Abstract
Silencing of O6‐methylguanine‐DNA methyltransferase (MGMT) protein expression because of MGMT gene promoter hypermethylation is considered to be associated with postoperative chemoradiotherapy benefits in glioblastoma multiforme (GBM) patients. The objective of this study was to clarify the usability of MGMT immunohistochemistry (IHC) as a clinical biomarker.
We immunostained a tissue microarray containing biopsy samples of 164 GBM patients from the European Organization for Research and Treatment of Cancer and the National Cancer Institute of Canada (EORTC/NCIC) trial 26981/22981 using two commercial anti‐MGMT antibodies (clones MT3.1 and MT23.2). Immunostaining results were semiquantitatively evaluated by four observers from three neuropathological laboratories using a predefined algorithm. We analyzed (i) inter‐ and intraobserver agreement on MGMT expression (kappa statistics); (ii) correlation of MGMT expression with MGMT promoter methylation status (kappa statistics); and (iii) correlation of MGMT expression with patient outcome (log‐rank test). Interobserver agreement on MGMT expression varied from slight to almost perfect, whereas intraobserver agreement ranged from substantial to almost perfect. MGMT expression showed poor to moderate correlation with MGMT promoter methylation status. We found no significant association of MGMT expression with patient outcome. In our hands, observer variability as well as lack of association with the MGMT promoter methylation status and patient survival impeded the use of anti‐MGMT immunohistochemistry as a clinical biomarker for routine diagnostic purposes.
Keywords: biomarker, glioblastoma, immunohistochemistry, MGMT, prognosis
INTRODUCTION
Glioblastoma multiforme (GBM) is the most common malignant type of primary brain tumor (33). A recent prospective multicenter study conducted through the collaboration of the European Organization for Research and Treatment of Cancer (EORTC) and the National Cancer Institute of Canada (NCIC) (EORTC/NCIC trial 26981/22981) showed that the addition of temozolomide to radiotherapy for newly diagnosed GBM results in a clinically meaningful and statistically significant survival benefit with minimal toxicity (52). Consequently, postoperative combined radio–chemotherapy followed by adjuvant chemotherapy with temozolomide is currently considered as standard adjuvant therapy for GBM patients. In a translational study conducted in parallel to the EORTC/NCIC trial 26981/22981, a strong correlation of the methylation status of the O6‐methylguanine‐methyltransferase (MGMT) gene promoter with temozolomide treatment effect and outcome was shown (26). MGMT promoter methylation results in transcriptional silencing, and therefore, inhibition of expression of MGMT, a DNA repair protein that removes methyl groups from the O6‐position of guanine, thus counteracting the effect of alkylating chemotherapy 17, 49, 55.
In the study by Hegi et al, MGMT promoter methylation status was assessed by methylation‐specific polymerase chain reaction (MSP) (26). However, MSP is a relatively complex and time‐consuming method not often available in the local treatment centers. In addition, the formalin fixation and paraffin embedding of tumor tissues deteriorates the DNA quality in the tissue, which may lead to failure of amplification by MSP, particularly in small samples (eg, stereotactic biopsies). MGMT protein can be visualized immunohistochemically, and commercial anti‐MGMT antibodies are available. There are several potential advantages of immunohistochemistry (IHC) as compared with MSP. IHC is a commonly used and reliable method in diagnostic histopathology and is available in most laboratories. Furthermore, IHC works on formalin‐fixed and paraffin‐embedded tissue and is less expensive than MSP. Several studies have reported significant associations of immunohistochemically assessed MGMT expression with patient outcome in glioma 2, 7, 12, 44, 48. A study by Friedman et al in 1998 indicated that pretherapy analysis of MGMT protein expression in malignant gliomas may help to identify patients in whom tumors are resistant to temozolomide (19). Some more recent studies on small patient series reported similar findings. Anda et al reported in a study on 18 patients that glioblastomas with strong immunohistochemical MGMT staining may show more resistance to alkylating chemotherapy (2). Chinot et al found in a study on 29 glioblastoma patients that MGMT expression correlated with response to temozolomide (12). Brell et al reported a correlation between MGMT protein expression and survival in patients with anaplastic gliomas who had received alkylating chemotherapy (7). Similar results were reported for pediatric patients with malignant gliomas (48). Therefore, there is broad interest in the clinical use of MGMT immunostaining in this tumor type. However, the clinical usability of MGMT IHC has never been systematically studied so far. Essential prerequisites for use of anti‐MGMT staining in the diagnostic setting are high observer agreement (analytical performance) and reproducible association with treatment response and patient outcome (clinical performance) (24).
The objective of the present study was to test whether MGMT IHC in GBM can be used as a clinical biomarker in the routine setting. To this end, we systematically assessed for the first time whether the analytical and clinical performances of MGMT immunostaining are adequate for routine diagnostic purposes.
MATERIALS AND METHODS
Patients
A total of 164 biopsies of glioblastoma patients were available for this study. All the cases belonged to a previously published glioblastoma cohort of 573 cases that have been prospectively recruited in a multicenter approach by collaboration of the EORTC and the NCIC (EORTC/NCIC trial 26981/22981) 26, 52. For our study, we extracted the data on MGMT promoter methylation status and patient outcome from the database of EORTC/NCIC trial 26981/22981.
In our total study cohort of 164 cases, the median age was 54 years (age range 25–70 years). Eighty‐one cases (49.4%) had been randomized to the “radiotherapy only” arm and 83 (50.6%) cases to the “radiotherapy plus temozolomide” arm. Median follow‐up time was 28 months (range 0–39 months). In 122 out of 164 (74.4%) cases, methylation status of the MGMT promoter as assessed by MSP was available from the database of the EORTC/NCIC trial 26981/22981. Of these 122 cases, 59 (48.4%) had a methylated MGMT promoter and 63 (51.6%) had an unmethylated MGMT promoter.
All patients provided written informed consent for molecular studies of their tumor, and the protocol was approved by the ethics committee at each study center.
Tissue microarray (TMA)
TMA (Figure 1) was constructed from paraffin blocks of glioblastoma specimens from 164 patients for which paraffin blocks comprising compact tumor tissue of adequate surface and 5 mm depth were available. The tissue array was constructed by retrieving tissue core biopsies of 0.6 mm diameter from selected tumor regions of the donor paraffin blocks and precisely arraying them on a new recipient block using an arrayer instrument (34). We included one tissue core per patient on the TMA. The representative area of the tumor on the block was chosen based on the respective hematoxylin‐ and eosin‐stained tissue section. All TMA cores were selected from tumor areas that were histologically representative for the entire tumor. These same blocks had been used for central pathology review and for assessment of the MGMT promoter methylation status by MSP 26, 52.
Figure 1.
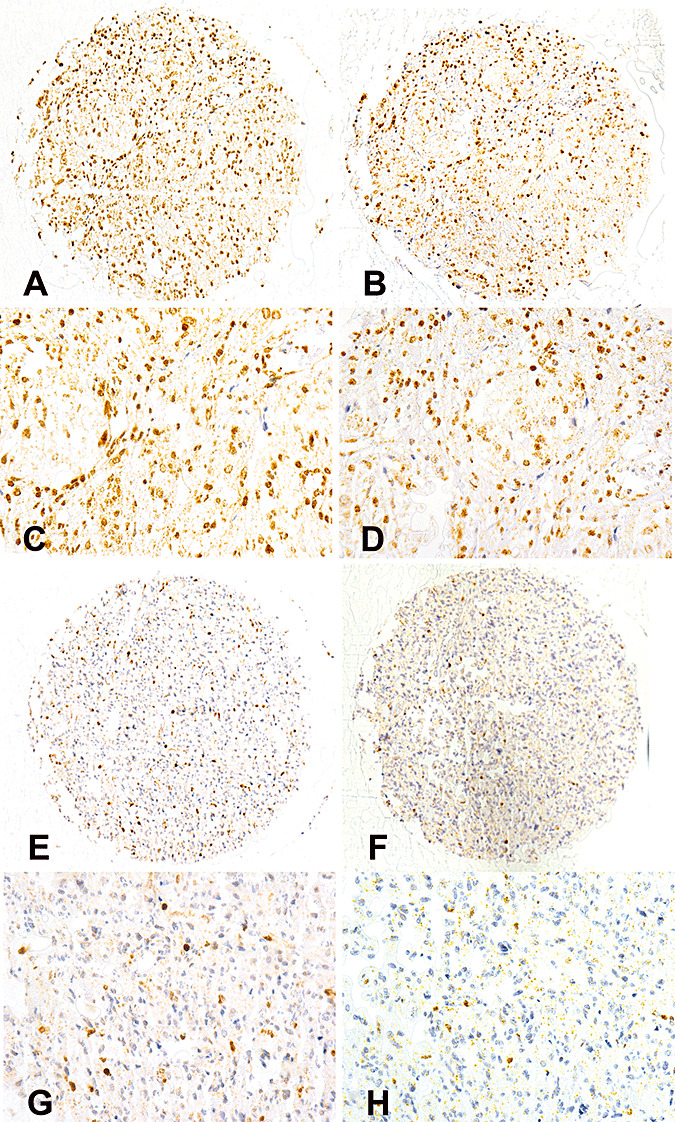
Immunohistochemical visualization of O6‐methylguanine‐methyltransferase (MGMT) protein expression on a tissue microarray (TMA) using two different primary antibodies (clones MT 3.1 and MT23.2). A–D. Sections of the same TMA tissue core stained with the two anti‐MGMT antibodies [antibody MT3.1 (A,C) and antibody MT23.2 (B,D); magnification: ×10 objective lens (A,B) and ×40 objective lens (C,D)]. Both antibodies (clones MT3.1 and MT23.2) show immunolabeling of the majority of tumor cell nuclei. E–H. Sections of another TMA tissue core stained with antibodies MT3.1 and MT23.2 [antibody MT3.1 (E,G) and antibody MT23.2 (F,H); magnification: ×10 objective lens (E,F) and ×40 objective lens (G,H)]. Both antibodies (clones MT3.1 and MT23.2) show immunolabeling of a minority of tumor cell nuclei.
Immunohistochemistry
TMA sections were immunostained using two different commercially available anti‐MGMT antibodies, namely clones MT3.1 (Dako, Copenhagen, Denmark) and MT23.2 (Zymed laboratories, Carlsbad, CA, USA). The TMA sections were deparaffinized with xylene for 30 minutes and rehydrated in decreasing concentrations of ethanol. The primary antibodies were used at final dilutions of 0.2 µg/mL (MT23.2) and 1.4 µg/mL (MT3.1), respectively, for an incubation period of 15 minutes at room temperature. Antibody binding was demonstrated with the DAKO‐catalyzed signal amplification horseradish peroxidase system® (Glostrup, Denmark), which was used according to the manufacturer's protocol. Immunoreactivity was visualized with 3′3′‐diaminobenzidine as the chromogen. All sections were counterstained with hematoxylin. Negative controls were carried out by omission of the respective primary antibody. In addition, tumor sections from cases with or without previously demonstrated MGMT expression were stained in parallel as positive and negative controls for the staining reaction.
Analysis
Evaluation of anti‐MGMT immunohistochemistry was performed by four observers from three neuropathology laboratories (JAH, JF, MP and RCJ). As a first step, an evaluation algorithm was circulated to all observers. After critical review and agreement on the algorithm by all observers (Table 1), assessment of both anti‐MGMT immunostained TMA sections (MT3.1 and MT23.2 antibodies) was performed. All observers reviewed the same TMA stained slides. Each observer evaluated both sections independently and was blinded to clinical data and MGMT promoter methylation status. For analysis of intraobserver agreement, each observer independently re‐evaluated both sections at least 3 weeks (range 3 to 6 weeks) after the first assessment. At reassessment, all observers were blinded to the results from the original first evaluation as well as to all clinical data and MGMT promoter methylation status as assessed by MSP.
Table 1.
Sequential questionnaire for evaluation of immunohistochemically visualized MGMT expression. For each biopsy sample (Figure 1) on both tissue microarray sections (one immunostained with MT3.1 antibody, one immunostained with MT23.2 antibody), each observer had to sequentially answer four questions as outlined in Table 1. For a given case, the “no” answer to question 1 terminated the evaluation. Semiquantitative assessment of MGMT expression in tumor cells was performed in two steps (questions 2a and 2b in Table 1). Abbreviation: MGMT = O6‐methylguanine‐methyltransferase.
| Question number | Question | Possible answers |
|---|---|---|
| 1 | Is assessable tumor tissue present? | No [no tumor tissue present/necrosis/too small (<30% of section)] |
| Yes | ||
| 2a | Is the tumor tissue MGMT positive? | No |
| Yes | ||
| 2b | How many tumor cells are positive? | Few (<10% of tumor cell nuclei) |
| Some (10%–50% of tumor cell nuclei) | ||
| Many (>50% of tumor cell nuclei) | ||
| 3 | Are there endothelial cells showing MGMT immunoreactivity? | No |
| Yes (only unequivocal endothelial cells) | ||
| 4 | Are there hematogenous cells showing MGMT immunoreactivity? | No |
| Yes (only unequivocal hematogenous cells; eg, lymphocytes) |
Statistics
The statistical software packages SAS® (SAS Institute Inc., Cary, NC, USA) and SPSS® (SPSS Inc., Chicago, IL, USA) were used for statistical calculations. A two‐tailed significance level of 5% was assumed.
Observer agreement
For each question of the evaluation algorithm (Table 1), Cohen's kappa and Cohen's weighted kappa were used to measure inter‐ and intraobserver agreement of MGMT IHC assessment. Kappa values were interpreted as follows: <0.2, poor observer agreement; 0.2–0.4, slight observer agreement; 0.4–0.6, moderate observer agreement; 0.6–0.8, substantial observer agreement; 0.8–1, almost perfect observer agreement (27). A two‐way ANOVA model was employed to compare differences between staining methods.
Correlation of MGMT IHC results with MSP results
Cohen's kappa was used to measure agreement between MGMT promoter methylation status (methylated or unmethylated) and immunohistochemically evaluated MGMT expression in tumor cells (semiquantitative MGMT values as assessed by question 2). For this purpose, we categorized the immunohistochemical MGMT values in three different ways: MGMT negative (“no” MGMT immunoreactive tumor cells) vs. MGMT positive (“few,”“some” or “many” MGMT immunoreactive tumor cells); low (“no” or “few” MGMT immunoreactive tumor cells) vs. high (“some” or “many” MGMT immunoreactive tumor cells) MGMT expression; and finally, MGMT expression in <50% of tumor cells (“no,”“few” or “some” immunoreactive tumor cells) vs. MGMT expression in ≥50% of tumor cells (“many” immunoreactive tumor cells).
Survival analysis
Overall survival was defined from the day of randomization until death of the patient. Patients not reported dead or lost to follow‐up were censored on the date of last visit. Survival probabilities were computed according to the Kaplan–Meier method. Log‐rank test was used to assess the prognostic effect of MGMT expression and MGMT promoter methylation status on overall survival. No correction for multiple testing was done as the statistical tests were performed for demonstration purposes only.
In order to explore the potential associations of the immunohistochemically evaluated MGMT expression in tumor cells (semiquantitative MGMT values as assessed by question 2) with patient survival, a four‐group comparison of MGMT expressions “no” vs. “few” vs. “some” vs. “many” MGMT immunoreactive tumor cells was tested (log‐rank test). In addition, in order to account for the semiquantitative nature of MGMT expressions, a linear trend across the four ordered MGMT categories was tested (log‐rank test).
RESULTS
Immunohistochemistry
Both anti‐MGMT antibodies (MT3.1 and MT23.2 antibodies) used in this study showed nuclear immunlabeling of variable extent and intensity (Figure 1). In addition to tumor cells, endothelial cells of tumor vasculature and tumor infiltrating lymphocytes (hematogenous cells) also showed labeling in a fraction of cases.
Interobserver agreement on MGMT IHC
Evaluation of both immunostained TMA sections (MT3.1 and MT23.2 antibodies) was performed at two different timepoints. For each antibody, timepoint and all six observer pairs (four observers = six pairs), kappa values were calculated as a measure of interobserver agreement. For the assessment of the sections, all observers used the same algorithm consisting of four predefined questions (Table 1). Ranges of kappa values for interobserver agreement are illustrated in Figure 2.
Figure 2.
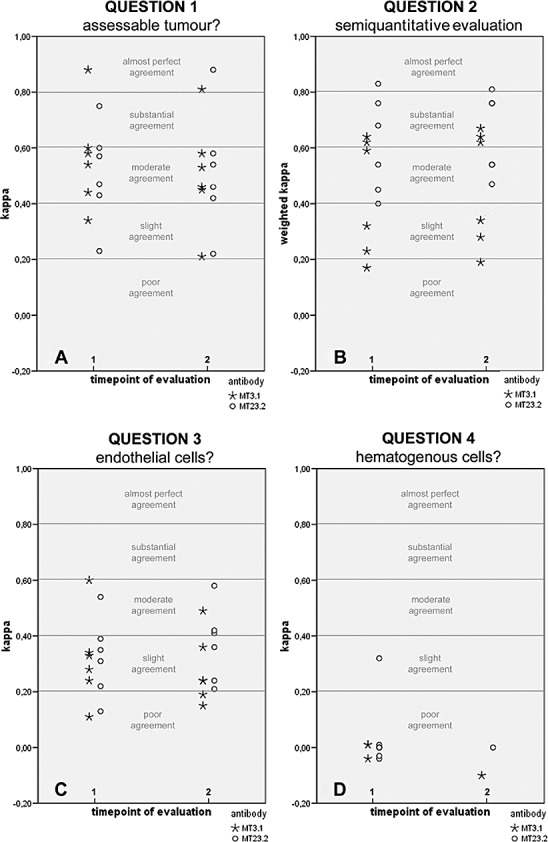
Illustration of kappa values for interobserver agreement on immunohistochemically visualized O6‐methylguanine‐methyltransferase protein expression in tissue microarray. (For a detailed description, refer to Results section.) Note that for question 2 (B), there are two kappa value ties at timepoint 2 with MT3.1 antibody. Therefore, only four instead of the expected six symbols (four observers = six observer pairs) are shown. For question 4 (D), less than six kappa values are shown for each antibody and timepoint because some observers showed no variation in an assessment run (ie, always voted solely “yes” or “no”). In such cases, no kappa values were computed.
-
•
Question 1: Is assessable tumor tissue present?
-
2
Kappa values calculated for both antibodies and for both timepoints showed variable interobserver agreement ranging from slight to almost perfect agreement (see Figure 2A).
-
•
Question 2: A. Is the tumor tissue MGMT positive (yes/no)? B. If positive, how many tumor cells are immunolabeled [few (<10%)/some (11%‐50%)/many (>50%)]?
-
4
As the assessment of the amount of positive tumor cells is a semiquantitative evaluation, weighted kappa calculation was used. Weighted kappa values calculated for both timepoints showed similar results (Figure 2B): The interobserver agreement was significantly higher on MT23.2 stained sections as compared with MT3.1 stained sections (P = 0.021). However, kappa values for both antibodies and both timepoints showed broad ranges (MT3.1: poor to substantial agreement; MT23.2: moderate to almost perfect agreement). Therefore, the degree of interobserver agreement seems to be associated with the type of anti‐MGMT antibody.
-
•
Question 3: Are there endothelial cells showing MGMT immunoreactivity (yes/no)?
-
6
Kappa calculation for both timepoints and both antibodies showed a similar, rather broad spectrum of interobserver agreement, ranging from poor to moderate (Figure 2C).
-
•
Question 4: Are there hematogenous cells showing MGMT immunoreactivity (yes/no)?
-
8
Kappa calculation for both timepoints and both antibodies showed only poor or slight interobserver agreement (Figure 2D).
Intraobserver agreement on MGMT IHC
To test intraobserver agreement, evaluation of both immunostained TMA sections (MT3.1 and MT23.2 antibodies) was performed twice by each observer at two different timepoints (time interval of 3 to 6 weeks). For each observer, kappa values were calculated between results of the first and second assessments. Ranges of kappa values for intraobserver agreement are illustrated in Figure 3.
Figure 3.
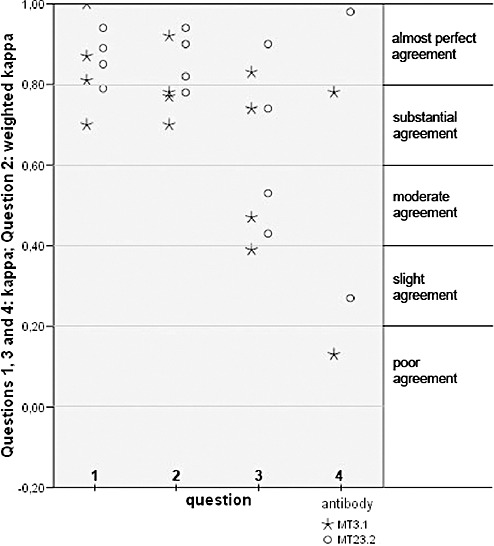
Illustration of kappa values for intraobserver agreement on immunohistochemically visualized O6‐methylguanine‐methyltransferase protein expression in tissue microarray. (For a detailed description, refer to Results section.) For question 4, less than four kappa values are shown for each antibody because some observers showed no variation in an assessment run (ie, always voted solely “yes” or “no”). In such cases, no kappa value was computed.
-
•
Question 1: Is assessable tumor tissue present?
-
2
Kappa calculation for both antibodies showed substantial or almost perfect intraobserver agreement (Figure 3). One observer reached perfect intraobserver agreement.
-
•
Question 2: A. Is the tumor tissue MGMT positive (yes/no)? B. If positive, how many tumor cells are immunolabeled [few (<10%)/some (11%‐50%)/many (>50%)]?
-
4
Kappa calculation for both antibodies showed substantial or almost perfect intraobserver agreement (Figure 3).
-
•
Question 3: Are there endothelial cells showing MGMT immunoreactivity (yes/no)?
-
6
Kappa calculation showed slight (MT3.1) or moderate (MT23.2) to almost perfect intraobserver agreement (Figure 3).
-
•
Question 4: Are there hematogenous cells showing MGMT immunoreactivity (yes/no)?
-
8
Kappa calculation showed poor to substantial (MT3.1) and slight to almost perfect (MT23.2) intraobserver agreement (Figure 3).
Correlation of MGMT MSP results with MGMT IHC results
For a detailed evaluation of the agreement between MGMT MSP (methylated or unmethylated) and MGMT IHC, we used the data of semiquantitative MGMT expression in tumor cells (MGMT values as assessed by question 2). We evaluated agreement using kappa analysis. For this purpose, we categorized the immunohistochemical MGMT values in three different ways (see also Materials and Methods):
-
(i)
MGMT negative vs. MGMT positive (Figure 4A): We found poor agreement between MSP and MGMT IHC for both antibodies (MT3.1 and MT23.2) and both timepoints of immunohistochemical MGMT evaluation.
-
(ii)
Low vs. high MGMT expression (Figure 4B): We found poor to slight (MT3.1) or poor to moderate (MT23.2) agreement between MSP and MGMT IHC for both timepoints of immunohistochemical MGMT evaluation.
-
(iii)
MGMT expression in <50% of tumor cells vs. MGMT expression in ≥50% of tumor cells (Figure 4C): For immunohistochemical MGMT evaluation utilizing the MT3.1 antibody, we found poor to slight agreement with MSP at timepoint 1 and poor agreement at timepoint 2. For the MT23.2 antibody, we found slight to moderate agreement with MSP results at both timepoints of immunohistochemical MGMT evaluation.
Figure 4.
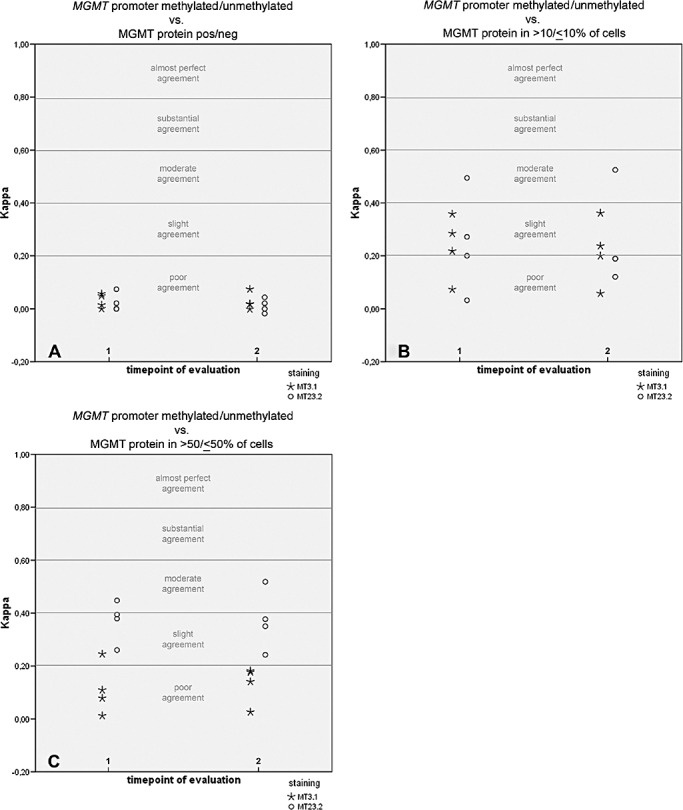
Illustration of kappa values for correlation between immunohistochemically visualized O6‐methylguanine‐methyltransferase (MGMT) protein expression and MGMT gene promoter status as assessed by methylation‐specific PCR. Immunohistochemical MGMT values were categorized in three different ways (for a detailed description, refer to Results section): MGMT negative vs. MGMT positive (A), low vs. high MGMT expression (cutoff 10%; B), MGMT expression in <50% of tumor cells vs. MGMT expression in ≥50% of tumor cells (C). Note that in A, there is one kappa value tie for the MT3.1 antibody at both timepoints, respectively, and one tie for the MT23.2 antibody at timepoint 1. In B, there is one kappa value tie at timepoint 2 with the MT23.2 antibody.
Survival analysis
We explored the potential impact of immunohistochemical MGMT expression in tumor cells (question 2; see also Materials and Methods) on patient survival on the basis of 16 assessments (four observers, two stainings, two timepoints; see also Table 2). None of the 16 assessments had a significant association of MGMT expression with patient outcome (Table 2). In contrast, presence of MGMT promoter methylation showed significant correlation with favorable patient survival (P = 0.0001, Figure 5), as reported previously in the larger original patient series (26).
Table 2.
Summary of results of survival analyses for all immunohistochemical assessments of MGMT expression in tumor cells (semiquantitative MGMT values as assessed by question 2, Table 1). The following situations were tested for all 16 assessments (four observers, two MGMT stainings, two timepoints): (i) MGMT qualitative—all four semiquantitative categories of MGMT expression (“no,”“few,”“some,”“many” MGMT immunoreactive tumor cells) were entered into log‐rank test as separate variables; (ii) MGMT linear trend—a linear trend across the four ordered MGMT categories (“no,”“few,”“some,”“many” MGMT immunoreactive tumor cells) was tested using the log‐rank test. We found no significant association of MGMT expression with patient outcome in any case. Abbreviation: MGMT = O6‐methylguanine‐methyltransferase.
| Assessment | Observer | Antibody | Timepoint | P‐values: MGMT qualitative | P‐values: MGMT linear trend |
|---|---|---|---|---|---|
| 1 | 1 | MT3.1 | 1 | 0.66 | 0.45 |
| 2 | 1 | MT3.1 | 2 | 0.98 | 0.66 |
| 3 | 2 | MT3.1 | 1 | 0.78 | 0.62 |
| 4 | 2 | MT3.1 | 2 | 0.80 | 0.66 |
| 5 | 3 | MT3.1 | 1 | 0.53 | 0.30 |
| 6 | 3 | MT3.1 | 2 | 0.19 | 0.11 |
| 7 | 4 | MT3.1 | 1 | 0.28 | 0.08 |
| 8 | 4 | MT3.1 | 2 | 0.30 | 0.14 |
| 9 | 1 | MT23.2 | 1 | 0.62 | 0.36 |
| 10 | 1 | MT23.2 | 2 | 0.62 | 0.37 |
| 11 | 2 | MT23.2 | 1 | 0.63 | 0.38 |
| 12 | 2 | MT23.2 | 2 | 0.42 | 0.17 |
| 13 | 3 | MT23.2 | 1 | 0.19 | 0.10 |
| 14 | 3 | MT23.2 | 2 | 0.18 | 0.14 |
| 15 | 4 | MT23.2 | 1 | 0.43 | 0.20 |
| 16 | 4 | MT23.2 | 2 | 0.37 | 0.36 |
Figure 5.
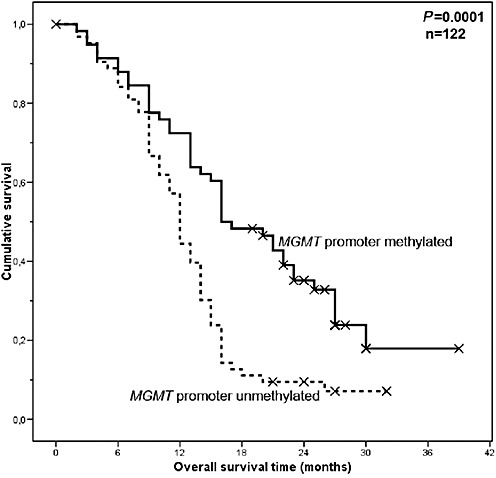
Kaplan–Meier curve showing a significant association of MGMT promoter methylation status with patient overall survival.
DISCUSSION
An association of MGMT promoter methylation status and clinical outcome of glioblastoma patients has been shown in independent investigations 14, 18, 25, 26, 28, 29, 35, 39, 47. Based on these findings, MGMT promoter methylation is considered a promising molecular factor predictive for chemotherapy response and longer survival in glioblastoma.
Some groups have also reported a prognostic significance of immunohistochemically assessed MGMT expression in glioblastoma (Table 3) 2, 11, 12, 44. Therefore, the use of anti‐MGMT IHC for diagnostic purposes is debated 2, 3, 4, 7, 11, 12, 13, 19, 21, 28, 30, 31, 36, 40, 41, 44, 48, 50. As a prerequisite for the routine diagnostic use of MGMT IHC in daily patient management, testing of its clinical usability is required 11, 40, 44, 51. In our study, we tested the clinical usability of MGMT IHC by analysis of observer agreement, its correlation with MGMT promoter methylation and its association with patient outcome. We used glioblastoma specimens from the patient cohort of the prospective randomized EORTC/NCIC trial 26981/22981 26, 52. We found variable observer agreement on immunohistochemical MGMT protein expression, insufficient correlation with MGMT promoter methylation status and no correlation with patient survival. Therefore, in our study, MGMT IHC does not prove to be a clinically usable tool in the diagnostic assessment of glioblastoma.
Table 3.
Summary of publications that have analyzed MGMT expression in glioblastoma by means of immunohistochemistry. Abbreviations: AA = anaplastic astrocytoma; AB = antibody; AOA = anaplastic oligoastrocytoma; AR‐10 = antigen retrieval 10; DA = diffuse astrocytoma; EDTA = ethylenediaminetetraacetic acid; GBM = glioblastoma multiforme; HGG = high grade glioma; MC = monoclonal; MIN = minutes; n.a. = not applicable; n.s. = not specified; O = oligodendroglioma; OA = oligoastrocytoma; o.n. = overnight; pAA = pediatric anaplastic astrocytoma; PBS = phosphate buffered saline; pGBM = pediatric glioblastoma multiforme; pHGG = pediatric high‐grade glioma; prGBM = primary glioblastoma multiforme; rGBM = relapsed glioblastoma multiforme; TEC = Tris‐ethylenediamine tetraacetic acid‐citrate.
| Reference number | Study cohort | MGMT AB clone (clonality) | MGMT AB supplier | Pretreatment, AB concentration, AB incubation time | Mode of evaluation of immunohistochemical MGMT expression | Categories used for semiquantitative evaluation of MGMT expression* | Observer agreement on MGMT expression | Correlation of MGMT expression with MGMT promoter methylation status | Cutoffs used for correlation of MGMT expression with patient survival | Significant correlation of MGMT expression with patient survival |
|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 18 GBM | MT3.1 (mc) | Neomarkers, CA, USA | 15 minutes microwave heating in citrate buffer (ph 6), 1:50, o.n. | Semiquantitative | 0, ≤19%, 20%–49%, ≥50% | Not done | Not done | 20% | Yes |
| 3 | 5 DA, 11 O, 6 AO, 12 AA/GBM | MT3.1 (mc) | Chemicon, CA, USA | Microwave heating in citrate buffer (ph 6), 1:200, o.n. | Semiquantitative | 0, <20%, 20%–40%, >40% | Not done | Not done | Not done | |
| 4 | 47 AA, 99 GBM | 3B8 (mc) | n.s. | Treatment with 0.1%Triton X‐l00 in PBS, 37 µg/mL, 60 minutes | Densitometry | Not done | Not done | 60 000 molecules/nucleus | Yes | |
| 8 | 21 O, 5 OA, 15 GBM | MT3.1 (mc) | Neomarkers, CA, USA | Microwave heating in TEC buffer (ph 7.8) for 36 minutes, 1:100, 120 minutes | Semiquantitative | ≤10%, 10%–50%, >50% | Not done | Not done | Not done | |
| 9 | 54 GBM | MT3.1 (mc) | Chemicon, CA, USA | 15 minutes incubation in citrate buffer (ph 6), 1:50, o.n. | Semiquantitative | <20%, >20% | Not done | Not done | 20% | No |
| 11 | 36 DA, 51 AA, 75 pGBM, 19 rGBM | MT3.1 (mc) | Chemicon, CA, USA | 20 minutes microwave boiling in 0.05% citraconic anhydride (ph 7.4) | Quantitative | Not done | Not done | 5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 50% | Yes | |
| 12 | 25 GBM | MT3.1 (mc) | Chemicon, CA, USA | n.s., 1:100, 60 minutes | Semiquantitative | <35%, >35% | Not done | Not done | 35% | Yes |
| 13 | 2 AA 1GBM | 3B8 (mc) | n.s. | n.s. | n.s. | Not done | Not done | Not done | ||
| 19 | 5 AA 33 GBM | MT3.1 (mc) | n.s. | Microwave heating in antigen retrieval buffer for 10 minutes, 5 µg/mL, o.n. | Semiquantitative | <20%, >20% | Not done | Not done | Not done | |
| 21 | 71 DA, 40 O 39 AA 259 GBM | MT3.1 (mc) | Chemicon, CA, USA | n.s. | Semiquantitative | <5%, >5% | Not done | Not done | Not done | |
| 22 | 6 AA, 17 GBM | MT3.1 (mc) | Dako, Copenhagen, Denmark | Heating in steamer, 1:500, 15 minutes | Semiquantitative | <10%, 10%–50%, >50% | Not done | No | Not done | |
| 30 | 24 AA, 40 GBM | 3B8 (mc) | n.s. | Treatment with 0.1%Triton X‐l00 in PBS, 37 µg/mL, 60 minutes | Densitometry | Not done | Not done | 60 000 molecules/nucleus | Yes | |
| 31 | 2 A, 2 O, 4 OA, 1 AA, 5 AO, 5 AOA, 14 GBM | n.s. | Chemicon, Hampshire, UK | 13 minutes microwave heating in sodium citrate, 1:1600, n.s. | n. s. | Not done | Poor | Not done | ||
| 36 | 3 DA, 15 AA, 8 GBM | Polyclonal | n.s. | n.s. | Semiquantitative | <33%, 34%–66%, ≥67% | Not done | Poor | Not done | |
| 40 | 1 DA, 1 XA, 3 AO, 5 AA, 22 GBM | MT3.1 (mc) | n.s. | Heating in AR‐10 buffer for 10 minutes, 4 µg/mL, o.n. | Semiquantitative | <20%, >20% | Not done | Poor | Not done | |
| 41 | 48 HGG | MT3.1 (mc) | Chemicon, CA, USA | Heating in AR‐10 buffer for 10 minutes, 5 µg/mL, o.n. | Quantitative | Not done | Not done | Not done | ||
| 44 | 69 HGG | MT3.1 (mc) | Neomarkers, CA, USA | Autoclave heating in citrate buffer (ph 6) for 10 minutes, 1:60, o.n. | Semiquantitative | <10%, >10% | Not done | Not done | 10% | Yes |
| 46 | 3 A, 14 AA, 17 GBM | MT3.1 (mc) | Lab Vision, CA, USA | 0.3% H2O2, 0.1% sodium azide for 20 minutes in PBS, 4 µg/mL, n.s. | n.s. | Not done | Not done | Not done | ||
| 48 | 8 pHGG, 31 pAA, 33 pGBM | MT23.2 (mc) | Zymed, CA, USA | 20 minutes heating in 10 mmol citrate buffer (ph 6) | Semiquantitative | Little or no expression, scattered positive cells, comparable to normal brain, overexpression | Not done | Not done | No, little or scattered positive cells vs. comparable to normal brain or over expression | Yes |
| 50 | 50 GBM | MT3.1 (mc) | Neomarkers, CA, USA | Steam disodium ethylene diamine‐tetraacetate (ph 8), 1:50, 30 minutes | Semiquantitative | 0, <10%, 10%–50%, >50% | Not done | No | Neg vs. pos | No |
| 57 | 10 A, 9 AA, 16 GBM | Polyclonal | n.s. | 3× microwave heating for 10 minutes in EDTA buffer (ph 8), 1:500, o.n. | Quantitative | Not done | Not done | Not done | ||
| Present study | 164 GBM | MT3.1 (mc) | Dako, Copenhagen, Denmark | 1.4 µg/mL, 15 minutes | Semiquantitative | 0, <10%, 10%–50%, >50% | Variable | Poor to moderate | Neg vs. pos, 10%, 50% | No |
| MT23.2 (mc) | Zymed, CA, USA | 0.2 µg/mL, 15 minutes |
Percent signifies fraction of immunoreactive cells.
Previously published studies on glioblastoma 2, 11, 12, 44 and other types of diffuse glioma 7, 11, 48 reported a significant association of immunohistochemically assessed MGMT expression and patient survival (see also Table 3). In glioblastoma, this association was confirmed neither by two other investigations in small retrospective series 9, 50 nor by our present study in a large prospective patient cohort. Possible explanations for the discrepancy of findings are sample sets composed of heterogeneous glioma types or small patient numbers in some of the studies (Table 3) (11). Other explanations may be methodological differences (eg, different pretreatment of sections prior to immunostaining; see Table 3). However, we used the same primary monoclonal antibodies that have been used in most of the previous studies in gliomas so far (Table 3).
Furthermore, different cutoff levels of semiquantitative MGMT assessment by immunostaining could be the cause for a significant association with patient outcome in some of the studies (11). In our study, we tested the potential association of MGMT expression with patient survival based on 16 immunohistochemical MGMT assessments using an overall group comparison of all four semiquantitative categories. In addition, a linear trend across the four ordered MGMT categories was also tested. In none of the cases was significance achieved. Therefore, evidence prevails that immunohistochemically detectable MGMT expression is not firmly associated with patient outcome. A possible explanation for this finding is that MGMT protein can be upregulated, and this upregulation may be induced by glucocorticoids, chemotherapy and radiotherapy. Thus, the expression of the protein at the time of diagnosis might not reflect the expression of the protein during therapy 1, 5, 20, 23, 37, 38, 51, 54.
In contrast to all but one (11) of the previous studies, we used TMA for immunohistochemical assessment of MGMT protein expression. The TMA contained all samples of the series in a single paraffin block allowing MGMT immunostaining of the whole series on one section, thus allowing for homogeneous staining conditions for the whole sample set, although differences in tissue handling prior to the TMA production (eg, tissue fixation, storage conditions) may introduce some heterogeneity. Furthermore, the TMA is particularly useful for inter‐ and intraobserver comparison of semiquantitative MGMT assessments, because all observers perform their assessments on the same restricted tissue area 15, 16, 32, 43. Under these conditions, intraobserver agreement was generally high, whereas interobserver agreement was highly variable. Possible explanations for the high interobserver variability could be interobserver differences in cutoff definition for intensity of the immunostaining signal (ie, differences in perception whether a given nucleus is stained darkly enough to be considered as positive) or interobserver differences in discrimination between specific immunostaining signal and background staining. Furthermore, high variability at identification of nonneoplastic cell elements within the tumor tissue (eg, endothelial cells, reactive astrocytes, microglial cells/macrophages and tumor‐infiltrating lymphocytes) may contribute to poor observer agreement. High intraobserver agreement may indicate that joint training of observers (eg, discussion of cases with discrepant results) could increase interobserver agreement. However, the results of immunohistochemical MGMT expression analysis did not reach a substantial correlation with MGMT promoter methylation status or a significant association with patient survival for any of the observers. Therefore, improvement of interobserver agreement on immunohistochemical MGMT expression by means of joint training is unlikely to provide additional analytical benefit.
Correlating semiquantitatively assessed MGMT expression with MGMT promoter methylation, we only found poor to moderate correlation. The best agreement (slight to moderate agreement) between MSP and MGMT IHC results was achieved using MT23.2 antibody and a cutoff of 50% immunolabeled cells for categorization of immunohistochemically evaluated MGMT expression. However, kappa values indicating substantial agreement between MGMT IHC and MSP were not achieved in any case. In our study, a potential methodological problem leading to insufficient correlation between MGMT IHC and MSP could be that the small tissue fraction viewed on the TMA may not be representative for the entire tumor in a given case. However, the TMA cores used in our study were specifically selected from tumor areas that were histologically classified as representative for the entire tumor. Moreover, a recently published paper systematically analyzed intratumoral distribution of MGMT promoter methylation and MGMT expression in anaplastic astrocytoma and glioblastoma (22). They found equal or highly similar MGMT expression in tissue samples taken from different sites of each individual tumor, thus clearly illustrating intratumoral homogeneity of immunohistochemical MGMT expression. However, major intratumoral regional variation in MGMT immunostaining would constitute another strong argument against the reliability of the immunohistochemical MGMT assessment for diagnostic purposes. Furthermore, in four previous studies, a comparably poor correlation between MGMT IHC and MSP was found 22, 31, 40, 50. Therefore, there is increasing evidence that immunohistochemically assessed MGMT expression is a poor indicator of MGMT promoter methylation status in glioblastoma. As discussed previously, a possible explanation for this finding is MGMT protein expression in entrapped pre‐existing, reactive or infiltrating cells 31, 44, 50, 51. MGMT expression has been shown in nonneoplastic endothelial cells, astrocytes, oligodendroglial cells and inflammatory cells 44, 50. Furthermore, thus far unknown control mechanisms of MGMT protein expression at transcriptional, posttranscriptional or translational level may exist 6, 8, 22, 57.
To conclude, in our study, observer variability and lack of association with MGMT promoter methylation status and with patient survival impede the use of MGMT IHC as a clinically useful biomarker for routine diagnostic purposes and clinical decision making. It remains to be seen whether novel anti‐MGMT antibodies directed against other epitopes are associated with better clinical and analytical performances.
Analysis of MGMT promoter methylation status (MSP‐PCR, real‐time MSP‐PCR, multiplex ligation‐dependent probe amplification, pyrosequencing, methylation‐sensitive high resolution melting) in glioblastoma remain promising tools for diagnostic purposes 10, 31, 42, 45, 46, 53, 56. However, the clinical and analytical performance of these molecular methods remain to be validated in prospective trials and by means of interlaboratory comparisons 28, 40, 51.
ACKNOWLEDGMENTS
We thank the EORTC for permission to use data from the EORTC/NCIC trial 26981/22981 for this research and Dr. Martin Van Den Bent for critical evaluation of the manuscript.
A translational research project of the European Organization for Research and Treatment of Cancer Brain Tumor Group.
REFERENCES
- 1. Alvino E, Castiglia D, Caporali S, Pepponi R, Caporaso P, Lacal PM et al (2006) A single cycle of treatment with temozolomide, alone or combined with O(6)‐benzylguanine, induces strong chemoresistance in melanoma cell clones in vitro: role of O(6)‐methylguanine‐DNA methyltransferase and the mismatch repair system. Int J Oncol 29:785–797. [PubMed] [Google Scholar]
- 2. Anda T, Shabani HK, Tsunoda K, Tokunaga Y, Kaminogo M, Shibata S et al (2003) Relationship between expression of O6‐methylguanine‐DNA methyltransferase, glutathione‐S‐transferase pi in glioblastoma and the survival of the patients treated with nimustine hydrochloride: an immunohistochemical analysis. Neurol Res 25:241–248. [DOI] [PubMed] [Google Scholar]
- 3. Andersson U, Malmer B, Bergenheim AT, Brannstrom T, Henriksson R (2004) Heterogeneity in the expression of markers for drug resistance in brain tumors. Clin Neuropathol 23:21–27. [PubMed] [Google Scholar]
- 4. Belanich M, Pastor M, Randall T, Guerra D, Kibitel J, Alas L et al (1996) Retrospective study of the correlation between the DNA repair protein alkyltransferase and survival of brain tumor patients treated with carmustine. Cancer Res 56:783–788. [PubMed] [Google Scholar]
- 5. Biswas T, Ramana CV, Srinivasan G, Boldogh I, Hazra TK, Chen Z et al (1999) Activation of human O6‐methylguanine‐DNA methyltransferase gene by glucocorticoid hormone. Oncogene 18:525–532. [DOI] [PubMed] [Google Scholar]
- 6. Blough MD, Zlatescu MC, Cairncross JG (2007) O6‐methylguanine‐DNA methyltransferase regulation by p53 in astrocytic cells. Cancer Res 67:580–584. [DOI] [PubMed] [Google Scholar]
- 7. Brell M, Tortosa A, Verger E, Gil JM, Vinolas N, Villa S et al (2005) Prognostic significance of O6‐methylguanine‐DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clin Cancer Res 11:5167–5174. [DOI] [PubMed] [Google Scholar]
- 8. Buccoliero AM, Arganini L, Ammannati F, Gallina P, Di Lorenzo N, Mennonna P, Taddei GL (2005) Oligodendrogliomas lacking O6‐methylguanine‐DNA‐methyltransferase expression. J Chemother 17:321–326. [DOI] [PubMed] [Google Scholar]
- 9. Cahill DP, Levine KK, Betensky RA, Codd PJ, Romany CA, Reavie LB et al (2007) Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res 13:2038–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cankovic M, Mikkelsen T, Rosenblum ML, Zarbo RJ (2007) A simplified laboratory validated assay for MGMT promoter hypermethylation analysis of glioma specimens from formalin‐fixed paraffin‐embedded tissue. Lab Invest 87:392–397. [DOI] [PubMed] [Google Scholar]
- 11. Capper D, Mittelbronn M, Meyermann R, Schittenhelm J (2007) Pitfalls in the assessment of MGMT expression and in its correlation with survival in diffuse astrocytomas: proposal of a feasible immunohistochemical approach. Acta Neuropathol (Berl) 115: 249–259. [DOI] [PubMed] [Google Scholar]
- 12. Chinot OL, Barrie M, Fuentes S, Eudes N, Lancelot S, Metellus P et al (2007) Correlation between O6‐methylguanine‐DNA methyltransferase and survival in inoperable newly diagnosed glioblastoma patients treated with neoadjuvant temozolomide. J Clin Oncol 25:1470–1475. [DOI] [PubMed] [Google Scholar]
- 13. Citron M, White A, Decker R, Wasserman P, Li B, Randall T et al (1995) O6‐methylguanine‐DNA methyltransferase in human brain tumors detected by activity assay and monoclonal antibodies. Oncol Res 7:49–55. [PubMed] [Google Scholar]
- 14. Criniere E, Kaloshi G, Laigle‐Donadey F, Lejeune J, Auger N, Benouaich‐Amiel A et al (2007) MGMT prognostic impact on glioblastoma is dependent on therapeutic modalities. J Neurooncol 83:173–179. [DOI] [PubMed] [Google Scholar]
- 15. De la Taille A, Viellefond A, Berger N, Boucher E, De Fromont M, Fondimare A et al (2003) Evaluation of the interobserver reproducibility of Gleason grading of prostatic adenocarcinoma using tissue microarrays. Hum Pathol 34:444–449. [DOI] [PubMed] [Google Scholar]
- 16. Diaz LK, Gupta R, Kidwai N, Sneige N, Wiley EL (2004) The use of TMA for interlaboratory validation of FISH testing for detection of HER2 gene amplification in breast cancer. J Histochem Cytochem 52:501–507. [DOI] [PubMed] [Google Scholar]
- 17. Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG (1999) Inactivation of the DNA repair gene O6‐methylguanine‐DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res 59:793–797. [PubMed] [Google Scholar]
- 18. Esteller M, Garcia‐Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V et al (2000) Inactivation of the DNA‐repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343:1350–1354. [DOI] [PubMed] [Google Scholar]
- 19. Friedman HS, McLendon RE, Kerby T, Dugan M, Bigner SH, Henry AJ et al (1998) DNA mismatch repair and O6‐alkylguanine‐DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant glioma. J Clin Oncol 16:3851–3857. [DOI] [PubMed] [Google Scholar]
- 20. Fritz G, Tano K, Mitra S, Kaina B (1991) Inducibility of the DNA repair gene encoding O6‐methylguanine‐DNA methyltransferase in mammalian cells by DNA‐damaging treatments. Mol Cell Biol 11:4660–4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fruehauf JP, Brem H, Brem S, Sloan A, Barger G, Huang W, Parker R (2006) In vitro drug response and molecular markers associated with drug resistance in malignant gliomas. Clin Cancer Res 12:4523–4532. [DOI] [PubMed] [Google Scholar]
- 22. Grasbon‐Frodl EM, Kreth FW, Ruiter M, Schnell O, Bise K, Felsberg J et al (2007) Intratumoral homogeneity of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas. Int J Cancer 121:2458–2464. [DOI] [PubMed] [Google Scholar]
- 23. Grombacher T, Mitra S, Kaina B (1996) Induction of the alkyltransferase (MGMT) gene by DNA damaging agents and the glucocorticoid dexamethasone and comparison with the response of base excision repair genes. Carcinogenesis 17:2329–2336. [DOI] [PubMed] [Google Scholar]
- 24. Gutman S, Kessler LG (2006) The US Food and Drug Administration perspective on cancer biomarker development. Nat Rev Cancer 6:565–571. [DOI] [PubMed] [Google Scholar]
- 25. Hegi ME, Diserens AC, Godard S, Dietrich PY, Regli L, Ostermann S et al (2004) Clinical trial substantiates the predictive value of O‐6‐methylguanine‐DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res 10:1871–1874. [DOI] [PubMed] [Google Scholar]
- 26. Hegi ME, Diserens AC, Gorlia T, Hamou MF, De Tribolet N, Weller M et al (2005) MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352:997–1003. [DOI] [PubMed] [Google Scholar]
- 27. Hsu CY, Ho DM, Yang CF, Chiang H (2003) Interobserver reproducibility of MIB‐1 labeling index in astrocytic tumors using different counting methods. Mod Pathol 16:951–957. [DOI] [PubMed] [Google Scholar]
- 28. Idbaih A, Omuro A, Ducray F, Hoang‐Xuan K (2007) Molecular genetic markers as predictors of response to chemotherapy in gliomas. Curr Opin Oncol 19:606–611. [DOI] [PubMed] [Google Scholar]
- 29. Ishii D, Natsume A, Wakabayashi T, Hatano H, Asano Y, Takeuchi H et al (2007) Efficacy of temozolomide is correlated with 1p loss and methylation of the deoxyribonucleic acid repair gene MGMT in malignant gliomas. Neurol Med Chir (Tokyo) 47:341–349. [DOI] [PubMed] [Google Scholar]
- 30. Jaeckle KA, Eyre HJ, Townsend JJ, Schulman S, Knudson HM, Belanich M et al (1998) Correlation of tumor O6 methylguanine‐DNA methyltransferase levels with survival of malignant astrocytoma patients treated with bis‐chloroethylnitrosourea: a southwest oncology group study. J Clin Oncol 16:3310–3315. [DOI] [PubMed] [Google Scholar]
- 31. Jeuken JW, Cornelissen SJ, Vriezen M, Dekkers MM, Errami A, Sijben A et al (2007) MS‐MLPA: an attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest 87:1055–1065. [DOI] [PubMed] [Google Scholar]
- 32. Kay E, O'Grady A, Morgan JM, Wozniak S, Jasani B (2004) Use of tissue microarray for interlaboratory validation of HER2 immunocytochemical and FISH testing. J Clin Pathol 57:1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kleihues P, Burger PC, Aldape KD, Brat DJ, Biernat W, Bigner DD et al (2007) Glioblastoma. In: WHO Classification of Tumours of the Central Nervous System. Louis N, Ohgaki H, Wiestler OD, Cavanee WK (eds), pp. 33–47. IARC Press: Lyon. [Google Scholar]
- 34. Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S et al (1998) Tissue microarrays for high‐throughput molecular profiling of tumor specimens. Nat Med 4:844–847. [DOI] [PubMed] [Google Scholar]
- 35. Krex D, Klink B, Hartmann C, Von Deimling A, Pietsch T, Simon M et al (2007) Long‐term survival with glioblastoma multiforme. Brain 130:2596–2606. [DOI] [PubMed] [Google Scholar]
- 36. Lee SM, Reid H, Elder RH, Thatcher N, Margison GP (1996) Inter‐ and intracellular heterogeneity of O6‐alkylguanine‐DNA alkyltransferase expression in human brain tumors: possible significance in nitrosourea therapy. Carcinogenesis 17:637–641. [DOI] [PubMed] [Google Scholar]
- 37. Lefebvre P, Zak P, Laval F (1993) Induction of O6‐methylguanine‐DNA‐methyltransferase and N3‐methyladenine‐DNA‐glycosylase in human cells exposed to DNA‐damaging agents. DNA Cell Biol 12:233–241. [DOI] [PubMed] [Google Scholar]
- 38. Margison GP, Povey AC, Kaina B, Santibanez Koref MF (2003) Variability and regulation of O6‐alkylguanine‐DNA alkyltransferase. Carcinogenesis 24:625–635. [DOI] [PubMed] [Google Scholar]
- 39. Martinez R, Schackert G, Yaya‐Tur R, Rojas‐Marcos I, Herman JG, Esteller M (2007) Frequent hypermethylation of the DNA repair gene MGMT in long‐term survivors of glioblastoma multiforme. J Neurooncol 83:91–93. [DOI] [PubMed] [Google Scholar]
- 40. Maxwell JA, Johnson SP, Quinn JA, McLendon RE, Ali‐Osman F, Friedman AH et al (2006) Quantitative analysis of O6‐alkylguanine‐DNA alkyltransferase in malignant glioma. Mol Cancer Ther 5:2531–2539. [DOI] [PubMed] [Google Scholar]
- 41. McLendon RE, Cleveland L, Pegram C, Bigner SH, Bigner DD, Friedman HS (1998) Immunohistochemical detection of the DNA repair enzyme O6‐methylguanine‐DNA methyltransferase in formalin‐fixed, paraffin‐embedded astrocytomas. Lab Invest 78:643–644. [PubMed] [Google Scholar]
- 42. Mikeska T, Bock C, El‐Maarri O, Hubner A, Ehrentraut D, Schramm J et al (2007) Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn 9:368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Milanes‐Yearsley M, Hammond ME, Pajak TF, Cooper JS, Chang C, Griffin T et al (2002) Tissue micro‐array: a cost and time‐effective method for correlative studies by regional and national cancer study groups. Mod Pathol 15:1366–1373. [DOI] [PubMed] [Google Scholar]
- 44. Nakasu S, Fukami T, Baba K, Matsuda M (2004) Immunohistochemical study for O6‐methylguanine‐DNA methyltransferase in the non‐neoplastic and neoplastic components of gliomas. J Neurooncol 70:333–340. [DOI] [PubMed] [Google Scholar]
- 45. Ogino S, Kawasaki T, Brahmandam M, Cantor M, Kirkner GJ, Spiegelman D et al (2006) Precision and performance characteristics of bisulfite conversion and real‐time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn 8:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ohe N, Saio M, Kijima M, Tamakawa N, Suwa T, Kojima Y et al (2003) In situ detection of O6‐methylguanine‐DNA methyltransferase messenger RNA in paraffin‐embedded human astrocytic tumor tissues by nested in situ RT‐PCR is useful in predicting chemotherapy‐resistance of tumors. Int J Oncol 22:543–549. [PubMed] [Google Scholar]
- 47. Paz MF, Yaya‐Tur R, Rojas‐Marcos I, Reynes G, Pollan M, Aguirre‐Cruz L et al (2004) CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res 10:4933–4938. [DOI] [PubMed] [Google Scholar]
- 48. Pollack IF, Hamilton RL, Sobol RW, Burnham J, Yates AJ, Holmes EJ et al (2006) O6‐methylguanine‐DNA methyltransferase expression strongly correlates with outcome in childhood malignant gliomas: results from the CCG‐945 Ciohort. J Clin Oncol 24:3431–3437. [DOI] [PubMed] [Google Scholar]
- 49. Qian XC, Brent TP (1997) Methylation hot spots in the 5′ flanking region denote silencing of the O6‐methylguanine‐DNA methyltransferase gene. Cancer Res 57:3672–3677. [PubMed] [Google Scholar]
- 50. Rodriguez FJ, Thibodeau SN, Jenkins RB, Schowalter KV, Caron BL, O'Neill BP et al (2007) MGMT immunohistochemical expression and promoter methylation in human glioblastoma. Appl Immunohistochem Mol Morphol 16:59–65. [DOI] [PubMed] [Google Scholar]
- 51. Stupp R, Hegi ME (2007) Methylguanine methyltransferase testing in glioblastoma: when and how? J Clin Oncol 25:1459–1460. [DOI] [PubMed] [Google Scholar]
- 52. Stupp R, Mason WP, Van Den Bent MJ, Weller M, Fisher B, Taphoorn MJ et al (2005) Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 352:987–996. [DOI] [PubMed] [Google Scholar]
- 53. Tanaka S, Oka H, Fujii K, Watanabe K, Nagao K, Kakimoto A (2005) Quantitation of O6‐methylguanine‐DNA methyltransferase gene messenger RNA in gliomas by means of real‐time RT‐PCR and clinical response to nitrosoureas. Cell Mol Neurobiol 25:1067–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ueda S, Mineta T, Nakahara Y, Okamoto H, Shiraishi T, Tabuchi K (2004) Induction of the DNA repair gene O6‐methylguanine‐DNA methyltransferase by dexamethasone in glioblastomas. J Neurosurg 101:659–663. [DOI] [PubMed] [Google Scholar]
- 55. Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW (1997) Methylation of discrete regions of the O6‐methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol 17:5612–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wojdacz TK, Dobrovic A (2007) Methylation‐sensitive high resolution melting (MS‐HRM): a new approach for sensitive and high‐throughput assessment of methylation. Nucleic Acids Res 35:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yuan Q, Matsumoto K, Nakabeppu Y, Iwaki T (2003) A comparative immunohistochemistry of O6‐methylguanine‐DNA methyltransferase and p53 in diffusely infiltrating astrocytomas. Neuropathology 23:203–209. [DOI] [PubMed] [Google Scholar]


