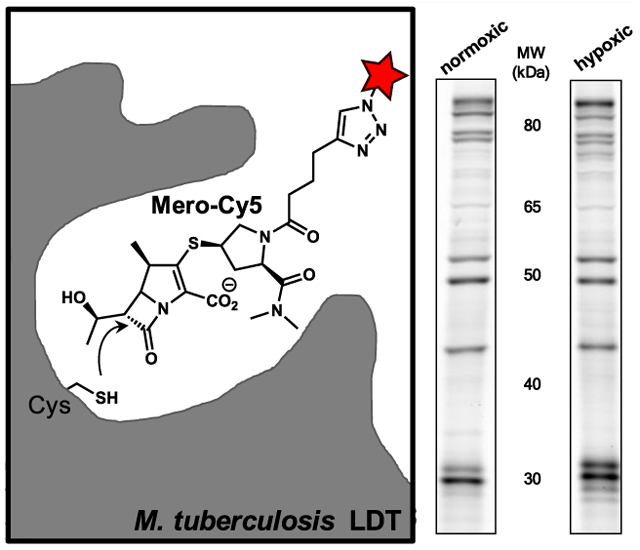
Abstract
Tuberculosis (TB), caused by the bacterial pathogen Mycobacterium tuberculosis (Mtb), infects 10 million people a year. An estimated 25% of humans harbor latent TB infections, an asymptomatic form of the disease. In both active and latent infections, Mtb relies on cell wall peptidoglycan for viability. In the current work, we synthesized fluorescent analogues of β-lactam antibiotics to study two classes of enzymes that maintain Mtb’s peptidoglycan: penicillin-binding proteins (PBPs) and l,d-transpeptidases (LDTs). This set of activity-based probes included analogues of three classes of β-lactams: a monobactam (aztreonam-Cy5), a cephalosporin (cephalexin-Cy5), and a carbapenem (meropenem-Cy5). We used these probes to profile enzyme activity in protein gel-resolved lysates of Mtb. All three out-performed the commercial reagent Bocillin-FL, a penam. Meropenem-Cy5 was used to identify β-lactam targets by mass spectrometry, including PBPs, LDTs, and the β-lactamase BlaC. New probes were also used to compare PBP and LDT activity in two metabolic states: dormancy and active replication. We provide the first direct evidence that Mtb dynamically regulates the enzymes responsible for maintaining peptidoglycan in dormancy. Lastly, we profiled drug susceptibility in lysates and found that meropenem inhibits PBPs, LDTs, and BlaC.
Keywords: antibiotic, tuberculosis, activity-based probe, fluorescent, carbapenem, proteomics
Graphical Abstract
Mycobacterium tuberculosis (Mtb) has the ignominious distinction of being the deadliest bacterial pathogen in the world. It causes both active tuberculosis (TB) and asymptomatic, latent disease.1,2 Latent TB infections (LTBIs) are common and widespread, creating a substantial barrier to eradication. In the human host, Mtb can transition to a dormant, non-replicating state in response to hypoxia, nutrient starvation, or other harsh conditions.3–5 Dormant Mtb is resistant to treatment with most front-line drugs.6,7 While dormant Mtb is associated with LTBIs, active TB patients also harbor metabolically varied Mtb.1,8,9 This suggests that the most effective drugs for curing TB will target both actively replicating and dormant Mtb.
Regardless of the metabolic state, maintenance of the cell wall peptidoglycan (PG) is required for Mtb viability. PG is composed of glycan chains linked to peptide stems that are cross-linked to form a mesh that surrounds bacterium.10 Two of the earliest classes of β-lactam drugs, the penicillins and cephalosporins, inhibit d,d-transpeptidases (DDTs). These enzymes are aptly called penicillin-binding proteins (PBPs), and they covalently modify PG.11 DDTs form 4 → 3 cross-links between a stem peptide’s fourth amino acid, d-alanine, and the third amino acid on an adjacent stem: meso-diaminopimelic acid (mDAP, Figure 1). In most bacteria, the PG is composed of 4 → 3 linkages. Yet in Mtb, 3 → 3 cross-links compose up to 80% of linkages.12,13 This covalent modification, bonding between mDAP3 residues, is made by l,d-transpeptidases (LDTs). Both LDTs and PBPs are critical to survival because they maintain the structure and rigidity of PG. The Mtb LDTs and PBPs are summarized in Table 1.
Figure 1.
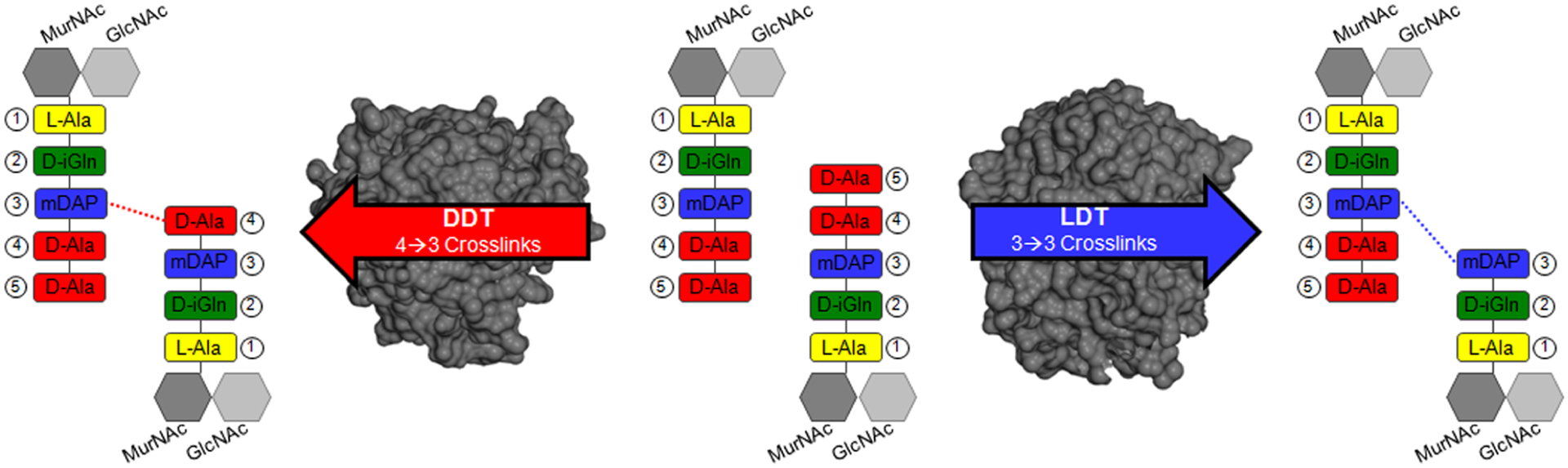
PBPs and LDTs catalyze transpeptidase reactions between different amino acid substrates. A subset of PBPs is called d,d-transpeptidases (DDTs), and they cross-link a position 4 d-Ala to a neighboring mDAP residue. Mtb also uses l,d-transpeptidases (LDTs), which form 3 → 3 cross-links (mDAP to mDAP). The DDT protein depicted in dark gray (left) is PonA1 (PDB ID: 5CXW) and the LDT structure (right) is LdtMt2 (PDB ID: 5K69). (Dark gray hexagon) MurNAc, N-acetylmuramic acid; (Light gray hexagon) GlcNAc, N-acetylglucosamine; l-Ala, l-alanine; d-iGln, d-isoglutamine; mDAP, meso-diaminopimelic acid; d-Ala, d-alanine.
Table 1.
LDTs and PBPs in M. tuberculosis (Mtb)
| name | RV # | enzyme type | MW (kDa) | |
|---|---|---|---|---|
| l,d-transpeptidases (LDTs) | LdtMt1 | RvO116c | LDT | 26.9 |
| LdtMt2 | Rv2518c | LDT | 43.4 | |
| LdtMt3 | Rvl433 | LDT | 28.7 | |
| LdtMt4 | RvO192 | LDT | 38.9 | |
| LdtMt5 | RvO483 | LDT | 47.9 | |
| penicillin-binding proteins (PBPs) | PonAl | Rv0050 | transglycosylase and DDT | 71.1 |
| PonA2 | Rv3682 | transglycosylase and DDT | 84.6 | |
| PBPA | Rv0016c | DDT | 51.6 | |
| PBPB | Rv2163c | DDT | 72.5 | |
| PBP-lipo | Rv2864 | DDT | 63.0 | |
| PBP4 | Rv3627c | carboxypeptidase | 46.8 | |
| DacB1 | Rv3330 | carboxypeptidase | 41.7 | |
| DacB2 | Rv2911 | carboxypeptidase | 29.7 | |
| BlaC | Rv2068c | β-lactamase | 32.6 | |
| Rv0907 | β-lactamase | 56.7 | ||
| Rvl367 | β-Mactamase | 41.3 |
Mtb uses PBPs, LDTs, and other enzymes to modify cell wall PG to survive environmental changes.14,15 Transcriptional analyses support that the PBPs and LDTs are dynamically regulated during these metabolic transitions. The LdtMt1 transcript is up-regulated 17-fold under nutrient starvation, while PBPA is down-regulated.3 DacB1’s transcript is up-regulated 5-fold under conditions that induce dormancy.16 The corresponding enzymes likely have non-redundant roles in dormancy, and PBPs and LDTs are biochemically distinct enzyme classes.11 For example, PBPA is an essential enzyme,17 and both DacB1 and PonA2 are required for infectivity.18,19 PonA2, a bifunctional transglycosylase and DDT, is associated with non-replicating mycobacteria20 and survival under hypoxia.21 The loss of PonA2 confers β-lactam drug-resistance and makes LdtMt2 essential.22 LdtMt1 and LdtMt2 are required for virulence,23,24 and LdtMt1 is believed to remodel PG for survival in dormancy.3,12 There is recent evidence that LDTs mediate PG maturation and confer cell shape in mycobacteria.25,26
The central role of LDTs and PBPs in Mtb PG biosynthesis and infection makes them candidate drug targets for treating TB. β-lactam antibiotics target PG biosynthesis and are widely used, safe, and effective drugs. However, they are not part of the standard treatment regimen for TB. One reason for this is likely historical.27 A study from 1949 described a “penicillinase” that made Mtb resistant to β-lactam antibiotics.28 The enzyme responsible is BlaC, a β-lactamase, and its discovery indicated that those first β-lactam antibiotics would be unsuccessful in treating TB. BlaC also inactivated the cephalosporin class, which was introduced in the 1960s.29 Yet, we now know that BlaC has limited activity against carbapenems30 and is inactivated by clavulanate,31 a clinically approved β-lactamase inhibitor. Moreover, as reviewed by Story-Roller and Lamichhane,27 the carbapenem class inhibits LDTs, which may provide distinct (non-PBP) drug targets in Mtb.
To summarize, the discovery of the mycobacterial LDTs12,32 has renewed interest in understanding how PBPs and LDTs maintain PG integrity and how these enzymes interact with β-lactam antibiotics. A central motivation for this work is the potential for using clinically approved β-lactams to treat TB.22,27,33 However, the scarcity of information on protein targets of β-lactams in Mtb makes the development of new treatment regimens challenging. Furthermore, there is almost no information on which PBPs and LDTs are active in dormant Mtb.
Evaluating PBPs and LDTs together would be challenging using conventional biochemical or genetic approaches. A different option is to use activity-based probes (ABPs), which facilitate monitoring of entire classes of enzymes at once in lysates or tissues. ABPs consist of a reporter linked to an irreversible inhibitor, which provides specificity for the enzyme target.34,35 There is an established precedence for using ABPs to assign peptidase functions in other bacterial species.36–41 Although both the PBPs and LDTs are susceptible to inhibition by β-lactam antibiotics, their substrate preferences are distinct. For example, PBPs are inactivated by covalent modification at a catalytic serine by various β-lactam antibiotics (e.g., penams, monobactams, cephems, and carbapenems).42–44 In contrast, LDTs are inhibited by compounds in the carbapenem class, which bind to an active-site cysteine by thioester bond formation (Figure 2).12,32,45–47 Characterizing the targets of β-lactam antibiotics in Mtb would therefore require ABPs based on carbapenems and other scaffolds.
Figure 2.

Active-site labeling of PBPs and LDTs with β-lactam probes. (A) In PBPs, an active-site serine binds to a reactive β-lactam probe, as shown for Bocillin-FL. (B) In contrast, the LDT’s active-site cysteine binds to carbapenems (e.g., meropenem-Cy5). Insets: Active-site covalently bound to the β-lactam probe.
In this work, we used ABPs to profile the activity and regulation of Mtb PBPs and LDTs. We produced a small set of structurally diverse β-lactam probes, including a new fluorescent meropenem. We used these probes to analyze PBPs and LDTs in protein gel-resolved mycobacterial lysates. We identified enzymes associated with both dormant and actively replicating Mtb. Lastly, our results show that carbapenems, including meropenem, inhibit many of the Mtb PBPs and LDTs, suggesting that this class of β-lactam could be useful for treating latent or active TB infections.
RESULTS AND DISCUSSION
Identification of β-Lactam Targets Encoded in the Mtb Genome.
We initiated this project by generating a comprehensive list of Mtb enzymes with active sites likely to interact with a β-lactam. We searched the Mtb H37Rv genome for genes with protein family (i.e., Pfam) annotation indicating PBP, LDT, or β-lactamase activity.48 This search gave a total of 43 proteins with either validated or theoretical abilities to interact with β-lactams (see Supporting Information, Table S1). The abbreviated version of this list, Table 1, contains only the most likely β-lactam targets and includes 11 PBPs and five LDTs. This list was generated by combining the PBPs identified by Sauvage et al.,49 the five known LDTs,11 and BlaC.
Synthesis and Validation of Fluorescent β-Lactam Probes.
Next, we assembled our chemical toolkit for analyzing β-lactam targets in Mtb (Figure 3). For our studies, all ABPs contained a β-lactam antibiotic as the enzyme inhibitor. We obtained Bocillin-FL to represent the penam class, which is a commercially available green fluorescent penicillin.36 We additionally selected monobactam, cephalosporin, and carbapenem scaffolds for structural diversity. An alkyne handle was included in these ABPs to enable conjugation to fluorescent probes via a copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC). In designing the probes, we modified parent compounds at a position amenable to the synthetic introduction of an alkyne. Additionally, we selected a location that would be unlikely to disrupt binding. For example, we introduced the alkyne at a position in meropenem that was expected to orient the reporter out of the LDT active site.47 Lastly, we opted for probes derived from clinically approved β-lactam antibiotics.
Figure 3.
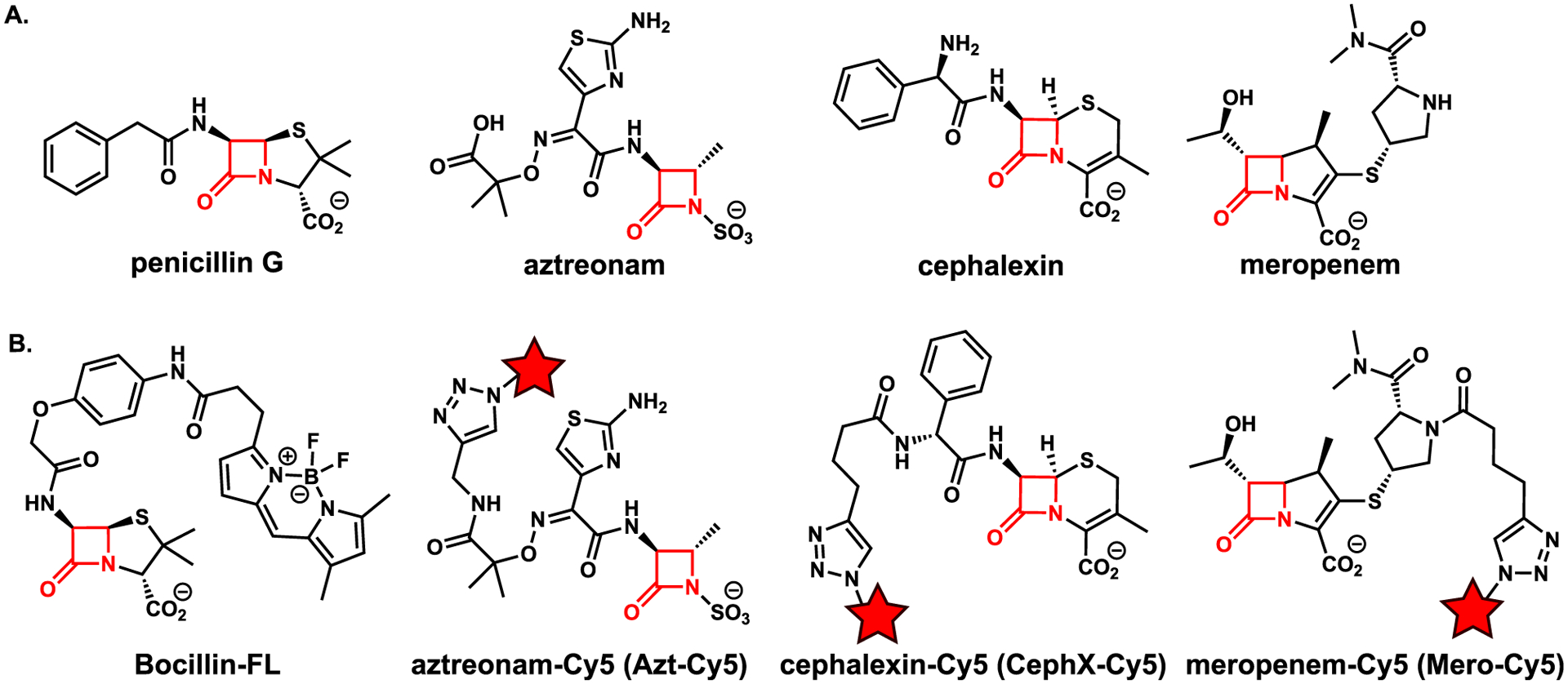
Structures of (A) β-lactam antibiotics and (B) activity-based probes used in the current work. The Cy5-conjugated derivatives were obtained by reacting the alkyne probe with sulfo-cyanine5 (Cy5) azide to produce Azt-Cy5, CephX-Cy5, and Mero-Cy5. The red star denotes the Cy5 fluorophore, and the β-lactam ring is highlighted in red.
For the monobactam compound, we synthesized the known aztreonam-N-alkyne.40 We found that increasing the reaction time resulted in a substantially higher yield than was previously reported (>95% yield in 3 d vs 19% in 30 min).40 For the cephalosporin and carbapenem scaffolds, we synthesized two new compounds, cephalexin-alkyne and meropenem-alkyne,50 respectively. For the cephalosporin probe, we used the readily available cephalexin as the starting material, providing a cost benefit over the more traditional cephalosporin C.38,40 We synthesized cephalexin-alkyne under basic conditions similar to ones used by Carlson and co-workers to synthesize cephalosporin C probes.38 A modification of these conditions to use a heterogeneous base (Cs2CO3) generated meropenem-alkyne from meropenem. We selected meropenem as our carbapenem-class probe because of its clinical significance for treating TB.51 Also, it was straightforward to modify, unlike faropenem and imipenem. These alkynes readily underwent CuAAC reactions with sulfo-cyanine5 (Cy5) azide to generate far-red fluorescent probes (Figure 3B): aztreonam-Cy5 (Azt-Cy5), cephalexin-Cy5 (CephX-Cy5), and meropenem-Cy5 (Mero-Cy5). The fluorescent probes were made in small quantities (e.g., low microgram scale) and used without further purification.
We used Bocillin-FL for the initial evaluation of enzyme targets in Mtb lysates (Figure 4). Prior work used this probe to detect PBPs—including PonA2—in Mycobacterium smegmatis (Msmeg), a rapidly growing mycobacterial species.20,52 We analyzed lysates prepared from mid log phase cultures of Mtb (mc26020), a ΔlysA ΔpanCD auxotroph of Mtb H37Rv.53 Bocillin-FL-labeled lysates were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and imaged on a fluorescence scanner to detect green fluorescence from the target-bound probe (Figure 4). Labeled proteins were nearly all over 50 kDa, despite 10 of the targets in Table 1 being of a lower molecular weight. Labeling was reduced by pretreatment with penicillin G and other β-lactam antibiotics (see Supporting Information; Figure S1). This competition experiment demonstrated that Bocillin-FL labeling was specific for proteins that bind to β-lactams. The fluorescent band intensity corresponded with activity not relative protein abundance (see Figure S1B). We observed three autofluorescent bands in the green channel. Additionally, there were three major drawbacks to using Bocillin-FL: (1) Bocillin-FL labeled less than half of the potential targets; (2) it labeled only two proteins under 50 kDa; (3) penicillins are not inhibitors of LDTs.
Figure 4.
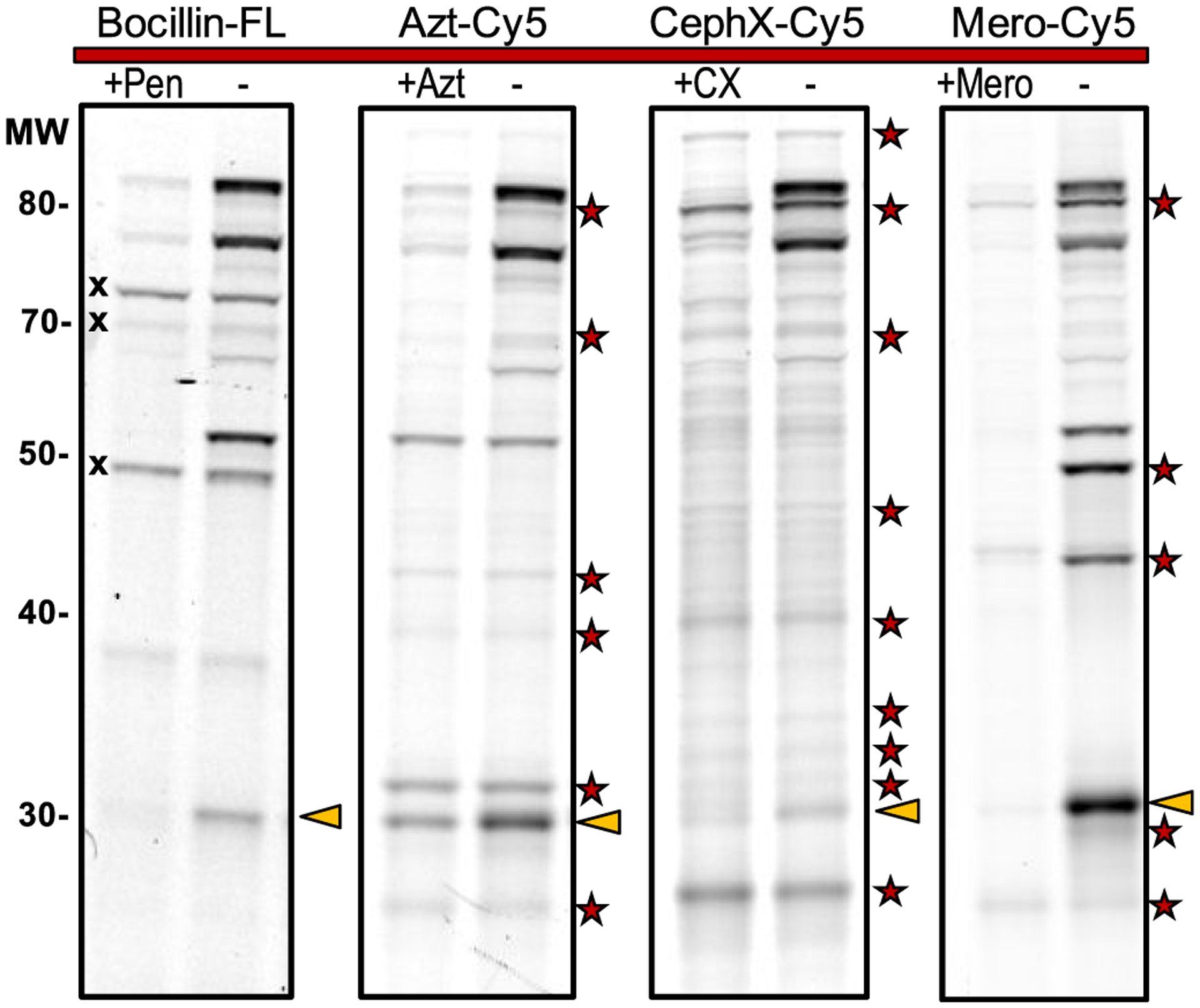
β-Lactam probes covalently bind to Mtb proteins, enabling fluorescent detection. Lysates were treated with a probe, resolved by SDS-PAGE, and imaged. Three autofluorescent bands (X) were observed with Bocillin-FL but not with Cy5 probes. Labeling was reduced by pretreatment with the parent antibiotic (left lanes). Pen, penicillin G; Azt, aztreonam; CX, cephalexin; Mero, meropenem. Red star: Targets bound and detected by Azt-Cy5, CephX-Cy5, or Mero-Cy5 (but not Bocillin-FL). BlaC migrates at 30 kDa and is denoted by gold triangles.
We overcame those drawbacks by using our far-red fluorescent ABPs: Azt-Cy5, CephX-Cy5, and Mero-Cy5. Mtb lysates were probe-treated, resolved by SDS-PAGE, and imaged to detect Cy5-labeled targets (Figure 4). All of these probes out-performed Bocillin-FL by revealing more fluorescent bands (i.e., enzymes inhibited by β-lactams). They also labeled multiple proteins with an apparent molecular weight of less than 50 kDa. This is further evidence that Bocillin-FL is a poor substrate for targets below 50 kDa because the lysis, treatment, and gel conditions were similar among the four fluorescent probes. This finding is consistent also with prior work that found that penams do not covalently inhibit the active site of LDTs,11 which have a molecular weight range of 26.9–47.9 kDa.
We performed competition experiments with the parent antibiotic, and labeling was reduced under these conditions. Some bands remained unchanged in the cephalexin-treated lysates, suggesting that CephX-Cy5 may have some non-specific interactions with proteins. However, pretreatment with aztreonam or meropenem significantly reduced labeling by the corresponding probe. This indicates that the labeling with Azt-Cy5 and Mero-Cy5 was specific for proteins that bind β-lactams. Notably, these new probes each revealed distinct target selectivity plus bands that were undetected by Bocillin-FL. All four probes detected BlaC activity at 30 kDa (see next section).
Identification of Protein Targets of Mero-Cy5.
We initiated the process of identifying proteins that were labeled by the carbapenem probe Mero-Cy5. In these studies, we used lysates from actively replicating cultures and dormant cultures of Mtb. We induced dormancy by incubation under hypoxic conditions, as described.5,54 Mtb lysates were fractionated by centrifugation (into pellet and supernatant), labeled with Mero-Cy5, and resolved by SDS-PAGE. The pellet was enriched in proteins associated with the cell wall and cell membrane (Figure S2).55 Fluorescent bands were excised and submitted for identification by liquid chromatography tandem mass spectrometry (LC–MS/MS).
We focused our analysis on proteins with putative PBP, LDT, or β-lactamase activity (see Table S1). Of the 43 proteins in Table S1, 32 were identified in our MS analysis (Table S2). Experimental details and the full data set are provided in the Supporting Information (SI), including annotated gels from which we excised bands (Figures S2–S6). A summary of the results from these studies is provided in Figure 5. Specifically, we identified DDTs (PonA1, PonA2, PBPA), LDTs (LdtMt2, LdtMt3, LdtMt5), carboxypeptidases (DacB1, DacB2, PBP4), and the β-lactamase BlaC. Most of these enzyme hits were identified in more than one sample.
Figure 5.
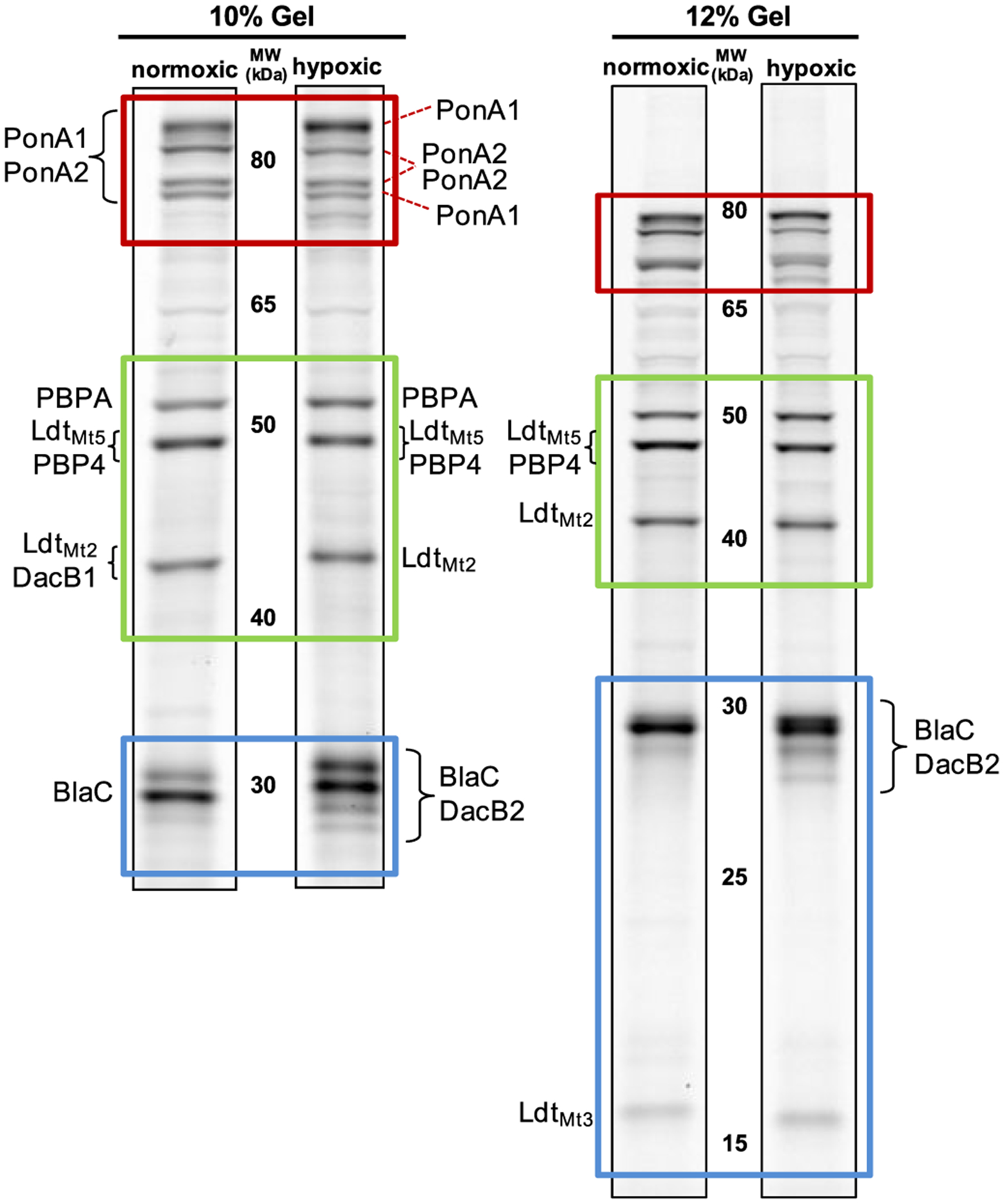
Analysis of Mtb lysates for MS-based identification of proteins labeled with Mero-Cy5. Lysates from normoxic and hypoxic cultures were resolved on 10% (left) or 12% (right) SDS-PAGE protein gels. Based on the data provided in the SI, the gels have been annotated to indicate the most likely PBP or LDT source of fluorescent bands. To simplify comparisons, three regions of the gels are boxed: High MW (~80 kDa; red); Middle MW (40–50 kDa; green); Low MW (15–30 kDa; blue). Figures S3, S5, and S6 include more information for each excised band, including protein identities, molecular weight, % coverage, and the number of peptides found per protein.
We further investigated one of the identified enzymes: BlaC. BlaC’s activity most likely contributes much of the signal at 30 kDa. In normoxic samples, we identified 18 unique peptides from BlaC with 69.7% sequence coverage (see Figure S3). We validated our assignment by comparing lysates from Mtb (mc26020) with Mycobacterium marinum, a close genetic relative, and with Msmeg (mc2155), a species that lacks a BlaC ortholog. We observed a clavulanate-sensitive band at 30 kDa for lysates from Mtb and M. marinum but not Msmeg (Figure S4). The major β-lactamase in Msmeg, BlaS (31 kDa), is not a target of clavulanate.56 This comparative analysis provides compelling support for our assignment of the fluorescent band at 30 kDa to BlaC activity.
Interestingly, we observed a different banding pattern near 30 kDa for samples analyzed from normoxic versus hypoxic cultures. We identified peptides from BlaC and carboxypeptidase DacB2; other β-lactamases were identified in considerably lower abundance in these excised bands. This made the source of the signal ambiguous because it could be from any combination of BlaC, DacB2, or the putative β-lactamases. While it has been shown that BlaC represents the majority of β-lactamase activity under regular growth conditions,38 similar studies have not been conducted with non-replicating bacteria. Further studies will be needed to learn more about the source of this up-regulated activity.
Next, we analyzed the middle region of the gel, which contained three fluorescent bands between 40 and 50 kDa (Figure S5). The fluorescent band directly above 50 kDa was identified as PBPA (51.6 kDa), on the basis of the MS data (77.4% sequence coverage, 28 peptide hits). The band just below 50 kDa contained peptides from PBP4 (46.8 kDa) and LdtMt5 (47.9 kDa). The analysis of the band just above 40 kDa resulted in masses corresponding to 14 proteins with relevant Pfam notation, including eight putative β-lactamases, four PBPs (PBP4, DacB1, PBPA, and PonA2) and two LDTs (LdtMt5 and LdtMt2). By molecular weight, DacB1 (41.7 kDa) and LdtMt2 (43.4 kDa) are the most likely to be present in this region. LdtMt2 was identified from three separate samples, supporting its contribution to the fluorescent band near 40 kDa.
We obtained peptide hits from both PonA1 and PonA2 in the bands excised near 80 kDa. We believe that the signal from the top and bottom of the four bands is from PonA1 because only that protein was identified in bands excised from hypoxic samples (Figure S6). The appearance of PonA1 in two locations is unsurprising because it is regulated by phosphorylation,52 which can influence protein migration by SDS-PAGE. We verified the presence of phosphorylated proteins in those bands by staining the gel with Molecular Probe’s Pro-Q Diamond stain (Figure S7). We believe that the two middle bands contain predominantly PonA2, with the lower band resulting from cleavage of a 3 kDa signal sequence, which was only identified in the upper of those two bands.
These studies enabled us to identify a variety of targets in gel-resolved lysates, including DDTs (PonA1, PonA1, PBPA), carboxypeptidases (DacB1, DacB2, PBP4), LDTs (LdtMt2, LdtMt3, LdtMt5), and the β-lactamase BlaC. Additionally, we identified 22 other potentially relevant proteins (see Table S2), which are worth investigating in future work. Our findings are in stark contrast to a 2020 report by Lopez Quezada and coworkers.57 In their work, non-replicating Mtb was treated with a cephalosporin-based ABP. Labeled protein targets were enriched and identified by MS-based proteomics. No PBPs or LDTs were identified, supporting their claim that those compounds only target enzymes that are not transpeptidases. Clearly there is more to learn about how different classes of β-lactam drugs exert their effect in Mtb.
Patterns of Enzyme Activity from Hypoxic and Normoxic Mtb.
Our MS analysis suggested that the activity patterns of PBPs and LDTs change in response to hypoxia. This fits prior observations that Mtb increases LDT-mediated (3 → 3) cross-links in dormancy.14,15,23 To better understand these differences, we analyzed ABP-labeled lysates resolved by SDS-PAGE (Figure 6). In these studies, we normalized by total protein to enable a direct comparison between band intensities observed under different culture conditions (i.e., normoxic versus hypoxic). We found that many targets were present in both metabolic states. For example, PonA1, PonA2, PBPA, LdtMt2, LdtMt3, and LdtMt5 maintained activity in dormancy. Other targets, particularly between 40 and 80 kDa, were less active in dormancy, and at least three bands were absent. Unfortunately, we were unable to determine which enzymes lost activity in dormancy, highlighting a challenge of identifying low abundance proteins from gel-excised bands.
Figure 6.

Mero-Cy5 reveals relative enzyme activities and drug targets in lysates obtained from normoxic and hypoxic Mtb cultures. Lane 1: no drug pretreatment. Lanes 2–10: drug pretreatment. 2, cephalexin; 3, ceftriaxone; 4, aztreonam; 5, penicillin G; 6, ampicillin; 7, carbenicillin; 8, meropenem; 9, faropenem; 10, clavulanate. Lane 11: lysates not treated with Mero-Cy5. Lane 12: lysates treated with a Cy5-(triazole)-butanoic acid. Putative protein identities were obtained by MS analysis. Legend: Gold stars denote enzymes that retain activity under hypoxic conditions. Red arrows indicate bands with enhanced activity in hypoxia. Black dash-marks indicate bands that were absent in hypoxia but present in normoxia.
The cephalexin probe indicated some up-regulation of faint bands in the 80 kDa region of the gel. These changes were also visible with Azt-Cy5, Mero-Cy5, and Bocillin-FL (Figure S8). It would be interesting to know if these changes are a result of post-translational modifications, such as glycosylation or phosphorylation.52 Bocillin-FL offered little insight on enzyme targets below 50 kDa, as expected.
As noted in the previous section, we observed up-regulated enzyme activity near 30 kDa from hypoxic lysates. In particular, the ~28 kDa band was only distinct in hypoxic samples. This region was also notable in lysates treated with Azt-Cy5, which showed similar up-regulation of bands from 25 to 30 kDa under hypoxia (Figure S8). Our proteomics results indicate that DacB2 migrates in this region of the gel. This suggests that DacB2 could have enhanced activity under hypoxic conditions, a notable result considering that the related carboxypeptidase DacB1 is transcriptionally up-regulated in dormancy.16 To summarize, our in-gel analysis provides evidence that the PBPs and LDTs are dynamically regulated in response to hypoxia.
Drug Susceptibility of PBPs and LDTs in Mtb.
There is evidence that β-lactams are therapeutically relevant drugs for TB patients.27,51,58 For example, there have been occasional reports of successfully treating TB patients with the coadministration of β-lactams with clavulanate, a BlaC inhibitor.51,59–65 A 1998 report described using amoxicillin/clavulanate to cure multidrug-resistant TB.64 Drug-resistant TB has also been treated with meropenem/clavulanate.59–61,63,65 That treatment helped to cure extensively drug-resistant TB patients (83% cure rate).63 This drug combination was also effective against dormant Mtb.66,67
Inspired by those findings, we set out to investigate the targets of β-lactam drugs in Mtb using ABPs. We tested cephalexin, ceftriaxone, aztreonam, penicillin G, ampicillin, carbenicillin, meropenem, faropenem, and clavulanate. All of these drugs, except faropenem, are prescribed for treating bacterial infections in the United States. For these studies, we analyzed ABP-labeled lysates alongside lysates pretreated with a selection of β-lactam antibiotics as inhibitors (Figure 6). Again, we studied lysates from dormant (hypoxic) and actively replicating cultures. After drug treatment, lysates were labeled with Mero-Cy5. As an additional control, we exposed lysates to a sulfo-Cy5 conjugated to 5-hexynoic acid instead of a β-lactam (Lane 12). The absence of labeling with this compound indicated that the β-lactam, not the Cy5 fluorophore, dictated target-binding specificity.
We were most interested in identifying enzymes that are targets of the carbapenems (i.e., meropenem and faropenem) because these drugs inhibit LDTs27 and undergo slower hydrolysis by BlaC compared with other β-lactam classes.31 As expected, pretreatment with meropenem (Lane 8), the closest structural analogue to Mero-Cy5, eliminated most of the fluorescent labeling. We observed that meropenem completely inhibited a band that corresponds to LdtMt5, which is consistent with the finding from Lamichhane et al. that meropenem acylates the LdtMt5 active site.47 Similar results were found with faropenem (Lane 9).
Ceftriaxone (Lane 3), a clinically approved cephem, was more effective than cephalexin (Lane 2) at blocking Mero-Cy5 binding, although neither antibiotic was an effective inhibitor of BlaC, LdtMt5, or LdtMt3. The penams (penicillin G and ampicillin) and carbenicillin (Lane 7) were less effective at inhibiting targets than other drugs but reduced the Mero-Cy5 labeling of PBPA. BlaC, which retained some activity in dormancy, was inhibited by several drugs (Lanes 7–10), including clavulanate (Lane 10) and meropenem (Lane 8).
Antibiotic susceptibility was also analyzed using Azt-Cy5 and CephX-Cy5, and we observed similar inhibition profiles (Figure S9). Overall, our findings indicate that PBP and LDT activities can be reduced or eliminated in vitro using clinically approved β-lactams.
CONCLUSION
In conclusion, we used fluorescent β-lactam probes to identify active PBPs, LDTs, and β-lactamases in Mtb. In a direct comparison with Bocillin-FL, we found that Azt-Cy5, CephX-Cy5, and Mero-Cy5 enabled more β-lactam targets to be detected in protein gel-resolved lysates. Although we collected data with Cy5-modified probes, we reported the synthesis of the alkyne derivatives because they enable facile modification with an assortment of reporters. For example, a biotinylated probe would enable the enrichment of rare or low-abundance β-lactam targets before proteomic analysis.68
We used Mero-Cy5, a new probe, to identify 32 proteins that bind β-lactams, including transpeptidases (PonA1, PonA2, PBPA, LdtMt2, LdtMt3, LdtMt5), carboxypeptidases (DacB1, DacB2, PBP4), BlaC, and numerous putative β-lactamases (see Table S2). We did not identify LdtMt1 or LdtMt4, although there is structural evidence that both LDTs bind carbapenems.32,46,69 Future studies might use purified proteins or genetic knockout strains to more definitively assign activities identified here.
Mero-Cy5 was used to investigate enzyme regulation in dormant and actively replicating samples. We found numerous changes in fluorescent banding patterns and intensity between the two metabolic states. These findings provide direct evidence for the first time that the PBP and LDT activities are different between dormant and actively replicating cultures. For example, we found that some bands (25–30 kDa) were up-regulated under hypoxia. A priority for future work is to identify the exact source of that activity because it is unclear if those bands are attributable to changes in the activity of BlaC, DacB2, or an uncharacterized protein. We are in the early stages of confirming these findings under other conditions that induce dormancy (e.g., carbon starvation3 or a multistress model57). Also, it would be interesting to study in more detail how various post-translational modifications alter PBP and LDT activities during different stages of infection.
Lastly, we treated lysates with a set of β-lactam drugs to identify their protein targets. We focused on a set of antibiotics in clinical use today and found that meropenem and faropenem were both good inhibitors of BlaC, PBPs, and LDTs. Overall, we demonstrated that Mero-Cy5 is a useful probe for target identification, the analysis of enzyme regulation, and determining enzyme inhibition by various β-lactam drugs.
It is worth noting that the far-red fluorescent probes described herein were developed with fluorescence imaging in mind. Prior studies have demonstrated that peptidoglycan biosynthesis occurs at both the poles and sidewall of mycobacteria.25,26,70 Mycobacteria have been labeled using various small molecule probes compatible with fluorescence microscopy, including fluorescent peptides,70 trehalose analogues,25 and fluorescent d-amino acids.25,26 In future work, we intend to examine LDT and PBP activity in the Mtb cell wall using fluorescent β-lactams, including Mero-Cy5. Alone or in combination with existing probes, Mero-Cy5 could be used to highlight the portions of the cell wall that have the requisite machinery for cross-linking peptidoglycan. These small molecule probes are compatible with live cell labeling and would enable new investigations into how PBPs or LDTs are regulated during cell division, pathogenesis, or dormancy.
METHODS
Synthesis of Activity-Based Probes.
Bocillin FL was obtained from ThermoFisher Scientific. Azt-alkyne, CephX-alkyne, and Mero-alkyne were synthesized from the parent β-lactam, as described in the Supporting Information. The red-fluorescent probes were generated in vitro via a CuAAC reaction (1 h, room temperature) of the alkyne (100 μM) with sulfo-Cyanine5-azide (100 μM), 1.25 mM THTPA/0.25 mM CuSO4, and sodium ascorbate (15 mM) in HEPES buffer (pH 7.3); the fluorescent probes were used without purification.
Mtb Culture Conditions.
Mtb mc26020 was grown in Middlebrook 7H9-OADC medium supplemented with lysine, pantothenate, and casamino acids. For normoxic cultures, the flasks were maintained at 37 °C with 100 rpm shaking under atmospheric conditions. For hypoxic cultures, mid log phase cultures were diluted to OD600 0.4 and grown as standing cultures under hypoxic conditions (1% O2 and 5% CO2) at 37 °C until a control flask containing methylene blue was decolorized.71
Preparation of Lysates.
Mtb grown under normoxic conditions was harvested at mid log phase (OD600 = 0.9–1.2). Mtb grown under hypoxic conditions was harvested at fixed time-points (OD600 = 0.4–0.8). All cells were collected by centrifugation (10 min, 4000g, 4 °C) and washed twice with PBS containing 0.05% Tween 80. Pellets were resuspended in detergent-free lysis buffer [50 mM Tris (pH 7.5 at 4 °C), 50 mM NaCl, 0.5 mM CaCl2, 0.5 mM MgCl2], lysed by mechanical disruption, and pelleted by centrifugation (10 min, 4000g, 4 °C). The supernatant was transferred to a separate tube. The beads and cell debris were resuspended in an equivalent volume of double detergent containing lysis buffer [50 mM Tris (pH 7.5 at 4 °C), 50 mM NaCl, 0.5 mM CaCl2, 0.5 mM MgCl2, 0.4% triton X-100], and the mechanical disruption and centrifugation steps were repeated. The combined supernatants were pelleted by centrifugation (10 min, 4000g, 4 °C) to remove insoluble debris and sterilized by filtration (0.2 μm PES membrane).
Lysate Labeling and Gel Imaging.
Lysates were diluted with detergent containing buffer [50 mM Tris (pH 7.5 at 4 °C), 50 mM NaCl, 0.5 mM CaCl2, 0.5 mM MgCl2, 0.2% triton X-100] to normalize the total protein concentration. For samples pretreated with antibiotics, the antibiotic was added for 15 min prior to probe labeling. Lysates were labeled with the Cy5-modified probe (5 μM) for 1 h at room temperature. Lysates were resolved by SDS-PAGE, washed, fixed, and imaged on a Typhoon multimode imager.
Band Excision and LC–MS/MS Proteomics.
All bands were excised from an SDS-PAGE gel on the basis of the fluorescent image and stored frozen. Gel slices were processed to remove SDS, reduced, and methylated. Proteins were trypsin-digested, and the resulting peptides were concentrated. LC–MS/MS-based proteomics was performed by the OHSU Proteomics Shared Resource Facility. Peptides were considered positively identified if their probability of identification was ≥99% with a false-discovery rate (FDR) of 0.67%. Proteins were considered positively identified if two or more unique peptides were detected (probability of identification ≥95%; FDR 0.0%).
Supplementary Material
ACKNOWLEDGMENTS
Funding for this research was provided by NIH (R01 AI149737), the Knight Cancer Institute, and the OHSU School of Medicine. S.R.L. was supported by an NIH T32 training grant (T32-AI07472). The authors are grateful to Dr. Gyanu Lamichhane (JHU) and Dr. Clifton Barry (NIH) for helpful discussions, Scotland Farley (OHSU) for synthesizing the aztreonam probe, and Dr. Kyle Gee (ThermoFisher Scientific) for providing Bocillin FL. Before publication we learned that Prof. Erin Carlson’s group (UMinn) had independently synthesized the Meropenem-alkyne compound. Therefore, we coordinated with her to deposit both papers simultaneously online (BioRxiv, Dec. 201950,72).
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acsinfecdis.0c00809
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsinfecdis.0c00809.
Tables of enzyme list and band excision data (XLSX)
Discussions of characterization and synthetic methods used, Cu-catalyzed azide-alkyne click reaction, mycobacterial culture conditions, hypoxia induced dormancy, preparation of lysates, analysis of labeled lysates by SDS-PAGE, detection of phosphoproteins in SDS-PAGE gels, and band excision for LC–MS/MS-based proteomics and figures of SDS-PAGE gels, Bocillin-FL labeling, Images of gels used for band excision experiments, visualization of the division of protein gels, protein identifications for bands, analysis of lysates, identification of proteins associated with fluorescent bands, analysis of the phosphorylation state of proteins labeled with Mero-Cy5, β-lactam probes show differences in transpeptidase activity, Azt-Cy5 and CephX-Cy5 selectively label proteins, LC trace and LC–MS results, and 1H NMR spectra (PDF)
Raw data files, results, peak lists, and search engine files for proteomics experiments will be deposited in an online database (i.e., ProteomeXchange via MassIVE).
The authors declare no competing financial interest.
REFERENCES
- (1).Barry CE, Boshoff HI, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, and Young D (2009) The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol 7, 845–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).WHO (2018) Global Tuberculosis Report, 20th ed., Geneva. [Google Scholar]
- (3).Betts JC, Lukey PT, Robb LC, McAdam RA, and Duncan K (2002) Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol 43, 717–731. [DOI] [PubMed] [Google Scholar]
- (4).Wayne LG, and Hayes LG (1996) An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun 64, 2062–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Wayne LG (2001) In Vitro Model of Hypoxically Induced Nonreplicating Persistence of Mycobacterium tuberculosis. Methods Mol. Med 54, 247–269. [DOI] [PubMed] [Google Scholar]
- (6).Deb C, Lee CM, Dubey VS, Daniel J, Abomoelak B, Sirakova TD, Pawar S, Rogers L, and Kolattukudy PE (2009) A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One 4, e6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Nathan C, and Barry CE 3rd. (2015) TB drug development: immunology at the table. Immunol. Rev 264, 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Bentrup K. H. z., and Russell DG (2001) Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 9, 597–605. [DOI] [PubMed] [Google Scholar]
- (9).Esmail H, Barry CE, and Wilkinson RJ (2012) Understanding latent tuberculosis: the key to improved diagnostic and novel treatment strategies. Drug Discovery Today 17, 514–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Hett EC, and Rubin EJ (2008) Bacterial Growth and Cell Division: a Mycobacterial Perspective. Microbiol. Mol. Biol. Rev 72, 126–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Tolufashe GF, Sabe VT, Ibeji CU, Ntombela T, Govender T, Maguire GEM, Kruger HG, Lamichhane G, and Honarparvar B (2018) Structure and function of L,D- and D,D-transpeptidase family enzymes from Mycobacterium tuberculosis. Curr. Med. Chem 27, 3250. [DOI] [PubMed] [Google Scholar]
- (12).Lavollay M, Arthur M, Fourgeaud M, Dubost L, Marie A, Veziris N, Blanot D, Gutmann L, and Mainardi J-L (2008) The Peptidoglycan of Stationary-Phase Mycobacterium tuberculosis Predominantly Contains Cross-Links Generated by l,d-Transpeptidation. J. Bacteriol 190, 4360–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wietzerbin J, Das BC, Petit JF, Lederer E, Leyh-Bouille M, and Ghuysen JM (1974) Occurrence of D-alanyl-(D)-meso-diaminopimelic acid and meso-diaminopimelyl-meso-diaminopimelic acid interpeptide linkages in the peptidoglycan of Mycobacteria. Biochemistry 13, 3471–3476. [DOI] [PubMed] [Google Scholar]
- (14).Seiler P, Ulrichs T, Bandermann S, Pradl L, Jörg S, Krenn V, Morawietz L, Kaufmann SHE, and Aichele P (2003) Cell-Wall Alterations as an Attribute of Mycobacterium tuberculosis in Latent Infection. J. Infect. Dis 188, 1326–1331. [DOI] [PubMed] [Google Scholar]
- (15).Bacon J, Alderwick LJ, Allnutt JA, Gabasova E, Watson R, Hatch KA, Clark SO, Jeeves RE, Marriott A, Rayner E, Tolley H, Pearson G, Hall G, Besra GS, Wernisch L, Williams A, and Marsh PD (2014) Non-Replicating Mycobacterium tuberculosis Elicits a Reduced Infectivity Profile with Corresponding Modifications to the Cell Wall and Extracellular Matrix. PLoS One 9, e87329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Cho SH, Goodlett D, and Franzblau S (2006) ICAT-based comparative proteomic analysis of non-replicating persistent Mycobacterium tuberculosis. Tuberculosis 86, 445–460. [DOI] [PubMed] [Google Scholar]
- (17).Griffin JE, Gawronski JD, DeJesus MA, Ioerger TR, Akerley BJ, and Sassetti CM (2011) High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism. PLoS Pathog. 7, e1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, and Ehrt S (2009) Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J. Bacteriol 191, 625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Dutta NK, Mehra S, Didier PJ, Roy CJ, Doyle LA, Alvarez X, Ratterree M, Be NA, Lamichhane G, Jain SK, Lacey MR, Lackner AA, and Kaushal D (2010) Genetic requirements for the survival of tubercle bacilli in primates. J. Infect. Dis 201, 1743–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Patru M-M, and Pavelka MS (2010) A Role for the Class A Penicillin-Binding Protein PonA2 in the Survival of Mycobacterium smegmatis under Conditions of Nonreplication. J. Bacteriol 192, 3043–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Saxena A, Srivastava V, Srivastava R, and Srivastava BS (2008) Identification of genes of Mycobacterium tuberculosis upregulated during anaerobic persistence by fluorescence and kanamycin resistance selection. Tuberculosis 88, 518–525. [DOI] [PubMed] [Google Scholar]
- (22).Wivagg CN, Bhattacharyya RP, and Hung DT (2014) Mechanisms of [beta]-lactam killing and resistance in the context of Mycobacterium tuberculosis. J. Antibiot 67, 645–654. [DOI] [PubMed] [Google Scholar]
- (23).Gupta R, Lavollay M, Mainardi J-L, Arthur M, Bishai WR, and Lamichhane G (2010) The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat. Med 16, 466–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Schoonmaker MK, Bishai WR, and Lamichhane G (2014) Nonclassical transpeptidases of Mycobacterium tuberculosis alter cell size, morphology, the cytosolic matrix, protein localization, virulence, and resistance to beta-lactams. J. Bacteriol 196, 1394–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).García-Heredia A, Pohane AA, Melzer ES, Carr CR, Fiolek TJ, Rundell SR, Lim HC, Wagner JC, Morita YS, Swarts BM, and Siegrist MS (2018) Peptidoglycan precursor synthesis along the sidewall of pole-growing mycobacteria. eLife 7, e37243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Baranowski C, Welsh MA, Sham L-T, Eskandarian HA, Lim HC, Kieser KJ, Wagner JC, McKinney JD, Fantner GE, Ioerger TR, Walker S, Bernhardt TG, Rubin EJ, and Rego EH (2018) Maturing Mycobacterium smegmatis peptidoglycan requires non-canonical crosslinks to maintain shape. eLife 7, e37516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Story-Roller E, and Lamichhane G (2018) Have we realized the full potential of beta-lactams for treating drug-resistant TB? IUBMB Life 70, 881–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Iland CN, and Baines S (1949) The effect of penicillin on the tubercle bacillus: Tubercle penicillinase. J. Pathol. Bacteriol 61, 329–335. [Google Scholar]
- (29).Wang F, Cassidy C, and Sacchettini JC (2006) Crystal Structure and Activity Studies of the Mycobacterium tuberculosis β-Lactamase Reveal Its Critical Role in Resistance to β-Lactam Antibiotics. Antimicrob. Agents Chemother 50, 2762–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Tremblay LW, Fan F, and Blanchard JS (2010) Biochemical and Structural Characterization of Mycobacterium tuberculosis β-Lactamase with the Carbapenems Ertapenem and Doripenem. Biochemistry 49, 3766–3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Hugonnet J-E, and Blanchard JS (2007) Irreversible Inhibition of the Mycobacterium tuberculosis β-Lactamase by Clavulanate. Biochemistry 46, 11998–12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Cordillot M, Dubée V, Triboulet S, Dubost L, Marie A, Hugonnet J-E, Arthur M, and Mainardi J-L (2013) In Vitro Cross-Linking of Mycobacterium tuberculosis Peptidoglycan by l,d-Transpeptidases and Inactivation of These Enzymes by Carbapenems. Antimicrob. Agents Chemother 57, 5940–5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Keener AB (2014) Oldie but goodie: Repurposing penicillin for tuberculosis. Nat. Med 20, 976. [DOI] [PubMed] [Google Scholar]
- (34).Puri AW, and Bogyo M (2009) Using Small Molecules To Dissect Mechanisms of Microbial Pathogenesis. ACS Chem. Biol 4, 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kocaoglu O, and Carlson EE (2016) Progress and prospects for small-molecule probes of bacterial imaging. Nat. Chem. Biol 12, 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Gee KR, Kang HC, Meier TI, Zhao G, and Blaszcak LC (2001) Fluorescent Bocillins: Synthesis and application in the detection of penicillin-binding proteins. Electrophoresis 22, 960–965. [DOI] [PubMed] [Google Scholar]
- (37).June CM, Vaughan RM, Ulberg LS, Bonomo RA, Witucki LA, and Leonard DA (2014) A fluorescent carbapenem for structure function studies of penicillin-binding proteins, β-lactamases, and β-lactam sensors. Anal. Biochem 463, 70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Kocaoglu O, Calvo RA, Sham L-T, Cozy LM, Lanning BR, Francis S, Winkler ME, Kearns DB, and Carlson EE (2012) Selective Penicillin-Binding Protein Imaging Probes Reveal Substructure in Bacterial Cell Division. ACS Chem. Biol 7, 1746–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Kocaoglu O, and Carlson EE (2015) Profiling of β-Lactam Selectivity for Penicillin-Binding Proteins in Escherichia coli Strain DC2. Antimicrob. Agents Chemother 59, 2785–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Staub I, and Sieber SA (2008) β-Lactams as Selective Chemical Probes for the in Vivo Labeling of Bacterial Enzymes Involved in Cell Wall Biosynthesis, Antibiotic Resistance, and Virulence. J. Am. Chem. Soc 130, 13400–13409. [DOI] [PubMed] [Google Scholar]
- (41).Staub I, and Sieber SA (2009) β-Lactam Probes As Selective Chemical-Proteomic Tools for the Identification and Functional Characterization of Resistance Associated Enzymes in MRSA. J. Am. Chem. Soc 131, 6271–6276. [DOI] [PubMed] [Google Scholar]
- (42).Filippova EV, Kieser KJ, Luan CH, Wawrzak Z, Kiryukhina O, Rubin EJ, and Anderson WF (2016) Crystal structures of the transpeptidase domain of the Mycobacterium tuberculosis penicillin-binding protein PonA1 reveal potential mechanisms of antibiotic resistance. FEBS J. 283, 2206–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Arora D, Chawla Y, Malakar B, Singh A, and Nandicoori VK (2018) The transpeptidase PbpA and noncanonical transglycosylase RodA of Mycobacterium tuberculosis play important roles in regulating bacterial cell lengths. J. Biol. Chem 293, 6497–6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Wivagg CN, Wellington S, Gomez JE, and Hung DT (2016) Loss of a Class A Penicillin-Binding Protein Alters β-Lactam Susceptibilities in Mycobacterium tuberculosis. ACS Infect. Dis 2, 104–110. [DOI] [PubMed] [Google Scholar]
- (45).Kumar P, Kaushik A, Lloyd EP, Li SG, Mattoo R, Ammerman NC, Bell DT, Perryman AL, Zandi TA, Ekins S, Ginell SL, Townsend CA, Freundlich JS, and Lamichhane G (2017) Non-classical transpeptidases yield insight into new antibacterials. Nat. Chem. Biol 13, 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Dubée V, Triboulet S, Mainardi J-L, Ethève-Quelquejeu M, Gutmann L, Marie A, Dubost L, Hugonnet J-E, and Arthur M (2012) Inactivation of Mycobacterium tuberculosis l,d-Transpeptidase LdtMt1 by Carbapenems and Cephalosporins. Antimicrob. Agents Chemother 56, 4189–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Brammer Basta LA, Ghosh A, Pan Y, Jakoncic J, Lloyd EP, Townsend CA, Lamichhane G, and Bianchet MA (2015) Loss of a Functionally and Structurally Distinct ld-Transpeptidase, LdtMt5, Compromises Cell Wall Integrity in Mycobacterium tuberculosis. J. Biol. Chem 290, 25670–25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).The UniProt Consortium (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Sauvage E, Kerff F, Terrak M, Ayala JA, and Charlier P (2008) The penicillin-binding proteins: structure and role in peptidoglycan biosynthesis. FEMS Microbiol Rev. 32, 234–258. [DOI] [PubMed] [Google Scholar]
- (50).Sharifzadeh S, Dempwolff F, Kearns DB, and Carlson EE (2020) Harnessing β-Lactam Antibiotics for Illumination of the Activity of Penicillin-Binding Proteins in Bacillus subtilis. ACS Chem. Biol 15, 1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Jaganath D, Lamichhane G, and Shah M (2016) Carbapenems against Mycobacterium tuberculosis: a review of the evidence. Int. J. Tuberc. Lung Dis 20, 1436–1447. [DOI] [PubMed] [Google Scholar]
- (52).Kieser KJ, Boutte CC, Kester JC, Baer CE, Barczak AK, Meniche X, Chao MC, Rego EH, Sassetti CM, Fortune SM, and Rubin EJ (2015) Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria. PLoS Pathog. 11, e1005010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Sambandamurthy VK, Derrick SC, Jalapathy KV, Chen B, Russell RG, Morris SL, and Jacobs WR (2005) Long-Term Protection against Tuberculosis following Vaccination with a Severely Attenuated Double Lysine and Pantothenate Auxotroph of Mycobacterium tuberculosis. Infect. Immun 73, 1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Tallman KR, Levine SR, and Beatty KE (2016) Profiling Esterases in Mycobacterium tuberculosis Using Far-Red Fluorogenic Substrates. ACS Chem. Biol 11, 1810–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Rezwan M, Lanéelle M-A, Sander P, and Daffé M (2007) Breaking down the wall: Fractionation of mycobacteria. J. Microbiol. Methods 68, 32–39. [DOI] [PubMed] [Google Scholar]
- (56).Flores AR, Parsons LM, and Pavelka MS (2005) Genetic analysis of the β-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to β-lactam antibiotics. Microbiology 151, 521–532. [DOI] [PubMed] [Google Scholar]
- (57).Lopez Quezada L, Smith R, Lupoli TJ, Edoo Z, Li X, Gold B, Roberts J, Ling Y, Park SW, Nguyen Q, Schoenen FJ, Li K, Hugonnet J-E, Arthur M, Sacchettini JC, Nathan C, and Aubé J (2020) Activity-Based Protein Profiling Reveals That Cephalosporins Selectively Active on Non-replicating Mycobacterium tuberculosis Bind Multiple Protein Families and Spare Peptidoglycan Transpeptidases. Front. Microbiol 11, 1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Catalao MJ, Filipe SR, and Pimentel M (2019) Revisiting Anti-tuberculosis Therapeutic Strategies That Target the Peptidoglycan Structure and Synthesis. Front. Microbiol 10, 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Dauby N, Muylle I, Mouchet F, Sergysels R, and Payen MC (2011) Meropenem/clavulanate and linezolid treatment for extensively drug-resistant tuberculosis. Pediatr. Infect. Dis. J 30, 812–813. [DOI] [PubMed] [Google Scholar]
- (60).De Lorenzo S, Alffenaar JW, Sotgiu G, Centis R, D’Ambrosio L, Tiberi S, Bolhuis MS, van Altena R, Viggiani P, Piana A, Spanevello A, and Migliori GB (2013) Efficacy and safety of Meropenem-clavulanate added to linezolid-containing regimens in the treatment of MDR-/XDR-TB. Eur. Respir. J 41, 1386–1392. [DOI] [PubMed] [Google Scholar]
- (61).Payen MC, De Wit S, Martin C, Sergysels R, Muylle I, Van Laethem Y, and Clumeck N (2012) Clinical use of the Meropenem-clavulanate combination for extensively drug-resistant tuberculosis. Int. J. Tuberc. Lung Dis 16, 558–560. [DOI] [PubMed] [Google Scholar]
- (62).England K, Boshoff HIM, Arora K, Weiner D, Dayao E, Schimel D, Via LE, and Barry CE (2012) Meropenem-Clavulanic Acid Shows Activity against Mycobacterium tuberculosis In Vivo. Antimicrob. Agents Chemother 56, 3384–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Payen MC, Muylle I, Vandenberg O, Mathys V, Delforge M, Van den Wijngaert S, Clumeck N, and De Wit S (2018) Meropenem-clavulanate for drug-resistant tuberculosis: a follow-up of relapse-free cases. Int. J. Tuberc. Lung Dis 22, 34–39. [DOI] [PubMed] [Google Scholar]
- (64).Chambers HF, Kocagöz T, Sipit T, Turner J, and Hopewell PC (1998) Activity of Amoxicillin/Clavulanate in Patients with Tuberculosis. Clin. Infect. Dis 26, 874–877. [DOI] [PubMed] [Google Scholar]
- (65).Tiberi S, Payen M-C, Sotgiu G, D’Ambrosio L, Alarcon Guizado V, Alffenaar JW, Abdo Arbex M, Caminero JA, Centis R, De Lorenzo S, Gaga M, Gualano G, Roby Arias AJ, Scardigli A, Skrahina A, Solovic I, Sulis G, Tadolini M, Akkerman OW, Alarcon Arrascue E, Aleska A, Avchinko V, Bonini EH, Chong Marín FA, Collahuazo López L, de Vries G, Dore S, Kunst H, Matteelli A, Moschos C, Palmieri F, Papavasileiou A, Spanevello A, Vargas Vasquez D, Viggiani P, White V, Zumla A, and Migliori GB (2016) Effectiveness and safety of Meropenem/clavulanate-containing regimens in the treatment of MDR- and XDR-TB. Eur. Respir. J 47, 1235–1243. [DOI] [PubMed] [Google Scholar]
- (66).Hugonnet J-E, Tremblay LW, Boshoff HI, Barry CE, and Blanchard JS (2009) Meropenem-Clavulanate Is Effective Against Extensively Drug-Resistant Mycobacterium tuberculosis. Science 323, 1215–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Solapure S, Dinesh N, Shandil R, Ramachandran V, Sharma S, Bhattacharjee D, Ganguly S, Reddy J, Ahuja V, Panduga V, Parab M, Vishwas KG, Kumar N, Balganesh M, and Balasubramanian V (2013) In Vitro and In Vivo Efficacy of β-Lactams against Replicating and Slowly Growing/Nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother 57, 2506–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Tallman KR, Levine SR, and Beatty KE (2016) Small Molecule Probes Reveal Esterases with Persistent Activity in Dormant and Reactivating Mycobacterium tuberculosis. ACS Infect. Dis 2, 936–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Correale S, Ruggiero A, Capparelli R, Pedone E, and Berisio R (2013) Structures of free and inhibited forms of the l,d-transpeptidase LdtMt1 from Mycobacterium tuberculosis. Acta Crystallogr., Sect. D: Biol. Crystallogr 69, 1697–1706. [DOI] [PubMed] [Google Scholar]
- (70).Pidgeon SE, Apostolos AJ, Nelson JM, Shaku M, Rimal B, Islam MN, Crick DC, Kim SJ, Pavelka MS, Kana BD, and Pires MM (2019) L,D-Transpeptidase Specific Probe Reveals Spatial Activity of Peptidoglycan Cross-Linking. ACS Chem. Biol 14, 2185–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Devasundaram S, Gopalan A, Das SD, and Raja A (2016) Proteomics Analysis of Three Different Strains of Mycobacterium tuberculosis under In vitro Hypoxia and Evaluation of Hypoxia Associated Antigen’s Specific Memory T Cells in Healthy Household Contacts. Front. Microbiol 7, 1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Levine SR, and Beatty KE (2019) Investigating β-lactam drug targets in Mycobacterium tuberculosis using chemical probes. bioRxiv 12, 881631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


