ABSTRACT
DNA replication is essential for the growth and development of Chlamydia trachomatis; however, it is unclear how this process contributes to and is controlled by the pathogen’s biphasic life cycle. While inhibitors of transcription, translation, cell division, and glucose-6-phosphate transport all negatively affect chlamydial intracellular development, the effects of directly inhibiting DNA polymerase have never been examined. We isolated a temperature-sensitive dnaE mutant (the dnaEts mutant) that exhibits an ∼100-fold reduction in genome copy number at the nonpermissive temperature (40°C) but replicates similarly to the parent at the permissive temperature of 37°C. We measured higher ratios of genomic DNA nearer the origin of replication than the terminus in the dnaEts mutant at 40°C, indicating that this replication deficiency is due to a defect in DNA polymerase processivity. The dnaEts mutant formed fewer and smaller pathogenic vacuoles (inclusions) at 40°C, and the bacteria appeared enlarged and exhibited defects in cell division. The bacteria also lacked both discernible peptidoglycan and polymerized MreB, the major cell division-organizing protein in Chlamydia responsible for nascent peptidoglycan biosynthesis. We also found that the absolute genome copy number, rather than active genome replication, was sufficient for infectious progeny production. Deficiencies in both genome replication and inclusion expansion were reversed when the dnaEts mutant was shifted from 40°C to 37°C early in infection, and intragenic suppressor mutations in dnaEts also restored genome replication and inclusion expansion in the dnaEts mutant at 40°C. Overall, our results show that genome replication in C. trachomatis is required for inclusion expansion, septum formation, and the transition between the microbe’s replicative and infectious forms.
IMPORTANCE Chlamydiae transition between infectious, extracellular elementary bodies (EBs) and noninfectious, intracellular reticulate bodies (RBs). Some checkpoints that govern transitions in chlamydial development have been identified, but the extent to which genome replication plays a role in regulating the pathogen’s infectious cycle has not been characterized. We show that genome replication is dispensable for EB-to-RB conversion but is necessary for RB proliferation, division septum formation, and inclusion expansion. We use new methods to investigate developmental checkpoints and dependencies in Chlamydia that facilitate the ordering of events in the microbe’s biphasic life cycle. Our findings suggest that Chlamydia utilizes feedback inhibition to regulate core metabolic processes during development, likely an adaptation to intracellular stress and a nutrient-limiting environment.
KEYWORDS: DNA replication, cell division, Chlamydia trachomatis, peptidoglycan
INTRODUCTION
The phylum Chlamydiae contains obligately intracellular bacteria that replicate inside eukaryotic host cells; multiple species are important human and animal pathogens (1, 2). Chlamydiae transition between extracellular infectious, but non-metabolically active, elementary bodies (EBs) and intracellular replicative, but noninfectious, reticulate bodies (RBs). How they transition between these developmental forms is unknown, but inhibiting these transitions prevents the production of infectious progeny. Determining the sequence of events during chlamydial development could identify checkpoints that could be targets for highly discriminating antichlamydial antibiotics.
Chlamydial development and pathogenesis are intertwined. EBs are preloaded with proteins and other molecules that promote entry into host cells (3), evasion of innate immunity (4), and disengagement from progression along the host cell endocytic pathway (5). EBs display minimal metabolic activity until they transition into RBs inside nascent chlamydial parasitophorous vacuoles (inclusions), although they can passively take up glucose-6-phosphate (6, 7) and incorporate amino acids into newly synthesized proteins (6, 8). Several EB characteristics may restrain metabolic activity. DNA supercoiling (9), histone-like proteins (10), and regulation of the transcriptional machinery by type three secretion system (T3SS) chaperones (11, 12) block transcription in EBs. Disulfide cross-linked envelope proteins must be reduced to relax EB membranes and initiate EB-to-RB conversion (13). A preloaded T3SS in EBs may sequentially secrete effectors into the host cytoplasm that drive early chlamydial development (14–16). Some early T3SS effectors that are secreted into the host cytosol facilitate EB entry, while others (Inc proteins) localize to the inclusion membrane, where they prevent fusion of the inclusion with lysosomes, promote fusion of the inclusion with exocytic host vesicles, and facilitate the acquisition of host nutrients (17, 18).
Genome replication begins within 4 h of Chlamydia trachomatis entry (19, 20). The stimuli that trigger genome replication and chlamydial cell division are unknown, but these processes require de novo protein synthesis (21). The first replication of a C. trachomatis genome takes as long as 6 h, whereas subsequent doublings during RB proliferation take 1.5 to 3 h, depending on the stage of the developmental cycle (19, 20). The rate of RB division does not approach logarithmic growth until midway through the developmental cycle (22), so additional rounds of genome replication must begin prior to the completion of early cell division events. While the processes of replication and cell division are decoupled in other well-studied bacteria (23), the observed delay between genome replication and RB division in Chlamydia suggests that these processes are linked by a common factor, such as cell size (22) or a signal that affects the population as a whole.
Chlamydial genome replication proceeds in the absence of cell division when these pathogens enter a viable but not cultivatable (persistent) state (24–26), which can be triggered by nutrient restriction and/or immune stress in vitro. Persistence may be a pathoadaptation that permits atypical RBs (also known as aberrant bodies) to acquire host resources during stressful periods and prepare for rapid reentry into the canonical developmental cycle when favorable growth conditions return (24, 27, 28). In this context, persistence may represent the exploitation of separate, compartmentalized DNA replication and division mechanisms that are employed by Chlamydia during normal development, since DNA replication continues in the absence of cell division.
An unresolved question in chlamydial development is how Chlamydia regulates the expansion of its inclusion. Over the course of development, the inclusion membrane expands in size as a result of the redirection of vesicles by Chlamydia from the host cell exocytic pathway to the inclusion and the subsequent fusion of these vesicles with the inclusion membrane. Previous efforts to unravel the role of genome replication in regulating inclusion expansion have yielded contradictory results. DNA gyrase inhibitors cause C. trachomatis to enter persistence, effectively blocking both cell division and inclusion expansion (29). Antibiotics that block de novo chlamydial transcription and translation block genome replication, cell division and inclusion expansion (30–32). In contrast, inhibiting the transport of glucose-6-phosphate, a major energy source for EBs, blocks genome replication and cell division but not inclusion expansion (21, 33). Thus, it is unclear if inclusion expansion is contingent upon genome replication. Determining which of these observations show the effects of indirect versus direct inhibition of DNA replication on bacterial and inclusion size is necessary. Unfortunately, at the time of earlier studies, there were no tools available to directly address the dependence of inclusion expansion on DNA replication due to a lack of specific inhibitors and/or a mutant of the DNA polymerase (21).
We isolated a temperature-sensitive (TS) C. trachomatis mutant, which we hereafter refer to as the dnaEts mutant for simplicity, that has an amino acid substitution in the catalytic subunit of DNA polymerase III (DnaE), the primary replicative polymerase (34). We show here that the dnaEts mutant can initiate, but not complete, genome replication and that this defect causes time-dependent “pauses” in physiological processes associated with chlamydial development. The ability of the dnaEts mutant to form inclusions and to transition into RBs was unaffected at the nonpermissive temperature. In contrast, dnaEts mutant inclusions did not enlarge and contained aberrant RBs, and these aberrant RBs had cell division defects at the nonpermissive temperature, suggesting that DNA replication is required for RB division and proportional expansion of inclusions. We exploit the utility of temperature-sensitive mutants for exploring essential gene function in Chlamydia and demonstrate the power of this approach for dissecting the events of the chlamydial developmental cycle. Here, we use the dnaEts mutant to directly analyze chlamydial DNA replication during the microbe’s progression through its developmental cycle, providing fundamental insights into chlamydial biology.
RESULTS
A point mutation in dnaE blocks C. trachomatis genome replication.
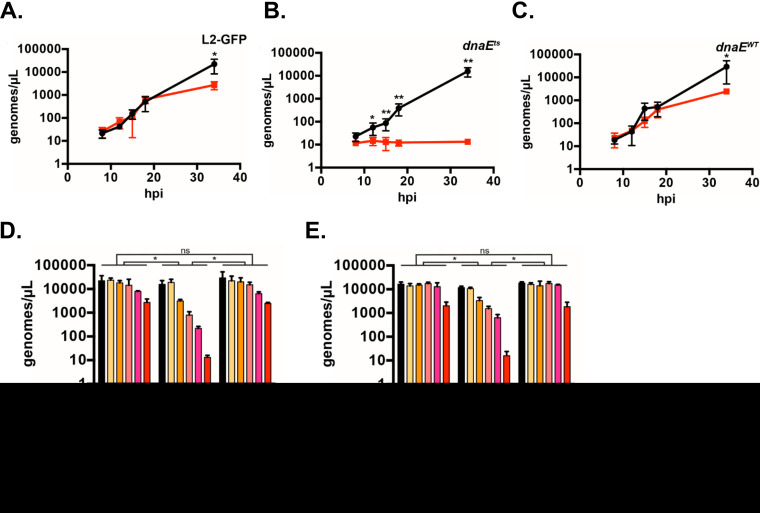
DnaE is the catalytic core subunit of the DNA polymerase III holoenzyme, the major replicative DNA polymerase in bacteria (35). While screening ethyl methanesulfonate (EMS)-mutagenized C. trachomatis isolates (34), we identified a temperature-sensitive mutant (the dnaEts mutant) that cannot proliferate at 40°C but develops normally at 37°C. The temperature-sensitive phenotype of the dnaEts mutant mapped to a single nucleotide change in dnaE (dnaEC2554T) that causes a proline-to-serine amino acid substitution in DnaE (DnaEP852S) (34). We compared the genome copy accumulation of the dnaEts mutant, an isogenic recombinant of the dnaEts mutant with wild-type dnaE (hereafter referred to as the dnaEWT recombinant), and the parent strain, C. trachomatis L2-GFP (green fluorescent protein [GFP]-expressing C. trachomatis L2), using quantitative PCR (qPCR) (Fig. 1A to C). The genome accumulation in the strains was similar at 37°C. Incubation at 40°C reduced the genome accumulation of the parent and the complemented mutant (the dnaEWT recombinant) approximately 10-fold from that at 37°C, but their genome copies still increased more than 100-fold between 8 h postinfection (hpi) and 34 hpi at 40°C. The genome copy number of the dnaEts mutant did not increase when it was incubated at 40°C. Thus, genome replication is impaired in the dnaEts mutant at nonpermissive, but not permissive, temperatures, which is consistent with the temperature-sensitive phenotypes of Escherichia coli dnaEts mutants (36).
FIG 1.
Genome replication in the dnaEts mutant is impaired at 40°C and rescued by shifting to 37°C. (A to C) HeLa cells were infected at an MOI of 0.1 with the parent (A), dnaEts (B), or isogenic recombinant (dnaEWT) (C) strain and were incubated at 37°C (black lines) or 40°C (red lines). Bacteria were harvested at 8, 12, 15, 18, or 34 hpi, and the genome copy number was determined by qPCR. The means of results from three experiments performed in duplicate ± standard deviations are shown. Data were analyzed by one-way ANOVA followed by Dunnett’s test. *, P < 0.05; **, P < 0.01. (D, E) HeLa cells were infected at an MOI of 0.2 with the parent, dnaEts, or dnaEWT strain and were incubated at 37°C or 40°C or were shifted from 40°C to 37°C at the times indicated in the legend. The genome copy number at 34 hpi total growth was determined by qPCR (D). The temperature shift assay was modified to determine if the time spent at 37°C affected the number of genome copies in cultures. Cultures were shifted from 40°C at the times indicated in the legend and incubated at 37°C for 26 h—regardless of the prior time spent at 40°C—and the genome copy number was determined by qPCR (E). Samples incubated at 37°C or 40°C only in both panels D and E were harvested at 34 hpi. Bars show the means for three experiments ± standard deviations. Data were analyzed by two-way ANOVA and Tukey’s test to assess statistical significance. *, P < 0.05; ns, not significant.
Shifting dnaEts mutant cultures from 40°C to 37°C by 8 hpi restored genome accumulation in the dnaEts mutant at 34 hpi to levels comparable to those of L2-GFP and the dnaEWT recombinant, suggesting that DnaE is dispensable during the EB-to-RB transition (Fig. 1D). The dnaEts mutant’s genome copies increased approximately 10-fold following shifting from 40°C to 37°C at 18 hpi over levels with continued incubation at 40°C. We hypothesized that the low genome copy number of the dnaEts mutant incubated continuously at 40°C could either reflect less time spent at 37°C or reveal that extended inhibition of genome replication is lethal. To differentiate between these possibilities, the assay was modified so that the infected cells were incubated for 26 h at 37°C regardless of the length of prior incubation at 40°C; both the 37°C and 40°C control infections were still stopped at 34 hpi. A linear decline in genome numbers was observed for bacteria incubated at 37°C for the same amount of time (Fig. 1E). Thus, inhibition of genome replication is reversible during the EB-to-RB transition but is lethal during RB proliferation.
Intragenic suppressor mutations in the dnaEts mutant rescue genome replication.
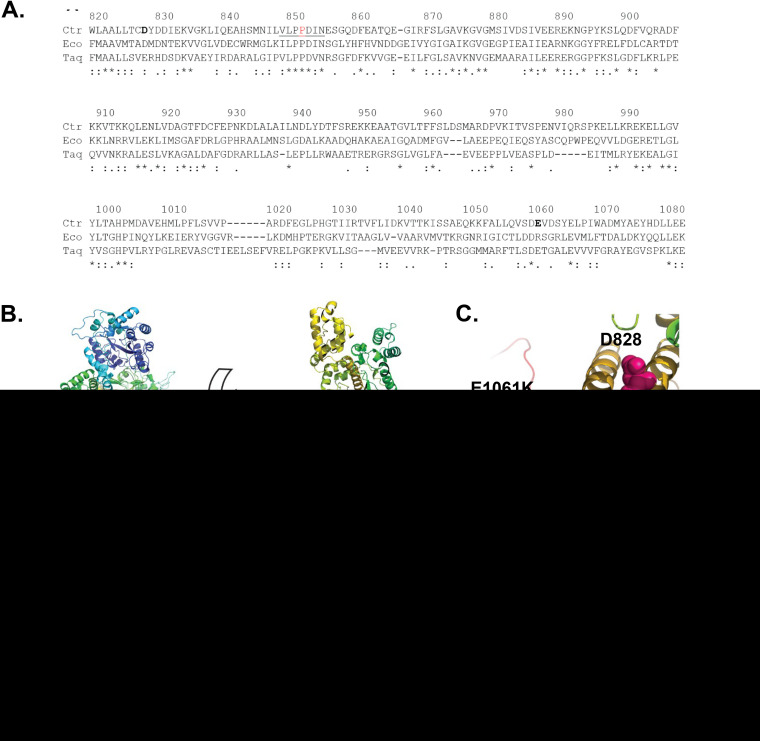
Sequence alignments and tertiary-structure modeling (37) of chlamydial DnaE with DnaE from E. coli and Thermus aquaticus (Fig. 2A and B) showed that the altered residue in the dnaEts mutant (P852) is in a conserved (V/I)LPPD(I/V)N motif in the β-clamp binding domain. The β-clamp gives the holoenzyme its high processivity during replication; however, the β-clamp is predicted to bind DnaE distal to the site of the mutation (38–40). Proline 852 is located in a predicted three-stranded β-sheet of unknown function that is conserved in E. coli DnaE (EcDnaE) and T. aquaticus DnaE (TaDnaE) (PDB IDs 2HQA and 3E0D, respectively).
FIG 2.
Intragenic suppressor mutations restore the function of the dnaEts mutant. (A) Alignments of DnaE from C. trachomatis (Ctr), E. coli (Eco), and Thermus aquaticus (Taq). Numbering is based on CtDnaE residue positions. (B) Phyre2 structural model of CtDnaE colored from blue (N terminus) to red (C terminus), with D828 (pink), P852 (blue), and E1061K (green) shown in space fill. (Left) View looking directly at the DNA template channel; (right) view looking down at the bottom of the channel. (C) Zoomed-in view of the right side of panel B. (D to F) HeLa cells were infected at an MOI of 0.2 with the parent, dnaEts, or dnaEWT strain, or with one of the two suppressor mutants (referred to as D828Y and E1061K for simplicity), and were incubated at 37°C (black bars) or 40°C (red bars) (D). The cells were fixed at 34 hpi, labeled with an anti-LPS antibody, and imaged at ×10. The 37°C/40°C ratio of progeny (E) and genome copies (F) for each mutant was normalized to that for L2-GFP at 34 hpi (gray bars) or 48 hpi (black bars). Bars show the means for three experiments ± standard deviations. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; ns, not significant.
To understand how the P852S substitution might impair DnaE function, we selected for strains with restored DnaE function (41). Serial passage of 15 populations (totaling ∼1 × 1010 EBs) of the dnaEts mutant at 40°C yielded 12 temperature-resistant mutant populations (Table 1). Sequencing of dnaE from these populations both revealed that the original C2554T mutation causing the TS P852S substitution was retained and identified additional mutations that caused E1061K or D828Y amino acid substitutions in DnaE. Isolates were plaque-cloned from the original populations, and resequencing of dnaE confirmed that the isolates retained dnaEC2254T, indicating that the isolates were suppressor mutants (data not shown). The individual suppressor mutants encoded either the E1061K or the D828Y change but not both. Genome sequencing of two representative suppressor mutant isolates identified no other mutations in the E1061K suppressor mutant; however, a synonymous mutation for G250 in the thiol:disulfide interchange protein DsdD was detected in the D828Y suppressor mutant.
TABLE 1.
Summary of the dnaEts mutant suppressor screen
| Flask | Passage when positive | Suppressor(s) present |
|---|---|---|
| 1 | None | |
| 2 | 3 | E1061K |
| 3 | 2 | D828Y, E1061K |
| 4 | 2 | D828Y, E1061K |
| 5 | 2 | E1061K |
| 6 | None | |
| 7 | 4 | D828Y, E1061K |
| 8 | 3 | E1061K |
| 9 | 4 | E1061K |
| 10 | 3 | E1061K |
| 11 | 4 | E1061K |
| 12 | 4 | E1061K |
| 13 | None | |
| 14 | 4 | E1061K |
| 15 | 2 | E1061K |
C. trachomatis DnaE (CtDnaE) E1061 and D828 are located in less-conserved regions than P852. CtDnaE D828 aligns with an aspartate in EcDnaE and a glutamate in TaDnaE. CtDnaE E1061 is preceded by a conserved aspartate; however, the charge state of the residue at position 1061 differs between EcDnaE and TaDnaE. Structural modeling predicted that P852S does not directly interact with either of the altered amino acids in the DnaE suppressor mutants (Fig. 2B and C). D828 falls in the helix-hairpin-helix (HhH) motif at the helix-hairpin transition, whereas E1061 is proximal to the oligonucleotide-binding (OB)-fold domain. Both of these features interact with the DNA template during DNA polymerization (42, 43). Therefore, we hypothesized that DnaEP852S has reduced contact with the DNA template at 40°C, which can be ameliorated by either D828Y or E1061K.
We tested if the suppressor mutations could rescue dnaEts mutant phenotypes. The parent, dnaEts, and dnaEWT strains and the two suppressor mutants (hereafter referred to as the DnaED828Y P852S and DnaEP852S E1061K suppressors) formed similar-sized inclusions at 37°C. At 40°C, dnaEts mutant inclusions were much smaller (Fig. 2D). DnaEP852S E1061K inclusions were similar in size to the inclusions of the parent and dnaEWT recombinant strains at 40°C, whereas the size of DnaED828Y P852S mutant inclusions was intermediate between those of the dnaEWT recombinant and dnaEts mutant strains. The DnaED828Y P852S and DnaEP852S E1061K suppressors produced ∼2-fold fewer progeny than the parent and dnaEWT recombinant strains at 40°C but produced many more progeny than the dnaEts mutant (Fig. 2E). The genome accumulation of the parent, dnaEWT recombinant, DnaED828Y P852S suppressor, and DnaEP852S E1061K suppressor strains did not differ and was ∼100-fold higher than that of the dnaEts mutant at 40°C (Fig. 2F). Thus, DNA replication and progeny yield are largely restored in the suppressor mutants.
DNA replication processivity is reduced in the dnaEts mutant.
DNA replication can be divided into three general steps. Initiation consists of loading the polymerase onto the single-stranded template; elongation is characterized by template-dependent addition of nucleotides; and termination is the release of the template by the polymerase. Although dnaEts mutant genomes did not accumulate at 40°C (Fig. 1), we could not distinguish if the dnaEts mutant failed to initiate replication or had insufficient processivity.
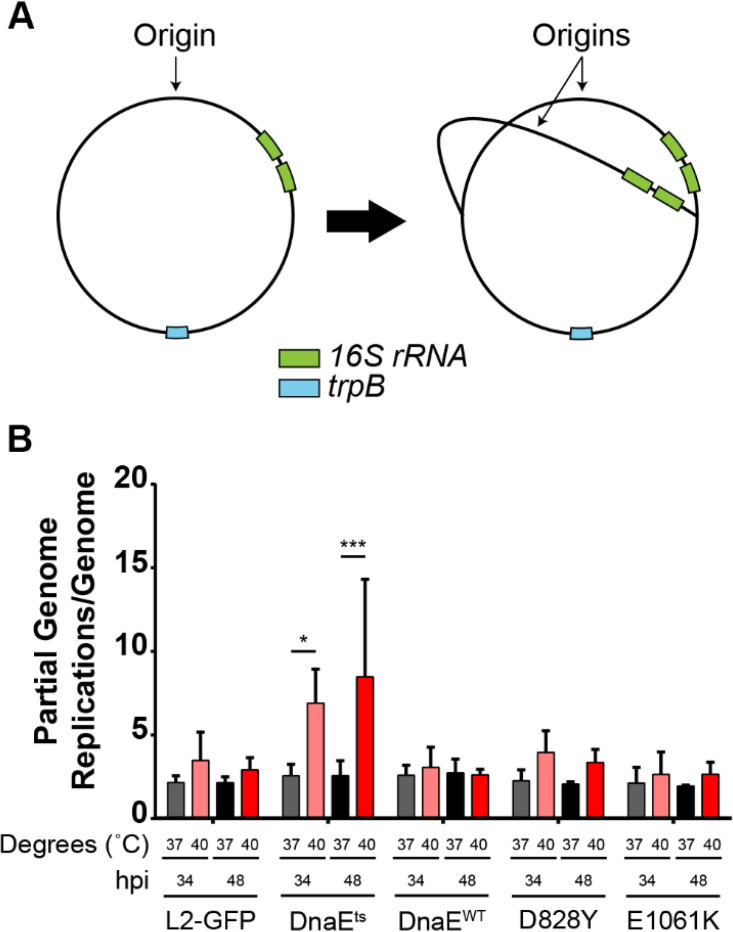
Modeling suggested that DnaE-DNA template interactions in the dnaEts mutant might be impaired (Fig. 2A to C). We designed a duplex qPCR assay to quantify copies of the 16S rRNA and trp genes. Tandem copies of the rRNA operon are located approximately 200 kbp from the chromosomal origin of replication in C. trachomatis, whereas trpB is located near the replication terminus (Fig. 3A). We reasoned that a ratio of the 16S rRNA gene to the trpB gene that was similar to the ratio in the wild type could indicate a replication initiation defect, whereas a higher ratio in the mutant would indicate an inability to complete replication. The parent, dnaEts mutant, and dnaEWT recombinant strains and the suppressor mutants had 16S rRNA gene/trpB ratios of ∼3 at 37°C (Fig. 3B). In contrast, the 16S rRNA gene/trpB ratio was ∼8 when the dnaEts mutant was incubated at 40°C for 34 hpi. Incubation of the dnaEts mutant for 48 hpi at 40°C increased this ratio further. For all other strains, the 16S rRNA gene/trpB ratio at 40°C was similar to that at 37°C (Fig. 3B). Since the dnaEts mutant cannot replicate its genome at 40°C (Fig. 1), and more genome counts were measured closer to the origin than to the terminus, these results indicate that the P852S substitution in the dnaEts mutant inhibits DnaE processivity.
FIG 3.
DNA replication is altered in the dnaEts mutant. (A) Approximate locations of the 16S rRNA (green) and trpB (blue) genes. As the replication forks move away from the origin, the copy number of genes near the origin, such as the 16S rRNA gene, increases before that of genes near the terminus, such as trpB. (B) Ratio of the 16S rRNA gene to trpB determined by qPCR. HeLa cells were infected at an MOI of 0.1 with the parent, dnaEts mutant, dnaEWT recombinant strain, or one of the two suppressor mutants (D828Y or E1061K) and were incubated at 37°C or 40°C for 34 or 48 hpi. Bars show the means for three experiments ± standard deviations. *, P < 0.05; ***, P < 0.001.
Disruption of DnaE activity prevents inclusion expansion but not maturation.
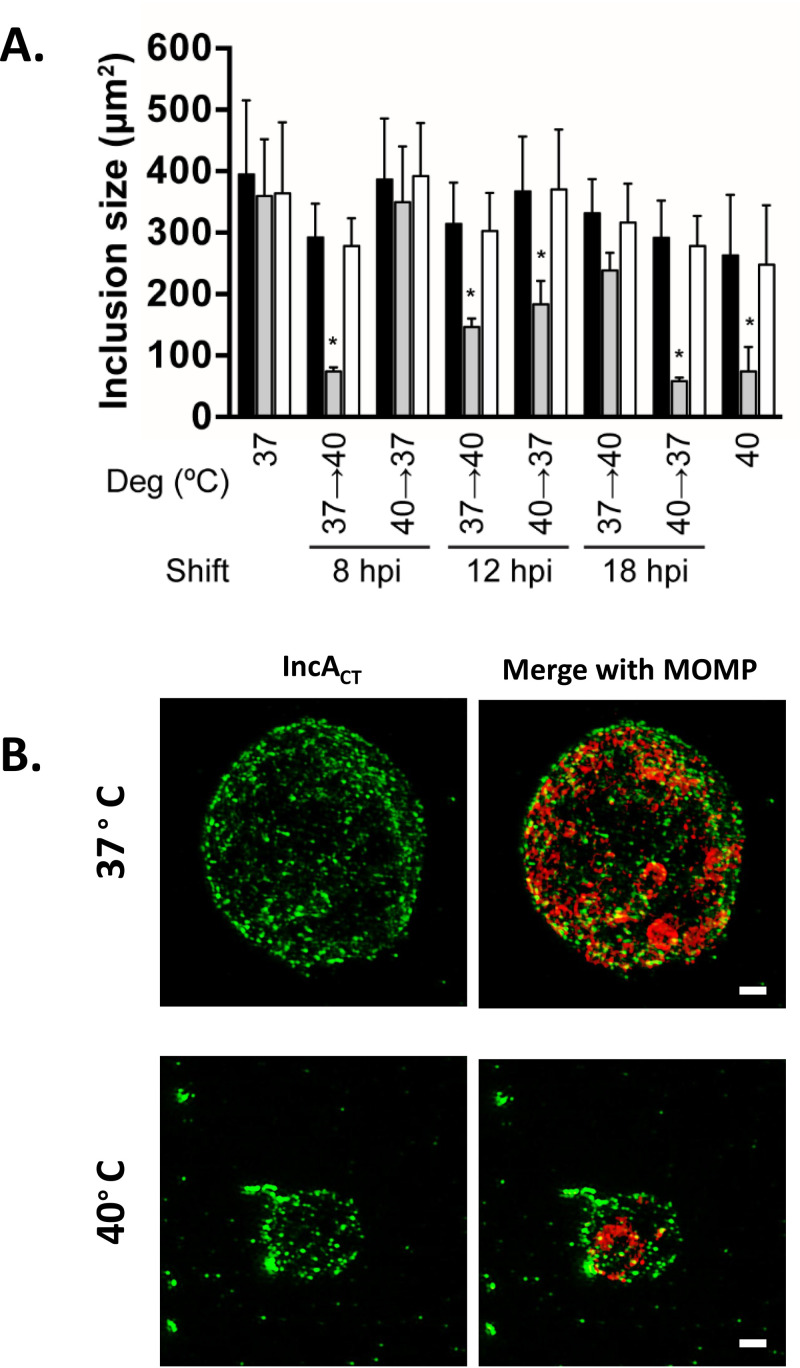
The relationship between chlamydial genome replication and inclusion expansion is unclear (21, 30, 31). To test if genome replication is required for inclusion expansion, the cross-sectional areas of the parent, the dnaEts mutant, and the dnaEWT recombinant strain inclusions in infected HeLa cells were compared after shifting from 37°C to 40°C and from 40°C to 37°C. Shifting from 37°C to 40°C prior to 18 hpi significantly reduced the inclusion size in the dnaEts mutant at 34 hpi (Fig. 4A). Shifting the dnaEts mutant from 40°C to 37°C at 12 hpi or later had a similar effect. Both shifts produced inclusions comparable in size to inclusions in the dnaEts mutant that was continuously incubated at 40°C for 34 hpi. Therefore, genome replication is essential for inclusion expansion during RB proliferation but dispensable for inclusion expansion later in chlamydial development.
FIG 4.
Inclusion size, but not maturation, is dependent on functional DnaE. (A) HeLa cells were infected at an MOI of 0.2 with the parent (filled bars), the dnaEts mutant (gray bars), or dnaEWT recombinant (white bars) strain and either were continuously incubated at 37°C or 40°C or were shifted between 37°C and 40°C at the times indicated. The cells were fixed at 34 hpi, labeled with an anti-LPS antibody, and imaged. Bars show the means for three experiments ± standard deviations. Two-way ANOVA and Tukey’s test were used to determine statistical significance. *, P < 0.05. (B) Cells infected with the dnaEts mutant were incubated at 37°C or 40°C for 24 h, fixed, and labeled with anti-MOMP and anti-IncA. Bars, 1 μm.
Inclusion expansion requires the recruitment of host vesicles by secreted chlamydial effectors. The chlamydial “secretome” is temporally regulated, and the array of Inc proteins in the inclusion membrane increases as the developmental cycle progresses (15). Thus, different Inc proteins can be used as a proxy for inclusion maturation (19). The small size of the dnaEts mutant inclusions suggested that they might be immature. However, the dnaEts mutant inclusions from 37°C and 40°C cultures both displayed IncA, a mid-to-late (∼18 to 24 hpi)-developmental cycle effector protein which mediates homotypic inclusion fusion (14, 15, 44, 45) (Fig. 4B). Thus, the dnaEts mutant can transcribe, translate, and secrete a midstage effector protein in the absence of de novo genome synthesis. In contrast, many persistence-inducing conditions that inhibit cell division in C. trachomatis directly affect transcription (24, 46, 47) and translation (27, 48), which can result in the failure of mid- and late-stage Inc proteins to accumulate in inclusion membranes (G. W. Liechti, unpublished data).
Disrupting DNA replication elicits the formation of aberrant bodies.
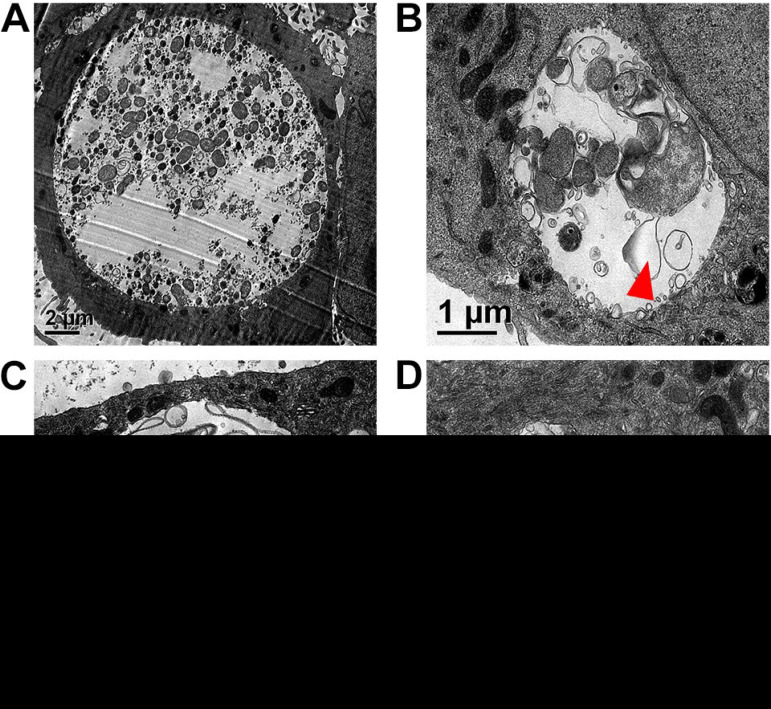
Since the dnaEts mutant does not replicate its genome at 40°C (Fig. 1), we determined whether DNA replication is required for the EB-to-RB and/or RB-to-EB transition by using transmission electron microscopy (TEM). At 37°C, the dnaEts mutant inclusions contained many small, electron-dense EBs and some larger RBs (Fig. 5A). At 40°C, dnaEts mutant inclusions contained enlarged and misshapen RBs or aberrant bodies but no EBs. Most of these RBs had enlarged periplasms (Fig. 5B and C), and empty vacuoles and cytoplasmic protrusions were prominent in some inclusions (Fig. 5D). No dividing RBs were observed. Similar RB morphologies have been observed following treatment with peptidoglycan (PG)-targeting antibiotics (49–58), gamma interferon (24, 59, 60), or high levels of nonessential amino acids (61). These observations suggest that septation is inhibited in the dnaEts mutant at 40°C and that inclusion integrity may be somewhat compromised, although the inclusions appear to mature normally (Fig. 4B).
FIG 5.
Loss of DnaE activity halts cell division and induces aberrant bodies. HeLa cells were infected at an MOI of 0.5 with the dnaEts mutant and were incubated at 37°C (A) or 40°C (B to D) for 32 hpi. Cells were processed for TEM. The dnaEts mutant inclusions contained numerous RBs and EBs at 37°C (A) but were smaller and contained aberrant RBs with enlarged periplasms (arrowheads) at 40°C (B and C) or had altered inclusion morphologies (D) but no EBs.
Inhibiting DNA replication causes degradation of the peptidoglycan ring.
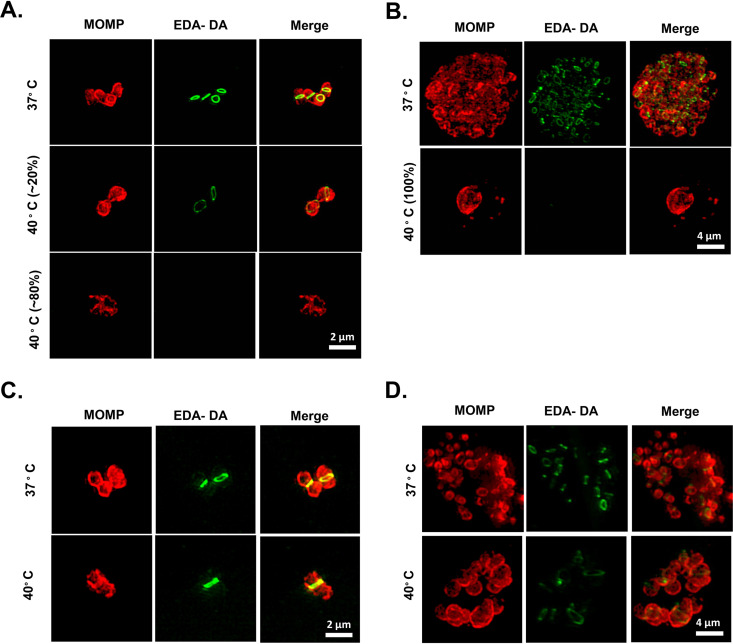
Chlamydial cell division requires peptidoglycan biosynthesis (52, 62, 63), and aberrant bodies exhibit altered peptidoglycan localization and/or the gradual loss of peptidoglycan over time (58, 64). To determine if defects in DNA replication block cell division and induce aberrant-body formation by altering peptidoglycan synthesis, we analyzed peptidoglycan localization in the dnaEts mutant by using a dipeptide labeling strategy (63, 65). When the dnaEts mutant was grown at 40°C, the vast majority of visible RBs at 12 hpi lacked peptidoglycan labeling (Fig. 6A). Most of the inclusions contained more than one RB or an RB with a discernible PG septum, further confirming that dnaEts mutant RBs are capable of limited cell division. However, by 24 hpi, all of the dnaEts mutant inclusions contained only peptidoglycan-negative RBs, the majority of which were enlarged and aberrant (Fig. 6B). We also examined peptidoglycan localization in the DnaEP852S E1061K suppressor. In contrast to the findings for the dnaEts mutant, peptidoglycan labeling was detected in DnaEP852S E1061K suppressor inclusions at 12 and 24 hpi at both 37°C and 40°C (Fig. 6C and D).
FIG 6.
Inhibition of DnaE blocks peptidoglycan biosynthesis. HeLa cells were infected at an MOI of 1 with the dnaEts (A, B) or dnaEts E1061K (C, D) strain and were incubated in 1 mM EDA-DA at 37°C or 40°C for 12 (A, C) or 24 (B, D) hours. Peptidoglycan (green) was labeled via a click chemistry reaction. The chlamydial outer membrane was labeled with anti-MOMP (red). Images are representative of >100 inclusions from at least 12 microscopic fields for each mutant/condition, and the percentages indicated in panel A represent approximate estimates.
Chlamydia and Chlamydia-related bacteria lack FtsZ, a filament-forming cytoskeletal protein and major organizer of the bacterial division complex (66). In its place, chlamydiae utilize another filament-forming protein, MreB, in order to synthesize septal peptidoglycan and initiate septation (64, 67–69). Inhibiting MreB polymerization causes degradation of the peptidoglycan ring and blocks cell division (23). We labeled chlamydial MreB with a polyclonal antibody (70) and determined that while filamentous MreB was easily discernible in the dnaEts mutant at 37°C, it was almost completely absent at 40°C (see Fig. S1 in the supplemental material). Since peptidoglycan biosynthesis is dependent on MreB polymerization in Chlamydia trachomatis (64), our data suggest that the loss of DNA replication inhibits peptidoglycan biosynthesis by inhibiting MreB polymerization.
The genome copy number is a checkpoint for the RB-to-EB transition.
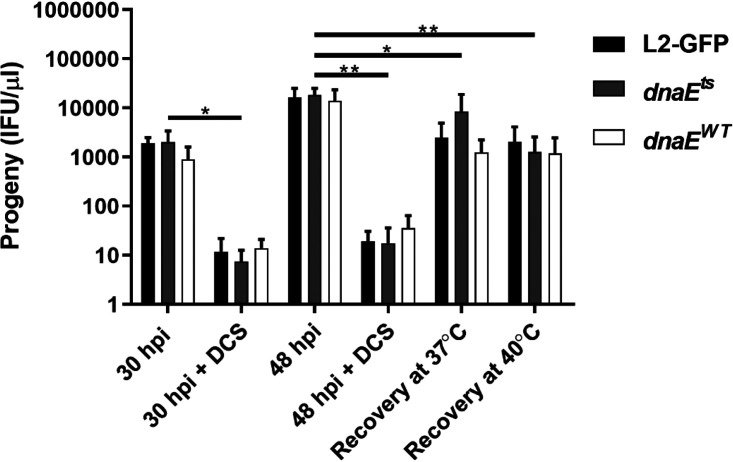
Loss of active DNA polymerization or a reduced chromosome copy number could explain the peptidoglycan biosynthesis and MreB localization defects of the dnaEts mutant. To better understand whether active DNA replication is required for septation, we used the peptidoglycan-targeting antibiotic d-cycloserine (DCS) to induce aberrant bodies, which continue to replicate their genomes but are unable to septate and thus divide (62, 71–73). DCS treatment caused reversible aberrant-body formation (Fig. S2) and reduced EB production by the parent, the dnaEts mutant, and dnaEWT recombinant strains at 30 hpi (Fig. 7). When the cultures recovered at 37°C or 40°C for 18 h after DCS removal, all of the strains formed similar numbers of progeny (Fig. 7). Thus, even though the dnaEts mutant cannot replicate its genome at 40°C, cell division and EB production proceed normally. These results indicate that the chromosomal copy number, rather than active DNA replication, is a checkpoint for the RB-to-EB transition and support a model where DNA replication continues during d-cycloserine-mediated persistence (24, 28).
FIG 7.
DnaE is dispensable for septation and EB formation if DNA replication has been completed. HeLa cells were infected at an MOI of 1 with L2-GFP, the dnaEts mutant, or the dnaEWT recombinant and were incubated with or without d-cycloserine (DCS) for 30 hpi or 48 hpi at 37°C, or at 37°C and 40°C after the removal of DCS at 30 hpi. Progeny from each condition were counted in HeLa cells. Bars show the means for three experiments performed in triplicate ± standard deviations. Significance was assessed by two-way ANOVA followed by Tukey’s post hoc test. *, P < 0.05; **, P < 0.01.
DISCUSSION
A TS DnaE mutant fails to replicate its genome.
Overall, our results show that the dnaEts mutant is able to undergo EB-to-RB conversion but that cell division by this mutant is impaired beginning in middevelopment. Determining if increased genome accumulation in the dnaEts mutant incubated at 40°C for 18 hpi prior to shifting to 37°C is due to a Chlamydia subpopulation escaping the normal stress response (Fig. 5) could clarify some of these observations. Active genome replication was not required for cell division to proceed if preexisting genomes were present, since dnaEts mutant progeny production following incubation at either 37°C or 40°C was indistinguishable (Fig. 7). This suggests that at the population level, most RB-to-EB conversions occur after genome replication has been completed. Thus, our data suggest that complete DNA replication is needed for proper septum assembly and that conversion between RBs and EBs can occur as long as the chromosome has been duplicated.
Elongation of the DNA template is impaired with DnaEP852S.
Reduced initiation complex formation, nucleotide addition, and/or processivity of DnaE could have all potentially been causes of the dnaEts mutant phenotypes that we observed. However, structural modeling did not differentiate between these possibilities (Fig. 2). A TS screen for E. coli mutants that could not replicate DNA (74) identified a TS E. coli dnaE allele (dnaE486) that encodes EcDnaES885P (75). The analogous residue in CtDnaE (A922) is predicted to contact P852 in our model, so the mechanisms of temperature sensitivity may be similar in these two mutant proteins, and the residues may be in a critical, uncharacterized DnaE motif.
We also identified two intragenic suppressor mutations that restored temperature resistance in the dnaEts mutant. In E. coli, similar suppressor screens identified several extragenic suppressor mutations of two different DnaEts proteins (76). These suppressor mutations were located primarily in genes that mediate core metabolic processes, suggesting a common mechanism of suppression via a reduced growth rate (75). Since our dnaEts suppressor mutants acquired intragenic mutations and developed normally, a reduced growth rate is unlikely to be the relevant mechanism here. Another group showed that E. coli DnaAts and DnaCts mutants that denatured at a nonpermissive temperature could renature at the permissive temperature (77). Similarly, since the function of the DnaEP852S mutant is comparable to that of the DnaEWT recombinant strain at 37°C, we hypothesize that this mutant does not have a discrete defect but is simply unstable at higher temperatures. The effects of specific amino acid substitutions in EcDnaE have been evaluated using reconstituted in vitro DNA replication assays (36, 43). Studies of DnaEP852S that employ these approaches could be warranted, because they could provide new structure-function insights into what appears to be an important and uncharacterized DnaE motif.
While we hypothesize that the defects in replication that we observed arise from a reduction in the processivity of the DnaEts mutant due to its instability at 40°C, we cannot exclude the possibility that the dnaEts mutant could be a temperature-sensitive synthesis (TSS) mutant. TSS mutants cannot generate new functional protein at the nonpermissive temperature; however, any temperature-sensitive protein synthesized in a TSS mutant prior to shifting to the nonpermissive temperature retains its activity. Results similar to what we observed upon the temperature shift to 40°C have been obtained using protein synthesis inhibitors on C. trachomatis; DNA replication is halted with no detectable residual activity (21). Our temperature shift assays demonstrate that longer incubations at 40°C before shifting to 37°C result in fewer genome copies, even when the time spent at 37°C is kept constant (Fig. 1E), suggesting that any residual activity of the DnaEts mutant is insufficient to maintain genome replication at 40°C.
DnaE inhibition affects inclusion size and stability.
Although there are indirect ways to block DNA replication in Chlamydia spp., including blocking transcription and translation or introducing double-stranded DNA breaks, in this study we are able to determine the impact on DNA replication when specifically inhibiting DNA polymerase activity. Cell growth and survival must balance DNA and protein synthesis with nutrient availability, and multiple feedback loops likely determine if and when the chlamydial genome is replicated (76). Conversely, inhibition of DNA replication through transcriptional, translational, or gyrase inhibition blocks key stages in Chlamydia development, including inclusion expansion. Inhibition of the glucose-6-phosphate transporter, UhpC, inhibits DNA replication but does not prevent inclusion expansion (21), suggesting that carbon sources or the energy derived from them drives inclusion expansion and that DNA replication is independent of inclusion expansion. In contrast, the size of dnaEts mutant inclusions at 40°C correlated with the genome copy number, and changes in inclusion size during temperature shifts also reflected the genome copy number (Fig. 2 and 3). The UhpC inhibitor may indirectly cause inhibition of DNA replication due to the absence of a primary carbon source but may still allow for inclusion maturation. However, for the dnaEts mutant, any feedback systems that regulate DNA replication would still license initiation; the lack of DNA replication is a direct effect of a faulty polymerase. Since the dnaEts mutant inclusion grows and acquires mid-to-late effectors (Inc proteins) at the expected points in development (Fig. 4B), it appears that inclusion maturation is genome replication independent. Meanwhile, in the absence of functional DnaE, the bacteria repeatedly initiate replication in a futile attempt to replicate the genome in preparation for cell division.
The integrity of dnaEts mutant inclusions at 40°C appears to be compromised (Fig. 5); however, to confirm this, additional experiments will be needed to establish whether mechanisms of clearance are upregulated in the host cells and the full repertoire of inclusion membrane proteins in these inclusions. Inclusion membrane stability is established early in development, and expression of some inclusion membrane proteins is associated with stability peaks within 2 h of EB entry (78). Whether other T3SS proteins are dependent on DNA replication to localize to the inclusion membrane remains to be evaluated.
The chromosomal copy number regulates cell division and EB formation in C. trachomatis.
Our observation that some dnaEts mutant inclusions contained multiple RBs when this mutant was continuously incubated at the nonpermissive temperature was surprising. Several possibilities could explain the observation of multiple RBs per inclusion in the dnaEts mutant at 40°C. The explanation that DnaEP852S retains some residual function seems unlikely, because we did not detect any increase in dnaEts mutant genomes when this mutant was continuously incubated at 40°C. Cell division requires at least two chromosomes, and we detected multiple replication forks per chromosome during normal chlamydial development (Fig. 3). Barring the possibility that the dnaEts mutant is a TSS mutant, another potential explanation is that other, minor DNA polymerases, such as PolA (DNA polymerase I), could compensate for the lack of DnaE if the majority of the genome was already replicated, which would be sufficient for a single cell division event. While DNA replication does not occur in EBs, an intriguing possibility is that DNA replication in RBs outpaces RB-to-EB transition events. This would give rise to EBs containing multiple chromosomal copies, thereby bypassing the need to replicate the genome before the first round of division upon transitioning back to RBs. While the genome copy number is a common measure of chlamydial growth in the chlamydia field, the number of full and/or partial genomes has never been determined at the single-cell level, and advances in microscopy will be required to address this possibility.
Replication and septation are decoupled in Chlamydia spp. (24), so there is precedent for the idea that cell division could proceed in the absence of genome replication. It is also clear that both RBs and persistent chlamydiae can maintain multiple chromosome copies. Finally, de novo transcription of DNA replication genes begins after immediate-early genes, which likely establish the inclusion niche (14). In either scenario, having near-complete or fully replicated genomes could increase gene dosage and enhance transcription early in infection, when few bacteria are present and an immediate need to generate proteins in order to avoid immune detection and acquire nutrients exists. Fluorescent in situ hybridization experiments may be able to elucidate the degree of genome replication in single EBs.
Nucleoid occlusion: a replication-dependent model for the regulation of cell division in Chlamydia.
The loss of both MreB and peptidoglycan at the division septum in the dnaEts mutant at 40°C suggests that incomplete genome duplication in C. trachomatis acts as a form of negative feedback inhibition for cell division. We hypothesize that septum formation may be inhibited via nucleoid occlusion, which is a common regulatory process in bacteria that prevents cell division until after genome replication is complete (79). Most nucleoid occlusion systems function by inhibiting FtsZ (80–82), which chlamydiae lack (66). The only known nucleoid occlusion system that does not rely directly on FtsZ is the Noc/ParB system. In this system, DNA is recruited to the inner membrane by Noc, which is initially located at midcell and eventually moves to the cell poles (83). While at midcell, Noc outcompetes FtsZ at the inner membrane, preventing FtsZ-dependent septum formation until after DNA replication is nearly complete. Most Chlamydia spp. encode Noc/ParB homologs (84), and, since MreB replaces FtsZ in the septal peptidoglycan synthase complex in Chlamydia (64) this system may function by limiting MreB localization. Future studies should address whether chlamydial Noc inhibits MreB localization at midcell until the completion of replication.
In conclusion, inhibiting replication completion is more detrimental than inhibiting replication initiation in Chlamydia. Defects in replication completion affected not only the genome copy number, cell division, and developmental form conversion but also the expansion and stability of the inclusion. Our results suggest that (i) the completion of genome replication serves as a regulatory checkpoint in cell division in Chlamydia and that (ii) these pathogens lack a negative regulatory feedback mechanism for responding to defects in replication completion. These two conclusions provide an appealing resolution to what were otherwise contradictory reports linking replication to other aspects of chlamydial development and offer a new avenue for therapeutic development that targets replication completion rather than initiation.
MATERIALS AND METHODS
Chlamydia strains and cultivation.
McCoy and HeLa 229 (HeLa) cells were obtained from the American Type Culture Collection. HeLa USU cells were obtained from the laboratory of Anthony Maurelli (University of Florida). All cells were maintained in high-glucose Dulbecco’s modified Eagle medium (DMEM) (HyClone) supplemented with nonessential amino acids, 10 mM HEPES, and 10% fetal bovine serum (DMEM-10; Atlanta Biologicals). C. trachomatis L2 434/Bu was transformed with pGFP::SW2 (a gift from Ian Clarke), which constitutively expresses green fluorescent protein (GFP) and encodes a β-lactamase (85). The dnaEts mutant (sucCG603A pmpGG1629A dnaEC2554T copBG1283A) and its isogenic recombinant (dnaEWT [sucCG603A pmpGG1629A copBG1283A]) have been described previously (34). All strains used in this study were plaque purified twice (86) before purification over a 30% MD-76R (Mallinckrodt Pharmaceuticals) cushion (87). Cell culture was performed in a 5% CO2 humidified incubator at 37°C unless stated otherwise.
Whole-genome sequencing.
MD-76R-purified EBs were treated with DNase I (Promega) prior to whole-genome amplification using a REPLI-g kit (Qiagen) as described previously (34). Samples were multiplexed using the NexteraXT Dual Index primer kit, combined in equimolar quantities, and sequenced at the Tufts University Genomics Core Facility in Boston, MA. Paired-end 100-bp reads were performed on an Illumina MiSeq system.
Genome sequence data were analyzed as described elsewhere (88). Ambiguous sequences and mutation calls with low quality scores were resolved by PCR and Sanger sequencing.
Temperature shift assays.
HeLa cells in 96-well plates (Eppendorf) were infected at a multiplicity of infection (MOI) of 0.2 in triplicate for each strain. Replicate experimental plates were initially incubated at 40°C and then moved to 37°C at either 8, 12, 15, or 18 hpi, or vice versa. Control plates were incubated at 37°C or 40°C throughout. At the times indicated in the figure legends, cells were fixed with 3.7% formaldehyde. Cells were subsequently blocked and permeabilized with blocking buffer (1% bovine serum albumin [BSA] and 0.1% saponin in phosphate-buffered saline [PBS]) and were labeled with conditioned EVI-H1 hybridoma supernatant, which contains a chlamydial lipopolysaccharide (LPS)-specific monoclonal antibody, and a DyLight 594-conjugated goat anti-mouse IgG secondary antibody (Thermo Scientific) in blocking buffer. Four images per well at ×10 were acquired on an EVOS FL Auto microscope (Thermo Scientific). Inclusion cross-sectional areas were measured in CellProfiler (89) using a modified pipeline (90).
For persistence assays, infections were fixed with 3.7% formaldehyde to preserve the GFP fluorescence of the chlamydiae. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired on a Nikon TiE fluorescence microscope with a 100× oil immersion objective. Images are representative of >50 inclusions with at least 10 fields of view for each condition examined and were processed with ImageJ.
Progeny assays.
HeLa cells were infected at an MOI of 0.1 in sucrose-phosphate-glutamic acid buffer (SPG) with 30% MD-76R-purified (87) EBs by centrifugation at 1,600 × g for 1 h at room temperature and rocking at 37°C for 30 min. The SPG was then aspirated, and DMEM-10 plus 1 µg/ml cycloheximide were added. The infected cultures were incubated at 37°C or 40°C. The medium was aspirated at 34 hpi and replaced with SPG. The infected plates were then frozen at −80°C. EBs were harvested by thawing the plates and were titrated on fresh HeLa cell monolayers (88). A parallel 96-well plate was infected to determine the titer of the inoculum, incubated at 37°C, and fixed with 3.7% formaldehyde at 34 hpi to normalize fold change differences between strains.
Infections were performed as described above for persistence assays, except that after the inoculum was removed, DMEM-10 with or without 400 μM (41 mg/ml) d-cycloserine (Sigma-Aldrich) was used, and the infected cultures were incubated at 37°C for 30 hpi. At 30 hpi, either the infections were stopped or the medium was replaced with fresh DMEM-10, and the cultures were incubated for an additional 18 h at 37°C or 40°C.
Genome copy number.
Genomic DNA was extracted from aliquots of samples generated for progeny formation assays using a gMax kit according to the manufacturer’s instructions (IBI Scientific). The FastStart TaqMan Probe master mix (Roche) was used to quantitate the genome copies in 1 μl of purified genomic DNA (gDNA) using primers 5′-GTAGCGGTGAAATGCGTAGA-3′ and 5′-CGCCTTAGCGTCAGGTATAAA-3′ with a TaqMan probe specific to the 16S rRNA gene of Chlamydia (5′-/56-FAM/ATGTGGAAG/ZEN/ACCACCAGT-3′). Genome copy numbers were determined using a standard curve generated from serial dilutions of a plasmid containing a cloned copy of a chlamydial 16S rRNA gene. Reaction mixtures underwent 40 cycles of amplification on an Eppendorf Realplex4 system using the following conditions: 20 s at 95°C, 1 min at 60°C, and 20 s at 68°C. Copy number analysis was performed using Realplex software, and final copy number values were divided by 2 to account for the two copies of the 16S rRNA gene on the genome.
Duplex qPCR experiments also included the trpB-specific primers 5′-CACTCCATTTCCGCTGGA-3′ and 5′-GCTCGTCCTGACTCATGCATT-3′ and TaqMan probe 5′-/HEX/CTTCAGTTGGGCCAGATCATGCCG/BQ1/-3′ (91). Copy numbers were determined using a standard curve generated from a plasmid containing a cloned copy of the L2 trpB gene.
Transmission electron microscopy.
HeLa cells were infected at an MOI of 0.5 as described above. Infections were fixed at 32 hpi for 1 h on ice using 4% paraformaldehyde–2.5% glutaraldehyde. Sample processing was performed essentially as described elsewhere (92). The cells were washed with sodium cacodylate, embedded in resin, and stained with osmium tetroxide. Ultrathin sections were further stained with uranyl acetate and lead citrate. Images were captured using a JEOL JEM 1011 microscope with a Gatan 890 4k × 4k digital camera at the Indiana University Microscopy Center, Bloomington, IN.
DnaE suppressor screen.
HeLa cell monolayers in 15 T-175 flasks were infected at an MOI of 30 (∼1 × 1010 EBs total) by rocking for 2 h at 37°C. The inoculum was replaced with fresh DMEM-10 with 1 µg/ml cycloheximide, and the infected cultures were incubated at 40°C for 34 hpi. The monolayers were harvested into SPG using glass beads. Host cell debris was removed by centrifugation at 500 × g for 5 min. Supernatants were used to infect fresh HeLa cell monolayers in 2 wells of a 6-well plate and were centrifuged at 1,600 × g to enhance infection. Selection at 40°C for 34 h was repeated five times. A portion of the resulting progeny was scaled up at 37°C to generate a stock and to isolate genomic DNA by alkaline lysis for Sanger sequencing of dnaE using the primers listed in Table 2.
TABLE 2.
Primers used for dnaE Sanger sequencing
| Position (bp) | Direction | Primer |
|---|---|---|
| –200 | Forward | CGGCTTTACAGGATGGTTCG |
| 312 | Forward | CTCTCCTCTCTTGCTTACACAG |
| 843 | Forward | CTATTTGCAGCACATCCAGAGAC |
| 1379 | Forward | CAGAGAGCGCGTTATTAACTATGC |
| 1894 | Forward | CCACGATTCCTCTAGATGACC |
| 2265 | Forward | GAACAGATGGTCAAGGAACG |
| 2808 | Forward | GCTATCCTTAATGACCTTTACGACAC |
| +297 | Reverse | GTTCGTGTTGAGTATATTAATGCTATTACAGG |
Genome sequencing was performed as described above for the two clonal intragenic suppressors referred to in the text as the D828Y (sucCG603A pmpGG1629A dnaEG2482T C2554T copBG1283A dsdDG750A) and E1061K (sucCG603A pmpGG1629A dnaEC2554T G3181A copBG1283A) suppressors.
Protein alignment and structural modeling.
ClustalW was used for primary sequence alignment. The Phyre2 server was used to generate a tertiary structure of CtDnaE (37). CtDnaE was modeled with >90% confidence due to the high similarity of the EcDnaE and TaDnaE crystal structures (PDB accession codes 2HQA and 3E0D, respectively). Relevant residues were identified in the PyMOL Molecular Graphics System, version 2.0 (Schrödinger, LLC).
Peptidoglycan and immunofluorescence labeling and fluorescence imaging.
HeLa cells were infected with the dnaEts mutant or dnaEts E1061K suppressor at an MOI of 1 and were grown in the presence of a 1 mM concentration of the peptidoglycan-incorporating dipeptide ethynyl-d-alanine–d-alanine (EDA-DA) (63) (generously provided to us by Michael VanNieuwenhze). At the indicated times, the cells were washed three times with 1× PBS and were permeabilized with methanol and 0.5% Triton X-100 (5 min each) prior to blocking with 3% BSA for 1 h. Cells were then incubated in the presence of cupric sulfate and azide-modified Alexa Fluor 488, resulting in the fusing of the Alexa Fluor dye and PG-integrated dipeptide via a click chemistry reaction, as described previously (63, 64). The major outer membrane protein (MOMP) was labeled with an anti-MOMP primary antibody (goat) and a secondary antibody (Alexa Fluor 594-conjugated chicken anti-goat antibody) at 1:500 and 1:1,000 dilutions, respectively. MreB was labeled utilizing a rabbit anti-MreB primary antibody (70) (generously provided to us by Scott Hefty of the University of Kansas) and an anti-rabbit secondary antibody conjugated to Alexa Fluor 488. IncA was labeled utilizing mouse polyclonal anti-IncA antibodies (generously provided to us by Dan Rockey of Oregon State University).
Fluorescence imaging was conducted utilizing a Zeiss 700 confocal microscope or a Zeiss ELYRA super resolution imaging suite set to SIM (structural imaging microscopy) mode. Laser intensities and settings were optimized for samples grown at the permissive temperature (37°C) for 12 hpi. z-stracks were used to generate maximum-intensity projections. Images were processed with Zen software (Zeiss). Images are representative of >20 fields of view per strain/condition/time point examined.
Statistics.
Statistical analyses were performed using GraphPad Prism, version 6.0. Data from growth curves and genome accumulation experiments were log10 transformed prior to analysis. All data were analyzed by either one- or two-way analysis of variance (ANOVA) followed by the post hoc tests specified in the figure legends to correct for multiple comparisons.
Data availability.
Unique research resources, including mutant bacterial strains, will be made available to others in the private and public sectors as soon as appropriate agreements covering such transfers can be executed, or, in the case of project data, shall be published as soon as possible or otherwise shared with other researchers pursuant to NIH policies regarding data sharing. All raw imaging data generated during the course of this work will be stored in perpetuity on both external drives as well as our internal networked server, was made available to reviewers prior to publication, and will be made available to all interested parties subsequent to publication, according to NIH guidelines. The Uniformed Services University of the Health Sciences (USUHS)-Henry Jackson Foundation (HJF) Joint Office of Technology Transfer (JOTT) manages all technology transfer activities for data and resources generated by USUHS and HJF researchers. The JOTT will adhere to all applicable policies, principles, guidelines, and procurement rules in making unique research resources readily available for research purposes to qualified individuals and entities and in ensuring that there is no more than a short-term restriction on publication or public dissemination of data in order for the JOTT to evaluate and file for patent protection on any subject invention that may arise from the funded study. Dissemination of unpublished data that are considered to be proprietary or confidential shall occur pursuant to a suitable confidential disclosure agreement prepared by the JOTT. Material transfer agreements (MTAs), license agreements, and Cooperative Research and Development Agreements (CRADAs), as appropriate, will be used to transfer material property. MTAs will be established to share resources among members of the research community for noncommercial research use.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael VanNieuwenhze (Indiana University) for providing us with the peptidoglycan-labeling reagent EDA-DA, as well as Scott Hefty (University of Kansas) and Dan Rockey (Oregon State University) for providing us with MreBCT and IncACT-specific antisera, respectively. We thank Rick Morrison (University of Arkansas), Stephen Jordan (Indiana University), and Tony Maurelli (University of Florida) for helpful comments in preparing this article.
This work was made possible by grant R01AI099278 to D.E.N., grant R35 GM138202 to G.W.L., and a faculty start-up award to G.W.L. J.A.B. was supported by grant T32AI007637.
The views and opinions expressed here are our own and should not be construed as being “official” or representing the views or policy of the Uniformed Services University or the Department of Defense.
Author contributions are as follows: Conceptualization and Design, J.A.B., D.E.N., and G.W.L.; Data Curation and Formal Analysis, J.A.B., D.E.N., and G.W.L.; Investigation, Methodology, Validation, Visualization, J.A.B., M.B., A.B., B.D.S., D.E.N., G.W.L.; Writing — Original Draft, J.A.B. and G.W.L.; Writing — Review & Editing, J.A.B., M.B., A.B., B.D.S., D.E.N., and G.W.L.; Funding Acquisition, Project Administration, and Supervision, D.E.N. and G.W.L. All authors read and approved the final manuscript.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Horn M. 2008. Chlamydiae as symbionts in eukaryotes. Annu Rev Microbiol 62:113–131. doi: 10.1146/annurev.micro.62.081307.162818. [DOI] [PubMed] [Google Scholar]
- 2.Taylor-Brown A, Vaughan L, Greub G, Timms P, Polkinghorne A. 2015. Twenty years of research into Chlamydia-like organisms: a revolution in our understanding of the biology and pathogenicity of members of the phylum Chlamydiae. Pathog Dis 73:1–15. doi: 10.1093/femspd/ftu009. [DOI] [PubMed] [Google Scholar]
- 3.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. 2004. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A 101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson DE, Taylor LD, Shannon JG, Whitmire WM, Crane DD, McClarty G, Su H, Kari L, Caldwell HD. 2007. Phenotypic rescue of Chlamydia trachomatis growth in IFN-γ treated mouse cells by irradiated Chlamydia muridarum. Cell Microbiol 9:2289–2298. doi: 10.1111/j.1462-5822.2007.00959.x. [DOI] [PubMed] [Google Scholar]
- 5.Fields KA, Hackstadt T. 2002. The chlamydial inclusion: escape from the endocytic pathway. Annu Rev Cell Dev Biol 18:221–245. doi: 10.1146/annurev.cellbio.18.012502.105845. [DOI] [PubMed] [Google Scholar]
- 6.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. 2012. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc Natl Acad Sci U S A 109:19781–19785. doi: 10.1073/pnas.1212831109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sixt BS, Siegl A, Muller C, Watzka M, Wultsch A, Tziotis D, Montanaro J, Richter A, Schmitt-Kopplin P, Horn M. 2013. Metabolic features of Protochlamydia amoebophila elementary bodies—a link between activity and infectivity in Chlamydiae. PLoS Pathog 9:e1003553. doi: 10.1371/journal.ppat.1003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grieshaber S, Grieshaber N, Yang H, Baxter B, Hackstadt T, Omsland A. 2018. Impact of active metabolism on Chlamydia trachomatis elementary body transcript profile and infectivity. J Bacteriol 200:e00065-18. doi: 10.1128/JB.00065-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niehus E, Cheng E, Tan M. 2008. DNA supercoiling-dependent gene regulation in Chlamydia. J Bacteriol 190:6419–6427. doi: 10.1128/JB.00431-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackstadt T, Baehr W, Ying Y. 1991. Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1. Proc Natl Acad Sci U S A 88:3937–3941. doi: 10.1073/pnas.88.9.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields KA, Fischer ER, Mead DJ, Hackstadt T. 2005. Analysis of putative Chlamydia trachomatis chaperones Scc2 and Scc3 and their use in the identification of type III secretion substrates. J Bacteriol 187:6466–6478. doi: 10.1128/JB.187.18.6466-6478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shen L, Macnaughtan MA, Frohlich KM, Cong Y, Goodwin OY, Chou CW, LeCour L, Jr, Krup K, Luo M, Worthylake DK. 2015. Multipart chaperone-effector recognition in the type III secretion system of Chlamydia trachomatis. J Biol Chem 290:28141–28155. doi: 10.1074/jbc.M115.670232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen S, McMahon RM, Martin JL, Huston WM. 2019. Life inside and out: making and breaking protein disulfide bonds in Chlamydia. Crit Rev Microbiol 45:33–50. doi: 10.1080/1040841X.2018.1538933. [DOI] [PubMed] [Google Scholar]
- 14.Belland RJ, Zhong G, Crane DD, Hogan D, Sturdevant D, Sharma J, Beatty WL, Caldwell HD. 2003. Genomic transcriptional profiling of the developmental cycle of Chlamydia trachomatis. Proc Natl Acad Sci U S A 100:8478–8483. doi: 10.1073/pnas.1331135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Almeida F, Borges V, Ferreira R, Borrego MJ, Gomes JP, Mota LJ. 2012. Polymorphisms in Inc proteins and differential expression of inc genes among Chlamydia trachomatis strains correlate with invasiveness and tropism of lymphogranuloma venereum isolates. J Bacteriol 194:6574–6585. doi: 10.1128/JB.01428-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mueller KE, Plano GV, Fields KA. 2014. New frontiers in type III secretion biology: the Chlamydia perspective. Infect Immun 82:2–9. doi: 10.1128/IAI.00917-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore ER, Ouellette SP. 2014. Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins. Front Cell Infect Microbiol 4:157. doi: 10.3389/fcimb.2014.00157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber MM, Bauler LD, Lam J, Hackstadt T. 2015. Expression and localization of predicted inclusion membrane proteins in Chlamydia trachomatis. Infect Immun 83:4710–4718. doi: 10.1128/IAI.01075-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaw EI, Dooley CA, Fischer ER, Scidmore MA, Fields KA, Hackstadt T. 2000. Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle. Mol Microbiol 37:913–925. doi: 10.1046/j.1365-2958.2000.02057.x. [DOI] [PubMed] [Google Scholar]
- 20.Mathews SA, Volp KM, Timms P. 1999. Development of a quantitative gene expression assay for Chlamydia trachomatis identified temporal expression of sigma factors. FEBS Lett 458:354–358. doi: 10.1016/s0014-5793(99)01182-5. [DOI] [PubMed] [Google Scholar]
- 21.Engstrom P, Bergstrom M, Alfaro AC, Syam Krishnan K, Bahnan W, Almqvist F, Bergstrom S. 2015. Expansion of the Chlamydia trachomatis inclusion does not require bacterial replication. Int J Med Microbiol 305:378–382. doi: 10.1016/j.ijmm.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Lee JK, Enciso GA, Boassa D, Chander CN, Lou TH, Pairawan SS, Guo MC, Wan FYM, Ellisman MH, Sutterlin C, Tan M. 2018. Replication-dependent size reduction precedes differentiation in Chlamydia trachomatis. Nat Commun 9:45. doi: 10.1038/s41467-017-02432-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang JD, Levin PA. 2009. Metabolism, cell growth and the bacterial cell cycle. Nat Rev Microbiol 7:822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Belland RJ, Nelson DE, Virok D, Crane DD, Hogan D, Sturdevant D, Beatty WL, Caldwell HD. 2003. Transcriptome analysis of chlamydial growth during IFN-gamma-mediated persistence and reactivation. Proc Natl Acad Sci U S A 100:15971–15976. doi: 10.1073/pnas.2535394100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lambden PR, Pickett MA, Clarke IN. 2006. The effect of penicillin on Chlamydia trachomatis DNA replication. Microbiology (Reading) 152:2573–2578. doi: 10.1099/mic.0.29032-0. [DOI] [PubMed] [Google Scholar]
- 26.Thompson CC, Carabeo RA. 2011. An optimal method of iron starvation of the obligate intracellular pathogen, Chlamydia trachomatis. Front Microbiol 2:20. doi: 10.3389/fmicb.2011.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinkworth AJ, Wildung MR, Carabeo RA. 2018. Genomewide transcriptional responses of iron-starved Chlamydia trachomatis reveal prioritization of metabolic precursor synthesis over protein translation. mSystems 3:e00184-17. doi: 10.1128/mSystems.00184-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beatty WL, Morrison RP, Byrne GI. 1994. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol Rev 58:686–699. doi: 10.1128/MR.58.4.686-699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dreses-Werringloer U, Padubrin I, Jurgens-Saathoff B, Hudson AP, Zeidler H, Kohler L. 2000. Persistence of Chlamydia trachomatis is induced by ciprofloxacin and ofloxacin in vitro. Antimicrob Agents Chemother 44:3288–3297. doi: 10.1128/aac.44.12.3288-3297.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scidmore MA, Fischer ER, Hackstadt T. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect Immun 71:973–984. doi: 10.1128/iai.71.2.973-984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scidmore MA, Rockey DD, Fischer ER, Heinzen RA, Hackstadt T. 1996. Vesicular interactions of the Chlamydia trachomatis inclusion are determined by chlamydial early protein synthesis rather than route of entry. Infect Immun 64:5366–5372. doi: 10.1128/IAI.64.12.5366-5372.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scherler A, Jacquier N, Kebbi-Beghdadi C, Greub G. 2020. Diverse stress-inducing treatments cause distinct aberrant body morphologies in the Chlamydia-related bacterium, Waddlia chondrophila. Microorganisms 8:89. doi: 10.3390/microorganisms8010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engstrom P, Krishnan KS, Ngyuen BD, Chorell E, Normark J, Silver J, Bastidas RJ, Welch MD, Hultgren SJ, Wolf-Watz H, Valdivia RH, Almqvist F, Bergstrom S. 2014. A 2-pyridone-amide inhibitor targets the glucose metabolism pathway of Chlamydia trachomatis. mBio 6:e02304-14. doi: 10.1128/mBio.02304-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brothwell JA, Muramatsu MK, Toh E, Rockey DD, Putman TE, Barta ML, Hefty PS, Suchland RJ, Nelson DE. 2016. Interrogating genes that mediate Chlamydia trachomatis survival in cell culture using conditional mutants and recombination. J Bacteriol 198:2131–2139. doi: 10.1128/JB.00161-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson DL, Cox MM. 2008. Lehninger principles of biochemistry, 5th ed. W H Freeman and Company, New York, NY. [Google Scholar]
- 36.Gefter ML, Hirota Y, Kornberg T, Wechsler JA, Barnoux C. 1971. Analysis of DNA polymerases II and 3 in mutants of Escherichia coli thermosensitive for DNA synthesis. Proc Natl Acad Sci U S A 68:3150–3153. doi: 10.1073/pnas.68.12.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McHenry CS. 2011. Bacterial replicases and related polymerases. Curr Opin Chem Biol 15:587–594. doi: 10.1016/j.cbpa.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Georgescu RE, Yurieva O, Kim SS, Kuriyan J, Kong XP, O'Donnell M. 2008. Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp. Proc Natl Acad Sci U S A 105:11116–11121. doi: 10.1073/pnas.0804754105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stukenberg PT, Studwell-Vaughan PS, O'Donnell M. 1991. Mechanism of the sliding beta-clamp of DNA polymerase III holoenzyme. J Biol Chem 266:11328–11334. [PubMed] [Google Scholar]
- 41.Giebel AM, Hu S, Rajaram K, Finethy R, Toh E, Brothwell JA, Morrison SG, Suchland RJ, Stein BD, Coers J, Morrison RP, Nelson DE. 2019. Genetic screen in Chlamydia muridarum reveals role for an interferon-induced host cell death program in antimicrobial inclusion rupture. mBio 10:e00385-19. doi: 10.1128/mBio.00385-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wing RA, Bailey S, Steitz TA. 2008. Insights into the replisome from the structure of a ternary complex of the DNA polymerase III alpha-subunit. J Mol Biol 382:859–869. doi: 10.1016/j.jmb.2008.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pavlov AR, Belova GI, Kozyavkin SA, Slesarev AI. 2002. Helix-hairpin-helix motifs confer salt resistance and processivity on chimeric DNA polymerases. Proc Natl Acad Sci U S A 99:13510–13515. doi: 10.1073/pnas.202127199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hackstadt T, Scidmore-Carlson MA, Shaw EI, Fischer ER. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol 1:119–130. doi: 10.1046/j.1462-5822.1999.00012.x. [DOI] [PubMed] [Google Scholar]
- 45.Suchland RJ, Rockey DD, Bannantine JP, Stamm WE. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect Immun 68:360–367. doi: 10.1128/iai.68.1.360-367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muramatsu MK, Brothwell JA, Stein BD, Putman TE, Rockey DD, Nelson DE. 2016. Beyond tryptophan synthase: identification of genes that contribute to Chlamydia trachomatis survival during gamma interferon-induced persistence and reactivation. Infect Immun 84:2791–2801. doi: 10.1128/IAI.00356-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ouellette SP, Rueden KJ, Rucks EA. 2016. Tryptophan codon-dependent transcription in Chlamydia pneumoniae during gamma interferon-mediated tryptophan limitation. Infect Immun 84:2703–2713. doi: 10.1128/IAI.00377-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonner CA, Byrne GI, Jensen RA. 2014. Chlamydia exploit the mammalian tryptophan-depletion defense strategy as a counter-defensive cue to trigger a survival state of persistence. Front Cell Infect Microbiol 4:17. doi: 10.3389/fcimb.2014.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hurst EW, Landquist JK, Melvin P, Peters JM, Senior N, Silk JA, Stacy GJ. 1953. The therapy of experimental psittacosis and lymphogranuloma venereum (inguinale) II. The activity of quinoxaline-1:4-dioxide and substituted and related compounds, with a note on the morphological changes induced in lymphogranuloma virus by these compounds and by antibiotics. Br J Pharmacol Chemother 8:297–305. doi: 10.1111/j.1476-5381.1953.tb00798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moulder JW, Novosel DL, Officer JE. 1963. Inhibition of the growth of agents of the psittacosis group by d-cycloserine and its specific reversal by d-alanine. J Bacteriol 85:707–711. doi: 10.1128/JB.85.3.707-711.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura A, Manire GP. 1968. Effect of penicillin on the multiplication of meningopneumonitis organisms (Chlamydia psittaci). J Bacteriol 96:875–880. doi: 10.1128/JB.96.4.875-880.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matsumoto A, Manire GP. 1970. Electron microscopic observations on the effects of penicillin on the morphology of Chlamydia psittaci. J Bacteriol 101:278–285. doi: 10.1128/JB.101.1.278-285.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyrick PB. 2010. Chlamydia trachomatis persistence in vitro: an overview. J Infect Dis 201(Suppl 2):S88–S95. doi: 10.1086/652394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kintner J, Lajoie D, Hall J, Whittimore J, Schoborg RV. 2014. Commonly prescribed beta-lactam antibiotics induce C. trachomatis persistence/stress in culture at physiologically relevant concentrations. Front Cell Infect Microbiol 4:44. doi: 10.3389/fcimb.2014.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dlugosz A, Muschiol S, Zakikhany K, Assadi G, D'Amato M, Lindberg G. 2014. Human enteroendocrine cell responses to infection with Chlamydia trachomatis: a microarray study. Gut Pathog 6:24. doi: 10.1186/1757-4749-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frohlich KM, Hua Z, Quayle AJ, Wang J, Lewis ME, Chou CW, Luo M, Buckner LR, Shen L. 2014. Membrane vesicle production by Chlamydia trachomatis as an adaptive response. Front Cell Infect Microbiol 4:73. doi: 10.3389/fcimb.2014.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hai J, Serradji N, Mouton L, Redeker V, Cornu D, El Hage Chahine JM, Verbeke P, Hemadi M. 2016. Targeted delivery of amoxicillin to C. trachomatis by the transferrin iron acquisition pathway. PLoS One 11:e0150031. doi: 10.1371/journal.pone.0150031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Slade JA, Brockett M, Singh R, Liechti GW, Maurelli AT. 2019. Fosmidomycin, an inhibitor of isoprenoid synthesis, induces persistence in Chlamydia by inhibiting peptidoglycan assembly. PLoS Pathog 15:e1008078. doi: 10.1371/journal.ppat.1008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beatty WL, Morrison RP, Byrne GI. 1994. Immunoelectron-microscopic quantitation of differential levels of chlamydial proteins in a cell culture model of persistent Chlamydia trachomatis infection. Infect Immun 62:4059–4062. doi: 10.1128/IAI.62.9.4059-4062.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Frohlich KM, Buckner L, Quayle AJ, Luo M, Feng X, Beatty W, Hua Z, Rao X, Lewis ME, Sorrells K, Santiago K, Zhong G, Shen L. 2011. Altered protein secretion of Chlamydia trachomatis in persistently infected human endocervical epithelial cells. Microbiology (Reading) 157:2759–2771. doi: 10.1099/mic.0.044917-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gussmann J, Al-Younes HM, Braun PR, Brinkmann V, Meyer TF. 2008. Long-term effects of natural amino acids on infection with Chlamydia trachomatis. Microb Pathog 44:438–447. doi: 10.1016/j.micpath.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 62.Gordon FB, Quan AL. 1972. Susceptibility of Chlamydia to antibacterial drugs: test in cell cultures. Antimicrob Agents Chemother 2:242–244. doi: 10.1128/aac.2.3.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liechti GW, Kuru E, Hall E, Kalinda A, Brun YV, VanNieuwenhze M, Maurelli AT. 2014. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506:507–510. doi: 10.1038/nature12892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liechti G, Kuru E, Packiam M, Hsu Y-P, Tekkam S, Hall E, Rittichier JT, VanNieuwenhze M, Brun YV, Maurelli AT. 2016. Pathogenic Chlamydia lack a classical sacculus but synthesize a narrow, mid-cell peptidoglycan ring, regulated by MreB, for cell division. PLoS Pathog 12:e1005590. doi: 10.1371/journal.ppat.1005590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuru E, Radkov A, Meng X, Egan A, Alvarez L, Dowson A, Booher G, Breukink E, Roper DI, Cava F, Vollmer W, Brun Y, VanNieuwenhze MS. 2019. Mechanisms of incorporation for d-amino acid probes that target peptidoglycan biosynthesis. ACS Chem Biol 14:2745–2756. doi: 10.1021/acschembio.9b00664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rivas-Marin E, Canosa I, Devos DP. 2016. Evolutionary cell biology of division mode in the bacterial Planctomycetes-Verrucomicrobia-Chlamydiae superphylum. Front Microbiol 7:1964. doi: 10.3389/fmicb.2016.01964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ouellette SP, Karimova G, Subtil A, Ladant D. 2012. Chlamydia co-opts the rod shape-determining proteins MreB and Pbp2 for cell division. Mol Microbiol 85:164–178. doi: 10.1111/j.1365-2958.2012.08100.x. [DOI] [PubMed] [Google Scholar]
- 68.Ranjit DK, Liechti GW, Maurelli AT. 2020. Chlamydial MreB directs cell division and peptidoglycan synthesis in Escherichia coli in the absence of FtsZ activity. mBio 11:e03222-19. doi: 10.1128/mBio.03222-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee J, Cox JV, Ouellette SP. 2020. Critical role for the extended N terminus of chlamydial MreB in directing its membrane association and potential interaction with divisome proteins. J Bacteriol 202:e00034-20. doi: 10.1128/JB.00034-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kemege KE, Hickey JM, Barta ML, Wickstrum J, Balwalli N, Lovell S, Battaile KP, Hefty PS. 2015. Chlamydia trachomatis protein CT009 is a structural and functional homolog to the key morphogenesis component RodZ and interacts with division septal plane localized MreB. Mol Microbiol 95:365–382. doi: 10.1111/mmi.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerard HC, Branigan PJ, Schumacher HR, Jr, Hudson AP. 1998. Synovial Chlamydia trachomatis in patients with reactive arthritis/Reiter's syndrome are viable but show aberrant gene expression. J Rheumatol 25:734–742. [PubMed] [Google Scholar]
- 72.Gerard HC, Krausse-Opatz B, Wang Z, Rudy D, Rao JP, Zeidler H, Schumacher HR, Whittum-Hudson JA, Kohler L, Hudson AP. 2001. Expression of Chlamydia trachomatis genes encoding products required for DNA synthesis and cell division during active versus persistent infection. Mol Microbiol 41:731–741. doi: 10.1046/j.1365-2958.2001.02550.x. [DOI] [PubMed] [Google Scholar]
- 73.Pokorzynski ND, Brinkworth AJ, Carabeo R. 2019. A bipartite iron-dependent transcriptional regulation of the tryptophan salvage pathway in Chlamydia trachomatis. Elife 8:e42295. doi: 10.7554/eLife.42295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wechsler JA, Gross JD. 1971. Escherichia coli mutants temperature-sensitive for DNA synthesis. Mol Gen Genet 113:273–284. doi: 10.1007/BF00339547. [DOI] [PubMed] [Google Scholar]
- 75.Vandewiele D, Fernandez de Henestrosa AR, Timms AR, Bridges BA, Woodgate R. 2002. Sequence analysis and phenotypes of five temperature sensitive mutator alleles of dnaE, encoding modified alpha-catalytic subunits of Escherichia coli DNA polymerase III holoenzyme. Mutat Res 499:85–95. doi: 10.1016/s0027-5107(01)00268-8. [DOI] [PubMed] [Google Scholar]
- 76.Strauss B, Kelly K, Dincman T, Ekiert D, Biesieda T, Song R. 2004. Cell death in Escherichia coli dnaE(Ts) mutants incubated at a nonpermissive temperature is prevented by mutation in the cydA gene. J Bacteriol 186:2147–2155. doi: 10.1128/jb.186.7.2147-2155.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zyskind JW, Deen LT, Smith DW. 1977. Temporal sequence of events during the initiation process in Escherichia coli deoxyribonucleic acid replication: roles of the dnaA and dnaC gene products and ribonucleic acid polymerase. J Bacteriol 129:1466–1475. doi: 10.1128/JB.129.3.1466-1475.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weber MM, Lam JL, Dooley CA, Noriea NF, Hansen BT, Hoyt FH, Carmody AB, Sturdevant GL, Hackstadt T. 2017. Absence of Specific Chlamydia trachomatis inclusion membrane proteins triggers premature inclusion membrane lysis and host cell death. Cell Rep 19:1406–1417. doi: 10.1016/j.celrep.2017.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wu LJ, Errington J. 2011. Nucleoid occlusion and bacterial cell division. Nat Rev Microbiol 10:8–12. doi: 10.1038/nrmicro2671. [DOI] [PubMed] [Google Scholar]
- 80.Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Mannik J. 2014. Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet 10:e1004504. doi: 10.1371/journal.pgen.1004504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bernhardt TG, de Boer PA. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18:555–564. doi: 10.1016/j.molcel.2005.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu LJ, Errington J. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915–925. doi: 10.1016/j.cell.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 83.Adams DW, Wu LJ, Errington J. 2015. Nucleoid occlusion protein Noc recruits DNA to the bacterial cell membrane. EMBO J 34:491–501. doi: 10.15252/embj.201490177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 85.Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN. 2011. Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog 7:e1002258. doi: 10.1371/journal.ppat.1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matsumoto A, Izutsu H, Miyashita N, Ohuchi M. 1998. Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J Clin Microbiol 36:3013–3019. doi: 10.1128/JCM.36.10.3013-3019.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Caldwell HD, Kromhout J, Schachter J. 1981. Purification and partial characterization of the major outer membrane protein of Chlamydia trachomatis. Infect Immun 31:1161–1176. doi: 10.1128/IAI.31.3.1161-1176.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rajaram K, Giebel AM, Toh E, Hu S, Newman JH, Morrison SG, Kari L, Morrison RP, Nelson DE. 2015. Mutational analysis of the Chlamydia muridarum plasticity zone. Infect Immun 83:2870–2881. doi: 10.1128/IAI.00106-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamentsky L, Jones TR, Fraser A, Bray MA, Logan DJ, Madden KL, Ljosa V, Rueden C, Eliceiri KW, Carpenter AE. 2011. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics 27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Osaka I, Hills JM, Kieweg SL, Shinogle HE, Moore DS, Hefty PS. 2012. An automated image-based method for rapid analysis of Chlamydia infection as a tool for screening antichlamydial agents. Antimicrob Agents Chemother 56:4184–4188. doi: 10.1128/AAC.00427-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ibana JA, Belland RJ, Zea AH, Schust DJ, Nagamatsu T, AbdelRahman YM, Tate DJ, Beatty WL, Aiyar AA, Quayle AJ. 2011. Inhibition of indoleamine 2,3-dioxygenase activity by levo-1-methyl tryptophan blocks gamma interferon-induced Chlamydia trachomatis persistence in human epithelial cells. Infect Immun 79:4425–4437. doi: 10.1128/IAI.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yu J, Rossi R, Hale C, Goulding D, Dougan G. 2009. Interaction of enteric bacterial pathogens with murine embryonic stem cells. Infect Immun 77:585–597. doi: 10.1128/IAI.01003-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Unique research resources, including mutant bacterial strains, will be made available to others in the private and public sectors as soon as appropriate agreements covering such transfers can be executed, or, in the case of project data, shall be published as soon as possible or otherwise shared with other researchers pursuant to NIH policies regarding data sharing. All raw imaging data generated during the course of this work will be stored in perpetuity on both external drives as well as our internal networked server, was made available to reviewers prior to publication, and will be made available to all interested parties subsequent to publication, according to NIH guidelines. The Uniformed Services University of the Health Sciences (USUHS)-Henry Jackson Foundation (HJF) Joint Office of Technology Transfer (JOTT) manages all technology transfer activities for data and resources generated by USUHS and HJF researchers. The JOTT will adhere to all applicable policies, principles, guidelines, and procurement rules in making unique research resources readily available for research purposes to qualified individuals and entities and in ensuring that there is no more than a short-term restriction on publication or public dissemination of data in order for the JOTT to evaluate and file for patent protection on any subject invention that may arise from the funded study. Dissemination of unpublished data that are considered to be proprietary or confidential shall occur pursuant to a suitable confidential disclosure agreement prepared by the JOTT. Material transfer agreements (MTAs), license agreements, and Cooperative Research and Development Agreements (CRADAs), as appropriate, will be used to transfer material property. MTAs will be established to share resources among members of the research community for noncommercial research use.