Abstract
Subcellular localization of messenger RNAs (mRNAs), as a prevalent mechanism, gives precise and efficient control for the translation process. There is mounting evidence for the important roles of this process in a variety of cellular events. Computational methods for mRNA subcellular localization prediction provide a useful approach for studying mRNA functions. However, few computational methods were designed for mRNA subcellular localization prediction and their performance have room for improvement. Especially, there is still no available tool to predict for mRNAs that have multiple localization annotations. In this paper, we propose a multi-head self-attention method, DM3Loc, for multi-label mRNA subcellular localization prediction. Evaluation results show that DM3Loc outperforms existing methods and tools in general. Furthermore, DM3Loc has the interpretation ability to analyze RNA-binding protein motifs and key signals on mRNAs for subcellular localization. Our analyses found hundreds of instances of mRNA isoform-specific subcellular localizations and many significantly enriched gene functions for mRNAs in different subcellular localizations.
INTRODUCTION
Subcellular localization of messenger RNAs (mRNAs) has proven to be a prevalent mechanism used in a variety of cell types in animal development (1). Particularly in highly complex cells, mRNAs are not distributed homogeneously throughout cells but are localized in specific cellular compartments. Besides localized protection from degradation and diffusion-coupled local entrapment, the predominant mechanism of mRNA localization is the directed transport of transcripts along with a polarized cytoskeletal network (1,2). First, the cis-regulatory elements presented in RNA molecules are recognized by diverse trans-regulatory factors, called RNA-binding proteins (RBPs). These cis-regulatory elements, known as zipcodes, are often but not exclusively found in 3′ untranslated regions (UTRs), for mediating the asymmetric localization of specific transcripts. Then, with the recruitment of the destination-specific proteins and adaptor proteins, the localization-competent ribonucleoprotein (RNP) complex is assembled. With the help of molecular motors, mRNA is transported along the cytoskeleton to the ultimate destination and anchored. Finally, spatially restricted protein synthesis is achieved through translational regulation (1,3–5). mRNA localization to distinct cellular compartments gives precise and efficient control over the translation process (6). There are some advantages to transporting mRNAs rather than proteins, such as low transport costs, preventing proteins from acting ectopically during translocation, and rapid local responses to extrinsic cues (7–9). There is mounting evidence supporting the important roles of the subcellular localization of mRNAs in a variety of cellular events, and a growing number of studies suggest that the aberrant regulation of mRNA localization can contribute significantly to human disease pathogenesis (10–12). For instance, mutations in genes involved in mRNA localization at the synapse have been linked to several human neurological diseases, including Fragile X syndrome (13).
In recent years, studies of the complex spatial distribution of mRNA within cells have generated a number of methods, many of which have been applied to improve classical in situ hybridization (ISH) and high-throughput RNA sequencing to detect and track single RNA subcellular localization in living cells (14–17). All of these novel techniques have advantages, but also costly. Therefore, computational methods are in demand as a complementary approach. The first computational predictor for mRNA subcellular localization was the RNATracker (18), which applied deep recurrent neural networks for predictions from raw sequences and provided a method to detect the candidate zipcodes by masking 100 nt of the sequence each time to evaluate its impact for prediction. With the accumulation of RNA localization data, Zhang et al. (19) proposed a machine-learning-based tool, named iLoc-mRNA that focused on the mRNA subcellular localization prediction of Homo sapiens, where a support vector machine (SVM) was applied to a combination of optimally preselected features. Recently, mRNALoc was developed to provide predictions for five subcellular locations of eukaryotic mRNAs using SVM on the pseudo K-tuple nucleotide composition features (20). In the real transcriptome world, mRNAs are localized in multiple compartments, as shown by data in an RNA subcellular localization database (21), in a way similar to that of about half of the proteins each localized in multiple compartments (22). However, none of the computational methods were designed for the multi-label subcellular localization prediction at the mRNA level.
To meet the need for multiple mRNA subcellular localization predictions, we proposed DM3LOC, a deep-learning method with multi-head self-attention, to predict mRNA subcellular localizations in multiple compartments. Different from previous predictors, which only consider mRNA localization in a single compartment, our task is much more challenging. Compared with traditional machine-learning methods like SVM, the deep-learning based methods, such as RNATracker, are considered more advanced and can provide a more biologically meaningful interpretation (23–25). In the deep-learning application for classical sequence-based prediction, the combination of convolutional neural networks (CNNs) and bi-directional long short-term memory (BLSTM) is a popular architecture, which was applied in (26) for DNA function prediction. After applying the combined CNNs and BLSTM, the attention mechanism was introduced on top of this hybrid architecture in DeepLoc (27) for protein subcellular localization prediction. The RNATracker applied an architecture similar to DeepLoc. In such an architecture, CNN is used to automatically extract features or motifs from sequences, BLSTM is used to consider the orientations and spatial distances between the features, and the subsequent attention layer is used to make the predictor attend to prediction-related regions by assigning high weights to significant regions automatically. Such an attention mechanism is referred to as single-head attention since only one attention-weight vector is applied. The single-head attention mechanism gives the model some intelligence for model interpretation; however, in some cases, the prediction performance was negatively affected. For example, in a study of protein subcellular localization prediction (28), the single-head attention mechanism showed poorer prediction performance than the non-attention architecture. The single-head attention may also experience some drawbacks in the interpretation power, which may be induced by the weighted sum of hidden states derived from previous layers with its single attention-weight vector. For example, multiple elements, which may be separated by relatively long segments, in an mRNA sequence can work together for the localization, whereas such synergistic effects may be missed by single-head attention.
To address the issue of the single-head attention mechanism, we applied a combination of CNN and multi-head self-attention architecture. Multi-head self-attention was first introduced in (29) for sentence embedding, which was used to characterize the multiple components in a sentence that together forms the overall semantics of the whole sentence. Later, the multi-head self-attention presented powerful advantages in the state-of-the-art natural language processing architectures, such as the Transformer (30) and Bert (31). Because the multiple heads attend to multiple subcellular elements simultaneously and characterize the global features formed from multiple elements of a sequence (multi-body effects), BLSTM becomes less important and can be removed. In addition, the recurrent model is computationally expensive, while the multi-head self-attention, basically composed of fully connected neurons, is computationally effective and facilitates more parallelization during training computations, especially for long sequences. Notably, multi-head self-attention architecture showed good results in our study. Evaluations on two independent benchmark datasets showed that DM3Loc outperformed RNATracker in both accuracy and speed. And, after a comparison with all the existing tools, DM3Loc in general obtained superior performance.
Besides the prediction, we demonstrate that DM3Loc has some benefits in the biological interpretation. Our study shows that DM3Loc is capable of generating sequence motifs, many of which can be matched to existing motifs of RNA binding proteins (RBPs). DM3Loc also has the ability to assess the contribution of nucleotides in an input mRNA sequence to subcellular localization at a useful resolution, which is consistent with known zipcode regions. Furthermore, from the prediction of all human mRNAs, we found hundreds of mRNA isoform-specific subcellular localization instances, which demonstrated the concept of isoform-specific mRNA subcellular localization on a large scale. We also conducted gene enrichment analyses for the mRNA subcellular localization groups and found significantly enriched gene ontology (GO) terms, many of which are consistent with existing knowledge. We also believe that our proposed approach could be useful for other sequence-based analyses. All the services and the standalone tool of DM3Loc can be freely accessed at the webserver http://dm3loc.lin-group.cn.
MATERIALS AND METHODS
Benchmark dataset
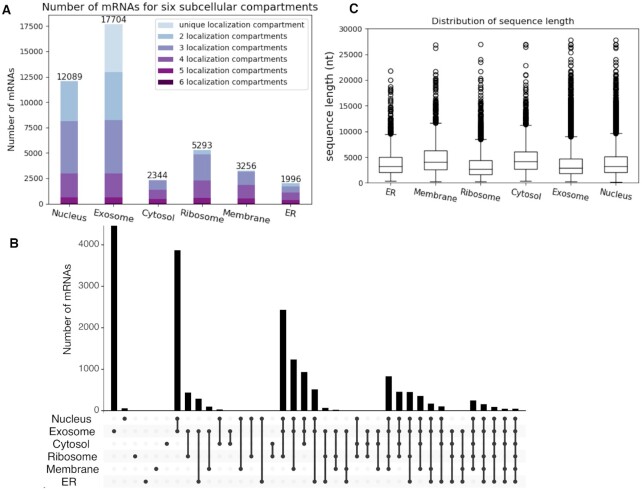
In this study, we assembled a benchmark dataset for mRNA subcellular localizations from the RNALocate database (21). Specifically, the RNALocate subcellular localization data in the EXCEL format were obtained from RNALocate in February 2020. The original complete sequences of mRNAs were downloaded from GenBank (https://www.ncbi.nlm.nih.gov/genbank/) (32), and the mRNA sequence data in the FASTA format were obtained from the NCBI in February 2020 (https://www.ncbi.nlm.nih.gov/sites/batchentrez). RNALocate collected the subcellular localization data from the literature, covering different cell types and conditions. In the RNALocate database, if there is any experimental evidence of one particular localization for one mRNA in the literature, the localization annotation will be assigned to the mRNA. For one mRNA, if it has multiple localization annotations, we combined them and assigned the mRNA with multi-labels. Our assembled benchmark dataset contains a total of 17 870 mRNAs and they are localized in six subcellular compartments: nucleus, exosome, cytosol, ribosome, membrane and endoplasmic reticulum (ER) which with experimentally verified multiple subcellular localizations in Homo sapiens. The distribution of this benchmark dataset is shown in Figure 1A. The number of mRNAs with localization at different compartments are 12 089 for nucleus, 17 704 for exosome, 2344 for cytosol, 5293 for ribosome, 3256 for membrane and 1996 for ER. Since an mRNA can localize at multiple compartments and we are considering six compartments, a total of 64 (26) possible combinations of mRNA localizations exist in theory; yet, only 40 localization combinations had mRNA samples in our data, as shown in Figure 1B. From this figure, we can say that the majority of the mRNAs in our dataset have multiple localization annotations. We analyzed the sequence length distribution for six subcellular localizations, which is shown in Figure 1C. The lengths of the mRNA sequences varied from ∼200 nt to ∼30 000 nt, in which the majority are shorter than 4000 nt. We also built a nonredundant dataset by removing the redundant sequences with a cut-off of 80% sequence similarity through CD-HIT-EST (33), which contains 17 023 mRNAs in total. For the performance evaluation, the nonredundant dataset was used to construct a 5-fold cross-validation dataset. Specifically, the nonredundant dataset was split into five folds and each fold had a similar distribution of subcellular localization categories. The 5-fold cross-validation dataset can be used directly to compare with other methods. All the data (the benchmark dataset with all the mRNAs, the nonredundant benchmark dataset, and the 5-fold cross-validation dataset) are freely available at the DM3Loc web server. In RNALocate, there are a large number (8846) of mRNAs localized in cytoplasm. However, because of ambiguous annotations of cytoplasm in early literature, which could include various compartments (such as ER), we only focus on the prediction of cytosol in this paper. Considering the number of cytoplasm mRNAs in RNALocate, although we did not provide a prediction for cytoplasm as the output of our tool, we still used the data to help with the training of the model by treating cytoplasm as an additional class.
Figure 1.
Statistics of mRNA subcellular localization benchmark data. (A) shows the number of mRNAs for six subcellular compartments. Each bar represents one compartment and the height represents the number of samples. For each bar, from the bottom to the top represent the number of mRNAs with six localization annotations to the number of mRNAs with a unique localization label, respectively. The total number of mRNAs in each compartment in (A) is labeled on the top. (B) An upset plot, which shows the detailed number of mRNA sequences in each intersection group. The intersection group is presented in the order of the compartment numbers (from left to right). The upper bar plot presents the number of mRNAs in each intersection group, while the bottom dots show the components of each group. (C) The distribution of sequence length for six subcellular localization compartments.
DM3Loc framework
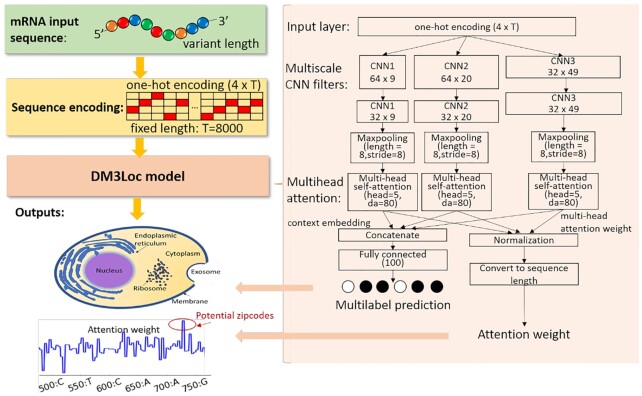
The framework of DM3Loc is presented in Figure 2. The model accepts mRNA sequences at variant lengths. We encoded the mRNA sequences using one-hot encoding for four types of nucleotides where T and U share the same coding. In our collected data, the length of the mRNA sequence varied from ∼200 nt to ∼30 000 nt, and the majority of mRNA sequences are shorter than ∼4000 nt. To take advantage of the mini-batch techniques for training and prediction, the length of input sequences was fixed at 8000 nt by extracting sequences from both ends or by padding. Specifically, for sequences longer than 8000 nt, we extracted 4000 nt from each end and concatenated these end sequences into 8000 nt; for sequences shorter than 8000 nt, we kept the whole sequences and right padded them with zero coding. Thus, the input sequences with variant lengths were encoded using a one-hot representation into a fixed-length matrix (4 × T, T = 8000) and fed into the input layer. To characterize the localization signals at variant lengths, a multiscale CNN was applied. The multiscale CNN consists of three different filter lengths, 9, 20 and 49, specifically, in three paths, each of which contains two convolutional layers followed by a max-pooling layer. Particularly, in the first convolutional layers, there are 64 9-length filters, 64 20-length filters and 32 49-length filters, while in the second convolutional layers, each type of filter contained 32 filters. After the max-pooling process, only the highest value of every eight continuous hidden neurons (pooling length = 8 and pooling stride = 8) from the last convolutional layer was kept, resulting in length T’ = T/8. In this framework, the multi-head self-attention mechanism is used to assess the contribution of sequence regions for localization by multiple heads (head = 5), which has the ability to further detect localization zipcodes during the prediction. The outputs of the multi-head self-attention are the multi-head attention weights and context embedding. To be consistent with the multi-scale CNN, three multi-head self-attention layers are separately applied to the output states of the three paths of CNN filters. Because the two convolutional layers and the following max-pooling layers, the nearby information of a nucleotide is convolved and pooled to some extent, our model cannot assess the contribution to localization at the single-nucleotide resolution per se, but it can reach a reasonable resolution. The filter length is 49 for each convolutional layer at most and the pooling stride is 8 for the max-pooling layer, as a result, the upper limit resolution of the attention weights is within 49 + 49 + 8 = 106 nt. Then, the outputs from the three multi-head self-attention layers are concatenated and fully connected to the output layer, which contains six neurons for six localization categories. We used the sigmoid activation function on the output layer, which makes the prediction values for each category in the range between 0 and 1. During the prediction, the attention weights of the CNN output layers are normalized and then converted back to the same length of the input sequence to analyze the localization zipcodes.
Figure 2.
DM3Loc framework. The input of the model is an mRNA sequence of any length; then, it is one-hot encoded into a fixed-length matrix (4 × T, T = 8000). There are two outputs of the DM3Loc model; one is the prediction scores for six localization categories and the other is the attention weights, which have the same length as the input sequence. The regions with high attention weights indicate the zipcode regions. The DM3Loc model mainly consists of multiscale CNN filters and multi-head self-attention layers. The multiscale CNN is used to capture the localization signals at variant lengths, and the multi-head self-attention is used to make the model attend more to the important sequence regions for localization.
Multi-head self-attention
Bengio et al. (29) proposed that the overall semantics of a sentence is formed by multiple components in it. Therefore, they proposed a multi-head self-attention mechanism to focus on different parts of a sentence. In this work, we borrowed this idea and made some modifications to the original mechanism, including a Mask operation for padding regions, smoothing the attention weights and normalizing the attention weights according to the effective length. To calculate the attention weights, we first need to calculate the energy score matrix E, as defined in Equation (1).
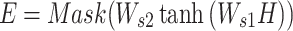 |
(1) |
Here, H is the 32-by-T’ hidden neurons output from each CNN path. 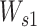 is a weight matrix with a shape of da-by-32, and da is the attention-dimension hyperparameter, which is 80.
is a weight matrix with a shape of da-by-32, and da is the attention-dimension hyperparameter, which is 80. 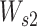 is a matrix of parameters with shape head-by-da, where head indicates the number of attention heads, which is 5. To overcome the overfitting and to get sparse energy scores, the L1-norm regularization is applied to
is a matrix of parameters with shape head-by-da, where head indicates the number of attention heads, which is 5. To overcome the overfitting and to get sparse energy scores, the L1-norm regularization is applied to 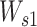 and
and 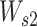 with the same loss weights of 0.001. Because we know where the paddings of the input sequences are during the model training, we can mask the padding regions for attention. The Mask operation works by adding an exceptionally large penalty (–10 000) to each position in the padding regions, which draws attention away from those regions.
with the same loss weights of 0.001. Because we know where the paddings of the input sequences are during the model training, we can mask the padding regions for attention. The Mask operation works by adding an exceptionally large penalty (–10 000) to each position in the padding regions, which draws attention away from those regions.
After the Mask operation, in the original multi-head self-attention paper, the softmax activation function was applied to the energy matrix to make the attention weights into probability values. We modified the original softmax function using a smoothing approach (34), which can aggregate selections from multiple top-scored regions. In the smoothing, the unbounded exponential function of the softmax function is replaced with the bounded logistic sigmoid function. The attention weight matrix A is calculated as:
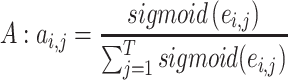 |
(2) |
where 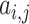 is the element of matrix A, and
is the element of matrix A, and 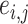 is the element of the energy matrix E.
is the element of the energy matrix E.
Because the Mask operation results in a variant effective length for the attention mechanism, the final attention weights should be normalized according to the ratio between the effective length without padding and the input length for the attention model, which is T’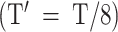 , as shown in Equation (3).
, as shown in Equation (3).
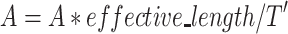 |
(3) |
The final context embedding M is calculated as the concatenation of each hidden neuron output from each CNN path, is defined as:
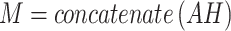 |
(4) |
where A is the head-by-T’ attention matrix from each CNN path, and H is the T’-by-32 hidden neurons output from each CNN path. The weighted sum was attained by multiplying A and H in each CNN model, which was concatenated along the second dimension (32 + 32 + 32), resulting in a head-by-96 embedding tensor.
To encourage the different attention heads to focus on different parts of an mRNA sequence, the penalization term is also applied and defined as:
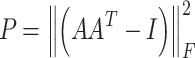 |
(5) |
where A is the attention matrix, I is an identity matrix, and 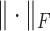 represents the Frobenius norm of a matrix. The more similarity between the two attention vectors, the higher the loss will be. The loss weight of the penalization term is 0.001.
represents the Frobenius norm of a matrix. The more similarity between the two attention vectors, the higher the loss will be. The loss weight of the penalization term is 0.001.
Extracting attention weights
From the multi-head self-attention model, the attention matrix A of each of the three CNN paths is generated, which yields three independent 5-by-T’ weight matrices. For real applications, we need an overall attention-weight vector that has the same length as the input sequence to assess the contribution of each base in the mRNA sequence. To get such overall attention weights, we first normalize the attention signals for each head in each CNN path through a z-score, respectively, and then we averaged all the z-score values along T’, resulting in an attention weight vector for each input sequence with length T’. Finally, the length T’ should be converted back to the original length T by first replicating eight times (because the pooling stride = 8 in the max-pooling layers) for each element in the normalized attention signals (resulted length T). We then remove the zero paddings for sequences shorter than T or add zero paddings for sequences longer than T to make the length the same as the original sequence length.
Extracting sequence binding motifs from CNN filters
The CNN filters can be used to build position-weight matrices (PWM) of sequence binding motifs. We applied a method similar to that of Alipanahi et al. (35) but modified the position-weight calculation. In particular, we scanned all the mRNA sequences in the benchmark data against the three types of CNN filters (the 9-length, 20-length and 49-length filters). For each of the CNN kernels, only one fragment was kept per sequence, which had to have the maximum response value (the value must > 0) of that CNN kernel. Thus, one fragment per sequence was needed to calculate the PWM of the CNN kernel. For each CNN kernel, we obtained a set of sequence fragments, which had the same length as the kernel after the scan. Instead of adding a value of 1 for all the effective positions, we calculated the PWM by adding the response values of the corresponding CNN feature maps for each position of the extracted fragments. Finally, for each of the CNN kernels, we obtained one PWM. Specifically, we obtained 64 PWMs for 9-length CNN, 64 PWMs for 20-length CNN filters, and 32 PWMs for 49-length CNN filters. We can use the PWMs to draw sequence logos to represent the sequence binding motifs.
Model training and testing
For a classification problem, the target of the training is to make the difference between the prediction vector and the true label vector as small as possible. We denote the prediction for the 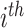 mRNA sample as
mRNA sample as 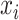 and its corresponding true label vector as
and its corresponding true label vector as 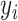 . Each element of
. Each element of 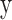 is a binary value, denoted as
is a binary value, denoted as 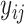 ,
, 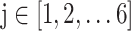 , which indicates whether that mRNA sample belongs to a certain localization category or not. If
, which indicates whether that mRNA sample belongs to a certain localization category or not. If 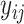 is 1, the
is 1, the 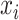 belongs to the class
belongs to the class  , 0 otherwise. The binary cross-entropy loss function was applied to treat each category independently. The loss of sample
, 0 otherwise. The binary cross-entropy loss function was applied to treat each category independently. The loss of sample 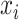 is defined as below:
is defined as below:
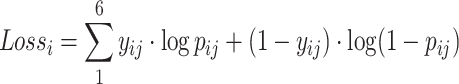 |
(6) |
where 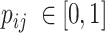 is the element of
is the element of 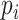 , which represents the prediction vector of the sample
, which represents the prediction vector of the sample  . Since there are six localization categories and we treated them independently, the loss of sample
. Since there are six localization categories and we treated them independently, the loss of sample 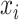 is the sum of the loss of individual localization categories.
is the sum of the loss of individual localization categories.
Because we treated cytoplasm as an additional class and used the cytoplasm data to help train the model, we actually have seven independent classes during the training. We applied different class weights for different localization categories to overcome the unbalanced training data problem. The class weight was calculated as the ratio of total training samples to the number of samples in each category. In particular, the class weights for nucleus, exosome, cytosol, ribosome, membrane, ER and cytoplasm are 1, 1, 7, 3, 5, 8 and 1, respectively. We used the Adam stochastic optimization method (36) with a learning rate of 0.001, and a learning rate decay of 5e–5. The deep-learning model was implemented using TensorFlow 1.12.0 and Keras 2.2.4. Model training and testing were performed with GPU Nvidia TITAN RTX. For the hyperparameter selection, such as the number of CNN filters and the number of CNN layers, we applied the GpyOpt package (https://sheffieldml.github.io/GPyOpt/), which implements one Bayesian optimization method on a small sample of data to find the hyperparameters with the global optimum solution.
For the performance evaluation, the model was trained and evaluated by a 5-fold cross-validation dataset. To use as much as possible training data, the prediction model used by the DM3Loc webserver was trained by a nested 8-fold cross-validation on the dataset before removing the redundant sequences. In particular, for each fold, we used 7 folds of the data to train the model and evaluated the last fold of data. We repeated this procedure eight times, before finally obtaining eight classifiers. When predicting the localization for an mRNA sequence, the average value obtained from the eight classifiers was used as the final prediction score. In the final prediction result, for each mRNA sequence, six prediction scores for the corresponding localization categories are provided (the cytoplasm category is not considered in the prediction) and the ones that have prediction scores higher than the pre-defined cutoffs are output as the localizations. We defined a series of cutoffs for each localization class according to their maximum MCC values with continuous cutoffs on the 8-fold cross-validation data. According to the MCC values along the continuous cutoffs (see Additional File1: Supplementary Figure S1), the default cutoffs were set as 0.68 for nucleus, 0.98 for exosome, 0.20 for cytosol, 0.39 for ribosome, 0.24 for membrane and 0.22 for ER.
RESULTS
Comparing DM3Loc with existing methods and tools
We compared the prediction performance of DM3Loc with existing methods and tools on the 5-fold cross-validation benchmark data. Here, we compared DM3Loc with the RNATracker from the deep-learning method perspective, where the same dataset was used to train and test these two methods, and both used the 8846 cytoplasm mRNAs to assist the training; and we compared DM3Loc with mRNALoc and iLoc-mRNA from the tool perspective, where the five folds of test data from the 5-fold cross-validation benchmark data were used to test these tools.
To make a fair comparison with the RNATracker on the multi-label classification problem, we modified the original code of the RNATracker to make all the settings identical to the DM3Loc. In particular, we replaced the softmax activation function and the Kullback-Leibler divergence loss function with the sigmoid activation function and the binary cross-entropy loss function in the RNATracker, respectively. Furthermore, since we also found localization signals in the 5′ end, we trained our model and the RNATracker model by truncating the whole sequences from both ends (4000 nt in each end). Although mRNALoc and iLoc-mRNA were not designed as multi-label predictors, both of them provide the prediction scores for each localization compartment; hence, we can extract the intermediate prediction scores for each compartment and evaluate their predictions of the localization on our dataset. Although different training sets were used, all the methods and tools were tested on the same five folds of test data for comparison. We used the area under the receiver operating characteristic (ROC), the precision-recall (PR) curves, and the Matthews correlation coefficient (MCC) to evaluate their performance. The average results of the five folds of test data are shown in Table 1, which shows that our method outperformed these methods and tools in most of the cases. It is worth mentioning that the test data has a performance bias towards mRNALoc and iLoc-mRNA since they both extracted the data from the same source (RNALocate) as we did for our methods; thus, some of our test data could be used to train their tools, while these test data were not seen during the training process of DM3Loc and RNATracker.
Table 1.
Performance comparison with existing methods and tools
| Method | Compartment | Average area under ROC | Average area under PR | MCC |
|---|---|---|---|---|
| DM3Loc | Nucleus | 0.7725 | 0.8765 | 0.3859 |
| Exosome | 0.7233 | 0.9964 | 0.0736 | |
| Cytosol | 0.7406 | 0.3193 | 0.2872 | |
| Ribosome | 0.7589 | 0.5478 | 0.3550 | |
| Membrane | 0.7558 | 0.4472 | 0.3115 | |
| ER | 0.6981 | 0.2502 | 0.2048 | |
| RNATracker | Nucleus | 0.7531 | 0.8601 | 0.3450 |
| Exosome | 0.7533 | 0.9970 | 0.0000 | |
| Cytosol | 0.7331 | 0.3176 | 0.1383 | |
| Ribosome | 0.7447 | 0.5365 | 0.2697 | |
| Membrane | 0.7386 | 0.4051 | 0.1927 | |
| ER | 0.6265 | 0.1880 | 0.0000 | |
| mRNALoc | Nucleus | 0.6075 | 0.7655 | 0.1501 |
| Exosome | 0.4065 | 0.9887 | -0.0294 | |
| Cytosol | 0.4529 | 0.1177 | -0.0134 | |
| ER | 0.3729 | 0.1402 | -0.1479 | |
| iLoc-mRNA | Nucleus | 0.5186 | 0.7200 | 0.0516 |
| Cytosol | 0.5310 | 0.1339 | 0.0253 | |
| Ribosome | 0.7940 | 0.6634 | 0.3899 | |
| ER | 0.8100 | 0.5702 | 0.3762 |
The results presented in Table 1 are the averages of the five folds of test data. To calculate the MCC, DM3Loc used the pre-defined default cutoffs; RNATracker used cutoff 0.5; mRNALoc used cutoff 0.1; iLoc-mRNA used cutoff 0.5.
To demonstrate the effect of the 5′ end, we retrained our model with sequences only extracted from the 5′ end and compared it with the RNATracker on the 5-fold cross-validation benchmark data. According to the length analyses in Figure 1B, 4000 nt can cover the majority of the mRNAs. Therefore, in this comparison, we only extracted 2000 nt at most to exclude the effect of the 5′ end region to some extent. The performance of the 5-fold cross-validation (see Additional File 1: Supplementary Table S1) is much lower than the current performance as shown in Table 1, which indicates the important roles of 5′ end in subcellular localization.
We conducted another comparison with RNATracker on the CeFra-Seq dataset provided by Yan et al. (18). Because the RNATracker used expression levels to estimate the localization likelihood, their model is a regression model. To make our model suitable for a regression problem, we replaced the output layer with the softmax activation function and the loss function with the Kullback-Leibler divergence, which is the same as the original RNATracker model. We evaluated our model and the RNATracker model by training and testing on 10-fold cross-validation data generated by us from the CeFra-Seq dataset. The performance was presented as the Pearson correlation coefficients between the experimental and predicted localization values of the combined folds. According to the performance (see Additional File 1: Supplementary Table S2), although our model was not tuned and designed for the regression problem, our performance was still superior to RNATracker, which demonstrated the benefit of the proposed architecture for sequence-based prediction. Another benefit of the proposed framework is the lower training time compared with RNATracker, which is 15s versus 107s per epoch. The increased training efficiency was mainly due to our omission of a recurrent model in our method. Recurrent models require much more computing time for such long sequences. Also, by utilizing this benefit, we could use advanced hyperparameter selection tools to tune more optimal model structures for this problem.
Application of DM3Loc on independent datasets
To further evaluate the performance of the DM3Loc predictor trained on the RNALocate dataset, we conducted the following two tests of DM3Loc on two independent datasets, i.e. the multiplexed error-robust FISH (MERFISH) dataset (37) and the APEX-Seq dataset (17). By combining the MERFISH approach with cellular structure imaging, Xia et al. (37) identified RNA species enriched in two subcellular compartments, which are the ER and nucleus from the ∼10 000 genes. The APEX-Seq dataset has 8 mRNA localization categories by the APEX-Seq techniques. By merging ERM and ER lumen as ER (used the maximum log2 fold-change as the merged value), there are three localization compartments in common to our categories, i.e., nucleus, cytosol, and ER. By matching the genes to the RefSeq database (38) and selecting one of the representative mRNAs (the mRNA isoform with the longest sequence) from the MERFISH dataset, we found 559 mRNA sequences enriched in ER and 864 mRNA sequences enriched in nucleus from 6526 mRNAs in total. By selecting one representative mRNA for a given ENSEMBL gene from the APEX-Seq dataset using the recommended log2 fold-change cutoff of 0.75 proposed by the authors, we found 1010 mRNAs enriched in the nucleus category, 1604 mRNAs enriched in the cytosol category and 173 mRNAs enriched in the ER category from 3219 mRNAs in total. We applied DM3Loc on the two mRNA datasets and showed the comparison between our predictions and the annotations of the two datasets in Venn plots (Additional File 1: Supplementary Figure S2). The statistical significance (P-value) of the observed overlapped mRNAs was calculated by a permutation test. Specifically, the distribution of the number of overlapping mRNAs was assumed to follow a hypergeometric distribution. The background distribution of the overlapping mRNAs was randomly drawn from the hypergeometric distribution for 10 000 times (left grey histograms in Supplementary Figure S2). The observed number of overlapping mRNAs of the corresponding localization compartment labelled from each dataset was shown as a red vertical line. According to the results, we found that all the observed numbers of overlapping mRNAs are significantly away from the random background distributions (P-values < 0.005). Please note that both MERFISH and APEX-Seq datasets were generated using only one cell line, while DM3Loc was trained on the data from different cell types under different conditions, which are unlikely to exactly match. However, our predictions still overlap well with the subcellular localization annotations from independent datasets.
Application of DM3Loc on the human transcriptome
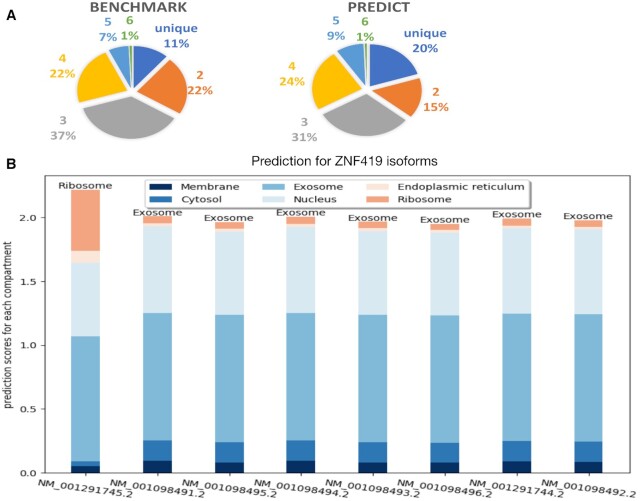
We applied DM3Loc to make localization predictions for all the human mRNAs. We collected all the mRNA sequences for the GRCh38 human reference genome, which was downloaded from the RefSeq database (38). There are 57 091 mRNAs in total from the 18 964 genes in the collected data after removing the predicted ones. The prediction results of DM3Loc for these mRNAs can be found in Additional File 2. We compared the distribution of mRNAs predicted as a single label, two labels, …, and up to six labels with the distribution in our benchmark dataset. The comparison is displayed in two pie charts, as shown in Figure 3A. From the result, we see that the predicted subcellular localization labels have a similar distribution to the one in the benchmark dataset. This collected mRNA data contains mRNA isoforms of the same genes. We analyzed the prediction results of these isoforms and found that a small number of isoforms of the same genes were predicted to have different subcellular localizations. In the analysis, for each isoform pair of the same gene, we considered that isoforms have significantly different subcellular localizations when for at least one compartment, it satisfies: (i) the predicted label is different (one isoform passes the default label cutoff and the other does not); (ii) the difference between their prediction scores is higher than 0.4. In total, 496 genes have at least one isoform pair satisfying this criterion, as shown in Additional File 3. For instance, as shown in Figure 3B, all the other isoforms of gene ZNF419, the zinc finger protein 419, were predicted to be localized in exosome, while the isoform NM_001291745 was predicted to be localized in ribosome. The concept of an isoform-specific mRNA subcellular localization is not new, i.e. it has been found in neural projections (39). In this paper, we demonstrate this phenomenon through the prediction method and provide a potential list of isoform-specific mRNA subcellular localizations.
Figure 3.
Applying DM3Loc to predict localization for all the human mRNAs (genome GRCh38). (A) The mRNA subcellular localization label distribution in benchmark and prediction pie charts. The left pie chart shows the label distribution of mRNAs in the benchmark dataset. The right pie chart shows the predicted label distribution of mRNAs in the GRCh38 human reference genome. The label distributions include a unique subcellular localization label, two labels, …, and six labels. (B) Prediction for isoforms of gene ZNF419. Each bar represents the prediction scores for the six subcellular localizations of each isoform; the predicted labels whose scores were higher than the default threshold are labels are on the top. The prediction scores of all the other isoforms are similar, except for isoform NM_001291745, which is predicted to be localized in the ribosome.
Analysis of CNN motifs
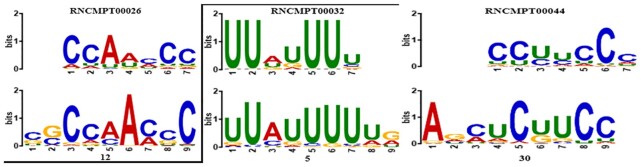
The CNN filters can be used to build position-weight matrices (PWM) of sequence binding motifs, which achieves the nucleotide-level resolution (see MATERIAL AND METHODS). Our model contains multiscale CNN filters, which include three different filter lengths, i.e. 9-length, 20-length and 49-length, resulting in independent PWMs from those three scales. The PWMs were used to represent the sequence binding motifs. We used TOMTOM (40) to map the learned motifs separately from each scale to the known RNA binding motifs of RBP (Homo sapiens has 102 in (41)). A total of 62 out of 64 9-length CNN motifs, 63 out of 20-length CNN motifs, and 32 out of 32 49-length CNN motifs were found to match the known motifs with P-value < 0.05, covering 99 out of 102 known RBP motifs. In contrast, only nine of the 30 CNN motifs were found to match in the RNATracker recently introduced by Yan et al. (18). The complete result can be found in Additional File 4. Some of the representative motifs were shown in Figure 4, where RNCMPT00026 (HNRNPK) matches to filter 12, RNCMPT00032 (HuR) matches to filter 5, and RNCMPT00044 (PCBP2) matches to filter 30 in the 9-length filter.
Figure 4.
Visualization of representative CNN motifs mapped to known RBPs. The known RBPs (top rows) are from (41). The CNN motifs (bottom rows) were generated from different CNN filters with the filter index labeled below.
Visualization of attention weights on sequences
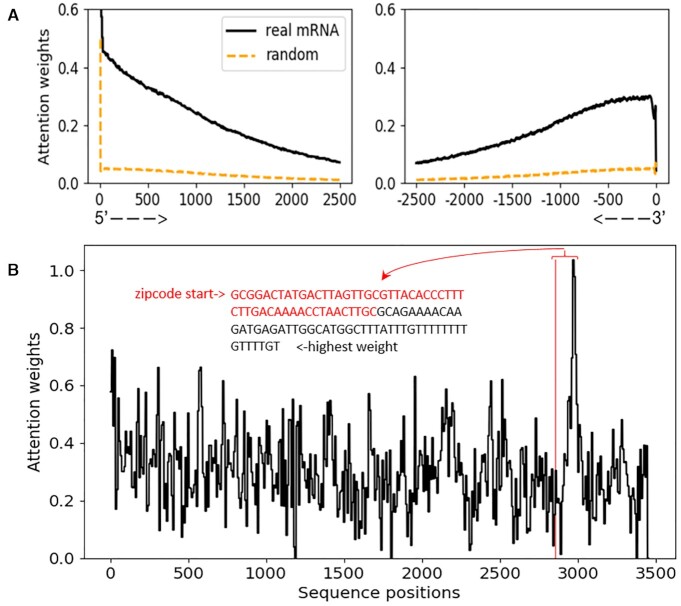
In this work, the attention weights generated along with the prediction can be used to monitor important mRNA sequence regions to the subcellular localization prediction. Here, we visualized the attention weights that vary along the sequences to investigate the impact of different sequence regions on subcellular localizations. In this experiment, we only used mRNAs longer than 5000 nt. Specifically, we truncated all the sequences from the center and aligned the left part of the attention weights to the 5′ end and the right part of the attention weights to the 3′ end, with each end's length set at 2500 nt. The resulting attention weights of length 2500 nt from each end is shown as the blue lines in Figure 5A. We also conducted an ablation test by randomly permuting the nucleotides of mRNAs to train a baseline model and present the attention weights of these permutated sequences, which is shown as the red lines in Figure 5A. We can clearly see from these results that both the 3′ end and 5′ end have high attention weights concentrated in real mRNAs, while all the attention weights are very low for the random sequences. The high peaks precisely located at the 5′ end and 3′ end may be introduced by artificial effects because of the end-effect of the CNN kernels. Except that, the attention weights may come from the localization regulatory elements located in both 5′ and 3′ UTR (42,43). A drop in the attention weights is near the 3′ end, which may be due to the poly(A) tails presented on most of the mRNA sequences; these poly(A) tails may not be useful for subcellular localization. Yan et al. (18) also drew attention weights for RNA localization; however, their method only focused on the 3′ end and used zero paddings at the 5′ end, making the 5′ end generally less informative; thus, the high attention weights were only presented in the 3′ end regions (Figure 5 in (18)). In our work, since all the mRNAs were longer than 5000 nt, the concentration of high attention weights on both ends of the mRNA is not introduced by the zero-padding of shorter sequences.
Figure 5.
Visualization of attention weights. (A) Attention weights for a set of mRNA sequences from the 5′ to the 3′ end. Only sequences longer than 5000 nt were used to draw this plot; therefore, all the positions have equal coverage of the bases. The black solid lines represent the attention weights for real mRNAs while the orange dashed lines represent the attention weights obtained from random models trained by permuted mRNA sequences. (B) Mapping attention weights to human β-actin localization zipcode. The x-axis represents the sequence position from the 5′ to 3′ end; the y-axis represents the attention weights, and the starting point of the zipcode region is indicated with a red vertical line. We designated the nucleotides’ location from the zipcode starting point to the position with the highest attention weight, where the zipcode region are in red and the non-zipcode nucleotides are in black.
Mapping attention weights to localization zipcode
To investigate the mapping of attention weights to a known localization zipcode, we conducted the following proof-of-concept experiment. In theory, the attention weights vary along sequences and can be used to assess the contributions of each region to a subcellular localization prediction, which can be directly used to infer localization zipcodes. However, to validate its effectiveness in real mRNA sequences is extremely difficult, since there are only dozens of well-characterized localization zipcodes and even fewer in the species (human) studied in this work. In addition, many localization signals operate at levels of both primary and secondary structures. We were able to find only one known zipcode that is markedly related to our data, which is the beta-actin mRNA in chicken embryo fibroblasts (CEFs), which localizes near an actin-rich region of cytoplasm. This localization is mediated by a 54-nt-long zipcode in the 3′-UTR sequence region and can be inhibited by anti-zipcode oligodeoxynucleotides (ODNs) (44). A homologous region of this zipcode is in the human β-actin sequence (52 nt). Because our model is trained on the human sequence, we used a human β-actin transcript sequence to test the mapping of attention weights in the human β-actin zipcode region. The human β-actin mRNA sequence is extracted from NC_000007.14, in which the localization zipcode starts at position 2855. Given the mRNA sequence of NC_000007.14, the attention weights are generated, as shown in Figure 5B. We can see from this figure that the position of the sequence with the highest attention weight is next to the starting point of the zipcode, which is within a reasonable distance under resolution 106 nt (see Materials and Methods) of our attention weights.
Gene enrichment analysis
To further understand the functional roles behind the subcellular localization of mRNAs, the functional enrichment analysis of gene ontology (GO) (45) was performed on the mRNA genes and their coding proteins for an analysis of their biological processes (BP), molecular functions (MFs), and cellular components (CCs) using the R package clusterProfiler (46). Since few mRNAs localize a single compartment in our study, we selected mRNAs that can transport to no more than two compartments to make the enrichment analyses more focused for each of the six localization compartments. We also conducted enrichment analyses for mRNAs that can localize in all six compartments (ALL-SIX). In total, we carried out functional enrichment analyses for seven compartment groups. From the enrichment results, each of the compartment groups had some significantly enriched GO terms, which are described in more detail in the following summary.
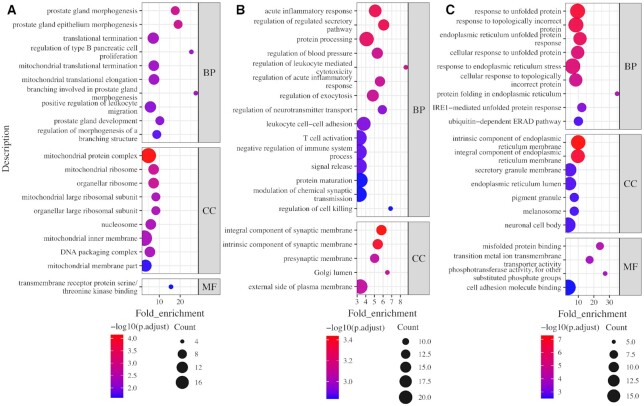
The mRNAs localize in ribosome are functionally enriched in GO terms ‘prostate gland development’ and ‘mitochondrial ribosome’ (Figure 6A). We found that the genes that were involved in GO term ‘prostate gland morphogenesis’ and encode the ribosome mRNAs are all related to different diseases or cancers, which is consistent with the known relationships between ribosome mRNAs and human maladies, including cancer (10,11). We investigated the genes that encode the mitochondrial ribosome, and found that all of these genes are located in the nuclear chromosome instead of the mitochondrial chromosome. Most of them are translated into mitochondrial ribosomal proteins, which help protein synthesis within mitochondrion. The widely accepted notion for protein transport to mitochondria is that the import occurs post-translationally after the protein is fully synthesized in the cytosol (47); however, mounting evidence also supports a co-translational import of some proteins into the mitochondria. Specifically, polysomes, a group of the mitochondrial ribosomes bound to an mRNA molecule were shown to be associated with the mitochondrial surface, and proteins synthesized from that polysome are imported co-translationally (48–50). The ribosome mRNAs enriched in the mitochondrial ribosome may have a similar mechanism where they form polysomes and are then imported into mitochondria co-translationally. mRNAs localized in ER are mainly enriched in GO terms ‘acute inflammatory response’, ‘immune-related processes’ and ‘synaptic membrane’ (Figure 6B), which is consistent with early observations noting that the transport of some ER mRNAs plays an important role in the inflammatory response and synaptic transmission (51,52). Taken together, these ER mRNAs reflect the important relationship between ER and inflammatory response within the central nervous system (12).
Figure 6.
Top 20 enrichment terms of mRNAs localized in ribosome, ER, and in all six of the compartments. (A) Ribosome, (B) ER, and (C) all six compartments.
Interestingly, mRNAs that can localize in all six compartments are mainly involved in GO terms ‘response to an unfolded protein,’ ‘ER stress,’ and ‘misfolded protein binding.’ We found that most of the gene functions of these ALL-SIX mRNAs are associated with ER. Some of the genes belong to the heat shock protein family, which perform chaperone functions, and some are associated with the regulation of apoptosis. It is known that the unfolded protein response (UPR) as a cellular stress response related to ER stress initially tries to restore normal functions of the proteins and then aims towards apoptosis if all attempts fail within a certain time span. Although the functions of the ALL-SIX mRNAs in these enriched terms were found to be associated with ER, none of these significantly enriched GO terms, i.e. ‘response to unfolded proteins,’ ‘response to topologically incorrect proteins,’ and ‘endoplasmic reticulum stress’ were found significantly enriched (P-adjust value ≤ 0.05) in mRNAs localized in ER specifically, which indicates the unique functions of ALL-SIX mRNAs. We assume that for an efficient response to UPR or ER stress, these mRNAs should be able to present anywhere in the cell.
mRNAs localized in other compartments are also significantly enriched with some GO terms. For example, mRNAs localized in the nucleus are mainly involved in the ribonucleotide catabolic process and channel activity (Additional File 1: Supplementary Figure S3), indicating the key roles of these mRNAs in ribonucleotide degradation and in the exporting of the nucleus. mRNAs localized in exosomes are mainly involved in cell adhesion, ion channel complex activity, receptor-ligand activity, and transporter activity (Additional File 1: Supplementary Figure S4), which exemplify the roles of exosome RNAs in intercellular communication (53). RNAs localized in cytosol are mainly involved in transport processes, such as the carboxylic acid transport and organic acid transport, as well as in the eating behavior and sialyltransferase activity (Additional File 1: Supplementary Figure S5). mRNAs localized in membranes are enriched to GO terms ‘NLS-bearing protein import into nucleus’, ‘lung epithelium development’, and ‘Ran GTPase binding’ (Additional File 1: Supplementary Figure S6). Altogether, these gene enrichment analyses suggest that the subcellular location of mRNA is tightly associated with its function. In this study, we provide a detailed gene list corresponding to the enriched terms, as shown in Additional File 5.
The DM3Loc web server
We developed a user-friendly web server, http://dm3loc.lin-group.cn, for easy access to the services of DM3Loc. Users can paste up to five mRNA sequences in the paste mode or upload a FASTA format file up to 5MB (∼1500 mRNA sequences) to the server through the upload mode. After the prediction, for each sequence, the prediction results and the attention weights along the sequence will be presented in the output panel. Users can visualize them or download them to local files. All the successfully submitted jobs will be saved in the users’ personal space on our server, allowing users to manage their jobs conveniently. Some of the existing mRNA localization methods also offer web servers, like the iLoc-mRNA. DM3Loc has some unique features, which compare with these other servers. In particular, this is the only web server that can predict mRNAs to multiple compartments; it is the only web server that provides visualization of the estimated contribution for each base to localization in the name of attention weights; and it provides larger sequence submission (up to a 5-MB file). We are confident that DM3Loc is a useful tool for studying mRNA subcellular localization.
DISCUSSION
Subcellular localization of mRNAs, as a prevalent mechanism, gives precise and efficient control over the translation process. Mounting evidence supports the important roles of this process in a variety of cellular events. Computational methods for mRNA subcellular localization prediction provide a useful approach to studying mRNA localization. However, few computational methods are designed for this purpose, and their performance has room for improvement. One notable deficiency is the lack of a tool to predict multiple localization annotations for mRNAs. In this work, we developed DM3Loc based on multi-head self-attention for multi-label subcellular localization prediction. DM3Loc provides prediction scores for all six of the subcellular localizations and gives the final predicted subcellular localizations that pass the pre-defined thresholds, which could be single or multiple localizations
The combination of CNN and BLSTM is still a popular architecture in the application of deep-learning for sequence-based prediction. Many attempts have been made to add attention mechanisms to this hybrid architecture. However, the single-head attention mechanism hardly improves the performance from the prediction perspective (28,54). We believe the bottleneck of the single-head attention is the reduced representation power by the compressed feature space. For example, before the regular attention, the dimension of the embedding layer is T (T = 8000) multiplied by the embedding size. After the single-head attention, the dimension becomes just the embedding size, which reduces the dimension of the context embedding layer by T times. The single-head attention only has one weight vector; thus, a single value is assigned for each base to assess its contribution for the subcellular localization that cannot well characterize the cases when the localization is determined by multiple elements combinatorically. In contrast, the multi-head self-attention introduces the multiple head design, which increases the dimension of the context embedding to an embedding size multiplied by the head number, making it possible to estimate the contribution of multiple elements for subcellular localization prediction independently and combinatorically. In summary, the advantage of applying the combination of CNN and multi-head self-attention in our DM3Loc model are 2-fold: (i) multiple heads can look into multiple regions simultaneously and independently, thereby enabling a more comprehensive assessment of the contributions for each element to subcellular localization; (ii) the replacement of the recurrent model facilitates more parallelization during training and prediction, which makes DM3Loc more computationally efficient. The advantages of such a multi-head self-attention have been demonstrated by the comparison of DM3Loc with the RNATracker method on two independent benchmark datasets.
To deal with the multi-label classification problem, DM3Loc treats the six subcellular localizations independently by applying a sigmoid activation function on the output layer and using the binary cross-entropy loss function (refer to Equation (6)). It is interesting to consider the relationship between different labels by designing a new loss function for the multi-label classification problem, as proposed by Zou et al. (55). In the future, if we have more reliable and diverse mRNA subcellular localization data, we could utilize this method to explore the relationship among different localization categories.
While most of the existing localization cis-regulatory elements are observed to be localized in the 3′ UTR, some regulatory elements also reside in the 5′ UTR or coding region of the mRNA (42,43). Thus, only using the sequences from 3′ UTR will miss some key information for localization prediction. From our experiment results (Additional File 1: Supplementary Table S1), the model trained by sequences extracted from the 3′ end only obtained poorer prediction performance than the model trained by sequences extracted from both ends. Also, from the visualization of the attention weights along sequences from the 5′ to 3′ end, we found both ends contain peak regions that are important for subcellular localization prediction (Figure 5A). All of these findings confirmed the important role of the elements on the 5′ end for subcellular localization.
Our study confirmed some early studies and suggested some interesting biological implications. We conducted functional enrichment analyses for seven compartment groups and from the results, each of the compartment groups has an amount of significantly enriched GO terms, most of which have consistent evidence for the putative functions to the localization of the corresponding compartment. Our study provides more mRNA lists for these functions. As an example, mRNA localizations play important roles in synapse formation and plasticity associated with neurological diseases (1,13,56). It is also known that ER stress has the potential to elicit aberrant inflammatory signaling and facilitate cell death within the central nervous system (12). Our GO enrichment analysis found that mRNAs that are localized in ER were mainly enriched to ‘modulation of chemical synaptic transmission,’ ‘component of synaptic membrane’ and ‘acute inflammatory response’. We hypothesize that for an acute inflammatory response and other stimuli, these mRNAs may rapidly accumulate in ER for proteins to synthesize locally and facilitate the synapse membrane insertion. Moreover, mutations in these mRNAs may result in human neurological disease, such as the Fragile X syndrome resulting from mutations in genes related to dendritic mRNA targeting (13). We also found that 65% of those ER mRNAs contain ‘membrane’ in their CC terms, indicating that the localization of these ER mRNAs is mainly for local synthesizing membrane proteins as a lower-cost transport than the post-translational transport. Another intriguing enrichment result is that the mRNAs can localize in all the compartments. They are mainly involved in the ‘response to unfolded protein,’ ‘endoplasmic reticulum stress,’ and ‘misfolded protein binding.’ Although most functions of the proteins translated from these mRNAs are found to be associated with ER, none of these GO terms were significantly enriched in ER mRNAs (P-adjust value < 0.05), which indicated a unique function for these ALL-SIX mRNAs. Our hypothesis for these mRNAs is that these mRNAs are present all over the cell and can be quickly translated into proteins locally upon ER stress. Some of these proteins function as molecular chaperones, such as the heat shock proteins, and some of these proteins can activate the apoptosis process when the chaperones fail.
CONCLUSIONS
In this study, we assembled a benchmark dataset for six mRNA subcellular localization compartments in Homo sapiens, in which each of the samples had single or multiple experimentally verified subcellular localization annotations. From the benchmark dataset, we built a nonredundant dataset and a 5-fold cross-validation dataset, which can be used directly to compare with other methods. We proposed DM3Loc, a multi-label deep-learning-based approach to predict mRNA subcellular localizations at multiple compartments from the raw sequence of mRNA. The DM3Loc applied a novel multi-head self-attention mechanism on top of the CNN models. The DM3Loc proved capable of generating sequence motifs, the majority of which can be matched to existing motifs of RNA binding proteins. DM3Loc has the ability to assess the segment-level contribution of the input sequence for subcellular localization directly through the multi-head self-attention at a useful resolution. Evaluations on independent benchmark datasets show that DM3Loc outperforms another deep-learning-based method RNATracker in both accuracy and speed, and in general, outperforms other existing tools for mRNA subcellular localization predictions. By applying DM3Loc to the human transcriptome, we found hundreds of mRNA isoform-specific subcellular localization predictions, which supports the existence of general isoform-specific mRNA subcellular localization from a computational perspective. The isoforms provided in this paper should be further validated through experimental methods, such as Fluorescence In Situ Hybridization (FISH). Through gene enrichment analyses, we found many significantly enriched gene ontology terms for mRNAs from different subcellular localization groups through functional enrichment analyses. These results extend existing knowledge about the functions of mRNAs involved in subcellular localization. All the services and the standalone tool of DM3Loc can be freely accessed at the webserver http://dm3loc.lin-group.cn. The proposed approach in this study provides a demonstration of how a deep-learning model with a multi-head self-attention mechanism facilitates the mRNA sequence analyses in the application of subcellular localization; our research also substantiated our line of reasoning, which presents the proposed deep-learning model as a useful tool for other sequence-based analyses.
The development of mRNA subcellular localization predictor is still at the early stage. The accuracy of DM3Loc is currently limited by the annotations in the RNALocate dataset. With mRNA subcellular localization becoming a more and more important topic, new techniques and diverse datasets will rapidly become available, and DM3Loc will have the ability to catch up with these new data for more accurate and comprehensive predictions.
DATA AVAILABILITY
The benchmark dataset with all the mRNAs, the nonredundant benchmark dataset, and the 5-fold cross-validation dataset are available on the DM3Loc webserver, http://dm3loc.lin-group.cn/ (under the menu Download/data). The 5-fold cross-validation dataset is also available in the GitHub repository, https://github.com/duolinwang/DM3Loc/tree/master/testdata. The DM3Loc web server can be accessed at http://dm3loc.lin-group.cn/. The stand-alone tool for locally using DM3Loc and source code are available in the GitHub repository, https://github.com/duolinwang/DM3Loc.
Supplementary Material
ACKNOWLEDGEMENTS
We like to thank Ms Carla Roberts for thoroughly proofreading this paper.
Contributor Information
Duolin Wang, Department of Electrical Engineering and Computer Science, Bond Life Sciences Center, University of Missouri, Columbia, MO 65203, USA.
Zhaoyue Zhang, Center for Information Biology, University of Electronic Science and Technology of China, Chengdu 610054, China.
Yuexu Jiang, Department of Electrical Engineering and Computer Science, Bond Life Sciences Center, University of Missouri, Columbia, MO 65203, USA.
Ziting Mao, Department of Electrical Engineering and Computer Science, Bond Life Sciences Center, University of Missouri, Columbia, MO 65203, USA.
Dong Wang, Department of Bioinformatics, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Hao Lin, Center for Information Biology, University of Electronic Science and Technology of China, Chengdu 610054, China.
Dong Xu, Department of Electrical Engineering and Computer Science, Bond Life Sciences Center, University of Missouri, Columbia, MO 65203, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Nature Scientific Foundation of China [61772119, 82070109]; Sichuan Provincial Science Fund for Distinguished Young Scholars [2020JDJQ0012]; Science Strength Promotion Program of UESTC; Paul K. and Diane Shumaker Endowment Fund at University of Missouri. Funding for open access charge: Paul K. and Diane Shumaker Endowment Fund at University of Missouri.
Conflict of interest statement. None declared.
REFERENCES
- 1. Medioni C., Mowry K., Besse F.. Principles and roles of mRNA localization in animal development. Development. 2012; 139:3263–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bullock S.L. Messengers, motors and mysteries: sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem. Soc. Trans. 2011; 39:1161–1165. [DOI] [PubMed] [Google Scholar]
- 3. Kloc M., Zearfoss N.R., Etkin L.D.. Mechanisms of subcellular mRNA localization. Cell. 2002; 108:533–544. [DOI] [PubMed] [Google Scholar]
- 4. Suter B. RNA localization and transport. Biochim. Biophys. Acta Gene Regul. Mech. 2018; 1861:938–951. [DOI] [PubMed] [Google Scholar]
- 5. Lewis R.A., Mowry K.L.. Ribonucleoprotein remodeling during RNA localization. Differentiation. 2007; 75:507–518. [DOI] [PubMed] [Google Scholar]
- 6. Holt C.E., Bullock S.L.. Subcellular mRNA localization in animal cells and why it matters. Science. 2009; 326:1212–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Di Liegro C.M., Schiera G., Di Liegro I.. Regulation of mRNA transport, localization and translation in the nervous system of mammals (Review). Int. J. Mol. Med. 2014; 33:747–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baj G., Leone E., Chao M.V., Tongiorgi E.. Spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments. PNAS. 2011; 108:16813–16818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mingle L.A., Okuhama N.N., Shi J., Singer R.H., Condeelis J., Liu G.. Localization of all seven messenger RNAs for the actin-polymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J. Cell Sci. 2005; 118:2425–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Uemura M., Zheng Q., Koh C.M., Nelson W.G., Yegnasubramanian S., De Marzo A.M.. Overexpression of ribosomal RNA in prostate cancer is common but not linked to rDNA promoter hypomethylation. Oncogene. 2012; 31:1254–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dolezal J.M., Dash A.P., Prochownik E.V.. Diagnostic and prognostic implications of ribosomal protein transcript expression patterns in human cancers. BMC Cancer. 2018; 18:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sprenkle N.T., Sims S.G., Sánchez C.L., Meares G.P.. Endoplasmic reticulum stress and inflammation in the central nervous system. Mol. Neurodegen. 2017; 12:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu-Yesucevitz L., Bassell G.J., Gitler A.D., Hart A.C., Klann E., Richter J.D., Warren S.T., Wolozin B.. Local RNA translation at the synapse and in disease. J. Neurosci. 2011; 31:16086–16093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang H., Nakamura M., Abbott T.R., Zhao D., Luo K., Yu C., Nguyen C.M., Lo A., Daley T.P., La Russa M.et al.. CRISPR-mediated live imaging of genome editing and transcription. Science. 2019; 365:1301–1305. [DOI] [PubMed] [Google Scholar]
- 15. Ren K., Wu R., Karunanayake Mudiyanselage A., Yu Q., Zhao B., Xie Y., Bagheri Y., Tian Q., You M.. In situ genetically cascaded amplification for imaging RNA subcellular locations. J. Am. Chem. Soc. 2020; 142:2968–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee J.H., Daugharthy E.R., Scheiman J., Kalhor R., Yang J.L., Ferrante T.C., Terry R., Jeanty S.S., Li C., Amamoto R.et al.. Highly multiplexed subcellular RNA sequencing in situ. Science. 2014; 343:1360–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fazal F.M., Han S., Parker K.R., Kaewsapsak P., Xu J., Boettiger A.N., Chang H.Y., Ting A.Y.. Atlas of subcellular RNA localization revealed by APEX-Seq. Cell. 2019; 178:473–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yan Z., Lécuyer E., Blanchette M.. Prediction of mRNA subcellular localization using deep recurrent neural networks. Bioinformatics. 2019; 35:i333–i342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Z.Y., Yang Y.H., Ding H., Wang D., Chen W., Lin H.. Design powerful predictor for mRNA subcellular location prediction in Homo sapiens. Brief. Bioinform. 2021; 22:526–535. [DOI] [PubMed] [Google Scholar]
- 20. Garg A., Singhal N., Kumar R., Kumar M.. mRNALoc: a novel machine-learning based in-silico tool to predict mRNA subcellular localization. Nucleic Acids Res. 2020; 48:W239–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang T., Tan P., Wang L., Jin N., Li Y., Zhang L., Yang H., Hu Z., Zhang L., Hu C.et al.. RNALocate: a resource for RNA subcellular localizations. Nucleic Acids Res. 2017; 45:D135–D138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thul P.J., Åkesson L., Wiking M., Mahdessian D., Geladaki A., Ait Blal H., Alm T., Asplund A., Björk L., Breckels L.M.et al.. A subcellular map of the human proteome. Science. 2017; 356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 23. Li H., Tian S., Li Y., Fang Q., Tan R., Pan Y., Huang C., Xu Y., Gao X.. Modern deep learning in bioinformatics. J. Mol. Cell Biol. 2020; doi:10.1093/jmcb/mjaa030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang B., Pan Z., Yin K., Khateeb A.. Recent advances of deep learning in bioinformatics and computational biology. Front. Genet. 2019; 10:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li Y., Huang C., Ding L., Li Z., Pan Y., Gao X.. Deep learning in bioinformatics: introduction, application, and perspective in the big data era. Methods. 2019; 166:4–21. [DOI] [PubMed] [Google Scholar]
- 26. Quang D., Xie X.. DanQ: a hybrid convolutional and recurrent deep neural network for quantifying the function of DNA sequences. Nucleic Acids Res. 2016; 44:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Almagro Armenteros J.J., Sønderby C.K., Sønderby S.K., Nielsen H., Winther O.. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics. 2017; 33:3387–3395. [DOI] [PubMed] [Google Scholar]
- 28. Sønderby S.K., Sønderby C.K., Nielsen H., Winther O.. International Conference on Algorithms for Computational Biology. 2015; Springer; 68–80. [Google Scholar]
- 29. Lin Z., Feng M., Santos C.N., Yu M., Xiang B., Zhou B., Bengio Y.. A structured self-attentive sentence embedding. 2017; arXiv doi:09 March 2017, preprint: not peer reviewedhttps://arxiv.org/abs/1703.03130.
- 30. Vaswani A., Shazeer N., Parmar N., Uszkoreit J., Jones L., Gomez A.N., Kaiser Ł., Polosukhin I.. Advances in Neural Information Processing Systems. 2017; 5998–6008. [Google Scholar]
- 31. Devlin J., Chang M.-W., Lee K., Toutanova K.. Bert: pre-training of deep bidirectional transformers for language understanding. 2018; arXiv doi:24 May 2019, preprint: not peer reviewedhttps://arxiv.org/abs/1810.04805v2.
- 32. Sayers E.W., Cavanaugh M., Clark K., Ostell J., Pruitt K.D., Karsch-Mizrachi I.. GenBank. Nucleic Acids Res. 2019; 47:D94–D99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fu L., Niu B., Zhu Z., Wu S., Li W.. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012; 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chorowski J.K., Bahdanau D., Serdyuk D., Cho K., Bengio Y.. Advances in Neural Information Processing Systems. 2015; 577–585. [Google Scholar]
- 35. Alipanahi B., Delong A., Weirauch M.T., Frey B.J.. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 2015; 33:831–838. [DOI] [PubMed] [Google Scholar]
- 36. Kingma D.P., Ba J.L.. Adam: a method for stochastic optimization. 2014; arXiv doi:30 January 2017, preprint: not peer reviewedhttps://arxiv.org/abs/1412.6980.
- 37. Xia C., Fan J., Emanuel G., Hao J., Zhuang X.. Spatial transcriptome profiling by MERFISH reveals subcellular RNA compartmentalization and cell cycle-dependent gene expression. PNAS. 2019; 116:19490–19499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D.et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taliaferro J.M., Vidaki M., Oliveira R., Olson S., Zhan L., Saxena T., Wang E.T., Graveley B.R., Gertler F.B., Swanson M.S.et al.. Distal alternative last exons localize mRNAs to neural projections. Mol. Cell. 2016; 61:821–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gupta S., Stamatoyannopoulos J.A., Bailey T.L., Noble W.S.. Quantifying similarity between motifs. Genome Biol. 2007; 8:R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A.et al.. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013; 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meer E.J., Wang D.O., Kim S., Barr I., Guo F., Martin K.C.. Identification of a cis-acting element that localizes mRNA to synapses. PNAS. 2012; 109:4639–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bergalet J., Lécuyer E.. The functions and regulatory principles of mRNA intracellular trafficking. Adv. Exp. Med. Biol. 2014; 825:57–96. [DOI] [PubMed] [Google Scholar]
- 44. Kislauskis E.H., Zhu X., Singer R.H.. beta-Actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 1997; 136:1263–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T.et al.. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000; 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Neupert W. Protein import into mitochondria. Annu. Rev. Biochem. 1997; 66:863–917. [DOI] [PubMed] [Google Scholar]
- 48. Kellems R.E., Allison V.F., Butow R.A.. Cytoplasmic type 80 S ribosomes associated with yeast mitochondria. II. Evidence for the association of cytoplasmic ribosomes with the outer mitochondrial membrane in situ. J. Biol. Chem. 1974; 249:3297–3303. [PubMed] [Google Scholar]
- 49. George R., Walsh P., Beddoe T., Lithgow T.. The nascent polypeptide-associated complex (NAC) promotes interaction of ribosomes with the mitochondrial surface in vivo. FEBS Lett. 2002; 516:213–216. [DOI] [PubMed] [Google Scholar]
- 50. Tsuboi T., Viana M.P., Xu F., Yu J., Chanchani R., Arceo X.G., Tutucci E., Choi J., Chen Y.S., Singer R.H.et al.. Mitochondrial volume fraction and translation speed impact mRNA localization and production of nuclear-encoded mitochondrial proteins. 2019; bioRxiv doi:10 October 2019, preprint: not peer reviewed 10.1101/529289. [DOI] [PMC free article] [PubMed]
- 51. Carpenter S., Fitzgerald K.A.. Transcription of inflammatory genes: long noncoding RNA and beyond. J. Interferon Cytokine Res. 2015; 35:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wickham L., Duchaîne T., Luo M., Nabi I.R., DesGroseillers L.. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999; 19:2220–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Batagov A.O., Kuznetsov V.A., Kurochkin I.V.. Identification of nucleotide patterns enriched in secreted RNAs as putative cis-acting elements targeting them to exosome nano-vesicles. BMC Genomics. 2011; 12(Suppl. 3):S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang D., Zeng S., Xu C., Qiu W., Liang Y., Joshi T., Xu D.. MusiteDeep: a deep-learning framework for general and kinase-specific phosphorylation site prediction. Bioinformatics. 2017; 33:3909–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zou Z., Tian S., Gao X., Li Y.. mlDEEPre: multi-functional enzyme function prediction with hierarchical multi-label deep learning. Frontiers in genetics. 2018; 9:714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mikl M., Vendra G., Doyle M., Kiebler M.A.. RNA localization in neurite morphogenesis and synaptic regulation: current evidence and novel approaches. J. Comp. Physiol. A Neuroethol. Sens. Neural Behav. Physiol. 2010; 196:321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The benchmark dataset with all the mRNAs, the nonredundant benchmark dataset, and the 5-fold cross-validation dataset are available on the DM3Loc webserver, http://dm3loc.lin-group.cn/ (under the menu Download/data). The 5-fold cross-validation dataset is also available in the GitHub repository, https://github.com/duolinwang/DM3Loc/tree/master/testdata. The DM3Loc web server can be accessed at http://dm3loc.lin-group.cn/. The stand-alone tool for locally using DM3Loc and source code are available in the GitHub repository, https://github.com/duolinwang/DM3Loc.