Abstract
Lysine acetylation (Kac) is well known to occur in histones for chromatin function and epigenetic regulation. In addition to histones, Kac is also detected in a large number of proteins with diverse biological functions. However, Kac function and regulatory mechanism for most proteins are unclear. In this work, we studied mutation effects of rice genes encoding cytoplasm-localized histone deacetylases (HDAC) on protein acetylome and found that the HDAC protein HDA714 was a major deacetylase of the rice non-histone proteins including many ribosomal proteins (r-proteins) and translation factors that were extensively acetylated. HDA714 loss-of-function mutations increased Kac levels but reduced abundance of r-proteins. In vitro and in vivo experiments showed that HDA714 interacted with r-proteins and reduced their Kac. Substitutions of lysine by arginine (depleting Kac) in several r-proteins enhance, while mutations of lysine to glutamine (mimicking Kac) decrease their stability in transient expression system. Ribo-seq analysis revealed that the hda714 mutations resulted in increased ribosome stalling frequency. Collectively, the results uncover Kac as a functional posttranslational modification of r-proteins which is controlled by histone deacetylases, extending the role of Kac in gene expression to protein translational regulation.
INTRODUCTION
Protein epsilon lysine acetylation (Kac) is a reversible post-translational protein modification that consumes acetyl-CoA. Histone Kac is well known for being important in the epigenetic regulation of gene expression in eukaryotic cells (1). Advancements in mass spectrometry (MS)-based proteomics have shown that, in addition to its occurrence on histone proteins, Kac is a widespread post-translational modification occurring at a large number of proteins of diverse biological function in different organisms (2–4). In particular, most enzymes of energetic metabolism such as glycolysis and the tricarboxylic acid (TCA) cycle, as well as photosynthesis in plants, are modified by Kac, and their acetylation status affects enzymatic activities and metabolic flux (2,5–7). However, the effect of Kac on activities of most proteins is generally unknown.
Regulation of histone Kac homeostasis by histone acetyltransferases (HAT) and histone deacetylases (HDAC) proteins is well documented (8–11). HAT and HDAC target specific genes or chromatin regions by interacting with other nuclear proteins or chromatin marks for epigenetic regulation of gene expression in responding to development and external signals (12). In addition, it has been shown that acetyl-CoA levels, which reflect cellular energy status, also regulate histone Kac. In yeast, unusually high levels of acetyl- CoA have a determinative role in histone acetylation and epigenetic regulation of gene expression (5,13). In plants, mutation of acetyl-CoA biosynthetic and catabolic enzymes respectively reduces and enhances genome-wide histone acetylation levels (14,15).
In addition to regulating histone Kac, several HATs or HDACs have been shown to also regulate Kac of non-histone proteins. For instance, a recent analysis finds that mammalian CBP/p300 acetylates many signaling proteins and enhancer-associated transcriptional regulators (16). It was shown that mammalian sirtuin proteins, which are NAD+-dependent HDACs, are involved in deacetylation of transcription regulators and/or metabolic enzymes in different cellular compartments (17–19). The plant sirtuin homolog, SRT1, was shown to deacetylate proteins with transcriptional activity in the nucleus (20,21). Arabidopsis HDAC protein, AtHDA14, found to reside in the chloroplasts, deacetylates many proteins having functions in photosynthesis (22). Recently, HDA9 was shown to deacetylate WRKY53, a transcription factor regulating stress responsive genes expression (23). However, for a vast majority of non-histone proteins, the responsible acetyltransferases or lysine deacetylases are not identified. Furthermore, non-enzymatic acetylation of proteins may occur when acetyl-CoA level is high in the cell (24). Whether and how non-histone protein Kac is regulated in response to cellular or metabolic signaling is generally unclear.
The rice genome encodes 18 HDACs from the three different families. The RPD3 family comprises 13 genes, the HDT and the Sir families contain two genes each (21,25,26). Previous studies showed that rice RPD3 members are involved in regulation of different developmental pathways including root growth and seed germination (27–30). In this work, we used quantitative proteomics to investigate the acetylomes regulated by three cytoplasm-localized and stress responsive RPD3 family members in rice seedling tissues. Our analysis revealed that HDA714 was a major deacetylase regulating the levels of a large number of Kac sites in hundreds of proteins of diverse functions including a large number of metabolic enzymes. In particular, we show that HDA714 was involved in deacetylation of ribosomal proteins which likely affected their stability and possibly translational efficiency of ribosome. The results point to a possible role of HDA714-regulated Kac in linking ribosome function and metabolic activity.
MATERIALS AND METHODS
Plant materials and growth conditions
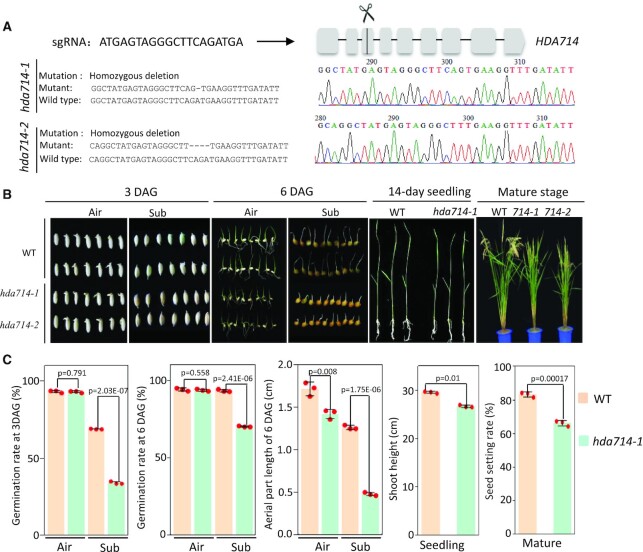
Rice (Oryza sativa) wild type and mutant plants were germinated and grown on hormone-free, one-half-strength Murashige and Skoog media under 16/8 h of light/dark at 30°C/25°C. For submergence treatment, rice seeds were submerged in distilled water for 6 days. The rice variety Zhonghua11 (Oryza sativa spp. Japonica) was used for transgenic plant production in this study. Tobacco (Nicotiana benthamiana) plants were grown at 20 ± 2°C, a photoperiod of 8 h, and an illumination of 100 μmol quanta/(m2 s) in soil for 6 weeks. Significance of differences between wild type and hda714–1 in germination rates (n = 120) under air or submergence at three or six DAG, seed setting rates (n = 9) at mature stage (number of filled grains to total number of reproductive sites and expressed as percentage), and seedling heights (14 days, n = 15) were tested by using the two-tail Student's t-test. For all data, the means ± SEM of three independent biological replicates were shown.
Vector construction for the CRISPR/Cas9 system
The single guide RNA (sgRNA) design for the CRISPR/Cas9 system and plasmid construction following the protocol as previously described (31). CRISPR-P (http://cbi.hzau.edu.cn/crispr) was used to select sgRNAs targeting exons of genes of interest. The Cas9 destination vector was driven by the maize ubiquitin promoter for expression in rice, and sgRNA expression was driven by the pol III type promoter of U3 sgRNA. Gibson Assembly Cloning method (32) was used to ligate the sgRNA to the Cas9 destination vector, then the binary T-DNA vectors for co-expression of Cas9 and sgRNA were transformed into rice calli. Genomic sequence of HDA705 (Os08g0344100), HDA706 (Os06g0571100), and HDA714 (Os12g0182700) were obtained from The Rice Annotation Project (https://rapdb.dna.affrc.go.jp/).
Subcellular localization assays
The full length cDNAs of rice HDAC genes were subcloned into the pCambia 1301 vector to create the GFP fusions. Protoplast isolation and transient expression were performed by following the AgI-PrI method (33). Briefly, the A. tumefaciens cells were infiltrated into abaxial epidermis of 6-week-old tobacco (N. benthamiana) leaves using a syringe without a needle. Then protoplasts were isolated from the transformed leaves, and photographed under a laser confocal microscope (Olympus FV1200).
Nuclear-Cytoplasmic protein fractionation
The nuclear fraction and cytoplasmic fraction were extracted from HDA714 overexpressing transgenic plants, which were generated by Agrobacterium-mediated transformation of HDA714 CDS fused to FLAG at the C-terminus under the control of the ubiquitin promoter. The HDA714 expression level in the overexpression lines OE1 and OE2 used in this study was respectively 32.0 and 59.7 times of wild type level. The OE lines showed no visible phenotype under normal growth conditions. 14 d seedlings were sampled and ground into powder with liquid nitrogen and extracted with lysis buffer (0.4 M of sucrose, 10 mM of Tris, pH 8.0, 10 mM of MgCl2, 0.1 mM of phenylmethylsulfonyl fluoride [PMSF], 5 mM of β-mercaptoethanol) supplemented with an EASYpack protease inhibitor cocktail (Roche). Then, the homogenate was filtered through a double layer of Miracloth (Millipore) followed by centrifuge at 1200g at 4°C for 10 min. The resultant pellet was purified by washing buffer (0.25 M of sucrose, 10 mM of Tris, pH 8.0, 10 mM of MgCl2, 1% Triton X-100, 0.1 mM of PMSF, 5 mM of β-mercaptoethanol, 1 protease inhibitor cocktail) and kept as the nuclear fraction. The supernatant was further centrifuged at 16 000g for 15 min to remove the cellular debris and kept as cytoplasmic fraction. The nuclear fraction was resuspended with 300 μl of loading buffer (25 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 10% [v/v] glycerol, 1% [w/v] SDS). The separated fractions were detected by immunoblotting using anti-H3 (Abcam, 1:1000 dilutions) and anti-actin (Sangon biotech, 1:1000 dilution).
RT-qPCR analysis
Total RNA and ribosome bound RNA were isolated using TRIzol reagent (Invitrogen). 0.05 micrograms of RNA were used to synthesize complementary DNA with reverse transcription kit (Vazyme). Realtime PCR was performed using SYBR Premix ExTaq(TaKaRa) on ABI 7500 real time PCR system. For the normalization of gene expression, rice ACTIN was used as an internal standard. Triplicate reactions were performed for each sample. The 2−△△CT method was used for quantification of relative expression (34). Primers used in RT-qPCR are listed in Supplementary Dataset S7.
Bimolecular fluorescence complementation (BiFC) assay
For BiFC assay, the fusions with split luciferase nLUC- and cLUC were first transformed into A. tumefaciens cells and then the indicated transformant pairs were infiltrated into abaxial epidermis of 6-week-old tobacco (N. benthamiana) leaves using a syringe without a needle. 48 h after the transfection, the transfected N. benthamiana leaves were infiltrated with 1 mM luciferin (Gold Biotechnology) and kept in dark for 10 min. Luciferase bioluminescence images were taken with the Chemi-Image System (Tanon 5200Multi).
In vivo degradation assay
For ribosomal protein stability analysis, ribosomal proteins and K to Q (to mimic Kac) or K–R (to inhibit Kac) substitution mutants were fused with the GFP tag and transformed into tobacco (N. benthamiana) leaf epidermal cells. 48 h after the transfection, the transfected N. benthamiana were treated with 100 μM cycloheximide (CHX), in the presence or absence of 50 μM MG132. Samples were then collected at 0 h, 12 h, and 24 h after the incubation.
Equal amounts of protein extractswere loaded and analyzed by immune-blotting using anti-GFP (Abmart, 1:1000 dilutions).
In vivo co-immunoprecipitation
To confirm the interaction between HDA714 and ribosomal proteins in vivo, the proteins were extracted from Nicotiana benthamiana leaves expressing 35Spro:HDA714-GFP/ 35Spro:RPS3-FLAG, 35Spro:HDA714-GFP/35Spro:RPS6-FLAG or 35Spro:HDA714-GFP/35Spro:RPL7a-FLAG constructs using lysis buffer (10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% Nonidet™ P40 Substitute) and then incubated with anti-GFP agarose beads (ChromoTek) for 5 h at 4°C. After three washes with washing/dilution buffer (10 mM Tris/Cl pH 7.5, 150 mM NaCl, 0.5 mM EDTA), the co-immunoprecipitated proteins were separated by SDS-PAGE and detected with anti-GFP (Abmart, 1:1000 dilution) and anti-FLAG (Sigma, 1:1000 dilution) antibodies, respectively. For co-immunoprecipitation, ten-day wild type rice seedlings were ground into power by liquid nitrogen and extracted by lysis buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% Nonidet™ P40 Substitute) and then incubated with anti-HDA714 (the full length HDA714 protein was used as antigen to produce polyclonal antibodies in rabbit) coated with protein-A magnetic beads (Thermo Fisher Scientific) for 5 h at 4°C. After four washes with PBST buffer, the co-immunoprecipitated proteins were separated by SDS-PAGE and detected with anti-RPS6 (Abcam, 1:1000 dilution) and anti-RPS3 (Abcam, 1:1000 dilution) antibodies.
Isolation of cytosolic ribosomes and polysomes from rice tissues
Cytosolic ribosome and polysome isolation were done as previously described (35,36). Briefly, 10 grams of 14 d rice seedlings were ground into a fined power with liquid nitrogen and suspended in the extraction buffer (200 mM Tris–HCl, pH 7.5, 200 mM KCl, 25 mM EGTA, 36 mM MgCl2, 1 mM sodium molybdate, 1 mM dithiothreitol, 50 μg/ml cycloheximide, 50 μg/ml chloramphenicol, 80 mM β-glycerophosphate, 1% (v/v) Triton X-100, 1% (v/v) Brij 35, 1% (v/v) Tween 40 and 1% (v/v) NP40). Then, the homogenate was filtered through four layers of Miracloth (Millipore) followed by centrifuge at 10 000g at 4°C for 15 min, two times. The supernatant was transferred onto the sucrose cushion buffer (1.3 M sucrose, 400 mM Tris–HCl, pH 7.5, 200 mM KCl, 5 mM EGTA, 36 mM MgCl2, 1 mM sodium molybdate, 1 mM dithiothreitol, 50 μg/ml cycloheximide, 50 μg/ml chloramphenicol, 80 mM β-glycerophosphate) and centrifuged at 149 000g for 18 h at 4°C. After centrifugation, the supernatant was removed and the ribosomal pellet was washed with ∼100 ul Staehelin A buffer (20 mM Tris–HCl, pH 7.5, 5 mM MgCl2, 1 mM sodium molybdate and 1 mM dithiothreitol) to remove any debris and kept for HDAC enzyme activity assays or polysomal RNA extraction.
In vitro and in vivo HDAC assays
In vitro deacetylation assay was done as previously described with modifications (37). Briefly, GST-tagged HDAC714 proteins were expressed and purified from E. coli cells. HDAC assays were carried out at 37°C for 12 h in reaction buffer (50 mM Tris pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 μM ZnCl2 and 1 mM DTT). Ribosomal proteins were prepared from wild type plants. The products were then subjected to Western blot analysis with anti-LysAc antibody (PTM Biolab, 1:1000 dilution). For in vivo RPS3 and RPS6 deacetylation assays, ten-day seedlings of wild type, hda714 mutants, and HDA714 over expression plants were collected. Samples were ground into power by liquid nitrogen and extracted by lysis buffer (10 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% Nonidet™ P40 Substitute) and then incubated with anti-RPS3/RPS6 antibodies coated with protein-A magnetic beads (Thermo Fisher Scientific) for 5 h at 4°C. After three washes with PBST buffer, the co-immunoprecipitated proteins were separated by SDS-PAGE and detected with anti-RPS6 (Abcam, 1:1000 dilution), anti-RPS3 (Abcam, 1:1000 dilution), and anti-LysAc antibody (PTM Biolab, 1:1000 dilution) antibodies. For in vitro RPS3 and RPS6 deacetylation assays, GST-tagged HDA714 and HIS-tagged RPS3, RPS6 proteins were expressed and purified from E. coli cells. The tagged r-proteins were acetylated in E. coli cells. Deacetylation assays were carried out at 37°C for 12 h in reaction buffer (50 mM Tris pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2, 1 μM ZnCl2 and 1 mM DTT). The products were then subjected to western blot analysis with anti-LysAc antibody (PTM Biolab, 1:1000 dilution).
Protein extraction and western Blot
Proteins were extracted as previously described (38,39). Briefly, 14-day seedlings were ground in liquid nitrogen with a mortar and pestle, and then homogenized in buffer containing 250 mM sucrose, 10 mM EGTA, 10 mM Tris–HCl, pH 7.5, 1% (v/v) TritonX-100, 1 mM PMSF and 1 mM DTT. After centrifugation at 15 000g for 15 min at 4°C, the supernatant was incubated in ice-cold acetone for more than 2 h at –20°C, and then centrifuged at 15 000g for 15 min at 4°C again. The obtained pellet was washed with cold acetone for three times, and then lyophilized and stored at –80°C for further use.
Protein pellet was dissolved in 100 mM NH4HCO3 (pH 8.0) and the protein concentration was measured by the Bradford method according to the manufacturer's instructions (Bio-Rad protein assay, USA). For immunoblotting, the procedure was as previously described (40,41). Briefly, protein was diluted with SDS loading buffer, and 20 ug protein of each sample was separated by 12% SDS-PAGE and electro-blotted onto PVDF. The blot was probed with anti-LysAc antibody (PTM Biolab, 1:1000 dilution) or anti-RPS3 (Abcam, 1:1000 dilution).
PROTEIN DIGESTION, ACETYLATED PEPTIDES ENRICHMENT, LC–MS/MS AND DATA ANALYSIS (SEE SUPPLEMENTAL METHODS)
RNA-seq, Ribo-seq and data analysis
Two-week-old rice plants grown under the same conditions as for acetylome analysis were used for RNA extraction and for isolation of ribosome protected RNA fragments. The RNA isolation procedures, libraries construction, high throughput sequencing, and data analysis are described in Supplemental methods.
RESULTS
Rice HDA714 KO results in defects in plant growth and development
Previous studies indicated that plant HDACs are localized in different cellular compartments (22,42,43). In rice, there are 13 RPD3-like HDAC genes that fall into the 3 classes of human HDAC proteins: class 1, class II and class IV (27,30) (Supplementary Figure S1A). The nomenclature of the plant HDACs (‘7’ for rice proteins) was based the ChromDB database (44). To study subcellular localization of rice HDACs, we fused GFP to seven of the rice RPD3-like HDAC proteins and tested their subcellular localization by transient expression in protoplasts. We found that HDA702, HDA704 and HDA711 were exclusively localized in the nucleus; HDA705 and HDA706 were detected in both the nucleus and cytoplasm, while HDA713 and HDA714 appeared to be only in the cytoplasm (Supplementary Figure S1B). Western blot analysis of cytoplasmic and nuclear fractions isolated from rice transgenic plants expressing the HDA714-FLAG fusion confirmed the cytoplasmic localization of the protein (Supplementary Figure S1C). To study functions of cytoplasm-localized HDACs, we knocked out (KO) the genes of HDA705, HDA706, HDA713 and HDA714 by CRISPR/Cas9. Rice HDA705 is a class I member, closely related to Arabidopsis HDA6. Rice HDA706 and Arabidopsis HDA2 belong to class IV, while rice HDA713 and HDA714 belong to class II. HDA714 is closely related to Arabidopsis HDA14 and HDA713 is orthologous to Arabidopsis HDA5 (Supplementary Figure S1A). The expression of HDA705, HDA706 and HDA714 is responsive to stresses (Supplementary Figure S1D). Except HDA713 for which we could not obtain any KO line, two independent KO lines for each of the other genes were characterized (Figure 1A, Supplementary Figure S2). The hda714 KO lines showed multiple phenotypes such as reduced growth at both seedling and mature stages and reduced fertility (Figure 1B, C). In addition, seed germination rates under water (submergence) were reduced in the mutants (Figure 1B, C). The hda705 KO lines also displayed reduced plant height at mature stage and reduced fertility (Supplementary Figure S2A, B). However, no clear developmental phenotype was observed for the hda706 KO lines (Supplementary Figure S2C, D).
Figure 1.
Phenotype of hda714 CRISPR/Cas9 mutants. (A) Designed HDA714 sgRNA sequence and position in the locus and the decoded mutations in two lines. (B) Phenotypes of hda714 mutant at germination, seedling and mature stages. Seeds were germinated in air or under submergence (sub) and photographed 3 and 6 days after germination (DAG). (C) Statistics of difference between wild type and hda714–1 germination rates (n = 120) under air or submergence at 3 or 6 DAG, seed setting rates (n = 9) at mature stage (Number of filled grains to total number of reproductive sites and expressed as percentage), and seedling heights (14 days, n = 15). For all the data, the means ± SEM of three independent biological replicates was shown. Significance of differences were analyzed using the two-tail Student's t-test (E is *10x).
Identification of a comprehensive protein acetylome in rice
To study whether the mutations affected the overall protein Kac levels in rice cells, we performed MS-based quantitative acetylproteomic studies of hda705, hda706 and hda714 (two independent KO lines per gene were combined) and wild type 14-day-old seedling green tissues (Supplementary Figure S3A). The data obtained from the two biological replicates were highly correlated, indicating a good reproducibility of the assays (Supplementary Figure S3B, C). In total, we detected 4868 acetylated lysine residues in 1952 proteins in wild type rice, among which 4091 Kac sites in 1729 proteins were quantifiable (Table 1, Supplemental Dataset S1). Calibration of the acetylomic data with the proteomes (Supplemental Dataset S1) revealed 3413 normalized Kac sites in 1333 proteins (Table 1). The numbers were 2–3 times higher than previously identified Kac sites and acetylated proteins in different rice tissues/organs (38–41,45–48) (Supplementary Figure S3D). The acetylated proteins identified in this work overlapped with 67% of previously identified protein acetylome in seedling tissues (46) (Supplementary Figure S3E). However, only 102 acetylated proteins were commonly found in rice seedling, seed, anther, and embryo acetylomes, suggesting existence of distinct protein acetylomes in different organs/tissues (Supplementary Figure S3F). Many acetylated proteins contained multiple Kac sites, of which 58 had more than 10 Kac sites (Supplementary Figure S4A, B). In addition, 66 Kac sites were detected in the core histones, revealing a number of novel sites such as H4K31 and several lysine residues in H2A and H2B (Supplementary Figure S4C, D). Furthermore, we found that 76 chromatin and transcription factors were acetylated (Supplemental Dataset S2), of which 18 have been functionally studied (Supplementary Table S1). Analysis of amino acid sequences flanking the 4868 Kac sites revealed an enrichment of phenylalanine (F), lysine (K) or histidine (H) at the +1 position, and aspartic acid (D), glutamic acid (E) or phenylalanine (F) at the -1 position (Supplementary Figure S4E), consistent with previous data (45,47,48). Acetylated proteins were found to be enriched for 20 of the 35 MapMan bins or functions (49), and especially for metabolic pathways such as photosynthesis, TCA cycle, and glycolysis (Supplementary Figure S4F). Together, our data identified a comprehensive protein acetylome in rice.
Table 1.
MS/MS spectra database search analysis summary
| Identified proteins | Identified sites | Quantifiable proteins | Quantifiable sites | Normalized proteins | Normalized sites | |
|---|---|---|---|---|---|---|
| Acetylome | 1952 | 4868 | 1729 | 4091 | 1333 | 3413 |
| Proteome | 6677 | NA | 5843 | NA | NA | NA |
Effect of the hda705, hda706 and hda714 mutations on proteome and protein acetylome
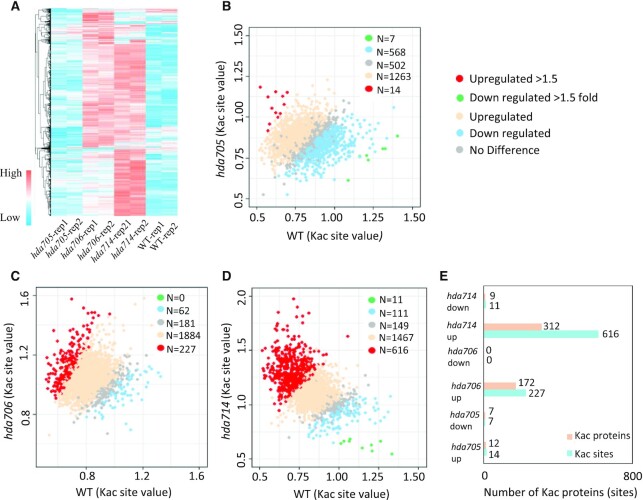
Proteome analysis revealed that the hda705, hda706 and hda714 mutations had no drastic effect on overall protein abundance (Supplementary Figure S5A-C, Supplemental Dataset S3). Relatively few proteins (82 up and 98 downregulated in hda705, 33 up and 71 downregulated in hda706, and 116 up and 93 downregulated in hda714) displayed >1.5 fold changes (Supplementary Figure S5A–C). The Kac changes in the mutants relative to wild type were correlated (r > 0.78) between the biological replicates (Figure 2A, Supplementary Figure S3B). The hda714 and hda706, but not hda705, mutations led to clear increases of Kac levels in many proteins (Figure 2A–D, Supplemental Dataset S4). In total, 227 (9.6%) Kac sites in 172 proteins and 616 (26.2%) Kac sites in 312 proteins showed higher (>1.5-fold) acetylation levels respectively in hda706 and hda714, while very few sites displayed lower Kac levels in the mutants (Figure 2E). In addition, a large number of Kac sites (1467 in hda714 and 1884 in hda706) showed <1.5-fold increases, while only 111 and 62 Kac sites showed <1.5-fold decreases in hda714 and hda706 respectively (Figure 2C, D). The data indicate that HDA706 and HDA714 are two major deacetylases required to maintain protein Kac homeostasis. The Kac sites regulated by HDA706 and HDA714 displayed similar motifs (Supplementary Figure S4E). More than 60% of upregulated Kac sites or protein in hda706 overlapped with those in hda714 (Supplementary Figure S5D), suggesting that the two HDACs may have redundant function in protein deacetylation.
Figure 2.
Relative protein lysine acetylation levels in hda705, hda706 and hda714 compared with wild type. (A) Heatmap of relative lysine acetylation levels in two replicates of hda705, hda706, hda714 and wild type. (B–D) Scattering plots of acetylation levels at individual Kac sites between wild type and the mutants. Significantly up and downregulated sites (>1.5-fold) in the mutants are indicated by red and green respectively. Up regulated (1–1.5-fold) and down regulated (1–1.5-fold) are indicated by light blue and tan respectively. (E) Numbers of Kac sites and proteins that were significantly up and downregulated (>1.5-fold) in hda705, hda706 and hda714 compared with wild type.
HDA714 regulates lysine acetylation of ribosomal proteins
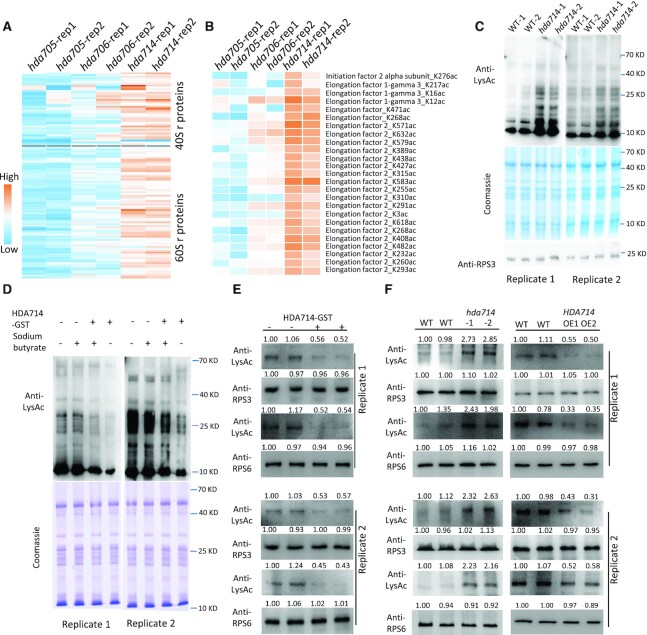
Analysis of published acetylomes in other organisms (22,50–52) revealed that Kac of r-proteins was a general phenomenon in eukaryotes (Supplementary Figure S6). At least105 annotated r-proteins (or isoforms) and 21 translational initiation and 12 elongation factors were largely acetylated in rice (Supplemental Dataset S5). Most of the acetylated ribosomal proteins and translational factors showed multiple Kac sites. In particular, 12 and 24 Kac sites were respectively found in r-protein RPS6 and elongation factor EF2, most of which showed higher acetylation in hda714 (Supplementary Figure S4B, Figure 3A, B, Supplemental Dataset S5). The HAD714 mutations led to increases (>1.5-fold) of Kac at 113 sites in 49 ribosomal proteins and 25 sites in four translational initiation and elongation factors (Figure 3A, B). To confirm the results, we isolated ribosomes from wild type and mutant seedling green tissues and analyzed by immunoblots using anti-LysAc antibody. The analysis revealed clear increases of ribosomal proteins Kac in hda714 mutant plants (Figure 3C). To confirm that HDA714 could deacetylate r-proteins, we used E. coli-produced GST-tagged HDA714 protein to incubate with r-proteins isolated from rice tissues in the presence or absence of sodium butyrate, an HDAC inhibitor. Kac levels of the r-proteins were tested by Western blots using anti-LysAc, which indicated that HDA714 reduced the Kac levels in the absence of sodium butyrate (Figure 3D). To further confirm in vitro deacetylation of individual r-proteins by HDA714, we produced His-tagged RPS3 and RPS6 in E. coli and incubated with or without HDA714-GST in deacetylation buffer. In the absence of HDA714, the tagged r- proteins showed high level of Kac indicating that the proteins were acetylated in E. coli cells (Figure 3E). The presence of HDA714 reduced their Kac levels (Figure 3E). To test in vivo deacetylation of RPS3 and RPS6 by HDA714, we immunoprecipitated RPS3 and RPS6 proteins from wild type, hda714 mutant and HDA714 overexpression plants and tested their Kac levels by Western blots. The analysis revealed higher Kac of the proteins in the mutant lines and lower Kac of the proteins in the over-expression plants compared with the wild type plants (Figure 3F).
Figure 3.
HDA714 loss-of-function resulted in increases of lysine acetylation of translational proteins. Relative acetylation levels at individual Kac sites of ribosomal proteins (A) and translational initiation and elongation factors (B) in hda705, hda706 and hda714 compared with wild type. (C) Analysis of Kac levels of ribosomal proteins isolated from hda714–1 (two biological replicates) and wild type (two biological replicates) plants by immunoblotting using anti-LysAc antibody. Coomassie staining and immunoblotting with anti-RPS3 antibody were used as loading control. Two replicates are shown. (D) In vitro r-protein deacetylation assays. Ribosomal proteins isolated from wild type plants were incubated with E. coli-produced GST-HDA714 protein (with or without addition of sodium butyrate) followed by immunoblotting using anti-LysAc antibody. Coomassie staining was used as loading control. Two independent replicates are shown. (E) In vitro deacetylation assays of RPS3 and RPS6. E. coli-produced His-tagged RPS3 and RPS6 proteins were incubated with (+) or without (−) GST-tagged-HDA714 in the deacetylation butter and analyzed by western blots with anti-RPS3, RPS6, and anti-LysAc (acetylated lysine residues). Replicates are shown. (F) Effect of HDA714 loss- or gain-of-function on Kac on RPS3 and RPS6 in vivo in rice plants. RPS3 and RPS6 were immunoprecipitated from HDA714 mutants (hda714–1, hda714–2), overexpression (OE1, OE2) and wild type plants (WT1, WT2) by anti-RPS3 or anti-RPS6 and analyzed by immuno-blots using the indicated antibodies. Two replicates are shown. Band intensities in (E) and (F) were measured using ImageJ with those WT1 set at 1.
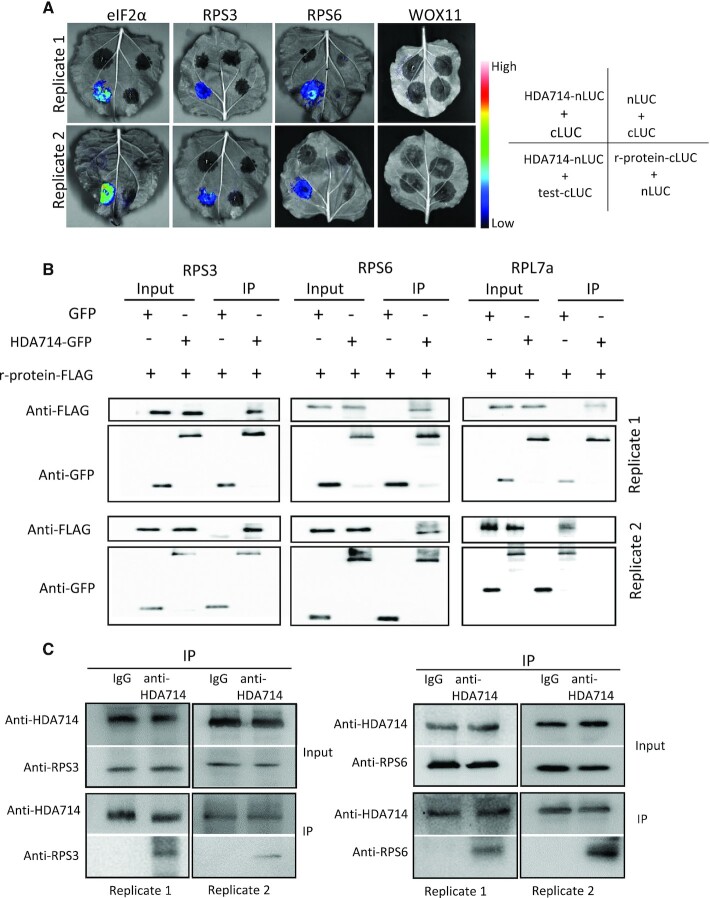
To test whether HDA714 could interact with r-proteins, we performed BiFC and co-immunoprecipitations. The assays revealed protein interaction between HDA714 and RPS3, RPS6, and RPL7a as well as eIF2α in tobacco cells (Figure 4A, B). Moreover, we performed co-immunoprecipitation assays with rice protein extracts using anti-HDA714 and analyzed the precipitated proteins by Western blots using anti-RPS3 and anti-RPS6. Results showed interaction between HDA714 and RPS3 and RPS6 in vivo in rice plants (Figure 4C). Collectively, the data indicate that HDA714 targets r-proteins for deacetylation.
Figure 4.
In vivo protein interaction between HDA714 and ribosomal and IF proteins. (A) Tobacco leaves co-infiltrated with Agrobacterium containing 35S-driven split luciferase (LUC) constructs as indicated were photographed with a charge-coupled device camera. BiFC visualization showing that interaction between HDA714-Nluc and tested proteins (RPS16-cLUC, RPS3-cLUC, eIF2α-cLUC) in the N. benthamiana epidermal cells was observed. For the negative controls (nLUC + cLUC, HDA714-nLUC+cLUC, RPS6-cLUC+nLUC, RPS3-cLUC+nLUC, and eIF2α-cLUC+nLUC, HDA714-nLUC+WOX11-cLUC) no interaction was observed. eIF2α: eukaryotic translation initiation factor 2 alpha subunit. WOX11: WUSCHEL-RELATED HOMEOBOX11 (WOX11) as a negative control. (B) Coimmunoprecipitation assay for the interaction between HDA714 and RPS3, RPS6 or RPL7a in vivo. Total proteins from N. benthamiana leaves co-expressing RPS3-, RPS6- or RPL7a-FLAG and HDA714-GFP were immunoprecipitated using anti-GFP agarose. The agarose-bound proteins were eluted and detected by immunoblotting using anti-GFP or anti-FLAG antibodies. IP, immunoprecipitation. (C) Tests of in vivo interaction between HDA714 and RPS3 and RPS6 in rice plants. Proteins extracted from wild type rice plants were precipitated with IgG or anti-HDA714 and analyzed by immunoblots with anti-RPS3, anti-RPS6 and anti-HDA714. IP, immunoprecipitation.
Lysine acetylation affects r-proteins stability
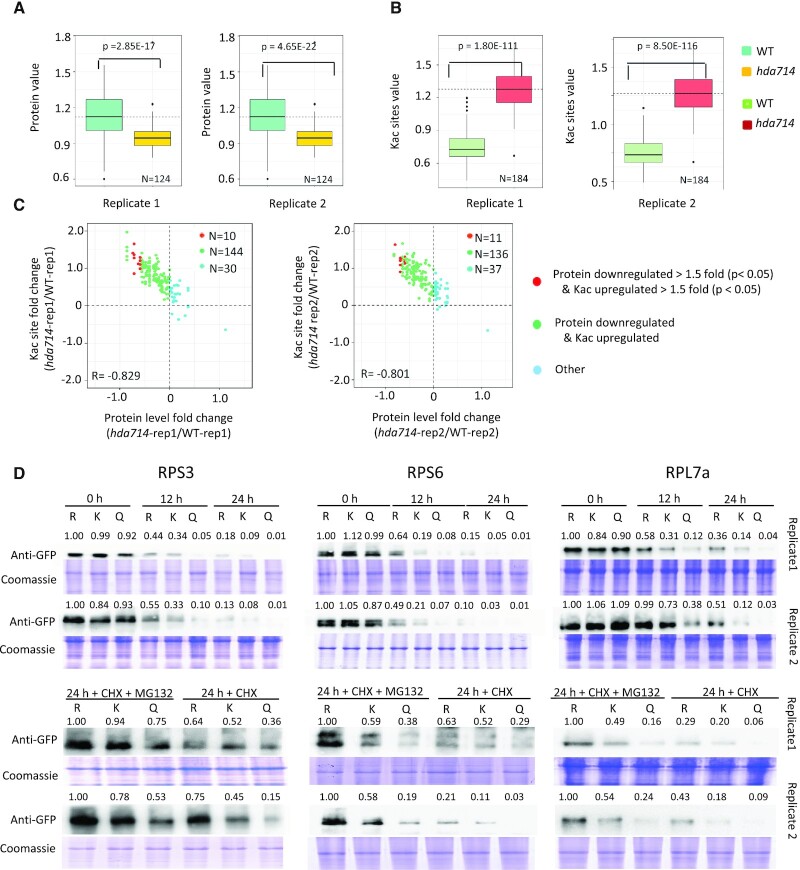
Interestingly, the 124 quantifiable r-proteins identified in the proteomic data showed significant decreases at protein levels but significant increases at Kac levels in hda714 relative to wild type (Figure 5A, B). The changes of r-proteins levels in hda714 were reversely correlated (r = –0.8) with that of Kac (Figure 5C). The lower abundance of r-proteins seemed to be unrelated to their transcript levels (Supplementary Figure S7, see below). The analysis suggested that increased Kac might reduce ribosomal protein abundance or stability.
Figure 5.
The hda714 mutation led to decreases of ribosomal proteins levels. (A) Box plots of abundance of ribosomal proteins detected in the hda714 and wild type proteomic data. (B) Box plots of acetylation levels of ribosomal proteins in hda714 and wild type plants. Horizontal lines in the boxes in (A) and (B) are medians; box limits indicate the 25th and 75th percentiles; whiskers extend to 5th and 95th percentiles. Significance of differences between wild type and the mutant indicated in the figures were analyzed using the two-tail Student's t-test (E is *10x). (C) Scattering plots of ribosomal proteins and acetylation changes in hda714 versus wild type. Correlation coefficients are indicated. (D) Ribosomal proteins (K and K to Q or R) were fused with the GFP tag and expressed in N. benthamiana by agroinfiltration. The transfected N. benthamiana were treated with cycloheximide (CHX) plus or minus MG132, and harvested at the indicated time points. Total protein was analyzed by Western blots with anti-GFP. Immunoblotting results were quantified using ImageJ (v1.6.0_24). Values in R mutations at 0 h are set as 1.
To test this hypothesis, we made point mutations to simultaneously substitute two to five lysine (K) residues in RPS3 (K62 and K75), RPS6 (K2, K14, K15, K23 and K157), and RPL7a (K118, K177 and K188) proteins by arginine (R) to inhibit acetylation or glutamine (Q) to mimic acetylation (Supplementary Figure S8A). The selected lysine residues showed >1.7-fold higher acetylation in hda714 than in wild type (Supplementary Figure S8B). The wild type (K) and the substitution (K to Q or R) proteins were fused with the GFP tag and expressed in tobacco (N. benthamiana) plants by infiltration. The transfected plants were treated with cycloheximide (CHX) to inhibit new protein synthesis and harvested at three time points for protein extraction. The levels of the tagged proteins were tested by Western blots using anti-GFP. The analysis revealed that 12 h after the CHX treatment, the K to R substitutions enhanced, while the K to Q mutations reduced the amount of the 3 tested r-proteins (Figure 5D). As many acetylated r-proteins are also polyubiquitinated (Supplementary Figure S8C) (53), we analyzed whether Kac affected polyubiquitination-mediated r-protein degradation by adding MG132, an inhibitor of ubiquitination E3 ligase in the assays. The presence of MG132 particularly slowed down the degradation of the K to Q substitution mutants compared with absence of the inhibitor (Figure 5D). The results suggest that Kac favors ubiquitination-mediated degradation of r-proteins.
The hda714 mutation increased ribosome stalling on mRNA
It has been shown that posttranslational modifications such as phosphorylation and ubiquitination of r-proteins affect ribosome function (54). Ribosome activity can be studied by investigating its binding frequency and binding profile to mRNAs by Ribo-Seq, which consists of sequencing mRNA segments protected by ribosomes from nuclease digestion (55). Translation output is also dependent on transcript steady-state abundance in the cytosol. To study whether increased Kac of r-proteins in hda714 altered the profile and frequency of ribosome binding to mRNAs, we performed both RNA-seq and Ribo-seq and analyzed the mRNA populations differentially bound by ribosome in wild type and hda714 mutants. Two biological replicates were performed for both RNA-seq and Ribo-seq analysis (Supplementary Tables S2 and S3, Supplementary Figure S9A, B). The RNA-seq revealed > 96% of the mapped reads in both wild type and mutant samples (Supplementary Table S2). There were a number of differentially expressed genes (Supplementary Figure S9C, Supplemental Dataset S6) in the mutant versus wild type. Among the highly repressed genes were Os11g0101200 (RNase P subunit p30) and Os01g0777400 (NB-ARC domain containing protein) (Supplemental Dataset S6).
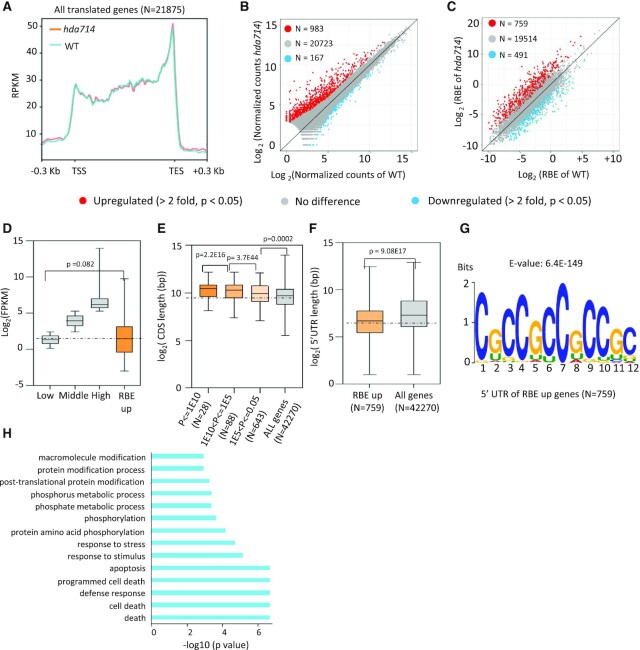
The lengths of Ribo-seq reads were about 26–29 nucleotides (nt) (Supplementary Figure S10A), consistent with the sizes protected by 80S ribosomes (55). However, the reads peak in hda714 appeared at 27-nt compared to 28-nt in the wild type (Supplementary Figure S10A). About 83% of the mapped reads were localized to annotated exons or coding DNA sequences (CDS) (Supplementary Figure S10B, C). The 5′ or 3′ ends of the reads in both wild type and mutants were accumulated at a 3-nucleotide periodicity from the start or stop codon which was not observed in the RNA-seq reads (Supplementary Figure S10D, E), indicating that most of the protected mRNAs were being translated by ribosomes. The distribution of aligned Ribo-seq reads over the translated CDS is dependent on the kinetics of the translation process (55). qRT-PCR were performed to validate the differential levels of several mRNA detected in hda714 versus wild type by Ribo-seq (using ribosome-bound mRNA) and RNA-seq (using total mRNA) (Supplementary Figure S11). Metagene analysis of ribosome binding profiling in all translating mRNA (detected by Ribo-seq, n = 21875) revealed that the binding frequency was high at the translation start site (TSS), and that after a slight and transient decrease the binding gradually increased and reached to the highest level at the translation end site (TES) (Figure 6A). This is consistent with previous results indicating that the assembly of the initiation complex is a relatively slow process, resulting in a pronounced accumulation of signal around the start codon (56–58). Ribosome accumulation at the last codon is caused by the slow kinetics of translation termination and peptide release (59). A similar profile of ribosome binding was found in hda714 (Figure 6A). In hda714 plants, 983 mRNA species showed significantly higher (>2-fold, P < 0.05) and 167 mRNA showed lower (>2-fold, P < 0.05) ribosomal binding compared with wild type (Figure 6B, Supplemental Dataset S6). Normalization of ribosome binding (Ribo-seq) with mRNA abundance (RNA-seq) identified 759 mRNA species with significantly higher (>2-fold, P < 0.05) and 491 with lower (>2-fold, P < 0.05) (Figure 6C, Supplemental Dataset S6) ribosomal binding efficiency (RBE). Among the RBE upregulated genes were Os06g0488200 (Similar to myosin heavy chain), Os08g0461600 (CCHC-type zinc finger domain containing protein), and Os04g0437500 (tetratricopeptide-like helical domain protein). Among the RBE downregulated genes were Os01g0586800 (WRKY5), Os07g0565100 (ribosomal RPS11) and Os03g0785900 (glutathione-S-transferase) (Supplemental Dataset S6). The transcripts that displayed higher RBE corresponded to most lowly expressed genes in the genome (Figure 6D). These genes are larger (longer transcripts (CDS) were more significantly downregulated for RBE, i.e. with lower P values) (Figure 6E), while their 5′-UTR (untranslated regions) are shorter than the genomic average (Figure 6F) and contained a cytidine (C)-rich sequence (Figure 6G). The genes are enriched for cell death and protein phosphorylation pathways (Figure 6H). The data suggested that the hda714 mutation resulted in a significant increase of ribosomal binding at a subset of low abundance mRNA with a C-rich sequence in the 5′-UTR.
Figure 6.
The hda714 mutation increased ribosomal binding frequency and characteristics of upregulated RBE mRNAs in the mutant. (A) Genome-wide metagene analysis of ribosomal footprinting density in gene coding region in wild type (blue) and hda714 (orange). RPKM: Reads Per Kilobase per Million mapped reads; TSS: translation start site; TES: translation end site. (B) Numbers of upregulated and downregulated (>2-fold, P < 0.05) genes detected by Ribo-seq in hda714 versus wild type. (C) Comparison of ribosome binding efficiency (RBE, Ribo-seq reads/RNA-seq reads) between wild type and hda714. Numbers of genes (mRNA) with higher or lower (>2-fold, P < 0.05) RBE are indicated. (D) Expression level analysis of genes with upregulated RBE compared with genome-wide gene expression levels which was divided into 3 intervals: the highest 25% (TPM > 39.33), the lowest 25% (1 ≤ TPM ≤ 5.8) and the middle (25–75%) (5.18 < TPM ≤ 39.3). Genes with TPM <1 were considered as unexpressed and were not included here. TPM: transcripts per kilobase of exon model per million mapped reads. Significance of differences was analyzed using the two-tail Student's t-test. For boxplots, horizontal lines show medians; box limits indicate the 25th and 75th percentiles. (E, F) Comparison of coding region and 5′ UTR (untranslated region) lengths between the genes with upregulated RBE in hda714 and all rice genes. CDS lengths are negatively correlated with p values of RBE up genes (E). Significance of differences was analyzed using the two-tail Student's t-test (E is *10−x). For boxplots, horizontal lines show medians; box limits indicate the 25th and 75th percentiles. (G) MEME motif of 5′UTR of genes with upregulated RBE in the mutant. E is *10−x. (H) GO pathway enrichment analysis of RBE up genes (N = 759).
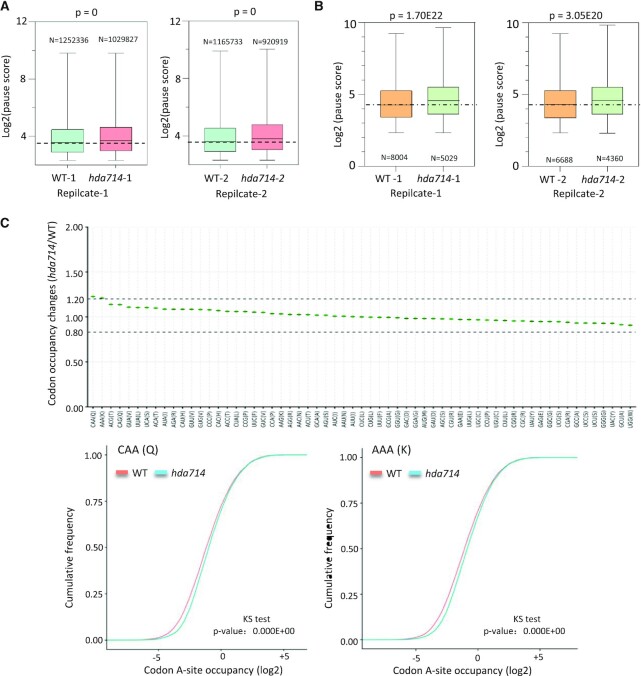
Previous studies have shown that higher ribosome binding might lead to translational pause (60,61). Analysis of ribosome binding frequency at individual positions relative to the average of the mRNA population (using the Pausepred software (62)) revealed an overall higher ribosome pausing score in hda714 compared with the wild type (Figure 7A). The 759 mRNA species that showed significantly higher RBE in the mutants also displayed a higher ribosomal pausing score (Figure 7B), suggesting that hyper Kac of r-proteins may lead to ribosome stalling, although an indirect effect is not excluded. To refine the analysis we used the recently developed RiboToolkit (63) to analyze codon occupancy profiling of the Ribo-seq dada. The analysis detected a clear increase (higher >1.2) of occupancy at codons within the A site encoding Gln and Lys in hda714 relative to wild type (Figure 7C), suggesting that the mutations resulted in ribosomal stalling or slower translation preferentially at these codons (64). Among the 759 mRNA with higher RBE, 32 had proteomic data, the protein levels of which appeared lower in hda714 than in wild type (Supplementary Figure S12), supporting the hypothesis that the increased RBE did not lead to a higher translation efficiency, but instead a lower one.
Figure 7.
Ribosomal pause score analysis in hda714 and wild type. (A) Overall ribosome pause scores in wild type and hda714. (B) Higher pause scores in hda714 upregulated RBE (N = 759) genes. Two replicates are shown in (A) and (B). Significances of differences were analyzed using the two-tail Student t test. For boxplots, horizontal lines show medians; box limits indicate the 25th and 75th percentiles. E is *10−x. (C) RiboToolkit analysis showed significant ribosome pausing at CAA (Gln) and AAA (Lys) codons in hda714 mutants in comparison to wild types (WT). Upper part: Codon occupancy changes between hda714 mutant and wild type. Lower part: Cumulative occupancy distribution of CAA (Q) and AAA (K) codons. P-values are from a one-sample Kolmogorov–Smirnov test.
DISCUSSION
In this work, we characterized a comprehensive protein acetylome in rice, with 4868 Kac sites and 1952 acetylated proteins detected and identified HDA706 and HDA714 as two major and redundant HDACs required for maintaining Kac homeostasis of a large number of rice proteins. HDA714 is closely related to Arabidopsis HDA14 and belongs to the same class as human HDAC4, 5, 6, 7, 9 and 10 (Supplementary Figure S1A). Arabidopsis HDA14 was shown to be involved in deacetylation of tubulin and some chloroplast proteins (65,66). Our data showed that the mutation of HDA714 also increased Kac of chloroplast proteins, suggesting some conserved functions between the two proteins. HDA706 is related to Arabidopsis HDA2 whose function is presently unknown. The limited effects of the HDA705 mutation on the rice seedling acetylome suggest that this HDAC may have a different function. As HDA705 is found within the same clade as Arabidopsis HDA6 that has been shown to play important roles in chromatin control of gene expression, HDA705 may function primarily in chromatin modification.
In particular, the present work identified HDA714 as a deacetylase of r-proteins. Ribosomes have traditionally been considered as housekeeping structures, invariable in composition within the same species. However, it has been shown that post-translational modifications such as phosphorylation and ubiquitylation of ribosomal proteins may have functional consequences for translational control (54). The data shown in this work indicate that Kac is an important posttranslational modification of r-proteins, which, in addition to reducing r-protein stability, may increase the combinatorial diversity of ribosome activities (54), analogously to the histone modifications that make up the so-called histone code (67). Our results are consistent with the results obtained in mammalian cells where treatment with HDAC6-selective inhibitor or HDAC6 knockdown induced r-protein RPL24 acetylation and affected ribosomal assembly (68). However, the functional consequences of ribosomal protein posttranslational modifications are generally unknown. It was shown the many ribosomal and translational elongation proteins are modified by non-degradative ubiquitylation under oxidative stress in yeast (69), and is important for resolving ribosomes stalled at the poly(A) sequences (70,71). Our data showing that the hda714 mutation increases overall ribosome stalling particularly at CAA and AAA codons within the A sites and reduced abundance of proteins encoded by mRNAs with higher RBE suggest that hyper Kac may have an opposite effect of non-degradative ubiquitylation to this regard. However, direct evidence of Kac effect on ribosome activity is lacking. Nevertheless, our data are consistent with recent results obtained in E. coli showing that acetylation of r-proteins significantly reduced the relative translation rate, and deacetylation partially restored the translation activity (72). Likely, Kac modifies not only histones for gene transcriptional regulation but also translational machinery.
Many r-proteins display both lysine acetylation and polyubiquitination. In many cases, a direct competition between the two lysine modifications results in inhibition of proteasome-mediated protein degradation by lysine acetylation (73). The observations that the presence of MG132 slowed down degradation of the K to Q (mimicking Kac) mutants of r-proteins in transient expression system suggest that Kac could stimulate polyubiquitination- proteasome-mediated r-protein degradation and that more complex and indirect processes also connect the two pathways. This is consistent with previous data showing that Kac enhances polyubiquitylation and degradation of target proteins. For instance, deacetylation of Arabidopsis MBP-1 (AtMBP-1), an unstable transcription factor, by a NAD+-dependent HDAC, namely SRT1, increases its stability (21). Similarly, Kac reduces the stability thus activity of phosphoenolpyruvate carboxykinase (PEPCK) in animal cells (74). PEPCK1 acetylation stimulates the interaction between PEPCK1 and the E3 ubiquitin ligase UBR5 thereby promoting PEPCK1 ubiquitylation and degradation. Conversely, SIRT2 deacetylates and stabilizes PEPCK1(74). Interaction of RING finger E3 ligases with r-proteins has been reported in rice (75). Whether Kac stimulates E3 ligase binding to r-proteins awaits further analysis. However, as MG123 did not efficiently inhibit degradation of K-Q substitution mutants (Figure 5D), it is possible that additional mechanisms may be involved in Kac-mediated stability of r-proteins. For instance, lysine acetylation/deacetylation controls local protein conformation by influencing proline isomerization (76,77).
Previous studies indicate that protein Kac is modulated by acetyl-CoA availability in the cell (78). In yeast, several regulatory and metabolic proteins are dynamically acetylated precisely in phase with the observed acetyl-CoA oscillations or in response to glucose levels in the media (13). The identification of a large number of Kac sites in r-proteins and translation IF and EF factors suggests that r-protein Kac may be modulated by intracellular acetyl-CoA levels, providing a possible link between translational regulation and cellular metabolic activity. It is suggested that cellular acetyl-CoA level increases in plants under stress, as under stress glycolysis is activated and there is accumulation of acetyl-CoA precursors such as pyruvate and acetate (79). We speculate that under stress higher acetyl-CoA may increase r-proteins Kac to stimulate their degradation to support stress tolerance. This is consistent with recent finding that in nutrient-starved mammalian cells r-proteins are selectively degraded largely through non-autophagy pathways (80), as r-proteins are required for ribosomal function and are used as a source of basic amino acids remobilization under the stress (80). The hyper Kac can be balanced by HDA714 to maintain r-protein homeostasis, as HDA714 expression is induced by different stresses (Supplementary Figure S1C). It is well established that nutrition status is sensed by the target of rapamycin complex 1 (TORC1) kinase that functions as a positive regulator of translation initiation and protein synthesis to promote cell growth and proliferation (81). Whether the protein Kac and the TORC1 pathways crosstalk to regulate translational activity remains to be studied.
Protein Kac affects metabolic enzyme activity such as inhibition of glycolytic enzymes (20,82–84). It was shown that Kac can also act as a feedback response to high acetyl-CoA levels in the cell, as the acetylation of an active site lysine residue inhibits the activity of acetyl-CoA synthetase (85). The inhibitory effect of Kac on glycolytic enzyme activities (20,82–84) may also be a feedback response to high acetyl-CoA levels in cells under stress. Glycolysis is a predominant energy metabolic pathway during rice seed germination under submergence (86). The hda714 mutations also resulted in hyper Kac of glycolytic enzymes (Supplemental Dataset S4). Due to their sessile life style, plants have to cope with rapidly changing environmental conditions by fine-tuning cellular activities for optimal growth and development. Stress signaling-induced gene expression, in which histone Kac is largely involved, is a primary response in plants. The present and previous results (22,43,87) suggest that protein Kac homeostasis may represent an additional cellular responsive mechanism that allows plants to acclimate their cellular activity to the environment. Likely, protein Kac appears at the nexus linking stress and metabolism to regulation of other cellular activities such as translation in addition to epigenetic regulation of gene expression. The present study suggests that HDA714-controlled protein Kac homoeostasis may play a role in coordinating stress, metabolism and ribosome function.
DATA AVAILABILITY
The complete mass spectrometry data produced by this research can be found in ProteomeXchage Consortium via the PRIDE (88) partner repository with the dataset identifier PXD014063.
The high throughput RNA-sequencing data generated in this study are deposited into SRA databases with the accession number: PRJNA547943 and PRJNA545844.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr W.D. Liu for help in acetylome characterization, H.Z. Song for help in confocal experiments and Dr Z.Y. Cheng in proteomic and acetylomic analysis. Q.X. performed most of the experiments and analyzed the data. Q.L. analyzed the Ribo-seq and RNA-seq data, Z.C., Y.Y., Y.L. and Y.Z. contributed to the experimentation. D.X.Z. supervised the project, analyzed the data and wrote the paper with inputs from Q.X. All authors read and approved the final manuscript.
Contributor Information
Qiutao Xu, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, 430070 Wuhan, China.
Qian Liu, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, 430070 Wuhan, China.
Zhengting Chen, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, 430070 Wuhan, China.
Yaping Yue, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, 430070 Wuhan, China.
Yuan Liu, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, 430070 Wuhan, China.
Yu Zhao, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, 430070 Wuhan, China.
Dao-Xiu Zhou, National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, 430070 Wuhan, China; Institute of Plant Sciences Paris-Saclay (IPS2), CNRS, INRAE, University Paris-Saclay, 91405 Orsay, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31730049]; National Key Research and Development Program of China [2016YFD0100802]; Huazhong Agricultural University Scientific & Technological Self-innovation Foundation [2016RC003]; National Natural Science Foundation of China [32070563]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Verdin E., Ott M.. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015; 16:258–264. [DOI] [PubMed] [Google Scholar]
- 2. Narita T., Weinert B.T., Choudhary C.. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019; 20:156–174. [DOI] [PubMed] [Google Scholar]
- 3. Guan K.L., Xiong Y.. Regulation of intermediary metabolism by protein acetylation. Trends Biochem. Sci. 2011; 36:108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M.. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009; 325:834–840. [DOI] [PubMed] [Google Scholar]
- 5. Shen Y., Wei W., Zhou D.X.. Histone acetylation enzymes coordinate metabolism and gene expression. Trends Plant Sci. 2015; 20:614–621. [DOI] [PubMed] [Google Scholar]
- 6. Zhao S., Xu W., Jiang W., Yu W., Lin Y., Zhang T., Yao J., Zhou L., Zeng Y., Li H.et al.. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010; 327:1000–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Menzies K.J., Zhang H., Katsyuba E., Auwerx J.. Protein acetylation in metabolism - metabolites and cofactors. Nat. Rev. Endocrinol. 2016; 12:43–60. [DOI] [PubMed] [Google Scholar]
- 8. Berger S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002; 12:142–148. [DOI] [PubMed] [Google Scholar]
- 9. Dancy B.M., Cole P.A.. Protein lysine acetylation by p300/CBP. Chem. Rev. 2015; 115:2419–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pikaard C.S., Mittelsten Scheid O.. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014; 6:a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sabari B.R., Zhang D., Allis C.D., Zhao Y.M.. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017; 18:90–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sharma G., Giri J., Tyagi A.K.. Sub-functionalization in rice gene families with regulatory roles in abiotic stress responses. Crit. Rev. Plant Sci. 2016; 35:231–285. [Google Scholar]
- 13. Cai L., Sutter B.M., Li B., Tu B.P.. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell. 2011; 42:426–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen C., Li C.L., Wang Y., Renaud J., Tian G., Kambhampati S., Saatian B., Nguyen V., Hannoufa A., Marsolais F.et al.. Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat. Plants. 2017; 3:814–824. [DOI] [PubMed] [Google Scholar]
- 15. Wang L., Wang C., Liu X., Cheng J., Li S., Zhu J.K., Gong Z.. Peroxisomal beta-oxidation regulates histone acetylation and DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:10576–10585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weinert B.T., Narita T., Satpathy S., Srinivasan B., Hansen B.K., Scholz C., Hamilton W.B., Zucconi B.E., Wang W.W., Liu W.R.et al.. Time-resolved analysis reveals rapid dynamics and broad scope of the CBP/p300 acetylome. Cell. 2018; 174:231–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Westerheide S.D., Anckar J., Stevens S.M. Jr, Sistonen L., Morimoto R.I.. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009; 323:1063–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang Y.P., Zhou L.S., Zhao Y.Z., Wang S.W., Chen L.L., Liu L.X., Ling Z.Q., Hu F.J., Sun Y.P., Zhang J.Y.et al.. Regulation of G6PD acetylation by SIRT2 and KAT9 modulates NADPH homeostasis and cell survival during oxidative stress. EMBO J. 2014; 33:1304–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carrico C., Meyer J.G., He W., Gibson B.W., Verdin E.. The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications. Cell Metab. 2018; 27:497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang H., Zhao Y., Zhou D.X.. Rice NAD(+)-dependent histone deacetylase OsSRT1 represses glycolysis and regulates the moonlighting function of GAPDH as a transcriptional activator of glycolytic genes. Nucleic Acids Res. 2017; 45:12241–12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu X.Y., Wei W., Zhu W.J., Su L.F., Xiong Z.Y., Zhou M., Zheng Y., Zhou D.X.. Histone deacetylase AtSRT1 links metabolic flux and stress response in Arabidopsis. Mol. Plant. 2017; 10:1510–1522. [DOI] [PubMed] [Google Scholar]
- 22. Hartl M., Fussl M., Boersema P.J., Jost J.O., Kramer K., Bakirbas A., Sindlinger J., Plochinger M., Leister D., Uhrig G.et al.. Lysine acetylome profiling uncovers novel histone deacetylase substrate proteins in Arabidopsis. Mol. Syst. Biol. 2017; 13:949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng Y., Ge J., Bao C., Chang W., Liu J., Shao J., Liu X., Su L., Pan L., Zhou D.X.. Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plant. 2020; 13:598–611. [DOI] [PubMed] [Google Scholar]
- 24. Wagner G.R., Payne R.M.. Widespread and enzyme-independent Nepsilon-acetylation and Nepsilon-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J. Biol. Chem. 2013; 288:29036–29045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li C., Huang L.M., Xu C.G., Zhao Y., Zhou D.X.. Altered levels of histone deacetylase OsHDT1 affect differential gene expression patterns in hybrid rice. PLoS One. 2011; 6:e21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang L., Sun Q., Qin F., Li C., Zhao Y., Zhou D.X.. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007; 144:1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu Y., Qin F., Huang L., Sun Q., Li C., Zhao Y., Zhou D.X.. Rice histone deacetylase genes display specific expression patterns and developmental functions. Biochem. Biophys. Res. Commun. 2009; 388:266–271. [DOI] [PubMed] [Google Scholar]
- 28. Zhao J., Li M., Gu D., Liu X., Zhang J., Wu K., Zhang X., Teixeira da Silva J.A., Duan J.. Involvement of rice histone deacetylase HDA705 in seed germination and in response to ABA and abiotic stresses. Biochem. Biophys. Res. Commun. 2016; 470:439–444. [DOI] [PubMed] [Google Scholar]
- 29. Chung P.J., Kim Y.S., Jeong J.S., Park S.H., Nahm B.H., Kim J.K.. The histone deacetylase OsHDAC1 epigenetically regulates the OsNAC6 gene that controls seedling root growth in rice. Plant J. 2009; 59:764–776. [DOI] [PubMed] [Google Scholar]
- 30. Fu W., Wu K., Duan J.. Sequence and expression analysis of histone deacetylases in rice. Biochem. Biophys. Res. Commun. 2007; 356:843–850. [DOI] [PubMed] [Google Scholar]
- 31. He Y.B., Zhang T., Yang N., Xu M.L., Yan L., Wang L.H., Wang R.C., Zhao Y.D.. Self-cleaving ribozymes enable the production of guide RNAs from unlimited choices of promoters for CRISPR/Cas9 mediated genome editing. J. Genet. Genomics. 2017; 44:469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., Smith H.O.. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009; 6:343–U341. [DOI] [PubMed] [Google Scholar]
- 33. Berestovoy M., Tyurin A., Kabardaeva K., Sidorchuk Y., Fomenkov A., Nosov A., Goldenkova-Pavlovaa I.. Transient gene expression for the characteristic signal sequences and the estimation of the localization of target protein in plant cell. Bio-protocol. 2018; 8:doi:10.21769/BioProtoc.2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Livak K.J., Schmittgen T.D.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 35. Klang Arstrand H., Turkina M.V.. Isolation of cytosolic ribosomes. Methods Mol. Biol. 2017; 1511:241–247. [DOI] [PubMed] [Google Scholar]
- 36. Rivera M.C., Maguire B., Lake J.A.. Isolation of ribosomes and polysomes. Cold Spring Harb. Protoc. 2015; 2015:293–299. [DOI] [PubMed] [Google Scholar]
- 37. Lu Y., Xu Q., Liu Y., Yu Y., Cheng Z.Y., Zhao Y., Zhou D.X.. Dynamics and functional interplay of histone lysine butyrylation, crotonylation, and acetylation in rice under starvation and submergence. Genome Biol. 2018; 19:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang Y.F., Hou Y.X., Qiu J.H., Li Z.Y., Zhao J., Tong X.H., Zhang J.. A quantitative acetylomic analysis of early seed development in rice (Oryza sativa L.). Int. J. Mol. Sci. 2017; 18:1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nallamilli B.R.R., Edelmann M.J., Zhong X.X., Tan F., Mujahid H., Zhang J., Nanduri B., Peng Z.H.. Global analysis of lysine acetylation suggests the involvement of protein acetylation in diverse biological processes in rice (Oryza sativa). PLoS One. 2014; 9:e89283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. He D.L., Wang Q., Li M., Damaris R.N., Yi X.L., Cheng Z.Y., Yang P.F.. Global proteome analyses of lysine acetylation and succinylation reveal the widespread involvement of both modification in metabolism in the embryo of germinating rice seed. J. Proteome Res. 2016; 15:879–890. [DOI] [PubMed] [Google Scholar]
- 41. Xue C., Liu S., Chen C., Zhu J., Yang X., Zhou Y., Guo R., Liu X., Gong Z.. Global proteome analysis links lysine acetylation to diverse functions in Oryza sativa. Proteomics. 2018; 18:doi:10.1002/pmic.201700036. [DOI] [PubMed] [Google Scholar]
- 42. Chung P.J., Kim Y.S., Park S.H., Nahm B.H., Kim J.K.. Subcellular localization of rice histone deacetylases in organelles. FEBS Lett. 2009; 583:2249–2254. [DOI] [PubMed] [Google Scholar]
- 43. Konig A.C., Hartl M., Pham P.A., Laxa M., Boersema P.J., Orwat A., Kalitventseva I., Plochinger M., Braun H.P., Leister D.et al.. The Arabidopsis class II sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol. 2014; 164:1401–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gendler K., Paulsen T., Napoli C.. ChromDB: the chromatin database. Nucleic. Acids. Res. 2008; 36:D298–D302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xiong Y., Peng X., Cheng Z., Liu W., Wang G.L.. A comprehensive catalog of the lysine-acetylation targets in rice (Oryza sativa) based on proteomic analyses. J. Proteomics. 2016; 138:20–29. [DOI] [PubMed] [Google Scholar]
- 46. Zhou H., Finkemeier I., Guan W.X., Tossounian M.A., Wei B., Young D., Huang J.J., Messens J., Yang X.B., Zhu J.et al.. Oxidative stress-triggered interactions between the succinyl- and acetyl-proteomes of rice leaves. Plant Cell Environ. 2018; 41:1139–1153. [DOI] [PubMed] [Google Scholar]
- 47. Meng X., Lv Y., Mujahid H., Edelmann M.J., Zhao H., Peng X., Peng Z.. Proteome-wide lysine acetylation identification in developing rice (Oryza sativa) seeds and protein co-modification by acetylation, succinylation, ubiquitination, and phosphorylation. Biochim. Biophys. Acta Proteins Proteom. 2018; 1866:451–463. [DOI] [PubMed] [Google Scholar]
- 48. Li X., Ye J., Ma H., Lu P.. Proteomic analysis of lysine acetylation provides strong evidence for involvement of acetylated proteins in plant meiosis and tapetum function. Plant J. 2018; 93:142–154. [DOI] [PubMed] [Google Scholar]
- 49. Thimm O., Blasing O., Gibon Y., Nagel A., Meyer S., Kruger P., Selbig J., Muller L.A., Rhee S.Y., Stitt M.. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004; 37:914–939. [DOI] [PubMed] [Google Scholar]
- 50. Wang G., Guo L., Liang W., Chi Z., Liu L.. Systematic analysis of the lysine acetylome reveals diverse functions of lysine acetylation in the oleaginous yeast Yarrowia lipolytica. AMB Express. 2017; 7:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walley J.W., Shen Z., McReynolds M.R., Schmelz E.A., Briggs S.P.. Fungal-induced protein hyperacetylation in maize identified by acetylome profiling. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tatham M.H., Cole C., Scullion P., Wilkie R., Westwood N.J., Stark L.A., Hay R.T.. A proteomic approach to analyze the aspirin-mediated lysine acetylome. Mol. Cell. Proteomics. 2017; 16:310–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen X.L., Xie X., Wu L., Liu C., Zeng L., Zhou X., Luo F., Wang G.L., Liu W.. Proteomic analysis of ubiquitinated proteins in rice (Oryza sativa) after treatment with pathogen-associated molecular pattern (PAMP) elicitors. Front Plant Sci. 2018; 9:1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Simsek D., Barna M.. An emerging role for the ribosome as a nexus for post-translational modifications. Curr. Opin. Cell Biol. 2017; 45:92–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Andreev D.E., O’Connor P.B.F., Loughran G., Dmitriev S.E., Baranov P.V., Shatsky I.N.. Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Res. 2017; 45:513–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S.. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009; 324:218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Juntawong P., Girke T., Bazin J., Bailey-Serres J.. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2014; 111:E203–E212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van Heesch S., Witte F., Schneider-Lunitz V., Schulz J.F., Adami E., Faber A.B., Kirchner M., Maatz H., Blachut S., Sandmann C.L.et al.. The translational landscape of the human heart. Cell. 2019; 178:242–260. [DOI] [PubMed] [Google Scholar]
- 59. Calviello L., Ohler U.. Beyond read-counts: ribo-seq data analysis to understand the functions of the transcriptome. Trends Genet. 2017; 33:728–744. [DOI] [PubMed] [Google Scholar]
- 60. Das Sharma S., Metz J.B., Li H., Hobson B.D., Hornstein N., Sulzer D., Tang G., Sims P.A.. Widespread alterations in translation elongation in the brain of juvenile Fmr1 knockout mice. Cell Rep. 2019; 26:3313–3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li G.W., Oh E., Weissman J.S.. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 2012; 484:538–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kumari R., Michel A.M., Baranov P.V.. PausePred and Rfeet: webtools for inferring ribosome pauses and visualizing footprint density from ribosome profiling data. RNA. 2018; 24:1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu Q., Shvarts T., Sliz P., Gregory R.I.. RiboToolkit: an integrated platform for analysis and annotation of ribosome profiling data to decode mRNA translation at codon resolution. Nucleic. Acids. Res. 2020; 48:W218–W229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nedialkova D.D., Leidel S.A.. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 2015; 161:1606–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tran H.T., Nimick M., Uhrig R.G., Templeton G., Morrice N., Gourlay R., DeLong A., Moorhead G.B.. Arabidopsis thaliana histone deacetylase 14 (HDA14) is an alpha-tubulin deacetylase that associates with PP2A and enriches in the microtubule fraction with the putative histone acetyltransferase ELP3. Plant J. 2012; 71:263–272. [DOI] [PubMed] [Google Scholar]
- 66. Hartl M., Fussl M., Boersema P.J., Jost J.O., Kramer K., Bakirbas A., Sindlinger J., Plochinger M., Leister D., Uhrig G.et al.. Lysine acetylome profiling uncovers novel histone deacetylase substrate proteins in Arabidopsis. Mol. Syst. Biol. 2017; 13:949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jenuwein T., Allis C.D.. Translating the histone code. Science. 2001; 293:1074–1080. [DOI] [PubMed] [Google Scholar]
- 68. Wilson-Edell K.A., Kehasse A., Scott G.K., Yau C., Rothschild D.E., Schilling B., Gabriel B.S., Yevtushenko M.A., Hanson I.M., Held J.M.et al.. RPL24: a potential therapeutic target whose depletion or acetylation inhibits polysome assembly and cancer cell growth. Oncotarget. 2014; 5:5165–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Silva G.M., Finley D., Vogel C.. K63 polyubiquitination is a new modulator of the oxidative stress response. Nat. Struct. Mol. Biol. 2015; 22:116–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Juszkiewicz S., Hegde R.S.. Initiation of quality control during Poly(A) translation requires site-specific ribosome ubiquitination. Mol. Cell. 2017; 65:743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sundaramoorthy E., Leonard M., Mak R., Liao J., Fulzele A., Bennett E.J.. ZNF598 and RACK1 regulate mammalian ribosome-associated quality control function by mediating regulatory 40S ribosomal ubiquitylation. Mol. Cell. 2017; 65:751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang B.Q., Bu H.L., You D., Ye B.C.. Acetylation of translation machinery affected protein translation in E. coli. Appl. Microbiol. Biotechnol. 2020; 104:10697–10709. [DOI] [PubMed] [Google Scholar]
- 73. Caron C., Boyault C., Khochbin S.. Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. Bioessays. 2005; 27:408–415. [DOI] [PubMed] [Google Scholar]
- 74. Jiang W., Wang S., Xiao M., Lin Y., Zhou L., Lei Q., Xiong Y., Guan K.-L., Zhao S.. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol. Cell. 2011; 43:33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lim S.D., Hwang J.G., Jung C.G., Hwang S.G., Moon J.C., Jang C.S.. Comprehensive analysis of the rice RING E3 ligase family reveals their functional diversity in response to abiotic stress. DNA Res. 2013; 20:299–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Howe F.S., Mellor J.. Proline cis-trans isomerization is influenced by local lysine acetylation-deacetylation. Microbial. Cell. 2014; 1:390–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Howe F.S., Boubriak I., Sale M.J., Nair A., Clynes D., Grijzenhout A., Murray S.C., Woloszczuk R., Mellor J.. Lysine acetylation controls local protein conformation by influencing proline isomerization. Mol. Cell. 2014; 55:733–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shi L., Tu B.P.. Acetyl-CoA and the regulation of metabolism: mechanisms and consequences. Curr. Opin. Cell Biol. 2015; 33:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hu Y., Lu Y., Zhao Y., Zhou D.X.. Histone acetylation dynamics integrates metabolic activity to regulate plant response to stress. Front Plant Sci. 2019; 10:1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. An H., Ordureau A., Korner M., Paulo J.A., Harper J.W.. Systematic quantitative analysis of ribosome inventory during nutrient stress. Nature. 2020; 583:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wang X., Proud C.G.. Nutrient control of TORC1, a cell-cycle regulator. Trends Cell Biol. 2009; 19:260–267. [DOI] [PubMed] [Google Scholar]
- 82. Finkemeier I., Laxa M., Miguet L., Howden A.J., Sweetlove L.J.. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 2011; 155:1779–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Park S.H., Ozden O., Liu G., Song H.Y., Zhu Y., Yan Y., Zou X., Kang H.J., Jiang H., Principe D.R.et al.. SIRT2-mediated deacetylation and tetramerization of pyruvate kinase directs glycolysis and tumor growth. Cancer Res. 2016; 76:3802–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bontemps-Gallo S., Gaviard C., Richards C.L., Kentache T., Raffel S.J., Lawrence K.A., Schindler J.C., Lovelace J., Dulebohn D.P., Cluss R.G.et al.. Global profiling of lysine acetylation in Borrelia burgdorferi B31 reveals its role in central metabolism. Front. Microbiol. 2018; 9:2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Starai V.J., Celic I., Cole R.N., Boeke J.D., Escalante-Semerena J.C.. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002; 298:2390–2392. [DOI] [PubMed] [Google Scholar]
- 86. He D., Yang P.. Proteomics of rice seed germination. Front. Plant Sci. 2013; 4:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Gao X., Hong H., Li W.C., Yang L., Huang J., Xiao Y.L., Chen X.Y., Chen G.Y.. Downregulation of rubisco activity by non-enzymatic acetylation of RbcL. Mol Plant. 2016; 9:1018–1027. [DOI] [PubMed] [Google Scholar]
- 88. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M.et al.. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019; 47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The complete mass spectrometry data produced by this research can be found in ProteomeXchage Consortium via the PRIDE (88) partner repository with the dataset identifier PXD014063.
The high throughput RNA-sequencing data generated in this study are deposited into SRA databases with the accession number: PRJNA547943 and PRJNA545844.