoS Genet 10:e1004782. doi: 10.1371/journal.pgen.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hubbard TP, Chao MC, Abel S, Blondel CJ, Abel Zur Wiesch P, Zhou X, Davis BM, Waldor MK. 2016. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc Natl Acad Sci U S A 113:6283–6288. doi: 10.1073/pnas.1601718113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhuri RR, Morgan E, Peters SE, Pleasance SJ, Hudson DL, Davies HM, Wang J, van Diemen PM, Buckley AM, Bowen AJ, Pullinger GD, Turner DJ, Langridge GC, Turner AK, Parkhill J, Charles IG, Maskell DJ, Stevens MP. 2013. Comprehensive assignment of roles for Salmonella Typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 9:e1003456. doi: 10.1371/journal.pgen.1003456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Warr AR, Hubbard TP, Munera D, Blondel CJ, Zur Wiesch PA, Abel S, Wang X, Davis BM, Waldor MK. 2019. Transposon-insertion sequencing screens unveil requirements for EHEC growth and intestinal colonization. PLoS Pathog 15:e1007652. doi: 10.1371/journal.ppat.1007652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamner S, McInnerney K, Williamson K, Franklin MJ, Ford TE. 2013. Bile salts affect expression of Escherichia coli O157:H7 genes for virulence and iron acquisition, and promote growth under iron limiting conditions. PLoS One 8:e74647. doi: 10.1371/journal.pone.0074647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kus JV, Gebremedhin A, Dang V, Tran S-L, Serbanescu A, Foster DB. 2011. Bile salts induce resistance to polymyxin in enterohemorrhagic Escherichia coli O157:H7. J Bacteriol 193:4509–4515. doi: 10.1128/JB.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arenas-Hernández MMP, Rojas-López M, Medrano-López A, Nuñez-Reza KJ, Puente JL, Martínez-Laguna Y, Torres AG. 2014. Environmental regulation of the long polar fimbriae 2 of enterohemorrhagic Escherichia coli O157:H7. FEMS Microbiol Lett 357:105–114. doi: 10.1111/1574-6968.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol Microbiol 48:1609–1619. doi: 10.1046/j.1365-2958.2003.03531.x. [DOI] [PubMed] [Google Scholar]
- 27.Thanassi DG, Cheng LW, Nikaido H. 1997. Active efflux of bile salts by Escherichia coli. J Bacteriol 179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Froelich JM, Tran K, Wall D. 2006. A pmrA constitutive mutant sensitizes Escherichia coli to deoxycholic acid. J Bacteriol 188:1180–1183. doi: 10.1128/JB.188.3.1180-1183.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwan BW, Lord DM, Peti W, Page R, Benedik MJ, Wood TK. 2015. The MqsR/MqsA toxin/antitoxin system protects Escherichia coli during bile acid stress. Environ Microbiol 17:3168–3181. doi: 10.1111/1462-2920.12749. [DOI] [PubMed] [Google Scholar]
- 30.Fath MJ, Mahanty HK, Kolter R. 1989. Characterization of a purF operon mutation which affects colicin V production. J Bacteriol 171:3158–3161. doi: 10.1128/jb.171.6.3158-3161.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang LH, Fath MJ, Mahanty HK, Tai PC, Kolter R. 1995. Genetic analysis of the colicin V secretion pathway. Genetics 141:25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sueki A, Stein F, Savitski MM, Selkrig J, Typas A. 2020. Systematic localization of Escherichia coli membrane proteins. mSystems 5:e00808-19. doi: 10.1128/mSystems.00808-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jensen HM, Eng T, Chubukov V, Herbert RA, Mukhopadhyay A. 2017. Improving membrane protein expression and function using genomic edits. Sci Rep 7:13030. doi: 10.1038/s41598-017-12901-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dörr T, Delgado F, Umans BD, Gerding MA, Davis BM, Waldor MK. 2016. a transposon screen identifies genetic determinants of Vibrio cholerae resistance to high-molecular-weight antibiotics. Antimicrob Agents Chemother 60:4757–4763. doi: 10.1128/AAC.00576-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greene NG, Fumeaux C, Bernhardt TG. 2018. Conserved mechanism of cell-wall synthase regulation revealed by the identification of a new PBP activator in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 115:3150–3155. doi: 10.1073/pnas.1717925115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. 2010. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao MC, Abel S, Davis BM, Waldor MK. 2016. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol 14:119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fleurie A, Zoued A, Alvarez L, Hines KM, Cava F, Xu L, Davis BM, Waldor MK. 2019. A Vibrio cholerae BolA-like protein is required for proper cell shape and cell envelope integrity. mBio 10:e00790-19. doi: 10.1128/mBio.00790-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meeske AJ, Sham L-T, Kimsey H, Koo B-M, Gross CA, Bernhardt TG, Rudner DZ. 2015. MurJ and a novel lipid II flippase are required for cell wall biogenesis in Bacillus subtilis. Proc Natl Acad Sci U S A 112:6437–6442. doi: 10.1073/pnas.1504967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wivagg CN, Wellington S, Gomez JE, Hung DT. 2016. Loss of a class A penicillin-binding protein alters β-lactam susceptibilities in Mycobacterium tuberculosis. ACS Infect Dis 2:104–110. doi: 10.1021/acsinfecdis.5b00119. [DOI] [PubMed] [Google Scholar]
- 41.Klobucar K, Brown ED. 2018. Use of genetic and chemical synthetic lethality as probes of complexity in bacterial cell systems. FEMS Microbiol Rev 42. doi: 10.1093/femsre/fux054. [DOI] [PubMed] [Google Scholar]
- 42.Harms C, Domoto Y, Celik C, Rahe E, Stumpe S, Schmid R, Nakamura T, Bakker EP. 2001. Identification of the ABC protein SapD as the subunit that confers ATP dependence to the K+-uptake systems Trk(H) and Trk(G) from Escherichia coli K-12. Microbiology (Reading) 147:2991–3003. doi: 10.1099/00221287-147-11-2991. [DOI] [PubMed] [Google Scholar]
- 43.Edwards JC, Johnson MS, Taylor BL. 2006. Differentiation between electron transport sensing and proton motive force sensing by the Aer and Tsr receptors for aerotaxis. Mol Microbiol 62:823–837. doi: 10.1111/j.1365-2958.2006.05411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borisov VB, Murali R, Verkhovskaya ML, Bloch DA, Han H, Gennis RB, Verkhovsky MI. 2011. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc Natl Acad Sci U S A 108:17320–17324. doi: 10.1073/pnas.1108217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimada T, Tanaka K, Ishihama A. 2017. The whole set of the constitutive promoters recognized by four minor sigma subunits of Escherichia coli RNA polymerase. PLoS One 12:e0179181. doi: 10.1371/journal.pone.0179181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding Y, Davis BM, Waldor MK. 2004. Hfq is essential for Vibrio cholerae virulence and downregulates σE expression. Mol Microbiol 53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 47.Mecsas J, Rouviere PE, Erickson JW, Donohue TJ, Gross CA. 1993. The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev 7:2618–2628. doi: 10.1101/gad.7.12b.2618. [DOI] [PubMed] [Google Scholar]
- 48.Tam C, Missiakas D. 2005. Changes in lipopolysaccharide structure induce the σE-dependent response of Escherichia coli. Mol Microbiol 55:1403–1412. doi: 10.1111/j.1365-2958.2005.04497.x. [DOI] [PubMed] [Google Scholar]
- 49.Lima S, Guo MS, Chaba R, Gross CA, Sauer RT. 2013. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 340:837–841. doi: 10.1126/science.1235358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Testerman TL, Vazquez-Torres A, Xu Y, Jones-Carson J, Libby SJ, Fang FC. 2002. The alternative sigma factor σE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol Microbiol 43:771–782. doi: 10.1046/j.1365-2958.2002.02787.x. [DOI] [PubMed] [Google Scholar]
- 51.Missiakas D, Mayer MP, Lemaire M, Georgopoulos C, Raina S. 1997. Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol Microbiol 24:355–371. doi: 10.1046/j.1365-2958.1997.3601713.x. [DOI] [PubMed] [Google Scholar]
- 52.Fang FC, DeGroote MA, Foster JW, Bäumler AJ, Ochsner U, Testerman T, Bearson S, Giárd J-C, Xu Y, Campbell G, Laessig T. 1999. Virulent Salmonella Typhimurium has two periplasmic Cu, Zn-superoxide dismutases. Proc Natl Acad Sci U S A 96:7502–7507. doi: 10.1073/pnas.96.13.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mendler K, Chen H, Parks DH, Lobb B, Hug LA, Doxey AC. 2019. AnnoTree: visualization and exploration of a functionally annotated microbial tree of life. Nucleic Acids Res 47:4442–4448. doi: 10.1093/nar/gkz246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russ WP, Engelman DM. 2000. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol 296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- 55.Malinverni JC, Silhavy TJ. 2011. Assembly of outer membrane β-barrel proteins: the Bam complex. EcoSal Plus 4:10.1128/ecosalplus.4.3.8. doi: 10.1128/ecosalplus.4.3.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hayden JD, Ades SE. 2008. The extracytoplasmic stress factor, σE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One 3:e1573. doi: 10.1371/journal.pone.0001573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raivio TL, Leblanc SKD, Price NL. 2013. The Escherichia coli Cpx envelope stress response regulates genes of diverse function that impact antibiotic resistance and membrane integrity. J Bacteriol 195:2755–2767. doi: 10.1128/JB.00105-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guest RL, Wang J, Wong JL, Raivio TL. 2017. A bacterial stress response regulates respiratory protein complexes to control envelope stress adaptation. J Bacteriol 199. doi: 10.1128/JB.00153-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sousa T, Castro RE, Pinto SN, Coutinho A, Lucas SD, Moreira R, Rodrigues CMP, Prieto M, Fernandes F. 2015. Deoxycholic acid modulates cell death signaling through changes in mitochondrial membrane properties. J Lipid Res 56:2158–2171. doi: 10.1194/jlr.M062653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cabral DJ, Small DM, Lilly HS, Hamilton JA. 1987. Transbilayer movement of bile acids in model membranes. Biochemistry 26:1801–1804. doi: 10.1021/bi00381a002. [DOI] [PubMed] [Google Scholar]
- 61.Cremers CM, Knoefler D, Vitvitsky V, Banerjee R, Jakob U. 2014. Bile salts act as effective protein-unfolding agents and instigators of disulfide stress in vivo. Proc Natl Acad Sci U S A 111:E1610–E1619. doi: 10.1073/pnas.1401941111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang CC, Konisky J. 1984. Colicin V-treated Escherichia coli does not generate membrane potential. J Bacteriol 158:757–759. doi: 10.1128/JB.158.2.757-759.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gérard F, Pradel N, Wu L-F. 2005. Bactericidal activity of colicin V is mediated by an inner membrane protein, SdaC, of Escherichia coli. J Bacteriol 187:1945–1950. doi: 10.1128/JB.187.6.1945-1950.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun 59:4310–4317. doi: 10.1128/IAI.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lazarus JE, Warr AR, Kuehl CJ, Giorgio RT, Davis BM, Waldor MK. 2019. A new suite of allelic-exchange vectors for the scarless modification of proteobacterial genomes. Appl Environ Microbiol 85:e00990-19. doi: 10.1128/AEM.00990-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morales VM, Bäckman A, Bagdasarian M. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- 68.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S, Cava F, de Pedro MA, Brun YV, VanNieuwenhze MS. 2012. In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew Chem Int Ed Engl 51:12519–12523. doi: 10.1002/anie.201206749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dörr T, Möll A, Chao MC, Cava F, Lam H, Davis BM, Waldor MK. 2014. Differential requirement for PBP1a and PBP1b in in vivo and in vitro fitness of Vibrio cholerae. Infect Immun 82:2115–2124. doi: 10.1128/IAI.00012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chiang SL, Rubin EJ. 2002. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene 296:179–185. doi: 10.1016/s0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- 72.Bhasin M, Garg A, Raghava GPS. 2005. PSLpred: prediction of subcellular localization of bacterial proteins. Bioinformatics 21:2522–2524. doi: 10.1093/bioinformatics/bti309. [DOI] [PubMed] [Google Scholar]
- 73.Zimmermann L, Stephens A, Nam S-Z, Rau D, Kübler J, Lozajic M, Gabler F, Söding J, Lupas AN, Alva V. 2018. A completely reimplemented MPI Bioinformatics Toolkit with a new HHpred server at its core. J Mol Biol 430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 74.Käll L, Krogh A, Sonnhammer ELL. 2004. A combined transmembrane topology and signal peptide prediction method. J Mol Biol 338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 75.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Viklund H, Bernsel A, Skwark M, Elofsson A. 2008. SPOCTOPUS: a combined predictor of signal peptides and membrane protein topology. Bioinformatics 24:2928–2929. doi: 10.1093/bioinformatics/btn550. [DOI] [PubMed] [Google Scholar]
- 77.Omasits U, Ahrens CH, Müller S, Wollscheid B. 2014. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30:884–886. doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 78.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing reads and processed data for transposon insertion sequencing (Tn-Seq) experiments are available in the Gene Expression Omnibus (GEO) repository under accession number GSE162346.