Abstract
The health of a cell requires proper functioning, regulation, and quality control of its organelles, the membrane-enclosed compartments inside the cell that carry out its essential biochemical tasks. Aging commonly perturbs organelle homeostasis, causing problems to cellular health that can spur the initiation and progression of degenerative diseases and related pathologies. Here, we discuss emerging evidence indicating that age-related defects in organelle homeostasis stem in part from dysfunction of the autophagy-lysosome system, a pivotal player in cellular quality control and damage clearance. We also highlight natural examples from biology where enhanced activity of the autophagy-lysosome system might be harnessed to erase age-related organelle damage, raising potential implications for cellular rejuvenation.
Keywords: cell biology, autophagy, organelles, aging, rejuvenation
In eukaryotic cells, molecular waste and damaged materials can be delivered to lysosomes for enzymatic degradation via autophagy [1]. During this process, autophagic vesicles, termed autophagosomes, form around select cargo, then subsequently fuse with the lysosome to allow for targeted degradation. Though autophagosomes were first observed by electron microscopy in the mid-1950s [2], it was not until nearly 40 years later that the first autophagy genes were identified in yeast [3–5]. Since then, breakthroughs in live-cell imaging have enabled sophisticated, real-time imaging of the autophagic process in several eukaryotic species, including animals [6,7]. In addition, an expanding pharmacological toolkit of molecules that modify autophagic activity in vivo (Table 1) has facilitated manipulation of this system in live organisms and raised exciting therapeutic prospects.
Table 1.
Example drugs that modulate autophagy in vivo.
| Drug | Mode of action | Reference |
|---|---|---|
| Inducers | ||
| Rapamycin | Inhibits mTOR pathway | [15,71–73] |
| Torin1 | Inhibits mTOR pathway | [74,75] |
| PP242 | Inhibits mTOR pathway | [76] |
| Curcumin | Activates Transcription factor EB; Inhibits mTOR pathway; Activates ERK1/2 pathway | [77,78] |
| Metformin | Activates Sirtuin-1 | [79] |
| Resveratrol | Activates Sirtuin-1 | [80] |
| Trehalose | Inhibits SLC2a family of glucose transporters; Activates AMPK | [81] |
| Spermidine | Regulates acetylation and deacetylation of cellular proteins | [82,83] |
| Lithium | Reduces inositol triphosphate levels | [84,85] |
| Carbamazepine | Reduces inositol triphosphate levels | [86,87] |
| Valproic acid | Reduces inositol triphosphate levels | [88] |
| Inhibitors | ||
| Chloroquine | Impairs lysosomal acidification | [10,73,89] |
| Lys05 | Impairs lysosomal acidification | [90] |
| Wortmannin | Inhibits phosphatidylinositol 3-kinases | [91] |
| Bafilomycin A1 | Inhibits V-ATPase; Inhibits autophagosome-lysosome fusion | [19,92,93] |
| Spautin-1 | Inhibits USP10 and USP13, which regulate deubiquitination of Beclin-1 | [94,95] |
| DBeQ | Inhibits p97/VCP | [96] |
A defining feature of the autophagy-lysosome system is its unique ability to recalibrate cellular homeostasis in response to a cell’s needs. If a cell is under intrinsic or extrinsic stress, activation of autophagy can help to erase molecular damage and to recycle material needed to support basic biological functions [1]. When these mechanisms fail, the stress can amplify, leading to an irreparable collapse in cellular homeostasis. Notably, aging is accompanied by several molecular signs of stress. As cells get older, genetic instability increases, proteins cluster into non-functional aggregates, and organelles, the cellular mini-factories that execute distinct signaling and metabolic functions, become damaged and inefficient [8]. Is this age-related collapse in cellular health and homeostasis linked to defects in autophagy?
Remarkably, researchers have found that an early-age decrease in lysosome and autophagic activity may be an initiating “domino” in age-related cellular deterioration [9,10]. Consistent with this model, modifying autophagic activity has profound effects on the aging process; experimental inhibition of lysosomal and/or autophagic factors accelerates aging in various organisms [11–14], whereas interventions that boost autophagic activity delay the appearance of cellular signs of aging and extend lifespan [15–17]. Even human centenarians [18], like long-lived mutant animals [19], have been reported to display exceptionally high levels of autophagic activity. These and other findings highlight the autophagy-lysosome system as an emerging nexus in the control of aging and longevity (Figure 1). Still, molecular details of this regulation remain obscure.
Figure 1. Changes to autophagy of cellular organelles during the aging process.
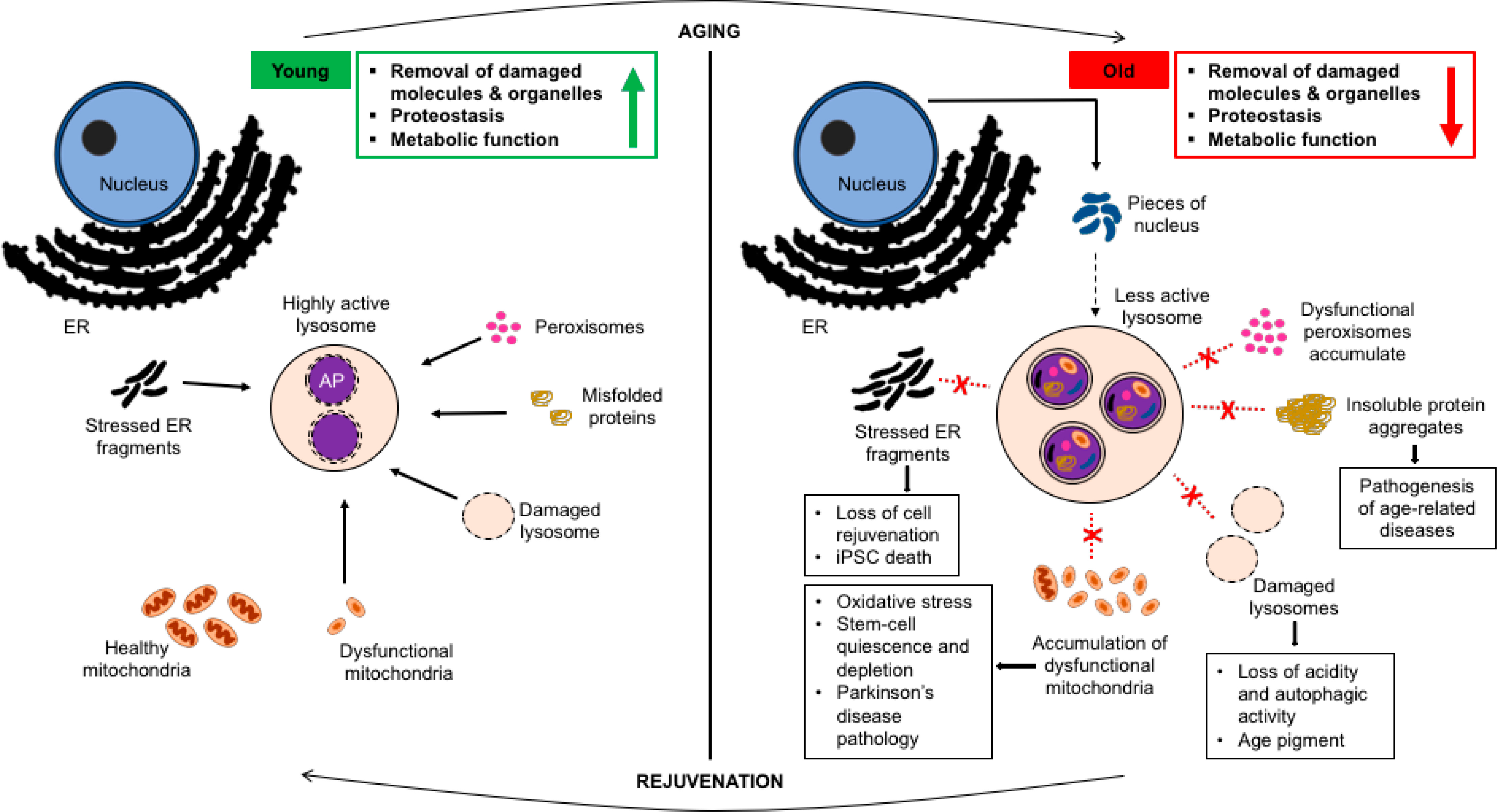
Lysosomes in young, healthy cells (on the left) are acidic and effectively degrade cellular waste, including organelles when necessary. This maintains robust homeostasis, which supports proper functioning not only of a cell but of a whole organism. However, in an old cell (on the right), lysosome dysfunction jeopardizes autophagic turnover, causing a build-up of damaged organelles along with protein aggregates; this leads to several age-related disease pathologies and brings about changes to organismal physiology. Re-establishing the correct dynamics of organelle turnover at lysosomes in old cells might provide one entry point to trigger a rejuvenation of cellular health and homeostasis. AP, autophagosome.
For one, how is different autophagic cargo handled in aging cells, and do changes to cargo turnover directly contribute to the aging process? Many studies have investigated how defective autophagy impedes protein-aggregate clearance in old cells [20]. This is an important line of research, given that impaired protein homeostasis (‘proteostasis’) is characteristic of many age-related diseases, including Alzheimer’s [21]. Yet, defective organelles are also common to age-related diseases [22–24], and their turnover is likewise sensitive to lysosome dysfunction [1,25]. To date, surprisingly little is known about the dynamics and control of organelle turnover in aging cells. Clarifying the regulation of organelle-specific autophagy during aging could provide novel clues on the biological basis of age-related disease, and might also hint at therapies for fighting the aging process.
Perhaps the most information is currently known regarding the age-related regulation of mitochondria, the energetic hubs of a cell. With age, mitochondrial function and homeostasis break down. Several proteins involved in oxidative phosphorylation and fatty-acid metabolism, two key cellular processes that occur at mitochondria, have been reported to decrease in abundance in old animals [26–28]. These molecular alterations, combined with other age-induced changes to mitochondrial protein levels and stoichiometry [29], are thought to impair mitochondrial activity and destabilize cellular bioenergetics and metabolism. As a consequence of this dysfunction, fragmented, oxidatively-damaged mitochondria are commonly seen in old cells of diverse eukaryotic species, ranging from yeasts to mammals [8,9,30–32]. Th ough healthy cells can effectively eliminate dysfunctional mitochondrial fragments by mitochondrial autophagy, or ‘mitophagy’ [33], mitochondrial-clearance mechanisms show signs of failure in old age [34,35]. This disrupts the balance between mitochondrial biogenesis and degradation, causing an age-dependent increase in damaged mitochondria that further exacerbates cell stress [34]. Mitophagy defects can predispose humans to degenerative disease; indeed, dysfunction of mitophagy factors, including Parkin and PINK1, is commonly seen in Parkinson’s disease patients [36,37]. Thus, impaired turnover of damaged organelles is at least partly to blame for some of the classic aging pathologies commonly seen in the clinic.
Importantly, impaired turnover with age does not appear to be limited to mitochondria. In cells, lysosomes are responsible for degrading additional types of organelles, including portions of the endoplasmic reticulum (ER), peroxisomes, and even other lysosomes. Like mitochondrial damage, ER stress accumulates in old cells [38]. Strikingly, genetic inhibition of ER-phagy causes progeric phenotypes and shortened lifespan in mice [39], hinting that ER turnover might be required to slow the pace of aging. Additionally, peroxisomes and lysosomes have been reported to increase in abundance in late age in some species and cell types [40,41]. In fact, uncleared lysosomes generate a non-degradable, autofluorescent ‘age pigment’, which has been used as a visual readout for biological age in multiple systems [42–44]. It will be important to clarify how directly these age-related changes in organelle number reflect impairment of the autophagy-lysosome system, and whether these changes bring about physiological effects on metabolic functioning in old animals.
While the general trend is that organelle turnover appears to decline with advanced age due to autophagy-lysosome dysfunction (Figure 1), this may not be true of all organelles, or for all stages of the aging process. For example, pieces of the nucleus are degraded at lysosomes in aging worms, even in the healthiest of individuals [45]. How nuclear autophagy (‘nucleophagy’) regulates organismal physiology, particularly during aging, is unclear, but it may be protective, as suggested in mouse models of laminopathies [46]. It remains to be seen whether other organelles likewise undergo regulated, active turnover in aging animals. Some organelles may even be degraded in early aging but start to accumulate later once lysosomes become dysfunctional. Understanding the dynamics and timing of organelle turnover at different stages of aging could reveal complexities that affect aging rate and/or stochasticity among different individuals in a population.
If organelle damage is generally characteristic of very old age, could harnessing organelle-specific autophagy help an old cell to regain its vitality and youthfulness? Germ (reproductive) cells provide a unique opportunity to study cellular rejuvenation, because age is naturally reset across generations. We and others have shown that cellular damage, including defective mitochondria, can be rapidly reversed as oocytes prepare for fertilization [47,48]. Removal of dysfunctional molecules and organelles is also seen during gametogenesis in single-celled yeast [49]. These findings imply that damage-clearance mechanisms may function centrally to the biological mechanisms of transgenerational rejuvenation. In support of this interpretation, lysosomes are activated in maturing oocytes prior to fertilization [47], and, once active, they could conceivably clear various forms of cellular damage, including dysfunctional organelles, to reset cellular health and homeostasis across generations. Though the specific cargo received by oocyte lysosomes awaits full description, identification of natural mechanisms that renew organelle health in the immortal germ-cell lineage could point the way to new strategies to counteract organelle damage in old somatic cells.
Lysosome induction has been reported to also occur during stem-cell activation and differentiation [50–52]. In these contexts, as in oocyte maturation, lysosome activation is linked to a developmental rewiring of cellular metabolism. Though, again, much attention has been paid to the role of lysosome activity in stem-cell proteostasis, there is recent evidence that organelle-specific autophagy plays a fundamental role in stem-cell and regenerative biology [53–57]. For one, impaired mitophagy leads to muscle stem-cell quiescence in old mice, and re-establishing autophagic flux is sufficient for old muscle stem cells to exit quiescence and regain stemness [58]. Importantly, defective mitophagy appears to cause oxidative stress and stem-cell depletion in other cell types as well [59,60]. These findings hint that mitochondrial turnover might be a pivotal determinant of regenerative capacity.
Notably, mitophagy also appears important in the generation of induced pluripotent stem cells (iPSCs) [57,61]. A number of rejuvenating events, including telomere re-lengthening and organelle renewal, have been associated with iPSC generation from differentiated cells [62–64]. Inhibiting mitochondrial fission, one of the early steps in mitophagy induction [33,65], prevents the conversion of fibroblasts to iPSCs [61]. Thus, it is exciting to speculate that organelle-specific autophagy may be integrated with other rejuvenating events involved in iPSC reprogramming, and that enhancing these activities might provide an entry point to improve the efficiency of this process.
Beyond mitophagy, other forms of organelle-specific autophagy are only beginning to be studied in the context of cellular regeneration and rejuvenation. Interestingly, elevated ER stress has been linked to iPSC death [66], and significant ER remodeling occurs as part of iPSC reprogramming [67]. In principle, ER quality control mechanisms, including ER-phagy, could aid regenerative capacity, particularly in old animals where persistent ER stress abounds [38]. As a compelling corollary, the ER has been shown to undergo dramatic rearrangements coincident with oocyte maturation and lysosome activation in the C. elegans germline [68]. How the ER and lysosomes are functionally and/or mechanically linked to support cellular rejuvenation is an important open question moving forward, as is the involvement of other organelle-turnover events in cellular-rejuvenation mechanisms.
In summary, dynamic changes to the landscape of the cell occur during aging, and several of these age-related changes can be traced to alterations in organelle homeostasis and turnover (Figure 1). Harnessing the natural rejuvenating capacities of the autophagy-lysosome system provides one possible means to reverse age-related organelle damage and re-establish a more youthful cellular environment (Figure 1). In fact, pharmacological tools that boost lysosome function (Table 1) are currently being tested as potential anti-aging therapies in old animals and humans [69,70]. Looking forward, it seems likely that growing knowledge on the mechanistic principles that govern organelle turnover at lysosomes, and the specific parts of these systems that fail with old age, will open new doors for aging-biology researchers in the quest to promote healthy aging, particularly at a cellular level.
FUNDING
We acknowledge funding from the LSU Office of Research and Economic Development, the LSU College of Science, and the LSU Department of Biological Sciences, as well as grants from the National Institutes of Health (R03AG067125; KAB) and the W. M. Keck Foundation (KAB). KAB is also a Glenn Foundation for Medical Research and AFAR Grant for Junior Faculty awardee.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
REFERENCES
- 1.Kaur J, Debnath J. Autophagy at the crossroads of catabolism an danabolism. Nat Rev Mol Cell Biol. 2015;16(8):461–72. [DOI] [PubMed] [Google Scholar]
- 2.Clark SL Jr. Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J Biophys Biochem Cytol. 1957;3(3):349–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119(2):301–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thumm M, Egner R, Koch B, Schlumpberger M, Straub M, Veenhuis M, et al. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 1994;349(2):275–80. [DOI] [PubMed] [Google Scholar]
- 5.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333(1–2):169–74. [DOI] [PubMed] [Google Scholar]
- 6.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3(5):452–60. [DOI] [PubMed] [Google Scholar]
- 7.Wang Y, Li Y, Wei F,Duan Y. Optical Imaging Paves the Way for Autophagy Research. Trends Biotechnol. 2017;35(12):1181–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes AL, Gottschling DE. An early age increase in vacuolar pH limits mitochondrial function and lifespan in yeast. Nature. 2012;492(7428):261–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baxi K, Ghavidel A, Waddell B, Harkness TA, de Carvalho CE. Regulation of Lysosomal Function by the DAF-16 Forkhead Transcription Factor Couples Reproduction to Aging in Caenorhabditis elegans. Genetics. 2017;207(1):83–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–4. [DOI] [PubMed] [Google Scholar]
- 12.Matecic M, Smith DL, Pan X, Maqani N, Bekiranov S, Boeke JD, et al. A microarray-based genetic screen for yeast chronological aging factors. PLoS Genet. 2010;6(4):e1000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simonsen A, Cumming RC, Brech A, Isakson P, Schubert DR, Finley KD. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4(2):176–84. [DOI] [PubMed] [Google Scholar]
- 14.Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4(3):330–8. [DOI] [PubMed] [Google Scholar]
- 15.Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, et al. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010;11(1):35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lapierre LR, De Magalhaes Filho CD, McQuary PR, Chu CC, Visvikis O, Chang JT, et al. The TFEB orthologue HLH-30 regulates autophagy and modulates longevity in Caenorhabditis elegans. Nat Commun. 2013;4:2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, et al. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiao FH, Chen XQ, Yu Q, Ye Y, Liu YW, Yan D, et al. Transcriptome evidence reveals enhanced autophagy-lysosomal function in centenarians. Genome Res. 2018;28(11):1601–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang JT, Kumsta C, Hellman AB, Adams LM, Hansen M. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife. 2017;6:e18459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hipp MS, Kasturi P, Hartl FU. The proteostasis network and its decline in ageing. Nat Rev Mol Cell Biol. 2019;20(7):421–35. [DOI] [PubMed] [Google Scholar]
- 21.Alzheimer A, Forstl H, Levy R. On certain peculiar diseases of old age. Hist Psychiatry. 1991;2(5 Pt 1):71–101. [DOI] [PubMed] [Google Scholar]
- 22.Cipolla CM, Lodhi IJ. Peroxisomal Dysfunction in Age-Related Diseases. Trends Endocrinol Metab. 2017;28(4):297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haas RH. Mitochondrial Dysfunction in Aging and Diseases of Aging. Biology (Basel). 2019;8(2):48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng W, Minakaki G, Nguyen M, Krainc D. Preserving Lysosomal Function in the Aging Brain: Insights from Neurodegeneration. Neurotherapeutics. 2019;16(3):611–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anding AL, Baehrecke EH. Cleaning House: Selective Autophagy of Organelles. Dev Cell. 2017;41(1):10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dai DF, Karunadharma PP, Chiao YA, Basisty N, Crispin D, Hsieh EJ, et al. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell. 2014;13(3):529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Narayan V, Ly T, Pourkarimi E, Murillo AB, Gartner A, Lamond AI, et al. Deep Proteome Analysis Identifies Age-Related Processes in C. elegans. Cell Syst. 2016;3(2):144–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popa-Wagner A, Sandu RE, Cristin C, Uzoni A, Welle KA, Hryhorenko JR, et al. Increased Degradation Rates in the Components of the Mitochondrial Oxidative Phosphorylation Chain in the Cerebellum of Old Mice. Front Aging Neurosci. 2018;10:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ingram T, Chakrabarti L. Proteomic profiling of mitochondria: what does it tell us about the ageing brain? Aging (Albany NY) 2016;8(12):3161–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leduc-Gaudet JP, Picard M, St-Jean Pelletier F, Sgarioto N, Auger MJ, Vallee J, et al. Mitochondrial morphology is altered in atrophied skeletal muscle of aged mice. Oncotarget. 2015;6(20):17923–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheckhuber CQ, Erjavec N, Tinazli A, Hamann A, Nystrom T, Osiewacz HD. Reducing mitochondrial fission results in increased life span and fitness of two fungal ageing models. Nat Cell Biol. 2007;9(1):99–105. [DOI] [PubMed] [Google Scholar]
- 32.Yasuda K, Ishii T, Suda H, Akatsuka A, Hartman PS, Goto S, et al. Age-related changes of mitochondrial structure and function in Caenorhabditis elegans. Mech Ageing Dev. 2006;127(10):763–70. [DOI] [PubMed] [Google Scholar]
- 33.Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20(9):1013–22. [DOI] [PubMed] [Google Scholar]
- 34.Palikaras K, Lionaki E, Tavernarakis N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature. 2015;521(7553):525–8. [DOI] [PubMed] [Google Scholar]
- 35.Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, et al. Measuring In Vivo Mitophagy. Mol Cell. 2015;60(4):685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–8. [DOI] [PubMed] [Google Scholar]
- 37.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304(5674):1158–60. [DOI] [PubMed] [Google Scholar]
- 38.Brown MK, Naidoo N. The endoplasmic reticulum stress response in aging and age-related diseases. Front Physiol. 2012;3:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng Y, Shapiro SL, Banduseela VC, Dieterich IA, Hewitt KJ, Bresnick EH, et al. Increased transport of acetyl-CoA into the endoplasmic reticulum causes a progeria-like phenotype. Aging Cell. 2018;17(5):e12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurz DJ, Decary S, Hong Y, Erusalimsky JD. Senescence-associated (beta)-galactosidase reflects an increase in lysosomal mass during replicative ageing of human endothelial cells. J Cell Sci. 2000;113(Pt 20):3613–22. [DOI] [PubMed] [Google Scholar]
- 41.Legakis JE, Koepke JI, Jedeszko C, Barlaskar F, Terlecky LJ, Edwards HJ, et al. Peroxisome senescence in human fibroblasts. Mol Biol Cell. 2002;13(12):4243–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Son HG, Altintas O, Kim EJE, Kwon S, Lee SV. Age-dependent changes and biomarkers of aging in Caenorhabditis elegans. Aging Cell. 2019;18(2):e12853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu H, Chen M, Manivannan A, Lois N, Forrester JV. Age-dependent accumulation of lipofuscin in perivascular and subretinal microglia in experimental mice. Aging Cell. 2008;7(1):58–68. [DOI] [PubMed] [Google Scholar]
- 44.Pincus Z, Smith-Vikos T, Slack FJ. MicroRNA predictors of longevity in Caenorhabditis elegans. PLoS Genet. 2011;7(9):e1002306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGee MD, Weber D, Day N, Vitelli C, Crippen D, Herndon LA, et al. Loss of intestinal nuclei and intestinal integrity in aging C. elegans. Aging Cell. 2011;10(4):699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park YE, Hayashi YK, Bonne G, Arimura T, Noguchi S, Nonaka I, et al. Autophagic degradation of nuclear components in mammalian cells. Autophagy. 2009;5(6):795–804. [DOI] [PubMed] [Google Scholar]
- 47.Bohnert KA, Kenyon C. A lysosomal switch triggers proteostasis renewal in the immortal C. elegans germ lineage. Nature. 2017;551(7682):629–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Goudeau J, Aguilaniu H. Carbonylated proteins are eliminated during reproduction in C. elegans. Aging Cell. 2010;9(6):991–1003. [DOI] [PubMed] [Google Scholar]
- 49.King GA, Goodman JS, Schick JG, Chetlapalli K, Jorgens DM, McDonald KL, et al. Meiotic cellular rejuvenation is coupled to nuclear remodeling in budding yeast. Elife. 2019;8:e47156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du G, Qiao Y, Zhuo Z, Zhou J, Li X, Liu Z, et al. Lipoic acid rejuvenates aged intestinal stem cells by preventing age-associated endosome reduction. EMBO Rep. 2020;21(8):e49583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leeman DS, Hebestreit K, Ruetz T, Webb AE, McKay A, Pollina EA, et al. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science. 2018;359(6381):1277–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie SZ, Garcia-Prat L, Voisin V, Ferrari R, Gan OI, Wagenblast E, et al. Sphingolipid Modulation Activates Proteostasis Programs to Govern Human Hematopoietic Stem Cell Self-Renewal. Cell Stem Cell. 2019;25(5):639–53.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Call JA, Wilson RJ, Laker RC, Zhang M, Kundu M, Yan Z. Ulk1-mediated autophagy plays an essential role in mitochondrial remodeling and functional regeneration of skeletal muscle. Am J Physiol Cell Physiol. 2017;312(6):C724–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, et al. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543(7644):205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Senos Demarco R, Uyemura BS, Jones DL. EGFR Signaling Stimulates Autophagy to Regulate Stem Cell Maintenance and Lipid Homeostasis in the Drosophila Testis. Cell Rep. 2020;30(4):1101–16.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song Q, Liu H, Zhen H, Zhao B. Autophagy and its role in regeneration and remodeling within invertebrate. Cell Biosci. 2020;10:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vazquez-Martin A, Van den Haute C, Cufi S, Corominas-Faja B, Cuyas E, Lopez-Bonet E, et al. Mitophagy-driven mitochondrial rejuvenation regulates stem cell fate. Aging (Albany NY). 2016;8(7):1330–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Prat L, Martinez-Vicente M, Perdiguero E, Ortet L, Rodriguez-Ubreva J, Rebollo E, et al. Autophagy maintains stemness by preventing senescence. Nature. 2016;529(7584):37–42. [DOI] [PubMed] [Google Scholar]
- 59.Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, Sadighi-Akha E, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208(3):455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Senos Demarco R, Jones DL. Mitochondrial fission regulates germ cell differentiation by suppressing ROS-mediated activation of Epidermal Growth Factor Signaling in the Drosophila larval testis. Sci Rep. 2019;9(1):19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vazquez-Martin A, Cufi S, Corominas-Faja B, Oliveras-Ferraros C, Vellon L, Menendez JA. Mitochondrial fusion by pharmacological manipulation impedes somatic cell reprogramming to pluripotency: new insight into the role of mitophagy in cell stemness. Aging (Albany NY). 2012;4(6):393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, Moreno R, et al. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 2010;28(4):661–73. [DOI] [PubMed] [Google Scholar]
- 63.Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4(2):141–54. [DOI] [PubMed] [Google Scholar]
- 64.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28(4):721–33. [DOI] [PubMed] [Google Scholar]
- 65.Pryde KR, Smith HL, Chau KY, Schapira AH. PINK1 disables the anti-fission machinery to segregate damaged mitochondria for mitophagy. J Cell Biol. 2016;213(2):163–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ha TW, Jeong JH, Shin H, Kim HK, Im JS, Song BH, et al. Characterization of Endoplasmic Reticulum (ER) in Human Pluripotent Stem Cells Revealed Increased Susceptibility to Cell Death upon ER Stress. Cells. 2020;9(5):1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simic MS, Moehle EA, Schinzel RT, Lorbeer FK, Halloran JJ, Heydari K, et al. Transient activation of the UPR(ER) is an essential step in the acquisition of pluripotency during reprogramming. Sci Adv. 2019;5(4):eaaw0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langerak S, Trombley A, Patterson JR, Leroux D, Couch A, Wood MP, et al. Remodeling of the endoplasmic reticulum in Caenorhabditis elegans oocytes is regulated by CGH-1. Genesis. 2019;57(2):e23267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Campisi J, Kapahi P, Lithgow GJ, Melov S, Newman JC, Verdin E. From discoveries in ageing research to therapeutics for healthy ageing. Nature. 2019;571(7764):183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Urfer SR, Kaeberlein TL, Mailheau S, Bergman PJ, Creevy KE, Promislow DEL, et al. A randomized controlled trial to establish effects of short-term rapamycin treatment in 24 middle-aged companion dogs. Geroscience. 2017;39(2):117–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, Oroz LG, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36(6):585–95. [DOI] [PubMed] [Google Scholar]
- 72.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273(7):3963–6. [DOI] [PubMed] [Google Scholar]
- 73.Nalbandian A, Llewellyn KJ, Nguyen C, Yazdi PG, Kimonis VE. Rapamycin and chloroquine: the in vitro and in vivo effects of autophagy-modifying drugs show promising results in valosin containing protein multisystem proteinopathy. PLoS One. 2015;10(4):e0122888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaizuka T, Morishita H, Hama Y, Tsukamoto S, Matsui T, Toyota Y, et al. An Autophagic Flux Probe that Releases an Internal Control. Mol Cell. 2016;64(4):835–49. [DOI] [PubMed] [Google Scholar]
- 75.Varga M, Fodor E, Vellai T. Autophagy in zebrafish. Methods. 2015;75:172–80. [DOI] [PubMed] [Google Scholar]
- 76.Feldman ME, Apsel B, Uotila A, Loewith R, Knight ZA, Ruggero D, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7(2):e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Song JX, Sun YR, Peluso I, Zeng Y, Yu X, Lu JH, et al. A novel curcumin analog binds to and activates TFEB in vitro and in vivo independent of mTOR inhibition. Autophagy. 2016;12(8):1372–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aoki H, Takada Y, Kondo S, Sawaya R, Aggarwal BB, Kondo Y. Evidence that curcumin suppresses the growth of malignant gliomas in vitro and in vivo through induction of autophagy: role of Akt and extracellular signal-regulated kinase signaling pathways. Mol Pharmacol. 2007;72(1):29–39. [DOI] [PubMed] [Google Scholar]
- 79.Song YM, Lee YH, Kim JW, Ham DS, Kang ES, Cha BS, et al. Metformin alleviates hepatosteatosis by restoring SIRT1-mediated autophagy induction via an AMP-activated protein kinase-independent pathway. Autophagy. 2015;11(1):46–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morselli E, Maiuri MC, Markaki M, Megalou E, Pasparaki A, Palikaras K, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.DeBosch BJ, Heitmeier MR, Mayer AL, Higgins CB, Crowley JR, Kraft TE, et al. Trehalose inhibits solute carrier 2A (SLC2A) proteins to induce autophagy and prevent hepatic steatosis. Sci Signal. 2016;9(416):ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Filfan M, Olaru A, Udristoiu I, Margaritescu C, Petcu E, Hermann DM, et al. Long-term treatment with spermidine increases health span of middle-aged Sprague-Dawley male rats. Geroscience. 2020;42(3):937–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Morselli E, Marino G, Bennetzen MV, Eisenberg T, Megalou E, Schroeder S, et al. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J Cell Biol. 2011;192(4):615–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, et al. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170(7):1101–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang D, Wang F, Zhai X, Li XH, He XJ. Lithium promotes recovery of neurological function after spinal cord injury by inducing autophagy. Neural Regen Res. 2018;13(12):2191–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329(5988):229–32. [DOI] [PubMed] [Google Scholar]
- 87.Schiebler M, Brown K, Hegyi K, Newton SM, Renna M, Hepburn L, et al. Functional drug screening reveals anticonvulsants as enhancers of mTOR-independent autophagic killing of Mycobacterium tuberculosis through inositol depletion. EMBO Mol Med. 2015;7(2):127–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ji MM, Wang L, Zhan Q, Xue W, Zhao Y, Zhao X, et al. Induction of autophagy by valproic acid enhanced lymphoma cell chemosensitivity through HDAC-independent and IP3-mediated PRKAA activation. Autophagy. 2015;11(12):2160–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Murakami N, Oyama F, Gu Y, McLennan IS, Nonaka I, Ihara Y. Accumulation of tau in autophagic vacuoles in chloroquine myopathy. J Neuropathol Exp Neurol. 1998;57(7):664–73. [DOI] [PubMed] [Google Scholar]
- 90.McAfee Q, Zhang Z, Samanta A, Levi SM, Ma XH, Piao S, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012;109(21):8253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang C, Tong Y, Ni W, Liu J, Xu W, Li L, et al. Inhibition of autophagy induced by overexpression of mda-7/interleukin-24 strongly augments the antileukemia activity in vitro and in vivo. Cancer Gene Ther. 2010;17(2):109–19. [DOI] [PubMed] [Google Scholar]
- 92.Zhang XD, Wang Y, Wang Y, Zhang X, Han R, Wu JC, et al. p53 mediates mitochondria dysfunction-triggered autophagy activation and cell death in rat striatum. Autophagy. 2009;5(3):339–50. [DOI] [PubMed] [Google Scholar]
- 93.Zhang H, Chang JT, Guo B, Hansen M, Jia K, Kovacs AL, et al. Guidelines for monitoring autophagy in Caenorhabditis elegans. Autophagy. 2015;11(1):9–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu J, Xia H, Kim M, Xu L, Li Y, Zhang L, et al. Beclin1 controls the levels of p53 by regulating the deubiquitination activity of USP10 and USP13. Cell. 2011;147(1):223–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melentijevic I, Toth ML, Arnold ML, Guasp RJ, Harinath G, Nguyen KC, et al. C. elegans neurons jettison protein aggregates and mitochondria under neurotoxic stress. Nature. 2017;542(7641):367–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Johnson AE, Shu H, Hauswirth AG, Tong A, Davis GW. VCP-dependent muscle degeneration is linked to defects in a dynamic tubular lysosomal network in vivo. Elife. 2015;4:e07366. [DOI] [PMC free article] [PubMed] [Google Scholar]


